Introduction
Renal cell carcinoma (RCC) represents a serious
health concern, with an estimated annual incidence of 69,330 new
cases and 14,400 deaths in the USA in 2017 (1). Worldwide kidney cancer causes
>100,000 deaths per year (2).
Clear cell RCC (ccRCC) is the most common subtype of renal cancer
(70-80%) (3), and is characterized
by the highest mortality rate compared with other RCC subtypes
(4). With a 50% risk of
metastasis, the prognosis of patients with ccRCC is poor, with a
5-year survival rate of 40% (5),
even with the administration of modern drugs (6). The development of ccRCC is associated
with extrinsic factors, including smoking, obesity and
hypertension. However, conditions such as hereditary von
Hippel-Lindau (VHL) disease or sporadic somatic inactivation of the
VHL gene have been found to have a very close genetic
association with ccRCC (4). The
VHL gene was mapped at chromosome 3p25.3 (7,8), and
the protein encoded by this gene (pVHL) was subsequently shown to
form a complex with elongin C, elongin B and cullin-2 (9), which possesses ubiquitin ligase E3
activity. Upon its formation, this complex specifically recognizes
the α subunits of two transcription factors, the hypoxia-inducible
factors (HIFs) 1A and 2A, and directs them for degradation along
the ubiquitin-proteasome pathway (8). Under normoxic conditions,
oxygen-dependent hydroxylation of HIFs at specific proline residues
by the enzyme prolyl hydroxylase (PHD) triggers binding of pVHL,
ubiquitination, and subsequent proteasomal degradation of either
HIF1A or HIF2A (2). Apart from
oxygen-dependent regulation, which requires pVHL for HIF
ubiquitination, another pathway also leads to the degradation of
the HIFs with the involvement of the P53 and mouse double minute 2
homolog (Mdm2) proteins (10).
Under hypoxic conditions, P53 binds to HIF1A and triggers its
removal through Mdm2-mediated ubiquitination and proteasomal
degradation, controlled by the phosphatase and tensin homolog
(PTEN)/phosphoinositide 3-kinase (PI3K)/AKT pathway (10-13).
Consequently, under normal physiological conditions, the HIF1A and
HIF2A proteins are degraded within minutes. However, during
hypoxia, the hydroxylation of HIFs is inhibited, and the two
proteins form a stable complex with the constitutively expressed
HIF-1β subunit (ARNT protein); this complex is subsequently
translocated to the cell nucleus. After binding with DNA at hypoxia
response elements, the HIF complex activates the expression of a
large variety of genes (14).
Although HIF1A and HIF2A represent different proteins, and they
partially overlap in the activation of target genes; for example,
the gene expression of C-X-C chemokine receptor type 4
(CXCR4) and solute carrier family 2 member 1 (SLC2A1)
has been shown to be activated by either HIF1A or HIF2A (15). One of the most well-characterized
genes activated by HIFs is vascular endothelial growth factor A
(VEGFA), which encodes heparin-binding protein. VEGFA
robustly induces the proliferation and migration of vascular
endothelial cells, and is essential for physiological and
pathological angiogenesis (16).
Currently, therapeutic approaches towards the treatment of advanced
ccRCC comprise various VEGF pathway-targeted agents, including
bevacizumab, sunitinib, sorafenib, pazopanib and axitinib (4). Sunitinib inhibits cellular signaling
by blocking the membrane receptors of platelet-derived growth
factor (PDGF), as well as VEGF, decreasing the rate of
neoangiogenesis, which is one of the most important pathological
mechanisms associated with RCC development (4). Although sunitinib was introduced as a
first-line post-operative adjuvant therapy for metastatic ccRCC due
to improvement of overall survival (OS) and progression-free
survival (PFS) rates (17), it was
reported that the outcome of ~15% of sunitinib-treated patients was
grim, due to intrinsic drug resistance and cancer recurrence
(6,18-20).
The mechanisms of the resistance to tyrosine kinase inhibitors
(TKIs) of the VEGF receptor (VEGFR) have yet to be fully
elucidated, although one research group previously observed
secondary resistance to angiogenesis inhibitors, reflected by
cancer progression following a period of clinical improvement
(21).
The deregulation of the VHL gene in ccRCC has
been widely reported (5,22,23).
Studies on the expression patterns of HIF1A (24-28),
HIF2A (25-28), VEGFA (5,24,27-29)
and P53 (30,31) in ccRCC have revealed variations in
terms of cancer progression and/or patient outcome. However, to the
best of our knowledge, to date, no comprehensive study of all the
above-mentioned genes in one cohort of ccRCC patients has been
performed with the use of quantitative and semi-quantitative
methods [i.e., reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) and western blot analysis]. Therefore, the first
aim of the present study was to identify the associations of the
mRNA and protein levels of VHL, HIF1A, HIF2A, VEGFA and
P53 with patient outcome. Furthermore, since the majority of
patients receiving sunitinib as first-line post-operative treatment
have a poor outcome, the possible associations between molecular
signatures and clinicopathological data were further analyzed.
Materials and methods
Patients and samples
Tissue samples were obtained from 36 patients with
ccRCC who underwent radical nephrectomy at the Department of
Urology, Medical University of Gdansk, Poland, between January,
2011 and September, 2013. The clinicopathological data of the
patients are presented in Table I.
A total of 11 patients received sunitinib following radical
nephrectomy. The present study was approved by the Independent
Bioethics Commission for Research of Medical University of Gdansk,
and written consent was obtained prior to surgery from all
patients.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
Patients
n=36 | Subgroups | No. |
|---|
| Age (years) | ≤62a | 19 |
| Median, 60.58±11.9
years | 62 | 17 |
| Range, 33-82
years | | |
| Sex | Female | 17 |
| Male | 19 |
| Body mass index
(BMI) | ≤25a | 14 |
| Median,
28.57±6.71 | >25 | 22 |
| Range,
19.82-45.52 | | |
| Creatinine
(mg/dl) | ≤1.21a | 32 |
| Median,
1.08±1.04 | >1.21 | 4 |
| Range,
0.65-7.06 | | |
| Blood urea nitrogen
(BUN) (mg/dl) | 7-20a | 26 |
| Median,
16.43±5.12 | >20 | 10 |
| Range,
7.4-28.2 | | |
| Estimated
glomerular filtration rate (eGFR) (ml/min/1.73 m2) | ≤60 | 8 |
| Range,
7.22-59.92 | |
| >60a | 28 |
| Hematocrit (HCT)
(%) | ≤40.55 | 18 |
| Median,
39.5±4.77 | >40.55 | 18 |
| Range,
28.6-46.9 | | |
| Hemoglobin (HGB)
(g/dl) | ≤11a | 5 |
| Median,
13.05±1.82 | >11 | 31 |
| Range, 9-15.7 | | |
| Glucose (GLC)
(mg/dl) | ≤99a | 20 |
| Median,
102.7±19.58 | >99 | 16 |
| Range, 77-167 | | |
| Sodium
(Na+) (mmol/l) | ≤145a | 36 |
| Median,
139.22±2.35 | >145 | 0 |
| Range, 133-144 | | |
| Potassium
(K+) (mmol/l) | ≤3.5 | 1 |
| Median,
4.33±0.46 | >3.5 | 35 |
| Range,
3.3-4.35 | | |
| Tumor location | Left kidney | 17 |
| Right kidney | 19 |
| Tumor size
(cm) | ≤7 cm | 17 |
| >7 cm | 19 |
| Fuhrman's | 1 | 2 |
| histological
grade | 2 | 16 |
| 3 | 11 |
| 4 | 7 |
| TNM stage | | |
| Non-metastatic,
n=21 | T1-2N0M0 | 21 |
| Metastatic,
n=15 | T1-2N1M0 | 0 |
| T3N0-1M0 | 7 |
| T1-4N2M0 | 6 |
| T1-4N0-2M1 | 2 |
| Sunitinib
(anti-VEGFR) | | |
| Yes | | 11 |
| No | | 25 |
Sample acquisition
Samples were obtained as previously described
(13,32,33).
In brief, dissected tissue samples of the primary ccRCC (n=36) and
corresponding normal kidney (n=36) were collected in the operating
theatre (by J.K.) and immediately placed in ~5 volumes of RNALater
(Ambion Inc., now a brand of Thermo Fisher Scientific, Inc.). Tumor
and normal kidney samples from 10 patients were placed in at least
10 volumes of 4% buffered formalin (pH 7.0-7.4; ChemPur).
RNALater-stored samples were subsequently used for RNA and protein
assessment; immunohistochemical (IHC) localization of proteins was
accomplished using the formalin-stored tissues.
Assessment of the mRNA expression of VHL,
HIF1A, HIF2A, VEGFA and P53
RNA isolation and cDNA synthesis were performed as
previously described (13,32,33).
Briefly, an ExtractMe RNA kit (Blirt) was used for RNA extraction
from tissue samples. Total RNA samples (2 µg) were reverse
transcribed with the use of RevertAid Reverse Transcriptase
(Fermentas; Thermo Fischer Scientific, Inc.). Details concerning
the RT-qPCR methodology are provided in Table II. All reactions were run in
duplicate; the measurement of glucuronidase beta (GUSB) gene
expression was used for the normalization of qPCR results (33) with Livak and Schmittgen's
2−∆∆Cq method (34).
 | Table IIDetails of qPCR assays. |
Table II
Details of qPCR assays.
| Gene name, GeneBank
ID | Primer
sequences | Amplicon size
(bp) | qPCR
efficiency | qPCR reaction
conditions | qPCR reaction
content |
|---|
| VHL,
NM_000551.3 |
5′-CGGACAGCCTATTTTTGCCAAT
5′-ATGTTTGCCCCTAAACATCACA | 400 | 96.4% | 95°C, 3 min; 37x
(95°C, 5 sec; 58°C, 10 sec; 72°C, 10 sec; 75°C, 10 sec - sample
reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C
reading every 0.3°C | 5 µl
SensiFast NoRox SYBR-Green (with SYBR-Green fluorophore) (BioLine,
London, GB), 200 nM each primer, Σ 10 µl |
| HIF1A,
NM_001243084.1 |
5′-ACCTGAAGAATTGGAAGAAATCAGA
5′-ATATCCAAATCACCAGCATCCA | 243 | 94.6% | | |
| HIF2A |
5′-CGTCCTGAGTGAGATTGAGAAG
5′-GACTCCTCGAAGTTCTGATTCC | 246 | 96.3% | | |
| VEGFA |
5′-GGGCTCATGGACGGGTGA
5′-ATCCATGAACTTCACCACTTCG | 328 | 92.7% | | |
| P53 |
5′-ACGACGGTGACACGCTTCCCTG
5′-CGCTAGGATCTGACTGCGGCTC | 84 | 99.1% | | |
| GUSB |
5′-ATGCAGGTGATGGAAGAAGTGGTG
5′-AGAGTTGCTCACAAAGGTCACAGG | 177 | 99.6% | | |
Western blot analysis
Western blot analysis was performed to compare the
protein levels of HIF1A, HIF2A, VEGFA, VHL and P53 in paired
tumor/unchanged kidney tissues of the 36 patients. Lysates were
extracted according to the method previously described in our
previous studies (13,32) with the use of a Mammalian Cell
Extraction kit (BioVision). Since the rapid degradation of the
HIF1A and HIF2A proteins occurs upon exposure to normal oxygen
pressure (35), all further steps
were performed immediately. Briefly, 10-µg protein samples
were loaded onto 10% polyacrylamide gels, resolved by 8% SDS-PAGE,
and transferred onto polyvinylidene difluoride membranes using the
Trans-Blot Turbo system (Bio-Rad Laboratories, Inc.). The membranes
were blocked by incubation with 3% albumin fraction V in
Tris-buffered saline (TBS) buffer at pH 7.4 (Sigma-Aldrich; now a
brand of Merck KGaA) with 0.1% Tween-20 (TBST) for 1 h at room
temperature, and were subsequently incubated overnight at 4°C with
specific primary antibodies dissolved in 2% albumin/TBS at a
dilution of 1:1,000. The following specific rabbit antibodies were
used: Polyclonal anti-HIF1A (cat. no. LS-B674), polyclonal
anti-HIF2A (EPAS1) (cat. no. LS-B4223), polyclonal anti-VEGFA (cat.
no. LS-B10263), polyclonal anti-VHL (cat. no. LS-C99277) and
polyclonal anti-P53 (cat. no. LS-B4558) (all purchased from
LifeSpan BioSciences). After washing 3 times with TBST, the blots
were incubated for 2 h at room temperature with horseradish
peroxidase-conjugated anti-rabbit IgG antibodies (1:10,000; cat.
no. A0545; Sigma-Aldrich; Merck KGaA). Anti-GAPDH
peroxidise-conjugated IgM antibodies (1:50,000; cat. no. G9295;
Sigma-Aldrich; Merck KGaA) were applied for 1 h at room temperature
as the loading control. Following triple washing with TBST,
immunoreactive bands were detected on medical X-ray films (Agfa
HealthCare) using Clarity Western ECL Blotting substrate (Bio-Rad
Laboratories, Inc.). Densitometric analysis of immunoreactive
protein bands was performed with Quantity One software (Bio-Rad
Laboratories, Inc.) and calculated as units =
Intensity/mm2. After normalizing the levels against
GAPDH for each sample, semi-quantitative results for the HIF1A,
HIF2A, VEGFA, VHL or P53 proteins extracted from tumor samples were
expressed according to the ratio: Mean no. of unitsTumor
/ mean no. of unitsControl.
Immunohistochemistry (IHC) for VHL,
HIF1A, HIF2A, VEGFA and P53 proteins
IHC staining was performed as previously described
(32,33). Formalin-fixed paraffin-embedded
tissue sections (6 µm) from the tumor and normal kidney
tissues of 10 patients with ccRCC were deparaffinized and hydrated
through xylenes and a graded alcohol series. Following antigen
retrieval in hot (90°C) acidic citrate buffer (Epitope Retrieval
Solution, pH 6.0; Leica Biosystems Ltd.), the samples were blocked
for endogenous peroxidase activity by incubation with 3% hydrogen
peroxide for 10 min, followed by incubation with 2.5% normal horse
serum [ImmPRESS™ Anti-Rabbit Ig (peroxidase) Polymer Detection kit,
Vector Laboratories Inc.; part of Maravai LifeSciences] to block
the non-specific binding of immunoglobulin. IHC staining was
performed using the same primary antibodies as those used for
western blot analysis at a 1:100 dilution (with the exception of
the anti-P53 antibody, which was used at a dilution of 1:50).
Following a 2-h incubation with primary antibodies at room
temperature, the slides were washed in PBS and incubated with an
appropriate secondary antibody [ImmPRESS™ Anti-Rabbit Ig
(peroxidase) Polymer Detection kit or ImmPRESS™ Anti-Mouse Ig
(peroxidase) Polymer Detection kit; Vector Laboratories, Inc.) for
30 min. The slides were rinsed in PBS, and immunoreactive cells
were visualized by the addition of 3,3′-diaminobenzidine solution
(DAB Peroxidase Substrate kit, Vector Laboratories, Inc.) and
counterstained with hematoxylin. The sections were subsequently
dehydrated, mounted in DPX Mountant, viewed under a Nikon Eclipse
E800 light microscope, and the acquisition of the microphotographs
was performed using NIS software (Nikon). The specificity of IHC
staining was determined by a negative control, which was prepared
under conditions identical to those described above; however, the
primary antibodies were replaced with 2.5% normal horse serum
[ImmPRESS™ Anti-Rabbit Ig (peroxidase) Polymer Detection kit;
Vector Laboratories, Inc.].
