Introduction
Pancreatic cancer (PC) is a highly aggressive
malignancy that is typically diagnosed at the advanced stage, when
few therapeutic options are viable (1). Despite decades of research and
clinical trials, the 5-year survival rate of PC is a poor 5%, and
the median survival duration is <6 months (2). Pancreatic ductal adenocarcinoma
(PDAC) is the most common histological subtype of pancreatic
neoplasms (3), and is predicted to
become the second leading cause of cancer-related death in the
United States by 2030 (4). The
poor clinical outcome of PDAC is due to its invasive progression
and distant metastasis, which is often non-resectable at the time
of diagnosis (1).
Currently, gemcitabine-based chemotherapy is the
standard treatment prescribed for patients with advanced PDAC.
However, its efficacy is limited due to intrinsic or acquired drug
resistance (5). Although the
recently approved FOLFIRINOX regimen (comprising fluorouracil,
leucovorin, irinotecan and oxaliplatin) elicited an improved
response from patients and increased overall survival compared to
gemcitabine mono-therapy, it is recommended only for a small group
of patients due to its side effects (6). Therefore, it is essential to
elucidate the underlying molecular mechanisms of PDAC, in order to
identify effective therapeutic targets. To that end, there is a
need for physiologically relevant models to simulate PDAC.
Cancer cell lines remain the primary in vitro
model for basic and translational cancer research. However, the
currently available pancreatic cancer cell lines were established
several decades ago, and may have been unintentionally
cross-contaminated with other cells (7). In addition, these decades-old cell
lines lack sufficient clinical information and reliable
certification, which has hindered research progression (8).
Accordingly, there is an urgent need for the
production of novel, well-established, fully characterized cancer
cell lines for research use. However, establishing cancer cell
lines directly from tumor samples is inefficient, while protocols
for deriving cancer cell lines from xenografts with a high success
rate have proved useful in other types of cancer research,
including for colon cancer and liver cancer (9,10).
The present study successfully established and fully
characterized a pancreatic cancer cell line derived from a PDAC
patient-derived xenograft (PDX). The genetic alterations and
phenotypes of the derived cell line were consistent with those of
the parent PDAC tumor. This well-established novel human pancreatic
cancer cell line is preferable for translational and molecular
studies, and can be a powerful tool to study the chemoresistance
patterns of tumors, and a subset of PDAC patients may benefit from
MEK inhibition treatment.
Materials and methods
Patient samples
The tissue sample was obtained from a patient with
PC who had undergone surgery at the Department of Surgical
Oncology, The First Affiliated Hospital of Zhejiang University
School of Medicine in August 2012. The tumor sample was confirmed
by at least two pathologists. The study was approved by the Ethical
Review Committee of The First Affiliated Hospital of Zhejiang
University School of Medicine, and written informed consent was
obtained from the patient.
Establishment of the PDX-derived cell
line
The patient-derived pancreatic cancer xenografts
were established as previously described (11). Briefly, patient tumor pieces were
subcutaneously implanted into five 4-6 week-old female BALB/c nude
mice obtained from the Model Animal Research Center of Nanjing, and
the tumors were measured every 5 days. The tumors were harvested
for second and third transplantation when they grew to
1,242.9±307.4 mm3 (largest diameter: 17.0±2.2 mm) and
1,196.6±136.4 mm3 (largest diameter: 17.3±1.4 mm),
respectively. The third generation PDXs were harvested when the
volume reached 157.5±17.2 mm3, and were then used for
establishing the cell line and other assays.
The cell line was established from the PDX using a
method reported in a previous study (12). The xenografts were enzymatically
digested, and then minced into small pieces (<1 mm3)
using sterile scissors. The homogenates were collected in DMEM
(HyClone; GE Healthcare Life Sciences) supplemented with 20% FBS
(Gibco; Thermo Fisher Scientific, Inc.), 1% penicillin and 1%
streptomycin (GENOM). The tumor cells were enriched with the
differential adhesion technique (13) in the initial generations at 37°C
under 5% CO2, with the medium replaced every 2-3 days.
The cells were passaged at 80-90% confluence and FBS was decreased
to 10% after 10 passages. The PDXPC1 cell line was obtained after
>80 serial passages over a period of 2 years, and aliquots were
frozen in liquid nitrogen.
Cell culture
The human pancreatic cancer cell line Panc-1 was
purchased from the American Type Culture Collection and validated
by comparing with a reference database of short tandem repeat (STR)
profiles. The cells were maintained in DMEM supplemented with 10%
FBS at 37°C under 5% CO2, and harvested using 0.25%
trypsin and 0.02% EDTA (Gino Biopharmaceutical Technology).
Spheroid formation
Panc-1 or PDXPC1 cells were suspended in serum-free
phenol red-free DMEM/F12 medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 1X B27 (Invitrogen; Thermo Fisher
Scientific, Inc.), 20 ng/ml human epidermal growth factor (Gibco;
Thermo Fisher Scientific, Inc.), 10 ng/ml basic fibroblast growth
factor (PeproTech, Inc.) and 5 µg/ml insulin (Novo Nordisk
A/S), and seeded at a density of 200 cells/well in an ultralow
attachment 6-well plate (Corning, Inc.). The cells were cultured
for 7 days and spheres >50 µm were counted under the
Nikon ECLIPSE Ti inverted microscope (Nikon Corporation)
(magnification, ×200).
STR analysis
The STR profile of the PDXPC1 cell line was verified
using the EX20 kit (AGCU ScienTech Incorporation) which allows for
co-amplification of 20 STR loci, along with the amelogenin gender
marker. The STR analysis of the PDXPC1 cell line was compared with
known cell lines in the Deutsche Sammlung von Mikroorganismen und
Zellkulturen (DSMZ) cell bank (https://www.dsmz.de).
Karyotyping
PDXPC1 cells at 80% confluence were incubated with
50 ng/ml Karyomax Colcemid (Thermo Fisher Scientific, Inc.) at 37°C
for 24 h. The cells were harvested and incubated with 0.075 M
hypotonic KCl solution at 37°C for 30 min, and fixed in a 3:1 (v/v)
mixture of methanol and acetic acid at 4°C for at least 1 h. After
dropping the fixed samples onto microscope slides, they were air
dried and viewed under the Ikaros system (MetaSystems) with a
bright-field microscope (Zeiss AG) (magnification, ×1,000). At
least five metaphases were counted per sample.
Generation of luciferase-overexpressing
cells
Briefly, 30-40% confluent PDXPC1 cells were
transduced with a lentiviral vector containing pBR322 (CMV/Firefly
luciferase/IRES/puromycin; Shanghai Genechem Co., Ltd.) at a
multiplicity of infection of 10, along with 40 µl HiTransG A
solution (Shanghai Genechem Co., Ltd.) per ml infective solution.
After 72 h, the transduced and control PDXPC1 cells were harvested
and re-seeded with 2 µg/ml puromycin (Shanghai Genechem Co.,
Ltd.) and expanded after >2 weeks. Luciferase-expressing PDXPC1
cells were confirmed by increased luminescence compared to control
cells in the presence of 150 µg/ml D-luciferin potassium
salt (MedChemExpress). The stably transduced PDXPC1 cells were
examined 1-2 weeks before every experiment.
Establishment of cell line-derived
xenograft model
All mouse experiments were approved by the Research
Ethics Committee of the First Affiliated Hospital, College of
Medicine, Zhejiang University. Panc-1 and PDXPC1 cells were
suspended in PBS at a density of 1×107 cells/ml, and 200
µl cell suspension was injected subcutaneously into the left
armpit of each mouse. Tumor growth was monitored every 5 days, and
tumor volumes were calculated using the formula [length (mm) x
width (mm)2]/2. A total of 45 days after inoculation,
the mice were sacrificed, and the tumors were harvested and fixed
in 4% paraformaldehyde (Sangon Biotech Co., Ltd.) at room
temperature for 24 h. Pancreatic orthotopic injection was performed
as previously described (14).
Briefly, the PDXPC1 cells were infected with lentiviral vectors
carrying the firefly luciferase reporter, and 50 µl stably
transduced cells (1×107 cells/ml) were injected. Tumor
growth was monitored twice a week for 6 weeks and the animals were
sacrificed. For in vivo bioluminescence imaging, the mice
were injected with D-luciferin potassium salt (150 mg/kg, i.p.;
MedChemExpress), and anesthetized with isoflurane 15 min later. The
concentrations of isoflurane used for inducing and maintaining the
anesthetic state were 3 and 1.5%, respectively. The intensity of
the bioluminescence signal was measured for 1-60 sec using the
In vivo Image system (PerkinElmer, Inc.), and analyzed using
the Living Imaging software (PerkinElmer, Inc.; version 4.3.1). All
mice were sacrificed by cervical dislocation during anesthesia (3%
isoflurane for induction, until the breathing had slowed) at the
end of the experiments, or upon reaching other ethical endpoints
(which did not occur throughout the study).
