Introduction
Choriocarcinoma is a highly malignant tumour that
develops from trophoblast cells and usually occurs in the uterus,
and it can cause severe local damage and metastasize to other areas
of the body (1). As the clinical
presentation of choriocarcinoma may vary, diagnosis may be
challenging and the prognosis of patients with choriocarcinoma is
related to the clinical stage and trophoblastic activity (1,2). It
is widely recognized that the regulatory process of trophoblast
invasion may be associated with growth factors, chemokines, protein
kinases and signaling pathways, and the changes in the regulation
of these factors may lead to various pathological changes (3). Therefore, a deeper understanding of
the mechanisms underlying cell proliferation and apoptosis in
choriocarcinoma is required to develop novel treatment strategies
and improve patient prognosis.
The disinterring and metalloprotease (ADAM) family
consists of several type I transmembrane proteins that have been
widely reported to be involved in various physiological functions,
such as cell-binding and intracellular signalling, related to human
tumour metastasis (4,5). Members of the ADAM family have two
major structural regions, the de-integrin and the metal matrix
protease regions, which degrade the extracellular matrix and
control cell adhesion and movement by regulating cell adhesion and
protease activity (5). Among the
members of the ADAM family, ADAM metallopeptidase domain 12
(ADAM12) expression is highly associated with several types of
epithelial cancer, including breast, skin, ovarian, stomach, lung,
prostate and brain cancer (6-10).
ADAM12 contributes to cell differentiation, tumour cell
proliferation, migration and invasion (8,11-18)
as well as apoptosis and endocrine resistance (19). Apoptosis is a well-known form of
programmed cell death and is a highly regulated and controlled
process. Autophagy allows the removal of unnecessary or
dysfunctional cellular components and allows the orderly
degradation and recycling of cellular components (20-22).
Both apoptosis and autophagy are known to play roles in several
diseases, including cancer (23-26).
However, the specific role of ADAM12 silencing in the apoptosis and
autophagy of choriocarcinoma cells, as well as the related
mechanisms, has not yet been described. Therefore, the present
study investigated the effects of ADAM12 silencing on the
proliferation and apoptosis of the human chorio-carcinoma JEG-3
cell line. Additionally, the potential mechanisms involved in
autophagy and other signalling pathways were explored in JEG-3
cells following ADAM12 silencing.
Materials and methods
Cell culture and transfection
The human choriocarcinoma JEG-3 cell line was
acquired from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences (27).
The cells were cultured in DMEM medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) and maintained in an incubator
containing 5% CO2 at 37˚C (28). ADAM12-small interfering RNAs
(ADAM12-siRNA; target 1, 5′-GCC TGA ATC GTC AAT GTC AAA-3′; target
2, 5′-CGC TCG AAA TTA CAC GGT AAT-3′; and target 3, 5′-GCG AGA TGA
GAG ATG CTA AAT-3′) were synthesized by Shanghai GeneChem Co., Ltd.
The siRNA targeted transcript variant 1 (NCBI Ref. Seq. NM_003474)
of ADAM12 was used. In addition, scrambled-siRNA (non-targeting
sequence, 5′-CCT AAG GTT AAG TCG CCC TCG-3′ (also synthesized by
Shanghai GeneChem Co., Ltd) was used as a negative control (si-NC).
A blank control (BC) group, consisting of untransfected JEG-3
cells, was also set up. Transfection was performed using
Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. JEG-3 cells culture were observed
using an optical microscope (magnification, ×200), and the
transfection efficiency at 48 h post-transfection was detected by
western blotting. In order to test whether ADAM12-knockout affected
autophagy, 10 mM 3-methyladenine (3MA; cat. no. M9281-100 mg;
Sigma-Aldrich) was co-applied with siRNA transfection and used for
blocking autophagy in JEG-3 cells, the ADAM12-siRNA-3MA group for
24 h.
Cell proliferation
JEG-3 cells in DMEM were seeded in a 96-well plate
(5×103 cells/well) and incubated for 24 h at 37˚C. In
addition, ADAM12-siRNA, ADAM12-siRNA-3MA and ascrambled-siRNA
groups were included. At 24 h post-transfection, 10 µl of
pre-warmed Cell Counting Kit-8 (CKK-8; Dojindo Molecular
Technologies, Inc.) solution were added to each well, and the cells
were incubated for 4 h at 37˚C and 5% CO2, and the
absorbance values of the cells in each group were measured at a
wavelength of 450 nm using a microplate reader.
Cell cycle analysis
JEG-3 cells were trypsinized to form a single cell
suspension and washed 2-3 times with PBS. The number of cells was
adjusted to 1×106 cells/ml. The cells were then
resuspended in 1 ml pre-cooled PBS and centrifuged at 250 × g for 5
min at 4˚C, and the supernatant was aspirated. The cells were
gently resuspended with 20 µl PBS and incubated with 600 µl
pre-cooled 100% ethanol (final concentration, 75%) overnight at
4˚C. The fixed cells were centrifuged at 250 × g for 5 min at 4˚C,
and the supernatant was aspirated. Next, the cells were resuspended
in 1 ml pre-cooled PBS, after which they were centrifuged at 250 ×
g for 5 min at 4˚C and collected. This process was repeated 1-2
times to remove the ethanol. Next, the cells were incubated with
150 µl propidium iodide (Sigma-Aldrich; Merck KGaA) for 30 min at
4˚C in the dark. The cells were subsequently analyzed using a
CytoFLEX S flow cytometer (Beckman Coulter, Inc.) and CytExpert
software (version 2.0; Beckman Coulter, Inc.). The percentage of
cells in each stage of cell cycle was analyzed.
Flow cytometry detection of the apoptosis
rate
The transfected and control JEG-3 cells were
collected by trypsin digestion without EDTA, washed twice with PBS
and centrifuged at 4˚C for 5 min at 520 × g. Approximately
1-5×105 cells were collected. Then, 500 µl binding
buffer was added to the cell suspension. The cells were incubated
with 5 µl Annexin V-FITC and 5 µl propidium iodide (eBioscience™
Annexin V Apoptosis Detection Kit FIFC; cat. no. 88-8005-72; Thermo
Fisher Scientific, Inc.) for 5-15 min at room temperature in the
dark. Apoptotic cells were detected within 1 h by a CytoFLEX S flow
cytometer (Beckman Coulter, Inc.) and analysed using CytExpert
software (version 2.0; Beckman Coulter, Inc.).
Western blotting
JEG-3 cells were treated with lysis buffer (50 mM
Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.5% sodium
deoxycholate, 0.1% sodium dodecylsulfate (SDS), 5 mM EDTA, 1 mM
4-(2-aminoethyl) benzenesul-fonyl fluoride hydrochloride, 5 µg/ml
pepstatin, 5 µg/ml leupeptin, 5 µg/ml aprotinin and 10 mM
1,10-phenanth-roline) for 30 min on ice. The samples were denatured
by boiling in SDS sample buffer (SDS-PAGE Gel Preparation kit;
CWBio). The protein concentration was determined using a BCA
Protein Concentration Assay kit (CWBio), and 50 µg protein/lane
were subjected to SDS-PAGE on 4-20% Tris-glycine gels and
transferred onto polyvinylidene difluoride membranes. The membranes
were dyed with 0.1% (w/v) ponceau S for 1-2 min at room temperature
and washed. Then, the membranes were blocked with 5% milk in
Tris-buffered saline and 0.1% Tween 20 (TBST) for 1 h at room
temperature. The membranes were washed with TBST for 5 min, and
incubated with primary antibodies (all used at a 1:1,000 dilution)
against ADAM12 (cat. no. 14139-1-AP; ProteinTech Group, Inc.),
autophagy related 5 (ATG5; cat. no. 10181-2-AP; ProteinTech Group,
Inc.), microtu-bule-associated protein-light-chain 3 [LC3B,
detected two bands LC3BI (upper bands) and LC3BII (lower bands);
cat. no. 18725-1-AP; ProteinTech Group, Inc.], caspase-3 (detected
pro- and cleaved caspase-3; cat. no ab13847; Abcam), caspase-9
(detected pro- and cleaved caspase-9; cat. no. ab202068; Abcam),
Bax (cat. no. 50599-2-Ig; ProteinTech Group, Inc.), p53 (cat. no.
10442-1-AP; ProteinTech Group, Inc.), phosphorylated-mTOR (Ser2448;
cat. no. ab109268; Abcam), mTOR (cat. no. ab2732; Abcam), p65-NF-κB
(cat. no. 10745-1-AP; ProteinTech Group, Inc.); proliferating cell
nuclear antigen (PCNA; cat. no. 10205-2-AP; ProteinTech Group,
Inc.) and β-actin (cat no. 60008-1-Ig; ProteinTech Group, Inc.)
overnight at 4˚C. The membranes were washed three times with TBST,
10 min/wash. Subsequently, the membranes were incubated with goat
anti-rabbit IgG horseradish peroxidase-conjugated secondary
antibodies (at 1:1,000 dilution; cat. no. SA00001-2; ProteinTech
Group, Inc.) at room temperature for 1 h, and washed three times
with TBST, 10 min/wash. The protein bands were visualized using the
SuperSignal West Pico Chemiluminescent Substrate (EMD Millipore).
ImageJ software (version 1.8.0; National Institutes of Health) was
used to analyze the greyscale values, with β-actin or PCNA as the
loading control. The calculation was performed as follows: The
amount of relevant protein expression=(the grey value of measured
protein/the grey value of internal parameter).
Immunofluorescence
JEG-3 cells were plated onto sterile coverslips and
allowed to attach overnight at 4˚C. JEG-3 cells were then washed
with PBS and fixed with 4% paraformalde-hyde in PBS (pH 7.4) for 20
min at room temperature. These cells were washed three times with
PBS and permeabilized with 0.1% Triton X-100 in PBS for 5 min. The
cells were then blocked with 1% bovine serum albumin (cat. no.
