Introduction
Neuroblastoma (NB) is one of the most common
extracranial solid tumors in children, accounting for 15% of all
childhood deaths from cancer (1).
It is almost exclusively a pediatric malignancy, and >90% of
patients are diagnosed at <10 years of age (2,3). NB
originates in neural crest cells of the sympathetic nervous system,
which are mainly found in the adrenal gland, neck, chest, abdomen
and pelvic cavity. Although the clinical diagnosis and treatment
for NB are continually improving, the 5-year survival rate for
children with high-risk NB remains <50% (4).
The mechanisms underlying NB pathogenesis and
development are complex, involving genetic and epigenetic
alterations, chromosomal changes, and the altered expression of
microRNAs (miRNAs/miRs) and long non-coding RNAs (lncRNAs)
(5,6). Among these genetic changes, the
Children's Oncology Group revealed that MYCN proto-oncogene, bHLH
transcription factor (MYCN) amplification is an independent
marker for NB prognosis and risk stratification (7,8).
However, MYCN amplification occurs in only 20-30% of patients with
primary NB (9). Due to the
diversity of clinical phenotypes and complex biological
characteristics, there is an urgent need to identify precise
biological markers for NB diagnosis and prognosis, as well as
potential molecular targets for chemotherapy.
lncRNAs are non-coding R NA transcripts >200
nucleotides long, which are involved in transcriptional and
post-transcriptional regulation (10), and have important value for the
diagnosis and treatment of tumors. They contribute to various
biological processes in tumorigenesis, including tumor
proliferation, metastasis, differentiation and cell death. lncRNAs
can be detected in urine, and prostate cancer-associated 3 was
recently approved by the US Food and Drug Administration to
identify prostate cancer (11,12).
In pediatric NB, various lncRNAs, including metastasis-associated
lung adenocarcinoma transcript 1 (MALAT1), HOXD antisense
growth-associated long non-coding RNA and
miR-100-let-7a-2-miR-125b-1 cluster host gene (linc-NeD125),
are involved in differentiation, tumor proliferation, invasion and
migration (13-16). Furthermore, in patients with
high-risk NB, small nucleolar RNA host gene 1 is highly expressed
and is strongly correlated with MYCN amplification (17,18).
As a member of the small nucleolar RNA host gene
family, small nucleolar RNA host gene 16 (SNHG16) is highly
expressed in several types of cancer (18-21).
It is regulated by the Wnt pathway in colorectal cancer and induced
breast cancer cell migration (19,20).
Although this suggests that SNHG16 may function as an
oncogene in cancer, its underlying molecular mechanisms are
unclear, particularly in pediatric NB. Therefore, the present study
further investigated the effects of SNHG16 on NB.
Materials and methods
Clinical patients
All patients with NB (aged between 7 months and 8
years) were clinically and histopathologically diagnosed at Beijing
Children's Hospital between May 2015 and December 2016 based on the
International Neuroblastoma Staging System (INSS) for clinical
staging of NB (22). The present
study was approved by the Ethics Committees of Beijing Children's
Hospital. A total of 40 surgical specimens were immediately
snap-frozen in liquid nitrogen prior to total RNA extraction.
Cell culture and transfection
The NB cell line SH-SY5Y (#CRL-2266; MYCN
non-amplified) was obtained from the American Type Culture
Collection (ATCC); this cell line is widely used in mechanistic and
drug development studies regarding NB (23,24).
Cells were cultured in Dulbecco's modified Eagle's medium (Corning,
Inc.) supplemented with 10% fetal bovine serum (FBS; Corning, Inc.)
in a humidified incubator containing 5% CO2 at 37°C.
According to the manufacturer's protocol, synthetic small
interfering (si)RNAs were transfected into cells at ~50% confluence
using the Lipofectamine RNAiMAX kit (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were further analyzed 8 h
post-transfection.
RNA interference
SH-SY5Y cells in the exponential growth phase were
seeded for 24 h and were then transfected with 100 nM siRNA at room
temperature using Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher
Scientific, Inc.). siRNA oligonucleotides were synthesized by
Sangon Biotech Co., Ltd., as follows: SNHG16
(siRNA1-SNHG16, 5′-CAGCAGUUGAGGGUUUGCUGUGUAUdTdT-3′,
siRNA2-SNHG16, 5′-GGACAACCUAGCUGUUGAAdTdT-3′); non-targeting
control (5′-UUCUCCGAACGUGUCACGUTT-3′). Cells were returned to the
incubator and were refreshed with normal medium 8 h
post-transfection, and further experiments were performed at
scheduled times.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
A total of 3×105 SH-SY5Y cells/well were
seeded in 6-well plates at ~50% confluence and transfected with
siRNA for 72 h. Tumor tissues and cells were homogenized in
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), and total RNA was extracted using the Direct-zol™ RNA
Miniprep kit (Zymo Research Corp.). The RevertAid™ H Minus First
Strand cDNA Synthesis kit (Invitrogen) was used for RT, and cDNA
templates were amplified with the SYBR Green Master mix (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Both RNA extraction
and RT procedures were performed according to the manufacturers'
protocols. qPCR thermal cycling was set as follows: Initial
denaturation at 95°C for 10 min; 40 cycles at 95°C for 15 sec and
60°C for 60 sec; followed by melt curve at 95°C for 15 sec, 60°C
for 60 sec and 95°C for 15 sec. For RNA expression detection,
GAPDH was used as a reference gene. The primer sequences
were as follows: SNHG16, forward 5′-CAGTCAGCCTCAGTTTCCAA-3′,
reverse 5′-AGGCAGGGCTGTGCTGAT-3′; and GAPDH, forward
5′-CGAGTCAACGGATTTGGTGGTAT-3′ and reverse
5′-AGCCTTCTCCATGGTGAAGAC-3′. The relative fold-change in mRNA
expression was calculated using the 2−ΔΔCq method
(25).
Real-time cell proliferation assay
Cell proliferation was measured by xCELLigence
real-time cell analysis (RTCA) (ACEA Biosciences, Inc.). Briefly, a
total of 4 ×103 SH-SY5Y cells were cultured in an
adaptive E-plate (ACEA Biosciences, Inc.) and transfected with
siRNA. The E-plate was then incubated at 37°C within the RTCA
Station inside the incubator, and device-defined cell index values
were recorded every 20 min for 72 h. Increases in cell numbers
altered the baseline impedance, which was monitored by gold
micro-electrodes located at the bottom of the E-plate. Data
analysis was performed using the RTCA Control Unit and preinstalled
RTCA software 2.0 (ACEA Biosciences, Inc.).
Colony formation assay
A total of 2×103 SH-SY5Y cells/well were
seeded in 6-well plates and transfected with siRNA. After 14 days,
cells were fixed with 4% paraformaldehyde for 10 min and stained
with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) for a further
10 min at room temperature. Images were captured using a light
microscope (IX73; Olympus Corporation), in order to observe cell
colony formation. Colonies containing >50 cells were counted and
recorded for statistical analysis. Independent assays were
conducted three times.
Wound healing assay
A total of 3×105 SH-SY5Y cells/well were
seeded in 6-well plates at ~50% confluence, and a 1-ml micropipette
tip was used to scratch the surface of the plate to create a
'wound'. After gentle washing with phosphate-buffered saline (PBS),
the attached cells were transfected with siRNA. Wound images were
captured under a light microscope (IX73; Olympus Corporation) at 0,
24, 48 and 72 h. The wound width was measured and analyzed by
AlphaView SA 3.4.0 software (ProteinSimple).
