Introduction
Generally, colorectal cancer (CRC) is the third most
common cause of morbidity worldwide and the fourth leading cause of
cancer-associated mortality (1).
Globally, CRC is the third most common type of cancer among men
(746,000 cases, 10.0% of total cancer cases) and the second among
women (614,000 cases, 9.2% of total cancer cases) (2,3). The
morbidity and mortality rates of CRC exhibit notable geographical
variation. CRC is also a main cause of cancer-associated mortality
among developed countries. In 2012, 1,4000,000 patients succumbed
to mortality due to cancer, of which 693,900 cases were associated
with CRC (4); however, the
morbidity and mortality rates of CRC were markedly lower among less
developed countries. In the USA, ~132,700 individuals were
diagnosed with CRC in 2015 and ~49,700 patients with CRC succumbed
in 2015 (5). CRC is a
multifactorial disease with a complex multistep process of
development associated with genetic background, environmental
factors and numerous genetic alterations (6). The risk factors of CRC include a high
fat diet, high adiposity index, smoking, alcoholism, stress,
inflammatory bowel diseases, and the predisposition to polyp
formation and hereditary components, which comprise hereditary
nonpolyposis CRC (6-10).
It is necessary to understand the molecular
mechanisms underlying the tumorigenesis of CRC, which may assist in
CRC characterization, the prognosis and stratification of patients,
and the development of personalized treatment (11,12).
The in vitro culture of tumor cells is a method used to
determine the biological characteristics and pathogenesis of a
tumor, and is important for tissue culture. At present, the
molecular mechanisms underlying the pathogenesis, recurrence and
metastasis of CRC remain to be elucidated; therefore, the
establishment of human CRC cell lines in vitro may provide
insight into the biological characteristics and development of CRC.
Additionally, the establishment of CRC cell lines provides a
convenient and relatively inexpensive cell model for anticancer
drug screening. China is a region with high CRC-associated
morbidity rates; however, few studies have established CRC cell
lines. CRC cell lines derived from China, including THC8908,
SIC-8101, HR-8348, Hce8693, HRC-99, DXH-1 and LST-R, have been
reported (13-19); the majority of these cell lines are
unsuitable for use due to cell contamination or improper storage.
Furthermore, other cells lines have not been recorded by
internationally recognized institutions for the standardized
identification of cell lines. At present, only one CRC cell line,
DXH-1, has been reported on the National Experimental Cell Resource
Sharing Platform. Therefore, it is important to establish CRC cell
lines with a Chinese genetic background to investigate the
biological characteristics and pathogenesis of CRC in China.
In the present study, the establishment and
characterization of the cc-006cpm8 cell line were conducted. The
cell line was analyzed with respect to growth properties, cellular
ultrastructure, neoplastic behavior in athymic BALB/c nude mice,
karyotype and the expression of tumor-associated antigens; the cell
line underwent authentication by short tandem repeat (STR)
profiling. Furthermore, the cell cultures were negative for
bacteria, fungi and mycoplasma. Exome capture DNA sequencing was
performed using the cc-006cpm8 cell line to efficiently identify
coding variants. The results revealed 20 hypermutated exons
comprising single nucleotide polymorphisms (SNPs), and insertion
and deletions (InDels) in this cell line, including single
nucleotide variants (SNVs) of mucin (MUC)19,
MUC16, MUC12, filaggrin (FLG) and AHNAK
nucleoprotein 2 (AHNAK2), and InDels of β defensin-126
(DEFB126), microRNA-3665 (MIR3665), WNK lysine
deficient protein kinase 1 and SLAIN motif-containing 1
(SLAIN1). Additionally, commonly mutated genes in CRC and
exon mutations of adenomatous polyposis coli (APC), tumor
protein p53 (TP53), Drosophila mothers against
decapentaplegic (SMAD)4,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α
(PIK3CA) and Kirsten rat sarcoma (KRAS), in addition
to genes of the DNA mismatch-repair (MMR) pathway were determined
(20).
Materials and methods
Tumor origin
Tumor specimens were derived from an 89-year-old
Chinese woman treated in the absence of chemotherapy and
radiotherapy, and had no family history. The patient had
moderately/poorly differentiated right colon invasive ulcerative
adenocarcinoma with liver metastasis as stage 4bN1bM1a, IV A. The
right hemicolectomy of this patient was performed at the Department
for Surgical Oncology of Nanjing First Hospital affiliated to
Nanjing Medical University (Nanjing, China) in March 16, 2015.
Following the surgical procedure, specimens were immediately
obtained from primary tumor tissues, and prepared for cell culture
and pathological analysis. Informed consent was obtained from the
patient and the experiments were approved by the Ethics Committee
of Nanjing First Hospital of Nanjing Medical University in
accordance with generally accepted guidelines for the use of human
material.
Tissue and cell culture
The tumor tissues were immersed in Dulbecco's PBS
(DPBS; Gibco; Thermo Fisher Scientific, Inc.) with 1,000 U/ml
penicillin G, 1,000 µg/ml streptomycin and 2.5 µg/ml
amphotericin B (DPBS with 10X Antibiotic-Antimycotic).
Subsequently, the tumor tissues were washed twice with transport
medium and immersed in DPBS with 10X Antibiotic-Antimycotic for 1-2
h at 4°C. Following two washes with transport medium, the tissues
were ground. The samples were plated in a sterile 6-cm culture dish
and cultured in RPMI-1640 supplemented with 10% heat inactivated
fetal bovine serum (both from Gibco; Thermo Fisher Scientific,
Inc.) and 1X Antibiotic-Antimycotic (RPMI-1640 complete medium),
and maintained in humidified incubators at 37°C in an atmosphere of
5% CO2 and 95% air. After 2 weeks, the initial cell
passage was performed when increased tumor cell growth was
observed. Differential trypsinization was used to remove
fibroblasts; a pure tumor cell population was obtained comprising
the cc-006cp cell line (21).
Subsequently, the monoclonal cell subline derived
from the cc-006cp cell line was plated onto semisolid medium. A
total of 103 viable cc-006cp cells were plated in 3 ml
of RPMI-1640 complete medium containing 0.3% agarose layered over
RPMI-1640 complete medium containing 0.5% agarose; the cells were
incubated at 37°C with 5% CO2. Colony formation was
observed after 12-15 days, following which single colonies were
selected and cultured with RPMI-1640 complete medium in an
incubator at 37°C with 5% CO2. Half of the media was
replaced every 2-3 days depending on the pH. Using this method, a
cell line of high purity and tumorigenicity was successfully
derived from human moderately/poorly differentiated colon cancer;
the cell line was stably passaged and was designated as cc-006cpm8,
which is one of eight sublines of the cc-006cp cell line.
Transmission electron microscopy
For electron microscopy, 1-5×105 of the
cc-006cpm8 cells plated in a 35-mm dish were fixed with 2.5%
glutaraldehyde in 0.1 mol/l phosphate buffer for 1 h at room
temperature, and post-fixed with 1% OsO4 in the
aforementioned buffer at 0°C for 30 min. The cells were rinsed with
the same buffer, dehydrated with ethanol, immersed twice in
absolute propylene oxide and embedded in Quetol 812. Sections were
cut at a thickness of 100 nm with a diamond knife and mounted onto
grids. Following staining with uranyl acetate and lead citrate, the
cells were observed using a JEM-1011 electron microscope at 80
kV.
Chromosome karyotype analysis
The cc-006cpm8 cells were treated with 0.075 mol/l
KCl solution for 20 min at 37°C, and then fixed with a solution of
methanol: Acetic acid (3:1) solution at 0°C for 10 min. Following
fixation, the slides were place in trypsin solution (0.02% trypsin
+3% Tris solution) in a water bath at 37°C for 3 min. The cells
were then stained with 10% Giemsa solution for 4 min at room
temperature. The karyotypes of 20 randomly selected cells were
analyzed using the GSL 120 Imaging System equipped with Zeiss.
H&E staining and Wright-Giemsa
staining
H&E staining
The patient's tumor tissues and the xenograft tumors
formed by cc-006cpm8 cells were fixed in 10% buffered formalin
(Thermo Fisher Scientific, Inc.) and processed for permanent
paraffin embedding on a Leica ASP 300 tissue processor (Leica
Microsystems); 5-µm paraffin sections were stained with
hematoxylin and eosin by using a Leica Autostainer XL. These
reagents of H&E staining (code no. 3801698) are manufatured by
Leica Biosystems Inc. The protocols of modified progressive H&E
stain are presented in Table
SI.
Wright-Giemsa staining
The chamber slides of the cc-006cpm8 cells were
washed with PBS (pH 6.5), and then placed 3-5 drops of
Wright-Giemsa Stain (code no. CO 135; Shanghai BOYAO Biotechnology
Inc.) upon the slides for 1-2 min at room temperature, and added
6-10 drops of PBS (pH 6.8) to mix Wright-Giemasa stain for 6-8 min,
and the stained slides were then rinsed with water and quickly
air-dried.
STR analysis
The cc-006cpm8 cell line, together with the positive
and negative controls, were amplified using the GenePrint 10 system
(Promega Corporation). The amplified products were then processed
using the ABI3730×l Genetic Analyzer (Applied Biosystems; Thermo
Fisher Scientific, Inc.). Data were analyzed using GeneMapper4.0
software (Applied Biosystems; Thermo Fisher Scientific, Inc.) and
then compared with the American Type Culture Collection (ATCC),
Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) or
Japanese Collection of Research Bioresources (JCRB) databases for
reference matching. The STR assay was conducted at the China Center
for Type Culture Collection (CCTCC).
In vitro growth properties of
cc-006cpm8
For growth curve and population doubling time (PDT)
analyses, 1×103 cells/well were plated onto 24-well
culture plates (Corning Incorporated) and cultured for 8 days in a
CO2 incubator. The cells were counted every hour in five
wells using a JuLi Stage real-time cell counter. Alterations in
cell confluence over time were presented as a growth curve. The PDT
of cells in the logarithmic phase was calculated as follows: PDT =
(t-t0) [lg2/(lgNt-lgN0)] (N0 and
Nt represent cell numbers at t0 and t separately).
To determine the plating efficiency, the cc-006cpm8
cells were plated at various densities, ranging between
1×102 and 5×102 cells in three replicate
60-mm dishes (Corning® Plastic Culture Dishes; Corning
Incorporated). Colonies comprising >50 cells were counted 9 days
later following staining with 0.5% crystal violet for 5 min at
25°C, and then counted under BX-4 Stereomicroscope (Shanghai
Optical Instrument Factory).
cc-006cpm8 single cell suspensions (103
cells in three replicate 60-mm dishes) with 0.3% agarose dissolved
in RPMI-1640 complete medium were placed in tissue culture dishes
(Corning® Plastic Culture Dishes; Corning Incorporated)
on a layer of 0.6% agarose (in the same medium), which had
previously set. The dishes were then transferred to a 37°C
incubator with 5% CO2. After 2 weeks, colonies
comprising >50 cells were counted and the number was divided by
1,000 to determine the colony-forming efficiency. Experiments were
conducted in triplicate.