V900933-100g; Sigma-Aldrich; Merck KGaA) and 22.52 mg/ml glycine in
PBST (PBS + 0.1% Tween 20) for 30 min at 37˚C to prevent
non-specific binding of the antibodies. The cells were incubated
with primary antibodies (diluted to 1:200 in 1% BSA in PBST)
against LC3B (cat. no. 18725-1-AP; ProteinTech Group, Inc.) in a
humidified incubator at 37˚C for 1 h. After washing three times in
PBS at 5 min for each wash, the cells were incubated for 1 h at
37˚C in the dark with a fluorescein-conjugated secondary IgG
antibody (1:200 in 1% BSA in PBS; Thermo Fisher Scientific, Inc.).
After washing three times with PBS for 5 min each in the dark, the
cells were incubated with 0.1-1 µg/ml DAPI at rom temperature for 1
min, washed with PBS and mounted with coverslips using a
fluorescent mounting medium. The coverslip was sealed with nail
polish to prevent drying and movement under the microscope. The
slides were stored in the dark at −20 or 4˚C. The slides were
imaged using a Zeiss Axiovert-200 inverted fluorescence microscope
(Carl Zeiss AG; magnification, ×400). ImageJ software (version
1.8.0; National Institutes of Health) was used to analyse the
immunofluorescence density (IFD). The formula used was as follows:
IFD=(the fluorescence intensity of the region of interest/the area
of the region of interest).
ELISA
A total of 50-100 µl of the prepared standard and
samples of JEG-3 cells supernatant were added to an antibody-coated
96-well microplate [human interleukin 1β (IL-1β) ELISA kit; cat.
no. CSB-E08053h; human interferon γ (IFNγ) ELISA kit; cat. no.
CSB-E04577h; and human tumor necrosis factor α (TNFα) ELISA kit;
cat. no. CSB-E04740h; all from CusaBio] The plate was covered and
incubated at room temperature for 2 h, and the solution was
thoroughly decanted from the wells. The wells were washed with TBST
four times using a squirt wash bottle or an automated 96-well plate
washer. Next, 100 µl of diluted detection antibodies was added to
the wells, and the plate was covered and incubated at room
temperature for 1 h, after which the solution was thoroughly
aspirated from the wells. After washing the wells four times, 100
µl of diluted HRP conjugate was added to each well and the plate
was covered and incubated at room temperature for 30 min, after
which the solution was thoroughly aspirated from the wells. After
washing the wells with TBST four times, 100 µl of the chromogenic
substrate TMB was added to each well, and the plate was then
developed at room temperature in the dark for 30 min. Next, 100 µl
of stop solution was added to each well, and then the solution in
the wells changed from blue to yellow. The plate was evaluated
within 30 min of stopping the reaction. The absorbance of each well
was read at 450 and 550 nm with a microplate reader, and the 550 nm
values were subtracted from the 450 nm values to correct for
optical imperfections in the microplate. MasterPlex ReaderFit
curve-fitting statistical software (version 2.0; Emerald Biotech
Co., Ltd.) was used to plot a four-parameter logistic curve fit to
the standards, and then the results for the test samples were
calculated.
Statistical analysis
Data are presented as the means ± standard error of
the mean of at least three independent experiments. Differences
among experimental groups were statistically analysed by SPSS
software (version 17.0; SPSS, Inc.) using the analysis of variance
followed by the least significant difference or Dunnett's post hoc
tests, as applicable. P<0.05 was considered to indicate a
statistically significant difference.
Results
Transfection and ADAM12 silencing in
JEG-3 cells
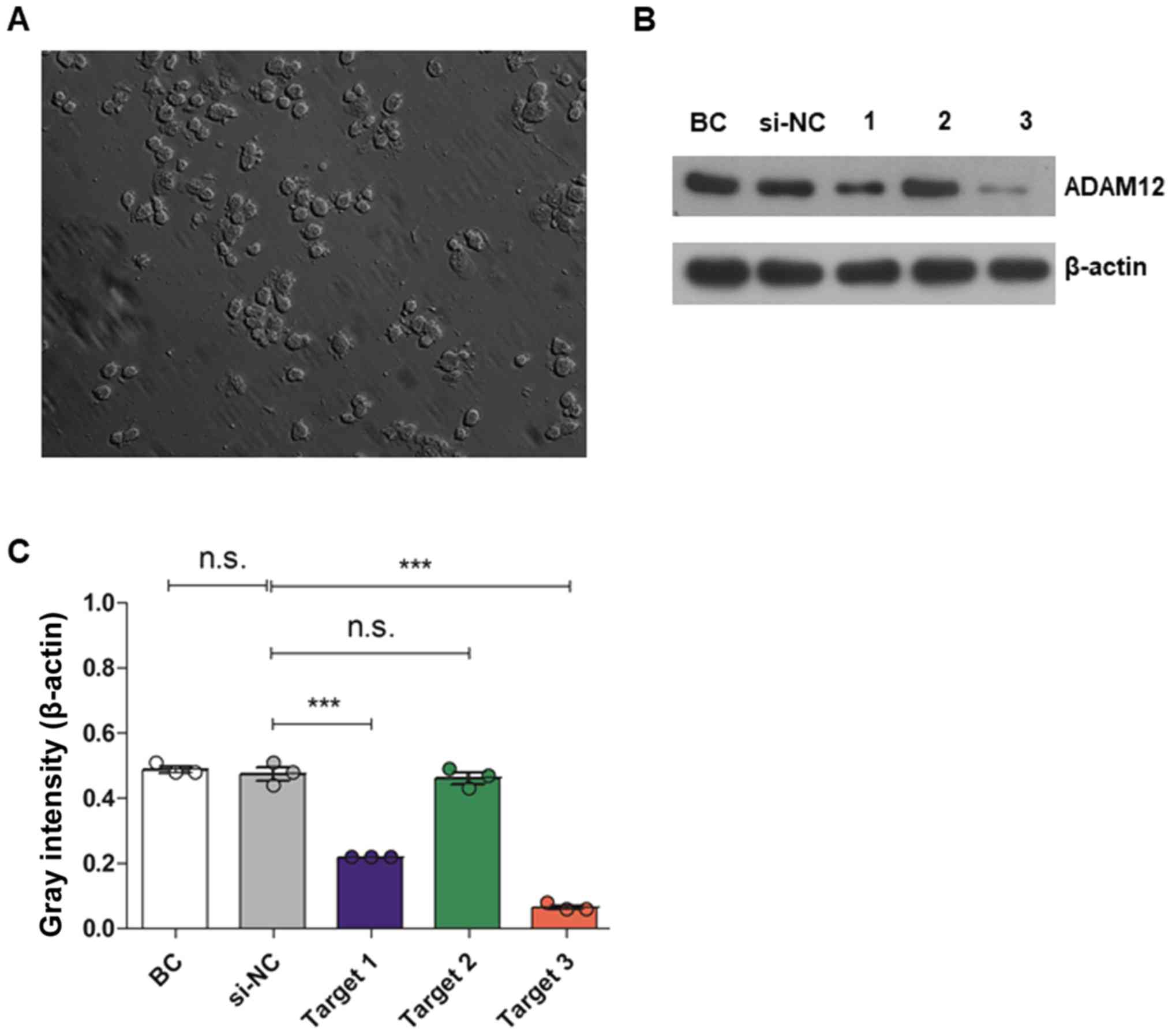
In the present study, untransfected JEG3 cells were
observed using an optical microscope (magnification, x200; Fig. 1A). JEG-3 cells were transfected
with three different targets of ADAM12-siRNA or scrambled-siRNA.
Western blotting revealed that at 48 h post-transfection, ADAM12
expression was decreased in cells transfected with ADAM12-siRNA
(target 1 and 3) but not in cells transfected with the si-NC) or
ADAM12-siRNA target 2, compared with the BC cells. ADAM12-siRNA
target 3 exhibited the best transfection efficiency compared with
the other two targets (P<0.05; Fig.
1B and C). Therefore, this siRNA was selected for subsequent
experimentation.
