Introduction
There are >200,000 newly-diagnosed cases of
kidney cancer each year globally according to a statistic
calculated in 2013, with the highest incidence in North America and
the lowest incidence in Asia and Africa (1). Histologically, kidney cancer can be
divided into two common types: Renal cell carcinoma (RCC) and
transitional cell carcinoma (2).
RCC is the most common type of kidney cancer, accounting for 90-95%
of cases in adults (3). The
incidence and mortality of kidney cancer have also increased over
the past decades at a rate of 2-3% per decade (3). Generally, RCC is asymptomatic, and
is, therefore, frequently diagnosed at an advanced stage (4), and up to 30% of patients with RCC
exhibit a metastatic tumor at diagnosis (3). To date, surgery remains the most
effective treatment for RCC, whereas it is usually resistant to
conventional chemoradiotherapy (5). Therefore, an improved understanding
of the biology of RCC can help us develop novel therapeutic
strategies and identify biomarkers for early detection and
prediction of prognosis and treatment responses, leading to
enhanced effectiveness of RCC control in clinical practice.
Previous studies have revealed that RCC development
is associated with gene mutations in chromosome 3p, which activate
oncogenes, such as c-Met, or inactivate tumor suppressor genes,
such as VHL (6,7). However, alterations of various genes
and gene pathways have also been associated with the development
and progression of RCC (8-12). The present study of the gene
alterations in RCC focused on eukaryotic initiation factors (eIFs).
Aberrant expression levels of eIFs have been observed in several
types of human cancer (13). eIFs
regulate the initiation of protein translation in eukaryotic cells
(14). For example, eIF3, the
largest initiation factor, binds to the 40S ribosomal subunit,
different initiation factors and mRNA to facilitate protein
translation in cells (15,16). Overexpression or underexpression of
a particular elF3 subunit is associated with the development and
progression of a number of tumors, including lung cancer, breast
cancer, hepatocellular cancer and intestinal cancer (17). During carcinogenesis and tumor
progression, gene transcription and protein translation are usually
upregulated in tumor cells (18).
This has been confirmed in various previous studies of eukaryotic
initiation factor 3c (eIF3C) in testicular seminoma (19), meningiomas (20), glioma (21,22),
colorectal cancer (22),
hepatocellular carcinoma (24,25)
and breast cancer (26).
Nevertheless, to the best of our knowledge, the role of eIF3C in
RCC has not been assessed.
In a preliminary experiment, the eIF3C mRNA level
was higher in RCC tissue than in adjacent normal tissues (Fig. S1). Therefore, the present study
aimed to evaluate whether eIF3C could be used as a potential
diagnostic marker or therapeutic target for RCC. To address this,
eIF3C expression was assessed in RCC and normal kidney tissues, and
the role of eIF3C in RCC malignant behavior was examined in
vitro and in vivo.
Materials and methods
Patients and tissue collection
A total of 16 pairs of tumor and matched distant
normal tissues were collected from patients with RCC (11 men and 5
women; median age at diagnosis, 54 years; age range, 37-74 years)
who underwent radical resection between February 2016 and July 2016
at the Third Affiliated Hospital of Soochow University. Distant
normal tissues were obtained >5 cm away from tumors to ensure
their normality. All patients were histologically diagnosed with
RCC and did not receive any pre-surgery chemotherapy or
radiotherapy. The present study was approved by the Ethics
Committee of the Third Affiliated Hospital of Soochow University
according to the principles of the Declaration of Helsinki. Written
informed consent was provided by all participants prior to
enrollment.
Immunohistochemistry
Tissues were fixed in 10% formalin at 4°C for 10 h.
Fixed and paraffin-embedded tissue blocks were retrieved from the
Pathology Department of The Third Affiliated Hospital of Soochow
University and cut into 4-µm thick sections. For
immunostaining of the eIF3C protein, the sections were
deparaffinized in xylene and rehydrated in descending alcohol
series. Antigens were repaired by heating the tissue sections at
100°C for 30 min in citrate (10 mmol/l; pH 6.0) (Beyotime). Then,
the sections were immersed in a 0.3% hydrogen peroxide solution for
30 min to block endogenous peroxidase activity, rinsed in PBS for 5
min, blocked with 3% BSA (Beyotime Institute of Biotechnology) at
room temperature for 30 min, and incubated with a primary antibody
at 4°C overnight according to the manufacturer's instructions.
Subsequently, the sections were incubated with the secondary
antibody at 37°C for 30 min. The antigen-antibody complex was
visualized after adding 3,3′-diaminobenzidine (DAB) for 2 min at
room temperature. The primary antibody against eIF3C (1:1,000
dilution; cat. no. ab170841) was purchased from Abcam, and the
secondary antibody and DAB were part of the MaxVision™ horseradish
peroxidase (HRP)-Polymer anti-Mouse/Rabbit IHC kit (cat. no. 5010;
Maxim Biotech, Inc.).
The immunostained sections were independently
reviewed by two experienced pathologists who were blinded to the
clinical parameters of the patients, and scores were evaluated from
five randomly selected ×20 microscopic fields under a light
microscope (Olympus Corporation), according to a previously
described H-score method (25).
The H-score = (% unstained tumor cells ×0) + (% weakly stained
tumor cells ×1) + (% moderately stained tumor cells ×2) + (%
strongly stained tumor cells ×3). The H-scores ranged between 0
(100% negative staining) and 300 (100% strong staining).
Cell lines and culture conditions
Four human RCC lines ACHN, 786-O, Caki-1 and A498,
were obtained from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences in September 2017 and maintained in
RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS; Biological
Industries) and 1% penicillin-streptomycin (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C in a humidified incubator with 5%
CO2. All cell lines were certified by Shanghai GeneChem
Co., Ltd. for short tandem repeat analysis on October 31, 2017, as
described in 2012 in American National Standards Institute Standard
by the American Type Culture Collection Standards Development
Organization (28) and in
Capes-Davis et al (29).
All cell lines were passaged <30 times.
Short hairpin RNA (shRNA) and cell
transfection
Lentiviruses carrying eIF3C shRNA (cat. no. PSC2752)
targeting the DNA sequence of 5′-GTC ACT AAA GGT CTG TTT A-3′, and
negative control shRNA (cat. no. PSC3741) with a targeting sequence
of 5′-TTC TCC GAA CGT GTC ACG T-3′ were obtained from Shanghai
GeneChem Co., Ltd. Both eIF3C and negative control shRNA were
designed and cloned into the GV115 vector (Shanghai GeneChem Co.,
Ltd.) double enzyme digested by AgeI/EcoRI.
Reconstructed vectors with eIF3C-shRNA and the negative control
shRNA were transformed into competent E. coli cells
(Shanghai GeneChem Co., Ltd.) and transformed cells in serum-free
LB solid medium (pH 7.0; Shanghai GeneChem Co., Ltd.) supplemented
with ampicillin (0.1 mg/ml; Genebase) were cultured at 37°C.
