Introduction
Synovial sarcoma (SS) is a rare tumour, with a
dismal survival when metastasis occurs. SS contains a
characteristic translocation (X;18)(p11;q11), representing the
fusion of SYT on chromosome 18 with either SSX1 or
SSX2, or rarely SSX4 on chromosome X. The resulting
fusion genes appear to be mutually exclusive and concordant in
primary and metastatic tumours. The 5-year survival rate is 55% for
axial SS and 84% for SS of the extremities. Currently, biological
factors are still unreliable for predicting the aggressiveness and
clinical evolution of the tumour (1).
A tumour can influence its microenvironment by
releasing extracellular signals, promoting tumour angiogenesis and
inducing the inflammatory response, while immune cells in the
microenvironment can affect the growth and evolution of cancer
cells. The association between cancer cells and their
microenvironment contributes to tumour heterogeneity (2) and is crucial for cancer progression.
C-X-C chemokine receptor 4 (CXCR4) is a seven-transmembrane G
protein-coupled chemokine receptor commonly expressed in tumour
cells and is involved in cell migration and invasion, as well as in
angiogenesis (3). The deregulated
expression of CXCR4 has been detected in several human cancers,
including melanoma, colon cancer (4), breast (5) and pancreatic cancer (6). CXCR4, by binding its ligand CXCL12
(SDF-1), activates cell signalling pathways promoting
proliferation, migration and the development of metastasis
(7,8), also playing a role in normal stem
cell homing (3). Previous studies
have demonstrated that high expression levels of CXCR4 activated by
CXCL12 are associated with a poor prognosis and a high rate of
metastasis in bone and soft tissue sarcomas (STS) (9,10),
also showing the pivotal role of CXCR4 in cancer cell migration
between the primary and metastatic site in SS. By multivariate
analysis, we recently reported that the nuclear protein expression
of CXCR4 was an independent adverse prognostic factor linked to the
use of chemotherapy for the survival of patients with SS,
suggesting a role of CXCR4 in drug resistance (11). MicroRNAs (miRNAs or miRs) are
involved in post-transcriptional gene expression regulation and
control important physiological processes, such as development,
cell differentiation and cell signalling (12,13).
The altered expression of miRNAs is strongly associated with the
malignant phenotype and there are data reporting a strong
association between miRNA expression, patient age and STS prognosis
(14,15). miR-494.3p is involved in tumour
progression and has been previously described as a CXCR4 regulator
in prostate (16) and breast
cancer cells, where its levels were significantly lower than those
in normal breast epithelial cells (17). In chondrosarcoma, miR-494 has been
shown to significantly inhibit cell proliferation, migration and
invasion in vitro and in vivo (18), whereas another study demonstrated
that it was downregulated in osteosarcoma when compared to normal
tissue. In osteosarcoma cell lines, miR-494 restoration has been
shown to inhibit cell proliferation, colony formation, migration
and invasion (19). In this study,
we analysed the expression of CXCR4 and miR-494.3p, in a series of
SS specimens and verified that the modulation of CXCR4 mediated by
the ectopic expression of miR-494.3p suppressed SS cell
proliferation and migration.
Materials and methods
Tumour specimens
Forty-two primary monophasic SS specimens were
selected from the Rizzoli Orthopedic Institute Biobank. All the
demographic and clinicopathological data of the patients were
collected (Table I). The specimens
were from 22 males and 20 females with a mean age of 43 years; 29
patients developed metastases and 24 succumbed to the disease. The
average follow-up time was 90 months (range, 13-70 months). All the
samples were treated in accordance with the authorizations issued
by the Ethics Committee of the Rizzoli Orthopedic Institute (no.
0033276) and written informed consent was obtained from all
patients. Fresh and paraffin-embedded non-necrotic tissue (≥90%
viable tumour cells) was used. Following the revision of the
histological slides according to histopathological and
immu-nohistochemical criteria and the presence of SS18 (SYT) gene
rearrangement and fusion transcripts (SSX1 and SSX2) the diagnosis
was confirmed by pathologists. A total of 20 non-paired normal
tissues were used as controls.
 | Table IClinicopathological features of
patients with SS. |
Table I
Clinicopathological features of
patients with SS.
| Case | Sex | Age | Site | Metastasis | Follow-up
(months) | Outcome |
|---|
| 1 | F | 17 | Elbow | x | 55 | DOD |
| 2 | F | 32 | Thigh | x | 24 | DOD |
| 3 | F | 51 | Thigh | x | 110 | DOD |
| 4 | M | 48 | Thigh | x | 16 | DOD |
| 5 | M | 63 | Paraspinal | x | 83 | DOD |
| 6 | F | 40 | Popliteus | x | 43 | DOD |
| 7 | F | 63 | Foot | x | 20 | DOD |
| 8 | M | 40 | Leg | x | 9 | DOD |
| 9 | F | 41 | Popliteus | x | 43 | DOD |
| 10 | M | 47 | Thigh | x | 49 | DOD |
| 11 | F | 51 | Thigh | x | 203 | DOD |
| 12 | F | 32 | Thigh | x | 24 | DOD |
| 13 | M | 49 | Forearm | x | 63 | NED |
| 14 | M | 29 | Popliteus | x | 120 | NED |
| 15 | M | 64 | Thigh | x | 50 | DOD |
| 16 | M | 70 | Scapular
girdle | x | 8 | DOD |
| 17 | M | 37 | Knee | x | 66 | DOD |
| 18 | F | 59 | Foot | | 72 | NED |
| 19 | M | 26 | Gluteus | x | 58 | DOD |
| 20 | F | 57 | Foot | x | 67 | DOD |
| 21 | M | 31 | Thigh | x | 12 | DOD |
| 22 | M | 14 | Foot | x | 93 | NED |
| 23 | F | 69 | Elbow | | 143 | NED |
| 24 | F | 16 | Gluteus | | 131 | NED |
| 25 | M | 61 | Wrist | | 153 | NED |
| 26 | M | 46 | Forearm | | 150 | NED |
| 27 | M | 39 | Leg | x | 23 | NED |
| 28 | F | 63 | Foot | x | 20 | DOD |
| 29 | M | 57 | Foot | x | 22 | DOD |
| 30 | F | 51 | Thigh | x | 54 | DOD |
| 31 | M | 36 | Leg | | 176 | NED |
| 32 | M | 57 | Popliteus | | 137 | NED |
| 33 | F | 13 | Gluteus | | 183 | NED |
| 34 | F | 46 | Leg | x | 88 | DOD |
| 35 | M | 29 | Leg | | 170 | NED |
| 36 | M | 38 | Thigh | | 138 | NED |
| 37 | M | 43 | Thigh | x | 310 | NED |
| 38 | F | 58 | Thigh | | 160 | NED |
| l39 | F | 58 | Foot | x | 44 | DOD |
| 40 | F | 21 | Elbow | | 116 | NED |
| 41 | M | 27 | Arm | | 260 | NED |
| 42 | F | 33 | Foot | x | 20 | DOD |
Synovial sarcoma cell lines
The human synovial sarcoma SW982 cell line (no.
