Introduction
To date, lung cancer remains as the leading type of
cancer according to its incidence and mortality rate worldwide
(1). It comprises two main
histological subtypes, non-small cell lung cancer (NSCLC) and
small-cell lung cancer (SCLC). Approximately 85% of lung cancer
cases are NSCLC, which includes squamous cell carcinomas,
adenocarcinomas and large cell carcinomas (2). The incidence of lung adenocarcinoma
continues to increase in developed countries. It has become the
most common subtype of lung cancer for both smokers and lifelong
non-smokers. However, relapse is commonly observed in 50 to 70% of
patients within 1 year following surgical therapy, which is closely
associated with the nature and quality of the surgery (3). Patients with NSCLC carrying mutated
epidermal growth factor receptor (EGFR) exhibit a good response to
tyrosine kinase inhibitor (TKI) (4). However, resistance to TKIs acquired
by cancer cells often limits the effects and corresponding benefits
(5). Therefore, intensive research
is required to shed light on the molecular and cellular machinery
which are ‘hijacked’ or acquired by the lung cancer cells during
tumor development and disease progression.
Ehm2, also known as erythrocyte membrane protein
band 4.1-like protein 4B (EPB41L4B), belongs to the Four.1 protein,
ezrin, radixin, moesin (FERM) superfamily. The overexpression of
Ehm2 has been observed in metastatic cancer cells (6). FERM proteins contain a highly
conserved FERM domain which mediates protein-protein interactions
through an interaction with the cytoplasmic tails of transmembrane
proteins (7,8). FERM proteins are
cytoskeletal-associated proteins mediating interactions between
transmembrane proteins and the cytoskeletal proteins. Upon an
interaction with Crb3 through its FERM-binding motif, Ehm2 can be
recruited to the plasma membrane, leading to an activation of
p114RhoGEF and the consequential control of the morphology and
cohesion of cancer cells (9).
Lulu2, a murine Ehm2 protein, has been shown to activate cortical
myosin II to alter contractile force in epithelial cells (10). In human fibrosarcoma cells, Ehm2
has been reported to be involved in steroid-regulated cytoskeletal
reorganization (11).
As a metastasis-associated protein, Ehm2 is
frequently dysregulated in human solid cancers (6,12).
The upregulation of Ehm2 has been observed in both tumor tissues
and cell lines of prostate cancer (13). The overexpression of Ehm2 can
promote prostate cancer progression and metastasis (13). The expression of Ehm2 is also
elevated in breast cancers, which is associated with metastasis and
a poor prognosis of patients with the disease (14). The knockdown of Ehm2 in MCF-7 cells
has been shown to not only increase apoptosis, but also to reduce
invasiveness through a regulation of matrix metalloproteinase
(MMP)9 (14).
Alternative splicing is a regulated process in gene
expression, which makes a single gene to encode different protein
products. It naturally occurs in approximately 95% of multi-exonic
genes in eukaryotes (15,16), and is also involved in various
disorders including cancers (17-21).
Through alternative splicing, the Ehm2 gene produces two
transcript variants, encoding two different isoforms, Ehm2/1 and
Ehm2/2. The isoform 1 has 382 amino acids missing in its C-terminal
region in comparison with Ehm2/2 (11). Although the involvement of Ehm2 in
disease progression is evident in both prostate and breast cancers,
as discussed above, the exact role and function played by the two
splice variants have yet to be elucidated.
In the present study, we examined the expression of
the two Ehm2 transcript variants in lung adenocarcinoma tissues.
The two variants exhibited distinct patterns of expression in lung
cancers. In vitro cell function assays revealed that Ehm2/1
exerted inhibitory effects, and that Ehm2/2 promoted both the
migration and invasion of A549 cells. We also examined the
association of the Ehm2 variants with epithelial-mesenchymal
transition (EMT) in lung cancer cells.
Materials and methods
Cells and cell culture
The A549 cell line was purchased from the American
Type Culture Collection (ATCC, Manassas, VA, USA). The cells were
cultured using Dulbecco’s modified Eagle’s medium (DMEM)-F12
(Thermo Fisher Scientific, Waltham, MA, USA) which was routinely
supplemented with 10% fetal bovine serum (Thermo Fisher Scientific)
and penicillin/streptomycin (KGY0023; KeyGEN BioTECH, Nanjing,
China) in an incubator at 37°C with 5% CO2 and 95%
humidity.
Human lung cancer specimens
Fresh-frozen NSCLC tissues (n=15) and matched
adjacent normal lung tissues from the same patients were collected
immediately following surgical resection at Xuanwu Hospital of
Capital Medical University (Beijing, China) after obtaining written
informed consent. These tissues were stored at −80°C until use.
Clinicopathological information was collected and recorded in a
database which includes sex, age, TNM stage, histological tumor
type and lymph node metastasis. The clinicopathological
characteristics of these tumor cases are presented in Table SI. All protocols were reviewed and
approved by the Ethics Committee of Xuanwu Hospital.
Immunohistochemistry using tissue
microarrays
Tumor tissue microarrays (TMA: HLug-Ade060PG-01)
were purchased from Shanghai Outdo Biotechnology Co., Ltd. Each
tissue microarray contained paraffin-embedded sections from 30 lung
adenocarcinoma tissues, as detailed in Table I, together with the paired normal
tissues. Pathological diagnosis for these lung adenocarcinomas was
available in the manufacturer’s instructions. Immunohistochemical
staining of tissue microarrays was performed as follows: After
being dewaxed for 2 h at 60°C and rehydrated by a gradient method,
antigen retrieval was performed using a citrate buffer (10 mM
citric acid, pH 6.0) followed by a blocking in a blocking buffer
containing 5% bovine serum albumin. Primary antibodies against
human Ehm2/1 (1:200; AP338037; OriGene Technologies, Inc.,
Rockville, MD, USA) and human Ehm2/2 (1:50; ab135616; Abcam,
Cambridge, UK) were used for probing (incubation for 20 min at room
temperature). A negative control was included without the addition
of any primary antibodies. The sections were subsequently incubated
with the relevant secondary antibody (ZSGB Biotechnology, Beijing,
China) for 20 min at room temperature. Microarrays were visualized
using diaminobenzidine (Cell Signal Technology, Inc., Danvers, MA,
USA) with a counterstaining of nuclei using hematoxylin. All these
processes were automatically carried out in a fully automated
immunohistochemistry and in situ hybridization system (Leica
BOND-MAX; Leica Biosystems, Richmond, IL, USA).
 | Table IClinicopathological characteristics
of the 30 tumor cases. |
Table I
Clinicopathological characteristics
of the 30 tumor cases.
| Parameters | Category | Cases, n=30 | Percentage (%) |
|---|
| Tissue paired
samples | Tumor | 30 | |
| Normal | 30 | |
| Lymph node
status | N0 | 4 | 13 |
| N>1 | 26 | 86 |
| Pathological
grades | I | 5 | 17 |
| II | 19 | 63 |
| III | 6 | 20 |
| TNM staging | TNM I | 3 | 10 |
| TNM II | 14 | 47 |
| TNM III | 13 | 43 |
| TNM IV | 0 | 0 |
| Age, years | ≤40 | 3 | 10 |
| 41-56 | 6 | 20 |
| ≥56 | 21 | 70 |
| Mean ± SD | 57.11±10.09 | | |
| Sex | Male | 11 | 37 |
| Female | 19 | 63 |
| Pathological
types | Adenocarcinoma | 30 | 100 |
| Squamous cell
carcinoma | 0 | 0 |
Semi-quantitative analysis of the
intensity of Ehm2/1 and Ehm2/2 immunochemical staining in tissue
microarrays
All immunochemical staining images in the tissue
microarrays were acquired using a microscope which equipped with a
digital camera (BX43; Olympus, Tokyo, Japan). The staining was
assessed by two independent investigators who were blinded to the
clinical information. The integrated optical density (IOD) of
Ehm2/1 and Ehm2/2 staining in the tissue microarrays was determined
using ImagePro® Plus software version 5.1 (Media
Cybernetics, Inc., Rockville, MD, USA). The average IOD/Ehm2
positively stained area (µm2) was calculated for
the positively stained tissues.
Cell transfection
Expression plasmids of Ehm2/1 (RC223085) and Ehm2/2
(RC212424) tagged with FLAG were purchased from OriGene
Technologies, Inc.. The shRNA targeting Ehm2/1 was purchased from
Genechem Co. (Shanghai, China). Plasmid transfections were
performed using jetPRIME® transfection reagent (Polyplus
Transfection, Illkirch, France) according to the manufacturer’s
instructions. The cells were harvested for an analysis of gene
expression approximately 48 h following transfection. A549
wild-type (A549 cells not transfected with any plasmid) and empty
vector transfected (pCMV-Entry) cells were used as controls.
