Introduction
Renal cell carcinoma is recognized as the third most
common urologic malignancy, accounting for ~3% of all malignant
diseases in adults globally in 2017 (1). Worldwide, clear cell renal cell
carcinoma (ccRCC) remain the most prevalent RCC subtype in 2017
(2). Although patients with
localized ccRCC usually have a satisfactory prognosis, numerous
patients are initially diagnosed at advanced stages, which are
characterized by a high degree of malignancy, high rates of local
invasion, and resistance to chemotherapy and radiotherapy (3,4).
Thus, an improved understanding of the mechanisms underlying ccRCC
progression and metastasis is urgently required to identify novel
biomarkers and develop more effective treatments for ccRCC.
MicroRNAs (miRNAs) are a class of small non-coding
RNA molecules ~22 nucleotides in length (5). A high level of a miRNAs in tissues
can result in degradation of target mRNA or translation blockage
(6,7). With the growing number of miRNA chips
in renal cancer, the miRNA expression data are frequently
inconsistent (8). Therefore,
comprehensive analysis of similar chips is of particular importance
(9,10). A number of studies used
bioinformatics approaches to identify novel diagnostic and
prognostic biomarkers; in particular, the analysis of miRNAs
expression datasets is a beneficial tool (11,12).
In the present study, four miRNA expression profiles
of ccRCC from the Gene Expression Omnibus (GEO) datasets were
analyzed, and then candidate miRNAs in The Cancer Genome Atlas
(TCGA)-Kidney Renal Clear Cell Carcinoma (KIRC) dataset were
validated, which revealed that miR-122 was highly expressed in
ccRCC and is associated with poor survival time. miR-122 was one of
the first examples of a tissue-specific miRNA of the liver that is
involved in multiple metabolic processes, including fatty acid
synthesis, cholesterol biosynthesis and β-oxidation (13-17).
Numerous studies demonstrated that miR-122 is dysregulated in a
number of cancer types, including liver, breast and renal cancer
(16-18). In liver cancer, loss of miR-122 may
result in the suppression of a hepatic phenotype and acquisition of
invasive properties (19). In
breast cancer, miR-122 has been determined to be rich in cancer
exosomes that assist with distant metastasis by affecting their
sugar metabolism (20). However,
the current knowledge of miR-122 regulation in renal cancer at a
molecular level remains limited.
Since predicted targets of miR-122 were enriched for
the Forkhead box O (FOXO) family signaling pathway (21), it was hypothesized that miR-122 may
function by targeting the FOXO3 mRNA FOXO3 belongs to the
Forkhead gene family, encoding the transcription factor FOXO3,
which exhibits consistent associations with longevity, apoptosis,
proteostasis and autophagy (22-25).
Notably, FOXO3 can suppress tumor growth (26). In leukemia and breast cancer cells,
anticancer drugs upregulate FOXO3 resulting in increased
B-cell lymphoma-2 (Bcl-2) like 11 (Bim) expression and
consequently inhibition of tumor growth (27-29).
Additionally, FOXO3 downregulates Myc, a stimulator of tumor
cell proliferation and survival (30,31).
The FOXO3 protein has been reported to be significantly
down-regulated in renal cancer, compared with adjacent tissues, but
the FOXO3 mRNA level was not notably changed, indicating a
post-transcriptional regulation may exist (32,33).
In the present study, it was demonstrated that
miR-122 is highly upregulated in ccRCC, and is associated with
ccRCC metastasis. miR-122 promotes cell proliferation and invasion
of ccRCC by targeting FOXO3 mRNA.
Materials and methods
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Chinese People’s Liberation Army (PLA) General Hospital (Beijing,
China). Each enrolled patient signed written informed consent prior
to sample collection. Animal experiments were approved by the
Experimental Animal Ethical Committee of Chinese PLA General
Hospital and were conducted in accordance with the Guide for the
Care and Use of Laboratory Animals (34).
Patients and tissue samples
The inclusion criteria included: i) Clinical and
imaging diagnosis of primary non-metastatic ccRCC; ii) aged 18-90
years old; and iii) underwent nephrectomy at Chinese PLA General
Hospital. The exclusion criteria included: i) Missing imaging data;
and ii) patients with severe liver and kidney disease,
cardiovascular disease, blood disease or other malignant tumor
types. A total of 46 ccRCC tissues, paired with adjacent non-tumor
renal tissues, were recruited randomly from patients who were
diagnosed with primary non-metastatic ccRCC and underwent
nephrectomy at the Department of Urology, Department of People’s
Liberation Army (PLA) General Hospital (Beijing, China) between
November 2013 and October 2015. The patient age range is 25-76
years, with a mean age of 54.2 years, and 50% were male and female.
All specimens were pathologically confirmed to be ccRCC by senior
pathologists of PLA General Hospital who were blind to the study.
The patients were followed up for 15-48 months (median, 35 months).
The nuclear grades and clinical stages were determined according to
the 2009 Fuhrman nuclear grading system and Tumor-Node-Metastasis
classification system, respectively, and 11 of them developed
metastasis at the endpoint (35,36).
Microarray data
The miRNA expression profiles were downloaded from
the GEO database (http://www.ncbi.nlm.nih.gov/geo/; accession numbers,
GSE24457, GSE23085, GSE71302 and GSE95385), which yielded 33 ccRCC
tissue samples and 33 paired normal samples in total. The data was
analyzed with the Morpheus Website (https://software.broadinstitute.org/morpheus/) using
P<0.05 and [logFC]>1 as cut-off criterion.
miRNA analysis of TCGA
OncoLnc online software (http://www.oncolnc.org) was used to validate the
expression of miRNAs. The TCGA-KIRC datasets (https://portal.gdc.cancer.gov/projects/TCGA-KIRC)
contains 506 ccRCC samples with clinical data.
miRNA target prediction and function
analysis
A total of four different target prediction datasets
were employed to identify the presumed targets of has-miR-122,
including TargetScan v7.1 (http://www.targetscan.org/), miRDB (http://www.mirdb.org/), miR-TarBase v7.0 (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and
PicTar (http://www.pictar.org/). Unique genes
with target sites in 3′-untranslated region (UTR) were
incorporated. Signaling pathway enrichment was performed using
Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/) PATHWAY websites.
Cell culture
Human ccRCC cell lines 769-P, 786-O, Caki-1, A498,
OS-RC-2 and SN12-PM6, the human renal proximal tubular epithelial
cell line HKC and the 293T cell line were purchased from the
Institute of Basic Medical Sciences Chinese Academy of Medical
Sciences, China Infrastructure of Cell Line Resources (Beijing,
China). 769-P, 786-O, Caki-1, A498, OS-RC-2 and SN12-PM6 cells were
used for RT-qPCR. 786-O and SN12-PM6 cells were used for western
blotting, immunofluorescence, MTS, colony formation, Transwell and
wound-healing assay. The 293T cell line was used for Luciferase
reporter assay. The cells were cultured in RPMI-1640 medium or
Dulbecco’s modified Eagle’s medium (both from HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) and maintained in a humidified atmosphere containing 5%
CO2 at 37°C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The total RNA was extracted from the cancer and
adjacent normal tissues of the selected patients using
TRIzol® reagent (Kang Wei Century Biological Technology
Co., Ltd., Beijing, China). Reverse transcription of mRNA was
performed using a One Step Realtime-PCR kit (Beijing Transgen
Biotech Co., Ltd., Beijing, China) and miRNA cDNA synthesis was
performed using a miRcute miRNA first-strand cDNA kit (Tiangen
Biotech, Beijing, China), according to the manufacturer’s
protocols. The miR-122 expression level was detected by using a
miRNA miRcute qPCR detection kit (Tiangen Biotech), according to
the manufacturer’s protocol, and miR-122 expression level was
normalized to small nucleolar RNA U6 and calculated using the
2-∆Cq method, where ∆Cq = Cq miR-122 - Cq U6 (37). FOXO3 mRNA expression level was
determined on the Applied Biosystems 7500 System using
SYBR® Green (Tiangen Biotech Co., Ltd.). The thermal
cycling conditions are 94°C for 30 sec, then 40 cycles for 94°C for
5 sec and 64°C for 34 sec. Peptidylprolyl isomerase A was used to
normalize the relative mRNA levels. The primers of FOXO3 are as
follows: 5′-CAAAGCAGACCCTCAAAC-3′ (sense) and
5′-CGGTATCACTGTCCACTT-3′ (antisense). The primer sequences of
peptidylprolyl isomerase A are as follows:
5′-ATGGTCAACCCCACCGTGT-3′ (sense) and 5′-TCTGCTGTCTTTGGGACCTTGTC-3′
(antisense).
