Introduction
Bcl2l10, also known as Diva, Bcl-b or Boo, is a
member of the Bcl-2 family of proteins, which are central mediators
of apoptosis and autophagy (1,2). The
Bcl-2 family proteins contain up to 4 conserved amino acid
sequences, known as Bcl-2 homology (BH) domains (1,3).
These proteins are grouped into 3 categories: i) Anti-apoptotic
proteins, including Bcl-2, Bcl-xL, Bcl-w, NR-13, A1 and Mcl-1; ii)
multi-domain pro-apoptotic proteins, such as Bax, and Bak; and iii)
BH3 domain-only proteins, such as Bad, Bim, Bid and Bik (1,4).
Interactions between pro-apoptotic and anti-apoptotic Bcl-2 family
proteins play important roles in controlling and promoting
apoptosis (1,3). However, Bcl2l10 reportedly has
contradictory functions in different apoptotic cells or tissues and
is recognized for its both pro-apoptotic (5-7) and
anti-apoptotic activities (8-10).
Previously, it was reported that Bcl2l10 was highly
expressed in mouse ovaries and oocytes and that it was associated
with cytoskeletal proteins (11,12).
It was demonstrated that Bcl2l10-depleted oocytes arrested at the
metaphase I stage, exhibiting abnormal spindle and chromosome
formation (12). These results
revealed that Bcl2l10 plays a crucial role in oocyte meiosis. By
using microarray analysis, it was found that Bcl2l10 was associated
with cytoskeletal proteins, such as targeting protein for xenopus
kinesin-like protein 2 (Tpx2) and centrosomal protein of 192 kDa
(Cep192), which are well-known cofactors of Aurora kinase A (Aurka)
(13). Notably, Bcl2l10 was
subsequently demonstrated to directly interact with Tpx2 in meiotic
spindles and to further regulate the expression and activity of
Aurka (14).
In somatic cells, Tpx2 directly interacts with Aurka
at spindle microtubules and protects Aurka from degradation
(15-17). Aurka controls multiple aspects of
mitotic cell division and plays important roles in centrosome
maturation and bipolar spindle formation in somatic cells (18). Aurka expression and activity are
upregulated in several types of solid tumors. Additionally, Aurka
overexpression has been shown to increase cancer cell malignancy
(19-22), promptly impair cell cycle
progression and cause chromosomal instability, supernumerary
centrosomes and multipolar spindles. These properties have led to
the consideration of Aurka as an oncogene and a target of cancer
therapy (23-25). Based on our previous finding that
Bcl2l10 is an upstream regulator of Aurka and considering that
Aurka is considered a crucial oncogenic factor (14), it was hypothesized that Bcl2l10
plays an important role in cancer cells. If Bcl2l10 is functionally
connected with Aurka in cancer cells, Bcl2l10 may be a novel
effective therapeutic target that can be used with Aurka inhibitors
as an anticancer drug.
The functions of Bcl2l10 in cancers have been
previously reported. Bcl2l10 plays roles as an oncogene and as a
tumor suppressor gene depending on the type of cancer. In gastric
and lung cancer cells, Bcl2l10 exhibits a low-level expression
relative to its expression in adjacent normal tissues, and the
overexpression of Bcl2l10 induces apoptosis and cell growth
inhibition (26-29). By contrast, Bcl2l10 acts as an
oncogene in myelodysplastic syndromes (MDS), acute myeloid leukemia
(AML), glioma and breast cancers (9,30,31).
However, although Bcl2l10 is highly expressed in human ovaries, the
biological functions of Bcl2l10 in ovarian cancer have yet to be
reported. Therefore, the aims of this study were to determine
whether Bcl2l10 regulates Aurka and to investigate the function of
Bcl2l10 in A2780 and SKOV3 ovarian cancer cells.
Materials and methods
Tissue microarray
Slides including 20 different types of human cancer
tissues (brain, esophagus, larynx, thymus, thyroid, breast,
stomach, pancreas, tongue, lung, lymph node, liver, kidney, ovary,
cervix, testis, colon, rectum, skin and soft tissue) were purchased
from AccuMax (ISU ABXIS). Slides made from formalin-fixed,
paraffin-embedded blocks and stained with hematoxylin and eosin
were used to define the most morphologically representative,
well-fixed cells. Single-tissue cores (1 mm in diameter) were
sampled from each paraffin block and assembled into a recipient
paraffin block using a tissue microarray instrument (AccuMax Array,
ISU ABXIS). To detect Bcl2l10 in cancer tissues,
immunohistochemical staining was performed with an anti-Bcl2l10
antibody. Briefly, antigen retrieval was conducted by boiling the
slide in 1X citrate buffer for 10 min, and blocking was then
performed in hydrogen peroxide solution for 10 min. The slide was
incubated with the anti-Bcl2l10 antibody (anti-BCL2L10, 1:100
dilution, ab151419, Abcam) for 1 h at room temperature and
incubated with a horseradish peroxidase (HRP) polymer solution (Lab
Vision; Thermo Fisher Scientific) for 15 min. Thereafter, the slide
was stained with a 3,3′-diaminobenzidine-tetrahydrochloride (DAB)
solution (Vector Laboratories) at room temperature for 3 min and
counterstained with hematoxylin (Sigma-Aldrich; Merck KGaA) for 5
min.
Cells and cell culture
Six ovarian cancer cell lines were used in this
study. The CAOV-3, SKOV3 and OVCAR-3 cells were obtained from the
Korean Cell Line Bank and the COV-318, A2780 cells were obtained
from Cellbank Australia; the TOV-21G cell line was purchased from
ATCC and the normal ovarian epithelia cell line, HOSE, was
purchased from Innoprot. The CAOV-3 and COV-318 cells were
maintained in DMEM (Gibco; Thermo Fisher Scientific) containing 10%
fetal bovine serum (FBS); the SKOV3 and OVCAR-3 cells were
maintained in RPMI-1640 (Gibco; Thermo Fisher Scientific)
containing 10% FBS, 25 mM HEPES (Gibco; Thermo Fisher Scientific),
25 mM NaHCO3 (Sigma-Aldrich; Merck KGaA); the TOV-21G
cells were maintained in MCDB and Medium 199 containing 15% FBS;
the A2780 cells were maintained in RPMI-1640 containing 10% FBS;
HOSE was maintained in medium provided from Innoprot.
Cell transfection
Bcl2l10 small-interfering RNA (siRNA) was
synthesized by Shanghai GenePharma Co., Ltd. and negative control
siRNA was purchased at Bioneer. One day prior to transfection, the
A2780 and SKOV3 cancer cells were seeded in 6-well plates. Before
transfection, the medium was removed and replaced with 1.5 ml of
fresh growth medium. All siRNAs were diluted in 0.25 ml of OPTI-MEM
to a final concentration of 100 nM, and Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific) was diluted in 0.25 ml of
OPTI-MEM. The solutions were incubated individually for 5 min at
room temperature and then combined and incubated for an additional
20 min at room temperature. The mixture was then added to each well
containing cells, which were maintained in a 5% CO2
atmosphere at 37°C for 48 h.
Total RNA extraction and cDNA
synthesis
After the cultured cells were washed twice with PBS,
1 ml of TRIzol reagent (Takara Bio) and 0.2 ml of chloroform were
added, and the mixture was incubated at room temperature for 10
min. The cells were centrifuged at 12,000 × g for 20 min at 4°C,
and the supernatants were transferred to new tubes and resuspended
in 0.5 ml of isopropanol. After the tubes were centrifuged at
12,000 × g for 10 min at 4°C, the supernatants were discarded, and
the precipitates were dried. The pellets were washed with 75%
ethanol, dried, and dissolved in 0.1% diethylpyrocarbonate
(DEPC)-treated water. To synthesize first-strand cDNA, total RNA (2
µg) was added to DNase I (New England Biolabs) and DNase I
buffer (New England Biolabs), and the total volume was adjusted to
11 ml with DEPC-treated water. The mixture was incubated at room
temperature for 15 min, 1 ml of 25 mM EDTA were added, and the
mixture was incubated at 65°C for 15 min. Subsequently, 1 ml of
oligo dT was added, and the mixture was incubated at 70°C for 10
min. M-MLV RNase (Promega), 5X buffer (Promega), RNase inhibitor
(Promega) and 10 mM dNTP were then added, and the reaction was
performed at 42°C for 1 h and 94°C for 2 min.
Western blot analysis
Proteins were extracted from the treated cells using
RIPA lysis buffer with a 1% protease inhibitor cocktail (Thermo
Fisher Scientific) and 0.5 M EDTA (pH 8.0). Protein concentrations
were estimated using the Bio-Rad protein assay reagent (Bio-Rad)
according to the manufacturer’s instructions. Protein extracts (30
µg) were separated using 10% SDS-PAGE and then transferred
onto PVDF membranes (Amersham Biosciences). The membranes were
blocked for 1 h in Tris-buffered saline/Tween [TBST; 0.2 M NaCl,
0.1% Tween-20, and 10 mM Tris (pH 7.4)] containing 5% non-fat dry
milk. Immunoblots were incubated overnight at 4°C with diluted
polyclonal antibodies against Bcl2l10 (#3869S, Cell Signaling
Technology), Aurka (#610939, BD Biosciences), cyclin-dependent
kinase (CDK)4 (sc-23896, Santa Cruz Biotechnology), cyclinD1
(#2978S, Cell Signaling Technology), p27 (#2552S, Cell Signaling
Technology), p21 (sc-6246, Santa Cruz Biotechnology), p16 (#80772S,
Cell Signaling Technology) and β-actin (PA1-183, Invitrogen; Thermo
Fisher Scientific) diluted 1:1,000, washed several times with TBST
and incubated with diluted solutions of goat anti-rabbit IgG
(#65-6120, Thermo Fisher Scientific) or goat anti-mouse IgG
(#62-6520, Thermo Fisher Scientific) diluted 1:2,000 for 1 h at
room temperature. After each step, the membranes were washed
several times with TBST, and the relative expression of the protein
bands was quantified using the chemiDoc XRS+ imaging system with
Image Lab software.
