Introduction
CRC has a high mortality rate in China (1,2),
accounting for >600,000 deaths annually, despite the advances in
treatment. The high CRC mortality rates are due to the high
frequency of tumor recurrence following surgical resection
(3,4). Hence, it is important to investigate
the molecular and cellular growth processes of CRC in order to
identify potential treatment targets.
MicroRNAs (miRNAs/miRs) are aberrantly expressed in
numerous types of cancers and can function either as tumor
suppressors or oncogenes. For example, Zhang et al (5) found miR-187 was downregulated in
colorectal cancer, while Zhang et al (6) verified miR-646 was downregulated in
gastric cancer (GC) tissues and Liu et al (7) found that miR-935 was upregulated in
liver cancer tissues and cells. miRNAs are a single-stranded class
of RNAs that cause inhibition of gene expression at the
post-transcriptional level by instantly binding to the
3′-untranslated regions (3′UTRs) of mRNAs. This inhibits their
translation process and causes degradation. miR-500a is located
within the p11 locus on the X chromosome and has two arms:
miR-500a-5p and miR-500a-3p. hsa-miR-500a-5p and hsa-miR-500a-3p
were previously referred to as hsa-miR-500a and
hsa-miR-500a*, respectively (http://mirdb.org/cgi-bin/search.cgi). miR-500a-5p was
previously reported to be an important factor in the development of
cancer. The expression of miR-500a-5p was found to be markedly
upregulated in human hepatocellular carcinoma (8), chronic lymphocytic leukemia (9), breast cancer (10) and lung cancer (11), while it was found to be
downregulated in CRC (12),
indicating miR-500a-5p has a role in tumorigenesis. However, the
role of miR-500a-5p has not been fully elucidated in CRC.
EMT is a key step in tumor metastasis (13,14).
EMT represents the loss of cell polarity and cell-cell adhesion by
epithelial cells, which results in transformation to a mesenchymal
phenotype, as well as migration and invasion, providing a mechanism
for cancer development (15,16).
Keratin and vimentin filaments regulate the intermediate filament
composition, while downregulation of E-cadherin reinforces the
destabilization of adhesion junctions and MMP9 enhances
extracellular matrix protein degradation and enables invasion
(17). Repression of epithelial
markers (cytokeratins and E-cadherin) and induction of mesenchymal
markers vimentin and matrix metallopeptidase (MMP)-9, constitute an
essential part of the EMT process. EMT is physiologically initiated
by certain autocrine factors, with TGF-β being the strongest
inducer that functions in the majority of epithelial cell types
tested in vivo (18).
Various studies have revealed that miRNAs affect the regulation of
EMT during cancer progression and metastasis (5,6,19).
For example, tumor suppressor miR-1271 binds to a particular target
mRNA in EMT-related genes, ZEB1 and TWIST1, and increased
expression of miR-1271 is considered to be an adverse prognostic
indicator in pancreatic cancer (19). Furthermore, miR-500a-5p has been
found to act as a tumor suppressor in CRC (12). However, the mechanism underlying
the tumor suppressor miR-500a-5p during EMT in CRC remains
unclear.
The Rhubarb plant roots (Rheum palmatum L.),
which are used as laxatives in Traditional Chinese Medicine,
contain rhein, aloe-emodin, and emodin (20-22).
Emodin is known for its antibacterial, anti-inflammatory and
immunosuppressive properties (23). The anticancer properties of emodin
have been investigated primarily in lung (24), liver (25) and stomach cancer (26), as well as CRC (27). It was previously demonstrated that
emodin can downregulate the expression of the mesenchymal marker,
vimentin, and upregulate the expression of epithelial markers, such
as E-cadherin, in gastric and pancreatic cancer cells by increasing
the content of miRNA (28,29). However, the mechanism by which
emodin regulates miR-500a-5p expression through EMT-like processes
in CRC cells has not been investigated.
