Introduction
Wilms’ tumor is one of the most common malignant
tumors of the abdomen in children. The prognosis of this cancer is
associated with age, tumor size and histological type. However, the
most important prognostic factor is local invasion and distant
metastasis of the tumor (1). The
mechanism underlying the pathogenesis and metastasis of Wilms’
tumor have yet to be fully elucidated, and further research is
required to design targeted therapies.
The role of lipid metabolism in cancer is attracting
increasing attention, and inhibition of lipid autophagy has been
shown to be of clinical value in other tumors (2). Due to the high metabolic demands of
tumor cells and their nutritional deficiency during
chemoradiotherapy, fatty acid metabolism can provide the necessary
energy for tumor cells. Furthermore, tumor cell division requires a
high energy supply. As phospho-lipids are an important part of the
cytoskeleton, phospholipid synthesis provides essential raw
materials for tumor cell proliferation (3-5).
Thus, recent publications have reported that lipid metabolism is
closely associated with tumor cell proliferation, invasion,
migration and chemosensitivity (6,7), and
it is emerging as an attractive target for cancer drug
discovery.
Fatty acid synthase (FASN) is essential for de
novo lipo-genesis and cellular substrate energy metabolism,
including the synthesis of long-chain fatty acids from acetyl-CoA,
malonyl-CoA and NADPH (8,9). The expression of FASN also has a
considerable impact on tumor progression: The expression of FASN in
a variety of solid tumors was found to be significantly increased
and was associated with poor prognosis (10), and the proliferation, invasion and
migration of tumor cells were significantly suppressed after its
downregulation (11-13). In particular, in renal cell
carcinoma, Albigeset al demonstrated that FASN played an
important role in the occurrence of tumors and that patients with
higher expression levels had a poor prognosis (14). However, only few studies (15) have addressed the role of FASN in
Wilms’ tumor.
Investigating the effect and mechanism of action of
FASN, a lipid-metabolizing protein, on the metastasis of Wilms’
tumors may provide a novel approach to the targeted therapy of this
disease. The aim of the present study was to examine the different
levels of lipid metabolism-related enzymes and determine whether
FASN is a key factor in the progression of Wilms’ tumor.
Materials and methods
Tissues
Between January 2007 and January 2012, a total of 65
patients with Wilms’ tumor who did not undergo radical or
palliative nephrectomy were selected for immunohistochemistry (IHC)
analysis. Clinical data, including sex, age, tumor size, stage,
histopathological type, metastasis and follow-up information, were
recorded. The patients included 35 males and 30 females, with a
mean age at diagnosis of 3.2 years (range, 0.25-11.8 years).
Detailed information may be found in Table I. The duration of the follow-up was
60 months. In addition, 20 cases of Wilms’ tumor and adjacent
tissues collected between March 2015 and December 2017 and stored
at -80°C were selected. The tumors were graded according to the
Fuhrman nuclear grade classification (16) as follows: G1, n=10; G2, n=7; and
G3, n=3. A total of 9 cases (G1, n=3; G2, n=3; and G3, n=3) were
used for label-free quantification. Radiotherapy, chemotherapy and
immunotherapy were not performed prior to surgery, and the samples
were verified by two pathologists postoperatively. The study
protocol was approved by the Ethics Committee of Shandong
Provincial Hospital Affiliated to Shandong University, and the
legal guardians of the patients provided informed consent regarding
the use of the patients’ tissues for research purposes.
 | Table ICorrelation of expression levels of
the FASN protein with the clinicopathological parameters of
patients with Wilms’ tumor. |
Table I
Correlation of expression levels of
the FASN protein with the clinicopathological parameters of
patients with Wilms’ tumor.
| Clinicopathological
parameters | n (%) | FASN expression
| P-value |
|---|
| High | Low |
|---|
| Sex | | | | 0.620 |
| Male | 35 (53.8) | 17 | 18 | |
| Female | 30 (46.2) | 17 | 13 | |
| Age (months) | | | | 0.083 |
| >24 | 28 (43.1) | 11 | 17 | |
| ≤24 | 37 (52.9) | 23 | 14 | |
| Stage | | | | |
| I | 32 (49.2) | 9 | 23 | |
| II | 21 (32.3) | 16 | 5 | 0.001 (II vs.
I) |
| III | 12 (18.5) | 9 | 3 | 0.007 (III vs.
I) |
| Histopathological
type | | | | 0.032 |
| Favorable | 46 (70.8) | 20 | 26 | |
| Unfavorable | 19 (29.2) | 14 | 5 | |
| Lymphatic
metastasis | | | | 0.086 |
| Yes | 10 (15.4) | 8 | 2 | |
| No | 55 (84.6) | 26 | 29 | |
Protein digestion
Proteins were re-dissolved in 500 mM
triethylammonium bicarbonate (TEAB). The protein concentration of
the supernatant was determined using the BCA protein assay, and 100
µg protein per condition was transferred into a new tube and
adjusted to a final volume of 100 µl with 8 M urea.
Subsequently, 11 µl of 1 M DTT was added, and the sample was
incubated at 37°C for 1 h and transferred into a 10 K
ultrafiltration tube (EMD Millipore). To remove the urea, the
samples were centrifuged after adding 100 mM TEAB three times at
14,000 × g at 4°C for 10 min, followed by the addition of 120
µl of 55 mM iodoacetamide to the sample and incubation for
20 min in the dark at room temperature. The proteins were then
digested with sequencing grade modified trypsin (Promega
Corporation).
Label-free quantification
First, 18 cases of in-solution digestion samples
(Wilms’ tumor and their adjacent tissues; G1, n=3; G2, n=3; and G3,
n=3) were acquired and analyzed by nanoflow liquid
chromatography-tandem mass spectrometry (nanoLC-MS/MS). The samples
were resuspended in 30 µl solvent C (C: water with 0.1%
formic acid; D: ACN with 0.1% formic acid), separated by nanoLC and
analyzed by online electrospray MS/MS. The experiments were
performed on an Easy-nLC 1000 system (Thermo Fisher Scientific,
Inc.) connected to a Q-Exactive mass spectrometer (Thermo Fisher
Scientific, Inc.) equipped with an online nano-electrospray ion
source. A 10-µl peptide sample was loaded onto the trap
column (Acclaim PepMap C18, 100 µm × 2 cm, Thermo Fisher
Scientific, Inc.) at a flow rate of 10 µl/min for 3 min, and
subsequently separated on the analytical column (Acclaim PepMap
C18, 75 µm × 15 cm) with a linear gradient from 3% D to 32%
D in 60 min. The column was re-equilibrated at the initial
conditions for 10 min. The column flow rate was maintained at 300
nl/min. The electrospray voltage of 2 kV vs. the inlet of the mass
spectrometer was used.
The mass spectrometer was run under data-dependent
acquisition mode and automatically switched under MS and MS/MS
mode. The MS1 mass resolution was set as 35 K with m/z 350-1,550,
and the MS/MS resolution was set as 17.5 K under HCD mode. The
dynamic exclusion time was set at 20 sec.
Volcano plot
The volcano plot, which plots significance vs.
fold-change on the y and x axes, respectively, is a type of scatter
plot that is used to quickly identify changes in large datasets
composed of replicate data. This plot is drawn by using the ggplot2
package (http://ggplot2.org).
Hierarchical cluster analysis (HCA)
HCA is an algorithmic approach to finding discrete
groups with varying degrees of (dis)similarity in a dataset
represented by a (dis)similarity matrix. This analysis is processed
with the pheatmap package (https://CRAN.R-project.org/package=pheatmap).
Gene Ontology (GO) analysis
Blast2GO version 4 (BioBam Bioinformatics S.L.) was
used for functional annotation. The whole protein sequence database
was analyzed by BlastP using a whole database and mapped and
annotated with a GO database. Statistically altered functions of
differentially expressed proteins were calculated by Fisher’s exact
test in Blast2GO.
Search Tool for the Retrieval of
Interacting Genes/Proteins (STRING) analysis
The protein-protein interaction network was analyzed
using STRING v.10 (http://string-db.org/).
Database for Annotation, Visualization
and Integrated Discovery (DAVID) functional annotation tool
The differentially expressed proteins were also
identified using DAVID functional annotation for functional
analysis (https://david.ncifcrf.gov/).
Kyoto Encyclopedia of Genes and Genomes
(KEGG) analysis
Pathway analysis was processed by KOBAS (http://kobas.cbi.pku.edu.cn/). Pathways with P<0.05
were recognized as significantly altered.
Ingenuity Pathway Analysis (IPA)
IPA is a common tool used to model, analyze and
understand complex biological systems, particularly to reveal
different signaling networks and biological processes (17). IPA is an integrated biological
pathway analysis software based on cloud computing. With the
support of the highly structured bioinformatics platform Ingenuity
Knowledge Base, it can search for various types of related
information on genes, proteins and drugs, as well as constructing
interaction models. In addition, this tool can also analyze data
obtained through such techniques as genomics, microRNA,
experimental data for single-nucleotide polymorphisms, chips,
metabolomes and proteomes.
Immunohistochemistry analysis
Tissues obtained from Wilms’ tumors were subjected
to fixation with 4% paraformaldehyde for at least 72 h at room
temperature, dehydration using 10% formalin for 3 h, followed by
immersion in a series of graded ethanol baths, washing in xylene,
immersion and final embedding in paraffin. The tissue blocks were
cut into 5-µm sections. After dewaxing and rehydration,
heat-induced epitope retrieval was performed with 1 mM EDTA (pH
9.0) in a microwave oven. Then, samples were immersed in 3%
H2O2 aqueous solution for 30 min to exhaust
endogenous peroxidase. After closure blocking with goat serum for
30 min, the sections were incubated overnight at 4°C with primary
anti-FASN antibody (dilution 1:100, cat. no. 10624-2-AP,
ProteinTech Group, Inc.). Peroxidase-labeled secondary antibodies
were applied, and diaminobenzidine (DAB; Zhongshan) staining was
performed at 4°C for 10 sec. The slides were then counterstained
with hematoxylin at room temperature for 4 min and mounted. As a
negative control, samples were analyzed without the primary
antibody.
