Introduction
At present, primary liver cancer is one of the most
common malignancies in the Chinese population and is the second
leading cause of cancer-associated mortality in males (1,2).
Chronic hepatitis B virus (HBV) infection is one of the most
critical risk factors for liver cancer (3). Effective treatment strategies have
been reported in the past few decades; however, clinical studies
have demonstrated that the morbidity and mortality of
HBV-associated liver cancer remains high, on account of an
increased incidence of cancer metastasis and invasion (4). Previously, HBV X protein (HBx),
encoded by HBV, which can alter the cell cycle, proliferation and
apoptosis-associated target gene expression of hepatocytes, was
considered to be an essential protein in the development of
HBV-related liver cancer (5,6).
Although, the specific mechanisms as to how HBx mediates
hepatocarcinogenesis remain unclear.
Long noncoding RNA (lncRNA) is a type of noncoding
RNA >200 bp in length, without the ability to encode proteins
(7). LncRNAs can affect cell
behavior; the abnormal expression or function of lncRNAs are firmly
associated with abnormalities in cell status, differentiation,
developmental diseases and cancer (8-10).
An increasing number of studies have reported that lncRNAs are
involved in the development of liver cancer, including Hox
antigenic intergenic RNA, hepatocellular carcinoma upregulated
lncRNA (HULC), hepatocellular carcinoma upregulated EZH2-associated
lncRNA, Dreh and UCF1, by affecting cell proliferation, apoptosis
and metastasis (11-14). Numerous lncRNAs have been reported
to be associated with HBx. For example, elevated lncRNA-HULC
expression was observed in HBx-overexpressed liver cancer cell
lines with upregulated cell proliferation, which could be due to
the enhanced inhibition of tumor suppressor gene p18 located near
HULC (15). In addition, HBx-long
interspersed nuclear elements (LINE1), produced by the
transcription of the HBx gene promoter region, was revealed to be
expressed in the tumor tissues of patients with HBV-related liver
cancer and was associated with poor prognosis (16). Cell experiments demonstrated that
HBx-LINE1 could promote tumor cell colony formation, migration and
the epithelial-mesenchymal transition (EMT) process by activating
the Wnt signaling pathway, in turn inducing hepatocarcinogenesis
(16,17). It has been reported that such
lncRNAs were associated with HBx; however, the precise role of
HBx-related lncRNAs in primary liver cancer remains unknown. In
addition, improved insight as to how HBx regulates the expression
of lncRNAs requires further investigation.
LncRNA-activated by transforming growth factor
(TGF)-β (lncRNA-ATB) is a recently identified oncogenic lncRNA,
which is highly expressed in primary liver cancer tissues and
several liver cancer cell lines. Yuan et al (18) revealed that lncRNA-ATB, induced by
exogenous TGF-β, could promote the invasion-metastasis cascade by
upregulating zinc finger E-box binding homeobox 1 and activating
signal transducer and activator of transcription 3 (STAT3)
signaling. Whether lncRNA-ATB is involved in the development of
HBV-associated liver cancer and the exact mechanisms of lncRNA-ATB
upregulation have not been elucidated.
Autophagy is an essential physiological process in
the eukaryotic cell that regulates the metabolic situation in cells
(19). Autophagy removes excess or
self-damaged organelles, nucleic acids, macromolecular proteins and
few degradation products to facilitate cell recycling, which in
turn maintains intracellular homeostasis (20,21).
Li et al (22) revealed
that autophagy-induced TGF-β signaling contributed to the EMT and
invasion of liver cancer cells; whether autophagy-induced TGF-β
signaling occurs in HBV-related liver cancer is unknown.
The aim of the present study was to clarify the
effects of lncRNA-ATB on the development of HBV-related liver
cancer and its regulatory mechanism mediated by HBx. The results
demonstrated that high levels of lncRNA-ATB were positively
associated with hepatitis B surface antigen and
tumor-node-metastasis (TNM) stage. Cell experiments revealed that
HBx promoted the cell invasion and migration of liver cancer by
upregulating lncRNA-ATB expression. Furthermore, TGF-β was
significantly overexpressed by HBx-induced autophagy and activated
lncRNA-ATB. These results suggested that the oncogenic effects of
lncRNA-ATB could be mediated by HBx protein and may provide novel
insight into the role of lncRNAs in the progression of HBV-related
liver cancer.
Materials and methods
Cell culture and liver cancer
tissues
The normal human hepatocyte cell line L02 and human
liver cancer cell line HepG2 were purchased from the Cell Bank of
the Shanghai Institute of Cell Biology, Chinese Academy of Sciences
(Shanghai, China). The two cell lines were cultured in Dulbecco's
Modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) containing 10% fetal bovine serum (Hyclone;
GE Healthcare Life Sciences, Logan, UT, USA), 100 IU/ml penicillin
and 100 µg/ml streptomycin) in 5% CO2 at 37°C,
for subsequent experimental research.
A total of 26 HBV-related liver cancer and non-HBV
infected liver cancer tissues were obtained from patients who
underwent radical resections for primary liver cancer in Nanjing
Drum Tower Hospital (Nanjing University, Nanjing, China) during
April 2014 to March 2015. These liver cancer samples were from 17
males and 9 females (37-80 years old). All patients were diagnosed
by histopathological examination and the detection of HBV markers.
In the subsequent experiments, all patients were separated into two
groups based on lncRNA-ATB expression, and the cut-off was the 50th
percentile. Tissues following the resection procedures were
immediately placed in liquid nitrogen, then stored at −80°C. The
present study was approved by the Ethics Committee of Nanjing Drum
Tower Hospital and written informed consent was provided by all
patients.
Lentivirus production and construction of
stable cell lines with HBx overexpression
For the development of lentiviral vectors expressing
the HBx gene, HBx cDNA was amplified by polymerase chain reaction
(PCR) and subcloned into the lentiviral vector
pHBLV-CMVIE-ZsGreen-T2A-Puro using the one step directed cloning
kit (Hanbio Biotechnology Co., Ltd., Shanghai, China); empty vector
and non-transfected cells served as the control. The restriction
enzyme used for HBx cDNA and vector were EcoRI/XhoI
and XbaI/BamHI, respectively (Takara Biotechnology
Co., Ltd., Dalian, China). The PCR procedure includes:
Pre-denaturation at 95°C for 5 min, 95°C for 45 sec, 60°C for 45
sec, 72°C for 1 min (35 cycles) and finally the last cycle 72°C for
10 min, using an ABI StepOne Plus system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). To produce lentiviruses containing
the HBx gene, the present study transfected 293 cells (Hanbio
Biotechnology Co., Ltd.) with the resulting vector described above,
pSPAX2 and pMD2G (Invitrogen; Thermo Fisher Scientific, Inc.) using
Lipofectamine 2000™ (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocols (23). Infectious lentiviruses were
harvested at 48 and 72 h post-transfection and filtered through
0.45 µm polyvinylidene fluoride (PVDF) filters, then
centrifuged at 4°C, 72,000 × g/min for 120 min in a 40 ml
ultracentrifuge tube, dissolved in DMEM and stored in liquid
nitrogen. These lentiviruses were termed LV-control and LV- HBx,
respectively. The two lentiviral vectors encoded green fluorescent
protein.
The respective recombinant lentiviruses were added
to HepG2 cells plated in a 6-well plate with a MOI of 20 to obtain
cell lines stably expressing HBx. The supernatant was replaced with
complete culture media (DMEM with 10% fetal bovine serum) following
24 h. Cells stably expressing HBx were screened out following the
addition of 2 µg/ml puromycin into the media. Reverse
transcription-quantitative PCR (RT-qPCR) and western blotting were
performed to confirm the expression of HBx in the infected cells
following infection.
