Introduction
Gastric cancer (GC) is the fifth most prevalent and
aggressive type of cancer and the third leading cause of
cancer-related mortality worldwide (1). The incidence and mortality rates
associated with GC are increasing in East Asia and Eastern Europe.
In China, the most recent statistical analysis has revealed that GC
is the second most common type of cancer with a 5-year survival
rate of ~25-30% (2). The
development of endoscopy and surgical techniques has reduced the
5-year mortality rate associated with GC; however, for patients
with late-stage GC, the 5-year mortality rate remains ~30-50%
(3). Although the effectiveness of
treatment strategies, such as surgery combined with radiotherapy
and chemotherapy has somewhat improved, the prognosis of patients
with advanced GC remains poor due to peritoneal dissemination,
hematogenous spread and lymph node metastasis (4). Thus, it is crucial to explore the
underlying molecular mechanisms responsible for the progression of
GC.
Recent studies have indicated that non-coding RNAs
(ncRNAs) are novel regulatory molecules that play essential roles
in tumorigenesis (5,6). ncRNAs, including long non-coding RNAs
(lncRNAs) and microRNAs (miRNAs or miRs) are not capable of coding
proteins (7). miRNAs interact with
associated proteins to form the active RNA-induced silencing
complex, consequently suppressing the translation or inducing the
degradation of target mRNAs (8).
Aberrantly expressed miRNAs are associated with the proliferation,
apoptosis, growth, migration and invasion of cancer cells (9). In recent years, the effects of
lncRNAs on tumor development have been investigated. Previous
studies have suggested that lncRNAs are involved in the regulation
of various cellular functions and disease pathogenesis, such as
cancer metastasis at the transcriptional and post-transcriptional
level (10,11). The potential function of lncRNAs is
to regulate the expression levels of mRNAs and miRNAs by altering
chromatin modification and transcription (12). Furthermore, lncRNAs are capable of
inhibiting the expression of miRNAs via direct binding,
consequently producing competing endogenous RNAs (ceRNAs) that can
induce protein destabilization at the post-transcriptional level
(13). Accumulating evidence has
suggested that the impaired expression of lncRNA H19 (hereon
referred to as H19) plays an essential role in carcinogenesis. For
example, H19 has been shown to promote the progression of GC
(14), breast cancer (15), lung cancer (16) and colorectal cancer (17). In addition, a previous study
revealed that the upregulated expression of H19 can affect the
proliferation of GC cells and is associated with the initiation and
development of GC (18).
miR-22-3p, as a tumor suppressor miRNA, has been shown to be
significantly downregulated in GC (19), colorectal cancer (20), hepatocellular carcinoma (21), cervical cancer (22), prostate cancer and lung cancer
(23). The downregulation of
miR-22-3p can promote tumor growth and metastasis. A recent study
suggested that H19 functions as a ceRNA for miR-22-3p by regulating
its downstream Wnt/β-catenin signaling in nucleus pulposus cells
(NPCs) (24). Although H19
functions as a molecular sponge of miR-22-3p in GC, the underlying
mechanisms of this function remain unknown. The association between
H19 and GC requires further investigation. Thus, the aim of the
present study was to identify the functions of H19 in GC and to
investigate whether miR-22-3p is the target of H19. The results of
the present study reveal the regulatory mechanisms of action of H19
in GC, which may provide novel insight into potential therapeutic
targets for the treatment of GC.
Materials and methods
Patient samples
A total of 40 pairs of primary tumor and
para-cancerous tissues were obtained from patients with GC (28
males and 12 females; average age, 58.93±9.17 years) undergoing
surgical resection at the Second Affiliated Hospital of Chongqing
Medical University (Chongqing, China). The study was approved by
the Ethics Committee of Chongqing Medical University. Informed
consent was obtained from each patient prior to surgery. The
clinicopathological characteristics of the patient samples were
confirmed by two pathologists according to the diagnostic criteria
of World Health Organization (2010-2012). Metastasis was found in
28 cases, and 25 patients were diagnosed with stage I or II gastric
cancer. All samples were immediately stored in liquid nitrogen
until further use.
Cells, cell culture and transfection
AGS (cat. no. CRL-1739) and normal gastric mucous
epithelium cells (GES-1; cat. no. 28200) were purchased from the
American Type Culture Collection (ATCC, Manassas, VA, USA). BGC-823
(cat. no. TCHu 11), MKN-45 (cat. no. TCHu130) and SGC-7901 (cat.
no. TCHu 46) were purchased from Chinese Academy of Sciences
(Shanghai, China). The cells were cultured in RPMI-1640 medium
(HyClone; GE Healthcare Life Science, Logan, UT, USA) supplemented
with 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life
Science) and incubated at 37°C in a humidified atmosphere of 5%
CO2. Short hairpin RNA (shRNA) sequences targeting H19
(sh-H19) and Snail1 (sh-Snail1), as well as the negative control
(sh-NC) were obtained from Genepharm Co. Ltd. (Shanghai, China).
Following annealing, shRNAs were integrated into the lentiviral
pU6-Luc-Puro vector (Genepharm Co. Ltd.). To generate the H19
overexpression model, the H19 fragment was amplified using PCR and
then subcloned into the pcDNA3.1 vector (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). pcDNA3.1 vectors containing
wild-type (WT) H19 or Snail1, mutant (MUT) H19 or Snail1 were
obtained from RioBio (Guangzhou, China). miR-22-3p mimics,
inhibitors and negative control (miR-NC) were synthesized by
Genepharma Co. Ltd. (Shanghai, China). All transfections were
performed using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer’s
instructions. The cells were harvested at 48 h post-transfection
for use in further experiments.
Luciferase reporter assay
Targetscan (www.targetscan.org) and LncBase Predicted v.2
(http://www.microrna.gr/LncBase) were
employed to predict the potential targets of miR-22-3p or H19.
Wild-type segments of the 3′UTR of H19/Snail1 containing potential
binding sites of miR-22-3p were cloned into the pMIR-REPORT firefly
luciferase vector (Applied Biosystems, Foster City, CA, USA). The
mutant fragment of H19/Snail1 was used as a control. 293 cells
(ATCC) were co-transfected with the luciferase reporter vector and
miR-22-3p mimics or miRNA negative control (miR-NC) using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At
48 h post-transfection, the luciferase activity was evaluated using
the Dual Luciferase Reporter Assay System (Promega, Madison, WI,
USA) according to the manufacturer’s protocols. The activity was
normalized to Renilla luciferase.
Cell proliferation assay
Transfected AGS and SGC-7901 cells were seeded at a
density of 1×104/well in 96-well plates. Proliferation
rates were determined by Cell Counting kit-8 assay (CCK-8 Assay;
Beyotime, Shanghai, China) at days 1, 2, 3 and 4 post-transfection.
The absorbance at 450 nm was measured using a microplate reader
(9200, Bio-Rad Laboratories, Hercules, CA, USA) according to the
manufacturer’s instructions.
Transwell assay
The migration and invasion of the cells were
evaluated by a Transwell assay. The AGS and SGC-7901 cells were
harvested at 48 h post-transfection. For the migration assay,
5×104 cells in serum-free medium were inoculated onto
the upper chamber (BD Biosciences, Franklin Lakes, NJ, USA) with an
8-µm membrane. For the invasion assay, 2×105
cells in serum-free medium were inoculated onto the upper chamber
pre-coated with Matrigel (Sigma-Aldrich, St. Louis, MO, USA).
Culture medium containing 10% FBS was added to the lower chamber.
