Introduction
Despite the standard of care, consisting of surgical
resection combined with chemotherapy and radiotherapy, the
prognosis of patients with glioma remains dismal (1). A number of factors promote the
malignant potential of glioma, including the capacity of
glioblastoma multiforme (GBM) to regulate immune responses both
locally and systemically (2).
Increasing evidence indicates that cancer cells can change the
normal cell phenotype by the secretion of growth factors,
chemokines and cytokines, which in turn promotes tumor growth,
invasion and metastases (3,4). In
the brain, chronic inflammation due to the presence of glioma cells
results in the infiltration of reactive astrocytes, which are major
components of the invasive niche at the interface of glioma cells
(5-7). Tumor-stroma interactions are in part
mediated by secreting soluble factors, such as growth factors.
However, the mechanisms underlying the communication between
astrocytes and glioma cells are complex, and exosomes are likely
involved.
Exosomes are small (30-100 nm) vesicles secreted
from cells into the extracellular space (8). Exosomes are formed by an inward
budding of the plasma membrane in the late endosomes, thus
enclosing within the internal vesicles to form multivesicular
bodies (MVBs) within the cytoplasm, which may fuse with the plasma
membrane and release its content of exosomes (9). Exosomes contain a set of specific
proteins, including tetraspanins (CD9, CD63 and CD81), TSG101, Alix
and flotillin (10). Exosomes are
highly enriched in sphingomyelin, hexosylceramides and cholesterol
at the expense of phosphatidylethanolamine and phosphatidylcholine
(11). Several crucial signals,
including small GTPases, the endosomal sorting complex required for
transport (ESCRT) and the SNARE complex are involved in exosome
release (12). Although they were
initially considered as removal of the garbage for cells, more
recently exosomes have been shown to mediate intercellular
communications (13,14). Exosomes are released by many cell
types into the extracellular environment and are found in a variety
of body fluids, such as saliva, urine, blood, ascites, breast milk
and cerebrospinal fluid (15).
Exosomes carry a specific payload of fully functional proteins,
lipids and nucleic acids when transferred into recipient cells
(16-18). Functionally, exosomes have been
implicated in the modulation of host immune responses and microbial
pathogenesis by the regulation of intercellular communications
(19,20). In cancer, for instance, exosomes
can transfer oncogenic proteins and nucleic acids to modulate the
activity of recipient cells, and then shape the microenvironment of
tumor (21-23).
Long non-coding RNA (lncRNA) activated by TGF-β
(lncRNA-ATB) was initially identified as an lncRNA in
hepatocellular carcinoma (HCC). LncRNA-ATB expression was
significantly increased hepatocellular carcinoma, which promotes
HCC cells invasion and tumor growth. Additionally, lncRNA-ATB can
induce epithelial-mesenchymal transition (EMT) and promote invasion
by competitively binding and sequestering the microRNA (miRNA or
miR)-200 family in HCC (24). Our
previous study revealed that lncRNA-ATB promoted the migration and
invasion of glioma cells by acting as a competitive endogenous RNA
(ceRNA) of miR-200a (25).
Recently, plasma lncRNA-ATB expression has been shown to be
increased in lung disease, suggesting that circular lncRNA-ATB may
play a key role in disease (26).
In this study, we aimed to investigate the
functional role of glioma cell-derived exosomal lncRNA-ATB in
astrocytes. We describe the effects of glioma cell-derived exosomes
on the activation of astrocytes. We demonstrate that lncRNA-ATB in
glioma cell-derived exosomes activates astrocytes through the
suppression of miR-204-3p and that reactive astrocytes stimulate
glioma cell invasion. These results suggest that lncRNA-ATB may
prove to be a novel therapeutic target for the treatment of
invasive glioma.
Materials and methods
Cell culture
The A172 and U251 glioma cells were purchased from
the Chinese Academy of Sciences (Shanghai, China) and cultured in
Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan, UT, USA)
with high glucose supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
streptomycin (100 µg/ml), penicillin (100 U/ml). Normal
human astrocytes (NHAs) were obtained from Sun Yat-Sen University
and cultured in astrocyte medium with low glucose supplemented with
10% fetal bovine serum. All cell lines were cultured at 37°C in a
humidified incubator containing 5% CO2.
Isolation and analysis of exosomes
For exosomes isolation, we first transplanted an
equal number of different cells into 10 cm plates and changed the
culture medium with fresh DMEM-supplemented serum, which was
depleted of exosomes. The culture medium was collected following
centrifugation at 3,000 × g for 15 min. Exosomes were extracted
from the cell culture medium using a Total Exosome Isolation kit
(Thermo Fisher Scientific) according to the manufacturer’s
instructions. The samples were then examined by transmission
electron microscopy (TEM) on a JEM 1010 transmission electron
microscope at an accelerating voltage of 80 Kv. Digital images were
obtained using the AMT Imaging System (Advanced Microscopy
Techniques Corp., Woburn, MA, USA).
Uptake of exosomes by astrocytes
Exosomes from the A172 and U251 glioma cells were
labeled with Dil (Sigma, St. Louis, MO, USA) according to the
supplier’s instructions, suspended in low serum medium (10
µg/ml), and incubated with the astrocytes for 24 h at 37°C.
Following incubation, the cells were processed as previously
described (27).
miRNA transfection and plasmid
construction
miR-204-3p mimic (sequence: GCUGGGAAGGCAAAGGGACGU)
and negative control (NC) were designed and synthesized by RiboBio
Co., Ltd. (Guangzhou, China). miRNA was trans-fected into the cells
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.). The transfection process was conducted according to
manufacturer’s guidelines. lncRNA ATB sequences (forward,
5′-CTCAAGCTTGGC CCTGGGGCTCTGCAA-3′ and reverse, 5′-GGAATTCTG
GTAAATGAGTCCAAAGTC-3′) were synthesized and subcloned into the
pCDNA3.1 vector (Sangon Biotech, Shanghai). The pCDNA constructs or
the empty vector were transfected into the cells cultured in 6-well
plates according to the manufacturer’s instructions (Invitrogen;
Thermo Fisher Scientific, Inc.).
Immunofluorescence staining
The NHAs were fixed with 4% paraformaldehyde, and
permeabilized with 0.2% Triton X-100 in 1% bovine serum albumin
(BSA) for 10 min and blocked with 5% BSA for 1 h at room
temperature. Incubation with anti-glial fibrillary acidic protein
(GFAP) antibodies (1:400, ab7260; Abcam, Cambridge, MA, USA) at 4°C
overnight was followed by incubation with rabbit IgG, Cy3 (1:100,
bs-0295P; Bioss Antibodies Inc., Beijing) at room temperature for 1
h. The cells were mounted with SlowFade Gold antifade reagent with
DAPI (Sigma, St. Louis, MO, USA) and images were acquired using
fluorescence microscopy (Olympus IX71; Olympus, Tokyo, Japan).
Counting the number of lipid droplets containing a specific amount
of pixels was carried out using Image J software.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the cultured cells and
exosomes using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer’s instructions.
