Over the past decade, several studies have
demonstrated that cancer initiation and progression are determined
not only by cancer cells, but also by the tumor microenvironment
(TME) (1-4), which includes fibroblasts, immune
cells and endothelial cells, among others (5). Mounting evidence suggests that
microRNAs (miRNAs) play a key role in shaping the biology and
function of tumor stromal cells (5-8). The
deregulation of miRNAs has been associated with almost every aspect
of cancer initiation and progression (9-11).
In 2006, Volinia et al first identified a miRNA signature in
human cancers and demonstrated that the predicted targets of these
deregulated miRNAs were classic oncogenes or tumor suppressors
(12). One of the oncogenic miRNAs
identified was miR-21, which was found to be highly expressed in
breast and colon cancer, and its overexpression was correlated with
poor patient survival. Furthermore, miR-21 was demonstrated to
target programmed cell death 4, an apoptosis-inducing protein, thus
promoting tumor growth (13,14).
Additionally, miR-34a was identified as a downstream target of the
p53 tumor suppressor gene (15).
With the recent advances in miRNA detection techniques, cancer
cell-derived miRNAs have emerged as promising diagnostic biomarkers
and therapeutic targets, as has been described in detail elsewhere
(16-18).
Fibroblasts are one of the major components of the
TME. At the primary tumor site, fibroblasts acquire distinct
phenotypic characteristics and become CAFs through a miRNA-mediated
regulation of multiple signaling pathways. CAFs differ from normal
fibroblasts (NFs) in their high expression of α-smooth muscle actin
and their pro-tumorigenic properties (28). CAFs secrete a wide range of
pro-inflammatory molecules, including interleukins, chemo-kines and
extracellular matrix (ECM) components, ultimately promoting tumor
growth by modulating tumor-associated inflammation or directing
cell-to-cell communication (29).
Broniszet al reported that miR-320 expression was
significantly reduced when phosphatase and tensin homolog
(PTEN) on chromosome 10 was ablated, which induced the
transition of NFs to CAFs in breast cancer (30). Loss of the PTEN gene in
stromal fibroblasts results in the activation of an oncogenic
secretome. The downregulation of miR-320 and upregulation of one of
its direct targets, ETS proto-oncogene 2, transcription factor, are
critical events in PTEN-deficient stromal fibroblasts that
induce the oncogenic secretome, which in turn promotes tumor
angiogenesis and tumor cell invasion (30).
In breast cancer, the pro-metastatic miRNA miR-9 was
shown to induce a switch in human breast fibroblasts from a normal
towards a CAF phenotype, thereby contributing to tumor progression
(31). In addition, the
downregulation of miR-200s was reported to play an important role
in reprogramming NFs into CAFs. miR-200s target friend leukemia
virus integration 1 (Fli-1) and transcription factor 12 (TCF12) in
the breast cancer microenvironment (32,33).
Moreover, decreased miR-205 was shown to convert breast NFs into
CAFs by promoting Yes-associated protein 1 (YAP1) expression, which
has been proven to be involved in angiogenesis (34).
In ovarian cancer, low expression of miR-214 and
high expression of miR-155 are involved in reprogramming quiescent
fibroblasts to CAFs. miR-214 directly targets C-C motif ligand 5
(CCL5), which is essential for CAF function. The over-expression of
miR-155 in NFs induces fibroblasts to develop CAF-like phenotypes.
The disruption of these miRNAs is sufficient to reverse the
functions of CAFs, thereby reducing ovarian cancer growth and
metastasis (32). Furthermore,
miR-155 was shown to convert NFs into CAFs in pancreatic cancer by
targeting p53-inducible nuclear protein 1 (35).
In melanoma, melanocytes are specialized in
producing pigment vesicles, referred to as melanosomes. Melanosomes
have been shown to carry miRNAs, including miR-211, into primary
fibroblasts, triggering changes such as increased proliferation,
migration and pro-inflammatory gene expression, all of which are
known characteristics of CAFs (36).
In lung cancer, downregulation of miR-1 and miR-206,
and upregulation of miR-31, may promote the NF-CAF switch by
stimulating CCL2/vascular endothelial growth factor A (VEGFA)
expression (37). Taken together,
these findings underline the importance of miRNAs in driving the
conversion of NFs into CAFs in several tumor entities.
MSCs are progenitor cells that are known to
participate in tumor stroma formation and tumor progression. Bone
marrow-derived MSCs (BM-MSCs) can migrate to tumor sites, where
they exert tumor-promoting and pro-metastatic effects. We
previously demonstrated that miR-155-5p downregulation induced
BM-MSCs to acquire the phenotype of GC-MSCs through the activation
of nuclear factor-κB p65 (NF-κB p65). In that study, NF-κB p65 and
the inhibitor of NF-κB kinase subunit ε were identified as the
targets of miR-155-5p (38).
miR-214, miR-221 and miR-222 were upregulated in GC-MSCs compared
with GCN-MSCs. GC-MSCs significantly promoted the proliferation and
migration of GC cells. Targeted inhibition of miR-221 in GC-MSCs
was found to inhibit its tumor-promoting effects (26).
Let-7 is downregulated in prostate cancer-associated
MSCs, which exhibit a stronger propensity for migration and
invasion compared with normal MSCs. Interleukin (IL)-6 was
identified as a direct target gene of let-7. The overexpression of
let-7 reduces IL-6 expression and represses the
metastasis-promoting activity of cancer-associated MSCs (39). miR-146a overexpression leads to
increased secretion of chemo-tactic proteins, including CXCL1,
interferon gamma-induced protein 10, CCL5 and IL-6, which are
crucial for the growth and migration of multiple myeloma cells. The
Notch pathway was also shown to be involved in the miR-146a-induced
cytokine and chemokine secretion in MSCs (40). In myeloid neoplasms, miR-7977 in
MSCs was found to induce aberrant reduction of hematopoietic growth
factors, including Jagged-1, stem cell factor and angiopoietin-1,
thereby reducing the hematopoietic-supporting capacity of bone
marrow CD34+ cells (41). This evidence indicates that the
aberrant expression of miRNAs can promote the evolution of MSCs in
the TME, facilitating tumor progression.