RNAi treatment
Small interfering RNAs (siRNAs) against FOXO3
(siFOXO3), small interfering negative controls (siNC), mimics
miR-122, mimics NC, inhibitor miR-122 and inhibitor NC were
synthesized by Tiangen Biotech Co., Ltd.. siFOXO3 and siNC were
being transfected into 786-O cells, and mimics miR-122 and
inhibitor miR-122 and the corresponding NCs were transfected into
786-O and SN12-PM6 cells. Transfection was conducted using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Essential experiments were conducted 48 h after
transfection. The sequences are as follows: siFOXO3
5′-CCAAUGUGUUUCAACUUUAAA-3′ (sense), and
5′-UAAAGUUGAAACACAUUGGAG-3′ (antisense); siNC
5′-UAGGAGUGGAGCAGAGUGAAG-3′ (sense), and
5′-GCUAUGGGUGUCGAUAAUGAG-3′ (antisense); miR-122 mimics
5′-UGGAGUGUGACAAUGGUGUUUG-3′ (sense), and
5′-AACACCAUUGUCACACUCCAUU-3′ (antisense); miR-122 NC
5′-UUCUCCGAACGUGUCACGUTT-3′ (sense), and
5′-ACGUGACACGUUCGGAGAATT-3′ (antisense); inhibitor miR-122
5′-CAAACACCAUUGUCACACUCCA-3′ (sense), and
5′-GAGUGUGUGACAAUGGUGUUGC-3′ (anti-sense); inhibitor NC
5′-CAGUACUUUUGUGUAGUACAA-3′ (sense), and 5′-GUACUACACAAAAGUACGA-3′
(antisense).
Western blotting
Total proteins of tissues or cells were extracted
with radioimmunoprecipitation assay lysis buffer (Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China) mixed with
EDTA-free Protease Inhibitor (Roche Applied Science, Penzberg,
Germany). Protein concentrations were detected using the
bicinchoninic acid method. The proteins (5 μg) were
separated using 12% SDS-PAGE, and then transferred to
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). The membranes were blocked with 5% bovine serum albumin
(Beijing Solarbio Science & Technology Co., Ltd.) for 1 h at
37°C and then incubated with primary antibodies against FOXO3
(1:1,000; cat. no. 2497; Cell Signaling Technology, Inc., Danvers,
MA, USA), E-cadherin (1:1,000; cat. no. 24E10; Cell Signaling
Technology, Inc.), N-cadherin (1:1,000; cat. no. D4R1H; Cell
Signaling Technology), α-smooth muscle actin (SMA; 1:200; cat. no.
ab7817; Abcam, Cambridge, UK) and β-actin (1:2,000; cat. no. TA-08;
OriGene Technologies, Inc., Rockville, MD, USA) overnight at 4°C
followed by horseradish peroxidase-conjugated secondary goat
anti-rabbit and anti-mouse IgG (H+L) antibodies (1:2,000; cat. no.
ab97051 and ab97023, respectively; Abcam) incubation for 1 h at
37°C. Immunoreactive bands of the proteins were normalized to
β-actin and visualized using enhanced chemiluminescence detection
reagent (Thermo Fisher Scientific, Inc.).
Immunofluorescence
After 24-h transfection the treated cells were
seeded on coverslips. Following fixation with 4%
paraformaldehyde-PBS for 10 min at room temperature, cells were
permeabilized with 0.5% Triton X-100 for 15 min and blocked with
bovine serum albumin (3%) for 30 min at room temperature. The cover
slips were incubated with primary anti-FOXO3 antibody at 1:200
dilution for 1 h at 37°C and then incubated with fluorescein
isothiocyanate-conjugated goat anti-rabbit secondary antibodies
(cat. no. TA130015; OriGene Technologies, Inc.) at 1:100 dilution
for 1 h at 37°C. Nuclei staining was counterstained with 0.2 mg/ml
DAPI for 15 min at 37°C. Stained cells were visualized under an
Olympus confocal microscope at ×200 magnification (Olympus
Corporation, Tokyo, Japan).
Construction of plasmids
During plasmid construction, the open reading frame
of FOXO3 (2.5 ng; 0.25 ng/μl) was cloned into the
PLV-EGFP(2A) lentiviral (25 ng; 2.5 ng/μl) vector (InovoGen
Tech. Co., Beijing, China) between the sites of XbaI and
EcoRI to generate PLV-EGFPFOXO3. The miR-122 segment (2.5
ng; 0.25 ng/μl; InovoGen Tech. Co) was inserted into
pLVshRNA-EGFP(2A) lentiviral vector (2.5 ng; 0.25 ng/μl;
InovoGen Tech. Co.) to generate PLV-EGFPmiR-122. The sequence of
miR-122 segment is
5′-CCTTAGCAGAGCTGTGGAGTGTGACAATGGTGTTTGTGTCTAAACTATCAAACGCCATTATCACACTAAATAGCTACTGCTAGGC-3′.
PLV-EGFPFOXO3, PLV-EGFPmiR-122 and PLV-EGFP empty vector (EV) were
transfected into cells using Lipofectamine 2000 to synthesize FOXO3
overexpressed, miR-122 overexpressed and control ccRCC cells
according to the aforementioned protocol. The subsequent
experimentations were conducted two weeks after the
transfection.
Luciferase reporter assay
The wild-type or mutated 3′-UTR of FOXO3 containing
the miR-122 binding site was cloned into a psiCHECK2 vector
(Promega Corporation, Madison, WI, USA) provided by Genewiz, Inc.
(Beijing, China). To test the function of miR-122 on luciferase
activity, 293T cells were co-transfected with luciferase reporter
of wild-type (WT) or mutated (MUT) 3′-UTR and miR-122 mimics or
control using Lipofectamine 2000. Cells were assayed using
Dual-Luciferase® Reporter Assay System (cat. no. E1910;
Promega Corporation) 24 h after transfection. Luciferase assays
were measured on the basis of ratio of Renilla/firefly
luciferase activities in accordance with the Dual-Luciferase Assay
Manual (38). The luciferase
activity of each control vector was set to 1.
MTS assay
The treated cells were seeded into a 96-well plate
(1×103 cells/well). At 0, 24, 48, 72, 96 and 120 h after
seeding in 10% FBS RPMI-1640 medium at 37°C, 20 μl Celltiter
96® AQueous One Solution (Promega Corporation) was added
to the cells and then co-incubated for 2 h at 37°C prior to
absorbance measurement. The absorbance was recorded at 490 nm using
an automatic enzyme-linked immunosorbent assay reader (BioTek
Instruments, Inc., Winooski, VT, USA) with a 96-well plate
reader.
Colony formation assay
The treated cells were seeded on a 6-well plate at a
density of 1×103 cells/well. After culturing in 10% FBS
RPMI-1640 medium at 37°C for 14 days, the treated cells were fixed
with 100% methanol for 15 min at room temperature and stained with
1% crystal violet for 20 min at room temperature prior to the
colony numbers being counted. Colonies consisting of ≥50 cells were
counted under a light microscope at ×200 magnification.
Transwell migration and Matrigel invasion
assays
Transwell assays were performed in Transwell
chambers (Corning Incorporated, Corning, NY, USA) containing
polycarbonate membrane filters with a pore size of 8 μm. The
membranes were coated with Matrigel (200 ng/ml; BD Biosciences,
Franklin Lakes, NJ, USA) for the invasion assay. A total of
1×104 cells in 200 μl serum-free RPMI-1640 medium
were seeded in the upper chamber, and 500 μl 10% FBS
RPMI-1640 medium was added into the lower chamber. After culturing
for 12 (migration) or 24 (invasion) h at 37°C, the cells that
adhered to the lower surface of the Transwell chambers were fixed
with 100% methanol for 15 min at room temperature and stained with
1% crystal violet for 20 min at room temperature. The cells were
counted in five random fields under a light microscope at ×200
magnification (Olympus Corporation; ×200), and the mean values were
then calculated.
Wound-healing assay
A wound-healing assay was performed in 6-well
plates. A total of 2×105 treated cells were seeded on
6-well plates. After culturing in RPMI-1640 medium at 37°C
overnight, the confluent monolayer of cells was serum-starved and
scratched using a sterile 200 μl pipette tip. Images of the
same position were captured using a light microscope at ×20
magnification at 0, 6 and 12 h after the scratching. The coverage
of the scratching area was measured at five random positions for
each well.
In vivo growth assay of xenograft
tumor
A total of 1×107 ccRCC cells stably
expressing plv-miR-122 or EV were subcutaneously injected into the
left armpit of male BALB/c nude mice (4 weeks old; weight, 14±1.2
g; 10 mice/group; 20 mice in total), which were supplied by Animal
Experiment Center of PLA General Hospital and maintained at a
specific pathogen free facility with a constant humidity and
temperature at 26°C, filtered air, and free access to food and
water at 12/12-h light/dark cycle. Following tumor formation, the
tumor volume was calculated weekly by using the following formula:
V (mm3) = 0.5 × length (mm) × width2
(mm2). All mice were sacrificed by cervical dislocation
for weight measurement of xenograft tumors eight weeks after the
injection.