Using a Nanodrop spectrophotometer (Implen GmbH, Munich, Germany),
the RNA concentration and quality were determined by the 260/280 nm
absorbance. Subsequently, the RNA was reverse transcribed into high
capacity cDNA using PrimeScriptTM RT Master Mix (Perfect Real-Time)
(Takara Biotechnology, Dalian, China). Maxima SYBR-Green/ROX qPCR
Master Mix (Thermo Fisher Scientific, Inc.) was used for
quantitative PCR. In brief, each PCR reaction mixture in a total
volume of 10 µl, containing 5 µl of 2X SYBR-Green
Master Mix, 1 µl of sense and antisense primers, 3 µl
of cDNA, was run for 45 cycles with denaturation at 95°C for 15
sec, annealing at 60°C for 30 sec, and extension at 72°C for 30
sec. The primers for genes were determined as follows: lncRNA-ATB
forward, 5′-ACAAGCTGTGCAGTCTCAGG-3′ and reverse,
5′-CTAGGCCCAAAGACAATGGA-3′; GFAP forward,
5′-AGGTCCATGTGGAGCTTGAC-3′ and reverse, 5′-GCCATTGCCTCATACTGCGT-3′;
and GAPDH forward, 5′-AGCAAGAGCACAAGAGGAAG-3′ and reverse, 5′-GGT
TGAGCACAGGGTACTTT-3′.
The All-in-One™ miRNA First-Strand cDNA Synthesis
kit (Genecopoeia, Guangzhou, China) was used for miRNA reverse
transcription and RT-qPCR was conducted using the All-in-One™ miRNA
qPCR kit (Genecopoeia) for miR-204-3p and U6
(miRQ0022693-1-1/MQP-0202, respectively; RiboBio Co., Ltd.),
respectively, using ABI 7100 (Applied Biosystems, Darmstadt,
Germany). For relative quantification, the 2−ΔΔCq value
was calculated and used as an indication of the relative expression
levels (28), which was calculated
by subtracting the CT values of the control gene from the CT values
of lncRNA-ATB, GFAP and miR-204-3p.
Cell migration and invasion assays
The cell migratory and invasive ability was examined
using 24-well chambers with an 8 μm pore size (Corning, Inc.,
Corning, NY, USA). A total of 5×103 cells were
resuspended in 100 µl serum-free medium and seeded into the
upper chamber with or without pre-coated with 500 ng/ml Matrigel
solution (BD Biosciences, San Jose, CA, USA), while
serum-containing medium was placed in the bottom chamber of
Transwell plates, following incubation at 37°C for 48 h for the
migration and invasion assays. Non-migrated and non-invaded cells
from the upper chamber were scraped off using a cotton swab. The
migrated and invaded cells on the lower chamber membrane were fixed
with 4% polyoxymethylene and stained with crystal violet (Sigma) at
room temperature for 5 min, and dried. Five predetermined fields
were counted under a microscope (Olympus IX71, (Olympus; ×200
magnification). All assays were performed in triplicate.
Luciferase reporter assays
The lncRNA-ATB fragment containing the predicted
miR-204-3p binding site (miRDB, http://mirdb.org/), and the putative sequences of the
binding site were cloned into a pmirGlO Dual-luciferase miRNA
Target Expression Vector (Promega, Madison, WI, USA) to form the
reporter vector pmiRGLO-lncRNA-ATB. PmiRGLO-lncRNA-ATB was
co-transfected with miR-204-3p mimics or NC into NHAs using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Following 48 h of transfection, luciferase assay was carried out
using a Dual-Luciferase Reporter Assay System (Promega) according
to the manufacturer’s instructions. All assays were independently
performed in triplicate.
RNA immunoprecipitation
The EZ-Magna RIP RNA-binding protein
immunoprecipitation kit (EMD Millipore, Billerica, MA, USA) was
used in RNA immunoprecipitation (RIP). RIP was implemented to pull
down endogenous miR-204-3p associated with lncRNA-ATB in NHAs, and
was performed following the manufacturer’s instructions. NHAs were
lysed with RIP lysis buffer, and 100 µl of cell lysate were
incubated with RIP immunoprecipitation buffer containing magnetic
beads conjugated with human anti-Argonaute 2 (Ago2) antibody (EMD
Millipore) and normal IgG (EMD Millipore) was used as negative
control. The samples were incubated with Proteinase K buffer and
then target RNA was extracted using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer’s
instructions. Purified RNA was subjected to RT-qPCR analysis.
Western blot analysis
Total proteins were extracted from the cells using
RIPA buffer with PMSF (Beyotime Institute of Biotechnology,
Shanghai, China) on ice, subjected to 10% SDS-PAGE gel and
electrophoretically transferred onto polyvinylidene difluoride
(PVDF) membranes. The membranes were incubated in 5% non-fat milk
dissolved in Tris-buffered saline (TBS) containing 0.1% Tween-20
for 1.5 h at room temperature and then incubated with primary
antibodies at 4°C overnight as follows: CD63, CD9 and GAPDH
(1:1,000, ab193349/ab223052/ab9484, respectively; Abcam). Following
incubation at room temperature for 2 h with secondary antibodies
(goat anti-rabbit or goat anti-mouse, 1:5,000, ZB-2301/ZB-2305,
respectively; ZSGB-BIO, Beijing, China), immune complexes were
visualized using the SuperSignal® West Femto Trial kit
(Thermo Fisher Scientific, Inc.) and blot bands were scanned using
Find-do ×6 Tanon (Tanon, Shanghai, China).
Statistical analysis
Experimental data are presented as the means ±
standard deviation (SD). GraphPad Prism V5.0 software (GraphPad
Software, Inc., La Jolla, CA, USA) was used for statistical
analysis. Differences were analyzed using SPSS 17.0 statistical
software with the Student’s t-test or one-way ANOVA. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
Isolation and characterization of glioma
cell-derived exosomes
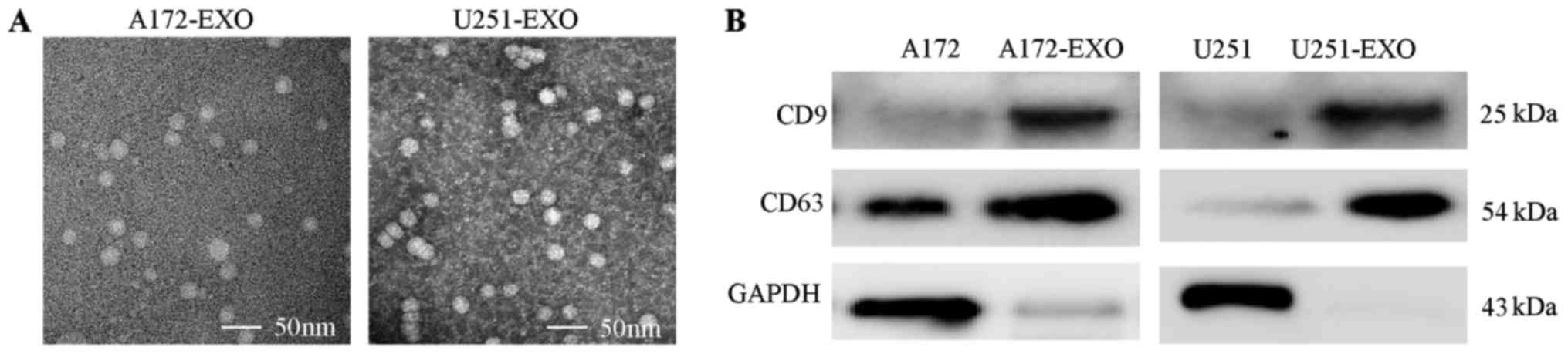
Exosomes purified from the media of the A172 and
U251 glioma cells are shown in Fig.
1A, as visualized by TEM. TEM revealed a relatively uniform
population of membrane-bound vesicles around 100 nm. Moreover,
western blot analyses revealed the isolated particles expressed
markers associated with exosomes (CD9 and CD63) (Fig. 1B). Thus, these nanovesicles have
the characteristics of exosomes and can be isolated in a consistent
manner.