Transwell assay
Transwell chambers (pore size, 8 μm; Costar;
Corning, Inc.) were used to conduct a migration assay. A total of
250 μl serum-free medium containing 4×104 cells
was added into the upper chamber, whereas 500 μl complete
medium was added into the bottom chamber. After 24 h at 37°C, the
cells on the upper chamber were discarded and the cells that had
migrated to the lower surfaces of the filters were fixed with 4%
para-formaldehyde for 10 min at room temperature. Images of the
cells were captured using a light microscope (IX73; Olympus
Corporation) following further staining with 0.1% crystal violet
(Sigma-Aldrich; Merck KGaA) at room temperature for 10 min.
Cell cycle analysis
A total of 3×105 SH-SY5Y cells/well were
cultured in 6-well plates; 72 h post-transfection, cells were
trypsinized, centrifuged at 110 × g for 5 min, and resuspended in
ice cold ethanol (70%) at 4°C overnight. PBS containing 2% FBS was
added to spin down the cells at 440 × g for 5 min. The cell pellet
was then incubated in PBS containing 2% FBS, 10 μl 1 mg/ml
propidium iodide solution and 2 μl 10 mg/ml RNAseA (Tiangen
Biotech Co., Ltd.) for 30 min at 37°C in the dark. Cell cycle
progression was assessed using a flow cytometer and CellQuest Pro
6.1 software (BD Biosciences); ≥1×104 cells were
analyzed for each sample.
Acridine orange (AO)/ethidium bromide
(EB) staining
AO and EB staining (Nanjing KeyGen Biotech Co.,
Ltd.) was used to visualize nuclear alterations characteristic of
apoptosis. AO is a vital dye that stains live and dead cells,
whereas EB only stains cells that have lost membrane integrity,
thus indicating apoptosis. Live cells appear uniformly green,
whereas apoptotic cells will incorporate EB and therefore will be
stained red-orange with condensed nuclei (26). A total of 3×105 SH-SY5Y
cells/well were cultured in 6-well plates, transfected with siRNA
for 72 h, and stained with AO (10 μg/ml) and EB (10
μg/ml) for 30 min at room temperature. Cellular apoptosis
was subsequently viewed and images were captured under a
fluorescence microscope (Olympus Corporation).
Caspase-3/7 activity detection
Caspase-3/7 activation was measured using the
Caspase-Glo 3/7 assay (Promega Corporation) according to the
manufacturer's protocol. Briefly, a total of 1×104
cells/well were seeded into 96-well plates, transfected with siRNA,
and cultured for 72 h. Subsequently, 100 μl Caspase-Glo 3/7
reagent was added to each well and the well contents were agitated
on a plate shaker. Finally, samples were incubated at room
temperature for 2 h and luminescence was measured using a
SpectraMax Microplate Luminometer (Molecular Devices, LLC).
RNA-binding protein (RBP) prediction
analysis
Potential RBPs that bind SNHG16 (27) were systematically identified using
starBase v2.0 software (starbase.sysu.edu.cn). The Cytoscape plug-in ClueGO
was then used to identify Gene Ontology (GO) terms and interpret
functions enriched for the predicted RBPs (28). Statistical parameters were set as
follows: Right-sided hypergeometric test, P<0.05 with
Benjamini-Hochberg correction; GO levels, 6-14; Kappa score
threshold, 0.4.
Statistical analysis
Publically available Gene Expression Omnibus (GEO)
datasets (www.ncbi.nlm.nih.gov/geo) GSE62564 (29-32)
and GSE16237 (33) were downloaded
for SNHG16 expression analysis. Kaplan-Meier survival
analysis and the log-rank test were performed based on survival
times collected from the GSE62564 dataset. All data were analyzed
using SPSS 19.0 software (IBM Corp.), and graphs were generated
using GraphPad Prism 5.0 (GraphPad Software, Inc.). All results are
expressed as the means ± standard deviations. Multiple comparisons
were assessed by one-way analysis of variance followed by
Bonferroni post hoc test. Student's t-test was performed to analyze
differences between two groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
SNHG16 expression is positively
associated with NB clinical characteristics
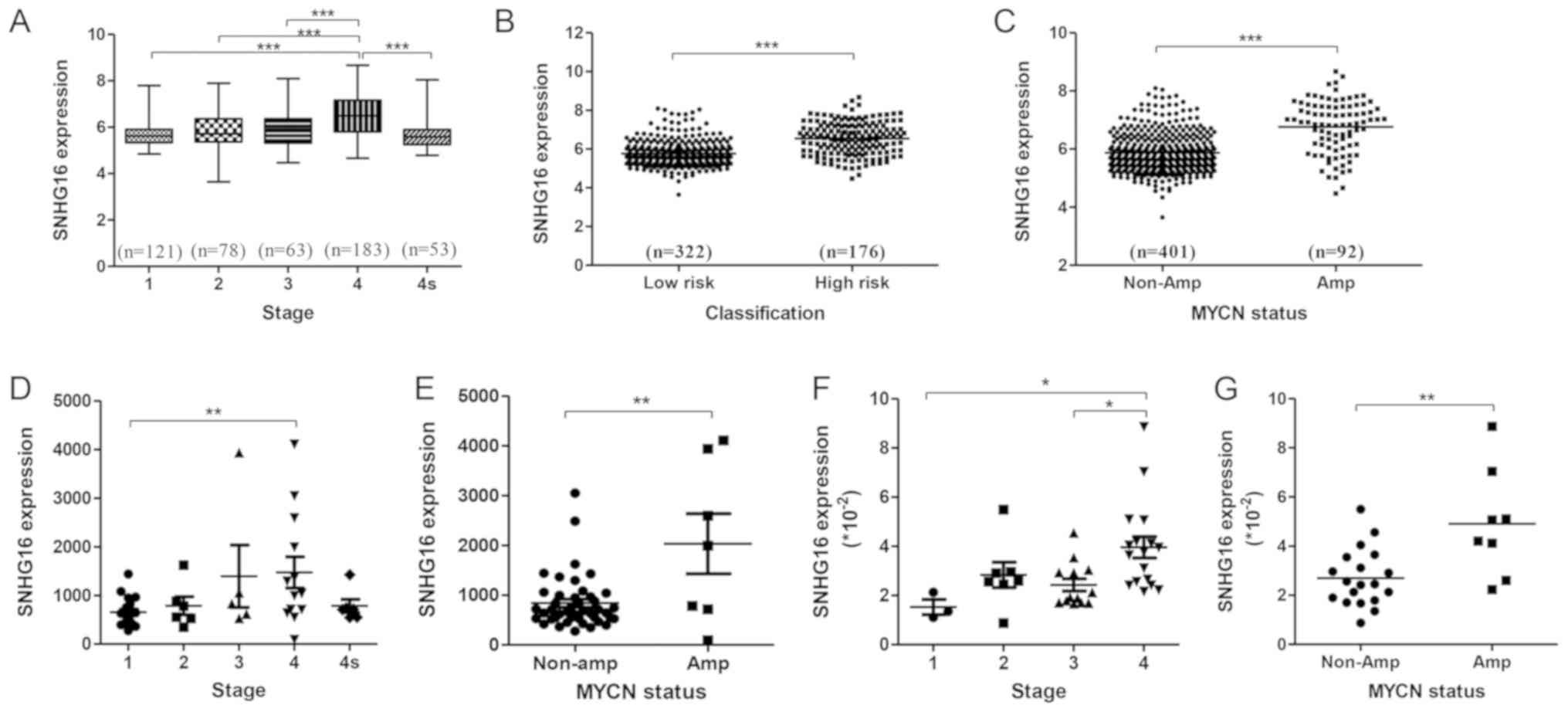
To explore the relationship between SNHG16
expression and the pathophysiological features of patients with NB,
the GEO dataset GSE62564 (498 samples) was analyzed by
stratification analysis based on INSS stage, risk group and
MYCN status. SNHG16 was expressed at significantly
higher levels in stage 4 tumors compared with in tumors at other
stages (Fig. 1A). In addition, it
was upregulated in high-risk NB and MYCN amplification
subtypes compared with in low-risk NB (Fig. 1B) and MYCN non-amplification
subtypes (Fig. 1C). Analysis of
another independent dataset, GSE16237 (51 samples), confirmed these
results (Fig. 1D and E). Analysis
of NB tissue samples (Table I)
revealed that SNHG16 expression was increased alongside
clinical staging of NB tumor progression (Fig. 1F) and in MYCN-amplified NB
(Fig. 1G). These findings
validated that SNHG16 expression was positively associated
with NB progression.
