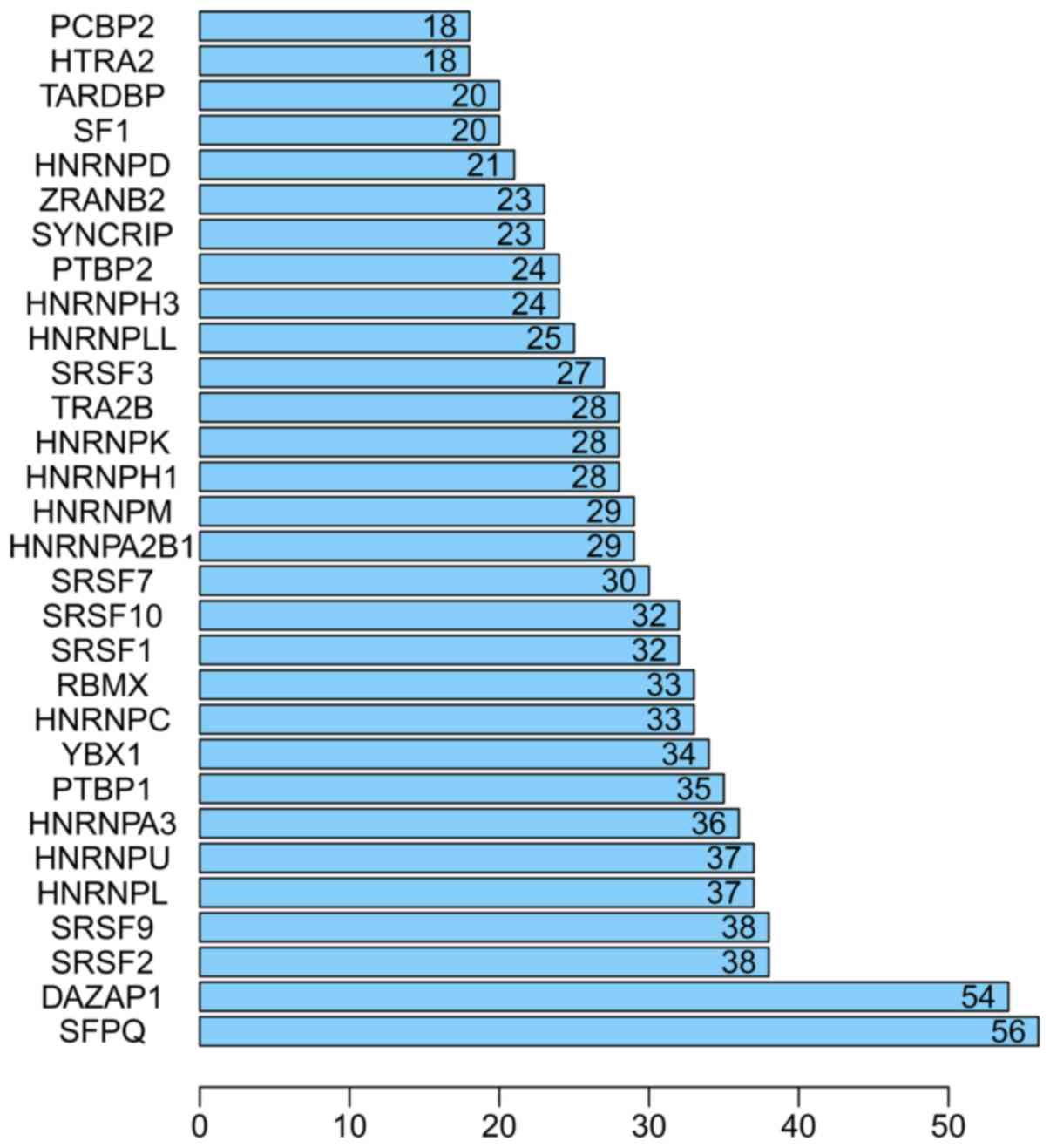
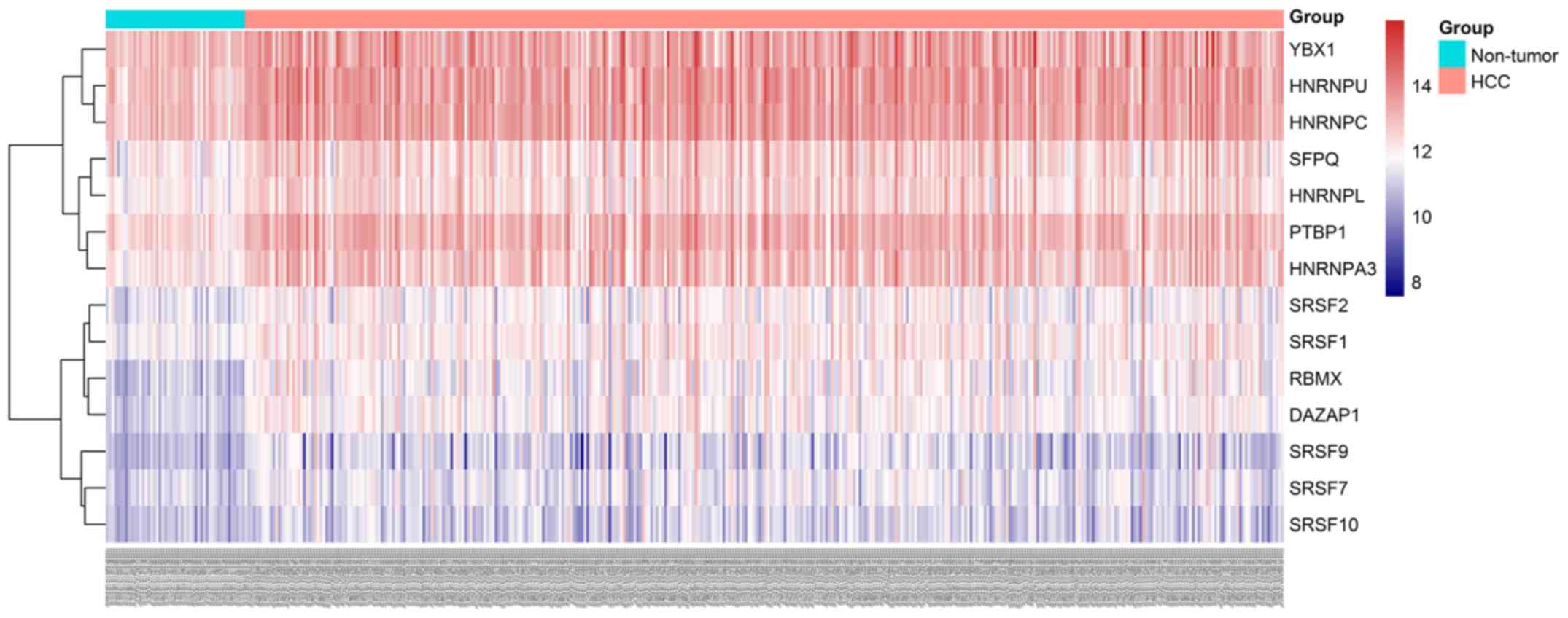
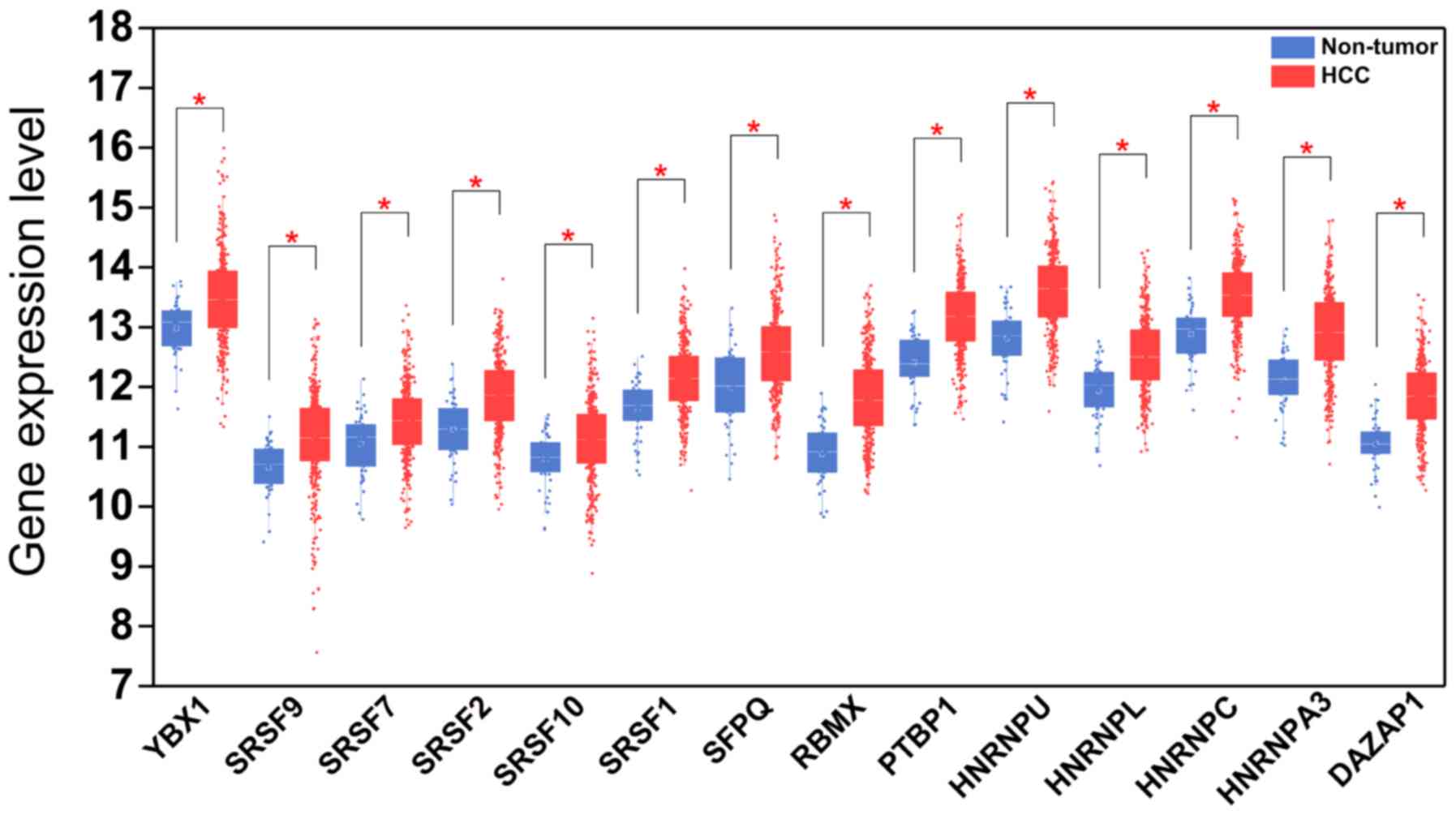
|
1
|
Breitbart RE, Andreadis A and Nadal-Ginard
B: Alternative splicing: A ubiquitous mechanism for the generation
of multiple protein isoforms from single genes. Annu Rev Biochem.
56:467–495. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Blencowe BJ: Alternative splicing: New
insights from global analyses. Cell. 126:37–47. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Takehara T, Liu X, Fujimoto J, Friedman SL
and Takahashi H: Expression and role of Bcl-xL in human
hepatocellular carcinomas. Hepatology. 34:55–61. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bingle CD, Craig RW, Swales BM, Singleton
V, Zhou P and Whyte MK: Exon skipping in Mcl-1 results in a bcl-2
homology domain 3 only gene product that promotes cell death. J
Biol Chem. 275:22136–22146. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Todaro M, Gaggianesi M, Catalano V,
Benfante A, Iovino F, Biffoni M, Apuzzo T, Sperduti I, Volpe S,
Cocorullo G, et al: CD44v6 is a marker of constitutive and
reprogrammed cancer stem cells driving colon cancer metastasis.
