Protein 4.1B/DAL-1 is encoded by erythrocyte
membrane protein band 4.1-like 3 (EPB41L3; also termed DAL-
1/ KIAA0987) and is important for cytoskeletal
rearrangements, intracellular transport and signal transduction,
with high levels of expression in the brain, intestine and kidney,
and lower levels in the pancreas, lung and liver (1-4).
Protein 4.1B/DAL-1 belongs to the 4.1 protein superfamily, which is
characterized by the presence of a highly conserved
four.one-ezrin-radixin-moesin (FERM) domain at the N-terminus
(3-6). Compared with 4.1 superfamily members,
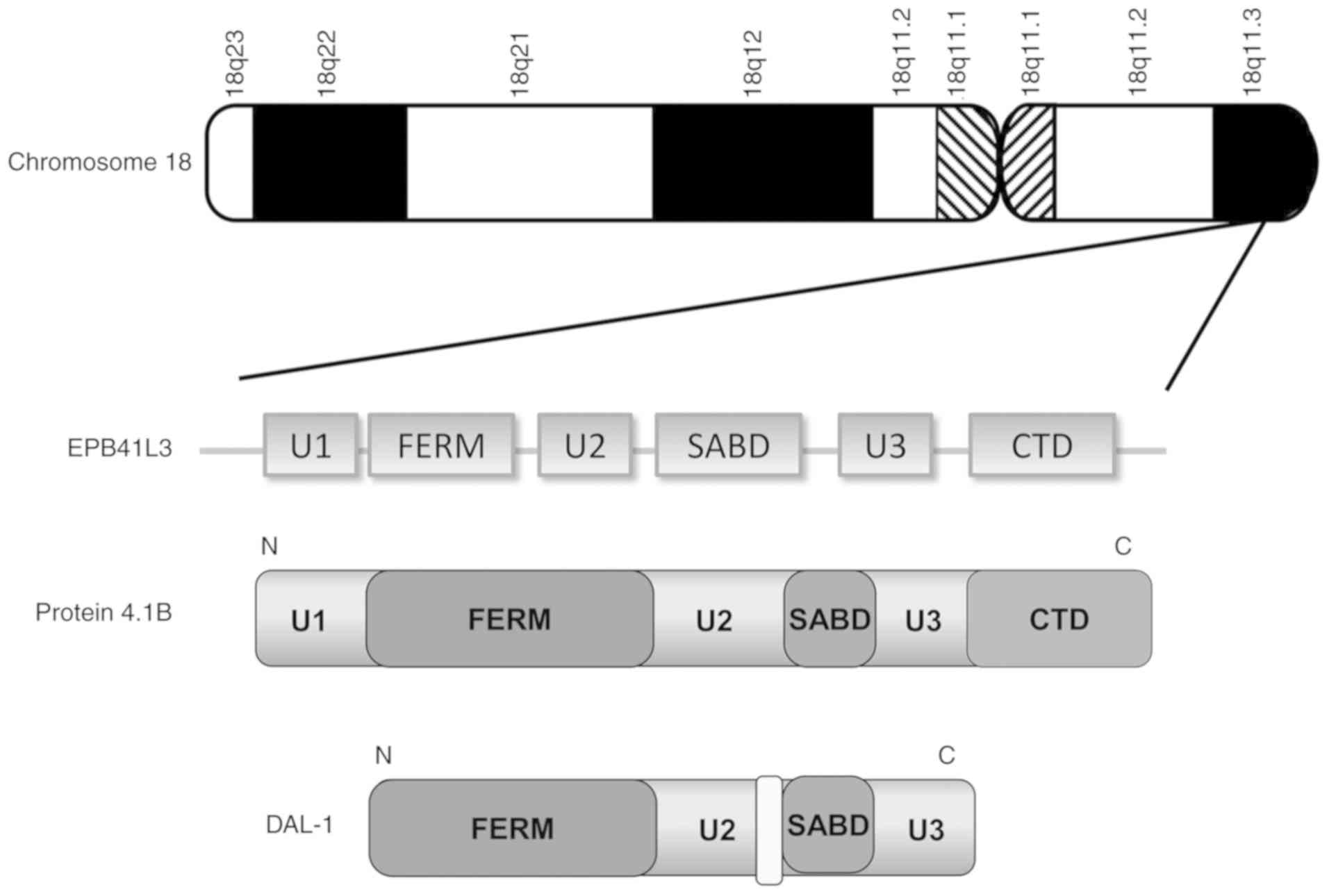
4.1B contains three conserved domains (FERM, SABD and CTD) and
three unique domains (U1, U2 and U3) (7). DAL-1, a short form of 4.1B, is also
located on human chromosome 18p11.3 and lacks the U1, parts of the
U2 and SABD, and full-length CTD (Fig.
1).
Protein 4.1B is a known tumor suppressor, and
inactivation contributes to the progression and development of
numerous cancer types, including lung adenocarcinoma (8-11),
meningiomas (12-16), breast cancer (17,18),
ovarian cancer (19) and prostate
cancer (20,21). A loss of protein 4.1B expression in
cancer is partly associated with a loss of heterozygosity (LOH) on
18p11.3 and/or the hypermethylation of CpG islands (17). Protein 4.1B, a pivotal regulator of
cytoskeletal structure, regulates various cytoskeletal-associated
processes in cancer, including cell migration and invasion
(22). Protein 4.1B has therefore
received attention as a potential therapeutic target in cancer.
Proteins isolated from human red blood cell
membranes were first recognized as members of the protein 4.1
superfamily. Since their discovery, over 40 proteins have been
demonstrated to belong to the protein 4.1 superfamily based on
sequence homology (3,23). In addition to the protein 4.1
subfamily, the protein 4.1 superfamily consists of four additional
subfamilies: i) Talin-related molecules; ii) the ezrin, radixin,
moesin protein family; iii) protein tyrosine phosphatases proteins;
and iv) novel band 4.1-like 4 proteins (4). Members of the protein 4.1
super-family possess diverse biological functions and share a
highly conserved FERM domain of 200-300 amino acids, which is
associated with the interaction with cytoplasmic domains of
specific transmembrane proteins. In addition to the FERM domain,
the protein 4.1 subfamily possesses two highly conserved
spectrin-actin-binding (SAB) and C-terminal (CT) domains (7). The protein 4.1 family includes four
homologues, 4.1R (erythrocyte-type), 4.1N (neuronal-type), 4.1G
(general-type) and 4.1B (brain-type), which are encoded by EPB41,
EPB41L1, EPB41L2 and EPB41L3, respectively (Fig. 2) (5,24).