Non-migratory cells were removed using a cotton swab following
overnight incubation at 37°C, while the migrated or invaded cells
in the lower chamber were fixed in 4% paraformaldehyde for 10 min
at room temperature, and stained using methanol and 0.1% crystal
violet. Finally, the number of migratory or invasive cells were
observed and counted using an inverted light microscope
(magnification, ×100; Olympus Corp., Tokyo, Japan).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR was used to determine the expression levels
of H19, miR-22-3p, Snail1, E-cadherin (E-Cad), α-smooth muscle
actin (α-SMA), vimentin (VI) and fibronectin (FN). Total RNA was
extracted from the tissues or cells using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA,
USA) according to the manufacturer’s instructions. miRNA was
extracted using the miRNeasy Mini kit (Qiagen, Shenzhen, China)
according to the manufacturer’s instructions. RNA was reverse
transcribed using a PrimeScript RT kit (Takara, Dalian, China)
according to the manufacturer’s instructions. For the assessment of
miR-22-3p, a TaqMan MicroRNA Assay kit (Applied Biosystems) was
used and qPCR was performed using the Applied Biosystem 7500
Real-Time PCR System; U6 was used for normalization. For H19,
Snail1, E-Cad, α-SMA, VI and FN, qPCR was performed using
SYBR-Green PCR Master Mix (Takara). Relative expression was
calculated and normalized to endogenous GAPDH. The forward and
reverse primer sequences were as follows: H19 forward, 5′-ATCGGTGC
CTCAGCGTTCGG-3′ and reverse, 5′-CTGTCCTCG CCGTC ACACCG-3′; Snail1
forward, 5′-GAAGATGCACATCCGA AGC-3′ and reverse,
5′-AGTGGGAGCGGAGAAAGG-3′; E-Cad forward, 5′-CAATGGTGTCCATGTGAACA-3′
and reverse, 5′-CCTCCTACCCTCCTGTTCG-3′; α-SMA forward,
5′-TCCCTTGAGAAGAGTTACGAGTTG-3′ and reverse,
5′-ATGATGCTGTTGTAGGTGGTTTC-3′; VI forward, 5′-CGCTTCGCCAACTACAT-3′
and reverse, 5′-AGGGCA TCCACTTCACAG-3′; FN forward,
5′-CCAAACCTCAAGC TCCCGTCA-3′ and reverse, 5′-GAGATTCTGCACATCAC
GGTCA-3′; and GAPDH forward, 5′-GCAAGAGCACAAG AGGAAGA-3′ and
reverse, 5′-ACTGTGAGGAGGGGAG ATTC-3′. The PCR program used for the
thermocycler was as follows: 95°C for 5 min, followed by 45 cycles
of 95°C for 15 sec, 60°C for 20 sec and 72°C for 10 sec. Relative
expression levels were evaluated via the 2−∆∆Cq method
(25). The results of significant
difference tests between the relative and raw numerical value are
the same.
Western blot analysis
Total protein was isolated from the tissues or cells
using radioimmunoprecipitation assay buffer (Beyotime). Protein
concentrations were evaluated by a BCA Protein Assay kit
(Beyotime). Equal amounts (40 µg) of extracted protein were
separated by 10% SDS-PAGE and transferred onto polyvinylidene
fluoride membranes (Millipore, Billerica, MA, USA). Subsequently,
the membranes were blocked with 5% (w/v) non-fat milk in TBST
buffer (Beyotime) for 1 h at 37°C. The membranes were then
incubated with anti-E-Cad (1:5,000; cat. no. ab40772), anti-α-SMA
(1:2,000; cat. no. ab124964), anti-VI (1:1,000; cat. no. ab20346),
anti-FN (1:1,000; cat. no. ab18265), anti-Snail1 (1:500; cat. no.
ab82846) and anti-GAPDH (1:1000; cat. no. ab8245) (all from Abcam,
Cambridge, MA, USA) at 4°C overnight. Following 3 washes in TBST
for 10 min, the membranes were incubated with horseradish
peroxidase-conjugated goat anti-mouse IgG (1:1,000; cat. no. 7076)
or goat anti-rabbit IgG (1:1,000; cat. no. 7074) (both from Cell
Signaling Technology, Danvers, MA, USA) for 1 h at 37°C. The
protein bands were visualized by an enhanced chemiluminescence kit
and quantified by densitometric analysis using ImageJ software
(NIH, Bethesda, MD, USA).
In vivo nude mouse xenograft and lung
metastasis assays
All animal experiments were ethically approved by
the Research Ethics Committee of Chongqing Medical University
(Chongqing, China). A total of 10 female BALB/C nude mice (5-6
weeks old) with a weight of 17-22 g were obtained from the
Experimental Animal Centre of the Third Military Medical University
(Chongqing, China). The mice were routinely housed in a humidity-
(80%); and temperature-controlled (22±2°C) environment for at least
3 days prior to the experiments. A total of 2×106
SGC-7901 cells transfected with sh-NC or sh-H19 were suspended in
200 µl phosphate-buffered saline and injected subcutaneously
into right side of the armpit regions of mice (5 mice/group. At 6
weeks post-injection, the mice were sacrificed, and the tumors were
isolated and measured. Tumor volume was calculated using the
following formula: V (mm3) = 0.5 × length ×
width2. The tumor tissues were snap-frozen in liquid
nitrogen until further use.
Statistical analysis
Data are presented as the means ± standard deviation
unless otherwise indicated and were analyzed using Graphpad Prism
v7.0 software (Graphpad Software Inc., La Jolla, CA, USA). A t-test
(two-sided) and one-way analysis of variance (ANOVA) were used for
statistical analysis. A student-Newman-Keuls test was performed as
a post hoc test following ANOVA. The correlation between the
expression levels of Snail1 and miR-22-3p was assessed using
Spearman’s correlation analysis, as appropriate. P<0.05 was
considered to indicate a statistically significant difference.
Results
H19 is upregulated and miR-22-3p is
downregulated in GC tissues and cells
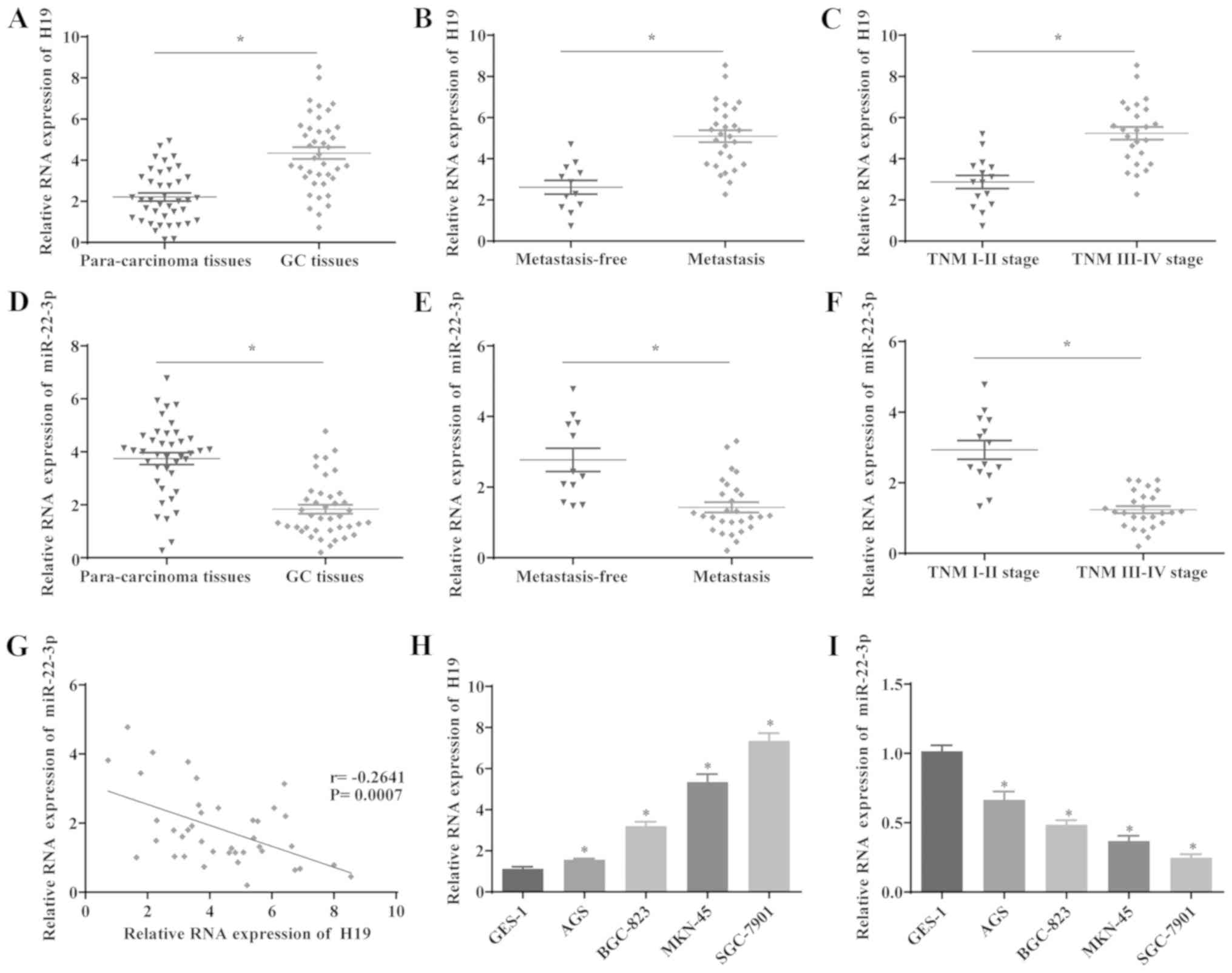
The expression levels of H19 and miR-22-3p in 40
paired GC and para-cancerous samples were determined by RT-qPCR.
