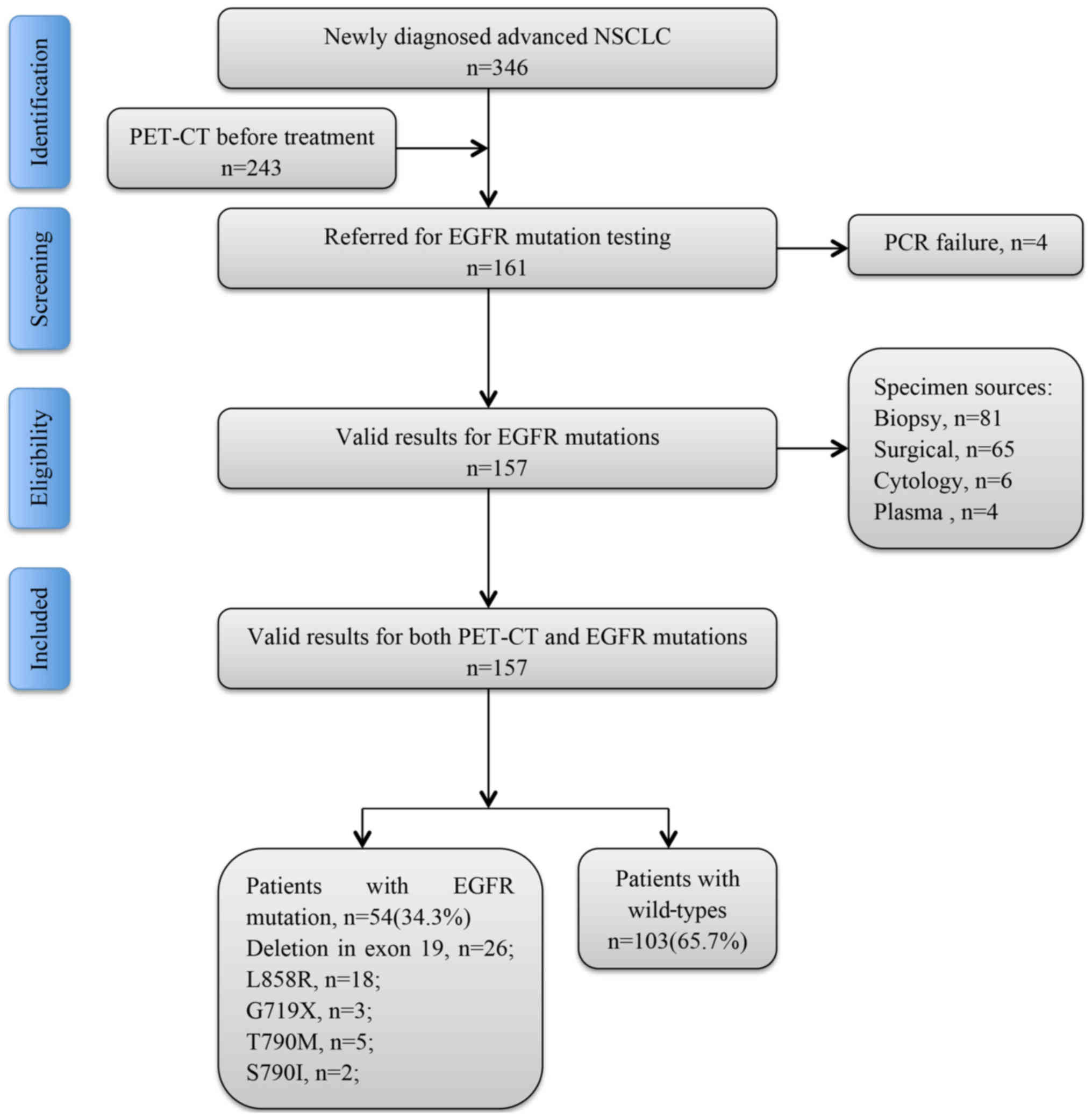
Introduction
Globally, lung cancer is the primary contributor to
cancer-associated mortality and the leading cause of mortality in
the majority of regions (1-4). An
estimated 222,500 novel lung cancer cases and 155,870 mortalities
were predicted to have occurred in 2017 in the USA (1). Of all patients with lung cancer,
those with non-small cell lung cancer (NSCLC) account for ~80%
(5). The identification and
investigation of genetic drivers such as epidermal growth factor
receptor (EGFR)-activating mutations, have contributed to a
gradual decrease in lung cancer-associated mortality (3,6,7).
EGFR is a member of a larger family of transmembrane
receptor tyrosine kinases (TKs) that activate cell proliferation
and survival (8). Mutations in the
TK domain of EGFR in NSCLC exhibit improved responses to
EGFR tyrosine kinase inhibitors (TKIs) such as gefitinib and
erlotinib (7), particularly exon
19 deletions and L858R in exon 21.
Previous studies have identified that female Asian
patients without a history of smoking and with adenocarcinoma
histology are more likely to exhibit EGFR mutations
(9). Therefore, validating the
EGFR genotype status in patients with NSCLC may help to
select those who will benefit from TKIs when making treatment
decisions. However, inaccessible tumor sites, insufficient tissues
for testing, heterogeneous tumors and a patient’s refusal to
undergo invasive detection all pose limitations to performing the
individual genotype test. Thus, developing non-invasive and
effective methods to help with identification of the status of the
EGFR gene is required.
[18F]fluorodeoxyglucose (FDG) positron
emission tomography (PET)-computed tomography (CT), which is based
on high glucose metabolism in lesions, serves an important function
in initial staging, evaluating the response following therapy and
radiation therapy planning during the management of NSCLC (10,11).
Therefore, as a non-invasive method, the quantification of glucose
metabolism using FDG-PET is one way to predict EGFR
mutations. The standard uptake value maximum (SUVmax), a
metabolic parameter from PET for FDG uptake, is associated with
prognosis in NSCLC and previous studies revealed that patients with
NSCLC with a low SUVmax for the primary lesion tend to
have better outcomes (12,13), indicating that a low
SUVmax may be associated with EGFR gene
mutations. However, in clinical practice, studies that aim to
reveal the FDG uptake and EGFR mutation status are
controversial, and the potential mechanisms by which EGFR
mutations alter FDG uptake remain largely unknown; therefore,
further clinical studies and investigations of the underlying
molecular mechanisms should be performed.
Glucose transporter 1 (GLUT1) serves crucial
functions in FDG uptake (14,15);
furthermore, GLUT1 expression can be altered by dysregulated
reactive oxygen species (ROS) activity (16,17),
in which NADPH oxidase 4 (NOX4) is primarily responsible for ROS
production (18). Considering the
aforementioned studies, we hypothesized that EGFR mutations
may regulate FDG uptake via the NOX4/ROS/GLUT1 axis in NSCLC.
In the present study, the association between
EGFR mutations and SUVmax was investigated, the
receiver operating characteristic (ROC) curve was analyzed to
identify the optimum cut-off value for SUVmax in
predicting EGFR mutation, GLUT1 expression and ROS activity
were determined in the A549 and PC-9 (EGFR mutation, 19del)
cell lines, and NOX4 mRNA and protein expression were investigated
to test the hypothesis. Subjects were recruited and enrolled in the
present study, and subjected to a battery of tests that included
FDG-PET-CT scanning and EGFR mutation testing. The study
flow chart is presented in Fig.
1.
Materials and methods
Patients and diagnosis
In total, 157 patients (median age 65.8 years;
range, 48-81 years) with NSCLC who were diagnosed at the Department
of Pathology of The Third Affiliated Hospital of Kunming Medical
University (Kunming, China) from June 2015 to October 2017 were
enrolled in the present study. All patients fulfilled the following
entry criteria: i) The diagnosis was made histologically, and the
patients underwent EGFR gene testing; ii) PET-CT was
performed prior to any therapy; iii) complete clinical information
was obtained; iv) histopathology was reviewed at Yunnan Cancer
Hospital (Yunnan, China); and v) written informed consent was
obtained from the patients. The study protocol was approved by the
Ethics Committee of The Third Affiliated Hospital of Kunming
Medical University. All procedures performed in the present study
that involved human participants were with the approval of the
Institutional Review Board of The Third Affiliated Hospital of
Kunming Medical University and in accordance with the 1964
Declaration of Helsinki and its later amendments or comparable
ethical standards.