Statistical analysis
Statistical analysis was performed using GraphPad
Prism version 6.07 (GraphPad Software) and Statistica version 13
(Dell Inc.) software. The following statistical tests were used:
Non-parametric Mann-Whitney U, Wilcoxon signed-rank and Fisher's
2×2 exact tests, Spearman's correlation, multivariate regression,
Kaplan-Meier survival tests with log-rank (Mantel-Cox) test, and
the Cox proportional hazard regression model. Survival associations
were presented as hazard ratios (HRs) with their 95% confidence
interval (CI) and P-values (36)
using Cox and Kaplan-Meier estimations. The OS and PFS rates were
calculated separately. In all analyses, a two-sided P<0.05 was
considered to indicate a statistically significant value, with a
95% confidence interval.
Results
Characteristics of the patients
The clinical, pathological and summary results of
the laboratory assessments of the 36 patients with ccRCC enrolled
in the present study are presented in Table I. Despite the malignancy, the
majority of the patients were in a relatively good condition, as
evidenced by the results of blood and urine laboratory tests and
physical examinations; none of the patients was diagnosed with
cachexia. The patients enrolled for sunitinib treatment had passed
the ESMO guidelines (37).
Of the 36 patients with ccRCC aged 60.6±11.9 years
[mean ± standard deviation (SD); Table
I], 21 were diagnosed [according to the
tumor-necrosis-metastasis (TNM) staging system] as stage I
(T1-2N0M0), 7 as stage III (T1-2N1M0 or T3N0-1M0) and 8 as stage IV
(T4N0-2M0 or T1-4N2M0 or T1-4N0-2M1), according to anatomic stage
and prognostic groups based on the 2010 TNM 7th classification of
RCC (38). At the time of surgery,
41.7% of the patients with ccRCC were diagnosed with local or
distant metastases. Histological nuclear staging was based on the
Fuhrman grading system (39),
which revealed that tumor tissues from 2 patients were classified
as grade 1, 16 were grade 2, 11 were grade 3, and 7 were classified
as grade 4. None of the patients had undergone chemo- or
radiotherapy prior to surgery. The mean follow-up period was 21
months (range, 3-48 months), and until the end of follow-up
collection, 23 patients were alive (64%); all deaths were
associated with ccRCC progression. The median OS rate was
undefined. During follow-up, metastases occurred in 14 (39%) of the
patients (data not shown), whereas the median PFS rate was
undefined. Post-operative treatment included sunitinib (an
anti-VEGFA agent), which was administered to 9 patients with
clinically advanced ccRCC (T1-2N1M0, T3N0-1M0, T1-4N2M0 and
T1-4N0-2M1), and to 2 patients with early ccRCC (T1-2N0M0),
according to a generally accepted schedule (40) of 1 cycle: 50 mg/day for 4 weeks,
followed by a 2-week interval. During follow-up, 9 and 10
sunitinib-treated patients succumbed to the disease or experienced
cancer recurrence, respectively.
Expression of the VHL, HIF1A, HIF2A,
VEGFA and P53 genes at the mRNA level
The mRNA levels of the selected genes were assessed
by RT-qPCR in the tumor tissues and corresponding normal kidney
samples of 36 patients with ccRCC. Different ratios of expression
for all genes between the cancerous tissues and microscopically
unaltered renal tissues were observed; the VHL mRNA ratio
was 30% lower in the cancer tissues (P<0.05; Fig. 1A), and its decreased levels were
observed in 20/36 (56%) of the ccRCC patients (Table III). The other analyzed genes
exhibited statistically significantly higher mRNA levels in the
ccRCC compared with the normal kidney tissue: HIF1A mRNA was
upregulated ~2-fold (P<0.05; Fig.
1A), and its level was increased in 56% of the cancer samples
(Table III); by comparison,
HIF2A mRNA was upregulated ~5-fold (P<0.01; Fig. 1A), and its higher expression was
observed in 58% (Table III) of
the patients. The VEGFA and P53 mRNA levels were
increased by 2- and 3-fold, respectively (P<0.01; Fig. 1A), and increased mRNA expression
rates of these genes were detected in 78 and 70% of the ccRCC
patients, respectively (Table
III). When the molecular assessment at the mRNA level was
compared with the clinicopathological data, it was observed that
the increased expression of the HIF2A and VEGFA genes
was associated with a higher Fuhrman's grade (3 and 4);
additionally, the mRNA levels increased by ~1.5- and 1.6-fold
(Fig. 1B) between grades 3+4 and
1+2, respectively (P<0.01). The mean expression rate of
VEGFA was also associated with clinically advanced ccRCC,
since its mRNA level was ~1.4-fold higher in the metastatic tumor
samples compared with early-stage tumors (P<0.01; Fig. 1C). When the expression levels for
each gene were divided according to their median levels in normal
samples (13,41) (Table
III), increased expression rates of HIF2A (42%) and
VEGFA (47%) were observed in samples with higher Furhman's
grades, whereas higher HIF2A (33%) and P53 (39%) mRNA
levels were observed in clinically advanced ccRCCs (Table III). No other associations were
observed between the laboratory or clinicopathological data of the
patients and the expression of the studied genes at the mRNA
level.
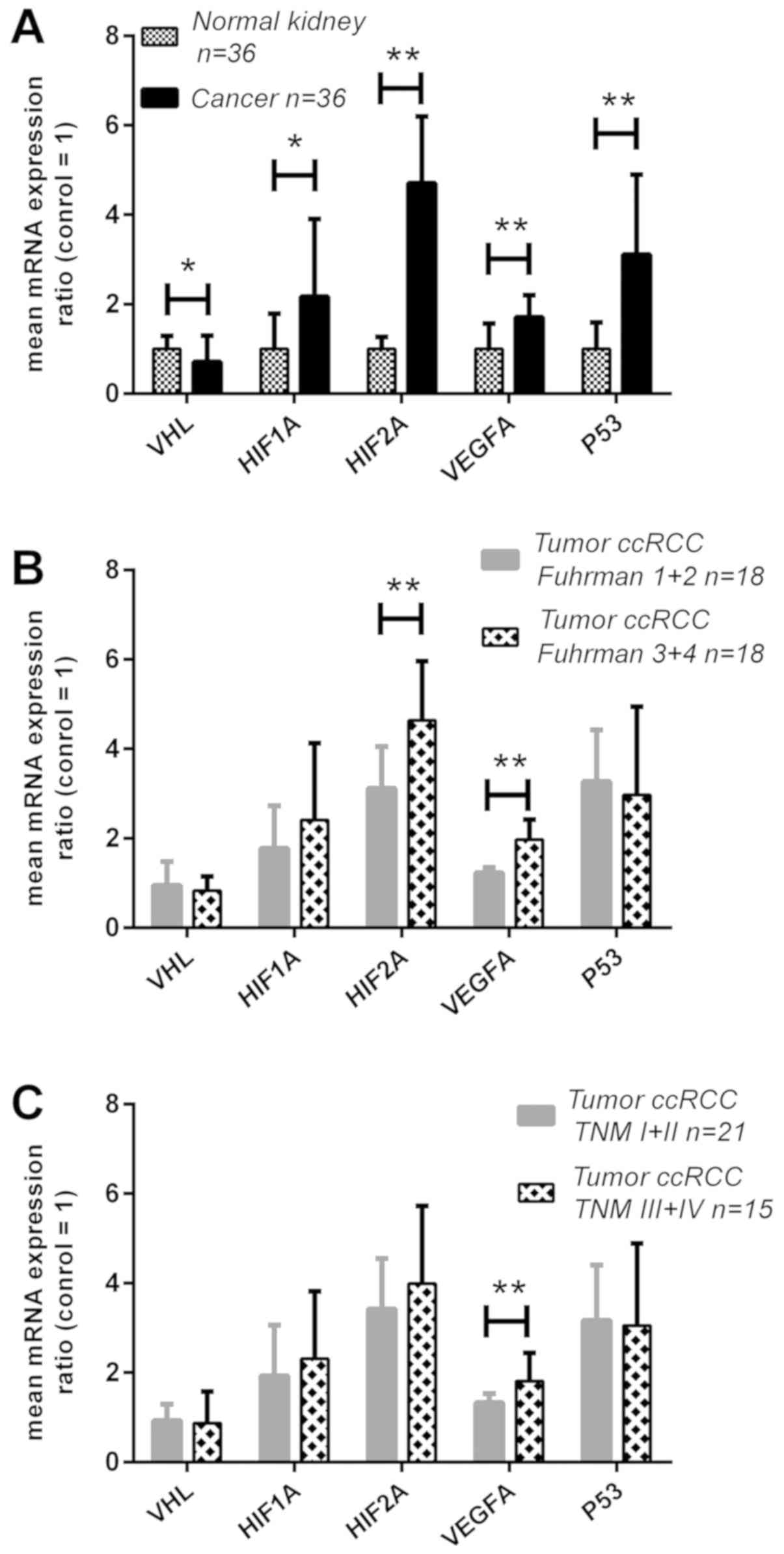 | Figure 1Expression analysis of the VHL,
HIF1A, HIF2A, VEGFA and P53 genes in ccRCC at the mRNA
level. Gene expression was assessed as described in the Materials
and methods. (A) Comparison between tumor and normal kidney
samples; (B) gene expression in tumor samples related to Fuhrman
grade; and (C) gene expression in tumor samples related to TNM
stage. Bars and whiskers represent mean ± standard deviation
normalized to control kidney samples. *P<0.05,
**P<0.01. (Wilcoxon signed-rank test for A,
Mann-Whitney U test for B and C). ccRCC, clear cell renal cell
carcinoma; VHL, Von Hippel-Lindau; HIF, hypoxia-inducible factor;
VEGF, vascular endothelial growth factor; qPCR, quantitative
polymerase chain reaction. |
 | Table IIIAssociation between VHL, HIF1A,
HIF2A, VEGFA and P53 mRNA levels and clinical data. |
Table III
Association between VHL, HIF1A,
HIF2A, VEGFA and P53 mRNA levels and clinical data.
| Patients n=36 | Subgroups | VHL qPCR
results (%)
| HIF1A qPCR
results (%)
| HIF2A qPCR
results (%)
| VEGFA qPCR
results (%)
| P53 qPCR
results (%)
|
|---|
| ↓ | ↑ | P-valuea | ↓ | ↑ | P-valuea | ↓ | ↑ | P-valuea | ↓ | ↑ | P-valuea | ↓ | ↑ | P-valuea |
|---|
| Age (years) | ≤6, n=19 | 10 (28) | 9 (25) | 0.74 | 8 (22) | 11 (31) | 1.0 | 9 (25) | 10 (28) | 0.52 | 5 (14) | 14 (39) | 0.69 | 8 (22) | 11 (31) | 0.15 |
| Median,
60.58±11.9, | >62, n=17 | 10 (28) | 7 (19) | | 8 (22) | 9 (25) | | 6 (17) | 11 (30) | | 3 (8) | 14 (39) | | 3 (8) | 14 (39) | |
| Range, 33-82, | | | | | | | | | | | | | | | | |
| Sex | Female, n=17 | 11 (31) | 6 (16) | 0.23 | 5 (14) | 12 (33) | 0.1 | 9 (25) | 8 (22) | 0.31 | 6 (17) | 11 (31) | 0.11 | 7 (19) | 10 (28) | 0.28 |
| Male, n=19 | 9 (25) | 10 (28) | | 11 (31) | 8 (22) | | 6 (17) | 13 (36) | | 2 (5) | 17 (47) | | 4 (11) | 15 (42) | |
| Tumor | ≤7 cm, n=17 | 7 (19) | 10 (28) | 0.18 | 9 (25) | 8 (22) | 0.5 | 7 (19) | 10 (28) | 1.00 | 3 (8) | 14 (39) | 0.69 | 5 (14) | 12 (33) | 1.00 |
| size (cm) | >7 cm, n=19 | 13 (36) | 6 (16) | | 7 (19) | 12 (33) | | 8 (22) | 11 (31) | | 5 (14) | 14 (39) | | 6 (17) | 13 (36) | |
| Fuhrman's | 1+2, n=18 | 10 (28) | 8 (22) | 1.00 | 5 (14) | 13 (36) | 0.09 | 12 (33) | 6 (17) | 0.006 | 7 (19) | 11 (31) | 0.04 | 8 (22) | 10 (28) | 0.15 |
| histological
grade, | 3+4, n=18 | 10 (28) | 8 (22) | | 11 (31) | 7 (19) | | 3 (8) | 15 (42) | | 1 (3) | 17 (47) | | 3 (8) | 15 (42) | |
| TNM stage | Non-metastatic,
n=21 | 12 (33) | 9 (25) | 0.9 | 7 (19) | 14 (39) | 0.17 | 12 (33) | 9 (25) | 0.04 | 7 (19) | 14 (39) | 0.10 | 10 (28) | 11 (31) | 0.011 |
| Metastatic,
n=15 | 8 (22) | 7 (19) | | 9 (25) | 6 (17) | | 3 (8) | 12 (33) | | 1 (3) | 14 (39) | | 1 (3) | 14 (39) | |
| Sunitinib
treatment | Yes, n=11 | 6 (17) | 5 (14) | 1.00 | 7 (19) | 4 (11) | 0.16 | 2 (6) | 9 (25) | 0.07 | 1 (3) | 10 (28) | 0.38 | 4 (11) | 7 (19) | 0.7 |
| No, n=25 | 14 (39) | 11 (31) | | 9 (25) | 16 (44) | | 13 (36) | 12 (33) | | 7 (19) | 18 (50) | | 7 (19) | 18 (50) | |
Expression of the VHL, HIF1A, HIF2A,
VEGFA and P53 genes at the protein level
Semi-quantitative assessment of the protein
expression levels was performed in paired tumor and normal kidney
samples of the 36 patients with ccRCC by western blot analysis and
the exemplary images are shown in Fig.
2. The expression levels of the analyzed proteins were
increased in the tumor samples: The HIF1A, HIF2A and P53 levels
were ~10- (P<0.001), 1.5- (P<0.05) and 4.8-fold (P<0.001)
higher in the cancer tissues, respectively, compared with those in
the normal kidney samples (Fig.
3A). No differences were observed in the pVHL and VEGFA protein
levels between the cancerous and normal tissues (Fig. 3A). However, the expression levels
of other proteins were increased in the tumor samples: When the
expression levels were divided by their median values in normal
tissues, higher levels of the HIF1A, HIF2A and P53 proteins in
cancer tissues were detected in 63, 64 and 75% of the patients,
respectively (Table IV). The
complete absence of the HIF1A protein was found in 10 normal and 3
cancer samples, whereas the absence of the HIF2A protein band was
found in 4 normal samples and 1 tumor sample (data not shown). Such
a result was not observed for the other proteins examined. When the
western blot analysis results were compared with the
clinicopathological data, no significant association between sex,
age, tumor progression or tumor size and VHL or P53 protein levels
was observed (Figs. 3B and C;
Table IV). On the other hand, the
protein ratios of HIF1A, HIF2A and VEGFA were markedly increased in
the tumors of patients with higher Fuhrman's grades; the HIF1A
levels were increased ~1.3-fold (P<0.05) in 22% of the samples
(Fig. 3B and Table IV), whereas those of HIF2A were
3.3-fold higher (P<0.001) in 44% of the samples (Fig. 3B and Table IV). The VEGFA protein level was
increased ~2-fold (44% of the samples) in samples with advanced
Fuhrman's grades in comparison with samples with Fuhrman's 1+2
grades (P<0.01; Fig. 3B and
Table IV). In addition, the
expression levels of the HIF2A and VEGFA proteins were associated
with metastatic ccRCC: The HIF2A protein level was ~2-fold higher
(P<0.001, Fig. 3C) in advanced
tumor samples (36% of the samples, Table IV), whereas that of VEGFA protein
was upregulated ~1.6-fold (P<0.01; Fig. 3C) in metastatic cases (39% of the
samples; Table IV). A higher
expression of HIF2A protein was also noted in larger tumors, with
17 of the 19 cases with tumors >7 cm exhibiting an increased
HIF2A level (Table IV).