Construction of the starvation-induced
autophagy model
The induction of cell autophagy was conducted
according to Klionsky et al (24). The cell medium of each group was
refreshed when the 6-well plate was covered with cells at 85%
confluence; the control cells were cultured with fresh DMEM, and
the experimental cells were washed three times with PBS to remove
the residual medium and then cultured with 1X Earle's balanced salt
solution (EBSS; Invitrogen; Thermo Fisher Scientific, Inc.) instead
of DMEM, to generate a starvation environment with sugar and amino
acid deficiency, and consequently induce autophagy. Cells were then
incubated at 37°C and 5% CO2 for 8 h and the optimal
duration for the induction of autophagy selected in this experiment
was 4 h post-starvation. Following the successful establishment of
the model, total RNA and protein in each group of cells were
extracted for subsequent analyses.
RNA interference and transfection
Small interfering (si)RNAs against lncRNA-ATB and
TGF-β (40 nmol/l) were transfected into HepG2 and HBx-HepG2 cells
respectively, which were plated in a 6-well plate at a density of
50%, using Lipofectamine 3000 according to manufacturer's
instructions. Scramble-control siRNA was used as the control, which
was transfected into the cells in the same manner as si-ATB and
si-TGF-β transfection. The catalogue numbers of
control/si-TGF-β/si-ATB are siN05815122147, siB09212165524 and
siB160908101333, respectively (Guangzhou RiboBio Co., Ltd.).
RT-qPCR confirmed the knockdown effect in HepG2 and HBx-HepG2 cells
at 48 h post-transfection.
RNA extraction and RT-qPCR analysis
Total RNA was isolated from cultured cells or tumor
tissues using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The first-strand cDNA was synthesized using
PrimeScript™ RT Master Mix (Takara Biotechnology Co., Ltd.); the
conditions for RT were: 37°C for 20 min and then heated at 85°C for
15 sec. cDNAs could be stored at -20°C. qPCR was conducted with
mixed cDNAs, gene primers and SYBR Green PCR master mix (Takara
Biotechnology Co., Ltd., Dalian, China) according to the
manufacturer's protocols, while the RNA expression levels were
measured using an ABI StepOne Plus system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). QPCR reactions were performed in
triplicate, while β-actin and 18S rRNA were selected as the
internal controls. The primers utilized for qPCR were as follows:
lncRNA-ATB forward, 5′-TCTGGCTGAGGCTGGTTGAC-3′, reverse,
5′-ATCTCTGGGTGCTGGTGAAGG-3′; HBx forward,
5′-CCCGTCTGTGCCTTCTCATC-3′, reverse, 5′-GTATGCCTC-AAGGTCGGTCG-3′;
β-actin forward, 5′-GGGAAATCGTGCGTGACATTAAG-3′, reverse,
5′-TGTGTTGGCGTACAGGTCTTTG-3′; Beclin-1 forward,
5′-CAGGAGAGACCCAGGAGGAA-3′, reverse, 5′-GCTGTTGGCACTTTCTGTGG3′;
microtubule-associated proteins 1A/1B light chain 3B (LC3b)
forward, 5′-CCGCACCTTCGAACAAAGAG-3′, reverse,
5′-TTGAGCTGTAAGCGCCTTC-T-3′; sequestosome 1 (SQSTM1) forward,
5′-TCTGGCTGAGGCTGGTTGAC-3′, reverse, 5′-ATCT-CTGGGTGCTGGTGAAGG-3′
and TGF-β forward, 5′-CACCATAAAGACAGGAACCTG-3′, reverse,
5′-GGAGGTGCCATCAATACCTGC3′. The relative expression of RNAs was
calculated using the comparative 2−ΔΔCq method (25,26).
Western blot analysis
Lysis buffer was used for the extraction of total
proteins of cells and tissues, which were quantified using a
Bicinchoninic Acid kit (Nanjing KeyGEN Biotech Co., Ltd., Nanjing,
China). Equal quantities of proteins (4 mg/ml) were separated by
SDS-PAGE (with 30% acrylamide), then transferred onto PVDF
membranes. Following incubation with antibodies (1:1,000) specific
for HBx (ab39716; Abcam, Cambridge, UK), LC3b (cat. no. 3868; Cell
Signaling Technology, Inc., Danvers, MA, USA), p62 (cat. no. 23214;
Cell Signaling Technology, Inc.), Beclin-1 (cat. no. 4122; Cell
Signaling Technology, Inc.), TGF-β (ab186838; Abcam) and β-actin
(ab179467; Abcam) for 12 h at 4°C; then the blots were incubated
with goat anti-rabbit (for HBx, LC3b and p62) or anti-mouse (for
TGF-β, β-actin and Beclin-1) IgG-horseradish peroxidase-conjugated
antibodies (1:5,000; Nanjing KeyGEN Biotech Co., Ltd.) for 2 h at
room temperature. The proteins were semi-quantified by
chemiluminescence method (Tanon Science & Technology Co., Ltd.,
Shanghai, China) and analyzed using ImageJ software (Version 1.5.1;
National Institutes of Health, Bethesda, MD, USA).
In vitro cell invasion and migration
assays
A Transwell assay was used to determine cell
invasion as described previously (27). Cells (~5×104) were
resuspended in 200 µl serum-free medium and added to the
upper layer of the Transwell chamber, while 500 µl of
complete medium was added into the lower chamber. Following 48 h of
culture at 37°C, the medium was discarded and noninvaded cells were
gently removed with a cotton swab and rinsed three times with PBS.
The remaining cells were then fixed using 3.7% paraformaldehyde at
room temperature for 20 min. The membranes were stained with 0.1%
crystal violet for 30 min at room temperature, and cell invasive
ability was analyzed by counting the number of stained cells in
random fields of view under a 400-fold inverted biological
microscope.
Cell migration ability was evaluated using a scratch
assay. A 200-µl pipette tip was used to scratch a straight
wound in cells seeded in 6-well plates and then cultured in
serum-free medium for 24 h at 37°C. Images of the wound width were
then captured at 0 and 24 h and compared under a 5-fold inverted
biological microscope. All experiments were independently repeated
in triplicate.
Statistical analysis
SPSS 19.0 software (IBM Corp., Armonk, NY, USA) was
used for statistical analysis. Each group of experiments was
repeated at least three times, and data are expressed as the mean ±
standard deviation. The χ2 test was used to assess the
association between lncRNA-ATB expression and the clinical
characteristics of patients. A two-tailed Student's t-test was used
for the comparison of independent variables. One-way analysis of
variance was used for the comparison of multiple groups, followed
by a Student-Newman-Keuls post-hoc test. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression of lncRNA-ATB in liver cancer
and its association with clinicopathological characteristics
To clarify the role of lncRNA-ATB in primary liver
cancer and whether HBx regulated lncRNA-ATB, the present study
selected 26 tumor tissues from patients with radical hepatectomy
and detected the expression of lncRNA-ATB by RT-qPCR. The
association between lncRNA-ATB content and the clinicopathological
characteristics of liver cancer, including the status of HBV
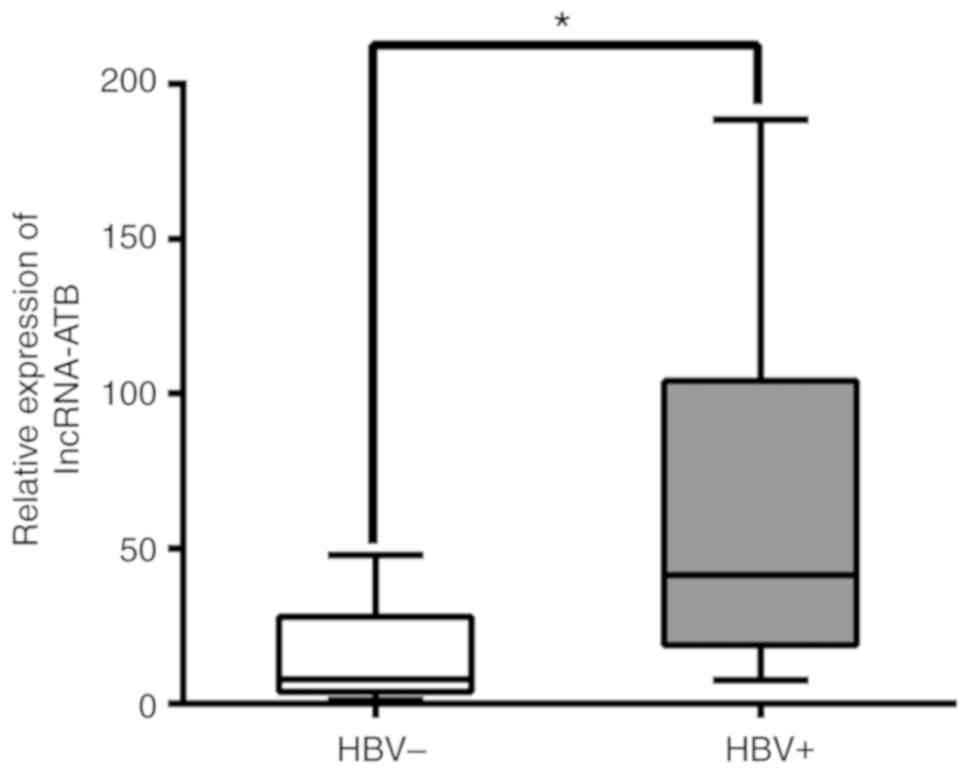
infection, was then evaluated. As presented in Fig. 1, lncRNA-ATB was significantly
associated with HBV. In addition, a significant association was
detected between more advanced TNM stage and a higher expression
levels of lncRNA-ATB, irrespective of patient age, gender, tumor
size, liver cirrhosis and histological differentiation (Table I). These results suggested that
increased expression of lncRNA-ATB in primary liver cancer may be
associated with HBV infection and advanced tumor development.