Immunofluorescence staining
The cells were seeded at density of 1×105
onto cover slides and incubated at 5% CO2 and 37°C.
After 24 h, the medium was removed from the dish, and prewarmed
(37°C) staining solution containing 100 nM MitoTracker (Invitrogen;
Thermo Fisher Scientific) stain was added. The cells were then
incubated for 15 min at 37°C, fixed with 4% formaldehyde (Electron
Microscopy Sciences) in medium for 20 min, and permeabilized with
0.2% Triton X-100 (Sigma-Aldrich; Merck KGaA) in PBS for 20 min.
The cells were then washed 3 times with PBS, blocked with 3% bovine
serum albumin in PBS for 1 h, and incubated overnight at 4°C with a
primary antibody against Bcl2l10 (1:100; #3869S, Cell Signaling
Technology). The cells were washed 3 times with PBS and then
incubated with Alexa Fluor 488-conjugated anti-rabbit IgG (1:100;
#A11034 Invitrogen; Thermo Fisher Scientific). Nuclei were stained
with 4′,6-diamidino-2-phenyl-indole (DAPI) (Invitrogen; Thermo
Fisher Scientific) at room temperature for 10 min, and the samples
were then cover-slipped with mounting solution (Dako). Observations
were performed under a confocal microscope (Leica).
Measurement of Aurka activity
Following transfection with Bcl2l10 siRNA, Aurka
activity was measured using the CycLex® Aurora-A Kinase
Assay/Inhibitor Screening kit (#CY-1165; MBL). According to
protocols provided by the manufacturer of this kit, cytoplasmic and
nuclear extracts were preferentially separated from whole cell
lysates using the Subcellular Protein Fraction kit (#78840; Thermo
Fisher Scientific). Each cell lysate was used to measure Aurka
activity according to manufacturer’s instruction.
Cycloheximide decay assay
Following siRNA transfection for 48 h, 50
µg/ml cycloheximide (Sigma-Aldrich; Merck KGaA) was added to
inhibit de novo protein synthesis. The cells were then
harvested at each indicated time point (0, 30, 60, 90, 120 and 150
min) and subjected to western blot analysis as described above.
Cell apoptosis assay
Apoptosis was evaluated by Annexin V-fluorescein
isothiocyanate (Annexin V-FITC) and propidium iodide (PI) staining
in transfected cells using an Annexin V-FITC Apoptosis Detection
kit (BD Biosciences) according to the manufacturer’s instructions.
The cells were harvested at 48 h after transfection. Cell pellets
were washed twice with PBS and suspended in 100 ml of 1X binding
buffer (10 mM HEPES/NaOH, 140 mM NaCl, 25 mM CaCl2, pH
7.4). The cells were then incubated with 5 ml of Annexin V-FITC and
10 ml of PI at room temperature for 15 min in the dark. Following
incubation, 400 ml of 1X binding buffer were added to each tube,
and the cells were analyzed immediately using a FACSCalibur flow
cytometer (BD Biosciences). Data analysis was performed using
FlowJo software (BD Biosciences), and FACS measurements were
performed for at least 3 independent experiments.
Cell cycle analysis
Cell cycle distribution was determined by staining
DNA with PI (Invitrogen; Thermo Fisher Scientific). At 48 h
following transfection, the cells were fixed with cooled 100%
ethanol and stained with 50 µg/ml PI and 1 mg/ml RNase A for
30 min at room temperature. Analytical cytometry was performed on a
FACSCalibur flow cytometer (BD Biosciences), and cell cycle
analysis was performed using FlowJo software (BD Biosciences). FACS
measurements were performed for at least 3 independent
experiments.
Cell Counting kit-8 (CCK-8) assay
Cell proliferation was determined using the CCK-8
kit (Dojindo Molecular Technologies). Briefly, 3×103
cells (A2780) and 2×103 cells (SKOV3) were seeded per
well in 96-well plates and allowed to adhere overnight prior to
transfection. At 0, 24, 48 and 72 h following transfection, 10
µl CCK-8 solution were added to each well for incubation at
5% CO2 and 37°C for an additional 4 h. Finally, the
optical density was determined at a wavelength of 450 nm using a
microplate reader (E-MAX, Molecular Devices).
In vitro Transwell invasion and migration
assays
The invasion assay was performed using Matrigel (BD
Biosciences). Briefly, chilled Matrigel was mixed to homogeneity
using a cooled pipette, diluted in serum-free media to a final
concentration of 1 mg/ml, and then used to coat a transwell (8-mm
pore size polycarbonate membrane) (Corning, Inc.). At 48 h
following transfection, the cells were resuspended in serum-free
medium and loaded into each upper well. Medium containing 10% FBS
was added to the lower chamber and served as a chemoattractant.
Following overnight incubation, cells were fixed with 4%
formaldehyde for 2 min, permeabilized with 100% methanol for 2 min
at room temperature, and then stained with 0.1% crystal violet
(Sigma-Aldrich; Merck KGaA) at room temperature for 10 min. Cells
on the upper surface of the Transwell were removed using cotton
swabs. A Transwell without Matrigel was used for the migration
assay, which was performed in the same manner as the invasion
assay. Subsequently, each chamber with the invaded or migrated
cells was soaked in 0.5 ml 10% acetic acid for 1 min to wash out
the crystal violet. Subsequently, 10% acetic acid was added at 200
µl/well to 96-well plates, and the absorbance was measured
at a wavelength of 562 nm using a microplate reader (Molecular
Devices).
In vitro wound healing assay
Cell motility was measured using the in vitro
wound healing assay using a Culture-Insert (Ibidi) according to the
manufacturer’s instructions. Briefly, at 48 h following
transection, the cells were resuspended and seeded on each side of
a culture insert. The inserts were then placed into wells of a
plate and incubated at 37°C and 5% CO2 to allow the
cells to grow to confluence. Subsequently, the inserts were removed
with sterile tweezers to create a cell-free area ('defined wound’)
of ~500 µm. The migration rate into this 'wound area’ was
measured after the indicated times (A2780: 0, 24 and 48 h; SKOV3:
0, 3 and 6 h).
Statistical analysis
Data are presented as the means ± SEM derived from 3
independent experiments. Differences among multiple groups were
analyzed using one-way ANOVA followed by Student-Newman-Keuls test.
Differences between negative control and Bcl2l10 siRNA-treated
groups were analyzed using the Student’s t-test. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
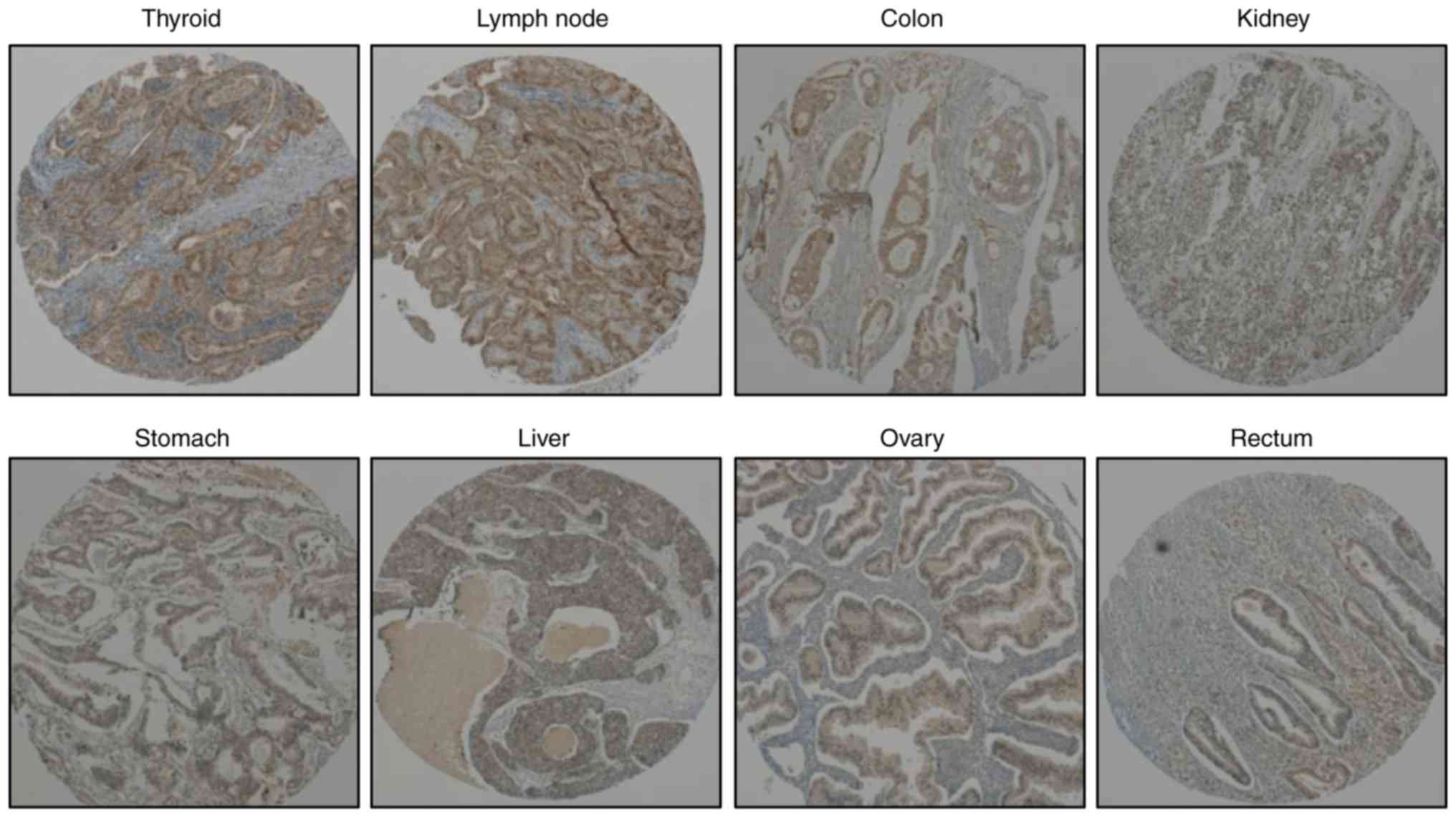
Bcl2l10 expression in various human
cancer tissues
In a previous study by the authors, it was
determined that Bcl2l10 was not expressed in mouse somatic cells,
such as ovarian cumulus and granulosa cells, but was detected in
oocytes (12). In the present
study, to determine the expression of Bcl2l10 in human cancer
tissues, a tissue microarray analysis was performed. The results
revealed that Bcl2l10 was expressed in various human cancer
tissues, including papillary thyroid carcinoma, lymph node
metastatic papillary carcinoma, colon adenocarcinoma, kidney renal
cell carcinoma, stomach adenocarcinoma, hepatocellular carcinoma,
ovarian clear cell carcinoma and rectum adenocarcinoma tissues.
