Introduction
Acute myeloid leukemia (AML) is a malignant and
aggressive disease that is sensitive to chemotherapy (1). Although current therapeutic
strategies provide a reasonable likelihood of achieving complete
remission for the majority of patients, the relapse rates and
subsequent disease-associated mortality remain unacceptably high
(1,2). There were an estimated 21,450 newly
diagnosed cases of AML and 10,920 AML-associated mortalities in the
USA in 2019 (2). Improvements in
chemotherapeutic regimens and supportive care have increased
overall survival rates in younger patients (<15 years of age) to
>60%; however, 40% ultimately relapse and require salvage
therapy (1). Therefore, there is
an urgent need to develop a novel therapeutic drug to enhance
chemosensitization. Diallyl disulfide (DADS), a major oil-soluble
compound derived from garlic, has multi-targeted antitumor
activities in diverse cancer types, which results in the induction
of cellular processes, including cell cycle arrest, growth
inhibition, differentiation and apoptosis (3). Our previous study (4) demonstrated that DADS could induce the
differentiation and inhibit the growth of HL-60 cells by increasing
the expression of acetylated histone H3, H4 and p21WAF1 in
vitro and in vivo. Using high-resolution mass
spectrometry, a total of 18 differentially expressed proteins were
identified after treatment with DADS, including four upregulated
and fourteen downregulated proteins, among which the protein
expression of cofilin 1 was downregulated (5).
Cofilin is an actin-binding protein universally
present in eukaryotes that has a molecular weight of 21 kD
(6). Two mammalian cofilin gene
subtypes exist, cofilin 1 and cofilin 2, which encode different
proteins. The former is expressed in a variety of tissues except
muscle, and the latter is mainly expressed in muscle tissue
(7,8). Cofilin can depolymerize actin
filaments and regulate the cytoskeleton (9). A basic function of cofilin is to
combine with and depolymerize F-actin intracellularly, thereby
inhibiting G-actin polymerization and accelerating actin filament
dynamic transformation (10). At
the same time, cofilin plays an important role in regulating cell
movement (11). The cofilin 1
pathway is regarded as a target to suppress the growth of breast
cancer cells (12) and also serves
an important role in inhibiting epithelial mesenchymal transition
in gastric adenocarcinoma cells (13).
Cofilin can be acted upon by two types of kinases,
LIM kinases (LIMKs) and testicular protein kinases, which induce
cofilin phosphorylation at the Ser3 site, inhibit the binding of
cofilin to F-actin and stabilize F-actin. Two types of cofilin
dephosphorylate, Slingshot and chronophin, which can
dephosphorylate cofilin, thereby causing the depolymerization and
removal of F-actin, promoting actin nucleation through Arp 2/3 and
inducing other actin polymerization events and directional movement
(14). Cofilin 1 helps regulate
glioblastoma cell migration by modulating the cytoskeleton via
multiple targets, including F-actin regulation and RhoGTPase
activity, such as small Rho-GTPases RhoA and Rac1 activity
(15). LIMK1 positively controls
the expression and phosphorylation of cofilin 1, leading to
rearrangement of the cellular actin cytoskeleton, which serves
roles in the growth and motility inhibition of breast cancer cells
(12).
Cofilin is associated with cell differentiation
in vivo and outside factors can control cofilin through
relative cell signaling pathways, thus regulating the actin
cytoskeleton to induce differentiation (16). Despite this information, no data
have indicated an association between the cofilin 1 pathway and
leukemia cell differentiation. The present study aimed to clarify
whether downregulation and inactivation of cofilin 1 by DADS
induces differentiation. In addition, the mechanism underlying the
inhibitory effects on the proliferation, migration and invasion of
human leukemia HL-60 cells, and the mechanisms by which DADS
mediates cofilin 1 downregulation and inactivation were
investigated.
Materials and methods
Reagents
DADS was obtained from Sigma-Aldrich; Merck KGaA.
DADS was dissolved in 0.1% Tween-80 (catalog no. E7034;
Sigma-Aldrich) at 8 g/l and stored at −20°C. Matrigel was obtained
from BD Biosciences. Transwell chambers (8-µm) were provided
by Corning, Inc. CCK-8 was obtained from Dojindo Molecular
Technologies, Inc. The total RNA kit II (catalog no. R6934) was
purchased from Omega Biotek, Inc., the PrimeScript™ RT reagent kit
(catalog no. RR037A) was purchased from Takara Bio, Inc., the PCR
Optimization kit (catalog no. D2381) was purchased from Promega
Corporation, and the BeyoECL Plus Hypersensitive ECL
Chemiluminescence kit (catalog no. P0018S), BCA Protein assay kit
(catalog no. P0012) and the β-actin mouse monoclonal antibody
(catalog no. AF5001) were purchased from Beyotime Institute of
Biotechnology. Nitrotetrazolium Blue chloride (NBT; catalog no.
N6876) was purchased from Sigma-Aldrich; Merck KGaA. The primary
antibodies against LIMK1 (catalog no. BS2016), phosphorylated
(p)-LIMK1 (catalog no. BS4115) and p-cofilin 1 (catalog no. BS4716)
were purchased from Bioword Technology, Inc. The primary antibody
against Rac1 (catalog no. 10485-2-AP), horseradish
peroxidase-conjugated affinipure goat anti-mouse IgG (H+L)
secondary antibody (catalog no. SA00001-1) and horseradish
peroxidase-conjugated affinipure goat anti-Rabbit IgG (H+L)
secondary antibody (catalog no. SA00001-2) were purchased from
ProteinTech Group, Inc. The primary antibodies against cofilin 1
(catalog no. S2056), Rho-associated protein kinase 1 (ROCK1;
catalog no. 1953-1), CD11b (catalog no. 1936-1) and CD33 (catalog
no. 3807-1) were provided by Epitomics; Abcam. The cofilin 1 high
expression vector (pcDNA3.1-cofilin 1-IRES2-EGFP eukaryotic
expression plasmid), empty vector pcDNA3.1 (control), cofilin 1
knockdown vector [pcDNA™6.2-GW/EmGFPmiR cofilin 1-microRNA
(miRNA)-expressing plasmid) and empty vector pcDNA™6.2 (control)
were constructed by Invitrogen; Thermo Fisher Scientific, Inc.
Cell culture and establishment of cells
lines stably overexpressing and silencing cofilin 1
The human leukemia cell line HL-60 was obtained from
the Cancer Research Institute, Xiangya Medical College, Central
South University, Changsha, China. Cells were maintained in
RPMI-1640 complete medium (Gibco; Thermo Fisher Scientific, Inc.)
containing 10% fetal bovine serum (catalog no. 11011-8611; Tianhang
of Sijiqing Hangzhou Technologies), 100 U/ml penicillin and 100
U/ml streptomycin in a humidified incubator at 37°C and 5%
CO2. Cells were transfected with 0.12 µg/l
pcDNA3.1-cofilin 1-IRES2-EGFP eukaryotic expression plasmid to
overexpress cofilin, 0.12 µg/l empty vector pcDNA3.1
(control), 0.05 µg/l pcDNA™6.2-GW/EmGFPmiR cofilin
1-microRNA (miRNA)-expressing plasmid to knockdown cofilin 1 and
0.05 µg/l empty vector pcDNA™6.2 (control) using Attractene
Transfection Reagent (catalog no. 301005; Qiangen GmbH) to generate
a HL-60 cell line stably overexpressing cofilin 1 and a cell line
in which cofilin 1 was silenced (termed cofilin-miR). G418 (catalog
no. 10131035; Gibco; Thermo Fisher Scientific, Inc.) was used to
screen positive cell clones. After screening for 10-14 days, the
concentration of G418 was halved to maintain the screening, and was
used in experiments after cell proliferation. Reverse
transcription-semi-quantitative PCR (RT-semi-qPCR) and western
blotting were performed to confirm the establishment of cofilin 1
stably overexpressing and silenced cell lines after transfection
for 48 h.
Cell proliferation analysis
The effect of DADS or cofilin 1 on HL-60 cell
proliferation was measured using the CCK-8 assay. Briefly,
4×103 cells were treated with 0.1% Tween-80 (control) or
DADS (8 µM) at room temperature for 24, 48 and 72 h after
being transfected or not and then exposed to CCK-8 (10
µl/well) for 4 h. The optical density (OD) values were
measured at 490 nm, and the inhibitory rate = (1 - OD value of
experience group/OD value of control group) x 100%. Each assay was
performed with six replicates.
RT-semi-qPCR
The total RNA kit II was used to extract total RNA,
according to the manufacturer's protocol, and complementary DNA was
generated using the PrimeScript™ RT reagent kit. Each gene primer
was synthesized by Invitrogen; Thermo Fisher Scientific, Inc., and
their sequences were as follows: Cofilin 1 (120 bp) forward,
5′-CAAGAAGGCGGTGCTCT-3′ and reverse, 5′-ACAAAGGTGGCGTAGGG-3′; Rac1
(100 bp) forward, 5′-CCCTATCCTATCCGCAAACA-3′ and reverse,
5′-CGCACCTCAGGATACCACTT-3′; ROCK1 (113 bp) forward,
5′-AAAACCTTATTTGTGCCTTCC-3′ and reverse, 5′-CGTTTCCCAAGCCCACT-3′;
LIMK1 (138 bp) forward, 5′-GGGGCATCATCAAGAGCA-3′ and reverse,
5′-GAGGACTAGGGTGGTTCAG-3′; and β-actin (367 bp) forward,
5′-ACACTGTGCCCATCTACGAGGGG-3′ and reverse,
5′-ATGATGGAGTTGAAGGTAGTTTCGTGGAT-3′. The reaction conditions were
as follows: β-actin, 94°C, 5 min, 30 cycles (94°C, 30 sec; 55.0°C,
30 sec; 72°C, 60 sec), 72°C, 5 min; and for all other primers,
94°C, 5 min; 30 cycles (94°C, 30 sec; 54.5°C, 30 sec; 72°C, 60
sec), 72°C, 5 min. The semi-qPCR products were separated by 2%
agarose gel electrophoresis. The Bio-Rad gel imaging system
(Bio-Rad Laboratories, Inc.) and AlphaImager 2200 software (version
5.0; Alpha Innotech Corporation) were used to photograph, scan and
calculate the relative OD values to demonstrate the gene expression
abundances. β-actin expression was regarded as the internal control
for determining the relative OD value.