Western blot analysis
The cells were lysed with a RIPA lysis buffer.
Protein extracts were prepared from frozen tissues as follows.
Tissues were homogenized using a protein isolation buffer [10 mM
Tris-HCl (pH 7.0), 160 mM NaCl, 1% Triton X-100, 1 mM EGTA, 1 mM
EDTA, and Roche complete protease inhibitors]. The samples were
incubated for 15 min on ice followed by a centrifugation at 16,000
× g for 30 min. The supernatants were stored at −80°C until use. A
BCA protein assay kit (Thermo Fisher Scientific) was used to
determine the protein concentration. A total of 25 µg of
protein samples were separated by sodiumdodecyl
sulfate-polyacrylamide gel electrophoresis followed by an electric
blotting onto polyvinylidenedifluoride membranes (Merck Millipore,
Billerica, MA, USA). Protein was probed with primary antibodies
against Ehm2/1 (1:1,000; AP33087; OriGene Technologies, Inc.),
Ehm2/2 (1:500; ab135616; Abcam), FLAG (1:1,000; F3165; Sigma, St.
Louis, MO, USA), E-cadherin (1:1,000; ab40772; Abcam), N-cadherin
(1:1,000; ab19348; Abcam), Snail1 (1:2,000; ab53519; Abcam), GAPDH
(1:3,000; sc-32233; Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA), at 4°C overnight. The horseradish peroxidase-conjugated goat
anti-mouse antibody (1:5,000; 115-035-003; Jackson ImmunoResearch
Laboratories, Inc., West Grove, PA, USA) and goat anti-rabbit
antibody (1:5,000; 111-035-03; Jackson ImmunoResearch Laboratories,
Inc.) were incubated with the membranes for 1 h at room
temperature. Protein bands were documented using a
chemiluminescence (ECL) detection system (FusionFx; VilberLourmat,
Collégien, France). Protein expression was determined by
densitometric analysis using Image J software (version 1.62;
National Institutes of Health, Bethesda, MD, USA). Quantitative
densitometric values of all proteins were normalized to GAPDH.
In vitro cell viability assay
The cells were seeded into 96-well plates at a
density of 3×103 cells per well. Cell viability was then
evaluated using a Cell Counting Kit-8 (CCK8; Dojindo, Kumamoto,
Japan). Absorbance was determined at a wavelength of 450 nm using a
spectrophotometer (BioTek, Winooski, VT, USA).
Wound healing assay
The cells were plated into 12-well plates at a
density of 6×105 cells per well and allowed to form a
monolayer. The cells were wounded with a fine gauge needle to
create a wound. The migration of the cells was recorded using an
inverted microscope (CKX41; Olympus Corp., Tokyo, Japan). Images
were captured using a camera (EOS600D; Canon, Tokyo, Japan) at the
indicated time point. The migration distances were determined and
analyzed using Image J software (version 1.62).
In vitro invasion assay
Matrigel (BD Biosciences, San Jose, CA, USA) was
used to pre-coat Transwell cell culture inserts (8 µm pore
size, 28617043; Costar, New York, NY, USA) and air dried. Following
rehydration, 20,000 cells were seeded into each insert. Following 2
days of incubation, the invaded cells were fixed in 4% formalin,
followed by staining with 0.5% crystal violet (0528; Amresco Inc.,
Solon, OH, USA) for 15 min at room temperature. The absorbance was
measured at 490 nm using a spectrophotometer (BioTek).
Gelatin zymography assay
The activities of MMPs were assayed as previously
described (14). The cells were
seeded into tissue culture flasks at a density of 1×106
cells. Once the cells reached 80% confluency, they were washed with
phosphate-buffered saline (PBS) and further cultured in serum-free
medium. After 6 h, conditioned medium was collected and
concentrated through an Amicon Ultra-0.5 ml centrifugal filter
(Merck Millipore, East Midlands, UK). Protein samples were prepared
using a non-reducing sample buffer. Protein samples were separated
via 10% SDS-PAGE containing 0.1% gelatin. Following
electrophoresis, the gels were washed and incubated overnight at
37°C. Following incubation, the gels were stained with Coomassie
blue and subsequently scanned on digital scanner images (HP Scanjet
G3110; HP Development Co., Beijing, China).
Immunofluorescence staining of cells
The cells were seeded on coverslips. Following
fixation with 4% paraformaldehyde, the cells were washed 3 times
with ice-cold phosphate-buffered saline (PBS), and then
permeabilized for 30 min. The cells were then incubated with
anti-FLAG (1:100; F3165; Sigma) antibody at room temperature for 1
h followed by further 1 h of incubation at room temperature with
Alexa Fluor® 488-conjugated goat anti-mouse secondary
antibody (1:200; A11029; Thermo Fisher Scientific). The slides were
mounted using ProLong® Gold Antifade Mountant with DAPI
(P36931; Thermo Fisher Scientific). A Leica SP8 confocal microscope
(Leica Microsystems, Wetzlar, Germany) was used to photograph the
slides.
RNA preparation and reverse
transcription-polymerase chain reaction (RT-PCR)
RNA extraction and RT-PCR were performed as
previously described (22). Total
RNA was isolated from frozen lung cancer tissues and lung
adenocarcinoma A549 cells using TRIzol® reagent
(15596018; Thermo Fisher Scientific). cDNA was generated from 1
µg RNA sample using an iScript™ cDNA Synthesis kit (1708891;
Bio-Rad Laboratories, Inc., Hercules, CA, USA). The reaction
conditions for reverse transcription were as follows: 25°C for 5
min, 42°C for 30 min, and 85°C for 5 min. PCR was carried out using
a REDTaq™ ReadyMix PCR reaction mix (R2523; Sigma) according to the
manufacturer’s instructions. The reaction conditions for PCR were
as follows: 94°C for 5 min for initial denaturation, followed by 30
cycles of 94°C for 10 sec, 56°C for 20 sec and 72°C for 30 sec. The
PCR primers were as follows: Ehm2/1 sense,
5′-CACTTTGAGAGACTGAAGCA-3′ and antisense,
5′-CAACTTCTACGACAGGAATATATGC-3′; Ehm2/2 sense,
5′-CCTGTTGCGGATCATGTGAAGTG-3′ and antisense,
5′-TATCAGGAAACGGGTTCATTGTATC-3′; and GAPDH sense,
5′-GGGAAGGTGAAGGTCGGAGT-3′ and antisense,
5′-TTGAGGTCAATGAAGGGGT-3′. The products were visualized on a 1.2%
agarose gel stained with GoldView (G8140; Solarbio, Beijing,
China).
Quantitative polymerase chain reaction
(qPCR)
qPCR was performed utilizing the ABI 7500 Fast
Real-time PCR system (Applied Biosystems, Foster City, CA, USA)
through the measurement of real-time SYBR-Green fluorescence
(1708880; Bio-Rad Laboratories, Inc.), and the results were
obtained by means of the comparative Cq method (2−ΔΔCq)
using GAPDH as an internal control (23). This experiment was performed in
triplicate. The following condition was used in the reaction: 94°C
for 5 min for initial denaturation, followed by 40 cycles of 94°C
for 10 sec, 56°C for 20 sec and 72°C for 30 sec. The PCR primers
were as follows: Ehm2/1 sense, 5′-TGAAGGTCGCATTGGAAAG-3′ and
antisense, 5′-CAT CATACTTTGCATCCCTCC-3′; Ehm2/2 sense, 5′-GTTGCG
GATCATGTGAAGTG-3′ and antisense, 5′-TGATTCCTC CTTTCCACCCT-3′; GAPDH
sense, 5′-CTGAGTACGTCG GGAGTC-3′ and antisense,
5′-GAGATGATGACCCTTTTG-3′.
Lung cancer survival analysis
The overall survival (OS), progression-free survival
(PFS) and post-progression survival (PPS) of the patients with lung
cancer was carried out using the web tool Kaplan-Meier Plotter
(http://kmplot.com/analysis/). The
Ehm2/1and Ehm2/2 probe sets were 220161_s_at and 233098_s_at,
respectively.
Comparision of sequences of Ehm2/1 and
Ehm2/2 in Ensemble and UCSC
Ensemble (http://asia.ensembl.org) is a genome browser for
vertebrate genomes that supports research in comparative genomics,
evolution, sequence variation, and so on. We queried Ehm2
(EPB41L4B) in human and found two transcripts for protein
coding. UCSC Genome Browser (http://genome.ucsc.edu) can also provide the
information of the Ehm2 transcript variants through the nearest
mirror-genome-asia.ucsc.edu
(http://genome-asia.ucsc.edu).