Glioma cell-derived exosomes activate
astrocytes
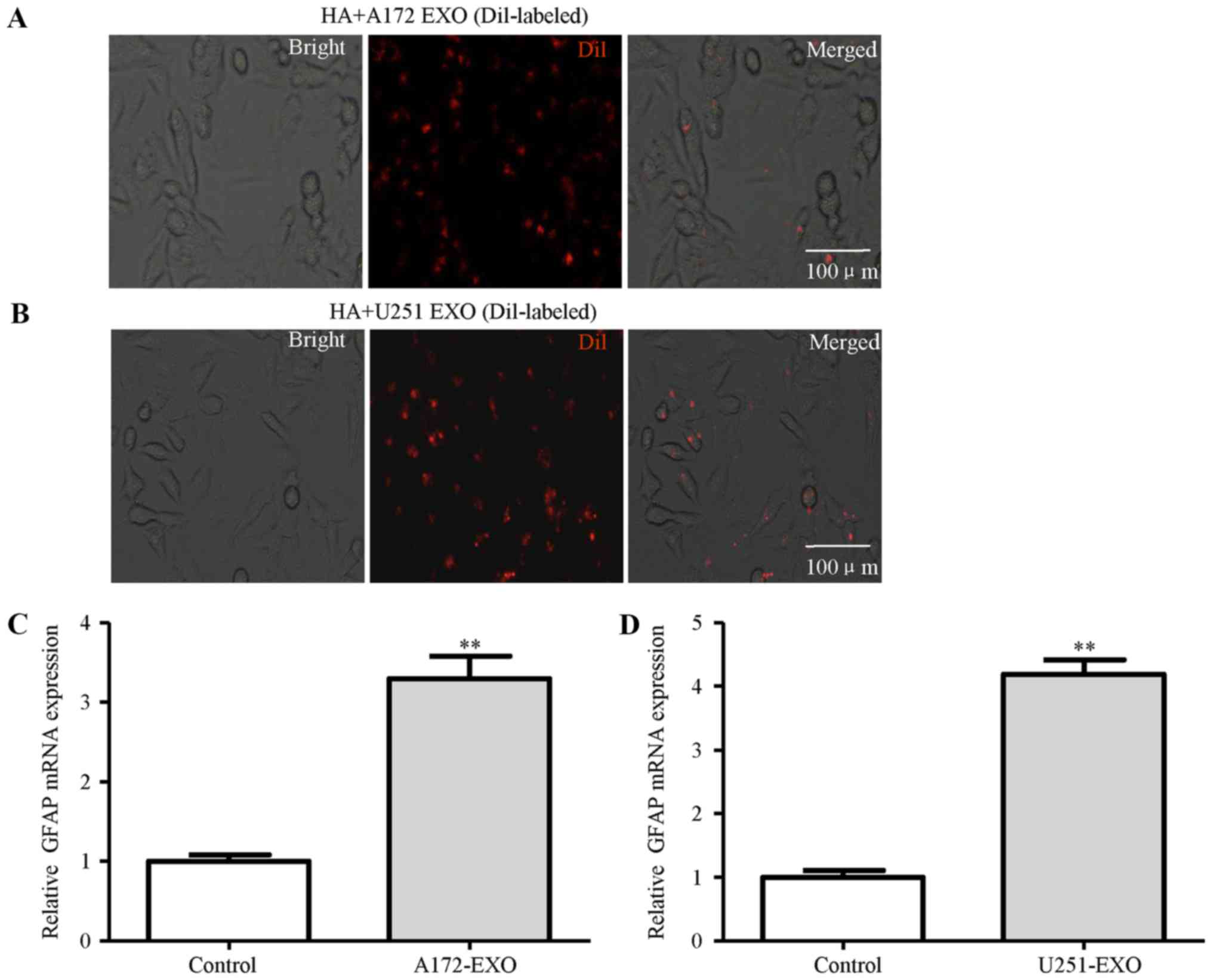
NHAs, a type of normal human brain astrocytes, were
selected for use as NHAs in our experimental model. The isolated
exosomes from the A172 and U251 glioma cells were labeled with Dil
dye (red), washed thoroughly and then added to the NHAs. The uptake
of the labeled exosomes by NHAs revealed a red signal under a
fluorescence microscope (Fig. 2A and
B). These results suggested that the uptake of exosomes by
these cells was very efficient. To gain insight into the effects of
exosomes derived from the glioma cells on astrocytes, the
activation of astrocytes was measured. The results of RT-qPCR
revealed that treatment with the A172 and U251 glioma cell-derived
exosomes induced the mRNA expression of GFAP, a marker of astrocyte
activation (Fig. 2C and D).
lncRNA-ATB in exosomes mediates astrocyte
activation
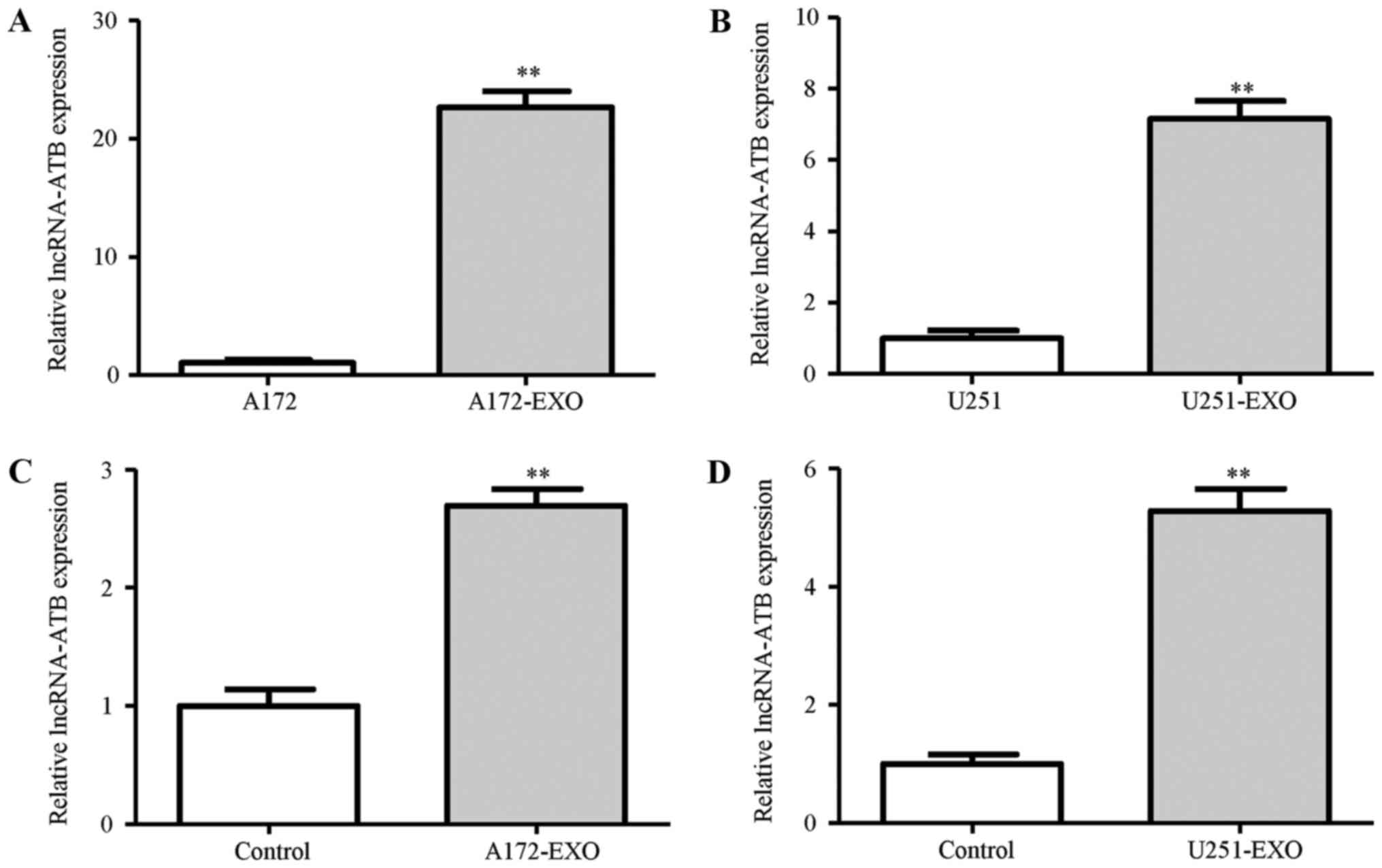
We then examined the mechanisms through which the
glioma cells-derived exosomes activate astrocytes. The results of
RT-qPCR demonstrated that the exosomes derived from the A172 and
U251 glioma cells contained higher levels of lncRNA-ATB when