 | Table IClinical characteristics of patients
with neuroblastoma enrolled in this study (n=40). |
Table I
Clinical characteristics of patients
with neuroblastoma enrolled in this study (n=40).
| Parameter | Number of patients
(%) |
|---|
| Age at diagnosis
(years) | |
| <1.5 | 10 (25) |
| 1.5-3 | 7 (17.5) |
| 3-7 | 18 (45) |
| >7 | 5 (12.5) |
| Sex | |
| Male | 21 (52.5) |
| Female | 19 (47.5) |
| MYCN
status | |
| Amplified | 8 (20) |
| Non-amplified | 20 (50) |
| Not clear | 12 (30) |
| Tumor stage | |
| I | 3 (7.5) |
| II | 7 (17.5) |
| III | 13 (32.5) |
| IV | 17 (42.5) |
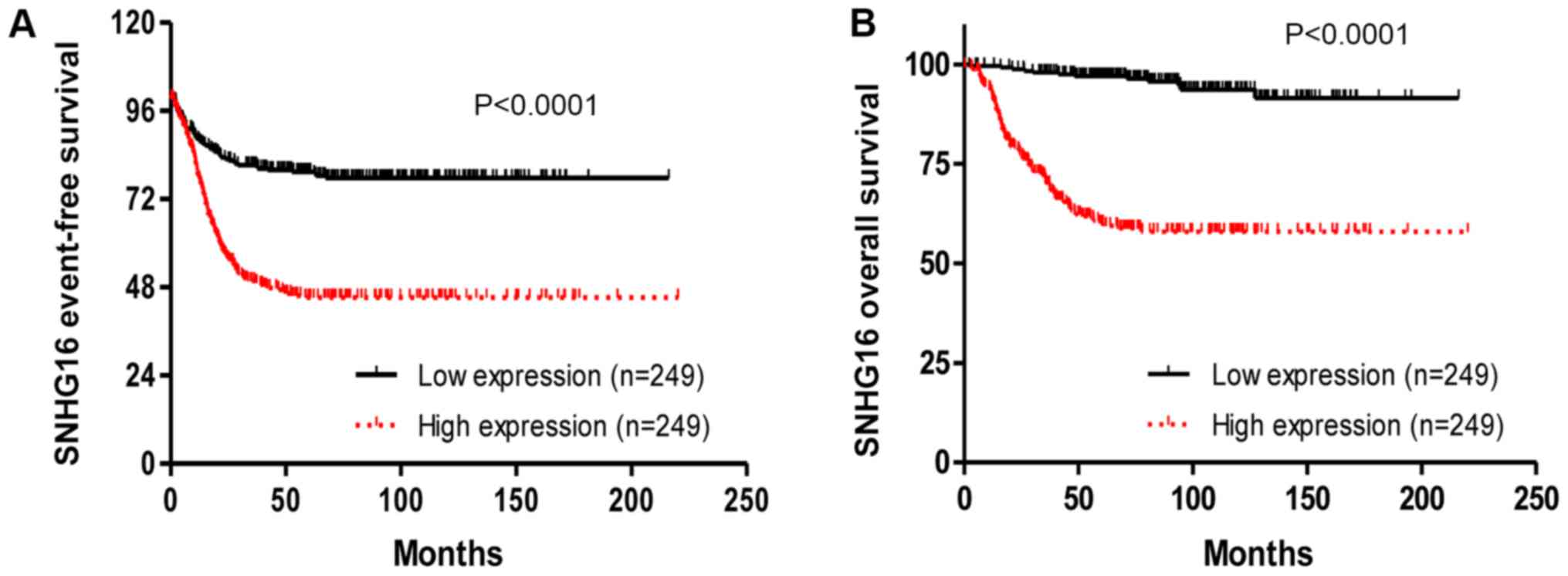
To explore the prognostic value of SNHG16,
Kaplan-Meier survival analysis was conducted based on GSE62564
data. Patients with low or high expression of SNHG16 were
grouped according to RNA sequencing data. Patients with high
expression had worse event-free survival (P<0.0001) and overall
survival (P<0.0001) than those with low expression (Fig. 2A and B), thus indicating that
SNHG16 may be a prognostic marker for patients with NB.
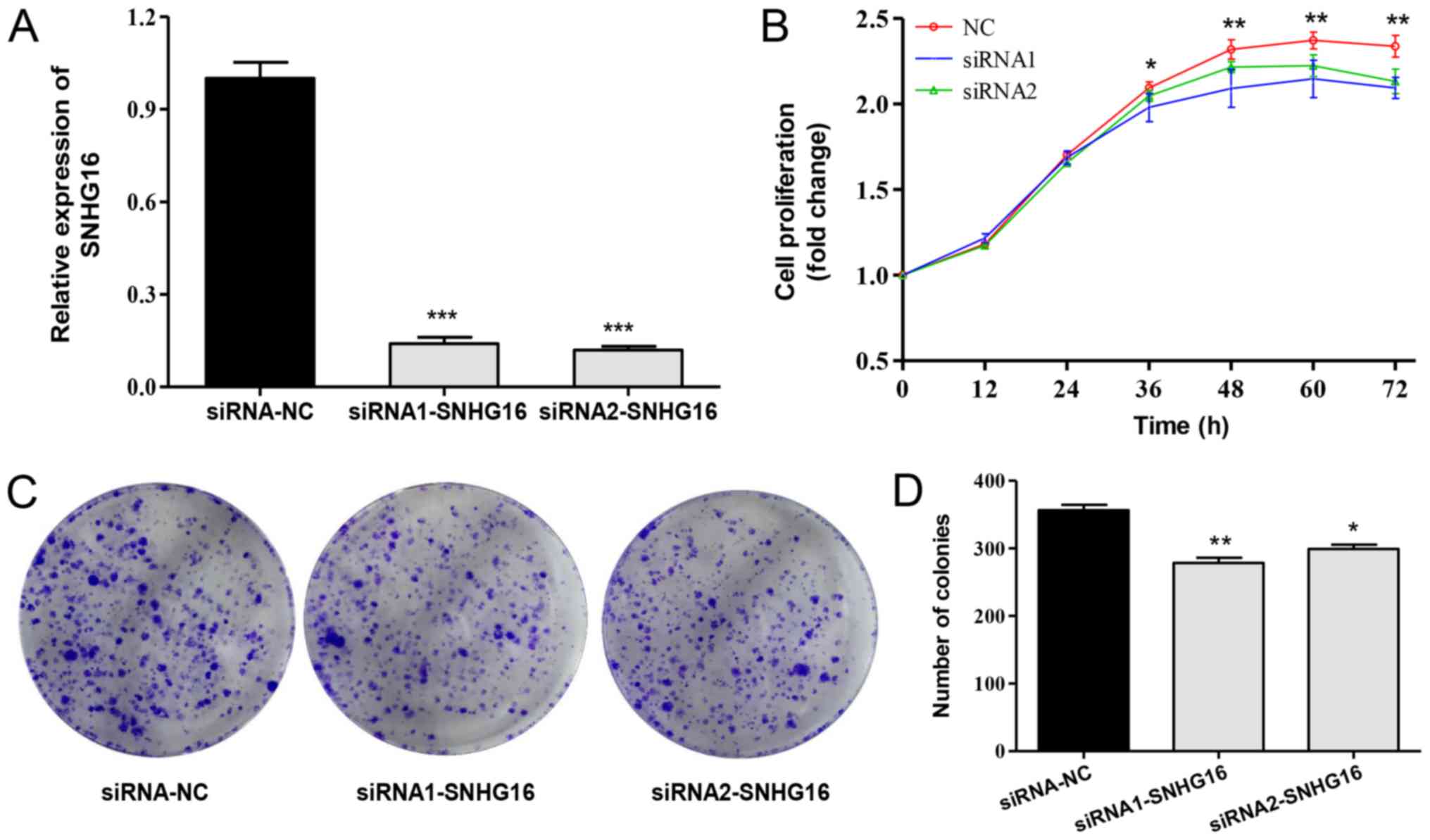
SNHG16 silencing inhibits proliferation
of SH-SY5Y cells
The gene knockdown efficiency of siRNAs was
validated by RT-qPCR. siRNA1-SNHG16 and siRNA2-SNHG16
effectively silenced SNHG16 expression (Fig. 3A). The real-time cell proliferation
assay revealed that cell growth in the SNHG16 silencing
group was significantly inhibited (Fig. 3B). This was confirmed by the colony
formation assay, which demonstrated that SNHG16 silencing
markedly reduced colony formation in SH-SY5Y cells (Fig. 3C and D). These findings indicated
that SNHG16 may contribute to NB cell proliferation. Both
siRNA1 and siRNA2 exhibited effective knockdown efficiency, and
were confirmed to inhibit cell proliferation; siRNA1 was used for
subsequent experiments.