Statistical analysis
The SPSS software package version 19.0 (SPSS, Inc.,
Chicago, IL, USA) was used for the statistical analyses.
Statistical analysis was performed using ANOVA and a Bonferroni
post hoc test for multiple group comparisons. P<0.05 was
considered to indicate a statistically significant difference.
Results
Differential expression of Ehm2/1
andEhm2/2 in human lung adenocarcinoma specimens
To analyze the protein levels of Ehm2 variants in
lung cancer, the immunohistochemical staining of Ehm2/1 and Ehm2/2
was performed on tissue microarrays which comprised 30 human lung
adenocarcinoma tissues paired with non-cancerous tissues. Two
tissue sections were invalid due to mounting issues and were
excluded from the analyses. All information regarding the
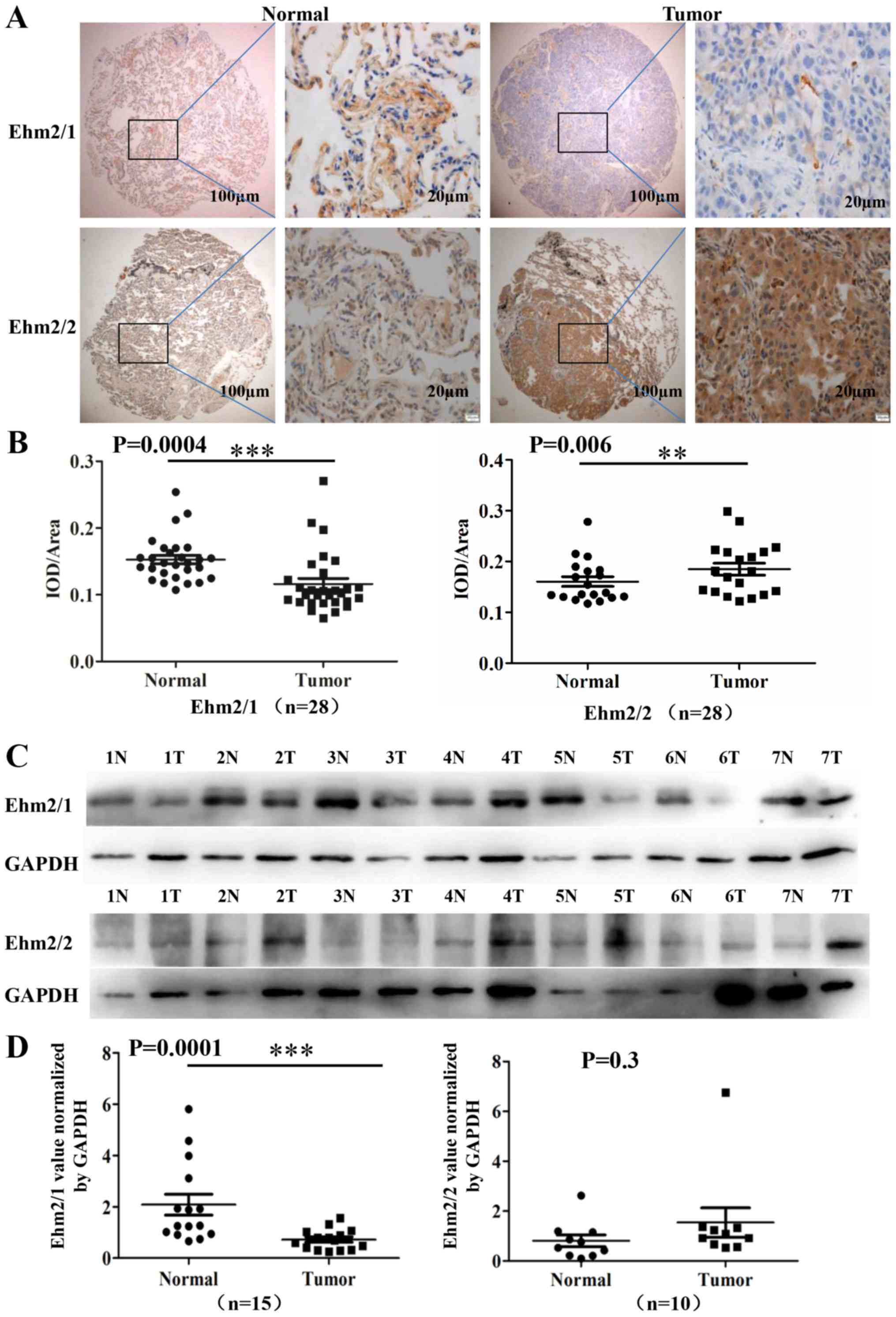
microarray tumor specimens is presented in Table I. The staining for Ehm2/1 was
significantly decreased in the lung adenocarcinoma tissues compared
with the paired normal tissues (P=0.0004, n=28; Fig. 1A and B), while the increased
expression of Ehm2/2 was observed in comparison to the paired
normal tissues (P=0.0060, n=28; Fig.
1A and B). We also determined the protein expression of these
two Ehm2 variants in lung cancer tissues by western blot analysis.
The normal lung tissues predominantly expressed Ehm2/1 (Fig. 1C). Similarly, Ehm2/1 expression was
markedly decreased in lung cancer tissues in comparison with the
controls (P<0.01, n=15; Fig.
1D). Ehm2/2 was found to be upregulated in the lung
adenocarcinoma tissues compared with the adjacent normal tissues,
but this did not reach a level of significance (P=0.3, n=10;
Fig. 1D). Our attempts to
quantitatively analyze the mRNA expression levels of Ehm2/1 and
Ehm2/2 in human samples using quantitative PCR were not successful
owing largely to the fact that an insufficient quantity of mRNA
isolated from these tissues was not adequate for the assay. Thus,
it is clear from the above-mentioned immunohistochemistry results
that the expression of Ehm2/2 was significantly increased in the
lung adenocarcinomas (P=0.006). Taken together, from the results of
immunohistochemistry and western blot analysis, it is clear that
the expression of the two Ehm2 transcript variants differs in human
lung adenocarcinoma.
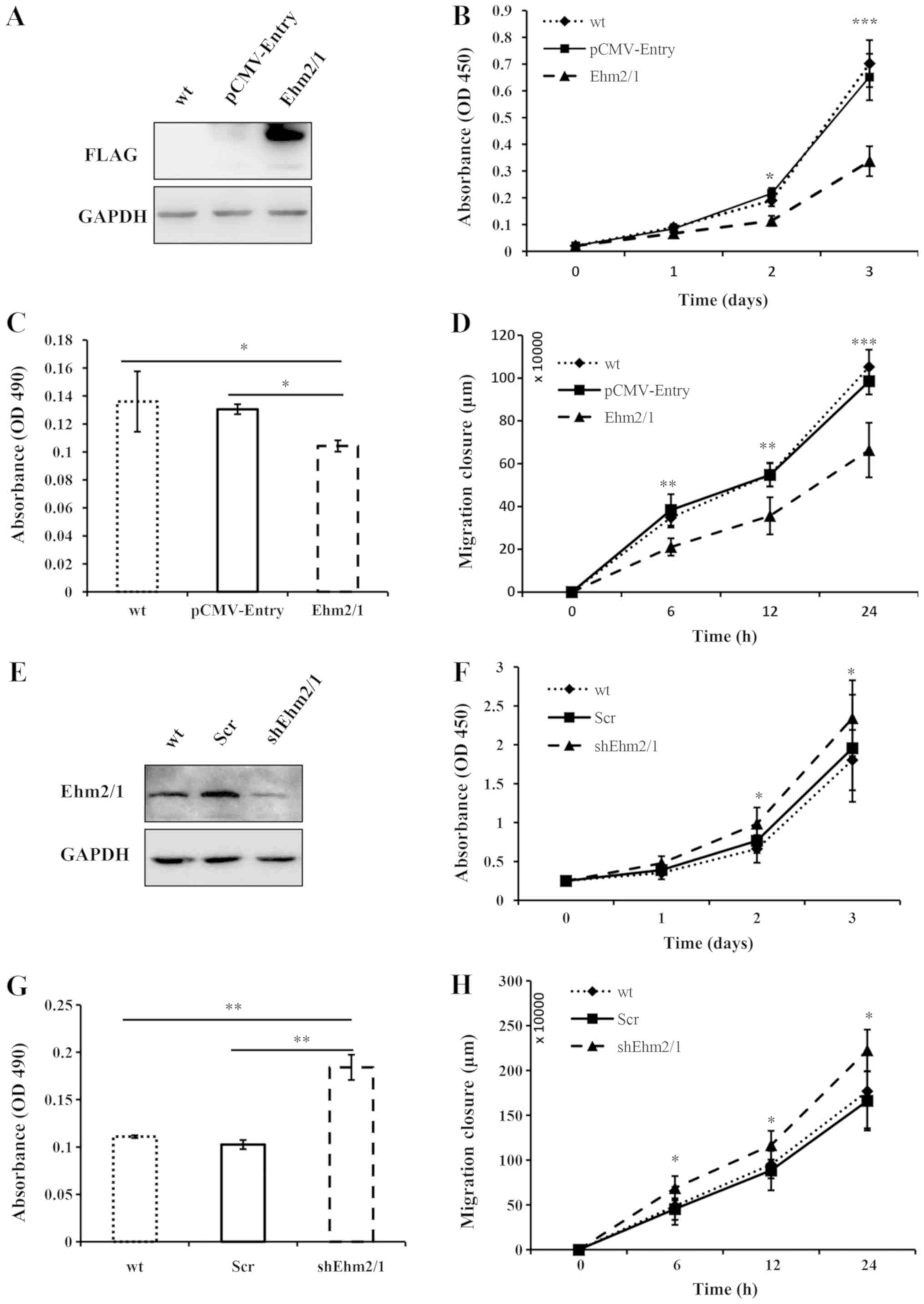
Ehm2/1 regulates the proliferation,
migration and invasion of A549 cells in vitro
To examine the function of Ehm2/1 in lung
adenocarcinoma, the human lung adenocarcinoma cell line, A549, was
used to create an overexpression cell model. The endogenous
expression of Ehm2 in the A549 cells is shown in Fig. S1. Western blot analysis was
performed to verify the expression of Ehm2/1 in the transfected
cells (Fig. 2A). Compared with the
A549 wild-type (A549 cells not transfected with any plasmid) and
empty vector-transfected (pCMV-Entry) control cells, the
overexpression of Ehm2/1 exerted an inhibitory effect on cell
proliferation in vitro (P<0.001, when compared with
pCMV-Entry control; Fig. 2B).