Tumor progression is intrinsically linked to immune
evasion. The activation of certain immune cells drives potent
antitumor responses, whereas the suppression of these cells
promotes tumor progression and metastasis. Cancer cells can
regulate miRNA expression in infiltrating immune cells, thereby
suppressing the local antitumor immune response and ultimately
leading to tumor progression (42-47).
Tumor-associated macrophages (TAMs) have both pro- and
anti-tumorigenic properties, depending on whether they belong to
the classical M1 or alternative M2 subtype. At the late stage of
human cancers, most of the infiltrating macrophages display an M2
phenotype, creating an immunosuppressive microenvironment that
favors tumor progression. Increased miR-21-5p delivery by
MSC-derived extracellular vesicles was recently reported to promote
the polarization of macrophages towards an M2 phenotype (48). miR-125b binds to the 3’
untranslated region of tumor necrosis factor-α (TNF-α), inhibiting
its production and sustaining an M1 phenotype (49). By contrast, miR-146 directly
inhibits the expression of the adaptor protein tumor necrosis
factor receptor-associated factor 6 and IL-1 receptor-associated
kinase-1 in the NF-κB pathway, thus reducing pro-inflammatory
cytokine production and promoting alternative M2 activation
(5).
In addition to regulating the polarization and
phenotype of TAMs, miRNAs have also been shown to regulate other
functionalities of TAMs, including infiltration, immune responses
and tumor-promoting effects. Ke et al reported that low
miR-148b levels in hepatocellular carcinoma (HCC) cells enhanced
the expression of colony-stimulating factor-1, promoting TAM
infiltration and HCC metastasis (50). Moreover, another crucial miRNA
involved in the modulation of immune responses is miR-155, which is
persistently downregulated in TAMs (51,52).
Soluble factors in HCC can downregulate miR-155 in TAMs. When the
expression of miR-155 is restored in macrophages, both direct and
indirect antitumor responses are promoted through the enhancement
of T-cell function (51). Using a
transgenic mouse model, Zhao et al demonstrated that
miR-125a, a downstream molecule of Notch signaling pathway, could
reprogram macrophages in the TME and restore their anti-tumor
potential by targeting certain factors, including factor inhibiting
hypoxia-inducible factor-1, interferon regulatory factor 4, and
RING1- and YY1-binding protein (53). A recent report indicated that
miR-511-3p exerted regulatory effects on TAMs, and that miR-511-3p
overexpression in TAMs inhibited tumor growth in vivo,
suggesting that miR-511-3p limits the tumor-promoting function of
TAMs (54). miR-21 and miR-29a are
delivered from cancer cells to macrophages via microvesicles and
subsequently bind to intracellular toll-like receptors (TLR7 or
TLR8). This miRNA-TLR interaction results in the activation of the
NF-κB pathway and increased production of the pro-inflammatory
cytokines IL-6 and TNF-α in TAMs, which in turn promotes cancer
metastasis (55). Unexpectedly,
apoptotic breast cancer cell-derived miR-375, which is released as
a non-exosome entity, is taken up by macrophages via the CD36
receptor and enhances the migration and infiltration of macrophages
towards tumor spheroids by targeting tensin 3 and paxillin
(56). Therefore, the miRNAs of
TME play a key role in regulating the phenotype and function of
TAMs through several pathways.
miRNAs play a key role in T-cell maturation,
activation and function. The immuno-suppressive FOXP3+
regulatory T cells (Tregs) are enriched in tumor tissues and are
associated with cancer development and progression (57,58).
Exosomal miR-24-3p inhibits T-cell proliferation and Th1 and Th17
differentiation, and promotes Treg development through the
repression of fibroblast growth factor 11 (59). Tumor-secreted miR-214 can induce
Tregs to secrete higher levels of IL-10 by downregulating
PTEN, leading to immune suppression and rapid tumor growth.
The inhibition of cancer cell-secreted miR-214 has been shown to
block Treg induction and tumor growth, suggesting that anti-miR-214
therapy can abolish tumor-induced Treg expansion and suppress tumor
growth (58). The silencing of
miR-21 has been reported to significantly reduce the proliferation
of chemokine receptor 6-positive Tregs by targeting PTEN and
the subsequently activated Akt pathway, which is crucial for the
induction and functional sustainability of CD4+
FOXP3+ Tregs (60).
miR-126 silencing was also shown to impair the expression of
FOXP3 and inhibit the expression of functional molecules in
Tregs.
Moreover, in a murine mammary cancer model, the
silencing of miR-126 in Tregs resulted in enhanced antitumor
activity of CD8+ T cells (61). Additionally, miR-149-3p was found
to enhance T-cell proliferation and secretion of cytokines,
indicative of increased T-cell activation, which may reverse
CD8+ T-cell exhaustion and promote CD8+
T-cell-mediated killing of 4T1 mouse breast tumor cells (62). Notably, in human colon cancer,
miR-448 could also suppress CD8+ T-cell apoptosis and
enhance CD8+ T-cell response by inhibiting indoleamine
2,3-dioxygenase 1 enzyme function (63). These results suggest that miRNAs
are key regulators of the functions of T cells in the TME.
Tumor growth is highly dependent on the
communication between tumor cells and cells of the TME. During
tumor progression, miRNAs participate in the intricate interactions
between tumor cells and tumor stromal cells, such as CAFs,
endothelial cells and infiltrating immune cells. For example,
fibroblasts provide a stromal framework for tumor cells during
early tumor growth (29,74). Recent studies have revealed that
the upregulated miR-7 suppresses RAS-association domain family
member 2 expression in CAFs, which in turn enhances the
proliferation of head and neck cancer cells (75). Wang et al also observed that
loss of CAF-derived exosomal miR-3188 may affect the proliferation
and apoptosis of tumor cells (76). Melanoma cell-derived miR-211 was
shown to induce CAF formation by directly targeting insulin-like
growth factor 2 and activating mitogen-activated protein kinase
signaling, which in turn enhances the growth of melanoma (36). TAMs play pivotal roles in tumor
growth (77-79). Yin et al reported that M2
macrophage-derived exosomal miR-501-3p facilitated tumor formation
in vivo via regulating the transforming growth factor
(TGF)-β signaling pathway (80).
