Introduction
Hypopharyngeal carcinoma (HPC) is one of the most
common head and neck squamous cell cancers; it is a relatively rare
but heterogeneous malignancy with a poor prognosis, and
hypopharyngeal squamous cell carcinoma (HSCC) is the most frequent
type of HPC (1). HPC is mainly
initiated in the pyriform sinus and the posterior wall of the
hypopharynx (2). Organ-preserving
chemoradiotherapy has been proposed as a practical choice for early
stage HPC, with surgical resection and reconstruction reserved for
patients with advanced and relapsed tumors (3). Despite recent advances in surgery and
chemotherapy, 5-year survival rates for patients diagnosed with HPC
have shown no significant improvement (4). Previous studies have suggested that
microRNAs (miRNAs), a large subgroup of short non-coding RNAs, are
aberrantly expressed in a wide range of human cancers, and serve
significant roles in the initiation, progression and metastasis of
various cancers, including head and neck squamous cell carcinoma
(5,6). The role of miRNAs in human cancer has
gained much attention (7), but how
these miRNAs may be involved in the onset and development of HPC
has yet to be elucidated.
miRNA (miR)-194-5p may serve as a diagnostic
indicator for myelodysplastic syndromes, in which downregulated
miR-194-5p expression levels were reported to be responsible for
the poor survival of patients with myelodysplastic syndromes
(8). Additionally, miR-194-5p
downregulation is correlated with drug resistance in metastatic
renal cell carcinoma (9). Notably,
the online prediction website TargetScan (http://www.targetscan.org/vert_71), used in the
present study, predicted that miR-194-5p may target Smad ubiquitin
regulatory factor 1 (SMURF1). SMURF1 is homologous to the
E6-associated protein C-terminus (HECT) domain ubiquitin ligase,
which is involved in several biological pathways that mediate
ubiquitination and degradation (10). SMURF1 silencing promotes the
reduction of the cancer stem cell-like population in head and neck
squamous cell carcinoma (11).
Furthermore, SMURF1 is reported to mediate the activation of the
phosphatidylinositol-3-kinase (PI3K)/protein kinase B
(AKT)/mammalian target of rapamycin (mTOR) pathway in clear cell
renal cell carcinoma (12). mTOR,
which is commonly deregulated in human cancers, has been reported
to play pivotal roles in cell-signaling pathways (13). A previous study found that the
inhibition of the mTOR/p70S6 kinase pathway could serve as a
potential target for the treatment of esophageal squamous cell
carcinoma owing to its activation in this disorder (14). On the basis of these findings, the
current study put forth a hypothesis that miR-194-5p may serve as a
tumor suppressor for HPC by regulating SMURF1, with the involvement
of the mTOR signaling pathway. To test this hypothesis, the
upregulation and depletion of miR-194-5p were used to determine the
effects of miR-194-5p on HPC cell viability, invasion and
migration, and SMURF1 silencing, together with mTOR inhibition, was
introduced to investigate the potential regulatory mechanisms.
Materials and methods
Ethics statement
The present study was approved by the Ethics
Committee of The First Hospital of China Medical University
(Shenyang, China), and written informed consent was obtained from
each participant prior to enrollment. The animal experiments were
conducted with approval of the Animal Ethics Committee of The First
Hospital of China Medical University, and all animal experimental
procedures conformed to the provisions of the Guide for the Care
and Use of Laboratory Animals by US Department of Agriculture
(15).
Study subjects
A total of 30 patients that were pathologically
diagnosed with HPC, and who underwent surgical resection at The
First Hospital of China Medical University between June 2011 and
December 2017, were enrolled into the present study. The enrolled
subjects included 28 males and 2 females, with a mean age of
63.1±8.3 years. None of the included patients underwent
chemotherapy or radiotherapy prior to enrollment. HPC tissues and
adjacent tissues (at least 2 cm away from the edge of cancer
tissues, with no cancer cells identified under a microscope) were
collected from these patients. All tissue specimens were fixed in
4% paraformaldehyde, embedded and sectioned, as described below.
The age, sex, primary tumor range (T) staging and regional lymph
node metastasis (N) staging of patients were recorded (Table I).
 | Table ILow miR-194-5p expression levels are
associated with hypopharyngeal carcinoma. |
Table I
Low miR-194-5p expression levels are
associated with hypopharyngeal carcinoma.
| Clinicopathological
feature | n | miR-194 -5p
expression
| P-valuea |
|---|
| Low | High |
|---|
| Age (years) | | | | 0.731 |
| ≤ 60 | 13 | 7 | 6 | |
| > 60 | 17 | 8 | 9 | |
| Sex | | | | 0.143 |
| Male | 28 | 15 | 13 | |
| Female | 2 | 0 | 2 | |
| T stage | | | | 0.046 |
| T1+T2 | 9 | 2 | 7 | |
| T3+T4 | 21 | 13 | 8 | |
| N stage | | | | 0.028 |
| N0+N1 | 14 | 4 | 10 | |
| N2+N3 | 16 | 11 | 5 | |
Hematoxylin and eosin (H&E)
staining
HPC and adjacent tissues were fixed in 10% neutral
formalin for 16-18 h at room temperature, dehydrated in gradient
ethanol [50, 70, 80, 95, 100 (I) and 100% (II)] for 15 min,
embedded in paraffin at 60°C for 30 min and sectioned (5
µm). The tissue sections were dewaxed by xylene I and xylene
II for 15 min, followed by rehydration at gradient ethanol (100,
95, 80, 70 and 50%) for 5 min, respectively, and then washed by
distilled water for 1 min. The sections were stained for 5 min with
hematoxylin, washed with water for 1 min, differentiated with 1%
hydrochloric acid ethanol for 15 sec and washed again with tap
water for 15 min. Subsequently, the sections were stained with 1%
eosin for 3 min and washed with tap water for 1 min. The sections
were dehydrated by gradient ethanol and then cleared with xylene I
and xylene II for 5 min each. Finally, the sections were sealed
with neutral gum and observed under an optical microscope.
Immunohistochemical staining
The sections were dewaxed and rehydrated as
aforementioned, followed by heat-induced antigen retrieval in a
microwave at a power of 800 W at 90°C for 5 min. Subsequently, the
sections were incubated with 3% H2O2 at room
temperature for 10 min, washed three times with PBS and blocked
with 10% goat serum (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) for 10 min at room temperature. Sections were
incubated with primary antibody against SMURF1 (1:100; catalog no.
ab38866; Abcam, Cambridge, MA, USA) at 4°C overnight, and
subsequently incubated with a horseradish peroxidase
(HRP)-conjugated goat anti-rabbit secondary immunoglobulin (Ig)G
antibody (1:2,000, catalog no. ab6721; Abcam) for 30 min at 37°C.
The sections were stained with diaminobenzidine at 37°C for 10 min,
counter-stained with hematoxylin for 30 sec at 37°C, differentiated
with hydrochloric acid ethanol, dehydrated with gradient ethanol,
cleared with xylene I and II, and sealed with neutral gum, as
aforementioned. The slides were examined under an optical light
microscope, and the positive cells were scored as previously
described (16). According to the
staining intensity and the percentage of positive cells, the cells
with cytoplasm or membranes stained with brown or brown-yellow
granules were considered as positive. A total of five visual fields
were observed in each section with an optical microscope
(magnification, ×400), and 200 cells were counted in each field.
The percentage of positive cells with brown/brown-yellow cytoplasm
or with brown/brown-yellow granules was calculated. The staining
intensity was scored as 3 points (dark brown), 2 points
(brown-yellow), 1 point (light yellow) or 0 points (unclear or
colorless); and the percentage of positive cells was graded as 3
points (>50% cells stained), 2 points (25-50% cells stained), 1
point (<25% cells stained) or 0 points (no cells stained). The
staining results of the tumors were expressed as the product of the
staining intensity and the percentage of positive cells, which were
classified similarly, with 0-1 for negative staining), 2-3 for
weakly positive, 4-5 for moderately positive), and >5 for strong
positive.