Ehm2/1 overexpression markedly reduced the malignant phenotype of
A549 cells, including invasiveness and migration (P<0.05 and
P<0.01, respectively, when compared with pCMV-Entry; Fig. 2C and D). A knockdown model of
Ehm2/1 in the A549 cells was also employed to verify the findings.
A decreased expression of Ehm2/1 was observed in the cells in which
Ehm2/1 was knocked down (shEhm2/1) compared with the A549 wild-type
and empty vector-transfected cells (Scr) (Fig. 2E). The knockdown of Ehm2/1
expression resulted in a significant increase in cell growth in
vitro (P<0.05, when compared with empty vector-transfected
cells; Fig. 2F), invasion
(P<0.01, when compared with empty vector-transfected cells;
Fig. 2G) and cell migration of the
A549 cells (P<0.05, when compared with empty vector-transfected
cells; Fig. 2H).
The invasive potential of the cancer cells is linked
to their ability to degrade the extracellular matrix, which to a
large degree is catalyzed by MMPs. Thus, in this study, we further
determined the activities of MMPs from the cells with the
differential expression of Ehm2 variants, using gelatin zymography
assay. There was a marked increase in the activities of MMPs in the
A549 cells in which Ehm2/1 was knocked down (Fig. S2). These findings indicate that
Ehm2/1 is a negative regulator of A549 cell viability, invasion and
migration.
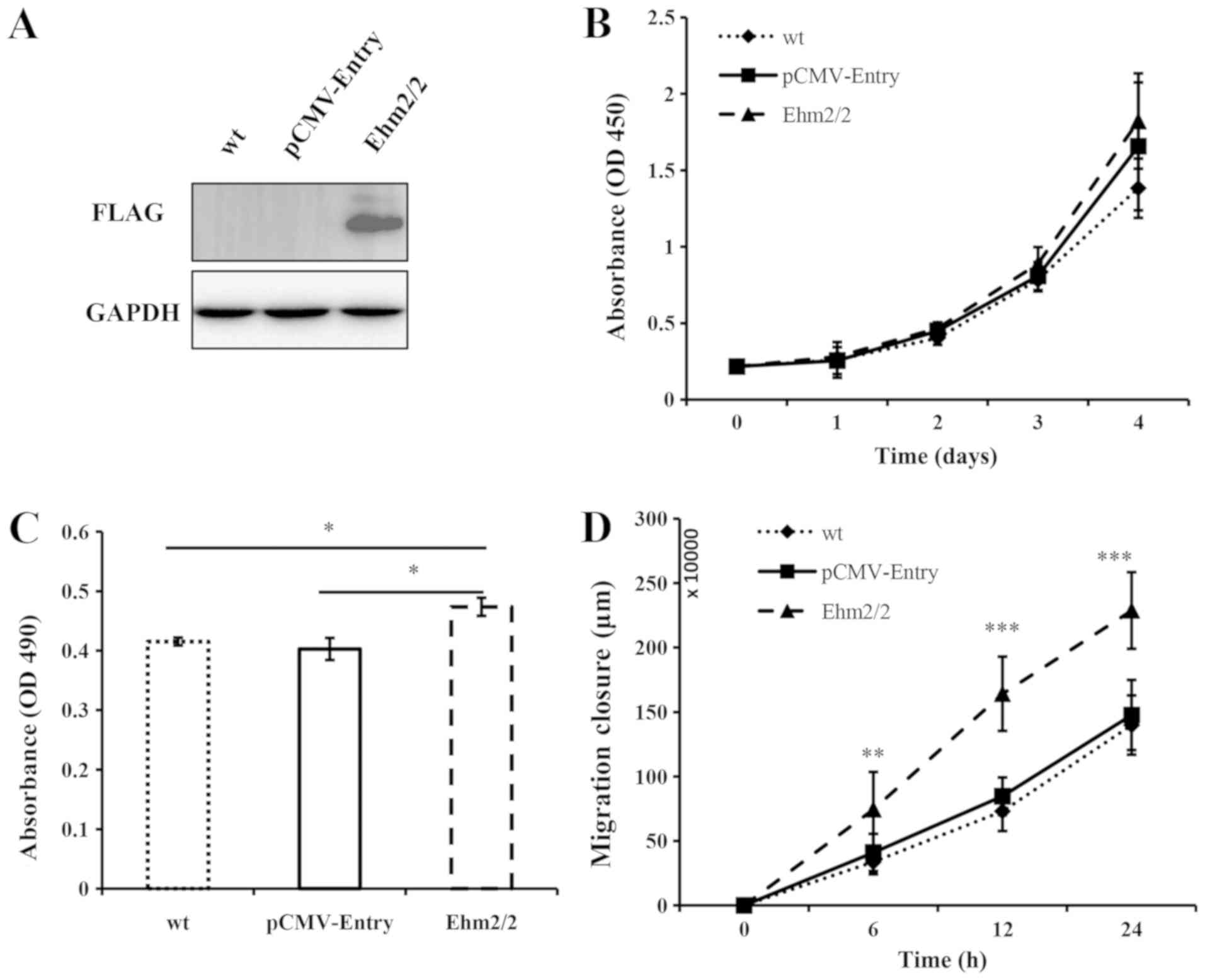
Ehm2/2 expression promotes A549 cell
invasion and migration
An Ehm2/2 overexpression cell line model was also
generated to examine the regulatory role of this molecule in lung
cancer cells. The expression level of Ehm2/2 was found to be
considerably elevated in the Ehm2/2-overexpressing cells (Fig. 3A). The overexpression of Ehm2/2 was
found to promote the proliferation of the A549 cells in comparison
with the wild-type cells and empty vector-transfected control
cells, although this difference was not statistically significant
(Fig. 3B). In comparison to the
control groups, the overexpression of Ehm2/2 markedly increased the
invasion of the A549 cells (P<0.05, when compared with
pCMV-Entry control; Fig. 3C). The
ectopic expression of Ehm2/2 also markedly promoted cell migration
(P<0.01, when compared with pCMV-Entry control; Fig. 3D). In addition, gelatin zymography
assay also revealed an increase in the activities of MMPs in the
Ehm2/2-overexpressing cells (Fig.
S2). Taken together, these data suggest that Ehm2/2 is a
promoter of A549 cell invasion and migration.