Western blot analysis
HL-60 cell lines were treated with or without DADS
(8 µM) at 37°C for 12, 24 and 48 h after being transfected
or not. Total protein was then extracted using lysis buffer
(catalog no. P0013; Beyotime Institute of Biotechnology) and 100
µg/ml PMSF (catalog no. ST505; Beyotime Institute of
Biotechnology, China), and quantified with a BCA protein assay kit.
Following electrophoresis, total protein (20-25 µg/lane) was
separated by 12% SDS-PAGE and transferred onto a PVDF membrane.
Primary antibodies, including anti-cofilin 1, anti-Rac1,
anti-ROCK1, anti-LIMK1, anti-p-cofilin 1, anti-p-T508-LIMK1 and
anti-β-actin, were used at 1:1,000, and incubated overnight at 4°C.
β-actin was employed as the loading control. The blots were washed
three times for 5 min in TBS-T (containing 1% Tween-20) and then
incubated with peroxidase-conjugated secondary antibody (1:1,000)
for 1 h at room temperature. Subsequently, the blots were washed
three times for 10 min in TBS-T and then developed using a BeyoECL
Plus Hypersensitive ECL Chemiluminescence kit. Alpha Imager 2200
(version 5.0; Alpha Innotech Corporation) was applied to evaluate
the ODs of the band densities.
NBT reduction assay
HL-60 cells were maintained at a logarithmic growth
rate and seeded at a density of 1×104 cells/well.
Following exposure to 8 µM DADS at 37°C for 72 h, cells were
collected by centrifugation at 201 × g at room temperature for 5
min. Differentiation of HL-60 cells was assayed by adding 200
µl NBT-TPA solution (2 mg/ml NBT and 0.24 mg/ml phorbol
12-myristate 13-acetate) in PBS to each well, incubation for 1 h at
37°C, and suspension in 0.4 ml cold 2 M HCl. Subsequently, the
formazan product was obtained by centrifugation at 700 × g at room
temperature for 10 min, and 200 µl DMSO was added to
dissolve the product. The absorbance of the solution was measured
at 570 nm.
Immunofluorescence of CD11b and CD33
Immunofluorescence analysis was performed to verify
the subcellular localization of the general myeloid differentiation
markers CD11b and CD33. HL-60 cells of each group were collected by
centrifugation at 201 × g at room temperature for 5 min. Cell
suspension was added to cover the slides and dried at room
temperature for 30 min. Subsequently, the cells were fixed with 4%
polyoxymethylene at room temperature for 30 min, rinsed with 0.5%
Triton X-100 in PBS for 15 min, and then blocked with goat serum at
room temperature for 30 min. The CD11b (1:100) and CD33 (1:100)
antibodies were incubated with the cells at 4°C overnight, followed
by 5-min washes in PBS three times. FITC-conjugated anti-rabbit IgG
(catalog no. ab6717; Abcam; 1:1,000) at room temperature for 1 h
was applied to assess CD11b and CD33. Images were obtained using an
inverted fluorescence microscope (Olympus Corporation;
magnification ×400) and assessed with PearlScope software (version
1.0.0.1202).
Cell migration and invasion assays
Cell migration and invasion were assessed as
previously described (17).
Invasion assays were performed using Transwell assays. For the
migration assay, cells were resuspended in serum-free RPMI-1640
medium and 100 µl cell suspension (1.0×106 cells)
was seeded into the upper chambers. RPMI-1640 (500 µl)
containing 10% FBS was added to the lower chambers. After
transfected and untransfected cells were exposed to 8 µM
DADS at 37°C for 24 h, hematoxylin was used to stain the cells at
room temperature for 30 min on the lower surface of the filter. The
cells were observed under an inverted microscope (Olympus
Corporation; magnification, ×400). The invasion assay protocol was
the same as that of the migration assay except that the upper
chambers were first coated with 1 mg/ml Matrigel. Invasion and
migration rates are presented as ratios between DADS-treated and
control group values.
Statistical analysis
All results are presented as the mean ± standard
deviation for three independent experiments. Comparisons between
groups were made by one-way ANOVA with Fisher's Least Significant
Difference for three groups and Tukey's test for >3 groups.
Statistical analyses were performed using SPSS 13.0 software (SPSS,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
Effect of DADS on the differentiation,
migration and invasion of HL-60 cells
Following incubation with 8 µM DADS for
different amounts of time, the proliferative ability of the human
leukemic cell line HL-60 was assayed by cell proliferation
analysis. Time-dependent cytotoxicity was determined after 8
µM DADS treatment, and the OD value of the treated cells was
0.311±0.012 at 24 h, 0.362±0.011 at 48 h and 0.389±0.020 at 72 h,
which were significantly lower than those of 0.540±0.009 at 24 h,
0.691±0.014 at 48 h and 0.791±0.019 at 72 h in the untreated
groups, respectively (Table
I).
 | Table IEffect of DADS on the proliferation
ability of HL-60 cells. |
Table I
Effect of DADS on the proliferation
ability of HL-60 cells.
| Time, h | Optical density at
490 nm
| P-value |
|---|
| Untreated
cells | DADS-treated
cells |
|---|
| 24 | 0.540±0.009 | 0.311±0.012 | 0.002 |
| 48 | 0.691±0.014 | 0.362±0.011 | <0.001 |
| 78 | 0.791±0.019 | 0.000±0.000 | <0.001 |
As presented in Table
II, the NBT reduction ability of the untreated HL-60 cells was
0.419±0.010 at 24 h, 0.446±0.016 at 48 h and 0.504±0.026 at 72 h,
and following treatment with 8 µM DADS, the NBT reduction
ability increased to 0.472±0.023 at 24 h (P=0.003), 0.536±0.006 at
48 h (P=0.001) and 0.620±0.013 at 72 h (P<0.001).
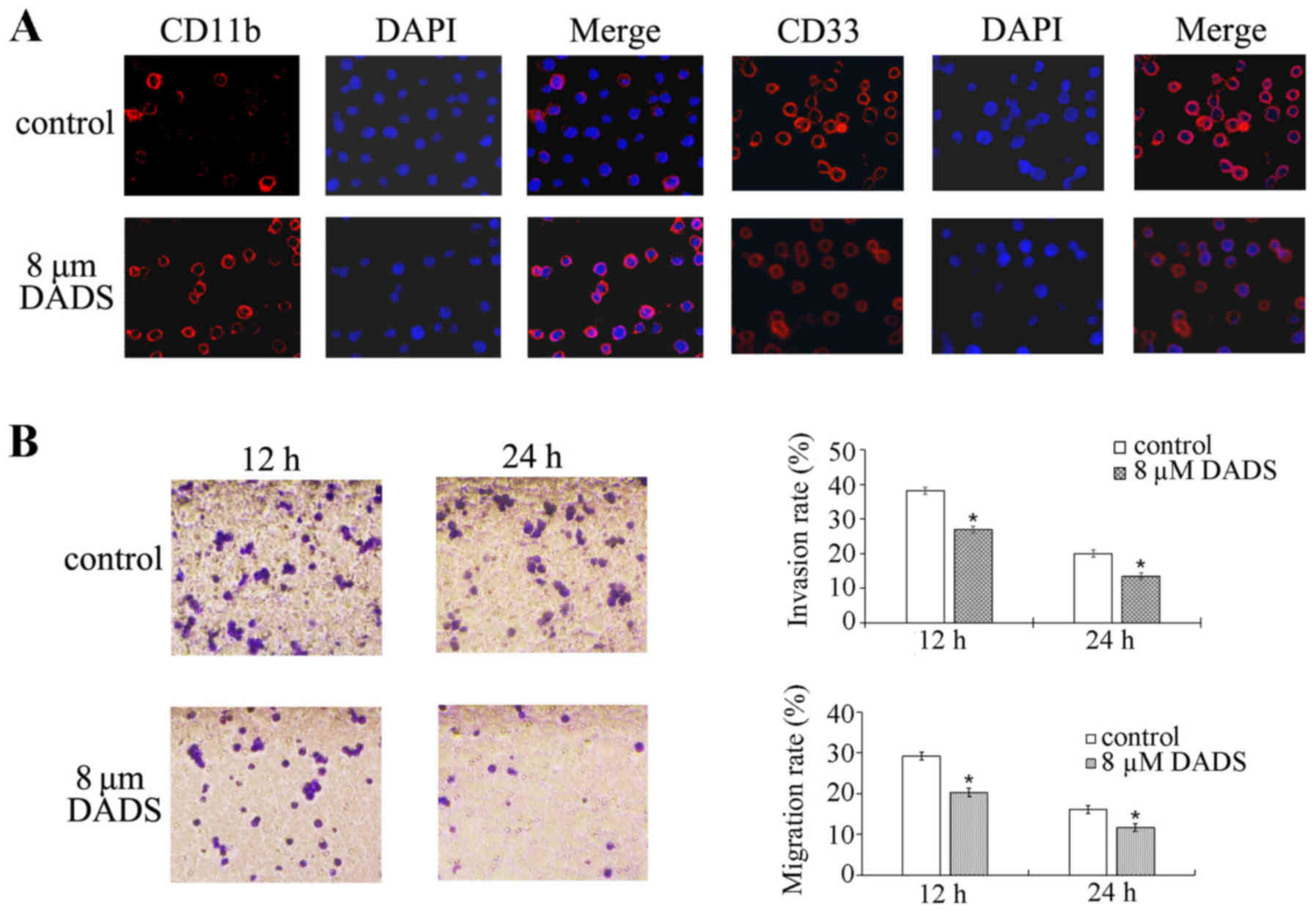
Immunofluorescence experiments demonstrated that the expression of
CD11b was markedly increased, while CD33 was markedly decreased, in
the DADS-treated group compared with the untreated group (Fig. 1A). These results suggest that DADS
can induce HL-60 cell differentiation.
