Introduction
Tongue squamous cell carcinoma (TSCC), one of the
most common type of oral squamous cell carcinoma (OSCC) that ranks
as the sixth leading cause of cancer-associated mortality
worldwide, presents with more aggressive characteristics compared
with other forms of OSCC; ~17,110 new cases and 2,510 mortalities
due to TSCC have been reported in the United States in 2018
(1,2). At present, surgical resection is
considered as the primary treatment option for patients with TSCC;
however, the majority of patients diagnosed with TSCC are of an
advanced stage, at which point surgery cannot be performed. For
patients who have undergone surgery, the inevitable oral
complications, including speech impediments and swallowing
dysfunction notably affect quality of life (3,4).
Although various treatments, including radiotherapy, chemotherapy
and targeted therapy have been applied to patients with TSCC,
unsatisfactory outcomes have been reported, partly due to distant
metastasis, tumor recurrence and drug resistance (4,5).
Therefore, it is important to identify additional reliable
biomarkers to accurately monitor the progression and predict the
prognosis of patients with TSCC.
Integrins, a large family of heterodimeric cell
surface receptors that participate in cell-extracellular matrix and
cell-cell interactions, are involved in a broad range of cellular
processes, such as cell growth, cell mobility and cell signaling
networks (6,7). As one member of the integrin family
of adhesion molecules, integrin a7 (ITGA7), located on chromosome
12p13 and comprising >27 exons spanning a region of ~22.5 kb,
has been reported to be a tumor suppressor in several carcinomas,
including prostate cancer (8),
breast cancer (9) and melanoma
(10). By contrast, ITGA7 was
determined to act as a tumor promoter in OSCC (11) and glioblastoma (12). In addition, ITGA7 has been reported
as a functional marker of cancer stem cells (CSCs) and serves a
critical role in the regulation of stem cell-like properties in a
variety of cancer cells (11,13).
Several previous studies investigated the potential role of ITGA7
in the pathogenesis of different types of carcinomas; however, the
specific function of ITGA7 in TSCC remains unknown.
Based on the aforementioned reports regarding the
regulatory roles of ITGA7 in tumor progression and the properties
of CSCs in carcinoma, we hypothesized that ITGA7 may act as a
critical regulator in the pathogenesis of TSCC. The present study
aimed to detect the expression of ITGA7 in tumor and paired
adjacent tissues, and to evaluate its association with the
clinicopathological characteristics and overall survival (OS) of
patients with TSCC. Additionally, the effects of ITGA7 knockdown
were investigated on the proliferation, apoptosis and sternness of
TSCC cells.
Materials and methods
Patients
A total of 60 patients with TSCC who underwent
surgical treatment in The Second Affiliated Hospital of Harbin
Medical University between January 2014 and December 2016 were
included in the present study. The inclusion criteria were: i)
Patients with a clinically and histopathologically confirmed
diagnosis of TSCC; ii) received surgical resection as initial
treatment; iii) archived tumor tissue and paired adjacent tissues
that were stored at the Pathological Department of the hospital;
and iv) clinicopathological and follow-up data were complete and
accessible. Patients were excluded if they underwent radiotherapy
or chemotherapy prior to surgery. The present study was approved by
the Institutional Review Board of The Second Affiliated Hospital of
Harbin Medical University, and written informed consent was
obtained from all patients or their guardians.
Data collection
The demographic data and clinicopathological
characteristics of patients, including age, gender, pathological
grade, T stage, N stage, and tumor-node-metastasis (TNM) stage,
which was evaluated according to the criteria of the 7th edition
American Joint Committee on Cancer, were obtained from medical
records. The survival data, which were used to calculate OS, were
obtained from follow-up records (last follow-up date was
30/06/2018). The OS was defined as the duration from surgical
treatment to patient mortality.
ITGA7 and CD133 expression in TSCC and
paired adjacent tissues
Immunohistochemistry and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) were
performed to detect ITGA7 expression levels; immuno- histochemistry
was also conducted to detect CD133 expression levels in TSCC and
paired adjacent tissues.
Immunohistochemistry
TSCC and paired adjacent tissue specimens from
enrolled patients were acquired from the Pathological Department of
the aforementioned hospital; the samples were paraffin-embedded.
Then, immunohistochemistry was performed to assess ITGA7 and CD133
expression in tumor and paired adjacent normal tissues. Briefly,
paraffin-embedded tissue sections (4-μm thick) were
deparaffinized with xylene and rehydrated with ethanol.
Heat-induced antigen retrieval was subsequently conducted in 0.01
mol/l sodium citrate buffer (pH 6.0) using a microwave, and
endogenous peroxidase activity was inhibited with freshly prepared
3% H2O2. Following blocking using 1.5% normal
goat serum (Shanghai Yeasen Biotechnology Co., Ltd.) at 37°C for 20
min, the sections were incubated at 4°C overnight with rabbit
polyclonal ITGA7 antibody (1:200; cat. no. ab203254; Abcam) and
rabbit polyclonal CD133 antibody (1:100; cat. no. ab19898; Abcam).
The next day, tissue sections were incubated with horseradish
peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody
(diluted 1:1,000 in 3% bovine serum albumin; cat. no. ab6721;
Abcam) at 37°C for 90 min. Subsequently, 3,3′-diaminobenzidine and
hematoxylin were applied for staining of the sections, which were
then sealed with neutral tree gum. ITGA7 and CD133 expression was
evaluated under a light microscope.
Assessment of ITGA7 and CD133 expression
in TSCC and paired adjacent tissues
ITGA7 and CD133 expression in TSCC and paired
adjacent tissues was assessed by scores based on the average
intensity and percentage of positively stained cells, as described
previously (14). The intensity
scores were graded as follows: 0, no staining; 1, weak staining,
light yellow; 2, moderate staining, yellow brown; and 3, strong
staining, brown. The percentage of stained cells was scored as: 0,
0%; 1, 1-25%; 2, 26-50%; 3, 51-75%; and 4, 76-100% positive cells.
The total score was calculated by multiplying the density score and
the percentage score; high expression was defined as a total score
of ≥3, while low expression was defined as a total score of
<3.
Cell sources and culture
Four human TSCC cell lines and one normal human oral
keratinocyte cell line were purchased through a third agent company
(Shanghai QeeJen Bio-Tech Co., Ltd). In detail, the CAL-27 cell
line was purchased from Deutsche Sammlung von Mikroorganismen und
Zellkulturen GmbH, the SCC-9 cell line was purchased from American
Type Culture Collection, the HSC-4 cell line was purchased from
Japanese Collection of Research Bioresources Cell Bank, the SCC-25
cell line was purchased from Cell Bank of Type Culture Collection
of Chinese Academy of Sciences, and the HOK cell line was purchased
from ScienCell Research Laboratories, Inc.
The CAL-27 and SCC-9 cell lines were cultured in 90%
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.).
The HSC-4 cell line was cultured in 90% Eagle's minimal essential
medium (MEM; Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS.
The SCC-25 cell line was cultured in 90% Dulbecco's modified
Eagle's medium/Ham's F12 medium (DMEM/F12; Gibco; Thermo Fisher
Scientific, Inc.) with 10% FBS. The HOK cell line was cultured in
90% oral keratinocyte medium (ScienCell Research Laboratories,
Inc.) with 10% FBS. All the cells were cultured in incubators at
37°C under 95% air and 5% CO2 conditions.
ITGA7 and CD133 expression in TSCC and
normal cell lines
qPCR, western blotting and immunofluorescence were
used for the detection of the mRNA and protein expression levels of
ITGA7, and western blotting and immunofluorescence were used for
the detection of the expression levels of CD133, in the human TSCC
cell lines CAL-27, SCC-9, HSC-4 and SCC-25 and the normal human
oral keratinocyte cell line HOK (as a normal control).
Lentivirus construction and transduction
into CAL-27 and HSC-4 cells
Control short hairpin RNA (shRNA) and
ITGA7-targeting shRNA shuttle plasmids were constructed using the
pGLV-U6 vector (Shanghai GenePharma Co., Ltd.), and transfected
into 293T cells (American Type Culture Collection) together with
envelope plasmids (Shanghai GenePharma Co., Ltd.) and packaging
plasmids (Shanghai GenePharma Co., Ltd.) using HilyMax (Dojindo
Molecular Technologies, Inc.). The cell supernatant was obtained at
48 and 72 h post-transfection. Following purification and
concentrating, the corresponding lentiviruses were collected. The
sequences used for the knockdown experiments were as follows: ITGA7
shRNA, forward
5′-CACCGCTGCCCACTCTACAGCTTTTCGAAAAAAGCTGTAGAGTGGGCAGC-3′ and
reverse, 5′-AAAAGCTGCCCACTCTACAGCTTTTTTCGAAAAGCTGTAGAGTGGGCAGC-3′;
control shRNA, forward
5′-CACCGTTCTCCGAACGTGTCACGTCGAAACGTGACACGTTCGGAGAA-3′ and reverse
5′-AAAATTCTCCGAACGTGTCACGTTTCGACGTGACACGTTCGGAGAAC-3′.
Subsequently, using a multiplicity of infection of 10, control
shRNA and ITGA7 shRNA lentiviruses were added to the medium for the
transduction of CAL-27 and HSC-4 cells with 6 μg/ml
polybrene (Sigma-Aldrich; Merck KGaA) for 24 h, followed by the
addition of fresh complete medium for another 48 h. Then, cells
were cultured with 8 μg/ml puromycin (Thermo Fisher
Scientific, Inc.) for 7 days to construct stably transduced CAL-27
and HSC-4 cells. Cells transduced with the control shRNA lentivirus
were termed as the negative control (NC) group, while the cells
transduced with the ITGA7shRNA lentivirus were termed as the
ITGA7(-) group.
ITGA7 expression in the NC and ITGA7(-)
groups
Following the construction of stably transduced
CAL-27 and HSC-4 cells, ITGA7 expression was detected by RT-qPCR,
western blotting and flow cytometry in the NC and ITGA7(-) groups,
in order to determine successful transduction.
Effects of ITGA7 knockdown on the
proliferation and apoptosis of CAL-27 and HSC-4 cells
In the NC and ITGA7(-) groups of stably transduced
CAL-27 and HSC-4 cells, the apoptotic rate was detected using a
fluorescein isothiocyanate (FITC) Annexin-V Apoptosis Detection kit
II (BD Biosciences). Expression levels of the cell apoptotic
markers cleaved (C)-Caspase 3 and Bcl-2 were detected via western
blotting. In addition, an equal quantity of cells of the NC and
ITGA7(-) groups were cultured for 72 h, and the proliferative
ability was detected at 0, 24, 48 and 72 h via a Cell Counting
Kit-8 (CCK-8) assay (Sigma-Aldrich; Merck KGaA).