Cell Stem Cell. 14:342–356. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng S, Kim H and Verhaak RGW: Silent
mutations make some noise. Cell. 156:1129–1131. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Salton M, Kasprzak WK, Voss T, Shapiro BA,
Poulikakos PI and Misteli T: Inhibition of vemurafenib-resistant
melanoma by interference with pre-mRNA splicing. Nat Commun.
6:71032015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bechara EG, Sebestyén E, Bernardis I,
Eyras E and Valcárcel J: RBM5, 6, and 10 differentially regulate
NUMB alternative splicing to control cancer cell proliferation. Mol
Cell. 52:720–733. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ghigna C, Giordano S, Shen H, Benvenuto F,
Castiglioni F, Comoglio PM, Green MR, Riva S and Biamonti G: Cell
motility is controlled by SF2/ASF through alternative splicing of
the Ron protooncogene. Mol Cell. 20:881–890. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jung H, Lee D, Lee J, Park D, Kim YJ, Park
WY, Hong D, Park PJ and Lee E: Intron retention is a widespread
mechanism of tumor-suppressor inactivation. Nat Genet.
47:1242–1248. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goldstein LD, Lee J, Gnad F, Klijn C,
Schaub A, Reeder J, Daemen A, Bakalarski CE, Holcomb T, Shames DS,
et al: Recurrent loss of NFE2L2 exon 2 is a mechanism for Nrf2
pathway activation in human cancers. Cell Rep. 16:2605–2617. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kornblihtt AR: Epigenetics at the base of
alternative splicing changes that promote colorectal cancer. J Clin
Invest. 127:3281–3283. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Poulikakos PI, Persaud Y, Janakiraman M,
Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al:
RAF inhibitor resistance is mediated by dimerization of aberrantly
spliced BRAF(V600E). Nature. 480:387–390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sotillo E, Barrett DM, Black KL, Bagashev
A, Oldridge D, Wu G, Sussman R, Lanauze C, Ruella M, Gazzara MR, et
al: Convergence of acquired mutations and alternative splicing of
CD19 enables resistance to CART-19 immunotherapy. Cancer Discov.
5:1282–1295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang BD, Ceniccola K, Hwang S, Andrawis R,
Horvath A, Freedman JA, Olender J, Knapp S, Ching T, Garmire L, et
al: Alternative splicing promotes tumour aggressiveness and drug
resistance in African American prostate cancer. Nat Commun.
8:159212017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sebestyén E, Zawisza M and Eyras E:
Detection of recurrent alternative splicing switches in tumor
samples reveals novel signatures of cancer. Nucleic Acids Res.
43:1345–1356. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trincado JL, Sebestyén E, Pagés A and
Eyras E: The prognostic potential of alternative transcript
isoforms across human tumors. Genome Med. 8:852016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen S, Wang Y, Wang C, Wu YN and Xing Y:
SURVIV for survival analysis of mRNA isoform variation. Nat Commun.
7:115482016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Parsons DW, Li M, Zhang X, Jones S, Leary
RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, et al: The
genetic landscape of the childhood cancer medulloblastoma. Science.