 | Table IIEffect of DADS on the reduction
ability of HL-60 cells. |
Table II
Effect of DADS on the reduction
ability of HL-60 cells.
| Time, h | NBT absorbance at
570 nm
| P-value |
|---|
| Untreated
cells | DADS-treated
cells |
|---|
| 24 | 0.419±0.010 | 0.472±0.023 | 0.003 |
| 48 | 0.446±0.016 | 0.536±0.006 | 0.001 |
| 72 | 0.504±0.026 | 0.620±0.013 | <0.001 |
Cells were exposed to 8 µM DADS for 12 and 24
h, and the effects of DADS on the migration and invasion of the
leukemia HL-60 cell line were detected using Transwell experiments.
As shown in Fig. 1B, the migration
and invasion abilities of HL-60 cells were decreased by 8 µM
DADS treatment for 12 and 24 h compared with the control cells
(migration at 12 h, P=0.001; migration at 24 h, P<0.001;
invasion at 12 h, P<0.001; invasion at 24 h, P<0.001). These
data demonstrate that DADS can inhibit the migration and invasion
of HL-60 cells.
DADS downregulates
Rac1/ROCK1/LIMK1/cofilin 1 signaling in HL-60 cells
Cofilin is reportedly associated with cell
differentiation (16); however, no
evidence of an association between the cofilin 1 pathway and
leukemia cell differentiation exists. Therefore, the present
investigated whether DADS affects the expression and activation of
cofilin 1 in leukemia HL-60 cells. As presented in Fig. 2, following treatment with 8
µM DADS for 12, 24 and 48 h, the mRNA and protein expression
levels of cofilin 1 in HL-60 cells were significantly inhibited in
a time-dependent manner. Similarly, the mRNA and protein expression
levels of Rac1, Rock1 and LIMK1 were also significantly lower in
HL-60 cells following exposure to DADS for 12, 24 or 48 h.
Additionally, both the phosphorylated/total cofilin 1 protein ratio
and the phosphorylated LIMK1/total LIMK1 protein ratio
significantly decreased at 12 h, 24 and 48 h after 8 µM DADS
treatment compared with the control.
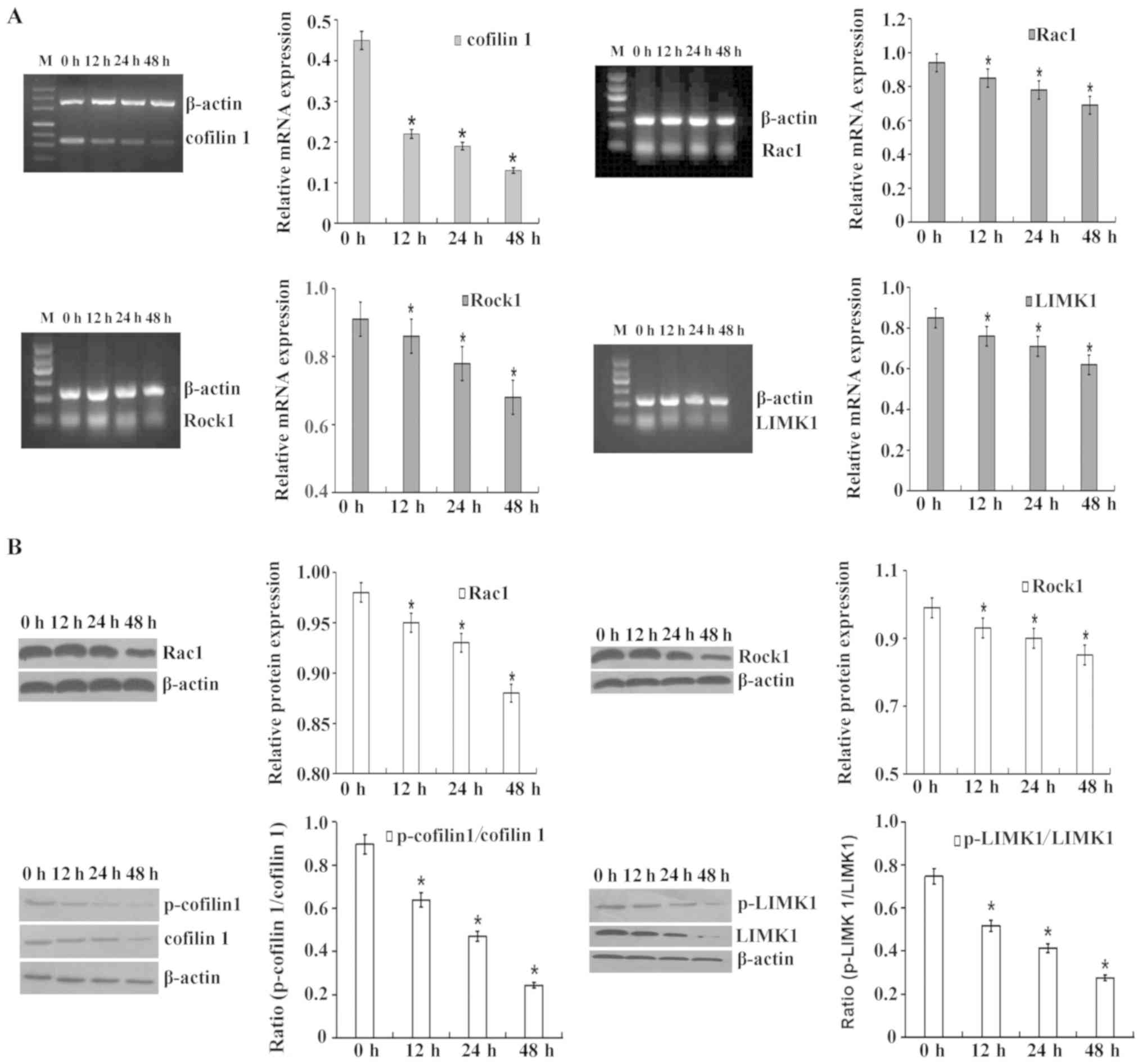 | Figure 2Inhibitory effect of DADS on
Rac1/ROCK1/LIMK1/cofilin 1 signaling in HL-60 cells. The cells were
treated with 8 µM DADS for the indicated amounts of time.
(A) Reverse transcription-semi-quantitative PCR was performed to
determine the mRNA expression levels of cofilin 1, Rac1, ROCK1 and
LIMK1. β-actin was used as an internal control for normalization.
(B) Western blotting was performed to determine cofilin 1,
p-cofilin 1, Rac1, ROCK1, LIMK1 and p-LIMK1 protein levels. β-actin
was used as a loading control. The relative fold changes in mRNA or
protein levels were compared with β-actin controls. The results are
presented as the mean ± standard error of the mean from three
independent experiments. *P<0.05 vs. the control (0 h
group). DADS, diallyl disulfide; ROCK1, Rho-associated protein
kinase 1; LIMK1, LIM domain kinase 1, p-, phosphorylated. |
Effect of DADS on the proliferation,
differentiation and invasion of cofilin 1-silenced HL-60 cells
Cell proliferation analysis indicated that the
proliferation inhibitory effect in cofilin 1-silenced cells
(cofilin-miR) was significantly enhanced by treatment with 8
µM DADS for 12, 24 and 48 h (Fig. 3A). However, 8 µM DADS could
significantly increase the NBT reduction ability in cofilin 1-miR
cells, demonstrating that DADS cooperates with cofilin 1 silencing
to affect the differentiation of HL-60 cells (Fig. 3B).