Effects of ITGA7 knockdown on CSC markers
of CAL-27 and HSC-4 cells
In the NC and ITGA7(-) groups of stably transduced
CAL-27 and HSC-4 cells, the CSC markers cluster of differentiation
(CD)24, CD44 and CD133 were detected by RT-qPCR and western
blotting. In addition, ITGA-positive cells, as well as
ITGA7-negative cells, were isolated by flow cytometry, and the
protein expression levels of ITGA7, CD24, CD44 and CD133 were
detected by western blotting.
Effects of ITGA7 knockdown on drug
resistance to cisplatin in CAL-27 and HSC-4 cells
Cisplatin (5 μM; Sigma-Aldrich; Merck KGaA)
was applied to the NC and ITGA7(-) groups of stably transduced
CAL-27 and HSC-4 cells for 24 h. Then, the drug resistance ability
was assessed by detecting cell viability and apoptosis. Briefly,
cell viability was detected via a CCK-8 assay and the relative cell
viability of the ITGA7 (-) group was calculated according to the
reference values of the NC group. The rate of cell apoptosis was
determined using an FITC Annexin-V Apoptosis Detection Kit II.
Effects of ITGA7 knockdown on the sphere
formation ability of CAL-27 and HSC-4 cells
The sphere formation ability of the NC and ITGA7(-)
groups of stably transduced CAL-27 and HSC-4 cells was analyzed by
a sphere formation assay. Briefly, transduced CAL-27 and HSC-4
cells were cultured in DMEM/F12 medium supplemented with 2% B27
(Gibco; Thermo Fisher Scientific, Inc.), 20 ng/ml epidermal growth
factor (EGF; Sigma-Aldrich; Merck KGaA), 20 ng/ml basic fibroblast
growth factor (bFGF; Gibco; Thermo Fisher Scientific, Inc.) and 4
μg/ml heparin (Sigma-Aldrich; Merck KGaA) for 10 days;
spheres of a diameter >50 μm were counted under a light
microscope (Olympus Corporation). The sphere formation ability was
calculated by dividing the number of these spheres by the total
number of seeded cells (200) ×1,000. In addition, the sphere
formation assay was conducted using the extreme limiting dilution
method. CAL-27 and HSC-4 cells were cultured in DMEM/F12 medium
supplemented with 2% B27, 20 ng/ml EGF (Sigma-Aldrich; Merck KGaA),
20 ng/ml bFGF and 4 μg/ml heparin (Sigma-Aldrich; Merck
KGaA) for 10 days in a 24 well-plate at densities of 1,000, 100 or
10 cells per well. Subsequently, spheres with a diameter >50
μm were counted under a light microscope (Olympus
Corporation) and the sphere formation ability was calculated using
an extreme limiting dilution analysis software (http://bioinf.wehi.edu.au/software/elda/) (15).
Expression of ITGA7 in CAL-27 and HSC-4
CSCs
CAL-27 and HSC-4 CSCs were generated by establishing
drug-resistant cells, followed by detection with a sphere formation
assay (16). Briefly, 5 μM
cisplatin was applied to CAL-27 and HSC-4 cells for 72 h, and then
cisplatin-free medium was applied to cells for another 72 h; these
processes were repeated until no effect of cisplatin on cell
viability/proliferation was observed by the CCK-8 assay. A sphere
formation assay was performed as aforementioned, and spheres of
CAL-27 and HSC-4 CSCs were isolated by centrifugation (300 × g at
room temperature for 3 min). To determine the successful
establishment of CSCs, the expression levels of CSC markers CD24,
CD44 and CD133, as well as ITGA7, were detected by RT-qPCR and
western blotting in CAL-27/HSC-4 CSCs and normal parental
CAL-27/HSC-4 cells.
RT-qPCR
TRIzol® reagent (Thermo Fisher Scientific, Inc.) was
used for the extraction of total RNA. The ReverTra Ace® qPCR RT kit
(Toyobo Life Science) was used to transcribe 1 μg RNA into
cDNA. SYBR® Green Realtime PCR Master Mix (Toyobo Life Science) was
used for qPCR, which was performed as follows: 95°C for 5 min,
followed by 40 cycles of 95°C for 5 sec and 61°C for 30 sec. The
sequences of primers employed for qPCR are listed in Table I. Relative fold changes in mRNA
expression were calculated using the 2–ΔΔCq method
(17,18). GAPDH was used as the internal
reference.
 | Table IPrimers used in quantitative PCR. |
Table I
Primers used in quantitative PCR.
| Gene | Forward primer
(5'-3') | Reverse primer (5'
- 3') |
|---|
| ITGA7 |
GCCACTCTGCCTGTCCAATG |
GGAGGTGCTAAGGATGAGGTAGA |
| CD24 |
GCTCCTACCCACGCAGATTT |
CACGAAGAGACTGGCTGTTGA |
| CD44 |
ACATCCTCACATCCAACACCTC |
CCTCCTGAAGTGCTGCTCCT |
| CD133 |
GCTGCTTGTGGAATAGACAGAATG |
GAAGGACTCGTTGCTGGTGAAT |
| GAPDH |
GAGTCCACTGGCGTCTTCAC |
ATCTTGAGGCTGTTGTCATACTTCT |
|
| ITGA7, integrin
α7. |
Western blotting
Radioimmunoprecipitation assay buffer (Thermo Fisher
Scientific, Inc.) was used for the extraction of total protein, and
a Pierce™ BCA Protein Assay kit (Thermo Fisher Scientific, Inc.)
was used to determine protein concentration; a standard curve was
used for adjustments. The protein sample (20 μg) was
fractionated via a NuPAGE™ 4-12% Bis-Tris Protein Gel (Invitrogen;
Thermo Fisher Scientific, Inc.), and then transferred to
nitrocellulose filter membranes (EMD Millipore). Subsequently, the
membranes were blocked using 5% skim milk for 2 h at 37°C, and were
incubated with primary antibodies overnight at 4°C. After the
secondary antibody was applied for 1 h at room temperature, Pierce™
Enhanced Chemiluminescence Plus Western Blotting Substrate (Thermo
Fisher Scientific, Inc.) was used for the visualization of bands,
which were then exposed using X-ray film (Kodak). GAPDH was used as
the internal reference. The detailed information regarding the
antibodies employed for western blot analysis are listed in
Table II.
 | Table IIAntibodies used in western
blotting. |
Table II
Antibodies used in western
blotting.
A, Primary
antibodies
|
|---|
| Target | Catalog no. | Company | Dilution |
|---|
| ITGA7 | ab203254 | Abcam | 1:2,000 |
| Caspase - 3 | #14220 | Cell Signaling
Technology, Inc. | 1:1,000 |
| Cleaved
caspase-3 | #9661 | Cell Signaling
Technology, Inc. | 1:1,000 |
| Bcl -2 | ab32124 | Abcam | 1:1,000 |
| CD24 | ab179821 | Abcam | 1:2,000 |
| CD44 | ab51037 | Abcam | 1:5,000 |
| CD133 | ab19898 | Abcam | 1:1,000 |
| GAPDH | #2118 | Cell Signaling
Technology, Inc. | 1:1,000 |
|
B, Secondary
antibody
|
| Target | Catalog no. | Company | Dilution |
|
| HRP conjugated goat
anti rabbit IgG | ab205718 | Abcam | 1:2,000 |
|
| ITGA7, integrin α7;
HRP, horseradish peroxidase; IgG, immunoglobulin G. |
Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 10 min
at room temperature, permeabilized with 0.2% Triton X-100 for 5 min
and blocked with PBS containing 2% BSA for 1 h at room temperature.
Cells were then incubated with primary antibodies targeting ITGA7
(1:200; cast. no. ab203254; Abcam) and anti-CD133 (1:100; cat. no.
ab19898; Abcam) overnight at 4°C. Following three washes with PBS,
cells were incubated with an FITC-conjugated goat anti-rabbit
immunoglobulin G antibody (1:1,000; cat. no. ab6717; Abcam) for 1 h
at 37°C. The nuclei were counterstained with DAPI. Images were
obtained using a BX41 fluorescence microscope (Olympus Corporation)
at x400 magnification.
Flow cytometry
Cells were harvested and washed with PBS, and then
incubated with rabbit polyclonal antibody against ITGA7 (1:50; cat.
no. ab203254; Abcam) in incubation buffer (PBS) for 1 h at 4°C.
Then, cells were washed with PBS and incubated with a
FITC-conjugated goat anti-rabbit immunoglobulin G antibody
(1:1,000; cat. no. ab6717; Abcam) for 1 h at 4°C in the dark.
Subsequently, the cells were washed again, resuspended in FACS
buffer (PBS) and analyzed by flow cytometry (Flowjo Vision 7.6; BD
Biosciences) using a FACS Canto II flow cytometer (BD
Biosciences).
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM Corp.) and GraphPad Prism 7.00 (GraphPad Software
Inc.). Normal distributed continuous variables were presented as
the mean + standard deviation, and categorized variables were
presented as a percentage. Comparisons of ITGA7 expression between
tumor and paired adjacent tissues were conducted via a McNemar
test. Associations between ITGA7 expression and patient
characteristics were determined by a χ2 or Wilcoxon rank
sum test. Differences between the OS of patients with low and high
ITGA7 expression were determined via Kaplan-Meier analysis,
followed a log-rank test. Univariate and multivariate Cox
proportional hazards regression analyses were performed to
determine the factors affecting OS. Comparisons among groups in
cell experiments were conducted with one-way ANOVA followed by
Dunnett's multiple comparisons test. Comparisons between two groups
in cell experiments were performed with unpaired parametric t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Baseline characteristics in patients with
TSCC
A total of 60 patients with TSCC were enrolled with
a mean age of 55.2+10.5 years, comprising 44 males and 16 females
(Table III). Regarding
pathological grade, the number of patients with TSCC in grade (G)1,
G2 and G3 were 8 (13.3%), 42 (70.0%) and 10 (16.7%), respectively.