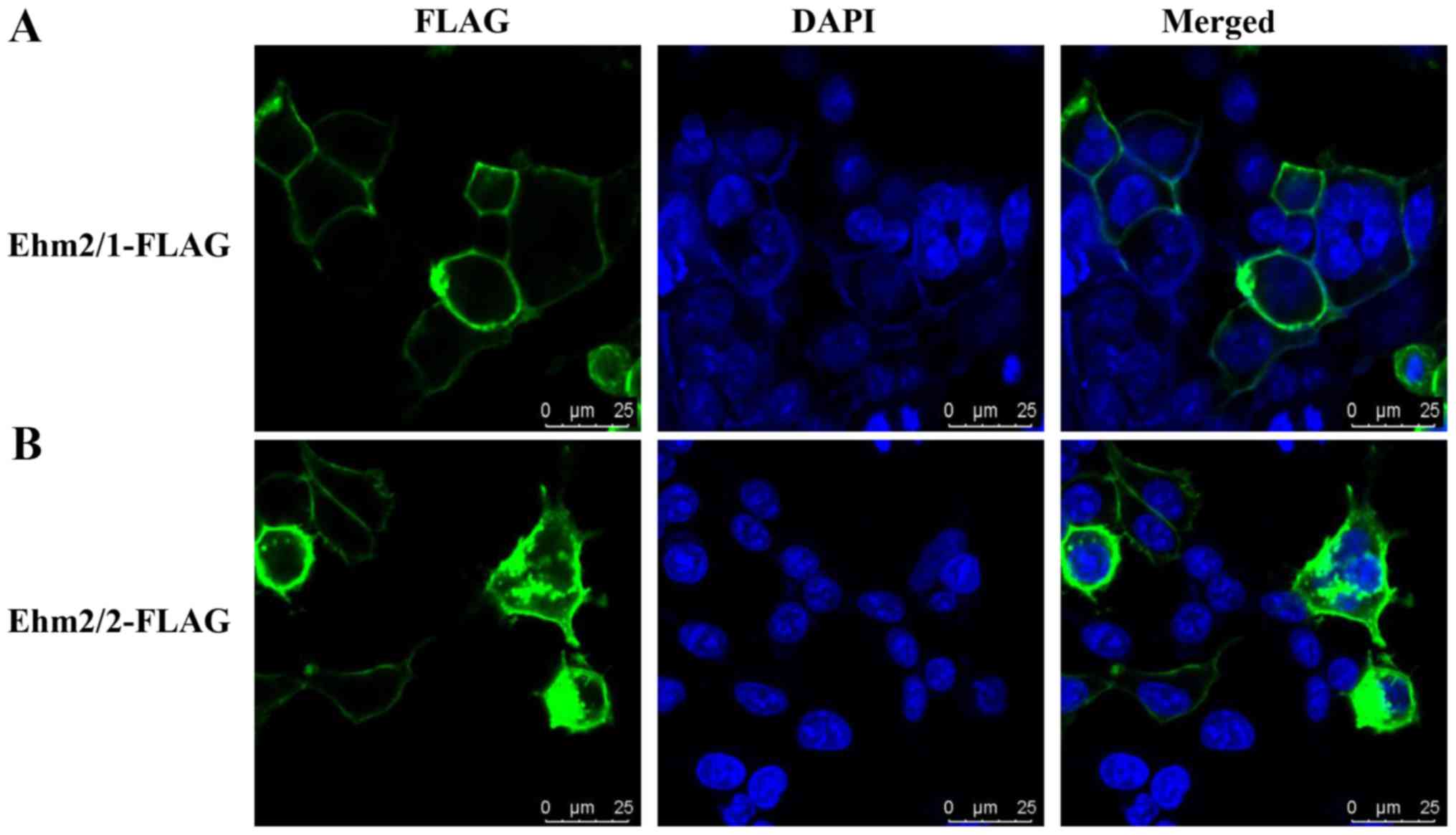
Subcellular distribution of Ehm2/1 and
Ehm2/2 differs in A549 cells
From the Ensemble and UCSC websites, we identified
that Ehm2/1 variant lacks several exons and has a different
3′-terminal exon (data not shown). This results in Ehm2/2 with a
long C-terminus and makes it longer than Ehm2/1. FERM proteins have
been reported to have additional protein functions in the
C-terminal region, including an autoregulatory function that
inhibits FERM domain activity (8,24).
Although thus far no functional domain has been found based on
protein sequence alignment in the C-terminal region of Ehm2/2, at
least to the best of our knowledge, we hypothesized that this
C-terminal region may be important for its function and subcellular
distribution through interacting with its FERM domain or with other
proteins. Combined with differential expression patterns and
differential effects on A549 cell function, we hypothesized that
Ehm2 variants have distinct subcellular distributions. To examine
this possibility, we performed immunofluorescence staining of the
A549 cells following the ectopic overexpression of Ehm2/1 and
Ehm2/2. As shown in Fig. 4, Ehm2/1
was mainly found in the cell membrane (Fig. 4A), while Ehm2/2 was more widely
distributed from the cell membrane, the cytoplasm and was even
found in the nuclei (Fig. 4B).
These results suggest that Ehm2 variants have different
distributions.
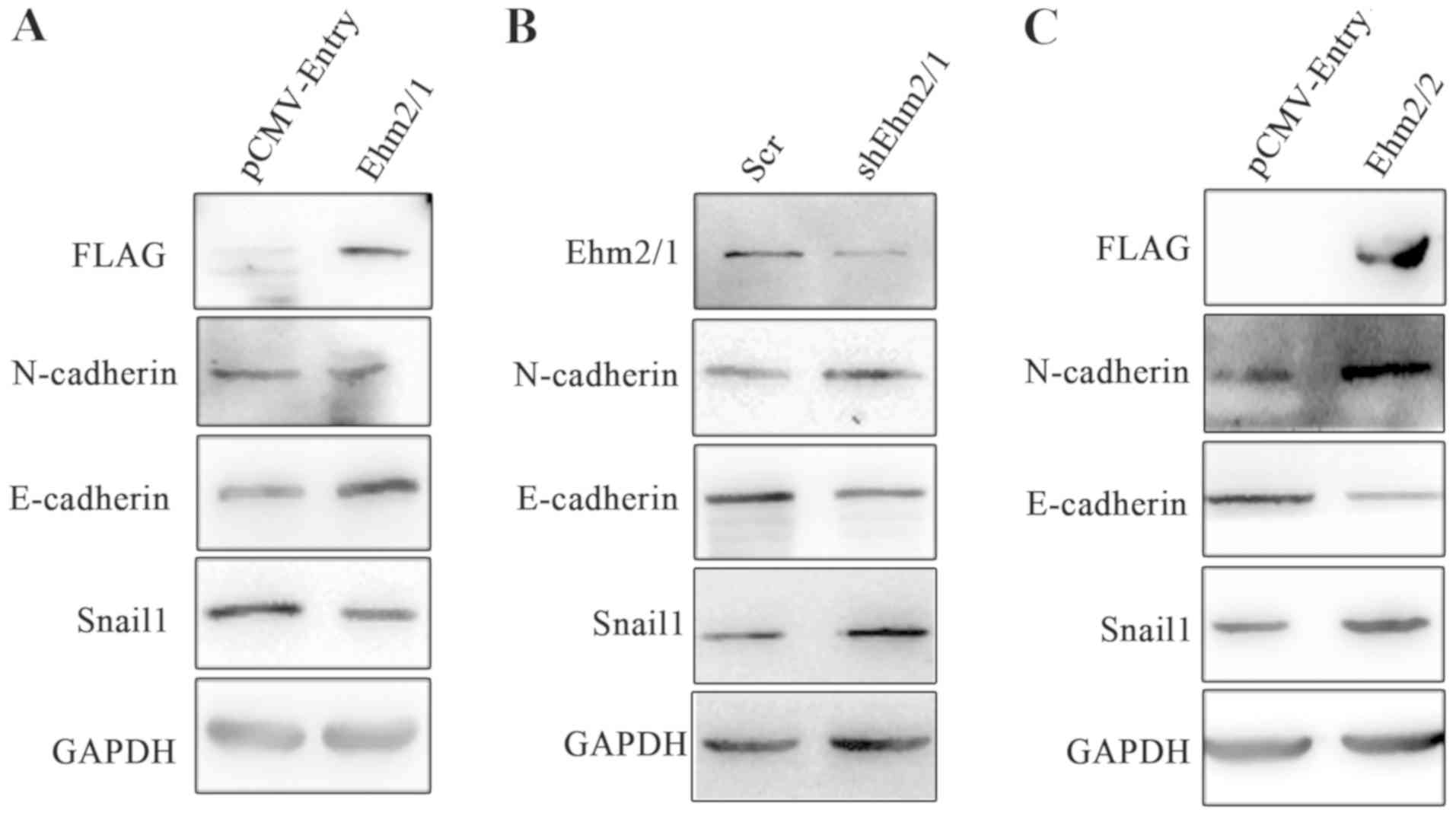
Ehm2/1 and Ehm2/2 influence EMT-marker
expression in an opposing manner
It is known that the process of EMT enables cell
migration and invasion. In consideration that Ehm2 variants
regulate the invasion and migration of A549 cells, in this study,
we detected the effects of Ehm2/1 and Ehm2/2 on EMT marker
expression. The ectopic expression of Ehm2/1 resulted in the
decreased expression of N-cadherin and Snail1, whereas the
expression of E-cadherin was elevated (Fig. 5A). In line with this result, the
knockdown of Ehm2/1 resulted in the downregulation of E-cadherin
expression and in the upregulated expression of both N-cadherin and
Snail1 (Fig. 5B). However, the
overexpression of Ehm2/2 resulted in the increased expression of
N-cadherin and Snail1, and in the decreased expression of
E-cadherin (Fig. 5C). These
results indicate that Ehm2/1 inhibits and Ehm2/2 promotes EMT in
lung cancer cells.