Dual-luciferase reporter gene assay
The online target prediction website TargetScan
(http://www.targetscan.org/vert_71)
was used to determine whether miR-194-5p targeted SMURF1. The
wild-type (wt) full length 3′-untranslated region (3′UTR) sequence
of SMURF1 mRNA was amplified by polymerase chain reaction (PCR) and
was ligated into the pGL3 dual-luciferase reporter gene vector
(Promega Corporation, Madison, WI, USA) following XhoI and
HindIII restriction enzyme digestion. Following
site-directed mutagenesis of the potential complementary binding
site, the pGL3 vector was double-enzyme digested by XhoI and
HindIII and the mut fragment was inserted into the
dual-luciferase reporter vector to construct mut recombinant dual
luciferase reporter vector. A total of 200 ng constructed vectors,
50 nmol/l miR-194-5p mimics (5′-UGCAGCAGCUUCTGCATGTCCT-3′) or 50
nmol/l mimics-NC (Guangzhou RiboBio Co., Ltd., Guangzhou, China)
and 5 μl of Lipofectamine® 2000 transfection reagents
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) were
dissolved together in the 250 µl Opti-MEM culture solution
(Invitrogen; Thermo Fisher Scientific, Inc.) to prepare the
transfection mixture, according to the manufacturer’s protocol, and
let stand at room temperature for 20 min. The transfection mixture
was inoculated into 6-well plates, gently mixed with the cell
culture solution by shaking and incubated in a 37°C CO2
incubator. Following 6 h incubation, the culture medium and
transfection reagents were replaced with 2 ml of RPMI-1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) for 72 h at 37°C. Cells
were collected and firefly and Renilla luciferase activities
were analyzed with the Luciferase Reporter Gene Assay kit (Promega
Corporation), according to the manufacturer’s protocol; firefly
luciferase activity was normalized to Renilla luciferase
activity.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Tissues (100 mg) or cells (5×106) were
used for total RNA extraction using TRIzol® reagent (Invitrogen,
Carlsbad, CA, USA), according to the manufacturer’s protocol. cDNA
was synthetized using the M-MLV Reverse Transcription kit
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer’s protocol; briefly, the reaction conditions were as
follows: 37°C for 60 min and 99°C for 5 min, and the reaction was
terminated at 4°C. The SYBR Prime Script miRNA RT-PCR kit (Takara
Biotechnology Co., Ltd., Dalian, China) was used to determine the
expressions of miR-194-5p in HPC and adjacent normal tissues, as
well as the human HPC cell lines. The 20 µl reaction was set
up as follows: 10 µl SYBR Premix Ex Taq II (2X), 0.8
µl of PCR forward primer (10 µmol/l), 0.8 µl
of Universal-miR qPCR primer (10 µmol/l), 0.4 µl of
ROX Reference Dye II (50X), 2 µl of cDNA template and 6
µl of ddH2O. The thermocycling conditions were as
follows: Initial denaturation at 95°C (10 min); followed by 40
cycles of denaturation at 95°C (10 sec), annealing at 60°C (10 sec)
and extension at 72°C (15 sec). The primers were synthesized by the
Shanghai Sangon Biotechnology Co. Ltd. (Shanghai, China); sequences
are indicated in Table II. U6 was
used as an internal reference and for normalization of miR-194-5p,
and GAPDH as an internal reference and for normalization for
SMURF1; the relative expressions of miR-194-5p and SMURF1 were
calculated using the 2−ΔΔCq method (17).
 | Table IIPrimer sequences for reverse
transcription quantitative polymerase chain reaction. |
Table II
Primer sequences for reverse
transcription quantitative polymerase chain reaction.
| Gene | Primer sequence
(5′-3′) |
|---|
| miR-194-5p | F:
5′-GCCGTCTGTAACAGCAACTCCA-3′ |
| R:
5′-GTGCAGGTCCGAGGTATTC-3′ |
| SMURF1 | F:
5′-CTCATCCCTCAACATCTGCTG-3′ |
| R:
5′-GCCCTCCTTTCTTCATCG-3′ |
| U6 | F:
5′-GCTTCGGCAGCACATATACT-3′ |
| R:
5′-GTGCAGGGTCCGAGGTATTC-3′ |
| GAPDH | F:
5′-GAAGGTCGGAGTCAACGGAT-3′ |
| R:
5′-CCTGGAAGATGGTGATGGGAT-3′ |
Western blot analysis
A total of 1×106 cells were collected and
washed two times with PBS with the supernatant being discarded.
Cells were lysed in radioimmunoprecipitation assay lysis buffer,
followed by an ice bath at 4°C for 30 min and centrifugation at
25,764 × g for 15 min. The supernatant was collected and
transferred to another clean Eppendorf tube. Protein concentration
was determined by a bicinchoninic acid assay. Protein samples
(20-30 µg) were separated by 12% SDS-PAGE. Proteins were
transferred to a polyvinylidene fluoride membrane, which was
subsequently blocked with 5% skimmed milk in tris-buffered saline +
0.05% Tween-20 (TBST) on a shaker at room temperature for 30 min.
The membranes were incubated at room temperature for 2 h with
primary rabbit polyclonal antibodies (diluted in blocking solution)
against SMURF1 (1:500; catalog no. ab38866), mTOR (1:2,000; catalog
no. ab2732), phosphorylated (p)-mTOR (1:2,000; catalog no.
ab109268), topoisomerase II (TOPO II; 1:2,000; catalog no.
ab52934), minichromosome maintenance 2 (MCM2; 1:1,000; catalog no.
ab108935), proliferating cell nuclear antigen (PCNA; 1:1,000;
catalog no. ab18197), Ki67 (1:5,000; catalog no. ab92742), matrix
metalloproteinase (MMP)-2 (1:2,000; catalog no. ab92536), MMP-9
(1:1,000; catalog no. ab38898) and GAPDH (1:2,500; catalog no.
ab9485), all purchased from Abcam. The membranes were subsequently
incubated with an HRP-conjugated secondary goat anti-rabbit IgG
antibody (1:2,000; catalog no. ab6721; Abcam) at room temperature
for 1 h. Protein bands were visualized using Enhanced
Chemiluminescence reagents (EMD Millipore, Billerica, MA, USA), and
washed three times with TBST, 10 min each. Images were captured
with a Gel Doc XR gel imaging instrument (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) and densitometric analysis was conducted
using ImageJ software (version 1.42, National Institutes of Health,
Bethesda, MD, USA); GAPDH was used as an internal control and for
normalization.
Cell culture and transfection
Human HPC cell lines FaDu and HSC-4 were provided by
the Cancer Institute of China Medical University (Beijing, China).
Cells were subcultured in Dulbecco’s modified Eagle’s medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1%
streptomycin at 37°C with 5% CO2 and saturated humidity.
The medium was replaced every 2 days, and the cells that grew well
were used for cell selection. The sequences of the transfected
components are as follows: miRNA mimic-negative control (mimic-NC),
5′-ACATTGTCGTTGAGGTACACCT-3′; miR-194-5p inhibitor-NC,
5′-AGGUTCAACUTGACGTACAGGA3′; miR-194-5p inhibitor,
5′-TCCACATGGAGTTGCTGTTACA-3′; and SMURF1 (XM_166483) small
interfering (si)RNA 5′-AAGAACCUUGCAAAGAAAGAC-3′. Nine experimental
groups were generated through liposome-mediated transfection and
included: i) Control, FaDu cells without any treatment; ii)
mimic-NC, FaDu cells transfected with mimic-NC; iii) miR-194-5p
mimics, FaDu cells transfected with miR-194-5p mimics; iv)
inhibitor-NC, FaDu cells transfected with inhibitor-NC; v)
miR-194-5p inhibitor, FaDu cells transfected with miR-194-5p
inhibitor; vi) miR-194-5p inhibitor + NC-siRNA, FaDu cells
co-transfected with miR-194-5p inhibitor and NC siRNA; vii)
miR-194-5p inhibitor + SMURF1 siRNA, FaDu cells transfected with
miR-194-5p inhibitor + SMURF1 siRNA; viii) miR-194-5p inhibitor +
0.01% dimethyl sulfoxide (DMSO), FaDu cells transfected with
miR-194-5p inhibitor a nd t reated with DMS; i x) a nd m i R-194
-5p inhibitor + rapamycin, FaDu cells transfected with miR-194-5p
inhibitor and treated with 100 ng/ml rapamycin. The mimics-NC,
inhibitor-NC, NC-siRNA, miR-194-5p mimics, miR-194-5p inhibitor and
SMURF1 siRNA were purchased from Guangzhou RiboBio Co., Ltd.; DMSO
and rapamycin was obtained from Beijing Propbs Biotechnology Co.,
Ltd. (Beijing, China). Cells were seeded (1×105
cells/well) into a 6-well plate; when the cells reached ~30%
confluency, a transfection mixture was prepared in accordance with
the instructions of the Lipofectamine 2000 transfection reagent: 10
µl miR-194-5p mimics or 50 nmol/l inhibitor, mimics-NC,
inhibitor-NC, NC-siRNA or SMURF1 siRNA and 5 µl of
Lipofectamine 2000 transfection reagents were dissolved together in
the Opti-MEM (Invitrogen; Thermo Fisher Scientific, Inc.) culture
solution to prepare the transfection mixture, and let stand at room
temperature for 20 min. The transfection mixture was inoculated
into 6-well plates, gently mixed with the cell culture solution by
shaking and incubated in a 37°C CO2 incubator for 6 h;
after which, the culture medium and transfection reagents were
replaced with 2 ml of RPMI-1640 medium containing 10% FBS and
incubated for 24 h prior to subsequent experimentation.
Cell Counting Kit-8 (CCK-8)
At 24 h post-transfection, FaDu cells were treated
with 0.25% trypsin and collected. Cells were centrifuged at 200 × g
for 5 min at 4°C and resuspended in fresh complete DMEM and
inoculated into 96-well plates at a density of 0.8×103
cells/well (l00 µl/well), with five duplicated wells for
each group. A CCK-8 kit (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) was used to examine cell viability. A total of 10
µl CCK-8 solution was gently added into each well along the
wall, gently shaken and mixed while avoiding bubbles. The mixture
was incubated at 37°C for 2 h, and the optical density was measured
at a wavelength of 450 nm on an automatic enzyme immunoassay
instrument after 0, 24, 48, 72 and 96 h of transfection, and the
growth curve was plotted.
Transwell cell migration and invasion
assay
Following 24 h transfection, FaDu cells were
detached with trypsin, and single cell suspensions were prepared.
Cells were washed two times with serum-free medium, and the number
of cells was counted and the concentration was adjusted to
5×106 cells/ml. A total of 100 µl of cell
suspension was slowly added to the upper chamber of each Transwell
chamber with or without Matrigel (for invasion and migration,
respectively), and 600 µl medium containing 10% FBS was
added to the lower chamber; a total of three duplicated wells were
used for each group, and the mixture was cultured in a 5%
CO2 incubator at 37°C for 24 h. The non-invading cells
in the upper membrane were removed with cotton swabs, and cells in
the lower chamber were fixed in methanol for 10 min and stained
with 0.1% crystal violet for 40 min. Five visual fields
(magnification, ×100) were randomly examined and the number of
invading cells in each group was counted; the average value was
recorded.