The results revealed that H19 expression was significantly
upregulated, whereas the expression level of miR-22-3p was
decreased in the GC tissues compared with the para-cancerous
tissues (Fig. 1A and D).
Furthermore, the effects of H19 and miR-22-3p expression on
metastasis and the prognosis of patients with GC were investigated.
The results indicated that H19 expression was significantly
elevated and that miR-22-3p expression was decreased in patients
with GC with lymph node metastasis compared with those without
metastasis (Fig. 1B and E). In
addition, the expression level of H19 was significantly increased
and that of miR-22-3p was decreased in aggressive GC tumors (TNM
stage III/IV), compared with stage I/II tumors, suggesting that the
upregulation of H19 and the downregulation of miR-22-3p are
associated with the development of GC (Fig. 1C and F). Furthermore, the
correlation between H19 and miR-22-3p expression was analyzed, and
the results indicated that the expression levels of H19 and
miR-22-3p inversely correlated in GC tissues (Fig. 1G). Additionally, H19 was
significantly upregulated and miR-22-3p was downregulated in GC
cells lines compared with GES-1 cells (Fig. 1H and I). These results suggested
that the expression of H19 and miR-22-3p is upregulated and
downregulated in GC respectively, which is also associated with
metastasis in patients with GC. Furthermore, the miR-22-3p level is
downregulated by H19 in GC cells, suggesting that H19 may exert its
regulatory function in GC via miR-22-3p.
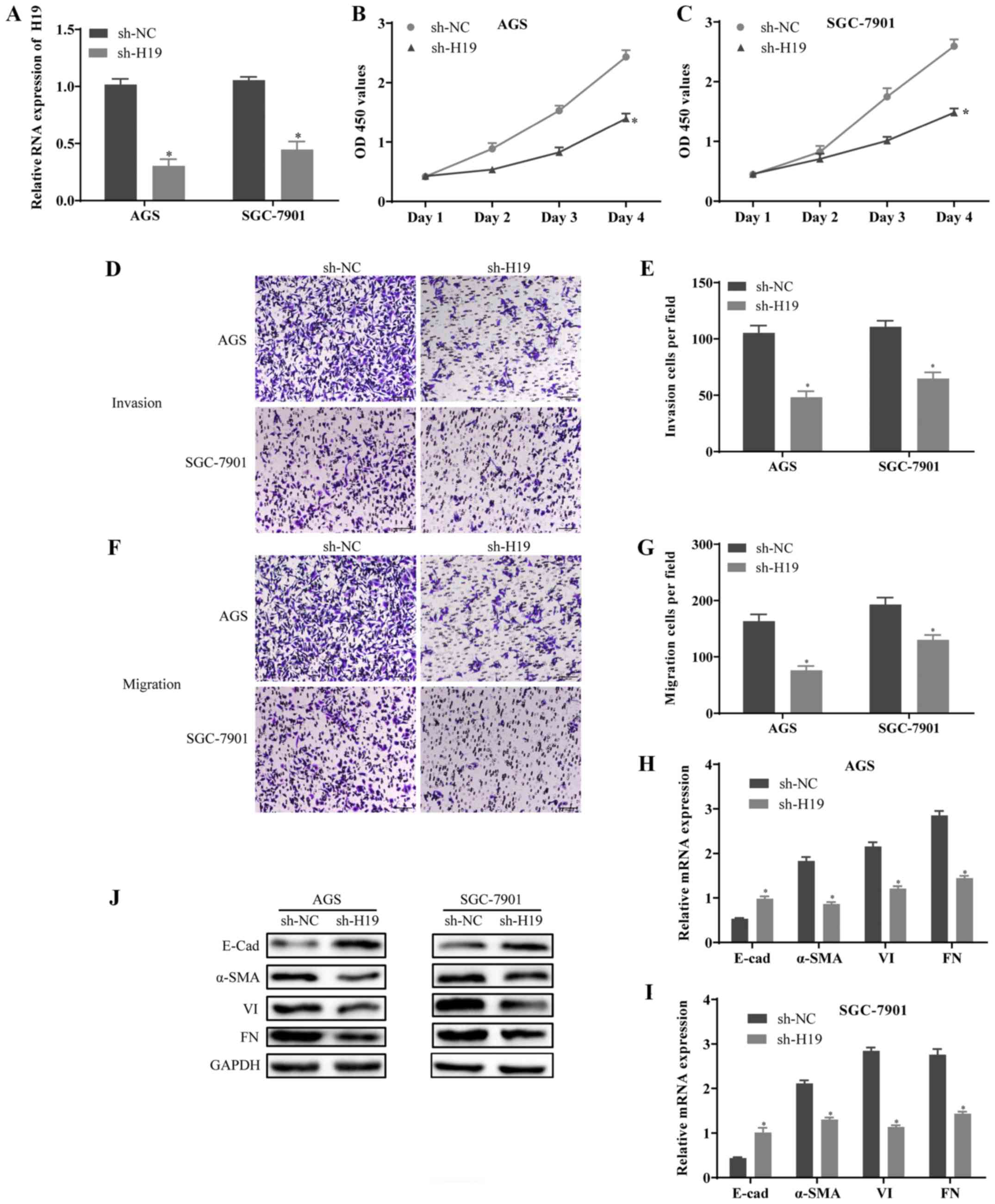
Downregulation of H19 suppresses cell
proliferation, invasion, migration and epithelial-mesenchymal
transition (EMT)
To examine the effects of H19 on the proliferation,
invasion and migration of GC cells, the expression of H19 was
silenced by sh-H19 in AGS (the lowest expression of H19) and
SGC-7901 (the highest expression of H19) cells. The knockdown
efficiency of H19 was determined by RT-qPCR (Fig. 2A). The results of CCK-8 assay
revealed that the proliferative ability of the AGS and SGC-7901
cells transfected with sh-H19 was decreased compared with the
controls (Fig. 2B and C). In
addition, Transwell assay indicated that the invasion and migration
of the AGS and SGC-7901 cells transfected with sh-H19 were
significantly suppressed (Fig.
2D-G). Furthermore, the results of RT-qPCR and western blot
analysis revealed that the expression level of E-Cad was notably
increased, whereas the expression levels of α-SMA, VI and FN were
decreased in the sh-H19-transfected cells compared with the
controls (Fig. 2H-J). On the
whole, these results suggested that the downregulation of H19
suppressed the growth, metastasis and EMT of GC cells.