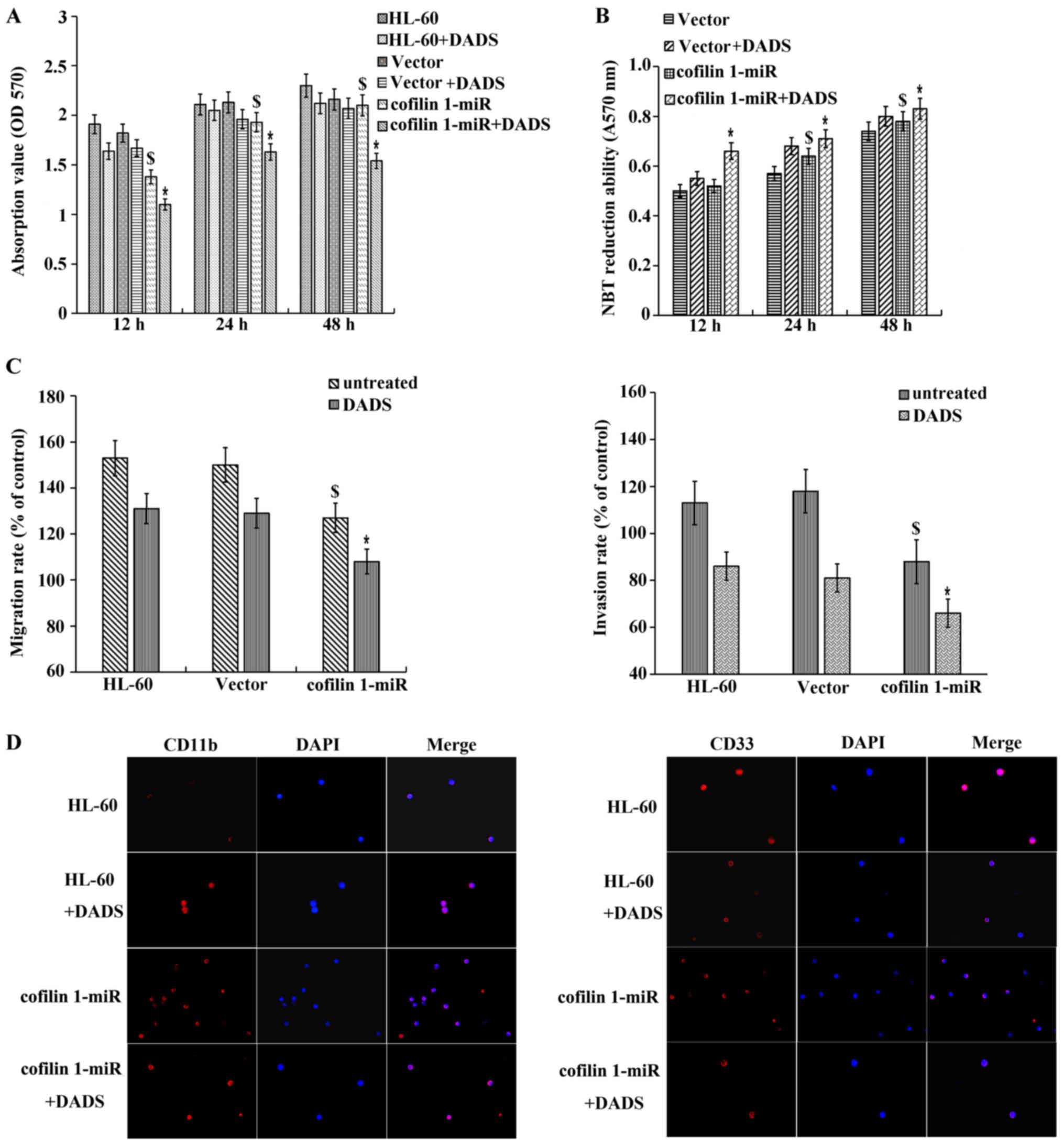 | Figure 3Effects of DADS and cofilin 1 miRNA
expression plasmid transfection on the proliferation,
differentiation and invasion of HL-60 cells. (A) Effect of DADS and
cofilin 1 miRNA expression plasmid transfection on the viability of
HL-60 cells. In total, 3×104 cells were treated with 8
µM DADS for 48 h, and the viable cells were analyzed for
cell proliferation. (B) Effect of DADS and cofilin 1 miRNA
expression plasmid transfection on NBT reduction in HL-60 cells.
HL-60 cells were treated with 8 µM DADS for 12, 24 and 48 h.
The differentiation of HL-60 cells was identified by the decrease
in NBT absorbance at 570 nm. $P<0.05 vs. the
untreated HL-60 group; *P<0.05 vs. the untreated
cofilin 1-miR group. (C) Effects of DADS and cofilin 1 miRNA
expression plasmid transfection on the migration and invasion of
HL-60 cells. Briefly, 8 µM DADS was added to HL-60 cells and
cells transfected with vector or a cofilin 1 miRNA expression
plasmid for 24 h. Cell migration and invasion were determined using
Transwell migration and invasion assays, respectively. The
migration rate is presented as the ratio of migrated cells between
treated and untreated HL-60 cells. The invasion rate is presented
as the ratio of the mean cell numbers between treated and untreated
cells. *P<0.05 vs. the untreated cofilin 1-miR group.
$P<0.05 vs. HL-60 cells (D) Expression of the cell
surface differentiation markers CD11b and CD33 following DADS
treatment and cofilin 1 miRNA expression plasmid transfection.
Magnification, ×200. CD11b and CD33 were identified by
immunofluorescence (red, left panel). The DNA-intercalating dye
DAPI was used to identify cell nuclei (blue, center panel). The
right panel displays a merged image to highlight the nuclear pool
of CD11b and CD33. Data are presented at the mean ± standard error
or the mean from three independent experiments. DADS, diallyl
disulfide; miRNA, microRNA; NBT, nitroblue tetrazolium; OD, optical
density; cofilin 1-miR, cells transfected with cofilin 1 miRNA. |
As shown in Fig.
3C, the numbers of cancer cells that migrated and invaded in
the Transwell assay were significantly decreased following
silencing of cofilin 1 compared with the control vector cells.
Furthermore, DADS could significantly strengthen this inhibitory
effect, suggesting that DADS negatively effects the invasive
ability of HL-60 cells via the downregulation of cofilin 1
expression.
Silencing of cofilin 1 enhanced CD11b expression and
inhibited CD33 expression. In addition, the expression of CD11b
increased, while the expression of CD33 decreased in the
DADS-treated cofilin 1-silenced cells compared with the untreated
cofilin 1-miR cells (Fig. 3D).
Effects of DADS on the Rac1-ROCK1-LIMK1
signaling pathway in cofilin 1-silenced HL-60 cells
To clarify the molecular mechanisms by which DADS
induces differentiation and exerts inhibitory effects, the present
study assessed whether silencing the cofilin 1 gene could interfere
with the effects of DADS on the Rac1-ROCK1-LIMK1 signaling pathway.
Silencing cofilin 1 significantly downregulated the protein
expression of Rac1, ROCK1 and LIMK1 compared with control cells,
and significantly increased the inhibitory effect of DADS on the
protein expression of cofilin 1, Rac1, ROCK1 and LIMK1 compared
with in the untreated cells (Fig.
4).
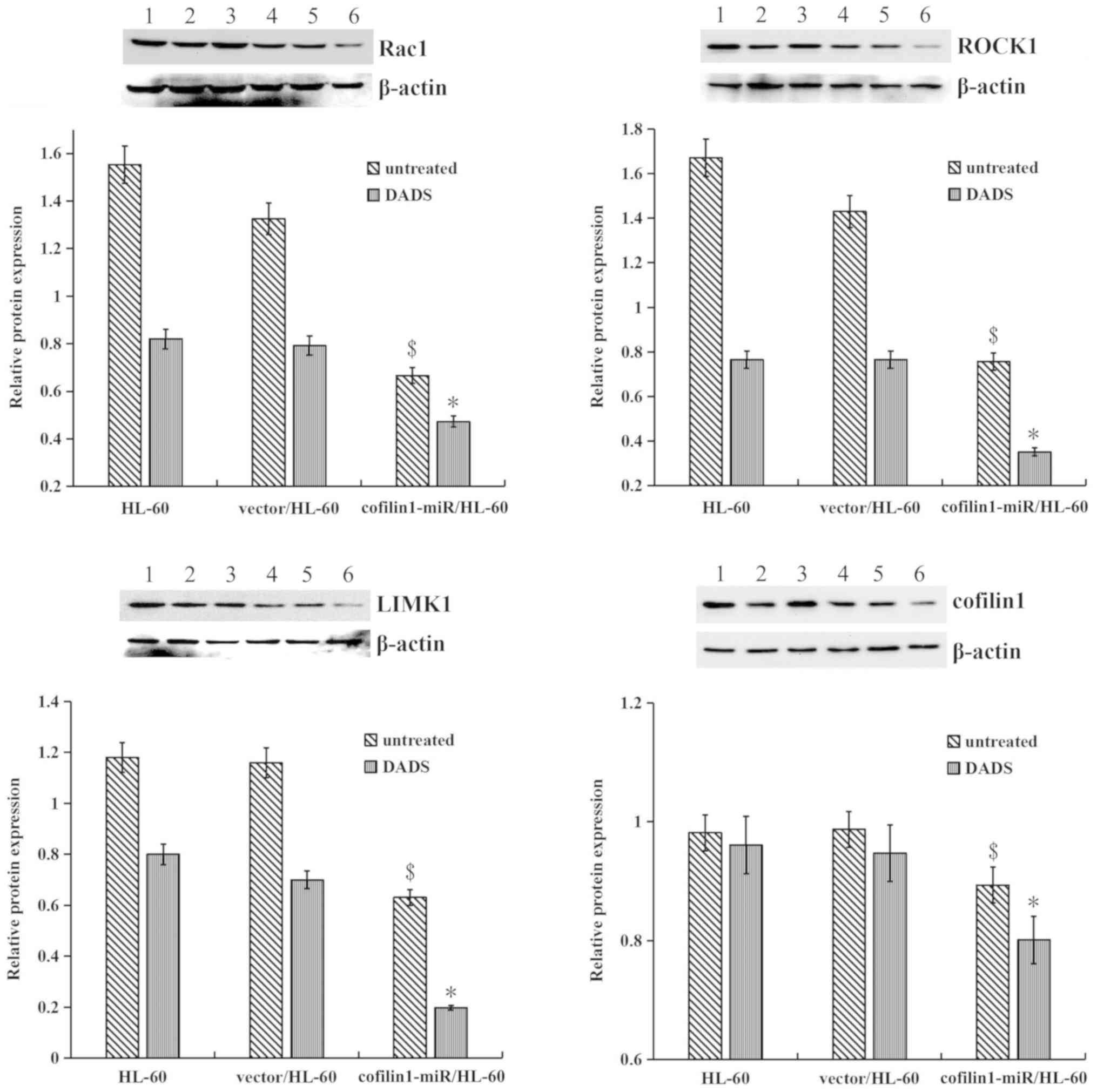 | Figure 4Effects of DADS on the
Rac1-ROCK1-LIMK1 signaling pathway in cofilin 1 gene-silenced HL-60
cells. Western blotting was used to detect Rac1, ROCK1, LIMK1 and
cofilin 1 protein levels. β-actin served as a loading control. The
relative fold changes in protein levels compared with the β-actin
control were analyzed. 1, untreated HL-60 cells; 2, HL-60 cells
treated with DADS; 3, vector/HL-60 cells; 4, vector/HL-60 cells
treated with DADS; 5, cofilin 1-miR/HL-60 cells; and 6, cofilin
1-miR/HL-60 cells treated with DADS. Data are presented at the mean
± standard error or the mean from three independent experiments.