Western blot analysis
To determine the FASN level, proteins were extracted
from the tissues by suspension in radioimmunoprecipitation assay
(RIPA) buffer (Solarbio). The samples were centrifuged at 13,523 ×
g at 4°C for 38 min, and the supernatants were recovered for
analysis. The protein concentration was determined using the
bicinchoninic acid (BCA) protein assay kit (Sigma-Aldrich; Merck
KGaA). Protein samples (40 µg) were electrophoresed on a
pre-cast bis-Tris polyacryl-amide gel (8-12%) and then transferred
onto a polyvinylidene difluoride membrane (EMD Millipore). The
membranes were blotted with rabbit anti-FASN (1:1,000) and mouse
anti-actin (all from ProteinTech Group, Inc.) followed by
horseradish peroxidase (HRP)-conjugated secondary antibodies
(1:5,000; ZsBio). Immunoblots were visualized using enhanced
chemiluminescence (LAS-4000; GE Healthcare).
mRNA expression analysis
The expression of the FASN gene was analyzed by
RT-qPCR. RNA was isolated from 40 frozen samples (20 from Wilms’
tumors and 20 from adjacent tissues) from which sufficient material
was obtained with the miRVana miRNA Isolation Kit (TaKaRa Bio,
Inc.). The quantity and quality of total RNA was determined with a
Nanodrop ND-2000 Spectrophotometer (Thermo Fisher Scientific,
Inc.). RT (37°C 15 min, 85°C 5 sec, and held at 4°C until use) was
performed using the TaqMan Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) in the GeneAmp PCR 9700
system and RT-qPCR amplification with the TaqMan Universal PCR
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.).
All RT-qPCR measurements were obtained in a 7900HT Fast Real Time
PCR System with the ExpressionSuite Software v1.0 (Applied
Biosystems; Thermo Fisher Scientific, Inc.). All siRNAs were
purchased from Personalbio, and their sequences were as follows:
5′-CTC ATCAAGTGGGACCACAG-3′ (forward) and 5′-CAGCGT
CTTCCACACTATGC-3′ (reverse) for FASN; 5′-AATCGT GCGTGACATTAAGG-3′
(forward) and 5′-TAGTTTCGTGGATGCCACAG-3′ (reverse) for actin.
RT-qPCR analysis was performed using SYBR Premix Ex Taq™ II
(Takara, Japan), according to the manufacturer′s protocol. The qPCR
reactions were performed on a Roche 480 Real-Time PCR System using
the standard procedure for two-step PCR amplification as follows:
Pre-denaturation at 95°C for 30 sec, PCR at 95°C for 5 sec, 60°C
for 30 sec, for 40 cycles; melting curve: 95°C for 5 sec, 60°C for
60 sec, 95°C for 15 sec; cooling: 50°C for 30 sec. After the
reaction, the amplification curve and melting curve of qPCR were
confirmed and data were analyzed (18). The FASN mRNA expression levels were
examined using the 2-ΔΔCq method and were compared
against the level of actin (19).
Immunofluorescence
Cell concentration smears were performed and the
cells were fixed with 4% paraformal-dehyde at room temperature for
20 min. The cells were thoroughly rinsed with 0.01 M PBS (5 min ×
3) and incubated in 0.3% Triton at room temperature for 20 min,
followed by washing in 0.01 M PBS (5 min × 3). Next, 30
µl/sample goat serum blocking solution was added at room
temperature for 60 min followed by primary antibody at 30
µl/sample (1:100 diluted in goat serum). The cells were
placed in a wet box at 4°C overnight and then washed in 0.01 M PBS
(5 min × 3). On the next day, fluorescent secondary antibody
(dilution 1:100) was added at 30 µl/sample (10% skimmed milk
powder) at room temperature in the dark for 60 min, followed by
addition of 4′,6-diamidino-2-phenylindole (DAPI) for 10 min. Next,
the cells were washed in 0.01 M PBS (5 min × 3) and mounted with an
anti-fluorescence quencher.
Statistical analysis
The data were statistically analyzed using Student’s
t-test, the χ2 test, or Fisher’s exact test using SPSS
version 19.0 (SPSS, Inc.). The survival curves were analyzed by the
Kaplan-Meier method. P<0.05 was considered to indicate
statistically significant differences. Tandem mass spectra were
processed by PEAKS Studio version 8.0 (Bioinformatics Solutions,
Inc.). Differentially expressed proteins were filtered if their
fold change was >2 and their P-value was <0.01 (significant
>20). The P-value for IPA data analysis was based on the
Right-Tailed Fisher’s Exact Test algorithm.
Results
MS revealed 19 differentially expressed
lipid metabolism-related enzymes in tumors and adjacent
tissues
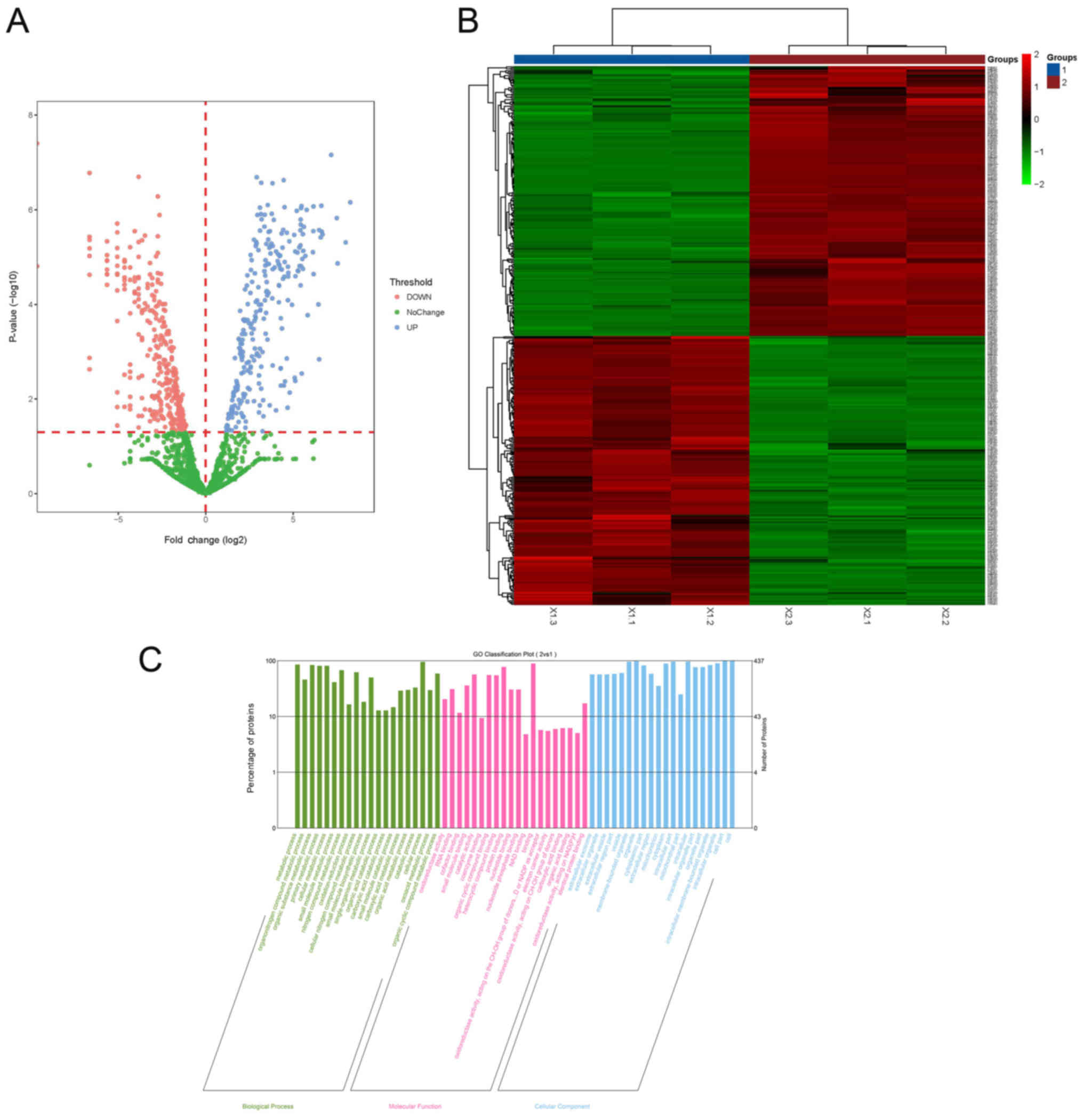
Label-free proteomics analysis revealed that there
were 219 upregulated and 218 downregulated proteins in the tumor
(Fig. 1). The HCA map indicates
the extent of the peptide or protein expression in terms of depth
of color, and the GO classification results can be visualized using
bar graphs. The GO classification bar graph uses the top 20
pathways with the smallest P-value (Fig. 1). The results were divided into
biological processes, molecular functions and cellular components.