Xenograft tumors in nude mice
A total of 54 male athymic BALB/c nude mice (age,
4-5 weeks; weight 18±2 g) were purchased from Beijing Vital River
Laboratory Animal Technology Co., Ltd. (Beijing, China). Cells in
the logarithmic growth phase collected and resuspended in PBS to a
concentration of 1×107 cells/ml. A total of 54 nude mice
were randomly divided into 9 groups (n=6 mice/group): i) Control
group, which were injected with untreated cells; ii) mimics-NC
group, which were injected with mimic-NC-transfected cells; iii)
miR-194-5p mimics group, which were injected with miR-194-5p
mimics-transfected cells; iv) inhibitor-NC group, which were
injected with miR-194-5p inhibitor-NC-transfected cells; v)
miR-194-5p inhibitor group, which were injected with miR-194-5p
inhibitor-transfected cells; vi) miR-194-5p inhibitor + NC siRNA
group, which were injected with cells co-transfected with
miR-194-5p inhibitor and NC siRNA; vii) miR-194-5p inhibitor +
SMURF1 siRNA group, which were injected with cells co-transfected
with miR-194-5p inhibitor and SMURF1 siRNA; viii) miR-194-5p
inhibitor + DMSO group, which were injected with miR-194-5p
inhibitor-transfected cells that were co-treated with DMSO); xi)
and miR-194-5p inhibitor + rapamycin group, which were injected
with miR-194-5p mimics-transfected cells that were co-treated with
rapamycin. The cell suspensions containing 1×107
transfected or treated cells were injected into the right armpit of
the nude mice using a disposable syringe. Experiments were
conducted in a specified pathogen-free barrier environment. Nude
mice were raised until the tumor was visible with the naked eye.
Tumor volume (volume = π /6 × short diameter2 × long
diameter) of nude mice was measured on days 7, 14, 21 and 28
following tumor transplantation. The nude mice were sacrificed on
day 28, and the weight of the tumors was measured.
Statistical analysis
Statistical analyses were conducted using SPSS 21.0
software (IBM Corp., Armonk, NY, USA). Data are expressed as the
mean ± standard deviation. Comparisons between two groups were
analyzed by independent-sample t-test or paired t-test. Comparisons
of miR-194-5p and clinicopathological features between two groups
were conducted using the χ2 test. The correlation
between miR-194-5p and SMURF1 expressions was analyzed by Pearson’s
correlation analysis. Comparisons of data among multiple groups
were tested using one-way analysis of variance (ANOVA) followed by
Tukey’s post hoc test. Comparisons of cell viability and tumor
volume at different time points were analyzed by two-way ANOVA.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Histopathological examination of HPC
tissues and adjacent normal tissues
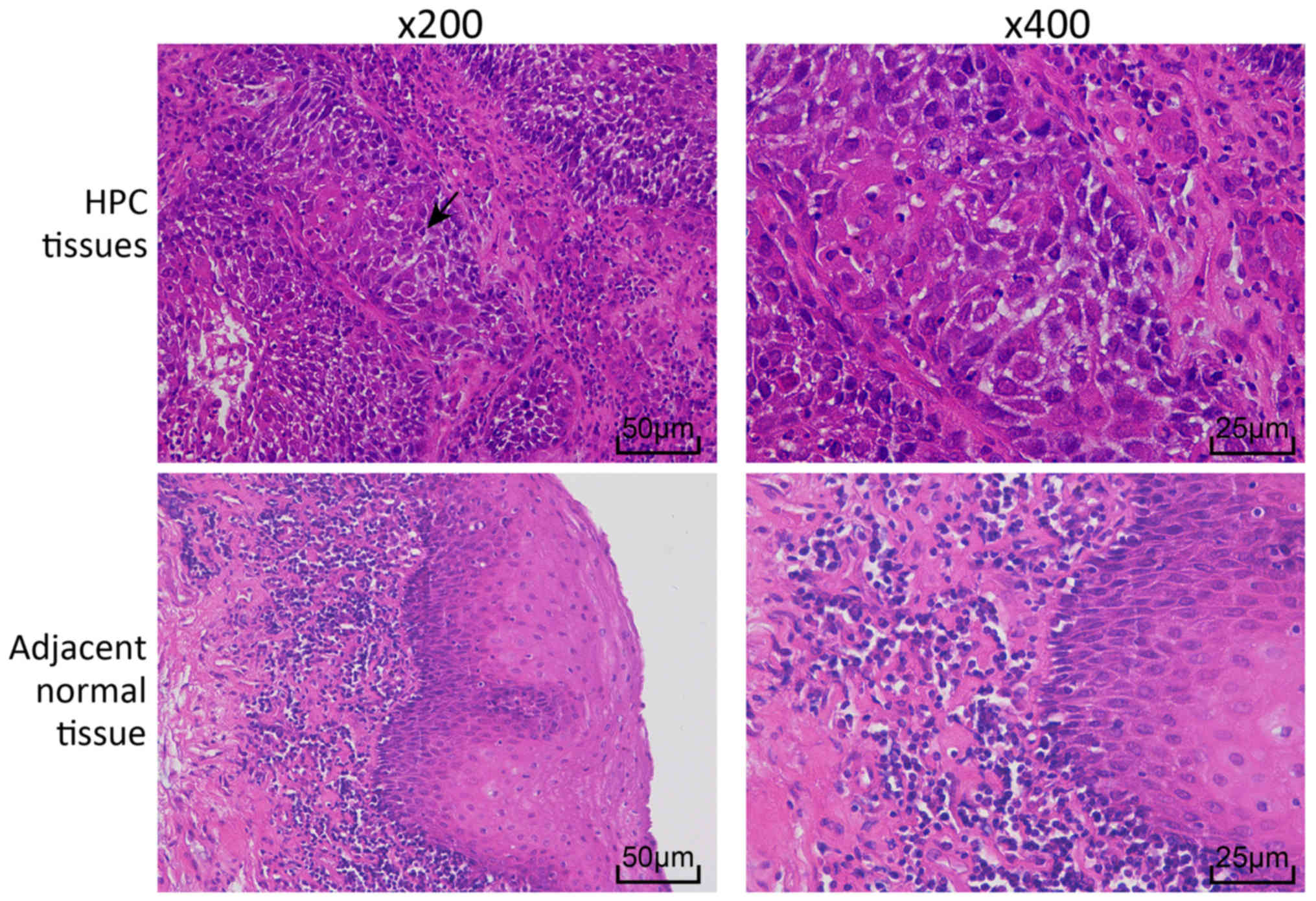
HPC and adjacent normal tissues were stained with
H&E to examine the histopathological changes (Fig. 1). In the HPC tissues, cells were
arranged in a disordered manner, with large nuclei that were deeply
stained, cancer nests were observed in the epithelial cells.
However, cells in the adjacent normal tissues were arranged in an
organized way.
Downregulated miR-194-5p is associated
with HPC progression
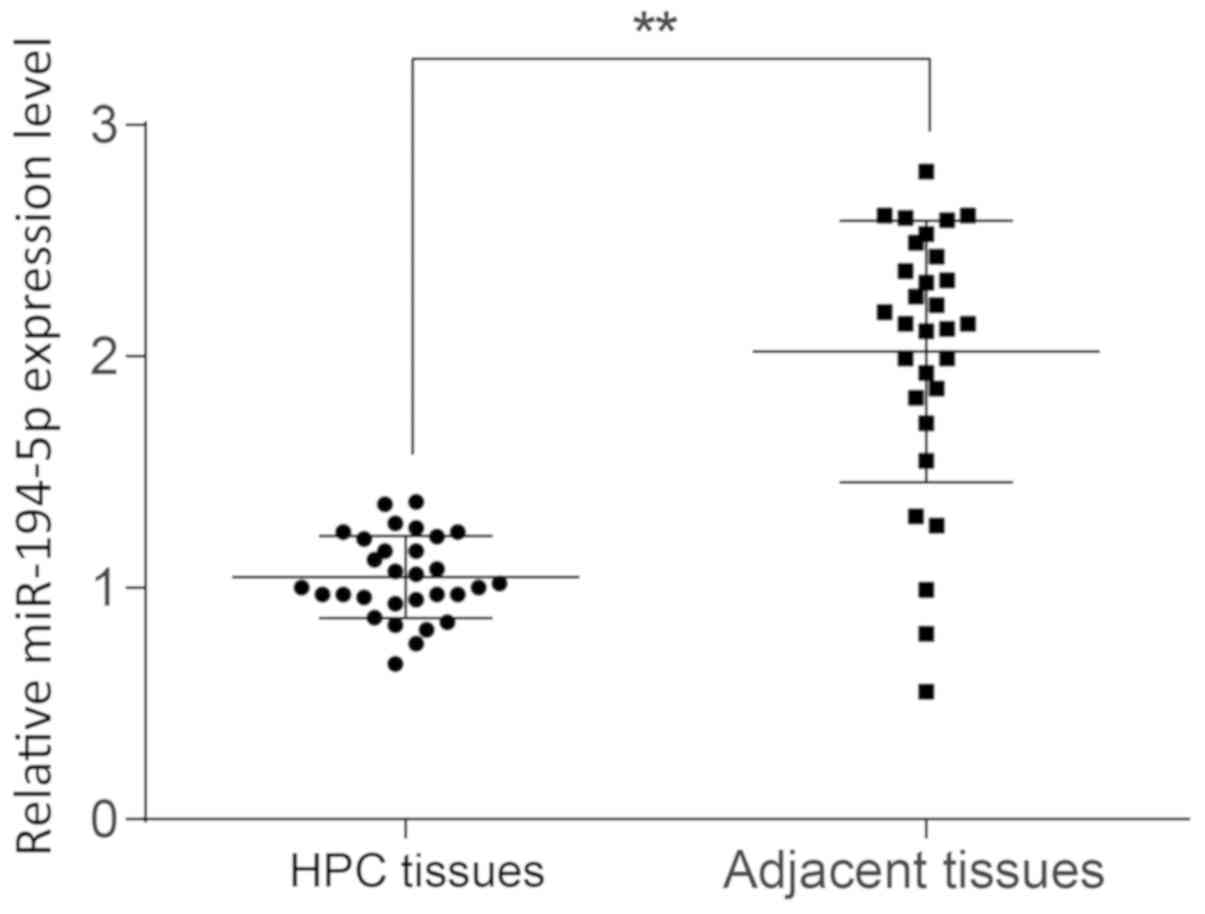
RT-qPCR was performed to examine the expression
levels of miR-194-5p in HPC tissues, and the results demonstrated
that miR-194-5p expression was significantly lower in HPC tissues
compared with expression in adjacent normal tissues (P<0.01;
Fig. 2). The median value of
miR-194-5p expression as set as the cut-off value, and the 30 HPC
tissues were divided into a high expression group (miR-194-5p
expression ≥1.02) and a low expression group (miR-194-5p expression
<1.02) to analyze the relationship between the expression level
of miR-194-5p and the clinicopathological features in patients with
HPC. The results indicated that there was no significant
relationship between the expression of miR-194-5p and age or sex
(P>0.05; Table I), whereas low
miR-194 expression levels were associated with T/N stages. These
results indicated that miR-194-5p expression was downregulated in
HPC, which may be involved in the progression and metastasis of
HPC.