$P<0.05 vs. the untreated HL-60 group;
*P<0.05 vs. the untreated cofilin 1-silenced/HL-60
group. DADS, diallyl disulfide; ROCK1, Rho-associated protein
kinase 1; LIMK1, LIM domain kinase 1; cofilin 1-miR, cells
transfection with cofilin 1 miRNA overexpressing vector. |
Effects of DADS on the proliferation,
differentiation and invasion of cofilin 1-overexpressing HL-60
cells
Next, it was observed whether cofilin 1
overexpression influences the effects of DADS on proliferation,
differentiation and invasion. As presented in Fig. 5A, following 24 h of DADS treatment,
the proliferative abilities of the cells in the untransfected,
negative control and high cofilin 1 expression groups were
significantly lower compared with that in the untreated groups. The
difference of the proliferative abilities between the
cofilin-overexpression group and untransfected group was
significant. The proliferation inhibition rate of DADS was
(37.42±3.67%) in the cofilin overexpression group, which was
significantly lower than that in the untransfected group
(44.78±4.19%) and the vector-transfected group (44.72±4.63%)
(Table III). This indicates that
cofilin 1 overexpression could reduce the inhibitory effect of DADS
on the proliferation of HL-60 cells.
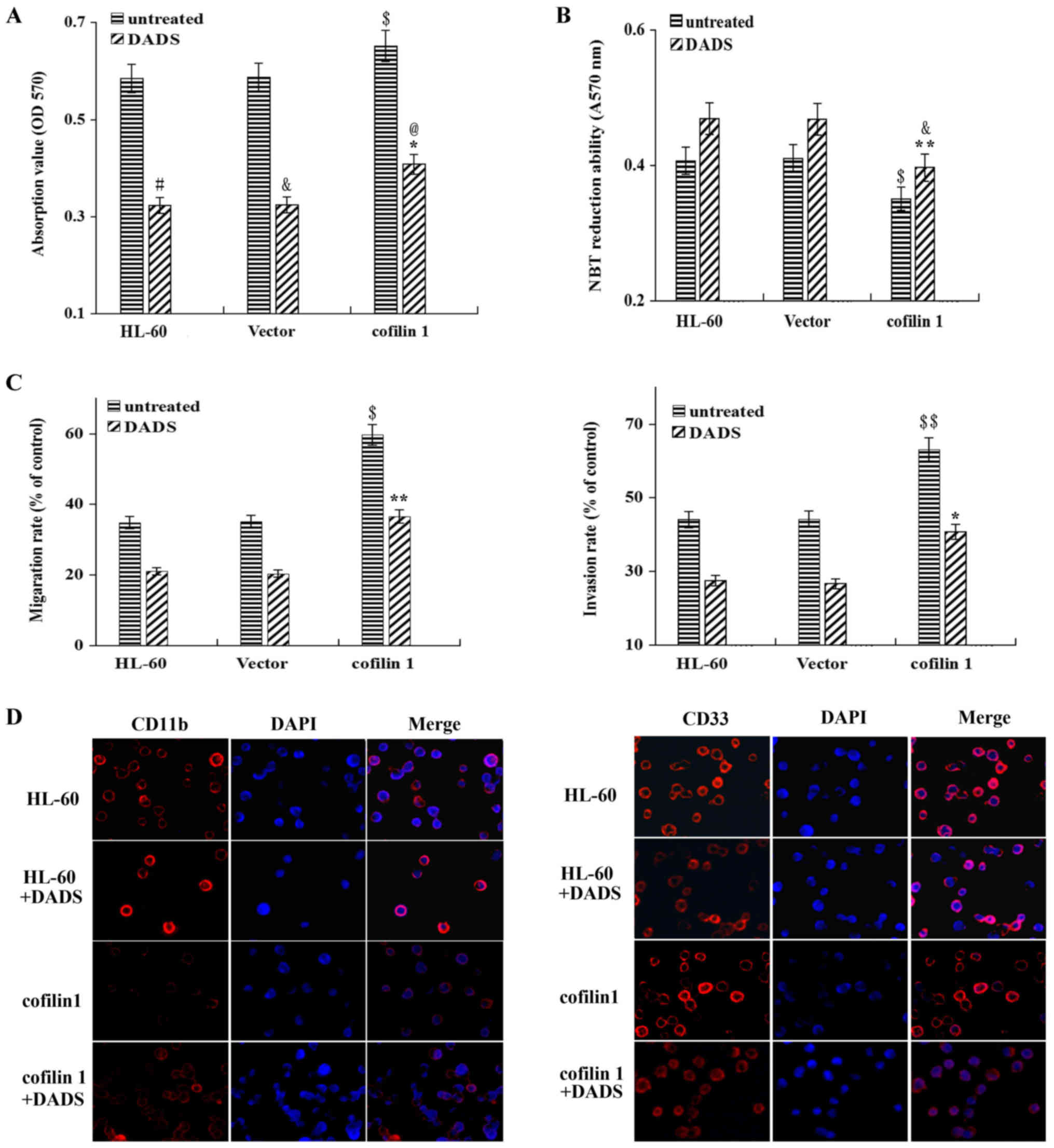 | Figure 5Effect of DADS on the proliferation,
differentiation and invasion of cofilin 1-transfected HL-60 cells.
(A) Effect of DADS on the viability of HL-60 cells,
vector-transfected cells and cofilin 1-transfected cells. In total,
3×104 cells were treated with 8 µM DADS for 48 h,
and the viable cells were analyzed for cell proliferation.
#P<0.01 vs. the untreated HL-60 group;
&P<0.01 vs. the untreated vector group;
$P<0.001 vs. the untreated HL-60 group and the
untreated vector-transfected group; *P<0.01 vs. the
untreated cofilin 1 group; @P<0.001 vs. the
untransfected cells treated with DADS and the vector-transfected
cells treated with DADS. (B) Effect of DADS on NBT reduction in
HL-60 cells, vector-transfected cells and cofilin 1-transfected
cells. HL-60 cells were treated with 8 µM DADS for 48 h. The
differentiation of HL-60 cells was determined by the decrease in
NBT absorbance at 570 nm. $P<0.05 vs. the untreated
HL-60 group; &P<0.05 vs. the HL-60 group with
DADS treatment; **P<0.01 vs. the vector group with
DADS treatment. (C) Effects of DADS on the migration and invasion
of HL-60 cells. Briefly, 8 µM DADS was added to HL-60 cells,
vector-transfected cells and cofilin 1-transfected cells for 24 h.
Cell migration and invasion were examined using Transwell migration
and invasion assays, respectively. The migration rate was
calculated as the ratio of migrated cells between DADS-treated and
untreated HL-60 cells. The invasion rate was estimated by the ratio
of mean cell numbers between treated and untreated cells.
$P<0.05, $$P<0.01 vs. the untreated HL-60 group;
*P<0.05, **P<0.01 vs. the untreated
cofilin 1 group. (D) Expression of the cell surface differentiation
markers CD11b and CD33 after DADS treatment and cofilin 1
transfection. Magnification, ×200. CD11b and CD33 were identified
by immunofluorescence (red, left panel). The DNA-intercalating dye
DAPI was used to identify cell nuclei (blue, center panel). The
right panel displays a merged image to highlight the nuclear pool
of CD11b and CD33. DADS, diallyl disulfide; OD, optical density;
NBT, nitroblue tetrazolium. |
 | Table IIIEffect of DADS on the proliferation
ability of HL-60 cells, vector-transfected cells and cofilin
1-transfected cells. |
Table III
Effect of DADS on the proliferation
ability of HL-60 cells, vector-transfected cells and cofilin
1-transfected cells.
| Group | Optical density
value
| P-value | Inhibitory rate,
% |
|---|
| Untreated
cells | DADS-treated
cells |
|---|
| Untransfected HL-60
cells | 0.585±0.0019 | 0.323±0.018a | 0.0069 | 44.78±4.19 |
| Vector-transfected
HL-60 cells | 0.587±0.021 | 0.324±0.017b | 0.0083 | 44.72±4.63 |
| Cofilin
1-transfected HL60 cells | 0.652±0.020c | 0.408±0.024d,e | 0.0011 | 37.42±3.67 |
NBT experiments demonstrated that cofilin 1
overexpression could significantly inhibit the reducing ability of
HL-60 cells. Furthermore, cofilin 1 overexpression significantly
reduced the DADS-induced cell reducing ability, compared with the
HL-60 cells treated with DADS (P=0.0133) and the vector group
treated with DADS (P=0.0049; Fig.
5B). Transwell migration and invasion experiments indicated
that overexpression of cofilin 1 significantly increased the
migration and invasion abilities of HL-60 cells (migration,
P=0.011; invasion, P=0.005) and significantly decreased the
DADS-induced migration and invasion inhibition (migration, P=0.004;
invasion, P=0.010; Fig. 5C).
Immunofluorescence experiments demonstrated that overexpression of
cofilin 1 decreased the expression of CD11b, while the expression
of CD33 had no obvious change, compared with the untreated HL-60
cells (Fig. 5D). CD11b expression
was increased and CD33 expression was decreased in the DADS-treated
cofilin 1-overexpressing cells compared with the cofilin
1-overexpressing cells without DADS treatment (Fig. 5D).