Positive colonies were selected by reverse
transcription-quantitative PCR (RT-qPCR) and sequencing. To
construct a stable eIF3C-depletion cell line, cells were seeded in
6-well plates at a density of 5×104 cells/well, grown
overnight, and transfected with eIF3C-shRNA (8×108
TU/ml) (sheIF3C) or negative control shRNA (1×109 TU/ml)
(shCtrl) lentivirus for 12 h. The culture medium was subsequently
replaced with fresh complete RPMI-1640 medium, and cells were grown
for an additional 72 h and subjected to fluorescence microscopy for
the visualization of the green fluorescence protein (GFP). The
efficiency of eIF3C depletion was evaluated through RT-qPCR and
western blotting.
RT-qPCR
Total cellular RNA was isolated from ACHN, 786-O,
A498 and Caki-1 cell lines using TRIzol® reagent
(Shanghai Pufei Biotechnology Co., Ltd.) and reverse transcribed
into cDNA using the M-MLV cDNA kit (Promega Corporation) according
to the manufacturer's instructions. The following steps were used:
Step 1, adding 2 µg total RNA, 1 µl Oligo(dT) and
RNase-Free H2O to 10 µl total volume, heating at
70°C for 5 sec; step 2, adding 4 µl 5X RT buffer, 2
µl 10 m MdNTPs, 0.4 µl Rnasin (40 U/µl), 1
µl M-MLV-RTase (200 U/µl) and 2.6 µl
RNase-Free H2O, heating at 42°C for 1 h and at 70°C for
10 min. Real-time quantitative polymerase chain reaction (RT-qPCR)
was performed to assess the expression levels of eIF3C using a SYBR
Master Mixture kit (cat. no. DRR041B) from Takara Biotechnology
Co., Ltd. according to the manufacturer's instructions. The
following thermocycling conditions were used: Stage 1, holding at
95°C for 30 sec, 1 cycle; stage 2, 2 steps PCR reacting at 95°C for
5 sec and at 60°C for 30 sec, respectively, 45 cycles; stage 3,
dissociating at 95°C for 15 sec, at 55°C for 30 sec and at 95°C for
15 sec, respectively, 1 cycle. GAPDH was used as an internal
control. The primers were designed as follows: Human eIF3C forward,
5′-AGA TGA GGA TGA GGA TGA GGA C-3′ and reverse, 5′-GGA ATC GGA AGA
TGT GGA ACC-3′; and human GAPDH forward, 5′-TGA CTT CAA CAG CGA CAC
CCA-3′ and reverse, 5′-CAC CCT GTT GCT GTA GCC AAA-3′. The relative
expression level of eIF3C was calculated using the
2−ΔΔCq method (30).
The experiments were performed in triplicate.
Celigo cell viability and proliferation
assay
Following depletion of elF3c expression, tumor cells
were subjected to a cell proliferation assay. Briefly, after 24 h
gene transfection, ACHN cells were seeded into 96-well plates at a
density of 2×103 cells/well and grown for up to 5 days.
On each day, the green fluorescence emission was observed and
recorded using a Celigo cytometer (Nexcelom Bioscience) to assess
the proliferation of tumor cells. The data are presented as the
mean ± SD (n=3).
The 96-well plates were subjected to an MTT assay
using the MTT reagent (Genview) and formazan was dissolved by DMSO
solution (Shiyi Corporation). An ELIASA microplate reader (Tecan
Group, Ltd.) was used and the optical density value at 490 nm was
used to estimate cell confluence. The data are presented as the
mean ± SD (n=3).
Cell cycle distribution assay
ACHN cells were cultured in 6-cm cell culture dishes
until they reached 80% confluence, and were then transfected for 5
days with a lentivirus targeting eIF3C or a negative control shRNA.
Cells were then harvested, washed with ice-cold D-Hanks solution,
and fixed with 75% ethyl alcohol at 4°C for at least 1 h.
Subsequently, cells were washed with ice-cold D-Hanks solution and
stained with 40X propidium iodide solution (2 mg/ml), 100X RNase
(10 mg/ml) and 1X D-Hanks solution according to the manufacturer's
instructions (Sigma-Aldrich; Merck KGaA). The cell cycle
distributions were then analyzed using Guava easyCyte HT flow
cytometry (EMD Millipore) and the results were analyzed by ModFit
LT 4.0 software (Verity Software House, Inc.).
Cell apoptosis assay
ACHN cells were cultured in 6-cm cell culture dishes
until they reached 80% confluence, and were transfected for 5 days
with a lentivirus targeting eIF3C or a negative control shRNA.
Transfected cells were harvested, washed with ice-cold D-Hanks
solution and fixed with 75% ethyl alcohol at 4°C for at least 1 h.
Staining was performed using Annexin V-APC apoptosis detection kit
(eBioscience; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Briefly, the cells were collected by
centrifugation at 265 × g at 4°C for 5 min, washed with D-Hanks
solution and then mixed with 1X binding buffer. Subsequently, 200
µl cell suspension was thoroughly mixed with 10 µl
Annexin V solution, followed by incubation in the dark at room
temperature for 10-15 min. Cells were then subjected to Guava
easyCyte HT flow cytometry (EMD Millipore) and the results were
analyzed by ModFit LT 4.0 software (Verity Software House,
Inc.).
Nude mouse tumor xenograft formation
assay
The animal protocol was approved by the
Institutional Animal Care and Use Committee of the Third Affiliated
Hospital of Soochow University (Changzhou, China). Female BALB/c
nude mice (n=14; 4 weeks old; average weight, 18.7±1.75 g) were
obtained from Shanghai SLAC Laboratory Animal Co., Ltd. All mice
were raised under specific pathogen-free conditions (23±3°C;
relative humidity, 40-70%) under a 12 h light/dark cycle. All mice
were adaptively fed with free access to water and standard mouse
chow. Animals were randomly divided into two experimental groups
and subcutaneously injected into the right forelimb armpit with
1×107 786-O cells (in ~200 µl PBS) following
transfection with a lentivirus carrying eIF3C- or scrambled shRNA.
Mice were regularly monitored for weight, health and xenograft size
for 7 weeks. Subsequently, all mice were euthanized by injection of
2% sodium pentobarbital (150 mg/kg of body weight), and after
complete coma, cervical dislocation was performed. The tumor volume
was measured for the greatest longitudinal diameter (length) and
the greatest transverse diameter (width) using a vernier caliper.
The tumor volume was calculated using the modified ellipsoidal
formula as follows: V=3.14/6 × L × W2, in which V
represents the whole volume of the tumor cell xenograft
(mm3), L indicates the length (mm), and W is the width
(mm).
Microarray and ingenuity pathway
analyses
Total cellular RNA was isolated from 786-O cells
after transfection with a lentivirus carrying eIF3C-shRNA (n=3) or
scrambled shRNA (n=3) using TRIzol® reagent (31). RNA quantity and quality were
evaluated using a NanoDrop 2000 spectrophotometer (Thermo Fisher
Scientific, Inc.) and RNA 2100 (Agilent Technologies, Inc.),
respectively, according to the manufacturers' instructions. The
genome-wide effects of eIF3C depletion were evaluated by GeneChip
PrimeView Human Affymetrix microarray (Affymetrix; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
resultant raw data and the differentially expressed genes (DEGs) in
the eIF3C-shRNA-infected RCC cell lines were identified based on
the criteria of an absolute fold change >1.3 and P<0.05. An
ingenuity pathway analysis (Ingenuity Systems; Qiagen, Inc.) was
performed to assess the functional and pathway annotations based on
all the DEGs.