The aim of the present study was to investigate
whether transfecting CRC cells with miR-500a-5p inhibitors would
result in EMT phenotypes, and to determine the role of transforming
growth factor (TGF)-β in the invasion of tumor cells, as well as
the effect of miR-500a-5p on TGF-β1-induced EMT. In addition, the
effect of emodin on miR-500a-5p inhibitor-induced EMT in CRC cells
was also investigated.
Materials and methods
Reagents and cell culture
The human SW1116 (Dukes′ type A, grade III), SW480
(Dukes' type B) and LoVo (Dukes′ type C, grade IV) CRC cell lines
were purchased from American Type Culture Collection (30). The cells were cultured using
RPMI-1640 medium (Thermo Fisher Scientific, Inc.) and supplemented
with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific,
Inc.), 100 mg/ml streptomycin and 100 mg/ml penicillin at 37°C in a
humidified incubator with 5% CO2.
Recombinant human TGF-β1 (cat. no. 240-B) was
purchased from R&D Systems, Inc. Emodin was purchased from
Sigma-Aldrich (Merck KGaA). Rabbit polyclonal anti-E-cadherin [cat.
no. 20874-1-AP; western blot analysis: 1:5,000 dilution;
immunohistochemistry (IHC), 1:1,000 dilution; immunofluorescence,
1:100 dilution] and mouse monoclonal anti-vimentin (cat. no.
60330-1-Ig; western blot analysis: 1:5,000 dilution; IHC, 1:2,000
dilution; immunofluorescence, 1:100 dilution) antibodies were
purchased from ProteinTech Group, Inc. Mouse monoclonal anti-GAPDH
antibody (cat. no. RM2002; western blot analysis: 1:5,000 dilution)
was purchased from Beijing Ray Antibody Biotech. Secondary
antibodies, including Alexa Fluor 594 (red)-conjugated goat
anti-rabbit IgG (cat. no. ZF-0516) and Alexa Fluor 488
(green)-conjugated goat anti-mouse IgG antibodies (cat. no.
ZF-0512) were purchased from OriGene Technologies, Inc.
Oligonucleotide transfection
miR-500a-5p mimics, miR-500a-5p mimics control
(m-NC), miR-500a-5p inhibitor and miR-500a-5p inhibitor control
(i-NC) were all purchased from Shanghai GenePharma Co., Ltd.. For
miRNA transfections, cells were seeded at a density of
5×104 cells/ml into 24-well plates 1-2 days prior to
transfection until the confluency reached 50-60%. miR-500a-5p
mimics (40 nM), miR-500a-5p inhibitor (100 nM) or their
corresponding controls miR (m-NC, 40 nM and i-NC, 100 nM) were
trans-fected into CRC cells using Lipofectamine® 3000
reagent for 36 h (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Following which,
transfection efficacy was determined using reverse
transcription-quantitative PCR (RT-qPCR). When m-NC or mimics were
combined with treatment of TGF-β1, the CRC cells were transfected
with miR-500a-5p mimics or m-NC for 24 h, then treated with 5 ng/ml
TGF-β1 for 24 h. When i-NC or miR-500a-5p inhibitor were combined
with treatment of emodin, 80 μM Emodin was used for 48 h
following transfection with i-NC or miR-500a-5p inhibitor for 24
h.
Treatment with TGF-β1
Recombinant human TGF-β1 (cat. no. 240-B) was
purchased from R&D Systems, Inc. The SW1116 and LoVo cells were
transfected with miR-500a-5p or m-NC for 24 h, and then treated
with 0, 1, 3, 5 ng/ml TGF-β1 for 48 h or 5 ng/ml TGF-β1 for 0, 12,
24 and 48 h. The expression of miR-500a-5p was subsequently
detected using qPCR.
