Introduction
Glioblastoma multiforme (GBM) is a lethal malignant
tumor accounting for 42% of the tumors of the central nervous
system, with a median survival of 15 months (1-3). At
present, no curative treatment is available and the most used
first-line drug, temozolomide (TMZ), only moderately increases the
life expectancy of the treated patients (4). In addition, a high proportion of
gliomas become TMZ-resistant with time. Therefore, novel drugs and
therapeutic protocols for combined treatments on TMZ-resistant
glioma cells are urgently needed.
In this respect, a novel strategy for therapeutic
protocols has been recently suggested, the targeting of microRNAs.
MicroRNAs (miRNAs) are short non-coding RNAs that function by
repressing translation or inducing the cleavage of the target mRNA
transcripts, thereby regulating gene expression at the
post-transcriptional level (5-7).
Altered miRNA expression has been firmly demonstrated to be
involved in cancer (8-13). Approaches based on the targeting of
oncomiRNAs and metastamiRNAs (associated with tumor progression and
metastasis, respectively) have been found to inhibit tumor cell
growth and metastasis, and in some examples to reverse the
resistance of tumor cells to anticancer drugs (14-17).
For instance, Chan et al (15) have reported that the inhibition of
oncomiR-138 prevents in vitro tumor sphere formation in
malignant gliomas and suppresses in vivo tumorigenesis.
Wagenaar et al (16) were
able to target the transcriptional network in hepatocellular
carcinomas cell lines with sequence-specific antagomiR targeting
miR-21, thereby inducing increased expression of miR-21-regulated
genes, associated with a loss of viability. In another example, Ma
et al (17) reported
inhibition of metastasis formation in a mouse model of mammary
tumor following silencing of the oncogenic miR-10a. In this
context, the use of peptide nucleic acids (PNAs) targeting
oncomiRNAs might be relevant (8).
PNAs are DNA analogues described for the first time
by Nielsen et al (18), in
which the sugar-phosphate backbone has been replaced by
N-(2-aminoethyl)glycine units (18-22).
PNAs are capable of forming Watson-Crick double helices following
efficient sequence-specific hybridization with complementary DNA
and RNA (23). Furthermore, they
are able to generate triple helices with double-stranded DNA and to
perform strand invasion (24-26).
In virtue of these biological activities, PNAs have been
demonstrated to be very efficient tools for
pharmacologically-mediated alteration of gene expression, both
in vitro and in vivo (27-29).
In summary, PNAs and PNA-based analogues were employed as antisense
molecules targeting mRNAs, triple-helix forming molecules targeting
eukaryotic gene promoters, artificial promoters, and decoy
molecules targeting transcription factors (26). Relevant to the present study, PNAs
have been demonstrated to be able of altering miRNA functions, both
in vitro and in vivo (30-37).
Cheng et al (37), for
instance, efficiently inhibited the function of oncomiR-155-5p in a
tumor mouse model by the design and synthesis of a
peptide-(anti-miR)-PNA construct able to target the tumor
microenvironment and to transport the anti-miR PNA across the
cellular plasma membranes under the acidic conditions which
characterize solid tumors. Recently, our group has reported that a
PNA targeting miR-221-3p (R8-PNA-a221) (38), bearing an oligoarginine peptide
(R8) enabling efficient uptake by glioma cells (30,31),
was able to strongly inhibit miR-221-3p in U251, U373 and T98G
glioma cells. The inhibition of miR-221-3p activity was associated
with an increased expression of the miR-221-3p target
p27Kip1, as analyzed by reverse
transcription-quantitative polymerase-chain reaction (RT-qPCR) and
by western blot analysis (38).
The present study determined the biological activity
of a combined treatment of glioma cell lines with two PNAs directed
against miRNAs regulating caspase-3 mRNA expression and conjugated
to the octaarginine R8 peptide, allowing efficient cellular uptake.
Effects on apoptosis were analyzed to determine whether additive
activity was were obtained by co-administration of the two PNAs to
the temozolomide-resistant T98G glioma cell line, and whether
combined treatments were associated with a reversion of the
drug-resistance phenotype.
Materials and methods
Synthesis and characterization of
PNAs
The protocols for the synthesis and the
characterization of the anti-miR-221 PNAs have been described in a
previously published study (38).
The synthesis of the new anti-miR-155 PNAs was performed using a
standard Fmoc-based automate peptide synthesizer (Syro I; Biotage,
Uppsala, Sweden), using a ChemMatrix-RinkAmide resin loaded with
Fmoc-Gly-OH (0.2 mmol/g) as first monomer, and using commercially
available monomers (Link Technologies, Bellshill, UK) with
HBTU/DIPEA coupling. After purification, the PNAs were
characterized by UPLC-MS on a Waters ACQUITY System equipped with a
ACQUITY UPLC BEH C18 Column(1.7 µm; 2.1×50 mm). Gradient:
100% A for 0.9 min, then 0-50% B in 5.7 min at 0.25 ml/min flow (A,
water + 0.2% formic acid; B, acetonitrile + 0.2% formic acid).
R8-PNA-a155: sequence H-R8-TAT CAC GAT TAG CAT
TAA-Gly-NH2; yield: 15.9% Rt=2.65 min;
calculated MW, 6184.3 g/mol; m/z found, 1238.2 [M+5H]5+,
1031.9 [M+6H]6+, 884.8 [M+7H]7+, 774.3
[M+8H]8+, 688.4 [M+9H]9+, 619.6
[M+10H]10+, 563.4 [M+11H]11+.
R8-PNA-a155-MUT: sequence H-R8-TAT TAC GGT TAA CAT
CAA-Gly-NH2; yield: 11.6% Rt=2.65 min; calculated mw,
6184.3 g/mol; m/z found, 1238.0 [M+5H]5+, 1032.0
[M+6H]6+, 884.7 [M+7H]7+, 774.2
[M+8H]8+, 688.4 [M+9H]9+, 619.6
[M+10H]10+, 563.3 [M+11H]11+.
Glioma cell lines and culture
conditions
T98G cells (39-41)
were grown in RPMI-1640 medium (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
EUROCLONE S.p.A., Pero, Italy), 100 U/ml penicillin and 100 mg/ml
streptomycin, in humidified atmosphere of 5% CO2/air. In
order to study possible anti-proliferative effects, the cell
number/ml was monitored using a Z2 Coulter Counter (Coulter
Electronics, Hialeah, FL, USA). Mycoplasma testing on T98G cells
was performed prior to each experiment.
RNA extraction
Cultured cells were trypsinized and collected by
centrifugation at 250 × g for 10 min at 4°C. Then, cells were lysed
with TRI Reagent (Sigma Aldrich; Merck KGaA, Darmstadt, Germany)
and the isolated RNA was washed once with cold 75% ethanol, dried
and dissolved in nuclease-free pure water prior to use (36).
Quantitative analyses of miRNAs
For miRNA quantification using RT-qPCR, reagents,
primers and probes for hsa-miR-221-3p and hsa-miR-155-5p were
obtained from Applied Biosystems (Thermo Fisher Scientific, Inc.).
Reverse transcription was performed using the TaqMan MicroRNA
Reverse Transcription kit, with 20 ng per sample used for the
assays. qPCR was performed as described elsewhere (36), using the CFX96 Touch Real-Time PCR
Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
The relative miRNA content was calculated using the comparative
cycle threshold method and U6 snRNA and has-let-7c were used as
references to normalize all RNA samples, as previously described
(38).
Analysis of caspase-3 gene
expression
Gene expression analysis was performed by RT-qPCR.
Total RNA (500 ng) was reverse transcribed by using random
hexamers. qPCR assays were performed using gene-specific
double-fluorescently labeled probes. Primers and probes used to
assay caspase-3 gene expression were purchased from Applied
Biosystems (Themo Fisher Scientific, Inc.). The relative expression
was calculated using the comparative cycle threshold method and the
human RPL13A (Assay ID: Hs04194366_g1) was used as a reference gene
(30,31,38).