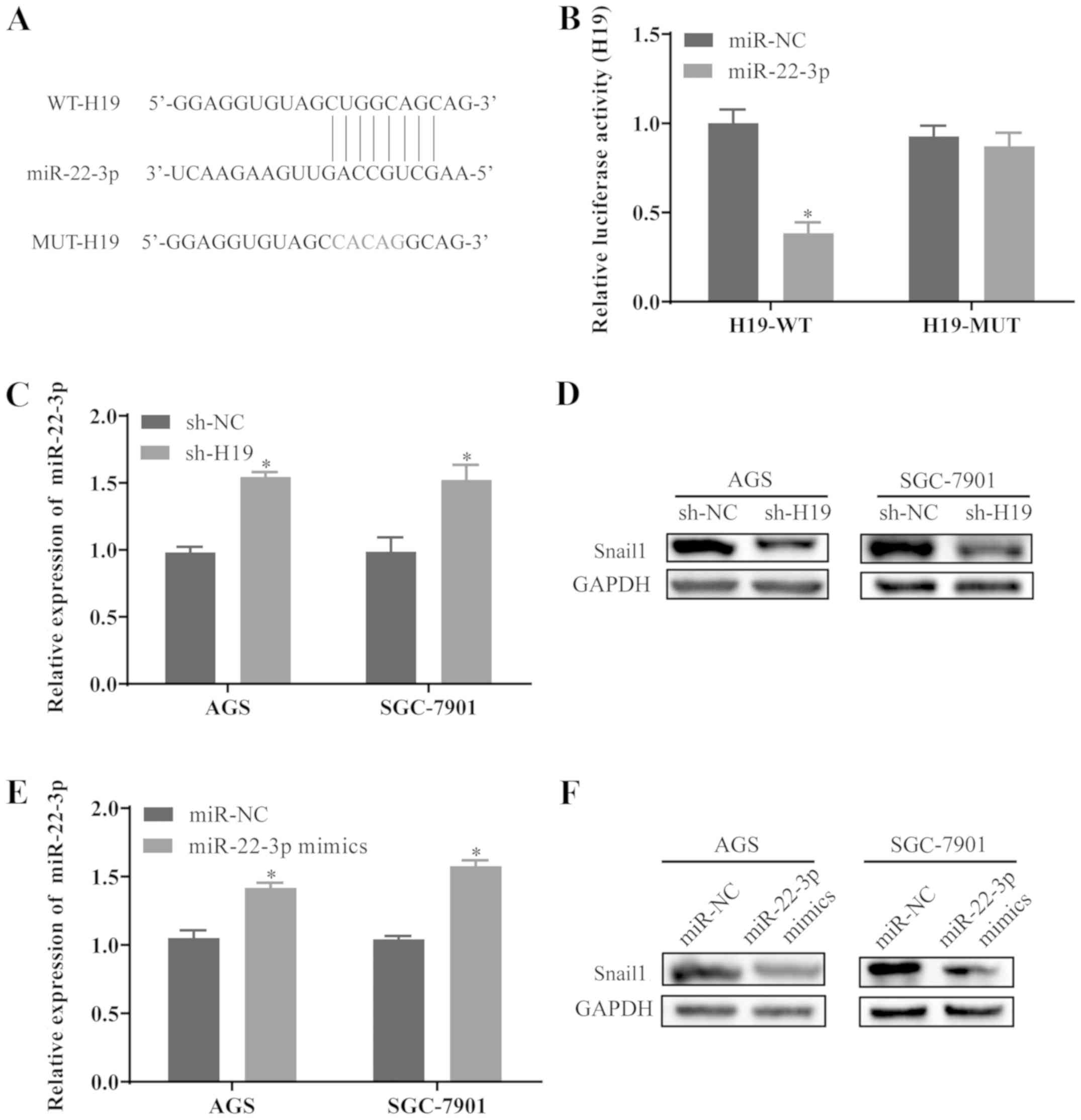
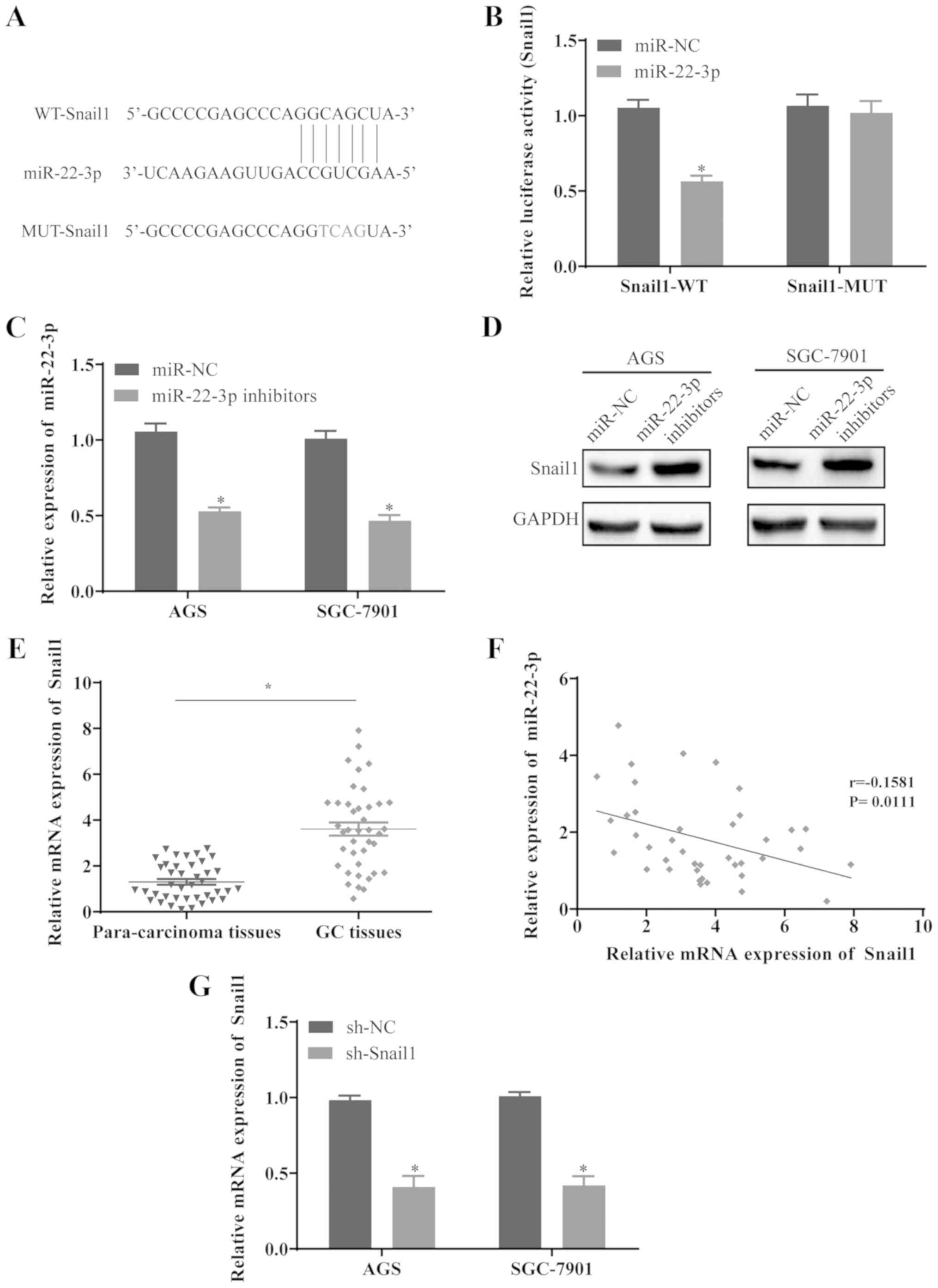
miR-22-3p is the potential target of H19
in GC cells
To determine whether H19 functions by suppressing
its target miRNA in GC, the potential binding sites of miR-22-3p in
H19 transcripts were predicted using LncBase Predicted v.2
(Fig. 3A). Luciferase reporter
plasmids containing the wild-type H19 (H19-WT) and mutant H19
(H19-MUT) of predicted miR-22-3p binding sites were constructed.
The results revealed that transfection with miR-22-3p mimics
significantly reduced the luciferase activity in H19-WT, while the
luciferase activity in 293 cells transfected with H19-MUT was not
affected by miR-22-3p mimics (Fig.
3B). In order to further examine the effects of H19 on the
expression level of miR-22-3p, the AGS and SGC-7901 cells were
transfected with sh-H19. The cells transfected with sh-H19
exhibited a marked increase in miR-22-3p levels (Fig. 3C). Furthermore, H19 knockdown
resulted in a prominent decrease in Snail1 protein expression in
the AGS and SGC-7901 cells (Fig.
3D). Additionally, a miR-22-3p overexpression model was
established by transfecting the AGS and SGC-7901 cells with
miR-22-3p mimics, and the transfection efficiency was confirmed by
RT-qPCR (Fig. 3E). Furthermore,
the overexpression of miR-22-3p inhibited the expression level of
Snail1 in the AGS and SGC-7901 cells (Fig. 3F), suggesting that the
miR-22-3p/Snail1 axis may be a potential target of H19 in GC
cells.