Macrophages transfected with a miR-125a mimic exhibited increased
phagocytic activity and repressed lung cancer growth in mice
(53).
Additionally, the increased levels of miR-27a
inhibited dendritic cell-mediated differentiation of Th1 and Th17
cells and enhanced the growth of melanoma cells (81). The downregulation of miR-638 was
demonstrated to promote angiogenesis and HCC growth by targeting
VEGF, whereas its overexpression inhibited tumor angiogenesis
(73). miR-143/145 in the lung
cancer microenvironment markedly promoted tumor growth by targeting
calcium/calmodulin-dependent protein kinase 1D, an inhibitory
kinase, the overexpression of which prevents the mitotic entry of
endothelial cells (68). Thus,
TME-associated miRNAs play key roles in tumor growth.
Resistance to chemotherapy and targeted therapy is a
major challenge in clinical practice. Accumulating evidence
suggests that miRNAs from the TME may affect drug resistance. In
HCC, exosomal miR-122 (122-Exo) from adipose tissue-derived MSCs
can be delivered into HCC cells, increasing the chemosensitivity of
HCC cells by inhibiting the expression of cyclin G1, a disintegrin
and metallopro-tease 10, and insulin-like growth factor receptor 1
(IGF1R). Intratumoral injection of 122-Exo significantly increased
the antitumor efficacy of sorafenib in HCC in vivo (88). Moreover, the chemotherapeutic agent
sorafenib can inhibit miR-101 expression and enhance
dual-specificity phosphatase 1 expression, leading to the
inhibition of macrophage-induced HCC growth (89). In GC, miR-21 was transferred from
macrophages to GC cells via exosomes and resulted in a significant
reduction in the sensitivity of GC cells to cisplatin chemotherapy
in vitro and in vivo, partially through the
regulation of the PTEN/phosphoinositide 3-kinase/AKT signaling
pathway (90). The levels of
miR-27a/b in the serum were significantly higher in patients with
esophageal cancer compared with those in healthy volunteers, and
high expression levels of miR-27a/b were shown to induce the
transformation of NFs into CAFs through upregulation of TGF-β,
leading to chemoresistance in esophageal cancer cells (91). These results suggest that
TME-derived miRNAs play a critical role in drug resistance.
According to the literature, miRNA-based antitumor
therapy can be broadly divided into two distinct approaches, namely
miRNA silencing by anti-miRNAs and miRNA restoration. Cubillos-Ruiz
et al reported that nanoparticles carrying oligonucleotide
duplexes markedly augmented miR-155 activity and transformed
tumor-associated dendritic cells from the immunosuppressive
phenotype to highly immunostimulatory cells, eliciting potent
antitumor responses that eliminate ovarian cancer cells (98). In a mouse breast cancer model,
let-7b delivered through a nucleic acid delivery system
expeditiously reprogrammed the functions of TAMs and
tumor-infiltrating dendritic cells, leading to the inhibition of
tumor growth. This strategy may represent a new approach to cancer
immunotherapy (99). Additionally,
miR-21 depletion in TAMs promoted tumori-cidal M1 polarization by
upregulating Janus kinase 2 and Signal transducer and activator of
transcription 1, and this, combined with programmed cell death
protein 1 blockade, may exert synergistic effects and exhibit
superior antitumor activity (100).
Moreover, the targeted interference of deregulated
miRNAs in cancer-associated endothelial cells was shown to decrease
vascular permeability. Pi et al reported that the elevated
expression of miR-302/367 significantly suppressed tumor growth by
restricting sprout angiogenesis and decreasing vascular
permeability (101). Schnittert
et al reported that anti-miR-199a, delivered via
peptide-based nanocomplexes, may inhibit human-derived pancreatic
stellate cell (hPSC) differentiation into CAFs and repress the size
of 3D hetero-spheroids, which consist of hPSCs and tumor cells
(102). In summary, targeting
miRNAs has emerged as a novel, promising approach to cancer
treatment.
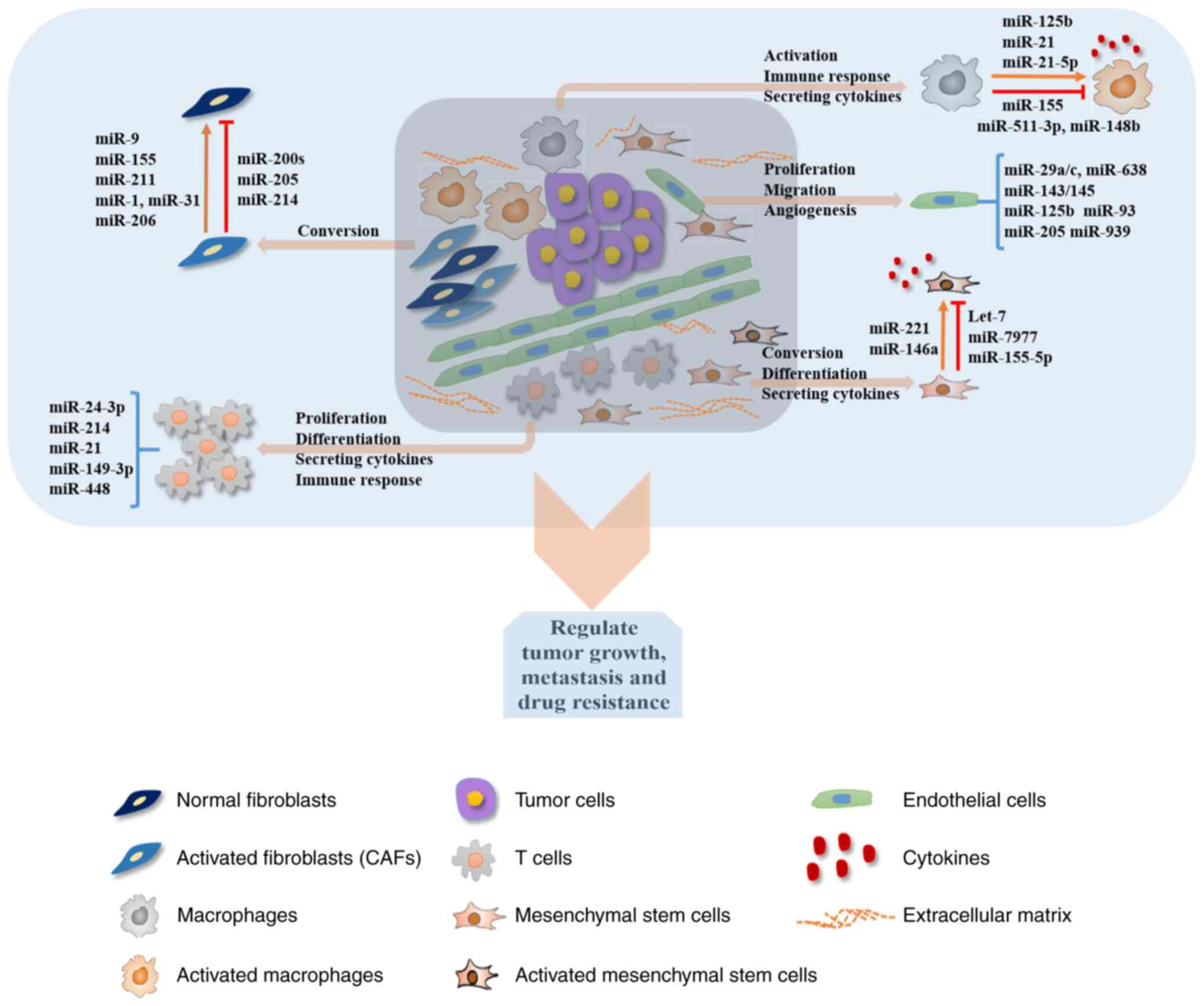
The present review summarized the roles of miRNAs in
the communication between tumor cells and tumor stromal cells.