Caspase-3 protein activity was analyzed using
Bio-PlexPro RBM Apoptosis Assays (Bio-Rad Laboratories, Inc.), an
immunoassay performed on magnetic beads. Glioblastoma cells were
seeded in 6-well plates, treated with the compounds and after 72 h
total cell extracts were prepared. Cells were washed with cold
sterile PBS, centrifuged, the pellet was suspended in LDB (lysate
dilution buffer) and after 8 thermal shock cycles using dry ice,
the suspension was centrifuged at 4°C for 10 min at 9000 × g. The
supernatant was carefully removed and transferred into new vials,
according to the manufacturer's instructions. Protein
quantification was performed using BCA protein assay (Thermo Fisher
Scientific, Inc.). For the analysis with Bio-PlexPro RBM Apoptosis
Panel 3, samples were diluted to a final concentration of 500
µg/ml with LDB. After the reconstitution of the standard,
seven serial 1:3 dilutions were prepared and the assay was
performed, according with the manufacturer's instructions. Briefly,
after reaction with 10 µl of Blocking buffer, 10 µl
of capture beads were added, the reaction incubated, washed three
times with 1X Assay buffer using Bio-Plex Pro Wash Station (Bio-Rad
Laboratories, Inc.) and, finally, detection antibodies (40
µl) were added for a second incubation. Afterwards, 20
µl of diluted streptavidin-phycoerythrin (SA-PE) were added
for 30 min at room temperature. After washing, the beads were
suspended in 100 µl 1X Assay buffer, incubated with shaking
for 30 sec and reading was performed at low PMT (photomultiplier
tube) with Bio-Plex 200 Array reader (Bio-Rad Laboratories, Inc.).
Data were analyzed with Bio-Plex Manager software (Bio-Rad
Laboratories, Inc.).
Analysis of apoptosis
Annexin V and Dead Cell assay were performed on T98G
cells using a Muse Cell Analyzer (Millipore Corporation, Billerica,
MA, USA), as described elsewhere (38). Cells were washed with sterile PBS,
trypsinized, suspended and diluted (1:2) with the one-step addition
of the Muse Annexin V and Dead Cell reagent. After incubation for
20 min at room temperature in the dark, samples were analyzed. Data
from prepared samples were acquired and recorded utilizing the
Annexin V and Dead Cell Software Module (Millipore Corporation)
(38).
Statistical analysis
Results are expressed as mean ± standard deviation.
GraphPad Prism software version 5 (GraphPad Software, Inc., La
Jolla, CA, USA) was used for statistical analysis. Comparison
between groups was made by two-way analysis of variance with
post-hoc Bonferroni comparison. P<0.05 was considered to
indicate a statistically significant difference.
Results
Identification of miRNA targets
First, it was confirmed that T98G are resistant to
TMZ treatment, by determining the cell proliferation rate and the
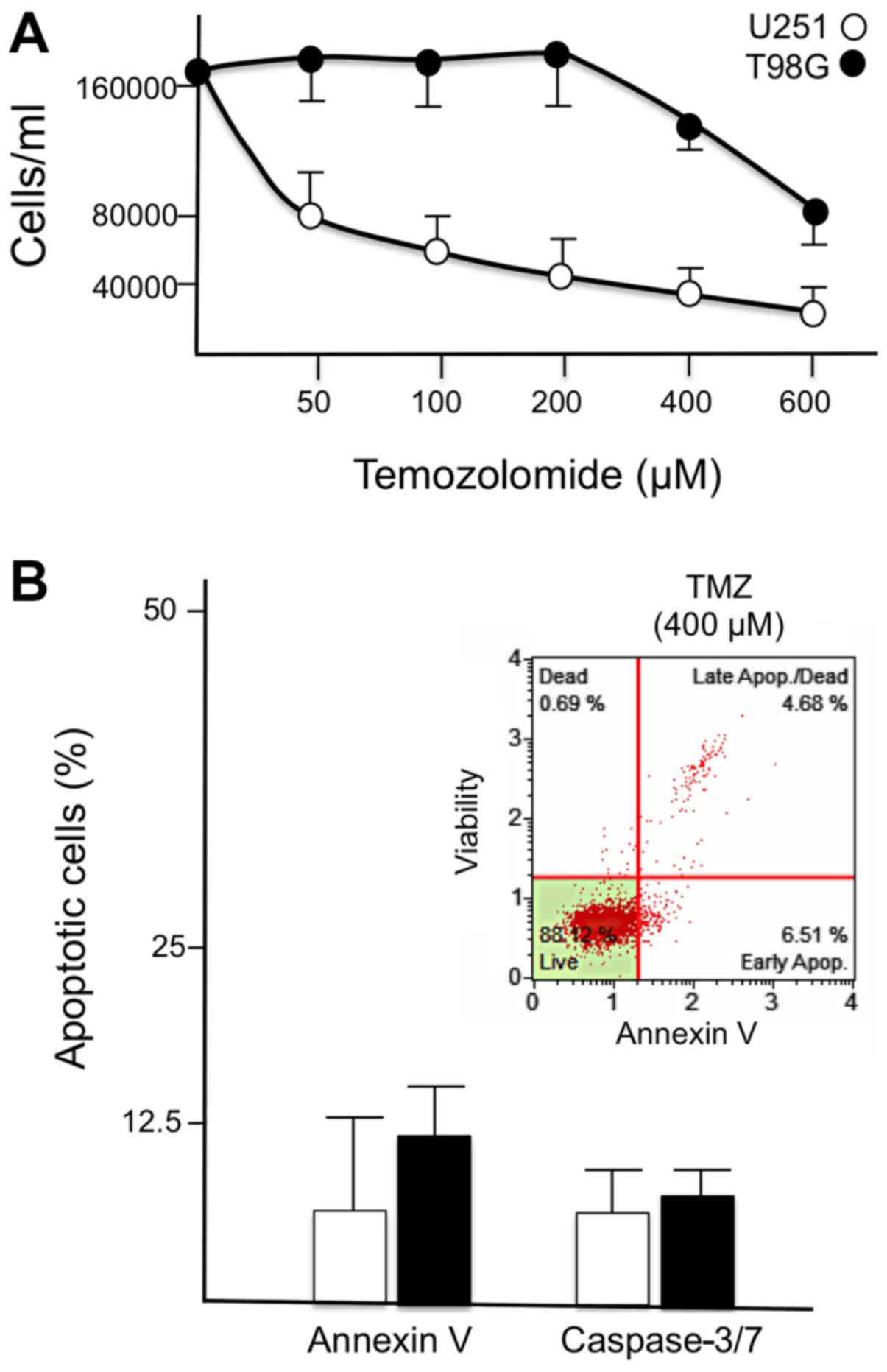
levels of induced apoptosis. As illustrated in Fig. 1A, the T98G cell line was resistant
to TMZ treatment, with only a slight inhibition of cell growth
observed when TMZ was used at very high concentrations (400 and 600
µM). By contrast, an IC50 inhibition was reached
at 50 µM, when TMZ-sensitive U251 glioma cells were treated
with TMZ (Fig. 1A). Furthermore,
TMZ failed to induce a major increase in the proportion of Annexin
V-positive cells when used in T98G cells (Fig. 1B).