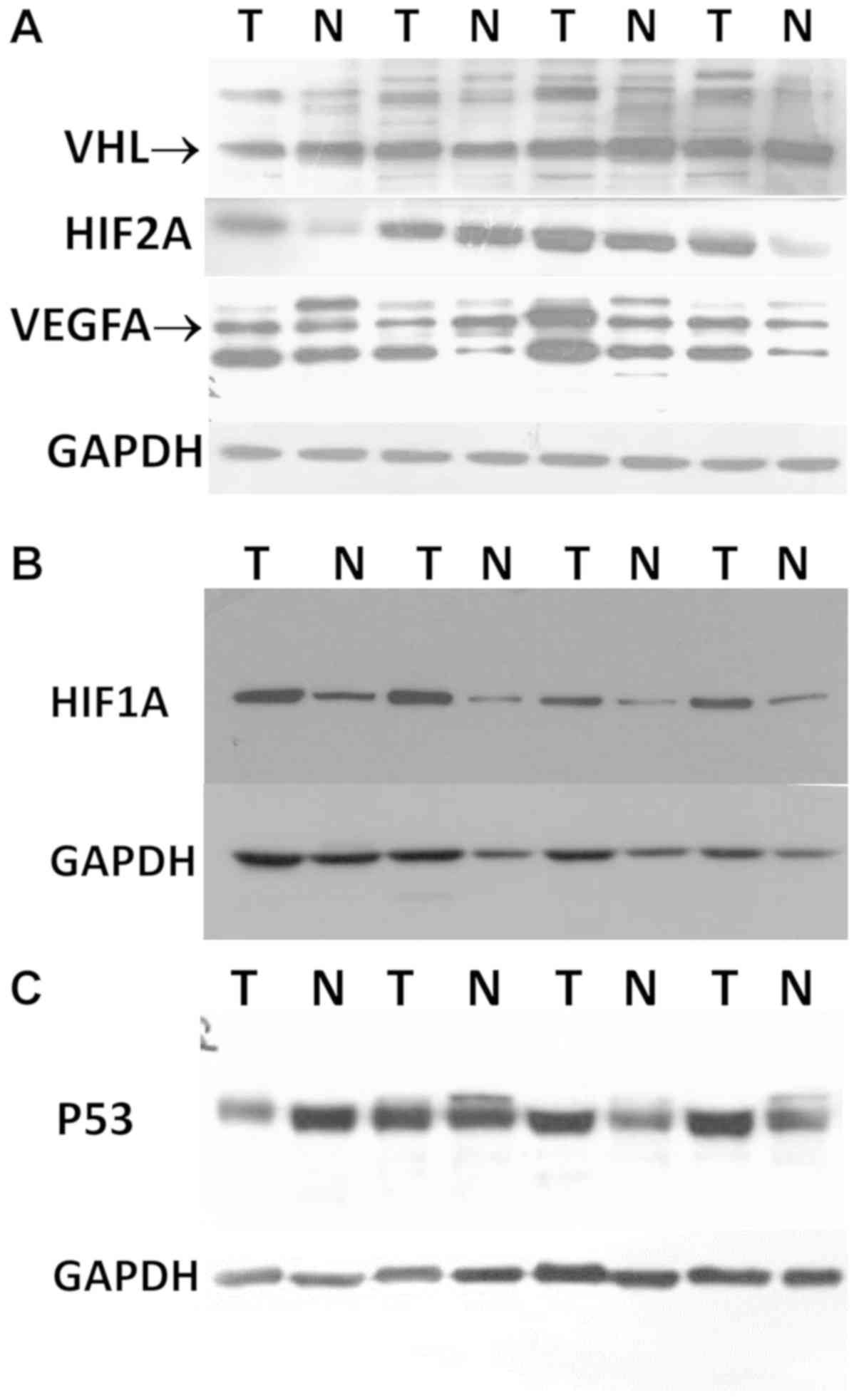 | Figure 2Western blot analysis images of VHL,
HIF1A, HIF2A, VEGFA and P53 proteins in ccRCC. Protein assessment
was performed as described in the Materials and methods. (A) Merged
image of VHL, HIF2A, VEGFA and GAPDH proteins as they were analyzed
in the same western blot membrane. (B and C) Western blot anlaysis
images of HIF1A and P53 proteins with corresponding GAPDH bands,
respectively. ccRCC, clear cell renal cell carcinoma; VHL, Von
Hippel-Lindau; HIF, hypoxia-inducible factor; VEGF, vascular
endothelial growth factor. |
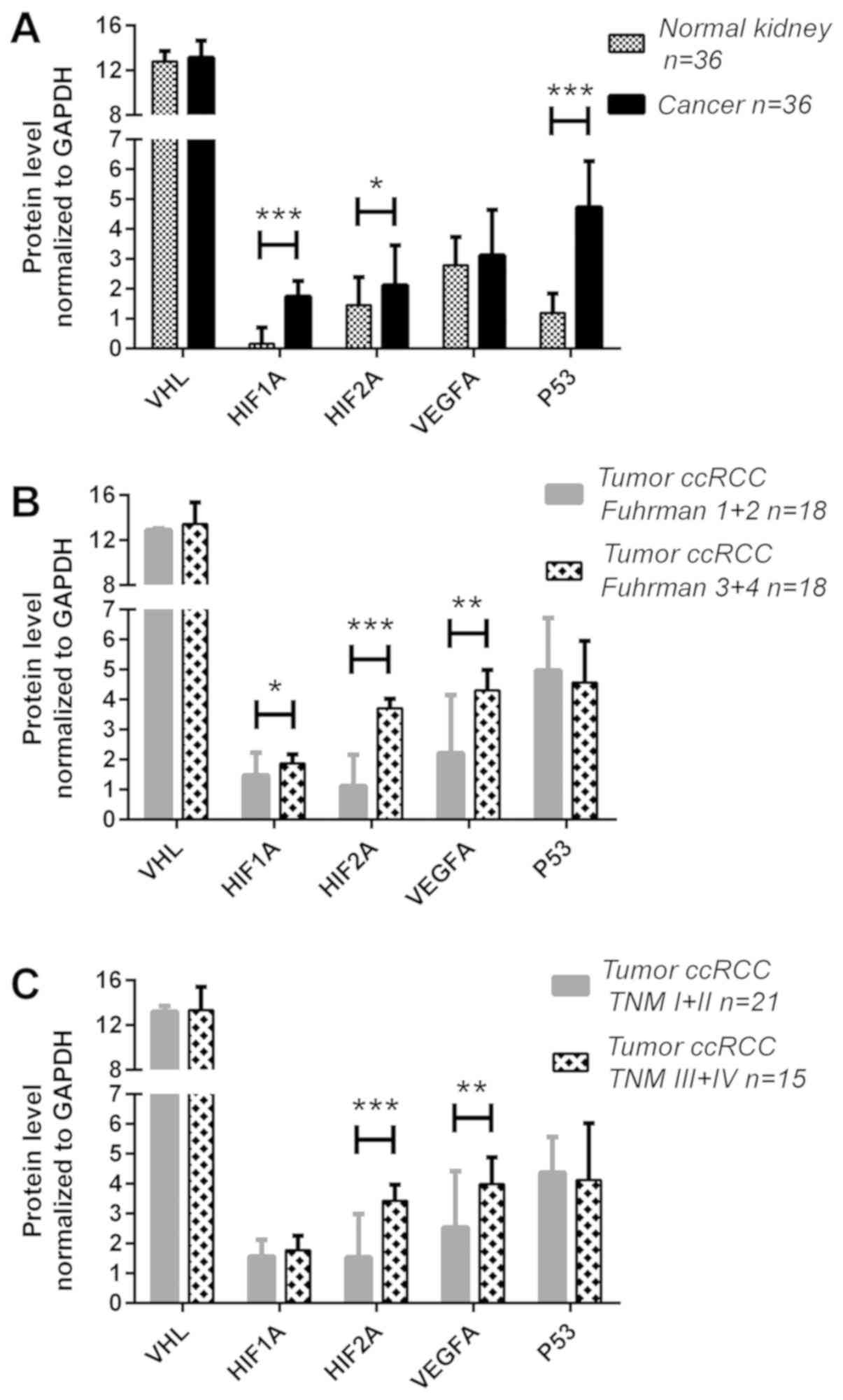 | Figure 3Expression analysis of the VHL,
HIF1A, HIF2A, VEGFA and P53 genes in ccRCC at the
protein level. Gene expression at protein level was assessed as
described in the Materials and methods. (A) Comparison between
tumor and normal kidney samples; (B) gene expression at protein
level in tumor samples related to Fuhrman grade; and (C) gene
expression at protein level in tumor samples related to TNM stage.
Bars and whiskers represent mean ± standard deviation normalized to
GAPDH level in each sample. *P<0.05,
**P<0.01, ***P<0.001. (Wilcoxon
signed-rank test for A, Mann-Whitney U test for B and C). ccRCC,
clear cell renal cell carcinoma; VHL, Von Hippel-Lindau; HIF,
hypoxia-inducible factor; VEGF, vascular endothelial growth factor;
qPCR, quantitative polymerase chain reaction. |
 | Table IVAssociation between VHL, HIF1A,
HIF2A, VEGFA and P53 protein levels and clinical data. |
Table IV
Association between VHL, HIF1A,
HIF2A, VEGFA and P53 protein levels and clinical data.
| Patients n=36 | Subgroups | pVHL WB results (%)
| HIF1A WB results
(%)
| HIF2A WB results
(%)
| VEGFA WB results
(%)
| P53 WB results (%)
|
|---|
| ↓ | ↑ | P-valuea | ↓ | ↑ | P-valuea | ↓ | ↑ P | -valuea | ↓ | ↑ P | -valuea | ↓ | ↑ | P-valuea |
|---|
| Age (years) | ≤62, n=19 | 13 (36) | 6 (17) | 0.73 | 5 (14) | 14 (39) | 0.72 | 5 (14) | 14 (39) | 0.29 | 8 (22) | 11 (31) | 0.15 | 6 (17) | 13 (36) | 0.45 |
| Median,
60.58±11.9 | >62, n=17 | 10 (28) | 7 (19) | | 6 (17) | 9 (31) | | 8 (22) | 9 (25) | | 3 (8) | 14 (39) | | 3 (8) | 14 (39) | |
| Range, 33-82 | | | | | | | | | | | | | | | | |
| Sex | Female, n=17 | 13 (36) | 4 (11) | 0.17 | 4 (11) | 13 (36) | 0.48 | 9 (25) | 8 (22) | 0.08 | 8 (22) | 9 (25) | 0.07 | 8 (22) | 9 (25) | 0.07 |
| Male, n=19 | 10 (28) | 9 (25) | | 7 (19) | 12 (33) | | 4 (11) | 15 (42) | | 3 (8) | 16 (44) | | 3 (8) | 16 (44) | |
| Tumor | ≤7 cm, n=17 | 10 (28) | 7 (19) | 0.73 | 4 (11) | 13 (36) | 0.48 | 11 (31) | 6 (17) | 0.001 | 6 (17) | 11 (31) | 0.72 | 5 (14) | 12 (33) | 0.71 |
| size (cm) | >7 cm, n=19 | 13 (36) | 6 (16) | | 7 (19) | 12 (33) | | 2 (5) | 17 (47) | | 5 (14) | 14 (39) | | 4 (11) | 15 (42) | |
| Fuhrman's | 1+2, n=18 | 10 (28) | 8 (22) | 1.00 | 1 (3) | 17 (47) | 0.003 | 12 (33) | 7 (19) | 0.005 | 9 (25) | 9 (25) | 0.02 | 7 (19) | 11 (31) | 0.12 |
| histological
grade | 3+4, n=18 | 10 (28) | 8 (22) | | 10 (28) | 8 (22) | | 2 (6) | 16 (44) | | 2 (6) | 16 (44) | | 2 (6) | 16 (44) | |
| TNM stage | Non-metastatic,
n=21 | 15 (42) | 6 (17) | 0.31 | 6 (17) | 15 (42) | 1.00 | 11 (31) | 10 (28) | 0.03 | 10 (28) | 11 (31) | 0.011 | 8 (22) | 13 (36) | 0.05 |
| Metastatic,
n=15 | 8 (22) | 7 (19) | | 5 (14) | 10 (28) | | 2 (5) | 13 (36) | | 1 (3) | 14 (39) | | 1 (3) | 14 (39) | |
| Sunitinib
treatment | Yes, n=11 | 7 (19) | 4 (11) | 1.00 | 6 (20) | 5 (17) | 0.23 | 1 (3) | 10 (31) | 0.01 | 0 (0) | 11 (31) | 0.015 | 4 (11) | 7 (19) | 0.4 |
| No, n=25 | 16 (44) | 9 (25) | | 5 (17) | 14 (47) | | 12 (38) | 9 (28) | | 11 (31) | 14 (38) | | 5 (14) | 20 (56) | |
Association between mRNA and protein
expression levels of the analyzed genes
The Spearman's correlation test was used to examine
the possible associations between VHL-HIF1A/HIF2A-VEGFA expression
at the mRNA and protein level, as well as P53 expression, in ccRCC
samples. The results are presented in Table V. First, medium-to-strong positive
correlations were observed between the mRNA and protein levels of
each gene [from Spearman's rank correlation coefficient (rs)=0.57
for P53 mRNA-protein to rs=0.74 for HIF1A mRNA-protein; P<0.05;
Table V]. Subsequently, it was
observed that the expression of P53 did not correlate with the
levels of any other analyzed genes. Notably, a weak negative
correlation was observed between the levels of the VHL protein and
the HIF1A and HIF2A proteins in ccRCC tissue (rs=−0.13 and =−0.19,
respectively; P<0.05; Table V).
