Introduction
For women, breast cancer is the most common type of
cancer with a high morbidity rate worldwide (1). According to the data of the National
Central Cancer Registry of China (NCCR), 4292,000 newly diagnosed
invasive breast cancer cases were reported in 2015 (2). Breast cancer accounts for 15% of all
new-onset malignant tumors in Chinese women (3). The increasing trend of breast cancer
reflects the changes in reproductive behavior, the prevalence of
obesity and physical inactivity over the past several decades in
China (4). Clinical studies have
indicated that there is very obvious heterogeneity in breast cancer
(5,6). However, the underlying mechanisms
related to the heterogeneity of breast cancer have not yet been
fully elucidated. Most importantly, a better mechanistic
understanding can reveal promising new therapeutic targets.
Fortunately, with the development of high-throughput sequencing
technologies, the classification of breast cancer extends from
pathological types to molecular types. The molecular typing of
breast cancer illustrates the heterogeneity of breast cancer in
part. Parker et al presented a molecular classification of
breast cancer, which was grouped into luminal A (Lum A), luminal B
(Lum B), Basal-like, human epidermal growth factor receptor
2-positive (HER2+), and Normal breast-like subtypes,
according to the PAM50 method (7).
The PAM50 breast cancer intrinsic classifier has lately been used
to assign the molecular subtypes, including Basal,
HER2+, Lum A, Lum B and Normal-like subtypes based on
the shrunken centroids of gene expression profiles (8). There is no doubt that precision
classification can enhance breast cancer diagnosis, treatment and
prognosis prediction in clinical practice.
Recently, the massive amount of data generated by
the sequencing of tumor samples have spawned a large amount of
bioinformatics databases, such as The Cancer Genome Atlas (TCGA,
https://cancergenome.nih.gov/) and Gene
Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), thus enabling us
to comprehensively understand tumors from large data analysis at
the multiple genetic level. Bioinformatics analysis provides an
alternative method with which to explore the genetic function, from
DNA to protein. In particular, bioinformatics analysis has an
absolute advantage over large quantities of data analysis.
The human Dickkopf (DKK) family members 1, 2, 3 and
4 are located within chromosomes 4, 8, 10, and 11 (DKK1 maps to
10q11, DKK2 maps to 4q25, DKK3 maps to 11p15.3, and DKK4 maps to
8p11). The DKK family encodes 4 types of secreted protein (DKK1,
DKK2, DKK3 and DKK4) that share 2 conserved cysteine-rich domains.
The cysteine-rich domain in the N-terminus is unique to each type
of DKKs, whereas the other one in the C-terminus is conserved among
the family members (9,10). A number of previous studies have
demonstrated changes in DKK expression in tumor cell lines or
tissues, such as colorectal cancer (11), ovarian carcinoma (12), gastric carcinoma (13) and renal cell carcinoma (14). DKK1 has been shown to be silenced
in colon cancer and to be associated with tumorigenesis (11). DKK2 contributes to tumorigenesis in
epithelial ovarian carcinoma through the Wnt/β-catenin signaling
pathway (12). Our group
previously found that a decreased DKK3 expression was closely
linked to an aggressive behavior and a poor prognosis of gastric
cancer (13). The expression of
DKK4 has also been shown to be significantly higher in renal cancer
tissues compared with adjacent normal kidney tissues (14).
In this study, we adopted cooperatively
bioinformatics and experimental methods to performed our analyses.
Firstly, we employed the Gene Expression-Based Outcome for Breast
Cancer Online (GOBO) platform that developed by Ringnér et
al (15) at Lund University
Canceromics Branch (Lund, Sweden) to perform the comprehensive
analysis of DKKs expression profiles and the corresponding
prognostic analysis of breast cancer. Secondly, we used our own
data to validate the data from bioinformatics analysis on the
expression profile of DKK2 in the normal-like subtype compared with
normal tissue by means of reverse transcription-quantitative PCR
(RT-qPCR). Thirdly, the breast cancer dataset from TCGA, Molecular
Taxonomy of Breast Cancer International Consortium (METABRIC,
http://molonc.bccrc.ca/aparicio-lab/research/metabric/)
and Kaplan-Meier plotter online platforms were used for the
analysis of the expression profiles and prognostic roles of DKKs in
breast cancer. Fourthly, a DKK2 protein-protein network was
constructed and enrichment analysis was conducted to explore the
underlying mechanisms of breast cancer. Finally, DKK2 mutation,
copy number variation and methylation were performed to explore the
genetic alterations in breast cancer. Taken together, the findings
of this study suggest that DKK2 may be a potential prognostic
biomarker for the normal-like subtype.
Materials and methods
DKKs mRNA expression profiles in breast
cancer and breast cancer subtypes
DKKs transcripts with molecular subtypes (PAM50) in
patients with breast cancer were examined using the GOBO platform,
which was developed by Ringnér et al at Lund University
Canceromics Branch (Lund, Sweden), which consists of three main
modules, including gene expression analysis, gene co-expression and
sample prediction. It is a publicly available platform, including
breast cancer subtype-related gene expression data and
corresponding clinical data from 1,881 subjects with breast cancer
based on Affymetrix microarray analysis (http://co.bmc.lu.se/gobo). The breast tumor set from
the 1,881 samples comprises 11 public datasets, including Basal
(n=280), HER2+ (n=214), Lum A (n=410), Lum B (n=414) and
Normal-like (n=255) subtypes according to the breast cancer PAM50
molecular subtype classification (15). The expression analyses of DKKs
transcripts in the subtypes and histological grade were performed
using one-way analysis of variance (one-way ANOVA). Besides, the
DKKs associated with the estrogen receptor (ER)-status were
analyzed using the Student's t-test. P-values <0.05 were
considered to indicate statistically significant differences.
Transcript levels of DKKs and clinical
outcomes in breast cancer subtypes
The transcript levels of DKKs associated with breast
cancer outcome, together with overall survival (OS) and
relapse-free survival (RFS), were analyzed accurately and in detail
using the Kaplan-Meier Plotter online platform, which integrates a
number of tumor-related microarrays and detailed clinical
prognostic information, as previously described (16). DKKs transcripts with molecular
subtypes and clinical outcome from patients with breast cancer were
analyzed using the GOBO outcome analysis module. The association of
clinical outcome with DKK2 gene expression levels in the subgroups
of breast cancer was analyzed through OS and RFS as the endpoint
and 10-year censoring. Breast cancer datasets were stratified into
3 quantiles based on DKK2 expression (lower, medium and upper
quartile), DKK2_low (log2 expression −5.749 to −0.305), DKK2_
medium (−0.305 to 0.502), and DKK2_high (0.502 to 5.422).
Subsequently, the Cox proportional hazards model was applied to
estimate the hazard ratio, and the ER status, node status, grade,
age and tumor size indicator were used as covariates. Logrank
P-values are shown as −log10 (P-value). P-values <0.05 were
considered to indicate statistically significant differences.
Breast cancer patient specimens and
RT-qPCR
We also used our data to further confirm the results
of bioinformatics analysis on DKKs and the subtypes. Tissue
specimens (17 Normal-like subtype and 28 ER-positive subtype) were
obtained from 45 patients (no chemotherapy or radiotherapy prior to
resection) at Shengjing Hospital of China Medical University
(Shenyang, China) from January, 2011 to September, 2013. Informed
written consent was obtained from all participants and the study
was approved by Ethics Committee of Shengjing Hospital. Total RNA
was extracted from these specimens using the RNeasy Mini kit
(Qiagen China, Shanghai, China) according to the manufacturer's
instructions. RT-qPCR was carried out using the ABI 7500 Real-Time
PCR system (Applied Biosystems, Foster City, CA, USA). The
RevertAid First Strand cDNA Synthesis kit (Thermo Fisher
Scientific, Burlington, ON, Canada) and SYBR® Premix
ExTaq™ II Kit (Takara Biomedical Technology, Dalian, China) were
used. Oligonucleotide primers for DKK2 were used as follows:
Forward, 5′-CCCCGTTCATTCCTGTTTG-3′ and reverse,
5′-TTCTCCACGGTCCAATCCT-3′. The primers for the internal control,
GAPDH, were forward, 5′-CAATGACCCCT TCATTGACC-3′ and reverse,
5′-TGGAAGATGGTGATGGG ATT-3′. The amplification of DKK2 and GAPDH
was performed with 1 cycle at 95°C for 10 min and 40 cycles of 95°C
for 15 sec and 60°C for 60 sec. Relative mRNA abundance was
normalized to the internal standard, GAPDH, using the
2−∆∆Cq method (17).