Histological and immunohistochemical
staining (IHC)
Fixed tumor tissues were embedded in paraffin, cut
into 4-µm sections, and stained with hematoxylin and eosin
(H&E) at room temperature (5 min and 3 min incubation times,
respectively) or further processed for IHC. The IHC was performed
as previously described (15).
Briefly, the tissue sections were first cleared with xylene and
rehydrated using an ethanol gradient, incubated with 3%
H2O2 to quench the endogenous peroxidases,
and then boiled in sodium citrate (pH 6) at 100°C for antigen
retrieval. After blocking with 3% goat serum (Fdbio Science) for 1
h to reduce nonspecific binding, the sections were incubated
overnight with the specific primary antibodies (Table S1) at 4°C. The immunopositive
signals were detected via a 20 min incubation at room temperature
with an out-of-the-box biotinylated secondary antibody (cat. no.
PV-8000; OriGene Technologies, Inc.) and 30 sec incubation with the
chromogen 3,3′-diaminobenzidine (cat. no. ZLI-9019; OriGene
Technologies, Inc.) at room temperature. After counterstaining with
hematoxylin for 3 min at room temperature, the slides were observed
under the Leica DM4000 light microscope (magnification, ×200; Leica
Microsystems GmbH).
Whole exome sequencing (WES)
DNA from PDXPC1 cells was extracted using QIAamp DNA
Blood Mini kit (Qiagen GmbH), according to the manufacturer's
protocol, and assessed by 1% agarose gel electrophoresis visualized
by gel-red (Sangon Biotech Co., Ltd.) staining. DNA libraries were
prepared using the SureSelect Human All Exon V5/V6 kit (Agilent
Technologies, Inc.) and were sequenced using the Illumina HiSeq X
Ten instrument (Illumina, Inc.). The generated fastq files were
matched with human reference genome version GRCh37 (hg19) using the
Burrows-Wheeler Aligner (version 0.7.13) and Samblaster (version
0.1.22) (16,17). The insertions/deletions and
single-nucleotide variations were detected using SAMtools
(http://www.htslib.org; version 1.0) and annotated
using ANNOVAR (http://annovar.openbio-informatics.org; version 2013,
August 23). The raw data were uploaded to the Sequence Read Archive
SRA database (https://www.ncbi.nlm.nih.gov/sra) in the National
Center for Biotechnology Information (accession no. SRR9312601). To
identify the cancer-related mutations, the filtering conditions
were as follows: i) Mutations reported in the Catalogue of Somatic
Mutations in Cancer (COSMIC) database (18), the rates of which in the 1,000
Genomes Project database (19)
were <0.1%, which were not reported in the Exome Aggregation
Consortium (ExAC) database (20);
or ii) mutations not reported in the COSMIC and dbSNP (21) databases, the rates of which were
<0.1% in the ExAC database and 0.3% in the 1000 Genomes Project
database.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from cell lines using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), and cDNAs were synthesized from 0.5 µg total RNA
using PrimeScript™ RT Master Mix (Takara Bio, Inc.), according to
the manufacturer's instructions as follows: 37°C for 15 min and
85°C for 5 sec. qPCR was performed with a StepOnePlus Real-Time PCR
system (Thermo Fisher Scientific, Inc.) using the TB Green™ Premix
Ex Taq™ kit (Takara Bio, Inc.). qPCR was performed as follows: 95°C
for 30 sec, then 40 cycles of 95°C for 5 sec and 60°C for 30 sec.
The primers used for qPCR are listed in Table S2. The gene expression level was
presented as the Cq value according to a previous study (22); ≥25 was defined as negative
expression and <25 as positive expression.
Western blot analysis
Cells were harvested and lysed using RIPA buffer
(Sigma-Aldrich; Merck KGaA), and quantified using the Pierce™ BCA
Protein Assay kit (Thermo Fisher Scientific, Inc.). A mass of 20
µg protein per lane was separated by SDS-PAGE on a 10% gel,
followed by immunoblotting (Immobilon P; EMD Millipore) with
specific antibodies. The primary antibodies were incubated at 4°C
overnight and secondary antibodies were incubated at room
temperature for 1 h. The specific bands were detected using an
enhanced chemiluminescence kit (Fdbio Science). The expression of
β-actin served as the internal reference throughout the study. The
antibodies are listed in Table
S1.
Invasion and migration assays
In vitro invasion and migration were assessed
by Transwell assays using membrane inserts of 8.0-µm pore
size (Millicell; EMD Millipore) with or without Matrigel (Corning,
Inc.) coating at 37°C for 1.5 h. The harvested cells were seeded in
the upper chamber at a density of 5×104 or
1×105 cells/well in 300 µl serum-free medium. The
lower chamber was filled with 700 µl complete DMEM (10%
FBS). The inserts were fixed with 95% ethanol at room temperature
for 15 min, stained with 0.1% crystal violet (Sigma-Aldrich; Merck
KGaA) at room temperature for 15 min and imaged after 24 h of
culture. The numbers of migratory/invasive cells were counted in
five random fields at ×200 magnification under a Nikon ECLIPSE Ti
inverted microscope (Nikon Corporation).
Flow cytometry analysis
Cell suspensions were harvested and stained with
anti-CD44-FITC (BD Biosciences; cat. no. 560977) and FITC-isotype
negative control (BD Biosciences; cat. no. 556655) for 30 min at
4°C in the dark. Flow cytometry was performed on a BD FACS Canto II
(BD Biosciences), and data were analyzed using FlowJo software
(FlowJo, LLC; version 10.0.7). Annexin V-FITC and propidium
iodidephycoerythrin staining was performed to detect apoptotic
cells, according to the manufacturer's protocol (BD Biosciences).
The apoptotic rate was calculated as the percentage of early and
late apoptotic cells in total cells.
Drug screening
Panc-1 and PDXPC1 cell lines were plated in 96-well
plates with 3,000 cells/well, and treated with a concentration
gradient of each compound for 24 h. After 48 or 72 h of incubation,
cell viability was assessed using the Cell Counting Kit-8 (Dojindo
Molecular Technologies, Inc.). The IC50 was calculated
via the Bliss method (23). Target
drugs for screening were found via the United States Food and Drug
Administration (USFDA; https://www.fda.gov/drugs/development-approval-process-drugs/drug-approvals-and-databases),
Drug Bank (https://www.drugbank.ca/) and
MycancerGenome (https://www.mycancergenome.org/) databases based on
somatic mutations of PDXPC1. The concentration ranges of each
compound are listed in Table S3,
and were selected for each compound based on published literature
(24) as well as the authors'
previous in vitro data (Du et al, unpublished).
Statistical methods
Statistical analyses were performed using GraphPad
Prism 7.0 Software (GraphPad Software, Inc.), and data are
presented as the mean ± SEM. Unpaired t-tests were used to evaluate
the difference between two groups, and multiple groups were
compared by one-way ANOVA with the Tukey post hoc test. P<0.05
was considered to indicate a statistically significant difference.
Each experiment was repeated at least three times.
Results
PDXPC1 cell line, established from PDX
models, retains the mutagenic signature of the original tumor
The primary culture of PDAC cells was from a PDAC
PDX model. The clinical features of the PDAC patient are listed in
Table I. The PDAC cells grew
consistently over a period of 2 years and maintained their
morphology (Fig. 1A). After 80
stable passages, this permanent PDAC cell line was designated
PDXPC1, and frozen for subsequent analysis. The spheroid morphology
of PDXPC1 is shown in Fig. 1B,
indicating a cancer stem cell phenotype. The STR analysis of this
cell line indicated no matches to any of the cell lines deposited
in the DSMZ cell bank (Fig. 1C).
The PDXPC1 cells exhibited aneuploidy resulting in chromosomal
numbers between 68 and 70 (Fig.
1D), which was consistent with the hypothesis that
tumorigenesis is typically accompanied by a high degree of
aneuploidy (25). To examine the
consistency with the original tumor tissue and PDX, an oncogenic
mutation screen of seven genes involved in PDAC progression
(26) was first performed, and the
results showed that the KRASG12V, tumor protein p53
(TP53)R155H and GNAS complex locus (GNAS)H69N
mutations were common between the primary patient tumor, PDX and
PDXPC1 (Fig. 1E), while the
remaining genes were wild-type. Thus, the PDXPC1 cell line retained
the mutagenic signature of the original tumor.