Statistical analysis
SPSS 23.0 (SPSS, Inc., Chicago, IL, USA) and Prism
5.0 (GraphPad Software, Inc., La Jolla, CA, USA) software were used
for all statistical analyses. Normally distributed variables were
presented as means ± standard deviation and compared using unpaired
Student’s t-test or one-way analysis of variance. Multiple
comparisons between the groups were performed using
Student’s-Newman-Keuls method.. Disease-free survival time was used
for prognostic evaluation of patients with ccRCC, which was defined
as the time from the date the patient underwent radical or partial
nephrectomy to the date of local recurrence, distant metastasis or
mortality. Association analysis between two variables were analyzed
by linear regression. Prognostic analysis was performed using the
Kaplan-Meier method with log-rank test. Pearson χ2 test
was used for Table I. P<0.05
was considered to indicate a statistically significant
difference.
 | Table IClinicopathological characteristics
of 46 patients with localized ccRCC. |
Table I
Clinicopathological characteristics
of 46 patients with localized ccRCC.
| Variables | Sex
| Age (years)
| BMI
(kg/m2)
| TNM staging
(34)
| Fuhrman grade
(35)
|
|---|
| Male | Female | <60 | ≥60 | <25 | ≥25 | I+II | III+IV | 1+2 | 3+4 |
|---|
| Total | 23 (50.00%) | 23 (50.00%) | 31 (67.39%) | 15 (32.61%) | 24 (52.17%) | 22 (47.83%) | 34 (73.91%) | 12 (26.09%) | 31 (67.39%) | 15 (32.61%) |
| High miR-122 | 14 (60.87%) | 9 (39.13%) | 17 (73.91%) | 6 (26.09%) | 10 (43.48%) | 13 (56.52%) | 15 (65.22%) | 8 (34.78%) | 17 (73.91%) | 6 (26.09%) |
| Low miR-122 | 9 (39.13%) | 14 (60.87%) | 14 (60.87%) | 9 (39.13%) | 14 (60.87%) | 9 (39.13%) | 19 (82.61%) | 4 (17.39%) | 14 (60.87%) | 9 (39.13%) |
| P-value | 1.0 | 0.35 | 0.24 | 0.17 | 0.34 |
Results
Integrated analysis identified
significantly upregulated miRNAs in ccRCC
miRNA expression profiles GSE24457, GSE23085,
GSE71302 and GSE95385 were downloaded from GEO. Upregulated miRNAs
(167, 195, 187 and 141) were extracted from the GSE24457, GSE23085,
GSE71302 and GSE95385 expression profile datasets, respectively.
Following integrated bioinformatic analysis, nine
consistently-upregulated miRNAs in ccRCC were identified from the
four profile datasets (Fig. 1A).
To further investigate whether the upregulated miRNAs associated
with ccRCC survival time, OncoLnc software was employed and
analyzed (506 ccRCC samples) survival data were analyzed for the
nine selected miRNAs. It was determined that high levels of miR-21,
miR-155 and miR-122 were significantly associated with poor overall
survival time (Fig. 1B). The other
six miRNAs had no significant effect on ccRCC prognosis.
Upregulation of miR-122 has been consistently observed in patients
with ccRCC with poor overall survival time, but few articles have
reported the functional importance of miR-122 in ccRCC pathogenesis
(39,40). The present study predicted the
target gene of miR-122 using TargetScan v7.1, miRDB, miR-TarBase
v7.0, and PicTar, and identified 87 consensus genes (Fig. 1C). Subsequently, KEGG pathway
analysis of the 87 consensus genes was performed to clarify the
potential biological functions of miR-122. Fig. 1D depicts the top six KEGG pathways
enriched of the miR-122 targets, which are: PI3K-FOXO3 pathway,
tumor necrosis factor-related apoptosis-inducing ligand signaling
pathway, ErbB receptor signaling network, sphingosine 1-phosphate
pathway, plasma membrane estrogen receptor signaling and the ADP
ribosylation factor 6 downstream pathway. Following screening, it
was determined that downregulation was only demonstrated for FOXO3
in ccRCC, so it was hypothesized that FOXO3 may be the direct
target of miR-122.
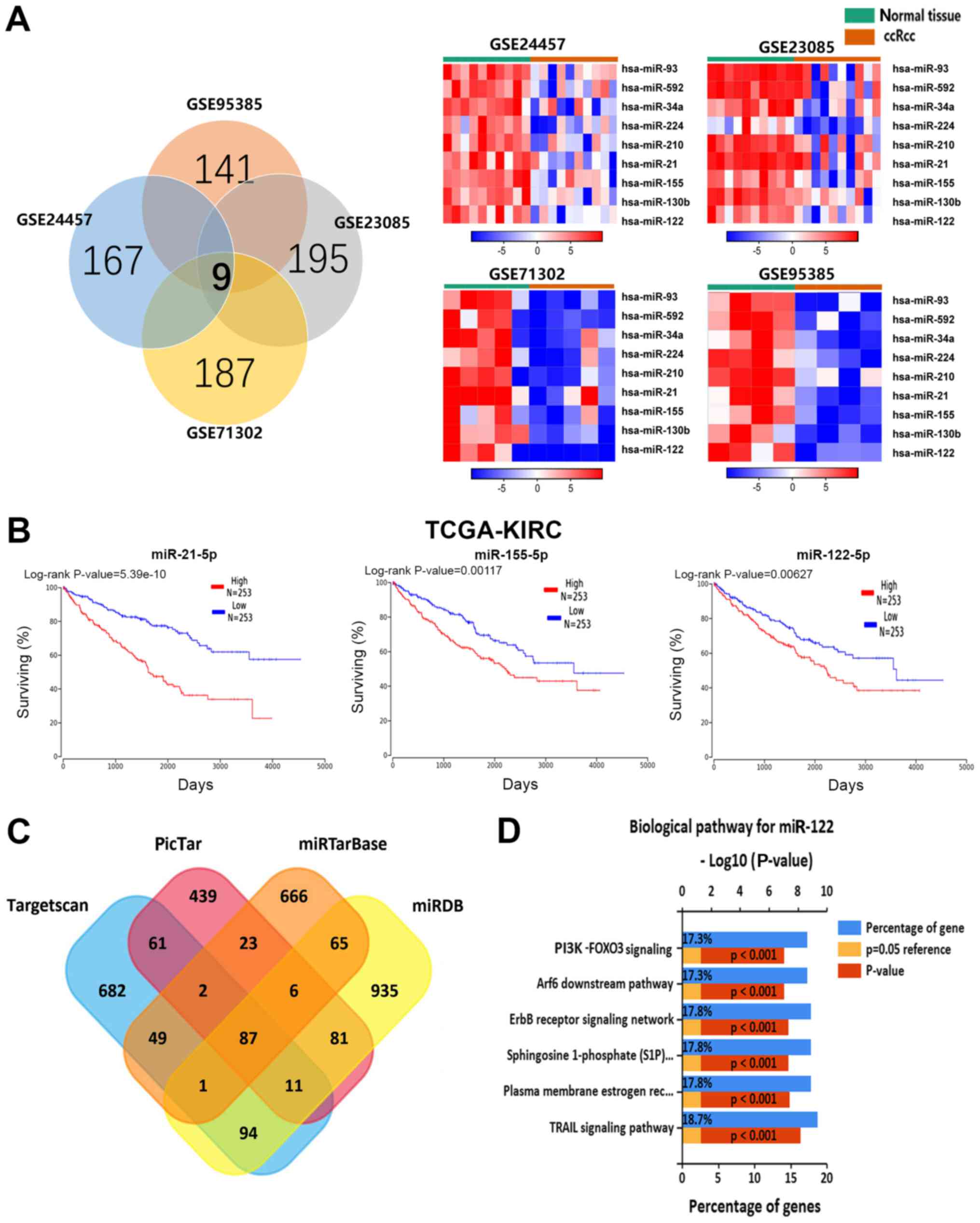 | Figure 1Integrated analysis identified
significantly upregulated miRNAs in ccRCC. (A) Identification of
nine upregulated miRNAs from the four cohort profile data sets
(GSE24457, GSE23085, GSE71302 and GSE95385) using the Morpheus
Website. Different color areas within the Venn diagram represent
different datasets, the number of overlapping parts in the figure
indicates the number of genes shared by the datasets, and the
number of non-overlapping parts indicates the total number of genes
in the dataset. (B) Kaplan-Meier analysis for overall survival time
for different miRNA expression levels from TCGA-KIRC sequencing
data. (C) Venn diagrams of putative miR-122 targets, which was
predicted by TargetScan, PicTar, miR-TarBase and miRDB. The number
of overlapping parts in the figure indicates the number of genes
shared by the datasets, and the number of non-overlapping parts
indicates the number of genes unique to the dataset. (D) The top
six Kyoto Encyclopedia of Genes and Genomes pathways are
cancer-specific pathways enriched for the miR-122 targets. miRNA,
microRNA; ccRCC, clear cell renal cell carcinoma; FOXO3, Forkhead
Box O3; PI3K, phosphoinositide 3-kinase; Arf6, ADP ribosylation
factor 6; TRAIL, tumor necrosis factor-related apoptosis-inducing
ligand; S1P, sphingosine 1-phosphate; TCGA-KIRC, The Cancer Genome
Atlas-Kidney Renal Clear Cell Carcinoma. |
Upregulation of miR-122 is associated
with downregulation of FOXO3
To investigate the role of miR-122 in ccRCC, miR-122
expression was examined in a number of ccRCC lines and 46 pairs of
ccRCC tissue samples (non-metastatic tumors and their adjacent
normal tissue specimens) (Table
I). Fig. 2A demonstrates that
miR-122 was significantly upregulated in ccRCC tissues, compared
with adjacent normal kidney tissues (P<0.001). Subsequently,
miR-122 expression level in different Fuhrman stages and T stages
of ccRCC tissues was analyzed. Compared with lower-Fuhrman-grade
group (Fuhrman I+II) and T1-stage group, significantly increased
levels of miR-122 expression were determined in the
higher-Fuhrman-grade group (Fuhrman III+IV) and later T-stage
groups (T2 and T3+T4) (Fig. 2B;
P<0.05).
