Introduction
Breast cancer is the most common type cancer
affecting women, and has an increasing worldwide incidence
(1,2). Breast cancer is divided into 5
subtypes, according to immunohistological and genetic
characteristics (3), and the
selection of therapeutic interventions, such as surgery,
radiotherapy, hormonal therapy, chemotherapy, molecular-targeted
therapy or combinations thereof, are dependent on these subtypes
(4). However, for aggressive
breast cancer phenotypes, such as triple-negative breast cancer,
the improvement of treatment strategies and the development of
novel therapies are warranted for more effective treatment.
Recently, the importance of immune checkpoints in
the tumor microenvironment has been elucidated, and the anti-tumor
effect of immune checkpoint inhibitors (ICIs) has been reported in
various types of cancer (5).
Anti-programmed cell death 1 (PD-1)/anti-programmed death ligand 1
(PD-L1) monoclonal antibodies (mAbs) have been shown to be
effective in several types of cancer, including lung, gastric and
kidney cancer (6-8). In addition, the outcomes of several
clinical trials with ICIs for patients with breast cancer have been
reported (9-11). As such, issues regarding patient
selection and biomarkers to predict responses to ICIs in breast
cancer patients remain debatable, and urgently need to be clarified
(9-11).
It has recently been reported that conditions within
the tumor microenvironment that are suitable for immunotherapy with
anti-PD-1/anti-PD-L1 mAbs include the following: The presence of
cytotoxic T lymphocytes (CTLs), the expression of human leukocyte
antigen (HLA) class I on the tumor cells, and a significant load of
neo-antigens and the expression of PD-L1 on tumor cells (12-15).
Previously, we reported that HLA class I and PD-L1 were mainly
upregulated by interferon-γ (IFN-γ) produced by CD8-positive T
lymphocytes within the tumor microenvironment in gastric cancer
(14). In the present study, using
breast cancer cell lines and clinical samples, we investigated the
mechanisms through which PD-L1 expression is regulated within the
tumor microenvironment in breast cancer, paying particular
attention to IFN-γ regulation, and discussed the clinical
implications of anti-PD-1/anti-PD-L1 mAbs in breast cancer
patients.
Materials and methods
Breast cancer cell lines and in vitro
drug treatment
A total of 4 breast cancer cell lines, MRK-nu-1,
BT-549, MCF-7 and MDA-MB-231, were used in the current study.
MRK-nu-1 was purchased from the Japanese Collection of Research
Bioresources Cell Bank (Osaka, Japan). The BT-549, MCF-7 and
MDA-MB-231 cells were purchased from the American Type Culture
Collection (ATCC, Manassas, VA, USA). All the cell lines, which
confirmed the absence of mycoplasma, were cultured in RPMI-1640
containing L-glutamine (Invitrogen/Thermo Fisher Scientific,
Waltham, MA, USA) with 5% fetal calf serum (Invitrogen/Thermo
Fisher Scientific) and 1% penicillin-streptomycin (Sigma-Aldrich,
St. Louis, MO, USA), and were verified as authentic through a short
tandem repeat analysis. The following reagents were used in in
vitro drug treatment: IFN-γ (R&D Systems, Minneapolis, MN,
USA), the ras/mitogen-activated protein kinase (MAPK) inhibitor,
PD98059 (Cell Signaling Technology, Danvers, MA, USA), the
phosphatidylinositol-3-kinase-protein kinase B (PI3K/AKT)
inhibitor, wortmannin (Cell Signaling Technology), and lapatinib
(GlaxoSmithKline, Brentford, UK). Lapatinib has been reported to be
a combined epidermal growth factor receptor/human epidermal growth
factor receptor 2 tyrosine kinase inhibitor and can inhibit both
the MAPK and PI3K/AKT pathways (13). DMSO (Sigma-Aldrich) was used as a
vehicle and a negative control for all treatments, and the cells
were cultured and treated in 12-well plates (Thermo Fisher
Scientific).
Clinical samples
Surgically-resected specimens were obtained from 111
patients who had undergone surgery for breast cancer at the First
Department of Surgery at Yamanashi University (Yamanashi, Japan)
between 2010 and 2014. Patients with stage II or III disease who
required adjuvant chemotherapy were enrolled into the study. No
patients had received pre-operative anti-tumor therapies, such as
radiotherapy or chemotherapy. Clinical and pathological information
were retrospectively obtained by reviewing the medical records.
Tumor grade and stage were defined in accordance with the UICC TNM
classification (7th edition) (16), and the histological classification
was defined in accordance with the criteria of the Japanese Breast
Cancer Society (17th edition) (17). This study was conducted in
accordance with the Declaration of Helsinki, and was approved by
the Institutional Ethical Committee of Yamanashi University
(Reference 1622). Written informed consent was obtained from all
participants.
Flow cytometry
The cells were stained with antibodies as previously
described (13), and the following
antibodies were used for staining: Annexin V-FITC (556420; BD
Biosciences, San Jose, CA, USA) at 1:20, 7-Aminoactinomycin D
(559925; BD Biosciences) at 1:20, and anti-human CD274 (PD-L1) PE
(12-5983-42; eBioscience, Santa Clara, CA, USA) at 1:20. An
isotype-matched immunoglobulin served as a negative control, and
dead and/or apoptotic cells were excluded using Annexin V and
7-Aminoactinomycin D. Staining was detected using an LSRII flow
cytometer (BD Biosciences).
Western blot analysis
All the samples were prepared, stained with the
antibodies and visualized as previously described (18). The following primary antibodies
were purchased from Cell Signaling Technology: STAT1 (14994S) at
1:1,000, p-STAT1 (8826S) at 1:1,000, p44/42 MAPK (ERK1/2) (4695S)
at 1:1,000, phospho-p44/42 MAPK (p-ERK1/2) (4370S) at 1:2,000 and
β-actin antibodies (4970S) at 1:2,000. A horseradish
peroxidase-linked anti-rabbit IgG (7074; Cell Signaling Technology)
at 1:2,000 was used as the secondary antibody.
Gene expression microarray analysis for
drug-treated cell lines
The isolation of total RNA and microarray gene
expression analysis were performed as previously described
(14).
IHC staining
Four-micron-thick sections were deparaffinized and
rehydrated. The slides were incubated with epitope retrieval
solution (Agilent Technologies, Santa Clara, CA, USA) at varying
conditions for different proteins: CD8, pH 9.0 for 20 min at
95-99°C in a water bath; p-STAT1, pH 6.0 for 10 min at 95-99°C in a
water bath; HLA class I-A, B and C, pH 6.0 for 20 min at 121°C in
an autoclave; PD-L1, pH 6.0 for 10 min at 110°C in an autoclave.
All slides were incubated with peroxidase blocking solution
(Agilent Technologies) for 10 min. Thereafter, the slides were
incubated at 37°C for 60 min or 4°C overnight with the following
primary antibodies: CD8 (clone C8/144B, M7103; Agilent
Technologies) at 1:100; p-STAT1 (clone D3B7, 8826S; Cell Signaling
Technology) at 1:800; HLA class I-A, B and C (clone EMR8-5, AB-46;
Hokudo, Sapporo, Japan) at 1:200; and PD-L1 (clone 28-8, ab205921;
Abcam, Cambridge, UK) at 1:400. Subsequently, for CD8, HLA class I
and PD-L1, the slides were incubated with an avidinbiotinylated
enzyme complex (Vector Laboratories, Burlingame, CA, USA), whereas
the slides for p-STAT1 were incubated with a horseradish
peroxidase-coupled anti-rabbit polymer (8114; Cell Signaling
Technology), which was a ready-to-use solution. The slides were
then incubated with diaminobenzidine (Agilent Technologies) at room
temperature for 5 min and counterstained with Mayer’s hematoxylin
solution, (131-09665; Wako/ Fujifilm, Tokyo, Japan) at room
temperature for 1 min.