ADAM12 silencing decreases cell
proliferation and increases apoptosis in JEG-3 cells
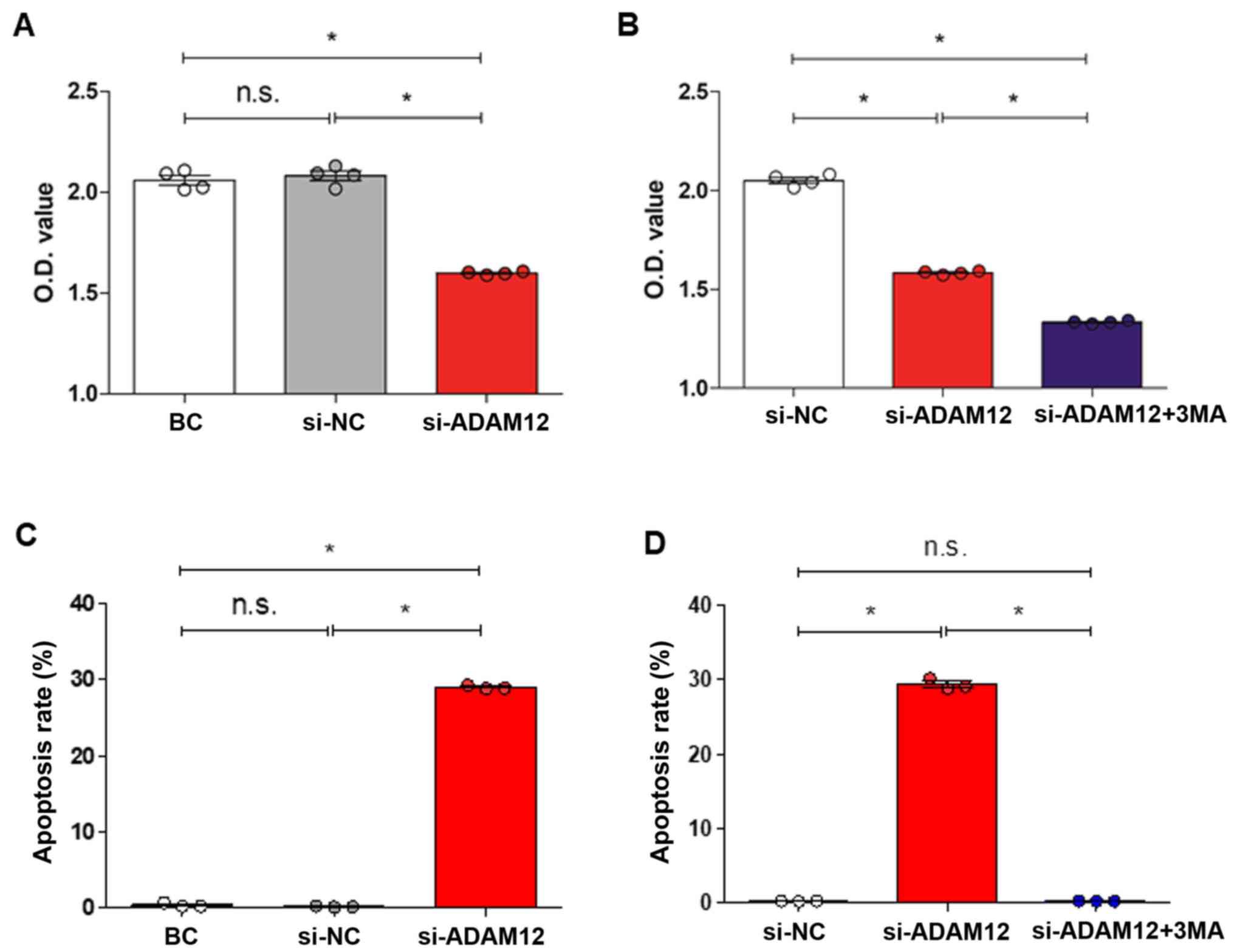
The CCK-8 assay showed that cell proliferation was
significantly decreased in the si-ADAM12 group compared with the
si-NC group (P<0.05; Fig. 2A).
Additionally, flow cytometry analysis revealed that the rate of
apoptosis in the si-ADAM12 group was significantly increased
compared with the si-NC group (P<0.05; Figs. 2C and S1A). Together, these findings confirmed
that cell apoptosis was increased after ADAM12 silencing.
The levels of apoptosis-associated proteins were
measured by western blotting following ADAM12-siRNA transfection
(Fig. 3A). The results revealed
that the levels of caspase-3 (Fig.
3G-I) and caspase-9 were not changed (P>0.05; Fig. 3J-L), but the levels of Bax and p53
levels were signifi-cantly increased in the si-ADAM12 group
compared with the si-NC group (Fig. 3C
and E; P<0.05).
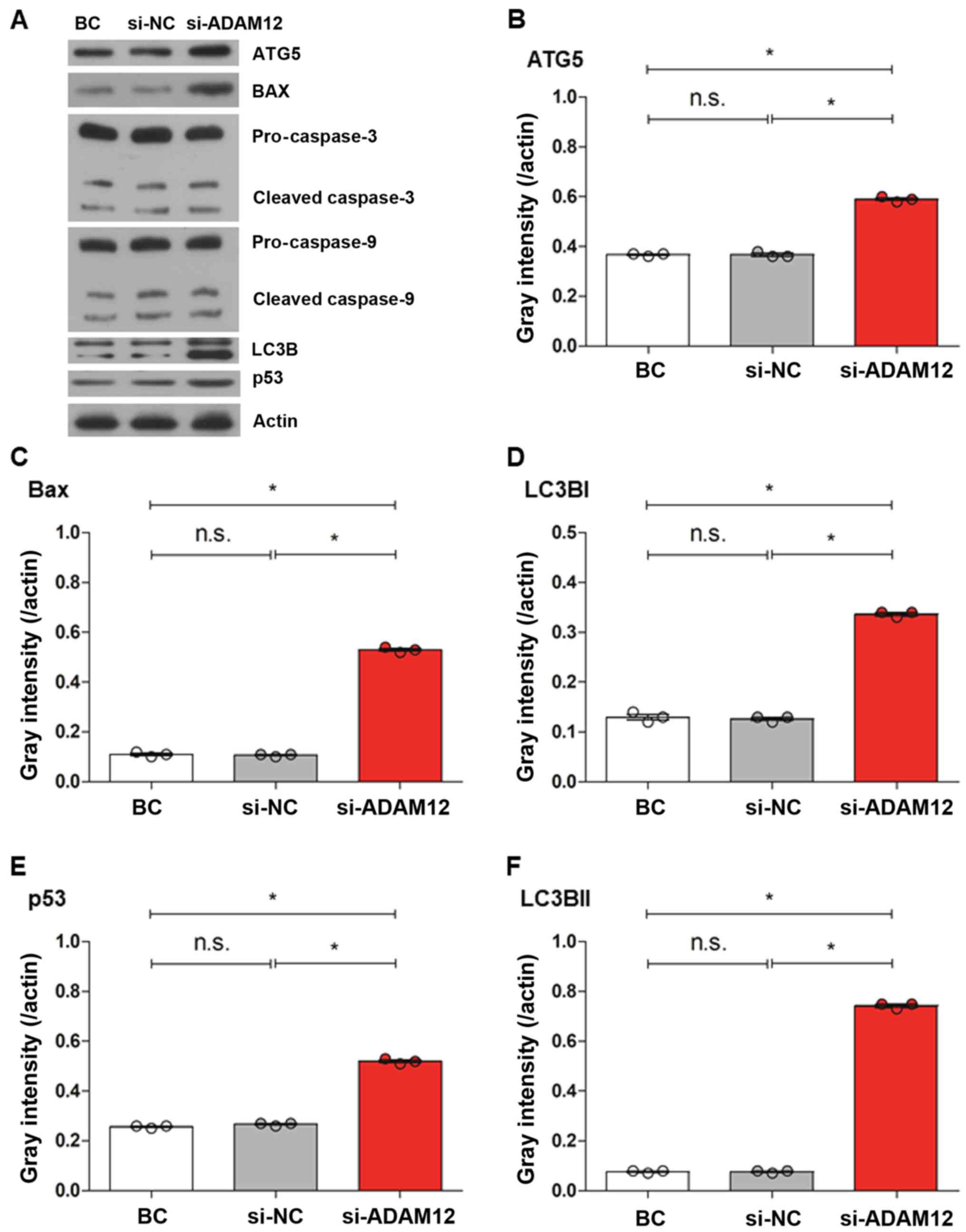 | Figure 3Western blotting was used to
investigate the expression levels of apoptosis- and
autophagy-assciacted proteins after ADAM12 silencing in JEG-3
cells. (A) Western blotting bands of autophagy-associated proteins
(ATG5 and LC3B) and apoptosis-associated proteins (caspase 3,
caspase 9, Bax and p53) in the BC, si-NC and si-ADAM12 groups. (B)
Quantification of western blotting showed that the level of ATG5
expression was increased in the si-ADAM12 group compared with the
BC and si-NC groups. (C) Quantification of western blotting showed
that the level of Bax expression was increased in the si-ADAM12
group compared with the BC and si-NC groups. (D) Quantification of
western blotting showed that the level of LC3BI expression was
increased in the si-ADAM12 group compared with the BC and si-NC
groups. (E) Quantification of western blotting showed that the
level of p53 expression was increased in the si-ADAM12 group
compared with the BC and si-NC groups. (F) Quantification of
western blotting showed that the level of LC3BII expression was
increased in the si-ADAM12 group compared with the BC and si-NC
groups. Western blotting was used to investigate the expression
levels of apoptosis- and autophagy-assciacted proteins after ADAM12
silencing in JEG-3 cells. Quantification of western blotting showed
that the level of (G) procaspase 3, (H) cleaved caspase 3 (19 kDa)
and (I) cleaved caspase 3 (17 kDa) expression in the si-ADAM12
groups was similar to the BC and si-NC groups. Quantification of
western blotting showed that the level of (J) procaspase 9, (K)
cleaved caspase 9 (37 kDa) and (L) cleaved caspase 9 (35 kDa) in
the si-ADAM12 group was similar to the BC and si-NC groups.
*P<0.05, as indicated. ADAM12, ADAM metallopeptidase
domain 12; ATG5, autophagy related 5; LC3B, microtubule-associated
protein-light-chain 3; BC, blank control; si, small interfering;
NC, negative control; n.s., not significant. |
ADAM12 silencing increases autophagy in
JEG-3 cells
The expression levels of the autophagy-associated
proteins LC3B and ATG5 in JEG-3 cells were detected by western
blotting after ADAM12 silencing. The results showed that the levels
of LC3BI and LC3BII in the si-ADAM12 cells were higher than those
in the control cells (P<0.05; Fig.
3D and F). Furthermore, the level of ATG5 expression was
increased in si-ADAM12 cells compared with the si-NC cells
(P<0.05; Fig. 3B).
In addition, the expression of the autophagy protein
LC3B was investigated using immunofluorescence analysis. The IFD of
LC3B in the si-ADAM12 group was significantly increased compared
with the BC and si-NC groups (P<0.05; Fig. 4A and B).