Analysis of tumorigenicity using nude
mice
Tumorigenicity was investigated by inoculating
5×106 cells/site cc-006cpm8 cells subcutaneously into
the shoulder and contralateral hip of five BALB/c male nude mice,
which were 4-6 weeks old and weighing 20-25 g, purchased from the
Comparative Medical Center of Yangzhou University (Yangzhou,
China). All mice were maintained in a HEPA-filtered environment at
24-25°C and humidity was maintained at 50-60%. Observations were
conducted at least twice a week via caliper measurements to analyze
progressive tumor growth. The study was performed in the Laboratory
Animal Center of Southeast University (Nanjing, China), and
followed the Constitution of the Laboratory Animal Ethics Committee
of Southeast University. The animal experimental ethical inspection
form was approved by the Animal Care and Welfare Committee of
Southeast University.
Mycoplasma assay
Mycoplasma contamination was assayed using the
Myco-PCR-Mix PCR Detection kit (Zhong Qiao). Cell lines should be
pre-cultured in the absence of mycoplasma active antibiotics for
several days to maximize test sensitivity. Samples should be
derived from cultures that are at 90-100% confluence. Cell culture
supernatants can be tested directly without prior preparation.
According to the reaction ratio, the sample/Positive Control
template/Negative Control template is added respectively to
Myco-PCR-Mix in the PCR tubes. The thermal cycler program was as
follows: the reaction is started with an initial denaturation for 5
min at 95°C; followed by 30 sec at 95°C, 45 sec at 55°C, 45 sec at
72°C for 36 cycles; and cooling down to 10°C. The PCR products were
then electrophoresed on 1.5% agar gel. The results of
electrophoresis can be observed directly under ultraviolet light
after staining with ethidium bromide (EB). If there is a 250 bp
band of sample same to positive control band size, it is indicated
that the sample was infected by mycoplasma. This method has been
reported to have high sensitivity in the detection of mycoplasma in
cell cultures and other cell culture-derived biologicals.
Immunohistochemical (IHC) analysis
The cell line and the original colon cancer tissues
were analyzed by immunohistochemistry. The cc-006cpm8 cells were
collected by centrifugation at 220 × g for 3 min; the pellets were
fixed in 10% formaldehyde, embedded in paraffin and then sliced
into 6-µm sections. The sections were stained using the
EnVision system. In the original colon cancer tissues, IHC staining
was performed on formalin-fixed paraffin-embedded tissues. The
paraffin sections were dewaxed in xylene and rehydrated with
distilled water. The slides were subsequently incubated with the
following antibodies for 60 min at room temperature: Anti-vimentin
(cat. no. MA3-745, Invitrogen; Thermo Fisher Scientific, Inc.;
1:20), anti-pan cytokeratin (cat. no. ab215838; 1:400),
anti-epithelial membrane antigen (EMA; cat. no. ab140355; 1:150),
anti-carcinoembryonic antigen (CEA; cat. no. ab4451; 1:100) (all
from Abcam), anti-p53 (cat. no. sc-47698, Santa Cruz Biotechnology,
Inc.; 1:250) and anti-Ki-67 (cat. no. ab8191; 1:50), anti-mucin 2
(cat. no. ab76774; 1:50), anti-caudal type homeobox transcription
factor 2 (CDX2; cat. no. ab70458; 1:1,000), anti-mutL homolog 1
(MLH1; cat. no. ab92312; 1:500), anti-mutS protein homolog (MSH2;
cat. no ab70270; 1:2,500), anti-mutS homolog 6 (MSH6; cat. no.
ab92471; 1:500), anti-postmeiotic segregation 2 (PMS2; cat. no.
ab110638; 1:200), anti-BRAF V600E (cat. no. ab200535; 5
µg/ml) (all from Abcam), anti-α-fetoprotein (AFP; cat. no.
PA5-11480, Invitrogen; Thermo Fisher Scientific, Inc.; 1:1,0000),
anti-glypican-3 (cat. no. ab207080; 1:500),
anti-hepatocyte-specific antigen (cat. no. ab75677; 1/1) (both from
Abcam). The reaction was performed using Autostainer Link 48 (Dako;
Agilent Technologies, Inc.). Secondary antibodies (Dako EnVision+,
HRP rab/mouse, K5007; Dako; Agilent Technologies, Inc.) were
incubated for 20 min and 3,3′-diaminobenzidene substrate chromogen
counterstaining for 4 min at room temperature.
For the cell line, the percentage of stained
positive cells was scored via a semiquantitative method according
to the following scheme: 0 (<5% of tumor cells); 1+ (5-25% of
tumor cells); 2+ (26-50% of tumor cells); 3+ (51-75% of tumor
cells) and 4+ (>75% of tumor cells). For the tumor tissues, the
slides for IHC were observed independently of the diagnosis
established from the HES-stained slides. For the slides of colon
cancer tissue, the positive index of IHC was determined in regions
of high proliferation. Automated detection was conducted using a
Dako AutoStainer Link 48. Normal expression was defined as nuclear
staining within tumor cells, using infiltrating lymphocytes as the
positive internal control. Negative protein expression was defined
as the complete absence of staining within tumor cells compared
with the positive labeling of internal non-neoplastic tissues.
Flow cytometric analysis
The cc-006cpm8 cells were cultured in a 100-mm plate
and then separated by trypsin-EDTA when the cells attained 90%
confluence. Subsequently, the cells were centrifuged at 300 × g for
10 min at room temperature and 107 cells/100 µl
were suspended (PBS pH 7.2, 2 mM EDTA and 0.5% bovine serum
albumin; Sigma-Aldrich). The cells were then divided into two
groups; one group was stained with human cluster of differentiation
44-fluorescein isothiocyanate (CD44-FITC; cat. no. 130-098-210) and
isotype control antibody mouse IgG1-FITC (cat. no. 130-098-847)
(both from Miltenyi Biotec) at 4°C for 10 min. The other group of
cells was stained with human EpCAM-phycoerythrin (CD326-PE; cat.
no. 130-098-115) and isotype control antibody mouse IgG1-PE (cat.
no. 130-098-845) (both from Miltenyi Biotec) under the same
conditions. Analysis was performed with a BD FACSCanto II flow
cytometer (BD Biosciences).
Detection of genetic mutations in
cc-006cpm8 cells
Exome capture sequencing was performed using the
Human Whole-Exon Capture (xGEN® Exome Research Panel)
kit of Integrated DNA Technologies and the Illumina Hiseq
sequencing platform. The High-Throughput Information Acquisition
Technique was used to collect the captured DNA fragments. Each
sample produced 10 Gb clean data on average via the PE150
sequencing strategy. The obtained data were annotated and counted
for SNVs and InDels.
Results
Patient specimens and primary
cultures
Dissociated cell suspensions from nine freshly
resected colorectal carcinoma tissues of various histological
grades were collected for the isolation of cancer cells as
aforementioned; two of the specimens were severely contaminated
with bacteria and were discarded. A total of five specimens were
primary cancer cells derived from a finite population of cells
(~5-6 passages) (22). Only two
specimens of resected colorectal tumor were used to establish cell
lines; one cell line was cc-006cpm8 and was established in the
present study for cellular characterization. In addition, the
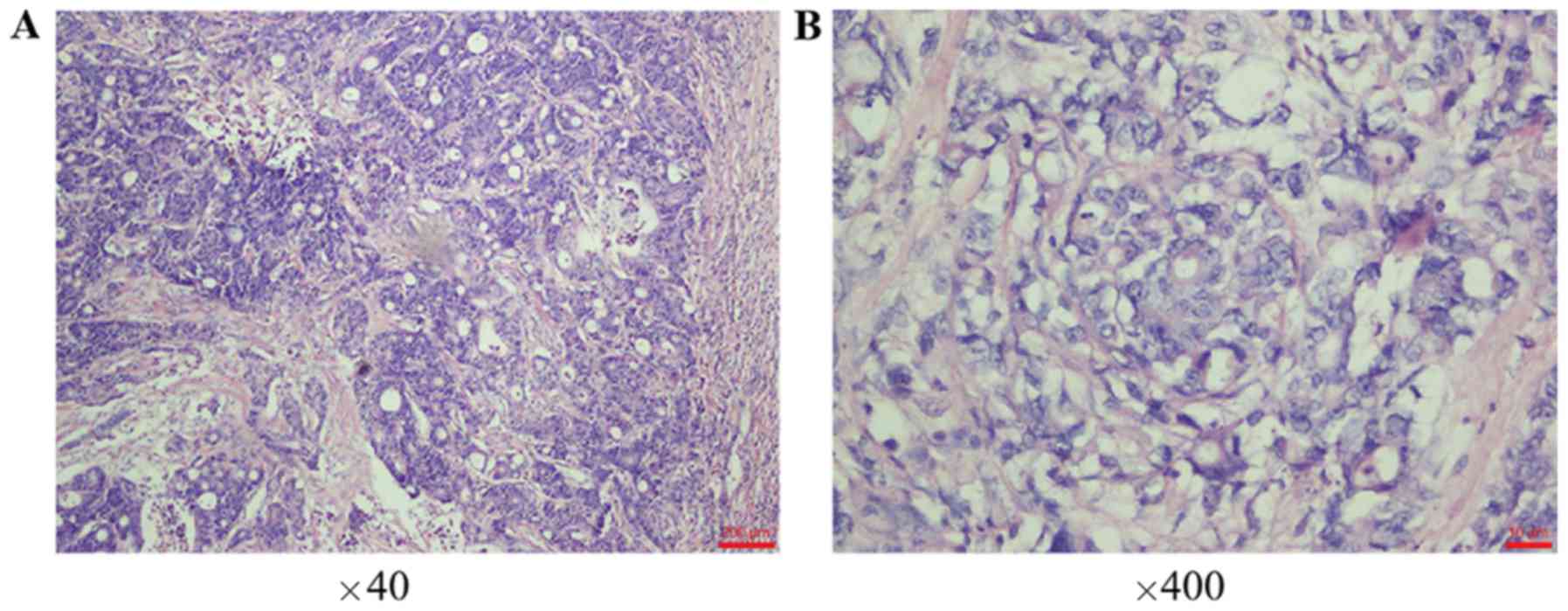
patient was diagnosed with invasive and ulcerative
moderately/poorly differentiated adenocarcinoma of the right colon
according to postoperative pathology, as demonstrated by H&E
staining of the primary tumor (Fig. 1A
and B).