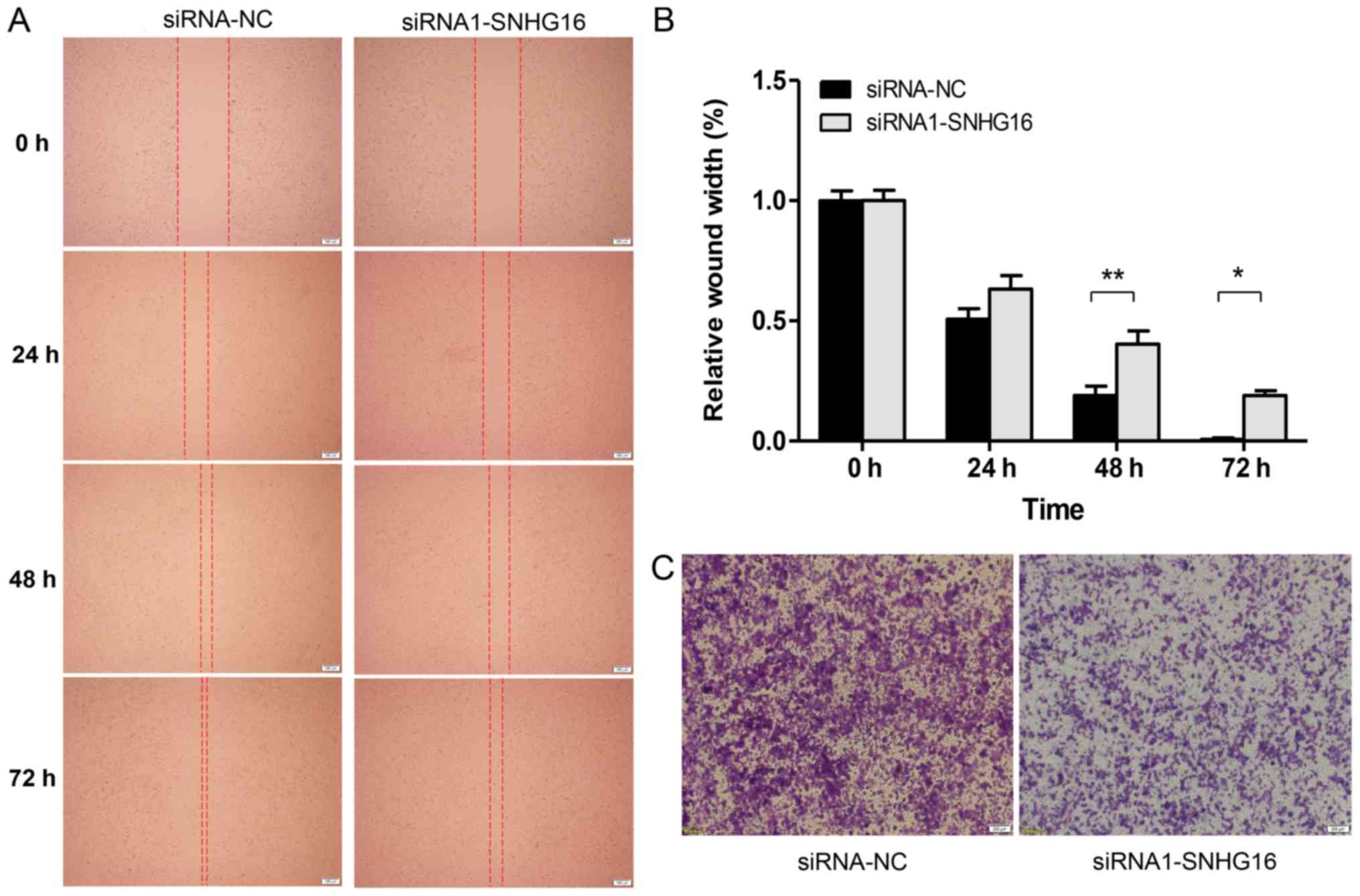
SNHG16 silencing inhibits migration of
SH-SY5Y cells
To explore whether SNHG16 affects cell
migration, a wound healing assay was conducted using SH-SY5Y cells
wounded with a micropipette tip. As shown in Fig. 4A, the wider width in the
SNHG16 silencing group suggested that SNHG16
knockdown significantly suppressed the migratory ability of SH-SY5Y
cells. Further quantification analysis revealed a significant
difference in the width between the SNHG16 silencing group
and the control group at 48 and 72 h after wounding (Fig. 4B). Furthermore, the results of a
Transwell assay further indicated that the migratory capacity of
SH-SY5Y cells transfected with siRNA-SNHG16 was markedly
decreased compared with the control group (Fig. 4C). These results suggested that
SNHG16 knockdown inhibited migration of SH-SY5Y cells.