 | Table IAssociation between lncRNA-ATB
expression and clinical characteristics in patients with liver
cancer (n=26). |
Table I
Association between lncRNA-ATB
expression and clinical characteristics in patients with liver
cancer (n=26).
| Factors | lncRNA-ATBa
| P-valueb |
|---|
| Low | High |
|---|
| All cases | 13 | 13 | |
| Age | | | |
| ≤55 | 6 | 8 | 0.430 |
| >55 | 7 | 5 | |
| Gender | | | |
| Male | 10 | 8 | 0.394 |
| Female | 3 | 5 | |
| HBsAg | | | |
| Positive | 4 | 12 | 0.001 |
| Negative | 9 | 1 | |
| Liver
cirrhosis | | | |
| With | 5 | 9 | 0.113 |
| Without | 8 | 4 | |
| Tumor size, cm | | | |
| ≤5 | 5 | 7 | 0.430 |
| >5 | 8 | 6 | |
| Histological
differentiation | | | |
| Well | 5 | 3 | 0.685 |
| Moderate | 7 | 9 | |
| Poor | 1 | 1 | |
| TNM stage | | | |
| I + II | 9 | 4 | 0.017 |
| III + IV | 2 | 11 | |
HBx upregulates the expression of
lncRNA-ATB in liver cancer cells
Previous studies have revealed that lncRNA-ATB can
promote the invasion and migration of various tumor cells including
primary liver cancer by activating the EMT and STAT3 pathways,
while lncRNA-ATB could induced by TGF-β (18,28-30).
However, whether abnormal expression of lncRNA-ATB is associated
with the invasive and migration abilities of HBV-associated liver
cancer cells remains unknown. To clarify this mechanism, the
present study constructed an HBx lentivirus transfected HepG2 cell
line (HBx-HepG2) which stably expressed HBx protein. The results of
western blotting and RT-qPCR revealed that the levels of HBx
protein and mRNA were markedly increased in HepG2 cells following
transfection, while the expression of HBx in the blank control and
empty plasmid groups (con-HepG2) was notable reduced (Fig. 2A and B).
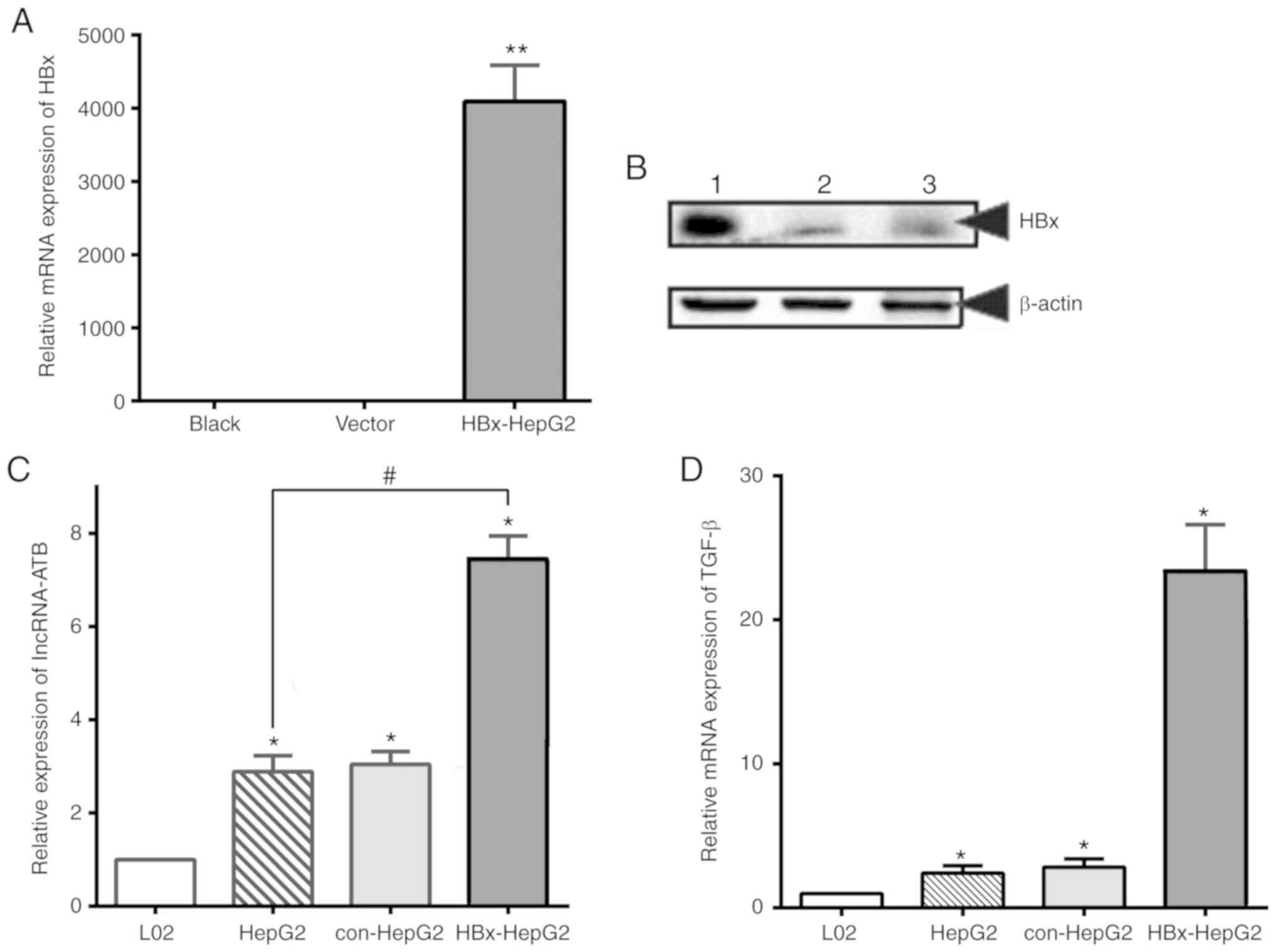 | Figure 2HBx upregulates the expression of
lncRNA-ATB and TGF-β in HepG2 cells. (A) RT-qPCR analysis and (B)
western blotting analysis of HBx expression in HepG2 cells
transfected with lentivirus. Lane 1, HBx lentivirus transfected
cells; lane 2, control vector and lane 3, blank. (C) RT-qPCR
analysis of lncRNA-ATB in L02, HepG2, con-HepG2 and HBx-HepG2
cells. (D) RT-qPCR analysis of TGF-β in HepG2 and HBx-HepG2 cells.