Bcl2l10 was particularly highly expressed in thyroid and lymph node
cancer tissues (Fig. 1). These
data support the possibility that Bcl2l10 is a cancer-related
gene.
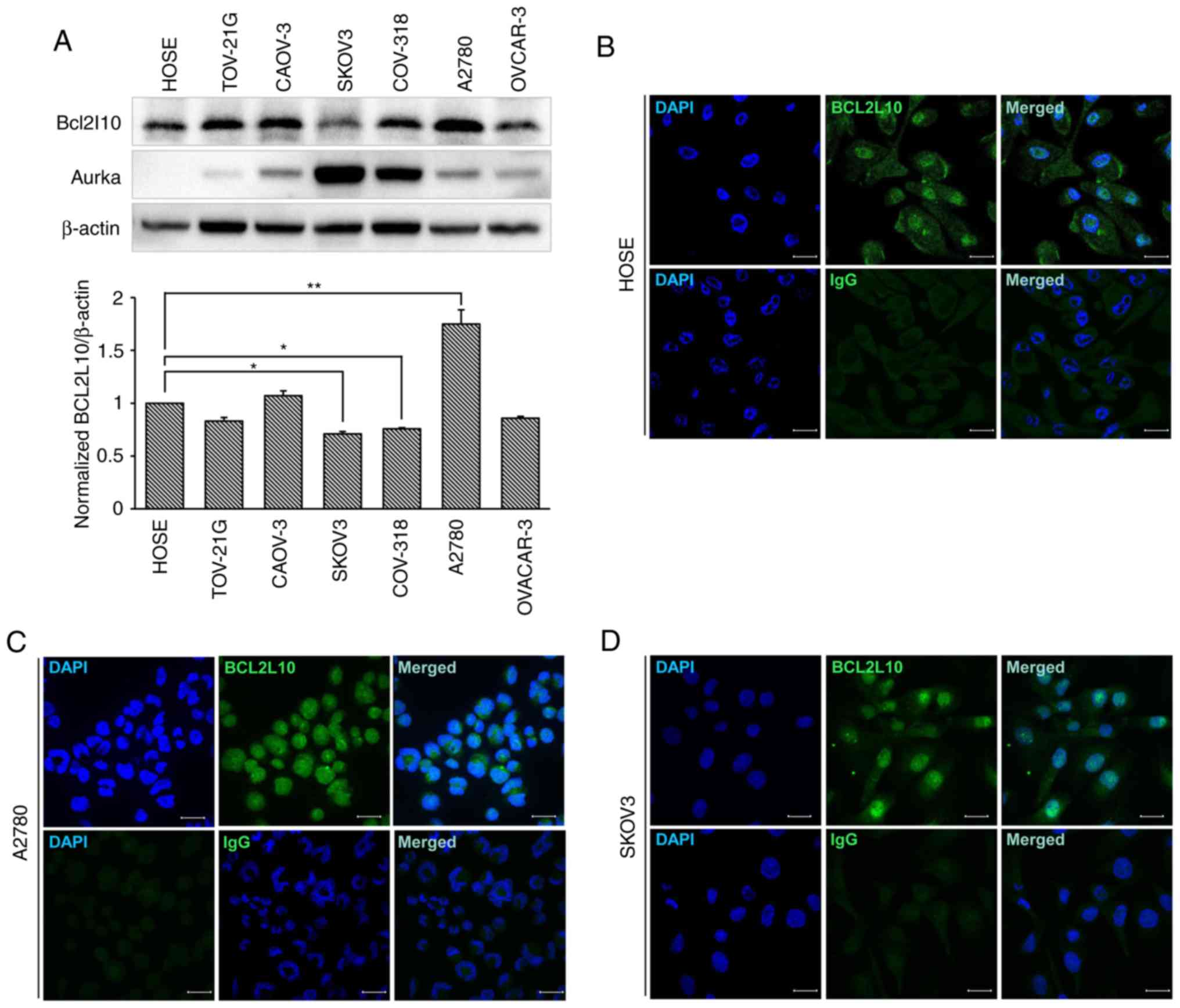
Expression and localization of Bcl2l10 in
human ovarian cancer cell lines
Prior to the analysis of the role of Bcl2l10 in
human ovarian cancer cells, the protein levels of Bcl2l10 were
examined in various human ovarian cancer cell lines (TOV-21G,
CAOV-3, SKOV3, COV-318, A2780 and OVCAR-3) and a normal primary
ovarian epithelial cell line (HOSE). The results indicated that
Bcl2l10 protein was expressed in all 6 ovarian cancer cell lines,
and the levels were significantly lower in the SKOV3 and COV-318
cells, and also tended to decrease in the TOV-21G and OVCAR-3 cells
compared to the levels in the normal ovarian cell line, HOSE cells
(Fig. 2A). The level of Bcl2l10 in
the A2780 cells was significantly higher than that in HOSE cells.
Since the A2780 cells expressed the highest level of Bcl2l10,
whereas the SKOV3 expressed the lowest level of Bcl2l10 among the 6
ovarian cancer cell lines, the A2780 and SKOV3 cells were selected
for use in further experiments to determine the role of Bcl2l10 in
ovarian cancer cells. Additionally, the expression of Aurka was
observed in normal ovarian cell and ovarian cancer cells. In the
HOSE cells, Aurka protein was not expressed as Aurka is a
well-known oncogene (32-34) (Fig.
2A). Of note, the expression pattern of Aurka protein in the 6
ovarian cancer cell lines was opposite to that of Bcl2l10 protein.
In other words, the TOV-21G, CAOV-3 and A2780 cells expressed high
levels of Bcl2l10 and expressed low levels of Aurka, whereas the
SKOV3 and COV-318 cells expressed low levels of Bcl2l10 and high
levels of Aurka (Fig. 2A).
To investigate the intracellular localization of
Bcl2l10 in ovarian cancer cells, immunofluorescence staining of the
HOSE, A2780 and SKOV3 cells was performed. The results revealed
Bcl2l10 that was mainly expressed in the nuclei of all 3 cell lines
(Fig. 2B-D).