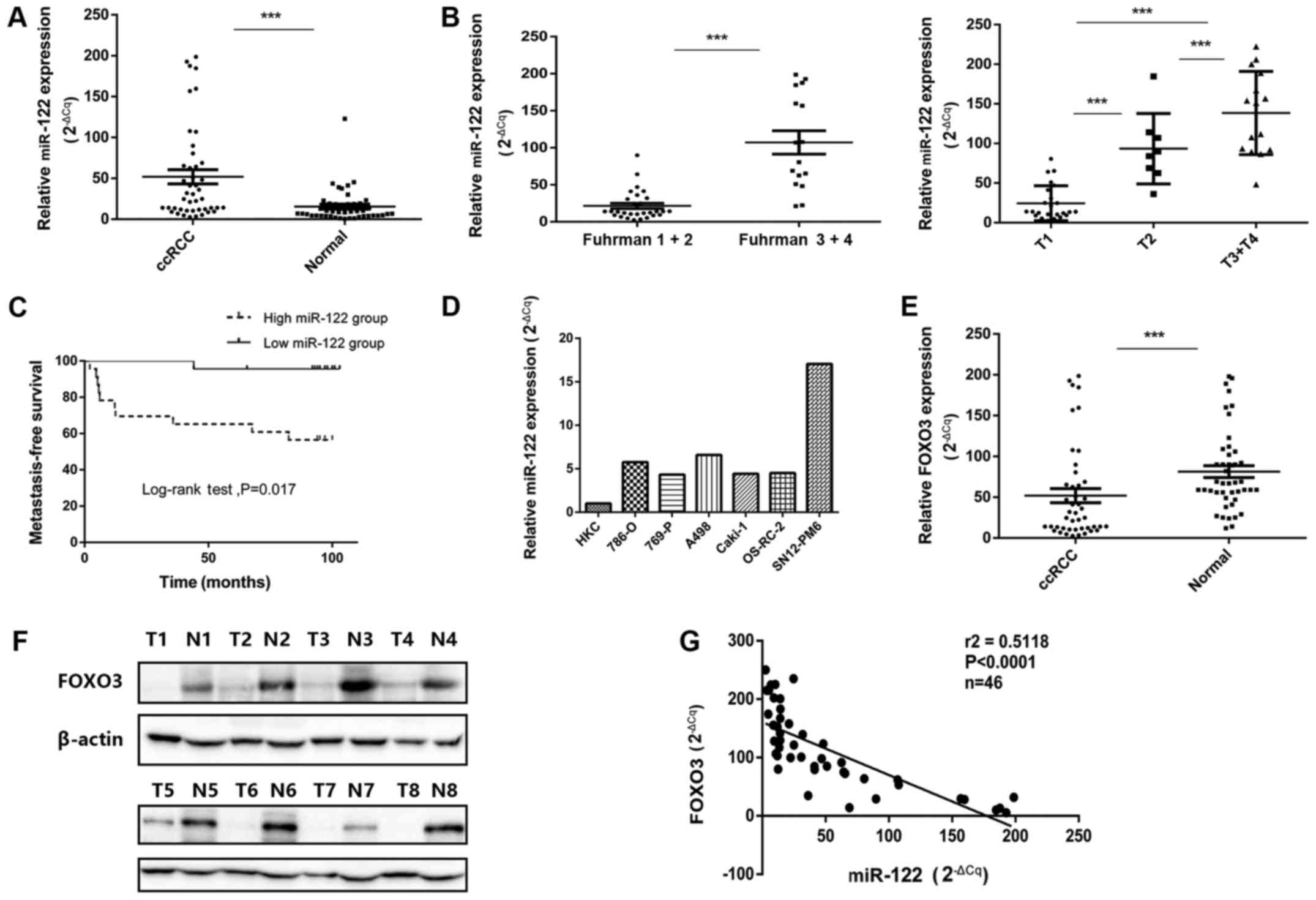 | Figure 2miR-122 expression levels in ccRCC
tissues and cell lines as well as correlation with FOXO3
expression. (A) Increased miR-122 expression levels in ccRCC
tissues, compared with adjacent normal tissues. (B) Association
between miR-122 expression levels and Fuhrman grades and T stage of
ccRCC. (C) Kaplan-Meier analysis of metastasis-free survival time
in patients with ccRCC with high (n=23) and low (n=23) miR-122
levels at initial diagnosis (P<0.01; log-rank test). The median
of the dataset served as the cutoff point. (D) miR-122 expression
levels in various ccRCC cell lines, compared with HKC cells. (E)
FOXO3 mRNA levels in ccRCC tissues, compared with normal adjacent
tissues. (F) Protein levels of FOXO3 in clinical samples. (G) The
negative correlation between FOXO3 mRNA levels and miR-122 levels
(n=46, r2=0.5118, P<0.0001). Data are presented as
the mean ± standard deviation (***P<0.001). miRNA,
microRNA; ccRCC, clear cell renal cell carcinoma; FOXO3, Forkhead
Box O3; T, tumor sample; N, normal sample. |
To investigate whether miR-122 expression is
associated with the prognosis of renal cancer, 46 selected patients
with primary non-metastatic ccRCC were followed up for 15-48 months
(median, 35 months) post-operatively. Subsequently, the ccRCC
dataset was ranked based on the miR-122 expression levels and the
median of the dataset was selected as the threshold between the
high miR-122 group and the low miR-122 group (n=23/group).
Kaplan-Meier analysis demonstrated that patients in the high
miR-122 group had a reduced metastasis-free survival time, compared
with those in the low miR-122 group (P<0.01; Fig. 2C). Furthermore, the expression of
miR-122 was measured in a number of renal cancer cell lines.
Compared with the HKC cell line, miR-122 expression was notably
increased in the SN12-PM6 and A498 cell lines and had increased
expression in 786-O, 769-P, Caki-1 and OS-RC-2 cell lines (Fig. 2D). These results were consistent
with the miR-122 expression levels detected in the ccRCC tissues.
Additionally, the FOXO3 expression in these specimens was examined,
and FOXO3 expression in ccRCC cancer tissues was significantly
reduced, compared with normal tissues (P<0.001; Fig. 2E). Western blot analysis also
demonstrated reduced protein levels of FOXO3 in ccRCC cancer
tissues, compared with adjacent normal tissues (Fig. 2F). Finally, miR-122 expression
demonstrated a significant inverse association with FOXO3 at
mRNA levels (r2=0.5118, P<0.001; Fig. 2G).
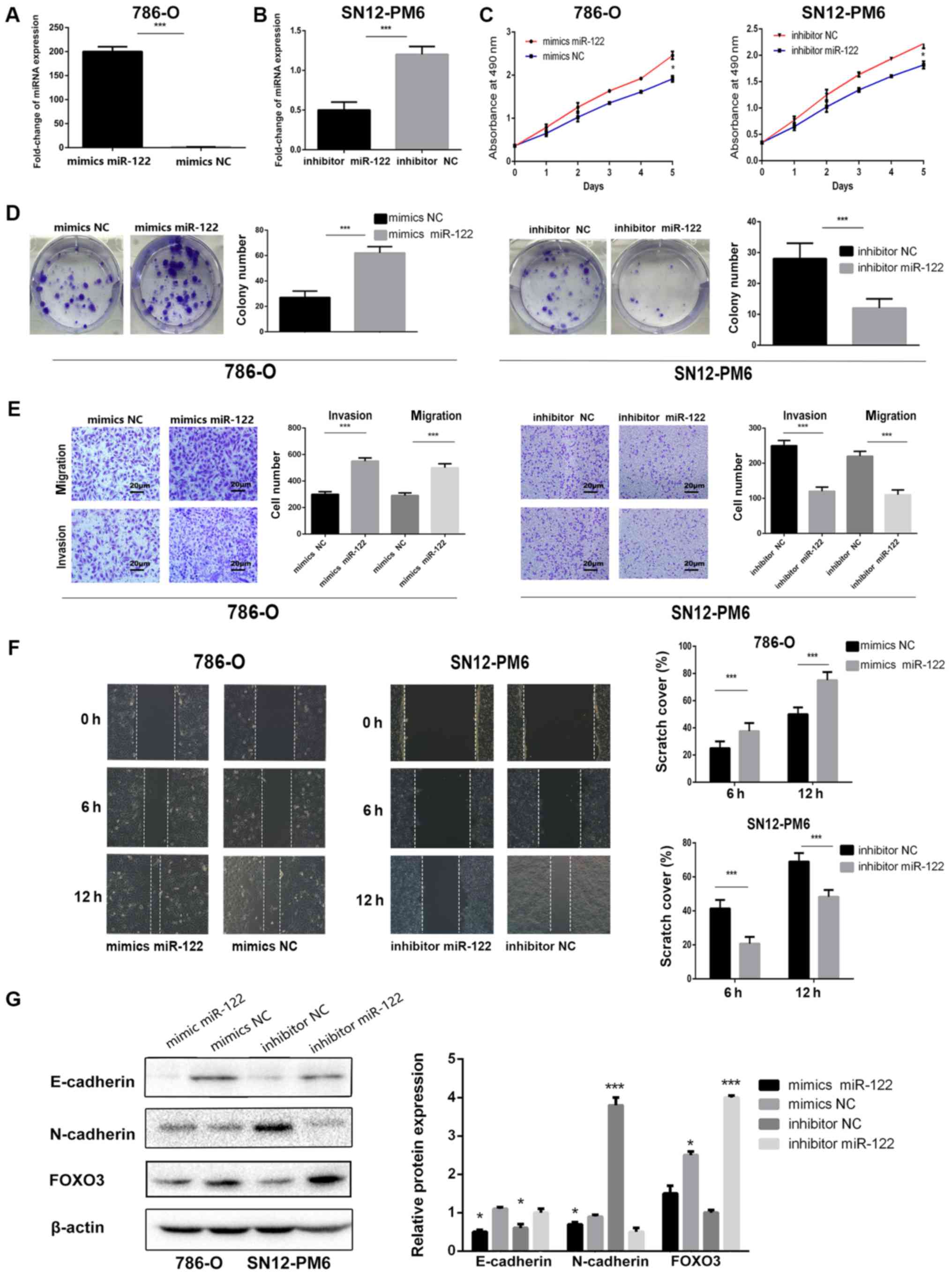
miR-122 promotes the proliferation and
migration of ccRCC cells in vitro
To investigate the biological roles of miR-122 in
ccRCC, miR-122 mimics were used to increase miR-122 expression and
a miR-122 inhibitor was used to decrease miR-122 expression. 786-O
cells, which have a low level of miR-122 expression (Fig. 2D), were transfected with miR-122
mimics to achieve significant miR-122 overexpression, compared with
mimics NC (P<0.001; Fig. 3A).