Morphological features and clonal
heterogeneity of cc-006cpm8 cells
The initial isolation of the primary tumor cells and
their propagation in vitro in RPMI-1640 complete medium for
8 days was conducted until those clones were observed. Numerous
different cellular shapes and clone morphologies were reported: i)
Carcinoma-associated fibroblasts and normal fibroblasts; ii) sparse
large semispheroid clusters with dichotomized processes; and iii)
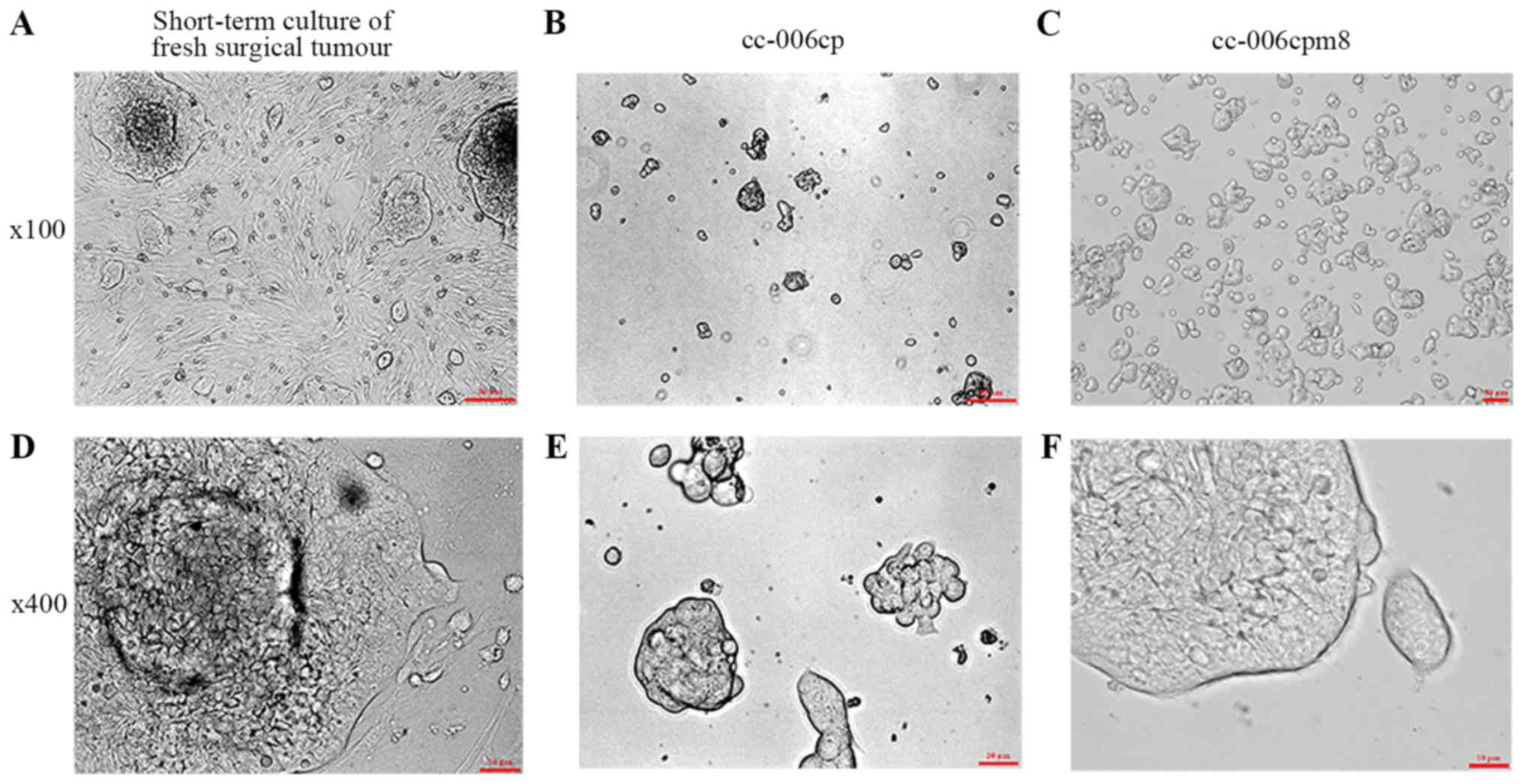
small closely-packed clones (Fig. 2A
and D). Following differential trypsinization, the cc-006cp
cell line possessed an epithelial phenotype without contaminating
fibroblasts; few clones were packed closely together, whereas
others were not (Fig. 2B and E),
and grew slowly compared with the cc-006cpm8 cell line (Fig. S1). From further subcloning,
cc-006cpm8 cells were selected as the monoclonal cell subline from
the cc-006cp cell line in semisolid medium (Fig. 2C and F).
In RPMI-1640 complete medium, the cc-006cpm8 cells
were cultured as several layers with tight adhesion to the plastic
surface and no contact suppression via microscopy (Fig. 3A). Following culture on chamber
slides and Wright-Giemsa staining, large nuclei, a thin layer of
cytoplasm and numerous nucleoli were observed in the cc-006cpm8
cells via microscopy (Fig. 3B).
Under a transmission electron microscope, the cc-006cpm8 cells
exhibited a prominent nucleolus, small irregular microvilli,
notable mitochondria and endoplasmic reticulum. Few lipid droplets
and vacuoles were observed (Fig. 3C
and D).
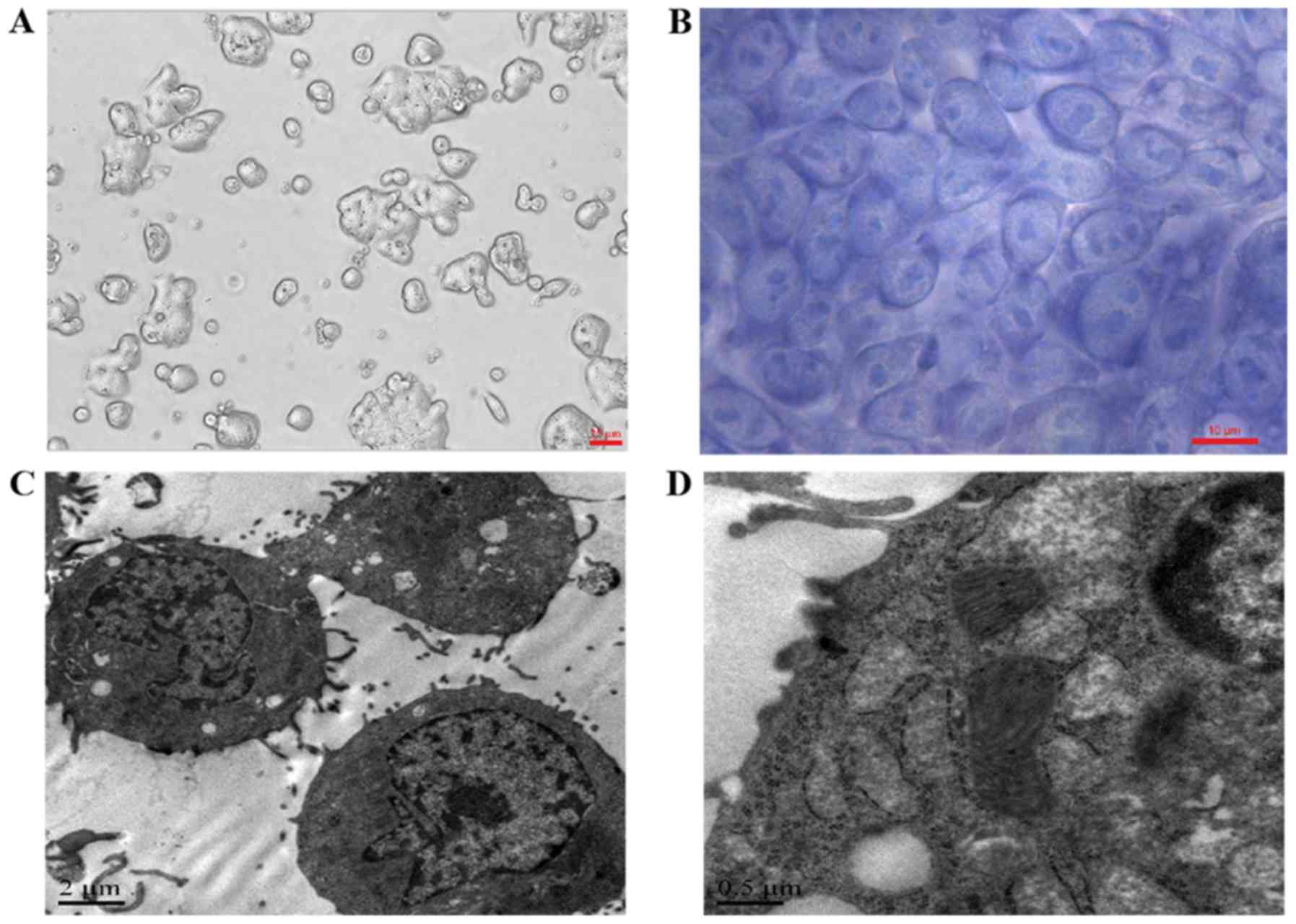 | Figure 3Morphological features of the
cc-006cpm8 cell line. (A) Cell morphology was observed under a
phase-contrast microscope. Original magnification, ×200. (B) In
situ Wright's staining of cc-006cpm8 cells. Original magnification,
×1,000. Below, electron micrographs show a cell possessing a
prominent nucleolus, small irregular microvilli, prominent
mitochondria, endoplasmic reticulum and few mucinous granules.
Original magnification, (C) ×5,000 and (D) 20,000. |
Growth properties of cc-006cpm8 cells in
vitro
The cc-006cpm8 cells were passaged continuously
>100 times over a period of 1 year; the cells exhibited
maintained vigorous growth. From the growth curve of the cc-006cpm8
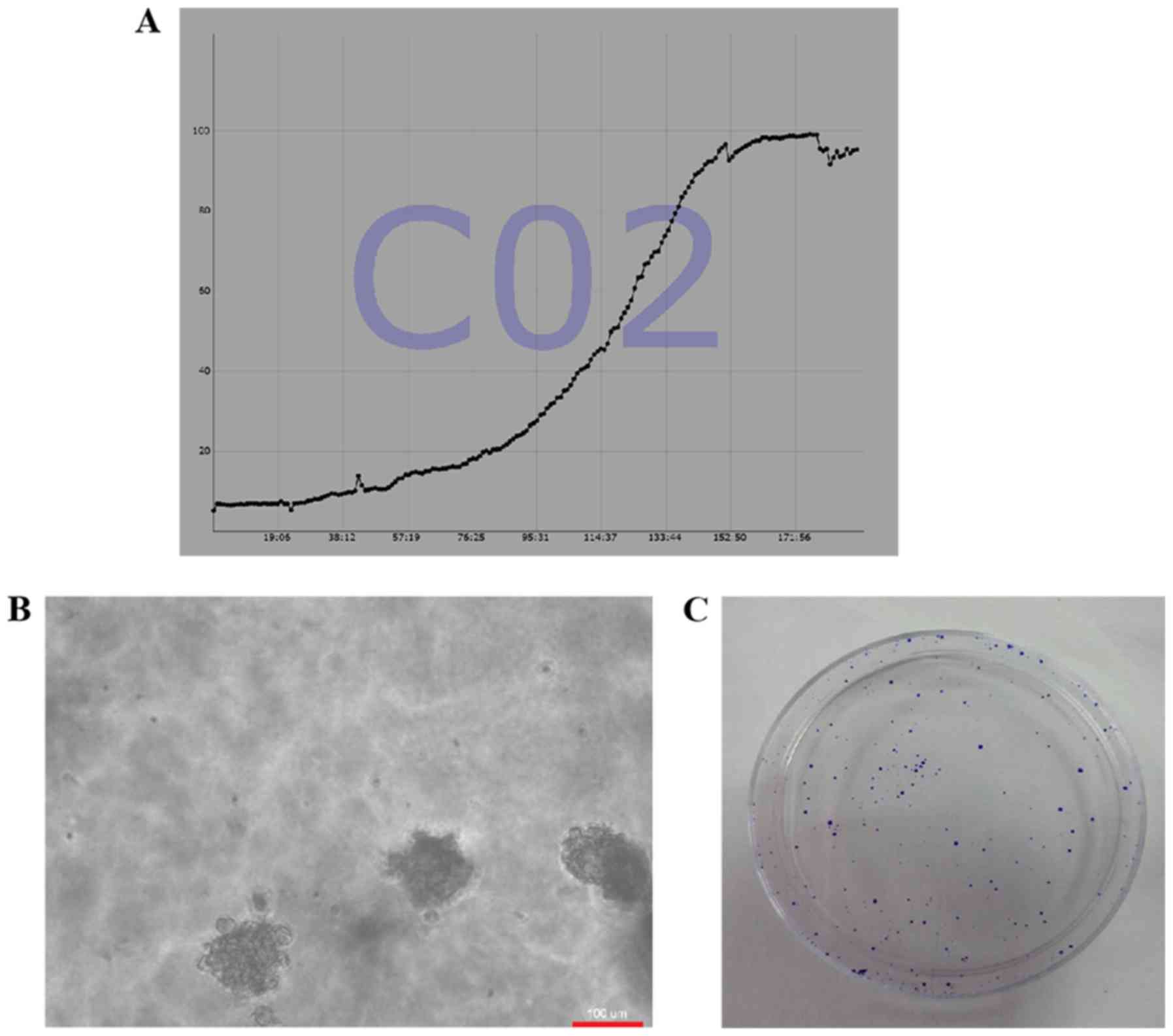
cells, the average PDT was determined to be 27 h (Fig. 4A). To analyze the proliferation of
this cell line, soft agar colony and clone formation assays were
performed. The former revealed that the colony formation rate of
the cc-006cpm8 cells was 73.2% (Fig.