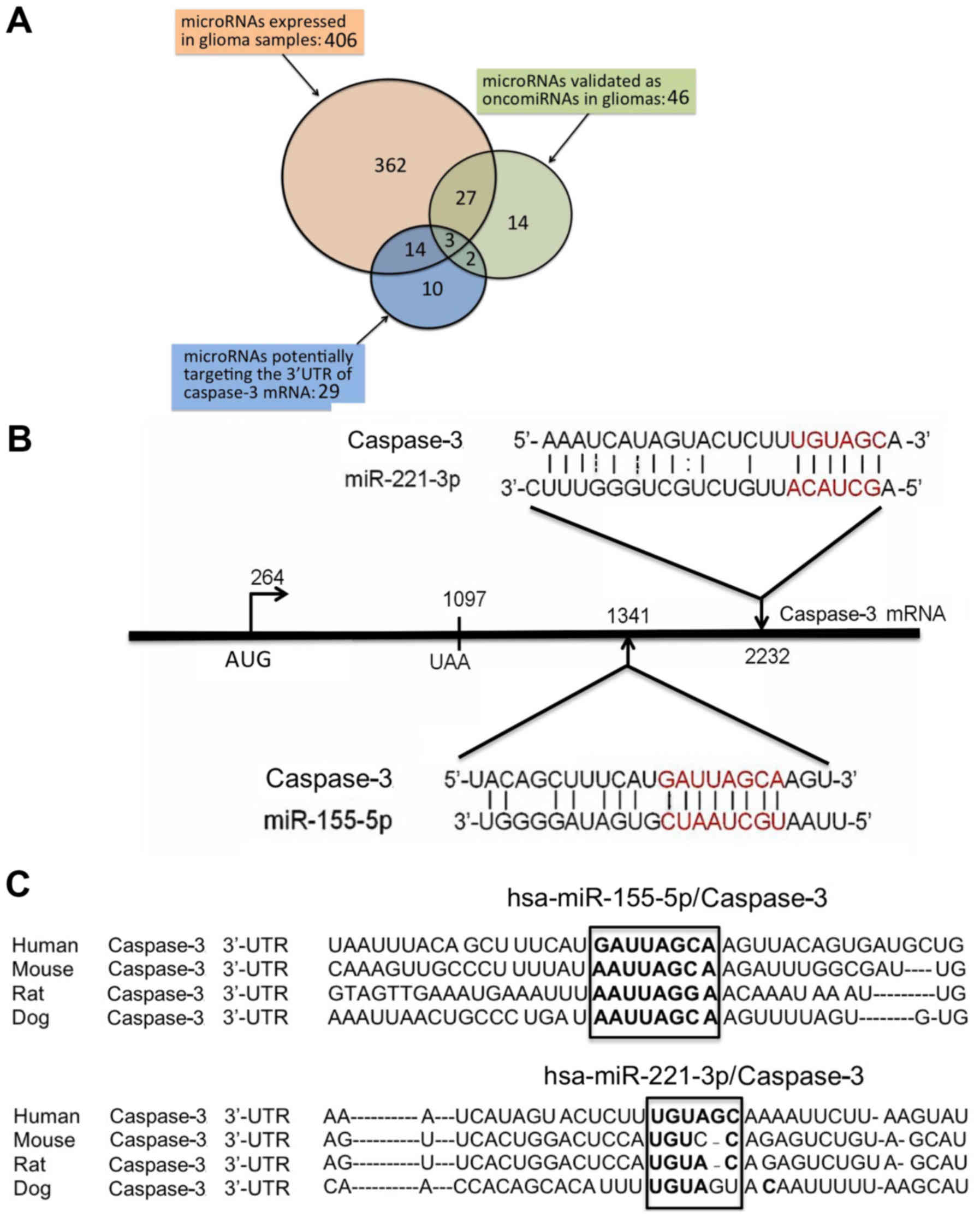
In order to identify possible miRNA targets for the
present study, three sets of miRNAs were compared: miRNAs highly
expressed in tissues from glioma patients; miRNAs validated in
gliomas for their oncogenic properties; and miRNAs putatively
interacting with the 3′ untranslated region (UTR) of caspase-3
mRNA, which is deeply involved in activation of the apoptotic
pathway. These lists of miRNAs are presented in the Tables SI-III. A Venn diagram for the
comparison of the three lists is illustrated in Fig. 2A. Only three miRNA sequences were
found in common within the three sets, miR-155-5p, miR-221-3p and
miR-30a. Of note, miR-221-3p has already been demonstrated to
exhibit anti-apoptotic effects in gliomas, and miR-221-3p targeting
induces apoptosis and reverses TMZ-resistance (42). Similar information is available on
the role of miR-155-5p, which regulates in the sensitization of
glioma cell lines to antitumor drugs (43). By contrast, no report is available
to date on the possible involvement of miR-30a to activation of
drug resistance in gliomas. For these reasons, the present study
focused on targeting miR-155-5p and miR-221-3p.
The location of the sites for these two miRNAs
within the 3′UTR sequences of the caspase-3 mRNA is shown in
Fig. 2B, also depicting the extent
of interactions between these two microRNAs and the caspase-3 mRNA
sequences. When this feature was compared with other miR-155-5p and
miR-221-3p validated targets, it was observed that the level of
interactions is similar and compatible with a true caspase-3 mRNA
regulation by miR-155-5p and miR-221-3p (data not shown). This is
further sustained by the finding that miR-155-5p and miR-221-3p
binding sites are of caspase-3 mRNA conserved through molecular
evolution (Fig. 2C).
R8-PNA-a155 and R8-PNA-a221 exhibit
inhibitory effects on miR-155-5p and miR-221-3p in glioma
cells
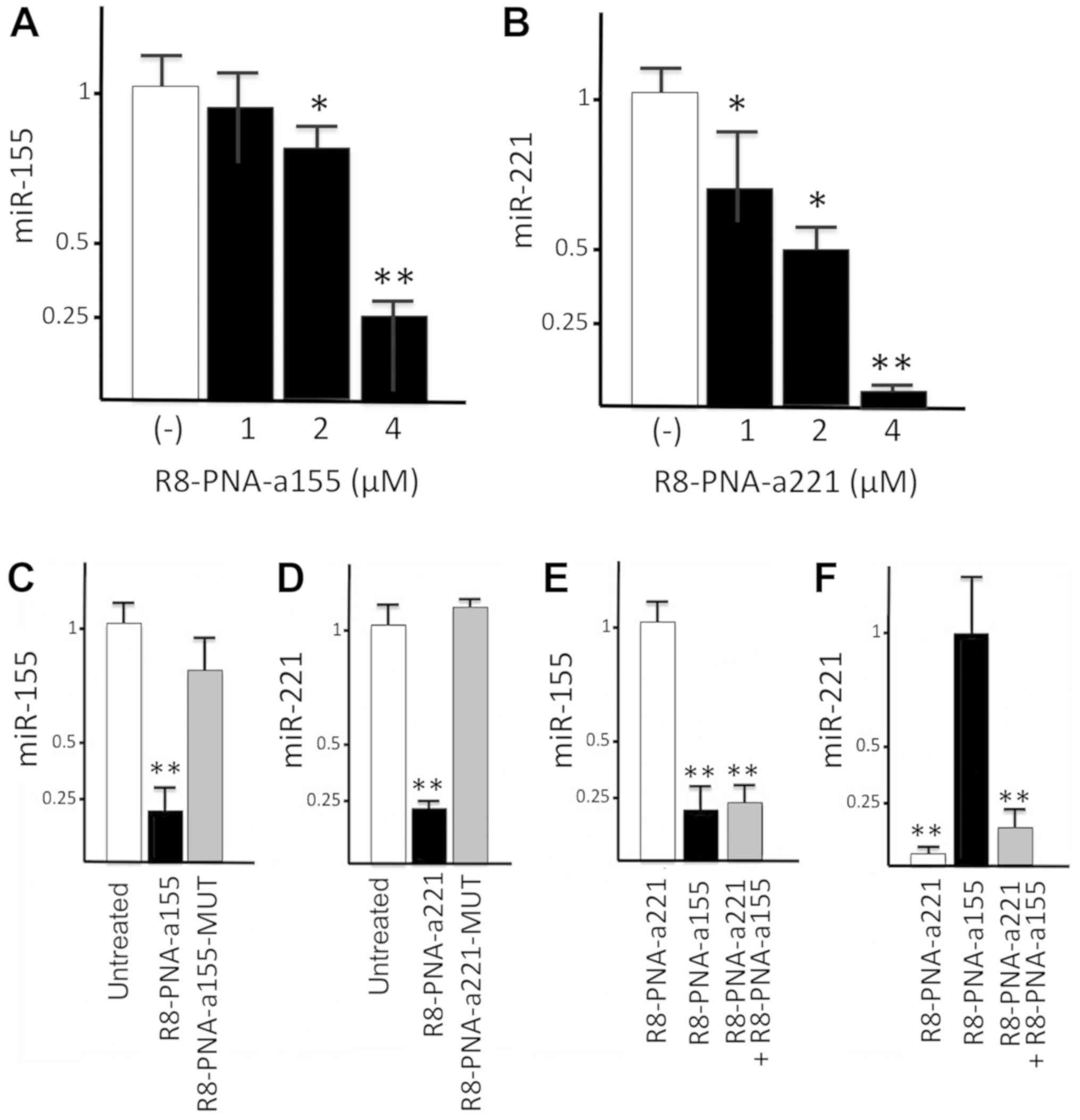
Firstly, glioma cells were treated with increasing
amounts of PNAs R8-PNA-a155 and R8-PNA-a221, targeting miR-155-5p
and miR-221-3p, respectively. The results of the experiments are
presented in Fig. 3A and B, and
clearly demonstrate that 4 µM PNAs was the optimal
concentration to significantly and reproducibly inhibit miR-155-5p
(Fig. 3A) and miR-221-3p (Fig. 3B) hybridization signals.