Effects of DADS on the Rac1-ROCK1-LIMK1
signaling pathway in cofilin 1-overexpressing HL-60 cells
After determining whether knockdown of cofilin 1
could enhance the suppressive effect of DADS on the
Rac1-ROCK1-LIMK1 signaling pathway, it was investigated whether
cofilin 1 over-expression serves a role in the effect of DADS on
HL-60 cells. RT-semi-qPCR and western blot results demonstrated
that DADS significantly lowered the mRNA and protein expression
levels of cofilin 1, Rac1, ROCK1 and LIMK1, and the ratios of
p-cofilin/cofilin 1 and p-LIMK1/LIMK1 in cofilin 1-overex-pressing
cells compared with those without DADS treatment (Fig. 6).
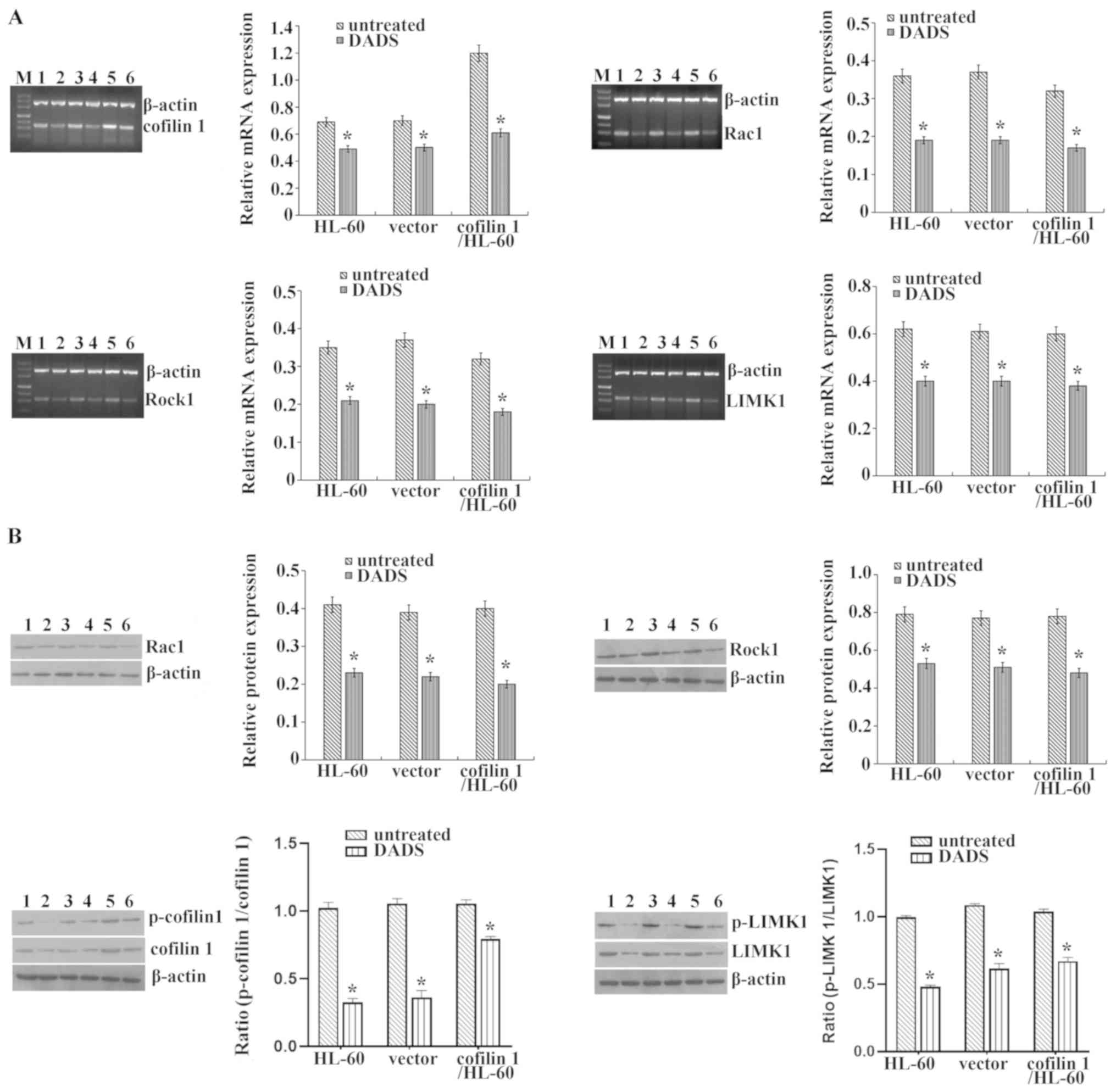 | Figure 6Effects of DADS on the
Rac1-ROCK1-LIMK1 signaling pathway in cofilin 1-transfected HL-60
cells. (A) Reverse transcription-semi-quantitative PCR was used to
determine the mRNA expression levels of Rac1, ROCK1, LIMK1 and
cofilin 1. β-actin was used as an internal control for
normalization. (B) Western blotting was employed to detect cofilin
1, p-cofilin 1, Rac1, ROCK1, LIMK1 and p-LIMK1 protein levels.
β-actin was regarded as a loading control. The relative fold
changes in mRNA or protein levels were compared with β-actin
controls. M, Maker; 1, untreated HL-60 cells; 2, HL-60 cells
treated with DADS; 3, vector/HL-60 cells; 4, vector/HL-60 cells
treated with DADS; 5, cofilin 1/HL-60 cells; and 6, cofilin 1/HL-60
cells treated with DADS. Data are presented at the mean ± standard
error or the mean from three independent experiments.
*P<0.05 vs. the untreated HL-60 group. DADS, diallyl
disulfide; ROCK1, Rho-associated protein kinase 1; LIMK1, LIM
domain kinase 1; p-, phosphorylated. |
Discussion
DADS, the main anticancer component in garlic, can
suppress the proliferation of numerous types of cancer cells, such
as leukemia, colon cancer and gastric cancer cells (4,5,18,19).
It has been suggested that DADS induces the differentiation of the
human gastric cancer cell line MGC803 (20) and the leukemia cell line HL-60
(5). Previously, we demonstrated
that 8 µM DADS could induce the differentiation of human
leukemia HL-60 cells to a granulocytic lineage, which is similar to
the effects of 1 µM all-trans retinoic acid (ATRA) (5). In the present study, DADS could
inhibit the proliferation, migration and invasion of HL-60 cells
and induce their differentiation, similar to ATRA. These results
suggest that DADS may be a potential compound for inducing
differentiation. However, the precise molecular mechanisms
underlying these anti-metastatic effects of DADS are not completely
understood.
In a previous study, DADS was shown to regulate the
expression of cofilin 1 (5). The
present study verified that DADS gradually decreased the mRNA and
protein expression, as well as the phosphorylation of cofilin 1 in
HL-60 cells in a time-dependent manner. Cofilin-1, an actin
depolymerizing factor (ADF)/cofilin superfamily member, has been
shown to be highly expressed in a number of cancer types and is
associated with proliferation, migration, invasion,
differentiation, metastasis and poor prognosis (16,21).
The expression level of cofilin and its subcellular distribution
play important roles in the high migratory ability of tumor cells
(22). The high expression of
cofilin-1 in malignant cells can enhance the speed of cell
migration, while silencing cofilin-1 can delay cancer cell
metastasis (15). Cofilin-1 can
depolymerize actin filaments by binding directly to actin and make
the actin filaments turn over quickly (23,24),
thus controlling cell movement, which serves an important role in
the proliferation and migration of tumor cells (11). Cofilin 1 is involved in tumor
invasion, metastasis and prognosis by regulating actin
reorganization and pseudopodia formation; sustained activation of
cofilin 1 promotes the formation and migration of filamentous
pseudopodia in prostate cancer cells and enhances the metastatic
ability of prostate cancer in vivo (25). O-(link)-N-acetylation glycosylation
modification of the cofilin 1 Ser108 site facilitates its correct
localization in the invasive pseudopod and promotes the invasion of
cancer cells (26). The high
expression and phosphorylation of cofilin 1 serves an important
role in the occurrence and development of cancer (27). The suppression of cofilin-1 and the
translocation of cofilin 1 from the cytoplasm to mitochondria can
induce cancer cell apoptosis (28,29).
High cofilin-1 expression is associated with cisplatin
chemoresistance in carcinomas (30). Serum cofilin-1 protein expression
is increased in carcinomas and may be a promising serum biomarker
for prognosis (31,32). Notably, the present study found
that silencing cofilin 1 by miRNA could markedly promote the
DADS-induced differentiation and inhibitory effect on the
proliferation and invasion of HL-60 cells. However, overexpression
of cofilin 1 significantly suppressed these effects induced by
DADS. These data provide evidence that cofilin 1 is involved in
DADS-induced differentiation and growth inhibition in human
leukemia HL-60 cells.