There was also a medium-to-strong positive correlation between
either HIF1A or HIF2A and VEGFA (at both the
mRNA and protein level); the associations were stronger for
HIF1A mRNA-VEGFA mRNA and HIF2A
mRNA-VEGFA mRNA (rs=0.71 and 0.73, respectively; P<0.05)
compared with HIF1A protein-VEGFA protein or HIF2A protein-VEGFA
protein (rs=0.58 and 0.69, respectively; Table V). A weak positive correlation was
also observed between the levels of HIF1A and HIF2A
mRNA (rs=0.42; P<0.05).
 | Table VSummary results of Spearman's
correlation tests between molecular data of VHL, HIF1A, HIF2A,
VEGFA and P53 gene expression. |
Table V
Summary results of Spearman's
correlation tests between molecular data of VHL, HIF1A, HIF2A,
VEGFA and P53 gene expression.
| Molecular
data/rsa | VHL
mRNA | VHL protein | HIF1A
mRNA | HIF1A protein | HIF2A
mRNA | HIF2A protein | VEGFA
mRNA | VEGFA protein | P53
mRNA | P53 protein |
|---|
| VHL
mRNA | | 0.63 | NS | NS | NS | NS | NS | NS | NS | NS |
| VHL protein | 0.63 | | NS | −0.13 | NS | −0.19 | NS | NS | NS | NS |
| HIF1A
mRNA | NS | NS | | 0.74 | 0.42 | NS | 0.71 | 0.49 | NS | NS |
| HIF1A protein | NS | −0.13 | 0.74 | | NS | 0.69 | 0.60 | 0.58 | NS | NS |
| HIF2A
mRNA | NS | NS | 0.42 | NS | | 0.68 | 0.73 | 0.64 | NS | NS |
| HIF2A protein | NS | −0.19 | NS | 0.69 | 0.68 | | 0.72 | 0.69 | NS | NS |
| VEGFA
mRNA | NS | NS | 0.71 | 0.60 | 0.73 | 0.72 | | 0.73 | NS | NS |
| VEGFA protein | NS | NS | 0.49 | 0.58 | 0.64 | 0.69 | 0.73 | | NS | NS |
| P53
mRNA | NS | NS | NS | NS | NS | NS | NS | NS | | 0.57 |
| P53 protein | NS | NS | NS | NS | NS | NS | NS | NS | 0.57 | |
Tissue localization of proteins
Immunohistochemical staining for VHL, HIF1A, HIF2A,
VEGFA and P53 was performed on paired tumor and normal kidney
samples of 10 patients with ccRCC (3 patients with non-metastatic
and 7 patients with metastatic ccRCC, of whom 3 were treated with
sunitinib). As shown in Fig. 4A,
in the unaffected part of the kidney, pVHL immunoreactivity was
strong in the cytoplasm and nuclei of the epithelial cells of the
proximal and distal tubules (PT - black arrows and DT - yellow
arrows in figures, respectively), whereas weaker pVHL
immunoreactivity was noted in the cytoplasm and nuclei of tumor
cells (Fig. 4B). HIF1A and HIF2A
immunoreactivity was predominantly present in the nuclei of cancer
cells (Fig. 4D and F), with a
particularly strong expression of HIF2A (Fig. 4F). In normal kidney tissue, HIF1A
protein expression was observed in the cytoplasm and nuclei of PT
and DT epithelial cells (black and yellow arrows, respectively,
Fig. 4C), whereas HIF2A protein
expression was mainly localized in the cytoplasm of PT and DT cells
(black and yellow arrows, respectively, Fig. 4E). The immunoreactivity of VEGFA
was moderate in the cytoplasm of PT and DT cells (black and yellow
arrows, respectively, Fig. 4G),
whereas in the tumor specimens, VEGFA protein expression was
strongly and homogeneously distributed in the cytoplasm of cancer
cells (Fig. 4H). Finally, we
observed very strong immunoreactivity of the P53 protein in the
cytoplasm and nuclei of both tumor (Fig. 4J) and normal kidney cells (stroma,
black arrows for PT and yellow arrows for DT; Fig. 4I).
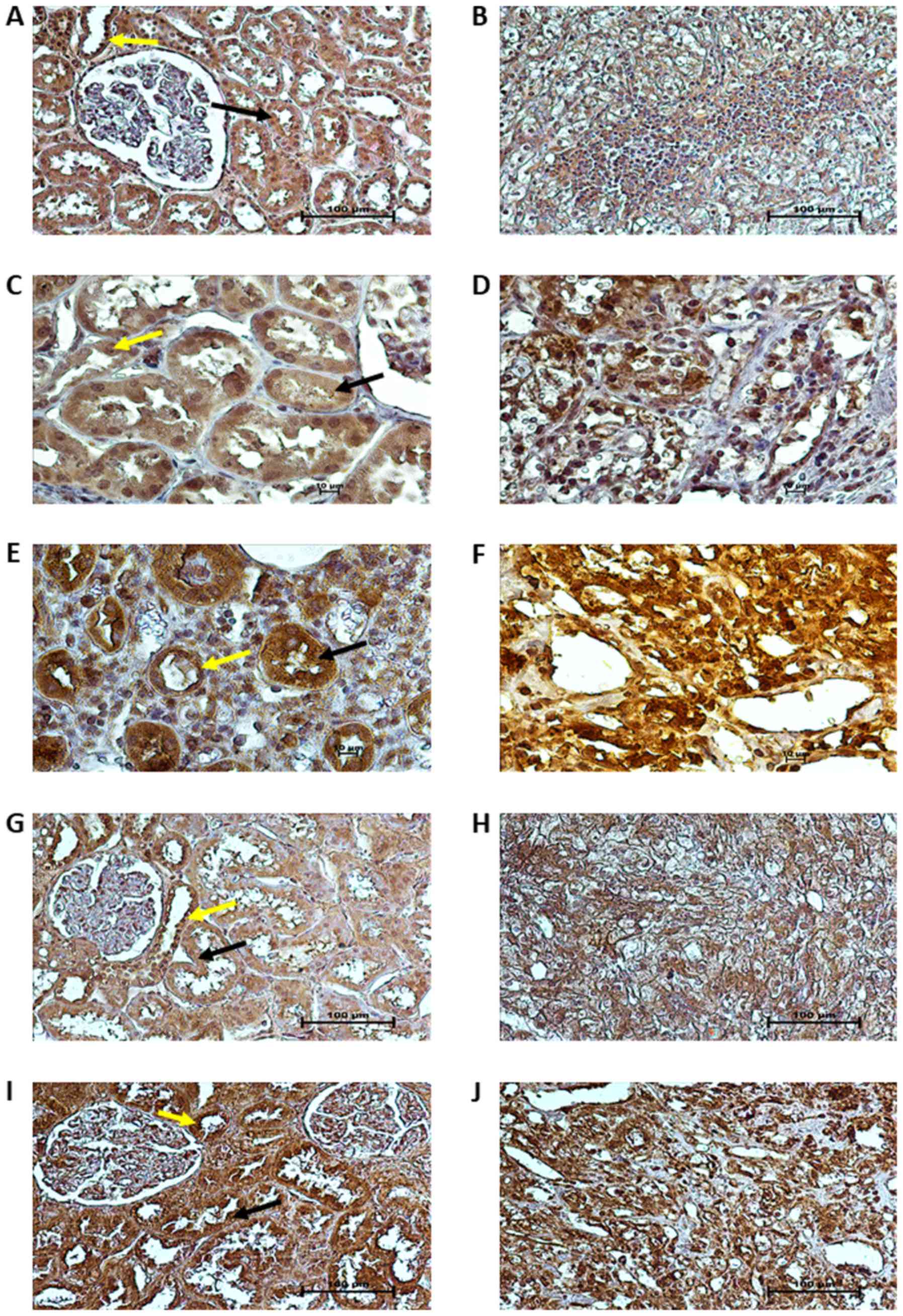 | Figure 4Localization of VHL, HIF1A, HIF2A,
VEGFA and P53 proteins in ccRCC and normal kidney. Immunoreactivity
for (A and B) VHL, (C and D) HIF1A, (E and F) HIF2A, (G and H)
VEGFA and (I and J) P53 proteins in normal kidney (A, C, E, G and
I) and TNM stage 3 and Fuhrman grade 3 ccRCC sections (B, D, F, H
and J) was demonstrated by immunohistochemical staining, as
described in the Materials and methods. Scale bars, 10 µm
(C-F) and 100 µm (A, B and G-J). Black and yellow arrows
arrows indicate proximal and distal tubules of unchanged kidney
morphological structure, respectively (A, C, E, G and I). ccRCC,
clear cell renal cell carcinoma; VHL, Von Hippel-Lindau; HIF,
hypoxia-inducible factor; VEGF, vascular endothelial growth
factor. |
Associations of clinicopathological and
molecular data with patient outcome
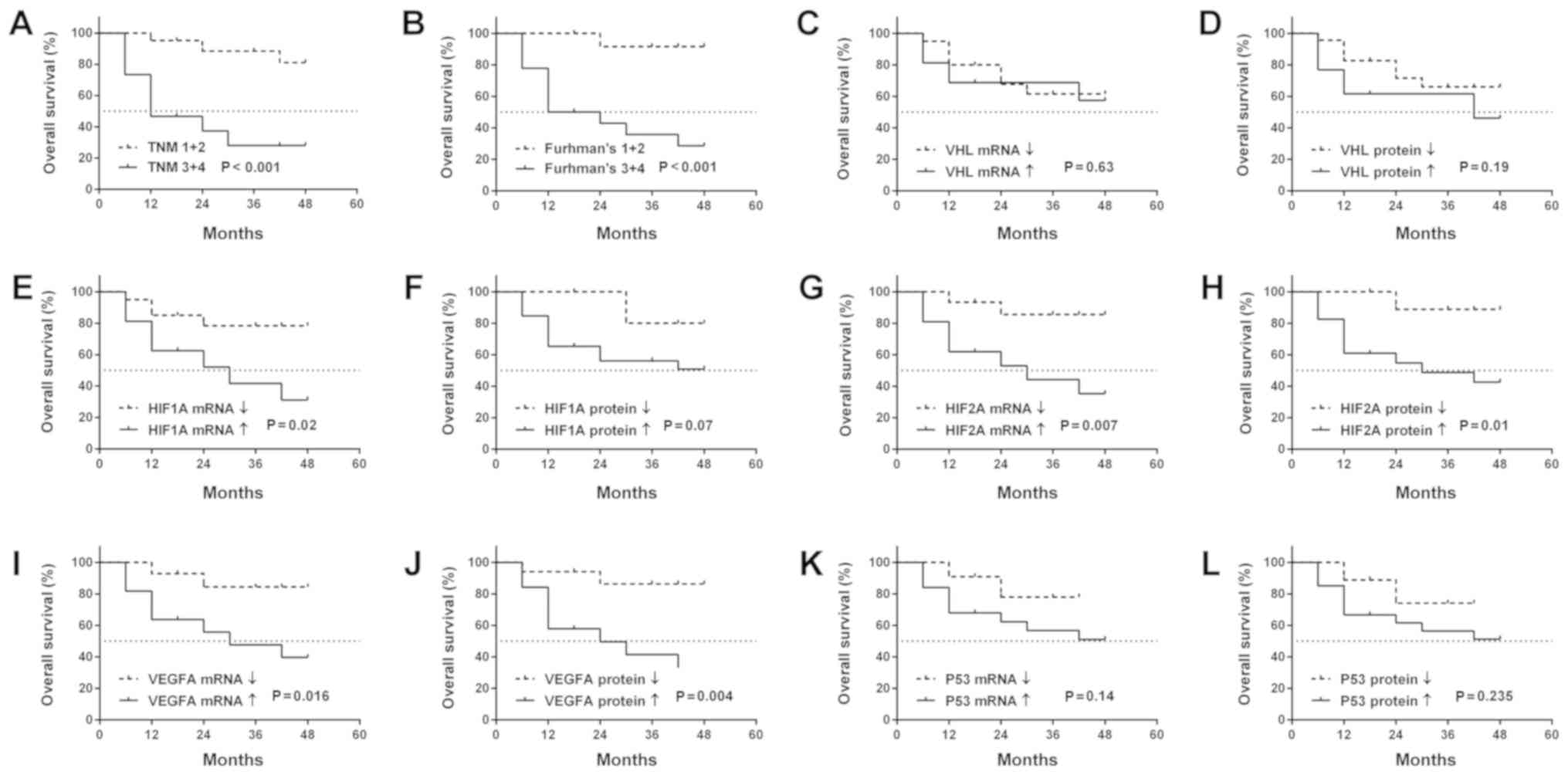
We found that higher Fuhrman's grade and TNM stage
were associated with OS (Fig. 5A and
B). No association was observed between the OS of patients and
other clinicopathological parameters (data not shown). It was
observed that patients with an increased expression of HIF1A
(mRNA), HIF2A (mRNA and protein) and VEGFA (mRNA and
protein) in ccRCC exhibited a shorter OS (Fig. 5E and G-J). There was no observed
association between OS and VHL or P53 gene
expression, neither at the mRNA nor the protein level (Fig. 5C, D, K and L).
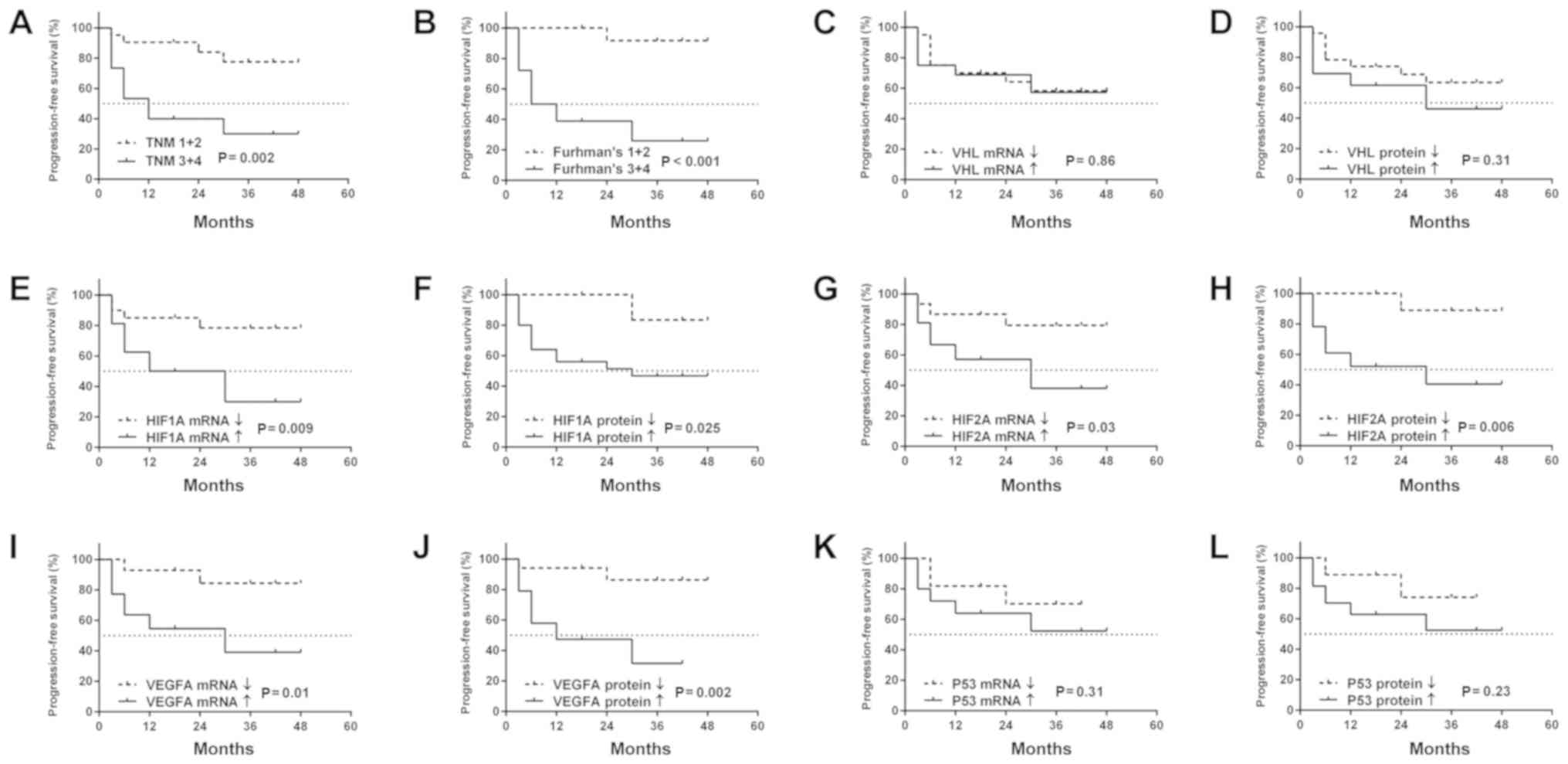
Similarly to OS, PFS was associated with a higher
Fuhrman's grade and TNM stage (Fig. 6A
and B), while no other clinicopathological values were related
to PFS (data not shown). Patients with increased levels of
HIF1A (mRNA and protein), HIF2A (mRNA and protein),
as well as VEGFA (mRNA and protein) were characterized by a
shorter PFS (Fig. 6E-J). There was
no connection between levels of mRNA or protein of the VHL
and P53 genes and the occurrence of cancer progression
(Fig. 6C, D, K and L).