HTB-93) was obtained from the American Type Culture Collection
(ATCC, Manassas, VA, USA) and the SYO-I cell line was kindly
provided by Dr Kawai (Department of Orthopaedic Surgery, National
Cancer Center Hospital, Tokyo, Japan). The cell lines were
routinely cultured using α-MEM medium supplemented with 10% FBS,
L-glutamine (2 mM), 100 U/ml penicillin and 100 µg/ml
streptomycin (Invitrogen/Thermo Fisher Scientific, Waltham, MA,
USA) at 37˚C in a 5% CO2 humidified incubator.
RNA extraction
Total RNA was extracted using TRizol reagent
(Invitrogen/Thermo Fisher Scientific) from the 42 SS tissue
samples, 20 non-tumour tissues and the two SS cell lines (SW982 and
SYO-I) following the manufacturer's instructions.
miRNA expression in clinical
specimens
Reverse transcription and quantitative PCR (qPCR) of
miR-494.3p (TaqMan miRNA assay no. 002365; Applied
Biosystems/Thermo Fisher Scientific) was performed in the 42 SS and
20 non-paired normal tissues following the TaqMan MicroRNA assay
protocol. miRNA expression was quantified by the ΔΔCq comparative
method using a pool of normal lymphocytes as a calibrator (20). Data were normalized to the
endogenous reference RNU44 (TaqMan miRNA assay no. 001094),
following Applied Biosystems indications (TaqMan miRNA assay no.
4373384; Applied Biosystems/Thermo Fisher Scientific).
CXCR4 gene expression in clinical
specimens
CXCR4 expression was quantified in 42 SS and in 20
non-tumour tissues by TaqMan Expression Assay (Hs00607978_sl;
Applied Biosystems/Thermo Fisher Scientific) according to
manufacturer's instructions following reverse transcription using
the SuperScript™ VILO™ cDNA Synthesis kit (Applied
Biosystems/Thermo Fisher Scientific). The expression of target
genes was calculated by the ΔΔCq comparative method and normalized
to the ACTB housekeeping gene (Hs99999903_m1 gene; TaqMan
Expression Assays; Applied Biosystems/Thermo Fisher Scientific). A
pool of normal lymphocytes was used as a calibrator.
Western blot analysis of CXCR4 protein
expression in clinical specimens
According to standard procedures, protein extracts
from the SS tissues and non-tumour tissues were prepared by mincing
and homogenizing fresh samples in extraction buffer (50 mM Tris-HCl
at pH 8.0, 150 mM NaCl, 1 mM DTT, 50 mM NaF, 0.5% sodium
deoxycholate, 0.1% SDS, 1% NP-40 and 0.1 mM PMFS) with complete
mixture of protease inhibitors (Roche Diagnostics, Laval, QC,
Canada). Protein extracts were obtained by lysis in 100-400
µl lysis buffer [20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2.5
mM sodium pyrophosphate, 1 mM glycerol phosphate, 1 mM
Na3VO4, 1 mM EDTA and 1% Triton] with
complete protease inhibitor mixture. A total of 50 µg of
protein extracts were prepared and analysed by 12% SDS-PAGE.
Following nitrocellulose blot transfer and blocking using 5% milk
in TBST 1X at room temperature for 1 h, the membranes were
incubated with a commercially available monoclonal antibody,
anti-CXCR4 (ab2074, Abcam, Cambridge, UK) diluted 1:1,000,
overnight in 4°C on a shaker. Anti-rabbit antibody conjugated to
horseradish peroxidase (cat. no. NA934V; GE Healthcare, Chicago,
IL, USA) was used as a secondary antibody at room temperature for 1
h. Proteins were visualized by incubation with ECL (EuroClone,
Milan, Italy) and quantified by densitometric analysis using a
GS-800 imaging densitometer and Quantity One Software (Bio-Rad,
Hercules, CA, USA). A rabbit anti-actin antibody (cat. no.
SAB5500001; Sigma Chemical Co, St. Louis, MO, USA) diluited
1:50,000 was used as a loading control.
miR-494.3p transfection in SS cell
lines
The SW982 and SYO-I cell lines were seeded at a
density of 2.5×105 cells/well in 6-well plates in 2 ml
of complete medium containing 10% FCS for 24 h. Transfection was
performed using Lipofectamine 2000 (Invitrogen/Thermo Fisher
Scientific) with 50 nM miR-494.3p precursor (cod. PM12409; Ambion
Inc., Austin, TX, USA) and miRNA negative precursor was used as a
negative control (scramble) (cod. AM17110; Ambion). The
transfection efficiency was monitored at 48 and 72 h
post-transfection by flow cytometry (FACSCalibur; BD Biosciences,
San Jose, CA, USA) using Cy™3 and FAM™ dye-labelled Pre-miR
Negative Controls (cod. AM17120; Ambion) and RT-PCR according to
the TaqMan MicroRNA assay protocol for the CXCR4 target gene. The
results were quantified by the ΔΔCq method using TaqMan Expression
Assays (Hs00607978_s1) and normalized to the ACTB housekeeping gene
(Hs99999903_m1) (Applied Biosystems/Thermo Fisher Scientific).
Flow cytometric analysis of CXCR4 in SS
cell lines
The SW982 and SYO-I cells were stained with
anti-CXCR4 antibody for 30 min at room temperature (ab2074; Abcam;
1:100 dilution in phosphate-buffered saline (PBS)]. Following
washes with wash buffer, the cells were incubated at room
temperature with a goat-anti-rabbit FITC-conjugated secondary
antibody (cat. no. 31635; Thermo Fisher Scientific) (1:80 dilution
in PBS) for 1 h in the dark. Following two additional washes in
wash buffer, flow cytometry was performed using a FACSCalibur flow
cytometer (BD Biosciences).
Cell growth assay
The number of adherent, viable cells was assessed
microscopically using an improved Neubauer cell counting chamber
(Incofar, Modena, Italy) in non-transfected and transfected cells
at 48 and 72 h from transfection. The viable cells were then
quantified by a Trypan blue exclusion assay. The cells were washed
once in PBS, harvested by trypsinization and stained with 1X trypan
blue solution (cat. no. 15250-061; Thermo Fisher Scientific). Cell
viability was assessed as the percentage of cells that excluded
trypan blue.
Apoptosis assay
Apoptotic cell death was analysed in non-transfected
and transfected cells at 48 and 72 h from transfection with the
Annexin V-FITC apoptosis detection kit (MEBCYTO Apoptosis kit, MBL
International, Woburn, MA, USA). Following the manufacturer's
instructions, adherent cells were trypsinized, suspended in 500
µl of staining solution containing FITC-conjugated Annexin V
antibody and propidium iodide (PI) and after 30 min of incubation
on ice were analysed by flow cytometry using a FACSCalibur flow
cytometer and CellQuest Software (BD Biosciences).