Deregulated expression of several miRNAs has been observed in both
types of cells, clearly highlighting the crucial roles of miRNAs in
cancer development and progression (Fig. 1; Table
I). Numerous examples in which miRNAs were demonstrated to
regulate the critical aspects of TME involved in tumor
angiogenesis, tumor cell growth and metastasis were herein
summarized. Previous studies elucidated some of the mechanisms by
which these small RNAs in the TME considerably affect tumor
biology. Undoubtedly, miRNA-related research has great potential
for the identification of important biomarkers and for promoting
the development of novel cancer therapies. As it has been reported
in previous studies, miRNA delivery to tumors is an attractive yet
challenging opportunity to improve therapeutic strategies for
cancer (103,104). However, more cancer-related
pathways must be identified as specific targets of miRNAs, and
additional efforts are required to ultimately design an optimal
delivery system for miRNAs. Several novel siRNA and miRNA delivery
systems are currently under development. However, despite the
enormous potential of microRNAs in cancer diagnosis and anticancer
clinical practice, microRNAs are not yet widely applied as cancer
biomarkers and therapy targets in the clinical setting, as numerous
obstacles must be overcome before their widespread application.
The present study was supported by the Special
Foundation for Young Scientists of Jiangsu Province (grant no.
QNRC2016379), Xuzhou Central Hospital and Xinyi People’s
Hospital.
The datasets used and analyzed during the present
study are available from the corresponding author on reasonable
request.
ZP and YT made substantial contributions to
conception and design and wrote the manuscript. GN and CC
critically revised the manuscript for important intellectual
content. ZP, YT, GN and CC reviewed and edited the manuscript. All
authors have read and approved the final version of the manuscript
for publication.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
The authors would like to thank Wenrong Xu and Xu
Zhang (Jiangsu Key Laboratory of Medical Science and Laboratory
Medicine, School of Medicine, Jiangsu University, Zhenjiang,
Jiangsu, China) for their ideas and support.
|
1
|
Naito Y, Yoshioka Y, Yamamoto Y and Ochiya
T: How cancer cells dictate their microenvironment: Present roles
of extracellular vesicles. Cell Mol Life Sci. 74:697–713. 2017.
View Article : Google Scholar :
|
|
2
|
Dachs GU and Chaplin DJ:
Microenvironmental control of gene expression: Implications for
tumor angiogenesis, progression, and metastasis. Semin Radiat
Oncol. 8:208–216. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Whiteside TL: Exosome and mesenchymal stem
cell cross-talk in the tumor microenvironment. Semin Immunol.
35:69–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chou J, Shahi P and Werb Z:
microRNA-mediated regulation of the tumor microenvironment. Cell
Cycle. 12:3262–3271. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li X, Wu Z, Fu X and Han W: A microRNA
component of the neoplastic Microenvironment: microregulators with
far-reaching impact. Biomed Res Int. 2013:7621832013.PubMed/NCBI
|
|
7
|
Zhang Y, Yang P and Wang XF:
Microenvironmental regulation of cancer metastasis by miRNAs.
Trends Cell Biol. 24:153–160. 2014. View Article : Google Scholar
|
|
8
|
Xu SJ, Hu HT, Li HL and Chang S: The role
of miRNAs in immune cell development, immune cell activation, and
tumor immunity: With a focus on macrophages and natural killer
cells. Cells. 8:pii: E1140. 2019. View Article : Google Scholar
|
|
9
|
Musumeci M, Coppola V, Addario A, Patrizii
M, Maugeri-Saccà M, Memeo L, Colarossi C, Francescangeli F, Biffoni
M, Collura D, et al: Control of tumor and microenvironment
cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene.
30:4231–4242. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review EMBO Mol Med. 9:8522017. View Article : Google Scholar
|
|
11
|
Que KT, Zhou Y, You Y, Zhang Z, Zhao XP,
Gong JP and Liu ZJ: MicroRNA-31-5p regulates chemosensitivity by
preventing the nuclear location of PARP1 in hepatocellular
carcinoma. J Exp Clin Cancer Res. 37:2682018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar
|
|
14
|
Frankel LB, Christoffersen NR, Jacobsen A,
Lindow M, Krogh A and Lund AH: Programmed cell death 4 (PDCD4) is
an important functional target of the microRNA miR-21 in breast
cancer cells. J Biol Chem. 283:1026–1033. 2008. View Article : Google Scholar
|
|
15
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Corsini LR, Bronte G, Terrasi M, Amodeo V,
Fanale D, Fiorentino E, Cicero G, Bazan V and Russo A: The role of
microRNAs in cancer: Diagnostic and prognostic biomarkers and
targets of therapies. Expert Opin Ther Targets. 16(Suppl 2):
S103–S109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nana-Sinkam SP and Croce CM: Clinical
applications for microRNAs in cancer. Clin Pharmacol Ther.