Cox proportional hazards test with multivariable
regression indicated that an increased HIF2A expression at
the mRNA and protein level, as well as increased VEGFA protein
levels, were independent prognostic factors of a worse outcome
(Table VI). Moreover, increased
levels of the HIF2A and VEGFA proteins were independent prognostic
factors of earlier cancer progression (Table VII). The administration of
sunitinib was not introduced to the Cox hazards test, since the
treatment was applied after the acquisition of the biological
material.
 | Table VIUnivariable and multivariable Cox
regression analysis of the overall survival rate of ccRCC
patients. |
Table VI
Univariable and multivariable Cox
regression analysis of the overall survival rate of ccRCC
patients.
| Parameters | Univariable
analysis
| Multivariable
analysis
|
|---|
| P-value | HR (95% CI) | P-value | HR (95% CI) |
|---|
| Sex | 0.38 | 1.63
(0.53-5.01) | | |
| Female vs.
male | | | | |
Age
>62 vs. ≤62 (years) | 0.91 | 0.93
(0.31-2.78) | | |
Tumor
size
>7 vs. ≤7 (cm) | 0.48 | 1.49
(0.49-4.41) | | |
Tumor
grade
T3+4 vs. T1+2) | 0.003 | 6.96
(1.89-25.68) | 0.11 | 3.04
(0.77-11.92) |
| Histological
grade | | | | |
| F3+4 vs. F1+2 | 0.008 | 16.25
(2.11-125.53) | 0.038 | 9.64
(1.12-82.32) |
VHL mRNA
levels
↓ vs. ↑ | 0.65 | 0.77
(0.26-2.32) | | |
VHL protein
levels
↓ vs. ↑ | 0.21 | 4.57
(1.35-15.37) | | |
HIF1A mRNA
levels
↑ vs. ↓ | 0.032 | 0.27
(0.08-0.89) | 0.09 | 0.99
(0.21-4.55) |
HIF1A protein
levels
↑ vs. ↓ | 0.089 | 5.85
(0.76-45.06) | | |
HIF2A mRNA
levels
↑ (>17.363) vs. ↓ (≤17.363) | 0.021 | 5.98
(1.31-27.44) | 0.03 | 3.21
(0.52-19.72) |
HIF2A protein
levels
↑ vs. ↓ | 0.031 | 8.51
(1.11-65.62) | 0.04 | 3.07
(0.22-42.19) |
VEGFA mRNA
levels
↑ vs. ↓ | 0.037 | 5.02
(1.11-22.94) | 0.09 | 1.09
(0.13-9.24) |
VEGFA protein
levels
↑ vs. ↓ | 0.014 | 6.58
(1.44-30.03) | 0.019 | 2.34
(0.34-16.06) |
P53 mRNA
levels
↑ vs. ↓ | 0.18 | 2.78
(0.61-12.57) | | |
P53 protein
levels
↑ vs. ↓ | 0.33 | 2.11
(0.46-9.53) | | |
 | Table VIIUnivariable and multivariable Cox
regression analysis of the progression-free survival rate of ccRCC
patients. |
Table VII
Univariable and multivariable Cox
regression analysis of the progression-free survival rate of ccRCC
patients.
| Parameters | Univariable
analysis
| Multivariable
analysis
|
|---|
| P-value | HR (95% CI) | P-value | HR (95% CI) |
|---|
| Sex | 0.29 | 1.81
(0.61-5.4) | | |
| Female vs.
male | | | | |
Age
>62 vs. ≤62 (years) | 0.71 | 0.82
(0.28-2.35) | | |
Tumor
size
>7 vs. ≤7 (cm) | 0.76 | 1.17
(0.41-3.35) | | |
Tumor
grade
T3+4 vs. T1+2 | 0.007 | 5.01
(1.54-16.18) | | |
| Histological
grade | | | | |
| F3+4 vs. F1+2 | 0.004 | 18.66
(2.42-143.35) | 0.028 | 6.64
(1.12-16.32) |
VHL mRNA
levels
↓vs. ↑ | 0.86 | 0.91
(0.31-2.65) | | |
VHL protein
levels
↓vs. ↑ | 0.34 | 0.59
(0.21-1.72) | | |
HIF1A mRNA
levels
↑ vs. ↓ | 0.02 | 0.25
(0.08-0.81) | 0.09 | 0.96
(0.21-4.55) |
HIF1A protein
levels
↑ vs. ↓ | 0.06 | 6.83
(0.98-52.27) | | |
HIF2A mRNA
levels
↑ (>17.363)vs. ↓(≤ 17.363) | 0.05 | 3.62
(0.99-13.22) | | |
HIF2A protein
levels
↑ vs. ↓ | 0.029 | 9.57
(1.25-73.44) | 0.031 | 3.75
(0.28-49.27) |
VEGFA mRNA
levels
↑ vs. ↓ | 0.028 | 5.37
(1.19-24.26) | 0.07 | 2.31
(0.33-15.69) |
VEGFA protein
levels
↑ vs. ↓ | 0.009 | 7.32
(1.62-32.97) | 0.04 | 2.34
(0.31-17.27) |
P53 mRNA
levels
↑ vs. ↓ | 0.34 | 1.86
(0.51-6.68) | | |
P53 protein
levels
↑ vs. ↓ | 0.26 | 2.34
(0.53-10.48) | | |
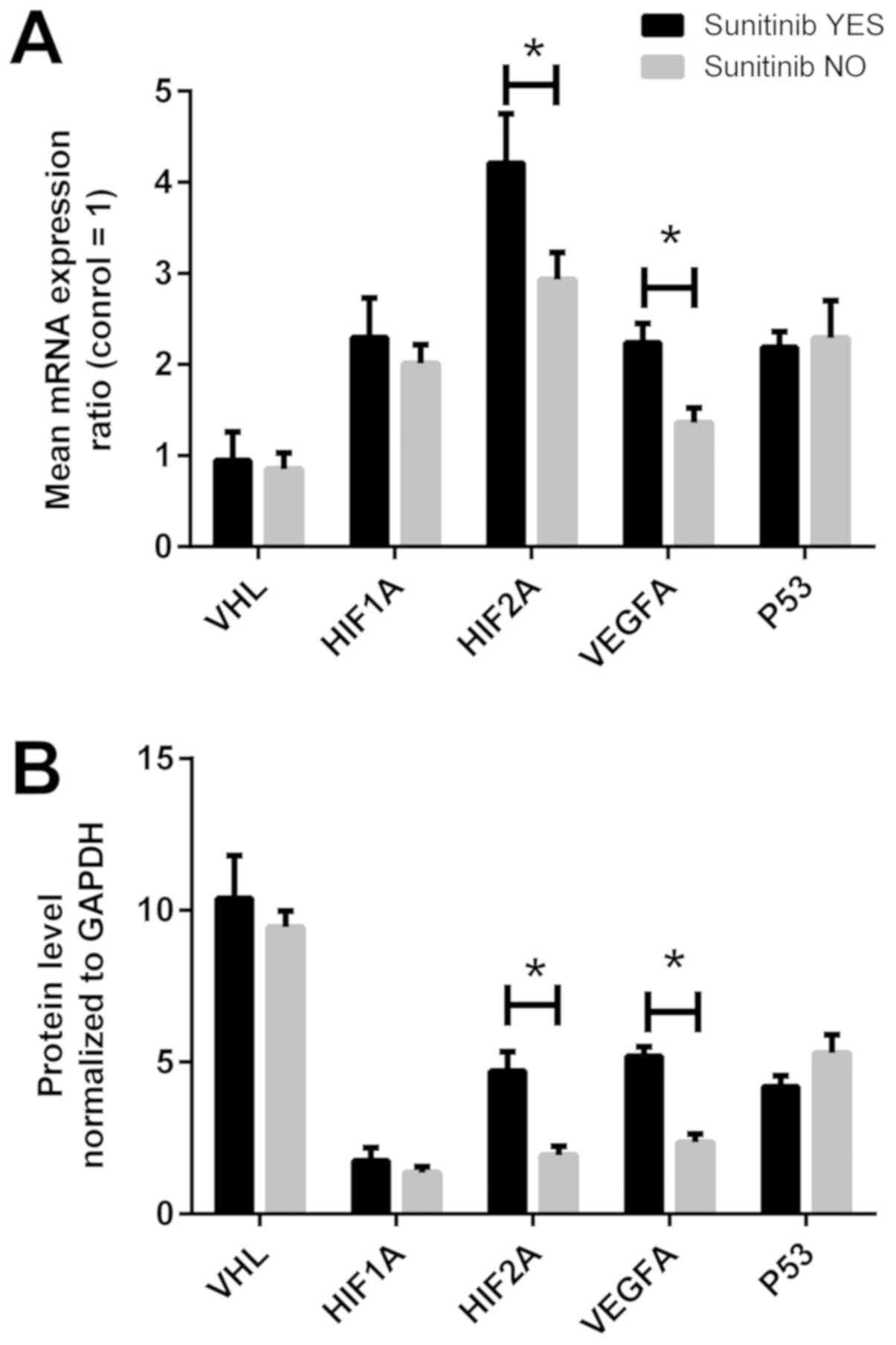
Gene expression in the tissues of
patients treated with sunitinib
The molecular data of the VHL, HIF1A, HIF2A,
VEGFA and P53 genes at the mRNA and protein level in
ccRCC tissues were analyzed according to adjuvant sunitinib
treatment. As shown in Fig. 7A,
the HIF2A and VEGFA mRNA levels were ~1.3- and
1.5-fold higher in the tissues of sunitinib-treated patients
compared with those in patients not receiving adjuvant treatment
(P<0.05). However, when the samples were divided according to
the median values of the gene expression in the controls, no
statistically significant differences were observed (Table III, bottom section). Unlike the
results observed at the mRNA level, parallel statistical
associations were observed at the protein level; the HIF2A protein
level was ~2-fold higher in 10 of the 11 sunitinib-treated patients
(P<0.05), whereas the VEGFA protein level was ~2.5-fold higher
in the cancer tissues of sunitinib-treated patients compared with
patients not receiving adjuvant treatment (Table IV, bottom section; Fig. 7B).
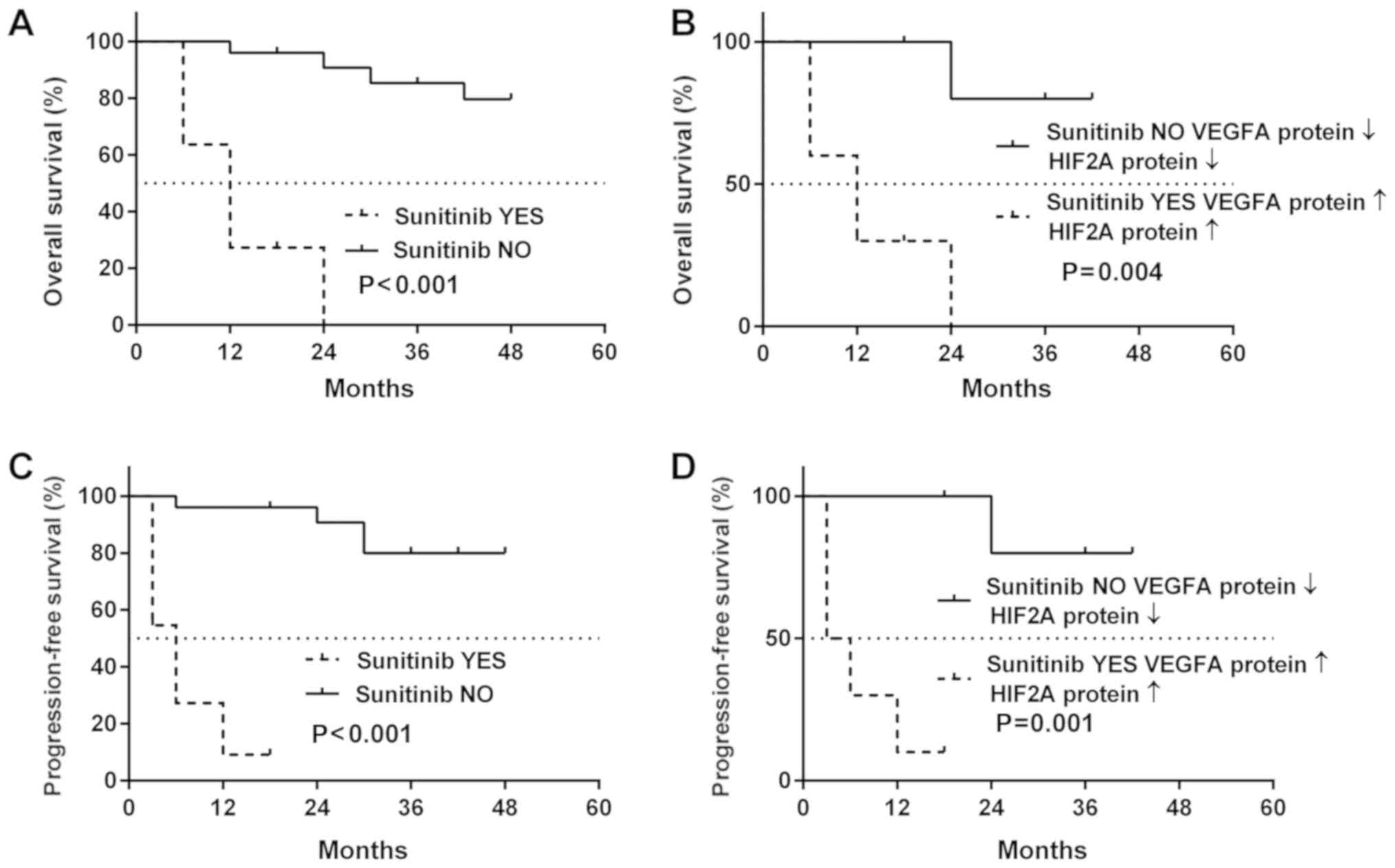
Effect of sunitinib treatment on patient
outcome
As shown in Fig.
8A, post-operative treatment with sunitinib had no positive
effect on the survival of patients with ccRCC, with a median
survival of sunitinib-treated patients of 12 months. On the
contrary, 80% of the patients with ccRCC who did not receive
sunitinib had a positive outcome (Fig.