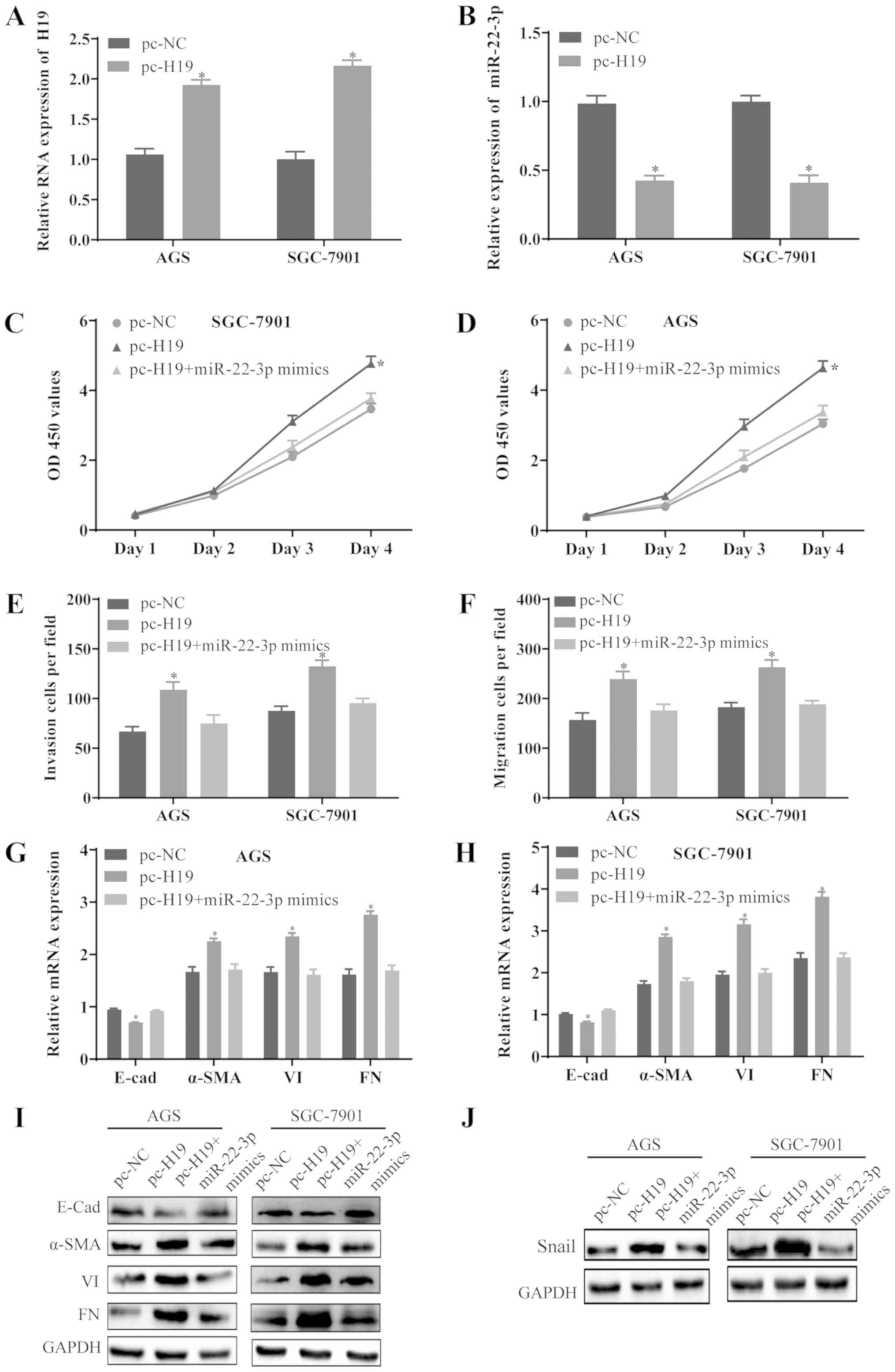
Overexpression of H19 promotes cell
proliferation, migration, invasion and EMT by regulating
miR-22-3p
To further investigate the association between H19
and miR-22-3p, the AGS and SGC-7901 cells were transfected with
pc-NC, pc-H19 or co-transfected with pc-H19 and miR-22-3p mimics.
The expression of H19 was significantly increased (Fig. 4A) and the level of miR-22-3p
decreased (Fig. 4B) in the AGS and
SGC-7901 cells transfected with pc-H19. Furthermore, the
overexpression of H19 promoted the proliferation (Fig. 4C and D), invasion (Fig. 4E), migration (Fig. 4F) and EMT (Fig. 4G-I) of the AGS and SGC-7901 cells,
whereas these effects were substantially abrogated by the
upregulation of miR-22-3p. In addition, the overexpression of H19
increased the level of Snail1, which was inhibited by the
upregulation of miR-22-3p (Fig.
4J). These results revealed that H19 may promote the
proliferation, invasion, migration and EMT of GC cells by
downregulating miR-22-3p and upregulating Snail1.
Snail1 is the target gene of miR-22-3p in
GC cells
Using the TargetScan database, the binding sites of
Snail1 on miR-22-3p was predicted (Fig. 5A). To investigate whether Snail1 is
the potential target of miR-22-3p, the WT and MUT fragments of
Snail1 were cloned into the downstream of the Firefly luciferase
coding domain in pGL-3 vector, and the results revealed that the
co-transfection of miR-22-3p and Snail1-WT reduced the relative
luciferase activity, whereas the activity remained unaltered in the
cells co-transfected with miR-22-3p and Snail1-MUT (Fig. 5B). To further determine whether
miR-22-3p can regulate the expression level of Snail1, the AGS and
SGC-7901 cells were transfected with miR-22-3p inhibitors. The
results confirmed that miR-22-3p was significantly downregulated in
the cells transfected with miR-22-3p inhibitors (Fig. 5C), whereas the protein level of
Snail1 was elevated (Fig. 5D).
Furthermore, Snail1 expression was upregulated in the GC tissues
compared with the paired para-cancerous tissues (Fig. 5E), and Snail1 expression inversely
correlated with miR-22-3p expression in GC tissues (Fig. 5F), suggesting that Snail1 may be a
potential target of miR-22-3p in GC. To further verify the role of
Snail1, the AGS and SGC-7901 cells were transfected with sh-Snail1
and Snail1 was significantly downregulated in the cells transfected
with sh-Snail1 (Fig. 5G).