Assessment of IHC staining
IHC analysis was performed by two independent
observers (TN and YN), who were blinded to all of the clinical
data. For assessment of CD8, PD-L1, HLA class I and p-STAT1,
hotspot areas of tumor infiltrating lymphocytes were reviewed in 4
randomly selected independent areas in marginal tumor regions at
×400 magnification. The expression levels were evaluated based on
IHC staining in 4 independent areas. CD8 was defined as the number
of stained lymphocytes, and calculated as the mean value of the 4
areas. p-STAT1 staining was evaluated via nuclei staining in the
tumor cells and tumor infiltrating immune cells (TIICs). The HLA
class I staining intensity was evaluated by the following criteria:
Positive, evidenced by dark brown staining in >30% of membrane
staining on tumor cells; and negative, evidenced by any lesser
degree of brown staining of appreciable or non-appreciable staining
on tumor cells. An H-score of membranous PD-L1 expression on the
tumor cells was calculated as previously described (14).
Gene expression microarray analysis using
The Cancer Genome Atlas (TCGA) dataset
We evaluated the mRNA expression levels of PD-L1
(CD274), the IFN-γ signature (19)
and CD8 T effector gene signature (20) in breast invasive carcinoma tissues.
The mRNA expression z-scores of genes, analyzed using the Illumina
Genome Analyzer RNA Sequencing Version 2, were obtained from the
TCGA breast invasive carcinoma tissue dataset through cBioPortal
(http://www.cbioportal.org/) (21,22).
The IFN-γ signature was calculated as the average expression level
of 6 IFN-γ-related genes: indoleamine 2,3-dioxygenase 1
(IDO1), C-X-C motif chemokine ligand 10 (CXCL10),
CXCL9, HLA-DRA, STAT1 and IFN-γ
(19). The CD8 T effector gene
signature was also calculated as the average expression level of 7
genes: CD8A, CD8B, eomesodermin (EOMES),
granzyme A (GZMA), granzyme B (GZMB), IFN-γ
and perforin 1 (PRF1) (20).
Statistical analysis
One-way analysis of variance followed by a Tukey’s
post hoc test were performed to determine the significance of the
flow cytometry results. Associations between the H-score of PD-L1
and the expression of p-STAT1 and HLA class I, as well as between
the number of PD-L1-positive TIICs and p-STAT1 expression in TIICs,
were also assessed using the Student’s t-test. Associations between
p-STAT1 and HLA class I expression levels were assessed using the
Chi-square test. Associations between the H-score of PD-L1 and the
number of CD8-positive cells, between PD-L1 mRNA expression
z-scores and the IFN-γ signature, and between PD-L1 mRNA expression
z-scores and the CD8 T effector gene signature were assessed using
a scatter diagram and Pearson’s product moment correlation
coefficient. Analyses were performed using SPSS Statistics Package
version 25 (IBM Corp., Armonk, NY, USA) and a value of P<0.05
was considered to indicate a statistically significant
difference.
Results
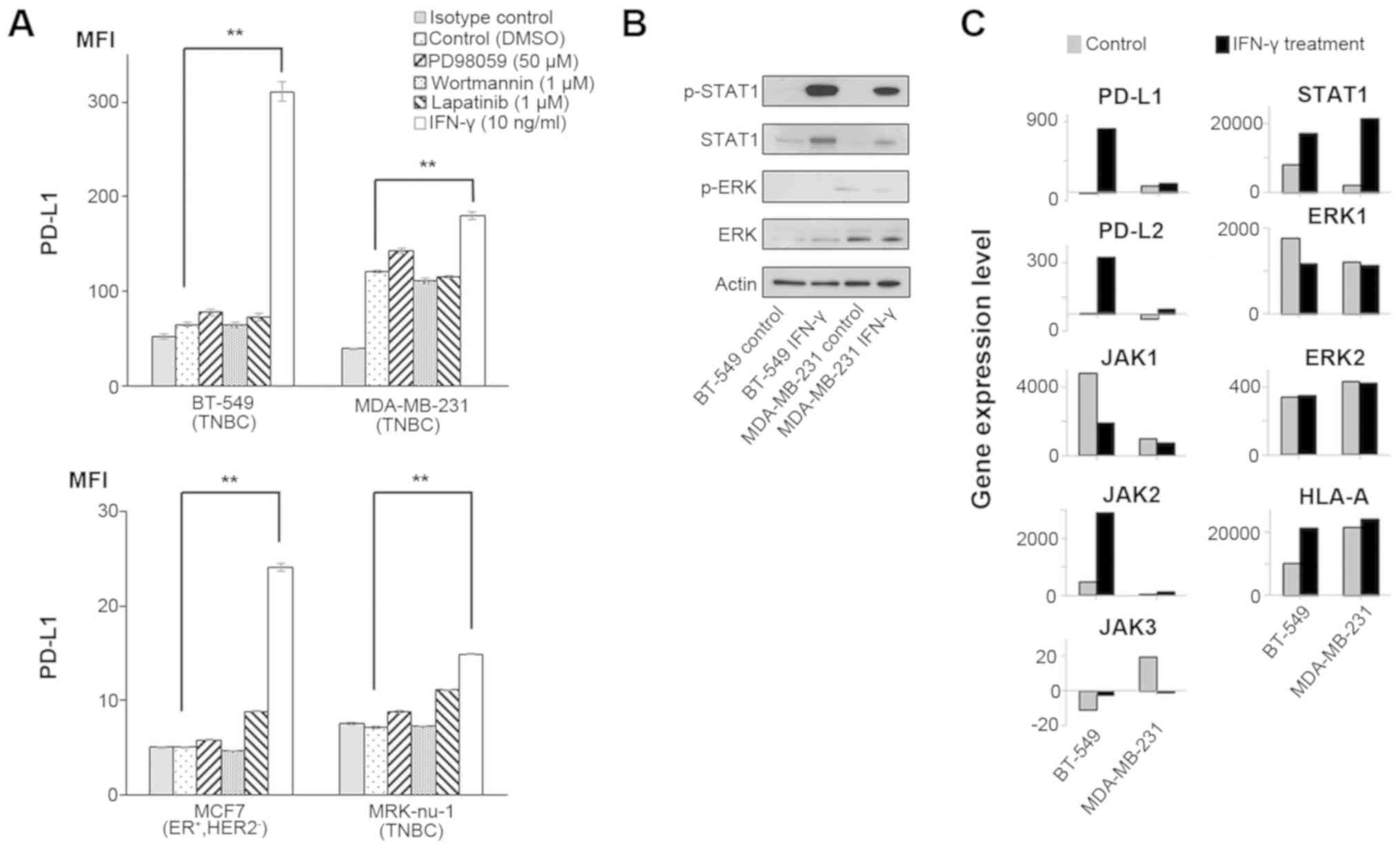
Upregulation of PD-L1 by IFN-γ in breast
cancer cell lines
We have recently reported that PD-L1 expression is
mainly regulated by IFN-γ via the JAK/STAT pathway, but not via the
MAPK and PI3K/AKT pathways in esophageal squamous cell carcinoma
cells and gastric cancer cells (14). In this study, in order to analyze
PD-L1 expression in breast cancer cells, 4 breast cancer cell lines
were treated with either IFN-γ (10 ng/ml), PD98059 (50 µM;
MAPK inhibitor), wortmannin (1 µM; PI3K-AKT inhibitor), or
lapatinib (1 µM) that can inhibit both the MAPK and PI3K/AKT
pathways for 48 h (13). We
assessed the optimal conditions of these reagents in previous
studies (13,14). As a result, PD-L1 expression was
significantly upregulated in all the tested cell lines only when
treated with IFN-γ (Fig. 1A).
Several previous studies have reported that IFN-γ
can upregulate PD-L1 expression through the JAK/STAT and MAPK
pathways in malignant tumors (23-25).