8A, solid line). Comparable results were obtained for cancer
recurrence: The median PFS for sunitinib-treated patients was 12
months, whereas the cancer-related deterioration of the health
status was not observed in >60% patients not receiving sunitinib
treatment (Fig. 8B). The molecular
data presented in Fig. 7 and
Tables III and IV revealed the possible association
between patient outcome, sunitinib treatment and the cancer tissue
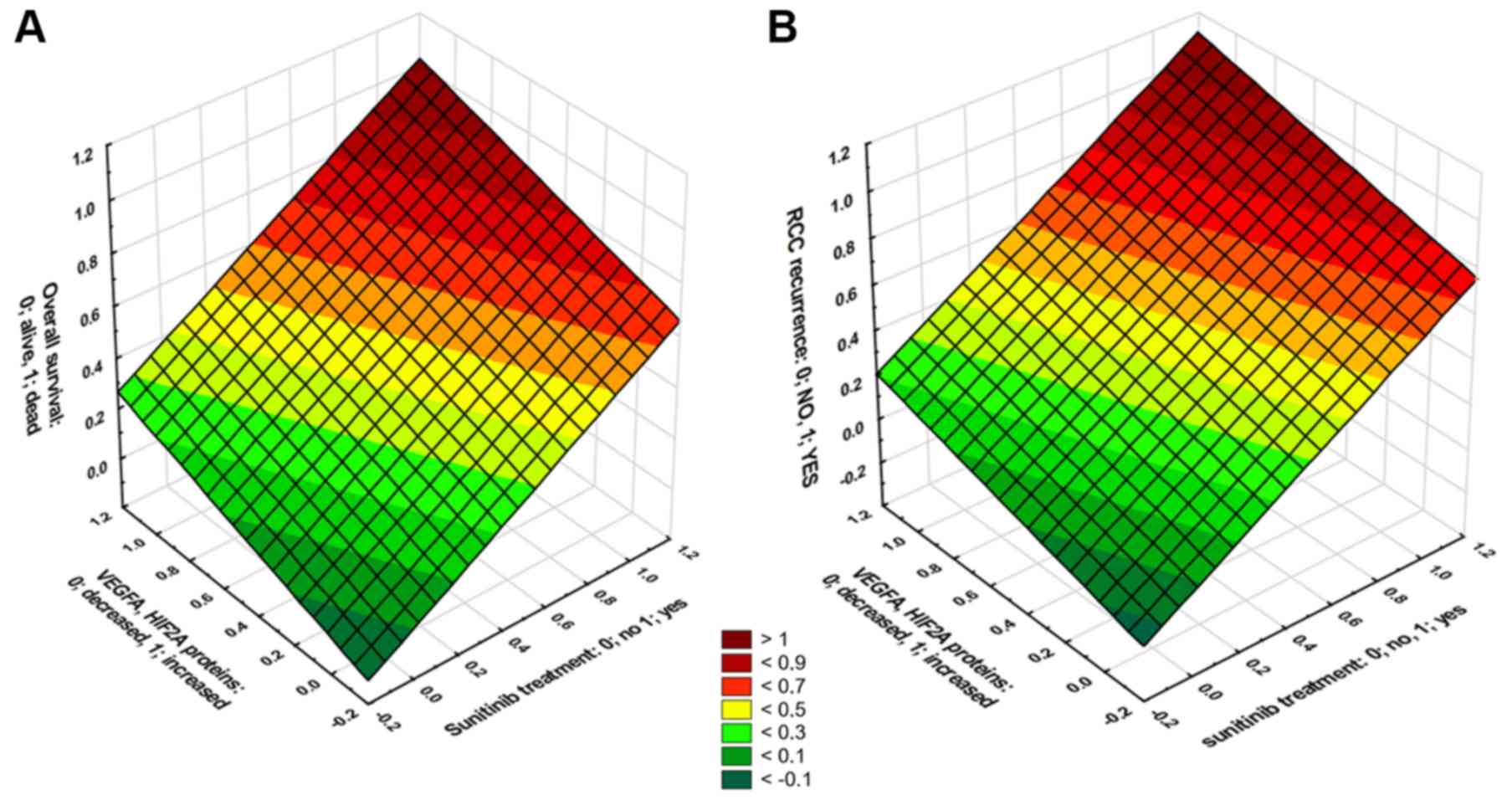
levels of the VEGFA and HIF2A proteins. In the sunitinib-treated
ccRCC patients who had high HIF2A and VEGFA protein levels in their
tumor tissues, the median OS was not altered (Fig. 8B); however, the PFS decreased to 3
months (Fig. 8D). The
multivariable regression revealed moderate-to-strong associations
between sunitinib treatment, increased levels of both VEGFA and
HIF2A proteins, and either death (b=0.57, P<0.001, Fig. 9A) or disease progression (b=0.69;
P<0.001, Fig. 9B).
Discussion
RCC is the 6th and 10th most common malignancy in
males and females, respectively, in the USA, according to the
estimated number of new cases in 2017 (1). More than 300,000 patients are
diagnosed annually, of whom ~143,000 succumb to the disease each
year, thus rendering RCC the 16th most common cause of
cancer-related mortality worldwide (1,22).
ccRCC is the most common histological subtype of RCC. At the
molecular level, deregulation of the expression of the
pVHL-HIFs-VEGFA pathway proteins is frequently observed in ccRCC
(42,43).
The aim of the present study was to simultaneously
assess the status of the VHL-HIF1/2A-VEGFA pathway in a group of
ccRCC patients from one clinical center. Similar to our previous
studies on ccRCC (13,33,41)
qPCR was selected to access gene expression at the mRNA level. At
the protein level, a semi-quantitative technique (western blot
analysis) was applied for all studied ccRCC samples and IHC
staining for the material available from 10 patients. The
statistical compatibility of the western blot analysis and IHC
methods has been noted by authors who analyzed human epidermal
growth factor receptor 2/neu (44)
or periostin (45) proteins in
human breast cancer tissues (44)
or cells (45), claudin-2 and −5
proteins in gastric carcinoma (46), or for the validation of antibody
clones for immunochemistry in non-small-cell lung cancer (47).
There is a common consensus that the majority of
ccRCCs are initiated by the somatic inactivation of the VHL tumor
suppressor gene (48,49). pVHL is a multifunctional factor
that acts as an adaptor protein recruiting different effector
proteins to different cellular targets, thereby regulating various
cellular processes (22). It has
been demonstrated that pVHL may trigger various processes, such as
glucose uptake and metabolism (50), angiogenesis (50,51),
the suppression of epithelial-to-mesenchymal transition (52), cell proliferation, survival or
apoptosis (50,51), or activation of the P53 pathway
(22,53). Since all the mentioned processes
may be involved in the mechanisms underlying carcinogenesis, the
expression of the VHL gene in ccRCC has been extensively
studied. Although the contribution of the mutated VHL gene
to ccRCC initiated in the course of Von Hippel-Lindau disease has
been described (22,54), further studies of this gene in
sporadic ccRCC are warranted. We have previously identified
molecular prognostic markers of ccRCC that were involved in the
Hippo pathway (13,41,55).
Although in the present study, a decreased VHL mRNA level
was observed in tumor samples, this was not found to be associated
with the clinicopathological data of the patients or with their
outcome or sunitinib treatment. Similar to our results, Xiao-Fen
et al reported the underexpression of VHL mRNA in
tumor samples from 75 RCC patients (the number of ccRCC cases was
not specified) in comparison to normal kidney tissues (23). However, in contrast to our
findings, Xiao-Fen et al observed that this decreased
expression was associated with cancer TNM progression and tumor
size, although the lack of data on the histological subtypes of RCC
precludes a direct comparison between their results (23) and ours. Högner et al
(56), using qPCR, observed a
strong underexpression of VHL mRNA in 80.6% of the studied
69 ccRCC patients; however, they investigated the associations
between clinicopathological and molecular data. On the contrary to
the listed studies, deep-genome and mRNA studies on 48 ccRCC cases
conducted by Girgis et al did not reveal any changes in
VHL mRNA levels, despite the observed hypermethylation of
the VHL genomic locus (57). Other studies on VHL gene
expression were mainly based on DNA analysis focusing on the
hypermethylation status of the VHL gene promoter locus or
mutational analysis of the VHL exons (5,22,23,57-59).
Since previous investigations of the tumor tissues from patients
with ccRCC treated with sunitinib focused on VHL mutations
(60-62), to the best of our knowledge, the
present study is the first to investigate the expression of
VHL at both the mRNA and protein level in patients with
ccRCC treated with first-line sunitinib. Finally, our findings of
weaker immunoreactivity of pVHL demonstrated by IHC staining are
only mentioned in a few studies (63,64)
in which the authors reported either markedly decreased pVHL
immunoreactivity in early ccRCC (63), or a reduction trend of pVHL
expression in association with ccRCC progression (64).
The most well-known function of the VHL protein is
the regulation of cellular response to oxygen depletion via
maintenance of the cellular lifespan of HIF transcription factors.
There is a rapid proteasomal degradation of HIFs during normoxia;
however, no association between pVHL and HIFs has been established
during tumor-associated hypoxia (65). In this study, although high levels
of HIF1A and HIF2A expression were observed at both
the mRNA and protein level in ccRCC samples, there was no observed
association between HIF1A/2A and VHL protein levels. Therefore, it
may be hypothesized that the control of pVHL upon HIF degradation
was incomplete in the analyzed ccRCC samples. This is supported by
Nyham et al (66), who
observed the overexpression of HIF1A and HIF2A at the protein
level, as assessed by western blot analysis in 88 and 100% of 17
ccRCC patients, respectively, regardless of the genetic mutations
of VHL that were detected in 43.5% of the tumors (66); therefore, they proposed that
control of the degradation of HIFs by an intact pVHL is not
effective during ccRCC progression (66). The results of this study,
demonstrating highly increased levels of HIF proteins in samples
with intact levels of pVHL, may support this hypothesis, although
our focus was not VHL genetic alterations.
The increased levels of HIF1A and
HIF2A at both the mRNA and protein level in ccRCC samples
observed in the present study confirm previous findings (24,26-28,67).
For example, Turner et al investigated a group of 34 ccRCC
patients and observed overexpression of HIFs at the mRNA and
protein level; the HIF1A mRNA level was ~2-fold and the
HIF2A mRNA was 5-fold higher in tumor samples compared with
normal kidneys (28). Using
western blot analysis, the authors revealed the upregulation of
HIF1A and HIF2A protein levels in 77 and 79% of the tumor cases,
respectively (28). The level of
HIF proteins were positively assocaited with the increased number
of blood vessels in tumor tissue, suggestive of neoangiogenesis
(28). Similar to our findings, a
positive correlation between HIFs and VEGFA expression at
both the mRNA and protein level was also reported (28).
The finding of a shorter OS in ccRCC patients
exhibiting increased levels of HIF1A, HIF2A and VEGFA in the
present study was consistent with the results reported by Ebru
et al (27). Using IHC in a
group of 72 ccRCC patients, they observed a strong association
between shorter OS and high expression of HIF1A, HIF2 and VEGFA, as
well as Ki-67 protein and microvessel density (27). Another study has also reported a
high immunoreactivity of HIF1A, HIF2 and VEGFA in the nuclei or/and
cytoplasm of ccRCC cells (68).
High levels of HIF2A, but not HIF1A, were found to
be associated with the risk of death and cancer recurrence,
independent of sunitinib treatment. The stronger impact of HIF2A,
rather than HIF1A, on ccRCC progression has also been previously
reported (65,69-75).
Maroto et al observed a similar pattern of shorter PFS in 10
sunitinib-treated patients, but only for both HIF2A- and
c-Myc-positive cases (at the protein level) (76). Philips and Atkins also reported
that HIF2A is more relevant in ccRCC development and progression
compared with HIF1A, assuming that HIF2A is the dominant oncogene
in RCC (75). Shen et al
further suggested the oncogenic role of HIF2A and the
tumor-suppressive role of HIF1A in the development of
pVHL-defective RCCs (69). The
dual role of HIF1A was also reported by Lindgren et al
(70); favorable prognosis of 92
RCC patients (including 66 ccRCC cases) was observed in association
with increased HIF1A protein levels, as determined by western blot
analysis (70). Furthermore,
studies on pre-neoplastic kidney lesions of patients with VHL
disease also support the key role of HIF2A in the transformation of
dysplastic cells, as the HIF2A expression was increased while that
of HIF1A was decreased, as assessed by IHC (71,72).
The association between high levels of HIF2A and ccRCC progression
has also been observed in cell lines representing various stages of
RCC progression (73,74) or in mouse xenograft models
(77). In a clinical study, Kamai
et al analyzed tumor samples obtained from 129 patients with
ccRCC, and found that increased expression of the HIF2A protein
(measured by western blot analysis) was associated with worse
clinical status, local and distant metastasis, and shorter OS
(78). The possibility of using
HIF2A inhibitors to block ccRCC progression and recurrence was
recently suggested (75,79,80).
Therefore, novel HIF2A antagonists were developed, such as PT2399
(80) and PT2385 (81), which block the PAS-B domain of the
HIF2A subunit. Furthermore, new-generation dual mammalian target of
rapamycin inhibitors (GDC-0980 and BKM120), which block both TORC1
and TORC2 activity, including HIF2A expression, have been
introduced to phase I trials of advanced RCC (75,82).
Whereas these data confirm the importance of HIF2A inhibition in
modern ccRCC chemotherapy, the results of the present study
strongly suggest that the measurement of HIF2A protein levels may
serve as independent prognostic marker in ccRCC, also in
sunitinib-treated patients (76).
However, it must be noted that Beuselinck et al (62) observed high levels of HIF2A
mRNA in sunitinib-sensitive ccRCCs. The differences in methodology
may explain the opposite observations, since Beuselinck et
al used β-actin as a reference gene (62), which may have affected the results
(33), whereas we used
β-glucuronidase, which was carefully normalized for ccRCC samples
in our previous study (33).
It has been widely confirmed that HIF1A, as well as
HIF2A, trigger transcription of the VEGFA gene to VEGFA, and
its receptors (VEGFRs) play pivotal roles in vasculogenesis and
angiogenesis under physiological conditions, as well as in cancer,
including ccRCC (10,24,50,54,65,74,80,83).
The present study also revealed that the expression of
VEGFA, either at the mRNA or protein level, was strongly
associated with ccRCC progression, and that patients with higher
VEGFA expression exhibited a poorer outcome and earlier recurrence
of cancer. Additionally, Cox analysis revealed that ccRCC patients
with high levels of VEGFA mRNA in tumor tissues had an increased
risk of cancer progression, while an increased risk of death was
associated with high levels of the VEGFA protein in tumor samples.
Other studies have also reported the important effect of VEGFA
expression on the progression of ccRCC (27,83-85),
while Wang et al also revealed that high levels of
VEGFA mRNA may serve as a prognostic marker in ccRCC
(83).
The IHC detection of VEGFA has been widely used for
the assessment of its expression in ccRCC tissues (5,27,83-85).
Our findings demonstrated an increased cytoplasmic presence of
VEGFA in cancer cells, consistent with previous reports (5,27,83-85).
For example, Veselaj et al (84) and Dagher et al (5) observed that increased VEGFA
immunoreactivity in tumor samples from ccRCC patients was
associated with cancer progression and disease-free survival
(5,84), as well as with an increased risk of
death (84).