EGFR mutation analysis
An AmoyDx® EGFR 29 Mutations
Detection kit (Amoy Diagnostics Co., Ltd., Xiamen, China) was used
to detect the EGFR mutation in DNA extracted from tissue and
plasma samples using a Qiagen DNA mini-kit (Qiagen GmbH, Hilden,
Germany), according to the manufacturer’s protocol. The kit
methodology is based on amplification refractory mutation system
(ARMS) technology, which was used to detect 29 mutations in exons
18 to 21 of the EGFR gene (19) All ARMS primer pairs (AmoyDx,
Super-ARMS, 19 del, forward, 5ʹ-GTTAAAATTCCCGTCGCTATCAAGACATCT-3ʹ,
and reverse, 5ʹ-CACAGCAAAGCAGAAACT CACAT-3ʹ; L858R, forward,
5ʹ-GCAGCATGTCAAGATCACAGATTTTGGGCG-3ʹ, and reverse,
5ʹ-GTCAGGAAAATGCTGGCTGACCTAAAG-3ʹ; T790M, forward,
5ʹ-CTCACCTCCACCGTGCARCTCATCAT-3ʹ, and reverse,
5ʹ-CAATATTGTCTTTGTGTTCCCGGACA-3ʹ; G719X, forward,
5ʹ-CTCACCTCCACCGTGCARCTCATCAT-3ʹ, and reverse,
5ʹ-CCGTGCCGAACGCACCGGAGCA-3ʹ; S790I, forward,
5ʹ-AGCGTGGACAACCCCCACCAC-3ʹ, and reverse,
5ʹ-CCGTGCCGAACGCACCGGAGCA-3ʹ) were used for polymerase chain
reaction (PCR), with the following criteria: Concentration of 1
mmol/l, control reaction primers at a concentration of 0.1 mmol/l.
PCR was performed with denaturation at 94°C for 30 sec, 30 cycles
of 95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec, and
another 72°C for 6 min.
Interpretation and image analysis of
FDG-PET-CT scans
FDG-PET-CT scan images were acquired in the
department of PET-CT Center of Yunnan Cancer Hospital using the
syngo. via platform (Siemens Healthineers, Erlangen, Germany)
(slice thickness, 3-5 mm). The patients fasted for a minimum of 6
h, an FDG dose of 12 mCi was administered, and the patients were
scanned from the skull base to the mid-thigh using multiple bed
positions (two or three bed positions; acquisition time, 2 min/bed
position) 1 h after injection. CT-attenuated data were
reconstructed using ordered subset expectation maximization for the
two scanner sites. Representative images are presented in Fig. 2A and B. The images were reviewed by
two board-certified nuclear medicine physicians with 2 and 10 years
of experience, respectively. A syngo MultiModality WorkPlace system
(Siemens Healthineers) was used to select and measure structures
throughout the body using the region-of-interest (ROI) tool within
the software. Circular ROIs with a diameter of 10 mm were drawn on
transaxial FDG-PET-CT images using the fusion CT scan as an
anatomical guide.
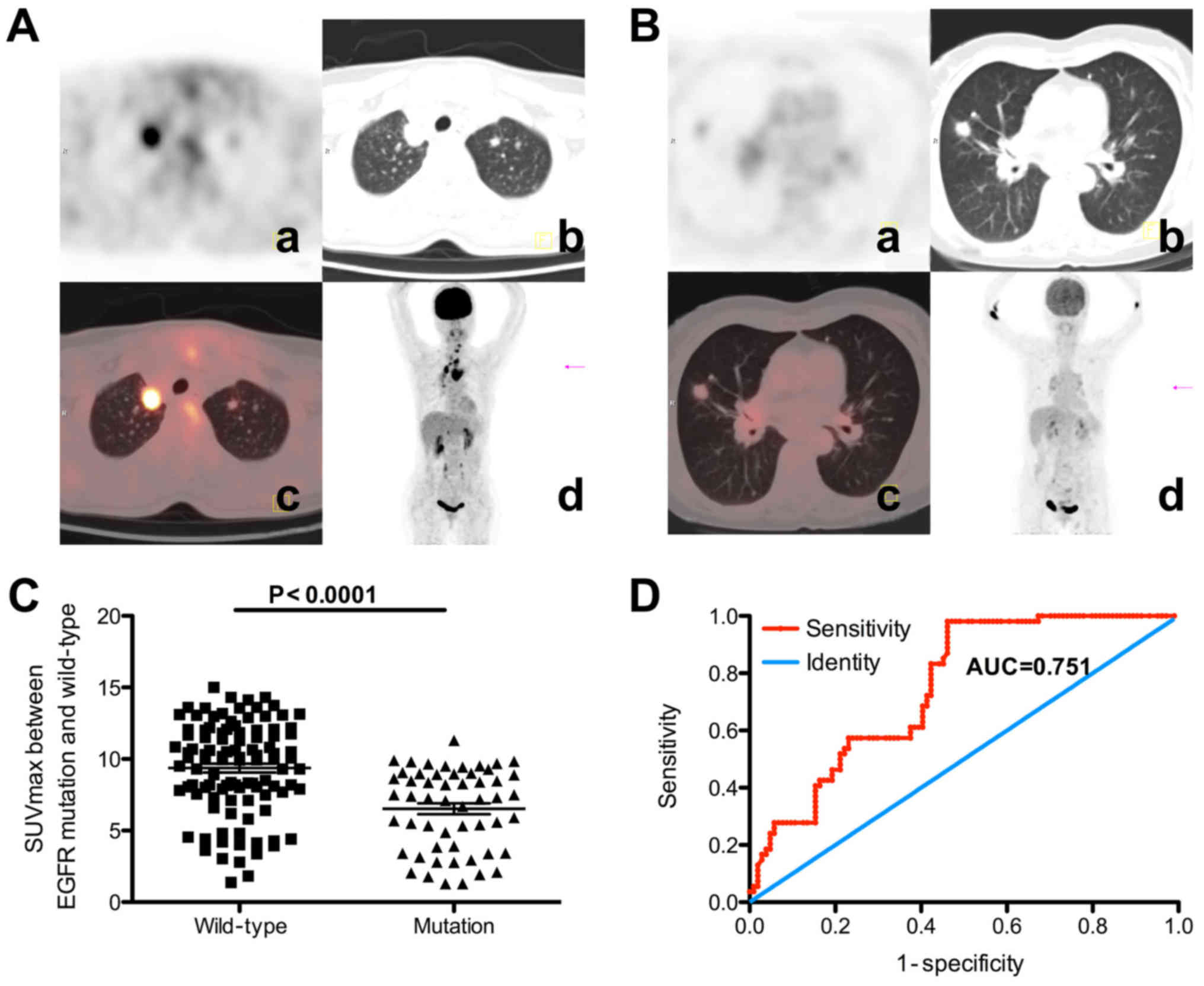 | Figure 2Representative FDG-PET-CT images and
SUVmax values for EGFR mutation and wild-type
patients with NSCLC. (A) A 53-year-old man underwent a PET-CT scan
to identify a nodule in the upper lobe of the right lung, which was
diagnosed pathologically as adenocarcinoma, and EGFR
detection revealed no positive mutation. Increased FDG uptake was
detected in the lesion, with an SUVmax of 11.7. (a) PET
portion of the PET-CT (transaxial); (b) CT portion of the PET-CT
(transaxial); (c) combined PET-CT images (transaxial); (d) MIP. (B)
A 64-year-old woman underwent a PET-CT test to identify a mass in
the upper lobe of the right lung, which was diagnosed
pathologically as adenocarcinoma, and EGFR detection
revealed an exon 19 deletion. Slight FDG uptake was observed in the
mass, with an SUVmax of 3.1. (a) PET portion of the
PET-CT (transaxial); (b) CT portion of the PET-CT (transaxial); (c)
combined PET-CT images (transaxial); (d) MIP. (C) Association
between SUVmax and EGFR mutation status. The
SUVmax was significantly lower in EGFR
mutation-positive patients (mean, 6.52±0.38) compared with in
wild-type EGFR patients (mean, 9.37±0.31; P<0.001). (D)
Receiver operating characteristic curve analysis of
SUVmax cut-off value. The SUVmax cut-off
point of 9.92 can best discriminate the EGFR mutation
status, with an AUC of 0.75 (95% confidence interval, 0.68-0.83).