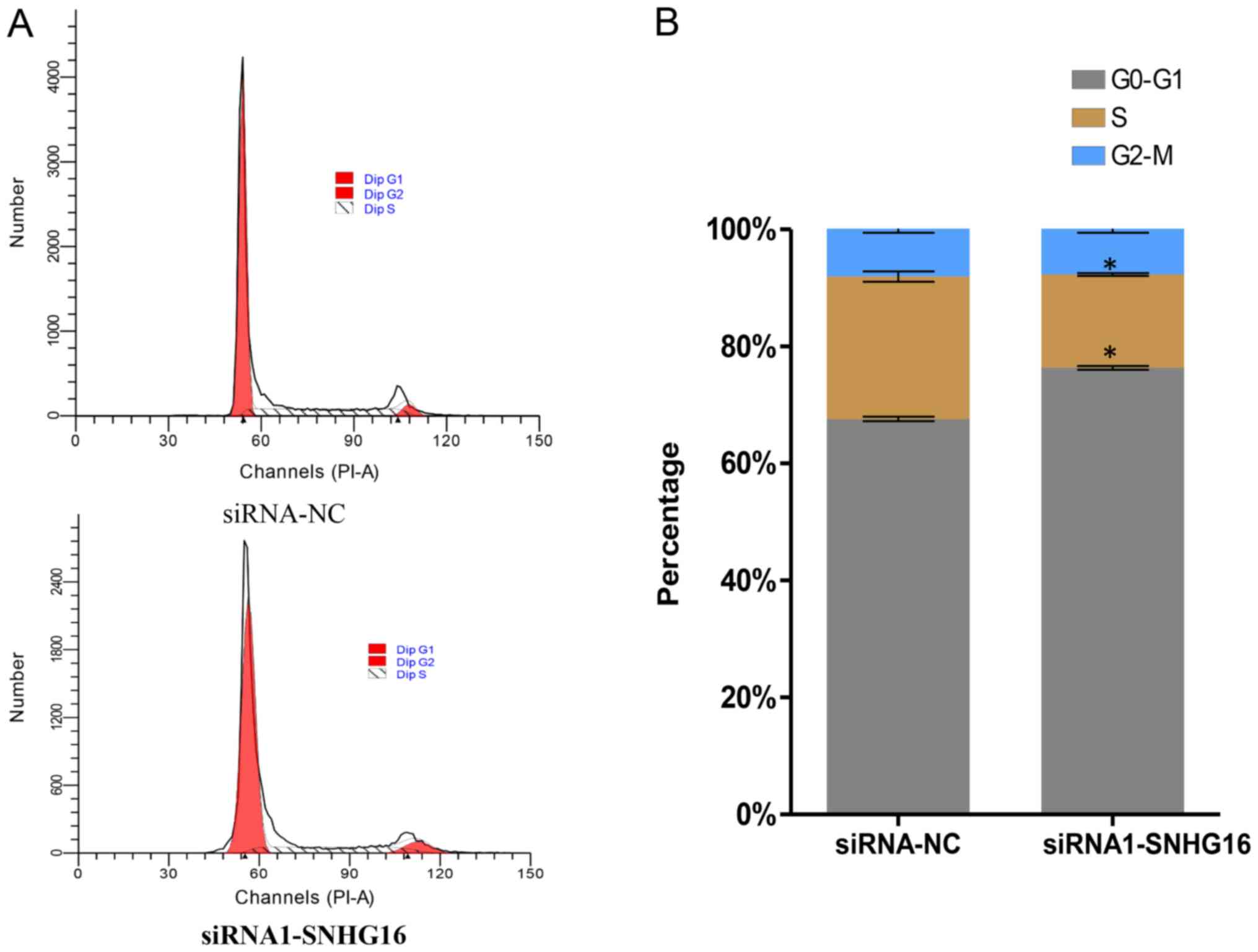
SNHG16 silencing induces cell cycle
arrest in SH-SY5Y cells
The cell cycle is closely associated with cell
proliferation; therefore, flow cytometry was used to investigate
the potential effects of SNHG16 on cell cycle alterations.
The percentage of cells in G0/G1 phase was
significantly increased, whereas the percentage in S phase was
decreased in the SNHG16-knockdown group compared with in the
control group (Fig. 5A and B;
Table II). These results
demonstrated that SNHG16 silencing in SH-SY5Y cells induced
cell cycle arrest at the G0/G1 phase.
 | Table IICell cycle arrest of SH-SY5Y cells
following siRNA-small nucleolar RNA host gene 16 transfection. |
Table II
Cell cycle arrest of SH-SY5Y cells
following siRNA-small nucleolar RNA host gene 16 transfection.
| Distribution of
cell cycle (%)
|
|---|
| Group |
G0/G1 | S |
G2/M |
|---|
| NC | 67.60±0.65 | 24.34±1.52 | 8.08±0.97 |
| siRNA | 76.35±0.59a | 15.94±0.41a | 7.72±0.98 |
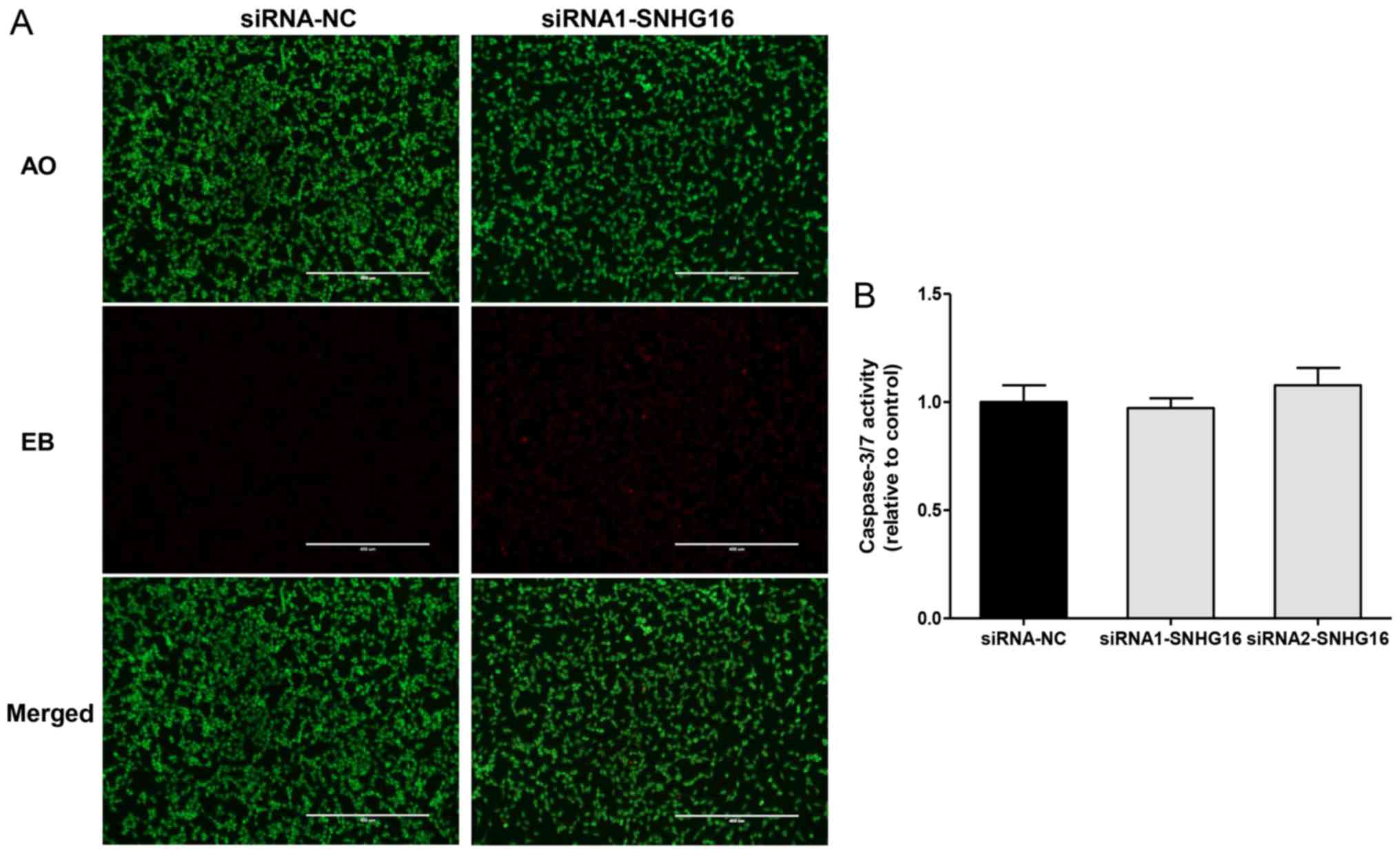
Apoptosis is undetectable in SH-SY5Y
cells following SNHG16 silencing
Cellular apoptosis was measured by AO/EB staining
following siRNA transfection. As shown in Fig. 6A, the cells in both groups appeared
uniformly green with no distinct red-orange staining, indicating no
detectable apoptotic cells. However, both cell number and density
were reduced in the SNHG16-knockdown group, suggesting that
SNHG16 silencing inhibited cell proliferation without
apoptosis. Detection of caspase-3/7 activity also revealed that
SNHG16 knockdown had no significant effect on caspase-3/7
activity (Fig. 6B). These results
indicated that SNHG16 knockdown did not induce apoptosis in
NB.