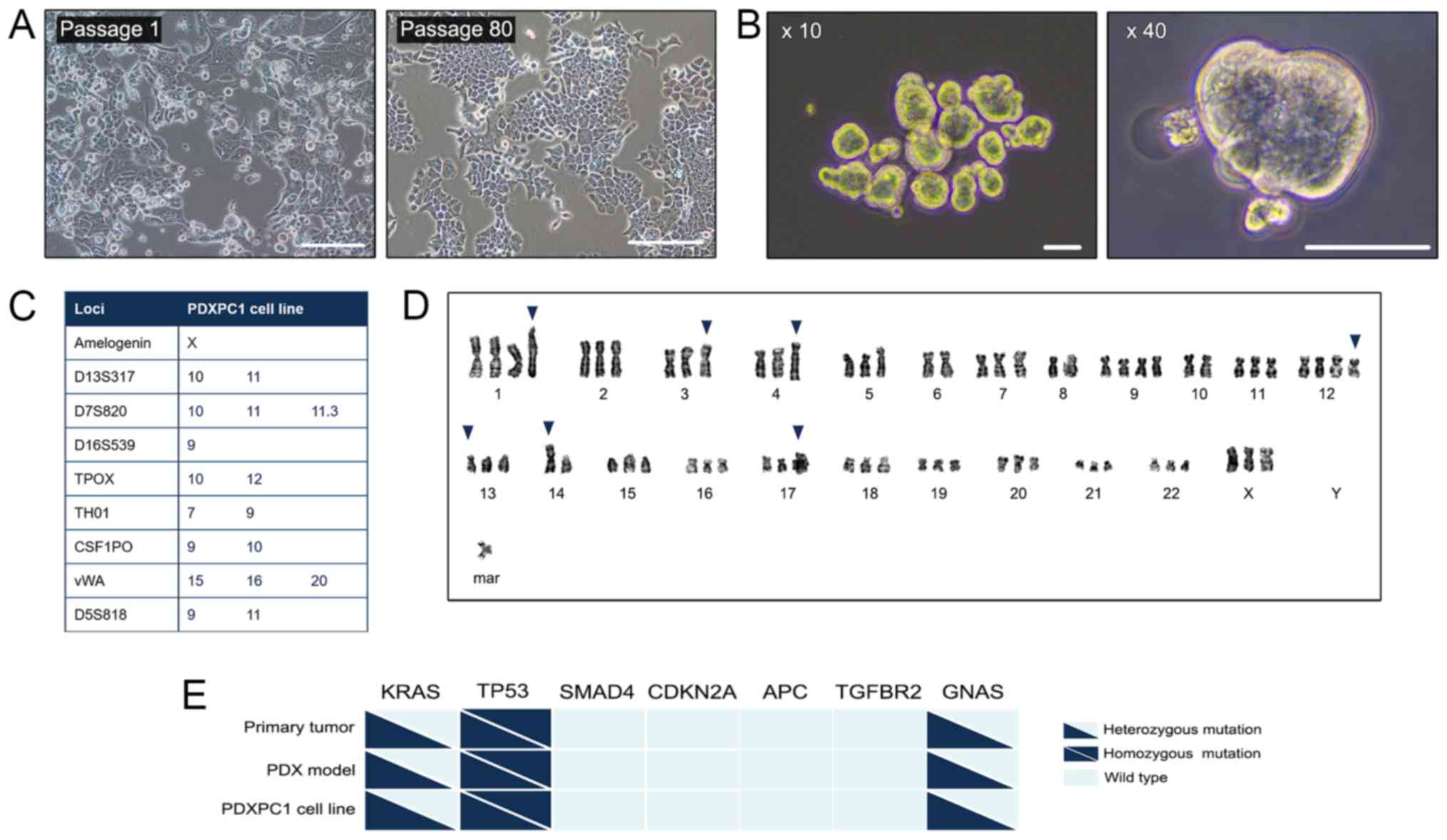 | Figure 1Establishment of the PDXPC1 cell
line. (A) Representative images of PDXPC1 cells at passage 1 (left)
and passage 80 (right). Scale bar, 200 µm. (B)
Representative images of PDXPC1 spheroids at ×10 (left) and ×40
(right) magnification. Scale bar, 50 µm. (C) The STR profile
of the PDXPC1 cell line. In order to protect the identity of the
donor, only listed the 8 core STR loci plus amelogenin are listed.
(D) Representative aneuploidy images of PDXPC1 metaphases with
chromosome number 69. The representative karyotype of PDXPC1 was:
69, XXX, +1, der(1)t(1;2)(p11;q11.1), der(3)t(3;10)(p13;p11.2), add(4) (q35), −6, −8, +9, −10, +12,
del(12)(q24.2), der(13)t(13;17)(p10;q10), −14,
rob(14;14)(q10;q10), der(17)t(17;?)(q12;?), +mar. The blue arrows
indicate the abnormalities. (E) Genetic mutations in the primary
tumor, PDX model and PDXPC1 cell line. Triangles and squares
indicate heterozygous and homozygous mutations respectively, and
wild type is shown as light blue. CDKNA2A, cyclin-dependent kinase
inhibitor 2A; APC, APC regulator of WNT signaling pathway; TGFBR2,
transforming growth factor receptor β2; GNAS, GNAS complex locus;
TP53, tumor protein p53. |
 | Table IClinical data for the donor
patient. |
Table I
Clinical data for the donor
patient.
|
Characteristics | Patient data |
|---|
| Sex | Female |
| Age (years) | 70 |
| Histological
diagnosis | Pancreatic ductal
adenocarcinoma |
| Tumor size
(cm) | 9.5×5.5 |
| Tumor number | 1 |
| TNM stagea | IIA |
| Metastasis | No |
| Location | Body-tail |
| Level of tumor
marker CA199 | 682.9 U/ml |
| History of
chemotherapy | No |
PDXPC1 cell line induces rapid in vivo
tumor growth and maintains a similar histomorphology and
immunophenotype to the original tumor
To evaluate the potential tumorigenic ability of the
PDXPC1 cells, PDXPC1 cells in comparison with a commercial
pancreatic cancer cell line, Panc-1 were subcutaneously injected
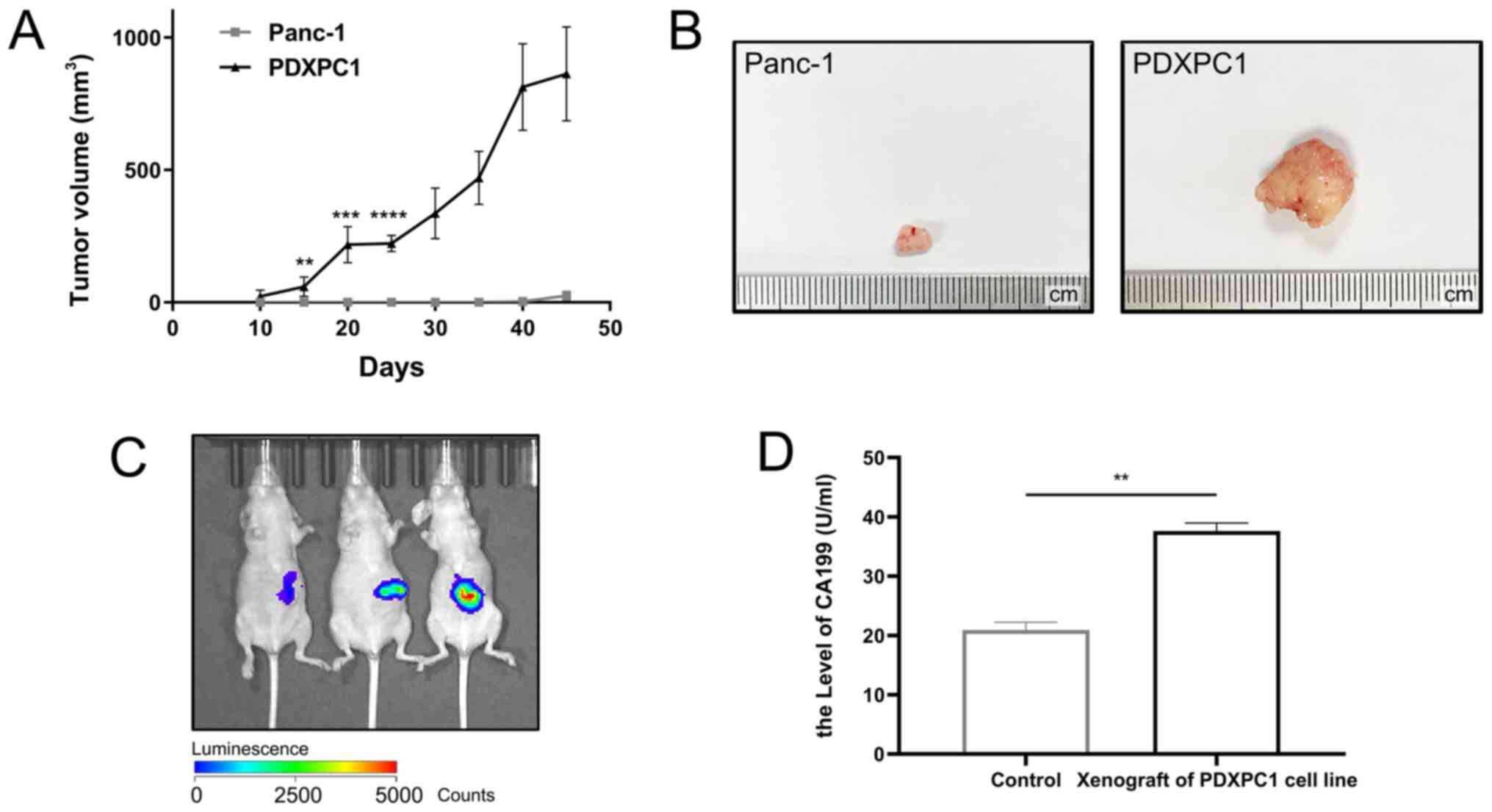
into immunodeficient mice. As shown in Fig. 2A, PDXPC1 cells resulted in
aggressive tumor growth that exceeded 10% of the body weight within
45 days of inoculation. By contrast, Panc-1 tumor growth was very