4B) and that of clone formation was ~66.5%; the values of which
were obtained from a small number of the 100 cells cultured in 6-mm
dishes for 9 days. The resulting clones were stained with crystal
violet and colonies constituting >50 cells were counted
(Fig. 4C). The findings of these
assays revealed the proliferative properties of cc-006cpm8
cells.
Tumorigenicity of cc-006cpm8 in vivo
Athymic nude xenograft mice were used in the present
study to investigate the tumorigenicity of cc-006cpm8 cells in
vivo. Tumor formation was analyzed at least twice a week via
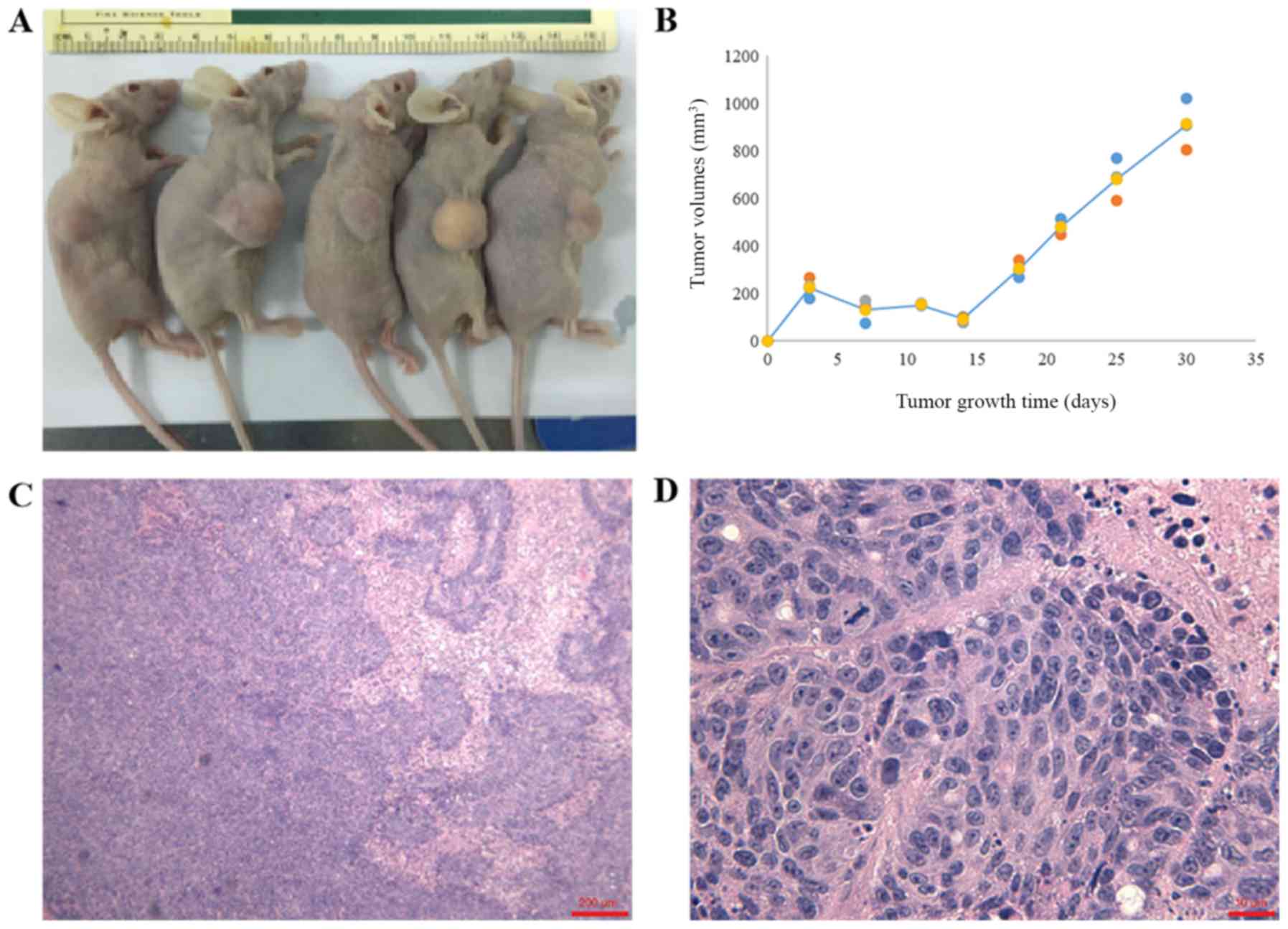
caliper measurements; tumors were detected in all mice after 4
weeks (Fig. 5A and B). These
xenograft tumors exhibited similar histology to the primary tumor
samples obtained from the patient, as determined by H&E
staining (Fig. 5C and D). The
xenograft tumor was nodular and adhered to the skin.
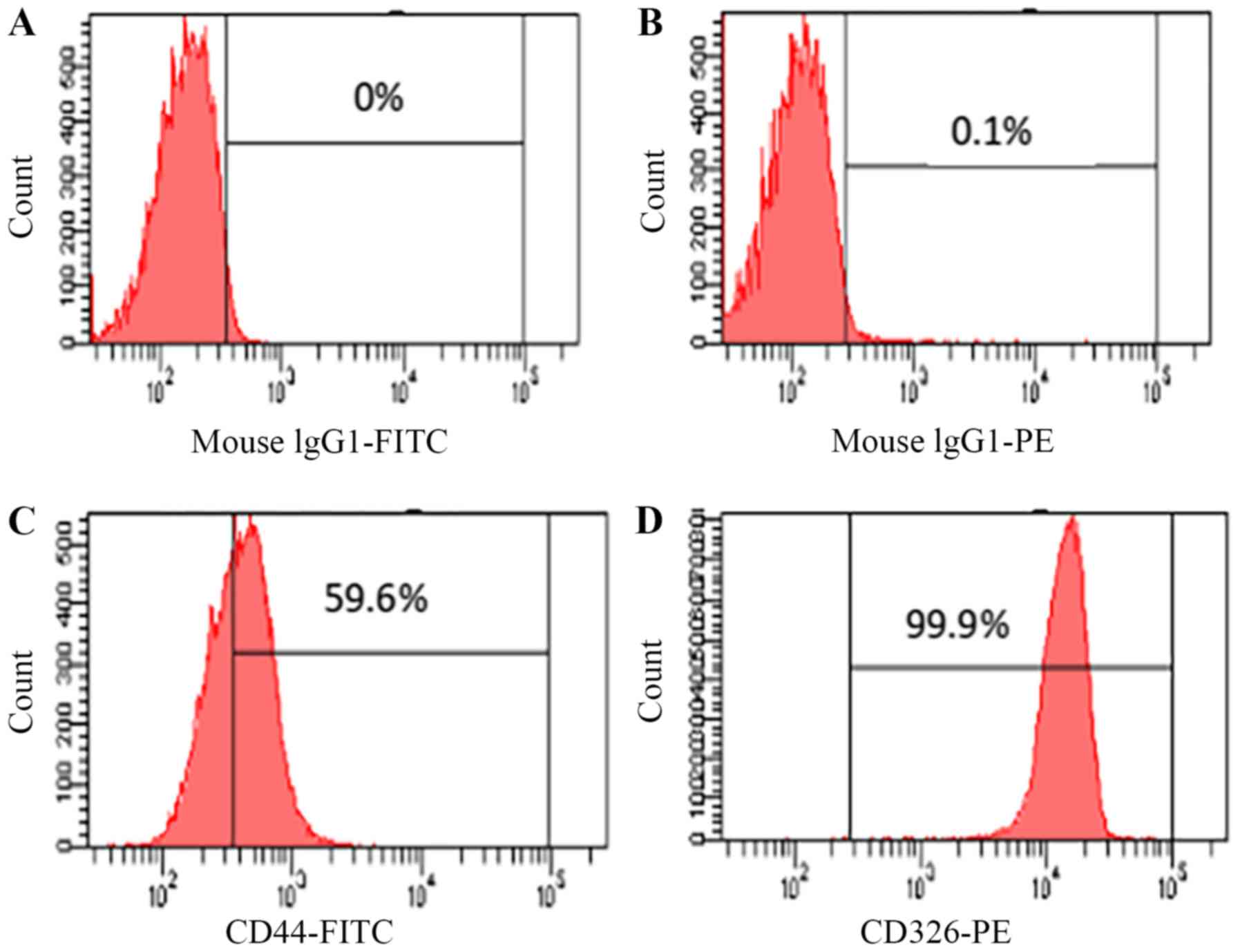
Expression of CD326 and CD44
To confirm the origin of the cc-006cpm8 cell line,
flow cytometric analysis was performed. The results revealed the
proportion of cancer stem cells (CSCs) in the subline. Compared
with the control, there were ~99.9% of EpCAM(+) cells in the
cc-006cpm8 population, which suggested that cc-006cpm8 cells were
of epithelial origin; however, compared with the control, there
were 59.6% of CD44(+) cells in the cc-006cpm8 population (Fig. 6). The results may indicate that
heterogeneous expression was also observed within the same
population of cc-006cpm8 cells.