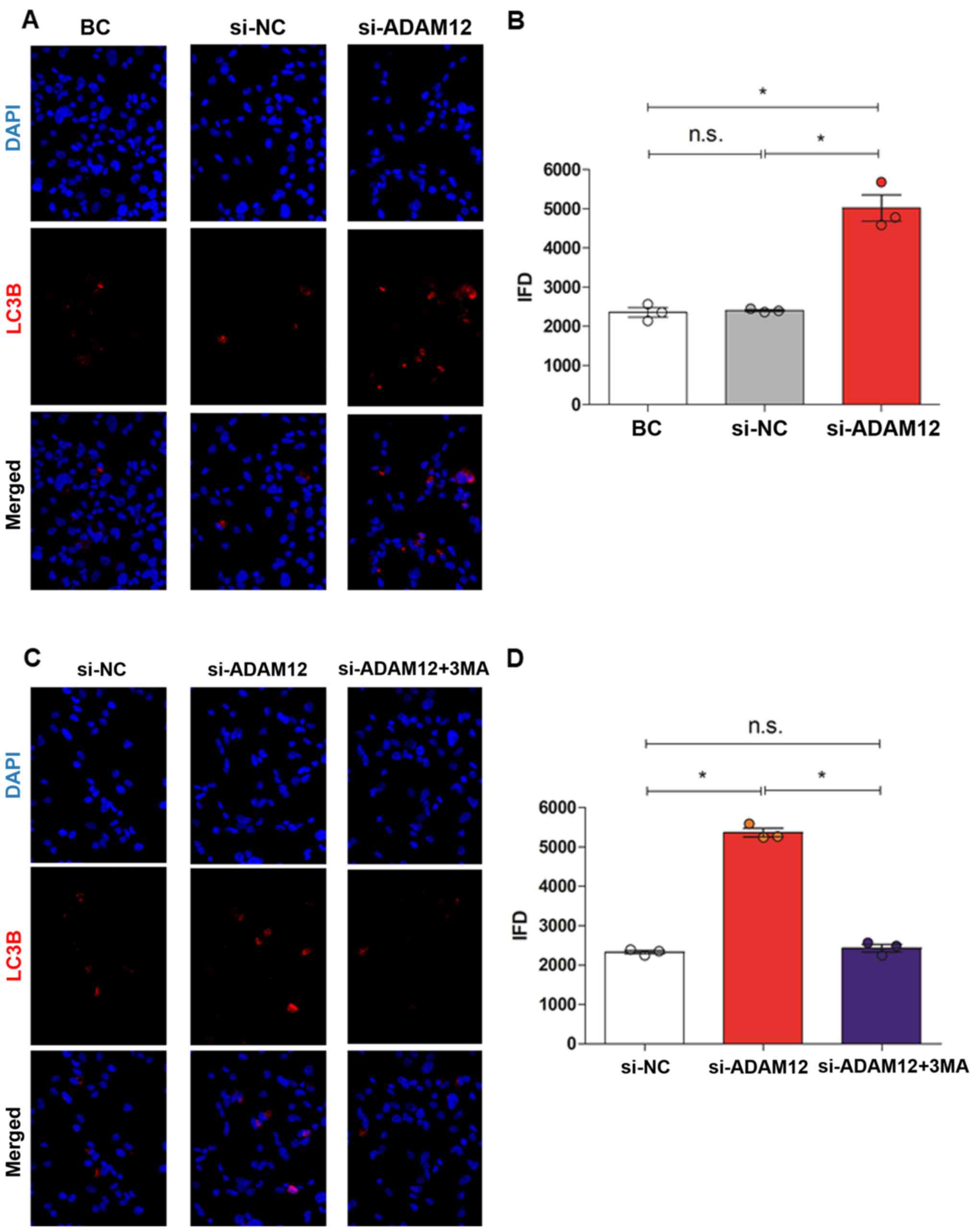 | Figure 4Immunofluorescence staining of the
autophagy-associated protein LC3B in JEG-3 cells after ADAM12
silencing. (A) Images of immunofluorescence staining
(magnification, x400). (B) Quantification of immunofluorescence
showed that the IFD of LC3B was increased in the si-ADAM12 group
compared with the BC and si-NC groups. (C) Images of
immunofluorescence staining (magnification, x400). (D)
Quantification of immunofluorescence showed that the IFD of LC3B
was decreased in the si-ADAM12 + 3MA group compared with the
si-ADAM12 group, but similar to the si-NC group.
*P<0.05, as indicated. LC3B, microtubule-associated
protein-light-chain 3; ADAM12, ADAM metallopeptidase domain 12;
IFD, immunofluorescence density; si, small interfering; 3MA,
3-methyladenine; NC, negative control; BC, blank control; n.s., not
significant. |
Autophagy mediates the effect of ADAM12
silencing on cell proliferation and apoptosis in JEG-3 cells
To investigate whether cell apoptosis was mediated
by autophagy in JEG-3 cells after ADAM12 silencing, the
ADAM12-siRNA trans-fected JEG-3 cells were treated with an
autophagy inhibitor 3-methyladenine (3MA). The cell cycle of the
3MA-treated JEG-3 cells after ADAM12 silencing was analyzed
(Figs. 5 and S2), and the results showed that the
number of cells in the si-ADAM12 group was reduced in the S and G2
phases (P<0.05; Fig. 5C-E) but
increased in the G1 phase, compared with the si-NC group
(P>0.05; Fig. 5B). However, no
differences were observed in all the cell cycle phases (G1, S and
G2) between the si-ADAM12 + 3MA and si-ADAM12 groups (P>0.05;
Fig. 5B-E).
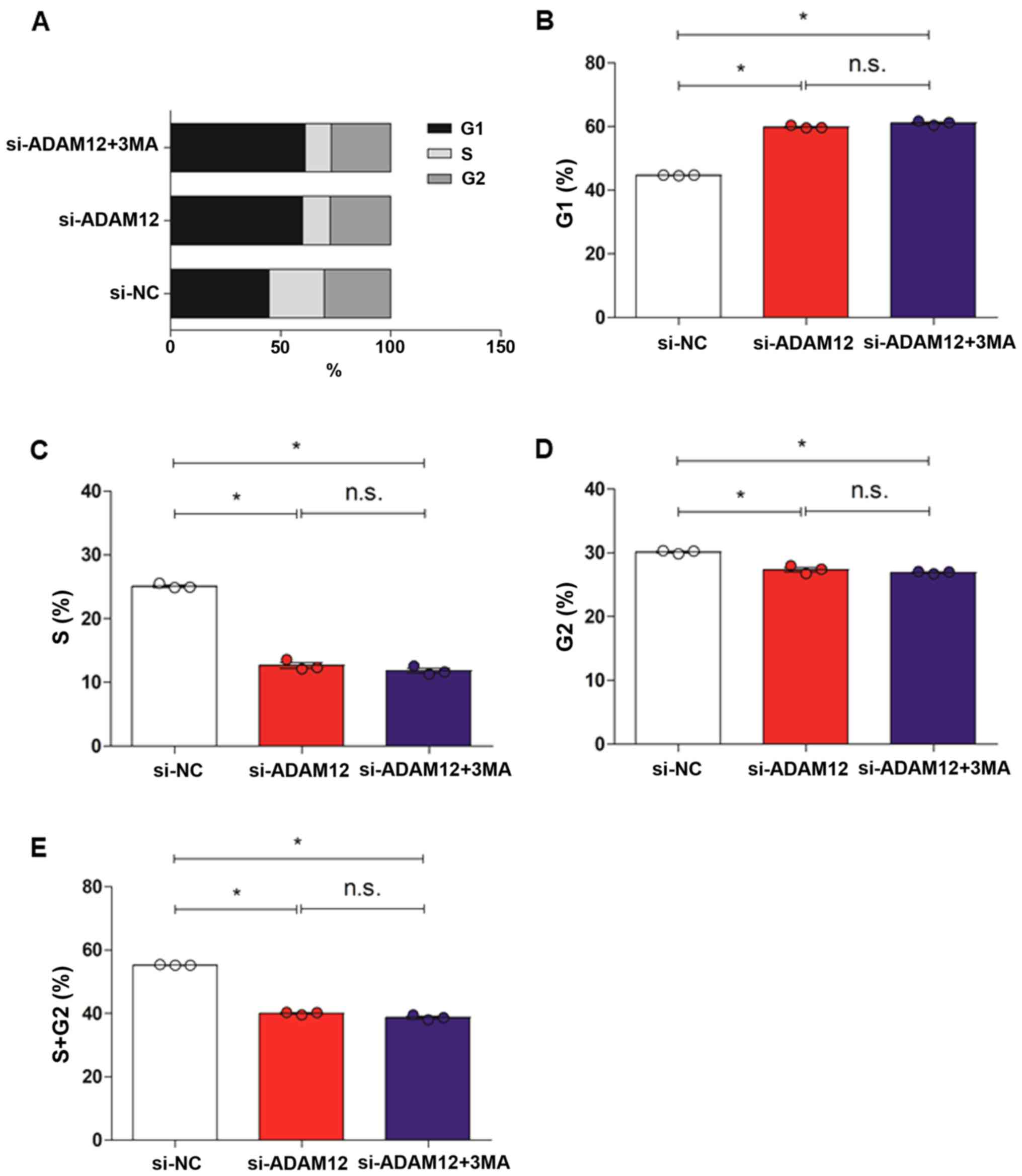 | Figure 5Cell cycle analysis of si-ADAM12
transfected JEG-3 cells after treatment with the autophagy
inhibitor 3MA. (A) Column plot of cell cycle (G1, S and G2 phases)
in JEG-3 cells in the si-NC, si-ADAM12 and si-ADAM12 + 3MA groups.
(B) Quantification of the G1 phase showed that the percentage of
cells in this phase was increased in the JEG-3 cells in the
si-ADAM12 + 3MA group compared with the si-NC group, but was
similar to the si-ADAM12 group. (C) Quantification of S phase
showed that the percentage of cells in this stage was decreased in
the JEG-3 cells in the si-ADAM12 + 3MA group compared with the
si-NC group, but was similar to the si-ADAM12 group. (D)
Quantification of the G2 phase showed that the percentage of cells
in this phase was decreased in the JEG-3 cells in the si-ADAM12 +
3MA group compared with the si-NC group, but was similar to the
si-ADAM12 group. (E) Quantification of the S and G2 phase showed
that the percentage of cells in this S+G phase was decreased in
JEG-3 cells in the si-ADAM12 + 3MA group compared with the si-NC
group, but was similar to the si-ADAM12
group.*P<0.05, as indicated. si, small interfering;
ADAM12, ADAM metallopeptidase domain 12; 3MA, 3-methyladenine; NC,
control; n.s., not significant. |
The CCK-8 assay showed that the cell proliferation
in the si-ADAMA12 + 3MA group was decreased compared with the of
si-ADAM12 and si-NC groups (Fig.