The FERM domain, first isolated from human
erythrocyte ghosts, is a unique module in the family of peripheral
membrane proteins that functions as a plasma membrane-cytoskeleton
linker (6,25). Structurally, the FERM domain has a
cloverleaf-like architecture with N-lobes, α-lobes and C-lobes
(also termed F1, F2 and F3). The N-terminal FERM domain of 4.1B has
a number of interacting partners. For example, 4.1B binds via its
FERM domain to the cytoplasmic domain of cell adhesion molecule
(CADM)1/tumor suppressor in lung cancer 1 (TSLC1) (11,26)
and CADM4 (27). In addition,
members of the membrane-associated guanylate kinase homologs,
including membrane palmitoylated protein (MPP)1/p55, MPP2/discs
large MAGUK scaffold protein 2 and MPP3, form a tripartite complex
through their interaction with 4.1B and CADM1 (28). Protein arginine N-methyltransferase
(PRMT)3 and PRMT5 also interact with 4.1B (29,30).
The interactions between 4.1B and contactin associated protein
(CNTNAP)1 or CNTNAP2 serve essential roles in the orga-nization of
myelinated axons (31). The FERM
domain of 4.1B also mediates binding to the 14-3-3 isoforms β, γ
and η (32).
Protein 4.1B/DAL-1 maintains the mechanical
integrity of cell membranes through its ability to form ternary
complexes with spectrin and actin via its SAB domain (33). Functional variation exists amongst
the different splice variants of SAB comprising of exons 16 and 17.
Strong spectrin and actin binding affinity requires both exons,
whilst exon 17 alone demonstrates weak affinity (34,35).
The SAB domain is also responsible for the interaction with
sarcomeric proteins, such as myosin, α-actin, and tropomyosin
(36). The association between
4.1B and αvβ8 integrin occurs via its CT domain (37). In addition to these domains,
protein 4.1B contains three unique regions (U1, U2 and U3).
Membrane localization of the U2 domain, which is located between
the FERM and SAB domains, is essential for 4.1B to function as a
growth suppressor in meningioma (14). Collectively, protein 4.1B/DAL-1
engages in a wide range of cellular functions through its
interactions with dynamic molecules via its specific FERM, SAB and
CT domains.
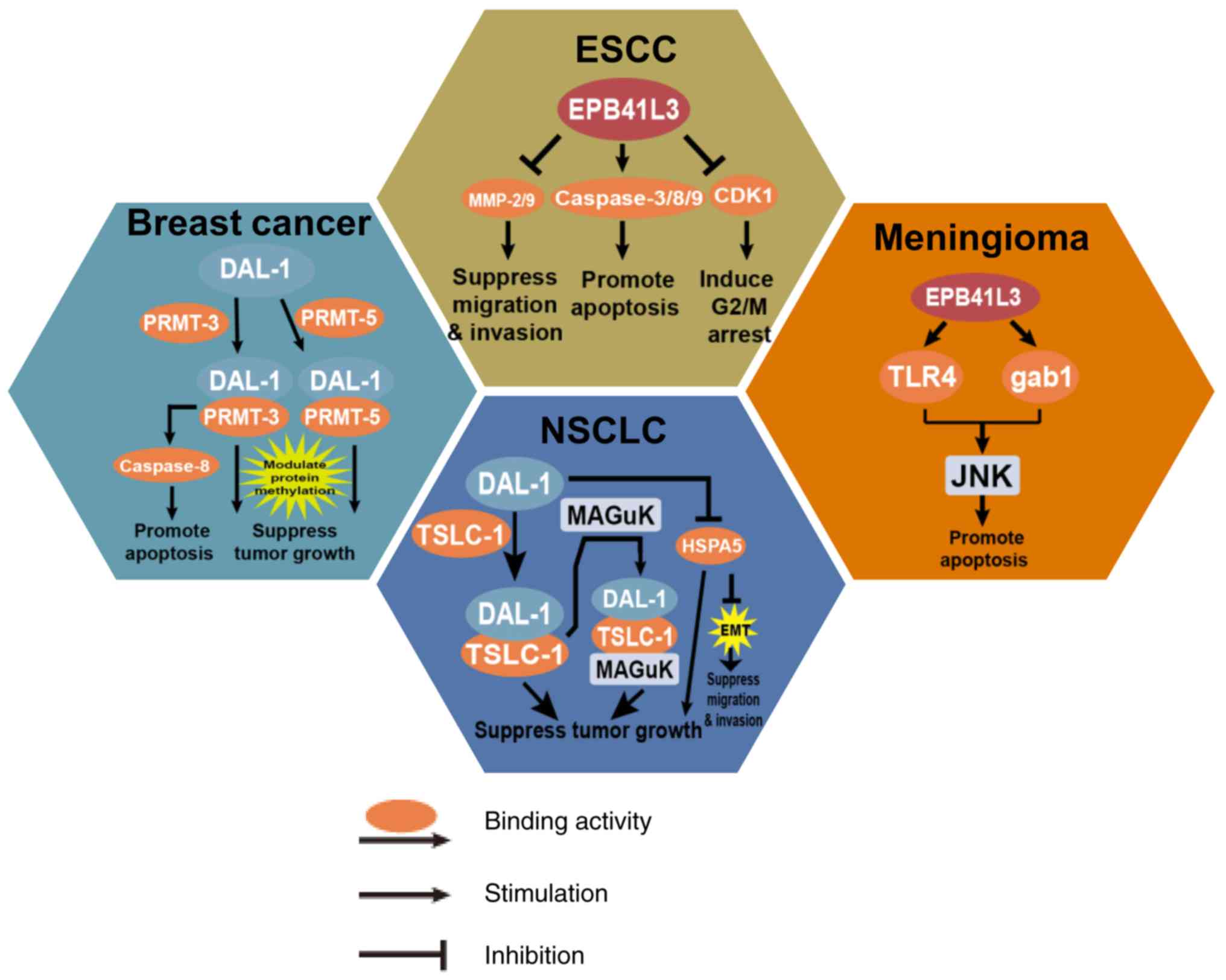
Aberrant protein 4.1B/DAL-1 expression occurs due to
a LOH and/or DNA hypermethylation, and is implicated in numerous
cancer types with a crucial role in tumor development and
progression (Fig. 3).
DAL-1, a short form of protein 4.1B possessing all
of its functional domains, was originally identified to be
downregulated in primary lung tumors and lung cancer cell lines
with a potential role in the suppression of tumor growth (8,38,39).
The hypermethylation of DAL-1 strongly correlates with a loss of
DAL-1, and predicts a short overall survival in patients with NSCLC
(9). In addition, EPB41L3
localizes within chromosomal region 18p11.3, which is influenced by
a LOH in NSCLC (13). These
studies indicate that both DNA methylation and LOH promote aberrant
DAL-1 expression in NSCLC.