Therefore, in this study, we assessed the effects of IFN-γ on both
the JAK/STAT and MAPK pathways by western blot analysis and gene
expression microarray analysis in the two breast cancer cell lines,
BT-549 and MDA-MB-231. Western blot analysis revealed that IFN-γ
treatment increased the p-STAT1 level, but not the p-ERK level in
both cell lines (Fig. 1B). Since
we performed a gene expression microarray analysis in both cell
lines, we did not perform statistical analysis for these data. Gene
expression microarray analysis also revealed that PD-L1, PD-L2,
HLA-A and the JAK/STAT pathway (JAK2 and STAT1) were concomitantly
increased by IFN-γ in both cell lines, although there was no
increase in ERK1 and ERK2 expression levels (Fig. 1C and Table I), similar to the findings of our
previous studies (14,18). Taken together, IFN-γ could induce
the up-regulation of PD-L1 mainly through the JAK-STAT pathway in
these breast cancer cell lines.
 | Table IMicroarray analysis in untreated and
IFN-γ-treated cell lines. |
Table I
Microarray analysis in untreated and
IFN-γ-treated cell lines.
| Molecules | BT-549 cells
| MDA-MB-231 cells
|
|---|
| Untreated | IFN-γ
treatment | Untreated | IFN-γ
treatment |
|---|
| PD-L1 | −11.7 | 815.5 | 77.0 | 110.2 |
| PD-L2 | 2.8 | 331.0 | −28.5 | 27.1 |
| JAK1 | 4,755.1 | 1,898.8 | 991.6 | 725.5 |
| JAK2 | 483.1 | 2,893.8 | 54.7 | 132.3 |
| JAK3 | −11.4 | −2.5 | 19.4 | −1.3 |
| STAT1 | 7,945.8 | 17,080.4 | 2,062.7 | 21,414.4 |
| ERK1 | 1,751.8 | 1,168.5 | 1,205.7 | 1,126.4 |
| ERK2 | 337.5 | 348.6 | 427.4 | 420.7 |
| AKT1 | 3,942.4 | 3,620.0 | 3,554.2 | 3,387.5 |
| AKT2 | −6.8 | 45.7 | 47.9 | 36.2 |
| AKT3 | 30.0 | 32.3 | 38.9 | 19.5 |
| HLA-A | 10,130.7 | 21,337.2 | 21,600.1 | 24,005.1 |
| HLA-B | 4,876.9 | 18,634.7 | 16,104.7 | 34,904.7 |
| HLA-C | 543.5 | 1,476.6 | 275.3 | 999.9 |
| HLA-E | 5,622.3 | 23,216.8 | 7,399.6 | 20,547.7 |
| HLA-F | 1,024.7 | 5,301.0 | 6,858.1 | 15,147.7 |
| HLA-G | 202.8 | 790.5 | 1,438.5 | 3,585.4 |
| HLA-H | 3,053.5 | 9,808.8 | 14,781.6 | 21,520.7 |
| HLA-DPA1 | −33.2 | 444.8 | 4,823.8 | 20,231.7 |
| HLA-DPB1 | −21.4 | −25.8 | 57.4 | 592.6 |
| HLA-DPB2 | 24.4 | 2.0 | 1.9 | 25.6 |
| HLA-DQA1 | −48.9 | −21.7 | 11.9 | 1,474.5 |
| HLA-DQA2 | 15.9 | −4.0 | 2.1 | 5.8 |
| HLA-DQB1 | −3.3 | 2.4 | 86.4 | 1,452.1 |
| HLA-DQB2 | −4.1 | 31.8 | 26.0 | 16.2 |
| HLA-DRA | 114.4 | 1,398.5 | 10,456.1 | 37,182.3 |
| HLA-DRB1 | −19.9 | −2.3 | 29.4 | 78.0 |
| HLA-DRB3 | 977.7 | 951.9 | 2,582.4 | 8,289.0 |
| HLA-DRB4 | 129.8 | 127.0 | 2,279.9 | 7,899.3 |
| HLA-DRB5 | −9.3 | −17.3 | −13.3 | 3.4 |
| HLA-DRB6 | 25.3 | 66.2 | 2,246.7 | 9,342.0 |
| TAP1 | 7,741.5 | 35,696.9 | 5,333.0 | 32,228.8 |
| TAP2 | 1,172.4 | 4,463.4 | 1,381.3 | 4,233.3 |
| Tapasin | 2,660.3 | 4,533.1 | 2,592.0 | 5,062.1 |
| β-2
microglobulin | 19,602.0 | 24,741.4 | 17,045.4 | 24,486.9 |
| LMP2 | 496.3 | 2,226.0 | 532.9 | 4,351.8 |
| LMP7 | 1,303.3 | 4,801.6 | 1,593.7 | 8,038.8 |
| LMP10 | 4,038.4 | 15,703.9 | 5,087.5 | 17,694.3 |
| PA28a | 8,930.9 | 16,073.7 | 8,037.7 | 16,002.5 |
| PA28b | 20,159.3 | 34,904.7 | 10,156.0 | 25,767.6 |
| Calnexin | 1,895.6 | 1,647.5 | 2,059.7 | 2,221.7 |
| Calreticulin | 11,055.8 | 7,468.7 | 6,234.9 | 9,945.8 |
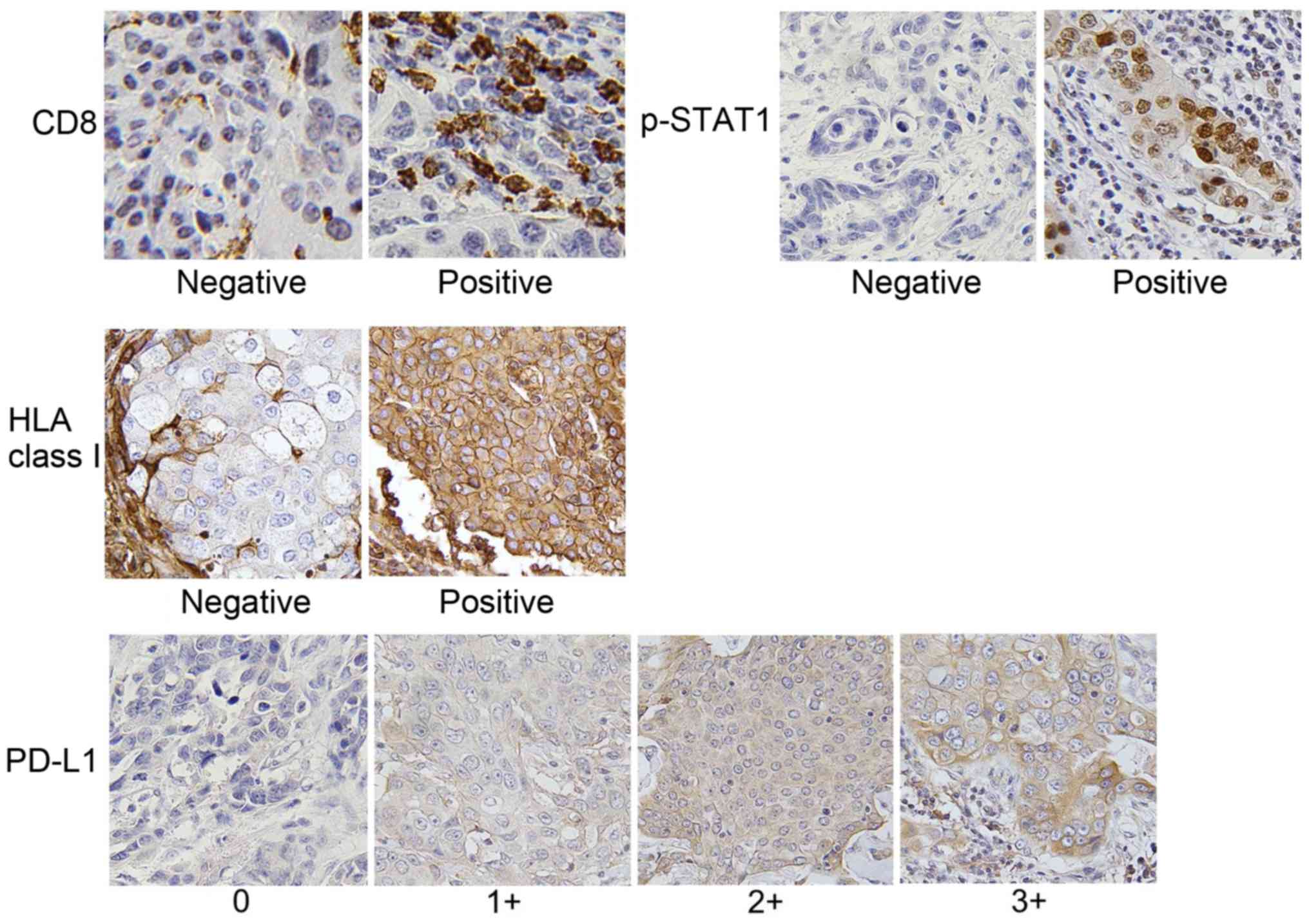
PD-L1 expression is upregulated in
p-STAT1-positive tumors
We then evaluated the expression levels of CD8,
p-STAT1, HLA class I and PD-L1 by IHC using the specimens of 111
patients with breast cancer. The clinicopathological data are
presented in Table II, and
representative IHC staining for each molecule is shown in Fig. 2. Furthermore, representative IHC
staining with serial sections for CD8, p-STAT1, HLA class I and
PD-L1 is illustrated in Fig.