Western blotting
Total cellular protein from tissue samples and cell
lines was extracted using an ice-cold RIPA (high) lysis buffer
(Beyotime Institute of Biotechnology) and centrifuged at 12,000 × g
at 4°C for 15 min. The protein concentration was determined using
the bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology). Equal amounts of protein (30 µg/µl)
were separated by 10% SDS-PAGE and then transferred onto PVDF
membranes (EMD Millipore). These membranes were blocked with 5%
fat-free milk in TBS-0.5% Tween-20 (TBS-T) at room temperature for
1 h and then incubated with primary antibodies at room temperature
for 2 h. The antibodies used in the present study were anti-c-JUN
(1:200 dilution; cat. no. ab32137; Abcam), anti-NFKB inhibitor α
(NFKBIA; 1:2,000 dilution; cat. no. ab7217; Abcam), anti-AKT
(1:1,000 dilution; cat. no. 9272; Cell Signaling Technology, Inc.),
anti-phosphorylated-(p-)AKT (1:1,000 dilution; cat. no. 13038; Cell
Signaling Technology, Inc.), anti-caspase-3 (1:1,000 dilution; cat.
no. 9662; Cell Signaling Technology, Inc.), anti-caspase-8 (1:1,000
dilution; cat. no. 4790; Cell Signaling Technology, Inc.),
anti-caspase-9 (1:1,000 dilution; cat. no. ab2324; Abcam) and
anti-GAPDH (1:2,000 dilution; cat. no. sc-32233; Santa Cruz
Biotechnology, Inc.). All aforementioned antibodies of caspases
could detect both the cleaved and total caspases. Blots were washed
three times in TBS-T and further incubated with a secondary
antibody goat anti-mouse immunoglobulin G (IgG)-HRP (1:5,000
dilution; cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) or goat
anti-rabbit IgG-HRP (1:5,000 dilution; cat. no. sc-2004; Santa Cruz
Biotechnology, Inc.) at room temperature for 90 min. Immunoreactive
protein bands were developed using the Pierce™ ECL Western Blotting
Substrate kit (Thermo Fisher Scientific, Inc.) and exposed to x-ray
films. The protein bands were semi-quantified using ImageJ v1.37
software (National Institutes of Health).
Statistical analysis
All data are presented as the mean ± SD of at least
three repeated experiments, and the difference between groups was
analyzed using Student's t-test by GraphPad Prism v6.0 software
(GraphPad Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
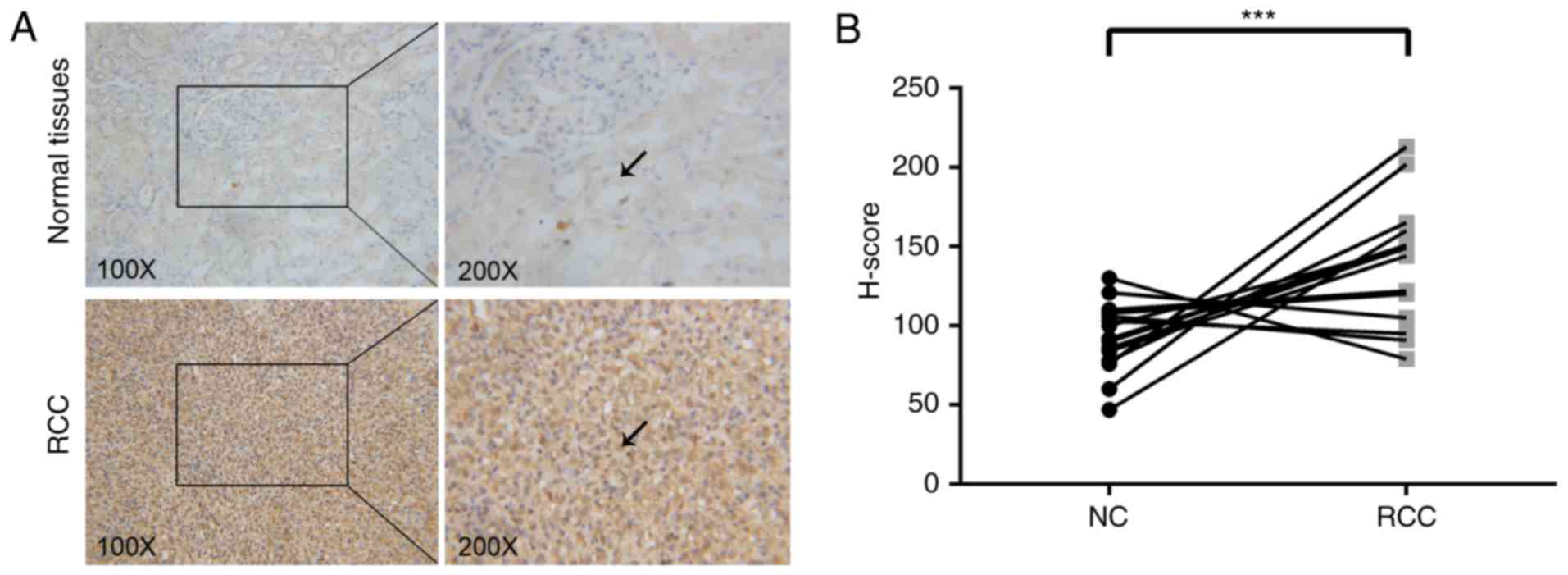
eIF3C expression is upregulated in RCC
tissues
The present study analyzed the expression levels of
eIF3C in 16 paired RCC and distant normal tissues using
immunohistochemistry. In almost all cases, eIF3C staining was
stronger in the RCC tissue compared with in the paired distant
normal tissue (Fig. 1A).
Subsequently, the immunostaining data was quantified using the
H-score. The H-score was significantly higher in the RCC tissue
compared with in the normal tissues (Fig. 1B), indicating that eIF3C may
contribute to the development or progression of RCC.