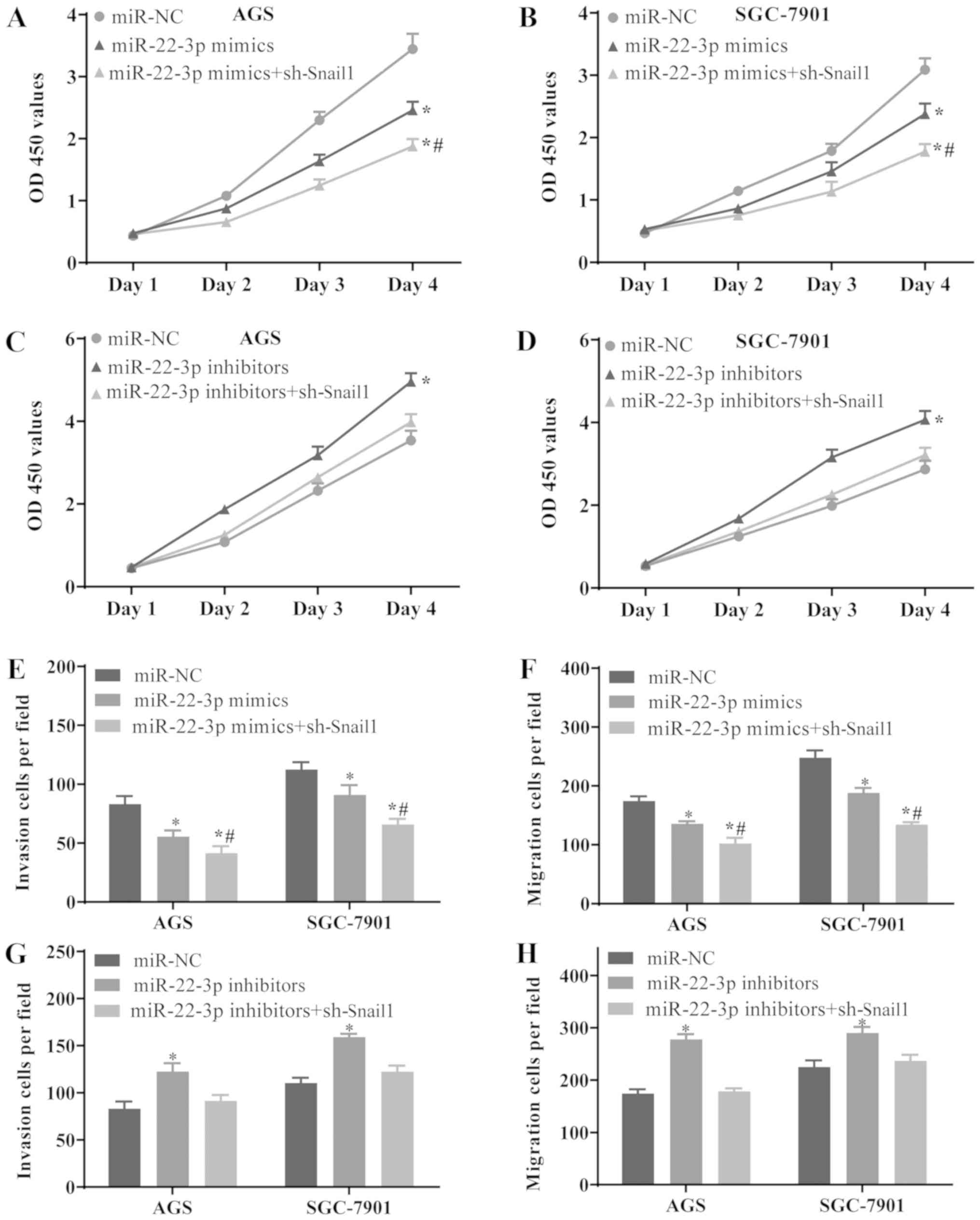
Downregulation of Snail1 enhances the
effects exerted by the overexpression of miR-22-3p and reverses the
effects exerted by the silencing of miR-22-3p in GC cells
To investigate whether the functions of Snail1 as
regards the growth, metastasis and EMT of GC cells are regulated by
miR-22-3p, the AGS and SGC-7901 cells were transfected with miR-NC,
miR-22-3p mimics or co-transfected with miR-22-3p mimics and
sh-Snail1, miR-22-3p inhibitors and sh-Snail1, respectively. The
results revealed that the overexpression of miR-22-3p suppressed
the proliferation (Fig. 6A and B),
invasion (Fig. 6E) and migration
(Fig. 6F) of the AGS and SGC-7901
cells, while these effects were enhanced by the silencing of Snail1
expression. Conversely, the downregulation of miR-22-3p promoted
the proliferation (Fig. 6C and D),
invasion (Fig. 6G) and migration
(Fig. 6H) of GC cells, whereas
these effects were abrogated by the silencing of Snail1 expression.
On the whole, the results of the present study suggest that
miR-22-3p may inhibit the growth of GC by downregulating Snail1. In
summary, H19 may regulate the proliferation, invasion, migration
and EMT of GC cells via the miR-22-3p/Snail1 signaling pathway.
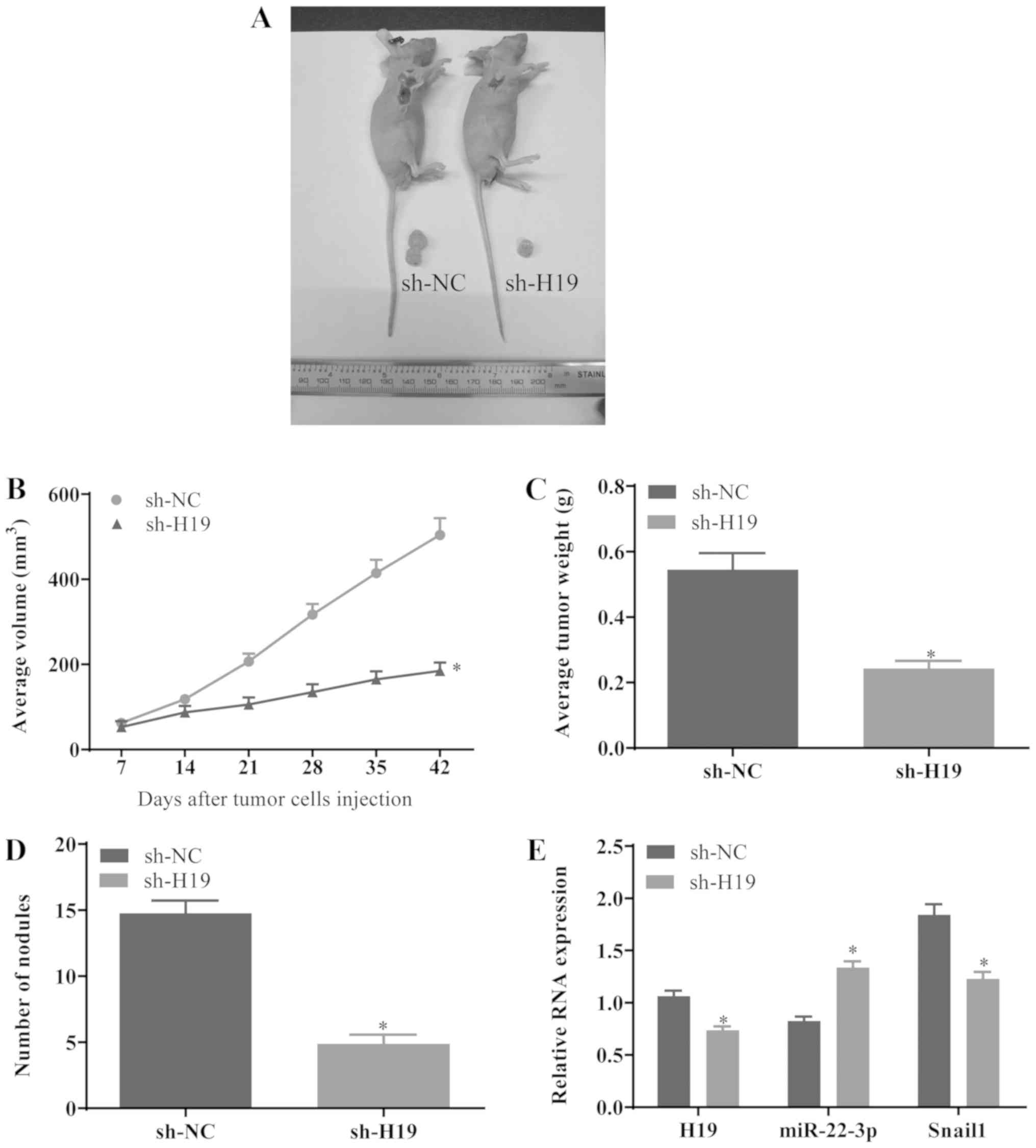
Downregulation of H19 suppresses the
development of GC in vivo
To investigate whether sh-H19 suppresses the growth
and metastasis of gastric tumors, GC cells transfected with sh-NC
or sh-H19 were subcutaneously injected into BALB/C nude mice. At 6
weeks post-injection, the mice were sacrificed and the isolated
tumors were measured (Fig. 7A).
The mean value of the tumor volume in the sh-H19 group was
significantly reduced compared with that in the sh-NC group
(Fig. 7B). In addition, the tumor
weight in the sh-H19 group was notably decreased compared with that
in the sh-NC group (Fig. 7C).
Furthermore, the numbers of macroscopic nodules were significantly
reduced in the sh-H19 group (Fig.
7D). Additionally, RT-qPCR revealed that the mRNA levels of H19
and Snail1 were downregulated in the sh-H19 group, whereas the
expression level of miR-22-3p was elevated (Fig. 7E) in these tumors, suggesting that
the downregulation of H19 may suppress the growth of gastric tumors
via the miR-22-3p/Snail1 axis in vivo.
Discussion
lncRNAs are RNA transcripts >200 nt in length,
with no capacity of coding proteins. Studies on lncRNAs have
revealed their significance over the past decade, and there is
adequate evidence to suggest that lncRNAs are important regulators
of the proliferation and migration of cancer cells; aberrant lncRNA
expression plays important roles in the progression of numerous
types of cancer; however, the regulatory functions and underlying
mechanisms of various lncRNAs remain unclear (26,27).