slow, and only palpable nodes were detected (Fig. 2B). To simulate the tumor
microenvironment more accurately, luciferase-labeled PDXPC1 cells
were transplanted into the murine pancreas, and tumors were
detected after 15 days using an in vivo imaging system
(Fig. 2C). Since the patient donor
of the PDXPC1 cell line had elevated serum levels of CA199, a
circulating diagnostic biomarker of PDAC (27), the peripheral sera of the
tumor-bearing and control mice was analyzed and high levels of
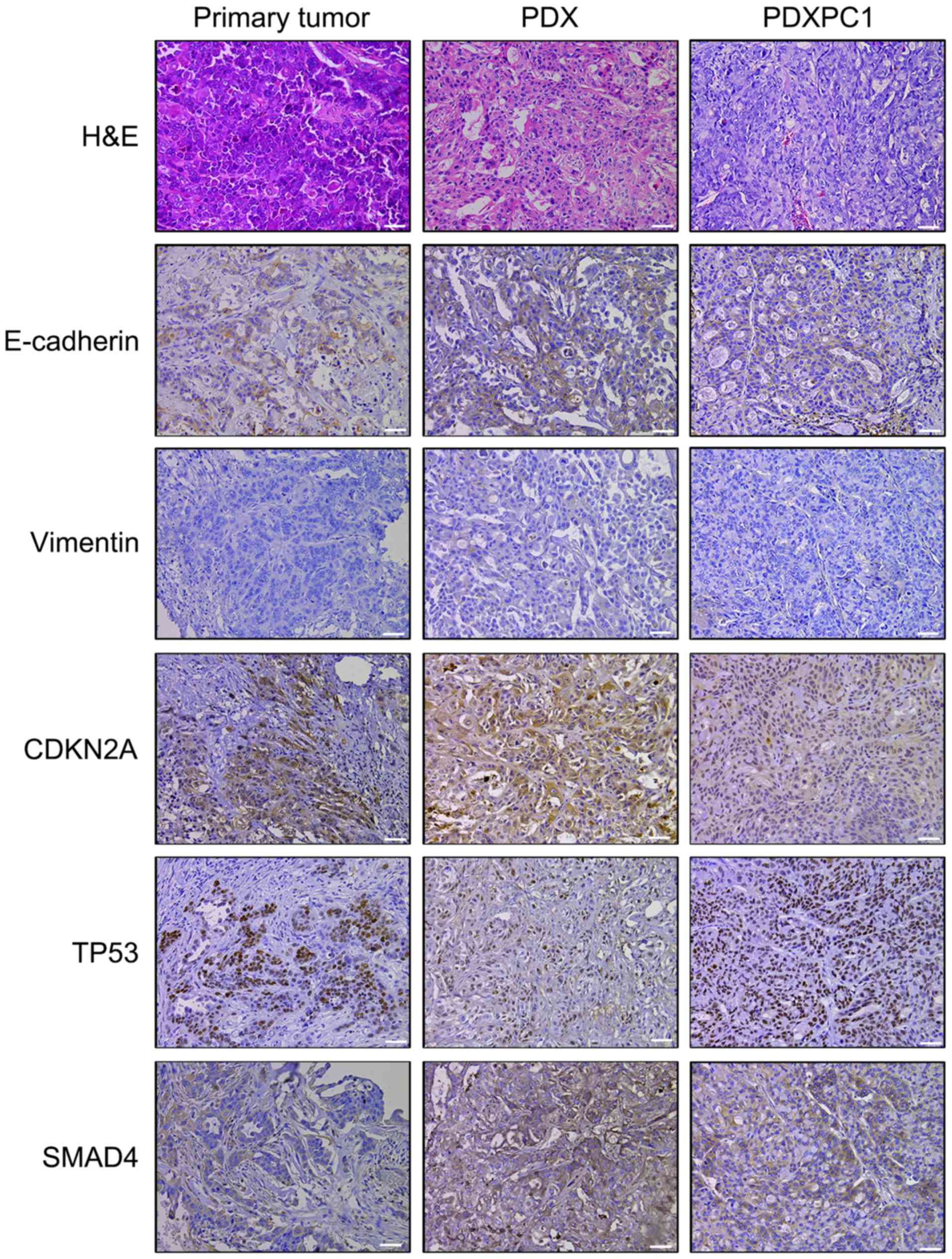
CA199 were detected in the former (Fig. 2D). H&E staining confirmed
similar histomorphology between the PDXPC1-derived xenografts,
patient-derived xenografts and the primary tumor (Fig. 3). In addition, E-cadherin and
vimentin immunostaining showed that PDXPC1 cells maintained their
epithelial nature. These three samples were also positive for
cyclin-dependent kinase inhibitor 2A (CDKN2A or p16), TP53 and
SMAD4, all of which are inactivated in early pancreatic
carcinogenesis (28). Taken
together, the PDXPC1 cell line maintained the key characteristics
of the original tumors.
PDXPC1 cell line maintains its epithelial
origin and carries overexpression of HER2, CDKN2A and programmed
cell death 1 ligand 1 (PD-L1)
WES of the PDXPC1 cell line was performed to fully
characterize the PDXPC1 cells. Since the mutational data of matched
normal tissues was lacking, the non-mutational polymorphisms were
first excluded using public databases, and the remaining
polymorphisms were filtered through the COSMIC database to identify
the cancer-related mutations. The driver mutations were
specifically searched for using publicly available databases, and
of the four commonly mutated genes in PDAC (29), KRASG12V was detected in
the PDXPC1 cell line. In addition, other genes that are mutated in
PDAC [such as GNAS, catenin β1 (CTNNB1), ring finger protein 43 and
fibroblast growth factor receptor 2] and some other cancer types
(e.g., chromodomain helicase DNA binding protein 8,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit-α,
myosin heavy chain 9 and stromal antigen 2) (30-33)
were also identified in the established cell lines (Fig. 4A).