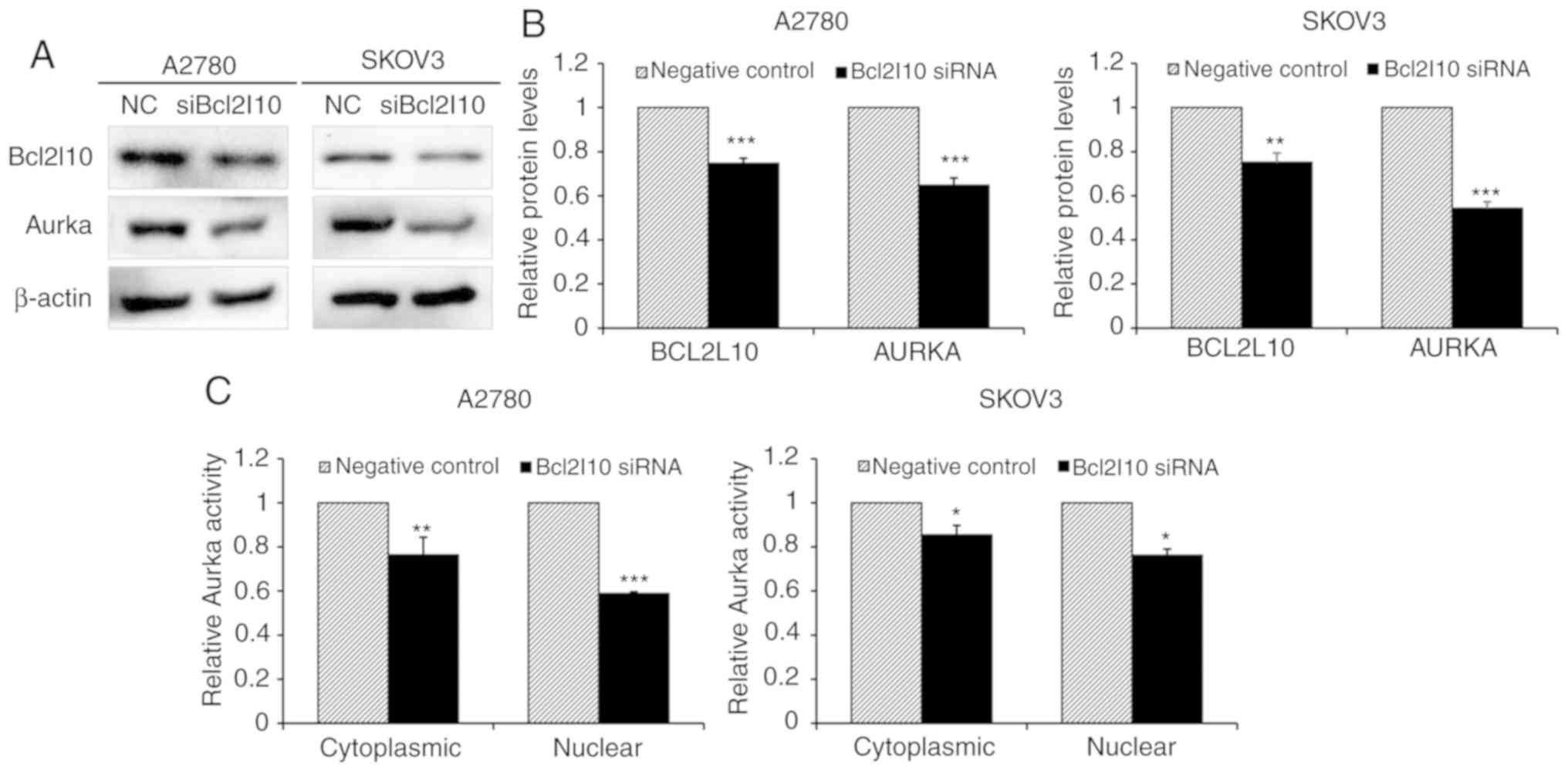
Bcl2l10 regulates the expression and
catalytic activity of Aurka in ovarian cancer cells
Based on a previous finding that Bcl2l10 regulates
the expression and activities of Aurka in mouse oocytes as a master
regulator of Aurka (14), this
study investigated whether Bcl2l10 also affects Aurka expression in
ovarian cancer cells by western blot analysis. The protein levels
of Bcl2l10 and Aurka were decreased following the knockdown of
Bcl2l10 by siRNA in the A2780 and SKOV3 cells (Fig. 3A and B). In addition, to determine
whether Aurka activity was affected by Bcl2l10, cytoplasmic and
nuclear protein fractions were harvested from negative control- and
Bcl2l10 siRNA-transfected cells, and the kinase activity of Aurka
was measured by the colorimetric method. The results revealed that
both the cytoplasmic and nuclear Aurka activity were significantly
reduced in the Bcl2l10 siRNA-transfected A2780 and SKOV3 cells
(Fig. 3C). These results are
consistent with our previous findings (14) using mouse oocytes and indicated
that Bcl2l10 regulates the expression and activities of Aurka in
ovarian cancer cells.
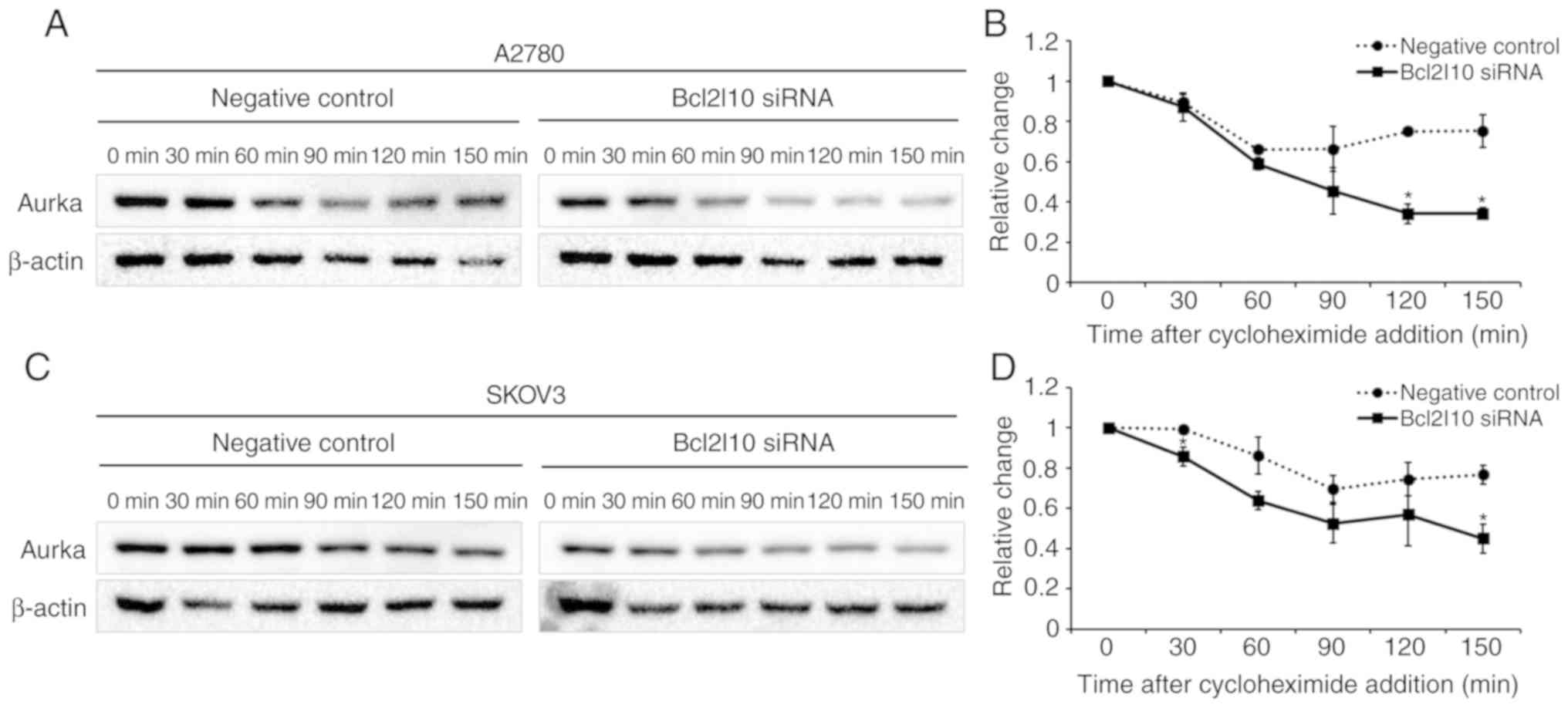
Bcl2l10 knockdown affects Aurka protein
stability in ovarian cancer cells
As a decrease in Aurka expression was observed after
Bcl2l10 knockdown, we then determined whether Bcl2l10 regulates
Aurka stability in ovarian cancer cells. Following transfection
with negative control or Bcl2l10 siRNA into the A2780 and SKOV3
cells, the cells were treated with cycloheximide to inhibit de
novo protein synthesis. Protein lysates were harvested at
30-min intervals following cycloheximide treatment, and western
blot analysis was performed to investigate Aurka stability. The
expression of Aurka declined rapidly in both cell lines following
trans-fection with Bcl2l10 siRNA (Fig.
4). These results indicate that Bcl2l10 contributes to Aurka
protein stability, which explains the decrease in Aurka protein
levels by Bcl2l10 knockdown.
Bcl2l10 knockdown does not affect the
apoptotic death of ovarian cancer cells
As Bcl2l10 is a member of the apoptotic gene family,
it was hypothesized that Bcl2l10 may affect the apoptosis of
ovarian cancer cells. To examine this hypothesis, the A2780 and
SKOV3 cells were transfected with negative control or Bcl2l10
siRNA. The percentage of apoptotic cells was then evaluated using
flow cytometric analysis with Annexin V/PI double staining. Annexin
V-/PI- staining indicates viable cells, which
are not permeable to PI due to their intact cell membranes.