compared with those derived from the parent glioma cells (Fig. 3A and B). To determine whether
lncRNA-ATB secreted from the A172 and U251 glioma cells can be
transferred to astrocytes via exosomes, we measured the lncRNA-ATB
levels in astrocytes treated with exosomes derived from the glioma
cells. Similar to that observed for exosome uptake, an increase in
the expression of lncRNA-ATB was observed in the NHAs following
treatment with the glioma cell-derived exosomes at 24 h (Fig. 3C and D). To explore the possible
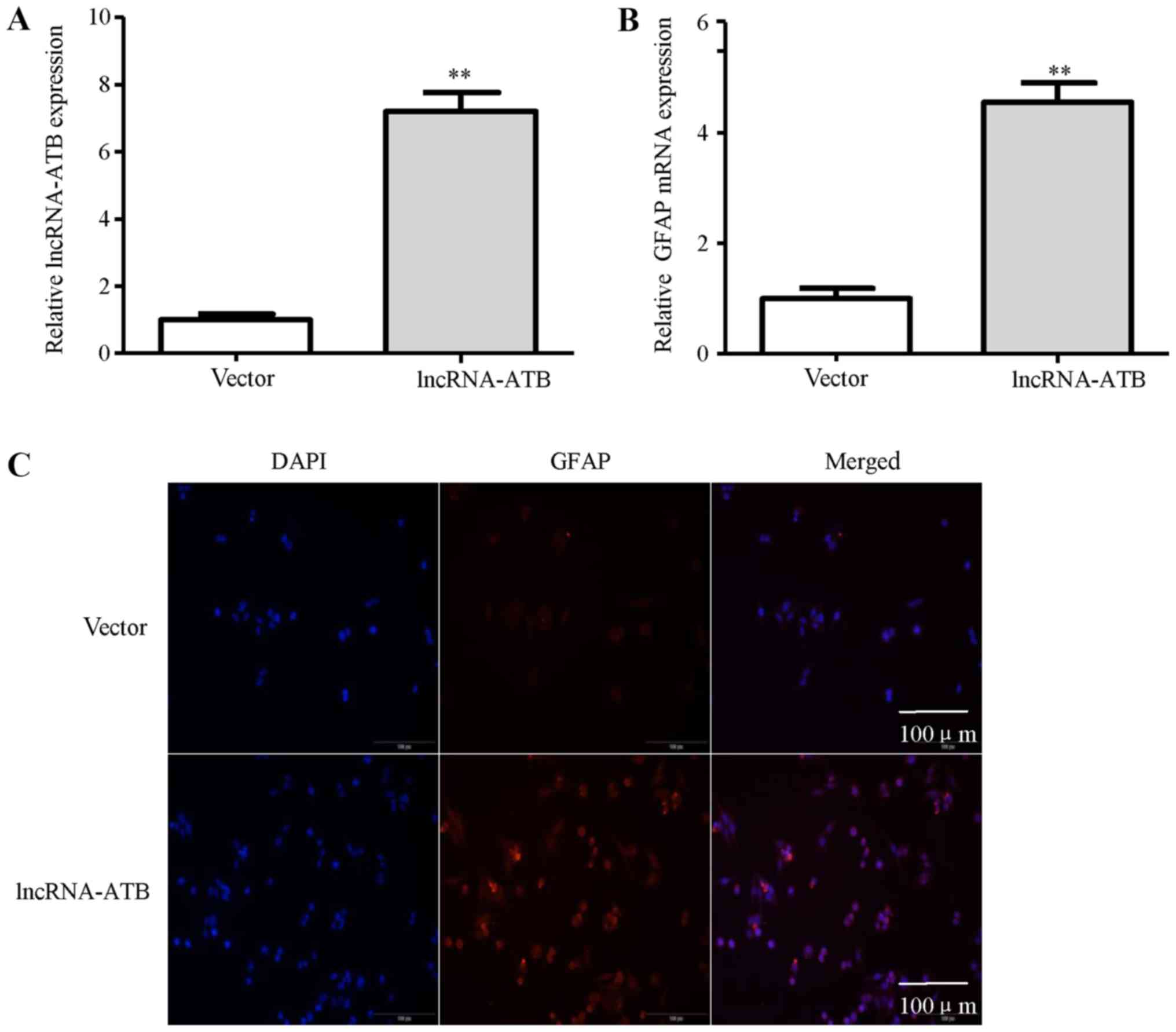
functional role of lncRNA-ATB in astrocytes, lncRNA-ATB was
transfected into astrocytes. The results of RT-qPCR analysis
revealed the elevated expression of lncRNA-ATB in the NHAs
(Fig. 4A). The enhanced expression
of lncRNA-ATB in the NHAs significantly increased GFAP mRNA
expression (Fig. 4B). In addition,
the increase in the number of GFAP-positive cells shown by
immunofluorescence staining was observed in the NHAs transfected
with lncRNA-ATB compared with the empty vector-transfected cells
(Fig. 4C). Collectively, these
findings indicate that tumor-derived exosomal lncRNA-ATB mediates
the activation of astrocytes.