Finally, in this study, we investigated whether the
P53 gene expression pattern was associated with either ccRCC
progression or the expression of VHL-HIFs-VEGFA axis components,
since such an approach has not yet been reported, at least to the
best of our knowledge. Although higher P53 mRNA and protein levels
were detected in tumor samples, there was no obvious association
between P53 expression and clinicopathological or molecular data of
the other analyzed genes and proteins. Our finding of the strong
immunoreactivity of the P53 protein in tumor cells is consistent
with other reports on P53 in ccRCC (27). Shi et al observed a higher
expression of P53 at the mRNA and protein level, as assessed by
qPCR, western blot analysis and IHC, in tumor samples, but without
any association with clinical variables (86). Ebru et al did not identify a
connection between P53 protein expression, as determined by IHC,
and the outcome of 62 ccRCC patients (27). Furthermore, the authors did not
observe any associations between P53 and HIF1A, HIF2A or VEGFA
protein immunoreactivity in the same samples (27). As regards first-line sunitinib
treatment, Zhu et al observed that high levels of P53, as
determined by IHC, were associated with favorable OS; however, they
did not include sunitinib-resistant cases in their study (87). Based on the results of the present
and previous studies, it may be suggested that P53 gene
expression at the mRNA or protein level is not associated with
ccRCC progression. Moreover, the present study is, to the best of
our knowledge, the first to analyze P53 expression in
sunitinib-resistant ccRCC.
Sunitinib is selective inhibitor of multiple
receptor tyrosine kinases, including VEGFRs (1-3),
PDGFRs and c-kit, and it was approved by FDA in 2006 (2009 in
Poland) for the treatment of metastatic RCC (17). It has been demonstrated that
patients treated with sunitinib had longer median PFS compared with
those treated with interferon (11.0 vs. 5.1 months, respectively)
(17). However, later studies did
not yield such optimistic values, since most of the advanced RCCs
treated with sunitinib developed intrinsic drug resistance
(18-20). The results of this study
demonstrated that patients who were treated with sunitinib
post-operatively had significantly shorter OS and PFS compared with
ccRCC patients who did not receive such treatment. Since there was
no difference in the pre-operative clinical status between
sunitinib-treated and non-treated patient groups, it may be
hypothesized that the poor outcome may be due to the drug
treatment. In addition, Busch et al analyzed the outcome of
35 metastatic RCC cases (29 ccRCCs) who received first-line
sunitinib treatment, and reported a poor prognosis due to intrinsic
drug resistance (88). Lim et
al observed a shorter OS and PFS in 33 out of 134 metastatic
ccRCC patients treated with first-line sunitinib (21). A recent meta-analysis of adjuvant
therapy in metastatic RCC did not reveal an increase of OS or DFS
in association with sunitinib treatment; however, such therapy was
associated with severe adverse events (20). Duran et al stated that,
eventually, all patients with advanced RCC will become resistant to
first-line TKIs, suggesting that second-line treatment should be
introduced (43). The molecular
mechanism underlying intrinsic resistance to first-line sunitinib
treatment remains elusive (21);
however, molecular studies on sunitinib resistance in RCC are
currently in progress. Giuliano et al observed
overexpression of the ABCB1 gene, which participates in the
accumulation of the drug in autolysosomes of 786-O and RCC10 cell
lines, favoring cellular efflux of sunitinib (89). Butz et al observed
downregulation of miR-1 and miR-663a targeting FRAS1 and MDGA1 gene
expression in a sunitinib-resistant ccRCC xenograft model (90). The results of the present study,
showing higher HIF2A and VEGFA mRNA and protein expression, with no
underexpression of pVHL, in sunitinib-treated ccRCC patients
compared with those receiving no adjuvant treatment, must be
verified by further studies, since only Beuselinck et al
observed the opposite pattern of high HIF2A and VEGFA levels in
sunitinib-sensitive tumor samples (62). Notably, recent results on
sunitinib-resistant RCC cell lines (786-0, Caki-1, Caki-2 and
SN12K1) reported by Kamli et al partially support our
findings, as Caki-2 and SN12K1 cells exhibited overexpression of
VEGFA (91).
In conclusion, based on the observation that
sunitinib-treated ccRCC patients with high levels of VEGFA and
HIF2A protein expression and unchanged levels of pVHL in tumor
samples are characterized by higher risk of death and cancer
recurrence, we recommend that cRCC patients with this molecular
profile are not administered sunitinib as first-line treatment.
However, since only 11 of the 36 analyzed ccRCC patients were
treated with sunitinib, this conclusion is merely a hypothesis, and
large-scale replication studies in independent subject panels are
required to validate the results. Moreover, the assessment of VHL,
VEGFA and HIF2A protein levels in ccRCC tissues in the future may
prove to be helpful in selecting an effective drug treatment.
Acknowledgments
Not applicable.
Funding
The present study was funded by the ST-12 and
ST-02-0117/07 internal funds of the Medical University of Gdańsk,
Poland.
Availability of data and materials
The datasets used and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
PMW performed the statistical analyses and drafted
the manuscript. PMW and JK conceived and designed the study. JK
collected tissue samples and patient data and revised the
manuscript. AKC, AW, MS, AR and AL performed molecular analyses and
statistical tests. ZK substantially contributed in the
interpretation of the results, as well as revised the manuscript
and provided funds, and MM collected tissue samples and provided
funds. All the authors have read and approved the final version of
this manuscript.
Ethics approval and consent to
participate
The study was approved by the Independent Bioethics
Commission at the Medical University of Gdańsk (permission no.
NKEBN/4/2011) and written consent was obtained prior to surgery
from all patients. All experimental procedures were performed
according to the regulations and internal biosafety and bioethics
guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests to disclose.
Authors' information
The ORCID numbers for the authors on this study are
as follows: PMW: 0000-0002-4310-1616; JK, 0000-0001-5010-9336; AKC:
0000-0002-2942-6270; AW, 0000-0003-1792-8975; MS,
0000-0001-7176-0937; AR, 0000-0001-9459-2489; AL,
0000-0003-4405-8266; ZK, 0000-0002-9801-8166; MM,
0000-0002-7799-685X.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schödel J, Grampp S, Maher ER, Moch H,
Ratcliffe PJ, Russo P and Mole DR: Hypoxia, hypoxia-inducible
transcription factors, and renal cancer. Eur Urol. 69:646–657.
2016. View Article : Google Scholar :
|
|
3
|
Weinstock M and McDermott D: Targeting
PD-1/PD-L1 in the treatment of metastatic renal cell carcinoma.
Ther Adv Urol. 7:365–377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu SS, Quinn DI and Dorff TB: Clinical use
of cabozantinib in the treatment of advanced kidney cancer:
Efficacy, safety, and patient selection. OncoTargets Ther.
9:5825–5837. 2016. View Article : Google Scholar
|
|
5
|
Dagher J, Kammerer-Jacquet SF, Dugay F,
Beaumont M, Lespagnol A, Cornevin L, Verhoest G, Bensalah K,
Rioux-Leclercq N and Belaud-Rotureau MA: Clear cell renal cell
carcinoma: A comparative study of histological and chromosomal
characteristics between primary tumors and their corresponding
metastases. Virchows Arch. 471:107–115. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen YL, Ge GJ, Qi C, Wang H, Wang HL, Li
LY, Li GH and Xia LQ: A five-gene signature may predict sunitinib
sensitivity and serve as prognostic biomarkers for renal cell
carcinoma. J Cell Physiol. 233:6649–6660. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang JY, Peng SH, Li T, Ning XH, Liu SJ,
Hong BA, Liu JY, Wu PJ, Zhou BW, Zhou JC, et al: Risk factors for
survival in patients with von Hippel-Lindau disease. J Med Genet.
55:322–328. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hasumi H and Yao M: Hereditary kidney
cancer syndromes: Genetic disorders driven by alterations in
metabolism and epigenome regulation. Cancer Sci. 109:581–586. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kondo K, Klco J, Nakamura E, Lechpammer M
and Kaelin WG Jr: Inhibition of HIF is necessary for tumor
suppression by the von Hippel-Lindau protein. Cancer Cell.
1:237–246. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Singh D, Arora R, Kaur P, Singh B, Mannan
R and Arora S: Overexpression of hypoxia-inducible factor and
metabolic pathways: Possible targets of cancer. Cell Biosci.
7:622017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ravi R, Mookerjee B, Bhujwalla ZM, Sutter
CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL and Bedi A:
Regulation of tumor angiogenesis by p53-induced degradation of
hypoxia-inducible factor 1alpha. Genes Dev. 14:34–44.
2000.PubMed/NCBI
|
|
12
|
Joshi S, Singh AR and Durden DL: MDM2
regulates hypoxic hypoxia-inducible factor 1α stability in an E3
ligase, proteasome, and PTEN-phosphatidylinositol
3-kinase-AKT-dependent manner. J Biol Chem. 289:22785–22797. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rybarczyk A, Klacz J, Wronska A,
Matuszewski M, Kmiec Z and Wierzbicki PM: Overexpression of the
YAP1 oncogene in clear cell renal cell carcinoma is associated with
poor outcome. Oncol Rep. 38:427–439. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shenoy N and Pagliaro L: Sequential
pathogenesis of metastatic VHL mutant clear cell renal cell
carcinoma: Putting it together with a translational perspective.
Ann Oncol. 27:1685–1695. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Elvidge GP, Glenny L, Appelhoff RJ,
Ratcliffe PJ, Ragoussis J and Gleadle JM: Concordant regulation of
gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase
inhibition: The role of HIF-1alpha, HIF-2alpha, and other pathways.
J Biol Chem. 281:15215–15226. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hori Y, Ito K, Hamamichi S, Ozawa Y,
Matsui J, Umeda IO and Fujii H: Functional characterization of
VEGF- and FGF-induced tumor blood vessel models in human cancer
xenografts. Anticancer Res. 37:6629–6638. 2017.PubMed/NCBI
|
|
17
|
Hutson TE and Figlin RA: Evolving role of
novel targeted agents in renal cell carcinoma. Oncology (Williston
Park). 21:1175–1180; discussion 1184-1187, 1190. 2007.
|
|
18
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shibasaki N, Yamasaki T, Kanno T, Arakaki
R, Sakamoto H, Utsunomiya N, Inoue T, Tsuruyama T, Nakamura E,
Ogawa O, et al: Role of IL13RA2 in sunitinib resistance in clear
cell renal cell carcinoma. PLoS One. 10:e01309802015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun M, Marconi L, Eisen T, Escudier B,
Giles RH, Haas NB, Harshman LC, Quinn DI, Larkin J, Pal SK, et al:
Adjuvant vascular endothelial growth factor-targeted therapy in
renal cell carcinoma: A systematic review and pooled analysis. Eur
Urol. 74:611–620. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lim SH, Hwang IG, Ji JH, Oh SY, Yi JH, Lim
DH, Lim HY, Lee SJ and Park SH: Intrinsic resistance to sunitinib
in patients with metastatic renal cell carcinoma. Asia Pac J Clin
Oncol. 13:61–67. 2017. View Article : Google Scholar
|
|
22
|
Mehdi A and Riazalhosseini Y: Epigenome
aberrations: Emerging driving factors of the clear cell renal cell
carcinoma. Int J Mol Sci. 18:182017. View Article : Google Scholar
|
|
23
|
Xiao-Fen W, Ting C, Jie L, Deng-Yang M,
Qing-Feng Z and Xin L: Correlation analysis of VHL and Jade-1 gene
expression in human renal cell carcinoma. Open Med (Wars).
11:226–230. 2016.
|
|
24
|
Wan L, Huang J, Chen J, Wang R, Dong C, Lu
S and Wu X: Expression and significance of FOXP1, HIF-1a and VEGF
in renal clear cell carcinoma. J BUON. 20:188–195. 2015.PubMed/NCBI
|
|
25
|
Gstalder C, Ader I and Cuvillier O: FTY720
(fingolimod) inhibits HIF1 and HIF2 signaling, promotes vascular
remodeling, and chemosensitizes in renal cell carcinoma animal
model. Mol Cancer Ther. 15:2465–2474. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Du W, Zhang L, Brett-Morris A, Aguila B,
Kerner J, Hoppel CL, Puchowicz M, Serra D, Herrero L, Rini BI, et
al: HIF drives lipid deposition and cancer in ccRCC via repression
of fatty acid metabolism. Nat Commun. 8:17692017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ebru T, Fulya OP, Hakan A, Vuslat YC,
Necdet S, Nuray C and Filiz O: Analysis of various potential
prognostic markers and survival data in clear cell renal cell
carcinoma. Int Braz J Urol. 43:440–454. 2017. View Article : Google Scholar :
|
|
28
|
Turner KJ, Moore JW, Jones A, Taylor CF,
Cuthbert-Heavens D, Han C, Leek RD, Gatter KC, Maxwell PH,
Ratcliffe PJ, et al: Expression of hypoxia-inducible factors in
human renal cancer: Relationship to angiogenesis and to the von
Hippel-Lindau gene mutation. Cancer Res. 62:2957–2961.
2002.PubMed/NCBI
|
|
29
|
Qin C, Chen J, Li J, Ju X, Zhang S, Cao Q,
Han Z, Li P, Shao P, Wang M, et al: Variants in
angiogenesis-related genes and the risk of clear cell renal cell
carcinoma. Mutagenesis. 29:419–425. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Godlewski J, Krazinski BE, Kowalczyk AE,
Kiewisz J, Kiezun J, Kwiatkowski P, Sliwińska-Jewsiewicka A,
Wierzbicki PW and Kmieć Z: Expression and prognostic significance
of EP300, TP53 and BAX in clear cell renal cell carcinoma.
Anticancer Res. 37:2927–2937. 2017.PubMed/NCBI
|
|
31
|
Yuan L, Chen L, Qian K, Qian G, Wu CL,
Wang X and Xiao Y: Co-expression network analysis identified six
hub genes in association with progression and prognosis in human
clear cell renal cell carcinoma (ccRCC). Genom Data. 14:132–140.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Klacz J, Wierzbicki PM, Wronska A, et al:
Decreased expression of RASSF1A tumor suppressor gene is associated
with worse prognosis in clear cell renal cell carcinoma. Manuskrypt
wysłany do European Urology. 21.07.2015. 2015.
|
|
33
|
Klacz J, Wierzbicki PM, Wronska A,
Rybarczyk A, Stanislawowski M, Slebioda T, Olejniczak A,
Matuszewski M and Kmiec Z: Decreased expression of RASSF1A tumor
suppressor gene is associated with worse prognosis in clear cell
renal cell carcinoma. Int J Oncol. 48:55–66. 2016. View Article : Google Scholar :
|
|
34
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang LE, Arany Z, Livingston DM and Bunn
HF: Activation of hypoxia-inducible transcription factor depends
primarily upon redox-sensitive stabilization of its alpha subunit.
J Biol Chem. 271:32253–32259. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Avădănei ER, Wierzbicki PM, Giuşcă SE,
Grigoraş A, Amălinei C and Căruntu ID: Macrophage profile in
primary versus secondary liver tumors. Folia Histochem Cytobiol.
52:112–123. 2014. View Article : Google Scholar
|
|
37
|
Escudier B and Kataja V; ESMO Guidelines
Working Group: Renal cell carcinoma: ESMO Clinical Practice
Guidelines for diagnosis, treatment and follow-up. Ann Oncol.
21(Suppl 5): v137–v139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sobin LH, Gospodarowicz MK and Wittekind
C; International Union Against Cancer: TNM Classification of
Malignant Tumours Chichester. West Sussex, Hoboken, NJ:
Wiley-Blackwell; 2010
|
|
39
|
Delahunt B, Sika-Paotonu D, Bethwaite PB,
William Jordan T, Magi-Galluzzi C, Zhou M, Samaratunga H and
Srigley JR: Grading of clear cell renal cell carcinoma should be
based on nucleolar prominence. Am J Surg Pathol. 35:1134–1139.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dzik C, Reis ST, Viana NI, Brito G,
Paloppi I, Nahas W, Srougi M and Leite KRM: Gene expression profile
of renal cell carcinomas after neoadjuvant treatment with
sunitinib: New pathways revealed. Int J Biol Markers. 32:e210–e217.