93:98–104. 2013. View Article : Google Scholar
|
|
18
|
Pichler M and Calin GA: MicroRNAs in
cancer: From developmental genes in worms to their clinical
application in patients. Br J Cancer. 113:569–573. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Soon P and Kiaris H: MicroRNAs in the
tumour microenvironment: Big role for small players. Endocr Relat
Cancer. 20:R257–R267. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvi-ronment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kuninty PR, Schnittert J, Storm G and
Prakash J: MicroRNA targeting to modulate tumor microenvironment.
Front Oncol. 6:32016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fanini F and Fabbri M: Cancer-derived
exosomic microRNAs shape the immune system within the tumor
microenvironment: State of the art. Semin Cell Dev Biol. 67:23–28.
2017. View Article : Google Scholar :
|
|
23
|
Curtale G: MiRNAs at the crossroads
between innate immunity and cancer: Focus on macrophages. Cells.
7:pii: E12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bandini E, Rossi T, Gallerani G and Fabbri
F: Adipocytes and microRNAs crosstalk: A key tile in the mosaic of
breast cancer microenvironment. Cancers (Basel). 11:pii: E1451.
2019. View Article : Google Scholar
|
|
25
|
Aprelikova O, Yu X, Palla J, Wei BR, John
S, Yi M, Stephens R, Simpson RM, Risinger JI, Jazaeri A and
Niederhuber J: The role of miR-31 and its target gene SATB2 in
cancer-associated fibroblasts. Cell Cycle. 9:4387–4398. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang M, Zhao C, Shi H, Zhang B, Zhang L,
Zhang X, Wang S, Wu X, Yang T, Huang F, et al: Deregulated
microRNAs in gastric cancer tissue-derived mesenchymal stem cells:
Novel biomarkers and a mechanism for gastric cancer. Br J Cancer.
110:1199–1210. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Y, Lai L, Chen Q, Song Y, Xu S, Ma F,
Wang X, Wang J, Yu H, Cao X and Wang Q: MicroRNA-494 is required
for the accumulation and functions of tumor-expanded
myeloid-derived suppressor cells via targeting of PTEN. J Immunol.
188:5500–5510. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Erez N, Truitt M, Olson P, Arron ST and
Hanahan D: Cancer-associated fibroblasts are activated in incipient
neoplasia to orchestrate tumor-promoting inflammation in an
NF-kappaB-dependent manner. Cancer Cell. 17:135–147. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kalluri R and Zeisberg M: Fibroblasts in
cancer. Nat Rev Cancer. 6:392–401. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bronisz A, Godlewski J, Wallace JA,
Merchant AS, Nowicki MO, Mathsyaraja H, Srinivasan R, Trimboli AJ,
Martin CK, Li F, et al: Reprogramming of the tumour
microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol.
14:159–167. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baroni S, Romero-Cordoba S, Plantamura I,
Dugo M, D’Ippolito E, Cataldo A, Cosentino G, Angeloni V, Rossini
A, Daidone MG and Iorio MV: Exosome-mediated delivery of miR-9
induces cancer-associated fibroblast-like properties in human
breast fibroblasts. Cell Death Dis. 7:pp. e23122016, View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mitra AK, Zillhardt M, Hua Y, Tiwari P,
Murmann AE, Peter ME and Lengyel E: MicroRNAs reprogram normal
fibroblasts into cancer-associated fibroblasts in ovarian cancer.
Cancer Discov. 2:1100–1108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tang X, Hou Y, Yang G, Wang X, Tang S, Du
YE, Yang L, Yu T, Zhang H, Zhou M, et al: Stromal miR-200s
contribute to breast cancer cell invasion through CAF activation
and ECM remod-eling. Cell Death Differ. 23:132–145. 2016.
View Article : Google Scholar
|
|
34
|
Du YE, Tu G, Yang G, Li G, Yang D, Lang L,
Xi L, Sun K, Chen Y, Shu K, et al: MiR-205/YAP1 in activated
fibroblasts of breast tumor promotes VEGF-independent angiogenesis
through STAT3 signaling. Theranostics. 7:3972–3988. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pang W, Su J, Wang Y, Feng H, Dai X, Yuan
Y, Chen X and Yao W: Pancreatic cancer-secreted miR-155 implicates
in the conversion from normal fibroblasts to cancer-associated
fibroblasts. Cancer Sci. 106:1362–1369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dror S, Sander L, Schwartz H, Sheinboim D,
Barzilai A, Dishon Y, Apcher S, Golan T, Greenberger S, Barshack I,
et al: Melanoma miRNA trafficking controls tumour primary niche
formation. Nat Cell Biol. 18:1006–1017. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shen H, Yu X, Yang F, Zhang Z, Shen J, Sun
J, Choksi S, Jitkaew S and Shu Y: Reprogramming of normal
fibroblasts into cancer-associated fibroblasts by miRNAs-mediated
CCL2/VEGFA signaling. PLoS Genet. 12:e10062442016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu M, Wang M, Yang F, Tian Y, Cai J, Yang
H, Fu H, Mao F, Zhu W, Qian H and Xu W: miR-155-5p inhibition
promotes the transition of bone marrow mesenchymal stem cells to
gastric cancer tissue derived MSC-like cells via NF-κB p65
activation. Oncotarget. 7:16567–16580. 2016.PubMed/NCBI
|
|
39
|
Sung SY, Liao CH, Wu HP, Hsiao WC, Wu IH,
Jinpu Yu, Lin SH and Hsieh CL: Loss of let-7 microRNA upregulates
IL-6 in bone marrow-derived mesenchymal stem cells triggering a
reactive stromal response to prostate cancer. PLoS One. 8:pp.
e716372013, View Article : Google Scholar : PubMed/NCBI
|
|
40
|
De Veirman K, Wang J, Xu S, Leleu X, Himpe
E, Maes K, De Bruyne E, Van Valckenborgh E, Vanderkerken K, Menu E
and Van Riet I: Induction of miR-146a by multiple myeloma cells in
mesenchymal stromal cells stimulates their protumoral activity.