The samples were divided into 2 groups at the DKK2 median and
classified as high or low DKK2.
Validation of the DKK2 expression profile
via TCGA and METABRIC in breast cancer subtypes
The TCGA project, which was developed for cancer
research, integrates gene expression profiles, methylation, copy
number variation and mutation information in 33 cancer types, which
helps us to understand multi-dimensional genetic alterations in
tumors (18). TCGA breast cancer
dataset (19) is available at the
TCGA data portal, such as cBioPortal for Cancer Genomics
(http://www.cbioportal.org/) (20). According to the PAM50
classification, TCGA breast cancer dataset (19) was divided into 5 subtypes,
including the Basal (n=98), HER2+ (n=58), Lum A (n=230),
Lum B (n=125) and Normal-like (n=8) subtypes. Moreover, METABRIC
breast cancer dataset covers several gene expression cohorts, and
is widely used for the investigation of breast cancer and subtypes.
METABRIC breast cancer dataset (21,22)
was grouped into the Basal (n=199), HER2+ (n=220), Lum A
(n=679), Lum B (n=461) and Normal-like (n=140) subtypes based on
the PAM50 classification.
As mentioned above, the GOBO database consists of 11
datasets, which contribute to instability in performance and
efficiency of the result. Therefore, we used the publicly available
breast cancer dataset with homogeneity and a large sample size to
verify the results of GOBO. Subsequently, we undertook breast
cancer from TCGA (19) and breast
cancer from METABRIC (21,22) to validate expression profile of
DKK2 in breast cancer subtypes (22). The expression of DKK2 in the PAM50
subtypes was examined using one-way ANOVA with Dunnett's test with
GraphPad prism V7.0 software. P-values <0.05 were considered to
indicate statistically significant differences.
Validation of the association between
DKK2 mRNA and clinical outcome using the METABRIC database and
Kaplan-Meier platform in breast cancer subtypes
We analyzed the survival outcomes of breast cancer
and the subtypes according to the differential expression levels of
DKK2. Kaplan-Meier survival curves with hazard ratio and logrank
P-value were calculated and plotted with the online platform, which
is available at Kaplan Meier plotter (http://kmplot.com/analyisis). Kaplan-Meier for breast
cancer assessed the relevance of the expression levels of various
genes with regards to the clinical outcome (23). Furthermore, we examined METABRIC
breast cancer samples (21,22)
to validate the clinical outcome of DKK2 in the Normal-like
subtype. Data were exported from the cBioPortal platform, and
GraphPad prism V7.0 software was then used to perform the survival
analysis. The breast cancer sample sets were stratified into 2
quantiles based on DKK2 expression as follows DDK2_medium (medium,
0.424), DKK2_low (−3.04 to 0.424) and DKK2_high (0.424 to
0.534).
Analysis of DKK2 and genes correlation
with breast cancer by protein-protein interaction network
To clearly elucidate the underlying mechanisms
through which DKK2 is involved in the occurrence and progression of
breast cancer at the protein level, we selected breast
cancer-related proteins by means of known literature retrieval.
Specifically, we conducted the following retrieval expression:
'breast cancer*' and 'gene*' or
'protein*'. Subsequently, we performed PPI network
analysis via the Search Tool for the Retrieval of Interacting Genes
(STRING v10.5, https://string-db.org/), which
provides protein interaction from multiple aspects of evidence,
including text-mining, experiments, database, gene co-expression,
gene neighborhood, gene-fusion and literature co-occurrence
(24). The necessary parameters
were set as follows: Meaning of network edges, confidence; minimum
required interaction score, medium confidence (0.400). After the
PPI network was constructed, Cytoscape software (v3.1.6) was used
to visualize the network (25).
Functional and pathway enrichment
analysis
Gene ontology (GO) and pathway analysis were
performed using the functional enrichment analysis tool (FunRich
v3.1.3) software (http://www.funrich.org/), which classifies the
biological functions, as well as the signaling pathway of the
various proteins involved in the protein networks via
hypergeometric distribution algorithm (26). GO analysis consists of molecular
function (GO_MF), biological process (GO_BP) and cellular component
(GO_CC) (27). Pathway analysis
was integrated from the Kyoto Encyclopedia of Genes and Genomes
database (KEGG). A threshold value of 0.05 was established for
P-values and false discovery rate (FDR) (P<0.05, FDR
<0.05).
COSMIC analysis for DKK2 mutations
DKK2 mutation analysis was carried out using the
Catalog of Somatic Mutations in Cancer database (COSMIC)
(https://cancer.sanger.ac.uk/cosmic).
The pie charts present detailed information about distribution of
mutations types and substitutions on the coding strand in breast
cancer. The alteration frequency of DKK2 mRNA in breast cancer was
performed using BioPortal. All operations were performed according
to the cBioPortal instructions. The database query was based on
mutation and altered expression of the DKK2 in TCGA breast cancer
dataset (19) and METABRIC breast
cancer dataset (21,22).
Analysis of DKK2 mRNA expression with DNA
methylation analysis and copy number analysis via cBioPortal
database
The analysis of DKK2 mRNA expression with DNA
methylation and copy number was conducted using the cBioPortal
database. DKK2 copy number was analyzed by GISTIC, which is the
tool to identify gene targeted by somatic copy-number alteration.
Moreover, DKK2 methylation was analyzed by Human Methylation 450
(HM450) BeadChip kit. cBioPortal has integrated DNA methylation
analysis and copy number analysis in a user-friendly manner.
Statistical analyses
The statistical significance of expression of DKKs
transcripts in the subtypes was examined by one-way ANOVA. In
statistics, Dunnett's test is a multiple comparison method
developed by Canadian statistician Charles Dunnett to compare each
of a number of treatments with a single control. Multiple
comparisons to a control are also referred to as many-to-one
comparisons. After ANOVA, Dunnett's test, a post hoc test, was used
to perform multiple comparison test. The association of the DKKs
with ER-status and grade was analyzed by the Student's t-test.
Survival curves were plotted using Graphpad Prism software v7.0
(GraphPad Software Inc., San Diego, CA, USA), according to the
method of Kaplan and Meier, and the (two-sided) log-rank test was
used to assess the statistical significance of differences in OS
and RFS among distinct groups of patients. Multivariate analysis of
prognostic factors for OS and RFS was performed using the Cox
stepwise regression model. P-values <0.05 and FDR <0.05 were
considered to indicate statistically significant differences.