Our previous study demonstrated that DADS reduces
LIMK1 expression in gastric cancer MGC-803 cells (19) and inhibits the migration and
invasion capabilities of colon cancer SW480 cells via
downregulation of the Rac1-ROCK1/PAK1-LIMK1-ADF/cofilin signaling
pathway (33). Thereafter, it was
suspected that the Rac1-ROCK1/LIMK1 pathway is involved in
regulating cofilin expression and activation, and consequently,
differentiation induction in HL-60 cells. In the present study, it
was identified that DADS can suppress the mRNA and protein
expression of Rac1, ROCK1 and LIMK1, as well as the phosphorylation
of LIMK1 in HL-60 cells in a time-dependent manner. Silencing
cofilin 1 by miRNA decreased the protein expression of Rac1, ROCK1
and LIMK1, and increased the inhibitory effect of DADS on cofilin 1
mRNA expression and the protein expression of cofilin 1, Rac1,
ROCK1 and LIMK1. By contrast, high expression of cofilin 1 reduced
the inhibitory effect of DADS on these molecules. Cofilin is
regulated by various upstream signals, predominantly RhoGTP enzyme
family members, which are involved in tumor occurrence and
development. RhoGTP enzyme family members, including Rho, Rac and
Cdc42, are closely associated with cytoskeleton reorganization and
serve an important role in cell motility, migration and invasion
(34). The degree of Rac signaling
plays a crucial role in the balance between differentiation and
proliferation; cellular Rac1 is indispensable for differentiation
(35), and RhoA/ROCK signaling
plays an important role in differentiation induction (36). The activation of Rho and Rac1 can
phosphorylate the kinase ROCK and activate LIMK1, as Rac can
activate LIMK1, which induces cofilin 1 phosphorylation at Ser3 and
thus regulates the actin cytoskeleton; this process indicates the
formation of the Rac1-ROCK1/LIMK1-cofilin signaling pathway by
regulating tumor cell migration and invasion (37,38).
The inhibition of the ROCK/PTEN pathway and cofilin-1 expression is
involved in the induction of cancer cell apoptosis (39). Cofilin cuts fibrous type F-actin
and accelerates free actin polymerization, and the phosphorylated
state of cofilin 1 is regulated by LIMK1 (40). In addition, LIMK1-mediated cofilin
phosphorylation has important effects on tumorigenesis, matrix
adhesion, transfer speed and direction of tumor cell invasion
(41). In the present study, the
protein expression levels of Rac1, ROCK1 and LIMK1 increased in
cofilin 1-silenced HL-60 cells, while the expressions of these
molecules were not significantly altered in cofilin
1-overexpressing HL-60 cells. This indicates that there may be a
signal interaction centered on cofilin 1, which participates in
other signal pathways, such as the Rac1-WAVE2-Arp2/3 pathway
(42), when activated, and this
conclusion needs to be confirmed by further research in the future.
However, DADS can regulate the expression of Rac1, Rock1, LIMK1 and
cofilin 1, whether cofilin 1 is overexpressed or silenced.
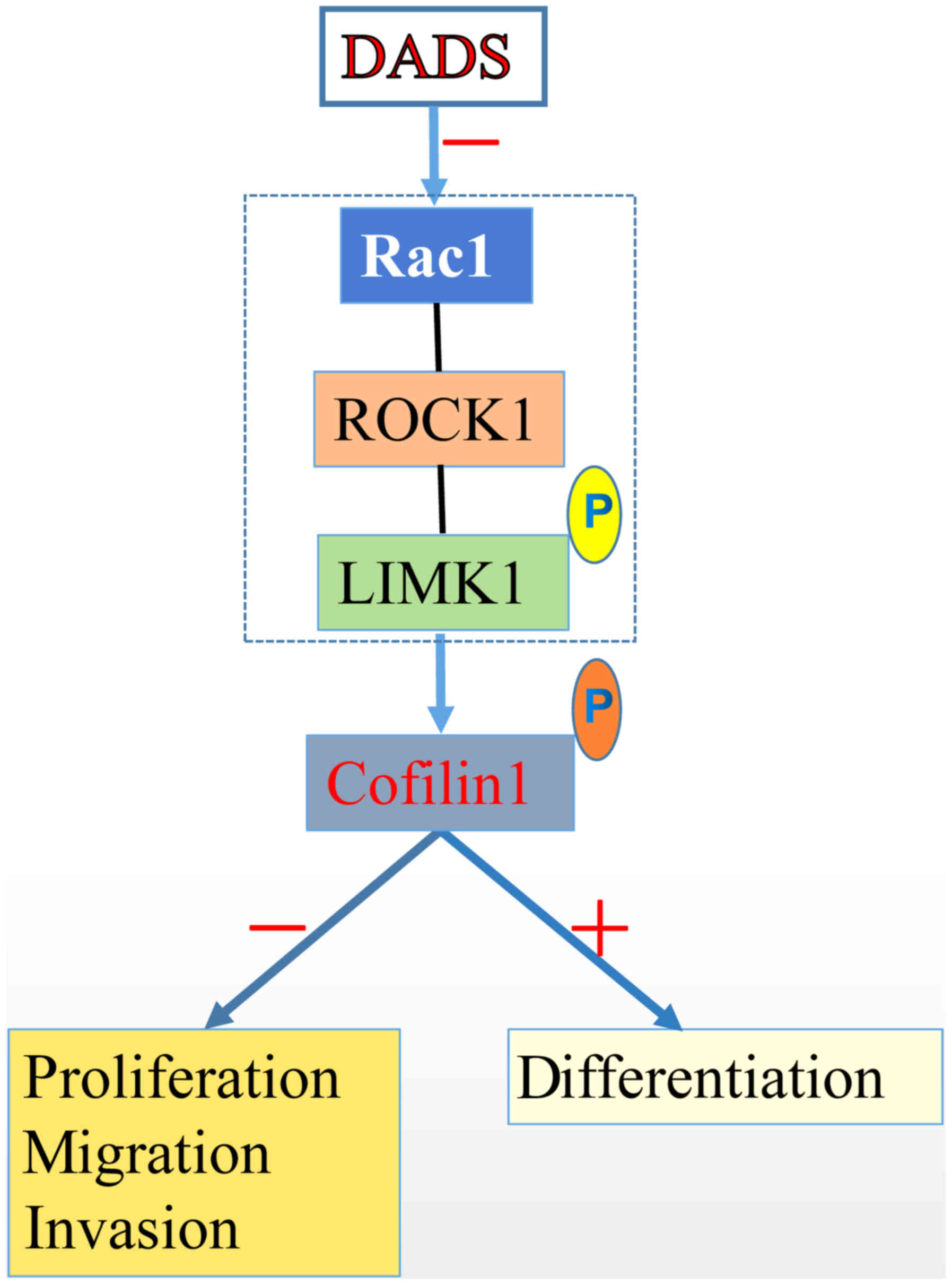
In summary, DADS could decrease cofilin 1 expression
and suppress its phosphorylation by negatively regulating the
Rac1-ROCK1-LIMK1 signaling pathway and lead to the inhibited
proliferation and induced differentiation of leukemia HL-60 cells
(Fig. 7). The present study helps
to identify targets for inducing leukemia differentiation and
provides a theoretical basis for leukemia differentiation induction
therapy.
Funding
This study was performed using equipment purchased
with funding from the construct program of the key discipline in
Hunan Province, China (Basic Medicine Sciences at University of
South China) [grant no. (2011)76]. This work was supported by the
National Natural Science Foundation of China (grant nos. 81100375,
31201027 and 81400117), the Patency Foundation of Innovation
Platform of Hunan Provincial University of China (grant no. 11K057)
and the Natural Science Foundation of Hunan Province (grant no.
2015JJ4042).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the corresponding author upon
reasonable request.
Authors' contributions
HL and QS conceived the idea and designed the study.
JH, XJ, YL and YJ performed all of the experiments. FL and HX
analyzed the data. HT, XZ and LY performed the statistical
analysis. HL wrote the manuscript. All authors have reviewed the
manuscript and read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
DADS
|
diallyl disulfide
|
|
NBT
|
nitroblue tetrazolium
|
|
BCA
|
bicinchoninic acid
|
|
ROCK1
|
Rho-associated protein kinase 1
|
|
RT-semi-qPCR
|
reverse
transcription-semi-quantitative polymerase chain reaction
|
|
OD
|
optical density
|
|
ATRA
|
all-trans retinoic acid
|
|
ADF
|
actin depolymerizing factor
|
|
LIMK1
|
LIM domain kinase 1
|
Acknowledgments
Not applicable.
References
|
1
|
Mahmud H, ter Elst A, Scherpen FJG, de
Boer TM, Kampen KR, de Haas V, Guryev V, Peppelenbosch MM, Kornblau
SM and de Bont ESJM: Peptide microarray of pediatric acute myeloid
leukemia is related to relapse and reveals involvement of DNA
damage response and repair. Oncotarget. 10:4679–4690.
2019.PubMed/NCBI
|
|
2
|
Surveillance, Epidemiology, and End
Results (SEER) Program, SEER*Stat Database: Incidence - SEER 9 Regs
Research Data, Nov 2017 Sub (1973-2015) - Linked To County
Attributes - Total U.S., 1969-2016 Counties. National Cancer
Institute, DCCPS, Surveillance Research Program. released April
2018, based on the November 2017 submission. https://www.seer.cancer.gov
|
|
3
|
Yi L and Su Q: Molecular mechanisms for
the anti-cancer effects of diallyl disulfide. Food Chem Toxicol.
57:362–370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao J, Huang WG, He J, Tan H, Liao QJ and
Su Q: Diallyl disulfide suppresses growth of HL-60 cell through
increasing histone acetylation and p21WAF1 expression in vivo and
in vitro. Acta Pharmacol Sin. 27:1459–1466. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ling H, He J, Tan H, Yi L, Liu F, Ji X, Wu
Y, Hu H, Zeng X, Ai X, et al: Identification of potential targets
for differentiation in human leukemia cells induced by diallyl
disulfide. Int J Oncol. 50:697–707. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nishida E, Maekawa S and Sakai H: Cofilin,
a protein in porcine brain that binds to actin filaments and
inhibits their interactions with myosin and tropomyosin.