3A.
 | Table IIClinical characteristics of the
patients with breast cancer (n=111). |
Table II
Clinical characteristics of the
patients with breast cancer (n=111).
|
Characteristics | No. of
patients |
|---|
| Age (years) | |
| Mean,
62.3±13.8 | |
| Range, 27-93 | |
| Primary
tumora | |
| T1 | 30 |
| T2-T4 | 81 |
| Stage
groupinga | |
| II | 88 |
| III | 23 |
| Molecular
subtypes | |
| Luminal A
likeb | 42 |
| Luminal B
like | 34 |
| Luminal-HER2 | 7 |
| HER2 | 12 |
| TNBC | 16 |
| ER | |
| Positive | 83 |
| Negative | 28 |
| PgR | |
| Positive | 72 |
| Negative | 39 |
| HER2 | |
| Positive | 20 |
| Negative | 91 |
| Ki67 | |
| <30% | 68 |
| ≤30% | 43 |
| Nuclear
gradea | |
| 1 | 24 |
| 2 | 45 |
| 3 | 42 |
| Histological
classificationc | |
| Invasive ductal
carcinoma (total) | 82 |
| Special
(total) | 29 |
| Mucinous
carcinoma | 4 |
| Medullary
carcinoma | 1 |
| Invasive lobular
carcinoma | 16 |
| Apocrine
carcinoma | 2 |
| Spindle cell
carcinoma | 2 |
| Invasive
micropapillary carcinoma | 1 |
| Carcinoma with
neuroendocrine features | 3 |
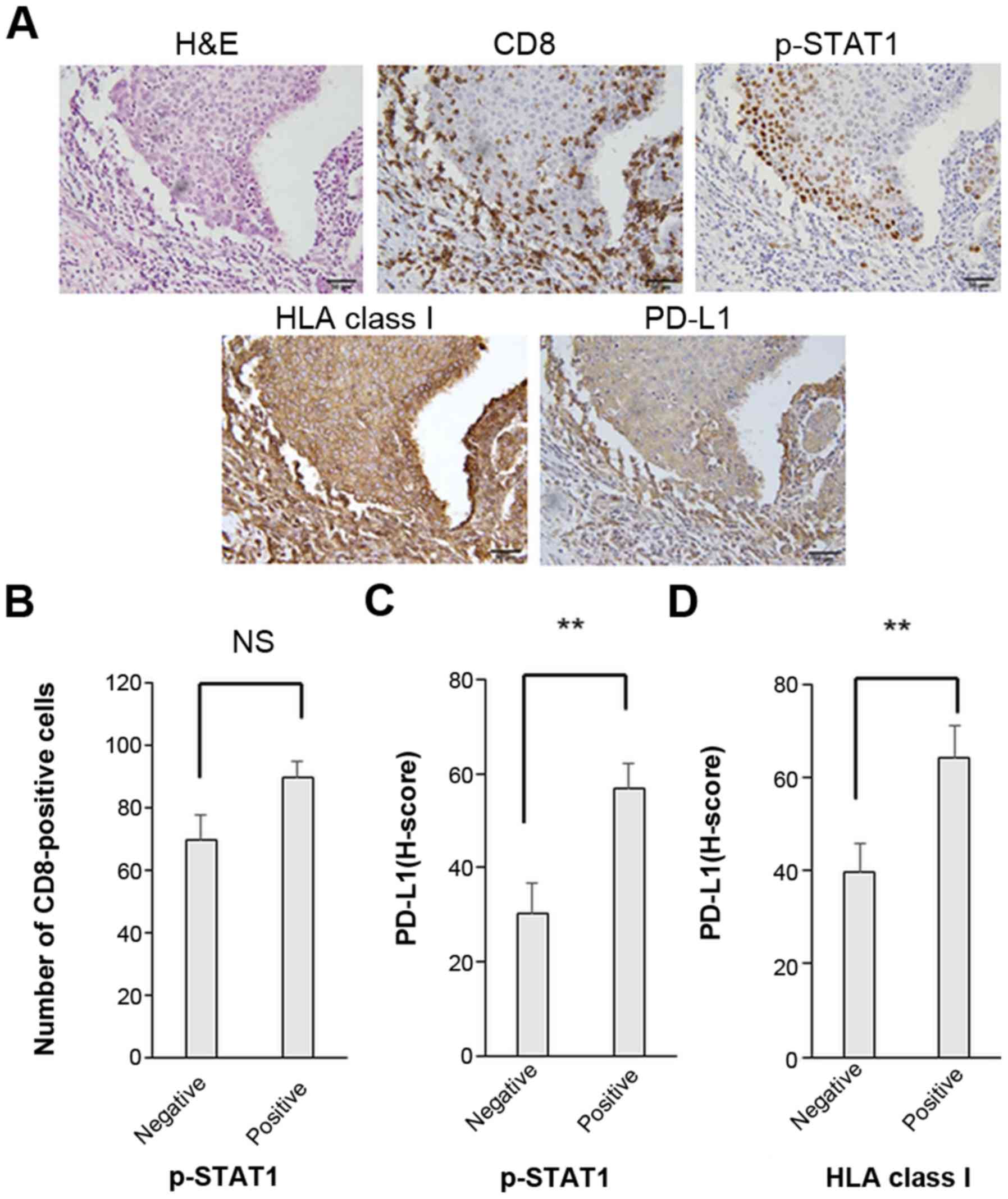
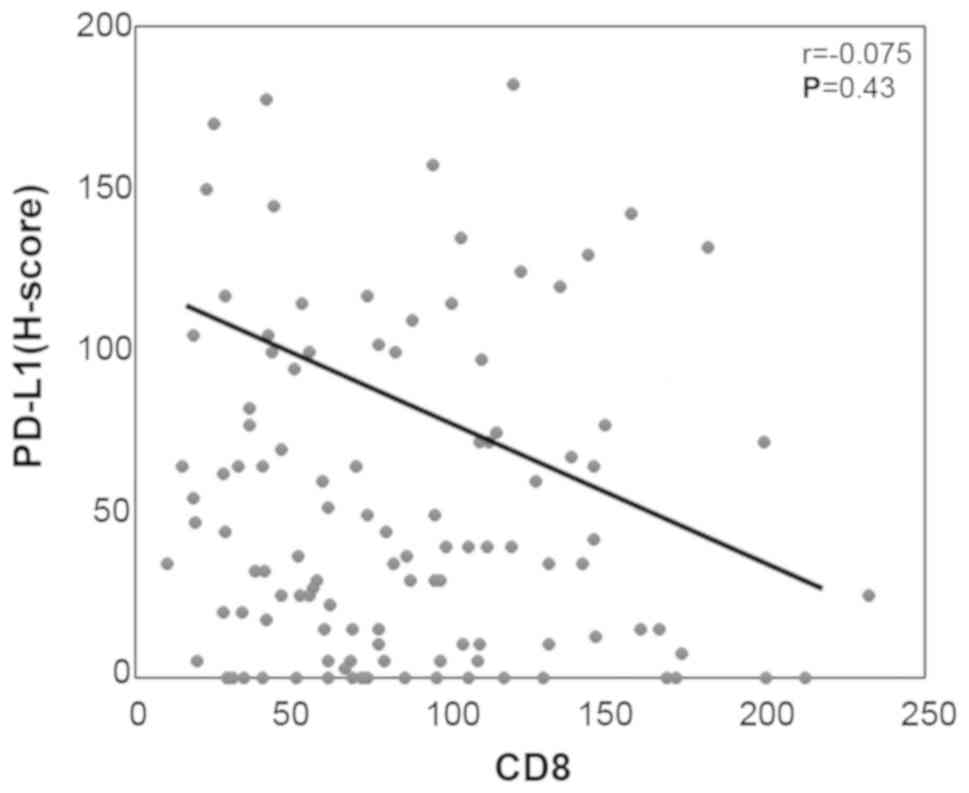
There were no significant associations or
correlations observed between the number of tumor infiltrating