18S rRNA was used as the internal control. *,
#P<0.05, **P<0.01. HBx, Hepatitis B
virus X protein; lncRNA-ATB, long noncoding RNA-activated by
transforming growth factor β; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; con-HepG2,
empty plasmid group; TGF-β, transforming growth factor-β. |
To further investigate the effects of HBx on
lncRNA-ATB in HBx-HepG2 cells, the present study compared the
expression of lncRNA-ATB in L02, HepG2, con-HepG2 and HBx-HepG2
cell lines. As presented in Fig.
2C, the content of lncRNA-ATB in HBx-HepG2 cells was
significantly increased following transfection with HBx lentivirus
compared with HepG2 cells. The results were similar in HepG2 and
con-HepG2 cells. In addition, the levels of TGF-β mRNA expression,
which has been reported to be an inducer of lncRNA-ATB expression,
were evaluated; a significant increase in HBx-HepG2 cells was
observed compared with HepG2 cells (Fig. 2D). These results suggested that HBx
may promote the expression of lncRNA-ATB in HepG2 cells by
upregulating the expression of TGF-β.
HBx-induced autophagy significantly
increases the expression of TGF-β and lncRNA-ATB
As aforementioned, alterations in cell behaviors,
including metabolism, secretion or degradation of functional
proteins, is one of the most critical underlying mechanisms as to
how HBx leads to the development of primary liver cancer (5,31).
In recent years, autophagy has been reported as an essential
mechanism for regulating the intracellular environment (20,32).
Whether HBx can affect the expression of intracellular lncRNA-ATB
by mediating the autophagy of liver cancer cells is unclear. To
investigate this, the present study detected the level of autophagy
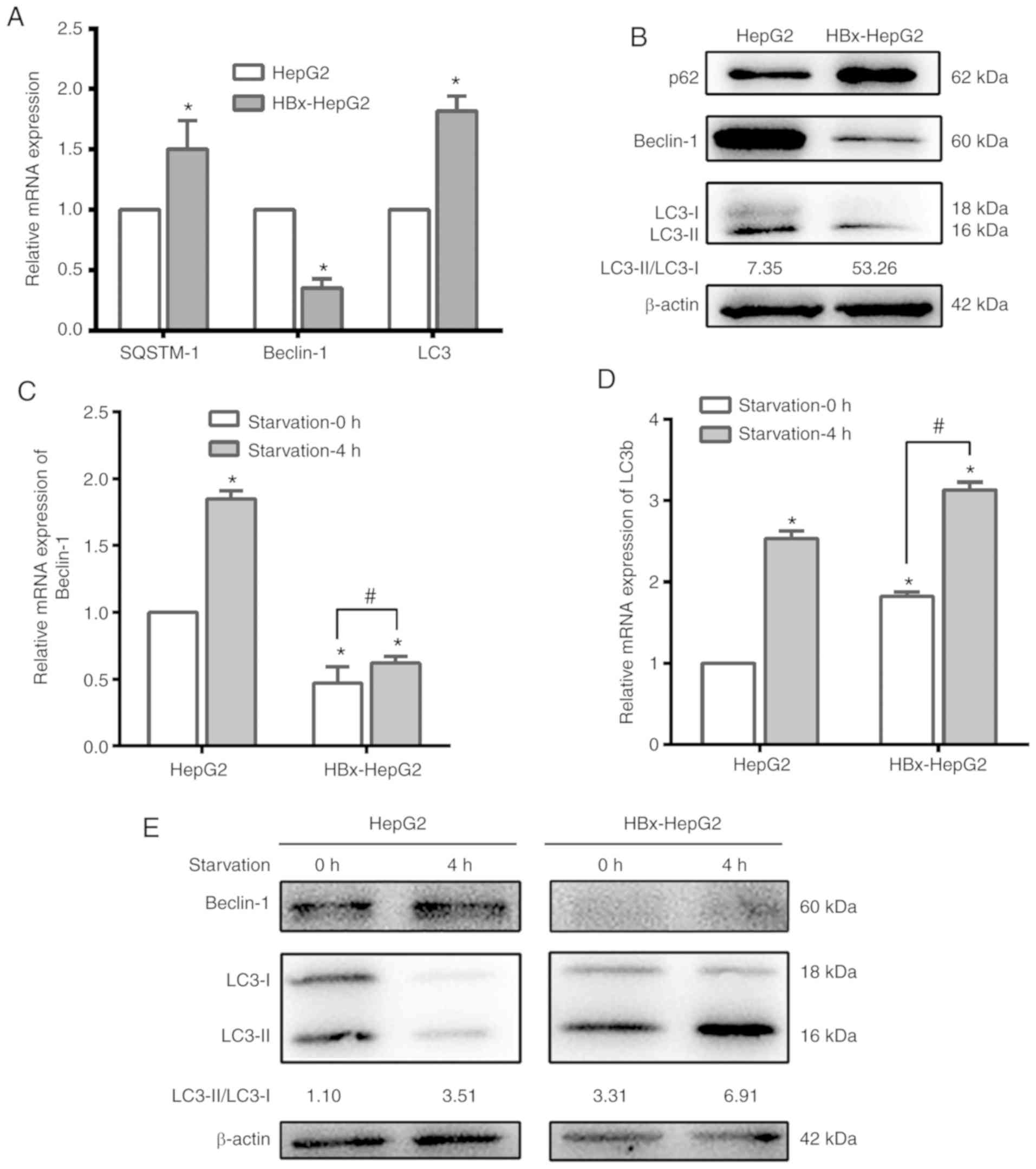
in HepG2 and HBx-HepG2 cells. As presented in Fig. 3A and B, the expression of
autophagy-associated markers SQSTM-1 and LC3 in HBx-HepG2 cells
were significantly increased following HBx transfection, while
Beclin-1, which reflects the successful synthesis of autophagic
precursors, exhibited a significant decrease compared with in HepG2
cells. HBx-HepG2 cells exhibited a marked increase in the
expression of p62 compared with in HepG2 cells. These results
indicated that HBx could promote autophagy in HepG2.
To further clarify whether lncRNA-ATB expression
levels in HBx-HepG2 are associated with high levels of autophagy,
the present study designed starvation-induced autophagy models
using EBSS, which has been widely used for the induction of
autophagy (33,34). Alterations in autophagy-associated
proteins in each group were detected once cells were cultured with
serum-free essential culture medium EBSS for 4 h. The expression
levels of Beclin-1 and LC3 were significantly increased in HepG2
and HBx-HepG2 cells following 4 h of starvation (Fig. 3C-E). The increase in LC3 content
and LC3 II/LC3 I indicated an increase in autophagy, while the
upregulated Beclin-1 suggests that autophagy may occur rapidly and
autophagic precursor protein may be synthesized in marked
quantities.