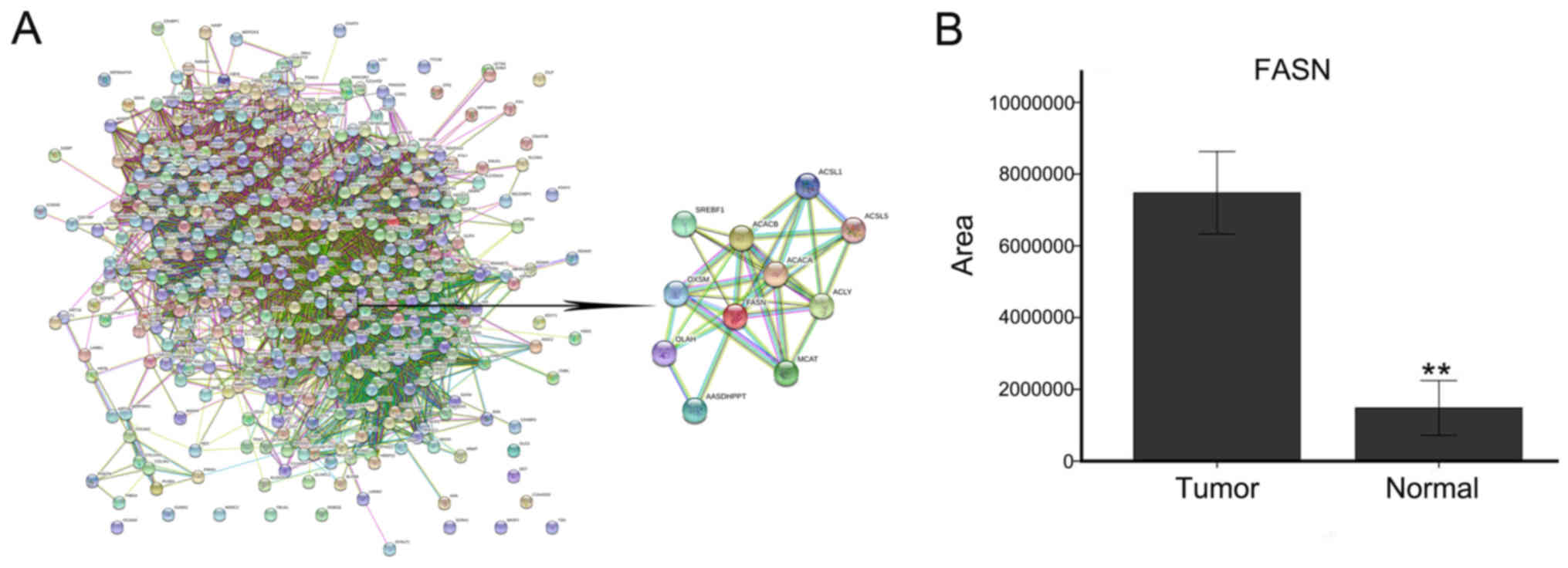
As shown in Fig. 2, 437 proteins
were input into STRING software, and there were associations among
418 of those proteins. Subsequently, the DAVID functional
annotation tool was used to perform protein functional analysis.
Among the 437 proteins, 19 were associated with lipid metabolism,
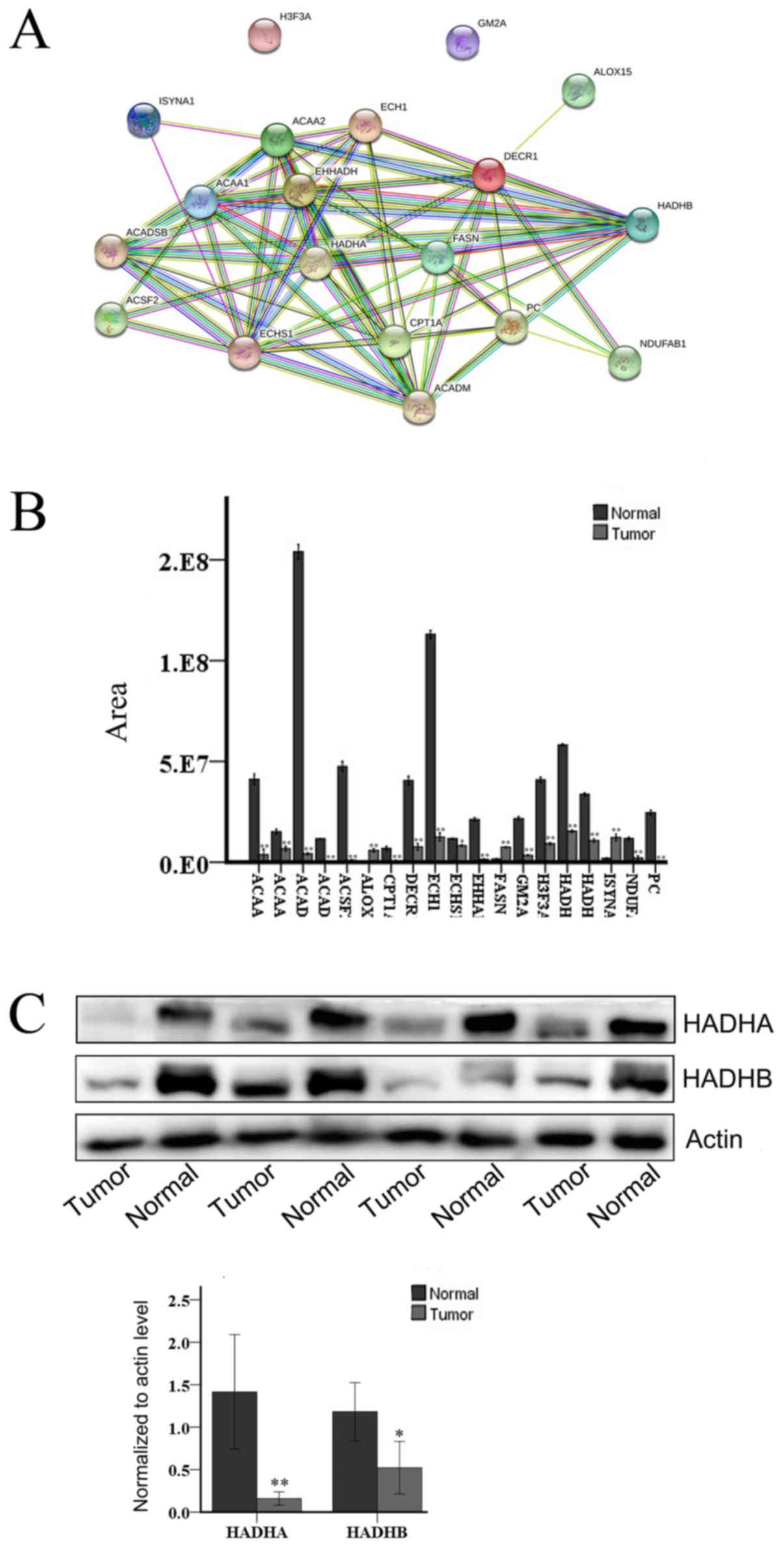
including lipid synthesis and decomposition (Fig. 3B). The upregulated proteins
included FASN, inositol-3-phosphate synthase 1 (ISYNA1), and
arachi-donate-15-lipoxygenase (ALOX15), and the downregulated
proteins included hydroxyacyl coenzyme A dehydrogenase beta [HADHB
(Fig. 3C)], acetyl coenzyme A
acyltransferase 1 (ACAA1), acetyl coenzyme A acyltransferase 2
(ACAA2), carnitine palmitoyltransferase 1A (CPT1A), enoylcoen-zyme
A hydratase (ECHS1), acyl-CoA synthetase family member 2 (ACSF2),
enoyl-coenzyme A, hydratase/3-hydroxy-acyl coenzyme A dehydrogenase
(EHHADH), pyruvate carboxylase (PC), acyl-CoA dehydrogenase
medium-chain (ACADM), short/branched chain acyl-CoA dehydrogenase
(ACADSB), enoyl coenzyme A hydratase 1 (ECH1), GM2 ganglioside
activator (GM2A), 2,4-dienoyl-CoA reductase 1 (DECR1), hydroxyacyl
coenzyme A dehydrogenase alpha [HADHA (Fig. 3C)], ubiquinone oxidoreductase
subunit AB1 (NDUFAB1) and histone H3.3 [H3F3A (Fig. 4)].