There were 10 (16.7%), 24 (40.0%), 24 (40.0%) and 2 (3.3%) patients
with TSCC of TNM stage I, II, III and IV, respectively. The
detailed information of other clinicopathological characteristics
of the patients is presented in Table III.
 | Table IIIClinical characteristics of patients
with TSCC. |
Table III
Clinical characteristics of patients
with TSCC.
|
Characteristics | TSCC patients
(n=60) |
|---|
| Age (years) | 55.2±10.5 |
| Gender
(male/female) | 44/16 |
| Pathological grade,
n (%) | |
| G1 | 8 (13.3) |
| G2 | 42 (70.0) |
| G3 | 10 (16.7) |
| T stage, n (%) | |
| T1 | 12 (20.0) |
| T2 | 30 (50.0) |
| T3 | 18 (30.0) |
| N stage, n (%) | |
| N0 | 38 (63.3) |
| N1 | 20 (33.3) |
| N2 | 2 (3.4) |
| TNM stage, n
(%) | |
| I | 10 (16.7) |
| II | 24 (40.0) |
| III | 24 (40.0) |
| IV | 2 (3.3) |
Comparison of ITGA7 expression between
tumor tissue and paired adjacent tissue
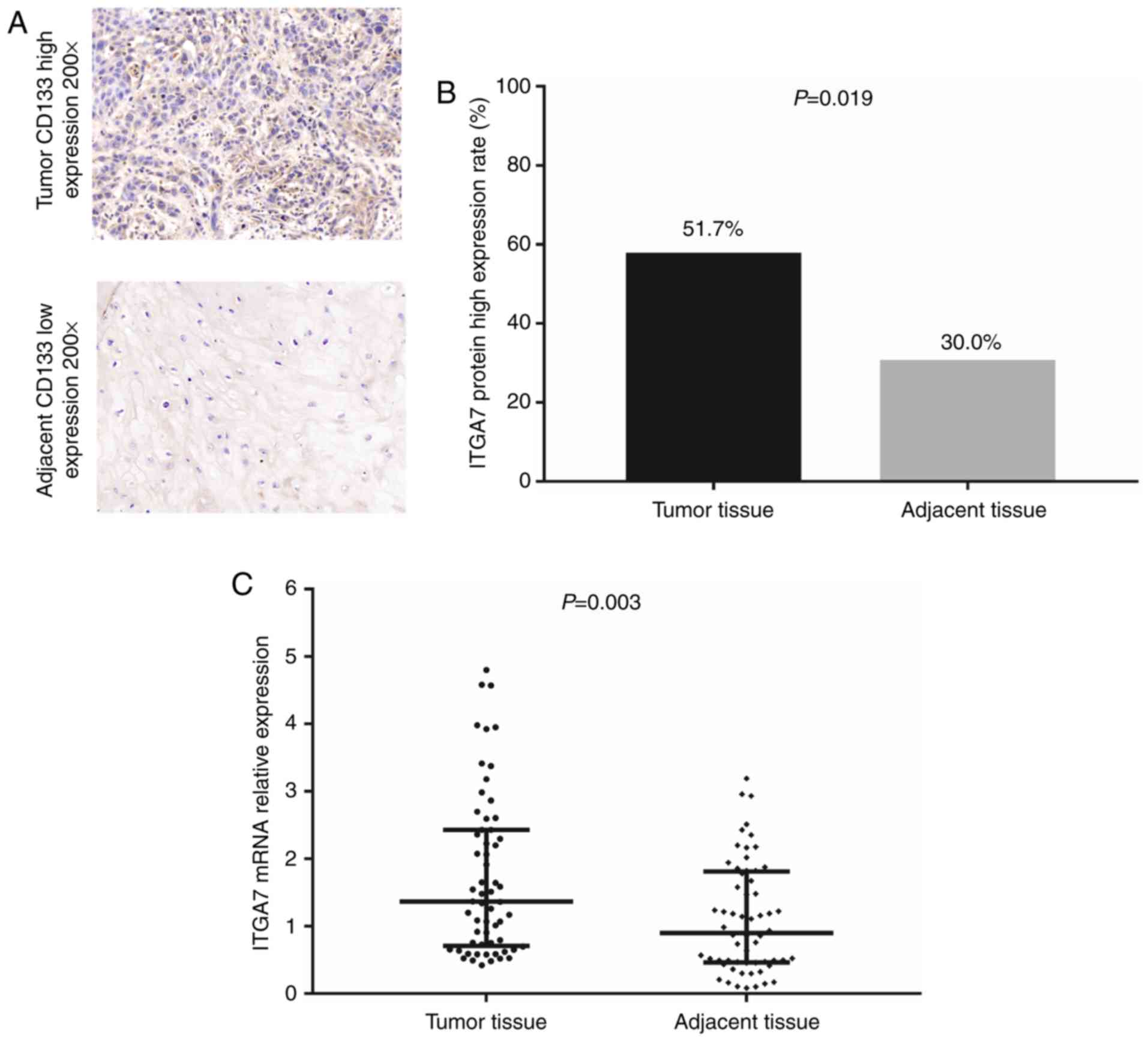
Immunohistochemistry was conducted for the detection
of ITGA7 expression in tumor and paired adjacent tissues;
representative images of high and low ITGA7 staining are presented
in Fig. 1A. The results from the
immunohistochemistry analysis demonstrated that the protein
expression levels of ITGA7 were increased in the tumor tissues
compared with paired adjacent tissues (P=0.019; Fig. 1B). Similar results were obtained by
RT-qPCR analysis for the ITGA7 mRNA expression levels (P<0.001;
Fig. 1C). In addition, the
expression of the CSC marker CD133 was detected by
immunohistochemistry in the patient tissues (Fig. S1A). The results revealed that
CD133 expression was upregulated in tumor tissues compared with
adjacent normal tissues (P=0.048; Fig. S1B).
Association between ITGA7 expression and
the clinicopathological characteristics of patients with TSCC
Increased protein expression of ITGA7 was associated
with advanced pathological grade (P=0.021), and higher N (P=0.011)
and TNM (P=0.005) stages. However, no association between ITGA7
protein expression and age (P=0.465 and 0.876) or gender (P= 0.876)
was observed in patients with TSCC (Table IV). In addition, high ITGA7 mRNA
expression was associated with advanced pathological grade
(P=0.003), increased T stage (P=0.002) and higher TNM stage
(P=0.010) in patients with TSCC (Table IV).
 | Table IVAssociation of ITGA7 expression with
patients' clinical characteristics. |
Table IV
Association of ITGA7 expression with
patients' clinical characteristics.
| Parameters | n | ITGA7 protein
expression
| P value | ITGA7 mRNA
expression | P value |
|---|
| Low | High | Low | High |
|---|
| Age, n (%) | | | | 0.465 | | | 0.584 |
| <60 years | 40 | 18 (45.0) | 22 (55.0) | | 21 (52.5) | 19 (47.5) | |
| ≥60 years | 20 | 11 (55.0) | 9 (45.0) | | 9 (45.0) | 11 (55.0) | |
| Gender, n (%) | | | | 0.876 | | | 0.243 |
| Male | 44 | 21 (47.8) | 23 (52.2) | | 20 (45.5) | 24 (54.5) | |
| Female | 16 | 8 (50.0) | 8 (50.0) | | 10 (62.5) | 6 (37.5) | |
| Pathological grade,
n (%) | | | | 0.021 | | | 0.003 |
| G1 | 8 | 7 (87.5) | 1 (12.5) | | 8 (100.0) | 0 (0.0) | |
| G2 | 42 | 19 (45.2) | 23 (54.8) | | 20 (47.6) | 22 (52.4) | |
| G3 | 10 | 3 (30.0) | 7 (70.0) | | 2 (20.0) | 8 (80.0) | |
| T stage, n (%) | | | | 0.073 | | | 0.002 |
| T1 | 12 | 8 (66.7) | 4 (33.3) | | 9 (75.0) | 3 (25.0) | |
| T2 | 30 | 15 (50.0) | 15 (50.0) | | 18 (60.0) | 12 (40.0) | |
| T3 | 18 | 6 (33.3) | 12 (66.7) | | 3 (16.7) | 15 (83.3) | |
| N stage, n (%) | | | | 0.011 | | | 0.064 |
| N0 | 38 | 23 (60.5) | 15 (39.5) | | 23 (60.5) | 15 (39.5) | |
| N1 | 20 | 6 (30.0) | 14 (70.0) | | 7 (35.0) | 13 (65.0) | |
| N2 | 2 | 0 (0.0) | 2 (100.0) | | 0 (0.0) | 2 (100.0) | |
| TNM stage, n
(%) | | | | 0.005 | | | 0.010 |
| I | 10 | 8 (80.0) | 2 (20.0) | | 8 (80.0) | 2 (20.0) | |
| II | 24 | 13 (54.2) | 11 (45.8) | | 15 (62.5) | 9 (37.5) | |
| III | 24 | 8 (33.3) | 16 (66.7) | | 7 (29.2) | 17 (70.8) | |
| IV | 2 | 0 (0.0) | 2 (100.0) | | 0 (0.0) | 2 (100.0) | |
Association between ITGA7 expression and
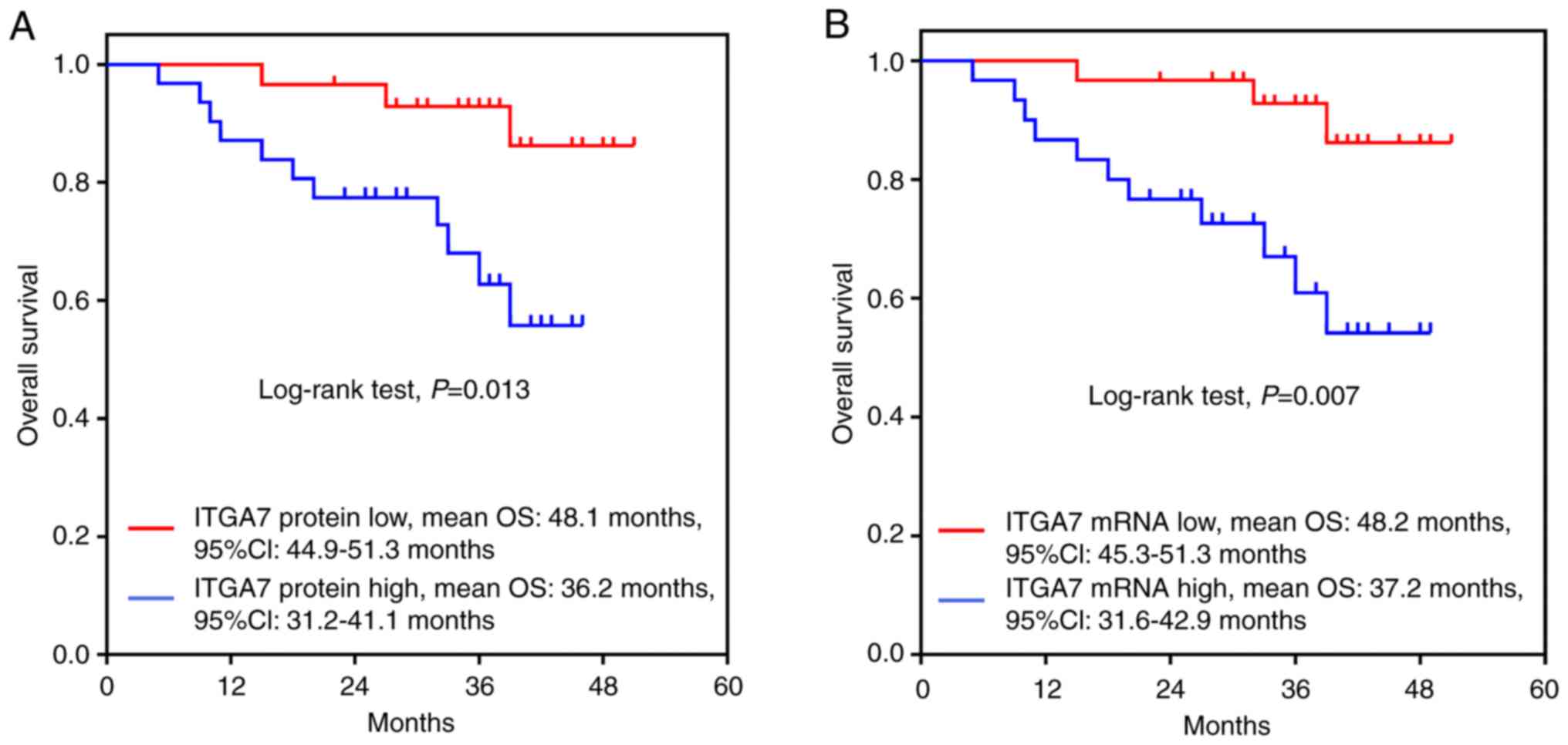
the OS of patients with TSCC
Kaplan-Meier analysis revealed that the OS was
shorter in patients with high ITGA7 protein expression levels [mean
OS: 36.2 months, 95% confidence interval (CI): 31.24-41.1 months]
compared with those possessing low ITGA7 protein expression (mean
OS: 48.1 months, 95% CI: 44.9-51.3 months; P=0.013; Fig. 2A). In addition, a similar trend was
reported for patients with high ITGA7 mRNA expression (mean OS:
37.2 months, 95% CI: 31.6-42.9 months) compared with those with low
ITGA7 mRNA expression (mean OS: 48.2 months, 95% CI: 45.3-51.3
months; P=0.007; Fig. 2B).