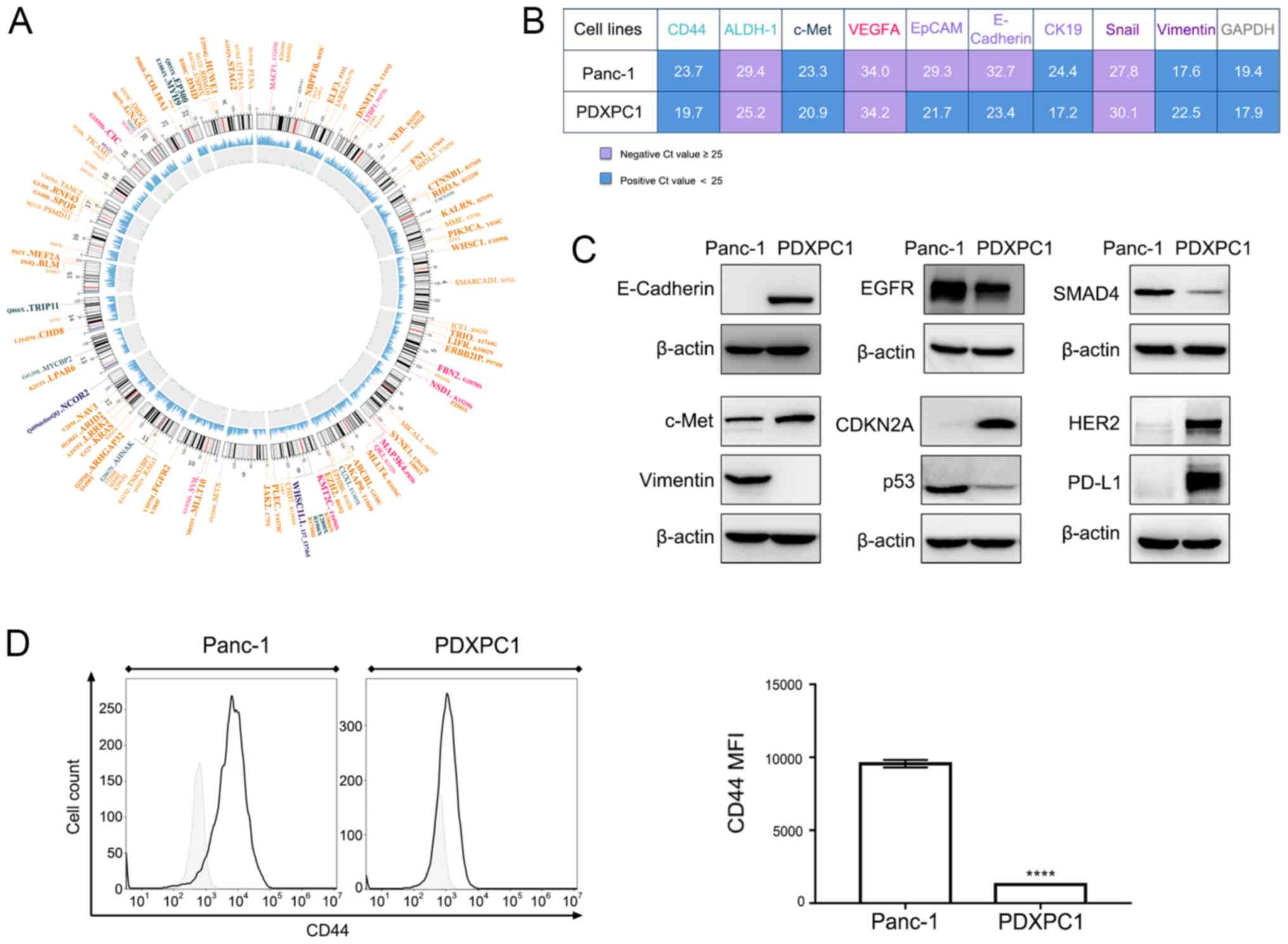 | Figure 4Molecular characterization of the
PDXPC1 cell line. (A) Circos plot of somatic mutations of PDXPC1
cells. The outer ring indicates the driver gene mutations (orange,
missense mutation; fuchsia, frame shift deletion; dark blue,
in-frame insertion; and dark green, nonsense mutation), and the
subsequent rings indicate the chromosomes, sequencing coverage and
the frequency of single-nucleotide variations and
insertions-deletions, each indicated by the size of the dots. (B)
Differential mRNA expression levels of PDXPC1 and Panc-1, with
GAPDH as the control. Cq values are presented as the mean ± SD of
three independent experiments. Cq <25 indicates positive
expression. (C) Phenotypic characterization of the PDXPC1 cell line
by western blotting. β-actin was used as the reference control. (D)
CD44 expression level in PDXPC1 and Panc-1 cell lines, as assessed
by flow cytometry. Left, Histogram of CD44 expression level in
Panc-1 and PDXPC1 cell lines. Right, rCD44 mean fluorescence index
of Panc-1 and PDXPC1 cell lines; ****P<0.0001 vs.
Panc-1. CDKNA2A, cyclin-dependent kinase inhibitor 2A; EGFR,
epidermal growth factor receptor; ALDH-1, aldehyde dehydrogenase 1;
VEGFA, vascular endothelial growth factor A; EpCAM, epithelial cell
adhesion molecule; CK19, keratin 19. |
Then, to fully characterize the PDXPC1 cell line,
the mRNA expression level of some genes of interest, shown in
Fig. 4B, was tested. The
expression levels of various epithelial and mesenchymal markers in
the PDXPC1 cell line were compared with the Panc-1 cell line, and
it was found that while the PDXPC1 cells expressed high levels of
the epithelial markers (E-cadherin, EpCAM and keratin 19) and low
levels of mesenchymal marker vimentin, Panc-1 expressed vimentin.
Furthermore, the PDXPC1 cells expressed the proto-oncogene c-Met
and the cancer stem cell markers CD44 and ALDH1, which have been
previously detected in PDAC (34).
Next, protein expression levels were examined using different
methods to identify the phenotype of the PDXPC1 cell line. The
epithelial nature of the PDXPC1 cell line, as well as highly
expressed c-Met, were consistent with the mRNA expression level
results (Fig. 4B). HER2 and EGFR,
which are related to pancreatic tumor progression (35,36)
were also highly expressed (Fig.
4C). The presence of CDKN2A, TP53 and SMAD4 was consistent with
the IHC results of the PDXPC1-derived xenograft. Since the PDXPC1
cells expressed higher levels of CD44 mRNA compared to Panc-1, the
relative CD44 protein levels were subsequently examined by flow
cytometry. CD44 protein levels were lower in the PDXPC1 cell line
(Fig. 4D). Since early trials of
single-agent immune checkpoint blockade therapy have not shown
encouraging results (37,38), the expression of PD-L1 in PDXPC1
cells was also examined, and aberrantly high levels were observed.
Taken together, the fully characterized PDXPC1 cell line is a
suitable model to study PDAC genesis.
PDXPC1 cell line exhibits relatively mild
biological behaviors
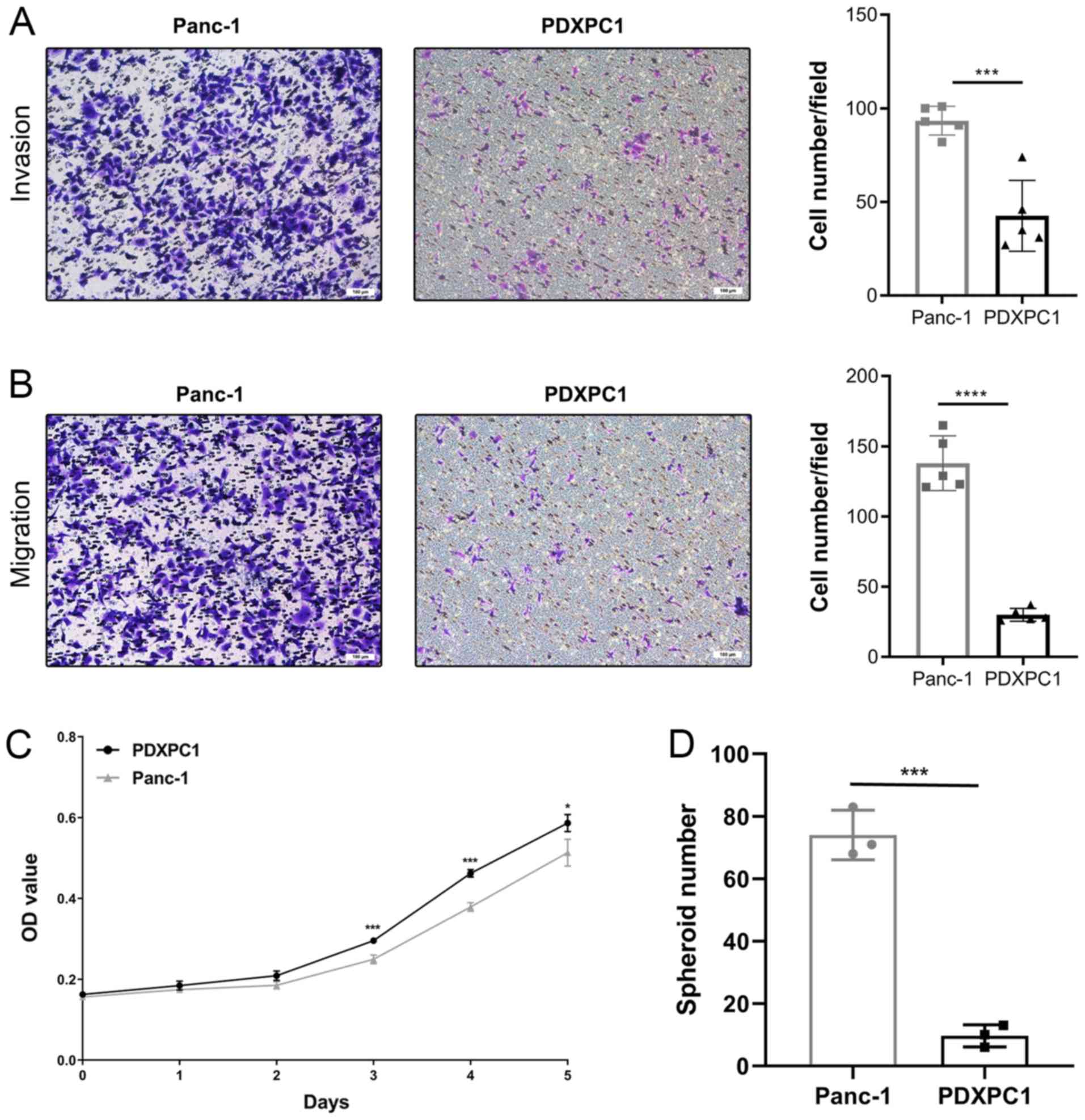
Next, the biological behaviors of the PDXPC1 cell
line were analyzed. The migration and invasion ability of PDXPC1
cells were evaluated, and the results indicated that the PDXPC1
cell line exhibited a lower migration and invasion potential than
the Panc-1 cell line (Fig. 5A and
B). Fig. 5C shows the in
vitro growth curve of PDXPC1 cells. Different from the in
vivo tumorigenesis study, the growth rate was similar between
PDXPC1 and Panc-1 cell lines. The doubling time of the PDXPC1 and
Panc-1 cell lines was 37.2 and 39.9 h, respectively. Considering
the different expression pattern of CD44, the cancer stem cell
marker, at the transcriptional and protein level the spheroid assay
was performed on the PDXPC1 and Panc-1 cell lines. The result
showed a lower spheroid formation rate of PDXPC1 cells compared to
that of Panc-1 cells, which was consistent with the protein
expression level data (Fig.