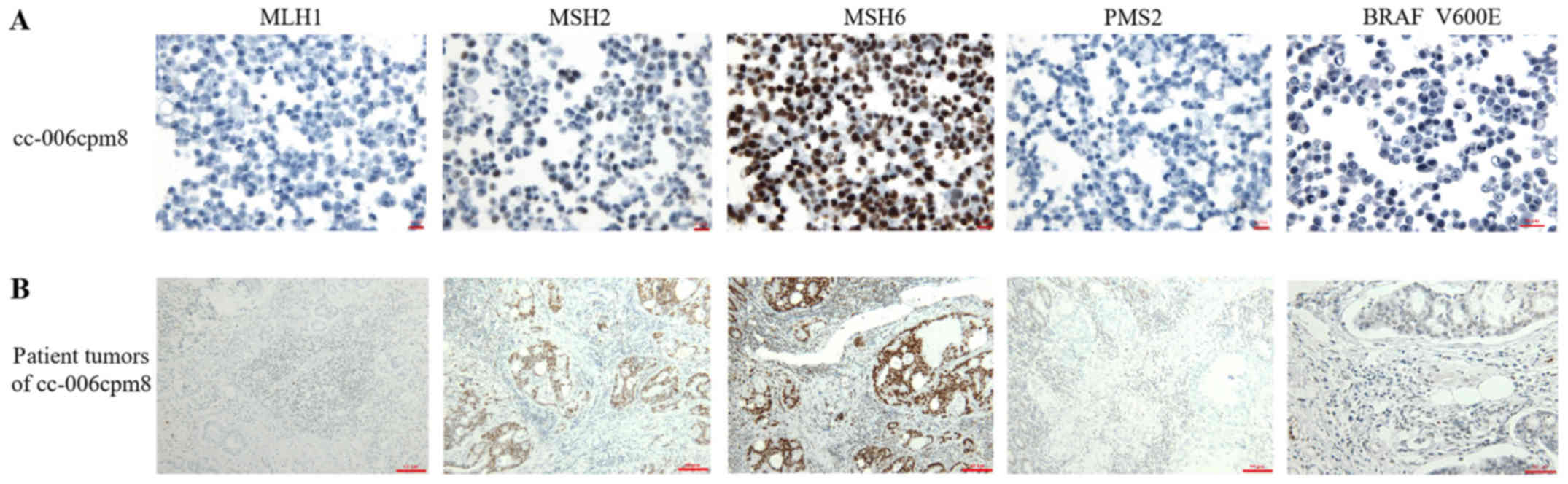
Antigen expression
To confirm the epithelial origin of the cc-006cpm8
cell line associated with the original tumor sections, the
expression of keratin and vimentin were investigated via
immunocytochemistry. These results demonstrated the positive
expression of pan-keratin (Fig.
7A) and negative expression of vimentin (Fig. 7B). These findings suggested the
epithelial origin of the cc-006cpm8 cell line. On further
observation of the cytokeratin (CK)7/CK20 profile,
CK7+/CK20− expression was present in this
cell line and the corresponding original tumor (Fig. S2). Additionally, the expression of
certain colon cancer markers in cc-006cpm8 cells and the original
tumor sections were determined. Notable positive expression of CDX2
and CEA (Fig. 7C and D), and mucin
2 (Fig. E) was observed. Furthermore 50% of the cells were
positively stained for Ki-67 (Fig.
7F); however, negative staining for p53 in the cell line and
the tumor sections from the patient was reported (Fig. 7G). In addition, positive staining
for EMA was detected in the original tumor sections from the
patient. By contrast, the cc-006cpm8 cell line exhibited 1%
positive staining (Fig. 7H). The
results further suggested the biological characteristics and cancer
heterogeneity in this cell line. The expression of DNA MMR proteins
was also determined by immunocytochemistry in the cell line and the
patient tumor sections, respectively. The results revealed the
negative expression of MLH1 and PMS2 and notable positive
expression of MSH2 and MSH6 (Fig.
8). Within MSI-H colorectal cancer, the BRAF V600E mutation is
associated with sporadic tumorigenesis (23). IHC analysis of BRAF V600E in the
cell line and the patient tumor sections was performed, and
positive expression was reported in the two sample types (Fig. 8). As the specimens were derived
from a patient with right colon adenocarcinoma and liver
metastasis, the expression of AFP, HepPar-1 and glypican-3 were
determined via IHC in the cc-006cpm8 cell and the tumor sections;
the antibodies used are those used in the diagnosis of
hepatocellular carcinoma. Therefore, this may eliminate the
possibility that the cc-006cpm8 cells and patient tumor sections
were of hepatic origin as negative expression was reported
(Fig. S3).
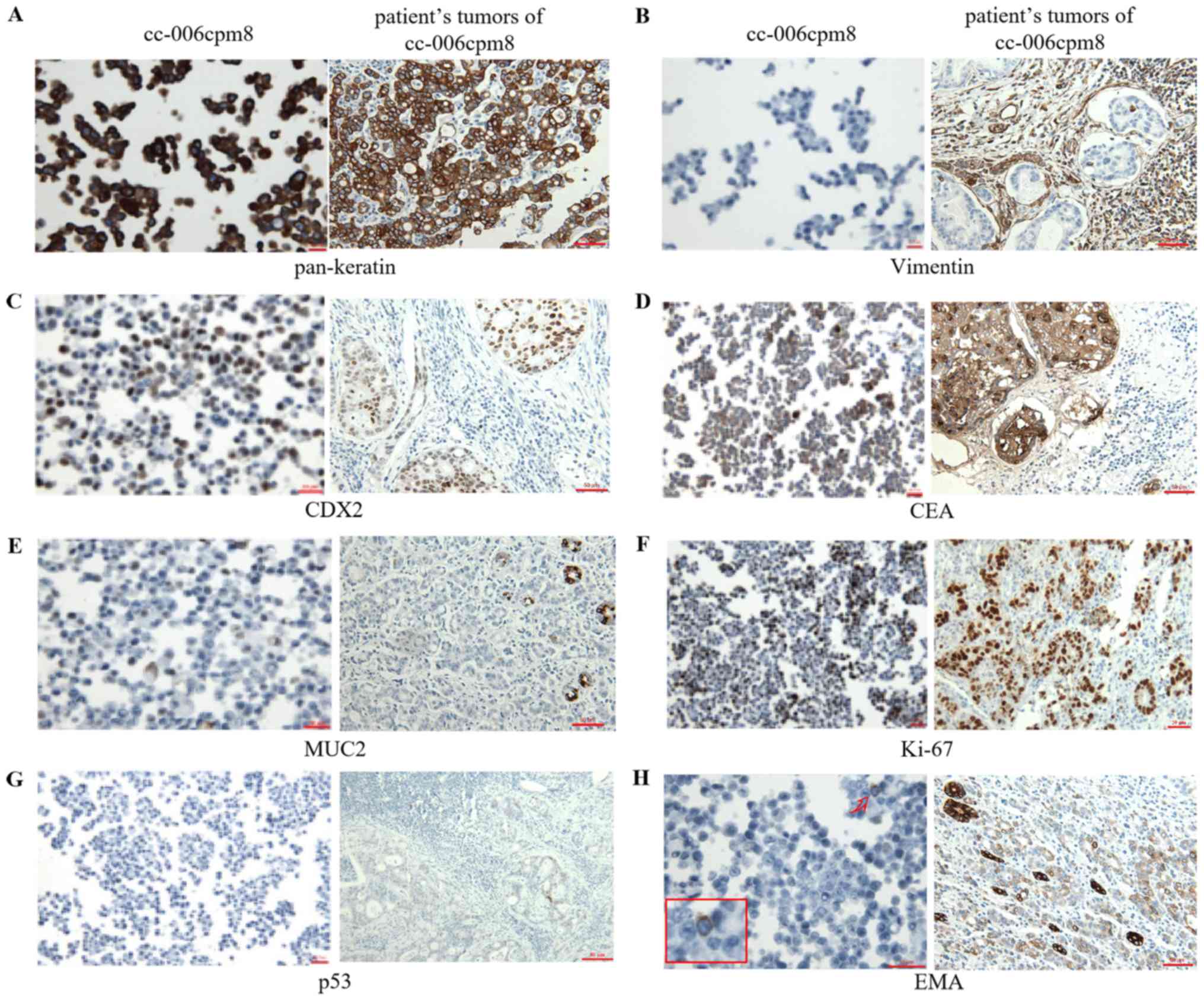 | Figure 7Expression of key colon cancer
markers in cc-006cpm8 cells and the original patient tumor
sections. Immunohistochemical analysis revealed the expression
profile (A) pan-keratin(+++), (C) CDX2(+), (B) vimentin(−), (D)
CEA(+++), (E) MUC2(+), (F) Ki-67(++), (G) p53(−) and (H) EMA(−) in
cc-006cpm8 cells. 1% positive staining of EMA (arrows) are shown in
high power view in the inset (magnification, ×1,000). Protein
expression in cc-006cpm8 cells was compared with that of the
original patient tumor sections, with the exception of the
expression of EMA. Magnification, ×400 (cell line) and 200
(original patient tumor sections). CDX2, caudal type homeobox
transcription factor 2; CEA, carcinoem-bryonic antigen; EMA,
epithelial membrane antigen; MUC2, mucin 2. |
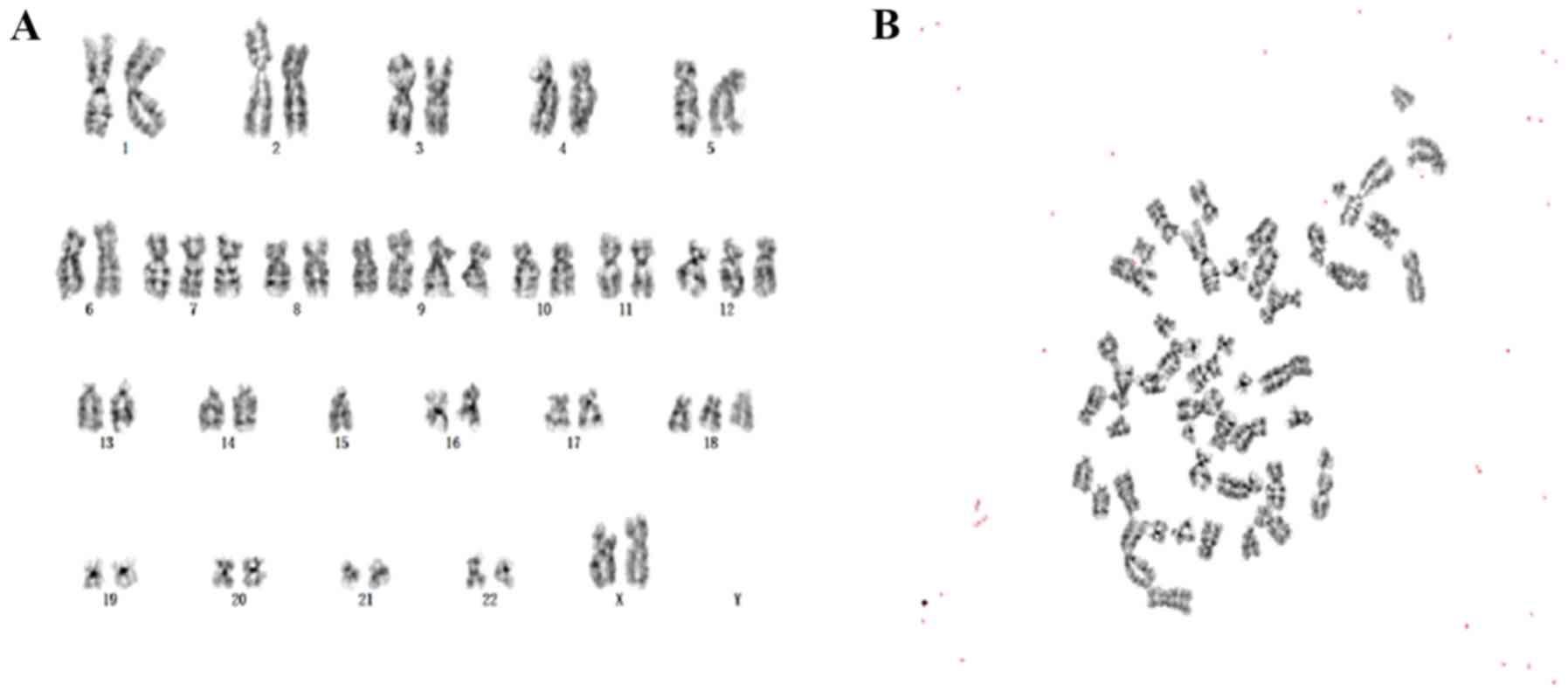
Chromosomal analysis
Chromosomal analysis of the cc-006cpm8 cell line was
performed at passages 5, 10 and 25, in which 20 karyotypes were
investigated using Giemsa banding. The results revealed that the
modal number of chromosomes in three different passages was
similar, indicating chromosomal stability. The modal number of
chromosomes in this cell line was 50, which accounted for 84.2% of
cells; the number of chromosomes ranged between 47 and 53.