331:435–439. 2011. View Article : Google Scholar :
|
|
20
|
Ghigna C, De Toledo M, Bonomi S, Valacca
C, Gallo S, Apicella M, Eperon I, Tazi J and Biamonti G:
Pro-metastatic splicing of Ron proto-oncogene mRNA can be reversed:
Therapeutic potential of bifunctional oligonucleotides and indole
derivatives. RNA Biol. 7:495–503. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee SC and Abdel-Wahab O: Therapeutic
targeting of splicing in cancer. Nat Med. 22:976–986. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Koh CM, Bezzi M, Low DH, Ang WX, Teo SX,
Gay FP, Al-Haddawi M, Tan SY, Osato M, Sabò A, et al: MYC regulates
the core pre-mRNA splicing machinery as an essential step in
lymphomagenesis. Nature. 523:96–100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Havens MA and Hastings ML:
Splice-switching antisense oligonucleotides as therapeutic drugs.
Nucleic Acids Res. 44:6549–6563. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Salton M and Misteli T: Small molecule
modulators of Pre-mRNA splicing in cancer therapy. Trends Mol Med.
22:28–37. 2016. View Article : Google Scholar :
|
|
25
|
Dvinge H, Kim E, Abdel-Wahab O and Bradley
RK: RNA splicing factors as oncoproteins and tumour suppressors.
Nat Rev Cancer. 16:413–430. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bonnal S, Vigevani L and Valcárcel J: The
spliceosome as a target of novel antitumour drugs. Nat Rev Drug
Discov. 11:847–859. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Webb TR, Joyner AS and Potter PM: The
development and application of small molecule modulators of SF3b as
therapeutic agents for cancer. Drug Discov Today. 18:43–49. 2013.
View Article : Google Scholar :
|
|
28
|
Yokoi A, Kotake Y, Takahashi K, Kadowaki
T, Matsumoto Y, Minoshima Y, Sugi NH, Sagane K, Hamaguchi M, Iwata
M, et al: Biological validation that SF3b is a target of the
antitumor macrolide pladienolide. FEBS J. 278:4870–4880. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kotake Y, Sagane K, Owa T, Mimori-Kiyosue
Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M and Mizui Y: Splicing
factor SF3b as a target of the antitumor natural product
pladienolide. Nat Chem Biol. 3:570–575. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fan L, Lagisetti C, Edwards CC, Webb TR
and Potter PM: Sudemycins, novel small molecule analogues of
FR901464, induce alternative gene splicing. ACS Chem Biol.
6:582–589. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xargay-Torrent S, López-Guerra M, Rosich
L, Montraveta A, Roldán J, Rodríguez V, Villamor N, Aymerich M,
Lagisetti C, Webb TR, et al: The splicing modulator sudemycin
induces a specific antitumor response and cooperates with ibrutinib
in chronic lymphocytic leukemia. Oncotarget. 6:22734–22749. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wan L, Yu W, Shen E, Sun W, Liu Y, Kong J,
Wu Y, Han F, Zhang L, Yu T, et al: SRSF6-regulated alternative
splicing that promotes tumour progression offers a therapy target
for colorectal cancer. Gut. 68:118–129. 2019. View Article : Google Scholar
|
|
33
|
Dvinge H and Bradley RK: Widespread intron
retention diversifies most cancer transcriptomes. Genome Med.
7:452015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hashemikhabir S, Budak G and Janga SC:
ExSurv: A web resource for prognostic analyses of exons across
human cancers using clinical transcriptomes. Cancer Inform.
15(Suppl 2): 17–24. 2016.PubMed/NCBI
|
|
35
|
Neelamraju Y, Gonzalez-Perez A,
Bhat-Nakshatri P, Nakshatri H and Janga SC: Mutational landscape of
RNA-binding proteins in human cancers. RNA Biol. 15:115–129. 2018.
View Article : Google Scholar :
|
|
36
|
Wang ZL, Li B, Luo YX, Lin Q, Liu SR,
Zhang XQ, Zhou H, Yang JH and Qu LH: Comprehensive Genomic
Characterization of RNA-Binding Proteins across Human Cancers. Cell
Rep. 22:286–298. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kechavarzi B and Janga SC: Dissecting the
expression landscape of RNA-binding proteins in human cancers.