Upregulated miR-194-5p reduces viability,
migration and invasion of FaDu cells and inhibits tumor growth
The expression levels of miR-194-5p in the HPC cell
lines FaDu and HSC-4 were examined using RT-qPCR. The results
demonstrated that the expression of miR-194-5p in the HSC-4 cell
line was significantly higher compared with expression in the FaDu
cells (P<0.05; Fig. 3A).
Therefore, the FaDu cell line was selected for subsequent
experimentation. FaDu cells were transfected with miR-194-5p mimics
or inhibitor and successful transfections were confirmed by
RT-qPCR, which demonstrated a significantly increased or reduced
level of miR-194-5p expression, respectively, compared with the
NC-transfected groups (P<0.01; Fig.
3B).
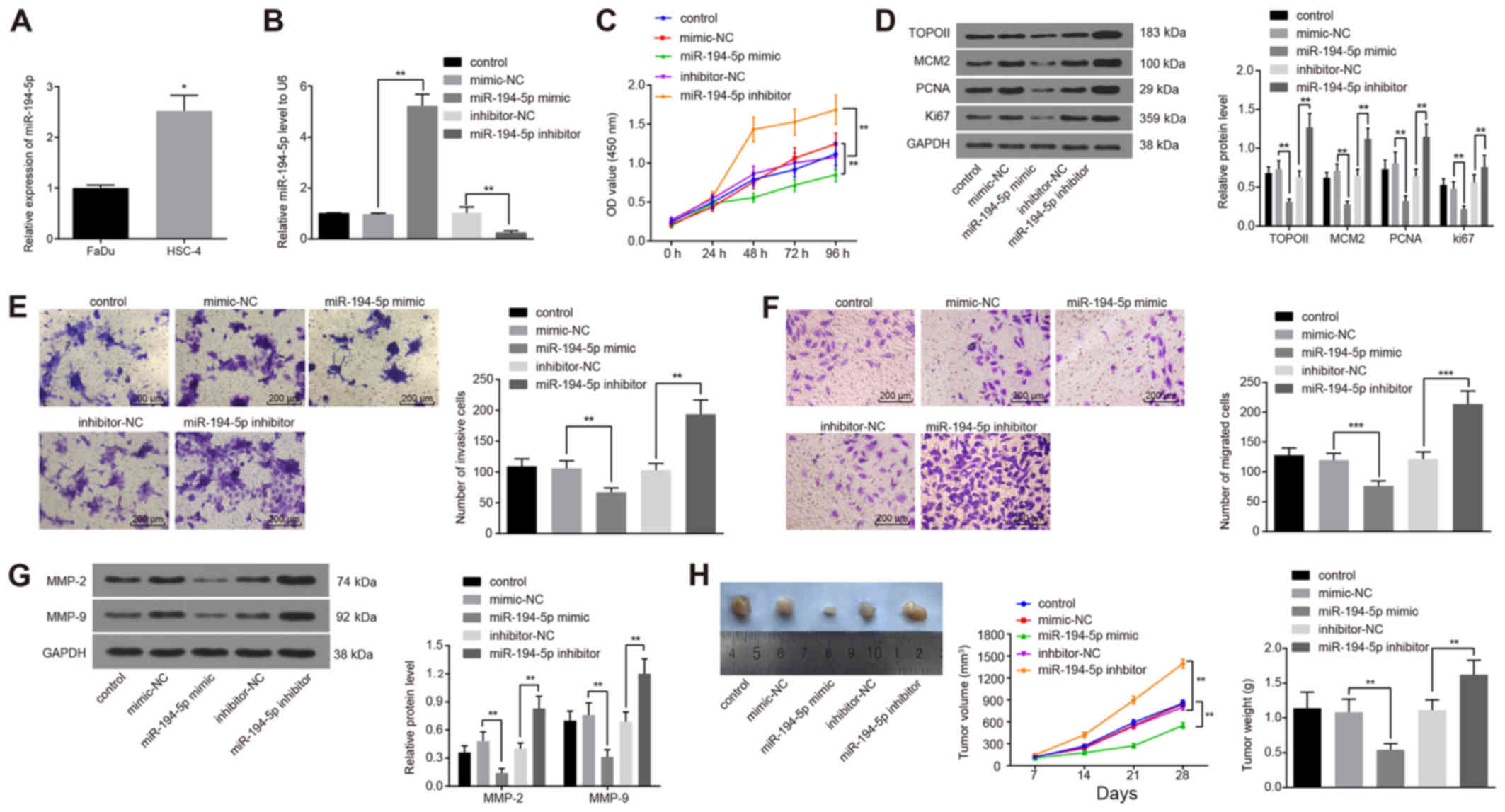 | Figure 3High miR-194-5p expression reduces
FaDu cell viability, invasion and migration. (A) miR-194-5p
expression in two HPC cell lines FaDu and HSC-4 detected by
RT-qPCR. (B) miR-194-5p expression detected by RT-qPCR following
miR-194-5p mimics or inhibitor transfection into FaDu cells. (C) OD
values were detected by Cell Counting Kit-8 at different time
points following miR-194-5p mimics or inhibitor transfections. (D)
Protein expression levels of proliferation-related genes TOPO II,
MCM2, PCNA and Ki67 were determined by western blot analysis. (E)
Invasive ability of transfected FaDu cells was measured by Matrigel
assay; magnification, ×400. (F) Migratory ability of transfected
FaDu cells was measured by Transwell assay; magnification, ×400.
(G) Protein expression levels of invasion-related factors MMP-2 and
MMP-9 were detected by western blot analysis. (H) Nude mice were
injected with cells treated with miR-194-5p mimics or inhibitor,
and tumor volumes and weights were examined. Experiments were
repeated three times, and data are presented as the mean ± standard
deviation; *P<0.05; **P<0.01;
***P<0.001. HPC, hypopharyngeal carcinoma; MCM2,
minichromosome maintenance 2; miR, microRNA; MMP, matrix
metalloproteinase; NC, negative control; OD, optical density; PCNA,
proliferating cell nuclear antigen; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; TOPO II,
topoisomerase II. |
CCK-8 was used to assess the effects of miR-194-5p
on the viability of FaDu cells. When the expression of miR-194-5p
was upregulated, the viability of FaDu cells decreased
significantly, whereas viability was significantly increased
following the downregulation of miR-194-5p expression (both
P<0.01; Fig. 3C), which
indicated that miR-194-5p may inhibit the viability of FaDu cells.
In addition, the protein expression levels of the
proliferation-related factors TOPO II, MCM2, PCNA and Ki67 were
examined by western blot analysis following miR-194-5p mimics or
inhibitor transfection, all of which were significantly decreased
following miR-194-5p mimics transfections and significantly
increased following transfection with the miR-194-5p inhibitor,
compare with the respective controls (P<0.01; Fig. 3D).
The effects of miR-194-5p on the invasion and
migration of FaDu cells was evaluated by Matrigel and Transwell
assays, respectively. When the expression of miR-194-5p was
upregulated, the number of invasive FaDu cells was significantly
decreased, whereas the downregulation of miR-194-5p resulted in an
increase in the number of invasive cells (P<0.01; Fig. 3E); these results suggested that
miR-194-5p may negatively regulate the invasive ability of FaDu
cells. Similarly, miR-194-5p mimics transfection resulted in a
significant decrease in the number of migrating FaDu cells, whereas
the number of migrating cells were significantly increased
following miR-194-5p inhibitor transfection (P<0.001; Fig. 3F); these data suggested that high
expression levels of miR-194-5p may inhibit the migratory ability
of FaDu cells. In addition, the expression of invasion-related
factors MMP-2 and MMP-9 were detected by western blot analysis
following transfection, and the results demonstrated that the
protein expression levels of MMP-2 and MMP-9 were significantly
decreased by miR-194-5p upregulation and significantly increased by
miR-194-5p depletion (P<0.01; Fig.