Additionally, SN12-PM6 cells, which have a high level of miR-122
expression, were transfected with a miR-122 inhibitor to achieve a
relatively low miR-122 expression, compared with inhibitor NC
(Fig. 3B).
Subsequently, the two cell lines were then analyzed
using an MTS assay. As depicted in Fig. 3C, overexpressing miR-122 in 786-O
cells significantly enhanced growth, compared with those
transfected with mimics NC (P<0.05). Additionally, miR-122
expression was inhibited in SN12-PM6 cells significantly attenuated
cell growth, compared with those transfected with inhibitor NC
(P<0.05). In the colony formation assay, the colony-formation
ability was significantly increased in 786-O cells following
transfection with the miR-122 mimics and significantly decreased in
SN12-PM6 cells transfected with the miR-122 inhibitor, compared
with NC (P<0.001; Fig. 3D).
The effect of miR-122 on invasion and migration were
examined by Transwell, Matrigel and wound healing assays. Fig. 3E depicts that the invasive and
migratory abilities of 786-O cells were significantly enhanced
following treatment with miR-122 mimics, compared with cells
treated with miR-122 mimics NC (P<0.001), and the migration and
invasion of SN12-PM6 cells were significantly attenuated following
treatment with the miR-122 inhibitor, compared with inhibitor NC
(P<0.001). Wound healing assays demonstrated that miR-122
promoted 786-O cell migration following transfection with miR-122
mimics, and SN12-PM6 cells displayed decreased migratory capacity
following treatment with the miR-122 inhibitor (Fig. 3F).
Additionally, two epithelial-mesenchymal transition
(EMT)-associated genes (N-cadherin and E-cadherin)
were examined. The present results collectively indicated that
mimics miR-122 significantly promotes the proliferation and
migration of ccRCC cells, compared with mimics NC (P<0.05;
Fig. 3G).
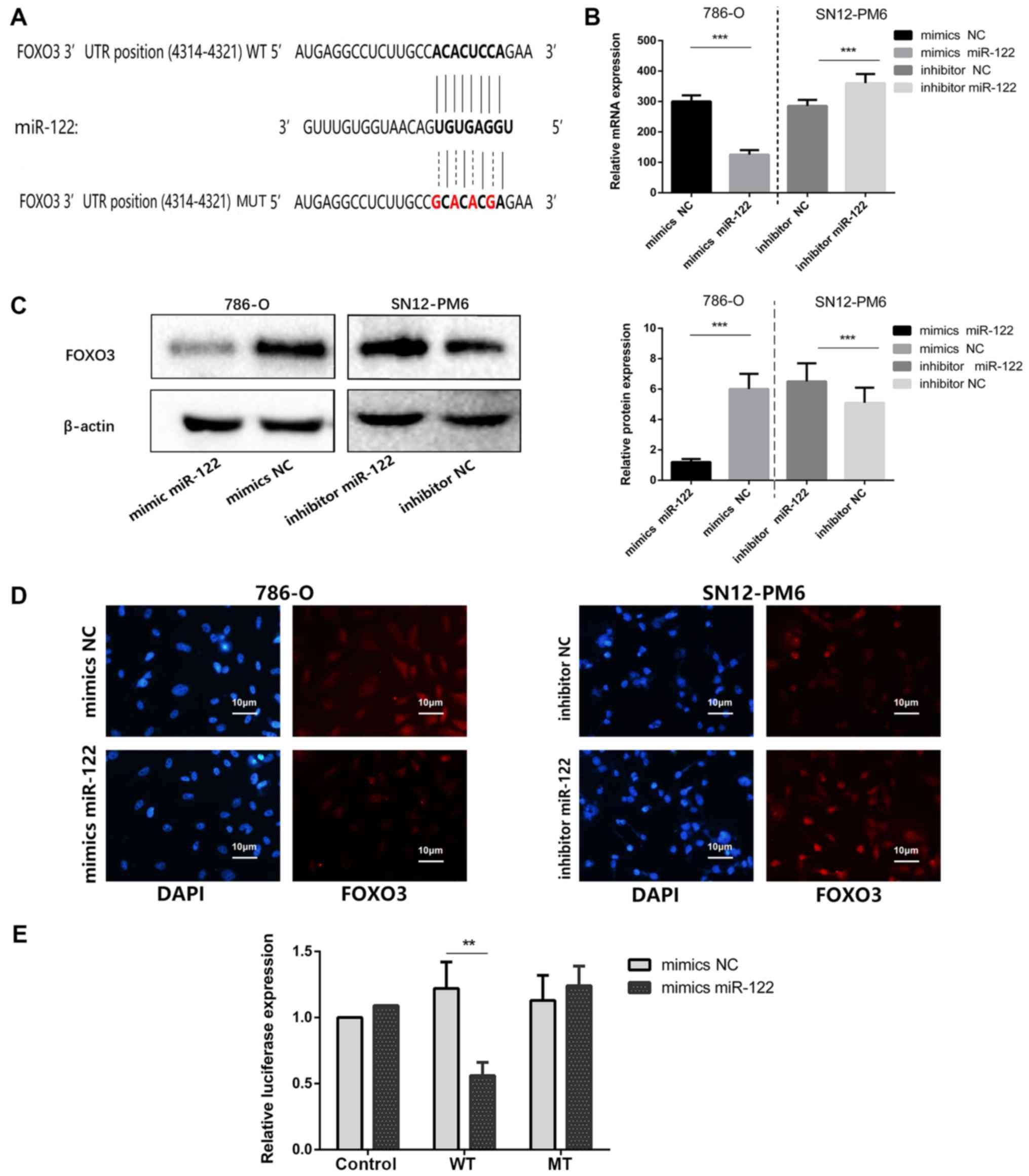
FOXO3 is a direct target of miR-122
As FOXO3 was predicted to be the target of
miR-122 and has been demonstrated to be downregulated in ccRCC. It
was hypothesized that upregulation of miR-122 may induce ccRCC
malignancy by attenuating FOXO3 expression. Fig. 4A depicts the putative miR-122
targeting sites in FOXO3 3′-UTR. As depicted by Fig. 4B and C, FOXO3 mRNA and protein
expression are significantly decreased in 786-O cells following
transfection with miR-122 mimics, compared with mimics NC
(P<0.001); however, FOXO3 expression is significantly increased
in SN12-PM6 cells following transfection with miR-122 inhibitor,
compared with inhibitor NC (P<0.001). Immunofluorescence assays
demonstrated that FOXO3 protein levels were decreased in 786-O
cells treated by miR-122 mimics, compared with cells transfected
with miR-122 mimics NC, and FOXO3 protein levels were increased in
SN12-PM6 cells following transfection with the miR-122 inhibitor,
compared with NC (Fig. 4D). These
data reveal that FOXO3 protein expression is negatively regulated
by miR-122. Bioinformatic predictions validated one conserved
miR-122 binding site on the 3′-UTR of FOXO3 mRNA. Subsequently, a
456-bp fragment was cloned from the FOXO3 3′-UTR containing the
miR-122 bonding site into a luciferase reporter plasmid. The WT
luciferase reporter plasmid or mutant MUT reporter plasmid was
separately co-transfected with miR-122 mimics or mimics NC. The
results revealed that miR-122 significantly repressed luciferase
activity of WT reporter, compared with MUT reporter (P<0.01;
Fig. 4E), indicating that miR-122
directly binds to the predicted site in the FOXO3 3′-UTR and
negatively regulates FOXO3 expression.