5D).
PDXPC1 cell line is resistant to multiple
therapeutic drugs excluding trametinib, a MEK inhibitor
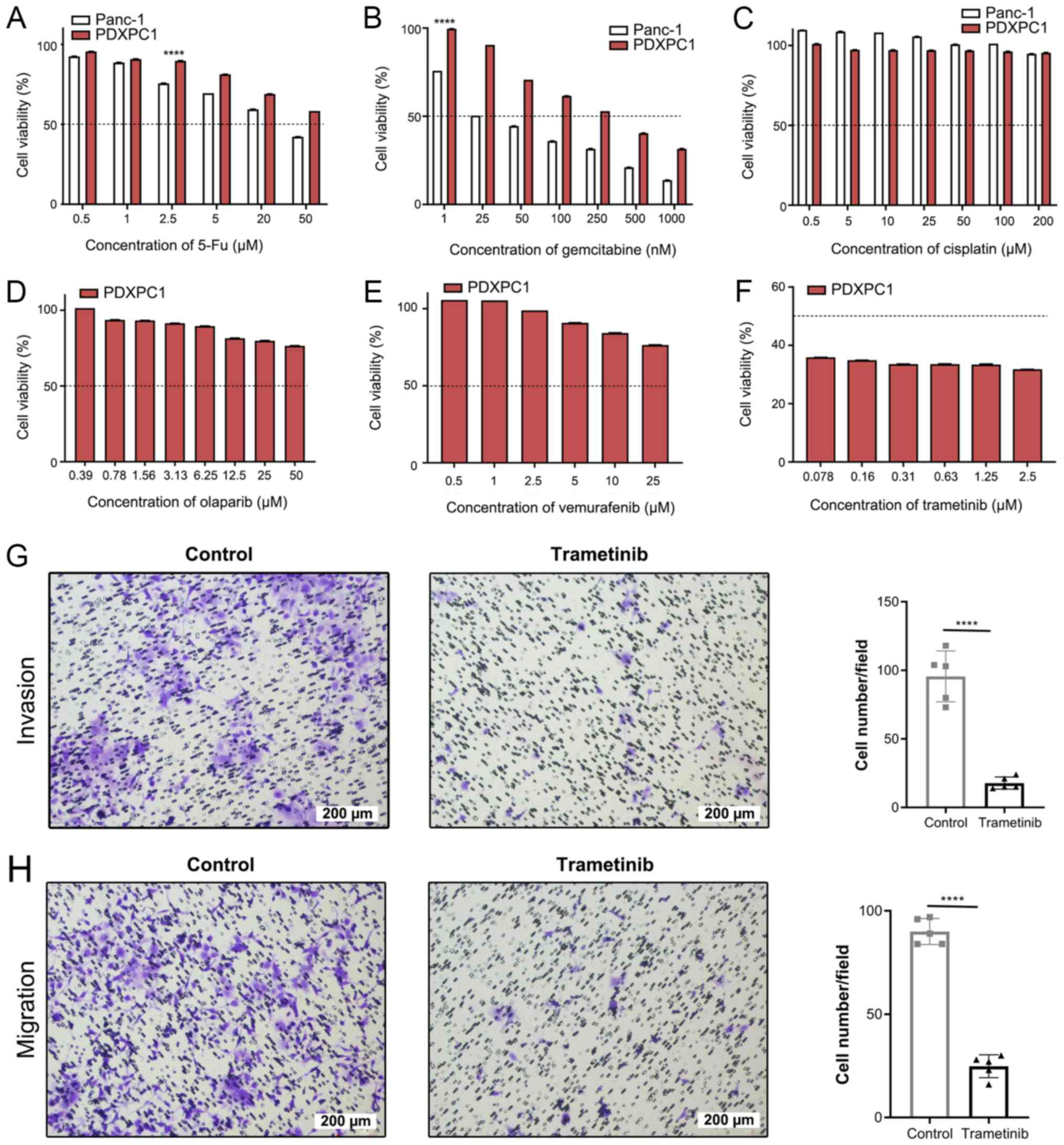
To determine the utility of PDXPC1 as a suitable
model for screening anti-PDAC drugs, the cells were treated with
gemcitabine, 5-fluoruracil (5-Fu) and cisplatin (Fig. 6A-C), which are routinely used in
the clinical management of PC. Both cell lines were resistant to
the DNA cross-linker cisplatin. While Panc-1 was sensitive to
gemcitabine and 5-Fu within the doses used, PDXPC1 only responded
to gemcitabine at high concentration levels. The PDXPC1 cell line
showed a drug-resistant phenotype according the IC50 of
gemcitabine (275.1±1.50 vs. 22.2±1.25 nM; 12.39-fold
difference).
Therefore, other target drugs were screened based on
the somatic mutational profile of PDXPC1 on online drug databases,
including USFDA, Drug Bank and MycancerGenome. The database of
MycancerGenome indicated that poly-ADP ribose polymerase (PARP)
inhibitors, BRAF inhibitors and MEK inhibitors may be the target
drugs for PDXPC1 cells with CTNNB1 mutations. Further drug
screening showed that PDXPC1 cell line was resistant to the PARP
inhibitor olaparib (Fig. 6D) and
the BRAF inhibitor vemurafenib (Fig
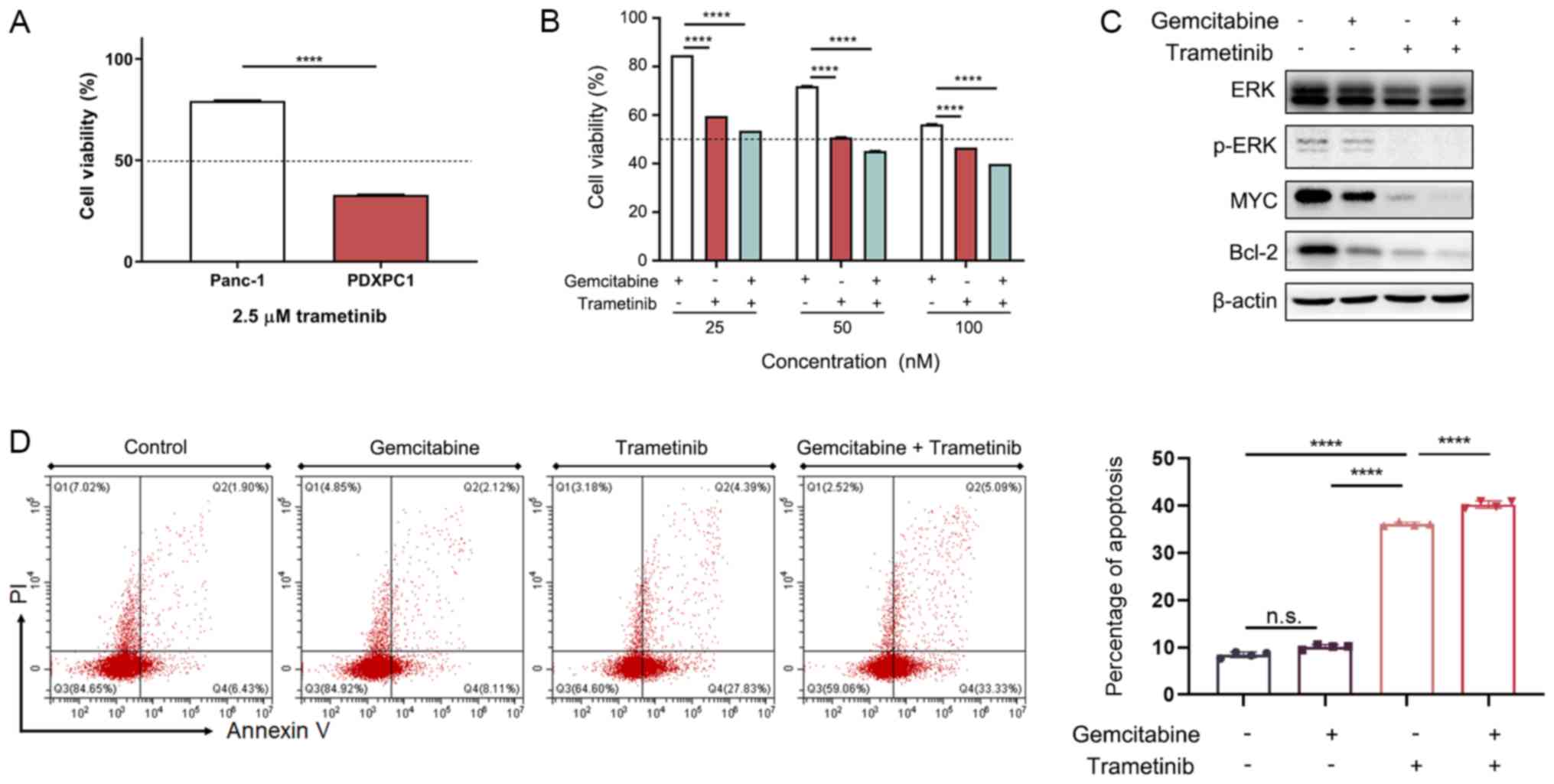
6E), but responded to the MEK inhibitor trametinib (Fig. 6F). Furthermore, trametinib-treated
PDXPC1 cells showed significantly decreased invasion and migration
(P<0.0001; Fig. 6G and H).