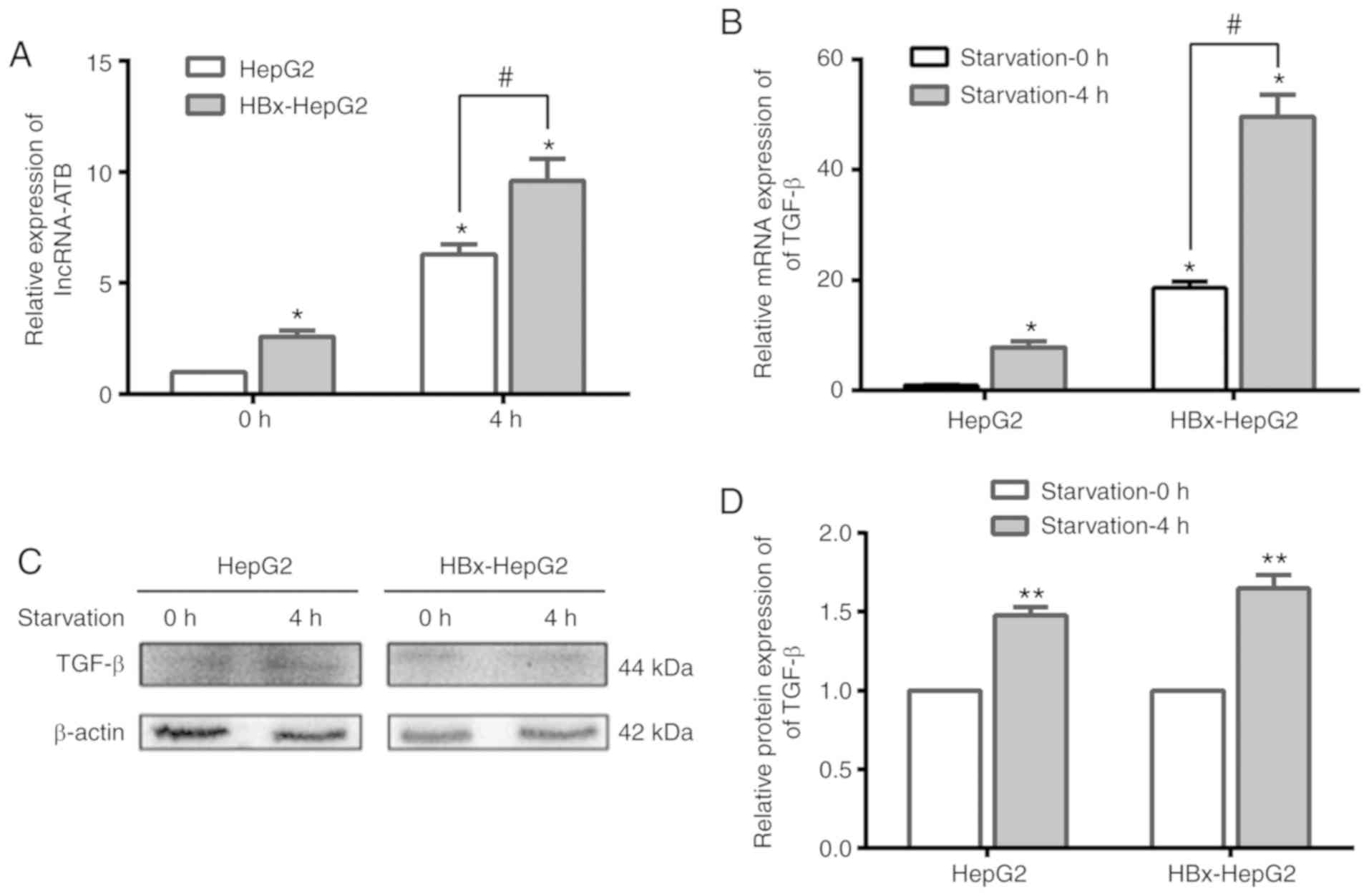
Following the successful induction of autophagy, the
content of lncRNA-ATB in HepG2 and HBx-HepG2 cells increased
significantly at 4 h compared with 0 h of starvation (Fig. 4A). Analysis of the lncRNA-ATB
inducer, TGF-β, revealed that the mRNA and protein expression
levels of TGF-β were upregulated in the two cell lines at 4 h of
starvation than at 0 h (Fig.
4B-D). These results indicated that starvation-induced
autophagy could significantly increase the levels of lncRNA-ATB and
TGF-β in HepG2 cells transfected with HBx.
Knockdown of lncRNA-ATB/TGF-β could
inhibit the level of autophagy in HepG2 cells
The aforementioned findings indicated an association
between autophagy and TGF-β/lncRNA-ATB expression in HBx-HepG2
cells. To further verify this association, the present study
employed siRNAs against lncRNA-ATB and TGF-β to observe their
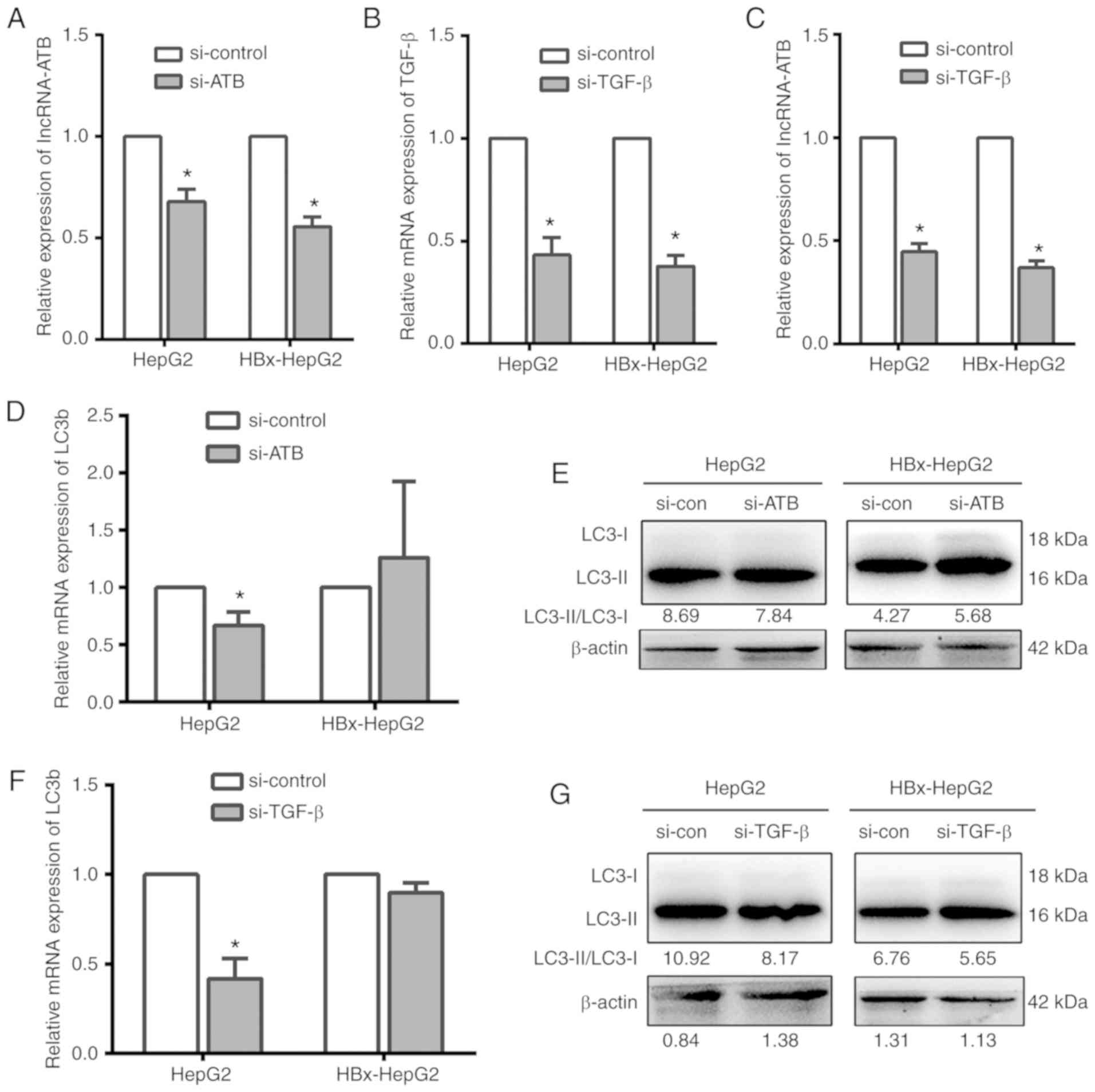
effects on cell autophagy following knockdown of lncRNA-ATB/TGF-β.