Results
Expression profiles of DKKs across breast
cancer subtypes
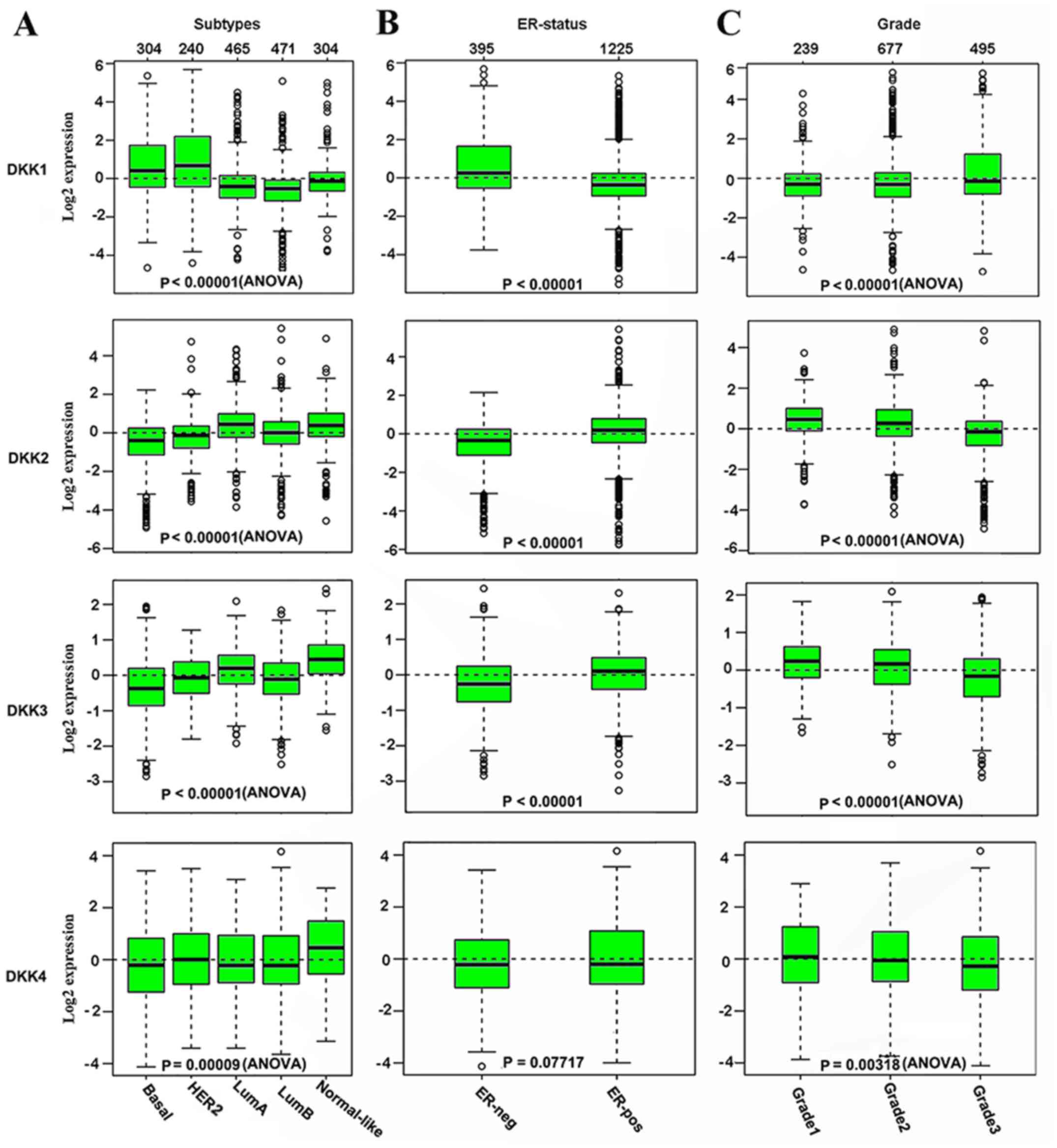
Gene expression analysis derived from the GOBO
expression module revealed that DKK1 expression was significantly
higher in the HER2-enriched, Basal and Normal-like subtype, while
it was downregulated in the Lum A and Lum B subtypes (P<0.00001,
Fig. 1A). The patients with an
ER-negative status exhibited a higher DKK1 expression than those
with an ER-positive status (P<0.00001, Fig. 1B). The highest DKK1 transcript
expression was observed in Grade 3 tumors, compared with Grade 1
and Grade 2 tumors (P<0.00001, Fig.
1C). The expression of DKK2 and DKK3 was significantly higher
in the Lum A and Normal-like subtypes (P<0.00001, Fig. 1A). However, the downregulation of
DKK2 and DKK3 was observed in the Basal, HER2+ and Lum B
subtypes. Unlike DKK1, the patients with an ER-positive status
exhibited a higher DKK2 and DKK3 expression than those with an
ER-negative status (P<0.00001, Fig.
1B). DKK4 expression was higher in the normal-like subtype and
lower in the Lum A subtype (P<0.00001, Fig. 1A). A higher expression level of
DKK2, DKK3 and DKK4 was observed in the lower grade and ER-positive
tumors (P<0.00001, Fig. 1B).
These observations provide further evidence of the DKK1-4 mRNA
levels associated with the subtypes, ER status and Grade in breast
cancer. Notably, we noted a consistent increase in the expression
of DKK family members, with the exception of DKK1, in the
normal-like subtypes.
DKKs are associated with the clinical
outcome of breast cancer patients
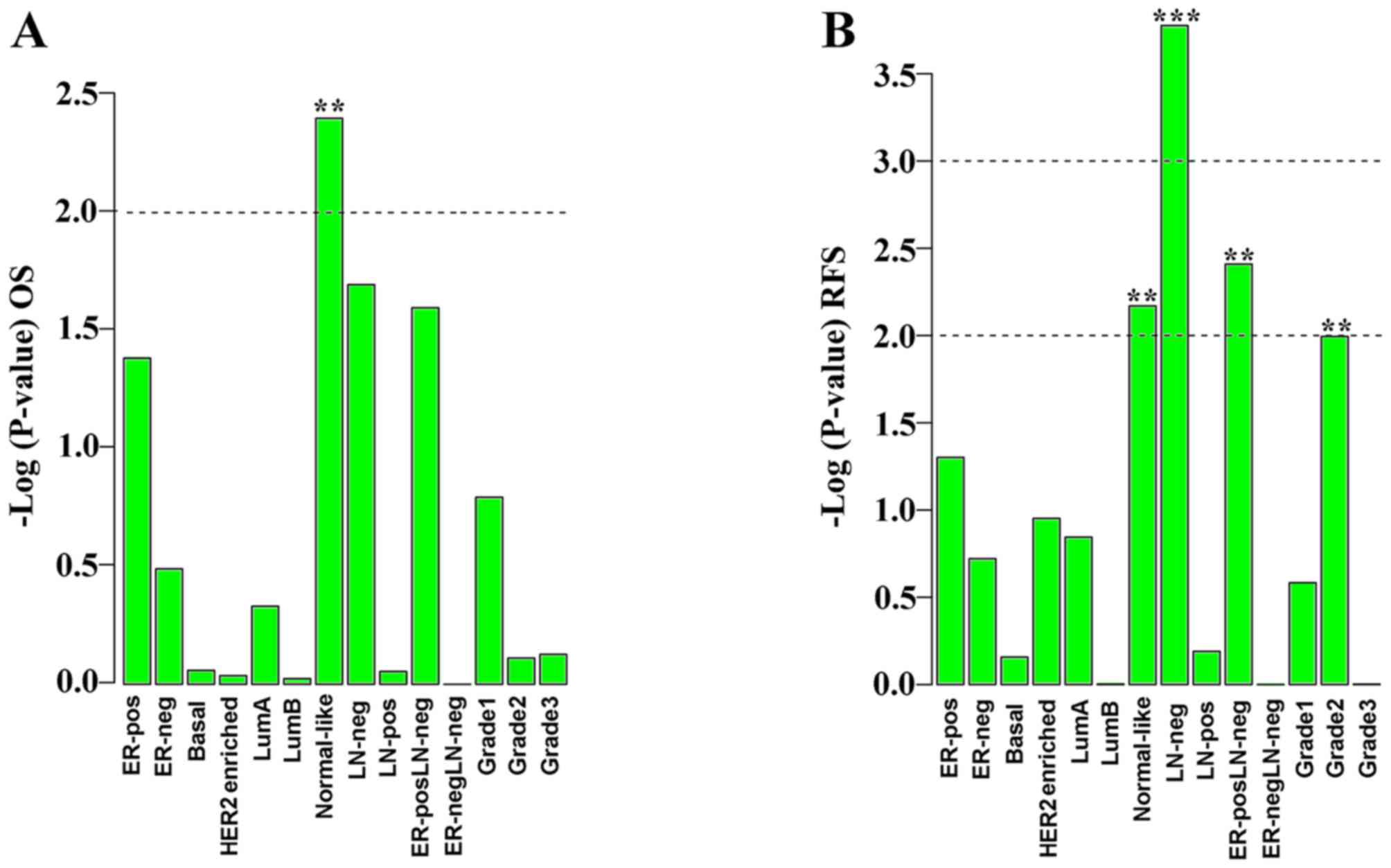
The GOBO outcome module suggested that the DKK2
level was associated with lymph node (LN)-negative (P<0.01)
status, ER-positive/LN-negative status (P<0.01), Grade 2
(P<0.01) and Normal-like subtype (P<0.001) (Fig. 2). The mRNA expression levels of
DKK1, DKK3 and DKK4 were not associated with the clinical prognosis
of the breast cancer subtypes (PAM50 classification) and the
results were statistically significant. A high DKK2 expression was
associated with a good OS of patients with all subtypes of breast
cancer (P=0.0477, Fig. 3A).