Cancer Lett. 377:17–24. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Horiguchi H, Kobune M, Kikuchi S, Yoshida
M, Murata M, Murase K, Iyama S, Takada K, Sato T, Ono K, et al:
Extracellular vesicle miR-7977 is involved in hematopoietic
dysfunction of mesenchymal stromal cells via poly(rC) binding
protein 1 reduction in myeloid neoplasms. Haematologica.
101:437–447. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jang JY, Lee JK, Jeon YK and Kim CW:
Exosome derived from epigallocatechin gallate treated breast cancer
cells suppresses tumor growth by inhibiting tumor-associated
macrophage infiltration and M2 polarization. BMC Cancer.
13:4212013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu Z, Zhao L, Zhu LY, He M, Zheng L and Wu
Y: MicroRNA-17, 20a regulates the proangiogenic function of
tumor-associated macrophages via targeting hypoxia-inducible factor
2α. PLoS One. 8:pp. e778902013, View Article : Google Scholar
|
|
44
|
Lagrange B, Martin RZ, Droin N, Aucagne R,
Paggetti J, Largeot A, Itzykson R, Solary E, Delva L and Bastie JN:
A role for miR-142-3p in colony-stimulating factor 1-induced
monocyte differentiation into macrophages. Biochim Biophys Acta.
1833:1936–1946. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Baj-Krzyworzeka M, Mytar B, Szatanek R,
Surmiak M, Węglarczyk K, Baran J and Siedlar M: Colorectal
cancer-derived microvesicles modulate differentiation of human
monocytes to macrophages. J Transl Med. 14:362016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li Y, Zhao L, Shi B, Ma S, Xu Z, Ge Y, Liu
Y, Zheng D and Shi J: Functions of miR-146a and miR-222 in
tumor-associated macrophages in breast cancer. Sci Rep.
5:186482015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Huber V, Vallacchi V, Fleming V, Hu X,
Cova A, Dugo M, Shahaj E, Sulsenti R, Vergani E, Filipazzi P, et
al: Tumor-derived microRNAs induce myeloid suppressor cells and
predict immunotherapy resistance in melanoma. J Clin Invest.
128:5505–5516. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ren W, Hou J, Yang C, Wang H, Wu S, Wu Y,
Zhao X and Lu C: Extracellular vesicles secreted by hypoxia
pre-challenged mesenchymal stem cells promote non-small cell lung
cancer cell growth and mobility as well as macrophage M2
polarization via miR-21-5p delivery. J Exp Clin Cancer Res.
38:622019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chaudhuri AA, So AY, Sinha N, Gibson WS,
Taganov KD, O’Connell RM and Baltimore D: MicroRNA-125b potentiates
macrophage activation. J Immunol. 187:5062–5068. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ke M, Zhang Z, Cong L, Zhao S, Li Y, Wang
X, Lv Y, Zhu Y and Dong J: MicroRNA-148b-colony-stimulating
factor-1 signaling-induced tumor-associated macrophage infiltration
promotes hepatocellular carcinoma metastasis. Biomed Pharmacother.
120:1095232019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
He M, Xu Z, Ding T, Kuang DM and Zheng L:
MicroRNA-155 regulates inflammatory cytokine production in
tumor-associated macrophages via targeting C/EBPbeta. Cell Mol
Immunol. 6:343–352. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zonari E, Pucci F, Saini M, Mazzieri R,
Politi LS, Gentner B and Naldini L: A role for miR-155 in enabling
tumor-infiltrating innate immune cells to mount effective antitumor
responses in mice. Blood. 122:243–252. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhao JL, Huang F, He F, Gao CC, Liang SQ,
Ma PF, Dong GY, Han H and Qin HY: Forced activation of notch in
macrophages represses tumor growth by upregulating miR-125a and
disabling tumor-associated macrophages. Cancer Res. 76:1403–1415.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Squadrito ML, Pucci F, Magri L, Moi D,
Gilfillan GD, Ranghetti A, Casazza A, Mazzone M, Lyle R, Naldini L
and De Palma M: miR-511-3p modulates genetic programs of
tumor-associated macrophages. Cell Rep. 1:141–154. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fabbri M, Paone A, Calore F, Galli R,
Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al:
MicroRNAs bind to Toll-like receptors to induce prometastatic
inflammatory response. Proc Natl Acad Sci USA. 109:E2110–2116.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Frank AC, Ebersberger S, Fink AF, Lampe S,
Weigert A, Schmid T, Ebersberger I, Syed SN and Brüne B: Apoptotic
tumor cell-derived microRNA-375 uses CD36 to alter the
tumor-associated macrophage phenotype. Nat Commun. 10:11352019.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Xu L, Xu W, Wen Z and Xiong S: In situ
prior proliferation of CD4+ CCR6+ regulatory T cells facilitated by
TGF-β secreting DCs is crucial for their enrichment and suppression
in tumor immunity. PLoS One. 6:e202822011. View Article : Google Scholar
|
|
58
|
Yin Y, Cai X, Chen X, Liang H, Zhang Y, Li
J, Wang Z, Chen X, Zhang W, Yokoyama S, et al: Tumor-secreted
miR-214 induces regulatory T cells: A major link between immune
evasion and tumor growth. Cell Res. 24:1164–1180. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ye SB, Zhang H, Cai TT, Liu YN, Ni JJ, He
J, Peng JY, Chen QY, Mo HY, Jun-Cui, et al: Exosomal miR-24-3p
impedes T-cell function by targeting FGF11 and serves as a
potential prognostic biomarker for nasopharyngeal carcinoma. J
Pathol. 240:329–340. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hu Y, Wang C, Li Y, Zhao J, Chen C, Zhou
Y, Tao Y, Guo M, Qin N, Ren T, et al: MiR-21 controls in situ
expansion of CCR6+ regulatory T cells through PTEN/AKT
pathway in breast cancer. Immunol Cell Biol. 93:753–764. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Qin A, Wen Z, Zhou Y, Li Y, Li Y, Luo J,
Ren T and Xu L: MicroRNA-126 regulates the induction and function
of CD4(+) Foxp3(+) regulatory T cells through PI3K/AKT pathway. J
Cell Mol Med. 17:252–264. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang M, Gao D, Shi Y, Wang Y, Joshi R, Yu
Q, Liu D, Alotaibi F, Zhang Y, Wang H, et al: miR-149-3p reverses
CD8+ T-cell exhaustion by reducing inhibitory receptors and
promoting cyto-kine secretion in breast cancer cells. Open Biol.