AV+/PI- staining indicates early apoptotic
cells; such staining occurs due to the loss of plasma membrane
asymmetry and the strong affinity of AV-FITC with
phosphatidylserine. AV+/PI+ staining is
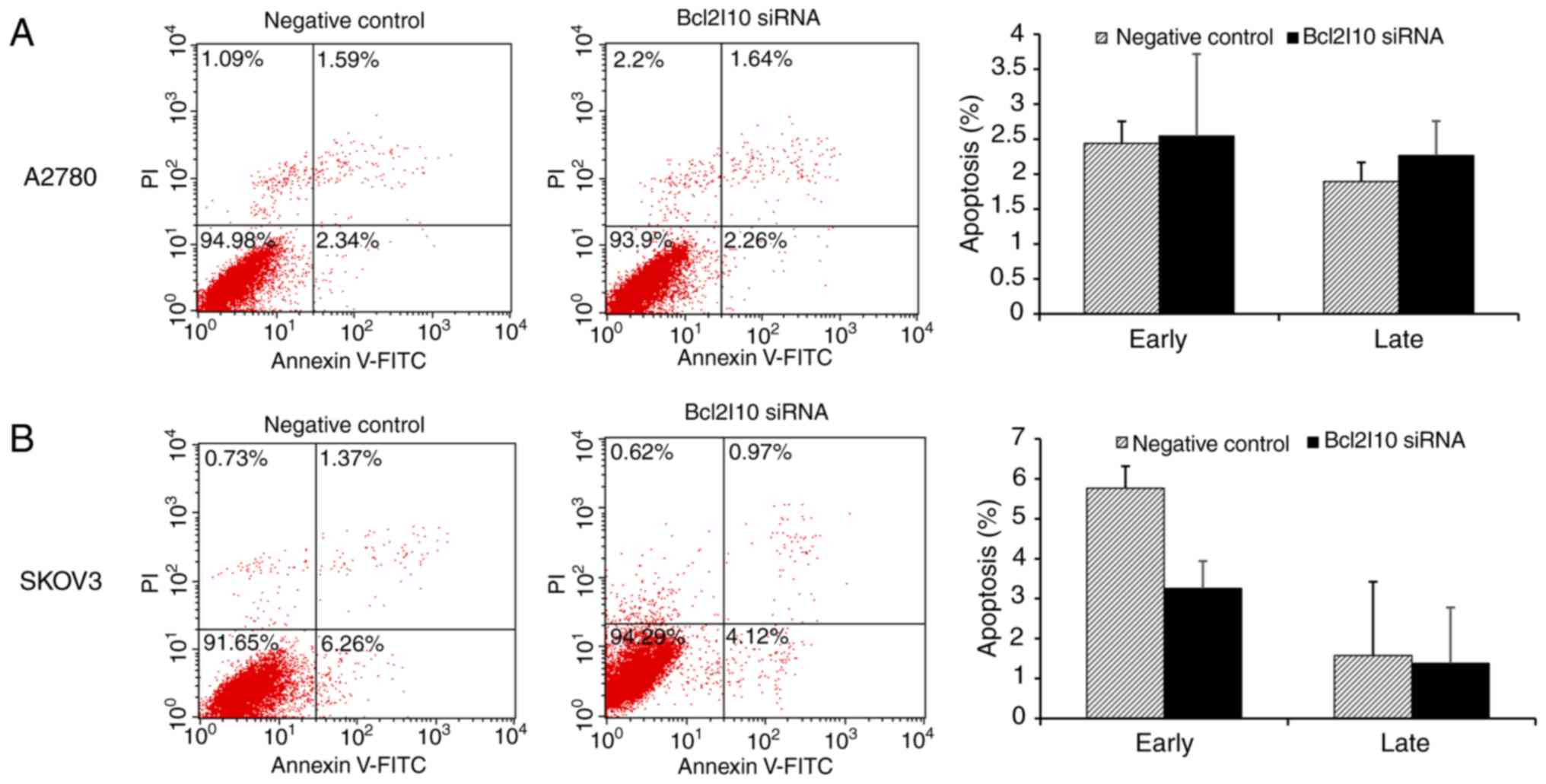
indicative of late apoptotic cells. As shown in Fig. 5, no significant difference was
observed in either early or late apoptosis between the negative
control- and Bcl2l10 siRNA-transfected A2780 cells (Fig. 5A). In the SKOV3 cells, the
knockdown of Bcl2l10 seemed to decrease the percentage of cells in
early apoptosis, although this was not statistically significant.
The percentage of cells in late apoptosis was not affected by
treatment with Bcl2l10 siRNA (Fig.
5B). Almost all of the A2780 and SKOV3 cells transfected with
Bcl2l10 siRNA were viable cells, as evidenced by
AV-/PI- staining (93.9 and 94.5%,
respectively). The results suggest that Bcl2l10 does not affect the
apoptosis of ovarian cancer cells.
Bcl2l10 knockdown promotes the
proliferation of ovarian cancer cells
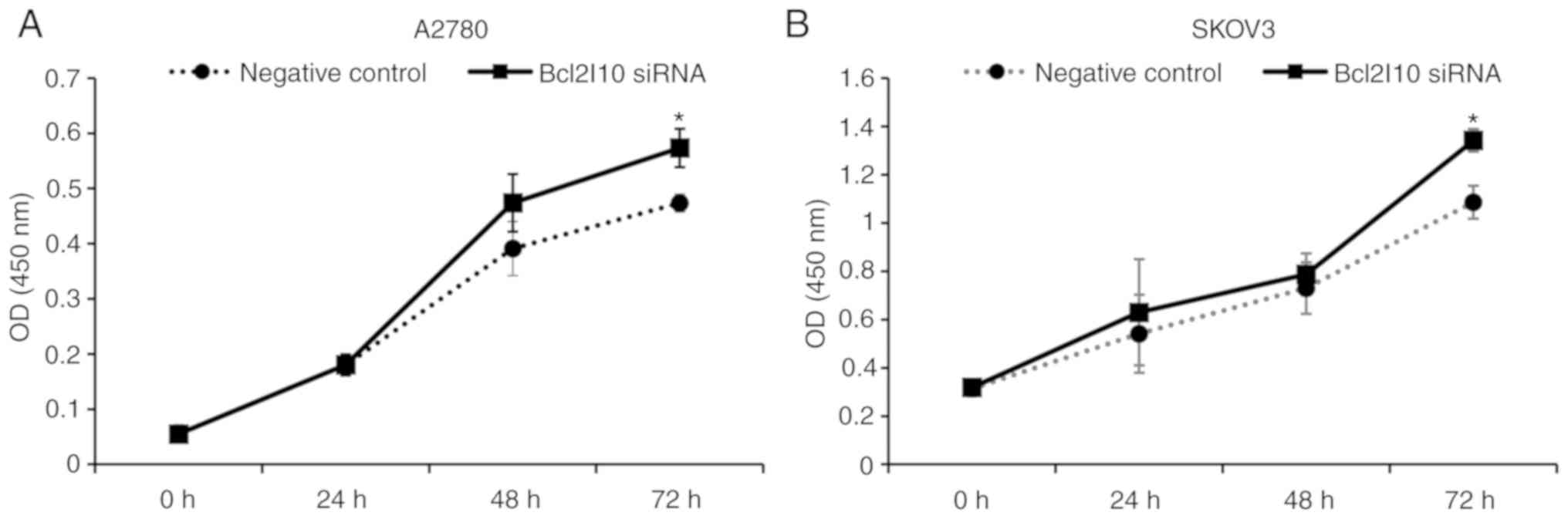
To determine the effects of Bcl2l10 on the viability
of ovarian cancer cells, aCCK-8 assay was performed for 72 h
following siRNA transfection. As shown in Fig. 6, the proliferation of the A2780 and
SKOV3 cells transfected with Bcl2l10 siRNA increased in a
time-dependent manner relative to the rate of negative
control-siRNA-transfected cells. At 72 h after Bcl2l10 siRNA
transfection, Bcl2l10 knockdown led to an increase in the
proliferation rate of 21 and 24% in the A2780 and SKOV3 cells,
respectively (Fig. 6). These
results suggest that endogenous Bcl2l10 suppresses the
proliferation of ovarian cancer cells.