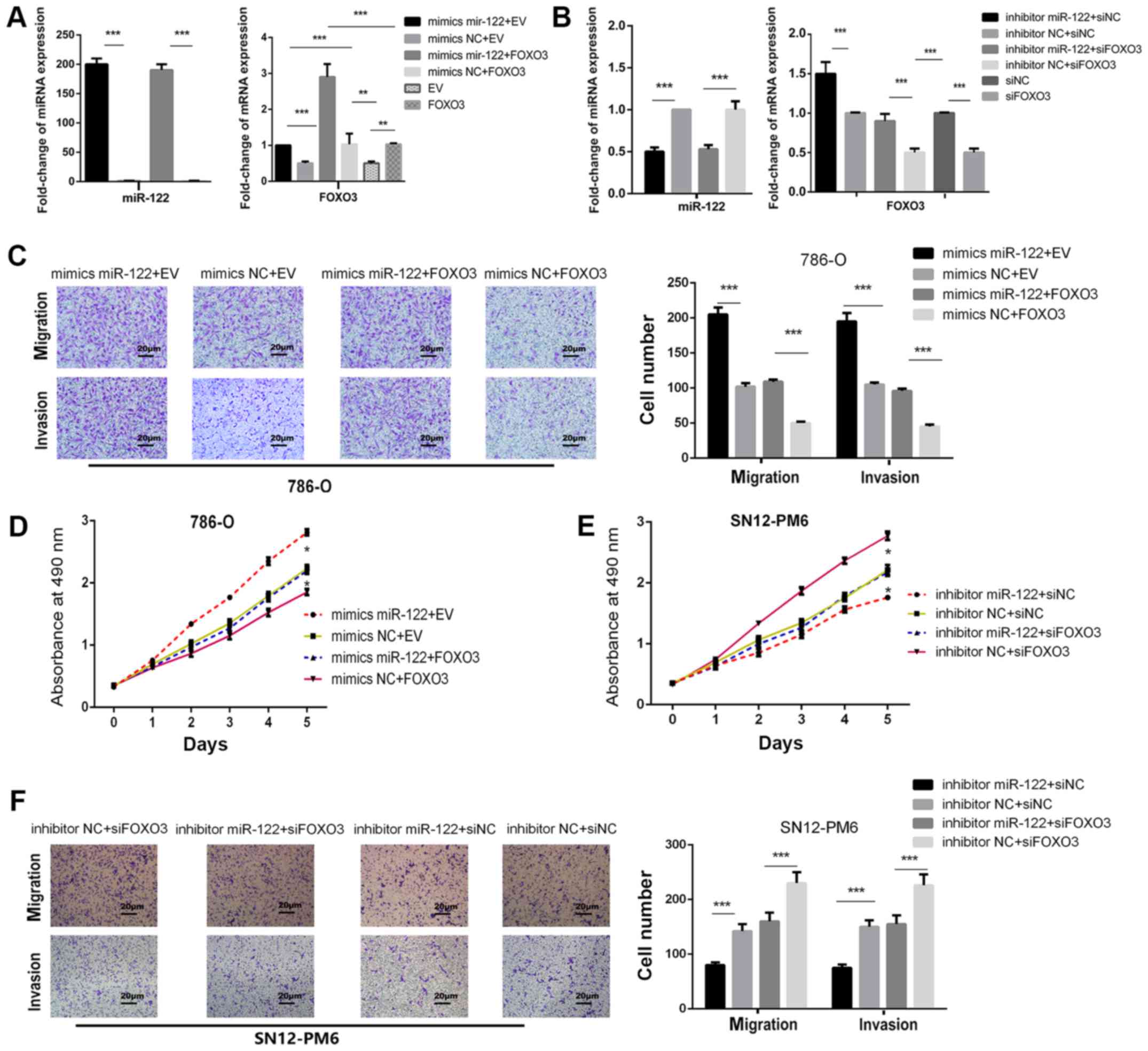
miR-122 promotes cell proliferation and
invasion by downregulating FOXO3
The present study examined whether FOXO3 reversed
the oncogenic effects of miR-122 in ccRCC cells. Firstly,
lentiviral FOXO3 particles (empty vector) were co-transfected with
miR-122 mimics (mimics NC) in 786-O cells. RT-qPCR analysis
confirmed that miR-122 mimics reduced FOXO3 expression, compared
with mimics NC groups (P<0.001; Fig. 5A). Additionally, transfecting
miR-122 inhibitor caused significant downregulation of miR-122 and
significantly upregulation of FOXO3, compared with inhibitor NC
groups (P<0.001; Fig. 5B). In
FOXO3 groups, the FOXO3 plasmid significantly attenuated the
proliferative and invasive abilities of 786-O cells transfected
with miR-122 mimics, compared with EV groups (P<0.05; Fig. 5C and D). Subsequently, a rescue
experiment was performed by co-transfecting FOXO3 siRNA or the siNC
and the miR-122 inhibitor or inhibitor NC into SN12-PM6 cells. In
siFOXO3 groups, FOXO3 downregulation effectively reversed the
attenuation of SN12-PM6 cell invasion and proliferation induced by
the miR-122 inhibitor, compared with siNC groups (P<0.05;
Fig. 5E and F). These data reveal
that miR-122 promotes proliferation and invasion of ccRCC by
downregulating FOXO3.
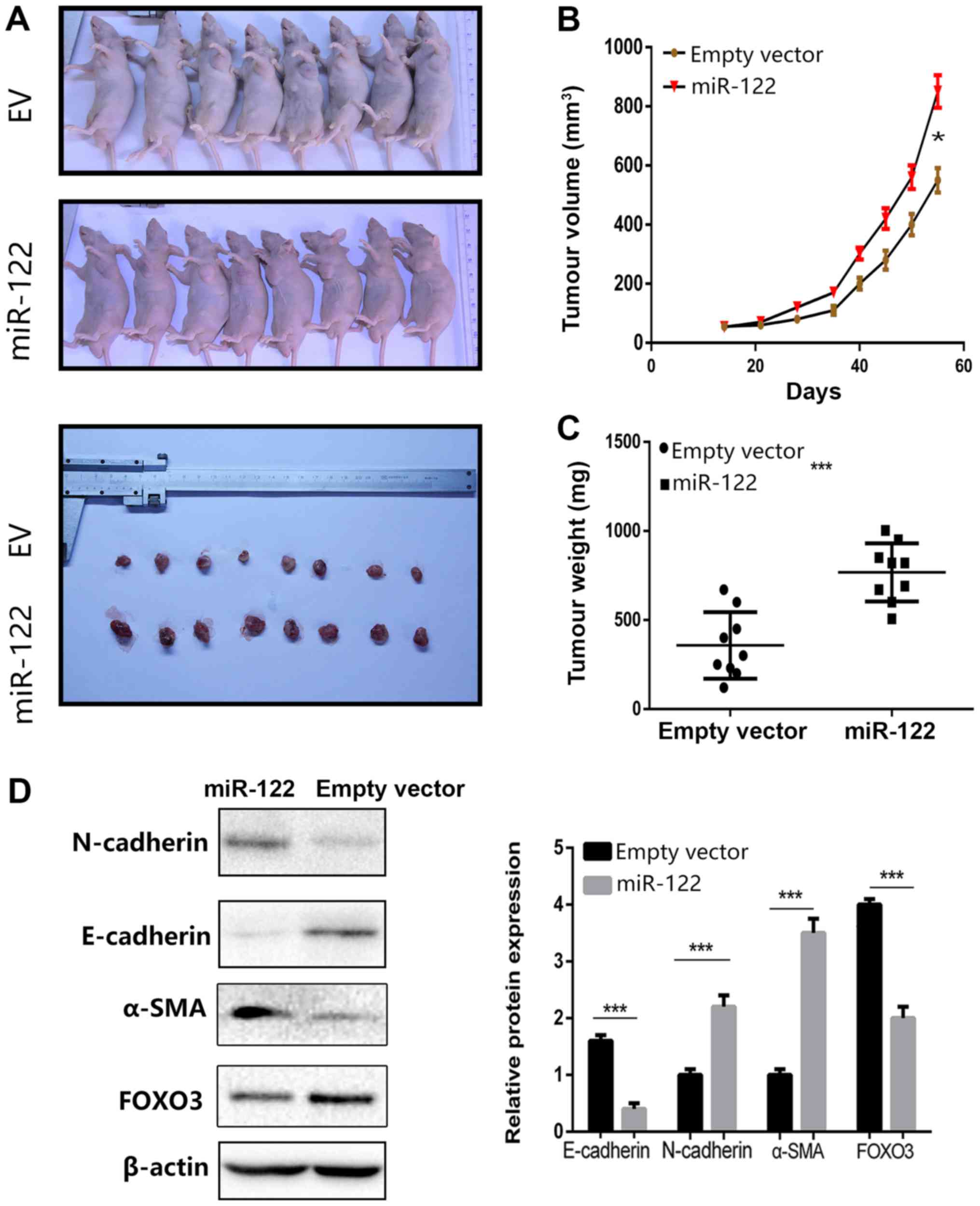
miR-122 promotes tumor growth and cell
invasion in vivo
A BALB/c nude mouse xenograft model and 786-O cells
were employed to verify the function of miR-122 in ccRCC. 786-O
cells stably transfected with lentiviral miR-122 particles or empty
vector were injected subcutaneously into mice. All mice were
sacrificed and dissected for tumor collection 8 weeks after
injection. The results revealed that miR-122 overexpression in
786-O cells significantly promoted tumor growth and significantly
boosted tumor size, compared with the controls (P<0.001;
Fig. 6A-C). Because EMT is a
crucial mechanism, through which tumors acquire malignancy, the
alteration of a number of EMT-association markers upon the
overexpression of miR-122, including E-cadherin, N-cadherin and
α-SMA, were further examined. miR-122 significantly enhanced the
expression of N-cadherin and α-SMA (P<0.001) and significantly
decreased the expression of E-cadherin (P<0.001), compared with
the EV cells (Fig. 6D).