The results of RT-qPCR demonstrated that the expression of target
RNA in cells was significantly inhibited following siRNA-ATB or
siRNA-TGF-β transfection in HepG2 and HBx-HepG2 cells compared with
the control, indicating successful transfection (Fig. 5A and B). In addition, the
expression of lncRNA-ATB was significantly decreased following
knockdown of TGF-β compared with the control (Fig. 5C). Conversely, the expression
levels of TGF-β were markedly altered following the knockdown of
lncRNA-ATB (data not shown), which suggested that TGF-β also
regulates the expression of lncRNA-ATB in HBx-HepG2 cells.
Further investigation demonstrated that LC3
expression was suppressed following the knockdown of lncRNA-ATB or
TGF-β in HepG2 cells; however, these observations were not noted in
HBx-HepG2 cells (Fig. 5D-G). These
results demonstrated that, at least in part, autophagy was
inhibited in the case of lncRNA-ATB knockdown in liver cancer
cells, while the expression of HBx could partially alleviate this
inhibition.
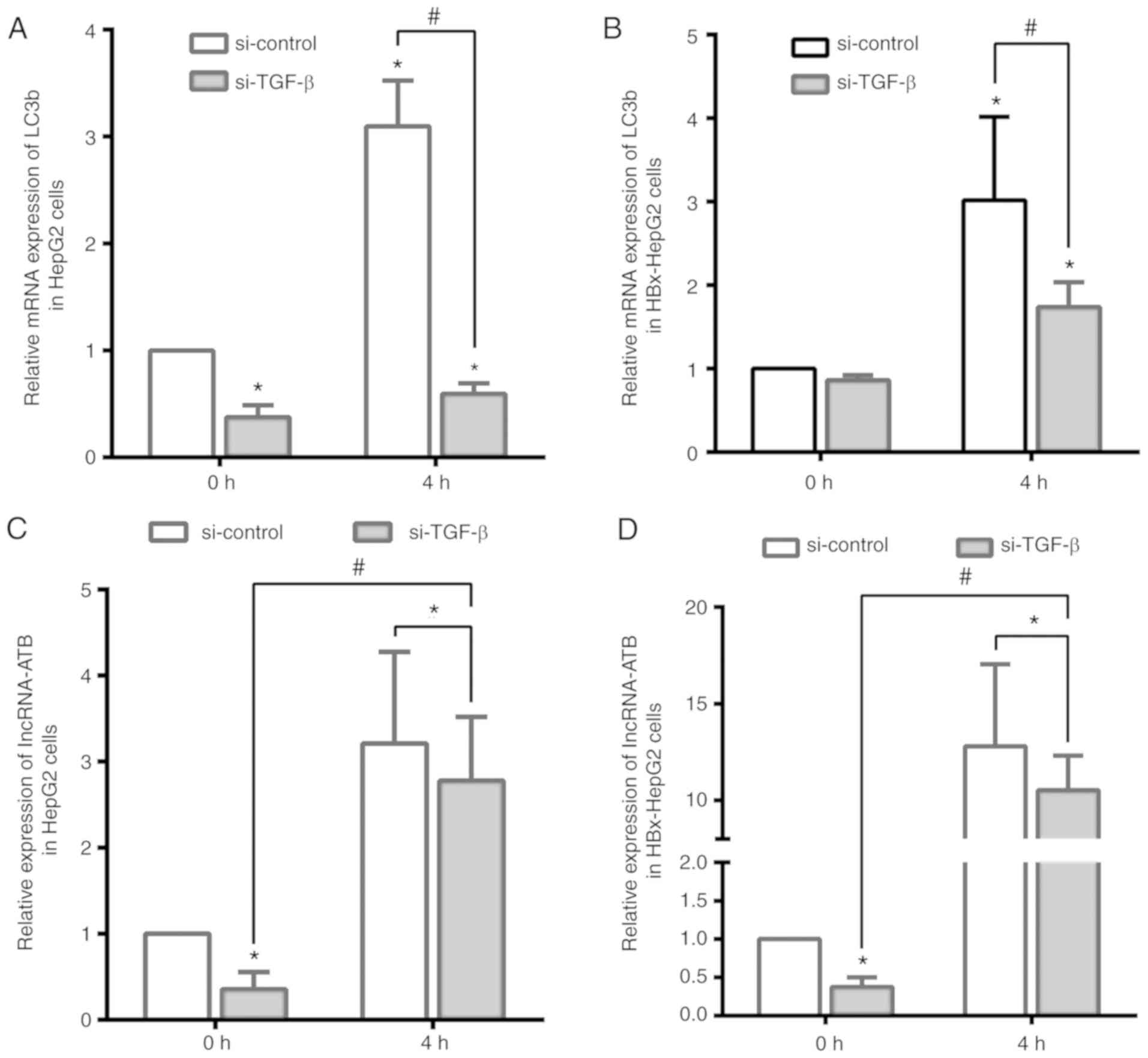
Starvation-induced autophagy upregulates
the expression of lncRNA-ATB following TGF-β knockdown
To further detect the association between
HBx-induced autophagy and lncRNA-ATB, the present study determined
lncRNA-ATB expression in starvation-induced autophagy and the
suppression of TGF-β in HepG2 and HBx-HepG2 cells. The mRNA levels
of the autophagy-associated protein LC3b were significantly
increased following starvation for 4 h, while such an increase was
observed in HBx-HepG2 and HepG2 cells transfected with si-TGF-β
(Fig. 6A and B). Combined with
previous experiments, this suggested that HBx-induced autophagy
could promote the expression of TGF-β but was not dependent on it.
The present study then investigated the expression levels of
lncRNA-ATB under these conditions. As presented in Fig. 6C and D, si-TGF-β significantly
inhibited the expression of lncRNA-ATB compared with the control at
0 h, but was significantly upregulated by starvation-induced
autophagy in HepG2 and HBx-HepG2 cells. Thus, it was concluded that
HBx-induced autophagy led to the abnormal expression of TGF-β, and
upregulated that of lncRNA-ATB in liver cancer cells.