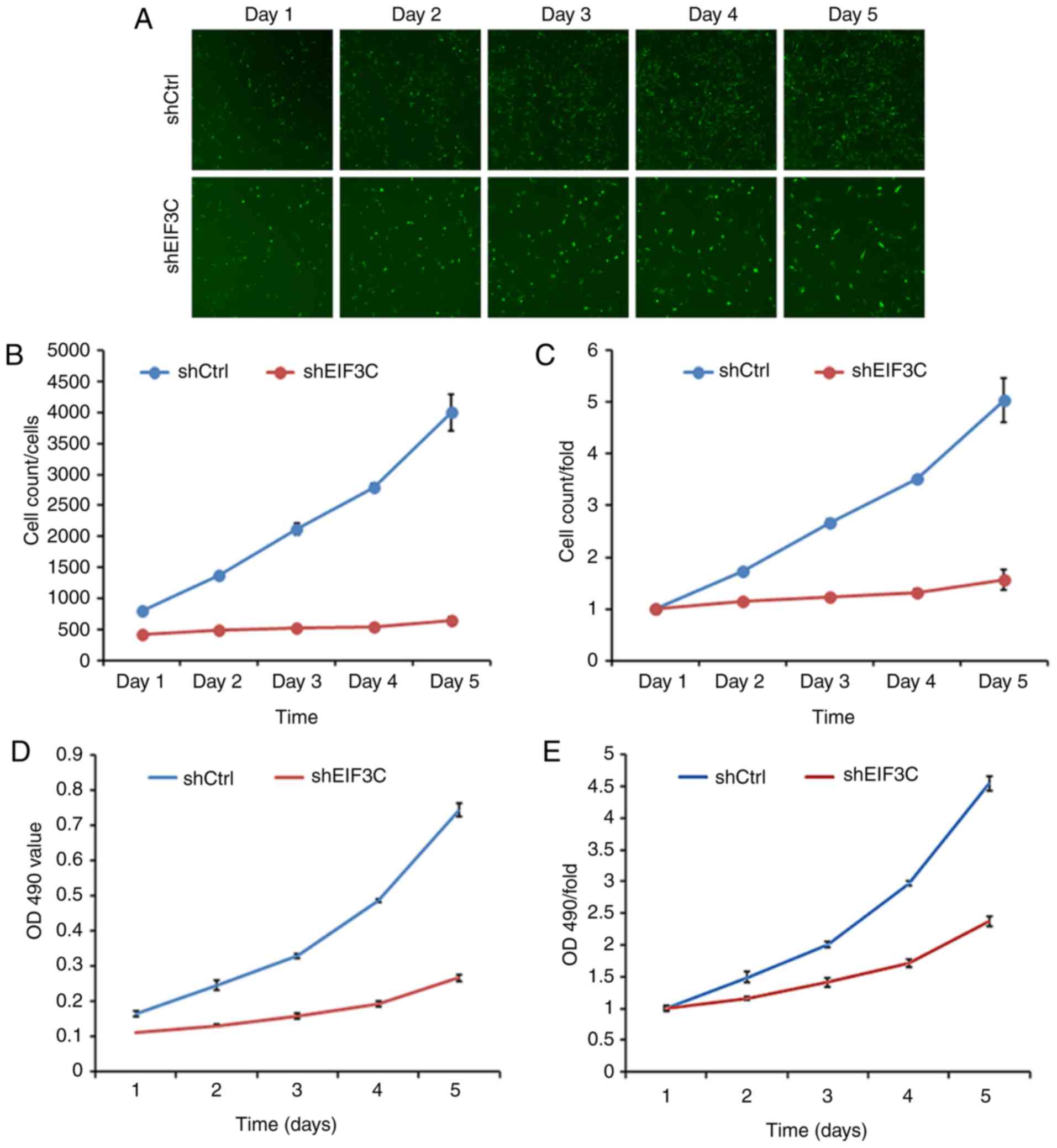
RCC cell proliferation is reduced and
cell cycle distribution is altered following eIF3C depletion
The present study assessed the mRNA expression
levels of eIF3C in four different RCC cell lines using RT-qPCR.
eIF3C expression was high in all four tested cell lines (Fig. 2F). Among them, eIF3C mRNA exhibited
the highest expression levels in 786-O cells and the lowest
expression levels in ACHN cells. Therefore, ACHN and 786-O cell
lines were selected for subsequent experiments in which eIF3C was
depleted (Fig. 2A), and
alterations in the tumor cell malignant phenotypes were explored.
Fig. 2G-I shows that transfection
with a lentivirus carrying eIF3C-shRNA resulted in significantly
decreased eIF3C mRNA and protein expression compared with
transfection with lentivirus carrying shCtrl.
Subsequently, the proliferation capacity of these
tumor cells was assessed using cell proliferation and viability
assays. The present study revealed that depletion of eIF3C
significantly reduced the viability and proliferation rate of ACHN
and 786-O cells compared with the control group (Fig. 2B-E). Additionally, the depletion of
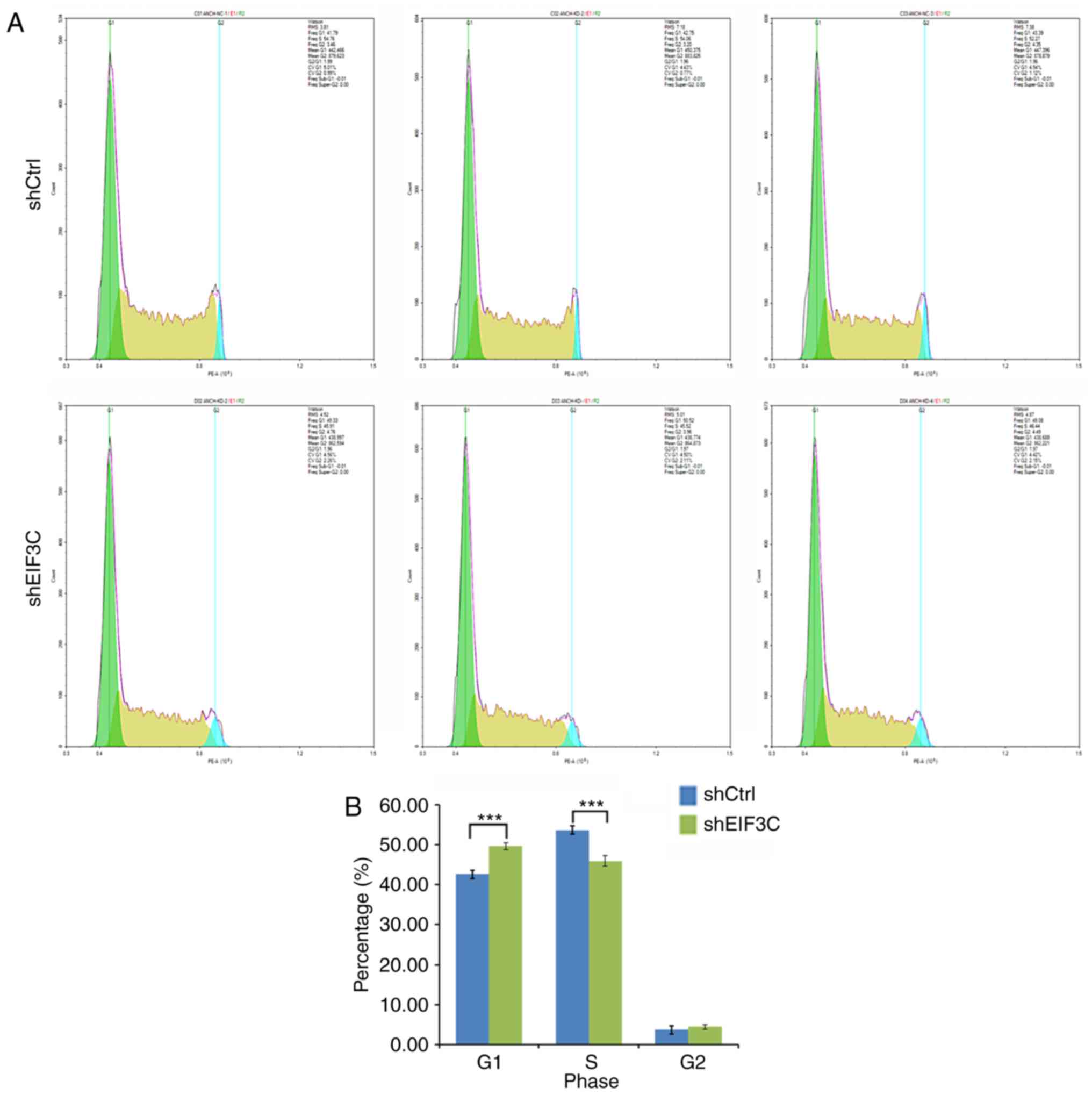
eIF3C remarkably altered the cell cycle distribution. There were
42.64±0.81, 53.70±1.28 and 3.67±0.60% of tumor cells in
G1, S and G2 phases, respectively, following
transfection with shCtrl, whereas there were 49.64±0.77, 45.96±0.46
and 4.40±0.41% of tumor cells in G1, S and G2
phases, respectively, after transfection with eIF3C-shRNA (Fig. 3A and B). The number of
G1-phase cells was significantly higher in the sheIF3C
group compared with in the shCtrl group (P<0.001). However, the
numbers of S-phase cells were lower in the sheIF3C group compared
with in the shCtrl group (P<0.001).