lncRNA-ATB activates astrocytes
physically associated with miR-204-3p
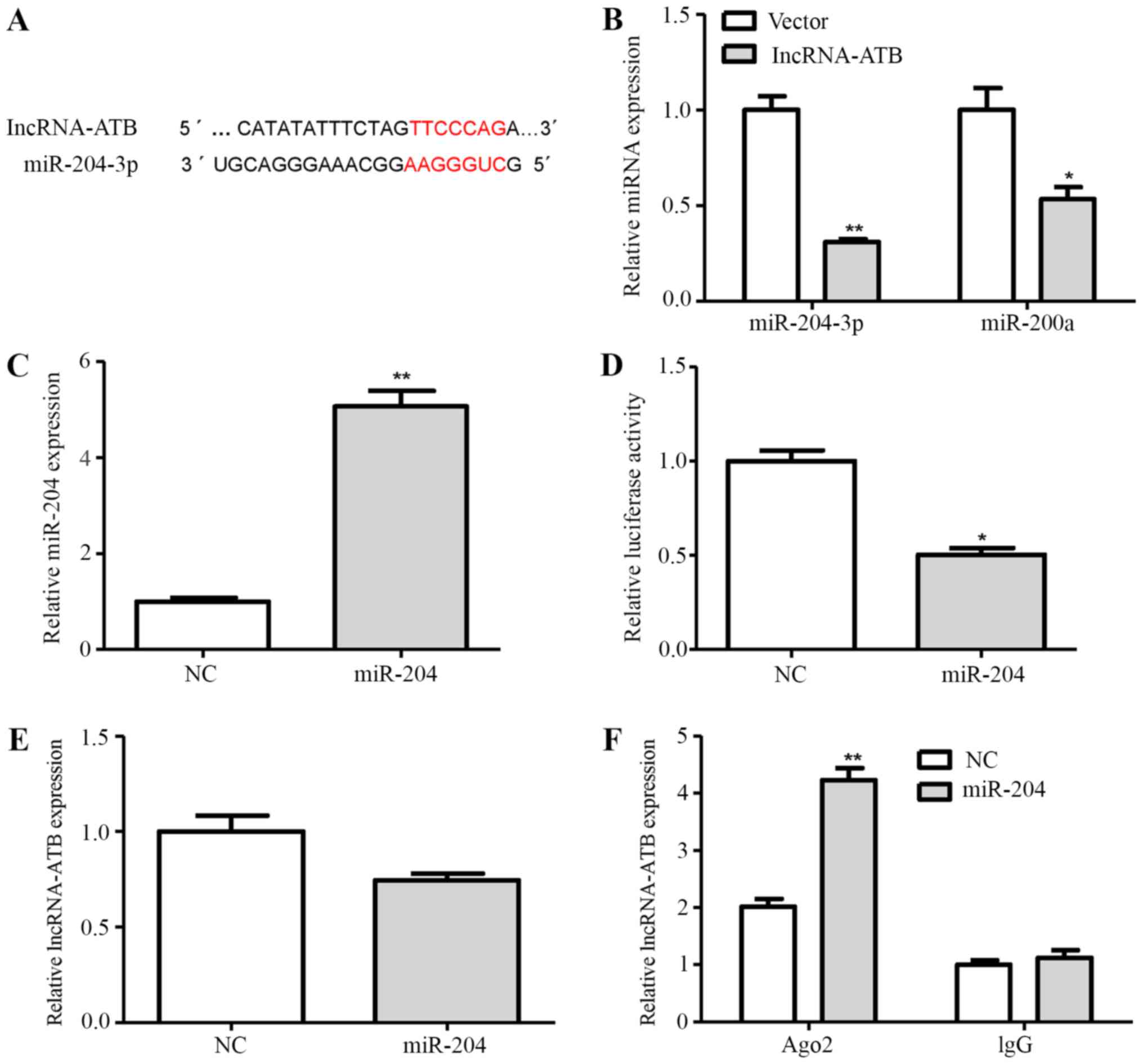
Bioinformatics analysis by miRDB revealed putative
targeting sites of miR-204-3p and miR-200a shared by lncRNA-ATB
(Fig. 5A). To further determine
whether lncRNA-ATB regulates the expression of miR-204-3p and
miR-200a, the NHAs were transfected with lncRNA-ATB or the empty
vector. The results revealed that the expression of miR-204-3p and
miR-200a was decreased by 69 and 46% in the lncRNA-ATB
vector-transfected cells compared with the empty vector
control-transfected cells, respectively (Fig. 5B). Therefore, in the subsequent
experiments, we mainly focused on the involvement of miR-204-3p in
the lncRNA-ATB-mediated activation of astrocytes.
To validate whether lncRNA-ATB is a target of
miR-204-3p in astrocytes, we constructed a luciferase reporter
plasmid of lncRNA-ATB containing miR-204-3p binding sites. As shown
in Fig. 5C and D, the ectopic
expression of miR-204-3p decreased lncRNA-ATB luciferase activity
compared with the negative control (NC). However, the
overexpression of miR-204-3p did not significantly affect
lncRNA-ATB expression in the NHAs (Fig. 5E).
miRNAs are known to bind their targets and cause
translational repression and/or RNA degradation in an
Ago2-dependent manner (25). Thus,
in this study, to determine whether lncRNA-ATB is regulated by
miR-204-3p in such a manner, we conducted anti-Ago2 RIP in NHAs
transiently overexpressing miR-204-3p. Endogenous lncRNA-ATB
pull-down was specifically enriched in the miR-204-3p-trans-fected
NHAs (Fig. 5F), suggesting that
miR-204-3p are bona fide lncRNA-ATB-targeting miRNAs. These data
demonstrated that miR-204-3p bound to lncRNA-ATB, but did not
induce the degradation of lncRNA-ATB. All these data suggested that
lncRNA-ATB was physically associated with miR-204-3p in the NHAs.
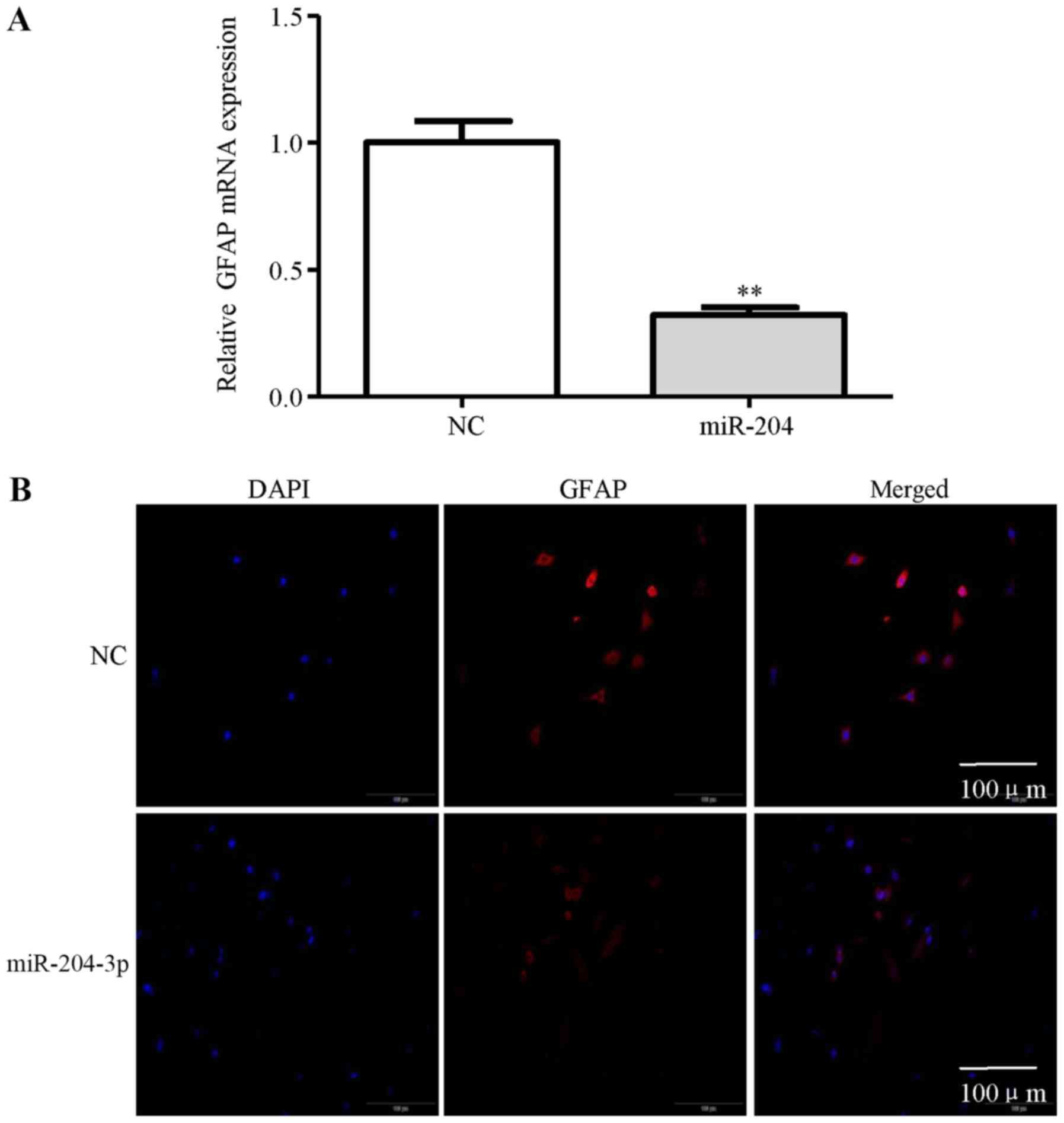
In addition, miR-204-3p inhibited GFAP expression in the NHAs
compared with the NC (Fig. 6A). As
shown in Fig. 6B, the decrease in
the number of GFAP-positive cells shown by immunofluorescence
staining was observed in the NHAs transfected with miR-204-3p
compared with the NC. These results suggest that lncRNA-ATB
activates astrocytes partly through the suppression of
miR-204-3p.