3G).
Finally, the data described above were verified with
in vivo experiments by means of the xenograft tumors in nude
mice (Fig. 3H). Compared with the
inhibitor-NC group, tumor volume in the nude mice transplanted with
the miR-194-5p inhibitor-treated cells was increased, and the
weight of tumors after 28 days was also significantly increased.
Compared with the mimics-NC group, tumor volume in the nude mice
was reduced and the tumor weight after 28 days was significantly
decreased in the miR-194-5p mimics group (P<0.05). These in
vivo experimental results indicated that elevated miR-194-5p
expression levels may contribute to the inhibition of tumor
growth.
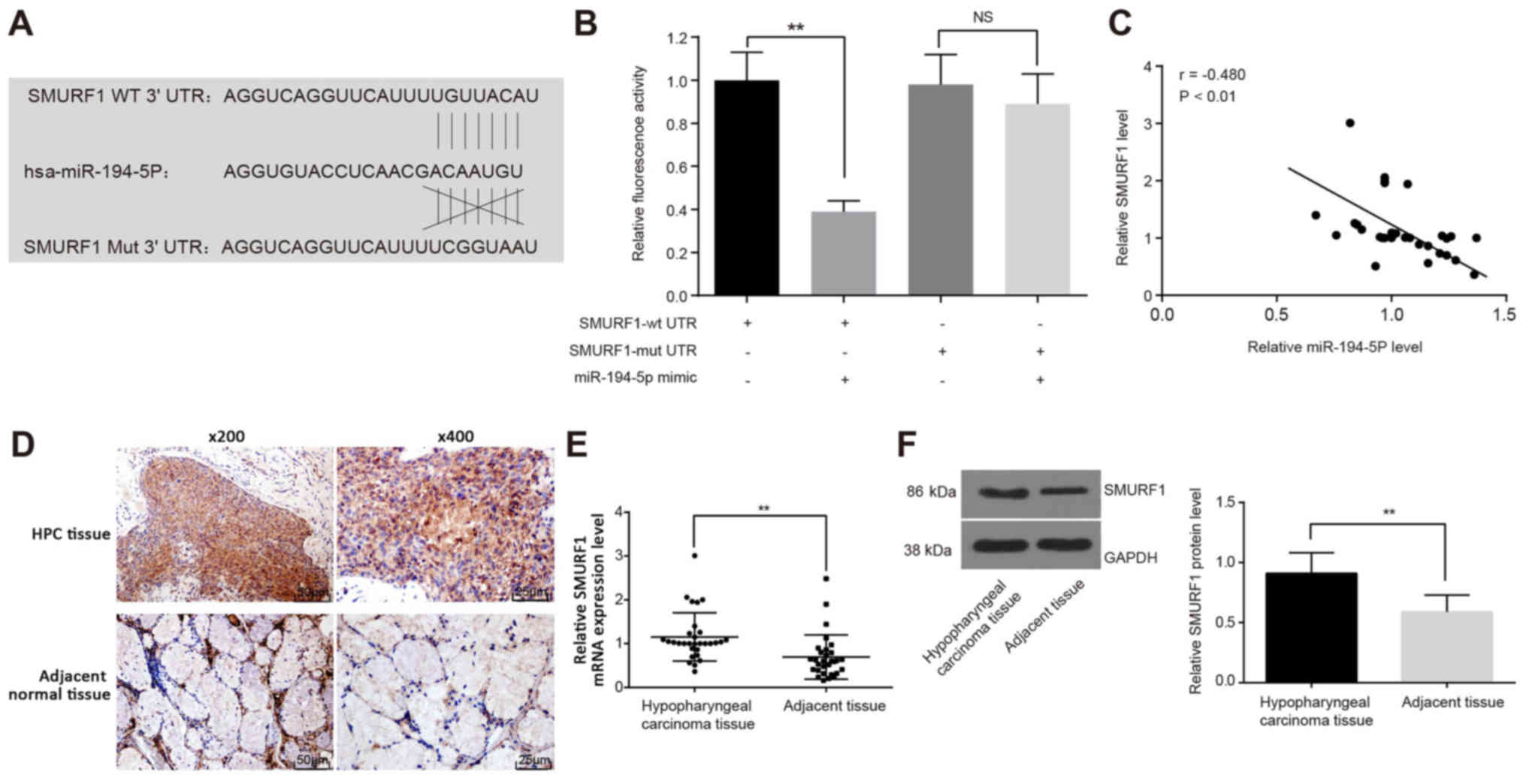
miR-194-5p binds to the SMURF1 3′UTR
miR-194-5 target genes were predicted using the
TargetScan online prediction website, which indicated that the seed
sequence of miR-194-5p targets the 3′UTR of SMURF1 mRNA (Fig. 4A). This potential interaction was
examined using luciferase assays in FaDu cells co-transfected with
either SMURF1-wtUTR or SMURF1-mutUTR and miR-194-5p mimics. The
luciferase activity of FaDu cells was significantly decreased in
SMURF1-wtUTR and miR-194-5p mimics co-treated cells (P<0.01;
Fig. 4B), which further
demonstrated that miR-194-5p can bind to and regulate SMURF1
expression. Pearson’s correlation analysis was used to verify the
correlation between miR-194-5p and SMURF1 mRNA, the results of
which indicated a negative correlation between SMURF1 and
miR-194-5p expression (r=-0.480; P<0.01; Fig. 4C). Subsequently,
immunohistochemical staining was performed to determine the
expression of SMURF1 in human HPC tissues and adjacent tissues,
which demonstrated that SMURF1 was mainly expressed in the
cytoplasm and cell membrane (Fig.
4D). The positive rate of SMURF1 protein in HPC tissues was
76.67% (23/30), which was significantly higher than that in the
adjacent tissues (16.67%; 5/30; P<0.01). The results of RT-qPCR
(Fig. 4E) and western blot
analysis (Fig. 4F) also revealed
that the mRNA and protein expression levels, respectively, of
SMURF1 were upregulated in HPC tissues compared with adjacent
tissues.
Upregulated miR-194-5p inhibits SMURF1
and mTOR signaling pathway activation
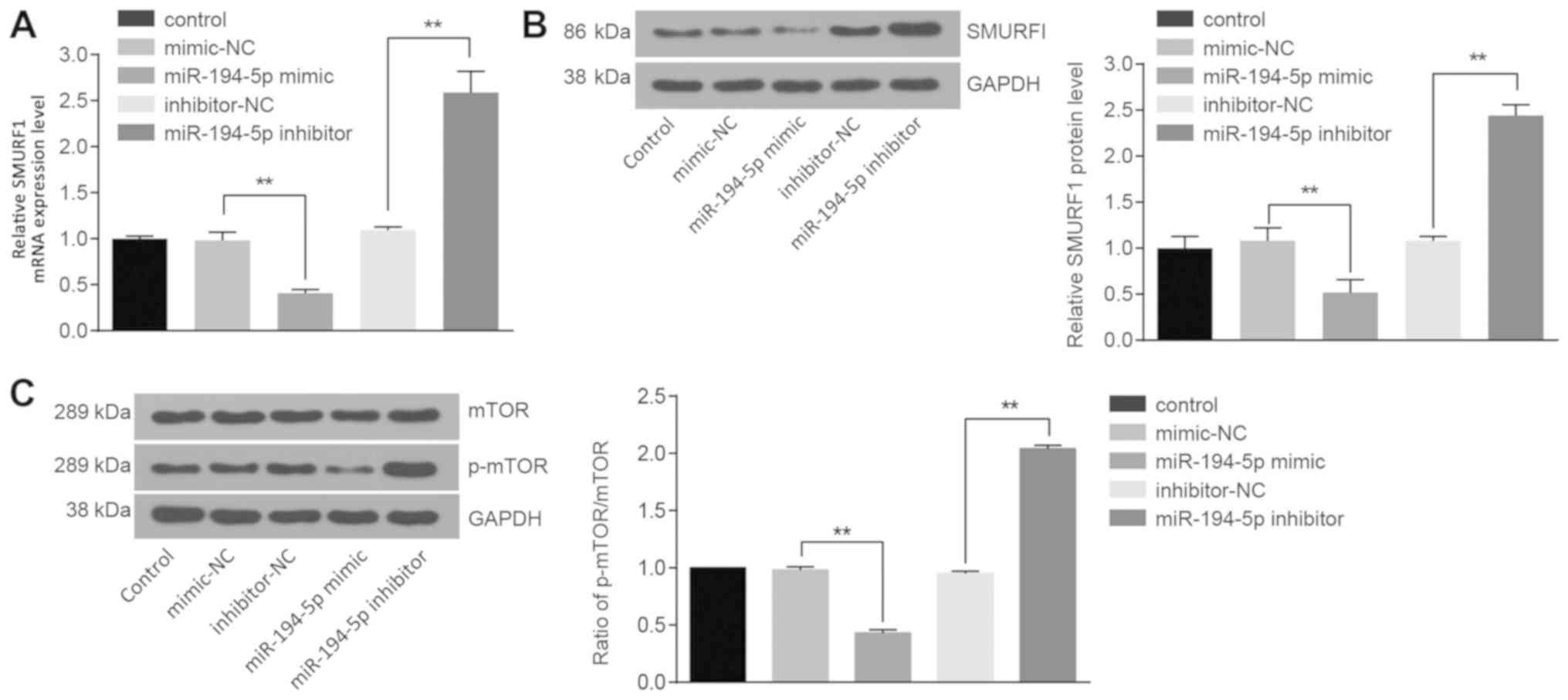
mRNA and protein expression levels of SMURF1 and
mTOR were examined, as well as the extent of mTOR phosphorylation.