2B). Flow cytometry was to analyse the apoptosis rate and
demonstrated that the rate of apoptosis in the si-ADAM12 group was
significantly increased (P<0.05; Figs. 2C, D and S1A) compared with the BC and si-NC
groups. The apoptosis rate was significantly decreased in the
si-ADAM12 + 3MA group compared with the si-ADAM12 group (P<0.05;
Figs. 2D and S1B). Moreover, the western blotting data
showed that the levels of the apoptotic proteins p53 and Bax were
significantly decreased in the si-ADAM12 + 3MA group compared with
the si-ADAM12 group (P<0.05; Fig.
6C and E). Additionally, the levels of Bcl-2 were significantly
decreased in the si-ADAM12 (P<0.05) but partly rescued in
si-ADAM + 3MA group, compared with the si-NC group (P<0.05;
Fig. 6I). The levels of the
autophagy-associate proteins LC3BI and LC3BII were decreased in the
si-ADAM12 + 3MA group compared with the si-ADAM12 group (P<0.05;
Fig. 6G and H). In addition, the
IFD of LC3B in the si-ADAM12 + 3MA group was consistent with that
of the si-NC (P>0.05), but the IFD in the si-ADAM12 + 3MA group
was significantly decreased compared with the si-ADAM12 group
(P<0.05; Fig. 4B and D).
Together, the findings confirmed that the pro-apoptotic effect of
ADAM12 silencing was blocked after the inhibition of autophagy in
JEG-3 cells.
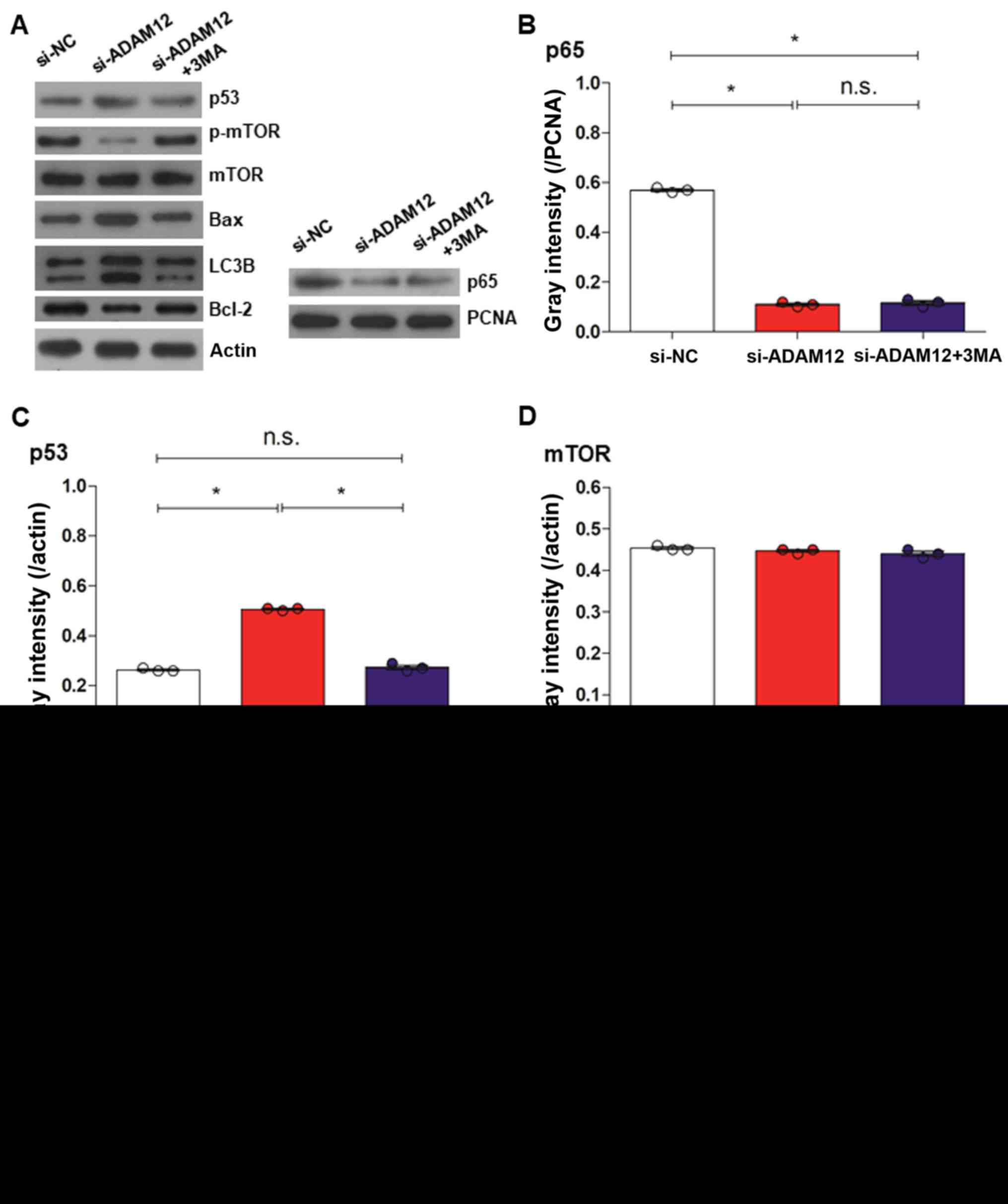 | Figure 6Western blotting was used to detect
the expression levels of apoptosis- and autophagy-associated
proteins, nuclear p65 and mTOR after 3MA treatment in si-ADAM12
transfected JEG-3 cells. (A) Western blotting bands of LC3B, Bax,
p53, nuclear p65 and m-TOR in the si-NC, si-ADAM12 and si-ADAM12 +
3MA groups. (B) Quantification of western blotting showed that the
level of nuclear p65 expression was decreased in the si-ADAM12 +
3MA group in compared with the si-NC group but was similar to the
si-ADAM12 group. (C) Quantification of western blotting showed that
the level of p53 expression was decreased in the si-ADAM12 + 3MA
group compared with the si-ADAM12 group but was similar to the
si-NC group. (D) Quantification of western blotting showed that the
level of mTOR expression in the si-ADAM12 + 3MA group was similar
to the si-ADAM12 and si-NC groups. (E) Quantification of western
blotting showed that the level of Bax expression was decreased in
the si-ADAM12 + 3MA group compared with the si-ADAM12 group but
similar to the si-NC group. (F) Quantification of western blotting
showed that the level of p-mTOR expression was increased in the
si-ADAM12 + 3MA group compared with the si-ADAM12 group but
slightly decreased compared with the si-NC group. Western blotting
was used to detect the expression levels of apoptosis- and
autophagy-associated proteins, nuclear p65 and mTOR after 3MA
treatment in si-ADAM12 transfected JEG-3 cells. (G) Quantification
of western blotting showed that the level of LC3BI expression was
decreased in the si-ADAM12 +3 MA group compared with si-ADAM12
group but was similar to the si-NC group. (H) Quantification of
western blotting showed that the level of LC3BII expression was
increased in the si-ADAM12 + 3MA group compared with the si-ADAM12
group but similar to the si-NC group. (I) Quantification of western
blotting showed that the level of Bcl-2 expression was decreased in
the si-ADAM12 group compared with the si-NC but partly rescued in
the si-ADAM12 + 3MA group. *P<0.05, as indicated.
3MA, 3-methyladenine; si, small interfering; ADAM12, ADAM
metallopeptidase domain 12; LC3B, microtubule-associated
protein-light-chain 3; NC, negative control; PCNA, proliferating
cell nuclear antigen; n.s., not significant. |
ADAM12 silencing decreases the
inflammatory response in JEG-3 cells
ELISA results showed that IL-1β (Fig. 7A), IFN-γ (Fig. 7B) and TNF-α (Fig. 7C) levels were significantly reduced
in the si-ADAM12 group compared with the BC and si-NC groups
(P<0.05).