CD8-positive T cells and the expression levels of p-STAT1 or PD-L1
(Figs. 3B and 4). Of note, in the p-STAT1-positive
cases, the tumor cells exhibited significantly higher expression
levels of both PD-L1 and HLA class I (P=0.003 and P=0.004,
respectively) (Fig. 3C and
Table III). Furthermore, in the
HLA class I-positive cases, PD-L1 expression was significantly
upregulated (P=0.007) (Fig. 3D).
These in vivo results suggest that the PD-L1 and HLA class I
expression levels are strongly related to the JAK/STAT pathway.
 | Table IIIAssociation between the p-STAT1
expression status and HLA class I positivity. |
Table III
Association between the p-STAT1
expression status and HLA class I positivity.
| p-STAT status | HLA class I
| Total |
|---|
| Negative | Positive |
|---|
|
p-STAT1-negative | 20 | 5 | 25 |
|
p-STAT1-positive | 41 | 45 | 86 |
| Total | 61 | 50 | 111 |
PD-L1 is upregulated in p-STAT1-positive
TIICs
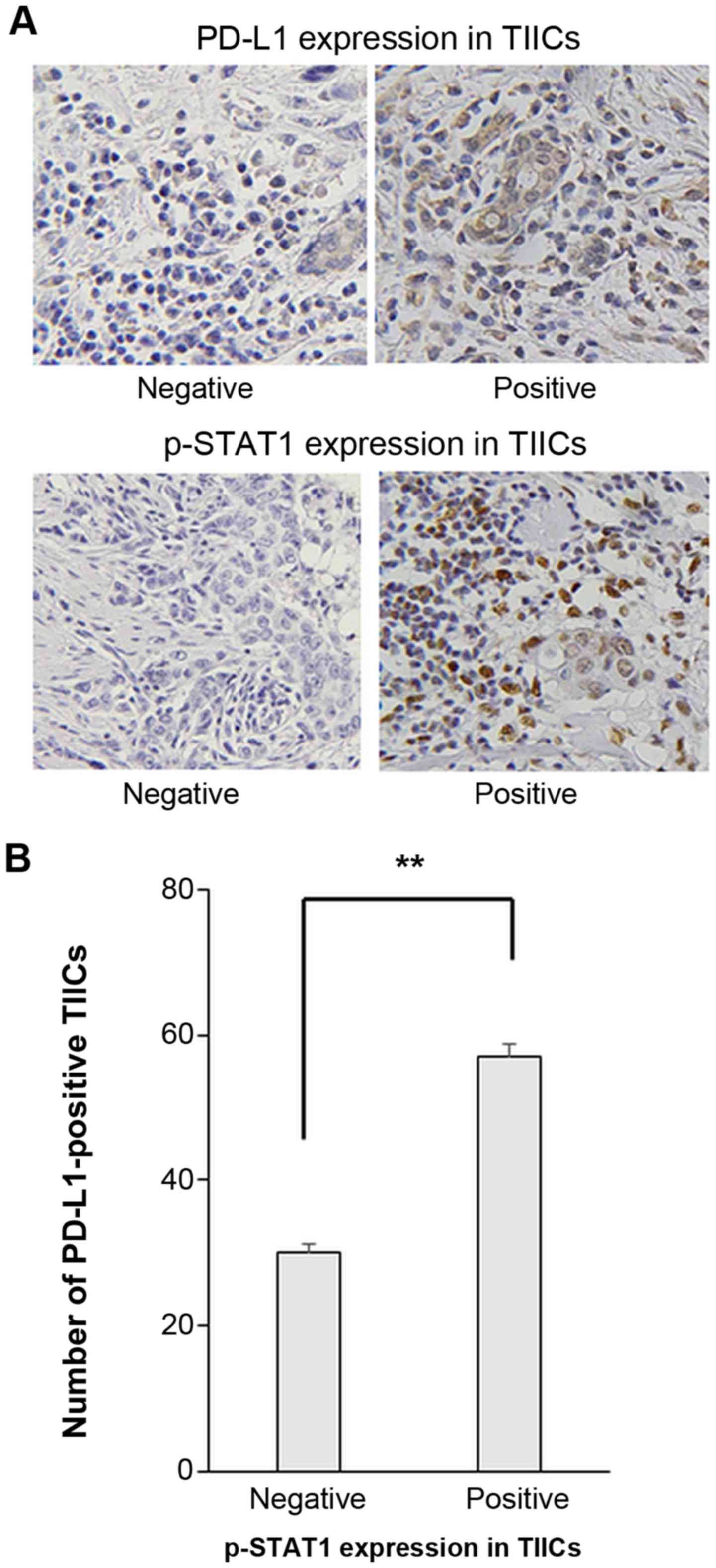
PD-L1 expression on TIICs was examined.
Representative IHC staining images of PD-L1 and p-STAT1 expression
on TIICs are presented in Fig. 5A.
The number of PD-L1-positive TIICs was significantly higher in
cases with p-STAT1 positively stained TIICs (Fig. 5B).