Compared with the cells transfected with mimics-NC, the mRNA and
protein expression levels of SMURF1 (P<0.01; Fig. 5A and B, respectively), the ratio of
p-mTOR to total mTOR in the cells treated with miR-194-5p mimics
were significantly decreased (P<0.01; Fig. 5C). However, the mRNA and protein
expression levels of SMURF1 (P<0.01; Fig. 5A and B, respectively), the protein
expression of mTOR and the extent of mTOR phosphorylation
significantly increased in cells treated with miR-194-5p inhibitor
(P<0.01; Fig. 5C). These data
indicated that miR-194-5p may downregulate SMURF1 expression and
may block the mTOR signaling pathway.
miR-194-5p inactivates the mTOR signaling
pathway by targeting SMURF1 to suppress FaDu cell viability,
migration and invasion and to inhibit tumor growth
The regulatory mechanisms of miR-194-5p in HPC and
the involvement of SMURF1 and the mTOR signaling pathway were
examined. First, the mRNA and protein expression levels of SMURF1
were detected to determine the siRNA-mediated silencing efficiency
following SMURF1-siRNA transfection into FaDu cells. Compared with
the NC siRNA group, the mRNA and protein expression levels of
SMURF1 in the SMURF1-siRNA group were decreased by 75 and 55%,
respectively (Fig. 6A and B,
respectively), which indicated that the silencing efficiency was
good and that SMURF1-siRNA could be used for subsequent
experiments. FaDu cells were treated with miR-194-5p inhibitor and
either co-transfected with SMURF1-siRNA or treated with the mTOR
inhibitor rapamycin, and the ratio of p-mTOR to total mTOR were
evaluated. The protein expression levels of mTOR and the extent of
mTOR phosphorylation were significantly decreased in the miR-194-5p
inhibitor + SMURF1 siRNA group compared with the miR-194-5p
inhibitor + NC siRNA group (P<0.01; Fig. 6C); similar results were observed in
the miR-194-5p inhibitor + rapamycin group compared with the
miR-194-5p inhibitor + DMSO group (P<0.01; Fig. 6C). These results suggested that
SMURF1 silencing may inactivate the mTOR signaling pathway.
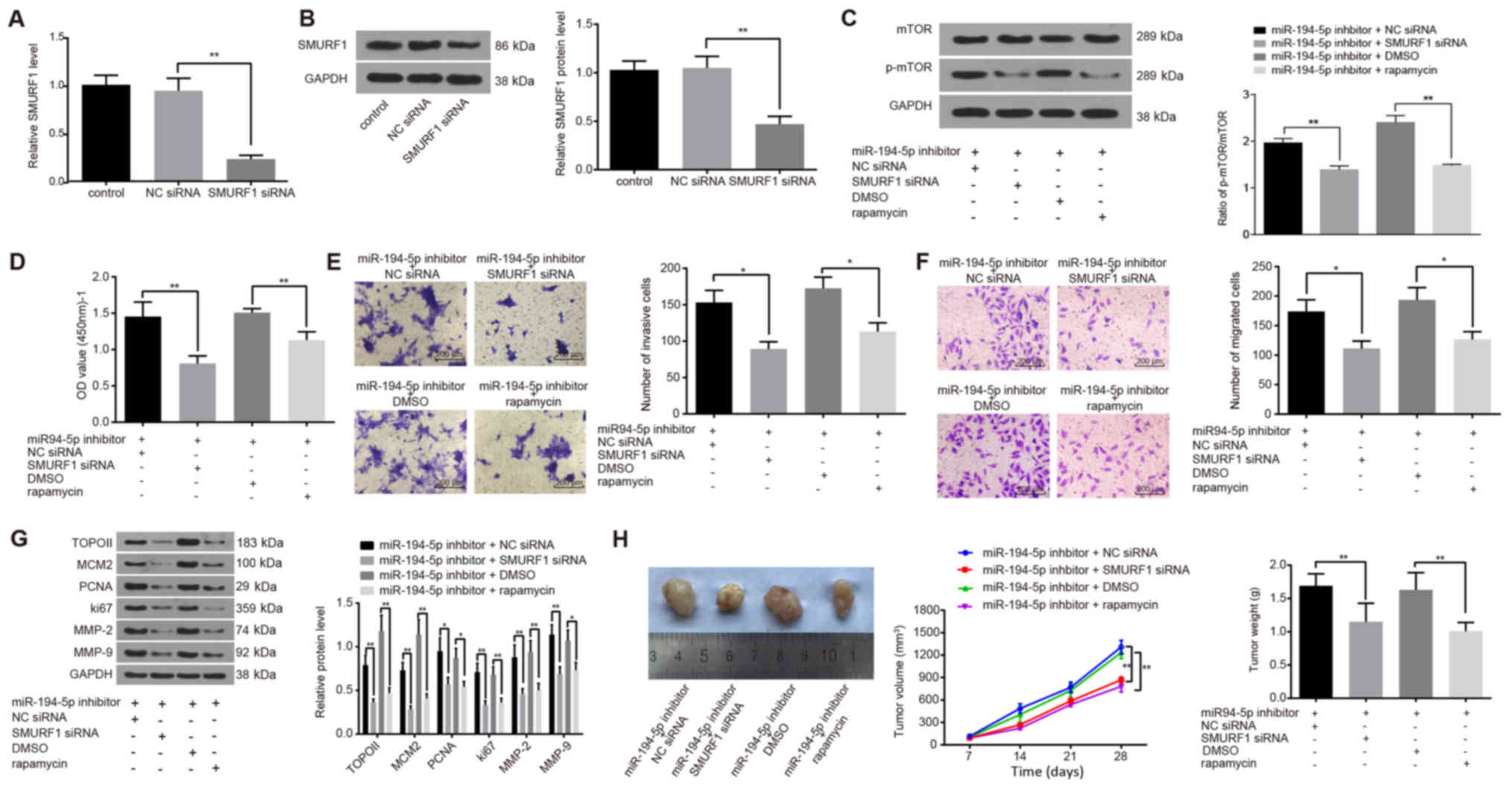 | Figure 6miR-194-5p inactivates the mTOR
signaling pathway by targeting SMURF1 to restrain the viability,
invasion and migration of FaDu cells, and to inhibit tumor growth.
(A and B) SMURF1-siRNA transfection successfully reduced SMURF1 (A)
mRNA and (B) protein expression, as detected by reverse
transcription-quantitative polymerase chain reaction and western
blot analysis, respectively. (C) Protein expression levels of mTOR
and the extent of mTOR phosphorylation in transfected FaDu cells
were detected by western blot analysis. (D) Viability of FaDu cells
at 24 h post-transfection was determined by Cell Counting Kit-8
assay. (E and F) Invasive and migratory abilities of FaDu cells at
24 h post-transfection detected by (E) Matrigel and (F) Transwell
assays. (G) Protein expression levels of proliferation-related
(TOPO II, MCM2, PCNA and Ki67) and invasion-related (MMP-2 and
MMP-9) factors in transfected FaDu cells were detected by western
blot analysis. (H) Nude mice were injected with cells treated with
miR-194-5p inhibitor and co-treated with SMURF1-siRNA or rapamycin,
and tumor volumes and weights were examined. Experiments were
repeated three times, and data are presented as the mean ± standard
deviation; *P<0.05; **P<0.01. MCM2,
minichromosome maintenance 2; miR, microRNA; MMP, matrix
metalloproteinase; mTOR, mammalian target of rapamycin; NC,
negative control; NS, no statistical significance; p,
phosphorylated; PCNA, proliferating cell nuclear antigen; siRNA,
small interfering RNA; SMURF1, Smad ubiquitin regulatory factor 1;
TOPO II, topoisomerase II. |
CCK-8, Matrigel and Transwell assays were used to
explore the effects of SMURF1 silencing and rapamycin on the
viability, invasion and migration of FaDu cells, respectively.
Compared with cells treated with miR-194-5p inhibitor alone, the
viability, invasion and migration of FaDu cells in the miR-194-5p
inhibitor + SMURF1 siRNA and in the miR-194-5p inhibitor +
rapamycin groups were significantly inhibited (all P<0.05;
Fig. 6D–F).
Western blot analysis was conducted to detect the
protein expression levels of proliferation- and invasion-related
factors (Fig. 6G). Compared with
cells treated with miR-194-5p inhibitor only, the expression levels
of the proliferation-related factors TOPO II, MCM2, PCNA and Ki67,
and of the invasion-related factors MMP-2 and MMP-9 were
significantly decreased in the miR-194-5p inhibitor + SMURF1 siRNA
group and in the miR-194-5p inhibitor + rapamycin group (P<0.05
or P<0.01; Fig. 6G). These
results suggested that SMURF1 silencing and rapamycin treatment
were able to antagonize the miR-194-5p-inhibitor-induced
upregulation of mTOR signaling pathway activation on the viability,
invasion and migration of FaDu cells and that miR-194-5p may
inactivate the mTOR signaling pathway by targeting SMURF1.
A tumorigenicity assay in nude mice was performed to
further confirm whether SMURF1 and the mTOR signaling pathway were
regulated by miR-194-5p (Fig. 6H).