The patients were divided into two groups according to this
threshold and it was identified that EGFR mutations were
more frequent in patients with a low SUVmax (≤9.92)
compared with in patients with a high SUVmax (>9.92)
(45.3 vs. 24.4%; P=0.007). FDG,
[18F]fluoro-2-deoxyglucose; PET, positron emission
tomography; CT, computer tomography; SUVmax, maximum
standardized uptake values; EGFR, epidermal growth factor
receptor; NSCLC, non-small cell lung cancer; MIP, maximum intensity
projection; AUC, area under the curve. |
Cell culture
Human NSCLC A549 and NCI-H1975 cells were purchased
from the American Type Culture Center (Manassas, VA, USA). PC-9
cells were purchased from RIKEN Cell Bank (Tsukuba, Japan) and is a
19del-positive cell line, whereas A549 is a cell line expressing
wild-type EGFR, and the NCI-H1975 cell line harbors the
L858R and T790M substitution EGFR mutations. NCI-H1975 and
PC-9 cells were grown in RPMI-1640 medium (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Hyclone; GE Healthcare, Logan, UT, USA), 2 mM
L-glutamine and 1% penicillin/streptomycin. A549 cells were
cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS, 2 mM L-glutamine and
1% penicillin/streptomycin. All cells were maintained and
propagated as monolayer cultures at 37°C in a humidified 5%
CO2 incubator.
NOX4 mRNA determination
Total RNA was extracted from A549 and PC-9 cells
using the TRIzol® reagent (Thermo Fisher Scientific,
Inc.), and was reverse-transcribed using a SuperScript II Reverse
Transcriptase kit (Takara Biotechnology Co., Ltd., Dalian, China),
according to the manufacturer’s protocol. Quantitative PCR (qPCR)
was performed using a SYBR Green Supermix kit (Takara Biotechnology
Co., Ltd.) and the ABI 7300 detection system (Thermo Fisher
Scientific, Inc.). Blank controls with no cDNA templates were
included to rule out contamination. The specificity of the PCR
product was confirmed by melting curve analysis and gel
electrophoresis. All gene expression levels were normalized to that
of the housekeeping gene U6. Relative expression levels of the
target gene normalized to U6 were calculated using the
2‐ΔΔCq method (20).
Each reaction was performed independently at least three times. The
following primer pairs were used: NOX4 primer set,
5ʹ-TGTTGGGCCTAGGATTGTGTT-3ʹ (forward) and
5ʹ-AGGGACCTTCTGTGATCCTCG-3ʹ (reverse); U6 primer set,
5ʹ-CTCGCTTCGGCAGCACA-3ʹ (forward) and 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ
(reverse). PCR was performed using the following parameters: 95°C
for 5 min, 30 cycles of 94°C for 30 sec, 58°C for 30 sec and 72°C
for 30 sec, and 72°C for 5 min.
Western blot analysis
The cells were solubilized in ice-cold
radioimmunoprecipitation assay lysis buffer. Amounts of 25 µg
protein (determined using a Bicinchoninic Acid Protein assay kit
from Abcam, Cambridge, UK) from the cytosolic fraction were
separated by SDS-PAGE (10% gel) and transferred onto a
polyvinylidene difluoride membrane. The membrane was incubated with
5% skimmed milk in Tris-buffered saline containing 0.2% Tween-20 at
37°C for 2 h. The membrane was then incubated with rabbit anti-NOX4
(cat. no. ab79971; 1:1,000 dilution) and mouse anti-β-actin (cat.
no. ab8226; 1:2,000 dilution) primary antibodies (both from Abcam)
at room temperature for 2 h. Following washing four times with PBS
containing 0.2% Tween-20 (PBST) each for 10 min, the membrane was
incubated with goat anti-rabbit (cat. no. sc-2030; 1:1,500
dilution) and goat anti-mouse (cat. no. sc-2005; 1:2,000 dilution)
secondary antibodies (Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) at 4°C overnight. Following washing four times with PBST each
for 10 min, proteins recognized by the antibody were visualized
with the Luminata Forte Western Blotting Substrate (EMD Millipore,
Billerica, MA, USA), according to the manufacturer’s protocol.
Image-Pro Plus software (version 6.0) was used to analyze the
relative protein expression, represented as the density ratio
against β-actin, which was used as an internal reference.
ROS detection
Intracellular ROS levels were determined using the
oxidative-sensitive fluorescent probe dihydroethidium (DHE;
Molecular Probes; Thermo Fisher Scientific, Inc.), as described
previously (21) with certain
modifications. Briefly, A549, PC-9 and NCI-H1975 cells
(2.5×105) in 6-well plates were incubated with 4 M DHE
at 37°C for 45 min. The cells were harvested and washed with PBS.
The fluorescence from oxidized DHE was detected at a wavelength of
630 nm and fluorescence images were captured using an Olympus BX51
fluorescence microscope (Olympus Corporation, Tokyo, Japan).
Statistical analysis
Categorical covariates were analyzed using Pearson’s
χ2 test or Fisher’s exact test as appropriate, and
continuous covariates were analyzed using Student’s t-test or
analysis of variance, as appropriate. A ROC curve was generated to
determine a cut-off for the SUVmax of the primary tumor.
Multivariate logistic regression analysis was performed to test the
variables that yielded predictors of EGFR mutations. The
area under the curve (AUC) was used for the predictive value.
P<0.05 was considered to indicate a statistically significant
difference. GraphPad Prism (version 6.0; GraphPad Software, Inc.,
La Jolla, CA, USA) was used for the analysis.
Results
Clinical features and EGFR mutations
The baseline characteristics of the patients are
listed in Table I. There were 157
patients (84 males and 73 females) that met the eligibility
criteria. Of those, 54 patients (34.3%) were EGFR
mutation-positive. Exon 19 deletion and L858R in exon 21 were the
most common mutations, accounting for 48% (26 patients, including 3
combined mutation types) and 33.3% (18 patients, all single
mutation), respectively. Other mutation types were single G719X (3
patients, 5.5%), single T790M (5 patients, 9.2%), single S768I (2
patients, 3.7%) and combined 19del+T790M (3 patients, 5.5%). The
EGFR mutations were more frequent in female patients
compared with in male patients (42.3 vs. 26.6%; P=0.045). The
median age was 58.3 years, and 96 patients (61.1%) had a history of
smoking. EGFR mutations were more frequent in non-smokers
compared with in smokers (49.1 vs. 19.1%; P=0.006). There were 144
patients (91.7%) with adenocarcinoma and the remaining 13 patients
were without adenocarcinoma (8.3%). EGFR mutation status was
more frequent in patients with adenocarcinoma compared with
patients without adenocarcinoma (36.8 vs. 7.7%; P=0.036). In
addition, patients harboring EGFR mutations had a lower
SUVmax compared with patients with wild-type EGFR
(63 vs. 40%) (Table I).
 | Table IEGFR mutation status among
various clinical characteristics. |
Table I
EGFR mutation status among
various clinical characteristics.