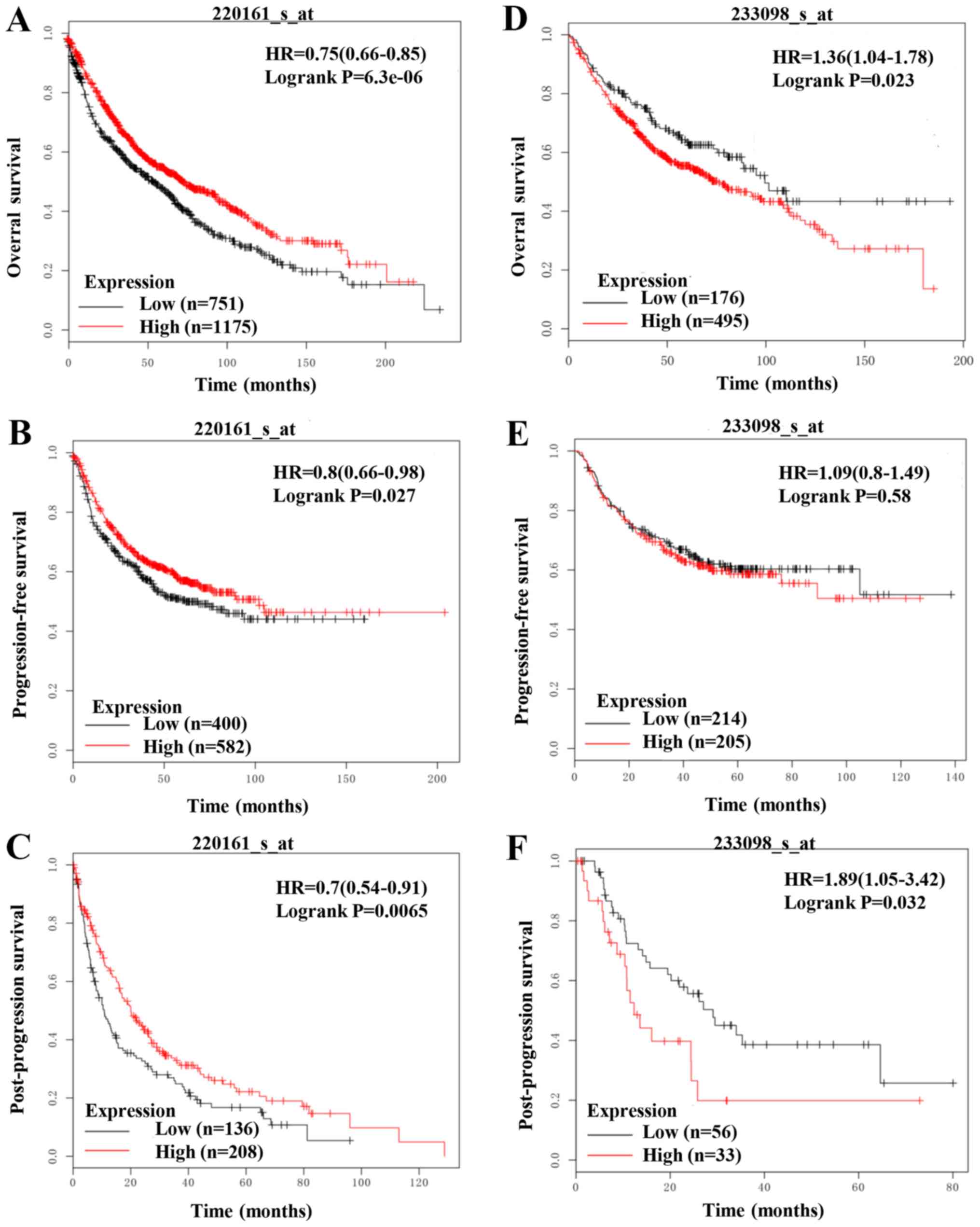
Ehm2/1 transcripts are positively
associated with and Ehm2/2 transcripts are negatively associated
with the longer survival of patients with lung cancer
In order to further elucidate the clinical
implication of Ehm2 variants in lung cancer, we analyzed the
association of Ehm2/1 (Affy ID 220161_s_at) and Ehm2/2 (Affy ID
233098_s_at) with patient survival using Kaplan-Meier survival
analysis (http://kmplot.com/analysis/). Lower
expression levels of Ehm2/1 were associated with a shorter OS
(P=6.3e−06; Fig. 6A) and PFS
(P=0.027; Fig. 6B). A lower
expression of Ehm2/1 was also associated with a poorer PPS
(P=0.0065; Fig. 6C). In contrast
to Ehm2/1, a higher Ehm2/2 expression was observed in patients who
had a poorer OS (P=0.023; Fig. 6D)
and PFS (P=0.58; Fig. 6E). The
elevated Ehm2/1 expression was also associated with a poor PPS
(P=0.032; Fig. 6F). Taken
together, these results indicate that Ehm2/1 inhibits and Ehm2/2
promotes disease progression and the relapse of lung cancer.
Discussion
In the current study, we demonstrated that two
transcript variants of Ehm2 had distinct expression patterns in
lung adenocarcinoma and had a different association with patient
survival in lung cancer. We further demonstrated that the two
transcript variants of Ehm2 had opposite effects on the functions
of A549 cells and EMT in A549 cells. Our data suggest that Ehm2/1
is a putative tumor suppressor during the disease progression and
metastasis of lung cancer, while Ehm2/2 may have an opposite
function.
The roles played by Ehm2 in cancer appear to be
controversial. As a metastasis-associated protein, Ehm2 has been
reported to be highly expressed in prostate and breast cancers,
which is associated with disease progression and metastasis
(13,14). On the other hand, Ehm2 has been
shown to exert an inhibitory effect on the migration of HeLa cells
by the regulation of cell morphology and cohesion as a downstream
molecule of CRB3A (25). In fact,
Ehm2 in these studies should be considered as total Ehm2, as the
anti-Ehm2 antibody used in these studies recognizes an internal
region of Ehm2, which is a sequence conserved in both Ehm2/1 and
Ehm2/2. However, which transcript variants dominate in these
processes has yet to be elucidated. In this study, we used
different antibodies specific to Ehm2/1 and Ehm2/2 to evaluate the
expression and functions of these transcript variants in lung
adenocarcinoma. In the present study, a decreased expression of
Ehm2/1 was found in lung cancers, while an upregulation of Ehm2/2
was observed in the tumors. Significant associations were found
between the reduced transcript levels of Ehm2/1 and poorer OS, PFS
and PPS, while higher transcript levels of Ehm2/2 were associated
with a poorer OS and PPS. Our results indicate that the two
transcript variants have differential expression patterns and
opposing functions in lung cancer development.
In line with their clinical implication, our in
vitro experiments revealed that Ehm2/1 and Ehm2/2 have opposite
effects on the functions of human lung adenocarcinoma A549 cells. A
significant reduction in cell growth, migration and invasion in
vitro was observed in Ehm2/1-overexpressing A549 cells. By
contrast, Ehm2/1 knockdown led to a significant increase in the
growth, migration and invasion of A549 cells. The overexpression of
Ehm2/2 markedly promoted the migration and invasion of A549 cells.