Next, we determined whether the PNA-mediated effects
were sequence-specific and selective for a given miRNA molecule. To
test this, glioma cells were treated with R8-anti-miR155 PNA and
R8-anti-miR221 PNA, and with PNAs including a mutated sequence
R8-PNA-a155-MUT and R8-PNA-a221-MUT (Fig. 3C and D). Treatment of T98G cells
with R8-PNA-a155 and R8-PNA-a221 resulted in a sharp and
significant inhibition of miR-155-5p and miR-221-3p hybridization
signals, respectively; by contrast, the mutant R8-anti-miR155-MUT
PNA and R8-anti-miR221-MUT PNA displayed only minor effects
(Fig. 3C and D). In a second set
of experiments, results supporting the concept that the effects
were specific were obtained. The results demonstrated that the
miR-155-5p hybridization signal was strongly reduced only when RNA
was isolated from glioma cells cultured for 48 h in the presence of
R8-PNA-a155, with no major effects on miR-221-3p levels (Fig. 3E and F). Conversely, the miR-221-3p
hybridization signal was significantly reduced only when RNA was
isolated from glioma cells cultured for 48 h in the presence of
R8-PNA-a221, while no major effects on miR-221-3p levels were
observed with R8-PNA-a155 (Fig. 3E and
F). Altogether these experiments support the concept that the
effects of R8-PNA-155 on miR-155-5p, and of R8-PNA-a221 on
miR-221-3p, are sequence-specific. As expected, miR-155-5p and
miR-221-3p hybridization signals were reduced by co-administrating
of R8-PNA-155 and R8-PNA-a221 (Fig. 3E
and F).
Effects of R8-PNA-a155 and R8-PNA-a221
treatment on caspase-3 expression
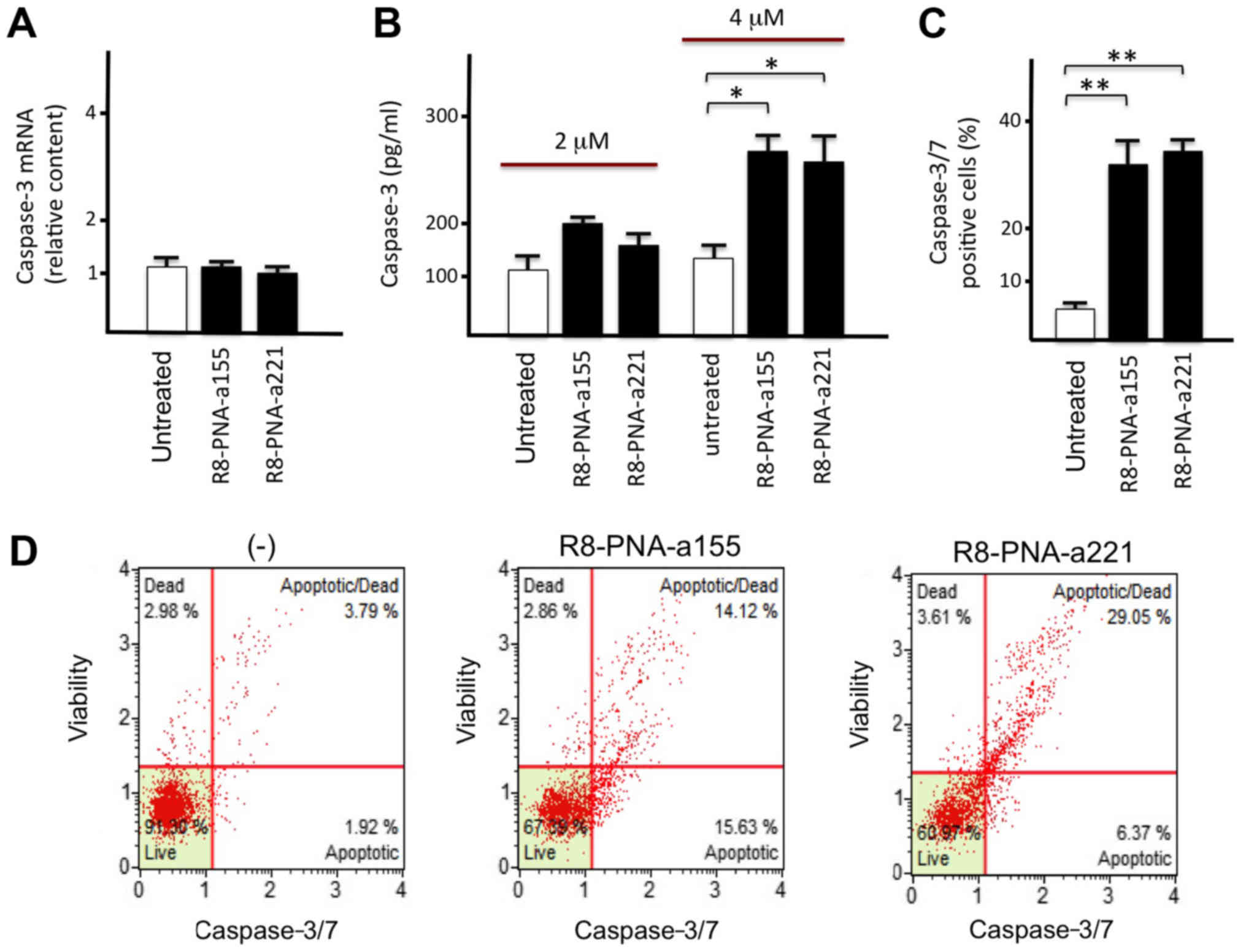
Since caspase-3 mRNA is a putative molecular target
of miR-155-5p and miR-221-3p (Fig.
2B), the glioma T98G cell line was treated with R8-PNA-a155 or
R8-PNA-a221 and caspase-3 mRNA and protein levels were determined.
To this aim, RNA was extracted for RT-qPCR analysis from an aliquot
of cells, while another aliquot was used for preparing protein
extracts for Bio-Plex analysis. The results demonstrated that
treatment with R8-PNA-a155 or R8-PNA-a221 did not affect caspase-3
mRNA levels (Fig. 4A). By
contrast, both treatments resulted in a significant
concentration-dependent increase of caspase-3 protein production,
as determined by Bio-Plex analysis (Fig. 4B). These results are compatible
with a regulation of caspase-3 expression by miR-155-5p and
miR-221-3p at the post-transcriptional level. Furthermore, analysis
of caspase 3/7 function confirmed the activation of the caspase-3/7
pathway in T98G cells treated with PNAs targeting miR-155-5p and
miR-221-3p (Fig. 4C and D).