Western blot analysis and protein
extraction
Protein was extracted from cells that reached 80-90%
confluency during the exponential growth phase and protein
expression was detected using western blot analysis. For emodin
treatment, cells were seeded in the 6-well plate and different
concentrations (0, 20, 40, 60, 80 μM) of emodin were added
to the medium until the cell density reached 80% for 48 h. Cells
were digested on the ice with RIPA lysis buffer (cat. no. P0013K;
Beyotime Institute of Biotechnology), including a protease
inhibitor. Protein were separated using 10% SDS-PAGE and proteins
were transferred onto Immobilon-P membranes (EMD Millipore; Merck
KGaA). After blocking with 5% skimmed milk in PBS-Tween-20 at room
temperature for 1 h, the membranes were incubated with
aforementioned primary antibodies overnight at 4°C. Subsequently, a
horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (cat.
no. ZB-2301) or goat anti-mouse IgG (cat. no. ZB-2305) secondary
antibodies (dilution 1:10,000; both OriGene Technologies, Inc.) was
used at room temperature for 1 h. The proteins were visualised
using ECL (PerkinElmer, Inc.).
RT-qPCR
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol and quantified with
a Nanodrop 2000 (Thermo Fisher Scientific, Inc.). RT was performed
using the Thermoscript RT System (Invitrogen; Thermo Fisher
Scientific, Inc.). The sequences of the primer used were as
follows: E-cadherin forward, 5′-TGC CCA GAA AAT GAA AAA GG-3′ and
reverse, 5′-GTG TAT GTG GCA ATG CG T TC-3′; vimentin forward,
5′-GGA GCT ACG TGA CTA CGT CCA-3′ and reverse, 5′-CTT GAA CTC GGT
GTT GAT GG-3′; GAPDH (used as internal control) forward, 5′-GTC AAC
GGA TTT GGT CGT ATTG-3′ and reverse, 5′-CTC CTG GAA GAT GGT GAT
GGG-3′.
For miR-500a-5p expression analysis, miRNA reverse
transcription reaction was performed using All-in-One™ miRNA
First-Strand cDNA Synthesis kit (cat. no. AMRT-0020;) and qPCR was
performed using All-in-OneTM miRNA RT-qPCR Detection kit
(cat. no. AOMD-Q020; both GeneCopoeia, Inc.). The miR-500a-5p
primer (cat. no. HmiRQP0545) and U6 primer (cat. no. HmiRQP9001)
were purchased from GeneCopoeia, Inc.. The thermocycling conditions
for qPCR was set as pre-denaturation at 95°C for 10 min, and 40
cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 20
sec, and extension at 72°C for 20 seconds. The small nuclear RNA U6
was used as the internal reference. The relative expression was
calculated using the 2-ΔΔCq method (31).
Immunofluorescence
SW1116 or LoVo cells were cultured at a density of
1x105 cells/ml on coverslips and cultured at normal
culture conditions (as aforementioned) until the cells reached
30-40% confluency and then were fixed with 4% parafor-maldehyde at
room temperature for 30 min. Then permeated with 0.2% Triton X-100
for 5 min. Non-specific binding was blocked using 1% bovine serum
albumin (Sigma-Aldrich; Merck KGaA). The cells were incubated with
aforementioned primary antibodies against E-cadherin and vimentin.
Following washing with PBS, the cells were incubated with Alexa
Fluor 594 (red)-conjugated goat anti-rabbit IgG and Alexa Fluor 488
(green)-conjugated goat anti-mouse IgG antibodies with dilution of
1:100. Cell nuclei were stained with 1 μg/ml Hoechst 33258
at room temperature for 30 min. The coverslips were then examined
using an Olympus CKX 41 fluorescence microscope (Olympus
Corporation) at x400 magnification.
F-actin cytoskeleton staining
The cells were cultured at a density of
1x105 cells/ml with 10% FBS at 37°C and 5%
CO2 on coverslips in a 24-well chamber until the cells
reached 30-40% confluency. PBS was used to wash the cells,
following which they were fixed in 4% paraformaldehyde at room
temperature for 30 min. Following treatment with 0.2% Triton X-100,
the cells were incubated with 5 U/ml phalloidin-FITC (Molecular
Probes; Thermo Fisher Scientific, Inc.) in PBS for 1 h at a 1:40
dilution. The nuclei were stained with 1 μg/ml Hoechst 33258
at room temperature for 30 min. F-actin cytoskeleton staining was
evaluated using a fluorescence microscope at ×400
magnification.