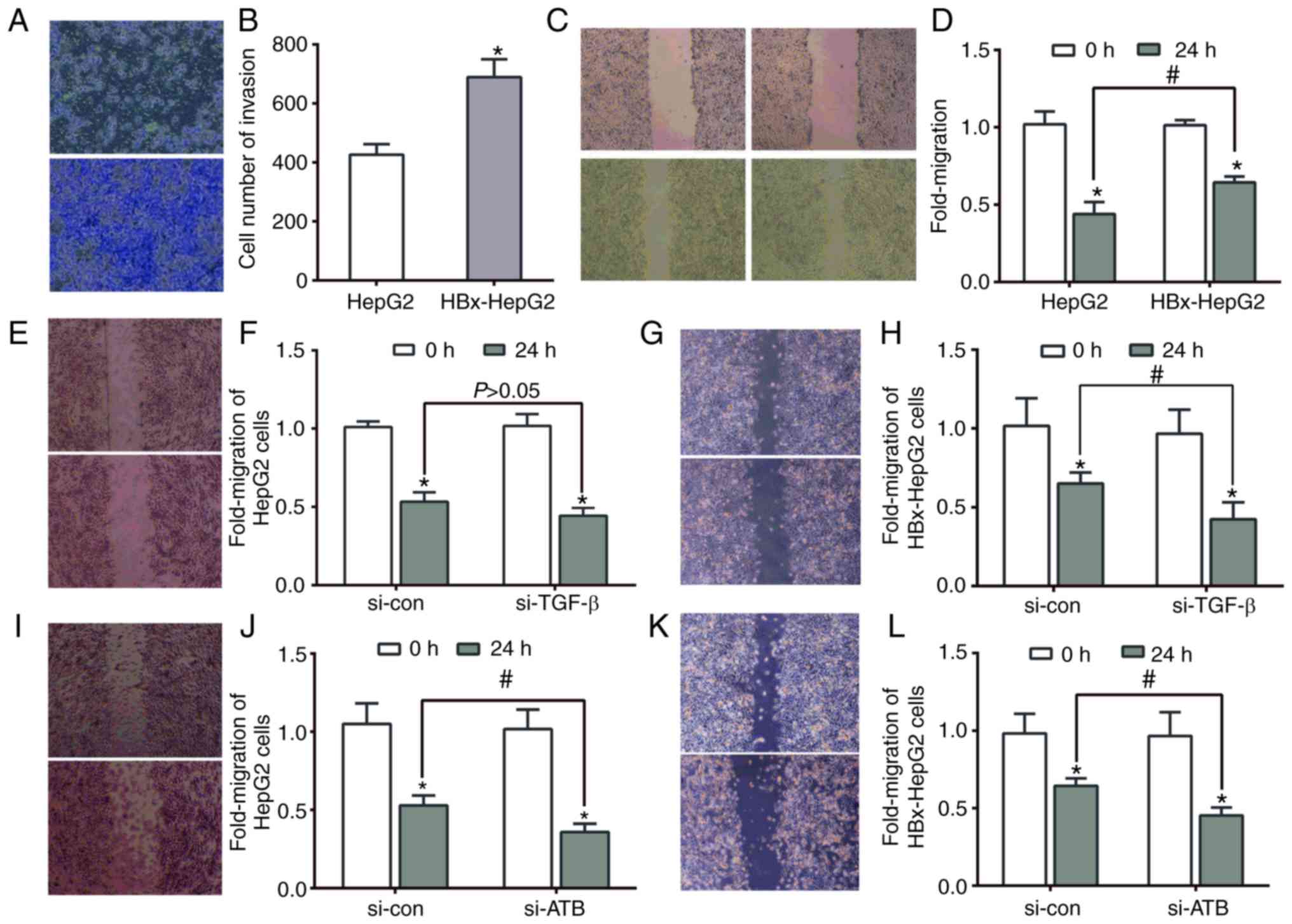
Effects of lncRNA-ATB on the invasion and
migration abilities of HBx-HepG2 cell lines
It has been reported that the invasion and migration
abilities of HBV-associated liver cancer cells were greater than
normal hepatocyte cell lines (35,36);
however, whether lncRNAs participate in the invasion and migration
of HBV-associated primary liver cancer is unclear. In the present
study, stably transfected HBx-vector significantly promoted cell
invasion and migration compared with HepG2 cells (Fig. 7A-D). A scratch-wound assay was then
conducted to investigate whether TGF-β or lncRNA-ATB affected the
migration ability of HBx-HepG2 cells. As presented in Fig. 7E-H, the migration ability of
HBx-HepG2 cells was significantly inhibited when TGF-β was
suppressed compared with the control group, which was not apparent
in HepG2 cells. In addition, such inhibition of migration was
significant in the two types of cells transfected with siRNA-ATB
(Fig. 7I-L). Therefore, it was
concluded that the invasion and migration of HBx-HepG2 may be
associated with the expression of lncRNA-ATB. Abnormal expression
of lncRNA-ATB may be an underlying mechanism of enhanced invasion
and migration of HBV-associated liver cancer cells.
Discussion
It has been reported that ~50% of primary liver
cancer cases are caused by HBV infection in China (37). HBV-associated liver cancer is more
prone to invasion and distant metastasis than cancer without HBV
infection, but the mechanism of HBV in primary liver cancer
development requires further investigation (17,38).
As HBV-related liver cancer is one of the most common subtypes of
liver cancer in China, the HBx protein encoded by X gene has been
reported to be the primary pathogenic factor of primary liver
cancer (31). Therefore, it is
necessary to improve the early diagnosis rate of HBV-related liver
cancer, in order to identify potential tumor markers with high
sensitivity and specificity. Following the discovery of lncRNA and
in-depth study of its function, increasing evidence has suggested
that lncRNAs may be involved in the invasion and metastasis of
liver cancer (12,13,39);
however, whether the abnormal expression of lncRNAs is associated
with the HBV infection in liver cancer remains unknown. LncRNA-ATB
is a novel lncRNA associated with liver cancer that can be
overexpressed via the addition of exogenous TGF-β; lncRNA-ATB
functions as a competing endogenous RNA by activating the EMT
pathway, and promotes the invasion and distant metastasis of tumor
cells (18,30). From these findings, the present
study hypothesized that HBx could upregulate the expression of
lncRNA-ATB, while overexpres-sion of lncRNA-ATB may promote the
invasion and migration of liver cancer cells. To the best of our
knowledge, the present study is the first to detect the expression
of lncRNA-ATB in primary liver cancer tissues with or without HBV
infection. The expression levels of lncRNA-ATB in HBV-related liver
cancer were significantly increased than in patients without HBV
infection; tissues with higher lncRNA-ATB levels were linked with
advanced TNM stage, which indicated that HBV could upregulate
lncRNA-ATB, a pathogenic factor associated with the development of
liver cancer.
However, whether the role of HBx protein in the
occurrence of liver cancer is associated with the abnormal
expression of lncRNA-ATB remains unknown. In the present study,
HBx-HepG2 cells, which stably expressed HBx was constructed and the
expression of lncRNA-ATB and TGF-β in HepG2 and HBx-HepG2 cells was
compared. The results revealed that HBx upregulated the expression
of lncRNA-ATB and TGF-β. Based on these observations, it was
proposed that this phenomenon may be due to the effects of
autophagy in HepG2 cells. Previous studies have demonstrated that
autophagy is an essential process in eukaryotes, that can affect or
reflect the synthesis and degradation of RNA and proteins,
including TGF-β (19,22,40).
To confirm our hypothesis, the present study investigated the
expression levels of the autophagy-associated proteins p62,
Beclin-1 and LC3b in HBx-HepG2 and HepG2 cells. The results
revealed that the expression of p62 and LC3b in HBx-HepG2 were
significantly higher than in HepG2, suggesting that HBx may be
associated with the induction of autophagy in HepG2. A model of
starvation-induced autophagy was constructed to simulate the
induction of autophagy by HBx, while the overexpression of
lncRNA-ATB and TGF-β were also detected in cells with or without
HBx overexpression at 4 h of starvation. Furthermore, it was
demonstrated that knockdown of TGF-β downregulated lncRNA-ATB in
HBx-HepG2 cells as determined by the RNA interference experiments
under conditions of starvation. Of note, knockdown of TGF-β or
lncRNA-ATB could inhibit autophagy in HepG2, but it did not affect
autophagy within HBx-HepG2 cells. This suggested that TGF-β may
affect autophagy in liver cancer cells, but the induction of
autophagy by HBx could compensate for this mechanism.
The present study also detected the effects of
lncRNA-ATB on the invasive and migration abilities of HepG2 cells
following transfection with HBx, to elucidate the role of
lncRNA-ATB in the progression of HBV-associated liver cancer. The
results revealed that the invasive and migration abilities of
HBx-HepG2 cells were promoted than in HepG2 cells, which were
further reduced following the knockdown of TGF-β or lncRNA-ATB.
This indicated that the invasive and metastatic potential of
HBV-related liver cancer may be associated with the upregulated
expression of TGF-β and lncRNA-ATB in liver cancer cells.
In conclusion, the present study demonstrated that
HBx could upregulate the expression of TGF-β by inducing autophagy.
Upregulated TGF-β may further stimulate the expression of
lncRNA-ATB in primary liver cancer, and finally promote the
invasion and migration of liver cancer cells. These results may
provide novel insight into the development of HBV-related liver
cancer. LncRNA-ATB may be considered as a therapeutic target in the
treatment of liver cancer; however, whether the altered invasion
and migration abilities induced by lncRNA-ATB affects normal
hepatocyte requires further investigation.