Factors affecting the OS of patients with
TSCC
Univariate Cox proportional hazards regression model
analysis was used to evaluate the effect of all clinicopathological
factors on the OS of patients with TSCC. The results revealed that
high ITGA7 protein (P=0.024) and mRNA expression (P=0.015), higher
pathological grade (P= 0.049), advanced T stage (P=0.012), higher N
stage (P=0.001), as well as advanced TNM stage (P<0.001) were
associated with poorer OS in patients with TSCC (Table V). However, when multivariate Cox
analysis was performed, only TNM stage could independently predict
OS, while high ITGA7 expression (protein or mRNA) was not an
independent factor for predicting shorter OS in patients with TSCC.
This suggested that ITGA7 may be indirectly associated with poor
prognosis by affecting tumor size, pathological grade or lymph node
metastasis in patients with TSCC.
 | Table VUnivariate and multivariate Cox
proportional hazards regression model analysis of factors affecting
overall survival. |
Table V
Univariate and multivariate Cox
proportional hazards regression model analysis of factors affecting
overall survival.
A, Univariate
analysis
|
|---|
| Parameters | Cox proportional
hazards regression model
|
|---|
| P-value | HR | 95% CI
|
|---|
| Lower | Higher |
|---|
| ITGA7 protein high
expression | 0.024 | 4.384 | 1.219 | 15.769 |
| ITGA7 mRNA high
expression | 0.015 | 4.927 | 1.367 | 17.756 |
| Age | 0.637 | 1.011 | 0.966 | 1.059 |
| Gender (male) | 0.102 | 5.460 | 0.713 | 41.790 |
| Higher pathological
grade | 0.049 | 2.499 | 0.999 | 6.248 |
| Higher T stage | 0.012 | 3.030 | 1.276 | 7.199 |
| Higher N stage | 0.001 | 5.990 | 2.084 | 17.216 |
| Higher TNM
stage |
<0.001 | 12.764 | 3.143 | 51.838 |
|
B, Multivariate
analysis
|
| Parameters | Cox proportional
hazards regression model
|
| P-value | HR | 95% CI
|
| Lower | Higher |
| ITGA7 protein high
expression | 0.643 | 1.416 | 0.326 | 6.140 |
| ITGA7 mRNA high
expression | 0.641 | 1.469 | 0.292 | 7.384 |
| Age | 0.908 | 1.073 | 0.326 | 3.528 |
| Gender (male) | 0.462 | 2.236 | 0.262 | 19.069 |
| Higher pathological
grade | 0.144 | 3.046 | 0.683 | 13.594 |
| Higher T stage | 0.219 | 0.502 | 0.167 | 1.507 |
| Higher N stage | 0.244 | 0.364 | 0.067 | 1.993 |
| Higher TNM
stage | 0.008 | 25.675 | 2.305 | 285.942 |
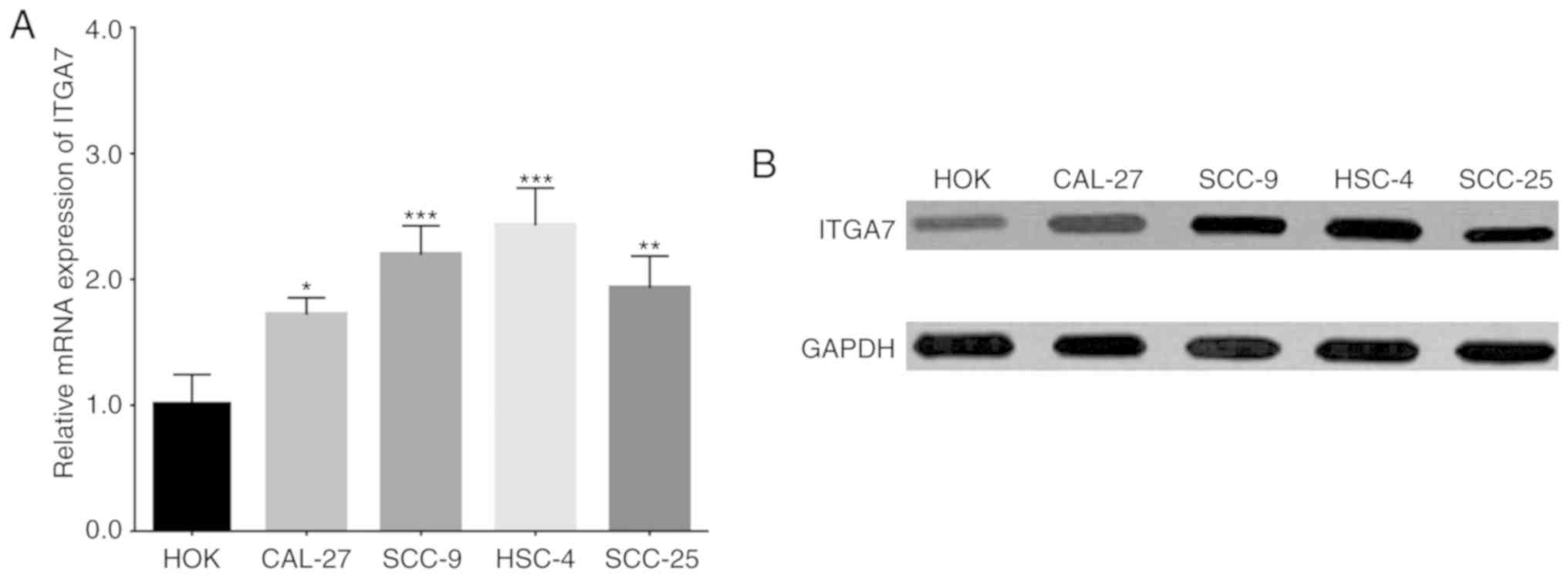
Comparison of ITGA7 expression in TSCC
and normal human oral keratinocyte cell lines
In order to investigate the function of ITGA7 in
TSCC cells, in vitro experiments were performed. Firstly,
the expression of ITGA7 was detected in several established TSCC
cell lines and a normal human oral keratinocyte cell line. Compared
to the normal HOK cells, both ITGA7 mRNA (Fig. 3A) and protein (Fig. 3B) expression levels were increased
in the human TSCC cell lines CAL-27, SCC-9, HSC-4 and SCC-25.