Astrocytes activated by lncRNA-ATB
regulate glioma cell invasion
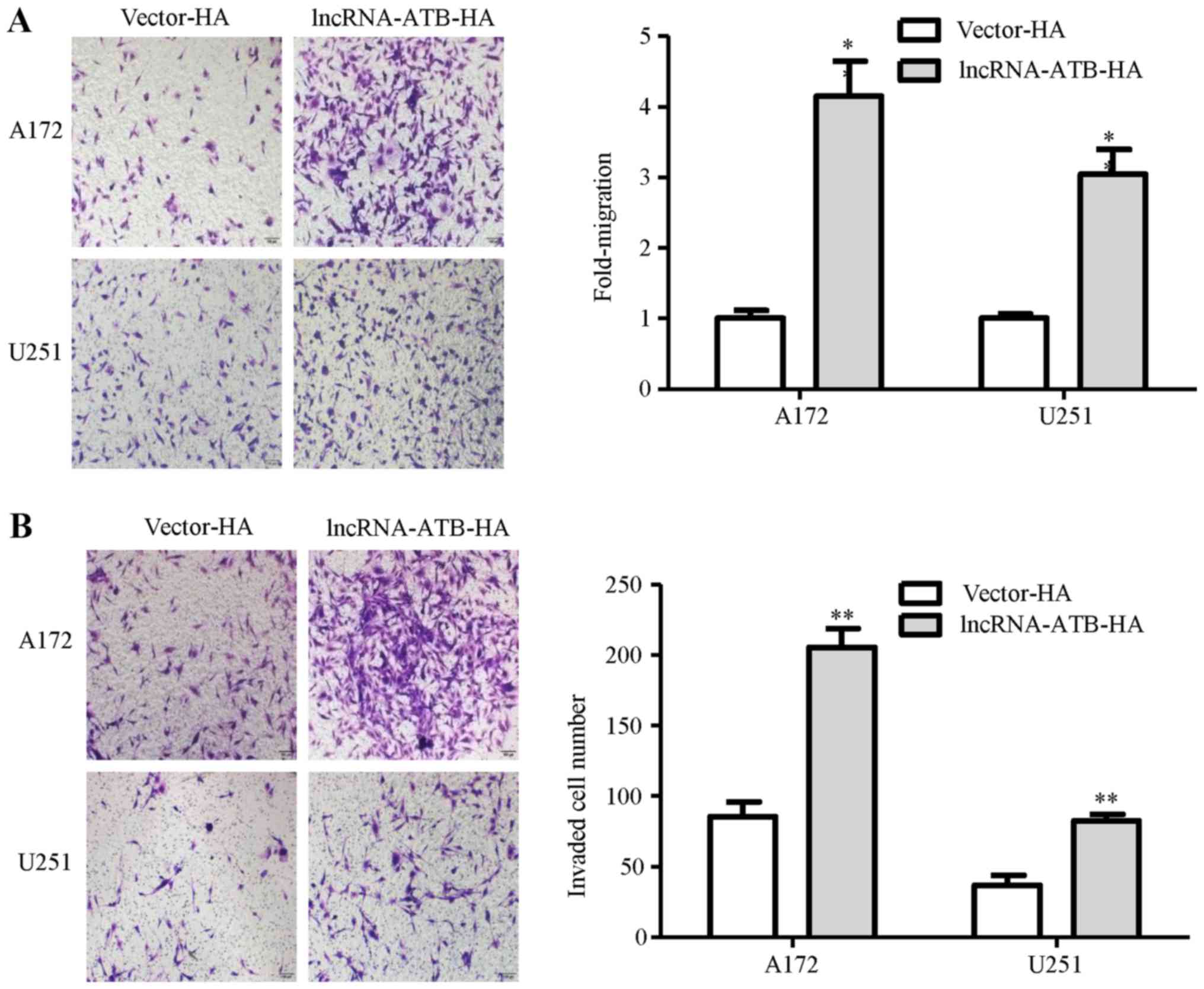
To determine the effects of astrocytes activated by
lncRNA-ATB on glioma cell migration and invasion, we investigated
the migration of the A172 and U251 glioma cells co-cultured with
vector-transfected NHAs or lncRNA-ATB-transfected NHAs. As a
result, the migration of the glioma cells was markedly increased
under lncRNA-ATB-NHA culture conditions (Fig. 7A). Similarly, the invasion of the
glioma cells was markedly increased when the cells were cultured
with lncRNA-ATB-NHAs in the bottom chamber (Fig. 7B).
Discussion
Gliomas are known to alter the phenotype of normal
cells in their environs to promote glioma invasion, and astrocytes
are key players in this process. A number of interactions between
gliomas and astrocytes are regulated through chemokines and
cytokines in the secretome (29).
This study supports an additional form of communication in which
lncRNAs are transferred from glioma cells into astrocytes via
exosomes. The transfer of lncRNA-ATB in glioma cell-derived
exosomes resulted in elevated levels in astrocytes and in the
suppression of miR-204-3p. Although other miRNAs and proteins exist
in these exsomes, the effects may be combinatorial.
Exosome transport is believed to be an effective
means for modulating cell signaling and biological function in
recipient cells (30). Several
studies have described the role of exosomes in the tumor immune
response, tumor invasiveness and metastasis (31-33).
Importantly, the ability of exosomes shed by tumor cells to
transfer the malignant phenotype to normal cells is mediated
through the delivery of nucleic acids and proteins, suggesting an
important mechanism for ‘dissemination’ of the tumor (34). Recently, GBM-derived exosomes have
been shown to promote angiogenesis during hypoxia, by the
transferring of mRNA transcripts and proteins to vascular
endothelial cells and pericytes (35,36).
However, the contribution of exosomes to the regulation of the
biological function of astrocytes is poorly understood. In the
present study, our data revealed that exosomes derived from glioma
cells were taken up by astrocytes. Moreover, the exosomes exerted a
promoting effect on the activation of astrocytes, supporting the
notion that exosomes released from malignant cells can affect the
functions of surrounding cells. A better characterization of glioma
cell-derived exosome contents and the mechanisms of astrocyte
interactions are necessary to reverse exosome-induced
dysfunction.
Increasing evidence indicates that tumor-derived
exosomes can regulate the functions of recipient cells probably
through delivering their carried non-coding RNAs. For example, high
levels of miR-451/miR-21 in GBM-EVs have been shown to be delivered
to microglia, presumably as a means for the tumor to manipulate its
environs (29). Qu et al
reported that lncARSR could be incorporated into exosomes and
transmitted to sensitive cells, thus disseminating sunitinib
resistance in renal cancer (37).
In this study, our results suggested that lncRNA-ATB embedded in
exosomes derived from glioma cells conferred the activation
phenotype to recipient astrocytes. Recently, plasma lncRNA-ATB
expression has been shown to be increased in lung disease (26). These findings suggest that circular
lncRNA-ATB may play a central role in glioma.