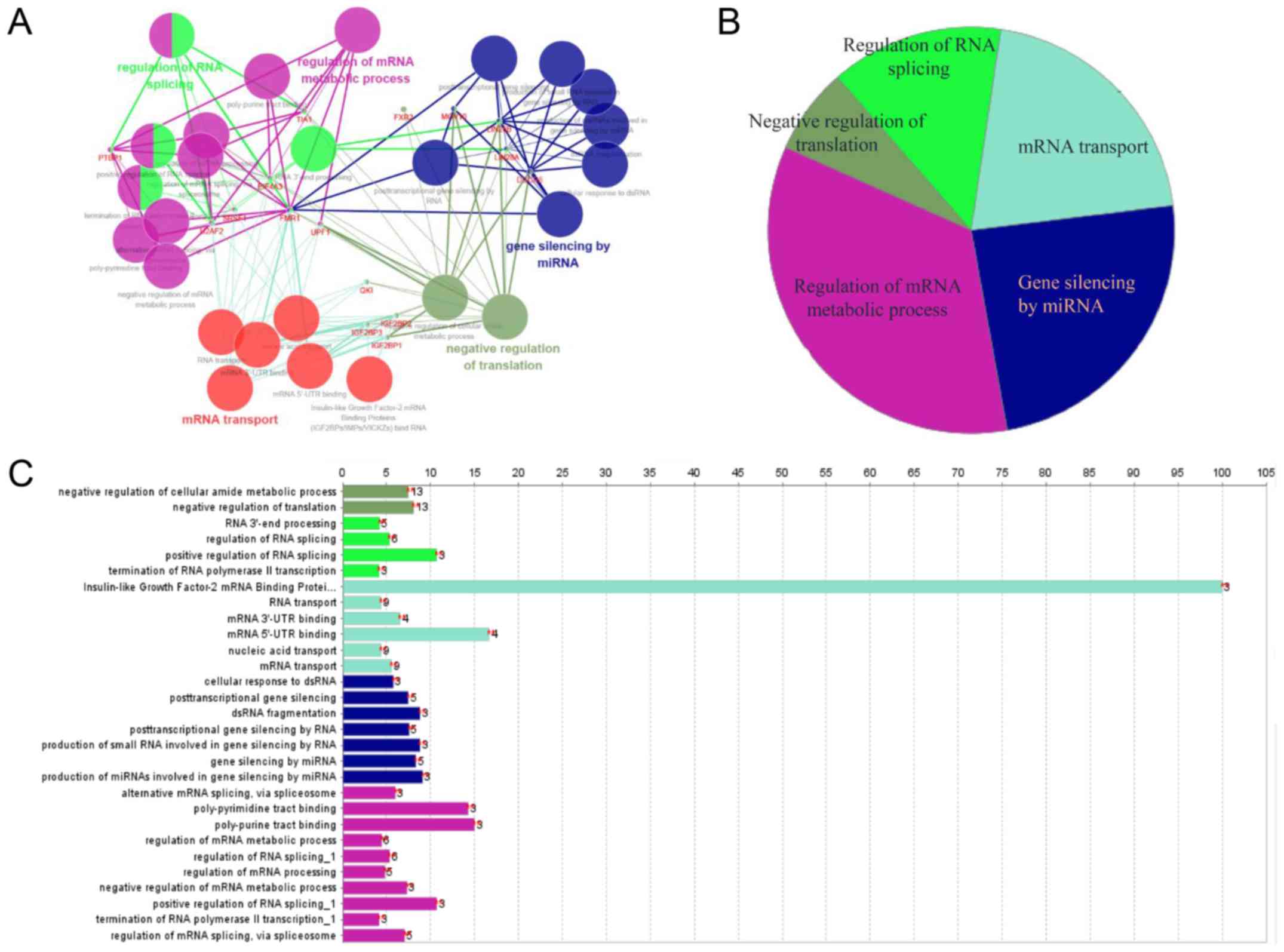
Implied RNA-related mechanism of SNHG16
function
To explore the underlying molecular mechanism of
SNHG16 function, starBase v2.0 was used to identify
potential RBPs that bind SNHG16 (34). Predicted target proteins of
SNHG16 are listed in Table
SI, and functional enrichment analysis was conducted by
Cytoscape. SNHG16-related RBPs were revealed to be mainly
involved in the regulation of mRNA metabolic processes, gene
silencing by miRNA, mRNA transport, RNA splicing and translation
(Fig. 7A-C), implying that the
molecular mechanism underlying the effects of SNHG16 is
associated with RNA-related processes.
Discussion
The results of the present study revealed that
SNHG16 expression was associated with the clinical
progression and prognosis of NB. In addition, SNHG16
silencing inhibited NB cell proliferation and migration, and
induced cell cycle arrest at G0/G1 phase in
SH-SY5Y cells.
lncRNAs have been reported to serve as signals of
specific cellular states, and may identify cellular pathologies,
provide prognostic value and predict therapeutic options for
patients with cancer (35). The
differential regulation of lncRNAs relative to coding genes may
underlie the high degree of tissue-specific lncRNA expression.
Furthermore, previous studies have suggested that the expression
and dysregulation of lncRNAs are cancer type-specific compared with
protein-coding genes (36,37). This suggests that lncRNAs should be
studied separately in different tumor types.
Sustaining proliferation and metastasis are the most
fundamental features of tumors (38). The present results indicated that
SNHG16 may contribute to NB progression. Using GEO datasets,
SNHG16 upregulation was revealed to be associated with
clinical staging of NB, and this was validated in a series of NB
samples. SNHG16 knockdown significantly inhibited
proliferation and migration of SH-SY5Y cells, whereas patients with
high SNHG16 expression had poor overall survival. Previous
studies have indicated that SNHG16 promotes tumor
proliferation via a diverse range of mechanisms. For example,
SNHG16 contributes to cervical cancer by directly targeting
miR-216-5p (39), and enhances
tumor proliferation through epigenetically silencing p21, which
acts as a tumor suppressor in bladder cancer (21,40).
SNHG16 has also been reported to be involved in the Wnt
pathway in colorectal cancer and affects genes involved in lipid
metabolism (19). Notably, lncRNAs
have both positive and negative effects on NB proliferation. For
example, high expression of lncRNAs, such as MYCN upstream
transcript, account for NB tumorigenesis (38), whereas lincNeD-125
negatively controls NB proliferation and activates the
anti-apoptotic factor BCL2 (15).
The primary aim of this study was to investigate the
effects and underlying mechanism of SNHG16 in NB
proliferation, and a series of experiments were performed. Notably,
MYCN amplification is an independent marker for NB prognosis
and risk stratification (7,8).
Existing evidence has suggested that SNHG16 is upregulated in a
MYCN amplification cell line and in NB samples (17,18).
Therefore, MYCN amplification may affect SNHG16
expression and subsequent function. In line with previous findings,
the present results demonstrated that the expression of
SNHG16 was increased in MYCN-amplified NB samples
compared with in MYCN non-amplified NB. A MYCN
non-amplified cell line was selected to examine the role of
SNHG16 in NB, because SH-SY5Y is a representative cell line
of NB, according to the ATCC, and it has been widely used in
mechanistic and drug development studies regarding NB (23,24).
The potential link between SNHG16 and MYCN
amplification requires independent research for further
clarification.