| Clinical
characteristic | EGFR status
| χ2 | P-value |
|---|
| Mutation
(n=54) | Wild-type
(n=103) |
|---|
| Age, years | | | 0.008 | 1.000 |
| ≤60 | 30 | 58 | | |
| >60 | 24 | 45 | | |
| Sex | | | 4.301 | 0.045 |
| Male | 33 | 45 | | |
| Female | 21 | 58 | | |
| Histopathology | | | 4.479 | 0.036 |
|
Adenocarcinoma | 53 | 91 | | |
|
Non-adenocarcinoma | 1 | 12 | | |
| Diameter, cm | | | 0.006 | 1.000 |
| ≤3 | 25 | 47 | | |
| >3 | 29 | 56 | | |
| AJCC stage | | | 1.205 | 0.752 |
| I | 12 | 20 | | |
| II | 12 | 29 | | |
| III | 18 | 28 | | |
| IV | 12 | 26 | | |
| Smoking status | | | 7.568 | 0.006 |
| Ever | 13 | 55 | | |
| Never | 41 | 48 | | |
| Location | | | 0.057 | 0.866 |
| Left | 32 | 59 | | |
| Right | 22 | 44 | | |
| Brain
metastasis | | | 0.656 | 0.498 |
| Yes | 21 | 47 | | |
| No | 33 | 56 | | |
| SUVmax
of tumor | | | 42.253
<0.001 | |
| ≤9.92 | 53 | 47 | | |
| >9.92 | 1 | 56 | | |
Association of SUVmax and EGFR
mutations
Using χ2 analysis, the EGFR
mutation status was identified to be significantly associated with
sex, smoking status, pathological type and the SUVmax of
the primary tumor (Table I). The
potential association between SUVmax and EGFR
mutation was investigated, and it was identified that the
SUVmax was significantly lower in patients with
EGFR mutations (mean, 6.52±0.38) compared with that in
patients with wild-type EGFR (mean, 9.37±0.31; P<0.001)
(Fig. 2C). ROC curve analysis
revealed an SUVmax cut-off point of 7.8 (Table I), with an AUC of 0.75 (95%
confidence interval, 0.68-0.83; Fig.
2D). Using χ2 analysis, EGFR mutation status
was identified to be significantly associated with sex, smoking
status, tumor histopathology and SUVmax of the primary
tumor (Table I). Using
multivariate analysis, smoking status (never-smoking),
histopathology (adenocarcinoma) and SUVmax (≤9.91) were
the statistically significant predictors of EGFR mutations
(Table II).
 | Table IIMultivariate analysis of potential
predictive factors for epidermal growth factor receptor gene
mutation. |
Table II
Multivariate analysis of potential
predictive factors for epidermal growth factor receptor gene
mutation.
| Predictive
factor | Univariate analysis
OR (95% CI) | P-value | Multivariate
analysis OR (95% CI) | P-value |
|---|
| Age | 0.97
(0.50-1.88) | 0.93 | | |
| Sex | 2.03
(1.04-3.96) | 0.04 | 1.30
(0.55-3.11) | 0.55 |
| Histopathology | 6.99
(0.88-55.27) | 0.03 | 11.87
(1.37-102.86) | 0.025 |
| Diameter | 1.03
(0.53-1.99) | 0.94 | | |
| AJCC stage | 0.97
(0.63-1.48) | 0.75 | | |
| Smoking status | 2.75
(1.32-5.74) | 0.006 | 3.31
(1.29-8.50) | 0.009 |
| Location | 1.09
(0.56-2.12) | 0.81 | | |
| Brain
metastasis | 0.76
(0.39-1.48) | 0.42 | | |
| SUVmax
≤9.92 | 63.15
(8.41-474.14) | <0.001 | 73.24
(9.52-563.63) | <0.001 |
Patients were divided into two groups according to
this threshold and it was identified that EGFR mutations were more
frequent in patients with a low SUVmax (≤9.92) compared
with in patients with a high SUVmax (>9.92) (53 vs.
1.8%; P<0.001).
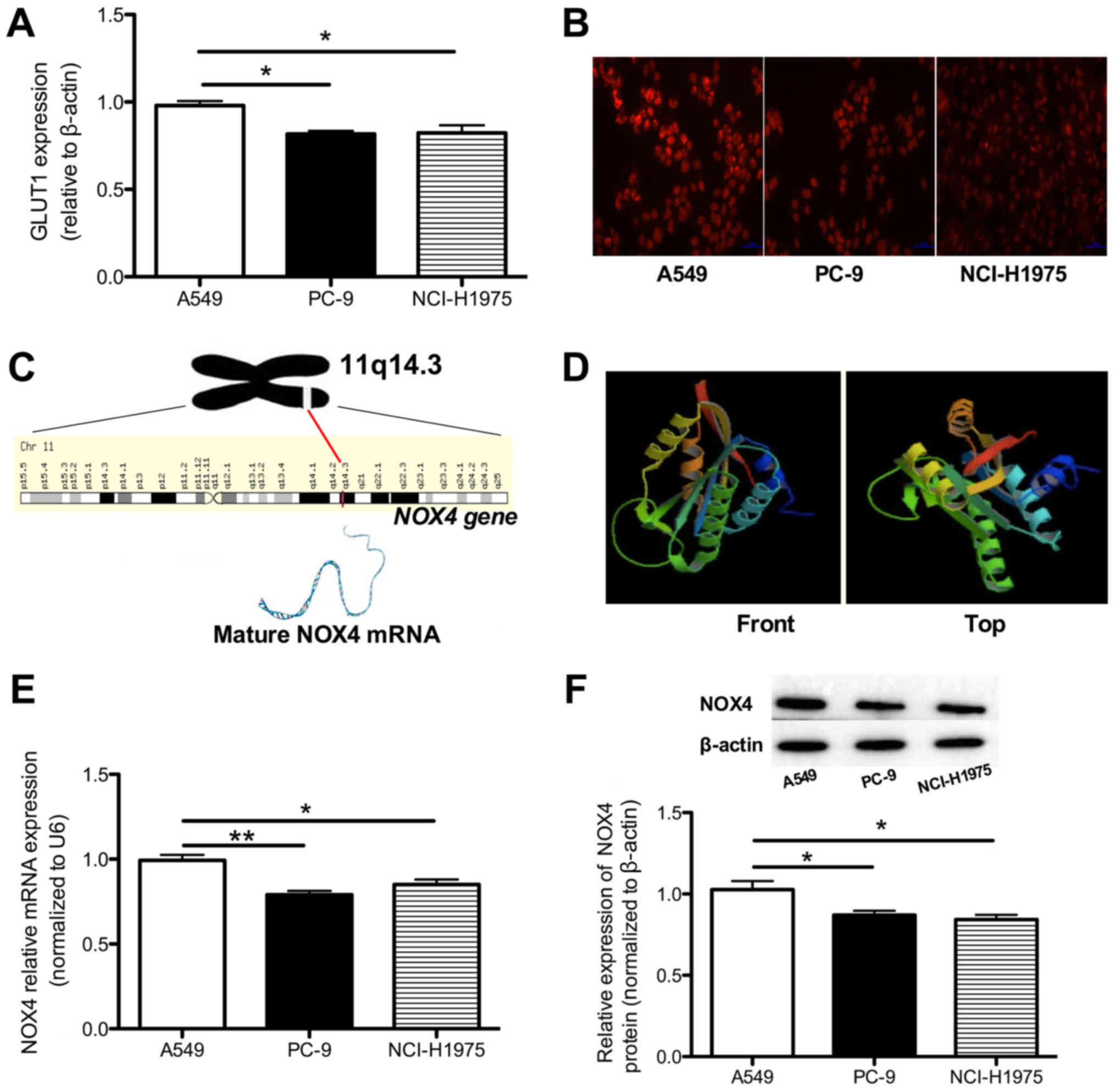
GLUT1 expression is downregulated in EGFR
mutated cell lines
Since GLUT1 has been investigated as an important
regulator of glucose transport, we hypothesized that the decreased
SUVmax associated with EGFR mutations may be
caused by downregulated GLUT1 expression. RT-qPCR revealed that
GLUT1 mRNA was decreased in the PC-9 and NCI-H1975 cell
lines compared with in the A549 cell line (0.82±0.07 and 0.72±0.04
vs. 0.98±0.04; P<0.05; Fig.
3A), indicating that decreased GLUT1 may be involved in the
downregulated FDG uptake in patients with an EGFR mutated
status.