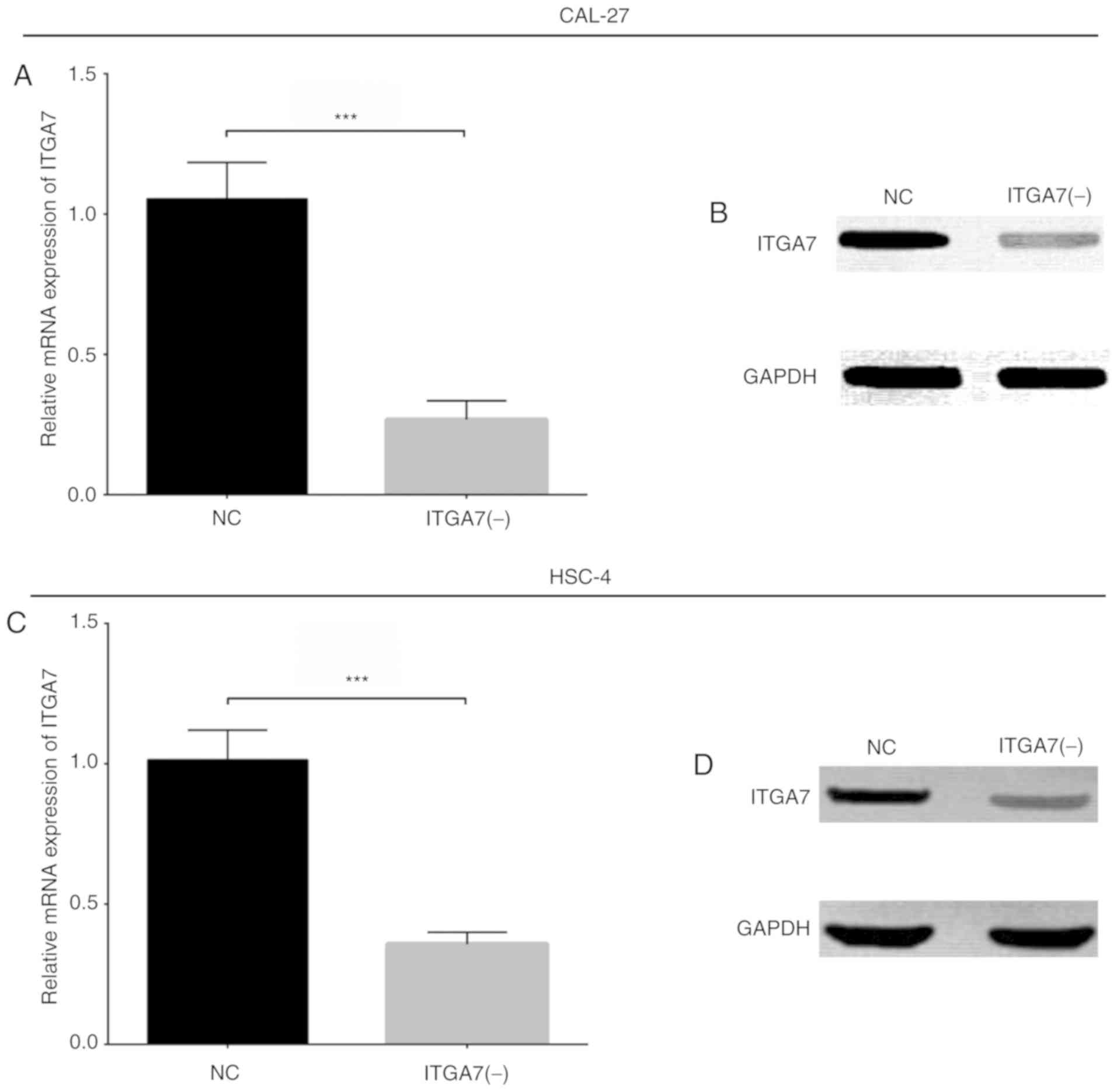
ITGA7 knockdown in CAL-27 and HSC-4
cells
In order to investigate the underlying mechanism of
ITGA7 in CAL-27 and HSC-4 cells, control NC shRNA and ITGA7 shRNA
lentiviruses were constructed and used to transduce these cell
lines, hence generating the NC and ITGA7(-) cell groups,
respectively. In CAL-27 cells, the mRNA (P<0.001; Fig. 4A) and protein (Fig. 4B) expression levels of ITGA7 were
down- regulated in the ITGA7(-) group compared with the NC group.
Additionally, a similar trend of ITGA7 expression at the mRNA
(P<0.001; Fig. 4C) and protein
(Fig. 4D) levels was observed
between the ITGA7(-) and NC groups of HSC-4 cells. These findings
suggested the successful construction of stably transduced
ITGA7-silenced TSCC cell lines. In addition, the results of flow
cytometry demonstrated that the percentage of ITGA7+ cells was
decreased in the ITGA7(-) group compared with the NC group, for
both the CAL-27 and HSC-4 cell lines (P<0.01; Fig. S2A-D).
Effects of ITGA7 knockdown on the
proliferation and apoptosis of CAL-27 and HSC-4 cells
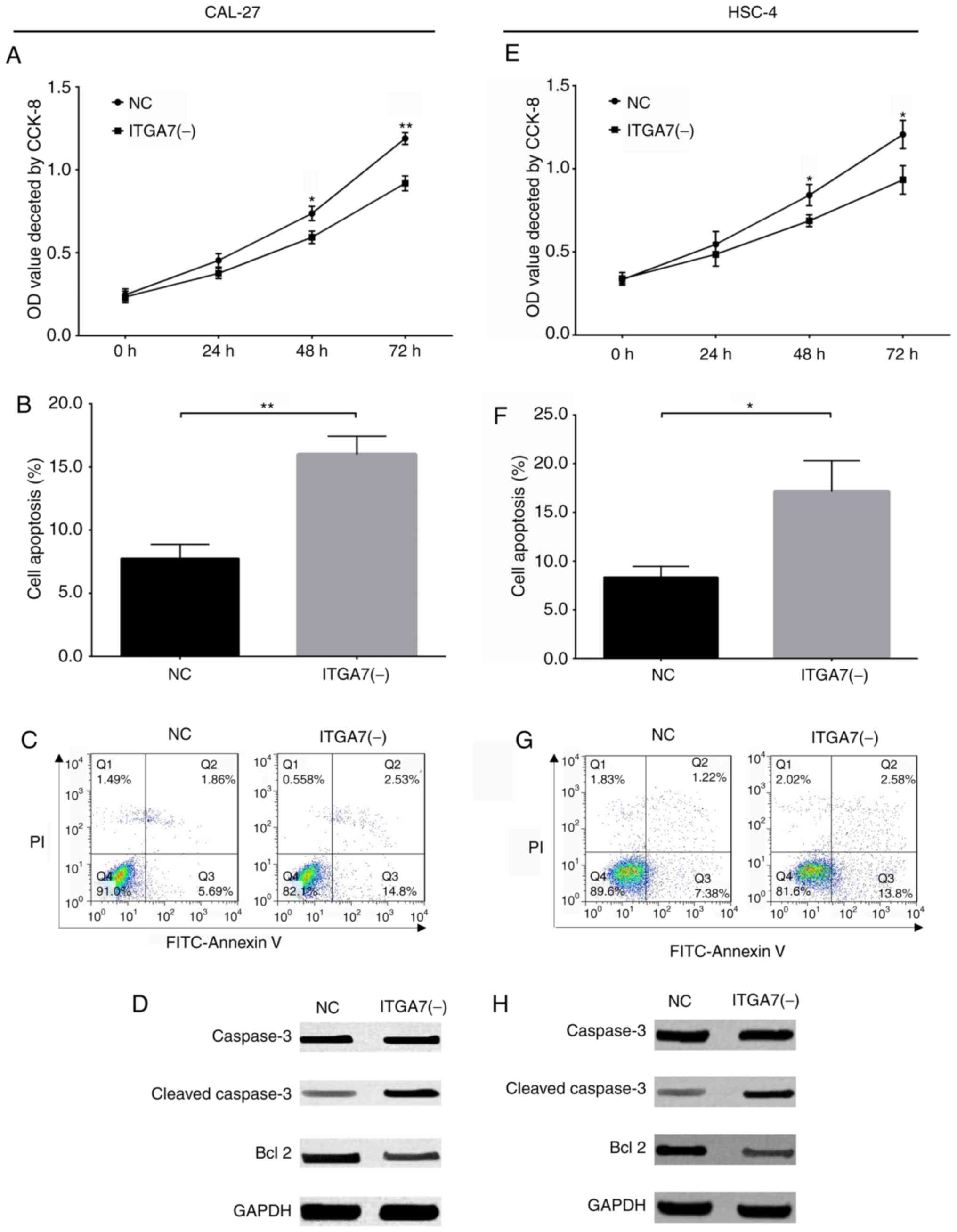
The present study investigated the effects of ITGA7
knockdown on the proliferation and apoptosis of CAL-27 and HSC-4
cells. A CCK-8 assay revealed that cell proliferation was decreased
in the ITGA7(-) group compared with the NC group at 48 (P<0.05)
and 72 h (P<0.01) for CAL-27 cells (Fig. 5A), and at 48 (P<0.05) and 72 h
(P<0.05) for HSC-4 cells (Fig.
5E). The rate of cell apoptosis was increased in the ITGA7(-)
group compared with the NC group for CAL-27 cells (P<0.01;
Fig. 5B and C) and HSC-4 cells
(P<0.05; Fig. 5F and G).
Western blot analysis revealed that the expression of the apoptotic
protein marker C-Caspase 3 was increased, but the expression of the
anti-apoptotic Bcl-2 was decreased, in the ITGA7(-) group compared
with the NC group for CAL-27 cells (Fig. 5D) and HSC-4 cells (Fig. 5H). These findings indicated that
ITGA7 knockdown inhibited cell proliferation, but promoted
apoptosis in CAL-27 and HSC-4 cells.
Effects of 1TGA7 knockdown on regulating
common CSC markers in CAL-27 and HSC-4 cells
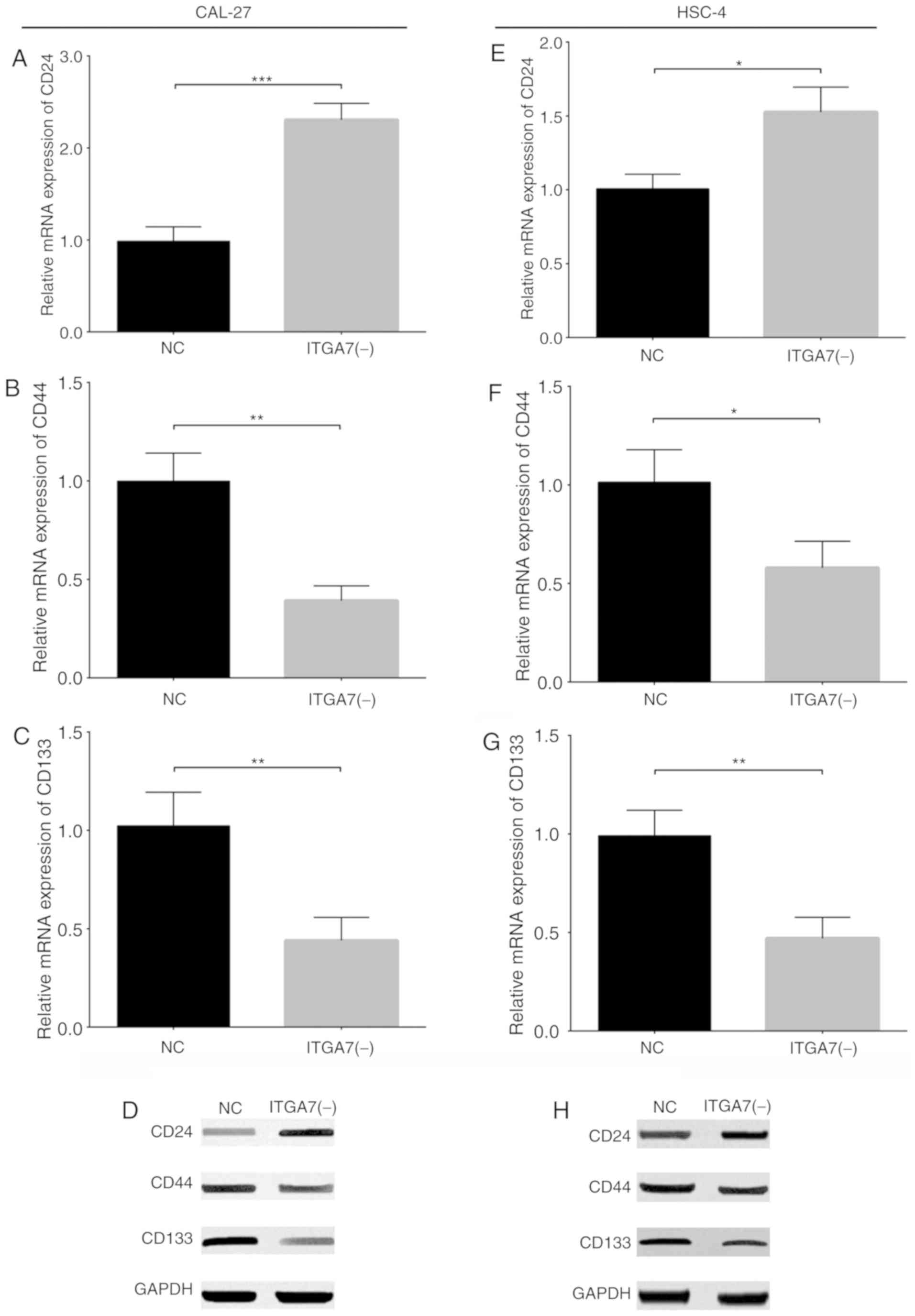
To investigate the effects of ITGA7 knockdown on the
sternness of TSCC cells, the expression of the common CSC markers
CD24, CD44 and CD133 was examined in CAL-27 and HSC-4 cells.