Therefore, the cell line was reported as hyper-diploid. In cells of
passage 10, the karyotypes of the clone with 50 chromosomes were
xx, +7, +9, +9, +12, −15 and +18 (Fig.
9A and B). Other karyotypes were also observed, including +8,
+10, +12, +13, +14, +16, +19, +21 and +x. Collectively, the
frequencies of +7, +8, +9, +12 and +18 were higher. The results
suggested that this cell line was characteristic of malignant tumor
cells.
STR analysis
The cc-006cpm8 cell line was subjected to STR
profiling by the CCTCC following the ANSI/ATCC ASN-0002-2011,
Authentication of Human Cell Lines: Standardization of STR
Profiling. The STR analysis of this cell line resulted in unique
human profiles, which did not completely match the profiles in the
ATCC, DSMZ or JCRB databases, or the National Experimental
Resources Cell Sharing Platform.
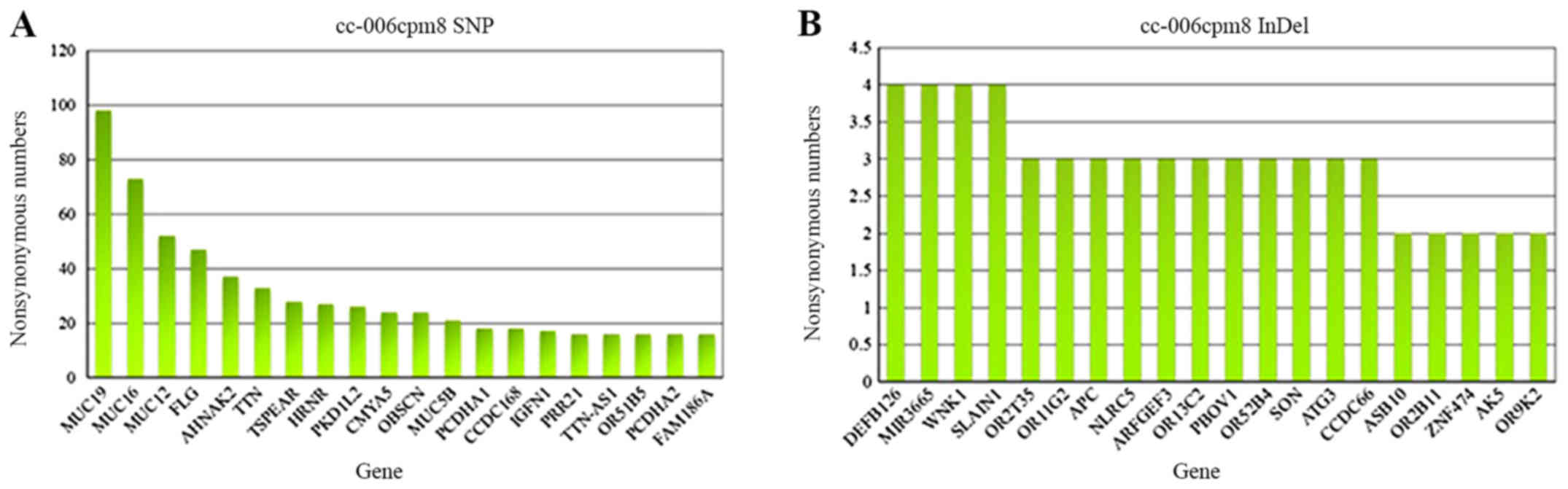
Detection of genetic mutations in
cc-006cpm8 cells
Exome capture DNA sequencing was performed on the
cell line; SNVs and InDels were counted from the data. As presented
in Table I, 27,134 total SNVs and
34,860 total InDels were reported in the cc-006cpm8 cell line.
Among the mutated genes, 1,510 novel SNVs and 32,658 novel InDels
were reported. In addition, 20 hypermutated exons containing SNPs
and InDels were detected in this cell line, including SNVs of
MUC19, MUC16, MUC12, FLG and
AHNAK2, and InDels of DEFB126, MIR3665, WNK
lysine deficient protein kinase 1 (WNK1) and SLAIN
motif-containing 1 (SLAIN1) (Fig. 10A and B). Additionally, 24
commonly mutated genes associated with CRC were reported; these
mutations may occur in the coding sequences of genes in this cell
line(Table II) (20). The genes associated with the DNA
MMR pathway were analyzed. MLH1 and MSH6 revealed no
mutations; however, MLH3, MSH2, MSH3 and
PMS2 exhibited missense mutations (24-26).
Furthermore, BRAF, APC, TP53, PIK3CA,
ERBB2 and TCF7L2 possessed missense mutations
comprising SNPs, and InDels of TGFBR2, ATM and
APC, which occurred via frameshift mutations. By contrast,
KRAS, NRAS, SMAD4, SMAD2,
ACVR2A, FBXW7, CTNNB1, ARID1A,
FAM123B and SOX9 exhibited no missense or frameshift
mutations in the coding regions.
 | Table ISummary of the whole exon sequencing
of the cc-006cpm8 cell line. |
Table I
Summary of the whole exon sequencing
of the cc-006cpm8 cell line.
| Term | SNPs (n) | InDels (n) |
|---|
| Total | 27,134 | 34,860 |
| Novel | 1,510 | 32,658 |
| Transitions | 20,192 | |
| Transeversions | 7,126 | |
| Homozygous | 10,729 | |
| Heterozygous | 16,362 | |
| Stopgain | 168 | |
| Stoploss | 11 | |
| Missense | 10,556 | |
| Frameshift | | 208 |
| Intronic | 7,969 | 1,621 |
| Intergenic | 3,142 | 375 |
| Splice3 | 10 | 9 |
| Splice5 | 19 | 12 |
| 5′-UTRs | 2,119 | 267 |
| 3′-UTRs | 2,318 | 670 |
 | Table IIExamples of alterations of coding
sequences in the cc-006cpm8 cell line. |
Table II
Examples of alterations of coding
sequences in the cc-006cpm8 cell line.
| Classification | Gene symbol | Status of coding
sequence
|
|---|
| SNPs | InDel |
|---|
| RTK/RAS
pathway | KRAS | No mutation | No mutation |
| NRAS | No mutation | No mutation |
| BRAF | rs9648696 (pE588G),
rs113488022 (pV600E) | No mutation |
| ERBB2 | rs1058808 | No mutation |
| TP53 pathway | TP53 | rs1042522,
rs2287499 (pR68G) | No mutation |
| ATM | No mutation | rs567403367 |
| TGF-β pathway | SMAD4 | No mutation | No mutation |
| SMAD2 | No mutation | No mutation |
| ACVR2A | No mutation | No mutation |
| TGFBR2 | No mutation | rs7937599 |
| IGF2/PI3K
pathway | PIK3CA | rs121913274 | No mutation |
| MMR pathway | MLH1 | No mutation | No mutation |
| MLH3 | rs17102999(pT942I),
rs175081(pN826D) | No mutation |
| MSH2 |
rs2303424(pQ915R) | No mutation |
| MSH3 | rs1650697(pI79V),
rs26279(pA1045T), rs184967(pQ949R) | No mutation |
| MSH6 | No mutation | No mutation |
| PMS2 | rs2228006 | No mutation |
| WNT pathway | APC | rs459552 | rs387906231,
rs121913334, rs387906234 |
| FBXW7 | No mutation | No mutation |
| TCF7L2 | rs77961654 | No mutation |
| CTNNB1 | No mutation | No mutation |
| ARID1A | No mutation | No mutation |
| FAM123B | No mutation | No mutation |
| SOX9 | No mutation | No mutation |
Discussion
CRC is a heterogeneous disease. In the present
study, a novel cell line (cc-006cpm8) originating from
moderately/poorly differentiated CRC with a Chinese genetic
background was established as few have been reported previously.
The cc-006cpm8 cells formed an immortal cell line that were
passaged >100 times in vitro with vigorous growth. This
cell line was examined at P0 and P100 respectively by STR analysis
and chromosome karyotype analysis. No significant changes were
found in either results (data not shown). In addition, this cell
line had a relatively high cloning formation rate and rapid growth.
Furthermore, the subline was negative for bacteria, fungi and
mycoplasma, as determined by PCR. However, further study data on
the phenotypic characteristics and the drug resistance of this cell
line, and comparison with another colorectal adenocarcinoma cell
line, are required. Despite this, the cc-006cpm8 cell line with
clinical biological characteristics of CRC may be useful in in
vitro and in vivo studies of this disease.
Cell lines can be separated by methods of infinite
dilution or plate cloning; however, the cc-006cpm8 cell line was
isolated via semisolid medium in the present study. The soft agar
colony formation method is well reported for the analysis of cell
proliferative ability (27-33).
Usually, the tumor cell colony formation rate is high (>10%),
whereas that of normal cells is weak (0 or 0.05-5%) in soft agar.
In addition, the adhesive and floating properties of tumor cells in
liquid culture may contribute to clone formation onto semisolid
medium. In addition, these properties inhibit contamination with
fibroblasts, which do not form colonies on soft agar. Therefore,
this technique may be conducive to the establishment of pure cells
lines with similar properties to that of tumor cells in
vivo.
Flow cytometric analysis demonstrated that the
expression of CD44 was 59.6% in cc-006cpm8 cells (a subline of
cc-006cp), and the expression of CD44 was 0.1% in the cc-006cp cell
line (data not shown). Further investigation is required to
determine the expression levels of additional colorectal CSCs
markers, including CD133, CD166 and ALDH1. The cell adhesion
molecule CD44 is a hyaluronic acid receptor, which has been
suggested as an alternative marker of CSCs. CD44 serves a major
role in cancer cell migration, and has been associated with the
initiation of tumor development of xenografts and colony formation,
in addition to tumor stage, lymph node infiltration, and patient
prognosis and survival (34,35).
To determine the epithelial origin, the expression of EpCAM cell
surface markers was analyzed and reported to be 99.9% in cc-006cpm8
cells. EpCAM, a similar protein to CD44, known for its role in cell
adhesion, may have versatile roles in signaling, cell migration,
proliferation and differentiation (36).