Decreased ROS activity is detected in the
PC-9 cell lines
Previous studies have identified that intracellular
ROS serve important functions in regulating GLUT1 expression. To
determine whether the different GLUT1 expression levels in A549,
PC-9 and NCI-H1975 cells are influenced by ROS levels, the
intracellular ROS level was determined in A549, PC-9 and NCI-H1975
cells. As presented in Fig. 3B, a
marked decrease in the intracellular concentration of ROS was
identified in PC-9 and NCI-H1975 cells, which confirmed our
hypothesis.
NOX4 mRNA and protein levels are
decreased in EGFR-mutated cell lines
NOX4 is a gene that maps to the 11q14.3
region and its sequence has been strictly conserved throughout
evolution. The NOX4 gene consists of 29 exons, and the NOX4
protein consists of 578 amino acids. This gene also encodes a
member of the NOX4 family of enzymes that functions as the
catalytic subunit of the NADPH oxidase complex (Fig. 3C and D). Previous studies have also
identified that NOX4 serves crucial functions in ROS production
(18,22). To investigate whether the altered
ROS activity was influenced by the NOX4 molecule, mRNA and protein
expression levels of NOX4 were determined in the A549, PC-9 and
NCI-H1975 cell lines. The NOX4 mRNA was decreased by 20% in
PC-9 (P<0.01) and by 14% in NCI-H1975 (P<0.05) cells,
respectively (Fig. 3E), whereas
the protein expression decreased by 13 and 16% in PC-9 and
NCI-H1975 cells, respectively (both P<0.05), compared with the
A549 cell line (Fig. 3F).
Discussion
In the present study, the association between
SUVmax and EGFR mutation status was investigated
in patients with NSCLC. The results revealed that patients who
harbored an EGFR mutation exhibited decreased
SUVmax values, and further studies revealed that the
EGFR mutation alters the SUVmax partially via the
NOX4/ROS/GLUT1 axis.
The aims of the present study were as follows: i) To
determine whether tumor metabolism can add significant value for
predicting EGFR gene mutation; and ii) to investigate the
molecular mechanisms by which lung lesions alter the metabolic
pathway.
The selection of a suitable therapeutic strategy for
a patient suffering from lung cancer is based on the gene status,
particularly EGFR. In 2009, Lara-Guerra et al
(23) carried out a Phase II study
that included 31 patients clinically diagnosed as stage I NSCLC,
who received pre-operative gefitinib. The results indicated that
tumor shrinkage was frequently seen in women who had never smoked,
and the EGFR mutation was the strongest predictor of
response. Apart from stage I patients, gefitinib is still useful in
patients with stage III/IV NSCLC (3 achieved complete response, 13
exhibited partial response, 3 had stable disease and 2 were
discontinued for side effects among the total 21 patients)
(24). All these studies indicate
the urgent requirement to validate the gene mutation, and a less
invasive test method is desirable. Although individual gene
detection has been recommended for advanced NSCLC, certain problems
(including tumor inaccessibility, insufficient sample tissue for
detection and unwillingness to perform invasive detection) have
hindered this potential benefit for patients with advanced NSCLC
(25). Consequently, a
non-invasive strategy for predicting EGFR gene mutation
status is advantageous, and the SUVmax, which represents
the most active metabolic location within the lesion, has been used
as the most convenient metabolic parameter in malignant diseases
including lung cancer. However, the association between EGFR
mutation and SUVmax differs markedly among studies, and
the data from previous association studies are summarized in
Table III. These differences are
observed because, first, the SUVmax, a semi-quantitative
index, varies with different PET scanners, fasting durations,
plasma levels and region of interest parameters, and, secondly,
different studies enrolled various sample sizes and disparate
pathology types, which may also contribute to variation. A
systematic meta-analysis should be performed to evaluate these
results (26). The results of the
present study indicated that never-smoking, female and lower
SUVmax were the most significant predictive factors for
the presence of the EGFR mutation, in accordance with
previous studies (26,27). Using a patient’s
clinicopathological and imaging data, which represents the
non-invasive examination, to diagnose EGFR mutation status
and other mutations is of marked importance. On the basis of the
results of the present study, with an SUVmax cut-off
value of 9.92, the sensitivity and specificity for our prediction
model were 98.15 and 53.85%, respectively.
 | Table IIISummary of published data on the
association between EGFR mutation and
[18F]fluoro-2-deoxyglucose uptake. |
Table III
Summary of published data on the
association between EGFR mutation and
[18F]fluoro-2-deoxyglucose uptake.
| Author, year | Primary
results | Pathology | No. of
patients | SUVmax
in EGFR mutation-positive | Ref. |
|---|
| Minamimoto et
al, 2017 | Lower
SUVmax was predictive for EGFR mutation | ADC | 131 | 4.2±3.8 | (27) |
| Liu et al,
2017 | No association
between SUVmax and EGFR mutation | ADC and others | 87 | Not shown | (25) |
| Takamochi et
al, 2017 | EGFR
mutations were more frequent with lower SUVmax | ADC | 734 | Median
SUVmax was 2.7 | (28) |
| Caicedo et
al, 2014 | No significant
differences were observed in SUVmax between
EGFR-positive and wild-type | ADC | 102 | Median
SUVmax was 5.7 | (29) |
| Yoshida et
al, 2016 | Lower levels of
SUVmax associated with T790M status | ADC | 34 | Median
SUVmax and SUVmean were 7.26 and 4.57,
respectively | (30) |
| Lee et al,
2015 | None of the
SUV-derived variables was significantly associated with EGFR
mutation | ADC and SCC | 206 | Not shown | (31) |
| Cho et al,
2016 | Lower
SUVmax was associated with EGFR mutation | ADC and SCC | 61 | SUVmax
9.6 exhibited highest sensitivity for EGFR mutation | (32) |
| Ko et al,
2014 | Patients with
higher SUVmax were more likely to exhibit EGFR
mutations | ADC | 132 | SUVmax
≥6 | (33) |
| Putora et
al, 2013 | No association
between SUVmax and EGFR status | ADC | 28 | SUVmax
10.7 vs. 9.9 in EGFR-positive and wild-type, respectively | (34) |
On the basis of the result that decreased FDG uptake
was identified in patients harboring an EGFR mutation
(9.37±0.31 vs. 6.52±0.38, wild-type vs. mutation), the ROC curve
was first analyzed, and it was identified that the
SUVmax cut-off point was 9.92 and the AUC was 0.75 (95%
confidence interval, 0.68-0.83). Next, we hypothesized that GLUT1,
which serves important functions in transporting glucose and is
expressed during all stages of embryonic development (35), may function as a key molecule in
regulating FDG uptake. Western blotting revealed that GLUT1
decreased markedly in PC-9 and NCI-H1975 cells compared with in
A549 cells, indicating that EGFR mutation status may
regulate FDG uptake by altering GLUT1 expression. Previous studies
have identified that GLUT1 expression may be regulated by ROS in
disparate pathways. Under normal conditions, ROS can be produced as
a product of normal mitochondrial energy metabolism, and slightly
increased ROS functions as a molecular signal to activate various
signaling pathways including glucose uptake. However, a
persistently high ROS level may reverse the traditional signaling
pathway (36). In the present
study, the ROS level was determined using DHE and it was revealed
that decreased ROS activity was detected in PC-9 and NCI-H1975 cell
lines, which is consistent with previous studies. Fiorentini et
al (37) identified that
decreasing ROS activity by adding the antioxidant EUK-134
downregulated total GLUT1 expression, partially indicating a
positive correlation between ROS activity and GLUT1 expression. The
dysregulation of the redox balance in cancer cells exerts crucial
functions in tumor development and the response to anticancer
therapies (38). Kawano et
al (39) transfected 293T
cells with a vector expressing an Ex19del mutant of human
EGFR and identified a marked increase in the intracellular
concentration of ROS, indicating a potential association between
EGFR mutation and ROS activity. In the present study, ROS
activity was also determined, and it was identified that PC-9 cells
and NCI-H1975 cells expressed lower ROS levels.