Cell invasion analysis
Transwell chambers (BD Biosciences) were used to
analyse the invasion ability of CRC cells. The LoVo or SW1116 cells
were transfected with miR-500a-5p mimics or inhibitor and suspended
in serum-free RPMI-1640 medium. A total of 200 μl RPMI-1640
medium (1×105 cells/ml) were placed into the upper
chamber of Matrigel-coated (at 37°C for 2 h) Transwell chambers (BD
Biosciences) with a PC membrane with a pore size of 8.0 μm.
RPMI-1640 (500 μl) supplemented with 20% FBS was placed in
the bottom chamber. After 1 complete day, residual cells on the
upper surface of the membrane were carefully detached with a cotton
swab. The cells that had migrated to the lower surface of the
membrane were fixed with methanol for 15 min at room temperature
and visualized by staining with 0.1% crystal violet solution at
room temperature for 30 min. Images of the invading cells were
obtained and were quantified using Axiovert 200 inverted microscope
(Carl Zeiss AG) at a magnification of x200, in at least five random
fields per filter. Each experiment was repeated independently,
three times.
Lentiviral vector construction and
transduction
The lentiviral vector was transducted and used for
subsequent experimentation after two weeks of puromycin (2
μg/ml) resistance screening. The miR-500a-5p lentivirus was
constructed by Shanghai GeneChem Co., Ltd. The red fluorescent
protein (RFP) and puromycin resistance genes (Shanghai GeneChem
Co., Ltd.) were cloned into a miR-500a-5p lentiviral expression
vector (Ubi-MCS-SV40-Cherry) and then transduced into the
lentiviral packaging cell line 293T. Polybrene (Shanghai GeneChem
Co., Ltd.) was added to the CRC cell lines to a final concentration
of 5 μg/ml for stable transduction. Screening for drug
resistance with puromycin was performed for 2 weeks to construct
stable expression cells. Fluorescence was observed under a
fluorescence microscope at ×100 magnification and the efficiency
was verified using qPCR.
Animal models
A total of 6 BALB/c-nu/nu female mice, (4-6 weeks
old; weight 16-20 g), were purchased from the Laboratory Animal
Unit, Southern Medical University. Mice were housed three per cage
in a specific-pathogen free laboratory and maintained with food and
water ad libitum at a constant ambient temperature with 12-h
light/dark cycle. To evaluate the miR-500a-5p in vivo
effects on the metastatic ability of CRC cells, nude mice were
randomly divided into two groups (m-NC-RFP and miR-500a-5p-RFP
group; n=3 per group). Subsequently, 5×106 m-NC-RFP/LoVo
or miR-500a-5p-RFP/LoVo cells that were transducted with
lentivirus, as aforementioned, (50 μl) were administered by
slow injection into the spleen via a 25-gauge needle. After 4
weeks, the mice were euthanized and their livers were removed.
In Vivo F Imaging System (Kodak) was used to evaluate the
fluorescence and number of tumor metastasis. Liver metastatic
tissues (4-μm) were fixed in 10% formalin for 24 h,
paraffin-embedded at room temperature (Wuhan Servicebio Technology
Co., Ltd.), and subsequently stained with hema-toxylin and eosin (H
& E) and IHC. Part of the liver metastatic tissues and adjacent
normal liver tissue, 0.5 cm away from the tumor tissue, were used
for RNA extraction and measurement of mRNA using qPCR. All animal
studies were conducted in accordance with the principles and
procedures outlined in the Southern Medical University of China
Guide for the Care and Use of Animals.