Depletion of eIF3C triggers apoptosis of
RCC cells
To assess the underlying mechanism of the reduced
cell viability following the depletion of eIF3C, an apoptosis assay
by flow cytometry as performed. The proportion of apoptotic cells
in eIF3C-depleted ACHN cells was ~2-fold higher compared with in
the cells transfected with scrambled shRNA (7.57±0.38 vs.
3.73±0.16%, P<0.01; Fig. 3C and
D). Additionally, the expression levels of caspase-3, caspase-8
and caspase-9 were detected, and it was ascertained that the
depletion of eIF3C could induce their upregulation with the
exception of caspase-8 (Fig. 3E and
F).
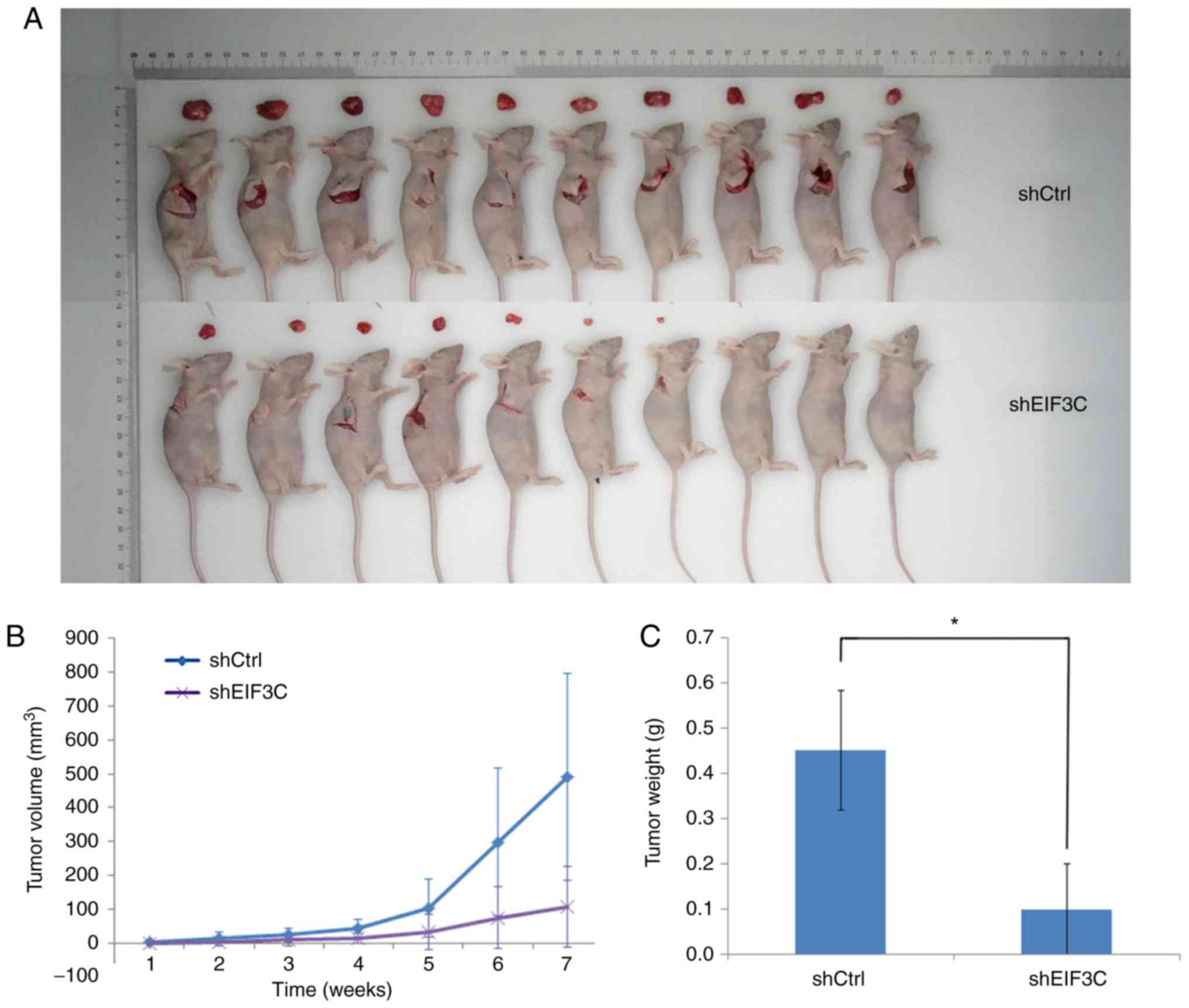
Effects of reduced eIF3C expression on
the formation and growth of nude mouse xenograft
The in vitro findings were further confirmed
using a nude mouse tumor cell xenograft assay. At the beginning of
the experiment, the average weights of the sheIF3C and shCtrl mice
were 18.5±2.32 and 18.9±0.99 g, respectively. At the end of the
experiment, the average body weights of the sheIF3C and shCtrl mice
were 22.9±2.30 and 23.0±0.96 g, respectively (data not shown). The
average tumor volume of the sheIF3C group was 106.72
mm3, whereas it was 491.61 mm3 in the shCtrl
group (Fig. 4). Additionally, the
average tumor xenograft weight in the sheIF3C group was
significantly lower compared with the shCtrl group (0.100±0.10 vs.
0.45±0.132 g; P<0.05; Fig.
4).