Previous studies have also identified that NOX4
serves crucial functions in ROS production (18,22).
NOX4 is a gene that maps to the 11q14.3 region, and its sequence
has been strictly conserved throughout evolution. The NOX4
gene consists of 29 exons, and the NOX4 protein consists of 578
amino acids. This gene encodes a member of the NOX family of
enzymes that functions as the catalytic subunit of the NADPH
oxidase complex. The encoded protein is localized to non-phagocytic
cells where it acts as an oxygen sensor and catalyzes the reduction
of molecular oxygen to various ROS. The ROS generated by this
protein have been implicated in numerous biological functions
including signal transduction, cell differentiation and tumor cell
growth (40,41). Furthermore, Prata et al
(16) identified that NOX4-derived
ROS could maintain a high glucose uptake rate by upregulating GLUT1
in a leukemic cell line (16). In
the present study, it was identified that NOX4 mRNA and protein
levels were significantly decreased in PC-9 and NCI-H1975 cells,
compared with in A549 cells, suggesting an underlying molecular
mechanism by which ROS activity is decreased. Previous studies have
identified that increased ROS activates hypoxia-inducible factor α
(HIF-α) and bind to HIF-α-response elements in the promoter regions
of target genes (including GLUT1), thereby increasing GLUT1
mRNA and protein levels (42,43).
Conversely, in patients with NSCLC harboring an EGFR
mutation, inhibited ROS activity may be responsible for the
downregulated GLUT1 protein level (Fig. 4). Indeed, it has been identified
previously that NOX4 is essential for EGFR TKI activity. Orcutt
et al (44) revealed that
the cytotoxicity of erlotinib, an EGFR TKI, was mediated by
induction of oxidative stress by inducing the expression of NOX4 in
human head and neck cancer. Sobhakumari et al (45) also revealed that erlotinib
increased NOX4 mRNA and protein expression by increasing its
promoter activity and mRNA stability in FaDu cells, which
potentially implied that the primary NOX4 expression is not enough
for erlotinib function and the relatively decreased NOX4 expression
possibly be a trigger which activates the erlotinib activity.
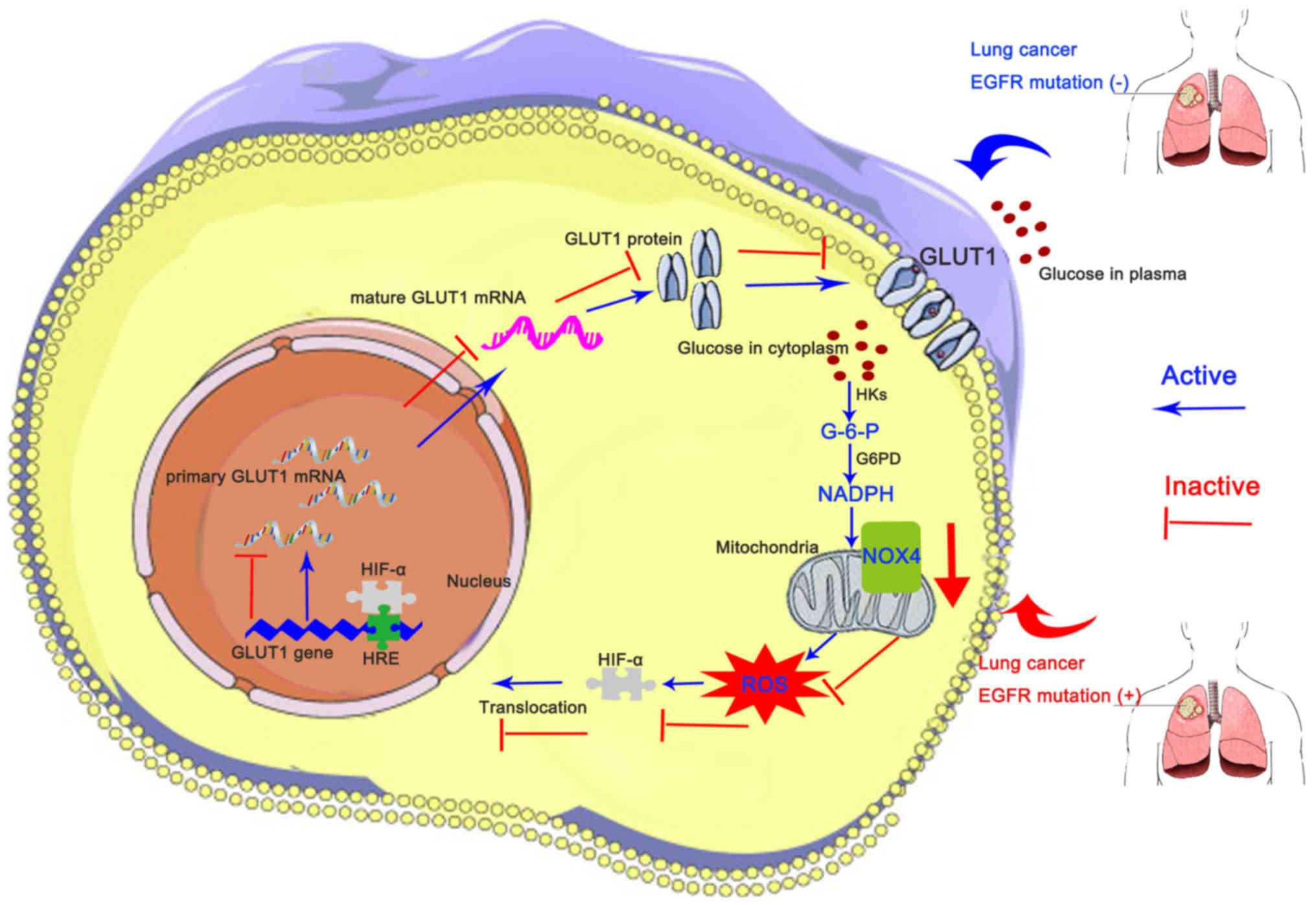 | Figure 4EGFR mutation alters FDG
uptake partially via the NOX4/ROS/GLUT1 axis. In patients with
EGFR mutation, NOX4 mRNA and protein levels are
downregulated, leading to decreased ROS activity, which inhibits
HIF-α translocation into the nucleus, resulting in decreased GLUT1
mRNA and protein expression and thereby hindering FDG uptake
(decreased maximum standardized uptake value). EGFR,
epidermal growth factor; FDG,
[18F]fluoro-2-deoxyglucose; NOX4, NAPDH oxidase 4; ROS,
reactive oxygen species; GLUT1, glucose transporter 1; HIF-α,
hypoxia-inducible factor α; HK, hexose kinase; G-6-P, glucose
6-phosphate; G6PD, glucose-6-phosphate dehydrogenase; HRE,
HIF-α-response element. |
The limitations of the present study should be
clarified. First, the study was designed retrospectively, with a
relatively small size (previous studies have ranged in size between
34 and 734 patients). With the accumulation of these small-sample
studies, a relatively objective and correct conclusion or opinion
may be drawn, for example, by meta-analysis. In addition, a
particular geographical issue should be considered. The patient
cohort in the present study was primarily from Yunnan Province, an
undeveloped, secluded and mountainous province of southwestern
China, therefore a number of individuals in this region are unable
to afford the relatively expensive cost of PET-CT and gene mutation
detection, directly leading to the small sample size. Secondly, a
bias could have existed in the process of the patient selection
process since the majority of the patients resided in Yunnan
Province that is known for high lung cancer rates (46-48).
Thirdly, differences in metabolic parameters among different
EGFR mutations, and between EGFR mutation and other
important mutations (e.g. KRAS) were not discussed, which we
intend to address in future studies. In the present study, although
direct evidence remains limited, patients with EGFR mutation
exhibited obviously decreased SUVmax compared with those
with no EGFR mutation. χ2 analysis revealed that
SUVmax is one predictor of EGFR mutation status
and univariate analysis indicated that SUVmax was the
only predictor of EGFR mutation. In the future, with the
requisite equipment, FDG uptake among different lung cancer cells
with various EGFR mutation status may be detected, which
will provide direct evidence. The sample size will be increased and
follow-up of patients assessed in the present study will be
continued, and it is intended to publish survival results in the
future. Finally, it should also be recog-nized that tissue testing
is the gold standard for judging EGFR mutation status.