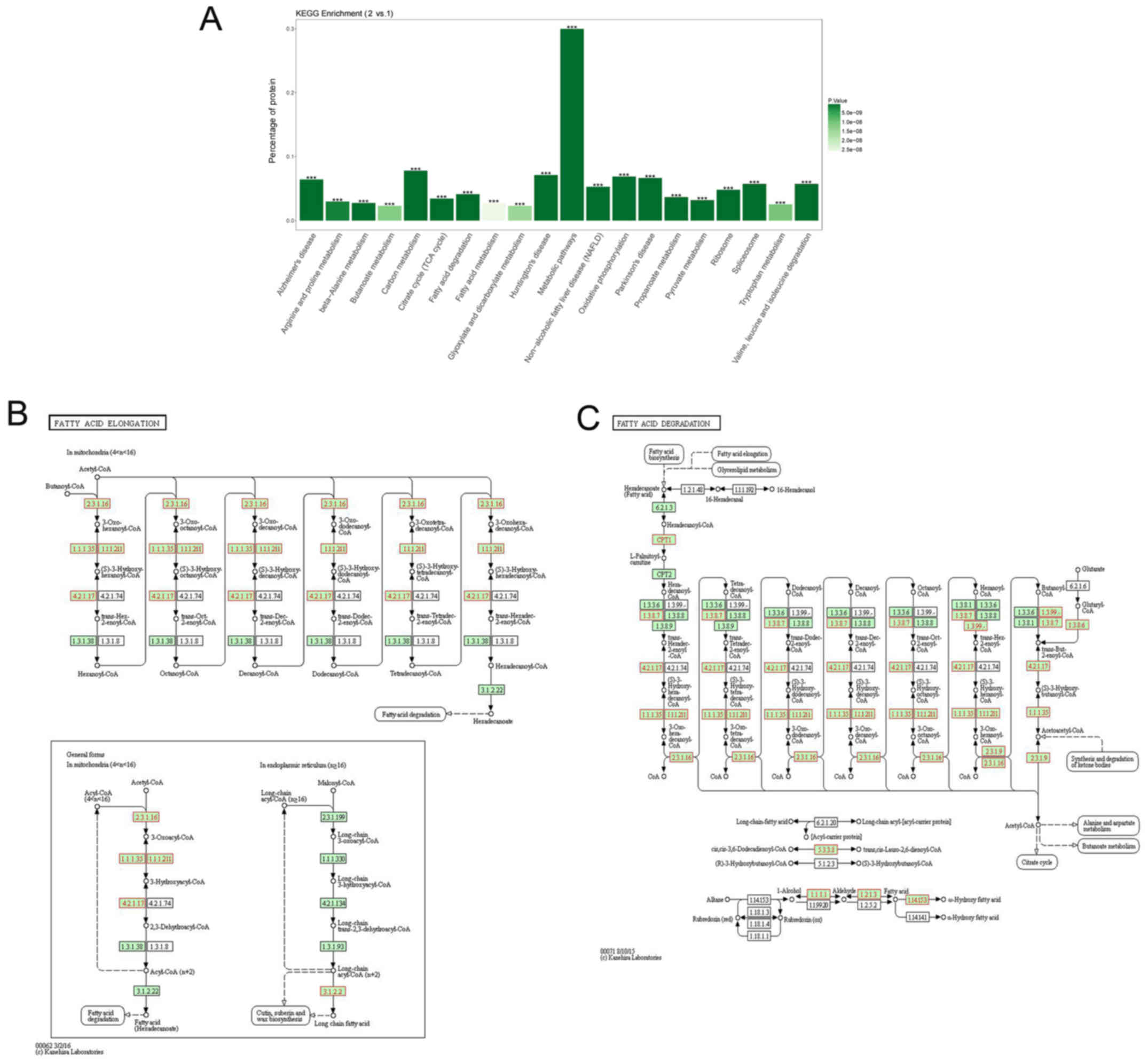 | Figure 3KEGG analysis of the 437
differentially expressed proteins. (A) The KEGG enrichment results
can be visualized by bar graphs, and the top 20 pathways with the
smallest P-value are plotted. The ordinate indicates the percentage
of total protein in the pathway. The darker the color, the smaller
the P-value. ***P<0.001. (B) The selected enzymes are
involved in the process of long-chain fatty acid extension. (C) The
selected enzymes are involved in the process of fatty acid
degradation. The EC code is concluded, such as ACAA [EC:2.3.1.16],
ECHS1 [EC:4.2.1.17], ACSF2 [EC:6.2.1.2], EHHADH
[EC:4.2.1.17&1.1.1.35&5.3.3.8], ACADM [EC:1.3.8.7], ACADSB
[EC:1.3.99.12], ECH1 [EC:5.3.3.21], DECR1 [EC:1.3.1.34], HADHA
[EC:4.2.1.17&1.1.1.211] (https://www.kegg.jp/). Group 1, tumor tissue; Group
2, adjacent tissue. (B and C) Green node, species-specific; blue
frame node, downregulation; and red frame node, upregulation. KEGG,
Kyoto Encyclopedia of Genes and Genomes. |
KEGG pathway analyses for lipid
metabolism
Fatty acid, triacylglycerol and ketone body
metabolism pathway analysis uncovered lipid metabolism-related
proteins or protein complexes, small molecules, and their
interactions (Fig. 3). KEGG
enrichment provides plotting of the top 20 pathways with the
smallest P-value. The ordinate indicates the percentage of total
protein in the pathway. Fatty acid metabolism and fatty acid
degradation are included among the top 20 most highly expressed
signaling pathways. The KEGG signaling pathway demonstrated the
pathways of fatty acid elongation and fatty acid degradation (the
EC code is used in the figure instead of the related enzyme). It
was observed that the HADH, HAHDB, ACAA1 and ACAA2 proteins all
hold important positions in the KEGG signaling pathway and
participate in this process.
Quantitative informatics analysis of
IPA
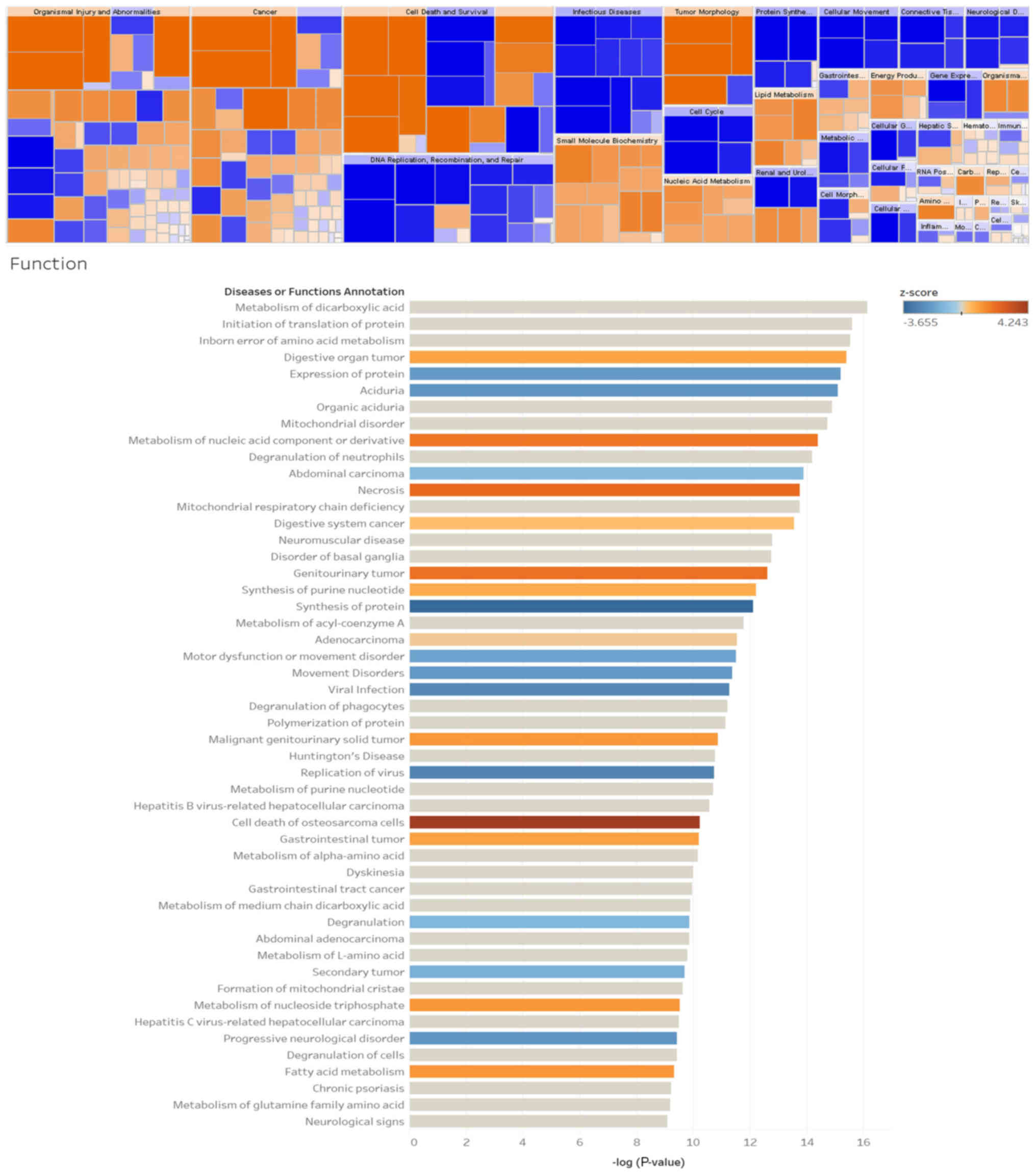
IPA analysis revealed that protein metabolism in
Wilms’ tumors is closely associated with tumor cell death, cell
cycle, and lipid metabolism (Fig.
5). Lipid metabolism plays an important role in tumors, and
signaling pathway analysis has demonstrated that dysregulated
proteins play an important role in fatty acid metabolism and are
involved in the classical pathways, apoptosis and autophagy of
multiple tumors (Fig. 5).