Effects of R8-PNA-a155 and R8-PNA-a221
treatment on apoptosis
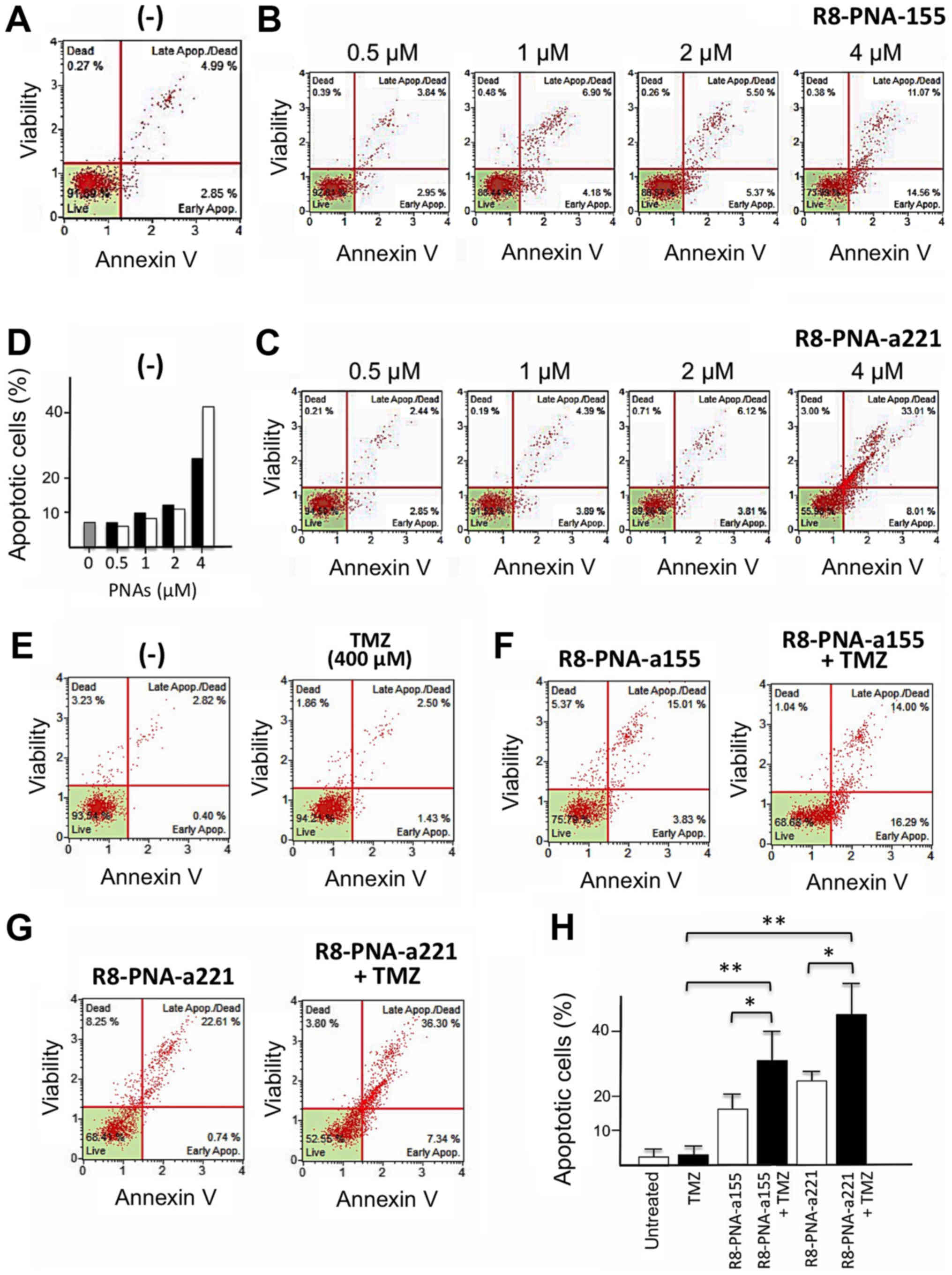
The proapoptotic effects of R8-PNA-a155 and
R8-PNA-a221 were confirmed to appear in a dose-dependent manner
(Fig. 5A-D). Cells were cultured
for 48 h in the absence (Fig. 5A)
or in the presence of 0.5, 1, 2 and 4 µM R8-PNA-a155
(Fig. 5B) or R8-PNA-a221 (Fig. 5C) PNAs and the Annexin-V assay was
performed. As clearly evident in Fig.
5D (which is in agreement with data in Fig. 3), only minor effects were observed
when 0.5-2 µM PNAs were employed, and the higher
proapoptotic effects were observed with 4 µM concentrations.
When the glioma cell line T98G was cultured in the presence of
singularly administered R8-PNA-a155 or R8-PNA-a221 (used at 4
µM concentration) a significant increase of early and late
apoptotic cells was found. Fig.
5E-G presents representative results obtained after treatment
of T98G glioma cell lines with 4 µM R8-PNA-a155, and 4
µM R8-PNA-a221, confirming the proapoptotic effects of
R8-PNA-a221 (as previously published by our group) (38), and demonstrating for the first time
the pro-apoptotic effects of R8-PNA-a155. Notably, when TMZ was
also administered in combination with R8-PNA-a155 or R8-PNA-a221, a
further significant increase of the proportion of apoptotic Annexin
V-positive cells was observed (Fig.
5F-H). These data suggest that treatment of the TMZ-resistant
T98G glioma cells with PNAs targeting miR-155-5p or miR-221-3p
resulted in a sensitization of glioma cells to TMZ.
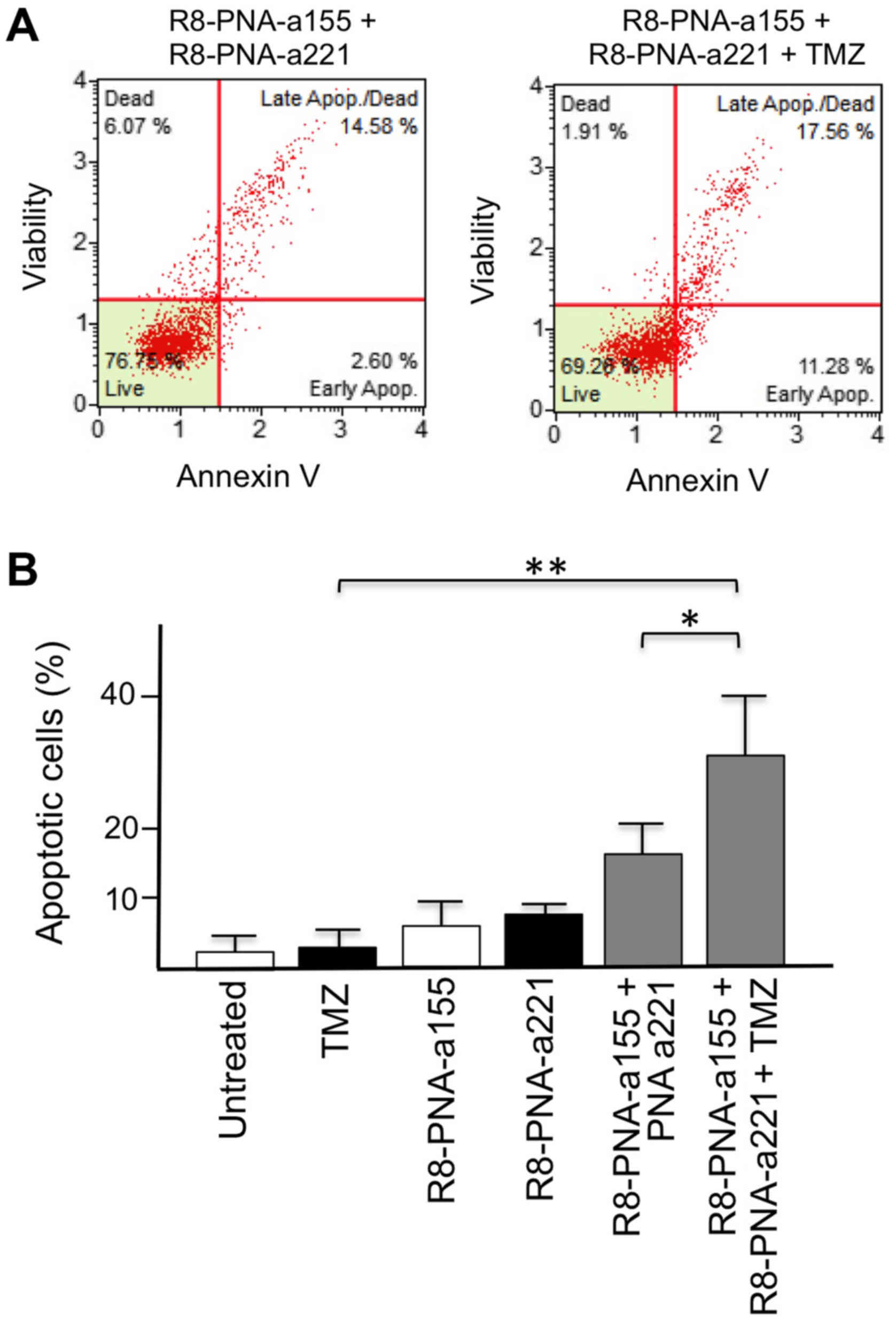
Effects of co-treatment with suboptimal
concentrations of R8-PNA-a155 or R8-PNA-a221 and TMZ on
apoptosis
Since dose-dependent off-target effects of antisense
molecules (including PNAs) is one of the major issues in this
potential therapeutic strategy, the effects of combined treatments
in the presence of 2 µM R8-PNA-a155 and R8-PNA-a221, which
is half of the dose used in the experiments depicted in Fig. 5, were determined. As illustrated in
Fig. 6, the levels of apoptosis
induction reached by the co-administration of 2 µM
R8-PNA-a155 and 2 µM R8-PNA-a221 in T98G cells are similar
to those obtained by singular administration of 4 µM
R8-PNA-a155 or R8-PNA-a221. This indicates that the use of
suboptimal concentrations of anti-miR-155 and anti-miR-221 PNAs
allows reaching high levels of apoptosis (Fig. 6A). This level was further increased
when TMZ was added (Fig. 6A and
B). These results demonstrated that co-administration of
R8-PNA-a155 and R8-PNA-a221 induced apoptosis in TMZ-treated T98G
cells at levels similar to those obtained following singular
administration of high doses of R8-PNA-a155 or R8-PNA-a221.