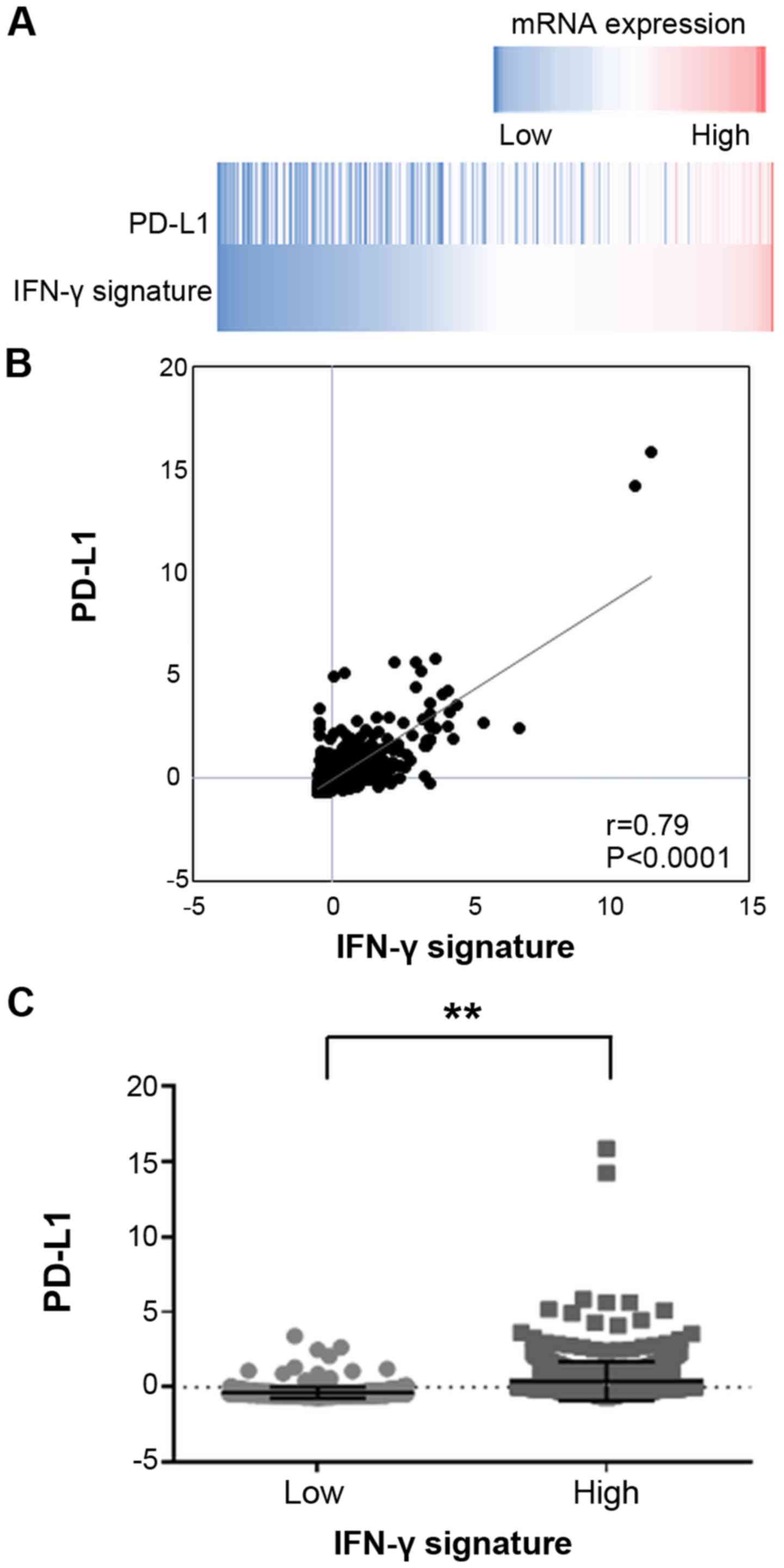
mRNA expression of PD-L1 is significantly
associated with the IFN-γ signature in breast cancer
The gene expression data of breast invasive
carcinoma tissues of 1,100 clinical cases were extracted from the
TCGA database. The data revealed a significantly positive
correlation between the mRNA expression levels of PD-L1 and the
IFN-γ signature (Fig. 6), and
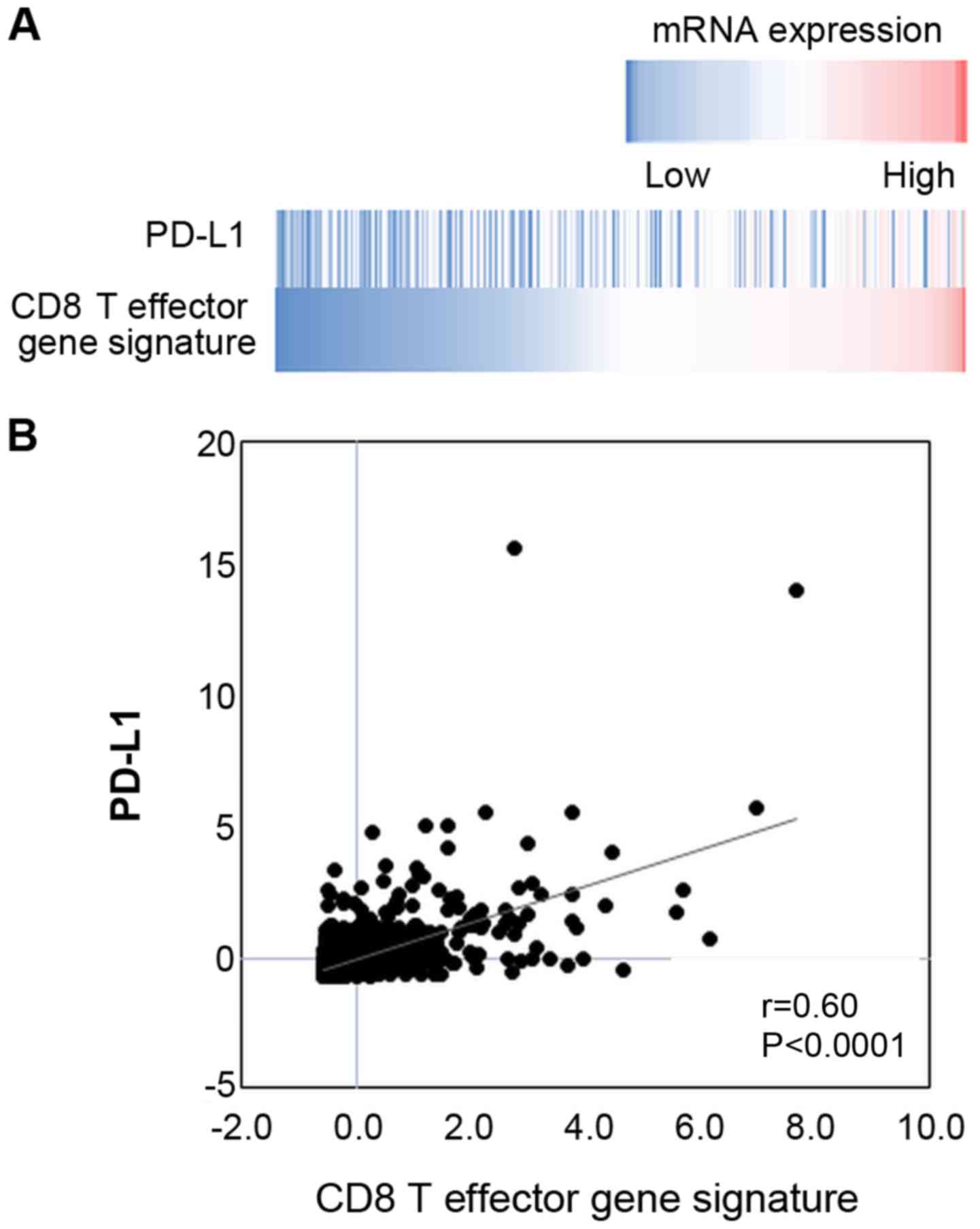
between those of PD-L1 and the CD8 T effector gene signature
(Fig. 7).
Discussion
In the present study, to the best of our knowledge,
we present novel and important findings relevant to anti-PD1/PD-L1
immunotherapy for breast cancer. First, the JAK/STAT pathway
stimulated by IFN-γ was shown to be involved in the expression of
PD-L1. Second, the expression levels of PD-L1 and HLA class I on
tumor cells were simultaneously upregulated in p-STAT1-positive
tumor cells, and PD-L1 expression was significantly upregulated in
p-STAT1-positive TIICs. Third, there was a positive correlation
between PD-L1 expression and the IFN-γ signature based on the gene
expression data from the TCGA database.