Our data suggest that Ehm2/1 is a putative suppressor of disease
progression in lung cancer, while Ehm2/2 may play an opposing role.
In fact, it is well known that different transcript variants of a
gene may have different or opposing biological functions. For
example, human osteopontin (OPN), has the 3 transcript variants of
OPN-a (full-length form), OPN-b (lacking exon 5) and OPN-c (lacking
exon 4). Chae et al demonstrated that OPN-a and OPN-b were
predominantly expressed in hepatocellular carcinoma tissues, while
OPN-c was expressed in normal liver tissues. The overexpression of
OPN-a and OPN-b promoted the migration of Hep3B cells, while OPN-c
exerted minimal effects. However, OPN-c inhibited the migrate of
SK-Hep1 cells, while OPN-a overexpression had no effect (26).
Ehm2 contains a conserved FERM domain that is
involved in the interaction between cytoplasmic proteins and
transmembrane proteins via its FERM domain (27). Laprise et al revealed that
Yurt, the Drosophila orthologue of Ehm2, can bind to Crb
through its FERM domain and be recruited to the apical membrane
(28). EPB41L5, a protein sharing
a high homology with Ehm2, has been shown to bind with
intracellular domain of Crumbs through its FERM domain, which is
involved in the regulation of cell polarity (29). A predominant expression of
endogenous Ehm2/1 at the cell membrane has been observed in a
breast cancer cell line (MCF-7) in our previous study (22). In accordance with these results,
the present study also revealed a predominant distribution of
Ehm2/1 at the cell membrane in theA549 cells. By contrast,
exogenous Ehm2/2 was widely distributed in the cells, including the
cell membrane, cytoplasm and even the nucleus. As mentioned above,
Ehm2/2 is longer than Ehm2/1 and has a longer C-terminal. Although
structural analysis identified no specific domains in the
C-terminus of Ehm2/2, we hypothesized that the C-terminus might be
related to the different subcellular localization of the molecule,
which is yet to be investigated. The present study clearly
demonstrated that the two Ehm2 transcript variants have different
expression patterns, different subcellular distributions and
opposing biological functions. It is thus highly probable that
different cellular distributions of Ehm2 variants may be one cause
leading to varying biological functions. In our previous study, we
reported that Ehm2/1 functions as a putative tumor suppressor in
the disease progression of breast cancer (14). In our ongoing studies, we found
that Ehm2/1 may stabilize E-cadherin by inhibiting its
ubiquitination and degradation (unpublished data). It is therefore
argued that different distributions of Ehm2 variants in different
cellular compartments enable them to exert biological functions via
different signaling pathway. This would be an exciting area to
explore in the future. The current research findings also raised
another question as to whether the two transcript variants interact
with each other. Presently, there is no direct and solid evidence
to suggest that the two variants do interact. However, in our
humble opinion, and from these protein sequences, the two
transcript variants of Ehm2 are unlikely to interact with each
other. Indeed, to the best of our knowledge, there has been no
report to date showing FERM proteins interacting with each other, a
notion worthwhile to explore in the future.
EMT is a highly regulated process, which is
associated with the enhanced migration and invasion of cancerous
cells, thus contributing to the local invasion and metastasis of
cancer cells. EMT is typically defined by the loss of the
epithelial phenotype and the acquiring of mesenchymal
characteristics, leading to reduced cell-cell adhesion, increased
motility and invasiveness and strengthened survival (30-32).
Our previous study demonstrated that Ehm2/1 overexpression
upregulated E-cadherin expression and decreased the migration of
MCF-7 cells (14). The results of
the present study also demonstrated that Ehm2/1 and Ehm2/2
regulated the migration and invasion of A549 cells, key hallmarks
of EMT. These findings led us to hypothesize that Ehm2/1 and Ehm2/2
may also regulate EMT in A549 cells. In line with this hypothesis,
we found that Ehm2/1 inhibited EMT, while Ehm2/2 promoted EMT. This
finding is indeed noteworthy and is in accordance to an ongoing
finding in breast cancer, where we demonstrated that Ehm2/1 may
stabilize E-cadherin by inhibiting its ubiquitination and
degradation. These two studies were on different types of cancer
and used different cell types, and suggest that Ehm2 is more widely
involved in EMT in different cancer types and that different
variants may have contrasting effects on EMT; these findings
warrant further investigation in the future.
MMPs are a family of enzymes involved in the
degradation of the extracellular matrix (ECM) and play important
roles in tumor invasion and metastasis (33). High levels of MMP2 expression are
often associated with metastasis (34). Studies have demonstrated that MMP2
leads to EMT via the proteolytic degradation of epithelial cell
junctional proteins in carcinomas (35). MMP2 and MMP9, also known as
gelatinases, play an important role for cleaving type I, IV
collagen and contribute to the process of metastasis (36). In this study, in accordance with
the data obtained from cell invasion assay, both the knockdown of
Ehm2/1 and the overexpression of Ehm2/2 increased the activity of
MMPs, including MMP2 and MMP9, suggesting that the regulation of
MMPs in a different manner by different Ehm2 variants is another
mechanism underlying their role in cancer metastasis. From this
point of view, it is very important to compare the two Ehm2
variants in primary and matched metastatic cancers. Unfortunately,
our collection was small and only had one metastatic cancer out of
the 15 lung cancer tissues making it difficult to draw a clear
conclusion. However, we are currently collecting a larger tissue
cohort, and hopefully with a longer follow-up of the patients, we
would be in a position to conduct the comparison in near
future.
In conclusion, the present study demonstrated that
the Ehm2 variant, Ehm2/1, was downregulated in lung adenocarcinoma
tissues and that these lower levels of Ehm2/1 were associated with
a poorer survival. In vitro cell function assays indicated
Ehm2/1 acted as an anti-oncogene in lung adenocarcinoma cells. This
study also demonstrated that the other variant, Ehm2/2, had a
contrasting pattern of expression from that of Ehm2/1, namely that
it was upregulated in lung adenocarcinoma cells and higher levels
of Ehm2/2 were associated with a poorer clinical outcome of
patients with lung cancer. Cell function assays indicated that
Ehm2/2 acted as an oncogene in lung adenocarcinoma cells. Moreover,
Ehm2/1 inhibited the invasion and migration of lung cancer cells
via the regulation of EMT, while Ehm2/2 increased the invasiveness
and motility of lung cancer cells by promoting EMT. Our research
provides new evidence that different transcript variants of a gene
can play different, even opposing, roles. It will be necessary to
validate these findings in in vivo models and accordingly we
are currently preparing recombinant lentivirus for overexpressing
Ehm2/2 and knocking down Ehm2/1 to order implement an orthotopic
xenografts tumor model in BALB/c nude mice. Hopefully, these future
investigation using in vivo models and indeed a larger
clinical cohort would assist in answering some of questions
relating to distant metastasis. Collectively, the current results
from clinical samples and cellular experiments are sufficient to
arrive at the conclusions that the two Ehm2 variants have
contrasting expression pattern in human lung adenocarcinoma and
exert contrasting biological functions in lung cancer cells. The
variants have important bearing to the disease progression of lung
adenocarcinoma.
Supplementary Materials
Funding
This study was supported by the National Natural
Science Foundation of the People’s Republic of China (grant no.
81672726).
Availability of data and materials
The corresponding experimental data of the present
study are available from the corresponding author on reasonable
request.
Authors’ contributions
SL performed most of the experiments with assistance
from JM, YS, SC and BL, and wrote the manuscript. XZ and MH
provided the lung cancer tissue samples from the Department of
Thoracic Surgery, Beijing Xuanwu Hospital. HY and WGJ conceived and
designed the study, and also provided supervision and critically
revised the manuscript. SL and HY performed the analysis of the
data and also wrote the manuscript. All authors have reviewed and
approved the final manuscript.
Ethics approval and consent to
participate
The procedures of the human tissues collection were
reviewed and approved by the Ethics Committee of Xuanwu Hospital,
Capital Medical University [approval no. (2014) 022].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would also like to thank Professor Gong
Liping of Capital Medical University for analyzing the results of
immunohistochemistry of the tissue microarrays.