Scratch wound-healing assay
Cell migration ability was determined using a
scratch wound-healing assay. The SW982 and SYO-I adherent cells
were rinsed with PBS to remove floating cells, and a vertical line
was scratched with a sterile 200 µl pipette (time 0) at 48 h
post-transfection. The wound closure was then monitored at 6, 18,
24, 36 and 72. The areas were measured using IMAGEJ v.1.45r
software (http://rsbweb.nih.gov/ij/).
Statistical analysis
miRNA expression is presented as the
2−ΔΔCq value for each sample. Values of the median (m)
and the 25th-75th percentile were calculated within the two
subsets. A non-parametric Mann-Whitney U test was performed to
compare miRNA median levels in the subsets and Spearman's rank
coefficient was used for correlation analysis. The non-parametric
Wilcoxon text was performed for protein analysis. In vitro
experiments were performed 3 times and statistical significance was
analysed by a Student's t-test. For all analyses, a value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
miRNA and CXCR4 expression in
specimens
miR-494.3p expression analysis performed in the 42
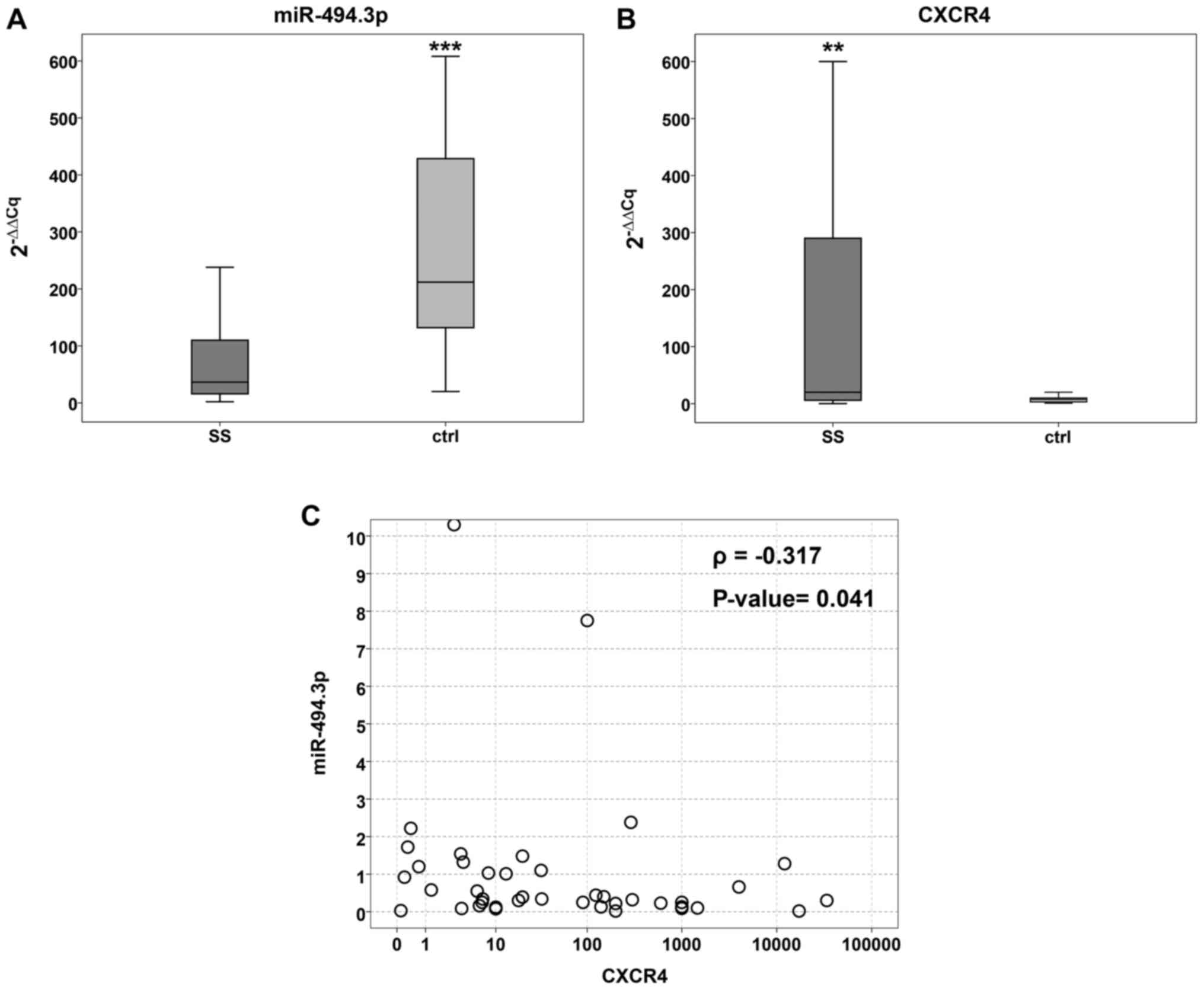
SS samples and 20 non-tumour tissues. The tumour tissues exhibited
significantly lower median values (2−ΔΔCq) in the
tumours compared to the controls (36.5; 25th-75th=15.2-112 and 212;
25th-75th=127-475, respectively) (P=0.001) (Fig. 1A).
Conversely, CXCR4 mRNA expression was significantly
higher in the tumour (median value 20; 25th- 75th=5.5-292) compared
to the non-tumour tissue (median value=7.5; 25th-75th=3-10)
(P=0.01) (Fig. 1B). Spearman's
correlation analysis performed on the 42 SS samples confirmed the
significant negative correlation between CXCR4 and miR-494.3p
expression (ρ=-0.317, P=0.041) (Fig.
1C).
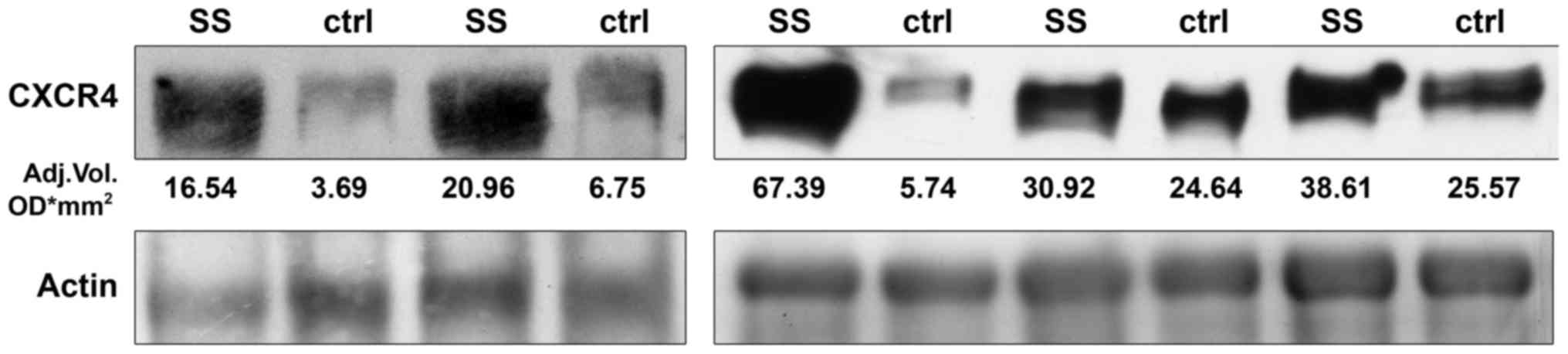
The high expression of the CXCR4 gene was
concomitant with the presence of a marked 44 kDa migration band
detected by western blot analysis. Densitometric analysis revealed
that CXCR4 protein levels were higher in the SS that in normal
adjacent tissue (30.92; 25th-75th=18.75-53 and 6.75;
25th-75th=4.71-25.10 respectively) with a difference at the limit
of statistical significance (P=0.063) (Fig. 2).