Bcl2l10 knockdown induces cell cycle
arrest at the G0/G1 phase by regulating G0/G1-related regulatory
genes in ovarian cancer cells
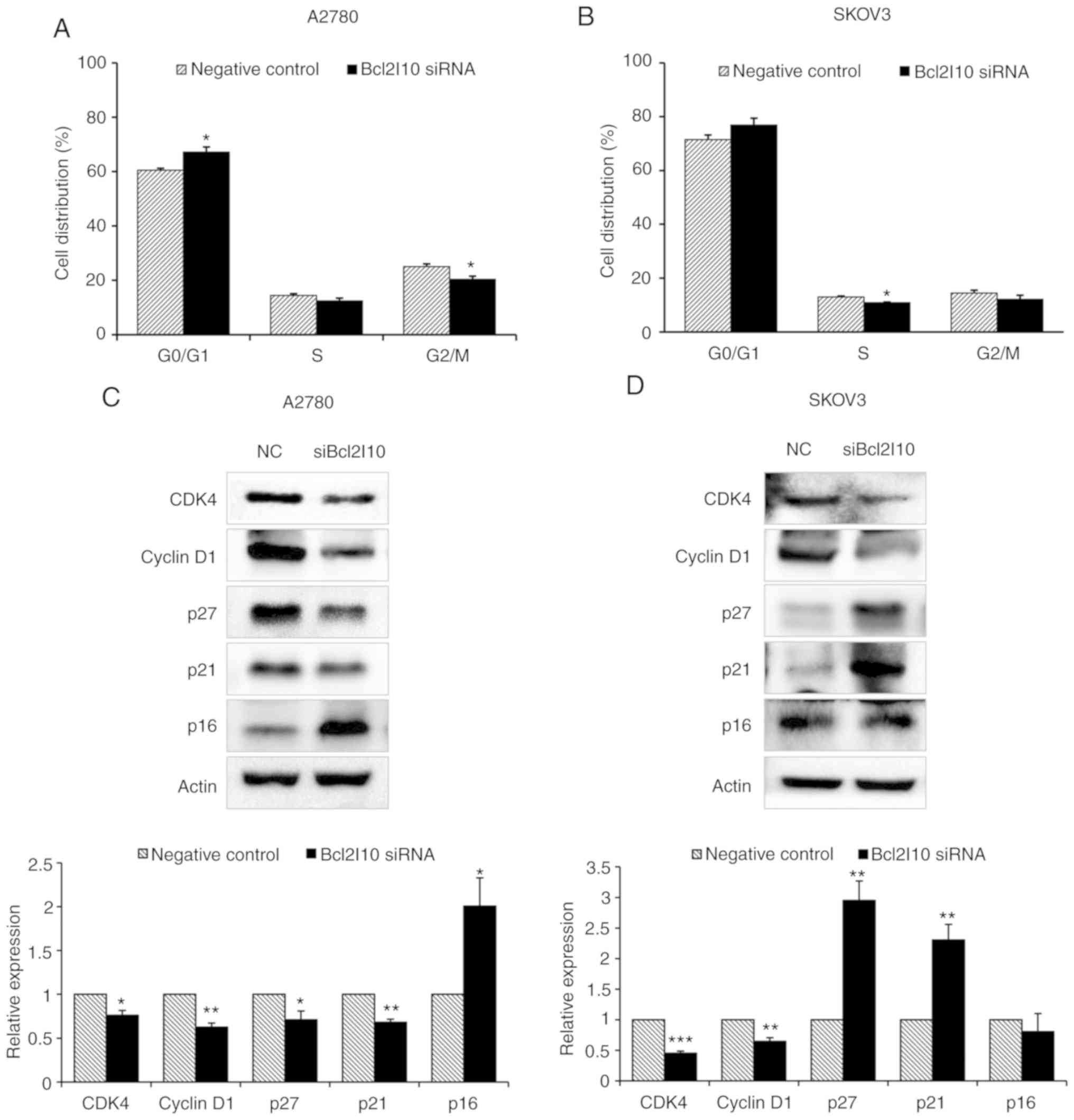
To examine whether the effects of Bcl2l10 on cell
proliferation are mediated via cell cycle regulation, cell cycle
changes were assessed using flow cytometry following Bcl2l10 siRNA
transfection. It was expected that Bcl2l10 knockdown would decrease
the cell population in the G0/G1 phase and would increase it in the
G2/M phase due to the above-mentioned finding that cell
proliferation was enhanced by Bcl2l10 suppression. However,
following Bcl2l10 knockdown in the A2780 and SKOV3 cells, the
percentage of cells in the G0/G1 phase was increased in the A2780
cells (60.5% in the control group vs. 67.2% in the Bcl2l10
siRNA-transfected group) and SKOV3 cells (71.5% in the control
group vs. 76.9% in the Bcl2l10 siRNA-transfected group), whereas
the percentages of cells in the S and G2/M phases were slightly
decreased (Fig. 7A and B). These
results indicated that Bcl2l10 knockdown induced cell cycle arrest
at the G0/G1 phase in ovarian cancer cells.
To investigate the mechanisms underlying the
induction of cell cycle arrest at the G0/G1 phase, the levels of
cyclins and CDKs that regulate the G0/G1 phase were examined by
western blot analysis. In both the A2780 and SKOV3 cells, the
levels of CDK4 and cyclin D1 were significantly decreased by
Bcl2l10 knockdown relative to the levels in the control group
(Fig. 7C and D). Subsequently, the
authors determined whether CDK inhibitors (CDKIs), such as p27, p21
and p16, which are known as inhibitors of CDK4, are affected by
Bcl2l10 knockdown (35). In the
Bcl2l10 siRNA-transfected group of A2780 cells, the levels of p27
and p21 were significantly decreased, whereas the level of p16 was
markedly increased (Fig. 7C). By
contrast, in the SKOV3 cells, Bcl2l10 knockdown markedly increased
the levels of p27 and p21, whereas the expression of p16 was not
markedly unaffected (Fig. 7D).
Taken together, these results suggest that Bcl2l10 mediates cell
cycle progression in the G0/G1 phase via the regulation of
G0/G1-related cyclins, CDKs and CDKIs in ovarian cancer cells.
Bcl2l10 knockdown promotes the migration
and invasion of ovarian cancer cells
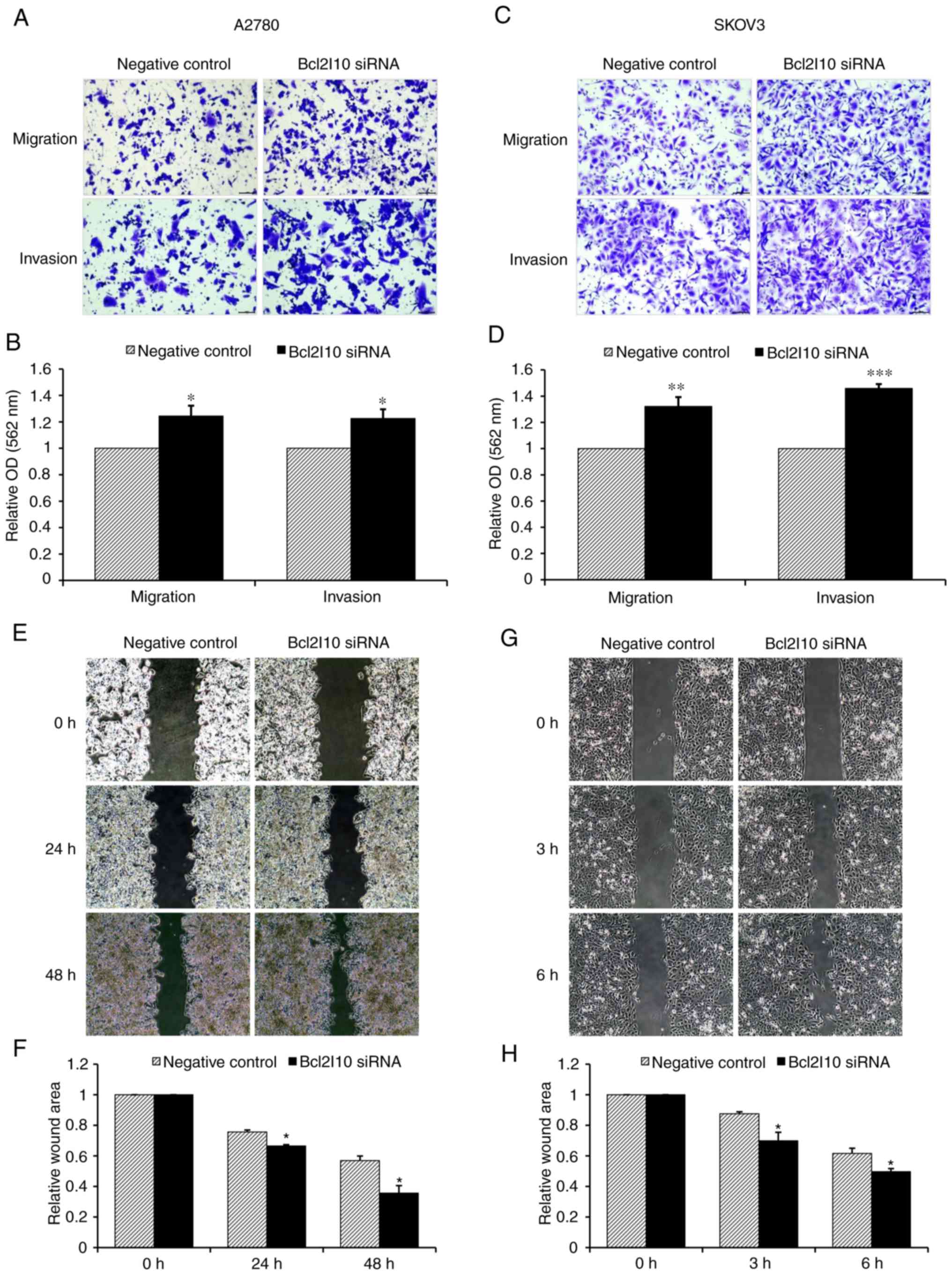
The roles of Bcl2l10 in cell migration and invasion
were then assessed using Transwell assays. The results revealed
that the numbers of migrated and invaded cells were increased by
Bcl2l10 suppression (Fig. 8A and
C). The rates of migration and invasion were quantified using
the dissolved crystal violet from cells passing through the
Transwell chamber. In the A2780 cells, the migratory and invasive
abilities were increased by 24.4 and 22.6%, respectively, in the
Bcl2l10 siRNA-transfected group relative to the control group
(Fig. 8B). Similarly, upon Bcl2l10
siRNA treatment, 32.2 and 45.9% of SKOV3 cells exhibited increased
migratory and invasive abilities, respectively, relative to the
abilities of the control cells (Fig.
8D). The migratory abilities were further confirmed by wound
healing assay. Bcl2l10 knockdown enhanced the motility of both the
A2780 and SKOV3 cells (Fig. 8E and
G). After 48 h, the A2780 cells filled 43.1 and 64.3% of the
wound area in the control and Bcl2l10 siRNA-transfected group,
respectively (Fig. 8F). After 6 h,
the SKOV3 cells filled 38.4 and 50.4% of the wound area in the
control and Bcl2l10 siRNA-transfected group, respectively (Fig. 8H). These data suggest that Bcl2l10
contributes to the regulation of the invasion and migration of
ovarian cancer cells in vitro.
Discussion
The present study investigated the functions of
Bcl2l10 in ovarian cancer cells. Previously, it was found that
Bcl2l10 is a master regulator of Aurka in mouse oocytes. Aurka is
reportedly overexpressed in a number of malignancies, including
breast, liver, pancreatic and ovarian cancers, inducing tumor
progression (32,36). As Aurka has been considered as an
important target for cancer therapy (37), it was hypothesized that if Bcl2l10
regulates tumor progression depending on Aurka, the targeting of
Bcl2l10 could be used in combination with Aurka inhibitor to
generate synergistic anticancer effects.