Moreover, a low DKK2 expression was also associated with a poor RFS
of patients with all subtypes of breast cancer (P=0.00493, Fig. 3B). On the contrary, a low DKK2
expression predicted a good OS for patients with the normal-like
subtype (P=0.00398, Fig. 3C). In
addition, a low DKK2 expression predicted a good RFS for patients
with the normal-like subtype (P=0.00677, Fig. 3D). On the whole, we comprehensively
analyzed the expression profiles and prognostic value of DKKs in
breast cancer subtypes. The main data indicated that a high DKK2
mRNA expression was associated with a better prognosis in breast
cancer, which is consistent with the findings of a previous study
(28). Nevertheless, we focused
our attention on the clinical prognostic value of DKK2 in the
normal-like subtype.
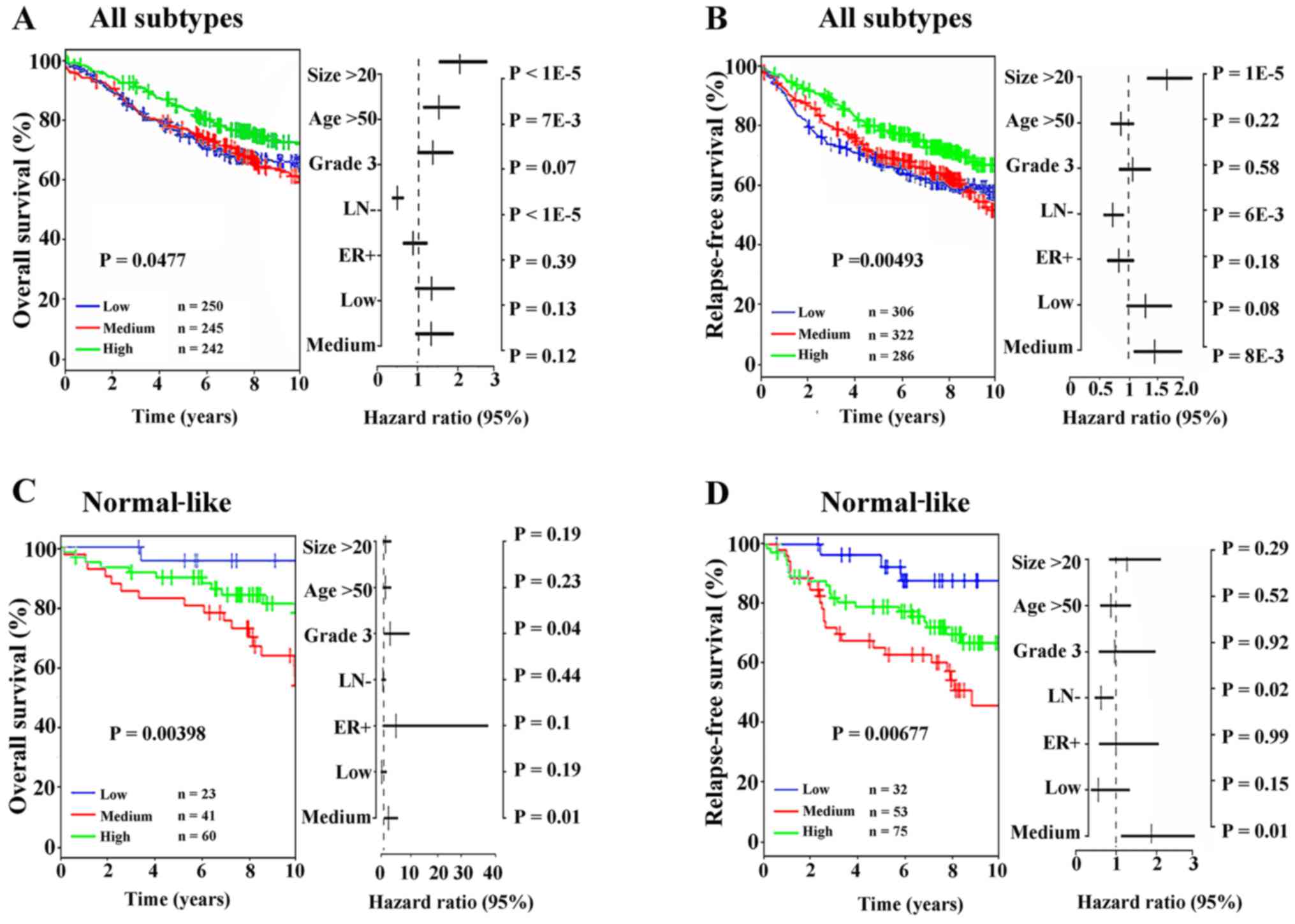 | Figure 3(A) All subtypes were stratified into
3 quantiles based on the DKK2 mRNA level followed by Cox
proportional hazards model using lymph node status and stratified
histological grade (histological grade 1, 2 and 3) and tumor size
as covariates, and using overall survival (OS) as the endpoint with
10-year censoring. (B) For all subtypes, the expression of DKK2
mRNA level was stratified into 3 quantiles based on DKK2 mRNA level
followed by Cox multivariable analysis using lymph node status and
stratified histological grade (histological grade 1 and 2 vs. 3)
and tumor size as covariates and using relapse-free survival (RFS)
as the endpoint with 10-year censoring. (C) The normal-like subtype
was stratified into 3 quantiles based on the DKK2 mRNA level
followed by Cox proportional hazards model using lymph node status
and stratified histological grade (histological grade 1, 2 and 3)
and tumor size as covariates, and using OS as the endpoint with
10-year censoring. (D) For the normal-like subtype, the mRNA
expression of DKK2 was stratified into 3 quantiles based on the
DKK2 mRNA level followed by Cox multivariable analysis using lymph
node status and stratified histological grade (histological grade 1
and 2 vs. 3) and tumor size as covariates and using RFS as the
endpoint with 10-year censoring. Samples in the 1,881-sample set
were stratified into 3 quantiles based on the DKK2 expression level
(lower quartile, −0.305; medium, 0.502; and upper quartile, 5.422)
for Kaplan-Meier analysis, including low (log2 expression −5.749 to
−0.305), medium (−0.305 to 0.502), and high (0.502 to 5.422). |
Validation of the DKK2 expression profile
via TCGA and METABRIC in breast cancer subtypes
To verify the accuracy of the expression profiles of
DKK2 in the PAM50-based molecular derived from GOBO in subtypes,
the TCGA breast cancer dataset (19) and METABRIC breast cancer dataset
(21,22) were used to validate the expression
profiles of DKK2 in breast cancer subtypes. The METABRIC breast
cancer datasets also indicated that the Lum A, Lum B and
Normal-like subtypes the expression of DKK2 exhibited an increasing
trend (Fig. 4A). Moreover, DKK2
expression was downregulated in the basal and HER2+
subtypes (P<0.0001; Fig. 4A).
The TCGA breast cancer dataset (19) also indicated that the transcript
level of DKK2 was upregulated in the Lum A, Lum B and normal-like
subtypes, and downregulated in the basal and HER2+
subtypes (P<0.0001) (Fig. 4B).