When the tumour population was divided according to
clinical follow-up, we found lower miR-494.3p median levels in the
metastatic compared to the non-metastatic subset (34.0;
25th-75th=13-115 and 56.0; 25th-75th=30-103, respectively) (P=0.4),
while the CXCR4 mRNA levels were higher in the first group (100;
6.2-1000 and 18.1; 0.8-32.5, respectively), revealing a difference
at the limit of statistical significance (P=0.06). No significant
differences were related to patient outcome (Table II).
 | Table IImiR-494.3p and CXCR4 median
expression level. |
Table II
miR-494.3p and CXCR4 median
expression level.
| 25 P | Median
2−ΔΔCT | 75 P | P-value |
|---|
| miR-494.3p | | | | |
|
Non-metastatic | 30 | 56 | 103 | 0.4 |
| Metastatic | 13 | 34 | 115 | |
| miR-494.3p | | | | |
| Alive
patients | 28.7 | 51 | 114.2 | >0.05 |
| Deceased
patients | 12.7 | 32 | 112.5 | |
| CXCR4 | | | | |
|
Non-metastatic | 0.8 | 18.1 | 32.5 | 0.06 |
| Metastatic | 6.2 | 100 | 1,000 | |
| CXCR4 | | | | |
| Alive
patients | 1.0 | 19 | 165.1 | >0.05 |
| Deceased
patients | 5.5 | 26 | 100 | |
miR-494.3p transfection in SS cell
lines
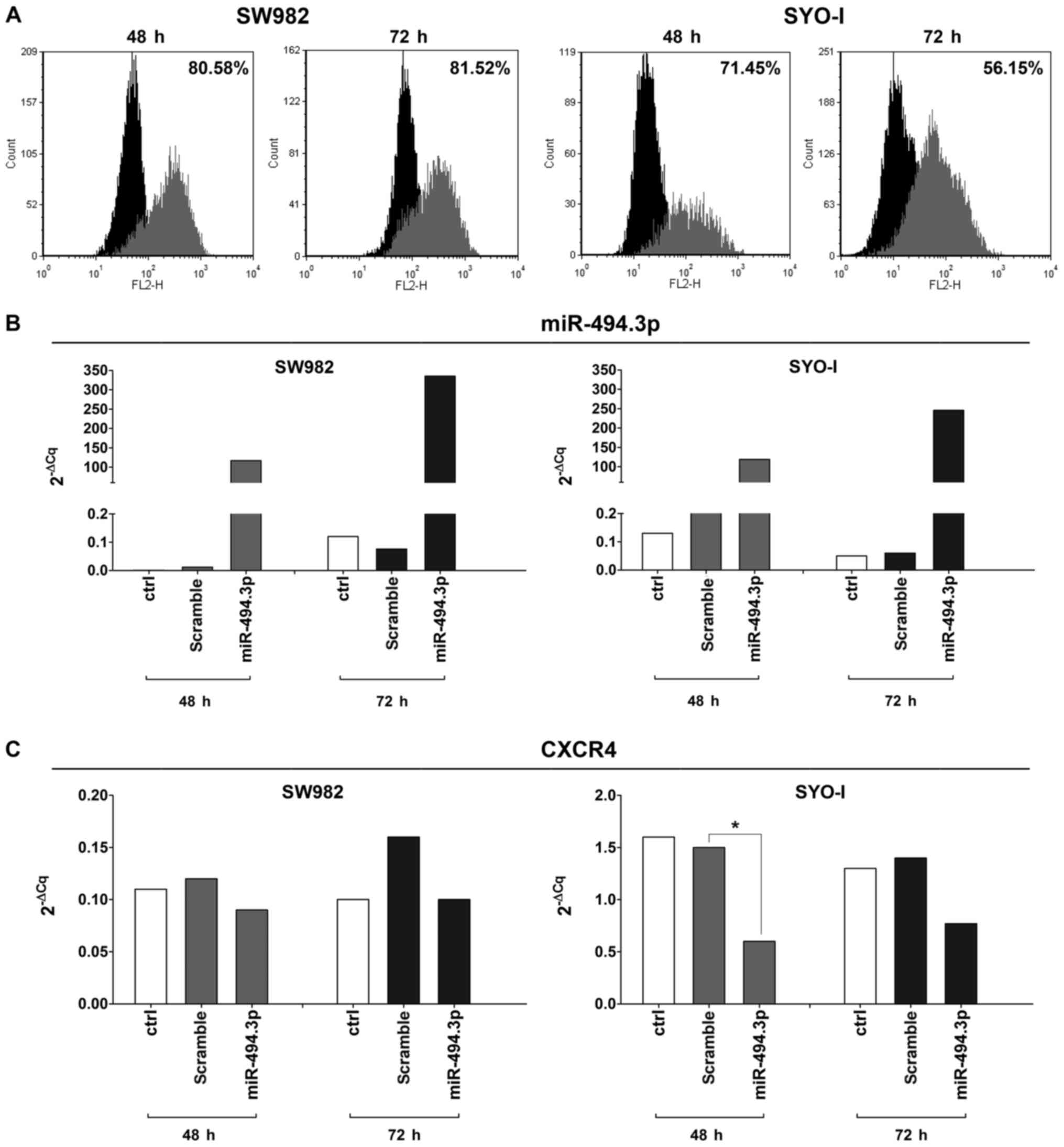
To verify the role of CXCR4 as a potential
miR-494.3p target, the SW982 and SYO-I cell lines with a minimal
expression of miR-494.3p (2−ΔCq values: 0.03 and 0.06,