2017. View Article : Google Scholar
|
|
41
|
Klacz J, Wierzbicki PM, Wronska A,
Rybarczyk A, Stanislawowski M, Slebioda T, Olejniczak A,
Matuszewski M and Kmiec Z: Decreased expression of RASSF1A tumor
suppressor gene is associated with worse prognosis in clear cell
renal cell carcinoma. Int J Oncol. 48:55–66. 2016. View Article : Google Scholar :
|
|
42
|
Akhtar M, Al-Bozom IA and Al Hussain T:
Molecular and metabolic basis of clear cell carcinoma of the
kidney. Adv Anat Pathol. 25:189–196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Duran I, Lambea J, Maroto P,
González-Larriba JL, Flores L, Granados-Principal S, Graupera M,
Sáez B, Vivancos A and Casanovas O: Resistance to targeted
therapies in renal cancer: The importance of changing the mechanism
of action. Target Oncol. 12:19–35. 2017. View Article : Google Scholar
|
|
44
|
Molina R, Ciocca DR, Tandon AK, Allred DC,
Clark GM, Chamness GC, Gullick WJ and McGuire WL: Expression of
HER-2/neu oncoprotein in human breast cancer: A comparison of
immunohistochemical and western blot techniques. Anticancer Res.
12B:B1965–B1971. 1992.
|
|
45
|
Ratajczak-Wielgomas K, Grzegrzolka J,
Piotrowska A, Matkowski R, Wojnar A, Rys J, Ugorski M and Dziegiel
P: Expression of periostin in breast cancer cells. Int J Oncol.
51:1300–1310. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang L, Sun X and Meng X: Differences in
the expression profiles of claudin proteins in human gastric
carcinoma compared with non-neoplastic mucosa. Mol Med Rep.
18:1271–1278. 2018.PubMed/NCBI
|
|
47
|
Parra ER, Villalobos P, Mino B and
Rodriguez-Canales J: Comparison of different antibody clones for
immunohistochemistry detection of programmed cell death ligand 1
(PD-L1) on non-small cell lung carcinoma. Appl Immunohistochem Mol
Morphol. 26:83–93. 2018.
|
|
48
|
Gao W, Li W, Xiao T, Liu XS and Kaelin WG
Jr: Inactivation of the PBRM1 tumor suppressor gene amplifies the
HIF-response in VHL−/− clear cell renal carcinoma. Proc
Natl Acad Sci USA. 114:1027–1032. 2017. View Article : Google Scholar
|
|
49
|
Tarade D and Ohh M: The HIF and other
quandaries in VHL disease. Oncogene. 37:139–147. 2018. View Article : Google Scholar
|
|
50
|
Morris MR and Latif F: The epigenetic
landscape of renal cancer. Nat Rev Nephrol. 13:47–60. 2017.
View Article : Google Scholar
|
|
51
|
Gossage L, Eisen T and Maher ER: VHL, the
story of a tumour suppressor gene. Nat Rev Cancer. 15:55–64. 2015.
View Article : Google Scholar
|
|
52
|
Pantuck AJ, An J, Liu H and Retting MB:
NF-kappaB-dependent plasticity of the epithelial to mesenchymal
transition induced by von Hippel-Lindau inactivation in renal cell
carcinomas. Cancer Res. 70:752–761. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Roe JS, Kim H, Lee SM, Kim ST, Cho EJ and
Youn HD: p53 stabilization and transactivation by a von
Hippel-Lindau protein. Mol Cell. 22:395–405. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang T, Niu X, Liao L, Cho EA and Yang H:
The contributions of HIF-target genes to tumor growth in RCC. PLoS
One. 8:e805442013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Godlewski J, Kiezun J, Krazinski BE,
Kozielec Z, Wierzbicki PM and Kmiec Z: The immunoexpression of YAP1
and LATS1 proteins in clear cell renal cell carcinoma: Impact on
patients' survival. BioMed Res Int. 2018:26536232018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Högner A, Krause H, Jandrig B, Kasim M,
Fuller TF, Schostak M, Erbersdobler A, Patzak A and Kilic E: PBRM1
and VHL expression correlate in human clear cell renal cell
carcinoma with differential association with patient's overall
survival. Urol Oncol. 36:94 e91–94 e14. 2018. View Article : Google Scholar
|
|
57
|
Girgis AH, Iakovlev VV, Beheshti B, Bayani
J, Squire JA, Bui A, Mankaruos M, Youssef Y, Khalil B, Khella H, et
al: Multilevel whole-genome analysis reveals candidate biomarkers
in clear cell renal cell carcinoma. Cancer Res. 72:5273–5284. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Patard JJ, Rioux-Leclercq N, Masson D,
Zerrouki S, Jouan F, Collet N, Dubourg C, Lobel B, Denis M and
Fergelot P: Absence of VHL gene alteration and high VEGF expression
are associated with tumour aggressiveness and poor survival of
renal-cell carcinoma. Br J Cancer. 101:1417–1424. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Beroukhim R, Brunet JP, Di Napoli A, Mertz
KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, et
al: Patterns of gene expression and copy-number alterations in
von-hippel lindau disease-associated and sporadic clear cell
carcinoma of the kidney. Cancer Res. 69:4674–4681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Song Y, Huang J, Shan L and Zhang HT:
Analyses of potential predictive markers and response to targeted
therapy in patients with advanced clear-cell renal cell carcinoma.
Chin Med J (Engl). 128:2026–2033. 2015. View Article : Google Scholar
|
|
61
|
Dornbusch J, Zacharis A, Meinhardt M,
Erdmann K, Wolff I, Froehner M, Wirth MP, Zastrow S and Fuessel S:
Analyses of potential predictive markers and survival data for a
response to sunitinib in patients with metastatic renal cell
carcinoma. PLoS One. 8:e763862013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Beuselinck B, Verbiest A, Couchy G, Job S,
de Reynies A, Meiller C, Albersen M, Verkarre V, Lerut E, Méjean A,
et al: Pro-angiogenic gene expression is associated with better
outcome on sunitinib in metastatic clear-cell renal cell carcinoma.
Acta Oncol. 57:498–508. 2018. View Article : Google Scholar
|
|
63
|
Sur S, Maurya AK, Roy A, Sharp TV, Pal DK
and Panda CK: Over expression of HIF1α is associated with
inactivation of both LimD1 and VHL in renal cell carcinoma:
Clinical importance. Pathol Res Pract. 213:1477–1481. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gervais ML, Henry PC, Saravanan A, Burry
TN, Gallie BL, Jewwet MA, Hill RP, Evans AJ and Ohh M: Nuclear
E-cadherin and VHL immunoreactivity are prognostic indicators of
clear-cell renal cell carcinoma. Lab Invest. 87:2152–1264. 2007.
View Article : Google Scholar
|
|
65
|
Baldewijns MM, van Vlodrop IJH, Vermeulen
PB, Soetekouw PMMB, van Engeland M and de Bruïne AP: VHL and HIF
signalling in renal cell carcinogenesis. J Pathol. 221:125–138.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Nyhan MJ, El Mashad SM, O'Donovan TR,
Ahmad S, Collins C, Sweeney P, Rogers E, O'Sullivan GC and McKenna
SL: VHL genetic alteration in CCRCC does not determine
de-regulation of HIF, CAIX, hnRNP A2/B1 and osteopontin. Cell Oncol
(Dordr). 34:225–234. 2011. View Article : Google Scholar
|
|
67
|
Damjanovic SS, Ilic BB, Beleslin Cokic BB,
Antic JA, Bankovic JZ, Milicevic IT, Rodic GS, Ilic DS, Todorovic
VN, Puskas N, et al: Tuberous sclerosis complex protein 1
expression is affected by VHL Gene alterations and HIF-1α
production in sporadic clear-cell renal cell carcinoma. Exp Mol
Pathol. 101:323–331. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Muriel López C, Esteban E, Berros JP,
Pardo P, Astudillo A, Izquierdo M, Crespo G, Sanmamed M, Fonseca PJ
and Martínez-Camblor P: Prognostic factors in patients with
advanced renal cell carcinoma. Clin Genitourin Cancer. 10:262–270.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Shen C, Beroukhim R, Schumacher SE, Zhou
J, Chang M, Signoretti S and Kaelin WG Jr: Genetic and functional
studies implicate HIF1α as a 14q kidney cancer suppressor gene.
Cancer Discov. 1:222–235. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lidgren A, Hedberg Y, Grankvist K,
Rasmuson T, Vasko J and Ljungberg B: The expression of
hypoxia-inducible factor 1alpha is a favorable independent
prognostic factor in renal cell carcinoma. Clin Cancer Res.
11:1129–1135. 2005.PubMed/NCBI
|
|
71
|
Mandriota SJ, Turner KJ, Davies DR, Murray
PG, Morgan NV, Sowter HM, Wykoff CC, Maher ER, Harris AL, Ratcliffe
PJ, et al: HIF activation identifies early lesions in VHL kidneys:
Evidence for site-specific tumor suppressor function in the
nephron. Cancer Cell. 1:459–468. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Martínez-Sáez O, Gajate Borau P,
Alonso-Gordoa T, Molina-Cerrillo J and Grande E: Targeting HIF-2 α
in clear cell renal cell carcinoma: A promising therapeutic
strategy. Crit Rev Oncol Hematol. 111:117–123. 2017. View Article : Google Scholar
|
|
73
|
Raval RR, Lau KW, Tran MGB, Sowter HM,
Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL and Ratcliffe
PJ: Contrasting properties of hypoxia-inducible factor 1 (HIF-1)
and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol
Cell Biol. 25:5675–5686. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zimmer M, Doucette D, Siddiqui N and
Iliopoulos O: Inhibition of hypoxia-inducible factor is sufficient
for growth suppression of VHL−/− tumors. Mol Cancer Res.
2:89–95. 2004.PubMed/NCBI
|
|
75
|
Philips GK and Atkins MB: New agents and
new targets for renal cell carcinoma. Am Soc Clin Oncol Educ Book.
34:e222–e227. 2014. View Article : Google Scholar
|
|
76
|
Maroto P, Esteban E, Parra EF,
Mendez-Vidal MJ, Domenech M, Pérez-Valderrama B, Calderero V,
Pérez-Gracia JL, Grande E and Algaba F: HIF pathway and c-Myc as
biomarkers for response to sunitinib in metastatic clear-cell renal
cell carcinoma. OncoTargets Ther. 10:4635–4643. 2017. View Article : Google Scholar
|
|
77
|
Wong SC, Cheng W, Hamilton H, Nicholas AL,
Wakefield DH, Almeida A, Blokhin AV, Carlson J, Neal ZC, Subbotin
V, et al: HIF2α-targeted RNAi therapeutic inhibits clear cell renal
cell carcinoma. Mol Cancer Ther. 17:140–149. 2018. View Article : Google Scholar
|
|
78
|
Kamai T, Tokura Y, Uematsu T, Sakamoto K,
Suzuki I, Takei K, Narimatsu T, Kambara T, Yuki H, Betsunoh H, et
al: Elevated serum levels of cardiovascular biomarkers are
associated with progression of renal cancer. Open Heart.
5:e0006662018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Cuvillier O: The therapeutic potential of
HIF-2 antagonism in renal cell carcinoma. Transl Androl Urol.
6:131–133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Chen W, Hill H, Christie A, Kim MS,
Holloman E, Pavia-Jimenez A, Homayoun F, Ma Y, Patel N, Yell P, et
al: Targeting renal cell carcinoma with a HIF-2 antagonist. Nature.
539:112–117. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wehn PM, Rizzi JP, Dixon DD, Grina JA,
Schlachter ST, Wang B, Xu R, Yang H, Du X, Han G, et al: Design and
activity of specific hypoxia-inducible factor-2α (HIF-2α)
inhibitors for the treatment of clear cell renal cell carcinoma:
Discovery of clinical candidate
(S)-3-((2,2-Difluoro-1-hydroxy-7-(methylsulfonyl)-
2,3-dihydro-1H-inden-4-yl)oxy)-5-fluorobenzonitrile (PT2385). J Med
Chem. 61:9691–9721. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Burris H, Rodon J, Sharma S, Herbst RS,
Tabernero J, Infante JR, Silva A, Demanse D, Hackl W and Baselga J:
First-in-human phase I study of the oral PI3K inhibitor BEZ235 in
patients (pts) with advanced solid tumors. J Clin Oncol. 28(Suppl
15): 282010. View Article : Google Scholar
|
|
83
|
Wang X, Zhang J, Wang Y, Tu M, Wang Y and
Shi G: Upregulated VEGFAand DLL4 act as potential prognostic genes
for clear cell renal cell carcinoma. OncoTargets Ther.
11:1697–1706. 2018. View Article : Google Scholar
|
|
84
|
Veselaj F, Manxhuka-Kerliu S, Neziri A,
Shahini L, Xharra S, Selmani L, Kerliu L and Kavaja F: Prognostic
value of vascular endothelial growth factor A in the prediction of
the tumor aggressiveness in clear cell renal cell carcinoma. Open
Access Maced J Med Sci. 5:167–172. 2017.PubMed/NCBI
|
|
85
|
Phuoc NB, Ehara H, Gotoh T, Nakano M,
Kamei S, Deguchi T and Hirose Y: Prognostic value of the
co-expression of carbonic anhydrase IX and vascular endothelial
growth factor in patients with clear cell renal cell carcinoma.
Oncol Rep. 20:525–530. 2008.PubMed/NCBI
|
|
86
|
Shi ZG, Li SQ, Li ZJ, Zhu XJ, Xu P and Liu
G: Expression of vimentin and survivin in clear cell renal cell
carcinoma and correlation with p53. Clin Transl Oncol. 17:65–73.
2015. View Article : Google Scholar
|
|
87
|
Zhu Y, Xu L, Zhang J, Hu X, Liu Y, Yin H,
Lv T, Zhang H, Liu L, An H, et al: Sunitinib induces cellular
senescence via p53/Dec1 activation in renal cell carcinoma cells.
Cancer Sci. 104:1052–1061. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Busch J, Seidel C, Weikert S, Wolff I,
Kempkensteffen C, Weinkauf L, Hinz S, Magheli A, Miller K and
Grünwald V: Intrinsic resistance to tyrosine kinase inhibitors is
associated with poor clinical outcome in metastatic renal cell
carcinoma. BMC Cancer. 11:2952011. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Giuliano S, Cormerais Y, Dufies M, Grépin
R, Colosetti P, Belaid A, Parola J, Martin A, Lacas-Gervais S,
Mazure NM, et al: Resistance to sunitinib in renal clear cell
carcinoma results from sequestration in lysosomes and inhibition of
the autophagic flux. Autophagy. 11:1891–1904. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Butz H, Ding Q, Nofech-Mozes R, Lichner Z,
Ni H and Yousef GM: Elucidating mechanisms of sunitinib resistance
in renal cancer: An integrated pathological-molecular analysis.
Oncotarget. 9:4661–4674. 2017.
|
|
91
|
Kamli H, Glenda GC, Li L, Vesey DA and
Morais C: Characterisation of the morphological, functional and
molecular changes in sunitinib-resistant renal cell carcinoma
cells. J Kidney Cancer VHL. 5:1–9. 2018. View Article : Google Scholar : PubMed/NCBI
|