A previous study revealed that a high expression of lncRNAs may
competitively bind to miRNAs, consequently inhibiting miRNA
expression and promoting the progression of GC; for example,
downregulated lncRNA-SNHG5 was shown to inhibit the proliferation
and migration of GC cells via the miR-32/KLF4 axis (28). Additionally, lncRNA-BC032469
competitively binds with miR-1207-5p and upregulates human
telomerase reverse transcriptase (hTERT), subsequently promoting
the proliferation of GC cells (29). Furthermore, lncRNAs regulate their
target genes by interacting with its mRNA directly. For example, Xu
et al (30) reported that
lncRNA-TINCR promoted the proliferation and apoptosis of GC cells
by binding to STAU1 protein and interfering with the stability of
Krüppel-like factor 2 (KLF2) mRNA. Recent studies have also
indicated that lncRNAs, such as H19 regulated the proliferation,
migration and invasion of GC cells by binding to their target
miRNAs (14,31).
Accumulating evidence indicates that H19 is
overexpressed in a variety of tumors and is associated with the
proliferation and metastasis of tumors by functioning as a ceRNA
(32,33). H19 regulates the migration and
invasion of breast cancer cells by targeting miR-675 (34). H19 also controls the expression
level of FADD via miR-675, consequently promoting the proliferation
and cell cycle progression and inhibiting the apoptosis of GC cells
(31). In addition, previous
studies have suggested that miR-22-3p is a potential tumor
suppressor, which is downregulated in various human tumors,
including GC (35,36). The aim of the present study was to
further investigate the underlying mechanisms of action of H19 and
miR-22-3p as regards the initiation and development of GC.
In the present study, H19 was significantly
upregulated and miR-22-3p was downregulated in GC tissues, and the
elevated expression of H19 and reduced level of miR-22-3p in GC
were associated with a poor prognosis of patients with GC.
Furthermore, the results of present study revealed negative
correlations between the expression of H19, miR-22-3p and Snail1 in
GC samples. Further experiments confirmed the association among the
H19, miR-22-3p and Snail1. Wang et al (24) reported that H19 could regulate
HO-induced deregulation in nucleus pulposus cell senescence,
proliferation via miR-22-3p. Additionally, the present study
indicated that miR-22-3p was the potential target gene of H19, and
that the downregulation of H19 suppressed the proliferation,
invasion, migration and EMT of GC cells. Furthermore, the
overexpression of H19 promoted the proliferation, invasion,
migration and EMT of GC cells, while these effects were
substantially abrogated by the upregulation of miR-22-3p,
suggesting that H19 may promote cell growth, metastasis and EMT in
GC via miR-22-3p.
It has been well established that EMT plays critical
roles in the development of tumor metastasis (37), and Snail is one of the major
activators of EMT signaling (38).
The upregulation of Snail may be a potential biomarker of EMT
(39). The activation of the EMT
program leads to the transformation of epithelial cells into
interstitial cells (40). E-Cad,
as a member of transmembrane glycoprotein family, is an EMT marker.
Numerous EMT inducers, including α-SMA, VI and FN suppress the
transcription of the E-Cad gene. The process of EMT is induced in
tumor development with the reduced expression of E-Cad and
increased levels of α-SMA, VI and FN (41). In the present study, the
downregulation of H19 and the overexpression of miR-22-3p
suppressed cell invasion and migration by inhibiting EMT, whereas
the silencing of Snail1 enhanced these effects.
Furthermore, luciferase reporter assay revealed that
Snail1 was the promising target gene of miR-22-3p, and the
overexpression of miR-22-3p suppressed cell proliferation, invasion
and migration by targeting Snail1. The downregulation of miR-22-3p
induced the proliferation, invasion and migration of GC cells,
while these effects were abolished by the silencing of Snail1
expression. In addition, Snail1 was significantly upregulated in GC
tissues compared with the paired para-cancerous tissues and its
expression inversely correlated with that of miR-22-3p in GC
tissues. Furthermore, Zuo et al (36) reported that miR-22-3p inhibited the
growth, migration and invasion of GC cells via Snail1. In addition,
the results of the present study revealed that the knockdown of H19
in a xenograft tumor model suppressed the growth of tumors in
vivo, suggesting that H19 may upregulate Snail1 expression by
inhibiting miR-22-3p and consequently promoting the development of
GC.
In conclusion, the present study revealed that H19
is a potential oncogene that may increase Snail1 mRNA transcripts
and promote the growth and metastasis of GC cells via miR-22-3p.
Snail1 induced EMT by regulating the expression levels of α-SMA,
VI, FN and E-Cad. Although there are some limitations to this
study, in that the migratory and invasive activity of the cells
could be further confirmed by examining cell proliferation- and
invasion-related genes, these findings revealed the essential role
of H19 and its functional mechanisms the proliferation, invasion,
migration and EMT of GC cells. The findings of this study suggest
that the H19/miR-22-3p/Snail1 axis may be a promising therapeutic
target for the treatment of GC.
Funding
This study was supported by grants from the
Municipal science and technology strategic cooperation project of
Nanchong (no. 18SXHZ0284) and National Natural Science Foundation
of China (grant no. 81101827).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
SL designed the present study. LG, SL and LL
performed the experiments. LG analyzed the data. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Chongqing Medical University. Informed consent was obtained from
each patient prior to surgery. All animal experiments were
ethically approved by the Research Ethics Committee of Chongqing
Medical University (Chongqing, China).
Patient consent for publication
Not applicable.
Competing interest
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Graham DY: Helicobacter pylori update:
Gastric cancer, reliable therapy, and possible benefits.
Gastroenterology. 148:719–31. e32015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hamashima C, Shabana M, Okada K, Okamoto M
and Osaki Y: Mortality reduction from gastric cancer by endoscopic
and radio-graphic screening. Cancer Sci. 106:1744–1749. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Allemani C, Weir HK, Carreira H, Harewood
R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A,
et al CONCORD Working Group: Global surveillance of cancer survival
1995-2009: Analysis of individual data for 25,676,887 patients from
279 population-based registries in 67 countries (CONCORD-2).
Lancet. 385:977–1010. 2015. View Article : Google Scholar
|
|
4
|
Oliveira C, Pinheiro H, Figueiredo J,
Seruca R and Carneiro F: Familial gastric cancer: Genetic
susceptibility, pathology, and implications for management. Lancet
Oncol. 16:e60–e70. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garajová I, Ferracin M, Porcellini E,
Palloni A, Abbati F, Biasco G and Brandi G: Non-coding RNAs as
predictive biomarkers to current treatment in metastatic colorectal
cancer. Int J Mol Sci. 18:15472017. View Article : Google Scholar :
|
|
6
|
Pop-Bica C, Gulei D, Cojocneanu-Petric R,
Braicu C, Petrut B and Berindan-Neagoe I: Understanding the role of
non-coding RNAs in bladder cancer: From dark matter to valuable
therapeutic targets. Int J Mol Sci. 18:15142017. View Article : Google Scholar :
|
|
7
|
Hao NB, He YF, Li XQ, Wang K and Wang RL:
The role of miRNA and lncRNA in gastric cancer. Oncotarget.
8:81572–81582. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chandra S, Vimal D, Sharma D, Rai V, Gupta
SC and Chowdhuri DK: Role of miRNAs in development and disease:
Lessons learnt from small organisms. Life Sci. 185:8–14. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhan JW, Jiao DM, Wang Y, Song J, Wu JH,
Wu LJ, Chen QY and Ma SL: Integrated microRNA and gene expression
profiling reveals the crucial miRNAs in curcumin anti-lung cancer
cell invasion. Thorac Cancer. 8:461–470. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang WC, Ren JL, Wong CW, Chan SO, Waye
MM, Fu WM and Zhang JF: lncRNA-NEF antagonized epithelial to
mesenchymal transition and cancer metastasis via cis-regulating
FOXA2 and inactivating Wnt/β-catenin signaling. Oncogene.
37:1445–1456. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Y, Huang W, Sun W, Zheng B, Wang C,
Luo Z, Wang J and Yan W: lncRNA MALAT1 promotes cancer metastasis
in osteosarcoma via activation of the PI3K-Akt signaling pathway.