Various biological processes are associated with
cell proliferation, including DNA damage, autophagy, apoptosis and
the cell cycle (41). In the
present study, SNHG16 silencing was revealed to affect cell
cycle progression and lead to G0/G1 arrest in
SH-SY5Y cells. This result supports a previous finding that
SNHG16 silencing is able to arrest gastric cancer cells at
G0/G1 (42),
as well as the reports that many lncRNAs are associated with
G0/G1 block in various tumor types. For
example, Yang et al (43)
reported that hepatocellular carcinoma up-regulated EZH2-associated
lncRNA is highly expressed in hepatitis B virus-related
hepatocellular carcinoma and negatively regulates the expression of
cyclin-dependent kinase inhibitors. Furthermore, Ye et al
(44) reported that lncRNA bladder
cancer-associated transcript 1 silencing induces
G0/G1 arrest through sponging miR-144.
Therefore, cell cycle control may be a potential mechanism
underlying SNHG16-mediated NB proliferation.
Sometimes cell cycle arrest is beneficial for DNA
repair to maintain homeostasis; however, it can also cause cellular
damage. Cells are equipped with cell cycle checkpoints to maintain
genome stability; when cells have DNA damage to be repaired, or DNA
replication is not complete, these checkpoints arrest the cell
cycle (45,46). This kind of cell cycle delay offers
more time for the repair of DNA damage and cellular homeostasis.
However, when cellular injuries are so severe that they exceed the
cellular repair capacity, apoptosis may occur. Blocking the cell
cycle is often associated with apoptosis; however, in this study,
apoptosis was not detected by AO/EB staining and caspase-3/7
activity assay following SNHG16 silencing. This finding
contradicts the previous findings of Christensen et al
(19); this previous study
reported that SNHG16-induced apoptosis is dependent on
caspase-3/7 activity in colorectal cancer cells. In addition, Lu
et al (47) demonstrated
that SNHG16 knockdown inhibits viability and induces
apoptosis in glioma cells. Because lncRNA function is highly cancer
type-specific compared with protein-coding genes (48,49),
this discrepancy may be accounted for by the different cell types
or regulatory mechanisms between these studies and the present
study.
The mechanisms underlying the effects of lncRNAs
are highly extensive and complex, and may involve protein or DNA
interactions (50). lncRNAs
typically function through RBPs, including the polycomb-group
proteins, heterochromatin protein 1 and DNA methyltransferases,
whose expression fluctuates in tumor samples and therefore may
provide prognostic clues (51).
Chen et al (52) reported
that the lncRNA MALAT1 regulates p53 through its interaction
with cell cycle and apoptosis regulator 2, whereas RNA pulldown and
RNA immunoprecipitation assays have demonstrated that the physical
association of colon cancer-associated transcript 1 with Livin
inhibits apoptosis in renal cell carcinoma (53). In the present study, bioinformatics
analysis predicted various RBPs that may interact with
SNHG16, and functional enrichment analysis demonstrated that
the potential RBPs regulated transcription and translation.
However, to confirm these predictions, in-depth study and molecular
experiments are urgently required. Nevertheless, the present
results suggested that SNHG16 regulated cell proliferation,
migration and the cell cycle in NB through multiple pathways
associated with transcription and translation.
In conclusion, the present study demonstrated that
high expression of SNHG16 was associated with NB progression
and poor clinical outcome. SNHG16 silencing inhibited cell
proliferation and migration, and induced cell cycle arrest at
G0/G1. Bioinformatics analysis revealed that
SNHG16 regulated the proliferation of NB cells through
multiple pathways associated with transcription and translation.
Therefore, SNHG16 may serve as a molecular marker for NB
therapy or prognosis.
Supplementary Materials
Funding
This work was supported by the National Natural
Science Foundation of China (grant nos. 81702463 and 81702787) and
the Beijing Health System Top Level Technical Personnel Training
Plan (grant no. 20153079).
Availability of data and materials
All data generated or analyzed in this study are
included in this published article.
Authors' contributions
YYu and FC performed most of the experiments and
wrote the manuscript. YYa, YJ JS and JL performed some experiments.
JT, SW and WY collected the clinical data and tissue samples. PC
and SH performed the statistical analysis. YG, XN and HW designed
the experiments and edited the manuscript. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committees of
Beijing Children's Hospital, and followed the principles of the
Helsinki Declaration II. Written informed consent was obtained from
the legal guardians of the patients.
Patient consent for publication
The legal guardians of the patients involved in
this study provided written informed consent prior to inclusion in
the study.
Competing interests
The authors declare they have no competing
interests.
Acknowledgments
The authors would like to thank Sarah Williams,
PhD, for language editing a draft of this manuscript.
References
|
1
|
Whittle SB, Smith V, Doherty E, Zhao S,
McCarty S and Zage PE: Overview and recent advances in the
treatment of neuroblastoma. Expert Rev Anticancer Ther. 17:369–386.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rogowitz E, Babiker HM, Kanaan M, Millius
RA, Ringenberg QS and Bishop M: Neuroblastoma of the elderly, an
oncologist's nightmare: Case presentation, literature review and
SEER database analysis. Exp Hematol Oncol. 3:202014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Esiashvili N, Goodman M, Ward K, Marcus RB
Jr and Johnstone PA: Neuroblastoma in adults: Incidence and
survival analysis based on SEER data. Pediatr Blood Cancer.
49:41–46. 2007. View Article : Google Scholar
|
|
4
|
Maris JM, Hogarty MD, Bagatell R and Cohn
SL: Neuroblastoma. Lancet. 369:2106–2120. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bosse KR and Maris JM: Advances in the
translational genomics of neuroblastoma: From improving risk
stratification and revealing novel biology to identifying
actionable genomic alterations. Cancer. 122:20–33. 2016. View Article : Google Scholar :
|
|
6
|
Salazar BM, Balczewski EA, Ung CY and Zhu
S: Neuroblastoma, a paradigm for big data science in pediatric
oncology. Int J Mol Sci. 18:182016. View Article : Google Scholar
|
|
7
|
Schneiderman J, London WB, Brodeur GM,
Castleberry RP, Look AT and Cohn SL: Clinical significance of MYCN
amplification and ploidy in favorable-stage neuroblastoma: A report
from the Children's Oncology Group. J Clin Oncol. 26:913–918. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
London WB, Bagatell R, Weigel BJ, Fox E,
Guo D, Van Ryn C, Naranjo A and Park JR: Historical time to disease
progression and progression-free survival in patients with
recurrent/refractory neuroblastoma treated in the modern era on
Children's Oncology Group early-phase trials. Cancer.
123:4914–4923. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peifer M, Hertwig F, Roels F, Dreidax D,
Gartlgruber M, Menon R, Krämer A, Roncaioli JL, Sand F, Heuckmann
JM, et al: Telomerase activation by genomic rearrangements in
high-risk neuroblastoma. Nature. 526:700–704. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar
|
|
11
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pandey GK and Kanduri C: Long noncoding
RNAs and neuro-blastoma. Oncotarget. 6:18265–18275. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bi S, Wang C, Li Y, Zhang W, Zhang J, Lv Z
and Wang J: lncRNA-MALAT1-mediated Axl promotes cell invasion and
migration in human neuroblastoma. Tumour Biol.