Effects of reduced eIF3C expression on
gene expression in vitro and in vivo
To explore the potential molecular events following
eIF3C depletion, a microarray analysis was conducted to identify
DEGs in eIF3C-depleted RCC 786-O cells. A total of 994 DEGs were
identified, including 516 upregulated and 478 downregulated genes
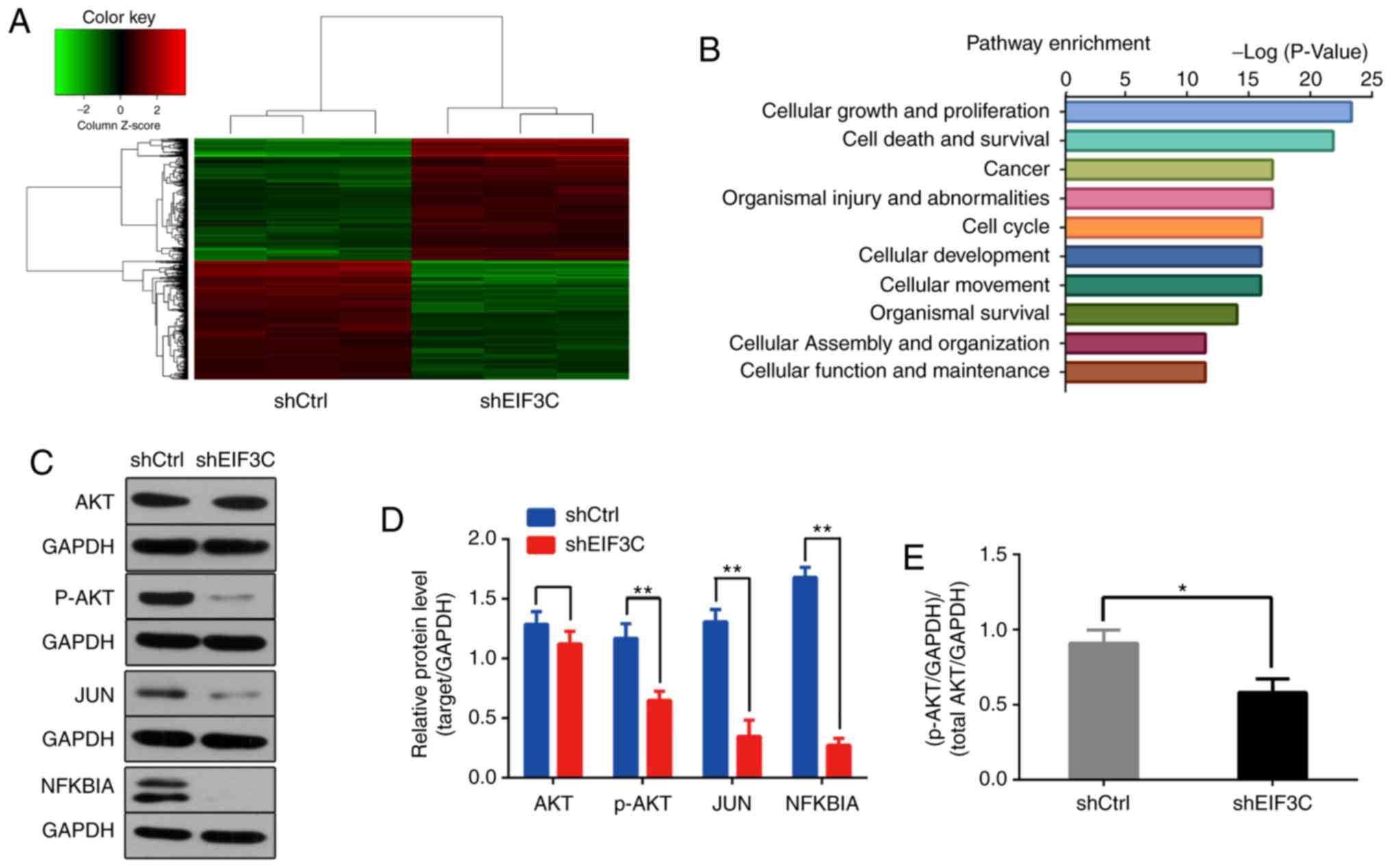
(Fig. 5A). The ingenuity pathway
analysis revealed that eIF3C-regulated genes were mainly involved
in the pathways of 'cell growth and proliferation', 'cell death and
survival', 'cancer', 'organismal injury and abnormalities', 'cell
cycle', 'cellular development' and 'cellular movement' (Fig. 5B). Common tumor-associated genes,
such as AKT, c-JUN and NFKBIA, whose expression levels changed
significantly, were selected. Western blotting results revealed
that all of them could be regulated by eIF3C (Fig. 5C and D).
Discussion
RCC is usually diagnosed at advanced stages of the
disease and is insensitive to chemoradiotherapy (5). Therefore, identification of novel
gene alterations and targets involved in this disease could help
medical oncologists effectively control RCC in clinical practice.
The present study first assessed eIF3C expression at the protein
level in RCC and normal kidney tissues and then investigated the
effects of eIF3C depletion on the regulation of malignant behaviors
of RCC cells in vitro and in nude mice. The data revealed
that eIF3C protein expression was significantly higher in RCC
tissues and cell lines compared with their corresponding controls.
The depletion of eIF3C reduced tumor cell proliferation, increased
the proportion of G1-phase tumor cells, and enhanced
tumor cell apoptosis compared with the controls. Furthermore,
depletion of eIF3C also inhibited the formation and growth of tumor
cell xenograft in nude mice. At the gene level, depletion of eIF3C
resulted in 516 upregulated and 478 downregulated genes. These
genes were involved in cell proliferation, survival and
cancer-related pathways, including the AKT, c-JUN and NFKBIA
signaling pathways. These findings were confirmed through western
blotting. Collectively, the data demonstrated that eIF3C exerted
oncogenic effects on the development and progression of RCC.
The proteins in the eIF family mainly function as
initiators of protein translation from their corresponding mRNAs in
eukaryotic cells (32). To date,
~11 eIF members have been reported to be involved in this process
(16). One of these protein family
members, eIF3, has 13 different subunits (eIF3a to eIF3 m)
(16) that can bind to the 40S
ribosome through the facilitation of methionyl-tRNA and mRNA
binding for protein translation (33). Previous studies have demonstrated
that the eIF3C subunit of eIF3 is a crucial binding partner for
eIF1 and eIF5 (34,35), and this subunit serves an essential
role in the selection of the translational start codon (36). During the development and
progression of cancer, protein synthesis is frequently upregulated,
and overexpression of eIF3C has been observed in various types of
cancer (19-26). However, to the best of our
knowledge, no studies have investigated the role of eIF3C in RCC.
The present study demonstrated that eIF3C expression was
upregulated in RCC tissues and cell lines. Zang et al
(37) have reported that
overexpression of eIF3b in tumors is associated with an aggressive
tumor phenotype and poor prognosis in patients with RCC. However,
as the present study had a small sample size, future studies with
large tissue sample sizes can further confirm the upregulation of
eIF3C and determine whether such upregulation is associated with
the prognosis or even the treatment response in RCC.
The data revealed that depletion of eIF3C inhibited
the malignant behavior in RCC, including a decrease in tumor cell
proliferation, increased apoptosis and altered cell cycle
progression. This corroborates the important role of eIF3C in the
development and progression of RCC. Indeed, a previous study has
demonstrated that eIF3C is able to induce exosome secretion and
promote angiogenesis and tumorigenesis in human hepatocellular
carcinoma (25). In breast cancer,
eIF3C targets the mTOR signaling pathway to inhibit cell
proliferation and induce apoptosis (26). Therefore, targeting eIF3C could be
a potential therapeutic approach for cancer treatment (13). Furthermore, the expression of other
eIF3 subunits could also be altered in RCC. For example, aberrant
eIF3e expression is essential for embryonic development and cell
proliferation (38), whereas
aberrant eIF3a expression occurs in non-small cell lung cancer,
which is associated with p27 expression and poor patient prognosis
(39). Additionally, eIF3b is able
to activate the β-catenin signaling pathway, leading to accelerated
progression of esophageal squamous cell carcinoma (40). Zhu et al (41) have reported that eIF3 h targets the
transforming growth factor-β and mitogen-activated protein kinase
signaling pathways in patients with hepatocellular carcinoma. Qi
et al (42) have reported
that overexpression of eIF3i can activate the synthesis of
prostaglandin-endoperoxide synthase 2 and β-catenin in colorectal
cancer. Therefore, it is necessary to further investigate the
functions of eIF3C in the development and progression of RCC.