respectively), were transfected with the miR-494.3p precursor. The
transfection efficiency was 80.58 and 81.52% at 48 and 72 h,
respectively for the SW982 cells, and 71.45 and 56.15% at 48 and 72
h, respectively for the SYO-1 cells (Fig. 3A). RT-PCR analysis confirmed the
increased expression of miR-494.3p in the SW982-transfected cells
compared to the scramble control (116 and 4,407-fold change at 48
and 72 h, respectively), as well as in the SYO-I-transfected cells
(519-fold at 48 h and 4,092-fold change at 72 h) (Fig. 3B). Accordingly, a decrease in the
levels of its potential target gene, CXCR4, was found in the
transfected cells compared to the scramble control, reaching
statistical significance in the SYO-I at 48 h (P=0.02) (Fig. 3C). A decreased protein expression
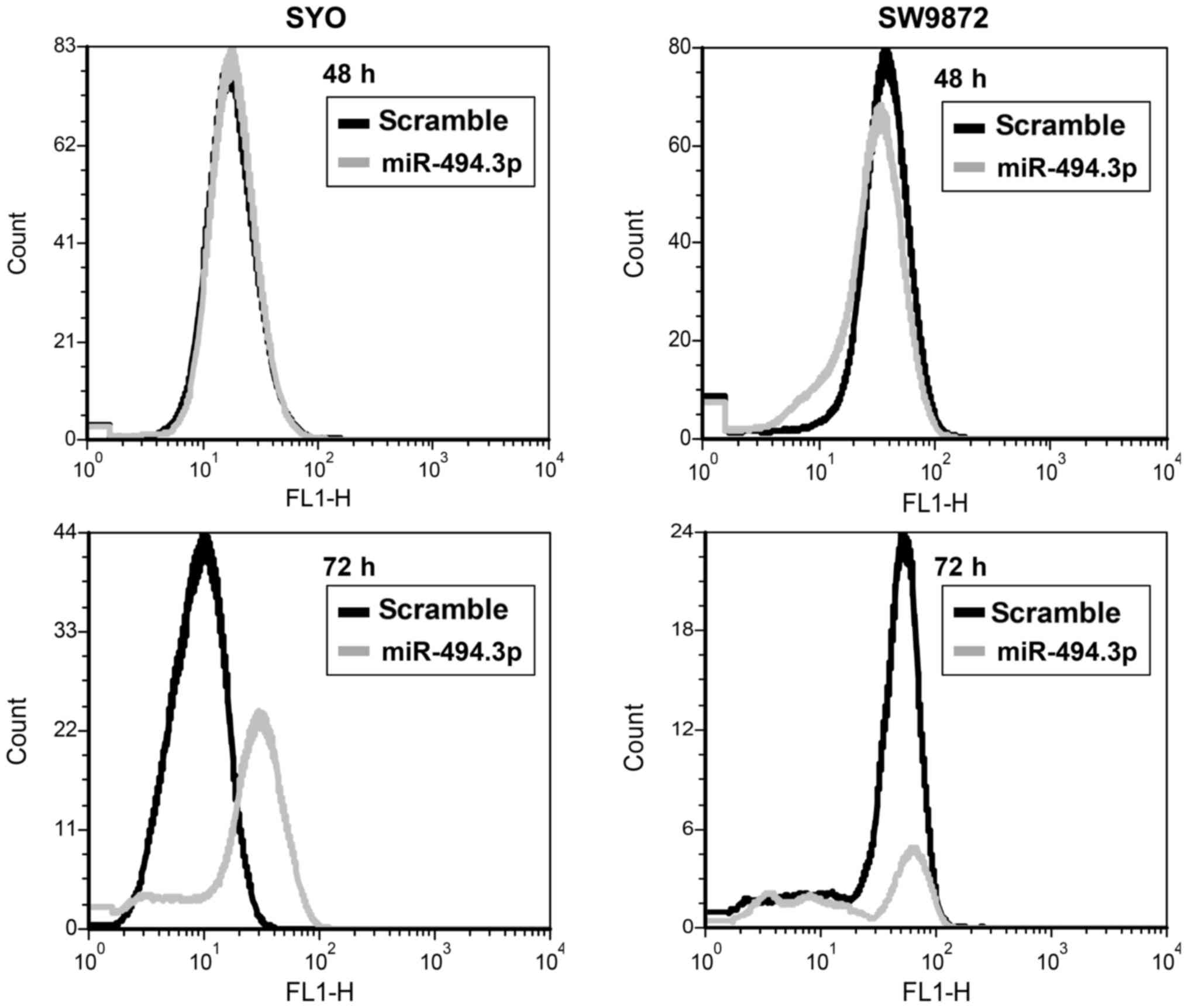
of CXCR4 was also observed by FACS analysis up to 72 h of
transfection (Fig. 4).
Effect of miR-494.3p ectopic expression
on SS cell lines
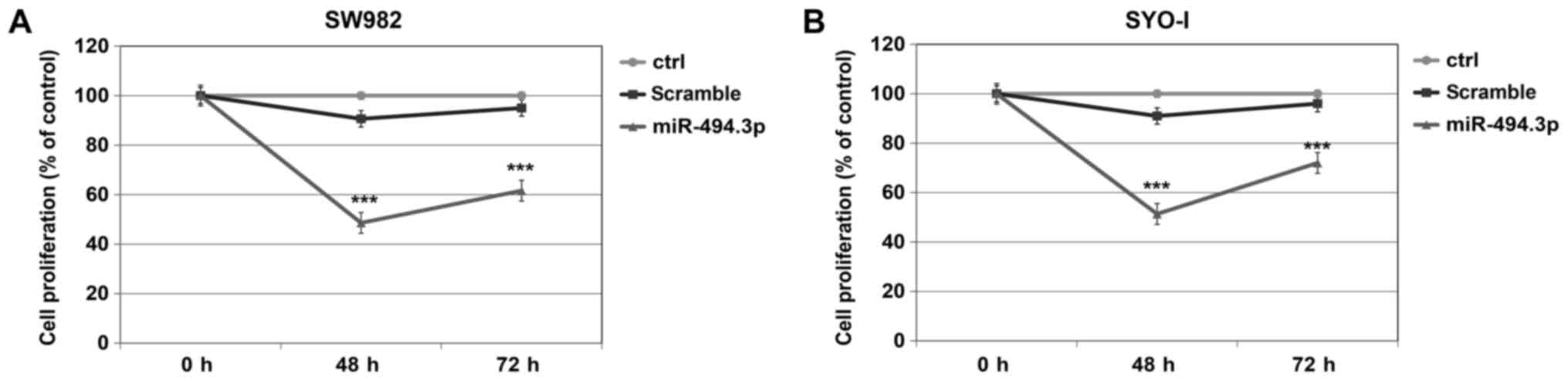
When compared to the scramble control, both the
SW982 and SYO-1 cells responded to miR-494.3p transfection with a
significant decrease in cell proliferation, of 86.42 and 77.21%,
respectively at 48 h (P=0.001) and of 54.22% and 33.33%,
respectively at 72 h (P=0.001) (Fig.
5).