39:10104283176997962017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yarmishyn AA, Batagov AO, Tan JZ, Sundaram
GM, Sampath P, Kuznetsov VA and Kurochkin IV: HOXD-AS1 is a novel
lncRNA encoded in HOXD cluster and a marker of neuroblastoma
progression revealed via integrative analysis of noncoding
tran-scriptome. BMC Genomics. 15(Suppl 9): S72014. View Article : Google Scholar
|
|
15
|
Bevilacqua V, Gioia U, Di Carlo V,
Tortorelli AF, Colombo T, Bozzoni I, Laneve P and Caffarelli E:
Identification of linc-NeD125, a novel long non coding RNA that
hosts miR-125b-1 and negatively controls proliferation of human
neuroblastoma cells. RNA Biol. 12:1323–1337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pandey GK, Mitra S, Subhash S, Hertwig F,
Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S,
et al: The risk-associated long noncoding RNA NBAT-1 controls
neuroblastoma progression by regulating cell proliferation and
neuronal differentiation. Cancer Cell. 26:722–737. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sahu D, Hsu CL, Lin CC, Yang TW, Hsu WM,
Ho SY, Juan HF and Huang HC: Co-expression analysis identifies long
noncoding RNA SNHG1 as a novel predictor for event-free survival in
neuroblastoma. Oncotarget. 7:58022–58037. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu M, Ohira M, Li Y, Niizuma H, Oo ML, Zhu
Y, Ozaki T, Isogai E, Nakamura Y, Koda T, et al: High expression of
ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is
associated with poor prognosis in neuroblastoma. Int J Oncol.
34:931–938. 2009.PubMed/NCBI
|
|
19
|
Christensen LL, True K, Hamilton MP,
Nielsen MM, Damas ND, Damgaard CK, Ongen H, Dermitzakis E, Bramsen
JB, Pedersen JS, et al: SNHG16 is regulated by the Wnt pathway in
colorectal cancer and affects genes involved in lipid metabolism.
Mol Oncol. 10:1266–1282. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cai C, Huo Q, Wang X, Chen B and Yang Q:
SNHG16 contributes to breast cancer cell migration by competitively
binding miR-98 with E2F5. Biochem Biophys Res Commun. 485:272–278.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duan W, Du L, Jiang X, Wang R, Yan S, Xie
Y, Yan K, Wang Q, Wang L, Zhang X, et al: Identification of a serum
circulating lncRNA panel for the diagnosis and recurrence
prediction of bladder cancer. Oncotarget. 7:78850–78858. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Brodeur GM, Pritchard J, Berthold F,
Carlsen NL, Castel V, Castelberry RP, De Bernardi B, Evans AE,
Favrot M and Hedborg F: Revisions of the international criteria for
neuro-blastoma diagnosis, staging, and response to treatment. J
Clin Oncol. 11:1466–1477. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Z, Yan S, Attayan N, Ramalingam S and
Thiele CJ: Combination of an allosteric Akt Inhibitor MK-2206 with
etoposide or rapamycin enhances the antitumor growth effect in
neuroblastoma. Clin Cancer Res. 18:3603–3615. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Radogna F, Cerella C, Gaigneaux A,
Christov C, Dicato M and Diederich M: Cell type-dependent ROS and
mitophagy response leads to apoptosis or necroptosis in
neuroblastoma. Oncogene. 35:3839–3853. 2016. View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Alarifi S, Ali D, Alkahtani S and Almeer
RS: ROS-mediated apoptosis and genotoxicity induced by palladium
nanoparticles in human skin malignant melanoma cells. Oxid Med Cell
Longev. 2017:84390982017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang JH, Li JH, Shao P, Zhou H, Chen YQ
and Qu LH: starBase: A database for exploring microRNA-mRNA
interaction maps from Argonaute CLIP-Seq and Degradome-Seq data.
Nucleic Acids Res. 39(Suppl 1): D202–D209. 2011. View Article : Google Scholar
|
|
28
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang C, Gong B, Bushel PR, Thierry-Mieg J,
Thierry-Mieg D, Xu J, Fang H, Hong H, Shen J, Su Z, et al: The
concordance between RNA-seq and microarray data depends on chemical
treatment and transcript abundance. Nat Biotechnol. 32:926–932.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
SEQC/MAQC-III Consortium: A comprehensive
assessment of RNA-seq accuracy, reproducibility and information
content by the Sequencing Quality Control Consortium. Nat
Biotechnol. 32:903–914. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Munro SA, Lund SP, Pine PS, Binder H,
Clevert DA, Conesa A, Dopazo J, Fasold M, Hochreiter S, Hong H, et
al: Assessing technical performance in differential gene expression
experiments with external spike-in RNA control ratio mixtures. Nat
Commun. 5:51252014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Su Z, Fang H, Hong H, Shi L, Zhang W,
Zhang W, Zhang Y, Dong Z, Lancashire LJ, Bessarabova M, et al: An
investigation of biomarkers derived from legacy microarray data for
their utility in the RNA-seq era. Genome Biol. 15:5232014.
View Article : Google Scholar
|
|
33
|
Ohtaki M, Otani K, Hiyama K, Kamei N,
Satoh K and Hiyama E: A robust method for estimating gene
expression states using Affymetrix microarray probe level data. BMC
Bioinformatics. 11:1832010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42D:D92–D97. 2014. View Article : Google Scholar
|
|
35
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao
SD, Zhang Y, Yang L, Shan W, He Q, et al: Comprehensive genomic
characterization of long non-coding RNAs across human cancers.
Cancer Cell. 28:529–540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu PY, Erriquez D, Marshall GM, Tee AE,
Polly P, Wong M, Liu B, Bell JL, Zhang XD, Milazzo G, et al:
Effects of a novel long noncoding RNA, lncUSMycN, on N-Myc
expression and neuroblastoma progression. J Natl Cancer Inst.
106:1062014. View Article : Google Scholar
|
|
39
|
Zhu H, Zeng Y, Zhou CC and Ye W:
SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the
tumorigenesis of cervical cancer cells. Arch Biochem Biophys.
637:1–8. 2018. View Article : Google Scholar
|
|
40
|
Cao X, Xu J and Yue D: lncRNA-SNHG-16
predicts poor prognosis and promotes tumor proliferation through
epigenetically silencing p21 in bladder cancer. Cancer Gene Ther.
25:10–17. 2018. View Article : Google Scholar
|
|
41
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lian D, Amin B, Du D and Yan W: Enhanced
expression of the long non-coding RNA SNHG16 contributes to gastric
cancer progression and metastasis. Cancer Biomark. 21:151–160.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ye JR, Liu L and Zheng F: Long noncoding
RNA bladder cancer associated transcript 1 promotes the
proliferation, migration, and invasion of nonsmall cell lung cancer
through sponging miR-144. DNA Cell Biol. 36:845–852. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Uryga A, Gray K and Bennett M: DNA damage
and repair in vascular disease. Annu Rev Physiol. 78:45–66. 2016.
View Article : Google Scholar
|
|
47
|
Lu YF, Cai XL, Li ZZ, Lv J, Xiang YA, Chen
JJ, Chen WJ, Sun WY, Liu XM and Chen JB: lncRNA SNHG16 functions as
an oncogene by sponging miR-4518 and up-regulating PRMT5 expression
in glioma. Cell Physiol Biochem. 45:1975–1985. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Winkle M, Kluiver JL, Diepstra A and van
den Berg A: Emerging roles for long noncoding RNAs in B-cell
development and malignancy. Crit Rev Oncol Hematol. 120:77–85.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar
|
|
50
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ferrè F, Colantoni A and Helmer-Citterich
M: Revealing protein-lncRNA interaction. Brief Bioinform.
17:106–116. 2016. View Article : Google Scholar
|
|
52
|
Chen R, Liu Y, Zhuang H, Yang B, Hei K,
Xiao M, Hou C, Gao H, Zhang X, Jia C, et al: Quantitative
proteomics reveals that long non-coding RNA MALAT1 interacts with
DBC1 to regulate p53 acetylation. Nucleic Acids Res. 45:9947–9959.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen S, Ma P, Li B, Zhu D, Chen X, Xiang
Y, Wang T, Ren X, Liu C and Jin X: lncRNA CCAT1 inhibits cell
apoptosis of renal cell carcinoma through up-regulation of Livin
protein. Mol Cell Biochem. 434:135–142. 2017. View Article : Google Scholar : PubMed/NCBI
|