Accumulating evidences indicates the common
existence of a widespread interaction network of ceRNAs. lncRNAs
may carry out its functions by targeting miRNAs and regulating
their functional roles. lncRNA-ATB promotes tumor cell invasion and
plays a key role in the distant metastasis of HCC by negatively
regulating the miR-200 family (24). Our previous study revealed that
lncRNA-ATB was abnormally upregulated in glioma, and patients with
glioma with a high lncRNA-ATB expression had a shorter overall
survival time. lncRNA-ATB functions as a ceRNA by decreasing
miR-200a expression, and upregulating TGF-β2 expression in human
glioma cells (25). In this study,
we demonstrated that lncRNA-ATB overexpression in astrocytes
significantly decreased miR-204-3p, and slightly (although still
significantly) decreased miR-200a expression, compared with the
empty vector-transfected cells. These results indicate that the
regulatory mechanism of lncRNA-ATB may be cell-specific. The
results of RNA-IP assay revealed that the expression of lncRNA-ATB
immunoprecipitated with in the miR-204-3p overexpression was
markedly increased. In addition, miR-204-3p inhibited astrocyte
activation. Therefore, these results suggest that lncRNA-ATB may
carry out its functions by the suppression of miR-204-3p in
astrocytes. Further studies are warranted in order to fully
elucidate the molecular mechanisms through which miR-204-3p
inhibits astrocyte activation.
Astrocytes are the most abundant cell type in the
brain and regulate the homeostasis of the brain microenvironment
(38). Astrocytes represent a
reactive phenotype when they contact tumor cells, expressing high
levels of GFAP (39). There is
evidence to indicate that reactive astrocytes promote the brain
metastasis of lung and breast cancer cells by secreting various
cytokines (40,41). Reactive astrocytes assist the
parenchymal infiltrative ability of glioma cells and further
augment glioma malignancy (42,43).
In this study, we demonstrated that astrocytes activated by
lncRNA-ATB promoted the migration and invasion of glioma cells.
Reactive astrocytes secrete chemokines or cytokines, such as TGF-β,
among others, to promote tumor cell migration and invasion
(44). Recently, studies have
reported that lncRNA-ATB governs the autocrine secretion of TGF-β
and increases the expression of by TGF-β via miR-200s (45,46).
This evidence suggests that exosomal lncRNA-ATB may activate
astrocytes to secrete TGF-β, which promotes the migration and
invasion of glioma cells. However, the mechanisms through which
NHAs activated by lncRNA-ATB plays its role in the regulation of
glioma cell migration/invasion is not yet clear, which is the field
we will focus on in subsequent study.
In conclusion, our results indicate that glioma
cell-derived exosomal lncRNA-ATB change phenotype of astrocytes. In
addition, exosomal lncRNA-ATB targeted and repressed of miR-204-3p
in astrocytes, which in turn promotes the invasion of glioma cells.
The findings of this study provide a novel molecular mechanism
underlying the crosstalk between glioma cells and astrocytes to
promote the invasion of glioma, which contributes to efficient
therapeutic strategies for the treatment of invasive glioma.
Funding
This study was supported by the National Natural
Science Foundation of China (nos. 81402078 and 81502149), the
Natural Science Foundation of Anhui Province (nos. 1608085MH225 and
1508085MH194), Key Research and Development Plan Project of Anhui
Province (no. 1804h08020270), the Academic Funding Project for Top
Talents in Colleges and Universities in Anhui Province (no.
gxbjZD10), and the Nova Pew Plan of the Second Affiliated Hospital
of Anhui Medical University (no. 2017KA01).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
EBB and EFC conceived and designed the experiments.
EBB, YDX, ZHY and FT, CCM performed the experiments and drafted the
manuscript. HLW and BZ contributed to the design of this study and
helped to draft the manuscript. BZ supervised the whole study and
revised the manuscript. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
GBM
|
glioblastoma multiforme
|
|
EVs
|
extracellular vesicles
|
|
MVBs
|
multivesicular bodies
|
|
lncRNAs
|
long non-coding RNAs
|
|
lncRNA-ATB
|
long non-coding RNA activated by
TGF-β
|
|
HCC
|
hepatocellular carcinoma
|
|
EMT
|
epithelial-mesenchymal transition
|
|
TEM
|
transmission electron microscopy
|
|
NHAs
|
normal human astrocytes
|
|
ceRNAs
|
competitive endogenous RNAs
|
Acknowledgments
Not applicable.
References
|
1
|
Johnson DR and O’Neill BP: Glioblastoma
survival in the United States before and during the temozolomide
era. J Neurooncol. 107:359–364. 2012. View Article : Google Scholar
|
|
2
|
Avril T, Vauleon E, Tanguy-Royer S, Mosser
J and Quillien V: Mechanisms of immunomodulation in human
glioblastoma. Immunotherapy. 3(Suppl 4): 42–44. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Coniglio SJ and Segall JE: Review:
Molecular mechanism of microglia stimulated glioblastoma invasion.
Matrix Biol. 32:372–380. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
D’Asti E, Garnier D, Lee TH, Montermini L,
Meehan B and Rak J: Oncogenic extracellular vesicles in brain tumor
progression. Front Physiol. 3:2942012. View Article : Google Scholar :
|
|
5
|
Fidler IJ, Balasubramanian K, Lin Q, Kim
SW and Kim SJ: The brain microenvironment and cancer metastasis.
Mol Cells. 30:93–98. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Katz AM, Amankulor NM, Pitter K, Helmy K,
Squatrito M and Holland EC: Astrocyte-specific expression patterns
associated with the PDGF-induced glioma microenvironment. PLoS One.
7:e324532012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee J, Borboa AK, Baird A and Eliceiri BP:
Non-invasive quantification of brain tumor-induced astrogliosis.
BMC Neurosci. 12:92011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang MK and Wong AS: Exosomes: Emerging
biomarkers and targets for ovarian cancer. Cancer Lett. 367:26–33.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Τhéry C: Exosomes: Secreted vesicles and
intercellular communications. F1000 Biol Rep. 3:152011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Batrakova EV and Kim MS: Using exosomes,
naturally-equipped nanocarriers, for drug delivery. J Control
Release. 219:396–405. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Aalberts M, van Dissel-Emiliani FM, van
Adrichem NP, van Wijnen M, Wauben MH, Stout TA and Stoorvogel W:
Identification of distinct populations of prostasomes that
differentially express prostate stem cell antigen, annexin A1, and
GLIPR2 in humans. Biol Reprod. 86:822012. View Article : Google Scholar
|
|
12
|
Friand V, David G and Zimmermann P:
Syntenin and syndecan in the biogenesis of exosomes. Biol Cell.
107:331–341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hagiwara K, Ochiya T and Kosaka N: A
paradigm shift for extracellular vesicles as small RNA carriers:
From cellular waste elimination to therapeutic applications. Drug
Deliv Transl Res. 4:31–37. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khalyfa A and Gozal D: Exosomal miRNAs as
potential biomarkers of cardiovascular risk in children. J Transl
Med. 12:1622014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Madison MN and Okeoma CM: Exosomes:
Implications in HIV-1 Pathogenesis. Viruses. 7:4093–4118. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hirsch E, Hilfiker-Kleiner D, Balligand
JL, Tarone G, De Windt L, Bauersachs J, Ferdinandy P, Davidson S,
Hausenloy DJ and Schulz R: Interaction of the heart and its close
and distant neighbours: Report of the Meeting of the ESC Working
Groups Myocardial Function and Cellular Biology. Cardiovasc Res.
99:595–599. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lugini L, Cecchetti S, Huber V, Luciani F,
Macchia G, Spadaro F, Paris L, Abalsamo L, Colone M, Molinari A, et
al: Immune surveillance properties of human NK cell-derived
exosomes. J Immunol. 189:2833–2842. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu H and Fan GC:
Extracellular/circulating microRNAs and their potential role in
cardiovascular disease. Am J Cardiovasc Dis. 1:138–149.