Genome Biol. 15:R142014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Luo C, Cheng Y, Liu Y, Chen L, Liu L, Wei
N, Xie Z, Wu W and Feng Y: SRSF2 regulates alternative splicing to
drive hepatocellular carcinoma development. Cancer Res.
77:1168–1178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li X, Qian X, Peng LX, Jiang Y, Hawke DH,
Zheng Y, Xia Y, Lee JH, Cote G, Wang H, et al: Corrigendum: A
splicing switch from ketohexokinase-C to ketohexokinase-A drives
hepatocellular carcinoma formation. Nat Cell Biol. 18:7092016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chiu YT, Wong JK, Choi SW, Sze KM, Ho DW,
Chan LK, Lee JM, Man K, Cherny S, Yang WL, et al: Novel pre-mRNA
splicing of intronically integrated HBV generates oncogenic chimera
in hepatocellular carcinoma. J Hepatol. 64:1256–1264. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Luo ZL, Cheng SQ, Shi J, Zhang HL, Zhang
CZ, Chen HY, Qiu BJ, Tang L, Hu CL, Wang HY, et al: A splicing
variant of Merlin promotes metastasis in hepatocellular carcinoma.
Nat Commun. 6:84572015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bayliss J, Lim L, Thompson AJ, Desmond P,
Angus P, Locarnini S and Revill PA: Hepatitis B virus splicing is
enhanced prior to development of hepatocellular carcinoma. J
Hepatol. 59:1022–1028. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yuan JH, Liu XN, Wang TT, Pan W, Tao QF,
Zhou WP, Wang F and Sun SH: The MBNL3 splicing factor promotes
hepatocellular carcinoma by increasing PXN expression through the
alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 19:820–832.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tremblay MP, Armero VE, Allaire A,
Boudreault S, Martenon-Brodeur C, Durand M, Lapointe E, Thibault P,
Tremblay-Létourneau M, Perreault JP, et al: Global profiling of
alternative RNA splicing events provides insights into molecular
differences between various types of hepatocellular carcinoma. BMC
Genomics. 17:6832016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ryan M, Wong WC, Brown R, Akbani R, Su X,
Broom B, Melott J and Weinstein J: TCGASpliceSeq a compendium of
alternative mRNA splicing in cancer. Nucleic Acids Res.
44:D1018–D1022. 2016. View Article : Google Scholar :
|
|
46
|
Goswami CP and Nakshatri H: PROGgeneV2:
Enhancements on the existing database. BMC Cancer. 14:9702014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xue L, Xie L and Song X and Song X:
Identification of potential tumor-educated platelets RNA biomarkers
in non-small-cell lung cancer by integrated bioinformatical
analysis. J Clin Lab Anal. 32:e224502018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhu J, Chen Z and Yong L: Systematic
profiling of alternative splicing signature reveals prognostic
predictor for ovarian cancer. Gynecol Oncol. 148:368–374. 2018.
View Article : Google Scholar
|
|
49
|
Ryan MC, Cleland J, Kim R, Wong WC and
Weinstein JN: SpliceSeq: A resource for analysis and visualization
of RNA-Seq data on alternative splicing and its functional impacts.
Bioinformatics. 28:2385–2387. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Boise LH, González-García M, Postema CE,
Ding L, Lindsten T, Turka LA, Mao X, Nuñez G and Thompson CB:
bcl-x, a bcl-2-related gene that functions as a dominant regulator
of apoptotic cell death. Cell. 74:597–608. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bae J, Leo CP, Hsu SY and Hsueh AJ:
MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member
MCL-1, encodes a proapoptotic protein possessing only the BH3
domain. J Biol Chem. 275:25255–25261. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li Y, Sun N, Lu Z, Sun S, Huang J, Chen Z
and He J: Prognostic alternative mRNA splicing signature in
non-small cell lung cancer. Cancer Lett. 393:40–51. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen L, Tovar-Corona JM and Urrutia AO:
Increased levels of noisy splicing in cancers, but not for
oncogene-derived transcripts. Hum Mol Genet. 20:4422–4429. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Qiao GJ, Chen L, Wu JC and Li ZR:
Identification of an eight-gene signature for survival prediction
for patients with hepatocellular carcinoma based on integrated
bioinformatics analysis. PeerJ. 7:e65482019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liao X, Zhu G, Huang R, Yang C, Wang X,
Huang K, Yu T, Han C, Su H and Peng T: Identification of potential
prognostic microRNA biomarkers for predicting survival in patients
with hepatocellular carcinoma. Cancer Manag Res. 10:787–803. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wahl MC, Will CL and Lührmann R: The
spliceosome: Design principles of a dynamic RNP machine. Cell.