The cc-006cpm8 cell line associated with the
original tumor sections exhibited a
CK7+/CK20− expression profile. In general,
the CK7−/CK20+ profile is known to be
characteristic of CRC (37-41);
however, not all CRCs exhibit this profile. A substantial
proportion of CRC are reported to be either CK7+ or
CK20−, particularly in right-sided and high-grade CRCs,
consisting of poorly differentiated adenocarcinoma (PDA) and
undifferentiated carcinoma, among others (42). Imai et al reported that
CK20− PDA significantly favored solid type (Por1) de
novo histogenesis, and a smaller proportion of stromal tissue.
By contrast, CK20+ PDA significantly favored non-solid
type (Por2) histogenesis in stepwise de-differentiation, and higher
levels of stromal tissue (43).
The expression of CDX2 may be a useful adjunct for the diagnosis of
intestinal adenocarcinomas. In the present study, notable positive
expression of CDX2 was shown by IHC staining of the cell line and
corresponding original tumor. CDX2 represents the latest in a
series of transcription factors that have been reported to have
important applications in diagnostic surgical pathology as specific
and sensitive markers of specific cell and tumor types (44). Among colorectal adenocarcinomas,
the association between tumor grade and CDX2 staining has been
controversial. Hinoi et al (45) demonstrated that a rare subset of
poorly differentiated colonic carcinomas, termed large cell
minimally differentiated carcinoma or medullary carcinoma, are
characterized by microsatellite instability and loss of CDX2
expression. Kaimaktchiev et al (46) examined 1,109 tissue microarray
samples of colorectal adenocarcinomas and found a lack of CDX2
reactivity in 14 (28%) of 50 poorly differentiated tumors.
The present study reported that the cc-006cpm8 cell
line exhibited negative expression of MLH1 and PMS2 via IHC;
MLH1 possessed no SNPs or InDel mutations in the coding
region, as determined by whole exome capture DNA sequencing. This
may be due to the abnormal hypermethylation of the MLH1
promoter, which prevents gene transcription (47). MLH1 is one of the DNA MMR
genes associated with Lynch syndrome. This disorder is caused by
germline mutations of the MMR genes, including MLH1,
MLH2, MLH6 and PMS2 (48). MSI-H CRC may exist in 2-3% Lynch
syndrome and 15% sporadic CRC cases. The patient enrolled in the
present study was 89 years old and had no family history. According
to the Bethesda and revised Bethesda criteria (49), the patient was determined to have
Lynch syndrome. In addition, within MSI-H CRC, the BRAF V600E
mutation has been associated with sporadic tumorigenesis (23). Therefore, IHC for BRAF V600E in the
cell line and patient tumor sections was conducted; the cells and
tissues exhibited notable positive expression. Collectively, the
aforementioned findings further support that the patient possessed
sporadic CRC rather than Lynch syndrome.
From the results of genetic mutation detection in
the present study, a frequency spectrum of mutated genes in the
cc-006cpm8 cell line was obtained (Fig. 10A and B). Numerous gene mutations
occurred with high frequencies, including SNPs of MUC19,
MUC16, MUC12, FLG and AHNAK2, and
InDels of DEFB126, MIR3665, WNK1 and
SLAIN1. Mucins are glycoproteins encoded by the MUC genes.
There are four categories of mucins: Transmembrane (TM), secreted
gel forming, secreted non-gel forming and unclassified mucins.
MUC19 is a secreted gel forming mucin gene (50). At present, investigations into the
association between MUC19 and CRC remain limited.
MUC12 is a TM mucin and has been reported as a novel mucin
gene associated with membranes (51). It has been suggested that TM mucins
promote tumor invasion (52).
MUC12 is widely known to be downregulated in CRC (51,53).
Matsuyama et al (54)
reported that the mRNA expression of MUC12 is an independent
prognostic marker of stage II and Ⅲ CRC. Patients with low
expression levels of MUC12 exhibited poor disease-free
survival. Therefore, the high-frequency mutation of MUC12
may be associated with the poor prognosis of patients with CRC.
With a low expression of MUC12, patients may present with
poorer disease-free survival. MUC16 was first identified
from the molecular cloning of CA125 in 2001 (55). MUC16 also belongs to the TM
class of mucins and is the largest of its type (56). In addition, MUC16 serves as
a biomarker for ovarian cancer worldwide (57); however, MUC16 has also been
detected in other types of cancer, including pancreatic and gastric
cancer, and colorectal adenocarcinomas. Zhang et al
(58) reported the detection of
CA125 (MUC16) as an effective diagnostic marker of CRC. For
FLG, few investigations have been conducted. Skaaby et
al (59) proposed that there
were no statistically significant associations between the
loss-of-function mutation of FLG and sporadic types of
cancer, such as CRC. Therefore, mutations of FLG in CRC
require further investigation. At present, the association between
AHNAK2 and CRC has not been determined; however,
AHNAK2 has been revealed to be a prognostic biomarker for
clear cell renal cell carcinoma and pancreatic ductal
adenocarcinoma (60,61). Additionally, Wen et al
(62) reported AHNAK2 as a
susceptibility gene for systemic lupus erythematosus; however,
mutations of DEFB126, MIR3665, WNK1 and
SLAIN1 were not associated with CRC according to previous
studies. Therefore, further investigation into the role of
high-frequency gene mutations in the cc-006cpm8 cell line for the
development of CRC is required.
In conclusion, a novel stable cell line of
moderately/poorly differentiated CRC (cc-006cpm8) of Chinese
genetic background was generated in the present study. The
cc-006cpm8 cells were well characterized as a cell line and may be
useful for the investigation of CRC in the future.
Supplementary Materials
Funding
The present study was partly supported by grants
from the National Natural Science Foundation of China (grant nos.
81572928 and 81772978) and the Jiangsu Provincial Special Program
of Medical Science (grant no. BE2017611) to JC.
Availability of data and materials
The datasets used and analyzed during this study
are available from the corresponding author on reasonable
request.
Authors' contributions
XC analyzed and interpreted the results of the
detection of this novel cell line, and wrote a part of the
manuscript. YX performed identification of the cell line, and was a
major contributor in writing the manuscript. XH was responsible for
specimen collection. JW wrote part of the manuscript and modified
the images and was involved in data processing. YC and RH performed
the cell culture. HC and XS produced the pathological sections. HZ
organized the data for all the experiments. YG provided useful
interpretation of the genetic mutations data. JC conceived and
designed and supervised the study. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
For the use of patient sample, the patient signed
informed consent forms for the use of their sample in scientific
research. The experiments were approved by the Ethics Committee of
Nanjing First Hospital of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
CRC
|
colorectal cancer
|
|
MMR
|
mismatch-repair
|
|
SNP
|
single nucleotide polymorphism
|
|
InDel
|
insertion-deletion
|
|
SNV
|
single nucleotide variant
|
|
STR
|
short tandem repeat
|
|
CEA
|
carcinoembryonic antigen
|
|
EMA
|
epithelial membrane antigen
|
|
CDX2
|
caudal type homeobox transcription
factor 2
|
|
PDT
|
population doubling time
|
|
PCR
|
polymerase chain reaction
|
|
IDT
|
Integrated DNA Technologies
|
|
CCTCC
|
China Center for Type Culture
Collection
|
|
TM
|
transmembrane
|
|
ccRCC
|
clear cell renal cell carcinoma
|
|
PDAC
|
pancreatic ductal adenocarcinoma
|
|
SLE
|
systemic lupus erythematosus
|
|
ANSI
|
American National Standards
Institute
|
|
ATCC
|
American Type Culture Collection
|
|
DSMZ
|
Deutsche Sammlung von Mikroorganismen
und Zellkulturen
|
Acknowledgments
The authors would like to thank Professor Wenbin
Huang from the Department of Pathology of Nanjing First Hospital
for IHC analysis and Professor Xiuqing Ji from the Genetic Medicine
Center of Maternity Hospital attached to Nanjing Medical University
for chromosomal analysis.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
2
|
GLOBOCAN: Estimated cancer incidence,
mortality and prevalence worldwide in 2012. 2012, http://globocan.iarc.fr/Default.aspx.
|
|
3
|
Bray F, Ferlay J, Laversanne M, Brewster
DH, Gombe Mbalawa C, Kohler B, Piñeros M, Steliarova-Foucher E,
Swaminathan R, Antoni S, et al: Cancer Incidence in Five
Continents: Inclusion criteria, highlights from Volume X and the
global status of cancer registration. Int J Cancer. 137:2060–2071.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends - an update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar
|
|
5
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Worthley DL, Whitehall VL, Spring KJ and
Leggett BA: Colorectal carcinogenesis: Road maps to cancer. World J
Gastroenterol. 13:3784–3791. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Arends MJ: Pathways of colorectal
carcinogenesis. Appl Immunohistochem Mol Morphol. 21:97–102.
2013.PubMed/NCBI
|
|
8
|
Barrasa JI, Olmo N, Lizarbe MA and Turnay
J: Bile acids in the colon, from healthy to cytotoxic molecules.
Toxicol In Vitro. 27:964–977. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tariq K and Ghias K: Colorectal cancer
carcinogenesis: A review of mechanisms. Cancer Biol Med.
13:120–135. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tejpar S and Van Cutsem E: Molecular and
genetic defects in colorectal tumorigenesis. Best Pract Res Clin
Gastroenterol. 16:171–185. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fakih MG: Metastatic colorectal cancer:
Current state and future directions. J Clin Oncol. 33:1809–1824.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nagahashi M, Wakai T, Shimada Y, Ichikawa
H, Kameyama H, Kobayashi T, Sakata J, Yagi R, Sato N, Kitagawa Y,
et al: Genomic landscape of colorectal cancer in Japan: Clinical
implications of comprehensive genomic sequencing for precision
medicine. Genome Med. 8:1362016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu HY, Li C, Wang LM, Yang Y, Zhan ZL and
Zhang TZ: Establishment and characterization of a cell line
THC-8908 from human well differentiated colon adenocarcinoma. Chin
J Clin Oncol. 20:330–333. 1993.
|
|
14
|
Fu ZM: Establishment and characteristics
of a human colon adenocarcinoma cell line (SIC-8101). Chin J Clin
Oncol. 13:47–49. 1986.
|
|
15
|
Zhang ZX, Xu SH and Qian LJ: Establishment
and characterization of a cell line HR-8348 derived from human
poorly differentiated adenocarcinoma of rectum. Sci Sin B.
30:522–532. 1987.PubMed/NCBI
|
|
16
|
Zhang XZ: Establishment and
characterization of a cell line Hce-8693 from human
undifferentiated caecal adenocarcinoma. Zhonghua Zhong Liu Za Zhi.
11:413–415. 1989.In Chinese. PubMed/NCBI
|
|
17
|
Zheng XL, Zhou ZG, Gu J, Li HG, Lin L and
Deng YL: Establishment and characterization of a cell line HRC-99
from human rectal adenocarcinoma. World Chin J Digestology.
13:1510–1513. 2005.
|
|
18
|
Ding KF, Hu YT, Xiao Q, He JJ and Zheng S:
A cell line DXH-1 from human colon carcinoma and its application.