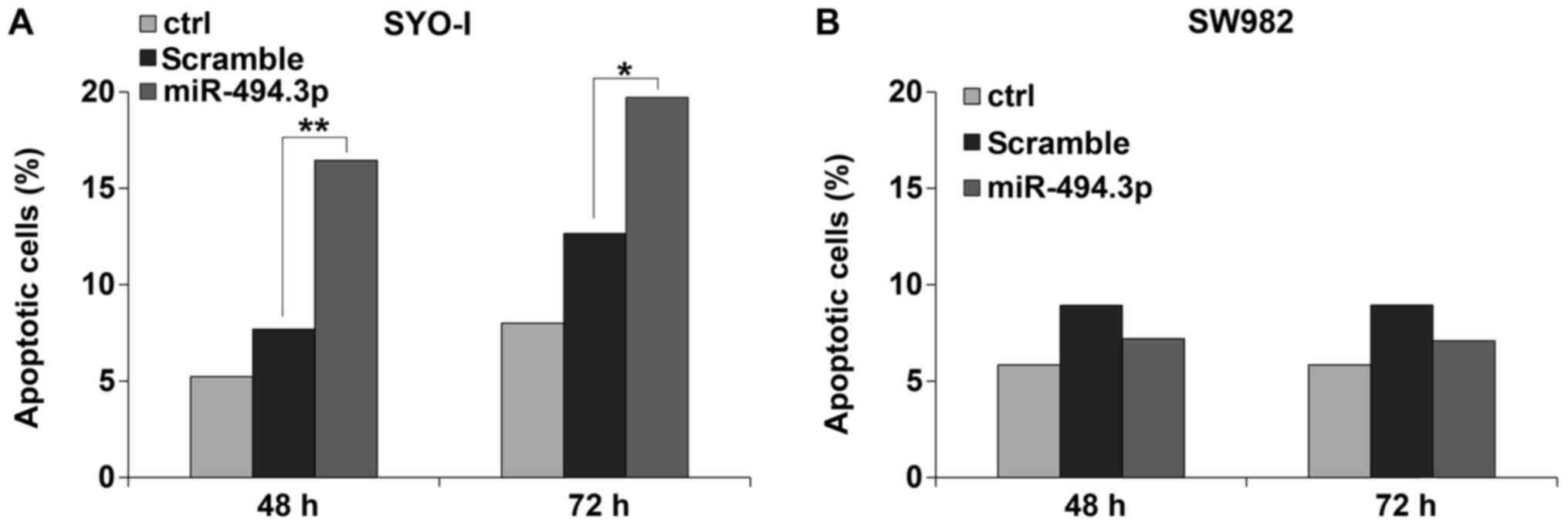
Concomitantly, the SYO-I cells responded with a
significant increase in the apoptotic fraction: 16.4% vs. 7.7% at
48 h (P=0.01) and 19.7% vs. 12.6% at 72 h (P=0.05) (Fig. 6A). No significant changes were
found for the SW982 cells (Fig.
6B).
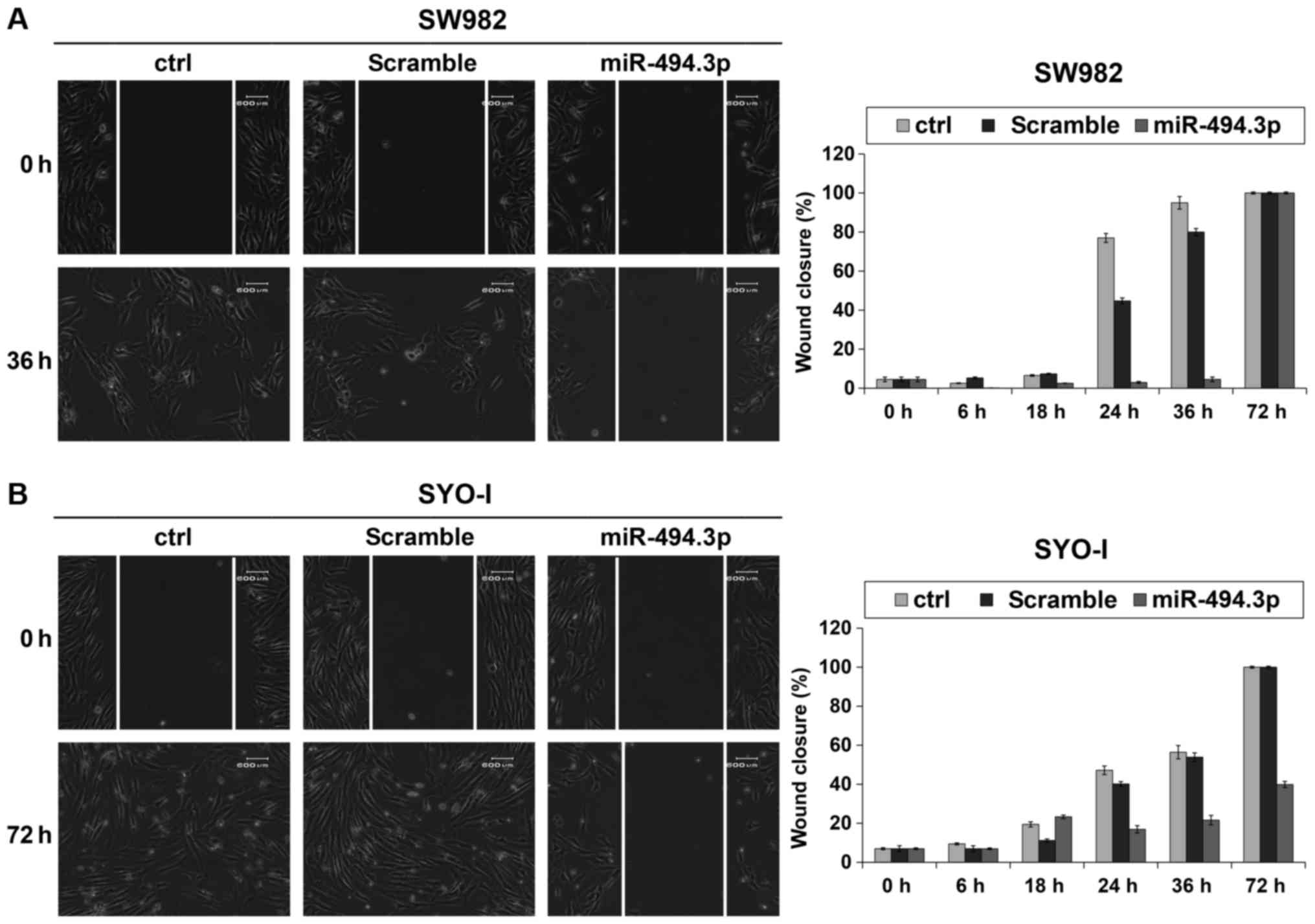
Migration assay revealed that the ectopic expression
of miR-494.3p attenuated the migration of the SW982 cells up to 36
h shifting to control values at 72 h (Fig. 7A), while the SYO-I cells also
exhibited a slower migration, reaching a complete coverage of the
scratch area only after 72 h(Fig.
7B).