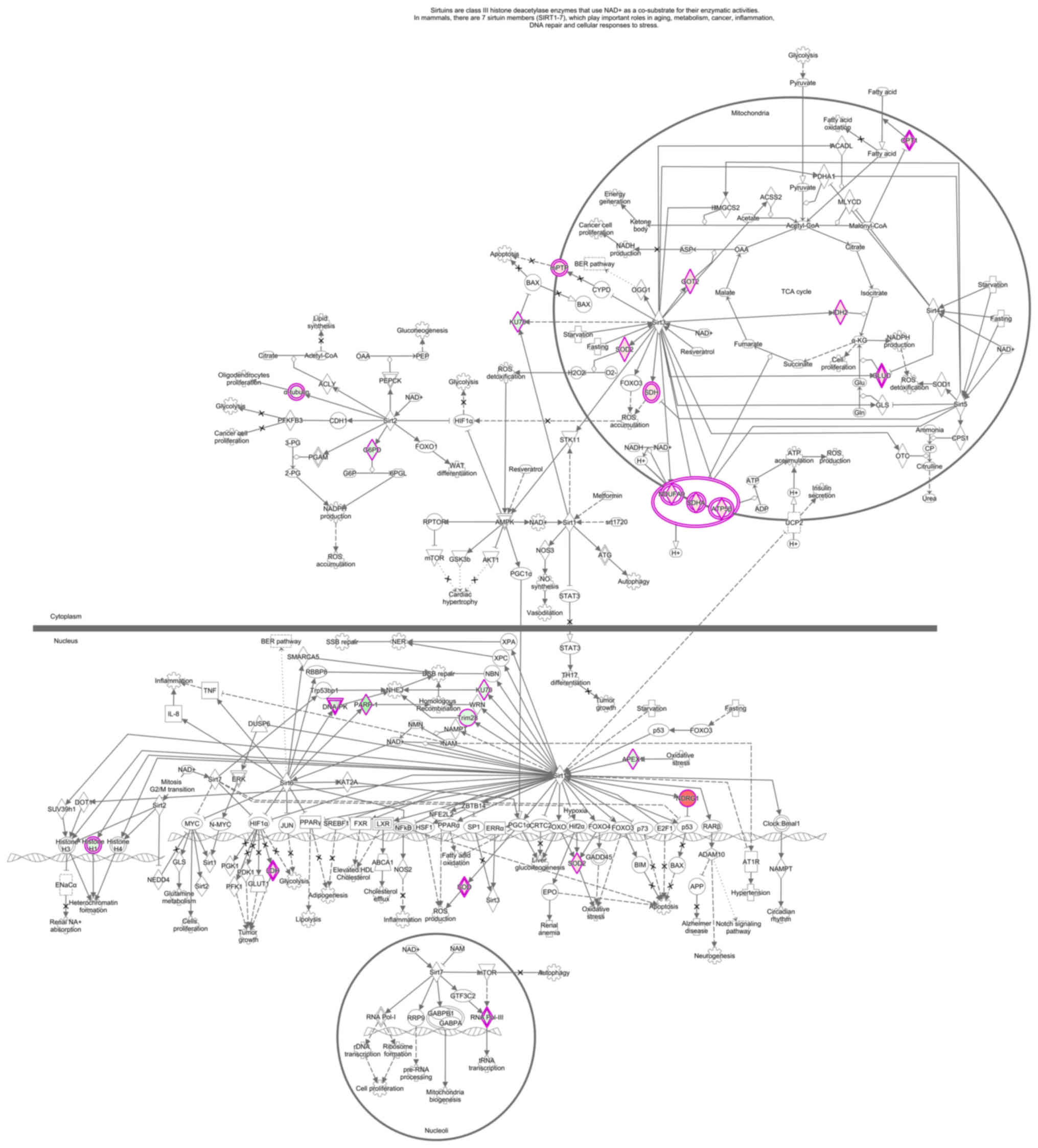
Signaling pathway analysis revealed that dysregulated proteins play
an important role in fatty acid metabolism pathways, and that
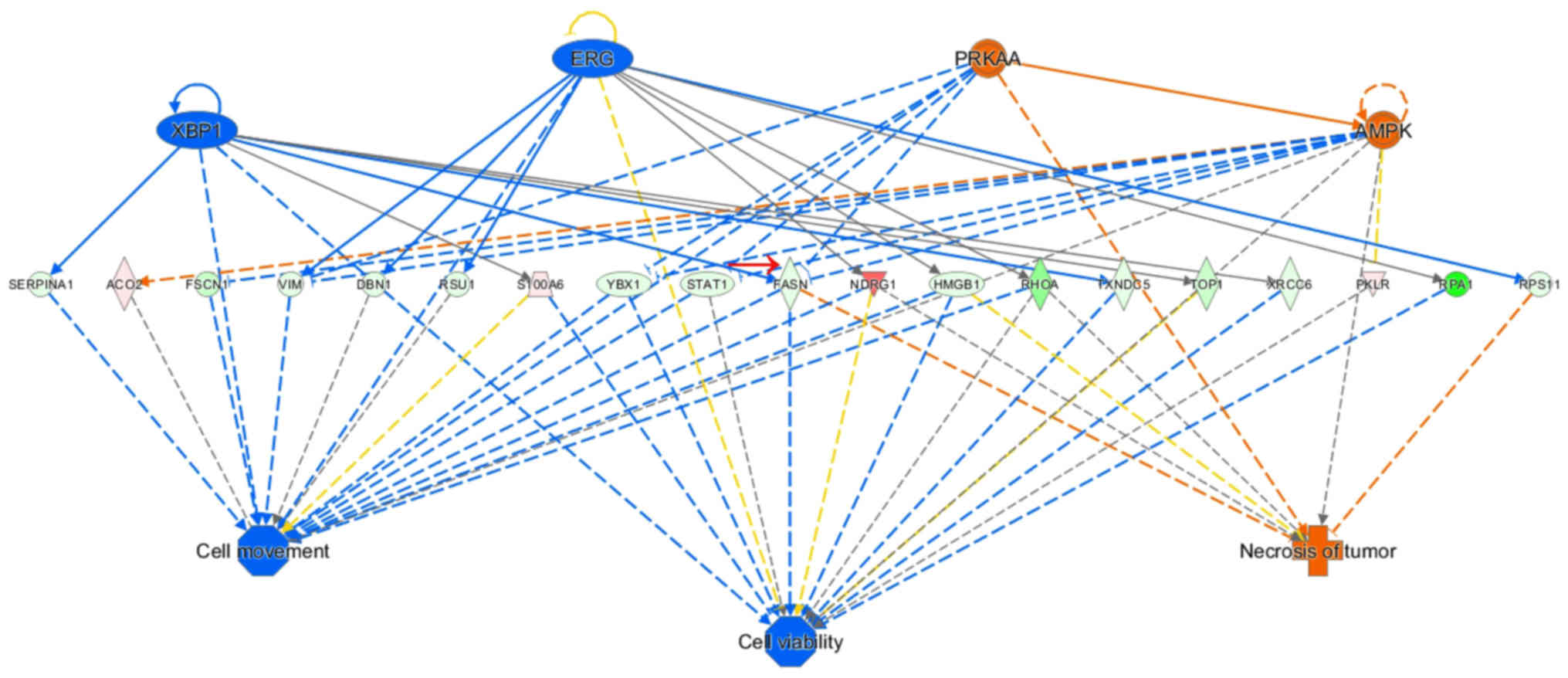
sirtuin plays a key role in multiple tumors (Fig. 6). It may be inferred from the
regulatory sub-effector network that FASN is involved in cell
migration, cell proliferation and tumor necrosis at important
positions in the total regulatory network and is implicated in
various signaling pathways (Fig.
7).
Validation of FASN expression between
Wilms’ tumor and adjacent tissues
Next, MS was used to determine whether the
expression of FASN in tumors was higher compared with that in
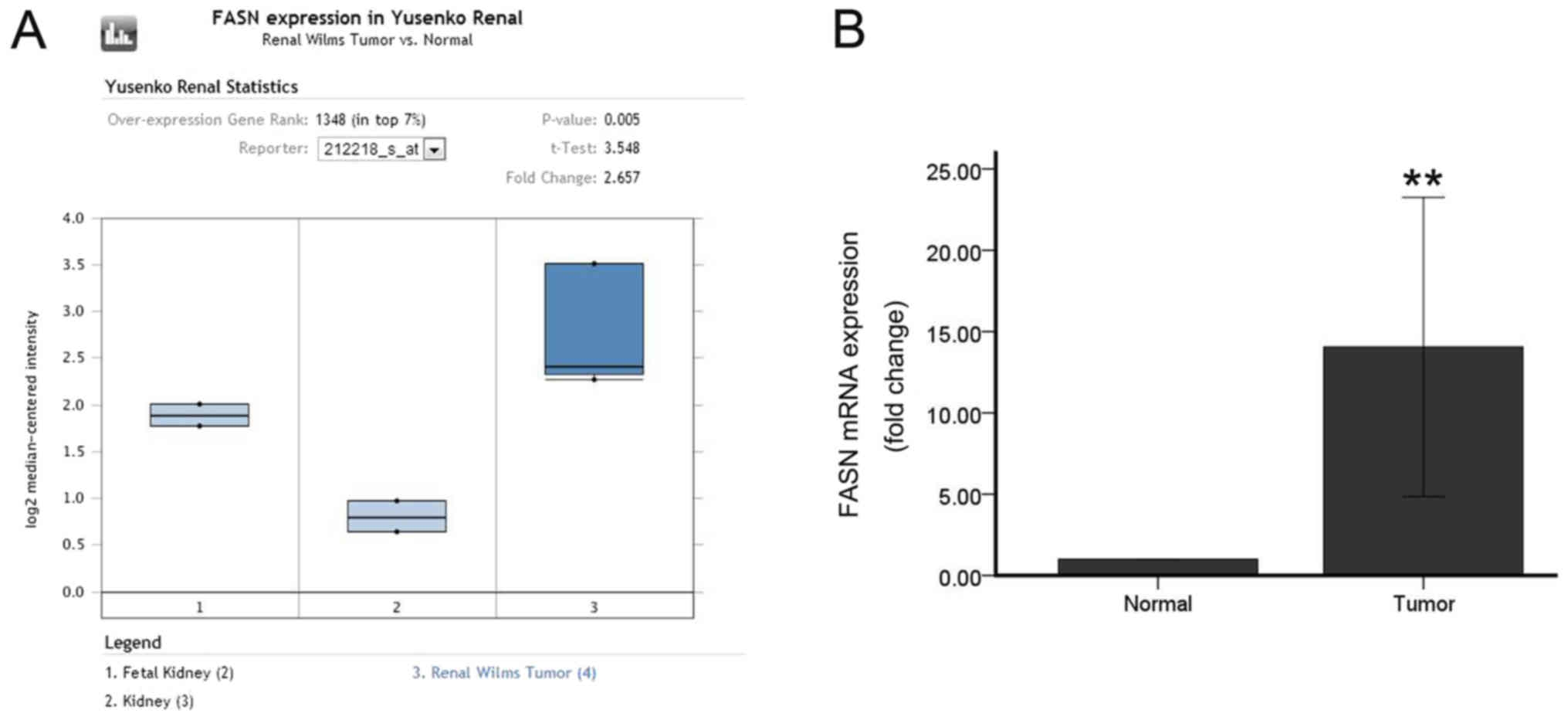
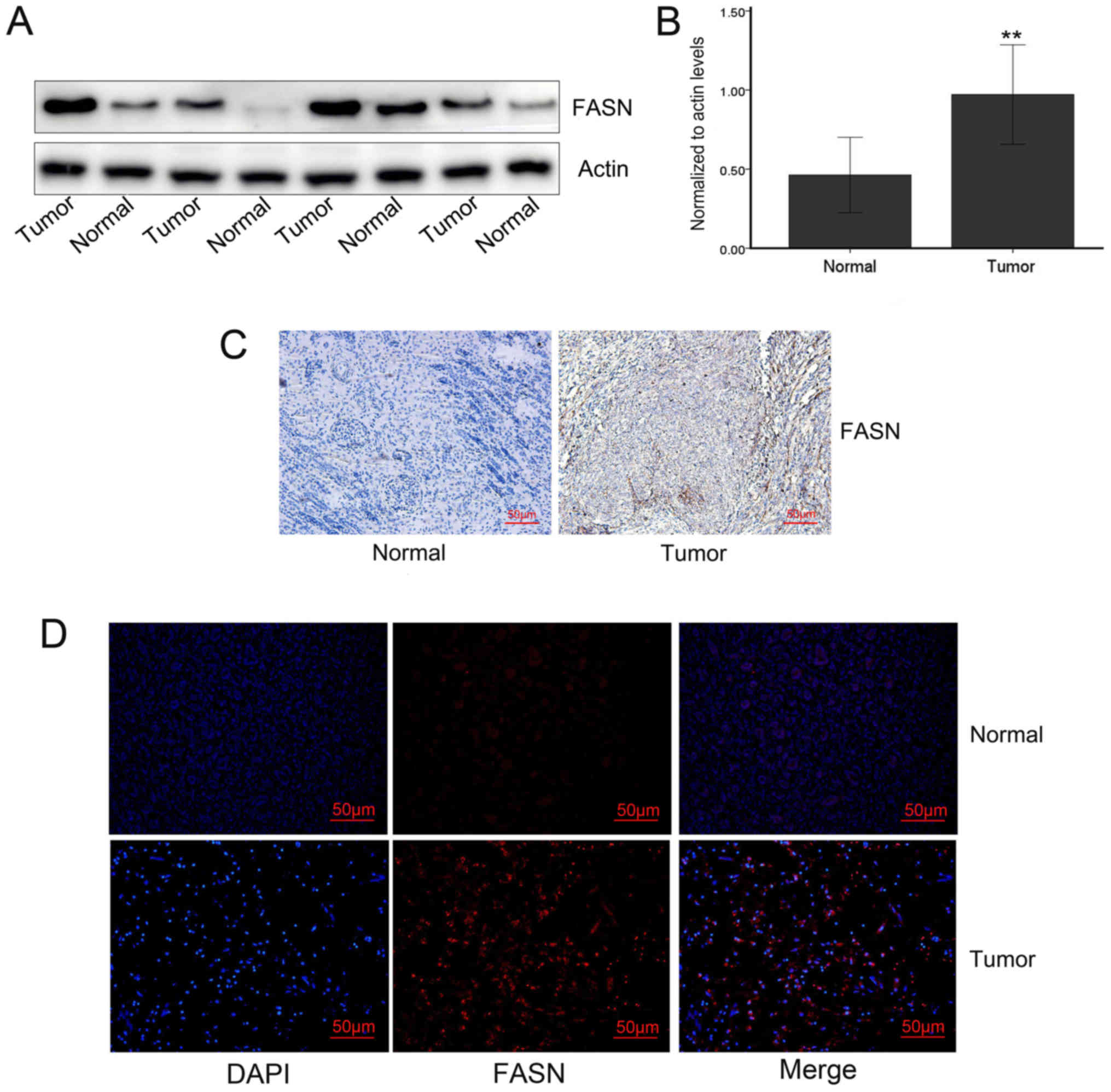
adjacent tissues (Fig. 2).
Analysis of the Oncomine database, RT-qPCR, immunohistochemistry,
western blotting and immunofluorescence were further performed to
verify the results (Figs. 8 and
9). Oncomine database analysis
demonstrated that FASN gene expression was significantly increased
in Wilms’ tumors (P=0.005 vs. normal tissue; Fig. 8A). Then, RT-PCR was used to detect
the expression of FASN in 20 pairs of tumors and adjacent tissues,
and the expression of FASN was found to be upregulated in Wilms’
tumors (P<0.01; Fig. 8B). The
results of immunohistochemistry, western blotting and
immunofluorescence all verified that the expression of the FASN
protein was significantly increased in tumors compared with that in
adjacent tissues.
FASN expression and its correlation with
clinicopathological parameters
The protein expression of FASN and the
clinicopathological parameters of the patients are summarized in
Table I. Among Wilms’ tumor
tissues, positive staining signals of FASN were detected in 34
samples (52.31%) with high immunoreactivity and 31 (47.69%) with
low immunoreactivity. The association between FASN expression and
tumor stage (III vs. I, P= 0.001; II vs. I, P=0.007) and