H & E staining
For H and E staining, 4-μm thick paraffin
embedded sections from liver metastatic tissues of nude mice were
prepared. The sections were washed in xylene twice for 20 min, then
in a descending alcohol series (100% ethanol for 20 min, 100%
ethanol for 5 min, 95% alcohol for 5 min, 90% alcohol for 5 min,
80% alcohol for 5 min, 70% alcohol for 5 min) then distilled water
for 5 min. The sections were stained with Harris hematoxylin (cat.
no. G1004-250; Tiengen Biotech Co., Ltd.) for 3-8 min, and washed
with distilled water for 5 min. The sections were subsequently
differentiated with 1% hydrochloric acid alcohol for 2 sec then
they were washed with distilled water for 5 min, 0.6% ammonia for 5
min, and lastly distilled water for 5 min. The sections were
further stained with eosin (cat. no. G1210-2; Tiengen Biotech Co.,
Ltd.) staining solution for 1-3 min, then washed in an ascending
alcohol series (95% alcohol twice for 5 min, 100% ethanol twice for
5 min), and then xylene twice for 5 min. Lastly the sections were
air dried and sealed with natural resin All the aforementioned
steps were conducted at room temperature.
IHC
Paraffin-embedded sections from liver metastatic
tissues of nude mice were cut into 4-μm sections and
transferred to glass slides. The slides were deparaffinised with
xylene, rehydrated in a descending ethanol series and washed, as
aforementioned. The sections were then immersed in 3% hydrogen
peroxide to block endogenous peroxidase activity at room
temperature for 20 min. The antigen retrieval step were conducted
using citrate buffer at 95°C for 30 min. Non-specific binding was
blocked using 5% bovine serum albumin (Sigma-Aldrich; Merck KGaA)
at room temperature for 1 h. The sections were subsequently
incubated with anti-E-cadherin or anti-vimentin primary antibody at
4°C overnight then incubated for 1 h at room temperature with a
HRP-conjugated goat anti-rabbit IgG (cat. no. PV-6001-6.0) or goat
anti-mouse IgG (cat. no. PV-6002-3.0; both OriGene Technologies,
Inc.) secondary antibodies. DAB staining solution (cat. no.
ZLI-9017; OriGene Technologies, Inc.) was added for 2-8 min and the
nuclei were counterstained with hematoxylin for 5 min at room
temperature.
Bioinformatics and statistical
analysis
The targets of miRNA were predicted using two
bioinformatics algorithms: mirwalk2 (http://mirdb.org/miRDB) and TargetScan (http://www.targetscan.org/vert_72). All
statistical analyses were performed using SPSS version 22.0 (IBM
Corp.). The numerical data with biological replicates are expressed
as mean ± standard error. Mann-Whitney U test was used to compare
data between two groups. One-way ANOVA was used when comparing the
means of multiple groups. Dunnett's post hoc test was used to
compare the experimental groups with the control group while
Bonferroni's post hoc test was used to compare multiple
experimental groups following one-way ANOVA. P<0.05 was
considered to indicate statistically significant difference. The
experimental procedures were performed three times.