A noticeable feature of protein 4.1B/DAL-1 in lung
cancer is its close association with epithelial-mesenchymal
transition (EMT). It has been reported that protein 4.1B/DAL-1
maintains the phenotype of epithelial cells and attenuates EMT by
inhibiting PI3K/Akt/Mdm2/p53 signaling as a result of the
suppression of heat shock protein 5 in NSCLC (40,41).
A loss of protein 4.1B/DAL-1 leads to a substantial decrease in the
expression of numerous EMT markers, including E-cadherin and
β-catenin in lung cancer cells (42). Additionally, through interaction
with the tumor suppressor gene TSLC-1, an immunoglobulin
superfamily cell-adhesion molecule possessing strong anti-tumor
ability, protein 4.1B/DAL-1 regulates cell motility and actin
rearrangements in cancer cells (11,26,43).
Several studies have evaluated the role of
4.1B/DAL-1 in breast cancer and have reported that its ability to
modulate the activity of protein arginine methyltransferases PRMT3
and PRMT5 is via direct binding to the catalytic core domain of
PRMTs in breast cancer cell lines (29,30).
Notably, protein 4.1B/DAL-1 can enhance and inhibit PRMT5 catalytic
activity in a substrate-specific manner (29,30).
Furthermore, protein 4.1B/DAL-1 and protein methylation mediated by
protein arginine methyltransferases cooperate to induce apoptosis
via a caspase-8-dependent pathway in MCF-7 breast cancer cells
(45). These findings suggest that
4.1B/DAL-1 influences breast cancer cell growth through the
modulation of PRMT-dependent post-translational methylation.
Bisulfite sequencing and methylation-specific PCR
revealed elevated levels of EPB41L3 hypermethylation in prostate
cancer tissues and cell lines compared with the low methylation
observed in normal prostate tissues (46-49).
In particular, EPB41L3 expression increased through co-treatment
with the DNA methyltransferase inhibitor (5-aza-2′-deoxycyti-dine)
and a histone deacetylase inhibitor (SAHA) in Du145 and 22Rv1
prostate cancer cell lines, respectively (46). These studies illustrate that the
loss of protein 4.1B/DAL-1 in prostate cancer is primarily
associated with aberrant DNA methylation. Decreased expression of
4.1B in highly metastatic prostate cancer cells indicates its role
as a negative modulator of cancer progression and metastasis
(21,47). Notably, 4.1B-deficient mice
demonstrate increased susceptibility for aggressive and spontaneous
prostate tumors in transgenic adenocarcinoma of the mouse prostate
tumor models (21). Based on these
findings, reduced 4.1B expression appears an important event in
prostate cancer that causally contributes to an aggressive tumor
phenotype.
Hypermethylated CpG sites in the promoter region of
EPB41L3 lead to loss of expression and are identified in ESCC,
which is consistent with studies in other cancer types (50). By contrast, it has been reported
that EPB41L3 is significantly upregulated in ESCC compared with
normal tissue. The enhanced expression of EPB41L3 is associated
with the response to neoadjuvant chemoradiation in ESCC (51). Hence, further studies investigating
the expression of EPB41L3 in ESCC are necessary to elucidate the
full spectrum of its functions in EPB41L3.
Protein 4.1B/DAL-1 plays a significant role in
suppressing cell migration and invasion via inhibiting matrix
metalloproteinase (MMP)2 and MMP9 in ESCC (52). EPB41L3 positively regulates
apoptosis through activating caspase-3/8/9, and inducing G2/M
arrest via the CDK1 pathway (53).
The specific functions of protein 4.1B/DAL-1 in the pathogenesis of
ESCC are relatively uncharacterized and require further
exploration.
The methylation of EPB41L3 can distinguish
precancerous lesions from invasive disease in cervical specimens.
High methylation of EPB41L3 in women with cervical intraepithelial
neoplasia grade 2/3 (CIN2/3) representing the pre-tumorigenic stage
implies a higher cancer risk (54-57).
Specifically, the methylation levels at CpG sites (438, 427 and
425) in EPB41L3 may distinguish CIN2/3 from negative
intraepithelial lesions or malignancy/CIN1 and cancer from CIN2/3
(58). Recent studies targeting
women living with HIV-1 (WLHIV) in Burkina Faso and South Africa
revealed that the hypermethylation of EPB41L3 is frequent among
WLHIV and occurs in conjunction with low CD4+ counts and
a poor efficacy of antiretroviral therapy (59). Indeed, when screening for HPV
infection, DNA methylation assessments of EPB41L3 offer promise as
an objective molecular-based approach for early detection and
diagnosis (60-69).
A loss of protein 4.1B/DAL-1 occurs in ≥50% of the
cases of sporadic meningiomas. As a major cause of downregulating
EPB41L3, a LOH on chromosome segment 18p11.3 is common in
meningiomas. However, meningioma with 18p11.3 LOH does not
correlate with the nucleotide inactivation of EPB41L3 (12-14).
EPB41L3 hypermethylation is a prognostic factor for poor survival
in diffuse gliomas, and treatment with the demethylating agent
5-aza-2′-deoxycytidine and the histone deacetylase inhibitor
trichostatin A have been shown to restore EPB41L3 expression in
glioma cells (70). A loss of
protein 4.1B/DAL-1 is reported as an early event in meningioma
tumorigenesis, suggesting it plays a crucial role in growth
regulation during meningioma pathogenesis (12). As a tumor suppressor, protein
4.1B/DAL-1 is associated with the activation of JNK in meningioma
(16). Protein 4.1B/DAL-1 mediated
growth suppression in meningioma requires the sequential activation
of Src, Rac1 and JNK (16).
Furthermore, the inhibition of Rac1 or JNK activation abrogates
protein 4.1B-growth suppression. However, it is poorly understood
how 4.1B/DAL-1 activates the JNK pathway.
In addition to esophageal carcinoma, abnormal
expression of EPB41L3 has been reported in other common digestive
system cancer types, including gastric cancer, intestinal
carcinoma, colorectal cancer, hepatocellular carcinoma and
pancreatic carcinoma (71-75). Consistent with previous findings
that Helicobacter pylori (HP)-induced inflammatory response
promotes EPB41L3 DNA methylation, the methylation of EPB41L3 is
significantly lower in remnant gastric cancers with the decline of
HP infection following distal gastrectomy (73,74).