histopathological type (P=0.032) was statistically significant.
There was no significant correlation between FASN expression and
patient sex or age (P=0.620 and P=0.083, respectively).
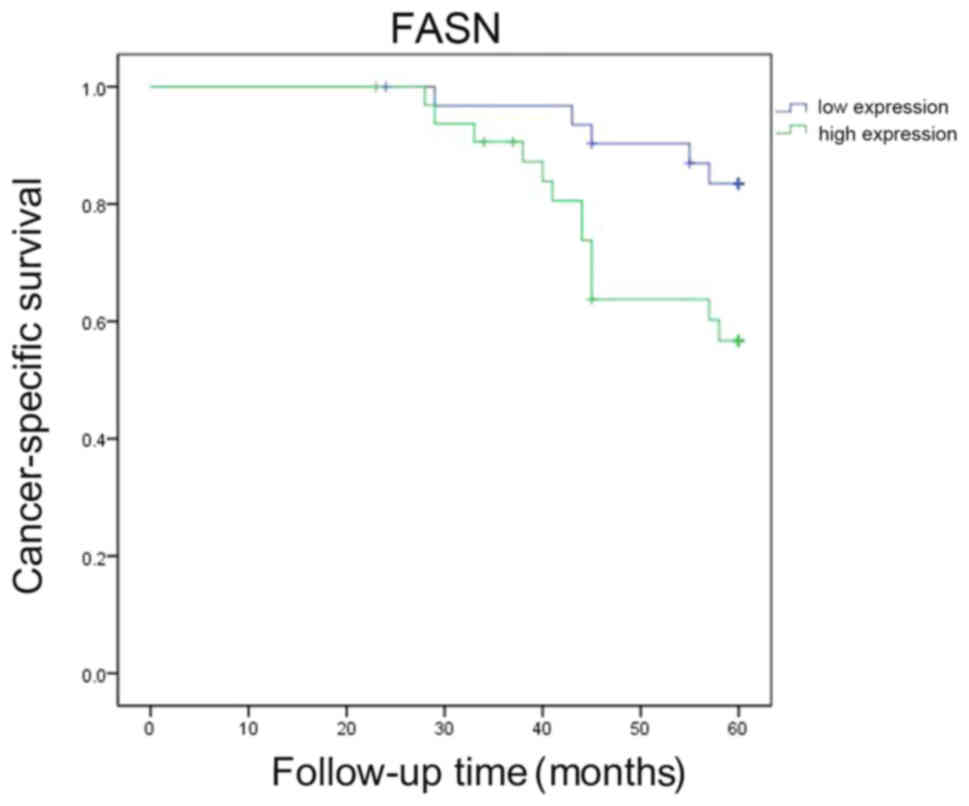
Survival analysis
Postoperative follow-up was performed for the 65
Wilms’ tumor patients (Fig. 10).
In the univariate analysis, the Kaplan-Meier survival curves
demonstrated that the survival of patients with high FASN
expression was significantly shorter compared with that of patients
with low FASN expression (P=0.016).
Discussion
Wilms’ tumor is the most common pediatric malignant
tumor of the urinary system, and its incidence rate is ~1/10,000
(20). As the tumor site is
concealed, the tumor is usually diagnosed at an advanced stage, and
there are currently no specific tumor markers. Wilms’ tumor poses a
serious threat to the patients’ wellbeing and the search for novel
anticancer targets to improve the clinical efficacy of Wilms’ tumor
treatment has been attracting increasing attention.
Lipid metabolism is an important and complex
biochemical reaction in the body involving numerous enzymes. This
process entails the digestion, absorption, synthesis and
decomposition of fat in living organisms. Fat is processed into the
substances required by the body to ensure the operation of normal
physiological functions (21).
Lipids are important substances that serve as energy storage and
energy supply, and phospholipids are important structural
components of biofilms. Currently, due to the high metabolic
properties of tumors, the study of energy metabolism in tumors has
been attracting increasing attention from researchers, as is the
interrelationship between lipid metabolism and the tumor. Lipid
metabolism is closely associated with the proliferation, invasion,
migration, radiosensitivity and chemosensitivity of various tumors,
such as lobular lung, breast and prostate cancer (6,22,23).