Discussion
Because of the poor prognosis of gliomas and of the
development of resistance to drugs commonly used in post-surgery
antitumor protocols, novel therapeutic strategies are highly needed
for GBM, some of which may already be tested in therapeutic
protocols for other tumors (44-47).
MicroRNAs are putative molecular targets, based on current
knowledge: glioma-associated oncomiRNAs have been described whose
expression is deeply impaired during onset and progression of these
tumors (48-50); the expression of some oncomiRNAs is
significantly associated with outcome, therefore rendering them a
very useful potential marker in the analysis of non-invasive liquid
biopsies (51); miRNA therapeutics
have been recently demonstrated to be useful in the treatment of a
variety of human pathologies, including gliomas (52-55);
miRNA pathways might exhibit a patient-to-patient variability,
allowing the design of personalized protocols (56,57);
and miRNA targeting might be employed to overcome drug-resistance,
an issue of great relevance in the management of patients with
glioma (58-61).
Gliomas express two microRNAs, miR-155-5p and
miR-221-3p, at high levels, and these are associated with oncogenic
activity and strong antiapoptotic effects (62-64).
These effects are mediated by a putative targeting by these miRNAs
of the 3′UTR sequence of the proapoptotic caspase-3 mRNA, possibly
leading to a downregulation of caspase-3 expression (42,43,65).
Therefore, miR-155-5p and miR-221-3p appear to be appealing targets
for the development of therapeutic protocols for gliomas.
The use of PNAs is very promising since, from their
introduction following the first description by Nielsen et
al (18), they have been
considered for therapeutic interventions on a variety of human
pathologies, including cancer. However, several issues should be
considered in proposing PNAs as therapeutic tools in miRNA
therapeutics: their delivery to target cells; possible off-target
effects due to the fact that a single miRNA might recognize several
mRNA targets; and the presence in the 3′UTR of each target mRNA of
several potential miRNA binding sites, which might require
co-administration of different anti-miRNA molecules, leading to
further complications of the off-targeting issue.
The major conclusion of the present study is that
two miRNAs (miR-155-5p and miR-221-3p), highly expressed in gliomas
and demonstrated to regulate caspase-3 gene expression (42,43,65),
were the target of PNA-based induction of apoptosis in the
TMZ-resistant T98G glioma cell line. Of note, TMZ was able to
further increase apoptosis induced by the PNAs targeting these two
miRNAs. Furthermore, combined treatment using low PNA doses and TMZ
resulted in higher pro-apoptotic effects, suggesting a
sensitization of glioma cells to TMZ.
The present results support the concept that
anti-miRNA strategy could lead to therapeutic relevant inhibition
of biological functions of miRNA-regulated mRNAs (8-11)
and that PNA-based anti-miRNA molecules are very promising reagents
as a tool for the development of therapeutic protocols for tumor
cell growth inhibition. In this context, and considering the low
uptake of PNAs (21), further
research on PNA analogues is necessary with the aim of increasing
delivery, improving stability, controlling the intracellular
distribution and the in vivo tissue targeting. In addition,
PNAs selectively interacting with specific mature miRNAs,
pre-miRNAs or pri-miRNAs might be compared as a further step for
the selection of the best candidate PNA-based drugs. The present
study strongly indicated that the combined treatment of target
glioma cells with PNAs inhibiting both miR-155-5p and miR-221-3p
was associated with a significant improvement of the efficacy of
the treatment, as evidenced by their effects on cell apoptosis.
This conclusion supports the strategy of designing multifunctional
PNA-containing systems or nanocarriers (67,68),
enabling to perform targeting of multiple miRNA sequences. Finally,
our data are compatible with a sensitization of T98G cells to TMZ,
supporting previous observations indicating anti-miRNA strategy may
be a potential tool to reverse drug resistance, which is one of the
major unresolved issues in the therapeutic management of patients
with glioma (58-60).
Supplementary Materials
Funding
This study was partially supported by the European
Union (EU) Horizon 2020 Research and Innovation Programme (grant
no. 633937; project ULTRAsensitive PLAsmonic devices for early
CAncer Diagnosis, ULTRAPLACAD). This work was also funded by CIB,
by COFIN-2009 and by AIRC (grant no. 13575; peptide nucleic acids
targeting oncomiR and tumor-suppressor miRNAs: cancer diagnosis and
therapy). GC was funded by the Verona Brain Research
Foundation.
Availability of data and materials
All data generated or analyzed during this study are
included within the manuscript.
Authors' contributions
RG and RC conceived and planned all the experiments.
Cell culture was performed by RM and EB. Bioinformatic analyses
were conducted by EF. Design and synthesis of PNAs were performed
by RC and AM. Treatments of the cells with PNAs were performed by
RM and LCC. Molecular analyses were performed by AF and JG.
Microarray-based analysis of miRNAs in gliomas has been conducted
by GC and MCD. Analysis of apoptosis was performed by IL and MCD.
RM, RG and GC contributed to the interpretation of the results. RG
wrote the manuscript. All authors provided critical feedback and
contributed to the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
PNA
|
peptide nucleic acid
|
|
miRNA
|
microRNA
|
|
GMB
|
glioblastoma multiforme
|
|
TMZ
|
temozolomide
|
|
Fl
|
fluorescein
|
|
FBS
|
fetal bovine serum
|
|
BCA
|
bicinchoninic acid
|
|
LDB
|
lysate dilution buffer
|
|
RT
|
reverse transcription
|
|
PCR
|
polymerase chain reaction
|
|
RT-PCR
|
reverse transcription polymerase-chain
reaction
|
|
SDS
|
sodium dodecylsulphate
|
|
SDS-PAGE
|
SDS-polyacrylamide-gel
electrophoresis
|
|
PBS
|
phosphate buffered saline
|
|
UPLC-MS
|
ultra-performance liquid
chromatography-mass spectrometry
|
Acknowledgments
We thank Nicoletta Bianchi for support and
suggestions.