In colorectal cancer, protein 4.1B/DAL-1 has been shown to be
downregulated through membrane proteomic analysis (72). In addition, a similar loss of
4.1B/DAL-1 occurs in intestinal epithelia malignant carcinomas of
mouse and humans (71). Protein
4.1B/DAL-1 is also downregulated in hepatocellular carcinoma (HCC),
and represses HCC cell migration and invasion (76). Using transgenic mouse models of
pancreatic b-cell carcinogenesis (Rip1Tag2), a loss of 4.1B has
been reported in the phenotypic transition from adenoma to
carcinoma (75). In addition,
EPB41L3 and HPV 16 methylation are markedly higher amongst
oropharyngeal cancer (OPC) with high sensitivity and specificity
for the detection of OPC from an oral gargle, suggesting that
measurements of HPV 16 and EPB41L3 methylation have utility in
identifying early OPC (77). High
methylation of EPB41L3 is also associated with anal intraepithelial
neoplasia (78).
Whilst protein 4.1B is expressed in the proximal
uriniferous tubules of the normal human kidney, its loss or
downregulation through EPB41L3 promoter hyper-methylation occurs is
~50% of renal clear cell carcinoma (RCCC) cases, implicating its
association with EPB41L3 methylation, tumor grade and
recurrence-free survival (79).
Furthermore, the protein 4.1B binding partner CADM4 serves an
important role in the adhesion of the proximal uriniferous tubules
that are the precursor cells of RCCC (27).
Similarly, promoter methylation of EPB41L3 is one of
the common mechanisms of genetic downregulation in ovarian cancer
(19). In addition, EPB41L3 is
up-regulated with age and contributes to a decreased proliferation
rate of human bone marrow-derived mesenchymal stem cells (80).
Protein 4.1B/DAL-1 encoded by EPB41L3, functions as
a tumor suppressor in human cancer (5,22).
The pathological functions of protein 4.1B/DAL-1 are versatile
through its regulation of various cellular processes during
carcinogenesis (Fig. 4).
Nevertheless, the functions and mechanisms of EPB41L3 in cancer
remain unknown.
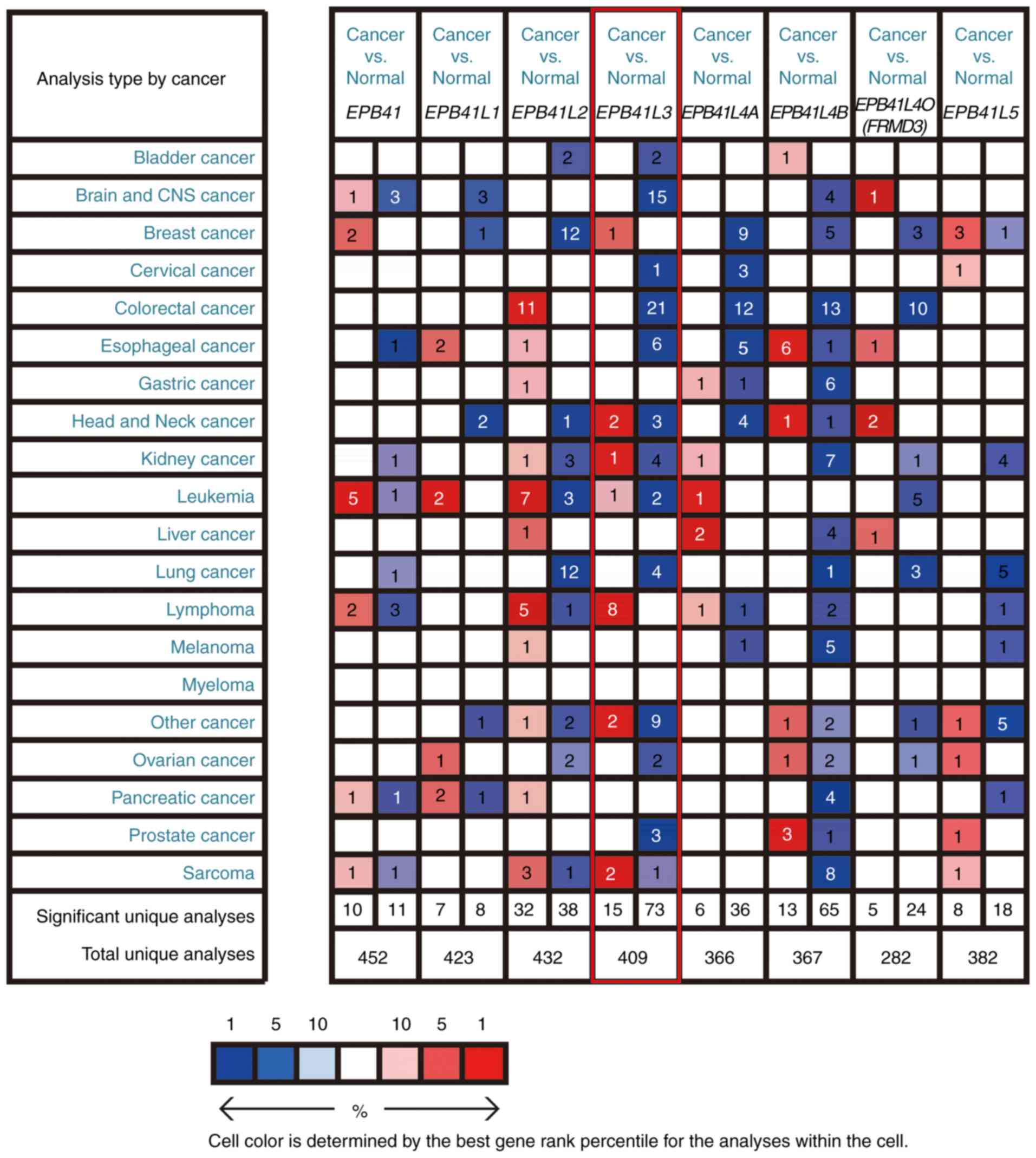
Promoter methylation is a major cause of EPB41L3
inactivation in cancer. Abnormal DNA methylation of EPB41L3 is
identified in numerous cancer types, including NSCLC and breast
cancer (9,44). Furthermore, aberrant EPB41L3 DNA
methylation possesses high diagnostic and prognostic significance
in cancer, indicating its potential use as a biomarker for cancer
diagnosis (Table I).
Re-expression of EPB41L3 results in significant
suppression of cell growth in ovarian cancer, lung cancer and
breast cancer (8,19,39).
Particularly, reintroduction of EPB41L3 into 4.1B/DAL-1-null lung,
breast and cervical cancer cell lines markedly suppresses cancer
cell growth and promotes apoptosis (8,39,64).
Hence, EPB41L3 represents an attractive therapeutic strategy.