Results
miR-500a-5p inhibits EMT in CRC
cells
Our previous research demonstrated that miR-500a-5p
is a tumor suppressor and inhibits CRC growth (12). To investigate the role of
miR-500a-5p in CRC progression, CRC cells were first transfected
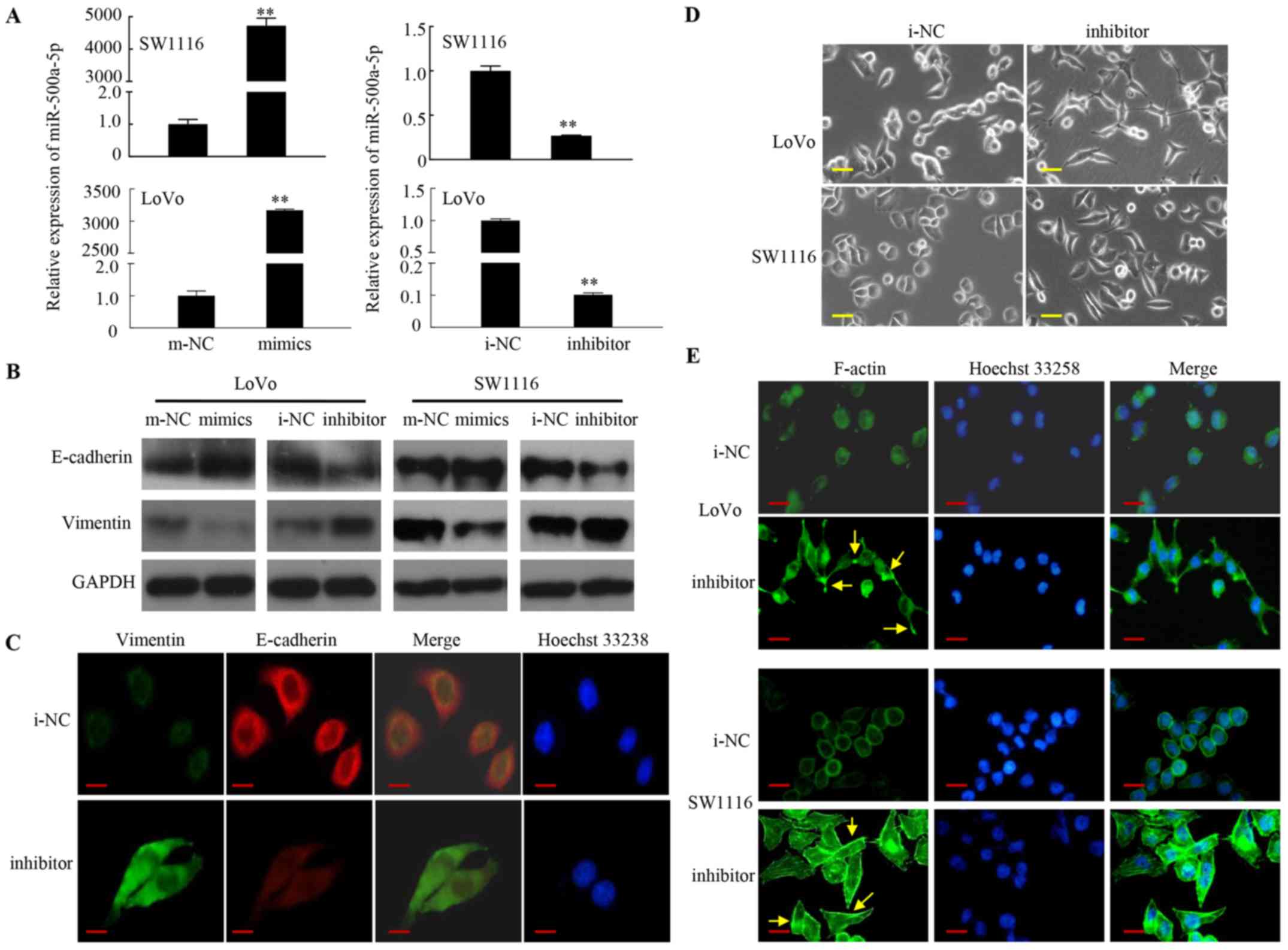
with miR-500a-5p mimics or inhibitor. As shown in Fig. 1A, miR-500a-5p was significantly
upregulated in LoVo-mimics or SW1116-mimics group and downregulated
in LoVo-inhibitor or SW1116-inhibitor group compared with that in
the negative control (NC) groups.
Second, the function of miR-500a-5p in CRC EMT was
investigated. The expression levels of the EMT biomarkers changed
in the CRC cell lines following transfection with mimics or with
m-NC. The results of the western blot analysis revealed that
vimentin expression was markedly reduced in the CRC cells
transfected with miR-500a-5p mimics compared with that in cells
transfected with m-NC, while E-cadherin expression was notably
upregulated. By contrast, the expression of vimentin, was
upregulated, whereas that of E-cadherin was downregulated in the
CRC cells transfected with miR-500a-5p inhibitor compared with that
in cells transfected with i-NC (Fig.