Funding
The present study was supported by a grant from the
National Natural Science Foundation of China (grant no. 81670566)
and Jiangsu Province's Key Provincial Talents Program (grant no.
ZDRCA2016066).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YHZ, JL, WZ and XLS made substantial contributions
to the design of the present study. YHZ, JL and FJY performed the
experiments. YHZ, JL and XLS wrote the manuscript. YZ, SW and YL
were involved in data analysis and produced initial figure drafts.
All authors have read and approved the final manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Nanjing Drum Tower Hospital (Nanjing, China) and
written informed consent was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
We thank Dr Jinglin Wang (Nanjing Drum Tower
Hospital, Nanjing, China) for technical assistance.
Abbreviations:
|
HBx
|
hepatitis B Virus protein x
|
|
lncRNA-ATB
|
long non-coding RNA activated by
TGF-β
|
|
TGF-β
|
transforming growth factor-β
|
|
LC3
|
microtubule-associated protein light
chain 3
|
|
siRNA
|
small interfering RNA
|
|
EBSS
|
Earle's balanced salt solution
|
|
DMEM
|
Dulbecco's modified Eagle media
|
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Han LL, Lv Y, Guo H, Ruan ZP and Nan KJ:
Implications of biomarkers in human hepatocellular carcinoma
pathogenesis and therapy. World J Gastroenterol. 20:10249–10261.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maucort-Boulch D, de Martel C, Franceschi
S and Plummer M: Fraction and incidence of liver cancer
attributable to hepatitis B and C viruses worldwide. Int J Cancer.
142:2471–2477. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
de Lope CR, Tremosini S, Forner A, Reig M
and Bruix J: Management of HCC. J Hepatol. 56(Suppl 1): S75–S87.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tang H, Oishi N, Kaneko S and Murakami S:
Molecular functions and biological roles of hepatitis B virus x
protein. Cancer Sci. 97:977–983. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martin-Vilchez S, Lara-Pezzi E,
Trapero-Marugán M, Moreno-Otero R and Sanz-Cameno P: The molecular
and pathophysiological implications of hepatitis B X antigen in
chronic hepatitis B virus infection. Rev Med Virol. 21:315–329.
2011.PubMed/NCBI
|
|
7
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Caley DP, Pink RC, Trujillano D and Carter
DR: Long noncoding RNAs, chromatin, and development.
ScientificWorldJournal. 10:90–102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang
H, Liang WC, Wang SS, Ko CH, Waye MM, et al: Hotair mediates
hepatocarcinogenesis through suppressing miRNA-218 expression and
activating P14 and P16 signaling. J Hepatol. 63:886–895. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang
Y, Tang GN, Zhou WP and Sun SH: Hepatitis B virus X protein
(HBx)-related long noncoding RNA (lncRNA) down-regulated expression
by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by
targeting the intermediate filament protein vimentin. Hepatology.
57:1882–1892. 2013. View Article : Google Scholar
|
|
15
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Heikenwalder M and Protzer U: LINE(1)s of
evidence in HBV-driven liver cancer. Cell Host Microbe. 15:249–250.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lau CC, Sun T, Ching AK, He M, Li JW, Wong
AM, Co NN, Chan AW, Li PS, Lung RW, et al: Viral-human chimeric
transcript predisposes risk to liver cancer development and
progression. Cancer Cell. 25:335–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Levine B and Kroemer G: Autophagy in the
pathogenesis of disease. Cell. 132:27–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ktistakis NT and Tooze SA: Digesting the
expanding mechanisms of autophagy. Trends Cell Biol. 26:624–635.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ponpuak M, Mandell MA, Kimura T, Chauhan S
and Cleyrat Deretic V: Secretory autophagy. Curr Opin Cell Biol.
35:1062015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li J, Yang B, Zhou Q, Wu Y, Shang D, Guo
Y, Song Z, Zheng Q and Xiong J: Autophagy promotes hepatocellular
carcinoma cell invasion through activation of
epithelial-mesenchymal transition. Carcinogenesis. 34:1343–1351.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stepanenko AA and Dmitrenko VV: HEK293 in
cell biology and cancer research: Phenotype, karyotype,
tumorigenicity, and stress-induced genome-phenotype evolution.
Gene. 569:1822015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Klionsky DJ, Abdalla FC, Abeliovich H,
Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello
Agostinis P, Aguirre-Ghiso JA, et al: Guidelines for the use and
interpretation of assays for monitoring autophagy. Autophagy.
445–544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Hou Z, Xu X, Fu X, Tao S, Zhou J, Liu S
and Tan D: HBx-long non-coding RNA MALAT1 promotes cell metastasis
via up-regulating LTBP3 in hepatocellular carcinoma. Am J Cancer
Res. 7:845–856. 2017.
|
|
28
|
Ke L, Xu SB, Wang J, Jiang XL and Xu MQ:
High expression of long non-coding RNA ATB indicates a poor
prognosis and regulates cell proliferation and metastasis in
non-small cell lung cancer. Clin Transl Oncol. 19:599–605. 2017.
View Article : Google Scholar
|
|
29
|
Saito T, Kurashige J, Nambara S, Komatsu
H, Hirata H, Ueda Sakimura S, Uchi R, Takano Y, Shinden Y, et al: A
long non-coding RNA activated by transforming growth factor-β an
independent prognostic marker of gastric cancer. Ann Surg Oncol.
22(Suppl 3): S915–S922. 2015. View Article : Google Scholar
|
|
30
|
Xiong J, Liu Y, Jiang L, Zeng Y and Tang
W: High expression of long non-coding RNA lncRNA-ATB is correlated
with metastases and promotes cell migration and invasion in renal
cell carcinoma. Jpn J Clin Oncol. 46:378–384. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Feitelson MA, Reis HM, Tufan NL, Sun B and
Pan J: Putative roles of hepatitis B x antigen in the pathogenesis
of chronic liver disease. Cancer Lett. 286:69–79. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Eitan E, Suire C, Zhang S and Mattson MP:
Impact of lysosome status on extracellular vesicle content and
release. Ageing Res Rev. 32:65–74. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tao H, Qian P, Lu J, Guo Y, Zhu H and Wang
F: Autophagy inhibition enhances radiosensitivity of Eca-109 cells
via the mitochondrial apoptosis pathway. Int J Oncol.
52:18532018.PubMed/NCBI
|
|
34
|
Barutcu SA, Girnius N, Vernia S and Davis
RJ: Role of the MAPK/cJun NH2-terminal kinase signaling pathway in
starvation-induced autophagy. Autophagy. 14:1586–1595. 2018.
View Article : Google Scholar :
|
|
35
|
Ringelhan M and Protzer U: Oncogenic
potential of hepatitis virus encoded proteins. Curr Opin Virol.
14:109–115. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ding D, Lou X, Hua D, Yu W, Li L, Wang J,
Gao F, Zhao Ren G, Li L and Lin B: Recurrent targeted genes of
hepatitis virus in the liver cancer genomes identified by a
next-generation sequencing-based approach. PLoS Genet.
8:e10030652012. View Article : Google Scholar
|
|
37
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273.e1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chan HL and Sung JJ: Hepatocellular
carcinoma and hepatitis virus. Semin Liver Dis. 26:153–161. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gu H, Guo X, Zou L, Zhu H and Zhang J:
Upregulation of microRNA-372 associates with tumor progression and
prognosis in hepatocellular carcinoma. Mol Cell Biochem. 375:23–30.
2013.PubMed/NCBI
|
|
40
|
Lamb CA, Yoshimori T and Tooze SA: The
autophagosome: Origins unknown, biogenesis complex. Nat Rev Mol
Cell Biol. 759–774. 2013. View Article : Google Scholar : PubMed/NCBI
|