References
|
1
|
von Neubeck C, Seidlitz A, Kitzler HH,
Beuthien-Baumann B and Krause M: Glioblastoma multiforme: Emerging
treatments and stratification markers beyond new drugs. Br J
Radiol. 88:201503542015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Buczkowicz P and Hawkins C: Pathology,
molecular genetics, and epigenetics of diffuse intrinsic pontine
glioma. Front Oncol. 5:1472015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pace A, Dirven L, Koekkoek JAF, Golla H,
Fleming J, Rudà R, Marosi C, Le Rhun E, Grant R, Oliver K, et al
European Association of Neuro-Oncology palliative care task force:
European Association for Neuro-Oncology (EANO) guidelines for
palliative care in adults with glioma. Lancet Oncol. 18:e330–e340.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alexander BM and Cloughesy TF: Adult
glioblastoma. J Clin Oncol. 35:2402–2409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Griffiths-Jones S: The microRNA Registry.
Nucleic Acids Res. 32:D109–D111. 2004. View Article : Google Scholar :
|
|
8
|
Piva R, Spandidos DA and Gambari R: From
microRNA functions to microRNA therapeutics: Novel targets and
novel drugs in breast cancer research and treatment (Review). Int J
Oncol. 43:985–994. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taylor MA and Schiemann WP: Therapeutic
opportunities for targeting microRNAs in cancer. Mol Cell Ther.
2:1–13. 2014. View Article : Google Scholar
|
|
10
|
Song MS and Rossi JJ: The anti-miR21
antagomir, a therapeutic tool for colorectal cancer, has a
potential synergistic effect by perturbing an
angiogenesis-associated miR30. Front Genet. 4:3012014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nana-Sinkam SP and Croce CM: Clinical
applications for microRNAs in cancer. Clin Pharmacol Ther.
93:98–104. 2013. View Article : Google Scholar
|
|
12
|
Hermansen SK and Kristensen BW: MicroRNA
biomarkers in glioblastoma. J Neurooncol. 114:13–23. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khalil S, Fabbri E, Santangelo A, Bezzerri
V, Cantù C, Di Gennaro G, Finotti A, Ghimenton C, Eccher A,
Dechecchi M, et al: miRNA array screening reveals cooperative
MGMT-regulation between miR-181d-5p and miR-409-3p in glioblastoma.
Oncotarget. 7:28195–28206. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shu M, Zheng X, Wu S, Lu H, Leng T, Zhu W,
Zhou Y, Ou Y, Lin X, Lin Y, et al: Targeting oncogenic miR-335
inhibits growth and invasion of malignant astrocytoma cells. Mol
Cancer. 10:592011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chan XH, Nama S, Gopal F, Rizk P, Ramasamy
S, Sundaram G, Ow GS, Ivshina AV, Tanavde V, Haybaeck J, et al:
Targeting glioma stem cells by functional inhibition of a
prosurvival oncomiR-138 in malignant gliomas. Cell Rep. 2:591–602.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wagenaar TR, Zabludoff S, Ahn SM, Allerson
C, Arlt H, Baffa R, Cao H, Davis S, Garcia-Echeverria C, Gaur R, et
al: Anti-miR-21 suppresses hepatocellular carcinoma growth via
broad transcriptional network de-regulation. Mol Cancer Res.
13:1009–1021. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma L, Reinhardt F, Pan E, Soutschek J,
Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW and Weinberg RA:
Therapeutic silencing of miR-10b inhibits metastasis in a mouse
mammary tumor model. Nat Biotechnol. 28:341–347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nielsen PE, Egholm M, Berg RH and Buchardt
O: Sequence-selective recognition of DNA by strand displacement
with a thymine-substituted polyamide. Science. 254:1497–1500. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nielsen PE: Targeting double stranded DNA
with peptide nucleic acid (PNA). Curr Med Chem. 8:545–550. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Borgatti M, Lampronti I, Romanelli A,
Pedone C, Saviano M, Bianchi N, Mischiati C and Gambari R:
Transcription factor decoy molecules based on a peptide nucleic
acid (PNA)-DNA chimera mimicking Sp1 binding sites. J Biol Chem.
278:7500–7509. 2003. View Article : Google Scholar
|
|
21
|
Gambari R: Peptide-nucleic acids (PNAs): A
tool for the development of gene expression modifiers. Curr Pharm
Des. 7:1839–1862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gambari R: Biological activity and
delivery of peptide nucleic acids (PNA)-DNA chimeras for
transcription factor decoy (TFD) pharmacotherapy. Curr Med Chem.
11:1253–1263. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nielsen PE: Peptide nucleic acids (PNA) in
chemical biology and drug discovery. Chem Biodivers. 7:786–804.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nielsen PE: Gene targeting and expression
modulation by peptide nucleic acids (PNA). Curr Pharm Des.
16:3118–3123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Krupnik OV, Guscho Y, Sluchanko K, Nielsen
P and Lazurkin Y: Thermodynamics of the melting of PNA(2)/DNA
triple helices. J Biomol Struct Dyn. 19:535–542. 2001. View Article : Google Scholar
|
|
26
|
Bentin T and Nielsen PE: Superior duplex
DNA strand invasion by acridine conjugated peptide nucleic acids. J
Am Chem Soc. 125:6378–6379. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hatamoto M, Ohashi A and Imachi H: Peptide
nucleic acids (PNAs) antisense effect to bacterial growth and their
application potentiality in biotechnology. Appl Microbiol
Biotechnol. 86:397–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gambari R, Borgatti M, Bezzerri V, Nicolis
E, Lampronti I, Dechecchi MC, Mancini I, Tamanini A and Cabrini G:
Decoy oligodeoxyribonucleotides and peptide nucleic acids-DNA
chimeras targeting nuclear factor kappa-B: Inhibition of IL-8 gene
expression in cystic fibrosis cells infected with Pseudomonas
aeruginosa. Biochem Pharmacol. 80:1887–1894. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pandey VN, Upadhyay A and Chaubey B:
Prospects for antisense peptide nucleic acid (PNA) therapies for
HIV. Expert Opin Biol Ther. 9:975–989. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Manicardi A, Fabbri E, Tedeschi T, Sforza
S, Bianchi N, Brognara E, Gambari R, Marchelli R and Corradini R:
Cellular uptakes, biostabilities and anti-miR-210 activities of
chiral arginine-PNAs in leukaemic K562 cells. ChemBioChem.
13:1327–1337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fabbri E, Manicardi A, Tedeschi T, Sforza
S, Bianchi N, Brognara E, Finotti A, Breveglieri G, Borgatti M,
Corradini R, et al: Modulation of the biological activity of
microRNA-210 with peptide nucleic acids (PNAs). ChemMedChem.
6:2192–2202. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gambari R, Fabbri E, Borgatti M, Lampronti
I, Finotti A, Brognara E, Bianchi N, Manicardi A, Marchelli R and
Corradini R: Targeting microRNAs involved in human diseases: A
novel approach for modification of gene expression and drug
development. Biochem Pharmacol. 82:1416–1429. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fabani MM and Gait MJ: miR-122 targeting
with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids
(PNA), and PNA-peptide conjugates. RNA. 14:336–346. 2008.
View Article : Google Scholar :
|
|
34
|
Fabani MM, Abreu-Goodger C, Williams D,
Lyons PA, Torres AG, Smith KG, Enright AJ, Gait MJ and Vigorito E:
Efficient inhibition of miR-155 function in vivo by peptide nucleic
acids. Nucleic Acids Res. 38:4466–4475. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Brown PN and Yin H: PNA-based microRNA
inhibitors elicit anti-inflammatory effects in microglia cells.
Chem Commun (Camb). 49:4415–4417. 2013. View Article : Google Scholar
|
|
36
|
Brognara E, Fabbri E, Aimi F, Manicardi A,
Bianchi N, Finotti A, Breveglieri G, Borgatti M, Corradini R,
Marchelli R, et al: Peptide nucleic acids targeting miR-221
modulate p27Kip1 expression in breast cancer MDA-MB-231
cells. Int J Oncol. 41:2119–2127. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar
|
|
38
|
Brognara E, Fabbri E, Bazzoli E, Montagner
G, Ghimenton C, Eccher A, Cantù C, Manicardi A, Bianchi N, Finotti
A, et al: Uptake by human glioma cell lines and biological effects
of a peptide-nucleic acids targeting miR-221. J Neurooncol.