1B).
Studies have shown that oncogenic miRNA promotes
tumor cell EMT and metastasis (32,33)
and it was found that miR-500a-5p acts as a tumor suppressor in CRC
(12). Third, it was determined
whether miR-500a-5p inhibitor has any effect on cell EMT. The
SW1116 cells transfected with miR-500a-5p inhibitor exhibited
decreased E-cadherin expression and increased vimentin expression
using immunofluorescence staining under fluorescence microscopy
(Fig. 1C).
Fourth, it was observed that the cell structure of
LoVo and SW1116 cells transfected with miR-500a-5p inhibitor was
transformed from a cubical shape via cellular bonding to a
spindle-like shape, and caused spreading of the cells. These
changes reflected EMT and were observed via a phase-contrast
microscope (Fig. 1D).
Traditionally, actin cytoskeleton transformation is
downstream of EMT reprogramming (34). Notably, it is raises questions on
how miR-500a-5p can reverse EMT. Finally, the results demonstrated
that F-actin filaments formed thick and regular subcortical bundles
in LoVo and SW1116 cells trans-fected miR-500-a-5p inhibitor
compared with that in cells transfected with i-NC (Fig. 1E). Therefore, these data indicate
that miR-500a-5p inhibits EMT in CRC cells.
TGF-β1-induced EMT is inhibited by
miR-500a-5p in CRC cells
TGFβ can activate EMT transcription factors through
SMAD-mediated changes in gene expression and signaling pathways
that are not mediated by SMADs (17). TGF-β regulates/induces EMT together
with other factors (e.g., FGF2 and EGF) (35) and miRNAs regulate TGF-β1-induced
EMT in multiple types of cancer. For example, miR-646 attenuates
the TGF-b1-induced EMT of gastric cancer cells and miR-141
suppresses EMT in laryngeal cancer through HOXC6-dependent TGF-β
signaling pathway (6,36). However, the role of miR-500a-5p in