References
|
1
|
Tomasini P, Greillier L and Barlesi F:
Advanced non-squamous non-small-cell lung cancer: Who and when
should be biologically screened today? Tomorrow? Curr Respir Care
Rep. 2:17–21. 2013.
|
|
2
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016.
|
|
3
|
Castells M, Thibault B, Delord JP and
Couderc B: Implication of tumor microenvironment in
chemoresistance: Tumor-associated stromal cells protect tumor cells
from cell death. Int J Mol Sci. 13:9545–9571. 2012.
|
|
4
|
Keedy VL, Temin S, Somerfield MR, Beasley
MB, Johnson DH, McShane LM, Milton DT, Strawn JR, Wakelee HA and
Giaccone G: American Society of Clinical Oncology provisional
clinical opinion: Epidermal growth factor receptor (EGFR) Mutation
testing for patients with advanced non-small-cell lung cancer
considering first-line EGFR tyrosine kinase inhibitor therapy. J
Clin Oncol. 29:2121–2127. 2011.
|
|
5
|
Neel DS and Bivona TG: Resistance is
futile: Overcoming resistance to targeted therapies in lung
adenocarcinoma. NPJ Precis Oncol. 1:1–6. 2017.
|
|
6
|
Shimizu K, Nagamachi Y, Tani M, Kimura K,
Shiroishi T, Wakana S and Yokota J: Molecular cloning of a novel
NF2/ERM/4.1 superfamily gene, ehm2, that is expressed in
high-metastatic K1735 murine melanoma cells. Genomics. 65:113–120.
2000.
|
|
7
|
Badouel C, Gardano L, Amin N, Garg A,
Rosenfeld R, Le Bihan T and McNeill H: The FERM-domain protein
Expanded regulates Hippo pathway activity via direct interactions
with the transcriptional activator Yorkie. Dev Cell. 16:411–420.
2009.
|
|
8
|
Pearson MA, Reczek D, Bretscher A and
Karplus PA: Structure of the ERM protein moesin reveals the FERM
domain fold masked by an extended actin binding tail domain. Cell.
101:259–270. 2000.
|
|
9
|
Marešová L, Vydarený T and Sychrová H:
Comparison of the influence of small GTPases Arl1 and Ypt6 on yeast
cells’ tolerance to various stress factors. FEMS Yeast Res.
12:332–340. 2012.
|
|
10
|
Nakajima H and Tanoue T: Lulu2 regulates
the circumferential actomyosin tensile system in epithelial cells
through p114RhoGEF. J Cell Biol. 195:245–261. 2011.
|
|
11
|
Chauhan S, Pandey R, Way JF, Sroka TC,
Demetriou MC, Kunz S, Cress AE, Mount DW and Miesfeld RL: Androgen
regulation of the human FERM domain encoding gene EHM2 in a cell
model of steroid-induced differentiation. Biochem Biophys Res
Commun. 310:421–432. 2003.
|
|
12
|
Hashimoto Y, Shindo-Okada N, Tani M,
Takeuchi K, Toma H and Yokota J: Identification of genes
differentially expressed in association with metastatic potential
of K-1735 murine melanoma by messenger RNA differential display.
Cancer Res. 56:5266–5271. 1996.
|
|
13
|
Wang J, Cai Y, Penland R, Chauhan S,
Miesfeld RL and Ittmann M: Increased expression of the
metastasis-associated gene Ehm2 in prostate cancer. Prostate.
66:1641–1652. 2006.
|
|
14
|
Yu H, Ye L, Mansel RE, Zhang Y and Jiang
WG: Clinical implications of the influence of Ehm2 on the
aggressiveness of breast cancer cells through regulation of matrix
metalloproteinase-9 expression. Mol Cancer Res. 8:1501–1512.
2010.
|
|
15
|
Stegle O, Drewe P, Bohnert R, Borgwardt K
and Rätsch G: Statistical Tests for Detecting Differential
RNA-Transcript Expression from Read Counts. Nat Preced.
10:4437–4448. 2010.
|
|
16
|
Lixia M, Zhijian C, Chao S, Chaojiang G
and Congyi Z: Alternative splicing of breast cancer associated gene
BRCA1 from breast cancer cell line. J Biochem Mol Biol. 40:15–21.
2007.
|
|
17
|
Black DL: Mechanisms of alternative
pre-messenger RNA splicing. Annu Rev Biochem. 72:291–336. 2003.
|
|
18
|
Ghigna C, Giordano S, Shen H, Benvenuto F,
Castiglioni F, Comoglio PM, Green MR, Riva S and Biamonti G: Cell
motility is controlled by SF2/ASF through alternative splicing of
the Ron protooncogene. Mol Cell. 20:881–890. 2005.
|
|
19
|
Moolenaar CE, Pieneman C, Walsh FS, Mooi
WJ and Michalides RJ: Alternative splicing of neural-cell-adhesion
molecule mRNA in human small-cell lung-cancer cell line H69. Int J
Cancer. 51:238–243. 1992.
|
|
20
|
Sato H, Hiyama K, Ishioka S, Maeda H and
Yamakido M: Alternative splicing, but not allelic loss, of the
FHIT gene increases with development of lung cancer. Int J
Oncol. 15:81–88. 1999.
|
|
21
|
Venables JP: Unbalanced alternative
splicing and its significance in cancer. BioEssays. 28:378–386.
2006.
|
|
22
|
Yu H, Ge Z, Si Y, Chen G, Zhang Y and
Jiang WG: The splice variant Ehm2/1 in breast cancer MCF-7 cells
interacted with β-catenin and increased its localization to plasma
membrane. RSC Advances. 6:78436–78444. 2016.
|
|
23
|
Yuan JS, Reed A, Feng C and Stewart CN:
Statistical analysis of real-time PCR data. BMC Bioinformatics.
7:852006.
|
|
24
|
Gary R and Bretscher A: Ezrin
self-association involves binding of an N-terminal domain to a
normally masked C-terminal domain that includes the F-actin binding
site. Mol Biol Cell. 6:1061–1075. 1995.
|
|
25
|
Loie E, Charrier LE, Sollier K, Masson JY
and Laprise P: CRB3A Controls the Morphology and Cohesion of Cancer
Cells through Ehm2/p114RhoGEF-Dependent Signaling. Mol Cell Biol.
35:3423–3435. 2015.
|
|
26
|
Chae S, Jun HO, Lee EG, Yang SJ, Lee DC,
Jung JK, Park KC, Yeom YI and Kim KW: Osteopontin splice variants
differentially modulate the migratory activity of hepatocellular
carcinoma cell lines. Int J Oncol. 35:1409–1416. 2009.
|
|
27
|
Mori T, Kitano K, Terawaki S, Maesaki R
and Hakoshima T: Crystallographic characterization of the radixin
FERM domain bound to the cytoplasmic tail of adhesion molecule
CD44. Acta Crystallogr Sect F Struct Biol Cryst Commun. 63:844–847.
2007.
|
|
28
|
Laprise P, Beronja S, Silva-Gagliardi NF,
Pellikka M, Jensen AM, McGlade CJ and Tepass U: The FERM protein
Yurt is a negative regulatory component of the Crumbs complex that
controls epithelial polarity and apical membrane size. Dev Cell.
11:363–374. 2006.
|
|
29
|
Gosens I, Sessa A, den Hollander AI,
Letteboer SJ, Belloni V, Arends ML, Le Bivic A, Cremers FP,
Broccoli V and Roepman R: FERM protein EPB41L5 is a novel member of
the mammalian CRB-MPP5 polarity complex. Exp Cell Res.
313:3959–3970. 2007.
|
|
30
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014.
|
|
31
|
Puisieux A, Brabletz T and Caramel J:
Oncogenic roles of EMT-inducing transcription factors. Nat Cell
Biol. 16:488–494. 2014.
|
|
32
|
Cervantes-Arias A, Pang LY and Argyle DJ:
Epithelial-mesenchymal transition as a fundamental mechanism
underlying the cancer phenotype. Vet Comp Oncol. 11:169–184.
2013.
|
|
33
|
Jezierska A and Motyl T: Matrix
metalloproteinase-2 involvement in breast cancer progression: A
mini-review. Med Sci Monit. 15:RA32–RA40. 2009.
|
|
34
|
Niemiec JA, Adamczyk A, Małecki K,
Majchrzyk K and Ryś J: Relationships between immunophenotype, Ki-67
index, microvascular density, Ep-CAM/P-cadherin, and MMP-2
expression in early-stage invasive ductal breast cancer. Appl
Immunohistochem Mol Morphol. 20:550–560. 2012.
|
|
35
|
Liu Y, Sun X, Feng J, Deng LL, Liu Y, Li
B, Zhu M, Lu C and Zhou L: MT2-MMP induces proteolysis and leads to
EMT in carcinomas. Oncotarget. 7:48193–48205. 2016.
|
|
36
|
John A and Tuszynski G: The role of matrix
metalloproteinases in tumor angiogenesis and tumor metastasis.
Pathol Oncol Res. 7:14–23. 2001.
|