Of note, it was found that cells expressing high
levels of Bcl2l10 expressed low levels of Aurka, whereas cells
expressing low levels of Bcl2l10 expressed high levels of Aurka. In
addition, there were also differences in baseline Aurka activity
between the A2780 and SKOV3 cells (data not shown). The Aurka
activities in the nuclei of SKOV3 cells were higher than those of
the A2780 cells, corresponding with the results that the expression
of Aurka was higher in the SKOV3 than in the A2780 cells. These
results provide evidence that Bcl2l10 regulates Aurka expression in
ovarian cancer cells. As was expected, the knockdown of Bcl2l10
decreased the protein expression and activity of Aurka in A2780 and
SKOV3 cells. It has been widely reported that the silencing of
Aurka expression by siRNA or treatment with the Aurka inhibitor,
MLN8237, blocks cancer progression by inducing apoptosis, cell
cycle arrest at the G2/M phase, autophagy, aneuploidy and mitotic
spindle failure in a number of types of cancer (38-41).
However, this study obtained contradictory results from
Bcl2l10-silenced cells, which exhibited phenotypic alterations
different from those of Aurka-silenced cells. The knockdown of
Bcl2l10 stimulated cell proliferation and mobility, whereas it
induced cell cycle arrest at the G0/G1 phase without affecting the
apoptosis of A2780 and SKOV3 cells.
Although there are contradictory data, relatively
lower expression levels of Bcl2l10 in ovarian cancer cells and
increased cell proliferation and motility by Bcl2l10 knockdown are
sufficient to conclude that Bcl2l10 is a tumor suppressor in
ovarian cancer cells. Genomic instability is a hallmark of
malignant transformation (42) and
the effects of the gain or loss of a single gene are likely to be
transmitted throughout the genome with the consequence that the
expression of other genes becomes secondarily modulated (43). Therefore, it was hypothesized that
the stimulation of cell proliferation by Bcl2l10 knockdown was
caused by changes in the expression or stability of other
Bcl2l10-related genes rather than Aurka downregulation. Although
Bcl2l10 knockdown decreased Aurka expression, the effects of other
genes whose expression was altered by Bcl2l10 knockdown may be
greater than that of decreased Aurka expression.
Bcl2l10 is a member of the Bcl-2 family that
regulates the balance between apoptosis and autophagy (2). In the present study, the authors
hypothesized that the depletion of Bcl2l10 may alter the apoptotic
rate of ovarian cancer cells; however, Bcl2l10 knockdown did not
affect the apoptotic rate. The apoptotic regulation of Bcl2l10 is
reportedly cell- or tissue-specific. The roles of proteins are
determined by their components based on cellular content, protein
interactions or external stimuli. However, the mechanisms
underlying the divergent cell- or tissue-specific activities of
Bcl2l10 have not yet been clarified (5-10).
Contributing to the conflicting results in a number of tissues,
this study demonstrated that Bcl2l10 had no effect on the apoptosis
of A2780 and SKOV3 cells. Thus, Bcl2l10 may not be a dominant
regulator of the apoptosis of ovarian cancer cells.
Cancer occurs due to inappropriate cell
proliferation (44,45). In normal cells, the cell cycle is
exquisitely controlled by signaling pathways for cell growth and
includes processes to correct errors that occur during cell
division (46). Cancer cells can
overcome these errors through the abnormal expression of cyclins
and CDKs, allowing them to undergo unregulated cell growth
(47). In the present study,
Bcl2l10 knockdown induced cell cycle arrest at the G0/G1 phase
through the upregulation of CDK inhibitors (p16, p21 and p27)
followed by the suppression of CDK4 and cyclin D1. Cell cycle
arrest represents a survival mechanism that provides cancer cells
the opportunity to repair their own damaged DNA, and cell cycle
retardation can activate the apoptotic cascade, leading to cancer
cell death (48). In this study,
it was found that the proliferation rate of Bcl2l10-silenced cells
was significantly increased relative to that of the control cells.
These results suggest that Bcl2l10 knockdown facilitates the
proliferation of ovarian cancer cells. Taken together, it can be
concluded that Bcl2l10 is involved in the cell cycle progression
and proliferation of ovarian cancer cells as a tumor suppressor
gene.
Metastasis, one of the most important issues in
cancer development (49),
comprises a series of events. epithelial- mesenchymal transition
and mesenchymalepithelial transition are important events for the
metastasis of carcinomas (50,51),
whereas cell invasion and migration are important processes in a
number of physiological events, such as embryo implantation
(52), angiogenesis (53) and inflammation (54). However, cell invasion and migration
are also involved in the pathophysiology of a number of diseases,
such as neuronal migration disorders, atherosclerosis and cancer
(55,56). In this study, it was found that
Bcl2l10 knockdown increased the number of invading or migrating
cells relative to the number among control cells. In the wound
healing assay, wounds were rapidly closed by the silencing of
Bcl2l10. These results suggest that Bcl2l10 may facilitate the
anti-metastatic functions in human ovarian cancer. However, the
mechanisms underlying this effect in the present study were not
addressed. Further studies are thus required to elucidate the
Bcl2l10 signaling network and to determine the mechanisms through
which it hinders the invasion and/or migration of ovarian cancer
cells.
In conclusion, this study demonstrated that Bcl2l10
regulates expression and activity of Aurka in ovarian cancer cells.
Importantly, the present study, to the best of our knowledge, is
the first to report that Bcl2l10 plays a role in tumorigenesis and
in the proliferation of ovarian cancer cells. To clarify the
molecular functions of Bcl2l10 in ovarian cancer cells, the authors
aim to explore novel Bcl2l10-related genes using RNA-Sequencing
analysis in future studies. In the future, our results may provide
a potential basis for developing ovarian anti-cancer agents
targeting Bcl2l10.
Funding
This research was supported by the Basic Science
Research Program through the National Research Foundation of Korea
(NRF) funded by the Ministry of Education
(NRF-2016R1A2B4013403).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article or are available from the
corresponding author upon reasonable request.
Authors’ contributions
JHW designed and performed experiments and wrote the
manuscript. SYL performed experiments and revised the manuscript.
JK provided experimental materials and analyzed the data. KHK was
involved in the conception and design of the study. KAL was
involved in the conception and design of the study and revised the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The tissues examined in this study were from a
tissue microarray; thus, no ethics approval was required.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
AURKA
|
Aurora kinase A
|
|
BH
|
Bcl-2 homology
|
|
RNAi
|
RNA interference
|
|
siRNA
|
small-interfering RNA
|
|
Tpx2
|
targeting protein for xenopus
kinesin-like protein 2
|
Acknowledgments
Not applicable.
References
|
1
|
Adams JM and Cory S: The Bcl-2 protein
family: Arbiters of cell survival. Science. 281:1322–1326. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Levine B, Sinha S and Kroemer G: Bcl-2
family members: Dual regulators of apoptosis and autophagy.
Autophagy. 4:600–606. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Opferman JT and Korsmeyer SJ: Apoptosis in
the development and maintenance of the immune system. Nat Immunol.
4:410–415. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zong WX, Lindsten T, Ross AJ, MacGregor GR
and Thompson CB: BH3-only proteins that bind pro-survival Bcl-2
family members fail to induce apoptosis in the absence of Bax and
Bak. Genes Dev. 15:1481–1486. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Inohara N, Gourley TS, Carrio R, Muñiz M,
Merino J, Garcia I, Koseki T, Hu Y, Chen S and Núñez G: Diva, a
Bcl-2 homologue that binds directly to Apaf-1 and induces
BH3-independent cell death. J Biol Chem. 273:32479–32486. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kang Y, Lee DC, Han J, Yoon S, Won M, Yeom
JH, Seong MJ, Ko JJ, Lee KA, Lee K and Bae J: NM23-H2 involves in
negative regulation of Diva and Bcl2L10 in apoptosis signaling.
Biochem Biophys Res Commun. 359:76–82. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mikata R, Yokosuka O, Fukai K, Imazeki F,
Arai M, Tada M, Kurihara T, Zhang K, Kanda T and Saisho H: Analysis
of genes upregulated by the demethylating agent
5-aza-2′-deoxycytidine in gastric cancer cell lines. Int J Cancer.
119:1616–1622. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ke N, Godzik A and Reed JC: Bcl-B, a novel
Bcl-2 family member that differentially binds and regulates Bax and
Bak. J Biol Chem. 276:12481–12484. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Naumann U, Weit S, Wischhusen J and Weller
M: Diva/Boo is a negative regulator of cell death in human glioma
cells. FEBS Lett. 505:23–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang H, Holzgreve W and De Geyter C:
Bcl2-L-10, a novel anti-apoptotic member of the Bcl-2 family,
blocks apoptosis in the mitochondria death pathway but not in the
death receptor pathway. Hum Mol Genet. 10:2329–2339. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yoon SJ, Kim JW, Choi KH, Lee SH and Lee
KA: Identification of oocyte-specific diva-associated proteins
using mass spectrometry. Korean J Fertil Steril DE. 33:189–198.