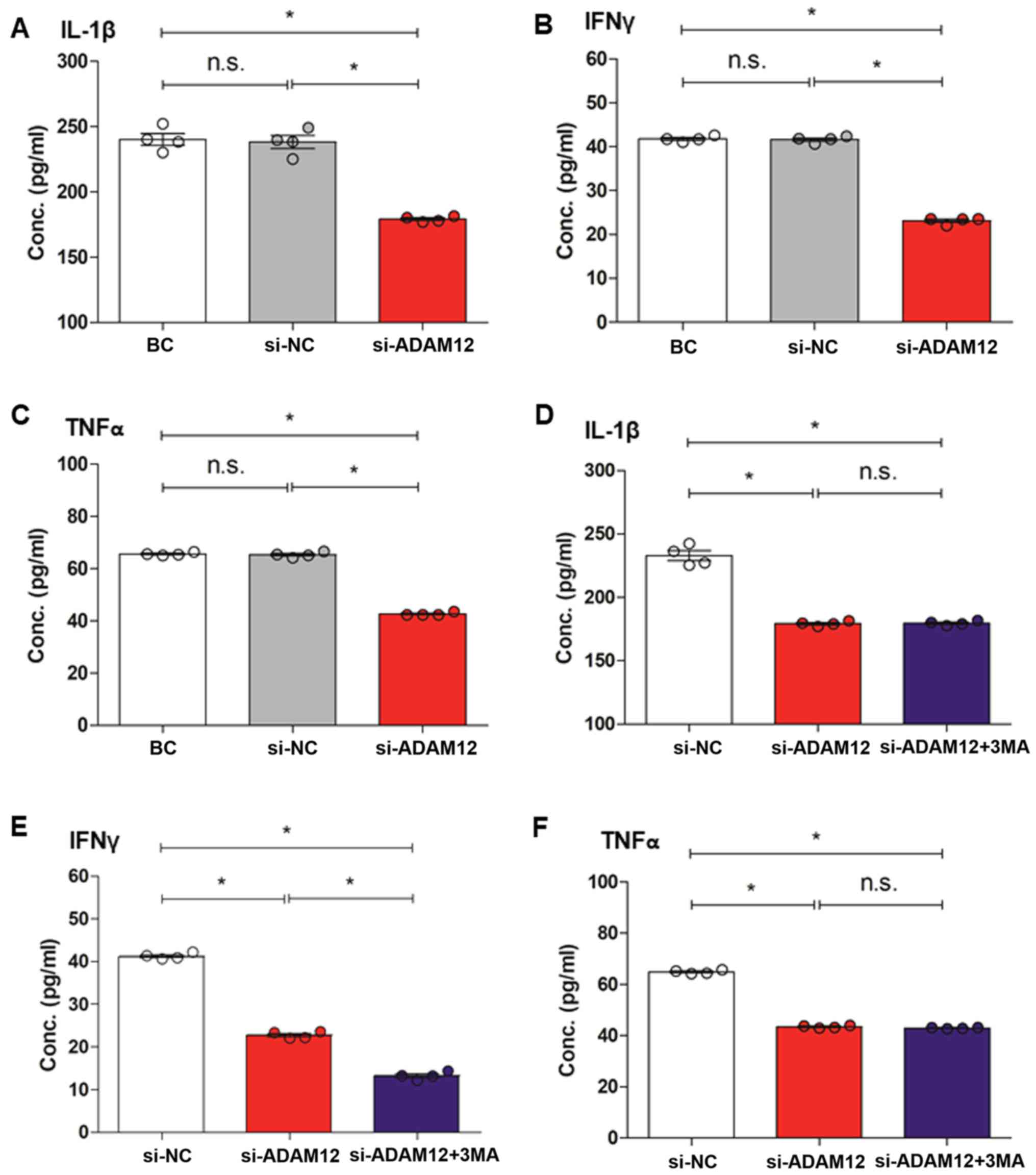 | Figure 7ELISA assay for the expression levels
of inflammatory factors after ADAM12 silencing in JEG-3 cells. (A)
Quantification of ELISA showed that the level of IL-1β expression
was decreased in the si-ADAM12 group compared with the BC and si-NC
groups. (B) Quantification of ELISA showed that the level of IFNγ
expression was decreased in the si-ADAM12 group compared with the
BC and si-NC groups. (C) Quantification of ELISA showed that the
level of TNFα expression was decreased in the si-ADAM12 group
compared with the BC and si-NC groups. (D) Quantification of ELISA
showed that the level of IL-1β expression was decreased in the
si-ADAM12 + 3MA group compared with si-NC group, but was similar to
the si-ADAM12 group. (E) Quantification of ELISA showed that the
level of IFNγ expression was greatly decreased in the si-ADAM12 +
3MA group compared with the si-NC group and slightly decreased
compared with the si-ADAM12 group. (F) Quantification of ELISA
showed that the level of TNFα expression was decreased in the
si-ADAM12 + 3MA group compared with the si-NC group but was similar
to the si-ADAM12 group. *P<0.05, as indicated.
ADAM12, ADAM metallopeptidase domain 12; IL-1β, interleukin-1β; BC,
blank control; si, small interfering; NC, negative control; IFNγ,
interferon γ; 3MA, 3-methyladenine; TNFα, tumour necrosis α; n.s.,
not significant. |
Autophagy fails to impact the effect of
ADAM12 silencing on the inflammatory response in JEG-3 cells
IL-1β (Fig. 7D),
IFN-γ (Fig. 7E) and TNF-α
(Fig. 7F) were significantly
reduced in the si-ADAM12 + 3MA group compared with the si-NC group
(P<0.05), while IL-1β (Fig. 7D)
and TNF-α concentrations (Fig. 7F)
exhibited little difference between the si-ADAM12 + 3MA and
si-ADAM12 groups (P>0.05). In addition, the level of IFN-γ was
significantly decreased in the si-ADAM12 + 3MA group compared with
the si-ADAM12 group (P<0.05; Fig.
7E).
NF-κB and mTOR signalling
To investigate the potential mechanism by which
ADAM12 silencing-mediated apoptosis in human choriocarcinoma cells,
the levels of p65-NF-κB (p65; Fig. 6A
and B), mTOR, and p-mTOR were measured in JEG-3 cells (Fig. 6A and F). ADAM12 silencing
significantly reduced p65 expression in the si-ADAM12 group
compared with the si-NC group (P<0.05; Fig. 6B), but the level of p65 protein was
similar in the si-ADAM12 + 3MA and si-ADAM12 groups (P>0.05;
Fig. 6B). Furthermore, mTOR
expression levels did not differ among the si-NC, si-ADAM12 and
si-ADAM12 + 3MA groups (Fig. 6D),
while p-mTOR expression was down-regulated in the si-ADAM12-treated
cells, compared with the si-ADAM12 + 3MA group, and this effect was
significantly rescued in the si-ADAM12 + 3MA group (P<0.05;
Fig. 6F).
Discussion
The prognosis of choriocarcinoma is related to
clinical stage and trophoblastic activity (1,2).
Treatment of chorio-carcinoma may include chemotherapy radiation
therapy or combination therapy (29,30).
Cancer-targeted therapies based on specific genes or functional
proteins are receiving increasing attention (31,32).
ADAM12 is a metalloprotein with cell adhesion and hydrolytic
activities and has been widely reported to mediate tumour cell
proliferation (16) and apoptosis
resitance (9). The present study
focused on the effect of ADAM12 silencing on cell apoptosis and
autophagy in choriocarcinoma cells. Multiple direct and indirect
interactions have been described suggesting mechanistic overlap and
interaction between the apoptosis machinery and autophagy proteins
(23-25). Although it was originally
identified as a cell survival mechanism, autophagy has highly
context-specific effects in mediating cell death (33,34).
In specific contexts, cell death can also be led by autophagy
(35-41). Studies in mammalian and other model
systems show that autophagy can paradoxically have pro-apoptosis or
pro-survival functions, depending on the context (23,42,43).
It has recently been shown that there are interplays between
autophagy-dependent apoptosis and other types of cell death
including apoptosis and necrosis (26).
The present study revealed that ADAM12 silencing
promoted cell apoptosis by activating autophagy in human
choriocarcinoma JEG-3 cells. Furthermore, ADAM12 silencing
decreased cell proliferation and increased the rate of apoptosis.
The levels of the apoptotic proteins Bax and p53 expression were
upregulated by ADAM12 silencing in JEG-3 cells but cleaved
caspase-3 levels were unchanged. Therefore, it was speculated that
ADAM12 silencing-induced apoptosis may occur through a
caspase-independent manner, as there are caspase-independent
apoptotic pathways, such as the apoptosis-inducing factor-mediated
pathway (44) and TNF receptor
superfamily member 1A-mediated apoptosis (45).
It was reported that the levels and activity of the
apoptotic proteins p53 and Bcl-2 can be regulated through diverse
mechanisms (46-48). The present study showed that ADAM12
silencing enhanced cell apoptosis but that after the inhibition of
autophagy, the rate of cell apoptosis did not change, which
suggested that autophagy indeed mediated the pro-apoptotic effect
of ADAM12 silencing. The autophagy-related protein ATG5 is a key
protein that is involved in the extension of the phagophore
membrane in autophagy vesicles and forms a complex with autophagy
related 12 and autophagy related 16 like 1 (49). These complexes are required for the
conjugation of LC3-I to phosphatidylethanolamine to form LC3-II
(50-54). Accordingly, the present study
provided evidence for the upregulation of autophagy by ADAM12
silencing by detecting autophagy markers, such as ATG5, LC3I and
LC3II proteins. The results showed that ADAM12 silencing
upregulated the expression levels of these autophagy proteins in
JEG-3 cells.
In the cellular network of signal integration,
autophagy and immunomodulation are interdependent (53-56).
Autophagy is involved in the induction and suppression of
inflammation and vice versa (55,57,58).
The proinflammatory cytokine TNFα has been shown to stimulate
autophagy, and autophagy also contributes to the secretion of this
cytokine (59-61). However, autophagy is regulated in
its response to inflammation, such as participating in the
regulation of inflammasome activation (62) and the clearance of protein
complexes, such as inflammasomes, through proteasomal degradation
(63). In the present study,
ADAM12 silencing not only increased autophagy but also exerted an
anti-inflammatory effect in JEG-3 cells, but this anti-inflammatory
effect of ADAM12 silencing was not blocked when autophagy was
inhibited in JEG-3 cells. At the molecular level, autophagy plays a
context-dependent pro-survival or pro-death role by regulating
different signalling pathways, such as p53, Bcl-2, and mTOR in
cancer (23,42,43,64).
The p53 gene is one of the target genes of the transcription factor
NF-κB (65), and the Bcl-2 gene is
transcriptionally regulated by NF-κB and directly links the
TNF-α/NF-κB signalling pathways (66,67).
NF-κB targets inflammation not only directly by increasing the
production of inflammatory cytokines, chemokines and adhesion
molecules, but it also regulates cell proliferation, apoptosis,
morphogenesis and differentiation (68-70).
In the present study, ADAM12 silencing reduced the expression level
of nuclear p65-NF-κB in JEG-3 cells. However, ADAM12 silencing did
not change the expression level of p65-NF-κB in JEG-3 cells, even
after autophagy was inhibited.
mTOR is a serine/threonine protein kinase and a key
factor serving as the convergence point for several upstream
stimuli and pathways to regulate cell growth, cell proliferation,
cell motility, cell survival, protein synthesis, translation and
autophagy (71-76). Growth factors, glucose and amino
acids also activate mTOR and suppress autophagy, whereas nutrient
deprivation will suppress mTOR, leading to the activation of
autophagy (77-80). mTOR and autophagy are closely
associated, and defects in signalling through either pathway are
known to drive the onset of a range of human diseases, such as
cancer and neurodegenerative diseases (80-82).