136:701–718. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Turunen JJ, Niemelä EH, Verma B and
Frilander MJ: The significant other: Splicing by the minor
spliceosome. Wiley Interdiscip Rev RNA. 4:61–76. 2013. View Article : Google Scholar :
|
|
58
|
Supek F, Miñana B, Valcárcel J, Gabaldón T
and Lehner B: Synonymous mutations frequently act as driver
mutations in human cancers. Cell. 156:1324–1335. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sterne-Weiler T and Sanford JR: Exon
identity crisis: Disease-causing mutations that disrupt the
splicing code. Genome Biol. 15:2012014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Diederichs S, Bartsch L, Berkmann JC,
Fröse K, Heitmann J, Hoppe C, Iggena D, Jazmati D, Karschnia P,
Linsenmeier M, et al: The dark matter of the cancer genome:
Aberrations in regulatory elements, untranslated regions, splice
sites, non-coding RNA and synonymous mutations. EMBO Mol Med.
8:442–457. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Singh S, Narayanan SP, Biswas K, Gupta A,
Ahuja N, Yadav S, Panday RK, Samaiya A, Sharan SK and Shukla S:
Intragenic DNA methylation and BORIS-mediated cancer-specific
splicing contribute to the Warburg effect. Proc Natl Acad Sci USA.
114:11440–11445. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gelfman S, Cohen N, Yearim A and Ast G:
DNA-methylation effect on cotranscriptional splicing is dependent
on GC architecture of the exon-intron structure. Genome Res.
23:789–799. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Shukla S, Kavak E, Gregory M, Imashimizu
M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R and
Oberdoerffer S: CTCF-promoted RNA polymerase II pausing links DNA
methylation to splicing. Nature. 479:74–79. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yuan H, Li N, Fu D, Ren J, Hui J, Peng J,
Liu Y, Qiu T, Jiang M, Pan Q, et al: Histone methyltransferase
SETD2 modulates alternative splicing to inhibit intestinal
tumorigenesis. J Clin Invest. 127:3375–3391. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ding X, Liu S, Tian M, Zhang W, Zhu T, Li
D, Wu J, Deng H, Jia Y, Xie W, et al: Activity-induced histone
modifications govern Neurexin-1 mRNA splicing and memory
preservation. Nat Neurosci. 20:690–699. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sharma A, Nguyen H, Geng C, Hinman MN, Luo
G and Lou H: Calcium-mediated histone modifications regulate
alternative splicing in cardiomyocytes. Proc Natl Acad Sci USA.
111:E4920–E4928. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kim S, Kim H, Fong N, Erickson B and
Bentley DL: Pre-mRNA splicing is a determinant of histone H3K36
methylation. Proc Natl Acad Sci USA. 108:13564–13569. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Feng J, Chen K, Dong X, Xu X, Jin Y, Zhang
X, Chen W, Han Y, Shao L, Gao Y, et al: Genome-wide identification
of cancer-specific alternative splicing in circRNA. Mol Cancer.
18:352019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kaida D, Motoyoshi H, Tashiro E, Nojima T,
Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima
H, et al: Spliceostatin A targets SF3b and inhibits both splicing
and nuclear retention of pre-mRNA. Nat Chem Biol. 3:576–583. 2007.
View Article : Google Scholar : PubMed/NCBI
|