RT-qPCR and western blotting demonstrated that the mRNA and protein
expression of CD24 (P<0.001; Fig.
6A and D) were increased, while that of CD44 (P<0.01;
Fig. 6B and D) and CD133
(P<0.01; Fig. 6C and D) were
decreased in the ITGA7(-) group compared with the NC group of
CAL-27 cells. Additionally, similar trends were observed in the
expression of CD24 (P<0.05; Fig. 6E
and H), CD44 (P<0.05; Fig. 6F
and H) and CD133 (P<0.01; Fig.
6G and H) between the ITGA7(-)and NC groups of HSC-4 cells.
These results suggested that ITGA7 knockdown regulated the
expression of common CSC markers in CAL-27 and HSC-4 cells.
In addition, immunofluorescence experiments were
performed to detect ITGA7 and CD133 expression in TSCC cell lines
and the normal human oral keratinocytes cell line, and the results
revealed that ITGA7 and CD133 expression levels were markedly
increased in human TSCC cell lines CAL-27, SCC-9, HSC-4 and SCC-25
compared with the normal HOK cells (Fig. S3). Furthermore, ITGA7-positive
cells and ITGA7-negative cells were sorted, and the protein
expressions of ITGA7, CD24, CD44 and CD133 were detected in the
separate cell populations by western blot analysis. The results
demonstrated that IT GA7, CD44 and CD133 protein expressions were
increased, while CD24 protein expression was decreased in
ITGA7-positive cells compared with ITGA7-negative cells in both
CAL-27 and HSC-4 cell lines (Fig.
S4). These findings indicated that high ITGA7 expression was
associated with high expression of CSC markers in TSCC cells.
Effects of 1TGA7 knockdown on drug
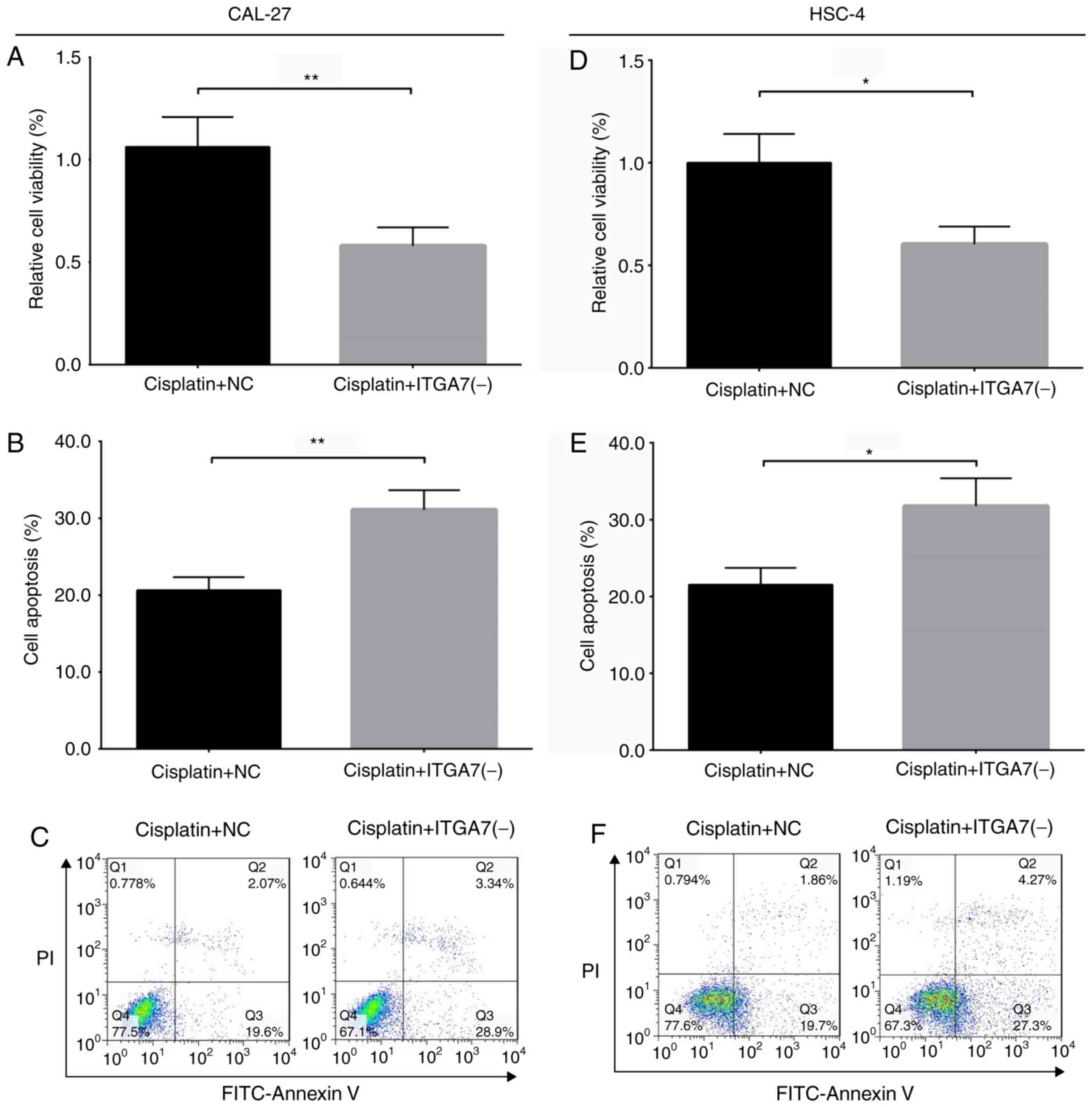
resistance to cisplatin in CAL-27 and HSC-4 cells
To further explore the role of ITGA7 in the
sternness of TSCC cells, the effect of ITGA7 knockdown on drug
resistance to cisplatin was investigated in CAL-27 and HSC-4 cells.
A CCK-8 assay revealed that the relative cell viability decreased
in the cisplatin + ITGA7(-) group compared with the cisplatin + NC
group for CAL-27 cells (P<0.01; Fig. 7A) and HSC-4 cells (P<0.05;
Fig. 7D). In addition, the rate of
apoptosis increased in the cisplatin + ITGA7(-) group compared with
the cisplatin + NC group of CAL-27 cells (P<0.01; Fig. 7B and C) and HSC-4 cells (P<0.05;
Fig. 7E and F). These results
suggested that ITGA7 knockdown decreased drug resistance to
cisplatin in these cell lines.
Effects of 1TGA7 knockdown on sphere
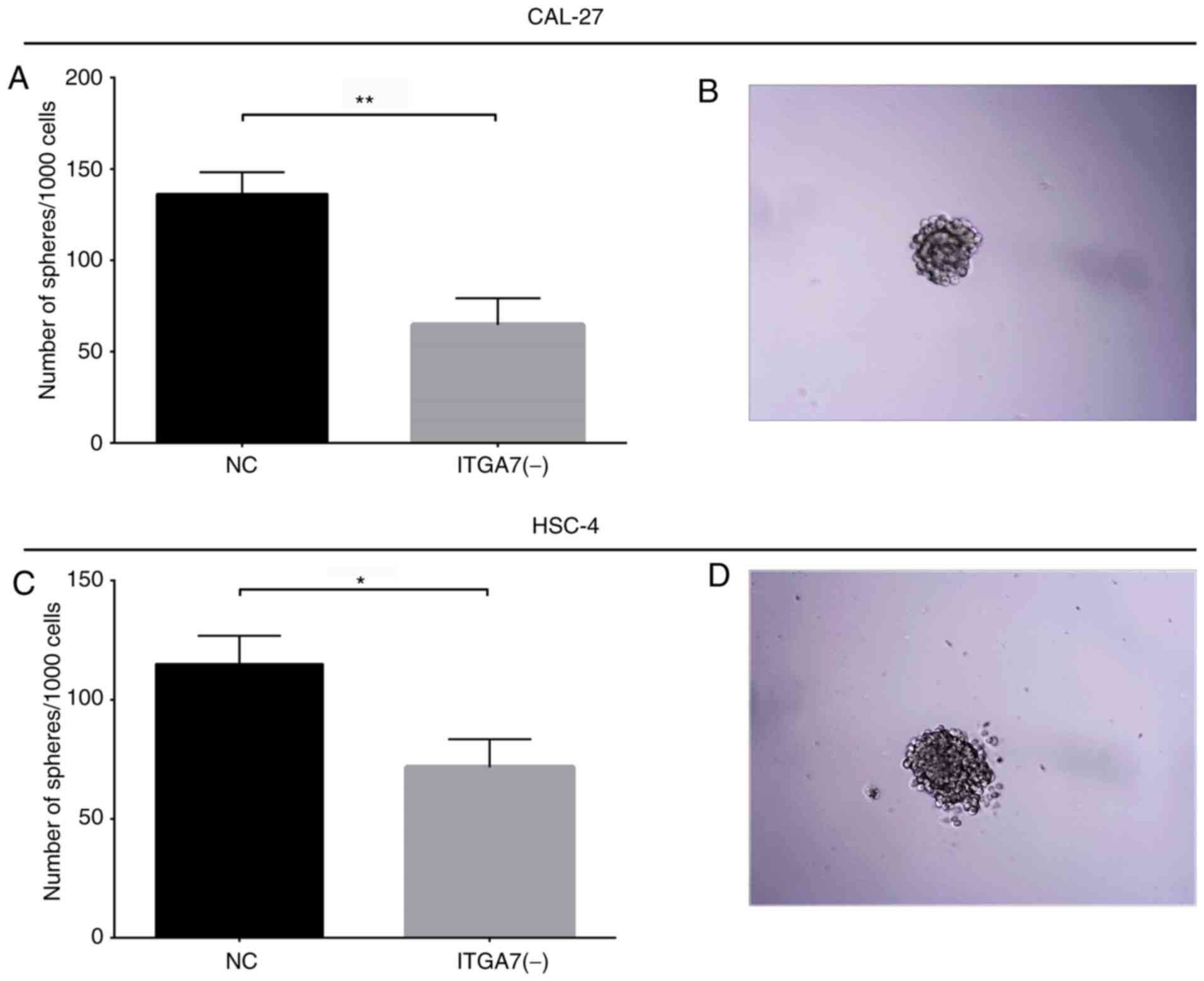
formation ability of CAL-27 and HSC-4 cells
To further validate the effects of ITGA7 knockdown
on the sternness of TSCC cells, sphere formation assays were
performed in CAL-27 and HSC-4 cells. The results demonstrated that
the sphere formation ability was reduced in the ITGA7(-) group
compared with the NC group for CAL-27 cells (P<0.01; Fig. 8A and B) and HSC-4 cells (P<0.05;
Fig. 8C and D). In addition, an
extreme limited dilution assay revealed that the sphere formation
ability was reduced in the ITGA7(-) group compared with the NC
group for CAL-27 and HSC-4 cells (both P<0.001; Table SI). These data indicated that
ITGA7 knockdown decreased the sphere formation ability of CAL-27
and HSC-4 cells. Therefore, the aforementioned findings that ITGA7
regulated the expression of CSC markers, decreased drug resistance
to cisplatin and suppressed sphere formation in CAL-27 and HSC-4
cells, indicated that ITGA7 knockdown may suppress TSCC cell
stemness.