These findings are mostly consistent with the results of the GOBO
dataset. Moreover, Dunnett's multiple test showed that the
expression of DKK2 in Normal-like was significantly higher than
Basal and HER2+ subtype (P<0.0001) (Fig. 4A and B). However, there are a few
samples for the normal-like subtype in the TCGA dataset, which may
account for the fact that the results shown in Fig. 4B are slightly inconsistent with the
results shown in Fig. 1A. However,
the prognostic role of DKK2 in the subtypes of breast cancer still
requires further investigations with larger sample sets in the
future.
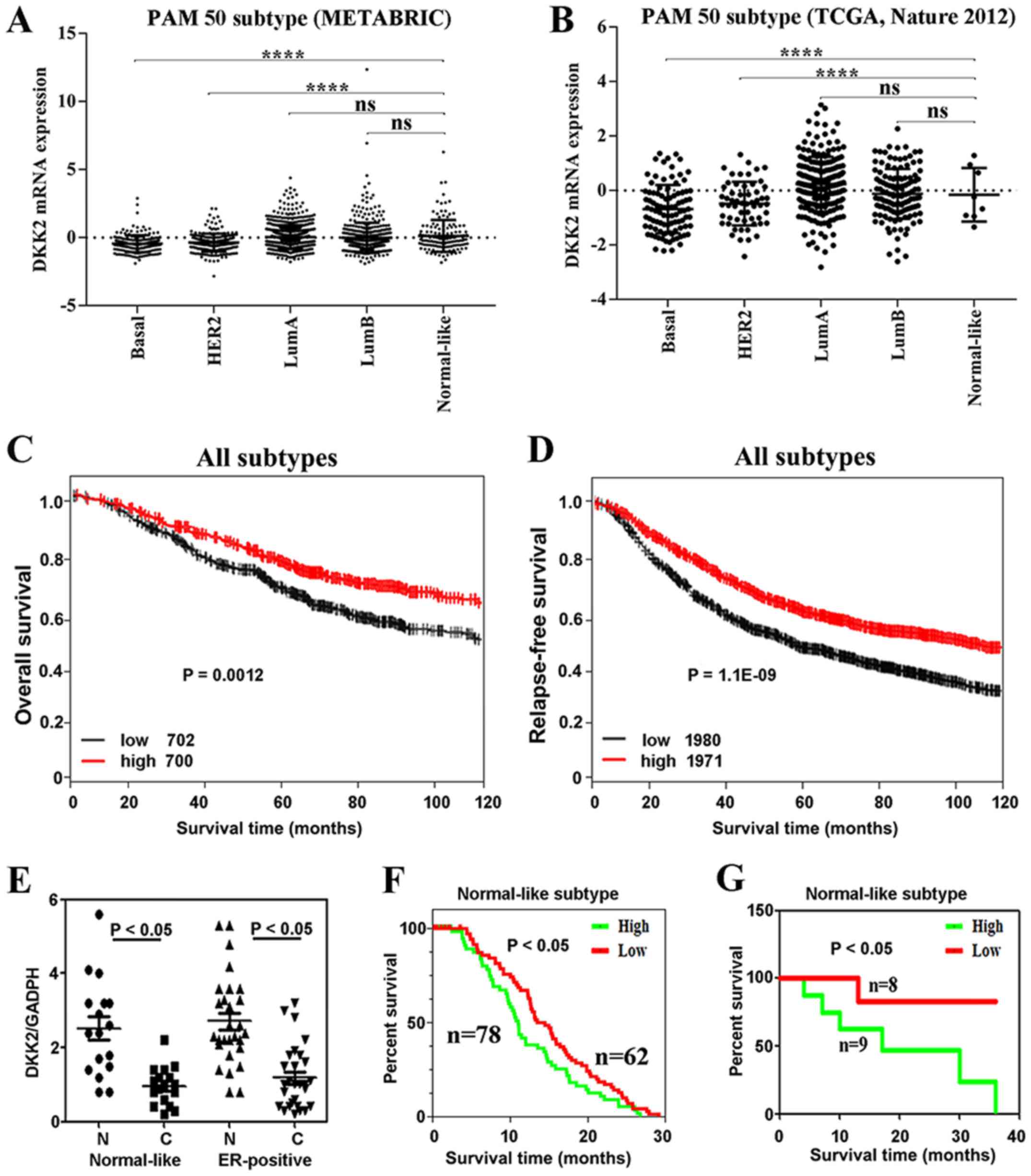 | Figure 4(A and B) METABRIC breast cancer
datasets (21,22) and TCGA breast cancer dataset
(19) validated the DKK2
expression level in breast cancer subtypes according to PAM50
classification by ANOVA and Dunnett's multiple comparison test. (C
and D) Kaplan-Meier online platform validated the association of
clinical outcome and DKK2 expression using overall survival (OS)
and relapse-free survival (RFS) as endpoints, respectively. The
breast cancer sample set was stratified into 2 quantiles based on
DKK2 medium (medium, 0.424) expression, including DKK2_low (−3.04
to 0.424) and DKK2_high (0.424 to 0.534). (E) RT-qPCR validated the
DKK2 mRNA expression level for normal-like and estrogen receptor
(ER+) subtypes versus normal control, respectively. (F
and G) METABRIC breast cancer dataset (21,22),
TCGA breast cancer dataset (22,43) and our data validated the
association of clinical outcome and DKK2 expression using OS,
respectively. The symbol **** and ns on top of the bars
stand for P<0.0001 and not significant, respectively; |
Validation of the association between
DKK2 mRNA expression and clinical outcome via the METABRIC database
and Kaplan-Meier platform in breast cancer subtypes
The Kaplan-Meier platform revealed that a high DKK2
expression was a good prognostic factor for OS in the all subtypes
(Fig. 4C). Moreover, an increased
DKK2 expression was also an excellent factor for RFS (Fig. 4D). METABRIC breast cancer (21,22)
indicated that a downregulated DKK2 mRNA expression was associated
with a better prognosis than the upregulation of its expression in
the Normal-like subtype (P<0.05; Fig. 4F). These findings are in line with
the results obtained from the GOBO platform.
Evaluation of DKK2 mRNA expression in 45
human breast cancer specimens
RT-qPCR analysis of DKK2 mRNA expression was carried
out in the Normal-like subtype and ER-positive subtype of breast
cancer. The results revealed that the mRNA level of DKK2 was lower
in both types than in the normal tissues (P<0.05, Fig. 4E). Kaplan-Meier analysis revealed
that an elevated DKK2 mRNA expression was associated with an
unfavorable prognosis of patients with the Normal-like subtype of
breast cancer (P<0.05, Fig.
4G). Through our own data validation, our results are
consistent with the results obtained from TCGA, METABRIC and
GOBO.