Collectively, the results in vivo indicate that miR-122
promotes EMT in ccRCC cells.
Discussion
Dysregulation of miRNAs is common in cancer, and
miRNAs can serve as a tumor promoter or suppresser (8,11,19).
Recent studies revealed that miRNAs may be secreted to circulation,
assisting cancer cells in metastasis and colonization (41,42)
Reports demonstrated that miR-122 is dysregulated in numerous
cancer types, including liver, breast and lung (15-18,43).
In hepatocellular carcinoma (HCC), overexpression of miR-122 can
induce cell cycle arrest and apoptosis by inhibiting Cyclin
G1 and Bcl-2 like 2 expression (19). Enhanced miR-122 expression can also
inhibit HCC cell growth and promote cell cycle arrest by activating
the Wnt/B-catenin-TCT signaling pathway (18). Additionally, overexpression of
miR-122 was reported to reduce the number of invasion and migration
cells in non-small-cell lung cancer (NSCLC) (44). These data support the hypothesis
that miR-122 acts as a tumor suppressor in HCC and NSCLC. However,
in colorectal cancer (CRC), increased miR-122 levels were
associated with a poor prognostic subtype, indicating that miR-122
may act as an oncogene in CRC (16). These data indicated that miR-122
serves an oncogenic or tumor-suppressing role in the progression of
cancer. Lian et al (40)
demonstrated that miR-122 can promote the proliferation and
invasion of renal cancer cell lines, but there was not further
verification in clinical samples and in vivo experiments.
Fan et al (39)
demonstrated that miR-122 can promote metastasis of ccRCC by
directly targeting Dicer, but they have not explained the screening
process of miR-122. In the present study, it was demonstrated that
miR-122 is overexpressed in ccRCC by the integrated analysis of
four miRNA expression profiles, and, to the best of our knowledge,
it was indicated for the first time that FOXO3 is a direct target
of miR-122 in ccRCC, which further clarified the mechanism of
miR-122 to promote renal cell proliferation and invasion.
The present study used integrated analysis strategy
and TCGA-KIRC validation to demonstrate that miR-122 is notably
upregulated in ccRCC. Further prognostic analysis of the present
ccRCC cohort indicated that high levels of miR-122 were associated
with poor metastasis-free survival time, demonstrating that miR-122
may serve an oncogenic role in ccRCC. Subsequently, in vitro
and in vivo experiments were conducted to confirm this
hypothesis.
786-O cells that were induced by overexpression of
miR-122 demonstrated aggressive growth; but the SN12-PM6 cells had
poor growth following miR-122 downregulation. The gene target of
miR-122 was predicted to investigate the mechanism underlying
miR-122 function in ccRCC. The data recognized FOXO3 as a direct
target of miR-122. The present analysis demonstrated a significant
inverse correlation between miR-122 and FOXO3 mRNA expression
levels in ccRCC (r2=0.05118, P<0.0001). Reduced
miR-122 level demonstrated a positive correlation with increased
FOXO3 mRNA level and vice versa. The present study established a
notable association between miR-122 and FOXO3. Knockdown of miR-122
in SN12-PM6 cells suppressed cell growth and attenuated cell
migration and invasion. The dual-luciferase reporter assay
indicated that miR-122 could bind to the WT sequence, rather than
the MUT sequence, of the target gene. Upregulation of miR-122 in
786-O cells downregulated FOXO3 levels by directly interacting with
the 3′-UTR of FOXO3. Therefore, it was concluded that miR-122
regulated FOXO3 expression by directly binding its 3′-UTR.
FOXO3 is a member of the FOXO family, which notably
regulates the proliferation of cells by modulating its downstream
genes, including cyclin-dependent kinases (26). FOXO3 is a critical gene to human
longevity, and it participates in numerous human physiology
processes, including glucose homeostasis, apoptosis, immunity, stem
cell homeostasis, autophagy and tumor suppression (45,46).
FOXO3 can suppress tumor growth by steering the induction of
p53-dependent apoptosis, increasing Bim expression or
downregulating Myc, a stimulator of tumor cell proliferation
and survival (47).
The present data reveal that miR-122 serves a vital
role in tumor progression of ccRCC by negatively regulating its
target FOXO3; thus, miR-122 may be considered as a potential
therapeutic target for ccRCC.
Funding
This work was financially supported by the National
Natural Science Foundation of China (grant no. 81402109) and
National Natural Science Foundation of China (grant no.
81702494).
Availability of data and materials
The datasets generated and analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors’ contributions
XZ and DN designed the studies. WN primarily
performed the experiments, wrote the manuscript and prepared the
figures. XM, YZ, YG and CP partially performed the experiments. All
authors have read and approved the final version of this
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Chinese People’s Liberation Army General Hospital (Beijing, China).
Each enrolled patient signed written informed consent prior to
sample collection. Animal experiments were approved by the
Experimental Animal Ethical Committee of Chinese PLA General
Hospital and were conducted in accordance with the Guide for the
Care and Use of Laboratory Animals.
Patient consent for publication
Patients included in the present study consented for
publication.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Abbreviations:
|
ccRCC
|
clear cell renal cell carcinoma
|
|
GEO
|
Gene Expression Omnibus
|
|
EMT
|
epithelial-mesenchymal transition
|
|
miRNAs
|
microRNAs
|
|
NC
|
negative control
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Capitanio U and Montorsi F: Renal cancer.
Lancet. 387:894–906. 2016. View Article : Google Scholar
|
|
3
|
Escudier B, Motzer RJ, Sharma P, Wagstaff
J, Plimack ER, Hammers HJ, Donskov F, Gurney H, Sosman JA, Zalewski
PG, et al: Treatment beyond progression in patients with advanced
renal cell carcinoma treated with nivolumab in CheckMate 025. Eur
Urol. 72:368–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wells JC, Stukalin I, Norton C, Srinivas
S, Lee JL, Donskov F, Bjarnason GA, Yamamoto H, Beuselinck B, Rini
BI, et al: Third-line targeted therapy in metastatic renal cell
carcinoma: Results from the International Metastatic Renal Cell
Carcinoma Database Consortium. Eur Urol. 71:204–209. 2017.
View Article : Google Scholar
|
|
5
|
Hu F, Xu P, Sun B and Xiao Z: Differences
in the MicroRNA profiles of subcutaneous adipose-derived stem cells
and omental adipose-derived stem cells. Gene. 625:55–63. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liang L, Lan F, Yin X, Ge S, Yu J and Yan
M: Metal-enhanced fluorescence/visual bimodal platform for
multiplexed ultrasensitive detection of microRNA with reusable
paper analytical devices. Biosens Bioelectron. 95:181–188. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tao Y, Yin D, Jin M, Fang J, Dai T, Li Y,
Li Y, Pu Q and Xie G: Double-loop hairpin probe and
doxorubicin-loaded gold nanoparticles for the ultrasensitive
electrochemical sensing of microRNA. Biosens Bioelectron.
96:99–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gattolliat CH, Couvé S, Meurice G, Oréar
C, Droin N, Chiquet M, Ferlicot S, Verkarre V, Vasiliu V, Molinié
V, et al: Integrative analysis of dysregulated microRNAs and mRNAs
in multiple recurrent synchronized renal tumors from patients with
von Hippel-Lindau disease. Int J Oncol. 53:1455–1468.
2018.PubMed/NCBI
|
|
9
|
Yi Z, Fu Y, Zhao S, Zhang X and Ma C:
Differential expression of miRNA patterns in renal cell carcinoma
and nontumorous tissues. J Cancer Res Clin Oncol. 136:855–862.
2010. View Article : Google Scholar
|
|
10
|
Lawrie CH, Larrea E, Larrinaga G,
Goicoechea I, Arestin M, Fernandez-Mercado M, Hes O, Cáceres F,
Manterola L and López JI: Targeted next-generation sequencing and
non-coding RNA expression analysis of clear cell papillary renal
cell carcinoma suggests distinct pathological mechanisms from other
renal tumour subtypes. J Pathol. 232:32–42. 2014. View Article : Google Scholar
|
|
11
|
Falzone L, Candido S, Salemi R, Basile MS,
Scalisi A, McCubrey JA, Torino F, Signorelli SS, Montella M and
Libra M: Computational identification of microRNAs associated to
both epithelial to mesenchymal transition and NGAL/MMP-9 pathways
in bladder cancer. Oncotarget. 7:72758–72766. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hafsi S, Candido S, Maestro R, Falzone L,
Soua Z, Bonavida B, Spandidos DA and Libra M: Correlation between
the overexpression of Yin Yang 1 and the expression levels of
miRNAs in Burkitt’s lymphoma: A computational study. Oncol Lett.