CN Patent CN201510631287.2. Filed September 29, 2015 issued
February 3, 2016.
|
|
19
|
Wang XY, Lai ZS, Yeung CM, Wang JD, Deng
W, Li HY, Han YJ, Kung HF, Jiang B and Lin MC: Establishment and
characterization of a new cell line derived from human colorectal
laterally spreading tumor. World J Gastroenterol. 14:1204–1211.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cancer Genome Atlas Network: Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brattain MG, Brattain DE, Fine WD, Khaled
FM, Marks ME, Kimball PM, Arcolano LA and Danbury BH: Initiation
and characterization of cultures of human colonic carcinoma with
different biological characteristics utilizing feeder layers of
confluent fibroblasts. Oncodev Biol Med. 2:355–366. 1981.PubMed/NCBI
|
|
22
|
Peeh DM: Are primary cultures realistic
models of prostate cancer? J Cell Biochem. 91:185–195. 2004.
View Article : Google Scholar
|
|
23
|
Capper D, Voigt A, Bozukova G, Ahadova A,
Kickingereder P, von Deimling A, von Knebel Doeberitz M and Kloor
M: BRAF V600E-specific immunohistochemistry for the exclusion of
Lynch syndrome in MSI-H colorectal cancer. Int J Cancer.
133:1624–1630. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aaltonen LA, Peltomäki P, Leach FS,
Sistonen P, Pylkkänen L, Mecklin JP, Järvinen H, Powell SM, Jen J,
Hamilton SR, et al: Clues to the pathogenesis of familial
colorectal cancer. Science. 260:812–816. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ionov Y, Peinado MA, Malkhosyan S, Shibata
D and Perucho M: Ubiquitous somatic mutations in simple repeated
sequences reveal a new mechanism for colonic carcinogenesis.
Nature. 363:558–561. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Parsons R, Li GM, Longley MJ, Fang WH,
Papadopoulos N, Jen J, de la Chapelle A, Kinzler KW, Vogelstein B
and Modrich P: Hypermutability and mismatch repair deficiency in
RER+ tumor cells. Cell. 75:1227–1236. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tsukamoto Y, Fumoto S, Noguchi T,
Yanagihara K, Hirashita Y, Nakada C, Hijiya N, Uchida T, Matsuura
K, Hamanaka R, et al: Expression of DDX27 contributes to
colony-forming ability of gastric cancer cells and correlates with
poor prognosis in gastric cancer. Am J Cancer Res. 5:2998–3014.
2015.PubMed/NCBI
|
|
28
|
Ercan D, Choi HG, Yun CH, Capelletti M,
Xie T, Eck MJ, Gray NS and Jänne PA: EGFR mutations and resistance
to irreversible pyrimidine based EGFR receptors. Clin Cancer Res.
21:3913–3923. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mayr C, Wagner A, Stoecklinger A, Jakab M,
Illig R, Berr F, Pichler M, Di Fazio P, Ocker M, Neureiter D, et
al: 3-Deazaneplanocin A may directly target putative cancer stem
cells in biliary tract cancer. Anticancer Res. 35:4697–4705.
2015.PubMed/NCBI
|
|
30
|
Hua G, Lv X, He C, Remmenga SW, Rodabough
KJ, Dong J, Yang L, Lele SM, Yang P, Zhou J, et al: YAP induces
high-grade serous carcinoma in fallopian tube secretory epithelial
cells. Oncogene. 35:2247–2265. 2016. View Article : Google Scholar :
|
|
31
|
Ukaji T, Lin Y, Banno K, Okada S and
Umezawa K: Inhibition of IGF-1 mediated cellular migration and
invasion by migracin A in ovarian clear cell carcinoma cells. PLoS
One. 10:e01376632015. View Article : Google Scholar
|
|
32
|
Bon H, Wadhwa K, Schreiner A, Osborne M,
Carroll T, Ramos-Montoya A, Ross-Adams H, Visser M, Hoffmann R,
Ahmed AA, et al: Salt-inducible kinase 2 regulates mitotic
progression and transcription in prostate cancer. Mol Cancer Res.
13:620–635. 2015. View Article : Google Scholar :
|
|
33
|
Kim T, Jeon YJ, Cui R, Lee JH, Peng Y, Kim
SH, Tili E, Alder H and Croce CM: Role of MYC-regulated long
noncoding RNAs in cell cycle regulation and tumorigenesis. J Natl
Cancer Inst. 107:dju5052015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dou J and Gu N: Emerging strategies for
the identification and targeting of cancer stem cells. Tumour Biol.
31:243–253. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Horst D, Kriegl L, Engel J, Kirchner T and
Jung A: Prognostic significance of the cancer stem cell markers
CD133, CD44, and CD166 in colorectal cancer. Cancer Invest.
27:844–850. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lugli A, Iezzi G, Hostettler I, Muraro MG,
Mele V, Tornillo L, Carafa V, Spagnoli G, Terracciano L and Zlobec
I: Prognostic impact of the expression of putative cancer stem cell
markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer.
Br J Cancer. 103:382–390. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Varadhachary GR, Abbruzzese JL and Lenzi
R: Diagnostic strategies for unknown primary cancer. Cancer.
100:1776–1785. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Moll R, Löwe A, Laufer J and Franke WW:
Cytokeratin 20 in human carcinomas. A new histodiagnostic marker
detected by monoclonal antibodies. Am J Pathol. 140:427–447.
1992.PubMed/NCBI
|
|
39
|
Loy TS and Calaluce RD: Utility of
cytokeratin immunostaining in separating pulmonary adenocarcinomas
from colonic adeno-carcinomas. Am J Clin Pathol. 102:764–767. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wauters CC, Smedts F, Gerrits LG, Bosman
FT and Ramaekers FC: Keratins 7 and 20 as diagnostic markers of
carcinomas metastatic to the ovary. Hum Pathol. 26:852–855. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chu P, Wu E and Weiss LM: Cytokeratin 7
and cytokeratin 20 expression in epithelial neoplasms: A survey of
435 cases. Mod Pathol. 13:962–972. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Park SY, Kim HS, Hong EK and Kim WH:
Expression of cytokeratins 7 and 20 in primary carcinomas of the
stomach and colorectum and their value in the differential
diagnosis of metastatic carcinomas to the ovary. Hum Pathol.
33:1078–1085. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Imai Y, Yamagishi H, Fukuda K, Okamura T,
Ono Y, Ban S, Inoue T and Ueda Y: Expression of cytokeratin 20
indicates invasive histological phenotype in poorly differentiated
colorectal adenocarcinoma. Anticancer Res. 34:159–167.
2014.PubMed/NCBI
|
|
44
|
Bayrak R, Haltas H and Yenidunya S: The
value of CDX2 and cytokeratins 7 and 20 expression in
differentiating colorectal adenocarcinomas from extraintestinal
gastrointestinal adeno-carcinomas: Cytokeratin
7−/20+ phenotype is more specific than CDX2
antibody. Diagn Pathol. 7:92012. View Article : Google Scholar
|
|
45
|
Hinoi T, Tani M, Lucas PC, Caca K, Dunn
RL, Macri E, Loda M, Appelman HD, Cho KR and Fearon ER: Loss of
CDX2 expression and microsatellite instability are prominent
features of large cell minimally differentiated carcinomas of the
colon. Am J Pathol. 159:2239–2248. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kaimaktchiev V, Terracciano L, Tornillo L,
Spichtin H, Stoios D, Bundi M, Korcheva V, Mirlacher M, Loda M,
Sauter G, et al: The homeobox intestinal differentiation factor
CDX2 is selectively expressed in gastrointestinal adenocarcinomas.
Mod Pathol. 17:1392–1399. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jasperson KW, Tuohy TM, Neklason DW and
Burt RW: Hereditary and familial colon cancer. Gastroenterology.
138:2044–2058. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Strate LL and Syngal S: Hereditary
colorectal cancer syndromes. Cancer Causes Control. 16:201–213.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Macrae F, Harris M, Massimo R and Alberto
M: Re: Revised Bethesda Guidelines for hereditary nonpolyposis
colorectal cancer (Lynch syndrome) and microsatellite instability.
J Natl Cancer Inst. 97:936–937; author reply 937-938. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Senapati S, Sharma P, Bafna S, Roy HK and
Batra SK: The MUC gene family: Their role in the diagnosis and
prognosis of gastric cancer. Histol Histopathol. 23:1541–1552.
2008.PubMed/NCBI
|
|
51
|
Williams SJ, McGuckin MA, Gotley DC, Eyre
HJ, Sutherland GR and Antalis TM: Two novel mucin genes
down-regulated in colorectal cancer identified by differential
display. Cancer Res. 59:4083–4089. 1999.PubMed/NCBI
|
|
52
|
Hollingsworth MA and Swanson BJ: Mucins in
cancer: Protection and control of the cell surface. Nat Rev Cancer.
4:45–60. 2004. View Article : Google Scholar
|
|
53
|
Packer LM, Williams SJ, Callaghan S,
Gotley DC and McGuckin MA: Expression of the cell surface mucin
gene family in adenocarcinomas. Int J Oncol. 25:1119–1126.
2004.PubMed/NCBI
|
|
54
|
Matsuyama T, Ishikawa T, Mogushi K,
Yoshida T, Iida S, Uetake H, Mizushima H, Tanaka H and Sugihara K:
MUC12 mRNA expression is an independent marker of prognosis in
stage II and stage III colorectal cancer. Int J Cancer.
127:2292–2299. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yin BW and Lloyd KO: Molecular cloning of
the CA125 ovarian cancer antigen: Identification as a new mucin,
MUC16. J Biol Chem. 276:27371–27375. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kufe DW: Mucins in cancer: Function,
prognosis and therapy. Nat Rev Cancer. 9:874–885. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yin BW, Dnistrian A and Lloyd KO: Ovarian
cancer antigen CA125 is encoded by the MUC16 mucin gene. Int J
Cancer. 98:737–740. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang B, Liang XL, Gao HY, Ye LS and Wang
YG: Models of logistic regression analysis, support vector machine,
and back-propagation neural network based on serum tumor markers in
colorectal cancer diagnosis. Genet Mol Res. May 13–2016.Epub ahead
of print. View Article : Google Scholar
|
|
59
|
Skaaby T, Husemoen LL, Thyssen JP,
Meldgaard M, Thuesen BH, Pisinger C, Jørgensen T, Carlsen K,
Johansen JD, Menné T, et al: Filaggrin loss-of-function mutations
and incident cancer: A population-based study. Br J Dermatol.
171:1407–1414. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang M, Li X, Zhang J, Yang Q, Chen W, Jin
W, Huang YR, Yang R and Gao WQ: AHNAK2 is a novel prognostic marker
and oncogenic protein for clear cell renal cell carcinoma.
Theranostics. 7:1100–1113. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lu D, Wang J, Shi X, Yue B and Hao J:
AHNAK2 is a potential prognostic biomarker in patients with PDAC.
Oncotarget. 8:31775–31784. 2017.PubMed/NCBI
|
|
62
|
Wen L, Zhu C, Zhu Z, Yang C, Zheng X, Liu
L, Zuo X, Sheng Y, Tang H, Liang B, et al: Exome-wide association
study identifies four novel loci for systemic lupus erythematosus
in Han Chinese population. Ann Rheum Dis. 77:4172018. View Article : Google Scholar
|