In conclusion, the results of the present study from
clinical samples and cell lines indicate that the FDG uptake was
decreased in patients with NSCLC with EGFR mutation. In
addition, with a cut-off value of 9.92, the SUVmax is
useful in predicting EGFR mutation, indicating that PET-CT
may be a useful non-invasive instrument for predicting EGFR
mutation in patients with NSCLC, thereby optimizing the clinical
treatment strategy. In addition, further experiments at the cell
and molecular levels validated that the NOX4/ROS/GLUT1 axis is
responsible for decreased FDG uptake in patients with NSCLC with
EGFR mutation, which may reveal potential treatment
targets.
Funding
The present study was supported by the Initiation
Foundation for Doctors of Yunnan Tumor Hospital (grant no.
BSKY201706) and Joint Special Fund from Yunnan Provincial Science
and Technology Department-Kunming Medical University for Applied
and Basic Research (grant no. 2018FE001-150).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors’ contributions
LC, YZ, XT and CY contributed to the design of the
study and wrote the manuscript. YT, RX, TC and JY performed the
experiments. MJ, FC, CW, HS and YH analyzed the data. CW, HS and YH
also revised and amended the manuscript. All authors have read and
approved this manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of The Third Affiliated Hospital of Kunming Medical
University. All procedures performed in the present study that
involved human participants were with the approval of the
Institutional Review Board of The Third Affiliated Hospital of
Kunming Medical University and in accordance with the 1964
Declaration of Helsinki and its later amendments or comparable
ethical standards. Written informed consent was obtained from all
patients.
Patient consent for publication
Written informed consent was obtained from the
patients for the publication of this the present paper.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
ARMS
|
amplification refractory mutation
system
|
|
AUC
|
area under the curve
|
|
CT
|
computed tomography
|
|
EGFR
|
epidermal growth factor receptor
|
|
FDG
|
[18F]fluoro-2-deoxyglucose
|
|
NSCLC
|
non-small cell lung cancer
|
|
PCR
|
polymerase chain reaction
|
|
PET
|
positron emission tomography
|
|
ROC
|
receiver operating characteristic
|
|
SUVmax
|
maximum standardized uptake value
|
|
TKI
|
tyrosine kinase inhibitor
|
Acknowledgments
Not applicable.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jung KW, Won YJ, Oh CM, Kong HJ, Lee DH
and Lee KH; Community of Population-Based Regional Cancer
Registries: Cancer statistics in Korea: Incidence, mortality,
survival, and prevalence in 2014. Cancer Res Treat. 49:292–305.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Fedewa SA, Miller KD,
Goding-Sauer A, Pinheiro PS, Martinez-Tyson D and Jemal A: Cancer
statistics for Hispanics/Latinos, 2015. CA Cancer J Clin.
65:457–480. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Janssen S, Käsmann L, Rudat V and Rades D:
Stereotactic body radiation therapy (SBRT) for recurrent non-small
cell lung cancer (NSCLC). Anticancer Res. 36:825–828.
2016.PubMed/NCBI
|
|
6
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Santarpia M, Altavilla G, Salazar MF,
Magri I, Pettineo G, Benecchi S and Rosell R: Tyrosine kinase
inhibitors for non-small-cell lung cancer: Finding patients who
will be responsive. Expert Rev Respir Med. 5:413–424. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo G, Gong K, Wohlfeld B, Hatanpaa KJ,
Zhao D and Habib AA: Ligand-independent EGFR signaling. Cancer Res.
75:3436–3441. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sholl LM, Yeap BY, Iafrate AJ,
Holmes-Tisch AJ, Chou YP, Wu MT, Goan YG, Su L, Benedettini E, Yu
J, et al: Lung adenocarcinoma with EGFR amplification has distinct
clinicopathologic and molecular features in never-smokers. Cancer
Res. 69:8341–8348. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kong FM, Ten Haken RK, Schipper M, Frey
KA, Hayman J, Gross M, Ramnath N, Hassan KA, Matuszak M, Ritter T,
et al: Effect of midtreatment PET/CT-adapted radiation therapy with
concurrent chemotherapy in patients with locally advanced
non-small-cell lung cancer: A phase 2 clinical trial. JAMA Oncol.
3:1358–1365. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Levine M and Julian J: Imaging: PET-CT
imaging in non-small-cell lung cancer. Nat Rev Clin Oncol.
6:619–620. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Na II, Byun BH, Kang HJ, Cheon GJ, Koh JS,
Kim CH, Choe DH, Ryoo BY, Lee JC, Lim SM, et al:
18F-fluoro-2-deoxyglucose uptake predicts clinical
outcome in patients with gefitinib-treated non-small cell lung
cancer. Clin Cancer Res. 14:2036–2041. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kobe C, Scheffler M, Holstein A, Zander T,
Nogova L, Lammertsma AA, Boellaard R, Neumaier B, Ullrich RT,
Dietlein M, et al: Predictive value of early and late residual
18F-fluorodeoxyglucose and
18F-fluorothymidine uptake using different SUV
measurements in patients with non-small-cell lung cancer treated
with erlotinib. Eur J Nucl Med Mol Imaging. 39:1117–1127. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Horiuchi C, Tsukuda M, Taguchi T, Ishiguro
Y, Okudera K and Inoue T: Correlation between FDG-PET findings and
GLUT1 expression in salivary gland pleomorphic adenomas. Ann Nucl
Med. 22:693–698. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Grabellus F, Nagarajah J, Bockisch A,
Schmid KW and Sheu SY: Glucose transporter 1 expression, tumor
proliferation, and iodine/glucose uptake in thyroid cancer with
emphasis on poorly differentiated thyroid carcinoma. Clin Nucl Med.
37:121–127. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Prata C, Maraldi T, Fiorentini D, Zambonin
L, Hakim G and Landi L: Nox-generated ROS modulate glucose uptake
in a leukaemic cell line. Free Radic Res. 42:405–414. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Prata C, Maraldi T, Zambonin L, Fiorentini
D, Hakim G and Landi L: ROS production and Glut1 activity in two
human megakaryocytic cell lines. Biofactors. 20:223–233. 2004.
View Article : Google Scholar
|
|
18
|
Lo YM: The amplification refractory
mutation system. Methods Mol Med. 16:61–69. 1998.PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
20
|
Weyemi U, Lagente-Chevallier O, Boufraqech
M, Prenois F, Courtin F, Caillou B, Talbot M, Dardalhon M, Al
Ghuzlan A, Bidart JM, et al: ROS-generating NADPH oxidase NOX4 is a
critical mediator in oncogenic H-Ras-induced DNA damage and
subsequent senescence. Oncogene. 31:1117–1129. 2012. View Article : Google Scholar :
|
|
21
|
Waris G, Turkson J, Hassanein T and
Siddiqui A: Hepatitis C virus (HCV) constitutively activates STAT-3
via oxidative stress: Role of STAT-3 in HCV replication. J Virol.
79:1569–1580. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kodama R, Kato M, Furuta S, Ueno S, Zhang
Y, Matsuno K, Yabe-Nishimura C, Tanaka E and Kamata T:
ROS-generating oxidases Nox1 and Nox4 contribute to oncogenic
Ras-induced premature senescence. Genes Cells. 18:32–41. 2013.
View Article : Google Scholar
|
|
23
|
Lara-Guerra H, Waddell TK, Salvarrey MA,
Joshua AM, Chung CT, Paul N, Boerner S, Sakurada A, Ludkovski O, Ma
C, et al: Phase II study of preoperative gefitinib in clinical
stage I non-small-cell lung cancer. J Clin Oncol. 27:6229–6236.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sunaga N, Tomizawa Y, Yanagitani N, Iijima
H, Kaira K, Shimizu K, Tanaka S, Suga T, Hisada T, Ishizuka T, et
al: II prospective study of the efficacy of gefitinib for the
treatment of stage III/IV non-small cell lung cancer with EGFR
mutations, irrespective of previous chemotherapy. Lung Cancer.