Discussion
SS is an aggressive tumour responsible of 7-8% of
malignant sarcomas. Localized SS has a high overall survival;
however, in relapsed patients the 5-year survival rate is 20-30%
with therapeutic strategies limited to surgical resection assisted
by radiotherapy, since chemotherapy is often ineffective and lacks
specific protocols (21).
Following the failure of anthracyclines, the multi-kinase
inhibitor, pazopanib, is the first targeted agent to be approved
for the treatment of advanced SS (22); however, other biomarkers are
required for prognosis and therapy.
On the bases of our previous results that identified
CXCR4 nuclear protein as a strong independent adverse prognostic
factor for SS patient survival (11), we analysed the CXCR4 mRNA levels in
a series of monophasic localized and metastatic SS specimens. The
significantly higher expression of the CXCR4 gene in the tumour
compared to normal tissue, also observed at protein level,
validated the role of the chemokine in tumour development (9,23).
Metastatic patients presented higher CXCR4 levels
than metastasis-free patients, with a difference at the limit of
statistical significance. Although no associations were found with
overall survival or other clinical parameters, probably requiring a
larger cohort, as in other STS (24), our data confirmed the association
between CXCR4 overexpression and poor prognosis in SS.
Biological data suggest that CXCR4 is a possible
candidate target of miR-494.3p that is often found downregulated in
tumours (16,17). Several studies have demonstrated a
deregulation of miRNAs in sarcoma, including SS (25) and Subramanian et al
suggested an association between miR-143 and the regulation of the
SYT/SSX-1 fusion gene (14). However, to date no evidence of a
correlation between miRNA expression and SS translocation fusion
genes has been found, at least to the best of our knowledge. In
chondrosarcoma, a low expression of miR-494.3p has been shown to be
associated with a poor prognosis (18).
In this study, miR-494.3p expression was
significantly lower in tumour compared to normal tissue. The
opposite trend of miR-494.3p and CXCR4 in SS emphasized the role of
miR-494.3p as a post-transcriptional regulator of CXCR4, confirming
previous data in prostate and breast cancer cells where this role
was observed by CXCR4 inhibition following miR-494.3p mimic
transfection (16,17).
In this study, concomitantly with higher CXCR4
levels, patients with SS with metastasis presented with lower
miR-494.3p levels when compared to those with no metastasis. In
order to demonstrate whether SS cell behaviour was modulated by
miR-494.3p through its potential target CXCR4, we transfected SW982
and SYO-I cell lines with miR-494-3p precursor. The significant
decrease in CXCR4 expression was associated with a decrease in cell
proliferation which was more evident in the SYO-I cells, that also
responded with a significant increase in the apoptotic cell
fraction in accordance with data on chronic myeloid leukemia and
prostate cancer (16,26).
In the SW982 cells we found that the significant
decrease in cell proliferation was not accompanied by an increased
apoptotic fraction probably due to a different apoptotic pathway
compared to the SYO-I cells (27,28).
The overexpression of miR-494.3p also inhibited ovarian cancer cell
proliferation by inducing apoptosis via FGFR2 (29), while in hepatocellular carcinoma it
conferred chemotherapy resistance by targeting PTEN and increasing
AKT expression (30).
By the evidence that the CXCL12/CXCR4 pathway may
play a role in the development of metastatic disease in STS
(9) we assessed the effect of
CXCR4 modulation on SS cell migration. Both the SW982 and SYO cells
responded to miR-494.3p overexpression by decreasing cell motility,
lasting longer in the SYO-I cells. Some data reported the role of
miR-494.3p as a promoter of cell migration in breast and lung
cancer (31,32), while by targeting different
pathways it also inhibited cell proliferation, migration and
invasion in ovarian cancer (29),
gastric cancer (33), osteosarcoma
(19) and chondrosarcoma (18).
In conclusion, the data of this study demonstrated a
negative significant correlation between CXCR4 and miR-494.3p
expression in SS surgical specimens and revealed that
non-metastatic patients presented a lower expression of CXCR4
concomitant with higher levels of miR-494.3p when compared to
metastatic patients. In vitro studies confirmed that
miR-494.3p transfection caused a reduction in its potential target
CXCR4, concomitant with a decrease in cell migration and
proliferation. Ongoing studies are required to better clarify the
clinical impact of CXCR4 in terms of prognostic and therapeutic
biomarker in SS.
Funding
This study was supported by 5‰ citizen income tax
contribution to the Rizzoli Orthopaedic Institute and Italian
Health Ministry.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LP performed the experiments, analysed data and
wrote the manuscript; SP performed the experiments and analysed the
data; MV and EB performed the experiments; SB and MG were in charge
of the pathological specimens; CF and EP were involved in patient
follow-up; PP conceived the study and approved the final version to
be published; MSB conceived the study, performed statistical
analysis, wrote the manuscript, and approved the final version to
be published. All authors have read and approved the contents of
this manuscript and declare that the work is original and had not
been submitted or published elsewhere.
Ethics approval and consent to
participate
All samples were treated in accordance with the
authorizations issued by the Ethics Committee of the the Rizzoli
Orthopedic Institute (no. 0033276) and written informed consent was
obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors wish to thank Dr A. Kawai for kindly
providing the SYO-I cell line, Dr Alba Balladelli for editing the
manuscript and Ms. Cristina Ghinelli for graphic work.
References
|
1
|
Picci P, Manfrini M, Fabbri N, Gambarotti
M and Vanel D: Synovial sarcoma In: Atlas of Musculoskeletal Tumors
and Tumor like Lesions. Springer; International Publishing, Cham;
pp. 359–364. 2014
|
|
2
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Furusato B, Mohamed A, Uhlén M and Rhim
JS: CXCR4 and cancer. Pathol Int. 60:497–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim J, Mori T, Chen SL, Amersi FF,
Martinez SR, Kuo C, Turner RR, Ye X, Bilchik AJ, Morton DL, et al:
Chemokine receptor CXCR4 expression in patients with melanoma and
colorectal cancer liver metastases and the association with disease
outcome. Ann Surg. 244:113–120. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Müller A, Homey B, Soto H, Ge N, Catron D,
Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al:
Involvement of chemokine receptors in breast cancer metastasis.
Nature. 410:50–56. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koshiba T, Hosotani R, Miyamoto Y, Ida J,
Tsuji S, Nakajima S, Kawaguchi M, Kobayashi H, Doi R, Hori T, et
al: Expression of stromal cell-derived factor 1 and CXCR4 ligand
receptor system in pancreatic cancer: A possible role for tumor
progression. Clin Cancer Res. 6:3530–3535. 2000.PubMed/NCBI
|
|
7
|
Schimanski CC, Schwald S, Simiantonaki N,
Jayasinghe C, Gönner U, Wilsberg V, Junginger T, Berger MR, Galle
PR and Moehler M: Effect of chemokine receptors CXCR4 and CCR7 on
the metastatic behavior of human colorectal cancer. Clin Cancer
Res. 11:1743–1750. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zeelenberg IS, Ruuls-Van Stalle L and Roos
E: The chemokine receptor CXCR4 is required for outgrowth of colon
carcinoma micrometastases. Cancer Res. 63:3833–3839.