9:1900612019. View Article : Google Scholar
|
|
63
|
Lou Q, Liu R, Yang X, Li W, Huang L, Wei
L, Tan H, Xiang N, Chan K, Chen J and Liu H: miR-448 targets IDO1
and regulates CD8+ T cell response in human colon cancer. J
Immunother Cancer. 7:2102019. View Article : Google Scholar :
|
|
64
|
Mangala LS, Wang H, Jiang D, Wu SY,
Somasunderam A, Volk DE, Lokesh GLR, Li X, Pradeep S, Yang X, et
al: Improving vascular maturation using noncoding RNAs increases
antitumor effect of chemotherapy. JCI Insight. 3:pii: 122387.
2018.PubMed/NCBI
|
|
65
|
Zhang H, Bai M, Deng T, Liu R, Wang X, Qu
Y, Duan J, Zhang L, Ning T, Ge S, et al: Cell-derived microvesicles
mediate the delivery of miR-29a/c to suppress angiogenesis in
gastric carcinoma. Cancer Lett. 375:331–339. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Di Modica M, Regondi V, Sandri M, Iorio
MV, Zanetti A, Tagliabue E, Casalini P and Triulzi T: Breast
cancer-secreted miR-939 downregulates VE-cadherin and destroys the
barrier function of endothelial monolayers. Cancer Lett.
384:94–100. 2017. View Article : Google Scholar
|
|
67
|
Guan B, Wu K, Zeng J, Xu S, Mu L, Gao Y,
Wang K, Ma Z, Tian J, Shi Q, et al: Tumor-suppressive microRNA-218
inhibits tumor angiogenesis via targeting the mTOR component RICTOR
in prostate cancer. Oncotarget. 8:8162–8172. 2017. View Article : Google Scholar :
|
|
68
|
Dimitrova N, Gocheva V, Bhutkar A, Resnick
R, Jong RM, Miller KM, Bendor J and Jacks T: Stromal expression of
miR-143/145 promotes neoangiogenesis in lung cancer development.
Cancer Discov. 6:188–201. 2016. View Article : Google Scholar :
|
|
69
|
Smits M, Wurdinger T, van het Hof B,
Drexhage JA, Geerts D, Wesseling P, Noske DP, Vandertop WP, de
Vries HE and Reijerkerk A: Myc-associated zinc finger protein (MAZ)
is regulated by miR-125b and mediates VEGF-induced angiogenesis in
glioblastoma. FASEB J. 26:2639–2647. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Fang L, Deng Z, Shatseva T, Yang J, Peng
C, Du WW, Yee AJ, Ang LC, He C, Shan SW and Yang BB: MicroRNA
miR-93 promotes tumor growth and angiogenesis by targeting
integrin-β8. Oncogene. 30:806–821. 2011. View Article : Google Scholar
|
|
71
|
Fang L, Du WW, Yang W, Rutnam ZJ, Peng C,
Li H, O'Malley YQ, Askeland RW, Sugg S, Liu M, et al: MiR-93
enhances angio-genesis and metastasis by targeting LATS2. Cell
Cycle. 11:4352–4365. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Szczyrba J, Nolte E, Hart M, Döll C, Wach
S, Taubert H, Keck B, Kremmer E, Stöhr R, Hartmann A, et al:
Identification of ZNF217, hnRNP-K, VEGF-A and IPO7 as targets for
microRNAs that are downregulated in prostate carcinoma. Int J
Cancer. 132:775–784. 2013. View Article : Google Scholar
|
|
73
|
Cheng J, Chen Y, Zhao P, Liu X, Dong J, Li
J, Huang C, Wu R and Lv Y: Downregulation of miRNA-638- promotes
angiogenesis and growth of hepatocellular carcinoma by targeting
VEGF. Oncotarget. 7:30702–30711. 2016.PubMed/NCBI
|
|
74
|
Schauer IG, Sood AK, Mok S and Liu J:
Cancer-associated fibroblasts and their putative role in
potentiating the initiation and development of epithelial ovarian
cancer. Neoplasia. 13:393–405. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Shen Z, Qin X, Yan M, Li R, Chen G, Zhang
J and Chen W: Cancer-associated fibroblasts promote cancer cell
growth through a miR-7-RASSF2-PAR-4 axis in the tumor
microenvi-ronment. Oncotarget. 8:1290–1303. 2017.
|
|
76
|
Wang X, Qin X, Yan M, Shi J, Xu Q, Li Z,
Yang W, Zhang J and Chen W: Loss of exosomal miR-3188 in
cancer-associated fibroblasts contributes to HNC progression. J Exp
Clin Cancer Res. 38:1512019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Biswas SK, Allavena P and Mantovani A:
Tumor-associated macrophages: Functional diversity, clinical
significance, and open questions. Semin Immunopathol. 35:585–600.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
De Palma M and Lewis CE: Macrophage
regulation of tumor responses to anticancer therapies. Cancer Cell.
23:277–286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Epelman S, Lavine KJ and Randolph GJ:
Origin and functions of tissue macrophages. Immunity. 41:21–35.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yin Z, Ma T, Huang B, Lin L, Zhou Y, Yan
J, Zou Y and Chen S: Macrophage-derived exosomal microRNA-501-3p
promotes progression of pancreatic ductal adenocarcinoma through
the TGFBR3-mediated TGF-β signaling pathway. J Exp Clin Cancer Res.
38:3102019. View Article : Google Scholar
|
|
81
|
Min S, Li L, Zhang M, Zhang Y, Liang X,
Xie Y, He Q, Li Y, Sun J, Liu Q, et al: TGF-β-associated miR-27a
inhibits dendritic cell-mediated differentiation of Th1 and Th17
cells by TAB3, p38 MAPK, MAP2K4 and MAP2K7. Genes Immun.