Treatment with DNA methyltransferase inhibitors can restore the
expression of EPB41L3 (81).
Engineered artificial transcription factors can reactivate EPB41L3
with tumor suppressive roles in breast, ovarian and cervical cancer
cell lines. Treatment of human amniotic fluid stem with CXCR4
promoters and adenovirus vector expressing DAL-1 permits the
targeting of lung cancer xenografts that overexpress DAL-1,
representing a strategy to restore EPB41L3 function (82). Such epigenetic reprogramming tools
to reintroduce silenced tumor suppressor genes represent an
emerging potential therapeutic strategy (64).
Protein 4.1B/DAL-1 is essential for
cytoskeleton-associated processes by interacting with a series of
protein molecules through its conserved FERM, SAB and CT domains. A
loss of protein 4.1B/DAL-1 expression, caused by aberrant DNA
methylation and/or LOH, is frequently observed in cancer. Thus,
protein 4.1B/DAL-1 can serve as a potential biomarker with high
sensitivity and specificity for cancer diagnosis. In addition,
reintroduction of protein 4.1B/DAL-1 by epigenetic inhibitors or
genome-editing tools significantly inhibits tumor progression,
indicating its potential as a novel promising target for cancer
therapy. Taken together, growing evidence highlights the importance
of protein 4.1B/DAL-1 in cancer progression, and further studies
will be required to elucidate the regulatory mechanisms of protein
4.1B/DAL-1 in human cancer.
This research was supported by Changzhou
Sci&Tech Program (grant no. 20180170).
Not applicable.
XY performed the original draft preparation. LP
performed most of the revision work. XY and LP participated in the
whole work. LW, XH and MZ contributed to figure preparation and
revision, as well as editing of the manuscript. ZL provided
financial support, edited the figures and contributed to editing of
the manuscript. All authors read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Cifuentes-Diaz C, Chareyre F, Garcia M,
Devaux J, Carnaud M, Levasseur G, Niwa-Kawakita M, Harroch S,
Girault JA, Giovannini M and Goutebroze L: Protein 4.1B contributes
to the organization of peripheral myelinated axons. PLoS One.
6:e250432011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hoover KB and Bryant PJ: The genetics of
the protein 4.1 family: Organizers of the membrane and
cytoskeleton. Curr Opin Cell Biol. 12:229–234. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Diakowski W, Grzybek M and Sikorski AF:
Protein 4.1, a component of the erythrocyte membrane skeleton and
its related homologue proteins forming the protein 4.1/FERM
superfamily. Folia Histochem Cytobiol. 44:231–248. 2006.
|
|
4
|
Takeuchi K, Kawashima A, Nagafuchi A and
Tsukita S: Structural diversity of band 4.1 superfamily members. J
Cell Sci. 107:1921–1928. 1994.PubMed/NCBI
|
|
5
|
Sun CX, Robb VA and Gutmann DH: Protein
4.1 tumor suppressors: Getting a FERM grip on growth regulation. J
Cell Sci. 115:3991–4000. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chishti AH, Kim AC, Marfatia SM, Lutchman
M, Hanspal M, Jindal H, Liu SC, Low PS, Rouleau GA, Mohandas N, et
al: The FERM domain: A unique module involved in the linkage of
cytoplasmic proteins to the membrane. Trends Biochem Sci.
23:281–282. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Parra M, Gee S, Chan N, Ryaboy D, Dubchak
I, Mohandas N, Gascard PD and Conboy JG: Differential domain
evolution and complex RNA processing in a family of paralogous
EPB41 (protein 4.1) genes facilitate expression of diverse
tissue-specific isoforms. Genomics. 84:637–646. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tran YK, Bögler O, Gorse KM, Wieland I,
Green MR and Newsham IF: A novel member of the NF2/ERM/4.1
superfamily with growth suppressing properties in lung cancer.
Cancer Res. 59:35–43. 1999.PubMed/NCBI
|
|
9
|
Kikuchi S, Yamada D, Fukami T, Masuda M,
Sakurai-Yageta M, Williams YN, Maruyama T, Asamura H, Matsuno Y,
Onizuka M and Murakami Y: Promoter methylation of DAL-1/4.1B
predicts poor prognosis in non-small cell lung cancer. Clin Cancer
Res. 11:2954–2961. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang H, Yan X, Pan Y, Wang Y, Wang N, Li
L, Liu Y, Chen X, Zhang CY, Gu H and Zen K: MicroRNA-223 delivered
by platelet-derived microvesicles promotes lung cancer cell
invasion via targeting tumor suppressor EPB41L3. Mol Cancer.
14:582015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yageta M, Kuramochi M, Masuda M, Fukami T,
Fukuhara H, Maruyama T, Shibuya M and Murakami Y: Direct
association of TSLC1 and DAL-1, two distinct tumor suppressor
proteins in lung cancer. Cancer Res. 62:5129–5133. 2002.PubMed/NCBI
|
|
12
|
Gutmann DH, Donahoe J, Perry A, Lemke N,
Gorse K, Kittiniyom K, Rempel SA, Gutierrez JA and Newsham IF: Loss
of DAL-1, a protein 4.1-related tumor suppressor, is an important
early event in the pathogenesis of meningiomas. Hum Mol Genet.
9:1495–1500. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tran Y, Benbatoul K, Gorse K, Rempel S,
Futreal A, Green M and Newsham I: Novel regions of allelic deletion
on chromosome 18p in tumors of the lung, brain and breast.
Oncogene. 17:3499–3505. 1998. View Article : Google Scholar
|
|
14
|
Nunes F, Shen Y, Niida Y, Beauchamp R,
Stemmer- Rachamimov AO, Ramesh V, Gusella J and MacCollin M:
Inactivation patterns of NF2 and DAL-1/4.1B (EPB41L3) in sporadic
meningioma. Cancer Genet Cytogenet. 162:135–139. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Robb VA, Gerber MA, Hart-Mahon EK and
Gutmann DH: Membrane localization of the U2 domain of Protein 4.1B
is necessary and sufficient for meningioma growth suppression.
Oncogene. 24:1946–1957. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gerber MA, Bahr SM and Gutmann DH: Protein
4.1B/differentially expressed in adenocarcinoma of the lung-1
functions as a growth suppressor in meningioma cells by activating
Rac1-dependent c-Jun-NH(2)-kinase signaling. Cancer Res.