As regards the energy metabolism of Wilms’ tumors, current research
mainly focuses on glucose metabolism, whereas lipid metabolism has
been less extensively studied. In our experiments, preliminary
screening was performed by MS of tumor and adjacent tissues. Among
the 437 proteins identified, 19 lipid metabolism-related proteins
were differentially expressed. When these 19 lipid metabolism
proteins were analyzed using KEGG, 11 proteins were found to be
involved in the process of long chain elongation and degradation of
fatty acids. Certain proteins exhibited synergy, such as EHHADH and
HAHDA, which are involved in the conversion of
(S)-3-hydroxy-dodecanoyl-CoA to 3-oxo-decanoyl-CoA. The 19
differentially expressed proteins were further analyzed: A total of
16 proteins exhibited low expression (HADHB, ACAA1, ACAA2, CPT1A,
ECHS1, ACSF2, EHHADH, PC, ACADM, ACADSB, ECH1, GM2A, DECR1, HADHA,
NDUFAB1 and H3F3A), and 3 proteins (FASN, ISYNA1 and ALOX15) were
highly expressed. In addition, these 16 proteins and ALOX15 are
associated with the metabolism of lipids. FASN and ISYNA1 are
involved in the process of lipid synthesis. To a certain extent,
lipid synthesis is active and degradation is inhibited in Wilms’
tumors. The reason for this finding may be that the majority of the
tissues we selected were from children who had not undergone
chemoradiotherapy and the tumor was not under hypoxic conditions.
Excessive glucose metabolism (24)
may inhibit lipid degradation to a certain extent. However,
phospholipids are the main constituents of cell membranes, and the
proliferation of tumors is a highly active process that requires
the support of lipid synthesis, which may explain the
overexpression of synthetase.
IPA is an integrated biological pathway analysis
software based on cloud computing. IPA analysis includes function,
disease correlation analysis, biological downstream effect
analysis, classical pathway correlation analysis, and pathway
activity effect prediction (25,26).
Through IPA analysis, the dysregulated proteins screened by MS were
found to be mainly involved in biological processes, such as cell
death, cell cycle progression and lipid metabolism. Although the
level of lipid metabolism is relatively lower in tumors compared
with that in the adjacent tissues, lipid metabolism plays an
important role in tumors; signaling pathway analysis demonstrated
that dysregulated proteins play an important role in fatty acid
metabolic pathways in multiple tumors. The classical pathway is
involved in tumor apoptosis and autophagy, and the dysregulated
protein is implicated in the process of post-translational
modification. Sirtuin (NAD-dependent deacetylase) plays a key role
in the epigenetic regulation of gene expression by altering
chromatin structure. This protein has been reported to deacetylate
histones and non-histones. It has been demonstrated that sirtuin
deacetylation of histones induces chromatin condensation, whereas
acetylation of histone acetyltransferase leads to chromatin
decondensation. This balance is crucial for maintaining normal
cellular function, and any disruption in this process may lead to
tumorigenesis (27).
Sirtuin-mediated protein deacetylation has been considered to play
an important role in cancer (28),
and there are reports of dylinin expression disorders in a variety
of human malignancies (29-31).
IPA results revealed that sirtuin may be dysregulated in Wilms’
tumors, and the fatty acid metabolism pathway may affect biological
behaviors, such as proliferation, apoptosis and autophagy of Wilms’
tumors by sirtuin. In addition, the dysregulated proteins can be
identified from the regulatory sub-effector network: FASN, one of
the lipid synthesis-related enzymes among the dysregulated
proteins, holds an important position in the total regulatory
network and is involved in processes such as cell migration, cell
proliferation and tumor necrosis. In addition, FASN is associated
with a variety of signaling pathways.
Several other metabolic proteins in Wilms’ tumors
have been found to be reduced compared with their levels in normal
tissues, such as aldehyde dehydrogenase and proteins associated
with propionic acid and butyrate metabolism and long-chain fatty
acid metabolism (24). FASN is a
multi-enzyme protein that catalyzes fatty acid synthesis. This
protein is not a single enzyme but a whole enzymatic system
composed of two identical 272-kDa multifunctional polypeptides
(32,33). Accumulating evidence shows that
FASN expression is upregulated in various human cancers and confers
a survival advantage to these cancer cells (12,34-36).
FASN overexpression has also been associated with resistance to
anticancer treatments, and FASN has been found to be closely
associated with the prognosis of a variety of tumors (37,38);
however, its role in Wilms’ tumors has not been extensively
investigated. The present study revealed high expression of FASN by
MS screening. This finding was further verified by Oncomine
database, RT-qPCR, western blot, immunofluorescence and
immunohistochemistry analyses, which confirmed the high expression
of FASN in Wilms’ tumors. Furthermore, the clinical case parameters
of 65 children with Wilms’ tumors were analyzed and it was observed
that the expression of FASN was closely associated with tumor stage
and pathological type, which confirmed that FASN may be implicated
in the malignant progression of Wilms’ tumors. In addition, the
results of the survival analysis revealed that the prognosis of
patients with high expression of FASN was relatively poor, which
may be used as a biomarker to predict the prognosis. Therefore, the
findings of the present study may provide a theoretical basis for
subsequent experiments in vivo and in vitro, and help
identify new targets for the treatment of Wilms’ tumors.
Reprogramming cellular energy metabolism is one of
the major markers of cancer, and cancer cells rely on fatty acids
as a building block for cell proliferation; thus, disrupting key
enzymes or modulators in cellular metabolism may be a promising new
approach to cancer therapy (39,40).
Targeting lipid metabolism has provided novel ideas for the
treatment of tumors (24). In
particular, there is an increasing number of studies on targeted
therapies for FASN, and inhibition of FASN expression may be of
therapeutic value in a variety of tumors. In addition to the
specific inhibitors of FASN, such as C75, Fasnall, and EGCG
(41-43), which may exert antitumor effects,
researchers committed to design specific drugs for FASN and
discovered a new antitumor drug, 'compound 34’ for treating various
tumors (44). Other studies have
also sought to treat tumors by identifying upstream regulators of
FASN or miRNAs (7,45). In other tumors, the study of FASN
is not limited to clinical specimens; through cell and animal
experiments, its mechanism of action has been investigated and was
found to include the AKT/mTOR signaling pathway, which is closely
associated with tumors (46,47).
To a certain extent, these previous studies may explain the
effectiveness of targeting FASN for tumor treatment. In addition,
the present study may also be of value as a guide for the treatment
of Wilms’ tumors, for which the targeted inhibition of FASN may
prove useful.
Although extensive studies on cancer and its
initiation are beginning to unravel the role of lipid metabolism in
this process, the lipid metabolism-related enzymes that are
implicated in Wilms’ tumor have not been determined. Due to the low
incidence of pediatric vs. adult tumors, there were relatively few
cases included in the present study. However, the abovementioned MS
screening may provide novel ideas for the study of lipid metabolism
in Wilms’ tumors, which may serve as a reference for subsequent
research. In addition, FASN appeared to be a useful biomarker for
Wilms’ tumors based on the analysis of the prognosis of 65
pediatric patients. This index provides theoretical support for the
prognosis assessment of children with Wilms’ tumors. Moreover, FASN
may prove to be useful in the targeted therapy of Wilms’ tumor.
Funding
The present study was supported by funding provided
by the National Natural Science Foundation (grant no. 81400575),
the Hunan Provincial Natural Science Foundation of China (grant no.
2017JJ4071), the Science and Technology Development Plan Project of
Shandong Province, China (grant nos. 2014GSF118144, 2018GSF118209
and 2019GSF108061), Jinan Science and Technology Bureau (grant no.
201602170) and the Shandong Provincial Natural Science Foundation
(grant nos. ZR2017MH091 and ZR2015HM048).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors’ contributions
XQW and GQD conceived and designed the study. XQW,
YDW and YFZ contributed to the work by designing and performing the
experiments, collecting and analyzing data, and drafting the
manuscript. FG collected patient samples and conceived the
experiments. WL and RDW read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Provincial Hospital affiliated to Shandong University (approval
no. 2019-124). All the included specimens were obtained following
informed consent by the patients’ guardians, and the patient data
were anonymized. The research protocol conformed to the principles
outlined in the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Jiang C, Li X, Zhao H and Liu H: Long
non-coding RNAs: Potential new biomarkers for predicting tumor
invasion and metastasis. Mol Cancer. 15:622016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Long J, Zhang CJ, Zhu N, Du K, Yin YF, Tan
X, Liao DF and Qin L: Lipid metabolism and carcinogenesis, cancer
development. Am J Cancer Res. 8:778–791. 2018.PubMed/NCBI
|
|
3
|
Gómez de Cedrón M and Ramírez de Molina A:
Microtargeting cancer metabolism: Opening new therapeutic windows
based on lipid metabolism. J Lipid Res. 57:193–206. 2016.
View Article : Google Scholar :
|
|
4
|
Huang C and Freter C: Lipid metabolism,
apoptosis and cancer therapy. Int J Mol Sci. 16:924–949. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Luo X, Cheng C, Tan Z, Li N, Tang M, Yang
L and Cao Y: Emerging roles of lipid metabolism in cancer
metastasis. Mol Cancer. 16:762017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Deep G and Schlaepfer IR: Aberrant Lipid
Metabolism Promotes Prostate Cancer: Role in Cell Survival under
Hypoxia and Extracellular Vesicles Biogenesis. Int J Mol Sci.