Potential of ITGA7 as a marker for CAL-27
and HSC-4 CSCs
In order to explore whether ITGA7 is a potential
marker for TSCC stem cells, the expression of ITGA7 in
drug-resistant (R-) CAL-27 and R-HSC-4 CSCs was detected, and
sphere formation assays were performed. A CCK-8 assay demonstrated
no notable differences between the R-CAL-27 + cisplatin (Fig. 9A) and R-HSC-4 + cisplatin groups
(Fig. 9I) at 0, 24, 48 and 72 h
(P>0.05), which indicated the successful generation of
drug-resistant cells. In addition, a sphere formation assay was
performed for CAL-27 (Fig. 9B) and
HSC-4 (Fig. 9J) cells. The number
of spheres/1,000 cells was increased in the R-CAL-27 (P<0.01;
Fig. 9C) and R-HSC-4 (P<0.01;
Fig. 9K) cells compared with the
corresponding parental control cell lines. Next, spheres from the
R-CAL-27 and R-HSC-4 groups were isolated by centrifugation,
serving as CAL-27 and HSC-4 CSCs. To further validate the
establishment of CAL-27 and HSC-4 CSCs, the expression of common
CSC markers was detected; the mRNA and protein expression levels of
CD24 (Fig. 9D and H) were
decreased, while those of CD44 (P<0.01; Fig. 9E and H) and CD133 (P<0.001;
Fig. 9F and H) were increased in
CAL-27 CSCs compared with parental CAL-27 cells. Additionally,
similar trends in the expression of CD24 (P<0.01; Fig. 9L and P), CD44 (P<0.01; and
Fig. 9M and P) and CD133
(P<0.001; Fig. 9N and P) were
observed for HSC-4 CSCs compared with parental HSC-4 cells, which
suggested that CAL-27 and HSC-4 CSCs were successfully obtained.
Following the successful establishment of CAL-27 and HSC-4 CSCs,
the expression of ITGA7 was detected in the TSCC stem cells. The
results revealed that the mRNA and protein expression levels of
ITGA7 were increased in CAL-27 CSCs compared with parental CAL-27
cells (P<0.001; Fig. 9G and H),
and in HSC-4 CSCs compared with parental HSC-4 cells (P<0.01;
Fig. 9O and P). These findings
indicated that ITGA7 may be a potential marker of TSCC stem
cells.
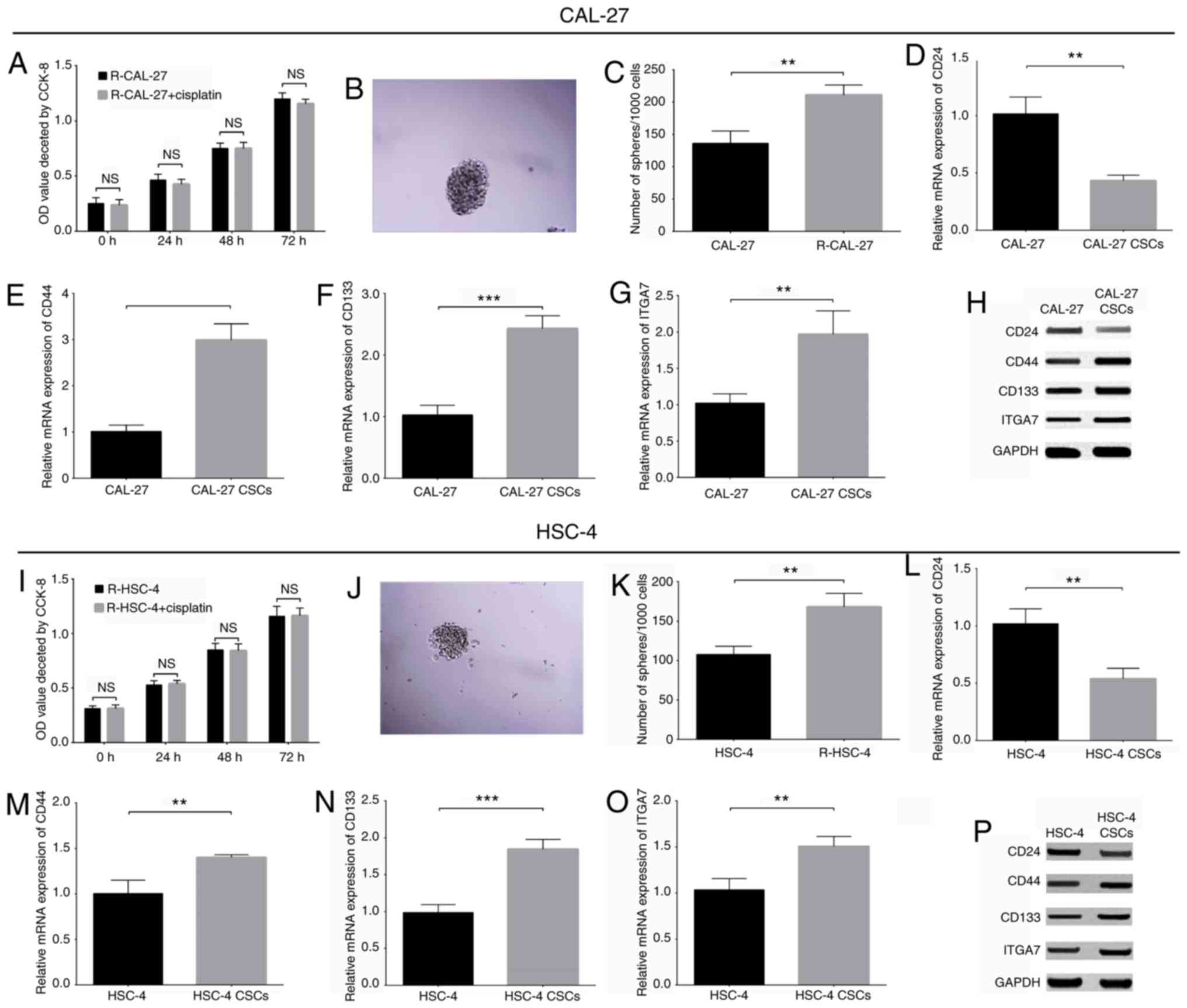 | Figure 9ITGA7 acts as a potential marker of
CSCs in CAL-27 and HSC-4 cell lines. (A) CCK-8 assay of untreated
and cisplatin treated R CAL-27 cells. (B) Representative
photomicrograph of a sphere (magnification, x200) after lentivirus
transfection in R-CAL-27 cells. (C) Number of spheres/1,000 cells
in R CAL-27 and parental CAL-27 cells. (D) CD24, (E) CD44, (F)
CD133 and (G) ITGA7 mRNA expression levels in CAL-27 CSCs and
CAL-27 parental cells. (H) Protein expression levels of CSC markers
in CAL-27 CSCs and CAL-27 parental cells. (I) CCK 8 assay of
untreated and cisplatin treated R HSC 4 cells. (J) Representative
photomicrograph of a sphere (magnification, ×200) after lentivirus
transfection in R-HSC-4 cells. (K) Number of spheres/1,000 cells in
R HSC-4 and parental HSC-4 cells. (L) CD24, (M) CD44, (N) CD133 and
(O) ITGA7 mRNA expression levels in HSC-4 CSCs and HSC-4 parental
cells. (P) Protein expression levels of CSC markers in HSC-4 CSCs
and HSC-4 parental cells. **P<0.01 and
***P<0.001. ITGA7, integrin α7; CSC, cancer stemness
cell; CCK-8, Cell Counting Kit-8; R-, resistant; OD, optical
density; NS, not significant. |
Discussion
Integrins are a type of heterodimeric cell-surface
adhesion molecule expressed in all nucleated cells (18). As one of the common integrins,
ITGA7, which forms a heterodimer with integrin p1, has been
reported to be critical for tumor propagation and the regulation of
CSC-associated properties (9-11).