It has been reported that conditions within the
tumor microenvironment that are favorable for immunotherapy with
anti-PD-1/anti-PD-L1 mAbs include greater T cell-infiltration, a
significant load of neo-antigens, IFN-γ signature and expression of
PD-L1 on the tumor cells (12-15),
although controversy remains depending on the tumor type. These
characteristics may serve as potential biomarkers for the
prediction of the responsiveness to ICIs. Furthermore, it is
generally accepted that reliable biomarkers should be simply and
easily measured in daily clinical practice. The findings of the
present study indicate that p-STAT1 expression within the tumor
microenvironment is a potential biomarker for immunotherapy with
anti-PD-1/anti-PD-L1 mAbs in breast cancer patients. This is based
on the fact that p-STAT1 expression significantly correlates with
PD-L1 and HLA class I expression on tumor cells. Both factors are
essential for anti-PD-1 immunotherapy (26,27),
and the results of the present study indicated that both molecules
were simultaneously upregulated by IFN-γ in the breast cancer
microenvironment.
Other underlying mechanisms may also contribute to
PD-L1 regulation in breast cancer. For instance, the involvement of
the loss of phosphatase and tensin homolog (PTEN) and the ensuing
activation of the PI3K pathway have been previously reported in the
expression of PD-L1 (28). In the
present study, our in vitro experiments and TCGA database analysis
revealed that PD-L1 expression was mainly regulated by IFN-γ via
the JAK/STAT pathway in breast cancer.
To date, there have been several reports describing
a positive correlation between the number of tumor-infiltrating
CD8-positive T cells and PD-L1 expression on tumor cells (29,30).
However, in the current study, we found no such correlation between
them (Figs. 3B and 4). By contrast, there was a significant
positive correlation between the mRNA expression levels of PD-L1
and the CD8 T effector gene signature from the TCGA dataset
(Fig. 7). We speculate that the
above discrepancy may be due to tumor-infiltrating CD8-positive T
cells detected in the IHC analysis, including both activated
effector CTLs and exhausted T cells. Furthermore, activated
effector CTLs can produce IFN-γ, resulting in PD-L1 upregulation,
while exhausted CD8-positive T cells do not produce IFN-γ.
According to the study by Huang et al, the T cell
invigoration to tumor burden ratio is associated with response to
anti-PD-1 immunotherapy (31);
therefore, the presence of activated effector CD8-positive T cells
in the tumor microenvironment is essential for immunotherapy with
anti-PD-1/anti-PD-L1 mAbs.
In addition to PD-L1 expression on tumor cells
(32-34), PD-L1 expression in TIICs has also
been reported to play an important role in immunotherapy with
anti-PD-1/anti-PD-L1 mAbs (35,36).
In the present study, we observed PD-L1 expression on TIICs
(Fig. 5A) and a greater number of
TIICs expressing PD-L1 on their membrane in cases with
p-STAT1-positive TIICs (Fig. 5B).
These observations suggest that the JAK/STAT pathway via IFN-γ may
also be involved in the PD-L1 expression in TIICs. Again,
p-STAT1-positivity in TIICs can possibly lead to the detection of
PD-L1-expressing TIICs, which can then be reinvigorated with
anti-PD-1/anti-PD-L1 mAbs, suggesting that p-STAT1-positivity
within the tumor microenvironment may be a biomarker for
immunotherapy with anti-PD-1/anti-PD-L1 mAbs.
Collectively, the results of the present study
indicate that p-STAT1 positivity is a potential biomarker for
patient selection for immunotherapy with anti-PD-1/anti-PD-L1 mAbs
in breast cancer, since p-STAT1 positivity significantly reflects
PD-L1 and HLA class I expressions on tumor cells as well as PD-L1
expression on TIICs.
Funding
This study was supported by the Japan Society for
the Promotion of Science Grants-in-Aid for Scientific Research
(Grant no. 17K10540), a Clinician Scientist Award (NMRC Grant no.
NMRC/CSA/0043/2012) and Clinician Scientist-Individual Research
Grant from the National Medical Research Council of Singapore (NMRC
Grant no. NMRC/CIRG/1364/2013), and the National Research
Foundation Singapore and the Singapore Ministry of Education under
its Research Centers of Excellence initiative.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
KM, YS and KK contributed to the study conception
and design. KM, KS, LFK, and VK performed the cell line
experiments. YN, MO, AK, SI and DI contributed to the acquisition
of the patient samples. YN and TN performed and evaluated the IHC
staining. YN, TT and KM analyzed the cell line and patient data.
YN, KM, and HO analyzed the TCGA dataset. YN, KM, TT, DI and KK
drafted the manuscript. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
Written informed consent forms were obtained from
all the participants in this study. All procedures were in
accordance with the ethical standards of the responsible committee
on human experimentation at Yamanashi University (Reference 1622)
and with the Helsinki Declaration. The study was approved by the
Domain Specific Review Board of the National Healthcare Group of
Singapore (Reference 2015/00209).
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
Abbreviations:
|
CTLs
|
cytotoxic T lymphocytes
|
|
HLA
|
human leukocyte antigen
|
|
ICIs
|
immune checkpoint inhibitors
|
|
IFN-γ
|
interferon-γ
|
|
IHC
|
immunohistochemistry
|
|
mAbs
|
monoclonal antibodies
|
|
PD-1
|
programmed cell death 1
|
|
PD-L1
|
programmed death ligand 1
|
|
TCGA
|
The Cancer Genome Atlas
|
|
TIICs
|
tumor infiltrating immune cells
|
Acknowledgments
The authors would like to thank Dr Haruka Nakada, Dr
Masayuki Inoue and Dr Hiroshi Nakagomi at Yamanashi Prefectural
Central Hospital (Yamanashi, Japan), and Dr Kazuyoshi Kunitomo at
Kofu Municipal Hospital (Kofu, Japan) for discussing the
interpretation of the data.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang J, Lv H, Xue Z, Wang L and Bai Z:
Temporal Trends of Common Female Malignances on Breast, Cervical,
and Ovarian Cancer Mortality in Japan, Republic of Korea, and
Singapore: Application of the Age-Period-Cohort Model. BioMed Res
Int. 2018:53074592018.PubMed/NCBI
|
|
3
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Coates AS, Winer EP, Goldhirsch A, Gelber
RD, Gnant M, Piccart-Gebhart M, Thürlimann B and Senn HJ; Panel
Members: Tailoring therapies - improving the management of early
breast cancer: St Gallen International Expert Consensus on the
Primary Therapy of Early Breast Cancer 2015. Ann Oncol.
26:1533–1546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Topalian SL, Drake CG and Pardoll DM:
Immune checkpoint blockade: A common denominator approach to cancer
therapy. Cancer Cell. 27:450–461. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Borghaei H, Paz-Ares L, Horn L, Spigel DR,
Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al:
Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell
Lung Cancer. N Engl J Med. 373:1627–1639. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kang YK, Boku N, Satoh T, Ryu MH, Chao Y,
Kato K, Chung HC, Chen JS, Muro K, Kang WK, et al: Nivolumab in
patients with advanced gastric or gastro-oesophageal junction
cancer refractory to, or intolerant of, at least two previous
chemotherapy regimens (ONO-4538-12 ATTRACTION-2): A randomised,
double-blind, placebo-controlled, phase 3 trial. Lancet.