66:5295–5303. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kittiniyom K, Mastronardi M, Roemer M,
Wells WA, Greenberg ER, Titus-Ernstoff L and Newsham IF:
Allele-specific loss of heterozygosity at the DAL-1/4.1B (EPB41L3)
tumor-suppressor gene locus in the absence of mutation. Genes
Chromosomes Cancer. 40:190–203. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kittiniyom K, Gorse KM, Dalbegue F, Lichy
JH, Taubenberger JK and Newsham IF: Allelic loss on chromosome band
18p11.3 occurs early and reveals heterogeneity in breast cancer
progression. Breast Cancer Res. 3:192–198. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dafou D, Grun B, Sinclair J, Lawrenson K,
Benjamin EC, Hogdall E, Kruger-Kjaer S, Christensen L, Sowter HM,
Al-Attar A, et al: Microcell-mediated chromosome transfer
identifies EPB41L3 as a functional suppressor of epithelial ovarian
cancers. Neoplasia. 12:579–589. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bernkopf DB and Williams ED: Potential
role of EPB41L3 (protein 4.1B/Dal-1) as a target for treatment of
advanced prostate cancer. Expert Opin Ther Targets. 12:845–853.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wong SY, Haack H, Kissil JL, Barry M,
Bronson RT, Shen SS, Whittaker CA, Crowley D and Hynes RO: Protein
4.1B suppresses prostate cancer progression and metastasis. Proc
Natl Acad Sci USA. 104:12784–12789. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Z, Zhang J, Ye M, Zhu M, Zhang B, Roy
M, Liu J and An X: Tumor suppressor role of protein 4.1B/DAL-1.
Cell Mol Life Sci. 71:4815–4830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Holzwarth G, Yu J and Steck TL:
Heterogeneity in the conformation of different protein fractions
from the human erythrocyte membrane. J Supramol Struct. 4:161–168.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Parra M, Gascard P, Walensky LD, Gimm JA,
Blackshaw S, Chan N, Takakuwa Y, Berger T, Lee G, Chasis JA, et al:
Molecular and functional characterization of protein 4.1B, a novel
member of the protein 4.1 family with high level, focal expression
in brain. J Biol Chem. 275:3247–3255. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Leto TL and Marchesi VT: A structural
model of human erythrocyte protein 4.1. J Biol Chem. 259:4603–4608.
1984.PubMed/NCBI
|
|
26
|
Busam RD, Thorsell AG, Flores A,
Hammarström M, Persson C, Öbrink B and Hallberg BM: Structural
basis of tumor suppressor in lung cancer 1 (TSLC1) binding to
differentially expressed in adenocarcinoma of the lung
(DAL-1/4.1B). J Biol Chem. 286:4511–4516. 2011. View Article : Google Scholar :
|
|
27
|
Nagata M, Sakurai-Yageta M, Yamada D, Goto
A, Ito A, Fukuhara H, Kume H, Morikawa T, Fukayama M, Homma Y and
Murakami Y: Aberrations of a cell adhesion molecule CADM4 in renal
clear cell carcinoma. Int J Cancer. 130:1329–1337. 2012. View Article : Google Scholar
|
|
28
|
Sakurai-Yageta M, Masuda M, Tsuboi Y, Ito
A and Murakami Y: Tumor suppressor CADM1 is involved in epithelial
cell structure. Biochem Biophys Res Commun. 390:977–982. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Singh V, Miranda TB, Jiang W, Frankel A,
Roemer ME, Robb VA, Gutmann DH, Herschman HR, Clarke S and Newsham
IF: DAL-1/4.1B tumor suppressor interacts with protein arginine
N-methyltransferase 3 (PRMT3) and inhibits its ability to methylate
substrates in vitro and in vivo. Oncogene. 23:7761–7771. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang W, Roemer ME and Newsham IF: The
tumor suppressor DAL-1/4.1B modulates protein arginine
N-methyltransferase 5 activity in a substrate-specific manner.
Biochem Biophys Res Commun. 329:522–530. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Horresh I, Bar V, Kissil JL and Peles E:
Organization of myelinated axons by Caspr and Caspr2 requires the
cytoskeletal adapter protein 4.1B. J Neurosci. 30:2480–2489. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu T, Robb VA, Singh V, Gutmann DH and
Newsham IF: The 4.1/ezrin/radixin/moesin domain of the
DAL-1/Protein 4.1B tumour suppressor interacts with 14-3-3
proteins. Biochem J. 365:783–789. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gimm JA, An X, Nunomura W and Mohandas N:
Functional characterization of spectrin-actin-binding domains in
4.1 family of proteins. Biochemistry. 41:7275–7282. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Discher DE, Winardi R, Schischmanoff PO,
Parra M, Conboy JG and Mohandas N: Mechanochemistry of protein
4.1's spectrin-actin-binding domain: Ternary complex interactions,
membrane binding, network integration, structural strengthening. J
Cell Biol. 130:897–907. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Discher D, Parra M, Conboy JG and Mohandas
N: Mechanochemistry of the alternatively spliced spectrin-actin
binding domain in membrane skeletal protein 4.1. J Biol Chem.
268:7186–7195. 1993.PubMed/NCBI
|
|
36
|
Kontrogianni-Konstantopoulos A, Huang SC
and Benz EJ Jr: A nonerythroid isoform of protein 4.1R interacts
with components of the contractile apparatus in skeletal myofibers.
Mol Biol Cell. 11:3805–3817. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McCarty JH, Cook AA and Hynes RO: An
interaction between {alpha}v{beta}8 integrin and Band 4.1B via a
highly conserved region of the Band 4.1 C-terminal domain. Proc
Natl Acad Sci USA. 102:13479–13483. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang Y, Xu R, Li G, Xie X, Long J and
Wang H: Loss of expression of the differentially expressed in
adenocarcinoma of the lung (DAL-1) protein is associated with
metastasis of non-small cell lung carcinoma cells. Tumour Biol.
33:1915–1925. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Charboneau AL, Singh V, Yu T and Newsham
IF: Suppression of growth and increased cellular attachment after
expression of DAL-1 in MCF-7 breast cancer cells. Int J Cancer.
100:181–188. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Qiu X, Guan X, Liu W and Zhang Y: DAL-1
attenuates epithelial to mesenchymal transition and metastasis by
suppressing HSPA5 expression in non-small cell lung cancer. Oncol
Rep. 38:3103–3113. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen X, Guan X, Zhang H, Xie X, Wang H,
Long J, Cai T, Li S, Liu Z and Zhang Y: DAL-1 attenuates
epithelial-to mesenchymal transition in lung cancer. J Exp Clin
Cancer Res. 34:32015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu F, Yang H, Zhang Z, Wang Z and Xiong J:
DAL-1/4.1B contributes to epithelial-mesenchymal transition via
regulation of transforming growth factor-β in lung cancer cell
lines. Mol Med Rep. 12:6072–6078. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Heller G, Fong KM, Girard L, Seidl S,
End-Pfützenreuter A, Lang G, Gazdar AF, Minna JD, Zielinski CC and
Zöchbauer-Müller S: Expression and methylation pattern of TSLC1
cascade genes in lung carcinomas. Oncogene. 25:959–968. 2006.