Compared with the Control nude mice transplanted with miR-194-5p
inhibitor + NC siRNA-treated or miR-194-5p inhibitor + DMSO-treated
cells, tumor growth in the nude mice transplanted with miR-194-5p
inhibitor + SMURF1-siRNA-treated or miR-194-5p inhibitor +
rapamycin-treated cells was inhibited and the weight of the tumors
decreased significantly at day 28 post-transplantation (all
P<0.01). These in vivo experiments demonstrated that
SMURF1 silencing or rapamycin inhibition of the mTOR signaling
pathway inhibited tumor growth and further verified that miR-194-5p
may inactivate the mTOR signaling pathway by targeting SMURF1.
Discussion
The prognosis of patients with HPC is often
unsatisfactory owing to perioperative complications, even though
many of these patients have undergone surgical excision following
definitive chemoradiotherapy (18). Consequently, it is of vital
importance to study other viable therapeutic strategies to prevent
the increasingly prevailing HPC. The results of the present study
suggested that miR-194-5p may serve as an anti-oncogene in HPC
through its effects on the SMURF1 expression and the mTOR signaling
pathway. Consequently, the present study demonstrated that
upregulated miR-194-5p may be able to disrupt HPC cell
proliferation and metastasis by suppressing SMURF1 and inactivating
the mTOR signaling pathway.
In the present study, low expression of miR-194-5p
was observed in HPC tissues, and increasing its expression led to
the inhibition of cancer cell viability, migration and invasion
in vitro. It has been demonstrated previously that miRNAs
are aberrantly expressed in various cancers and act as ‘tumor
suppressor genes’ or ‘oncogenes’ in carcinogenesis (19). Notably, miR-194-5p has been
proposed as a tumor suppressor; for example, in glioblastoma
multiforme, miR-194-5p inhibits cell growth and promotes apoptosis,
thus indicating a suppressive role in this tumor (20,21).
Another study suggested that miR-194-5p leads to the downregulation
of Bcl-2-associated transcription factor 1, a nuclear protein that
binds to Bcl-related proteins, to improve cell differentiation
(22). miR-194 was also reported
to be expressed at a low level in liver cancer, and its
upregulation contributed to the inhibition of cell metastasis in
mice (23). All these previous
studies have indicated the role of miR-194-5p upregulation in
cancer progression. Results from the present study demonstrated
that the viability, migration and invasion of the HPC cell line
FaDu are suppressed by overexpression of miR-194-5p, which
indicated a tumor-suppressive role of miR-194-5p. In addition, it
was demonstrated that overexpression of miR-194-5p decreased the
protein expression of TOPO II, MCM2, PCNA and Ki67, as well as
MMP-2 and MMP-9, in FaDu cells. PCNA and Ki67 are commonly applied
as markers for cell proliferation assessment (24). Regarding the role of TOPO II and
MCM2 in cell division, those two factors were also considered as
markers of cell proliferation or malignant transformation (25,26).
These data suggested that miR-194-5p may provide a therapeutic
target for HPC.
Results from the present study also revealed that
SMURF1 expression was upregulated in HPC tissues. As previously
reported, the degradation of ubiquitin-dependent protein is
correlated with various biological processes; for example, SMURF1
has been found to be involved in cancer progression (27). Another study reported that the
knock down of SMURF1 resulted in a restoration of bone
morphogenetic protein signaling and enhanced the induction of
differentiation of cancer stem cell-like cells, which may reduce
drug resistance and recurrence of head and neck squamous cell
carcinoma (11). The present study
also demonstrated that FaDu cells transfected with miR-194-5p
mimics exhibited reduced proliferation and migration, and
miR-194-5p was confirmed to target SMURF1. The gene expression
profile of a cell is determined by post-transcriptional processing
of mRNA transcripts (28). In the
present study, luciferase activity detection verified that
miR-194-5p negatively regulated the expression of SMURF1. The
results also revealed that SMURF1 silencing blocked the mTOR
signaling pathway and prevented HPC progression. Another study also
reported that mTOR-inhibiting therapy provided a novel therapeutic
approach in patients diagnosed with esophageal squamous cell
carcinoma (29). High levels of
mTOR phosphorylation were reported to be associated with poor
prognosis in esophageal squamous cell carcinoma, indicating that
mTOR inhibition may function as a target for the treatment of this
disease (30). As previously
reported, miR-99a and miR-100 suppress cell proliferation by
inhibiting the mTOR signaling pathway in human esophageal squamous
cell carcinoma (31). Another
study reported that high SMURF1 expression levels may result in
enhanced activation of the PI3K/AKT/mTOR signaling pathway
(31). Partly in line with the
present study results, Tao et al demonstrated that SMURF1
knockdown significantly suppressed the migration, invasion and
tumor growth of gastric cancer, whereas it was involved with a
mechanism involving the PI3K/Akt pathway and the SMURF1/DABIP axis
(32). These results indicated
that miR-194-5p may hinder HPC progression through inhibiting the
mTOR signaling pathway by downregulating SMURF1.
In summary, the present study data provided evidence
that upregulation of miR-194-5p may effectively repress cell
proliferation and invasion in HPC, whereas the expression of SMURF1
is downregulated and the mTOR signaling pathway is inactivated
(Fig. 7). These results indicate a
potential regulatory mechanism of miR-194-5p targeting SMURF1 to
block the mTOR signaling pathway. Therefore, it is suggested that
the miR-194-5p/SMURF1/mTOR axis may serve as an attractive
therapeutic target for HPC treatment, and miR-194-5p mimic
treatment might be a better alternative to the known treatment with
rapamycin. However, the results of this study were obtained using a
sample size. Therefore, further studies on the more detailed
mechanisms as well as the effectiveness and safety of miR-194-5p in
the treatment of this fatal malignancy should be analyzed with a
larger sample size.
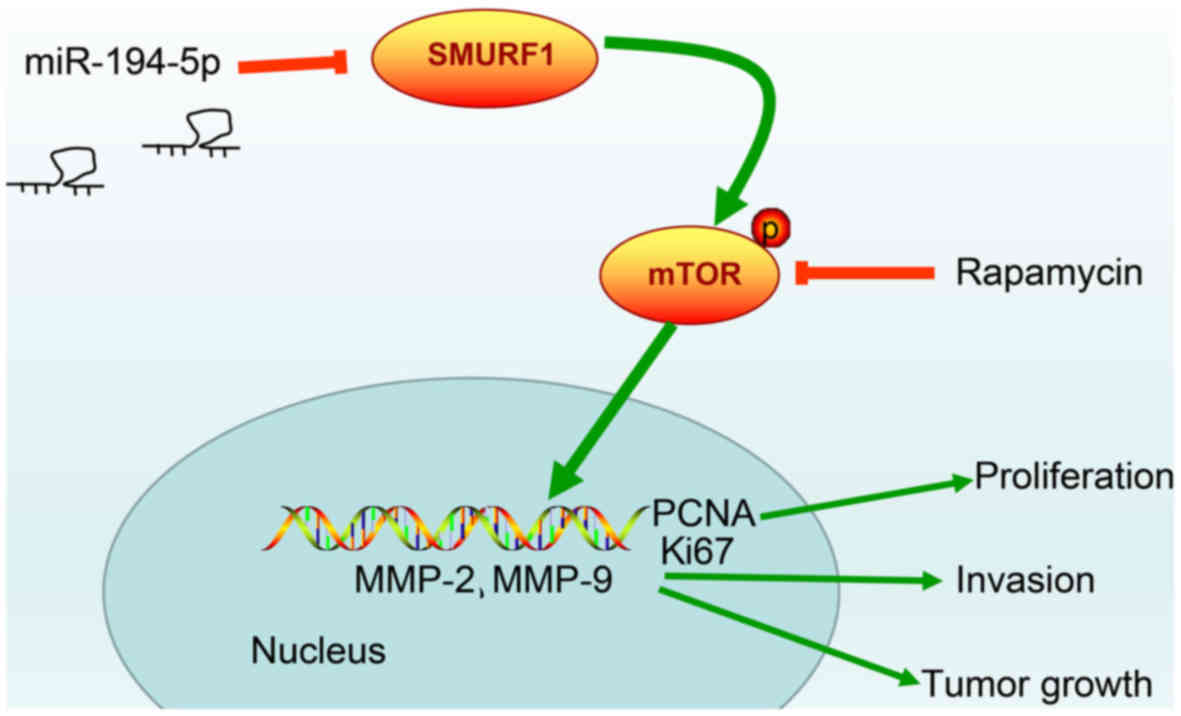 | Figure 7Molecular mechanism of miR-194-5p in
HPC involves the mTOR signaling pathway by targeting SMURF1.
miR-194-5p inactivates mTOR signaling pathway to downregulate the
proliferation-related genes, PCNA and Ki67, and to inhibit
invasion-related factors MMP-2 and MMP-9, thereby inhibiting HPC
cell proliferation, invasion and tumor growth. HPC, hypopharyngeal
carcinoma; MCM2, minichromosome maintenance 2; miR, microRNA; mTOR,
mammalian target of rapamycin; MMP, matrix metal-loproteinase;
PCNA, proliferating cell nuclear antigen; SMURF1, Smad ubiquitin
regulatory factor 1; TOPO II, topoisomerase II. |
Funding
This study was supported by The Natural Science
Foundation of Liaoning Province (grant nos. 20170541050 and
20170541026).