17:172016. View Article : Google Scholar
|
|
7
|
Duan J, Chen L, Zhou M, Zhang J, Sun L,
Huang N, Bin J, Liao Y and Liao W: MACC1 decreases the
chemosensitivity of gastric cancer cells to oxaliplatin by
regulating FASN expression. Oncol Rep. 37:2583–2592. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Smith S, Witkowski A and Joshi AK:
Structural and functional organization of the animal fatty acid
synthase. Prog Lipid Res. 42:289–317. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Szolkiewicz M, Nieweglowski T, Korczynska
J, Sucajtys E, Stelmanska E, Goyke E, Swierczynski J and Rutkowski
B: Upregulation of fatty acid synthase gene expression in
experimental chronic renal failure. Metabolism. 51:1605–1610. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu H, Liu JY, Wu X and Zhang JT:
Biochemistry, molecular biology, and pharmacology of fatty acid
synthase, an emerging therapeutic target and diagnosis/prognosis
marker. Int J Biochem Mol Biol. 1:69–89. 2010.PubMed/NCBI
|
|
11
|
Heuer TS, Ventura R, Mordec K, Lai J,
Fridlib M, Buckley D and Kemble G: FASN Inhibition and Taxane
Treatment Combine to Enhance Anti-tumor Efficacy in Diverse
Xenograft Tumor Models through Disruption of Tubulin Palmitoylation
and Microtubule Organization and FASN Inhibition-Mediated Effects
on Oncogenic Signaling and Gene Expression. EBioMedicine. 16:51–62.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Papaevangelou E, Almeida GS, Box C,
deSouza NM and Chung YL: The effect of FASN inhibition on the
growth and metabolism of a cisplatin-resistant ovarian carcinoma
model. Int J Cancer. 143:992–1002. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ventura R, Mordec K, Waszczuk J, Wang Z,
Lai J, Fridlib M, Buckley D, Kemble G and Heuer TS: Inhibition of
de novo Palmitate Synthesis by Fatty Acid Synthase Induces
Apoptosis in Tumor Cells by Remodeling Cell Membranes, Inhibiting
Signaling Pathways, and Reprogramming Gene Expression.
EBioMedicine. 2:808–824. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Albiges L, Hakimi AA, Xie W, McKay RR,
Simantov R, Lin X, Lee JL, Rini BI, Srinivas S, Bjarnason GA, et
al: Body Mass Index and Metastatic Renal Cell Carcinoma: Clinical
and Biological Correlations. J Clin Oncol. 34:3655–3663. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Camassei FD, Jenkner A, Ravà L, Bosman C,
Francalanci P, Donfrancesco A, Alò PL and Boldrini R: Expression of
the lipogenic enzyme fatty acid synthase (FAS) as a predictor of
poor outcome in nephroblastoma: An interinstitutional study. Med
Pediatr Oncol. 40:302–308. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fuhrman SA, Lasky LC and Limas C:
Prognostic significance of morphologic parameters in renal cell
carcinoma. Am J Surg Pathol. 6:655–663. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nagaprashantha LD, Talamantes T, Singhal
J, Guo J, Vatsyayan R, Rauniyar N, Awasthi S, Singhal SS and Prokai
L: Proteomic analysis of signaling network regulation in renal cell
carcinomas with differential hypoxia-inducible factor-2α
expression. PLoS One. 8:e716542013. View Article : Google Scholar
|
|
18
|
Wang Q, Geng F, Zhou H, Chen Y, Du J,
Zhang X, Song D and Zhao H: MDIG promotes cisplatin resistance of
lung adenocar-cinoma by regulating ABC transporter expression via
activation of the WNT/β-catenin signaling pathway. Oncol Lett.
18:4294–4307. 2019.PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Friedman AD: Wilms tumor. Pediatr Rev.
34:328–330; discussion 330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Werbrouck E, Van Gansbeke D, Vanreusel A
and De Troch M: Temperature Affects the Use of Storage Fatty Acids
as Energy Source in a Benthic Copepod (Platychelipus littoralis,
Harpacticoida). PLoS One. 11:e01517792016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cha YJ, Kim HM and Koo JS: Expression of
Lipid Metabolism-Related Proteins Differs between Invasive Lobular
Carcinoma and Invasive Ductal Carcinoma. Int J Mol Sci. 18:182017.
View Article : Google Scholar
|
|
23
|
Kim S, Lee Y and Koo JS: Differential
expression of lipid metabolism-related proteins in different breast
cancer subtypes. PLoS One. 10:e01194732015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aminzadeh S, Vidali S, Sperl W, Kofler B
and Feichtinger RG: Energy metabolism in neuroblastoma and Wilms
tumor. Transl Pediatr. 4:20–32. 2015.
|
|
25
|
Liu X, Wen F, Yang J, Chen L and Wei YQ: A
review of current applications of mass spectrometry for
neuroproteomics in epilepsy. Mass Spectrom Rev. 29:197–246.
2010.
|
|
26
|
Thomas S and Bonchev D: A survey of
current software for network analysis in molecular biology. Hum
Genomics. 4:353–360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Glozak MA, Sengupta N, Zhang X and Seto E:
Acetylation and deacetylation of non-histone proteins. Gene.
363:15–23. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Glozak MA and Seto E: Histone deacetylases
and cancer. Oncogene. 26:5420–5432. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen X, Sun K, Jiao S, Cai N, Zhao X, Zou
H, Xie Y, Wang Z, Zhong M and Wei L: High levels of SIRT1
expression enhance tumorigenesis and associate with a poor
prognosis of colorectal carcinoma patients. Sci Rep. 4:74812014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hao C, Zhu PX, Yang X, Han ZP, Jiang JH,
Zong C, Zhang XG, Liu WT, Zhao QD, Fan TT, et al: Overexpression of
SIRT1 promotes metastasis through epithelial-mesenchymal transition
in hepatocellular carcinoma. BMC Cancer. 14:9782014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang RH, Sengupta K, Li C, Kim HS, Cao L,
Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al: Impaired DNA damage
response, genome instability, and tumorigenesis in SIRT1 mutant
mice. Cancer Cell. 14:312–323. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Alberts AW, Strauss AW, Hennessy S and
Vagelos PR: Regulation of synthesis of hepatic fatty acid
synthetase: Binding of fatty acid synthetase antibodies to
polysomes. Proc Natl Acad Sci USA. 72:3956–3960. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stoops JK, Arslanian MJ, Oh YH, Aune KC,
Vanaman TC and Wakil SJ: Presence of two polypeptide chains
comprising fatty acid synthetase. Proc Natl Acad Sci USA.
72:1940–1944. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang Y, Yin X, Wu L, Qin Q and Xu J:
MAPK/P53-mediated FASN expression in bone tumors. Oncol Lett.
13:4035–4038. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
O’Malley J, Kumar R, Kuzmin AN, Pliss A,
Yadav N, Balachandar S, Wang J, Attwood K, Prasad PN and Chandra D:
Lipid quantification by Raman microspectroscopy as a potential
biomarker in prostate cancer. Cancer Lett. 397:52–60. 2017.
View Article : Google Scholar
|
|
36
|
Patel AV, Johansson G, Colbert MC,
Dasgupta B and Ratner N: Fatty acid synthase is a metabolic
oncogene targetable in malignant peripheral nerve sheath tumors.
Neuro-oncol. 17:1599–1608. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Diaz KP, Gondak R, Martins LL, de Almeida
OP, León JE, Mariano FV, Altemani A and Vargas PA: Fatty acid
synthase and Ki-67 immunoexpression can be useful for the
identification of malignant component in carcinoma ex-pleomorphic
adenoma. J Oral Pathol Med. 48:232–238. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sangeetha M, Deepa PR, Rishi P, Khetan V
and Krishnakumar S: Global gene deregulations in FASN silenced
retinoblastoma cancer cells: Molecular and clinico-pathological
correlations. J Cell Biochem. 116:2676–2694. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Currie E, Schulze A, Zechner R, Walther TC
and Farese RV Jr: Cellular fatty acid metabolism and cancer. Cell
Metab. 18:153–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ooi AT and Gomperts BN: Molecular
Pathways: Targeting Cellular Energy Metabolism in Cancer via
Inhibition of SLC2A1 and LDHA. Clin Cancer Res. 21:2440–2444. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Alwarawrah Y, Hughes P, Loiselle D,
Carlson DA, Darr DB, Jordan JL, Xiong J, Hunter LM, Dubois LG,
Thompson JW, et al: Fasnall, a Selective FASN Inhibitor, Shows
Potent Anti-tumor Activity in the MMTV-Neu Model of HER2(+) Breast
Cancer. Cell Chem Biol. 23:678–688. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wong A, Chen S, Yang LK, Kanagasundaram Y
and Crasta K: Lipid accumulation facilitates mitotic
slippage-induced adaptation to anti-mitotic drug treatment. Cell
Death Discov. 4:1092018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Giro-Perafita A, Palomeras S, Lum DH,
Blancafort A, Viñas G, Oliveras G, Pérez-Bueno F, Sarrats A, Welm
AL and Puig T: Preclinical Evaluation of Fatty Acid Synthase and
EGFR Inhibition in Triple-Negative Breast Cancer. Clin Cancer Res.
22:4687–4697. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lu T, Schubert C, Cummings MD, Bignan G,
Connolly PJ, Smans K, Ludovici D, Parker MH, Meyer C, Rocaboy C, et
al: Design and synthesis of a series of bioavailable fatty acid
synthase (FASN) KR domain inhibitors for cancer therapy. Bioorg Med
Chem Lett. 28:2159–2164. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Singh R, Yadav V, Kumar S and Saini N:
MicroRNA-195 inhibits proliferation, invasion and metastasis in
breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1.
Sci Rep. 5:174542015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shen M, Tsai Y, Zhu R, Keng PC, Chen Y,
Chen Y and Lee SO: FASN-TGF-β1-PD-L1 axis contributes to the
development of resistance to NK cell cytotoxicity of
cisplatin-resistant lung cancer cells. Biochim Biophys Acta Mol
Cell Biol Lipids. 1863:313–322. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tao T, Su Q, Xu S, Deng J, Zhou S, Zhuang
Y, Huang Y, He C, He S, Peng M, et al: Down-regulation of PKM2
decreases FASN expression in bladder cancer cells through
AKT/mTOR/SREBP-1c axis. J Cell Physiol. 234:3088–3104. 2019.
View Article : Google Scholar
|