118:19–28. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cao X, Gu Y, Jiang L, Wang Y, Liu F, Xu Y,
Deng J, Nan Y, Zhang L, Ye J, et al: A new approach to screening
cancer stem cells from the U251 human glioma cell line based on
cell growth state. Oncol Rep. 29:1013–1018. 2013. View Article : Google Scholar
|
|
40
|
Abdullah Thani NA, Sallis B, Nuttall R,
Schubert FR, Ahsan M, Davies D, Purewal S, Cooper A and Rooprai HK:
Induction of apoptosis and reduction of MMP gene expression in the
U373 cell line by polyphenolics in Aronia melanocarpa and by
curcumin. Oncol Rep. 28:1435–1442. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pen A, Durocher Y, Slinn J, Rukhlova M,
Charlebois C, Stanimirovic DB and Moreno MJ: Insulin-like growth
factor binding protein 7 exhibits tumor suppressive and vessel
stabilization properties in U87MG and T98G glioblastoma cell lines.
Cancer Biol Ther. 12:634–646. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang JK, Yang JP, Tong J, Jing SY, Fan B,
Wang F, Sun GZ and Jiao BH: Exosomal miR-221 targets DNM3 to induce
tumor progression and temozolomide resistance in glioma. J
Neurooncol. 131:255–265. 2017. View Article : Google Scholar
|
|
43
|
Liu Q, Zou R, Zhou R, Gong C, Wang Z, Cai
T, Tan C and Fang J: miR-155 regulates glioma cells invasion and
chemosensitivity by p38 isforms in vitro. J Cell Biochem.
116:1213–1221. 2015. View Article : Google Scholar
|
|
44
|
Jung J, Yeom C, Choi YS, Kim S, Lee E,
Park MJ, Kang SW, Kim SB and Chang S: Simultaneous inhibition of
multiple oncogenic miRNAs by a multi-potent microRNA sponge.
Oncotarget. 6:20370–20387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Anjum K, Shagufta BI, Abbas SQ, Patel S,
Khan I, Shah SAA, Akhter N and Hassan SSU: Current status and
future therapeutic perspectives of glioblastoma multiforme (GBM)
therapy: A review. Biomed Pharmacother. 92:681–689. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lozada-Delgado EL, Grafals-Ruiz N and
Vivas-Mejía PE: RNA interference for glioblastoma therapy:
Innovation ladder from the bench to clinical trials. Life Sci.
188:26–36. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Touat M, Idbaih A, Sanson M and Ligon KL:
Glioblastoma targeted therapy: Updated approaches from recent
biological insights. Ann Oncol. 28:1457–1472. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li C, Sun J, Xiang Q, Liang Y, Zhao N,
Zhang Z, Liu Q and Cui Y: Prognostic role of microRNA-21 expression
in gliomas: A meta-analysis. J Neurooncol. 130:11–17. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Beyer S, Fleming J, Meng W, Singh R, Haque
SJ and Chakravarti A: The role of miRNAs in angiogenesis, invasion
and metabolism and their therapeutic implications in gliomas.
Cancers (Basel). 9. pp. E852017, View Article : Google Scholar
|
|
50
|
Wang Y, Wang X, Zhang J, Sun G, Luo H,
Kang C, Pu P, Jiang T, Liu N and You Y: MicroRNAs involved in the
EGFR/PTEN/AKT pathway in gliomas. J Neurooncol. 106:217–224. 2012.
View Article : Google Scholar
|
|
51
|
Regazzo G, Terrenato I, Spagnuolo M,
Carosi M, Cognetti G, Cicchillitti L, Sperati F, Villani V,
Carapella C, Piaggio G, et al: A restricted signature of serum
miRNAs distinguishes glioblastoma from lower grade gliomas. J Exp
Clin Cancer Res. 35:1242016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen L and Kang C: miRNA interventions
serve as 'magic bullets' in the reversal of glioblastoma hallmarks.
Oncotarget. 6:38628–38642. 2015.PubMed/NCBI
|
|
53
|
Areeb Z, Stylli SS, Koldej R, Ritchie DS,
Siegal T, Morokoff AP, Kaye AH and Luwor RB: MicroRNA as potential
biomarkers in Glioblastoma. J Neurooncol. 125:237–248. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ouyang Q, Xu L, Cui H, Xu M and Yi L:
MicroRNAs and cell cycle of malignant glioma. Int J Neurosci.
126:1–9. 2016. View Article : Google Scholar
|
|
55
|
Wang H, Xu T, Jiang Y, Yan Y, Qin R and
Chen J: MicroRNAs in human glioblastoma: From bench to beside.
Front Biosci. 20:105–118. 2015. View
Article : Google Scholar
|
|
56
|
Gambari R, Brognara E, Spandidos DA and
Fabbri E: Targeting oncomiRNAs and mimicking tumor suppressor
miRNAs: New trends in the development of miRNA therapeutic
strategies in oncology (Review). Int J Oncol. 49:5–32. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Finotti A, Allegretti M, Gasparello J,
Giacomini P, Spandidos DA, Spoto G and Gambari R: Liquid biopsy and
PCR-free ultrasensitive detection systems in oncology (Review). Int
J Oncol. 53:1395–1434. 2018.PubMed/NCBI
|
|
58
|
Li W, Guo F, Wang P, Hong S and Zhang C:
miR-221/222 confers radioresistance in glioblastoma cells through
activating Akt independent of PTEN status. Curr Mol Med.
14:185–195. 2014. View Article : Google Scholar
|
|
59
|
Chen L, Zhang J, Han L, Zhang A, Zhang C,
Zheng Y, Jiang T, Pu P, Jiang C and Kang C: Downregulation of
miR-221/222 sensitizes glioma cells to temozolomide by regulating
apoptosis independently of p53 status. Oncol Rep. 27:854–860.
2012.
|
|
60
|
Xie Q, Yan Y, Huang Z, Zhong X and Huang
L: MicroRNA-221 targeting PI3-K/Akt signaling axis induces cell
proliferation and BCNU resistance in human glioblastoma.
Neuropathology. 34:455–464. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Costa PM, Cardoso AL, Mano M and de Lima
MC: MicroRNAs in glioblastoma: Role in pathogenesis and
opportunities for targeted therapies. CNS Neurol Disord Drug
Targets. 14:222–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yan Z, Che S, Wang J, Jiao Y, Wang C and
Meng Q: miR-155 contributes to the progression of glioma by
enhancing Wnt/β-catenin pathway. Tumour Biol. 36:5323–5331. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yang L, Li C, Liang F, Fan Y and Zhang S:
miRNA-155 promotes proliferation by targeting caudal-type homeobox
1 (CDX1) in glioma cells. Biomed Pharmacother. 95:1759–1764. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
De Santis R, Liepelt A, Mossanen JC, Dueck
A, Simons N, Mohs A, Trautwein C, Meister G, Marx G,
Ostareck-Lederer A, et al: miR-155 targets caspase-3 mRNA in
activated macrophages. RNA Biol. 13:43–58. 2016. View Article : Google Scholar :
|
|
66
|
Ergun S, Arman K, Temiz E, Bozgeyik I,
Yumrutaş Ö, Safdar M, Dağlı H, Arslan A and Oztuzcu S: Expression
patterns of miR-221 and its target caspase-3 in different cancer
cell lines. Mol Biol Rep. 41:5877–5881. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bertucci A, Lülf H, Septiadi D, Manicardi
A, Corradini R and De Cola L: Intracellular delivery of peptide
nucleic acid and organic molecules using zeolite-L nanocrystals.
Adv Healthc Mater. 3:1812–1817. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bertucci A, Prasetyanto EA, Septiadi D,
Manicardi A, Brognara E, Gambari R, Corradini R and De Cola L:
Combined delivery of temozolomide and anti-miR221 PNA using
mesoporous silica nanoparticles induces apoptosis in resistant
glioma Cells. Small. 11:5687–5695. 2015. View Article : Google Scholar : PubMed/NCBI
|