View Article : Google Scholar
|
|
44
|
Martín-Sánchez E, Pernaut-Leza E, Mendaza
S, Cordoba A, Vicente-Garcia F, Monreal-Santesteban I, Vizcaino JP,
De Cerio MJ, Perez-Janices N, Blanco-Luquin I, et al: Gene promoter
hypermethylation is found in sentinel lymph nodes of breast cancer
patients, in samples identified as positive by one-step nucleic
acid amplification of cytokeratin 19 mRNA. Virchows Arch.
469:51–59. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jiang W and Newsham IF: The tumor
suppressor DAL-1/4.1B and protein methylation cooperate in inducing
apoptosis in MCF-7 breast cancer cells. Mol Cancer. 5:42006.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schulz WA, Alexa A, Jung V, Hader C,
Hoffmann MJ, Yamanaka M, Fritzsche S, Wlazlinski A, Müller M,
Lengauer T, et al: Factor interaction analysis for chromosome 8 and
DNA methylation alterations highlights innate immune response
suppression and cytoskeletal changes in prostate cancer. Mol
Cancer. 6:142007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schulz WA, Ingenwerth M, Djuidje CE, Hader
C, Rahnenführer J and Engers R: Changes in cortical cytoskeletal
and extracellular matrix gene expression in prostate cancer are
related to oncogenic ERG deregulation. BMC Cancer. 10:5052010.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Vasiljević N, Ahmad AS, Carter PD, Fisher
G, Berney DM, Foster CS, Cuzick J and Lorincz AT: DNA methylation
of PITX2 predicts poor survival in men with prostate cancer.
Biomark Med. 8:1143–1150. 2014. View Article : Google Scholar
|
|
49
|
Schulz WA and Hoffmann MJ: Epigenetic
mechanisms in the biology of prostate cancer. Semin Cancer Biol.
19:172–180. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li X, Zhou F, Jiang C, Wang Y, Lu Y, Yang
F, Wang N, Yang H, Zheng Y and Zhang J: Identification of a DNA
methylome profile of esophageal squamous cell carcinoma and
potential plasma epigenetic biomarkers for early diagnosis. PLoS
One. 9:e1031622014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Maher SG, Gillham CM, Duggan SP, Smyth PC,
Miller N, Muldoon C, O'Byrne KJ, Sheils OM, Hollywood D and
Reynolds JV: Gene expression analysis of diagnostic biopsies
predicts pathological response to neoadjuvant chemoradiotherapy of
esophageal cancer. Ann Surg. 250:729–737. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zeng R, Huang JP, Li XF, Xiong WB, Wu G,
Jiang ZJ, Song SJ, Li JQ, Zheng YF and Zhang JR: Epb41l3 suppresses
esophageal squamous cell carcinoma invasion and inhibits MMP2 and
MMP9 expression. Cell Biochem Funct. 34:133–141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zeng R, Liu Y, Jiang ZJ, Huang JP, Wang Y,
Li XF, Xiong WB, Wu XC, Zhang JR, Wang QE and Zheng YF: EPB41L3 is
a potential tumor suppressor gene and prognostic indicator in
esophageal squamous cell carcinoma. Int J Oncol. Mar 14–2018.Epub
ahead of print.
|
|
54
|
Verlaat W, Van Leeuwen RW, Novianti PW,
Schuuring E, Meijer CJLM, Van Der Zee AGJ, Snijders PJF, Heideman
DAM, Steenbergen RDM and Wisman GBA: Host-cell DNA methylation
patterns during high-risk HPV-induced carcinogenesis reveal a
heterogeneous nature of cervical pre-cancer. Epigenetics.
13:769–778. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vasiljević N, Scibior-Bentkowska D,
Brentnall AR, Cuzick J and Lorincz AT: Credentialing of DNA
methylation assays for human genes as diagnostic biomarkers of
cervical intraepithelial neoplasia in high-risk HPV positive women.
Gynecol Oncol. 132:709–714. 2014. View Article : Google Scholar
|
|
56
|
Brentnall AR, Vasiljević N,
Scibior-Bentkowska D, Cadman L, Austin J, Szarewski A, Cuzick J and
Lorincz AT: A DNA methylation classifier of cervical precancer
based on human papillomavirus and human genes. Int J Cancer.
135:1425–1432. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Boers A, Wang R, van Leeuwen RW, Klip HG,
de Bock GH, Hollema H, van Criekinge W, de Meyer T, Denil S, van
der Zee AGJ, et al: Discovery of new methylation markers to improve
screening for cervical intraepithelial neoplasia grade 2/3. Clin
Epigenetics. 8:292016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Louvanto K, Franco EL, Ramanakumar AV,
Vasiljević N, Scibior-Bentkowska D, Koushik A, Cuzick J, Coutlée F
and Lorincz AT; Biomarkers of Cervical Cancer Risk Study Team:
Methylation of viral and host genes and severity of cervical
lesions associated with human papillomavirus type 16. Int J Cancer.
136:E638–E645. 2015. View Article : Google Scholar
|
|
59
|
Kelly HA, Chikandiwa A, Warman R, Segondy
M, Sawadogo B, Vasiljević N, Didelot MN, Meda N, Weiss HA,
Delany-Moretlwe S, et al: Associations of human gene EPB41L3 DNA
methylation and cervical intraepithelial neoplasia in women living
with HIV-1 in Africa. AIDS. 32:2227–2236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Clarke MA, Luhn P, Gage JC, Bodelon C,
Dunn ST, Walker J, Zuna R, Hewitt S, Killian JK, Yan L, et al:
Discovery and validation of candidate host DNA methylation markers
for detection of cervical precancer and cancer. Int J Cancer.
141:701–710. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cuschieri K, Ronco G, Lorincz A, Smith L,
Ogilvie G, Mirabello L, Carozzi F, Cubie H, Wentzensen N, Snijders
P, et al: Eurogin roadmap 2017: Triage strategies for the
management of HPV-positive women in cervical screening programs.
Int J Cancer. 143:735–745. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lorincz AT, Brentnall AR,
Scibior-Bentkowska D, Reuter C, Banwait R, Cadman L, Austin J,
Cuzick J and Vasiljević N: Validation of a DNA methylation HPV
triage classifier in a screening sample. Int J Cancer.