2011.PubMed/NCBI
|
|
19
|
Madison MN, Jones PH and Okeoma CM:
Exosomes in human semen restrict HIV-1 transmission by vaginal
cells and block intravaginal replication of LP-BM5 murine AIDS
virus complex. Virology. 482:189–201. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vojtech L, Woo S, Hughes S, Levy C,
Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R,
Tewari M, et al: Exosomes in human semen carry a distinctive
repertoire of small non-coding RNAs with potential regulatory
functions. Nucleic Acids Res. 42:7290–7304. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Clayton A: Cancer cells use exosomes as
tools to manipulate immunity and the microenvironment.
OncoImmunology. 1:78–80. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Epple LM, Griffiths SG, Dechkovskaia AM,
Dusto NL, White J, Ouellette RJ, Anchordoquy TJ, Bemis LT and
Graner MW: Medulloblastoma exosome proteomics yield functional
roles for extracellular vesicles. PLoS One. 7:e420642012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Taylor DD and Gercel-Taylor C:
Exosomes/microvesicles: Mediators of cancer-associated
immunosuppressive microenvironments. Semin Immunopathol.
33:441–454. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepato-cellular carcinoma. Cancer Cell. 25:666–681. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma CC, Xiong Z, Zhu GN, Wang C, Zong G,
Wang HL, Bian EB and Zhao B: Long non-coding RNA ATB promotes
glioma malignancy by negatively regulating miR-200a. J Exp Clin
Cancer Res. 35:902016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma J, Cui X, Rong Y, Zhou Y, Guo Y, Zhou
M, Xiao L and Chen W: Plasma LncRNA-ATB, a Potential Biomarker for
Diagnosis of Patients with Coal Workers’ Pneumoconiosis: A
Case-Control Study. Int J Mol Sci. 17:172016. View Article : Google Scholar
|
|
27
|
Zhou W, Fong MY, Min Y, Somlo G, Liu L,
Palomares MR, Yu Y, Chow A, O’Connor ST, Chin AR, et al:
Cancer-secreted miR-105 destroys vascular endothelial barriers to
promote metastasis. Cancer Cell. 25:501–515. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
van der Vos KE, Abels ER, Zhang X, Lai C,
Carrizosa E, Oakley D, Prabhakar S, Mardini O, Crommentuijn MH,
Skog J, et al: Directly visualized glioblastoma-derived
extracellular vesicles transfer RNA to microglia/macrophages in the
brain. Neurooncol. 18:58–69. 2016.
|
|
30
|
Melo SA, Sugimoto H, O’Connell JT, Kato N,
Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, et
al: Cancer exosomes perform cell-independent microRNA biogenesis
and promote tumorigenesis. Cancer Cell. 26:707–721. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Clayton A, Mitchell JP, Court J, Mason MD
and Tabi Z: Human tumor-derived exosomes selectively impair
lymphocyte responses to interleukin-2. Cancer Res. 67:7458–7466.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Janowska-Wieczorek A, Wysoczynski M,
Kijowski J, Marquez-Curtis L, Machalinski B, Ratajczak J and
Ratajczak MZ: Microvesicles derived from activated platelets induce
metastasis and angiogenesis in lung cancer. Int J Cancer.
113:752–760. 2005. View Article : Google Scholar
|
|
33
|
EL Andaloussi S, Mäger I, Breakefield XO
and Wood MJ: Extracellular vesicles: Biology and emerging
therapeutic opportunities. Nat Rev Drug Discov. 12:347–357. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Milane L, Singh A, Mattheolabakis G,
Suresh M and Amiji MM: Exosome mediated communication within the
tumor microenvironment. J Control Release. 219:278–294. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kucharzewska P, Christianson HC, Welch JE,
Svensson KJ, Fredlund E, Ringnér M, Mörgelin M, Bourseau-Guilmain
E, Bengzon J and Belting M: Exosomes reflect the hypoxic status of
glioma cells and mediate hypoxia-dependent activation of vascular
cells during tumor development. Proc Natl Acad Sci USA.
110:7312–7317. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Svensson KJ, Kucharzewska P, Christianson
HC, Sköld S, Löfstedt T, Johansson MC, Mörgelin M, Bengzon J, Ruf W
and Belting M: Hypoxia triggers a proangiogenic pathway involving
cancer cell microvesicles and PAR-2-mediated heparin-binding EGF
signaling in endothelial cells. Proc Natl Acad Sci USA.
108:13147–13152. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y,
Chen W, Liu F, Sun W, Li XF, et al: Exosome-Transmitted lncARSR
Promotes Sunitinib Resistance in Renal Cancer by Acting as a
Competing Endogenous RNA. Cancer Cell. 29:653–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Grosche J, Matyash V, Möller T,
Verkhratsky A, Reichenbach A and Kettenmann H: Microdomains for
neuron-glia interaction: Parallel fiber signaling to Bergmann glial
cells. Nat Neurosci. 2:139–143. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gagliano N, Costa F, Cossetti C, Pettinari
L, Bassi R, Chiriva-Internati M, Cobos E, Gioia M and Pluchino S:
Glioma-astrocyte interaction modifies the astrocyte phenotype in a
co-culture experimental model. Oncol Rep. 22:1349–1356. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fitzgerald DP, Palmieri D, Hua E, Hargrave
E, Herring JM, Qian Y, Vega-Valle E, Weil RJ, Stark AM, Vortmeyer
AO, et al: Reactive glia are recruited by highly proliferative
brain metastases of breast cancer and promote tumor cell
colonization. Clin Exp Metastasis. 25:799–810. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Seike T, Fujita K, Yamakawa Y, Kido MA,
Takiguchi S, Teramoto N, Iguchi H and Noda M: Interaction between
lung cancer cells and astrocytes via specific inflammatory
cytokines in the microenvironment of brain metastasis. Clin Exp
Metastasis. 28:13–25. 2011. View Article : Google Scholar :
|
|
42
|
Barbero S, Bajetto A, Bonavia R, Porcile
C, Piccioli P, Pirani P, Ravetti JL, Zona G, Spaziante R, Florio T,
et al: Expression of the chemokine receptor CXCR4 and its ligand
stromal cell-derived factor 1 in human brain tumors and their
involvement in glial proliferation in vitro. Ann N Y Acad Sci.
973:60–69. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Biasoli D, Sobrinho MF, da Fonseca AC, de
Matos DG, Romão L, de Moraes Maciel R, Rehen SK, Moura-Neto V,
Borges HL and Lima FR: Glioblastoma cells inhibit astrocytic
p53-expression favoring cancer malignancy. Oncogenesis. 3:e1232014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen W, Xia T, Wang D, Huang B, Zhao P,
Wang J, Qu X and Li X: Human astrocytes secrete IL-6 to promote
glioma migration and invasion through upregulation of cytomembrane
MMP14. Oncotarget. 7:62425–62438. 2016.PubMed/NCBI
|
|
45
|
Lei K, Liang X, Gao Y, Xu B, Xu Y, Li Y,
Tao Y, Shi W and Liu J: Lnc-ATB contributes to gastric cancer
growth through a MiR-141-3p/TGFβ2 feedback loop. Biochem Biophys
Res Commun. 484:514–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhu HY, Bai WD, Li C, Zheng Z, Guan H, Liu
JQ, Yang XK, Han SC, Gao JX, Wang HT, et al: Knockdown of
lncRNA-ATB suppresses autocrine secretion of TGF-β2 by targeting
ZNF217 via miR-200c in keloid fibroblasts. Sci Rep. 6:247282016.
View Article : Google Scholar
|