These results indicated that MEK is a therapeutic target in the
PDXPC1 cell line.
By contrast, the CTNNB1-wild type Panc-1 cells were
unresponsive to trametinib (Fig.
7A). The combination of low-dose trametinib and gemcitabine
significantly inhibited the PDXPC1 cells (Fig. 7B), suggesting the possibility of
other efficient combination treatments with trametinib.
Mechanistically, trametinib, but not gemcitabine, reduced the
expression of p-ERK, MYC and the pro-survival Bcl-2 protein
(Fig. 7C). Thus, trametinib
decreased PDXPC1 cell proliferation by activating the apoptotic
pathway via Bcl-2 degradation, which was confirmed by the
significantly increased apoptosis rates seen after treatment with
trametinib and trametinib together with gemcitabine, but not with
gemcitabine alone (Fig. 7D). Taken
together, the overall drug screening results indicated that a
subset of PDAC patients could benefit from a MEK inhibitor in the
therapeutic regimen.
Discussion
The poor prognosis of PDAC is partly attributed to
its chemo-resistance. Gemcitabine-based monotherapy or combination
treatments have been largely unsatisfactory in the majority of
patients with PDAC (5). PDXs
closely replicate the clinical characteristics of the tumors and
are therefore indispensable for accurate preclinical drug
evaluation. However, cancer cell lines are preferable for molecular
studies and high-throughput drug screenings. The present study
established a new stable pancreatic cancer cell line, PDXPC1, from
a PDAC PDX, since it was not possible to establish a cancer cell
line directly from a primary tumor sample. The results demonstrated
that the PDXPC1 cell line fully recapitulated the primary tumor and
provided a suitable tool for exploring the molecular basis of PDAC.
Furthermore, the specific responsiveness to the small molecular
inhibitors tested based on mutation information, and the
involvement of signaling pathways, indicated a potential novel
therapeutic approach for a subgroup of PDAC patients.
The target drug screening revealed that the PDXPC1
cell line, with a CTNNB1 mutation, may be susceptible to three
directed therapies, including BRAF, PARP and MEK inhibitors.
However, in vitro drug sensitivities showed that PDXPC1
cells were only response to trametinib, a small molecular compound
which specifically inhibits MEK1/2. CTNNB1, the gene encoding
β-catenin, is a key transcriptional activator of the Wnt/β-catenin
signaling pathway that is oncogenic in most human cancer types
(39). Mutant CTNNB1 has been
implicated in the pathogenesis of several cancer types, including
melanoma, colorectal cancer, hepatocellular carcinoma and
pancreatic solid-pseudopapillary tumor (40). Few relevant studies have been
conducted in pancreatic disorders, to the best of our knowledge.
The present study evaluated the inhibitory effect of trametinib in
the Panc-1 cell line, carrying wild-type CTNNB1, confirming that
the CTNNB1 mutation might be a target molecular marker for
trametinib treatment in PDAC. Plans for next stage may involve a
large number of PDAC cell lines and targeted CTNNB1 gene editing to
further confirm these conclusions.
The MEK-ERK pathway plays an important role in
cancer cell proliferation, migration and chemoresistance (41), and is constitutively activated in
>90% of PC cases, wherein it drives KRAS-dependent tumor growth
and decreases the survival rate (42). The intricate crosstalk between the
Wnt/β-catenin and mitogen-activated protein kinase (MAPK) pathways
has been reported in melanoma and colorectal cancer. In melanoma,
studies found that inhibition of the MAPK pathway resulted in
enhanced Wnt/β-catenin signaling, which induced the reduction of
tumor size due to apoptosis (43,44).
By contrast, Wnt/β-catenin and MAPK signaling synergistically
promoted cancer development in colon cancer (45). It appears that the relationship
between Wnt/β-catenin is dependent on the specific context, which
will be investigated in future work.
The poor prognosis of PDAC is partly attributed to
its chemoresistance. Gemcitabine based monotherapy or combination
treatments have been largely unsatisfactory in the majority of
patients with PDAC. The PDXPC1 cell line was susceptible to MEK
inhibitor-based monotherapy, resulting in ERK inactivation and
Bcl-2 degradation. Therefore, a subset of PDAC patients may benefit
from MEK inhibition treatment.
Supplementary Data
Funding
This research was funded by the National Natural
Science Foundation of China (grant no. 81772853), Zhejiang Science
and Technology Project (grant no. 2017C37171) and the Natural
Science Foundation of Zhejiang (grant no. LZ16H160001).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LST conceived the study. JZ and MK performed the
karyotyping and IHC experiments. KFH, XXD, YYH and ZZX performed
experiments. XXD analyzed the data and performed the statistical
analysis, as well as writing the original draft. KFH, JC, JZ and
LST reviewed and edited the manuscript. LST and KFH obtained
funding support.
Ethics approval and consent to
participate
The study was approved by the Ethical Review
Committee of The First Affiliated Hospital of Zhejiang University
School of Medicine, and written informed consent was obtained from
the patient. All mouse experiments were approved by the Research
Ethics Committee of the First Affiliated Hospital, College of
Medicine, Zhejiang University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Wolfgang CL, Herman JM, Laheru DA, Klein
AP, Erdek MA, Fishman EK and Hruban RH: Recent progress in
pancreatic cancer. CA Cancer J Clin. 63:318–348. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang G, He P, Tan H, Budhu A, Gaedcke J,
Ghadimi BM, Ried T, Yfantis HG, Lee DH, Maitra A, et al:
Integration of metabolomics and transcriptomics revealed a fatty
acid network exerting growth inhibitory effects in human pancreatic
cancer. Clin Cancer Res. 19:4983–4993. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kamisawa T, Wood LD, Itoi T and Takaori K:
Pancreatic cancer. Lancet. 388:73–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ryan DP, Hong TS and Bardeesy N:
Pancreatic adenocarcinoma. N Engl J Med. 371:1039–1049. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Heinemann V, Wilke H, Mergenthaler HG,
Clemens M, König H, Illiger HJ, Arning M, Schalhorn A, Possinger K
and Fink U: Gemcitabine and cisplatin in the treatment of advanced
or metastatic pancreatic cancer. Ann Oncol. 11:1399–1403. 2000.
View Article : Google Scholar
|
|
6
|
Michl P and Gress TM: Current concepts and
novel targets in advanced pancreatic cancer. Gut. 62:317–326. 2013.
View Article : Google Scholar
|
|
7
|
Fusenig NE, Capes-Davis A, Bianchini F,
Sundell S and Lichter P: The need for a worldwide consensus for
cell line authentication: Experience implementing a mandatory
requirement at the International Journal of Cancer. PLoS Biol.