13:621–631. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Nguyen DX, Bos PD and Massague J:
Metastasis: From dissemination to organ-specific colonization. Nat
Rev Cancer. 9:274–284. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y,
Li C, Chong M, Ibrahim T, Mercatali L, et al: miR-126 and miR-126*
repress recruitment of mesenchymal stem cells and inflammatory
monocytes to inhibit breast cancer metastasis. Nat Cell Biol.
15:284–294. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Li BL, Lu W, Qu JJ, Ye L, Du GQ and Wan
XP: Loss of exosomal miR-148b from cancer-associated fibroblasts
promotes endometrial cancer cell invasion and cancer metastasis. J
Cell Physiol. 234:2943–2953. 2019. View Article : Google Scholar
|
|
86
|
Cuiffo BG, Campagne A, Bell GW, Lembo A,
Orso F, Lien EC, Bhasin MK, Raimo M, Hanson SE, Marusyk A, et al:
MSC-regulated microRNAs converge on the transcription factor FOXP2
and promote breast cancer metastasis. Cell Stem Cell. 15:762–774.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhou SL, Hu ZQ, Zhou ZJ, Dai Z, Wang Z,
Cao Y, Fan J, Huang XW and Zhou J: miR-28-5p-IL-34-macrophage
feedback loop modulates hepatocellular carcinoma metastasis.
Hepatology. 63:1560–1575. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Lou G, Song X, Yang F, Wu S, Wang J, Chen
Z and Liu Y: Exosomes derived from miR-122-modified adipose
tissue-derived MSCs increase chemosensitivity of hepatocellular
carcinoma. J Hematol Oncol. 8:1222015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Wei X, Tang C, Lu X, Liu R, Zhou M, He D,
Zheng D, Sun C and Wu Z: MiR-101 targets DUSP1 to regulate the
TGF-β secretion in sorafenib inhibits macrophage-induced growth of
hepatocar-cinoma. Oncotarget. 6:18389–18405. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie
G, Ma Y and Shen L: Exosomal transfer of tumor-associated
macrophage-derived miR-21 confers cisplatin resistance in gastric
cancer cells. J Exp Clin Cancer Res. 36:532017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Tanaka K, Miyata H, Sugimura K, Fukuda S,
Kanemura T, Yamashita K, Miyazaki Y, Takahashi T, Kurokawa Y,
Yamasaki M, et al: miR-27 is associated with chemoresistance in
esophageal cancer through transformation of normal fibroblasts to
cancer-associated fibroblasts. Carcinogenesis. 36:894–903. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Bryant RJ, Pawlowski T, Catto JW, Marsden
G, Vessella RL, Rhees B, Kuslich C, Visakorpi T and Hamdy FC:
Changes in circulating microRNA levels associated with prostate
cancer. Br J Cancer. 106:768–774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Lee YS, Lim YS, Lee JC, Wang SG, Park HY,
Kim SY and Lee BJ: Differential expression levels of plasma-derived
miR-146b and miR-155 in papillary thyroid cancer. Oral Oncol.
51:77–83. 2015. View Article : Google Scholar
|
|
94
|
Yi SJ, Liu P, Chen BL, Ou-Yang L, Xiong WM
and Su JP: Circulating miR-31-5p may be a potential diagnostic
biomarker in nasopharyngeal carcinoma. Neoplasma. 66:825–829. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Kosaka N, Iguchi H and Ochiya T:
Circulating microRNA in body fluid: A new potential biomarker for
cancer diagnosis and prognosis. Cancer Sci. 101:2087–2092. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Hornick NI, Huan J, Doron B, Goloviznina
NA, Lapidus J, Chang BH and Kurre P: Serum exosome MicroRNA as a
minimally-invasive early biomarker of AML. Sci Rep. 5:112952015.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Donahue TR, Nguyen AH, Moughan J, Li L,
Tatishchev S, Toste P and Farrell JJ: Stromal microRNA-21 levels
predict response to 5-fluorouracil in patients with pancreatic
cancer. J Surg Oncol. 110:952–959. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Cubillos-Ruiz JR, Baird JR, Tesone AJ,
Rutkowski MR, Scarlett UK, Camposeco-Jacobs AL, Anadon-Arnillas J,
Harwood NM, Korc M, Fiering SN, et al: Reprogramming
tumor-associated dendritic cells in vivo using miRNA mimetics
triggers protective immunity against ovarian cancer. Cancer Res.
72:1683–1693. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Huang Z, Gan J, Long Z, Guo G, Shi X, Wang
C, Zang Y, Ding Z, Chen J, Zhang J and Dong L: Targeted delivery of
let-7b to reprogramme tumor-associated macrophages and tumor
infiltrating dendritic cells for tumor rejection. Biomaterials.
90:72–84. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Xi J, Huang Q, Wang L, Ma X, Deng Q, Kumar
M, Zhou Z, Li L, Zeng Z, Young KH, et al: miR-21 depletion in
macrophages promotes tumoricidal polarization and enhances PD-1
immuno-therapy. Oncogene. 37:3151–3165. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Pi J, Tao T, Zhuang T, Sun H, Chen X, Liu
J, Cheng Y, Yu Z, Zhu HH, Gao WQ, et al: A
MicroRNA302-367-Erk1/2-Klf2-S1pr1 pathway prevents tumor growth via
restricting angiogenesis and improving vascular stability. Circ
Res. 120:85–98. 2017. View Article : Google Scholar
|
|
102
|
Schnittert J, Kuninty PR, Bystry TF, Brock
R, Storm G and Prakash J: Anti-microRNA targeting using
peptide-based nano-complexes to inhibit differentiation of human
pancreatic stellate cells. Nanomedicine (Lond). 12:1369–1384. 2017.
View Article : Google Scholar
|
|
103
|
Pecot CV, Calin GA, Coleman RL,
Lopez-Berestein G and Sood AK: RNA interference in the clinic:
Challenges and future directions. Nat Rev Cancer. 11:59–67. 2011.
View Article : Google Scholar :
|
|
104
|
Ling H, Fabbri M and Calin GA: MicroRNAs
and other non-coding RNAs as targets for anticancer drug
development. Nat Rev Drug Discov. 12:847–865. 2013. View Article : Google Scholar : PubMed/NCBI
|