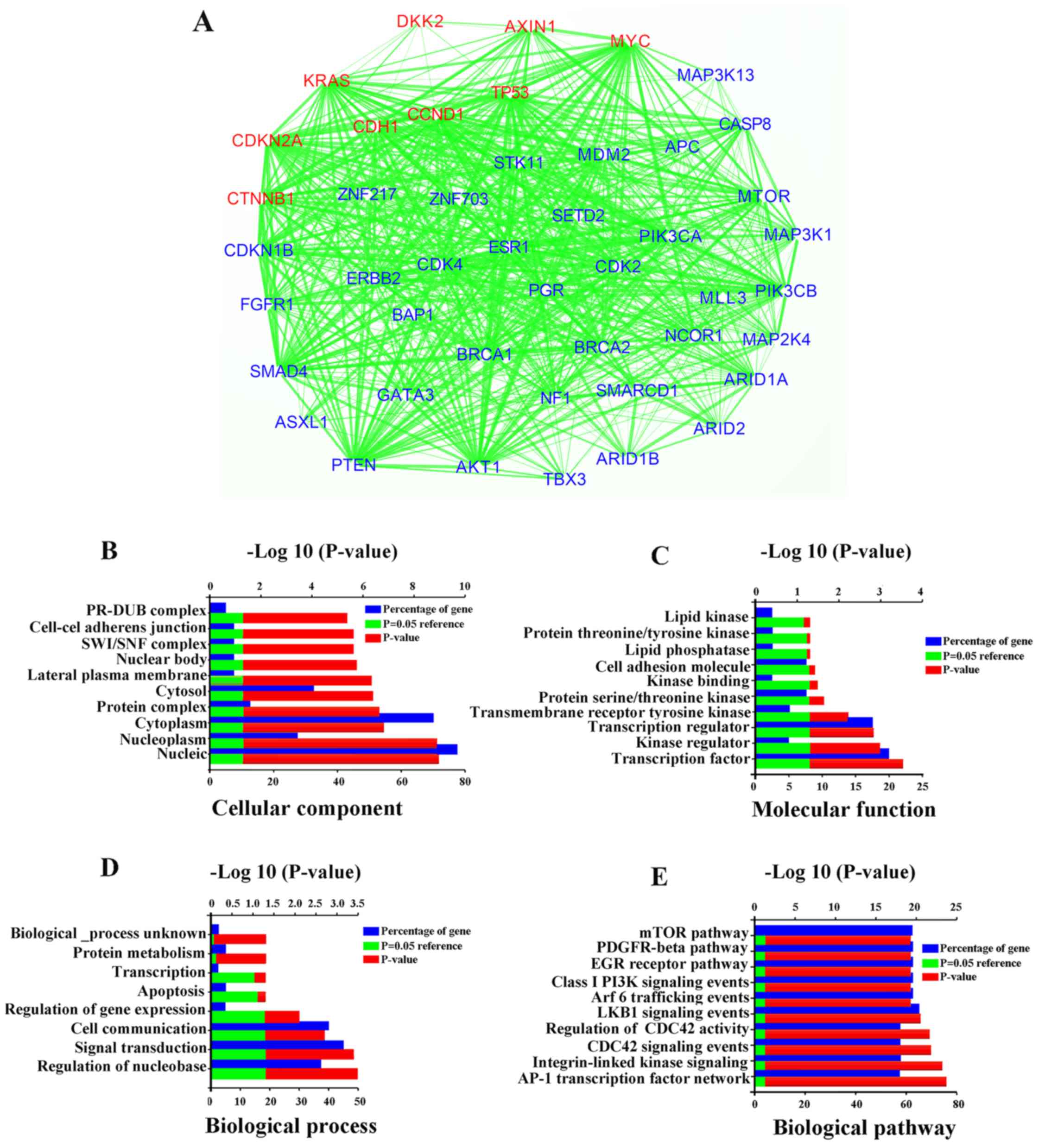
PPI network analysis
Based on the information from the literature
retrieval, we obtained 39 breast cancer-related proteins, including
TP53, ESR1, PGR, ERBB2, CTNNB1, AKT1, APC, ARID1A, ARID1B, ARID2,
ASXL1, BAP1, BRCA1, BRCA2, CASP8, CDH1, CDKN1B, CDKN2A, CCND1,
MDM2, ZNF217, MYC, FGFR1, ZNF703, GATA3, KRAS, MAP2K4, MAP3K1,
MAP3K13, KMT2C, NCOR1, NF1, PIK3CA, PTEN, SETD2, SMAD4, SMARCD1,
STK11 and TBX3. Furthermore, the PPI network consisted of 40 nodes
and 76 edges (average node degree of 1.15 and average local
clustering co-efficient of 0.313) (Fig. 5A). We were aware that DKK2 directly
interacted with AXIN1, KRAS, MYC, TP53, CCND1, CDH1, CDKN2A and
CTNNB1. The strength of the connection between DKK2 and CTNNB1 was
the strongest based on the STRING database. Among these genes,
TP53, APC and PTEN are typical tumor suppressor genes (29-31).
From the analytical results of PPI network, we also found that DKK2
interacted with SMAD4, MAP3K1 and MAP3K11, which referred to
TGF-β/Smad4 and MAPK signaling pathways through CTNNB1.
Additionally, high frequency mutations in the tumor suppressor
mentioned genes above have been detected in breast cancer (32). These findings suggest that DKK2 may
play a role in multiple signaling pathways. The tumor suppressive
effect of DKK2 depends on the synergistic effect of the tumor
suppressor genes.
GO and KEGG pathway enrichment
analysis
To further explore the protein functions derived
from the PPI network, FunRich was used to analyze functional and
pathway enrichment. The top significant terms of GO enrichment
analysis in FunRich were shown in context. Among these biological
process, protein metabolism, transcription, apoptosis, regulation
of gene expression, cell communication, signal transduction and
regulation of nucleobase. Besides, lipid kinase, cell adhesion,
kinase binding, and transcript factor activity were the major
molecular function. These proteins were related to several cellular
components, including protein complex, cytoplasm, cytosol,
nucleoplasm and nucleus (Fig.
5B–D). The top 10 significant terms of pathway enrichment
analysis in FunRich were mTOR, PDGFR-β, EGF-receptor, Class I PI3K,
Arf6 trafficking, LKB1, regulation of CDC42 activity, CDC42,
integrin-linked kinase and AP-1 transcription factor network
(Fig. 5E). The detailed results
are not shown in the text and are available at https://github.com/shaoyoucheng.
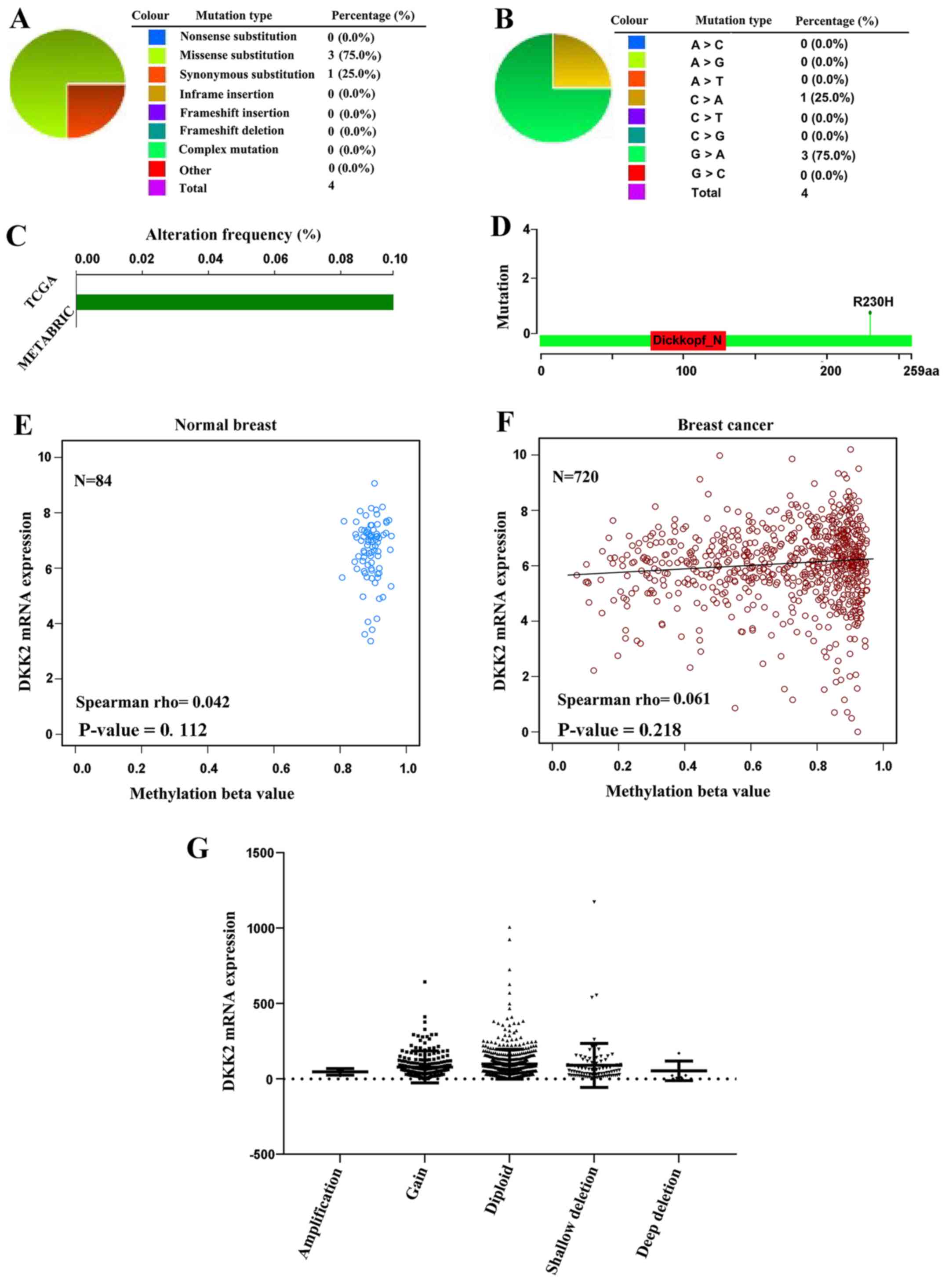
COSMIC analysis for DKK2 mutations and
cBioPortal analysis for alteration frequency of DKK2
The pie chart illustrated the information of
mutations of substitution missense, nonsense, synonymous, insertion
and deletion, which were generated by COSMIC. The substitution
missense rate was 75% and the substitution synonymous rate was 25%
of the mutant samples of breast cancer. G>A and C>A mutation
occupied 100% of the substitution mutation (Fig. 6A and B). The alteration frequency
of the DKK2 mutation in breast cancer was analyzed by BioPortal.