2003.PubMed/NCBI
|
|
9
|
Kim RH, Li BD and Chu QD: The role of
chemokine receptor CXCR4 in the biologic behavior of human soft
tissue sarcoma. Sarcoma. 2011.593708:2011.
|
|
10
|
Li YJ, Dai YL, Zhang WB, Li SJ and Tu CQ:
Clinicopathological and prognostic significance of chemokine
receptor CXCR4 in patients with bone and soft tissue sarcoma: A
meta-analysis. Clin Exp Med. 17:59–69. 2017. View Article : Google Scholar
|
|
11
|
Palmerini E, Benassi MS, Quattrini I,
Pazzaglia L, Donati D, Benini S, Gamberi G, Gambarotti M, Picci P
and Ferrari S: Prognostic and predictive role of CXCR4, IGF-1R and
Ezrin expression in localized synovial sarcoma: is chemotaxis
important to tumor response. Orphanet J Rare Dis. 10:62015.
View Article : Google Scholar
|
|
12
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Subramanian S, Lui WO, Lee CH, Espinosa I,
Nielsen TO, Heinrich MC, Corless CL, Fire AZ and van de Rijn M:
MicroRNA expression signature of human sarcomas. Oncogene.
27:2015–2026. 2008. View Article : Google Scholar
|
|
15
|
Fujiwara T, Kunisada T, Takeda K and Ozaki
T: MicroRNAs and soft tissue sarcomas. Adv Exp Med Biol.
889:179–199. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shen PF, Chen XQ, Liao YC, Chen N, Zhou Q,
Wei Q, Li X, Wang J and Zeng H: MicroRNA-494-3p targets CXCR4 to
suppress the proliferation, invasion, and migration of prostate
cancer. Prostate. 74:756–767. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song L, Liu D, Wang B, He J, Zhang S, Dai
Z, Ma X and Wang X: miR-494 suppresses the progression of breast
cancer in vitro by targeting CXCR4 through the Wnt/β-catenin
signaling pathway. Oncol Rep. 34:525–531. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Wang L, Liu Z, Zu C, Xing F, Yang P,
Yang Y, Dang X and Wang K: MicroRNA-494 inhibits cell proliferation
and invasion of chondrosarcoma cells in vivo and in vitro by
directly targeting SOX9. Oncotarget. 6:26216–26229. 2015.PubMed/NCBI
|
|
19
|
Zhi X, Wu K, Yu D, Wang Y, Yu Y, Yan P and
Lv G: MicroRNA-494 inhibits proliferation and metastasis of
osteosarcoma through repressing insulin receptor substrate-1. Am J
Transl Res. 8:3439–3447. 2016.PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Δ Δ C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
21
|
Krieg AH, Hefti F, Speth BM, Jundt G,
Guillou L, Exner UG, von Hochstetter AR, Cserhati MD, Fuchs B,
Mouhsine E, et al: Synovial sarcomas usually metastasize after
>5 years: A multi-center retrospective analysis with minimum
follow-up of 10 years for survivors. Ann Oncol. 22:458–467. 2011.
View Article : Google Scholar
|
|
22
|
van der Graaf WT, Blay JY, Chawla SP, Kim
DW, Bui-Nguyen B, Casali PG, Schöffski P, Aglietta M, Staddon AP,
Beppu Y, et al EORTC Soft Tissue and Bone Sarcoma Group: PALETTE
study group: Pazopanib for metastatic soft-tissue sarcoma
(PALETTE): A randomised, double-blind, placebo-controlled phase 3
trial. Lancet. 379:1879–1886. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Balkwill F: The significance of cancer
cell expression of the chemokine receptor CXCR4. Semin Cancer Biol.
14:171–179. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Oda Y, Tateishi N, Matono H, Matsuura S,
Yamamaoto H, Tamiya S, Yokoyama R, Matsuda S, Iwamoto Y and
Tsuneyoshi M: Chemokine receptor CXCR4 expression is correlated
with VEGF expression and poor survival in soft-tissue sarcoma. Int
J Cancer. 124:1852–1859. 2009. View Article : Google Scholar
|
|
25
|
Yu PY, Balkhi MY, Ladner KJ, Alder H, Yu
L, Mo X, Kraybill WG, Guttridge DC and Iwenofu OH: A selective
screening platform reveals unique global expression patterns of
microRNAs in a cohort of human soft-tissue sarcomas. Lab Invest.
96:481–491. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Salati S, Salvestrini V, Carretta C,
Genovese E, Rontauroli S, Zini R, Rossi C, Ruberti S, Bianchi E,
Barbieri G, et al: Deregulated expression of miR-29a-3p miR-494-3p
and miR-660-5p affects sensitivity to tyrosine kinase inhibitors in
CML leukemic stem cells. Oncotarget. 8:49451–49469. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jones KB, Su L, Jin H, Lenz C, Randall RL,
Underhill TM, Nielsen TO, Sharma S and Capecchi MR: SS18–SSX2 and
the mitochondrial apoptosis pathway in mouse and human synovial
sarcomas. Oncogene. 32:2365–2371. 2013. View Article : Google Scholar
|
|
28
|
Joyner DE, Albritton KH, Bastar JD and
Randall RL: G3139 antisense oligonucleotide directed against
antiapoptotic Bcl-2 enhances doxorubicin cytotoxicity in the
FU-SY-1 synovial sarcoma cell line. J Orthop Res. 24:474–480. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao X, Zhou Y, Chen YU and Yu F: miR-494
inhibits ovarian cancer cell proliferation and promotes apoptosis
by targeting FGFR2. Oncol Lett. 11:4245–4251. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu K, Liu S, Zhang W, Jia B, Tan L, Jin Z
and Liu Y: miR-494 promotes cell proliferation, migration and
invasion, and increased sorafenib resistance in hepatocellular
carcinoma by targeting PTEN. Oncol Rep. 34:1003–1010. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Macedo T, Silva-Oliveira RJ, Silva VAO,
Vidal DO, Evangelista AF and Marques MMC: Overexpression of mir-183
and mir-494 promotes proliferation and migration in human breast
cancer cell lines. Oncol Lett. 14:1054–1060. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Faversani A, Amatori S, Augello C, Colombo
F, Porretti L, Fanelli M, Ferrero S, Palleschi A, Pelicci PG,
Belloni E, et al: miR-494-3p is a novel tumor driver of lung
carcinogenesis. Oncotarget. 8:7231–7247. 2017. View Article : Google Scholar :
|
|
33
|
Zhao XQ, Liang TJ and Fu JW: miR-494
inhibits invasion and proliferation of gastric cancer by targeting
IGF-1R. Eur Rev Med Pharmacol Sci. 20:3818–3824. 2016.PubMed/NCBI
|