Biochemistry. 23:5307–5313. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Van Troys M, Huyck L, Leyman S, Dhaese S,
Vandekerkhove J and Ampe C: Ins and outs of ADF/cofilin activity
and regulation. Eur J Cell Biol. 87:649–667. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Verdoni AM, Aoyama N, Ikeda A and Ikeda S:
Effect of destrin mutations on the gene expression profile in vivo.
Physiol Genomics. 34:9–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bernstein BW and Bamburg JR: ADF/cofilin:
A functional node in cell biology. Trends Cell Biol. 20:187–195.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ostrowska Z and Moraczewska J: Cofilin - a
protein controlling dynamics of actin filaments. Postepy Hig Med
Dosw. 71:339–351. 2017. View Article : Google Scholar
|
|
11
|
Kapoor S: Cofilin-1 overexpression and its
role in tumor growth and progression in systemic malignancies. Int
J Radiat Biol. 90:1132014. View Article : Google Scholar
|
|
12
|
Li D, Wang H, Song H, Xu H, Zhao B, Wu C,
Hu J, Wu T, Xie D, Zhao J, et al: The microRNAs miR-200b-3p and
miR-429-5p target the LIMK1/CFL1 pathway to inhibit growth and
motility of breast cancer cells. Oncotarget. 8:85276–85289.
2017.PubMed/NCBI
|
|
13
|
Wang H, Gu H, Feng J, Qian Y, Yang L, Jin
F, Wang X, Chen J, Shi Y, Lu S, et al: Celastrus orbiculatus
extract suppresses the epithelial-mesenchymal transition by
mediating cytoskeleton rearrangement via inhibition of the Cofilin
1 signaling pathway in human gastric cancer. Oncol Lett.
14:2926–2932. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang TY, DerMardirossian C and Bokoch GM:
Cofilin phosphatases and regulation of actin dynamics. Curr Opin
Cell Biol. 18:26–31. 2006. View Article : Google Scholar
|
|
15
|
Schiapparelli P, Guerrero-Cazares H,
Magaña-Maldonado R, Hamilla SM, Ganaha S, Goulin Lippi Fernandes E,
Huang CH, Aranda-Espinoza H, Devreotes P and Quinones-Hinojosa A:
NKCC1 Regulates Migration Ability of Glioblastoma Cells by
Modulation of Actin Dynamics and Interacting with Cofilin.
EBioMedicine. 21:94–103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou J, Wang Y, Fei J and Zhang W:
Expression of cofilin 1 is positively correlated with the
differentiation of human epithelial ovarian cancer. Oncol Lett.
4:1187–1190. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Su B, Su J, He H, Wu Y, Xia H, Zeng X, Dai
W, Ai X, Ling H, Jiang H, et al: Identification of potential
targets for diallyl disulfide in human gastric cancer MGC-803 cells
using proteomics approaches. Oncol Rep. 33:2484–2494. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang YS, Xie N, Su Q, Su J, Huang C and
Liao QJ: Diallyl disulfide inhibits the proliferation of HT-29
human colon cancer cells by inducing differentially expressed
genes. Mol Med Rep. 4:553–559. 2011.PubMed/NCBI
|
|
19
|
Su B, Su J, Zeng Y, Liu F, Xia H, Ma YH,
Zhou ZG, Zhang S, Yang BM, Wu YH, et al: Diallyl disulfide
suppresses epithelial-mesenchymal transition, invasion and
proliferation by downregulation of LIMK1 in gastric cancer.
Oncotarget. 7:10498–10512. 2016.PubMed/NCBI
|
|
20
|
Ling H, Zhang LY, Su Q, Song Y, Luo ZY,
Zhou XT, Zeng X, He J, Tan H and Yuan JP: Erk is involved in the
differentiation induced by diallyl disulfide in the human gastric
cancer cell line MGC803. Cell Mol Biol Lett. 11:408–423. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang F, Wu D, Fu H, He F, Xu C, Zhou J, Li
D, Li G, Xu J, Wu Q, et al: Cofilin 1 promotes bladder cancer and
is regulated by TCF7L2. Oncotarget. 8:92043–92054. 2017.PubMed/NCBI
|
|
22
|
Nowak D, Mazur AJ, Popow-Woźniak A,
Radwańska A, Mannherz HG and Malicka-Błaszkiewicz M: Subcellular
distribution and expression of cofilin and ezrin in human colon
adenocarcinoma cell lines with different metastatic potential. Eur
J Histochem. 54:e142010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma M, Zhou L, Guo X, Lv Z, Yu Y, Ding C,
Zhang P, Bi Y, Xie J, Wang L, et al: Decreased cofilin1 expression
is important for compaction during early mouse embryo development.
Biochim Biophys Acta. 1793:1804–1810. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pfaendtner J, De La Cruz EM and Voth GA:
Actin filament remodeling by actin depolymerization factor/cofilin.
Proc Natl Acad Sci USA. 107:7299–7304. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Collazo J, Zhu B, Larkin S, Martin SK, Pu
H, Horbinski C, Koochekpour S and Kyprianou N: Cofilin drives
cell-invasive and metastatic responses to TGF-β in prostate cancer.
Cancer Res. 74:2362–2373. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang X, Pan Q, Sun D, Chen W, Shen A,
Huang M, Ding J and Geng M: O-GlcNAcylation of cofilin promotes
breast cancer cell invasion. J Biol Chem. 288:36418–36425. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chung H, Kim B, Jung SH, Won KJ, Jiang X,
Lee CK, Lim SD, Yang SK, Song KH and Kim HS: Does phosphorylation
of cofilin affect the progression of human bladder cancer? BMC
Cancer. 13:452013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Q, Ji Q, Tang Y, Chen T, Pan G, Hu S,
Bao Y, Peng W, Yin P, Shimizu H, et al: Mitochondrial translocation
of cofilin-1 promotes apoptosis of gastric cancer BGC-823 cells
induced by ursolic acid. Tumour Biol. 35:2451–2459. 2014.
View Article : Google Scholar
|
|
29
|
Xiao P, Ma T, Zhou C, Xu Y, Liu Y and
Zhang H: Anticancer effect of docetaxel induces apoptosis of
prostate cancer via the cofilin-1 and paxillin signaling pathway.
Mol Med Rep. 13:4079–4084. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Becker M, De Bastiani MA, Müller CB,
Markoski MM, Castro MA and Klamt F: High cofilin-1 levels correlate
with cisplatin resistance in lung adenocarcinomas. Tumour Biol.
35:1233–1238. 2014. View Article : Google Scholar
|
|
31
|
Satoh M, Takano S, Sogawa K, Noda K,
Yoshitomi H, Ishibashi M, Mogushi K, Takizawa H, Otsuka M, Shimizu
H, et al: Immune-complex level of cofilin-1 in sera is associated
with cancer progression and poor prognosis in pancreatic cancer.
Cancer Sci. 108:795–803. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng Y, Fang Y, Li S and Zheng B:
Detection of plasma cofilin protein for diagnosis of lung cancer.
Nan Fang Yi Ke Da Xue Xue Bao. 33:1551–1553. 2013.In Chinese.
PubMed/NCBI
|
|
33
|
Zhou Y, Su J, Shi L, Liao Q and Su Q: DADS
downregulates the Rac1-ROCK1/PAK1-LIMK1-ADF/cofilin signaling
pathway, inhibiting cell migration and invasion. Oncol Rep.
29:605–612. 2013. View Article : Google Scholar
|
|
34
|
Borensztajn K, Peppelenbosch MP and Spek
CA: Coagulation Factor Xa inhibits cancer cell migration via
LIMK1-mediated cofilin inactivation. Thromb Res. 125:e323–e328.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Niit M, Arulanandam R, Cass J, Geletu M,
Hoskin V, Côté G, Gunning P, Elliott B and Raptis L: Regulation of
HC11 mouse breast epithelial cell differentiation by the
E-cadherin/Rac axis. Exp Cell Res. 361:112–125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang L, Dai F, Tang L, Le Y and Yao W:
Macrophage differentiation induced by PMA is mediated by activation
of RhoA/ROCK signaling. J Toxicol Sci. 42:763–771. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Aggelou H, Chadla P, Nikou S, Karteri S,
Maroulis I, Kalofonos HP, Papadaki H and Bravou V: LIMK/cofilin
pathway and Slingshot are implicated in human colorectal cancer
progression and chemoresistance. Virchows Arch. 472:727–737. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Prunier C, Prudent R, Kapur R, Sadoul K
and Lafanechère L: LIM kinases: Cofilin and beyond. Oncotarget.
8:41749–41763. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gai WT, Yu DP, Wang XS and Wang PT:
Anti-cancer effect of ursolic acid activates apoptosis through
ROCK/PTEN mediated mitochondrial translocation of cofilin-1 in
prostate cancer. Oncol Lett. 12:2880–2885. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ishaq M, Lin BR, Bosche M, Zheng X, Yang
J, Huang D, Lempicki RA, Aguilera-Gutierrez A and Natarajan V: LIM
kinase 1 - dependent cofilin 1 pathway and actin dynamics mediate
nuclear retinoid receptor function in T lymphocytes. BMC Mol Biol.
12:412011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kaji N, Muramoto A and Mizuno K: LIM
kinase-mediated cofilin phosphorylation during mitosis is required
for precise spindle positioning. J Biol Chem. 283:4983–4992. 2008.
View Article : Google Scholar
|
|
42
|
Zhou T, Wang CH, Yan H, Zhang R, Zhao JB,
Qian CF, Xiao H and Liu HY: Inhibition of the Rac1-WAVE2-Arp2/3
signaling pathway promotes radiosensitivity via downregulation of
cofilin-1 in U251 human glioma cells. Mol Med Rep. 13:4414–4420.
2016. View Article : Google Scholar : PubMed/NCBI
|