In the present study, it was also observed that
depletion of eIF3C was able to suppress different signaling
pathways, including the Akt, c-JUN and NFKBIA singaling pathways.
This indicated that eIF3C promoted RCC malignant phenotypes in
vitro through these signaling pathways. Several eIFs family
members have been found to participate in these pathways indicated
above in various tumors, such as lung cancer, breast cancer,
hepatocellular cancer and intestinal cancer (17). A previous study has linked eIF1,
eIF5 and eIF6 to the PI3K/Akt/mTOR signaling pathway in colorectal
cancer (43), whereas Zang et
al (37) has demonstrated that
depletion of eIF3b suppresses the Akt pathway network in RCC cells,
including decreased expression of integrin/focal adhesion
kinase/Akt, Akt/mTOR/hypoxia-inducible factor/vascular endothelial
growth factor, Akt/mTOR/NF-κB, Akt/Bcl-2/Bax and Akt/glycogen
synthase kinase-3β pathway genes. Furthermore, Chen et al
(44) have demonstrated that eIF4b
may integrate the signals from the Pim and PI3K/Akt/mTOR signaling
pathways in Abl-expressing leukemic cells. Phosphorylated eIF2
exhibits translation-dependent control of the activation of NF-κB
(45). In order to review what
occurred in tumors, the aforementioned signaling pathways were
listed in Table SI. However,
there was insufficient data demonstrating eIF3C-related gene
regulation. The present study demonstrated that there were a total
of 994 differentially expressed genes in RCC cells following eIF3C
depletion. Future studies could characterize some of these genes in
RCC cells.
Overall, the findings provided initial evidence
regarding the role of eIF3C in the development and progression of
RCC. However, the underlying molecular mechanisms require further
investigation.
Supplementary Data
Abbreviations:
|
eIFs
|
eukaryotic initiation factors
|
|
eIF3C
|
eukaryotic initiation factor 3c
|
|
RCC
|
renal cell carcinoma
|
|
DAB
|
3,3′-diaminobenzidine
|
|
GFP
|
green fluorescence protein
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
Acknowledgments
Not applicable.
Funding
The present study was financially supported in part
by grants from the Natural Science Foundation of Jiangsu Province
(grant no. BK20151180), Applied Basic Research of Changzhou City
(grant no. CJ20159014) and Major Science and Technology Project of
Changzhou Health Bureau (grant no. ZD201405).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XH and ZC conceived and designed the project. KW, HY
and XW performed the experiments and acquired the data. MF analyzed
the data. KW and MF participated in writing the article. All
authors read and approved the final version of this manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from every
participant enrolled in the present study. The current study was
approved by the Ethics Committee of the Third Affliated Hospital of
Soochow University.
Patient consent for publication
All patients in the present study provided consent
for their data to be published.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Birkhäuser FD, Kroeger N and Pantuck AJ:
Etiology of renal cell carcinoma: Incidence, demographics, and
environmental factors. Renal Cell Carcinoma: Clinical Management.
Springer Science; New York, NY: pp. 3–22. 2013, View Article : Google Scholar
|
|
2
|
Zhou M and He H: Pathology of renal cell
carcinoma. Renal Cancer. 43:51–69. 2013.
|
|
3
|
Gupta K, Miller JD, Li JZ, Russell MW and
Charbonneau C: Epidemiologic and socioeconomic burden of metastatic
renal cell carcinoma (mRCC): A literature review. Cancer Treat Rev.
34:193–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Najjar YG and Rini BI: Novel agents in
renal carcinoma: A reality check. Ther Adv Med Oncol. 4:183–194.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rohan SM, Xiao Y, Liang Y, Dudas ME,
Al-Ahmadie HA, Fine SW, Gopalan A, Reuter VE, Rosenblum MK, Russo P
and Tickoo SK: Clear-cell papillary renal cell carcinoma: Molecular
and immunohistochemical analysis with emphasis on the von
Hippel-Lindau gene and hypoxia-inducible factor pathway-related
proteins. Mod Pathol. 24:1207–1220. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu CZ, Zheng JJ, Bai YH, Xia P, Zhang HC
and Guo Y: HMGB1/RAGE axis mediates the apoptosis, invasion,
autophagy, and angiogenesis of the renal cell carcinoma. Onco
Targets Ther. 11:4501–4510. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhai W, Ma J, Zhu R, Xu C, Zhang J, Chen
Y, Chen Z, Gong D, Zheng J, Chen C, et al: MiR-532-5p suppresses
renal cancer cell proliferation by disrupting the ETS1-mediated
positive feedback loop with the KRAS-NAP1L1/P-ERK axis. Br J
Cancer. 119:591–604. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Damayanti NP, Budka JA, Khella HWZ, Ferris
MW, Ku SY, Kauffman E, Wood AC, Ahmed K, Chintala VN,
Adelaiye-Ogala R, et al: Therapeutic targeting of
TFE3/IRS-1/PI3K/mTOR axis in translocation renal cell carcinoma.
Clin Cancer Res. 24:5977–5989. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Carlo MI, Mukherjee S, Mandelker D, Vijai
J, Kemel Y, Zhang L, Knezevic A, Patil S, Ceyhan-Birsoy O, Huang
KC, et al: Prevalence of germline mutations in cancer
susceptibility genes in patients with advanced renal cell
carcinoma. JAMA Oncol. 4:1228–1235. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mitchell TJ, Rossi SH, Klatte T and
Stewart GD: Genomics and clinical correlates of renal cell
carcinoma. World J Urol. 36:1899–1911. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Emmanuel R, Weinstein S, Landesman-Milo D
and Peer D: eIF3c: A potential therapeutic target for cancer.
Cancer Lett. 336:158–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sonenberg N and Hinnebusch AG: Regulation
of translation initiation in eukaryotes: Mechanisms and biological
targets. Cell. 136:731–745. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou M, Sandercock AM, Fraser CS, Ridlova
G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA,
Hershey JW, et al: Mass spectrometry reveals modularity and a
complete subunit interaction map of the eukaryotic translation
factor eIF3. Proc Natl Acad Sci USA. 105:18139–18144. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hinnebusch AG: eIF3: A versatile scaffold
for translation initiation complexes. Trends Biochem Sci.
31:553–562. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hershey JW: The role of eIF3 and its
individual subunits in cancer. Biochim Biophys Acta. 1849:792–800.