Controversial findings regarding the role of ITGA7 have been
reported for patients with cancer, and this may be partly due to
variations in the types of cancer and samples analyzed, or the
inclusion and exclusion criteria applied for the enrollment of
patients. However, the association between ITGA7 expression and the
clinicopathological characteristics and prognosis of patients with
TSCC was unknown (9,11). In the present study, ITGA7 was
determined to be upregulated in tumor tissues compared with in
paired adjacent tissues, and its high expression was associated
with increased pathological grade, higher N stage, and advanced TNM
stage in patients with TSCC. The potential explanations for these
observations are hypothesized as follows: i) ITGA7 promotes cell
migration and invasion, and inhibits cell apoptosis by interacting
with the epithelial-mesenchymal transition (EMT), focal adhesion
kinase (FAK)/Akt or other signaling pathways to enhance tumor
growth and metastasis, which may then be associated with increased
pathological grade, higher N stage and advanced TNM stage in
patients with TSCC (11,19); and ii) ITGA7 could induce the
stemness of TSCC cells to promote self-renewal and malignant
phenotypes in cancer cells, which may then contribute to the poor
prognosis of patients with TSCC. In addition, the potential of
ITGA7 as an indicator of prognosis for patients with TSCC was
investigated in the present study. The results demonstrated that
upregulated ITGA7 expression was associated with poor OS in
patients with TSCC, which was in accordance with previous studies
that reported a negative correlation of ITGA7 expression with OS in
patients with esophageal squamous cell carcinoma and glioma
(11,12). The possible reasons were as
follows: i) ITGA7 was associated with worse clinicopathological
characteristics, leading to poor prognosis and reduced OS in
patients with TSCC; and ii) ITGA7 may promote drug resistance by
inducing the sternness of TSCC cells. This could be associated with
the unsatisfactory treatment outcomes and poor OS in patients with
TSCC.
Recently, although limited data have been reported,
investigations into the molecular mechanism underlying the effects
of ITGA7 on the pathogenesis of carcinomas has been a novel
research focus. For example, ITGA7 was determined to induce EMT
promoting tumor metastasis, and to activate FAK/Akt signaling
suppressing cell apoptosis, in esophageal squamous cell carcinoma
(11). In lung cancer, ITGA7
induces cell migration and invasion by binding with S100 calcium
binding protein P (19). The role
of ITGA7 as a tumor promoter has been identified in several
carcinomas; however, few studies have determined the effects of
ITGA7 on the pathogenesis of TSCC. In the present study, RT-qPCR
and western blot analyses were conducted to detect the mRNA and
protein expression levels of ITGA7 in the human TSCC cell lines
CAL-27, SCC-9, HSC-4, SCC-25 and the normal human oral
keratinocytes cell line HOK. The present findings revealed that
ITGA7 was upregu- lated in human TSCC cell lines compared with the
normal HOK cell line. Subsequently, control shRNA and ITGA7-shRNA
lentiviruses were constructed and transduced into CAL-27 and HSC-4
cells; CCK-8 and Annexin-V/PI staining assays were performed to
investigate the effects of ITGA7 knockdown on the proliferation and
apoptosis of TSCC cells. The results revealed that ITGA7 knockdown
suppressed proliferation, but promoted apoptosis in CAL-27 and
HSC-4 cells.
According to previous studies, ITGA7 has been
reported as a functional CSC marker and is involved in the
regulation of stem cell-like properties in several types of cancer.
For instance, ITGA7 upregulates the expression of stemness-asso-
ciated genes (including octamer-binding transcription factor 4, sex
determining region Y-box 2, Nanog homeobox and CD90), and promotes
the self-renewal ability of esophageal squamous cell carcinoma
cells via the activation of the FAK-mediated signaling pathways
(11). Furthermore, in
vitro and in vivo experiments demonstrated that ITGA7
contributes to the growth and invasion of glioblastoma stem-like
cells, potentially through interacting with laminin-induced
outside-in signaling (12). These
previous data indicated that ITGA7 may serve a critical role in
regulating the stemness of cancer cells. In order to explore
whether ITGA7 affected TSCC cell stemness, the present study
investigated the effects of ITGA7 knockdown on the expression of
common CSC markers (CD24, CD44 and CD133), drug resistance to
cisplatin, as well as sphere formation ability. The findings
indicated that ITGA7 knockdown promoted the expression of CD24,
while it downregulated that of CD44 and CD133, compared with the NC
group of CAL-27 and HSC-4 cells. This suggested that ITGA7
knockdown regulated the expression of common CSC markers in TSCC
cells. In addition, ITGA7 knockdown reduced drug resistance to
cisplatin and decreased the sphere formation ability of CAL-27 and
HSC-4 cells. These observations supported the findings of a recent
study, which revealed that the apoptotic index of ITGA7-silenced
esophageal squamous cell carcinoma cells increased after 48 h
exposure to chemotherapeutic reagents (11). Furthermore, ITGA7-silenced KYSE180
and KYSE520 cells formed smaller and fewer spheroids compared with
control cells (11). Therefore,
the present results indicated that ITGA7 knockdown decreased TSCC
cell stemness.
Based on the findings that ITGA7 regulated TSCC cell
stemness, as evidenced by the regulation of CSC marker expression
and the reductions in drug resistance and sphere formation, it was
hypothesized that ITGA7 may be a novel potential marker of TSCC
stem cells. Thus, CAL-27 and HSC-4 CSCs were generated by
establishing drug-resistant cells, which were verfied via a sphere
formation assay and analysis of CSC marker expression. The present
study reported that the mRNA and protein expression levels of ITGA7
were increased in CAL-27 CSCs compared with parental CAL-27 cells;
similar findings were obtained for HSC-4 CSCs. These results
indicated that ITGA7 may act as a potential marker for TSCC stem
cells. Therefore, the findings of the present study provided novel
and comprehensive insight into the molecular mechanisms underlying
the role of ITGA7 in the pathogenesis of TSCC.
In summary, ITGA7 was determined to be upregulated
in tumor tissues, and its high expression was associated with worse
clinicopathological characteristics and poor overall survival in
patients with TSCC. In addition, ITGA7 knockdown suppressed
proliferation and stemness, but promoted apoptosis, in TSCC cells
in vitro. The present findings indicated that ITGA7 may
serve as a potential marker for TSCC stem cells.
Supplementary Data
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81672827).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CY contributed to study conception and design, and
reviewed the manuscript. ZL and CY provided the study materials and
revised the manuscript. ZL and YY were responsible for data
analysis and manuscript writing. All authors approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of The Second Affiliated Hospital of Harbin Medical
University, and written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Abbreviations:
|
ITGA7
|
integrin a7
|
|
OS
|
overall survival
|
|
TSCC
|
tongue squamous cell carcinoma
|
|
CSCs
|
cancer stem cells
|
|
OSCC
|
oral squamous cell carcinoma
|
|
TNM
|
tumor-node-metastasis
|
|
MOI
|
multiplicity of infection
|
References
|
1
|
Almangush A, Heikkinen I, Makitie AA,
Coletta RD, Laara E, Leivo I and Salo T: Prognostic biomarkers for
oral tongue squamous cell carcinoma: A systematic review and
meta-analysis. Br J Cancer. 117:856–866. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pfister DG, Ang KK, Brizel DM, Burtness
BA, Busse PM, Caudell JJ, Cmelak AJ, Colevas AD, Dunphy F, Eisele
DW, et al: Head and neck cancers, version 2.2013. Featured updates
to the NCCN guidelines. J Natl Compr Canc Netw. 11:917–923. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gore SM, Crombie AK, Batstone MD and Clark
JR: Concurrent chemoradiotherapy compared with surgery and adjuvant
radiotherapy for oral cavity squamous cell carcinoma. Head Neck.
37:518–523. 2015. View Article : Google Scholar
|
|
5
|
Iyer NG, Tan DS, Tan VK, Wang W, Hwang J,
Tan NC, Sivanandan R, Tan HK, Lim WT, Ang MK, et al: Randomized
trial comparing surgery and adjuvant radiotherapy versus concurrent
chemoradiotherapy in patients with advanced, nonmetastatic squamous
cell carcinoma of the head and neck: 10-year update and subset
analysis. Cancer. 121:1599–1607. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goel HL, Li J, Kogan S and Languino LR:
Integrins in prostate cancer progression. Endocr Relat Cancer.
15:657–664. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Juan-Rivera MC and Martinez-Ferrer M:
Integrin inhibitors in prostate cancer. Cancers (Basel).
10:E442018. View Article : Google Scholar
|
|
8
|
Tan LZ, Song Y, Nelson J, Yu YP and Luo
JH: Integrin alpha7 binds tissue inhibitor of metalloproteinase 3
to suppress growth of prostate cancer cells. Am J Pathol.
183:831–840. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bhandari A, Xia E, Zhou Y, Guan Y, Xiang
J, Kong L, Wang Y, Yang F, Wang O and Zhang X: ITGA7 functions as a
tumor suppressor and regulates migration and invasion in breast
cancer. Cancer Manag Res. 10:969–976. 2018. View Article : Google Scholar :
|
|
10
|
Ren B, Yu YP, Tseng GC, Wu C, Chen K, Rao
UN, Nelson J, Michalopoulos GK and Luo JH: Analysis of integrin
alpha7 mutations in prostate cancer, liver cancer, glioblastoma
multiforme, and leiomyosarcoma. J Natl Cancer Inst. 99:868–880.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ming XY, Fu L, Zhang LY, Qin YR, Cao TT,
Chan KW, Ma S, Xie D and Guan XY: Integrin a7 is a functional
cancer stem cell surface marker in oesophageal squamous cell
carcinoma. Nat Commun. 7:135682016. View Article : Google Scholar
|
|
12
|
Haas TL, Sciuto MR, Brunetto L, Valvo C,
Signore M, Fiori ME, di Martino S, Giannetti S, Morgante L, Boe A,
et al: Integrin a7 is a functional marker and potential therapeutic
target in glioblastoma. Cell Stem Cell. 21:35–50.e39. 2017.
View Article : Google Scholar
|
|
13
|
Carrasco-Garcia E, Auzmendi-Iriarte J and
Matheu A: Integrin a7: A novel promising target in glioblastoma
stem cells. Stem Cell Investig. 5:22018. View Article : Google Scholar
|
|
14
|
Abd El hafez A and El-Hadaad HA:
Immunohistochemical expression and prognostic relevance of Bmi-1, a
stem cell factor, in epithelial ovarian cancer. Ann Diagn Pathol.
18:58–62. 2014. View Article : Google Scholar
|
|
15
|
Hu Y and Smyth GK: ELDA: Extreme limiting
dilution analysis for comparing depleted and enriched populations
in stem cell and other assays. J Immunol Methods. 347:70–78. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dontu G, Abdallah WM, Foley JM, Jackson
KW, Clarke MF, Kawamura MJ and Wicha MS: In vitro propagation and
transcriptional profiling of human mammary stem/progenitor cells.
Genes Dev. 17:1253–1270. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Hynes RO: Integrins: Bidirectional,
allosteric signaling machines. Cell. 110:673–687. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hsu YL, Hung JY, Liang YY, Lin YS, Tsai
MJ, Chou SH, Lu CY and Kuo PL: S100P interacts with integrin a7 and
increases cancer cell migration and invasion in lung cancer.
Oncotarget. 6:29585–29598. 2015. View Article : Google Scholar : PubMed/NCBI
|