Availability of data and materials
The data sets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors’ contributions
SX and LH conceived and designed the experiments,
and wrote the paper. NY and YW obtained the and validated the
results. NZ and XJJ reviewed the results and discussions. SX, LH
and NY performed the statistical analysis and prepared figures. All
authors had final approval of the submitted and published
versions.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Hospital of China Medical University
(Shenyang, China), and written informed consent was obtained from
each participant prior to enrollment. The animal experiments were
conducted with approval of the Animal Ethics Committee of The First
Hospital of China Medical University, and all animal experimental
procedures conformed to the provisions of the Guide for the Care
and Use of Laboratory Animals by US National Institutes of
Health.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Zhang Y, Wang B, Chen X, Li W and Dong P:
AGO2 involves the malignant phenotypes and FAK/PI3K/AKT signaling
pathway in hypopharyngeal-derived FaDu cells. Oncotarget.
8:54735–54746. 2017.PubMed/NCBI
|
|
2
|
Wu P, Wu H, Tang Y, Luo S, Fang X, Xie C,
He J, Zhao S, Wang X, Xu J, et al: Whole-exome sequencing reveals
novel mutations and epigenetic regulation in hypopharyngeal
carcinoma. Oncotarget. 8:85326–85340. 2017.PubMed/NCBI
|
|
3
|
Chan JY and Wei WI: Current management
strategy of hypopharyngeal carcinoma. Auris Nasus Larynx. 40:2–6.
2013. View Article : Google Scholar
|
|
4
|
Zhang Y, Cong L, He J, Wang Y, Zou Y, Yang
Z, Hu Y, Zhang S and He X: Photothermal treatment with
EGFRmAb-AuNPs induces apoptosis in hypopharyngeal carcinoma cells
via PI3K/AKT/mTOR and DNA damage response pathways. Acta Biochim
Biophys Sin (Shanghai). 50:567–578. 2018. View Article : Google Scholar
|
|
5
|
Orenes-Piñero E, Montoro-García S, Patel
JV, Valdés M, Marín F and Lip GY: Role of microRNAs in cardiac
remodelling: New insights and future perspectives. Int J Cardiol.
167:1651–1659. 2013. View Article : Google Scholar
|
|
6
|
Nohata N, Hanazawa T, Kinoshita T, Okamoto
Y and Seki N: MicroRNAs function as tumor suppressors or oncogenes:
Aberrant expression of microRNAs in head and neck squamous cell
carcinoma. Auris Nasus Larynx. 40:143–149. 2013. View Article : Google Scholar
|
|
7
|
Han C, Shen JK, Hornicek FJ, Kan Q and
Duan Z: Regulation of microRNA-1 (miR-1) expression in human
cancer. Biochim Biophys Acta Gene Regul Mech. 1860.227–232.
2017.
|
|
8
|
Choi JS, Nam MH, Yoon SY and Kang SH:
MicroRNA-194-5p could serve as a diagnostic and prognostic
biomarker in myelo-dysplastic syndromes. Leuk Res. 39:763–768.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yumioka T, Osaki M, Sasaki R, Yamaguchi N,
Onuma K, Iwamoto H, Morizane S, Honda M, Takenaka A and Okada F:
Lysosome-associated membrane protein 2 (LAMP-2) expression induced
by miR-194-5p downregulation contributes to sunitinib resistance in
human renal cell carcinoma cells. Oncol Lett. 15:893–900.
2018.PubMed/NCBI
|
|
10
|
Wei R, Li B, Guo J, Li M, Zhu R, Yang X
and Gao R: Smurf1 targets Securin for ubiquitin-dependent
degradation and regulates the metaphase-to-anaphase transition.
Cell Signal. 38:60–66. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khammanivong A, Gopalakrishnan R and
Dickerson EB: SMURF1 silencing diminishes a CD44-high cancer stem
cell-like population in head and neck squamous cell carcinoma. Mol
Cancer. 13:2602014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ke M, Mo L, Li W, Zhang X, Li F and Yu H:
Ubiquitin ligase SMURF1 functions as a prognostic marker and
promotes growth and metastasis of clear cell renal cell carcinoma.
FEBS Open Bio. 7:577–586. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guertin DA and Sabatini DM: Defining the
role of mTOR in cancer. Cancer Cell. 12:9–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hou G, Xue L, Lu Z, Fan T, Tian F and Xue
Y: An activated mTOR/p70S6K signaling pathway in esophageal
squamous cell carcinoma cell lines and inhibition of the pathway by
rapamycin and siRNA against mTOR. Cancer Lett. 253:236–248. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
USDA Animal Care: Act Animal Welfare and
Regulations Animal Welfare. United States Department of
Agriculture; Washington, DC: 2003
|
|
16
|
Ito K, Liu Q, Salto-Tellez M, Yano T, Tada
K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, et al: RUNX3, a
novel tumor suppressor, is frequently inactivated in gastric cancer
by protein mislocalization. Cancer Res. 65:7743–7750. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔ C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
18
|
Kadota H, Fukushima J, Nakashima T,
Kumamoto Y, Yoshida S, Yasumatsu R, Shiratsuchi H, Morita M and
Komume S: Comparison of salvage and planned pharyngolaryngectomy
with jejunal transfer for hypopharyngeal carcinoma after
chemoradiotherapy. Laryngoscope. 120:1103–1108. 2010.PubMed/NCBI
|
|
19
|
Li Q, Qiu XM, Li QH, Wang XY, Li L, Xu M,
Dong M and Xiao YB: MicroRNA-424 may function as a tumor suppressor
in endometrial carcinoma cells by targeting E2F7. Oncol Rep.
33:2354–2360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Z, Lei B, Wu H, Zhang X and Zheng N:
Tumor suppressive role of miR-194-5p in glioblastoma multiforme.
Mol Med Rep. 16:9317–9322. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Su R, Cao S, Ma J, Liu Y, Liu X, Zheng J,
Chen J, Liu L, Cai H, Li Z, et al: Knockdown of SOX2OT inhibits the
malignant biological behaviors of glioblastoma stem cells via
up-regulating the expression of miR-194-5p and miR-122. Mol Cancer.
16:1712017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dell’Aversana C, Giorgio C, D’Amato L,
Lania G, Matarese F, Saeed S, Di Costanzo A, Belsito Petrizzi V,
Ingenito C, Martens JHA, et al: miR-194-5p/BCLAF1 deregulation in
AML tumorigenesis. Leukemia. 31:2315–2325. 2017. View Article : Google Scholar
|
|
23
|
Meng Z, Fu X, Chen X, Zeng S, Tian Y, Jove
R, Xu R and Huang W: miR-194 is a marker of hepatic epithelial
cells and suppresses metastasis of liver cancer cells in mice.
Hepatology. 52:2148–2157. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bologna-Molina R, Mosqueda-Taylor A,
Molina-Frechero N, Mori-Estevez AD and Sánchez-Acuña G: Comparison
of the value of PCNA and Ki-67 as markers of cell proliferation in
ameloblastic tumors. Med Oral Patol Oral Cir Bucal. 18:e174–e179.
2013. View Article : Google Scholar
|
|
25
|
Stromar IK and Jakic-Razumovic J: The
value of immunohistochemical determination of topoisomerase IIα and
Ki67 as markers of cell proliferation and malignant transformation
in colonic mucosa. Appl Immunohistochem Mol Morphol. 22:524–529.
2014. View Article : Google Scholar
|
|
26
|
Carreón-Burciaga RG, González-González R,
Molina-Frechero N and Bologna-Molina R: Immunoexpression of Ki-67,
MCM2, and MCM3 in Ameloblastoma and Ameloblastic Carcinoma and
Their Correlations with Clinical and Histopathological Patterns.
Dis Markers. 2015.683087:2015.
|
|
27
|
Fukunaga E, Inoue Y, Komiya S, Horiguchi
K, Goto K, Saitoh M, Miyazawa K, Koinuma D, Hanyu A and Imamura T:
Smurf2 induces ubiquitin-dependent degradation of Smurf1 to prevent
migration of breast cancer cells. J Biol Chem. 283:35660–35667.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wigington CP, Jung J, Rye EA, Belauret SL,
Philpot AM, Feng Y, Santangelo PJ and Corbett AH:
Post-transcriptional regulation of programmed cell death 4 (PDCD4)
mRNA by the RNA-binding proteins human antigen R (HuR) and T-cell
intracellular antigen 1 (TIA1). J Biol Chem. 290:3468–3487. 2015.
View Article : Google Scholar :
|
|
29
|
Boone J, Ten Kate FJ, Offerhaus GJ, van
Diest PJ, Rinkes IH and van Hillegersberg R: mTOR in squamous cell
carcinoma of the oesophagus: A potential target for molecular
therapy? J Clin Pathol. 61:909–913. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hirashima K, Baba Y, Watanabe M, Karashima
R, Sato N, Imamura Y, Hiyoshi Y, Nagai Y, Hayashi N, Iyama K, et
al: Phosphorylated mTOR expression is associated with poor
prognosis for patients with esophageal squamous cell carcinoma. Ann
Surg Oncol. 17:2486–2493. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun J, Chen Z, Tan X, Zhou F, Tan F, Gao
Y, Sun N, Xu X, Shao K and He J: MicroRNA-99a/100 promotes
apoptosis by targeting mTOR in human esophageal squamous cell
carcinoma. Med Oncol. 30:4112013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tao Y, Sun C, Zhang T and Song Y: SMURF1
promotes the proliferation, migration and invasion of gastric
cancer cells. Oncol Rep. 38:1806–1814. 2017. View Article : Google Scholar : PubMed/NCBI
|