In the present study, ADAM12 silencing increased the expression
level of autophagy proteins in JEG-3 cells and significantly
reduced the level of phosphorylated mTOR in JEG-3 cells, which is
responsible for the activation of the mTOR signalling pathway.
However, the downregulation of phosphorylated-mTOR expression was
rescued after the suppression of autophagy in JEG-3 cells, and the
pro-apoptotic effect of ADAM12 silencing was blocked after the
suppression of autophagy. These data confirmed that i) there might
be a relationship between the impeded nuclear enrichment of
p65-NF-κB by ADAM12 silencing and the anti-inflammatory process of
ADAM12 silencing; and ii) ADAM12 silencing activated autophagy via
the regulation of the mTOR signalling pathway.
The present study only investigated one
choriocarcinoma cell line, and further studies are warranted to
assess other choriocarcinoma cell lines and to validate the results
obtained. In conclusion, the present study revealed that ADAM12 may
serve as a potential anticancer agent due to its effects on
proliferation and apoptosis of choriocarcinoma cells.
Supplementary Data
Funding
The present study was supported by the Hunan
Provincial Natural Science Foundation of China (grant no.
2016JJ2169).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZHT designed the experiments. LW, ZHT, YZ, NKK, HNL
and YZ performed the experiments and analyzed the data. LW and ZHT
wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
References
|
1
|
Wreczycka-Cegielny P, Cegielny T, Oplawski
M, Sawicki W and Kojs ZC: Current treatment options for advanced
choriocar-cinoma on the basis of own case and review of the
literature. Ginekol Pol. 89:711–715. 2018. View Article : Google Scholar
|
|
2
|
Cheung AN, Zhang HJ, Xue WC and Siu MK:
Pathogenesis of choriocarcinoma: Clinical, genetic and stem cell
perspectives. Future Oncol. 5:217–231. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wells M: The pathology of gestational
trophoblastic disease: Recent advances. Pathology. 39:88–96. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Becherer JD and Blobel CP: Biochemical
properties and functions of membrane-anchored
metallprotease-disintegrin proteins (ADAMs). Curr Top Dev Biol.
54:101–123. 2003. View Article : Google Scholar
|
|
5
|
Klein T and Bischoff R: Active
metalloproteases of the A disin-tegrin and metalloprotease (ADAM)
family: Biological function and structure. J Proteome Res.
10:17–33. 2011. View Article : Google Scholar
|
|
6
|
Roy R, Wewer UM, Zurakowski D, Pories SE
and Moses MA: ADAM 12 cleaves extracellular matrix proteins and
correlates with cancer status and stage. J Biol Chem.
279:51323–51330. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fröhlich C, Nehammer C, Albrechtsen R,
Kronqvist P, Kveiborg M, Sehara-Fujisawa A, Mercurio AM and Wewer
UM: ADAM12 produced by tumor cells rather than stromal cells
accelerates breast tumor progression. Mol Cancer Res. 9:1449–1461.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roy R, Rodig S, Bielenberg D, Zurakowski D
and Moses MA: ADAM12 transmembrane and secreted isoforms promote
breast tumor growth: A distinct role for ADAM12-S protein in tumor
metastasis. J Biol Chem. 286:20758–20768. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kveiborg M, Fröhlich C, Albrechtsen R,
Tischler V, Dietrich N, Holck P, Kronqvist P, Rank F, Mercurio AM
and Wewer UM: A role for ADAM12 in breast tumor progression and
stromal cell apoptosis. Cancer Res. 65:4754–4761. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li H, Duhachek-Muggy S, Dubnicka S and
Zolkiewska A: Metalloproteinase-disintegrin ADAM12 is associated
with a breast tumor-initiating cell phenotype. Breast Cancer Res
Treat. 139:691–703. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rao VH, Vogel K, Yanagida JK, Marwaha N,
Kandel A, Trempus C, Repertinger SK and Hansen LA: Erbb2
up-regulation of ADAM12 expression accelerates skin cancer
progression. Mol Carcinog. 54:1026–1036. 2015. View Article : Google Scholar
|
|
12
|
Cheon DJ, Li AJ, Beach JA, Walts AE, Tran
H, Lester J, Karlan BY and Orsulic S: ADAM12 is a prognostic factor
associated with an aggressive molecular subtype of high-grade
serous ovarian carcinoma. Carcinogenesis. 36:739–747. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carl-McGrath S, Lendeckel U, Ebert M,
Roessner A and Röcken C: The disintegrin-metalloproteinases ADAM9,
ADAM12, and ADAM15 are upregulated in gastric cancer. Int J Oncol.
26:17–24. 2005.
|
|
14
|
Mino N, Miyahara R, Nakayama E, Takahashi
T, Takahashi A, Iwakiri S, Sonobe M, Okubo K, Hirata T, Sehara A
and Date H: A disintegrin and metalloprotease 12 (ADAM12) is a
prognostic factor in resected pathological stage I lung
adenocarcinoma. J Surg Oncol. 100:267–272. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peduto L, Reuter VE, Sehara-Fujisawa A,
Shaffer DR, Scher HI and Blobel CP: ADAM12 is highly expressed in
carcinoma-associated stroma and is required for mouse prostate
tumor progression. Oncogene. 25:5462–5466. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kodama T, Ikeda E, Okada A, Ohtsuka T,
Shimoda M, Shiomi T, Yoshida K, Nakada M, Ohuchi E and Okada Y:
ADAM12 is selectively overexpressed in human glioblastomas and is
associated with glioblastoma cell proliferation and shedding of
heparin-binding epidermal growth factor. Am J Pathol.
165:1743–1753. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rao VH, Kandel A, Lynch D, Pena Z, Marwaha
N, Deng C, Watson P and Hansen LA: A positive feedback loop between
HER2 and ADAM12 in human head and neck cancer cells increases
migration and invasion. Oncogene. 31:2888–2898. 2012. View Article : Google Scholar :
|
|
18
|
Georges S, Chesneau J, Hervouet S,
Taurelle J, Gouin F, Redini F, Padrines M, Heymann D, Fortun Y and
Verrecchia F: A disintegrin and metalloproteinase 12 produced by
tumour cells accelerates osteosarcoma tumour progression and
associated osteolysis. Eur J Cancer. 49:2253–2263. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Roy R and Moses MA: ADAM12 induces
estrogen-independence in breast cancer cells. Breast Cancer Res
Treat. 131:731–741. 2012. View Article : Google Scholar
|
|
20
|
Klionsky DJ: Autophagy revisited: A
conversation with Christian de Duve. Autophagy. 4:740–743. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mizushima N and Komatsu M: Autophagy:
Renovation of cells and tissues. Cell. 147:728–741. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kobayashi S: Choose delicately and reuse
adequately: The newly revealed process of autophagy. Biol Pharm
Bull. 38:1098–1103. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eisenberg-Lerner A, Bialik S, Simon HU and
Kimchi A: Life and death partners: apoptosis, autophagy and the
cross-talk between them. Cell Death Differ. 16:966–975. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
White E and DiPaola RS: The double-edged
sword of autophagy modulation in cancer. Clin Cancer Res.
15:5308–5316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thorburn A: Apoptosis and autophagy:
Regulatory connections between two supposedly different processes.
Apoptosis. 13:1–9. 2008. View Article : Google Scholar :
|
|
26
|
Doherty J and Baehrecke EH: Life, death
and autophagy. Nat Cell Biol. 20:1110–1117. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tan Z and Zhang Y, Deng J, Zeng G and
Zhang Y: Purified vitexin compound 1 suppresses tumor growth and
induces cell apoptosis in a mouse model of human choriocarcinoma.
Int J Gynecol Cancer. 22:360–366. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Deng J, Zhang Y and Tan Z: Proliferation
and apoptosis of choriocarcinoma cell JEG-3 induced by VB2 and its
in vitro mechanism. Zhong Nan Da Xue Xue Bao Yi Xue Ban.
38:476–782. 2013.In Chinese. PubMed/NCBI
|
|
29
|
Peng Z, Zhang C, Zhou W, Wu C and Zhang Y:
The STAT3/NFIL3 signaling axis-mediated chemotherapy resistance is
reversed by Raddeanin A via inducing apoptosis in choriocarcinoma
cells. J Cell Physiol. 233:5370–5382. 2018. View Article : Google Scholar :
|
|
30
|
Wu C, Yu S, Tan Q, Guo P and Liu H: Role
of AhR in regulating cancer stem cell-like characteristics in
choriocarcinoma. Cell Cycle. 17:2309–2320. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi D and Zhang Y, Lu R and Zhang Y: The
long non-coding RNA MALAT1 interacted with miR-218 modulates
choriocarcinoma growth by targeting Fbxw8. Biomed Pharmacother.
97:543–550. 2018. View Article : Google Scholar
|
|
32
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar
|
|
33
|
Denton D, Nicolson S and Kumar S: Cell
death by autophagy: Facts and apparent artefacts. Cell Death
Differ. 19:87–95. 2012. View Article : Google Scholar :
|
|
34
|
Anding AL and Baehrecke EH: Autophagy in
cell life and cell death. Curr Top Dev Biol. 114:67–91. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Arakawa S, Tsujioka M, Yoshida T,
Tajima-Sakurai H, Nishida Y, Matsuoka Y, Yoshino I, Tsujimoto Y and
Shimizu S: Role of Atg5-dependent cell death in the embryonic
development of Bax/Bak double-knockout mice. Cell Death Differ.