56:383–389. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu A, Han A, Zhu H, Ma L, Huang Y, Li M,
Jin F, Yang Q and Yu J: The role of metabolic tumor volume (MTV)
measured by [18F] FDG PET/CT in predicting EGFR gene
mutation status in non-small cell lung cancer. Oncotarget.
8:33736–33744. 2017.PubMed/NCBI
|
|
26
|
Guan J, Xiao NJ, Chen M, Zhou WL, Zhang
YW, Wang S, Dai YM, Li L, Zhang Y, Li QY, et al: 18F-FDG
uptake for prediction EGFR mutation status in non-small cell lung
cancer. Medicine (Baltimore). 95:e44212016. View Article : Google Scholar
|
|
27
|
Minamimoto R, Jamali M, Gevaert O,
Echegaray S, Khuong A, Hoang CD, Shrager JB, Plevritis SK, Rubin
DL, Leung AN, et al: Prediction of EGFR and KRAS mutation in
non-small cell lung cancer using quantitative 18F
FDG-PET/CT metrics. Oncotarget. 8:52792–52801. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Takamochi K, Mogushi K, Kawaji H,
Imashimizu K, Fukui M, Oh S, Itoh M, Hayashizaki Y, Ko W, Akeboshi
M, et al: Correlation of EGFR or KRAS mutation status with
18F-FDG uptake on PET-CT scan in lung adenocarcinoma.
PLoS One. 12:e01756222017. View Article : Google Scholar
|
|
29
|
Caicedo C, Garcia-Velloso MJ, Lozano MD,
Labiano T, Vigil Diaz C, Lopez-Picazo JM, Gurpide A, Zulueta JJ,
Richter Echevarria JA and Perez Gracia JL: Role of
[18F]FDG PET in prediction of KRAS and EGFR mutation
status in patients with advanced non-small-cell lung cancer. Eur J
Nucl Med Mol Imaging. 41:2058–2065. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoshida T, Tanaka H, Kuroda H, Shimizu J,
Horio Y, Sakao Y, Inaba Y, Iwata H, Hida T and Yatabe Y:
Standardized uptake value on (18)F-FDG-PET/CT is a predictor of
EGFR T790M mutation status in patients with acquired resistance to
EGFR-TKIs. Lung Cancer. 100:14–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee SM, Bae SK, Jung SJ and Kim CK: FDG
uptake in non-small cell lung cancer is not an independent
predictor of EGFR or KRAS mutation status: A retrospective analysis
of 206 patients. Clin Nucl Med. 40:950–958. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cho A, Hur J, Moon YW, Hong SR, Suh YJ,
Kim YJ, Im DJ, Hong YJ, Lee HJ, Kim YJ, et al: Correlation between
EGFR gene mutation, cytologic tumor markers, 18F-FDG
uptake in non-small cell lung cancer. BMC Cancer. 16:2242016.
View Article : Google Scholar
|
|
33
|
Ko KH, Hsu HH, Huang TW, Gao HW, Shen DH,
Chang WC, Hsu YC, Chang TH, Chu CM, Ho CL, et al: Value of
18F-FDG uptake on PET/CT and CEA level to predict
epidermal growth factor receptor mutations in pulmonary
adenocarcinoma. Eur J Nucl Med Mol Imaging. 41:1889–1897. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Putora PM, Früh M and Müller J: FDG-PET
SUV-max values do not correlate with epidermal growth factor
receptor mutation status in lung adenocarcinoma. Respirology.
18:734–735. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hogan A, Heyner S, Charron MJ, Copeland
NG, Gilbert DJ, Jenkins NA, Thorens B and Schultz GA: Glucose
transporter gene expression in early mouse embryos. Development.
113:363–372. 1991.PubMed/NCBI
|
|
36
|
Liemburg-Apers DC, Willems PH, Koopman WJ
and Grefte S: Interactions between mitochondrial reactive oxygen
species and cellular glucose metabolism. Arch Toxicol.
89:1209–1226. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fiorentini D, Prata C, Maraldi T, Zambonin
L, Bonsi L, Hakim G and Landi L: Contribution of reactive oxygen
species to the regulation of Glut1 in two hemopoietic cell lines
differing in cytokine sensitivity. Free Radic Biol Med.
37:1402–1411. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gorrini C, Harris IS and Mak TW:
Modulation of oxidative stress as an anticancer strategy. Nat Rev
Drug Discov. 12:931–947. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kawano Y, Iwama E, Tsuchihashi K,
Shibahara D, Harada T, Tanaka K, Nagano O, Saya H, Nakanishi Y and
Okamoto I: CD44 variant-dependent regulation of redox balance in
EGFR mutation-positive non-small cell lung cancer: A target for
treatment. Lung Cancer. 113:72–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jafari N, Kim H, Park R, Li L, Jang M,
Morris AJ, Park J and Huang C: CRISPR-Cas9 mediated NOX4 knockout
inhibits cell proliferation and invasion in HeLa cells. PLoS One.
12:e01703272017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Crosas-Molist E, Bertran E,
Rodriguez-Hernandez I, Herraiz C, Cantelli G, Fabra À, Sanz-Moreno
V and Fabregat I: The NADPH oxidase NOX4 represses epithelial to
amoeboid transition and efficient tumour dissemination. Oncogene.
36:3002–3014. 2017. View Article : Google Scholar :
|
|
42
|
Brunelle JK, Bell EL, Quesada NM,
Vercauteren K, Tiranti V, Zeviani M, Scarpulla RC and Chandel NS:
Oxygen sensing requires mitochondrial ROS but not oxidative
phosphorylation. Cell Metab. 1:409–414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jung KH, Lee JH, Thien Quach CH, Paik JY,
Oh H, Park JW, Lee EJ, Moon SH and Lee KH: Resveratrol suppresses
cancer cell glucose uptake by targeting reactive oxygen
species-mediated hypoxia-inducible factor-1alpha activation. J Nucl
Med. 54:2161–2167. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Orcutt KP, Parsons AD, Sibenaller ZA,
Scarbrough PM, Zhu Y, Sobhakumari A, Wilke WW, Kalen AL, Goswami P,
Miller FJ Jr, et al: Erlotinib-mediated inhibition of EGFR
signaling induces metabolic oxidative stress through NOX4. Cancer
Res. 71:3932–3940. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sobhakumari A, Schickling BM, Love-Homan
L, Raeburn A, Fletcher EV, Case AJ, Domann FE, Miller FJ Jr and
Simons AL: NOX4 mediates cytoprotective autophagy induced by the
EGFR inhibitor erlotinib in head and neck cancer cells. Toxicol
Appl Pharmacol. 272:736–745. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou Y, Yang Y, Yang C, Chen Y, Yang C, Du
Y, Zhao G, Guo Y, Ye L and Huang Y: Epidermal growth factor
receptor (EGFR) mutations in non-small cell lung cancer (NSCLC) of
Yunnan in southwestern China. Oncotarget. 8:15023–15033.
2017.PubMed/NCBI
|
|
47
|
Lan Q, He X, Shen M, Tian L, Liu LZ, Lai
H, Chen W, Berndt SI, Hosgood HD, Lee KM, et al: Variation in lung
cancer risk by smoky coal subtype in Xuanwei. China Int J Cancer.
123:2164–2169. 2008. View Article : Google Scholar
|
|
48
|
Mumford JL, He XZ, Chapman RS, Cao SR,
Harris DB, Li XM, Xian YL, Jiang WZ, Xu CW, Chuang JC, et al: Lung
cancer and indoor air pollution in Xuan Wei, China. Science.
235:217–220. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pieper U, Eswar N, Davis FP, Braberg H,
Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, Eramian
D, et al: MODBASE: A database of annotated comparative protein
structure models and associated resources. Nucleic Acids Res.
34:D291–D295. 2006. View Article : Google Scholar :
|