Less than 0.1% of the mutations in the patients with breast cancer
were screened (Fig. 6C). Moreover,
the event that genetic mutation for DKK2 results in nonsense
alteration in the 230th amino acid site was detected in breast
cancer (Fig. 6D).
mRNA expression with DNA methylation
analysis and copy number analysis
The scatter plot displayed that DKK2 methylation was
not associated with the DKK2 mRNA level in breast cancer by the
cBioPortal database (Fig. 6E and
F). In addition, the boxplot provided an overview of the DKK2
mRNA level associated with copy number of deep deletion, shallow
deletion, diploid, gain, and amplification, which was generated by
the cBioPortal database. Diploid was the cause for the decline in
the DKK2 mRNA level in breast cancer (Fig. 6G). On the whole, DKK2 copy number
alterations, particularly diploid, account for the elevated DKK2
level in breast cancer.
Discussion
DKK1 expression has been described to increase early
in the development of prostate cancer (33). However, in this study, we confirmed
a low DKK1 expression in the patients with grade 1 breast cancer.
Xu et al found that an elevated expression of DKK1 in
triple-negative cancers indicated a poor outcome for patients
(34). A relative increase in DKK1
expression has also been observed in ER-negative breast cancer
patients (35,36). Consistent with the findings of
previous studies, in this study, we also confirmed a high DKK1
expression in ER-negative breast cancer.
DKK3 has been reported to be frequently silenced, as
a valuable biomarker for breast cancer in the European population
(37,38). DKK3 methylation was previously
found in 78% of primary breast cancer tissues in an Asian
population (39). Patients
harboring DKK3 methylation have been shown to have a poor prognosis
in contrast to patients retaining an unmethylated DKK3 promoter
(37-39). In this study, we found that DKK3
expression was decreased with the progression of tumor stage. Veeck
et al found that the downregulation of DKK3 expression
predicted a poor OS and short RFS in breast cancer (37). However, the situation is more
complex when considering the subtypes of breast cancer. In this
study, we found that a high DKK3 expression was associated with a
poor Distant metastasis-free survival (DMFS) in ER-positive tumors,
ER-positive/LN-negative tumors, and Grade 3 tumors. However, a high
DKK3 expression was associated with a good RFS in both ER-negative
tumors and LN-positive tumors.
To the best of our knowledge, no previous studies
have examined the roles of DKK2 and DKK4 in breast cancer. As shown
in this study, we confirmed that DKK2 played significant and
various roles in different subtypes of breast cancer. However, we
did not find the expression of DKK4 transcripts and corresponding
outcomes in breast cancer (data not shown). The differential
expression of DKKs may be due to differences in tumor type, tumor
stage, or cellular subtypes of breast cancer.
In this study, we focused our attention on the
clinical prognostic value of DKKs in breast cancer subtypes
(PAM50-based). Our main finding was the different roles of DKK2 in
breast cancer and the subtypes. The overall trend was that a high
level of DKK2 indicated a poor survival of the Normal-like subtype,
while DKK2 was a good indicator for breast cancer (no
classification of breast cancer). The TCGA breast cancer dataset
(19) and METABRIC breast cancer
datasets (21,22) were used to validate the results of
expression profile of DKK2 from GOBO database in breast cancer
subtypes. We noted that the results shown in Fig. 4A and B were slightly inconsistent
with the results shown in Fig. 1A.
Furthermore, we found that the landscape of DKK2 expression in
breast cancer subtypes was consistent with that of GOBO, excluding
the Normal-like subtype. The limitation to a few samples for
normal-like subtypes in the TCGA dataset, may account for the
above-mentioned results. On the whole, our findings indicated that
DKK2 is a potential biomarker in the Normal-like subtype of breast
cancer.
PPI analysis suggested that DKK2 carries out its
functions through multiple signaling pathways, without being
confined to the Wnt/β-catenin pathway in breast cancer. Xiao et
al proposed that DKK2 secreted by tumor cells acts on cytotoxic
lymphocytes, inhibiting STAT5 signaling by impeding STAT5 nuclear
localization but independently of the Wnt/β-catenin pathway
(40). The results from the study
by Aravalli et al suggested that DKK2 was completely
silenced after MYC-induction in PICM-19-CSCs, suggesting that the
absence of DKK2 may be critical for inducing tumorigenesis
(41).
Therefore, it can be concluded that DKK2 plays a
role in multiple signaling pathways. In addition, we noticed that
DKK2 interacted with SMAD4, MAP3K1 and MAP3K11, which are involved
in the TGF-β/Smad4 and MAPK signaling pathways through CTNNB1, as
illustrated by the PPI network, respectively. Those findings
prompted that TGF-β/Smad4 pathway and MAPK pathway were altered
after DKK2 acting on CTNNB1, which was consistent with the findings
of a previous study (42).
Therefore, perhaps the following bold assumption can be made: DKK2
may play a role indirectly through the TGF-β/Smad4 and MAPK
pathways.
Furthermore, the genetic alterations in DKK2 in
breast cancer are mainly copy number variations and G>A
substitutions. However, we collected a low number of samples with
which to detect other breast cancer types. The prognostic roles of
DKKs in the different subtypes of breast cancer warrant further
investigations in the future. Experimental studies are required to
further explore the role and underlying mechanisms if action of
DKK2 in breast cancer.
Acknowledgments
Not applicable.
Funding
The present study was funded by National Natural
Scientific Foundation of China (no. 81572777), and Shenyang Science
and Technology Grant (17-230-9-41).
Availability of data and materials
The datasets generated and analyzed in the current
study are available at TCGA (https://cancergenome.nih.gov/) and cBioPortal for
Cancer Genomics (http://www.cbioportal.org/). Additional supplemental
results are available at https://github.com/shaoyoucheng.
Authors' contributions
YCS, XYX, PX and YW designed the study, analyzed the
bioinformatics data and drafted the manuscript. XYX, GQS, PX and
XCN collected the clinical data and tissue samples, and performed
the experiments and the statistical analysis on experiments. All
authors reviewed and approved the final manuscript.
Ethics approval and consent to
participate
Human breast cancer samples for our studies were
obtained from the Shengjing Hospital of China Medical University.