15:e20014382017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ye F, Chen C, Qin J, Liu J and Zheng C:
Genetic profiling reveals an alarming rate of cross-contamination
among human cell lines used in China. FASEB J. 29:4268–4272. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dangles-Marie V, Pocard M, Richon S,
Weiswald LB, Assayag F, Saulnier P, Judde JG, Janneau JL, Auger N,
Validire P, et al: Establishment of human colon cancer cell lines
from fresh tumors versus xenografts: Comparison of success rate and
cell line features. Cancer Res. 67:398–407. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bissig-Choisat B, Kettlun-Leyton C, Legras
XD, Zorman B, Barzi M, Chen LL, Amin MD, Huang YH, Pautler RG,
Hampton OA, et al: Novel patient-derived xenograft and cell line
models for therapeutic testing of pediatric liver cancer. J
Hepatol. 65:325–333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang H, Lu J, Tang J, Chen S, He K, Jiang
X, Jiang W and Teng L: Establishment of patient-derived gastric
cancer xenografts: A useful tool for preclinical evaluation of
targeted therapies involving alterations in HER-2, MET and FGFR2
signaling pathways. BMC Cancer. 17:1912017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu X, Qian L-J, Su X-Y, He KF, Jin KT, Gu
LH, Feng JG, Li GL, Zhou Q, Xu ZZ, et al: Establishment and
characterization of GCSR1, a multi-drug resistant signet ring cell
gastric cancer cell line. Int J Oncol. 46:2479–2487. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nikkhah M, Strobl JS, Peddi B and Agah M:
Cytoskeletal role in differential adhesion patterns of normal
fibroblasts and breast cancer cells inside silicon
microenvironments. Biomed Microdevices. 11:585–595. 2009.
View Article : Google Scholar
|
|
14
|
Kim MP, Evans DB, Wang H, Abbruzzese JL,
Fleming JB and Gallick GE: Generation of orthotopic and heterotopic
human pancreatic cancer xenografts in immunodeficient mice. Nat
Protoc. 4:1670–1680. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Du X, Wu L, Ur Rahman MS, Teng X, Teng L,
Ye J and Cao J: Promoter Hypomethylation Is Responsible for
Upregulated Expression of HAI-1 in Hepatocellular Carcinoma. Dis
Markers. 2019:91752152019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Faust GG and Hall IM: SAMBLASTER: Fast
duplicate marking and structural variant read extraction.
Bioinformatics. 30:2503–2505. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li MM, Datto M, Duncavage EJ, Kulkarni S,
Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ,
Younes A, et al: Standards and Guidelines for the Interpretation
and Reporting of Sequence Variants in Cancer: A Joint Consensus
Recommendation of the Association for Molecular Pathology, American
Society of Clinical Oncology, and College of American Pathologists.
J Mol Diagn. 19:4–23. 2017. View Article : Google Scholar :
|
|
19
|
Abecasis GR, Altshuler D, Auton A, Brooks
LD, Durbin RM, Gibbs RA, Hurles ME and McVean GA; 1000 Genomes
Project Consortium: A map of human genome variation from
population-scale sequencing. Nature. 467:1061–1073. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karczewski KJ, Weisburd B, Thomas B,
Solomonson M, Ruderfer DM, Kavanagh D, Hamamsy T, Lek M, Samocha
KE, Cummings BB, et al: The Exome Aggregation Consortium: The ExAC
browser: Displaying reference data information from over 60 000
exomes. Nucleic Acids Res. 45(D1): D840–D845. 2017. View Article : Google Scholar
|
|
21
|
National Center for Biotechnology
Information (US): General Information about dbSNP as a Database
Resource. SNP FAQ Archive [Internet]. National Center for
Biotechnology Information (US); Bethesda, MD: 2005
|
|
22
|
Cayrefourcq L, Mazard T, Joosse S,
Solassol J, Ramos J, Assenat E, Schumacher U, Costes V, Maudelonde
T, Pantel K, et al: Establishment and characterization of a cell
line from human circulating colon cancer cells. Cancer Res.
75:892–901. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bliss CI: The calculation of microbial
assays. Bacteriol Rev. 20:243–258. 1956.PubMed/NCBI
|
|
24
|
Tiriac H, Belleau P, Engle DD, Plenker D,
Deschênes A, Somerville TDD, Froeling FEM, Burkhart RA, Denroche
RE, Jang GH, et al: Organoid profiling identifies common responders
to chemotherapy in pancreatic cancer. Cancer Discov. 8:1112–1129.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Douville C, Springer S, Kinde I, Cohen JD,
Hruban RH, Lennon AM, Papadopoulos N, Kinzler KW, Vogelstein B and
Karchin R: Detection of aneuploidy in patients with cancer through
amplification of long interspersed nucleotide elements (LINEs).
Proc Natl Acad Sci USA. 115:1871–1876. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Seino T, Kawasaki S, Shimokawa M, Tamagawa
H, Toshimitsu K, Fujii M, Ohta Y, Matano M, Nanki K, Kawasaki K, et
al: Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche
Factor Dependence during Disease Progression. Cell Stem Cell.
22:454–467.e6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Duffy MJ, Sturgeon C, Lamerz R, Haglund C,
Holubec VL, Klapdor R, Nicolini A, Topolcan O and Heinemann V:
Tumor markers in pancreatic cancer: A European Group on Tumor
Markers (EGTM) status report. Ann Oncol. 21:441–447. 2010.
View Article : Google Scholar
|
|
28
|
Jones S, Zhang X, Parsons DW, Lin JC,
Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et
al: Core signaling pathways in human pancreatic cancers revealed by
global genomic analyses. Science. 321:1801–1806. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ellis MJ, Ding L, Shen D, Luo J, Suman VJ,
Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al:
Whole-genome analysis informs breast cancer response to aromatase
inhibition. Nature. 486:353–360. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wagner AH, Devarakonda S, Skidmore ZL,
Krysiak K, Ramu A, Trani L, Kunisaki J, Masood A, Waqar SN, Spies
NC, et al: Recurrent WNT pathway alterations are frequent in
relapsed small cell lung cancer. Nat Commun. 9:37872018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pietzak EJ, Bagrodia A, Cha EK, Drill EN,
Iyer G, Isharwal S, Ostrovnaya I, Baez P, Li Q, Berger MF, et al:
Next-generation Sequencing of Nonmuscle Invasive Bladder Cancer
Reveals Potential Biomarkers and Rational Therapeutic Targets. Eur
Urol. 72:952–959. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X,
Xiao Y, Yu KD, Liu YR, Yu Y, et al: Genomic and Transcriptomic
Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment
Strategies. Cancer Cell. 35:428–440.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Christen F, Hoyer K, Yoshida K, Hou HA,
Waldhueter N, Heuser M, Hills RK, Chan W, Hablesreiter R, Blau O,
et al: Genomic landscape and clonal evolution of acute myeloid
leukemia with t(8;21): An international study on 331 patients.
Blood. 133:1140–1151. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Penchev VR, Rasheed ZA, Maitra A and
Matsui W: Heterogeneity and targeting of pancreatic cancer stem
cells. Clin Cancer Res. 18:4277–4284. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tobita K, Kijima H, Dowaki S, Kashiwagi H,
Ohtani Y, Oida Y, Yamazaki H, Nakamura M, Ueyama Y, Tanaka M, et
al: Epidermal growth factor receptor expression in human pancreatic
cancer: Significance for liver metastasis. Int J Mol Med.
11:305–309. 2003.PubMed/NCBI
|
|
36
|
Dugan MC, Dergham ST, Kucway R, Singh K,
Biernat L, Du W, Vaitkevicius VK, Crissman JD and Sarkar FH:
HER-2/neu expression in pancreatic adenocarcinoma: Relation to
tumor differentiation and survival. Pancreas. 14:229–236. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Foley K, Kim V, Jaffee E and Zheng L:
Current progress in immunotherapy for pancreatic cancer. Cancer
Lett. 381:244–251. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Brahmer JR, Tykodi SS, Chow LQM, Hwu WJ,
Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al:
Safety and activity of anti-PD-L1 antibody in patients with
advanced cancer. N Engl J Med. 366:2455–2465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kongkanuntn R, Bubb VJ, Sansom OJ, Wyllie
AH, Harrison DJ and Clarke AR: Dysregulated expression of β-catenin
marks early neoplastic change in Apc mutant mice, but not all
lesions arising in Msh2 deficient mice. Oncogene. 18:7219–7225.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Giles RH, van Es JH and Clevers H: Caught
up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta.
1653:1–24. 2003.PubMed/NCBI
|
|
41
|
Balmanno K and Cook SJ: Tumour cell
survival signalling by the ERK1/2 pathway. Cell Death Differ.
16:368–377. 2009. View Article : Google Scholar
|
|
42
|
Hayes TK, Neel NF, Hu C, Gautam P, Chenard
M, Long B, Aziz M, Kassner M, Bryant KL, Pierobon M, et al:
Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is
Associated with MYC Degradation and Senescence-like Growth
Suppression. Cancer Cell. 29:75–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pollock PM, Harper UL, Hansen KS, Yudt LM,
Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J,
et al: High frequency of BRAF mutations in nevi. Nat Genet.
33:19–20. 2003. View
Article : Google Scholar
|
|
44
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jeong WJ, Yoon J, Park JC, Lee SH, Lee SH,
Kaduwal S, Kim H, Yoon JB and Choi KY: Ras stabilization through
aberrant activation of Wnt/β-catenin signaling promotes intestinal
tumorigenesis. Sci Signal. 5:ra302012. View Article : Google Scholar
|
|
46
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Manual. 7th edition.
American Joint Committee on Cancer; Chicago, IL: 2010
|