Cell Physiol Biochem. 51:1313–1326. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen G, Cao Y, Zhang L, Ma H, Shen C and
Zhao J: Analysis of long non-coding RNA expression profiles
identifies novel lncRNA biomarkers in the tumorigenesis and
malignant progression of gliomas. Oncotarget. 8:67744–67753.
2017.PubMed/NCBI
|
|
13
|
Cao MX, Jiang YP, Tang YL and Liang XH:
The crosstalk between lncRNA and microRNA in cancer metastasis:
Orchestrating the epithelial-mesenchymal plasticity. Oncotarget.
8:12472–12483. 2017.
|
|
14
|
Li P, Tong L, Song Y, Sun J, Shi J, Wu Z,
Diao Y, Li Y and Wang Z: Long noncoding RNA H19 participates in
metformin-mediated inhibition of gastric cancer cell invasion. J
Cell Physiol. 234:4515–4527. 2019. View Article : Google Scholar
|
|
15
|
Collette J, Le Bourhis X and Adriaenssens
E: Regulation of human breast cancer by the long non-coding RNA
H19. Int J Mol Sci. 18:23192017. View Article : Google Scholar :
|
|
16
|
Huang Z, Lei W, Hu HB, Zhang H and Zhu Y:
H19 promotes non-small-cell lung cancer (NSCLC) development through
STAT3 signaling via sponging miR-17. J Cell Physiol. 233:6768–6776.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li CF, Li YC, Wang Y and Sun LB: The
effect of lncRNA H19/miR-194-5p axis on the epithelial-mesenchymal
transition of colorectal adenocarcinoma. Cell Physiol Biochem.
50:196–213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhuang M, Gao W, Xu J, Wang P and Shu Y:
The long non-coding RNA H19-derived miR-675 modulates human gastric
cancer cell proliferation by targeting tumor suppressor RUNX1.
Biochem Biophys Res Commun. 448:315–322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jafarzadeh-Samani Z, Sohrabi S,
Shirmohammadi K, Effatpanah H, Yadegarazari R and Saidijam M:
Evaluation of miR-22 and miR-20a as diagnostic biomarkers for
gastric cancer. Linchuang Zhongliuxue Zazhi. 6:162017.
|
|
20
|
Li B, Li B, Sun H and Zhang H: The
predicted target gene validation, function, and prognosis studies
of miRNA-22 in colorectal cancer tissue. Tumour Biol.
39:10104283176922572017.PubMed/NCBI
|
|
21
|
Chen J, Wu FX, Luo HL, Liu JJ, Luo T, Bai
T, Li LQ and Fan XH: Berberine upregulates miR-22-3p to suppress
hepatocellular carcinoma cell proliferation by targeting Sp1. Am J
Transl Res. 8:4932–4941. 2016.PubMed/NCBI
|
|
22
|
Lv KT, Liu Z, Feng J, Zhao W, Hao T, Ding
WY, Chu JP and Gao LJ: miR-22-3p regulates cell proliferation and
inhibits cell apoptosis through targeting the eIF4EBP3 gene in
human cervical squamous carcinoma cells. Int J Med Sci. 15:142–152.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xin M, Qiao Z, Li J, Liu J, Song S, Zhao
X, Miao P, Tang T, Wang L, Liu W, et al: miR-22 inhibits tumor
growth and metastasis by targeting ATP citrate lyase: Evidence in
osteo-sarcoma, prostate cancer, cervical cancer and lung cancer.
Oncotarget. 7:44252–44265. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang X, Zou M, Li J, Wang B, Zhang Q, Liu
F and Lü G: lncRNA H19 targets miR-22 to modulate
H2O2-induced deregulation in nucleus pulposus
cell senescence, proliferation, and ECM synthesis through Wnt
signaling. J Cell Biochem. 119:4990–5002. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-∆ ∆ C (T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
26
|
Gooding AJ, Zhang B, Jahanbani FK, Gilmore
HL, Chang JC, Valadkhan S and Schiemann WP: The lncRNA BORG drives
breast cancer metastasis and disease recurrence. Sci Rep.
7:126982017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu H, Hu Y, Liu X, Song W, Gong P, Zhang
K, Chen Z, Zhou M, Shen X, Qian Y, et al: lncRNA TRERNA1 function
as an enhancer of SNAI1 promotes gastric cancer metastasis by
regulating epithelial-mesenchymal transition. Mol Ther Nucleic
Acids. 8:291–299. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao L, Han T, Li Y, Sun J, Zhang S, Liu
Y, Shan B, Zheng D and Shi J: The lncRNA SNHG5/miR-32 axis
regulates gastric cancer cell proliferation and migration by
targeting KLF4. FASEB J. 31:893–903. 2017. View Article : Google Scholar
|
|
29
|
Lü MH, Tang B, Zeng S, Hu CJ, Xie R, Wu
YY, Wang SM, He FT and Yang SM: Long noncoding RNA BC032469, a
novel competing endogenous RNA, upregulates hTERT expression by
sponging miR-1207-5p and promotes proliferation in gastric cancer.
Oncogene. 35:3524–3534. 2016. View Article : Google Scholar
|
|
30
|
Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen
WM, Huang MD and Shu YQ: SP1-induced upregulation of the long
noncoding RNA TINCR regulates cell proliferation and apoptosis by
affecting KLF2 mRNA stability in gastric cancer. Oncogene.
34:5648–5661. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan J, Zhang Y, She Q, Li X, Peng L, Wang
X, Liu S, Shen X, Zhang W, Dong Y, et al: Long Noncoding RNA
H19/miR-675 axis promotes gastric cancer via FADD/caspase 8/caspase
3 signaling pathway. Cell Physiol Biochem. 42:2364–2376. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu G, Xiang T, Wu QF and Wang WX: Long
noncoding RNA H19-derived miR-675 enhances proliferation and
invasion via RUNX1 in gastric cancer cells. Oncol Res. 23:99–107.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu Q, Yin J, Zeng A, Jin X, Zhang Z, Yan W
and You Y: H19 functions as a competing endogenous RNA to regulate
EMT by sponging miR-130a-3p in glioma. Cell Physiol Biochem.
50:233–245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vennin C, Spruyt N, Dahmani F, Julien S,
Bertucci F, Finetti P, Chassat T, Bourette RP, Le Bourhis X and
Adriaenssens E: H19 non coding RNA-derived miR-675 enhances
tumorigenesis and metastasis of breast cancer cells by
downregulating c-Cbl and Cbl-b. Oncotarget. 6:29209–29223. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu J, Huang Y, Wu Y, Liu F, Sun D, Wang K
and Qu H: NTRK2 is an oncogene and associated with microRNA-22
regulation in human gastric cancer cell lines. Tumour Biol.
37:15115–15123. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zuo QF, Cao LY, Yu T, Gong L, Wang LN,
Zhao YL, Xiao B and Zou QM: MicroRNA-22 inhibits tumor growth and
metastasis in gastric cancer by directly targeting MMP14 and Snail.
Cell Death Dis. 6:e20002015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhou LH, Yang YC, Zhang RY, Wang P, Pang
MH and Liang LQ: CircRNA_0023642 promotes migration and invasion of
gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci.
22:2297–2303. 2018.PubMed/NCBI
|
|
38
|
Xiang Z, Jiang DP, Xia GG, Wei ZW, Chen W,
He Y and Zhang CH: CXCL1 expression is correlated with Snail
expression and affects the prognosis of patients with gastric
cancer. Oncol Lett. 10:2458–2464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao J, Yang C, Guo S and Wu Y: GM130
regulates epithelial- to-mesenchymal transition and invasion of
gastric cancer cells via snail. Int J Clin Exp Pathol.
8:10784–10791. 2015.
|
|
40
|
Zhang J, Zhou Y and Yang Y: CCR7 pathway
induces epithelial-mesenchymal transition through up-regulation of
Snail signaling in gastric cancer. Med Oncol. 32:4672015.PubMed/NCBI
|
|
41
|
Feng Y, Sun T, Yu Y, Gao Y, Wang X and
Chen Z: MicroRNA-370 inhibits the proliferation, invasion and EMT
of gastric cancer cells by directly targeting PAQR4. J Pharmacol
Sci. 138:96–106. 2018. View Article : Google Scholar : PubMed/NCBI
|