Informed written consent was obtained from all participants and the
study was approved by Ethics Committee of Shengjing Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen JG, Chen HZ, Zhu J, Yang YL, Zhang
YH, Huang PX, Chen YS, Zhu CY, Yang LP, Shen K, et al: Cancer
survival in patients from a hospital-based cancer registry, China.
J Cancer. 9:851–860. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Varghese C and Shin HR: Strengthening
cancer control in China. Lancet Oncol. 15:484–485. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song JL, Chen C, Yuan JP and Sun SR:
Progress in the clinical detection of heterogeneity in breast
cancer. Cancer Med. 5:3475–3488. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zardavas D, Irrthum A, Swanton C and
Piccart M: Clinical management of breast cancer heterogeneity. Nat
Rev Clin Oncol. 12:381–394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Parker JS, Mullins M, Cheang MC, Leung S,
Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al:
Supervised risk predictor of breast cancer based on intrinsic
subtypes. J Clin Oncol. 27:1160–1167. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tibshirani R, Hastie T, Narasimhan B and
Chu G: Diagnosis of multiple cancer types by shrunken centroids of
gene expression. Proc Natl Acad Sci USA. 99:6567–6572. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brott BK and Sokol SY: Regulation of
Wnt/LRP signaling by distinct domains of Dickkopf proteins. Mol
Cell Biol. 22:6100–6110. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shao YC, Wei Y, Liu JF and Xu XY: The role
of Dickkopf family in cancers: From Bench to Bedside. Am J Cancer
Res. 7:1754–1768. 2017.PubMed/NCBI
|
|
11
|
Aguilera O, Fraga MF, Ballestar E, Paz MF,
Herranz M, Espada J, García JM, Muñoz A, Esteller M and
González-Sancho JM: Epigenetic inactivation of the Wnt antagonist
DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene.
25:4116–4121. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu J, Zhang S, Gu L and Di W: Epigenetic
silencing of DKK2 and Wnt signal pathway components in human
ovarian carcinoma. Carcinogenesis. 33:2334–2343. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu XY, Xia P, Yu M, Nie XC, Yang X, Xing
YN, Liu YP, Takano Y and Zheng HC: The roles of REIC gene and its
encoding product in gastric carcinoma. Cell Cycle. 11:1414–1431.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hirata H, Hinoda Y, Majid S, Chen Y, Zaman
MS, Ueno K, Nakajima K, Tabatabai ZL, Ishii N and Dahiya R:
DICKKOPF-4 activates the noncanonical c-Jun-NH2 kinase signaling
pathway while inhibiting the Wnt-canonical pathway in human renal
cell carcinoma. Cancer. 117:1649–1660. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ringnér M, Fredlund E, Häkkinen J, Borg Å
and Staaf J: GOBO: Gene expression-based outcome for breast cancer
online. PLoS One. 6:e179112011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The Cancer Genome Atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.
|
|
19
|
Koboldt DC, Fulton RS, McLellan MD,
Schmidt H, Kalicki-Veizer J, McMichael JF, Fulton LL, Dooling DJ,
Ding L, Mardis ER, et al: Cancer Genome Atlas Network:
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar
|
|
20
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBio-Portal. Sci Signal. 6:pl12013. View Article : Google Scholar
|
|
21
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: METABRIC Group: The genomic and transcriptomic architecture of
2,000 breast tumours reveals novel subgroups. Nature. 486:346–352.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pereira B, Chin SF, Rueda OM, Vollan HK,
Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ,
et al: The somatic mutation profiles of 2,433 breast cancers
refines their genomic and transcriptomic landscapes. Nat Commun.
7:114792016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar
|
|
24
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar
|
|
25
|
Agapito G, Guzzi PH and Cannataro M:
Visualization of protein interaction networks: Problems and
solutions. BMC Bioinformatics. 14(Suppl 1): S12013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Benito-Martin A and Peinado H: FunRich
proteomics software analysis, let the fun begin! Proteomics.
15:2555–2556. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
The Gene Ontology: Consortium: Expansion
of the Gene Ontology knowledgebase and resources. Nucleic Acids
Res. 45:D331–D338. 2017. View Article : Google Scholar
|
|
28
|
Mu J, Hui T, Shao B, Li L, Du Z, Lu L, Ye
L, Li S, Li Q, Xiao Q, et al: Dickkopf-related protein 2 induces
G0/G1 arrest and apoptosis through suppressing Wnt/β-catenin
signaling and is frequently methylated in breast cancer.
Oncotarget. 8:39443–39459. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duffy MJ, Synnott NC, McGowan PM, Crown J,
O'Connor D and Gallagher WM: p53 as a target for the treatment of
cancer. Cancer Treat Rev. 40:1153–1160. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu M, Liu X, Xu Y, Zhu S and Gao Y: Co
expression of Axin and APC gene fragments inhibits colorectal
cancer cell growth via regulation of the Wnt signaling pathway. Mol
Med Rep. 16:3783–3790. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim DH, Suh J, Surh YJ and Na HK:
Regulation of the tumor suppressor PTEN by natural anticancer
compounds. Ann NY Acad Sci. 1401:136–149. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stephens PJ, Tarpey PS, Davies H, Van Loo
P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell
GR, et al: Oslo Breast Cancer Consortium (OSBREAC): The landscape
of cancer genes and mutational processes in breast cancer. Nature.
486:400–404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hall CL, Daignault SD, Shah RB, Pienta KJ
and Keller ET: Dickkopf-1 expression increases early in prostate
cancer development and decreases during progression from primary
tumor to metastasis. Prostate. 68:1396–1404. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu WH, Liu ZB, Yang C, Qin W and Shao ZM:
Expression of dickkopf-1 and beta-catenin related to the prognosis
of breast cancer patients with triple negative phenotype. PLoS One.
7:e376242012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bu G, Lu W, Liu CC, Selander K, Yoneda T,
Hall C, Keller ET and Li Y: Breast cancer-derived Dickkopf1
inhibits osteoblast differentiation and osteoprotegerin expression:
Implication for breast cancer osteolytic bone metastases. Int J
Cancer. 123:1034–1042. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rachner TD, Göbel A, Thiele S, Rauner M,
Benad-Mehner P, Hadji P, Bauer T, Muders MH, Baretton GB, Jakob F,
et al: Dickkopf-1 is regulated by the mevalonate pathway in breast
cancer. Breast Cancer Res. 16:R202014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Veeck J, Bektas N, Hartmann A, Kristiansen
G, Heindrichs U, Knüchel R and Dahl E: Wnt signalling in human
breast cancer: Expression of the putative Wnt inhibitor Dickkopf-3
(DKK3) is frequently suppressed by promoter hypermethylation in
mammary tumours. Breast Cancer Res. 10:R822008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kloten V, Becker B, Winner K, Schrauder
MG, Fasching PA, Anzeneder T, Veeck J, Hartmann A, Knüchel R and
Dahl E: Promoter hypermethylation of the tumor-suppressor genes
ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast
cancer screening. Breast Cancer Res. 15:R42013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xiang T, Li L, Yin X, Zhong L, Peng W, Qiu
Z, Ren G and Tao Q: Epigenetic silencing of the WNT antagonist
Dickkopf 3 disrupts normal Wnt/β-catenin signalling and apoptosis
regulation in breast cancer cells. J Cell Mol Med. 17:1236–1246.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiao Q, Wu J, Wang WJ, Chen S, Zheng Y, Yu
X, Meeth K, Sahraei M, Bothwell ALM, Chen L, et al: DKK2 imparts
tumor immunity evasion through β-catenin-independent suppression of
cytotoxic immune-cell activation. Nat Med. 24:262–270. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Aravalli RN, Talbot NC and Steer CJ: Gene
expression profiling of MYC-driven tumor signatures in porcine
liver stem cells by transcriptome sequencing. World J
Gastroenterol. 21:2011–2029. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Meng Q, Mongan M, Wang J and Xia Y:
Repression of MAP3K1 expression and JNK activity by canonical Wnt
signaling. Dev Biol. 440:129–136. 2018. View Article : Google Scholar : PubMed/NCBI
|