390:2461–2471. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Escudier B, Motzer RJ, Sharma P, Wagstaff
J, Plimack ER, Hammers HJ, Donskov F, Gurney H, Sosman JA, Zalewski
PG, et al: Treatment Beyond Progression in Patients with Advanced
Renal Cell Carcinoma Treated with Nivolumab in CheckMate 025. Eur
Urol. 72:368–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nanda R, Chow LQ, Dees EC, Berger R, Gupta
S, Geva R, Pusztai L, Pathiraja K, Aktan G, Cheng JD, et al:
Pembrolizumab in Patients With Advanced Triple-Negative Breast
Cancer: Phase Ib KEYNOTE-012 Study. J Clin Oncol. 34:2460–2467.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Emens LA, Braiteh FS, Cassier P, Delord
JP, Eder JP, Fasso M, Xiao Y, Wang Y, Molinero L, Chen DS, et al:
Abstract 2859: Inhibition of PD-L1 by MPDL3280A leads to clinical
activity in patients with metastatic triple-negative breast cancer
(TNBC). Cancer Res. 75(Suppl 15): 28592015. View Article : Google Scholar
|
|
11
|
Schmid P, Cruz C, Braiteh FS, Eder JP,
Tolaney S, Kuter I, Nanda R, Chung C, Cassier P, Delord JP, et al:
Abstract 2986: Atezolizumab in metastatic TNBC (mTNBC): Long-term
clinical outcomes and biomarker analyses. Cancer Res. 77(Suppl 13):
29862017. View Article : Google Scholar
|
|
12
|
Mizukami Y, Kono K, Maruyama T, Watanabe
M, Kawaguchi Y, Kamimura K and Fujii H: Downregulation of HLA Class
I molecules in the tumour is associated with a poor prognosis in
patients with oesophageal squamous cell carcinoma. Br J Cancer.
99:1462–1467. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mimura K, Shiraishi K, Mueller A, Izawa S,
Kua LF, So J, Yong WP, Fujii H, Seliger B, Kiessling R and Kono K:
The MAPK pathway is a predominant regulator of HLA-A expression in
esophageal and gastric cancer. J Immunol. 191:6261–6272. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mimura K, Teh JL, Okayama H, Shiraishi K,
Kua LF, Koh V, Smoot DT, Ashktorab H, Oike T, Suzuki Y, et al:
PD-L1 expression is mainly regulated by interferon gamma associated
with JAK-STAT pathway in gastric cancer. Cancer Sci. 109:43–53.
2018. View Article : Google Scholar
|
|
15
|
Schumacher TN and Schreiber RD:
Neoantigens in cancer immunotherapy. Science. 348:69–74. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sobin LH, Gospodarowicz MK and Wittekind
C: International Union against Cancer. TNM Classification of
Malignant Tumours Wiley-Blackwell; West Sussex, UK; Hoboken, NJ:
2010
|
|
17
|
Japanese Breast Cancer Society: General
Rules for Clinical and Pathological Recording of Breast Cancer.
17th edition. Kanehara & Co. Ltd.; Tokyo: 2012
|
|
18
|
Mimura K, Kua LF, Shiraishi K, Kee Siang
L, Shabbir A, Komachi M, Suzuki Y, Nakano T, Yong WP, So J, et al:
Inhibition of mitogen-activated protein kinase pathway can induce
upregulation of human leukocyte antigen class I without
PD-L1-upregulation in contrast to interferon-γ treatment. Cancer
Sci. 105:1236–1244. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ayers M, Lunceford J, Nebozhyn M, Murphy
E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran
V, et al: IFN-γ-related mRNA profile predicts clinical response to
PD-1 blockade. J Clin Invest. 127:2930–2940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wallin JJ, Bendell JC, Funke R, Sznol M,
Korski K, Jones S, Hernandez G, Mier J, He X, Hodi FS, et al:
Atezolizumab in combination with bevacizumab enhances
antigen-specific T-cell migration in metastatic renal cell
carcinoma. Nat Commun. 7:126242016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBio-Portal. Sci Signal. 6:pl12013. View Article : Google Scholar
|
|
22
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun D and Ding A: MyD88-mediated
stabilization of interferon-gamma-induced cytokine and chemokine
mRNA. Nat Immunol. 7:375–381. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu J, Hamrouni A, Wolowiec D, Coiteux V,
Kuliczkowski K, Hetuin D, Saudemont A and Quesnel B: Plasma cells
from multiple myeloma patients express B7-H1 (PD-L1) and increase
expression after stimulation with IFN-{gamma} and TLR ligands via a
MyD88-, TRAF6-, and MEK-dependent pathway. Blood. 110:296–304.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamamoto R, Nishikori M, Tashima M, Sakai
T, Ichinohe T, Takaori-Kondo A, Ohmori K and Uchiyama T: B7-H1
expression is regulated by MEK/ERK signaling pathway in anaplastic
large cell lymphoma and Hodgkin lymphoma. Cancer Sci.
100:2093–2100. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Townsend A and Bodmer H: Antigen
recognition by class I-restricted T lymphocytes. Annu Rev Immunol.
7:601–624. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Šmahel M: PD-1/PD-L1 Blockade Therapy for
Tumors with Downregulated MHC Class I Expression. Int J Mol Sci.
18:182017. View Article : Google Scholar
|
|
28
|
Mittendorf EA, Philips AV, Meric-Bernstam
F, Qiao N, Wu Y, Harrington S, Su X, Wang Y, Gonzalez-Angulo AM,
Akcakanat A, et al: PD-L1 expression in triple-negative breast
cancer. Cancer Immunol Res. 2:361–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fourcade J, Sun Z, Benallaoua M, Guillaume
P, Luescher IF, Sander C, Kirkwood JM, Kuchroo V and Zarour HM:
Upregulation of Tim-3 and PD-1 expression is associated with tumor
antigen-specific CD8+ T cell dysfunction in melanoma patients. J
Exp Med. 207:2175–2186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu B, Arakawa Y, Yokogawa R, Tokunaga S,
Terada Y, Murata D, Matsui Y, Fujimoto KI, Fukui N, Tanji M, et al:
PD-1/PD-L1 expression in a series of intracranial germinoma and its
association with Foxp3+ and CD8+ infiltrating lymphocytes. PLoS
One. 13:e01945942018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang AC, Postow MA, Orlowski RJ, Mick R,
Bengsch B, Manne S, Xu W, Harmon S, Giles JR, Wenz B, et al: T-cell
invigoration to tumour burden ratio associated with anti-PD-1
response. Nature. 545:60–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang ZQ, Milne K, Derocher H, Webb JR,
Nelson BH and Watson PH: PD-L1 and intratumoral immune response in
breast cancer. Oncotarget. 8:51641–51651. 2017.PubMed/NCBI
|
|
33
|
Muenst S, Schaerli AR, Gao F, Däster S,
Trella E, Droeser RA, Muraro MG, Zajac P, Zanetti R, Gillanders WE,
et al: Expression of programmed death ligand 1 (PD-L1) is
associated with poor prognosis in human breast cancer. Breast
Cancer Res Treat. 146:15–24. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mori H, Kubo M, Yamaguchi R, Nishimura R,
Osako T, Arima N, Okumura Y, Okido M, Yamada M, Kai M, et al: The
combination of PD-L1 expression and decreased tumor-infiltrating
lymphocytes is associated with a poor prognosis in triple-negative
breast cancer. Oncotarget. 8:15584–15592. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yagi T, Baba Y, Ishimoto T, Iwatsuki M,
Miyamoto Y, Yoshida N, Watanabe M and Baba H: PD-L1 Expression,
Tumor-infiltrating Lymphocytes, and Clinical Outcome in Patients
With Surgically Resected Esophageal Cancer. Ann Surg. 269:471–478.
2019. View Article : Google Scholar
|
|
36
|
Kawazoe A, Kuwata T, Kuboki Y, Shitara K,
Nagatsuma AK, Aizawa M, Yoshino T, Doi T, Ohtsu A and Ochiai A:
Clinicopathological features of programmed death ligand 1
expression with tumor-infiltrating lymphocyte, mismatch repair, and
Epstein-Barr virus status in a large cohort of gastric cancer
patients. Gastric Cancer. 20:407–415. 2017. View Article : Google Scholar
|