2015. View Article : Google Scholar
|
|
18
|
Bhat M, Robichaud N, Hulea L, Sonenberg N,
Pelletier J and Topisirovic I: Targeting the translation machinery
in cancer. Nat Rev Drug Discov. 14:261–278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rothe M, Ko Y, Albers P and Wernert N:
Eukaryotic initiation factor 3 p110 mRNA is overexpressed in
testicular seminomas. Am J Pathol. 157:1597–1604. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Scoles DR, Yong WH, Qin Y, Wawrowsky K and
Pulst SM: Schwannomin inhibits tumorigenesis through direct
interaction with the eukaryotic initiation factor subunit c
(eIF3c). Hum Mol Genet. 15:1059–1070. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hao J, Liang C and Jiao B: Eukaryotic
translation initiation factor 3, subunit C is overexpressed and
promotes cell proliferation in human glioma U-87 MG cells. Oncol
Lett. 9:2525–2533. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hao J, Wang Z, Wang Y, Liang Z, Zhang X,
Zhao Z and Jiao B: Eukaryotic initiation factor 3C silencing
inhibits cell proliferation and promotes apoptosis in human glioma.
Oncol Rep. 33:2954–2962. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song N, Wang Y, Gu XD, Chen ZY and Shi LB:
Effect of siRNA-mediated knockdown of eIF3c gene on survival of
colon cancer cells. J Zhejiang Univ Sci B. 14:451–459. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li T, Li S, Chen D, Chen B, Yu T, Zhao F,
Wang Q, Yao M, Huang S, Chen Z and He X: Transcriptomic analyses of
RNA-binding proteins reveal eIF3c promotes cell proliferation in
hepatocellular carcinoma. Cancer Sci. 108:877–885. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee HY, Chen CK, Ho CM, Lee SS, Chang CY,
Chen KJ and Jou YS: eIF3C-enhanced exosome secretion promotes
angiogenesis and tumorigenesis of human hepatocellular carcinoma.
Oncotarget. 9:13193–13205. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao W, Li X, Wang J, Wang C, Jia Y, Yuan
S, Huang Y, Shi Y and Tong Z: Decreasing eukaryotic initiation
factor 3C (eIF3C) suppresses proliferation and stimulates apoptosis
in breast cancer cell lines through mammalian target of rapamycin
(mTOR) pathway. Med Sci Monit. 23:4182–4191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hammes LS, Tekmal RR, Naud P, Edelweiss
MI, Kirma N, Valente PT, Syrjänen KJ and Cunha-Filho JS:
Up-regulation of VEGF, c-fms and COX-2 expression correlates with
severity of cervical cancer precursor (CIN) lesions and invasive
disease. Gynecol Oncol. 110:445–451. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
ANSI/ATCC ASN-0002-2011: Authentication of
Human Cell Lines: Standardization of STR Profiling. ANSI eStandards
Store; 2012
|
|
29
|
Capes-Davis A, Reid YA, Kline MC, Storts
DR, Strauss E, Dirks WG, Drexler HG, MacLeod RA, Sykes G, Kohara A,
et al: Match criteria for human cell line authentication: Where do
we draw the line? Int J Cancer. 132:2510–2519. 2013. View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
31
|
Chomczynski P and Sacchi N: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dong Z and Zhang JT: Initiation factor
eIF3 and regulation of mRNA translation, cell growth, and cancer.
Crit Rev Oncol Hematol. 59:169–180. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Asano K, Vornlocher HP, Richter-Cook NJ,
Merrick WC, Hinnebusch AG and Hershey JW: Structure of cDNAs
encoding human eukaryotic initiation factor 3 subunits. Possible
roles in RNA binding and macromolecular assembly. J Biol Chem.
272:27042–27052. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Karásková M, Gunišová S, Herrmannová A,
Wagner S, Munzarová V and Valášek L: Functional characterization of
the role of the N-terminal domain of the c/Nip1 subunit of
eukaryotic initiation factor 3 (eIF3) in AUG recognition. J Biol
Chem. 287:28420–28434. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Erzberger JP, Stengel F, Pellarin R, Zhang
S, Schaefer T, Aylett CHS, Cimermančič P, Boehringer D, Sali A,
Aebersold R and Ban N: Molecular architecture of the 40SeIF1eIF3
translation initiation complex. Cell. 158:1123–1135. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Srivastava S, Verschoor A and Frank J:
Eukaryotic initiation factor 3 does not prevent association through
physical blockage of the ribosomal subunit-subunit interface. J Mol
Biol. 226:301–304. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zang Y, Zhang X, Yan L, Gu G, Li D, Zhang
Y, Fang L, Fu S, Ren J and Xu Z: Eukaryotic translation initiation
factor 3b is both a promising prognostic biomarker and a potential
therapeutic target for patients with clear cell renal cell
carcinoma. J Cancer. 8:3049–3061. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sadato D, Ono T, Gotoh-Saito S, Kajiwara
N, Nomura N, Ukaji M, Yang L, Sakimura K, Tajima Y, Oboki K and
Shibasaki F: Eukaryotic translation initiation factor 3 (eIF3)
subunit e is essential for embryonic development and cell
proliferation. FEBS Open Bio. 8:1188–1201. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shen J, Yin JY, Li XP, Liu ZQ, Wang Y,
Chen J, Qu J, Xu XJ, McLeod HL, He YJ, et al: The prognostic value
of altered eIF3a and its association with p27 in non-small cell
lung cancers. PLoS One. 9:e960082014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu F, Xu CZ, Gu J, Liu X, Liu R, Huang E,
Yuan Y, Zhao G, Jiang J, Xu C, et al: Eukaryotic translation
initiation factor 3B accelerates the progression of esophageal
squamous cell carcinoma by activating β-catenin signaling pathway.
Oncotarget. 7:43401–43411. 2016.PubMed/NCBI
|
|
41
|
Zhu Q, Qiao GL, Zeng XC, Li Y, Yan JJ,
Duan R and Du ZY: Elevated expression of eukaryotic translation
initiation factor 3H is associated with proliferation, invasion and
tumorigenicity in human hepatocellular carcinoma. Oncotarget.
7:49888–49901. 2016.PubMed/NCBI
|
|
42
|
Qi J, Dong Z, Liu J and Zhang JT: EIF3i
promotes colon onco-genesis by regulating COX-2 protein synthesis
and β-catenin activation. Oncogene. 33:4156–4163. 2014. View Article : Google Scholar
|
|
43
|
Golob-Schwarzl N, Schweiger C, Koller C,
Krassnig S, Gogg-Kamerer M, Gantenbein N, Toeglhofer AM, Wodlej C,
Bergler H, Pertschy B, et al: Separation of low and high grade
colon and rectum carcinoma by eukaryotic translation initiation
factors 1, 5 and 6. Oncotarget. 8:101224–101243. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen K, Yang J, Li J, Wang X, Chen Y,
Huang S and Chen JL: eIF4B is a convergent target and critical
effector of oncogenic Pim and PI3K/Akt/mTOR signaling pathways in
Abl transformants. Oncotarget. 7:10073–10089. 2016.PubMed/NCBI
|
|
45
|
Deng J, Lu PD, Zhang Y, Scheuner D,
Kaufman RJ, Sonenberg N, Harding HP and Ron D: Translational
repression mediates activation of nuclear factor kappa B by
phosphorylated translation initiation factor 2. Mol Cell Biol.
24:10161–10168. 2004. View Article : Google Scholar : PubMed/NCBI
|