2006.
|
|
12
|
Yoon SJ, Kim EY, Kim YS, Lee HS, Kim KH,
Bae J and Lee KA: Role of Bcl2-like 10 (Bcl2l10) in regulating
mouse oocyte maturation. Biol Reprod. 81:497–506. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim EA, Kim KS, Lee HS, Lee S, Kim EY, Seo
YM, Bae J and Lee KA: Downstream genes regulated by Bcl2l10 RNAi in
the mouse oocytes. Dev Reprod. 15:61–69. 2011.
|
|
14
|
Lee SY, Kim EY, Kim KH and Lee KA:
Bcl2l10, a new Tpx2 binding partner, is a master regulator of
Aurora kinase A in mouse oocytes. Cell Cycle. 15:3296–3305. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Joukov V: Aurora kinases and spindle
assembly: Variations on a common theme? Cell Cycle. 10:895–903.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kufer TA, Silljé HH, Körner R, Gruss OJ,
Meraldi P and Nigg EA: Human TPX2 is required for targeting
Aurora-A kinase to the spindle. J Cell Biol. 158:617–623. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Giubettini M, Asteriti IA, Scrofani J, De
Luca M, Lindon C, Lavia P and Guarguaglini G: Control of Aurora-A
stability through interaction with TPX2. J Cell Sci. 124:113–122.
2011. View Article : Google Scholar
|
|
18
|
Giet R, Petretti C and Prigent C: Aurora
kinases, aneuploidy and cancer, a coincidence or a real link?
Trends Cell Biol. 15:241–250. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Compérat E, Camparo P, Haus R,
Chartier-Kastler E, Radenen B, Richard F, Capron F and Paradis V:
Aurora-A/STK-15 is a predictive factor for recurrent behaviour in
non-invasive bladder carcinoma: A study of 128 cases of
non-invasive neoplasms. Virchows Arch. 450:419–424. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lassmann S, Shen Y, Jütting U, Wiehle P,
Walch A, Gitsch G, Hasenburg A and Werner M: Predictive value of
Aurora-A/STK15 expression for late stage epithelial ovarian cancer
patients treated by adjuvant chemotherapy. Clin Cancer Res.
13:4083–4091. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nishida N, Nagasaka T, Kashiwagi K, Boland
CR and Goel A: High copy amplification of the Aurora-A gene is
associated with chromosomal instability phenotype in human
colorectal cancers. Cancer Biol Ther. 6:525–533. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jeng YM, Peng SY, Lin CY and Hsu HC:
Overexpression and amplification of Aurora-A in hepatocellular
carcinoma. Clin Cancer Res. 10:2065–2071. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Anand S, Penrhyn-Lowe S and Venkitaraman
AR: AURORA-A amplification overrides the mitotic spindle assembly
checkpoint, inducing resistance to Taxol. Cancer Cell. 3:51–62.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Meraldi P, Honda R and Nigg EA: Aurora-A
overexpression reveals tetraploidization as a major route to
centrosome amplification in p53-/- cells. EMBO J.
21:483–492. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nikonova AS, Astsaturov I, Serebriiskii
IG, Dunbrack RL Jr and Golemis EA: Aurora A kinase (AURKA) in
normal and pathological cell division. Cell Mol Life Sci.
70:661–687. 2013. View Article : Google Scholar
|
|
26
|
Xu JD, Furuya T, Cao XX, Liu XL, Li QQ,
Wang WJ, Xu JW, Xu ZD, Sasaki K and Liu XP: Loss of BCL2L10 protein
expression as prognostic predictor for poor clinical outcome in
gastric carcinoma. Histopathology. 57:814–824. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu JD, Cao XX, Long ZW, Liu XP, Furuya T,
Xu JW, Liu XL, De Xu Z, Sasaki K and Li QQ: BCL2L10 protein
regulates apoptosis/proliferation through differential pathways in
gastric cancer cells. J Pathol. 223:400–409. 2011. View Article : Google Scholar
|
|
28
|
Mikata R, Fukai K, Imazeki F, Arai M,
Fujiwara K, Yonemitsu Y, Zhang K, Nabeya Y, Ochiai T and Yokosuka
O: BCL2L10 is frequently silenced by promoter hypermethylation in
gastric cancer. Oncol Rep. 23:1701–1708. 2010.PubMed/NCBI
|
|
29
|
Bai Y, Wang J, Han J, Xie XL, Ji CG, Yin
J, Chen L, Wang CK, Jiang XY, Qi W and Jiang HQ: BCL2L10 inhibits
growth and metastasis of hepatocellular carcinoma both in vitro and
in vivo. Mol Carcinog. 56:1137–1149. 2017. View Article : Google Scholar
|
|
30
|
Nougarede A, Popgeorgiev N, Kassem L,
Omarjee S, Borel S, Mikaelian I, Lopez J, Gadet R, Marcillat O,
Treilleux I, et al: Breast cancer targeting through inhibition of
the endoplasmic reticulum-based apoptosis regulator Nrh/BCL2L10.
Cancer Res. 78:1404–1417. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cluzeau T, Robert G, Mounier N, Karsenti
JM, Dufies M, Puissant A, Jacquel A, Renneville A, Preudhomme C,
Cassuto JP, et al: BCL2L10 is a predictive factor for resistance to
azacitidine in MDS and AML patients. Oncotarget. 3:490–501. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Carvajal RD, Tse A and Schwartz GK: Aurora
kinases: New targets for cancer therapy. Clin Cancer Res.
12:6869–6875. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fu J, Bian M, Jiang Q and Zhang C: Roles
of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res.
5:1–10. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW,
Sahin A, Brinkley BR and Sen S: Tumour amplified kinase STK15/BTAK
induces centrosome amplification, aneuploidy and transformation.
Nat Genet. 20:189–193. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
D’Andrilli G, Kumar C, Scambia G and
Giordano A: Cell cycle genes in ovarian cancer: Steps toward
earlier diagnosis and novel therapies. Clin Cancer Res.
10:8132–8141. 2004. View Article : Google Scholar
|
|
36
|
Gautschi O, Heighway J, Mack PC, Purnell
PR, Lara PN Jr and Gandara DR: Aurora kinases as anticancer drug
targets. Clin Cancer Res. 14:1639–1648. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Keen N and Taylor S: Aurora-kinase
inhibitors as anticancer agents. Nat Rev Cancer. 4:927–936. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou N, Singh K, Mir MC, Parker Y, Lindner
D, Dreicer R, Ecsedy JA, Zhang Z, Teh BT, Almasan A and Hansel DE:
The investigational Aurora kinase A inhibitor MLN8237 induces
defects in cell viability and cell-cycle progression in malignant
bladder cancer cells in vitro and in vivo. Clin Cancer Res.
19:1717–1728. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li JP, Yang YX, Liu QL, Pan ST, He ZX,
Zhang X, Yang T, Chen XW, Wang D, Qiu JX and Zhou SF: The
investigational Aurora kinase A inhibitor alisertib (MLN8237)
induces cell cycle G2/M arrest, apoptosis, and autophagy via p38
MAPK and Akt/mTOR signaling pathways in human breast cancer cells.
Drug Des Devel Ther. 9:1627–1652. 2015.PubMed/NCBI
|
|
40
|
Görgün G, Calabrese E, Hideshima T, Ecsedy
J, Perrone G, Mani M, Ikeda H, Bianchi G, Hu Y, Cirstea D, et al: A
novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and
cell-cycle arrest in multiple myeloma. Blood. 115:5202–5213. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Asteriti IA, Di Cesare E, De Mattia F,
Hilsenstein V, Neumann B, Cundari E, Lavia P and Guarguaglini G:
The Aurora-A inhibitor MLN8237 affects multiple mitotic processes
and induces dose-dependent mitotic abnormalities and aneuploidy.
Oncotarget. 5:6229–6242. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Komarova NL: Genomic instability in
cancer: Biological and mathematical approaches. Cell Cycle.
3:1081–1085. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang XD: Genome-wide screens for
effective siRNAs through assessing the size of siRNA effects. BMC
Res Notes. 1:332008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Collins K, Jacks T and Pavletich NP: The
cell cycle and cancer. Proc Natl Acad Sci USA. 94:2776–2778. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jones RG and Thompson CB: Tumor
suppressors and cell metabolism: A recipe for cancer growth. Genes
Dev. 23:537–548. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
del Rincón SV, Widschwendter M, Sun D,
Ekholm-Reed S, Tat J, Teixeira LK, Ellederova Z, Grolieres E, Reed
SI and Spruck C: Cks overexpression enhances chemotherapeutic
efficacy by overriding DNA damage checkpoints. Oncogene.
34:1961–1967. 2015. View Article : Google Scholar
|
|
48
|
Schwartz GK and Shah MA: Targeting the
cell cycle: A new approach to cancer therapy. J Clin Oncol.
23:9408–9421. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Spano D, Heck C, De Antonellis P,
Christofori G and Zollo M: Molecular networks that regulate cancer
metastasis. Semin Cancer Biol. 22:234–249. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kang Y and Massague J:
Epithelial-mesenchymal transitions: Twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yao D, Dai C and Peng S: Mechanism of the
mesenchymal-epithelial transition and its relationship with
metastatic tumor formation. Mol Cancer Res. 9:1608–1620. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Swanson WF, Roth TL and Wildt DE: In vivo
embryogenesis, embryo migration, and embryonic mortality in the
domestic cat. Biol Reprod. 51:452–464. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lamalice L, Le Boeuf F and Huot J:
Endothelial cell migration during angiogenesis. Circ Res.
100:782–794. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Luster AD, Alon R and von Andrian UH:
Immune cell migration in inflammation: Present and future
therapeutic targets. Nat Immunol. 6:1182–1190. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Friedl P and Alexander S: Cancer invasion
and the microenvironment: Plasticity and reciprocity. Cell.
147:992–1009. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Friedl P and Wolf K: Tumour-cell invasion
and migration: Diversity and escape mechanisms. Nat Rev Cancer.
3:362–374. 2003. View Article : Google Scholar : PubMed/NCBI
|