138:2745–2751. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Nedjai B, Reuter C, Ahmad A, Banwait R,
Warman R, Carton J, Boer S, Cuzick J and Lorincz AT: Molecular
progression to cervical precancer, epigenetic switch or sequential
model? Int J Cancer. Apr 21–2018.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Huisman C, van der Wijst MG, Falahi F,
Overkamp J, Karsten G, Terpstra MM, Kok K, van der Zee AG,
Schuuring E, Wisman GB and Rots MG: Prolonged re-expression of the
hypermethylated gene EPB41L3 using artificial transcription factors
and epigenetic drugs. Epigenetics. 10:384–396. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Brentnall AR, Vasiljević N,
Scibior-Bentkowska D, Cadman L, Austin J, Cuzick J and Lorincz AT:
HPV33 DNA methylation measurement improves cervical pre-cancer risk
estimation of an HPV16, HPV18, HPV31 and \textit{EPB41L3}
methylation classifier. Cancer Biomark. 15:669–675. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lorincz AT: Virtues and weaknesses of DNA
methylation as a test for cervical cancer prevention. Acta Cytol.
60:501–512. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
van Leeuwen RW, Ostrbenk A, Poljak M, van
der Zee AGJ, Schuuring E and Wisman GBA: DNA methylation markers as
a triage test for identification of cervical lesions in a high risk
human papillomavirus positive screening cohort. Int J Cancer.
144:746–754. 2019. View Article : Google Scholar :
|
|
68
|
Eijsink JJ, Lendvai Á, Deregowski V, Klip
HG, Verpooten G, Dehaspe L, de Bock GH, Hollema H, van Criekinge W,
Schuuring E, et al: A four-gene methylation marker panel as triage
test in high-risk human papillomavirus positive patients. Int J
Cancer. 130:1861–1869. 2012. View Article : Google Scholar
|
|
69
|
Lendvai Á, Johannes F, Grimm C, Eijsink
JJ, Wardenaar R, Volders HH, Klip HG, Hollema H, Jansen RC,
Schuuring E, et al: Genome-wide methylation profiling identifies
hypermethylated biomarkers in high-grade cervical intraepithelial
neoplasia. Epigenetics. 7:1268–1278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Perez-Janices N, Blanco-Luquin I, Tuñón
MT, Barba-Ramos E, Ibáñez B, Zazpe-Cenoz I, Martinez-Aguillo M,
Hernandez B, Martínez-Lopez E, Fernández AF, et al: EPB41L3, TSP-1
and RASSF2 as new clinically relevant prognostic biomarkers in
diffuse gliomas. Oncotarget. 6:368–380. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ohno N, Terada N, Murata S, Yamakawa H,
Newsham IF, Katoh R, Ohara O and Ohno S: Immunolocalization of
protein 4.1B/DAL-1 during neoplastic transformation of mouse and
human intestinal epithelium. Histochem Cell Biol. 122:579–586.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kume H, Muraoka S, Kuga T, Adachi J,
Narumi R, Watanabe S, Kuwano M, Kodera Y, Matsushita K, Fukuoka J,
et al: Discovery of colorectal cancer biomarker candidates by
membrane proteomic analysis and subsequent verification using
selected reaction monitoring (SRM) and tissue microarray (TMA)
analysis. Mol Cell Proteomics. 13:1471–1484. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Niwa T, Tsukamoto T, Toyoda T, Mori A,
Tanaka H, Maekita T, Ichinose M, Tatematsu M and Ushijima T:
Inflammatory processes triggered by Helicobacter pylori infection
cause aberrant DNA methylation in gastric epithelial cells. Cancer
Res. 70:1430–1440. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sugimoto K, Ito T, Hulbert A, Chen C,
Orita H, Maeda M, Moro H, Fukagawa T, Ushijima T, Katai H, et al:
DNA methylation genome-wide analysis in remnant and primary gastric
cancers. Gastric Cancer. Mar 12–2019. View Article : Google Scholar : Epub ahead of
print. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Terada N, Ohno N, Yamakawa H, Baba T,
Fujii Y, Christofori G, Ohara O and Ohno S: Protein 4.1B in mouse
islets of Langerhans and beta-cell tumorigenesis. Histochem Cell
Biol. 120:277–283. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhu L, Yang N, Chen J, Zeng T, Yan S, Liu
Y, Yu G, Chen Q, Du G, Pan W, et al: LINC00052 upregulates EPB41L3
to inhibit migration and invasion of hepatocellular carcinoma by
binding miR-452-5p. Oncotarget. 8:63724–63737. 2017.PubMed/NCBI
|
|
77
|
Giuliano AR, Nedjai B, Lorincz AT, Schell
MJ, Rahman S, Banwait R, Boulware D, Sirak B, Martin-Gomez L,
Abrahamsen M, et al: Methylation of HPV 16 and EPB41L3 in oral
gargles: Associations with oropharyngeal cancer detection and tumor
characteristics. Int J Cancer. Jul 15–2019.Epub ahead of print.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Lorincz AT, Nathan M, Reuter C, Warman R,
Thaha MA, Sheaff M, Vasiljević N, Ahmad A, Cuzick J and Sasieni P:
Methylation of HPV and a tumor suppressor gene reveals anal cancer
and precursor lesions. Oncotarget. 8:50510–50520. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Yamada D, Kikuchi S, Williams YN,
Sakurai-Yageta M, Masuda M, Maruyama T, Tomita K, Gutmann DH,
Kakizoe T, Kitamura T, et al: Promoter hypermethylation of the
potential tumor suppressor DAL-1/4.1B gene in renal clear cell
carcinoma. Int J Cancer. 118:916–923. 2006. View Article : Google Scholar
|
|
80
|
Jiang SS, Chen CH, Tseng KY, Tsai FY, Wang
MJ, Chang IS, Lin JL and Lin S: Gene expression profiling suggests
a pathological role of human bone marrow-derived mesenchymal stem
cells in aging-related skeletal diseases. Aging (Albany NY).
3:672–684. 2011. View Article : Google Scholar
|
|
81
|
Heller G, Geradts J, Ziegler B, Newsham I,
Filipits M, Markis-Ritzinger EM, Kandioler D, Berger W, Stiglbauer
W, Depisch D, et al: Downregulation of TSLC1 and DAL-1 expression
occurs frequently in breast cancer. Breast Cancer Res Treat.
103:283–291. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Li L, Li S, Cai T, Wang H, Xie X, Liu Z
and Zhang Y: The targeted inhibitory effects of human amniotic
fluid stem cells carrying CXCR4 promoter and DAL-1 on non-small
cell lung carcinoma growth. Gene Ther. 23:214–222. 2016. View Article : Google Scholar
|