24:1598–1608. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shimizu S, Kanaseki T, Mizushima N, Mizuta
T, Arakawa-Kobayashi S, Thompson CB and Tsujimoto Y: Role of Bcl-2
family proteins in a non-apoptotic programmed cell death dependent
on autophagy genes. Nat Cell Biol. 6:1221–1228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lamy L, Ngo VN, Emre NC, Shaffer AL III,
Yang Y, Tian E, Nair V, Kruhlak MJ, Zingone A, Landgren O and
Staudt LM: Control of autophagic cell death by caspase-10 in
multiple myeloma. Cancer Cell. 23:435–449. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu L, Alva A, Su H, Dutt P, Freundt E,
Welsh S, Baehrecke EH and Lenardo MJ: Regulation of an ATG7-beclin
1 program of autophagic cell death by caspase-8. Science.
304:1500–1502. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Elgendy M, Sheridan C, Brumatti G and
Martin SJ: Oncogenic Ras-induced expression of Noxa and Beclin-1
promotes autophagic cell death and limits clonogenic survival. Mol
Cell. 42:23–35. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Byun JY, Yoon CH, An S, Park IC, Kang CM,
Kim MJ and Lee SJ: The Rac1/MKK7/JNK pathway signals upregulation
of Atg5 and subsequent autophagic cell death in response to
onco-genic Ras. Carcinogenesis. 30:1880–1888. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dasari SK, Bialik S, Levin-Zaidman S,
Levin-Salomon V, Merrill AH Jr, Futerman AH and Kimchi A:
Signalome-wide RNAi screen identifies GBA1 as a positive mediator
of autoph-agic cell death. Cell Death Differ. 24:1288–1302. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Debnath J, Baehrecke EH and Kroemer G:
Does autophagy contribute to cell death? Autophagy. 1:66–74. 2005.
View Article : Google Scholar
|
|
43
|
Dalby KN, Tekedereli I, Lopez-Berestein G
and Ozpolat B: Targeting the prodeath and prosurvival functions of
autophagy as novel therapeutic strategies in cancer. Autophagy.
6:322–329. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Susin SA, Lorenzo HK, Zamzami N, Marzo I,
Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler
M, et al: Molecular characterization of mitochondrial
apoptosis-inducing factor. Nature. 397:441–446. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen W, Li N, Chen T, Han Y, Li C, Wang Y,
He W, Zhang L, Wan T and Cao X: The lysosome-associated
apoptosis-inducing protein containing the pleckstrin homology (PH)
and FYVE domains (LAPF), representative of a novel family of PH and
FYVE domain-containing proteins, induces caspase-independent
apoptosis via the lysosomal-mitochondrial pathway. J Biol Chem.
280:40985–40995. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen D, Zheng X, Kang D, Yan B, Liu X, Gao
Y and Zhang K: Apoptosis and expression of the Bcl-2 family of
proteins and P53 in human pancreatic ductal adenocarcinoma. Med
Princ Pract. 21:68–73. 2012. View Article : Google Scholar
|
|
47
|
Gursan N, Karakök M, Sari I and Gursan MS:
The relationship between expression of p53/Bcl-2 and
histopathological criteria in breast invasive ductal carcinomas.
Int J Clin Pract. 55:589–590. 2001.
|
|
48
|
Hemann MT and Lowe SW: The p53-Bcl-2
connection. Cell Death Differ. 13:1256–1259. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wesselborg S and Stork B: Autophagy signal
transduction by ATG proteins: From hierarchies to networks. Cell
Mol Life Sci. 72:4721–4757. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI,
Woo HN, Cho DH, Choi B, Lee H, Kim JH, et al: Essential roles of
Atg5 and FADD in autophagic cell death: Dissection of autophagic
cell death into vacuole formation and cell death. J Bio Chem.
280:20722–20729. 2005. View Article : Google Scholar
|
|
51
|
Mehrpour M, Esclatine A, Beau I and
Codogno P: Overview of macroautophagy regulation in mammalian
cells. Cell Res. 20:748–762. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Otomo C, Metlagel Z, Takaesu G and Otomo
T: Structure of the human ATG12~ATG5 conjugate required for LC3
lipidation in autophagy. Nat Struct Mol Biol. 20:59–66. 2013.
View Article : Google Scholar :
|
|
53
|
Cadwell K, Stappenbeck TS and Virgin HW:
Role of autophagy and autophagy genes in inflammatory bowel
disease. Curr Top Microbiol Immunol. 335:141–167. 2009.PubMed/NCBI
|
|
54
|
Fésüs L, Demény MÁ and Petrovski G:
Autophagy shapes inflammation. Antioxid Redox Signal. 14:2233–2243.
2011. View Article : Google Scholar
|
|
55
|
Levine B, Mizushima N and Virgin HW:
Autophagy in immunity and inflammation. Nature. 469:323–335. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen YM, Chang CY, Chen HH, Hsieh CW, Tang
KT, Yang MC, Lan JL and Chen DY: Association between autophagy and
inflammation in patients with rheumatoid arthritis receiving
biologic therapy. Arthritis Res Ther. 20:2682018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Saitoh T and Akira S: Regulation of innate
immune responses by autophagy-related proteins. J Cell Biol.
189:925–935. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Deretic V: Multiple regulatory and
effector roles of autophagy in immunity. Curr Opin Immunol.
21:53–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Harris J: Autophagy and cytokines.
Cytokine. 56:140–144. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Harris J and Keane J: How tumour necrosis
factor blockers interfere with tuberculosis immunity. Clin Exp
Immunol. 161:1–9. 2010.PubMed/NCBI
|
|
61
|
Yang R, Zhang Y, Wang L, Hu J, Wen J, Xue
L, Tang M, Liu Z and Fu J: Increased autophagy in fibroblast-like
synoviocytes leads to immune enhancement potential in rheumatoid
arthritis. Oncotarget. 8:15420–15430. 2017.PubMed/NCBI
|
|
62
|
Harris J, Lang T, Thomas JPW, Sukkar MB,
Nabar NR and Kehrl JH: Autophagy and inflammasomes. Mol Immunol.
86:10–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Shi CS, Shenderov K, Huang NN, Kabat J,
Abu-Asab M, Fitzgerald KA, Sher A and Kehrl JH: Activation of
autophagy by inflammatory signals limits IL-1β production by
targeting ubiquitinated inflammasomes for destruction. Nat Immunol.
13:255–263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Morselli E, Galluzzi L, Kepp O, Vicencio
JM, Criollo A, Maiuri MC and Kroemer G: Anti- and pro-tumor
functions of autophagy. Biochim Biophys Acta. 1793:1524–1532. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wu H and Lozano G: NF-kappa B activation
of p53. A potential mechanism for suppressing cell growth in
response to stress. J Biol Chem. 269:20067–20074. 1994.PubMed/NCBI
|
|
66
|
Catz SD and Johnson JL: Transcriptional
regulation of bcl-2 by nuclear factor kappa B and its significance
in prostate cancer. Oncogene. 20:7342–7351. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Grimm S, Bauer MK, Baeuerle PA and
Schulze-Osthoff K: Bcl-2 down-regulates the activity of
transcription factor NF-kappaB induced upon apoptosis. J Cell Biol.
134:13–23. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Boakye YD, Groyer L and Heiss EH: An
increased autophagic flux contributes to the antiinflammatory
potential of urolithin A in macrophages. Biochim Biophys Acta Gen
Subj. 1862:61–70. 2018. View Article : Google Scholar
|
|
69
|
Liu T, Zhang L, Joo D and Sun SC: NF-κB
signaling in inflammation. Signal Transduct Target Ther. 2:pii:
17023. 2017. View Article : Google Scholar
|
|
70
|
Davignon JL, Hayder M, Baron M, Boyer JF,
Constantin A, Apparailly F, Poupot R and Cantagrel A: Targeting
mono-cytes/macrophages in the treatment of rheumatoid arthritis.
Rheumatology (Oxford). 52:590–598. 2013. View Article : Google Scholar
|
|
71
|
Ip WKE, Hoshi N, Shouval DS, Snapper S and
Medzhitov R: Anti-inflammatory effect of IL-10 mediated by
metabolic reprogramming of macrophages. Science. 356:513–519. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ko JH, Yoon SO, Lee HJ and Oh JY:
Rapamycin regulates macrophage activation by inhibiting NLRP3
inflammasome-p38 MAPK-NFκB pathways in autophagy- and p62-dependent
manners. Oncotarget. 8:40817–40831. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Inoki K and Guan KL: Complexity of the TOR
signaling network. Trends Cell Biol. 16:206–212. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Reiling JH and Sabatini DM: Stress and
mTORture signaling. Oncogene. 25:6373–6383. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sarbassov DD, Ali SM and Sabatini DM:
Growing roles for the mTOR pathway. Curr Opin Cell Biol.
17:596–603. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wu YT, Tan HL, Shui G, Bauvy C, Huang Q,
Wenk MR, Ong CN, Codogno P and Shen HM: Dual role of
3-methylad-enine in modulation of autophagy via different temporal
patterns of inhibition on class I and III phosphoinositide
3-kinase. J Biol Chem. 285:10850–10861. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Jung CH, Ro SH, Cao J, Otto NM and Kim DH:
mTOR regulation of autophagy. FEBS Lett. 584:1287–1295. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Kim YC and Guan KL: mTOR: A pharmacologic
target for autophagy regulation. J Clin Invest. 125:25–32. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Paquette M, EI-Houjeiri L and Pause A:
mTOR pathways in cancer and autophagy. Cancers (Basel). 10:pii:
E18. 2018. View Article : Google Scholar
|
|
80
|
Dunlop EA and Tee AR: mTOR and autophagy:
A dynamic relationship governed by nutrients and energy. Semin Cell
Dev Biol. 36:121–129. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Nah J, Yuan J and Jung YK: Autophagy in
neurodegenerative diseases: From mechanism to therapeutic approach.
Mol Cells. 38:381–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Guo F, Liu X, Cai H and Le W: Autophagy in
neurodegenerative diseases: Pathogenesis and therapy. Brain Pathol.
28:3–13. 2018. View Article : Google Scholar
|