11:1021–1025. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ambade A, Satishchandran A and Szabo G:
Alcoholic hepatitis accelerates early hepatobiliary cancer by
increasing stemness and miR-122-mediated HIF-1α activation. Sci
Rep. 6:213402016. View Article : Google Scholar
|
|
14
|
Elhanati S, Ben-Hamo R, Kanfi Y, Varvak A,
Glazz R, Lerrer B, Efroni S and Cohen HY: Reciprocal regulation
between SIRT6 and miR-122 controls liver metabolism and predicts
hepatocarcinoma prognosis. Cell Reports. 14:234–242. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hopcraft SE, Azarm KD, Israelow B, Lévêque
N, Schwarz MC, Hsu TH, Chambers MT, Sourisseau M, Semler BL and
Evans MJ: Viral determinants of miR-122-independent hepatitis C
virus replication. mSphere. 1:12015.
|
|
16
|
Maierthaler M, Benner A, Hoffmeister M,
Surowy H, Jansen L, Knebel P, Chang-Claude J, Brenner H and
Burwinkel B: Plasma miR-122 and miR-200 family are prognostic
markers in colorectal cancer. Int J Cancer. 140:176–187. 2017.
View Article : Google Scholar
|
|
17
|
Pan C, Wang X, Shi K, Zheng Y, Li J, Chen
Y, Jin L and Pan Z: miR-122 reverses the doxorubicin-resistance in
hepatocellular carcinoma cells through regulating the tumor
metabolism. PLoS One. 11:e01520902016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ahsani Z, Mohammadi-Yeganeh S, Kia V,
Karimkhanloo H, Zarghami N and Paryan M: WNT1 gene from WNT
signaling pathway is a direct target of miR-122 in hepatocellular
carcinoma. Appl Biochem Biotechnol. 181:884–897. 2017. View Article : Google Scholar
|
|
19
|
von Felden J, Heim D, Schulze K, Krech T,
Ewald F, Nashan B, Lohse AW and Wege H: High expression of micro
RNA-135A in hepatocellular carcinoma is associated with recurrence
within 12 months after resection. BMC Cancer. 17:602017. View Article : Google Scholar
|
|
20
|
Fong MY, Zhou W, Liu L, Alontaga AY,
Chandra M, Ashby J, Chow A, O’Connor ST, Li S, Chin AR, et al:
Breast-cancer-secreted miR-122 reprograms glucose metabolism in
premetastatic niche to promote metastasis. Nat Cell Biol.
17:183–194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee CG, Kim JG, Kim HJ, Kwon HK, Cho IJ,
Choi DW, Lee WH, Kim WD, Hwang SJ, Choi S, et al: Discovery of an
integrative network of microRNAs and transcriptomics changes for
acute kidney injury. Kidney Int. 86:943–953. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Du WW, Yang W, Chen Y, Wu ZK, Foster FS,
Yang Z, Li X and Yang BB: Foxo3 circular RNA promotes cardiac
senescence by modulating multiple factors associated with stress
and senescence responses. Eur Heart J. 38:1402–1412. 2017.
|
|
23
|
Hagenbuchner J, Lungkofler L,
Kiechl-Kohlendorfer U, Viola G, Ferlin MG, Ausserlechner MJ and
Obexer P: The tubulin inhibitor MG-2477 induces autophagy-regulated
cell death, ROS accumulation and activation of FOXO3 in
neuroblastoma. Oncotarget. 8:32009–32026. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kumazoe M, Takai M, Bae J, Hiroi S, Huang
Y, Takamatsu K, Won Y, Yamashita M, Hidaka S, Yamashita S, et al:
FOXO3 is essential for CD44 expression in pancreatic cancer cells.
Oncogene. 36:2643–2654. 2017. View Article : Google Scholar
|
|
25
|
Kumazoe M, Takai M, Hiroi S, Takeuchi C,
Kadomatsu M, Nojiri T, Onda H, Bae J, Huang Y, Takamatsu K, et al:
The FOXO3/PGC-1β signaling axis is essential for cancer stem cell
properties of pancreatic ductal adenocarcinoma. J Biol Chem.
292:10813–10823. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Coomans de Brachène A and Demoulin JB:
FOXO transcription factors in cancer development and therapy. Cell
Mol Life Sci. 73:1159–1172. 2016. View Article : Google Scholar
|
|
27
|
Liang R and Ghaffari S: Mitochondria and
FOXO3 in stem cell homeostasis, a window into hematopoietic stem
cell fate determination. J Bioenerg Biomembr. 49:343–346. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Natarajan SK, Stringham BA, Mohr AM,
Wehrkamp CJ, Lu S, Phillippi MA, Harrison-Findik D and Mott JL:
FoxO3 increases miR-34a to cause palmitate-induced cholangiocyte
lipoapoptosis. J Lipid Res. 58:866–875. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Peng XL, So KK, He L, Zhao Y, Zhou J, Li
Y, Yao M, Xu B, Zhang S, Yao H, et al: MyoD- and FoxO3-mediated
hotspot interaction orchestrates super-enhancer activity during
myogenic differentiation. Nucleic Acids Res. 45:8785–8805. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Salcher S, Hermann M, Kiechl-Kohlendorfer
U, Ausserlechner MJ and Obexer P: C10ORF10/DEPP-mediated ROS
accumulation is a critical modulator of FOXO3-induced autophagy.
Mol Cancer. 16:952017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Song D, Ma J, Chen L, Guo C, Zhang Y, Chen
T, Zhang S, Zhu Z, Tian L and Niu P: FOXO3 promoted mitophagy via
nuclear retention induced by manganese chloride in SH-SY5Y cells.
Metallomics. 9:1251–1259. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee J and Park SH: Tumor-suppressive
activity of 1,25-dihydroxyvitamin D3 against kidney cancer cells
via up-regulation of FOXO3. Biosci Biotechnol Biochem.
80:1947–1953. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ni D, Ma X, Li HZ, Gao Y, Li XT, Zhang Y,
Ai Q, Zhang P, Song EL, Huang QB, et al: Downregulation of FOXO3a
promotes tumor metastasis and is associated with metastasis-free
survival of patients with clear cell renal cell carcinoma. Clin
Cancer Res. 20:1779–1790. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bloomsmith MA, Perlman JE, Hutchinson E
and Sharpless M: Behavioral management programs to promote
laboratory animal welfare. Management of Animal Care and Use
Programs in Research, Education, and Testing. Weichbrod RH,
Thompson GAH and Norton JN: CRC Press/Taylor & Francis; Boca
Raton, FL: pp. 63–82. 2018
|
|
35
|
Lee C, You D, Park J, Jeong IG, Song C,
Hong JH, Ahn H and Kim CS: Validation of the 2009 TNM
classification for renal cell carcinoma: Comparison with the 2002
TNM classification by concordance index. Korean J Urol. 52:524–530.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ficarra V, Novara G and Martignoni G: The
use of simplified versions of the Fuhrman nuclear grading system in
clinical practice requires the agreement of a multidisciplinary
panel of experts. Eur Urol. 56:782–784; discussion 784–785. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhou Y, Liu Y, Yan H, Li Y, Zhang H, Xu J,
Puthiyakunnon S and Chen X: miR-281, an abundant midgut-specific
miRNA of the vector mosquito Aedes albopictus enhances dengue virus
replication. Parasit Vectors. 7:4882014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wolter JM, Kotagama K, Pierre-Bez AC,
Firago M and Mangone M: 3′LIFE: A functional assay to detect miRNA
targets in high-throughput. Nucleic Acids Res. 42:e1322014.
View Article : Google Scholar
|
|
39
|
Fan Y, Ma X, Li H, Gao Y, Huang Q, Zhang
Y, Bao X, Du Q, Luo G, Liu K, et al: miR-122 promotes metastasis of
clear-cell renal cell carcinoma by downregulating Dicer. Int J
Cancer. 142:547–560. 2018. View Article : Google Scholar
|
|
40
|
Lian JH, Wang WH, Wang JQ, Zhang YH and Li
Y: MicroRNA-122 promotes proliferation, invasion and migration of
renal cell carcinoma cells through the PI3K/Akt signaling pathway.
Asian Pac J Cancer Prev. 14:5017–5021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Junker K, Heinzelmann J, Beckham C, Ochiya
T and Jenster G: Extracellular vesicles and their role in urologic
malignancies. Eur Urol. 70:323–331. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y,
Chen W, Liu F, Sun W, Li XF, et al: Exosome-transmitted lncARSR
promotes sunitinib resistance in renal cancer by acting as a
competing endogenous RNA. Cancer Cell. 29:653–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Akuta N, Kawamura Y, Suzuki F, Saitoh S,
Arase Y, Kunimoto H, Sorin Y, Fujiyama S, Sezaki H, Hosaka T, et
al: Impact of circulating miR-122 for histological features and
hepatocellular carcinoma of nonalcoholic fatty liver disease in
Japan. Hepatol Int. 10:647–656. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qin H, Sha J, Jiang C, Gao X, Qu L, Yan H,
Xu T, Jiang Q and Gao H: miR-122 inhibits metastasis and
epithelial-mesenchymal transition of non-small-cell lung cancer
cells. Onco Targets Ther. 8:3175–3184. 2015.PubMed/NCBI
|
|
45
|
Martins R, Lithgow GJ and Link W: Long
live FOXO: Unraveling the role of FOXO proteins in aging and
longevity. Aging Cell. 15:196–207. 2016. View Article : Google Scholar :
|
|
46
|
Lee S and Dong HH: FoxO integration of
insulin signaling with glucose and lipid metabolism. J Endocrinol.
233:R67–R79. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Morris BJ, Willcox DC, Donlon TA and
Willcox BJ: FOXO3: A major gene for human longevity - A
mini-review. Gerontology. 61:515–525. 2015. View Article : Google Scholar
|