TGF-β1-induced EMT in CRC cells remains unknown. It was found that
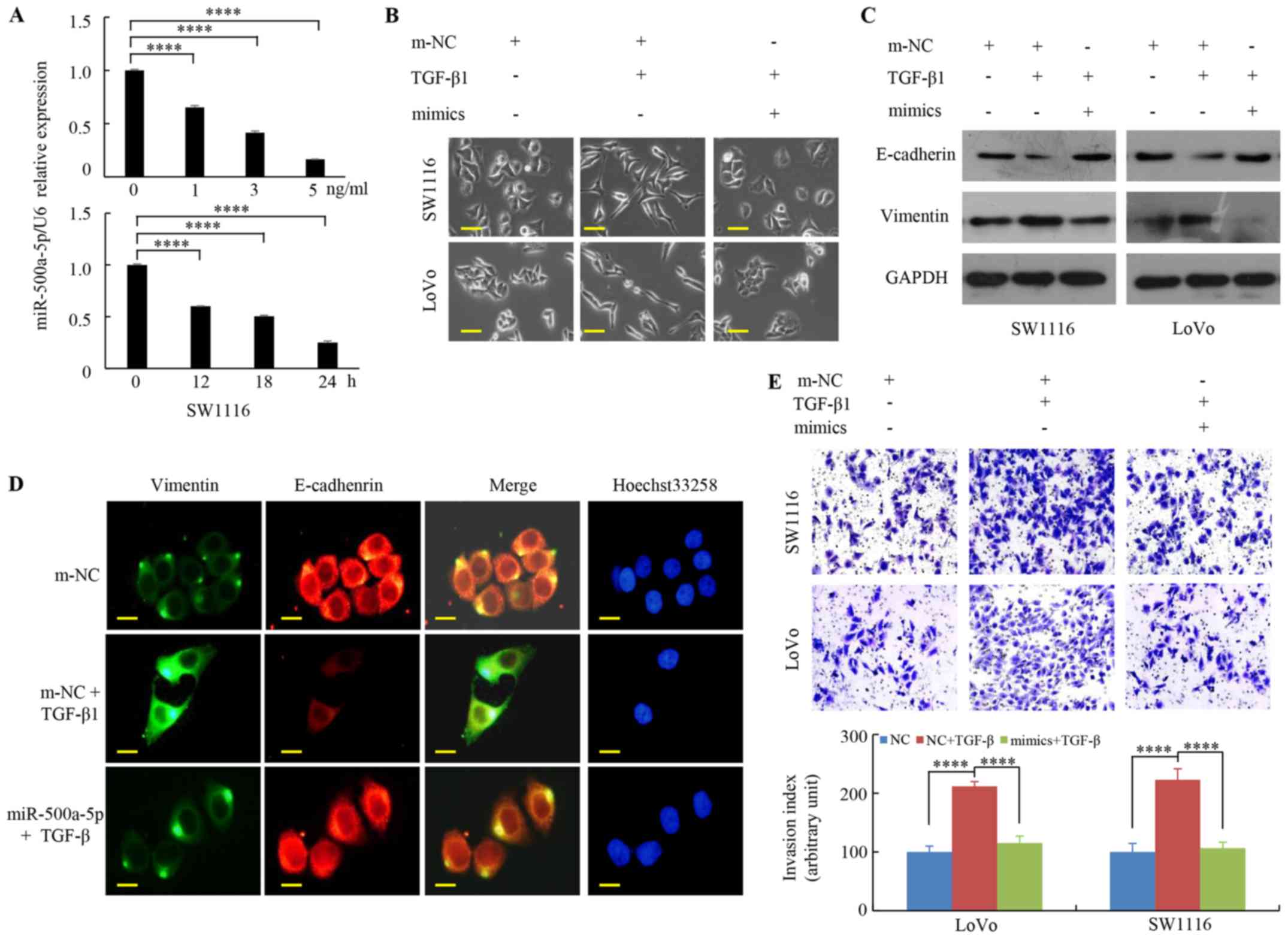
treatment with TGF-β1 significantly reduced miR-500a-5p expression
in a dose/time-dependent manner (Figs.
2A and S1). It was previously
demonstrated that
CRC cells exhibited a myofibroblast-like phenotype
caused by TGF-β1 (34). As shown
in Fig. 2B, enhancing miR-500a-5p
expression suppressed the transformation of CRC cells to a
myofibroblast structure when treated with TGF-β1. Moreover,
increased miR-500a-5p expression was associated with a markedly
decrease in the protein levels of vimentin and an increase in the
protein levels of E-cadherin when compared with that in the TGF-β1
group (Fig. 2C). The green signals
denote Vimentin staining, while the red signals denote E-cadherin
staining using immunofluorescence staining under a fluorescence
microscopy. The results revealed that TGF-β1 treatment decreased
the expression of E-cadherin and increased the expression of
vimentin; while miR-500a-5p overexpression reversed the
TGF-β1-mediated upregulation of vimentin and downregulation of
E-cadherin (Fig. 2D). Furthermore,
the Transwell experiments demonstrated that upregulated miR-500a-5p
expression inhibited the effects of TGF-β1 on tumor cell invasion
ability (Fig. 2E). The results
suggest that miR-500a-5p reduces TGF-β1-induced EMT in CRC
cells.
miR-500a-5p inhibitor-induced EMT is
suppressed by emodin in CRC cells
It was previously reported that emodin inhibits RKO
CRC cell invasion and migration by suppressing EMT (37). A total of three CRC cell lines at
Dukes′ stage A (SW1116), B (SW480) and C (LoVo) were selected from
the American Type Culture Collection to investigate whether the
effect of emodin is independent of the progression status in cells
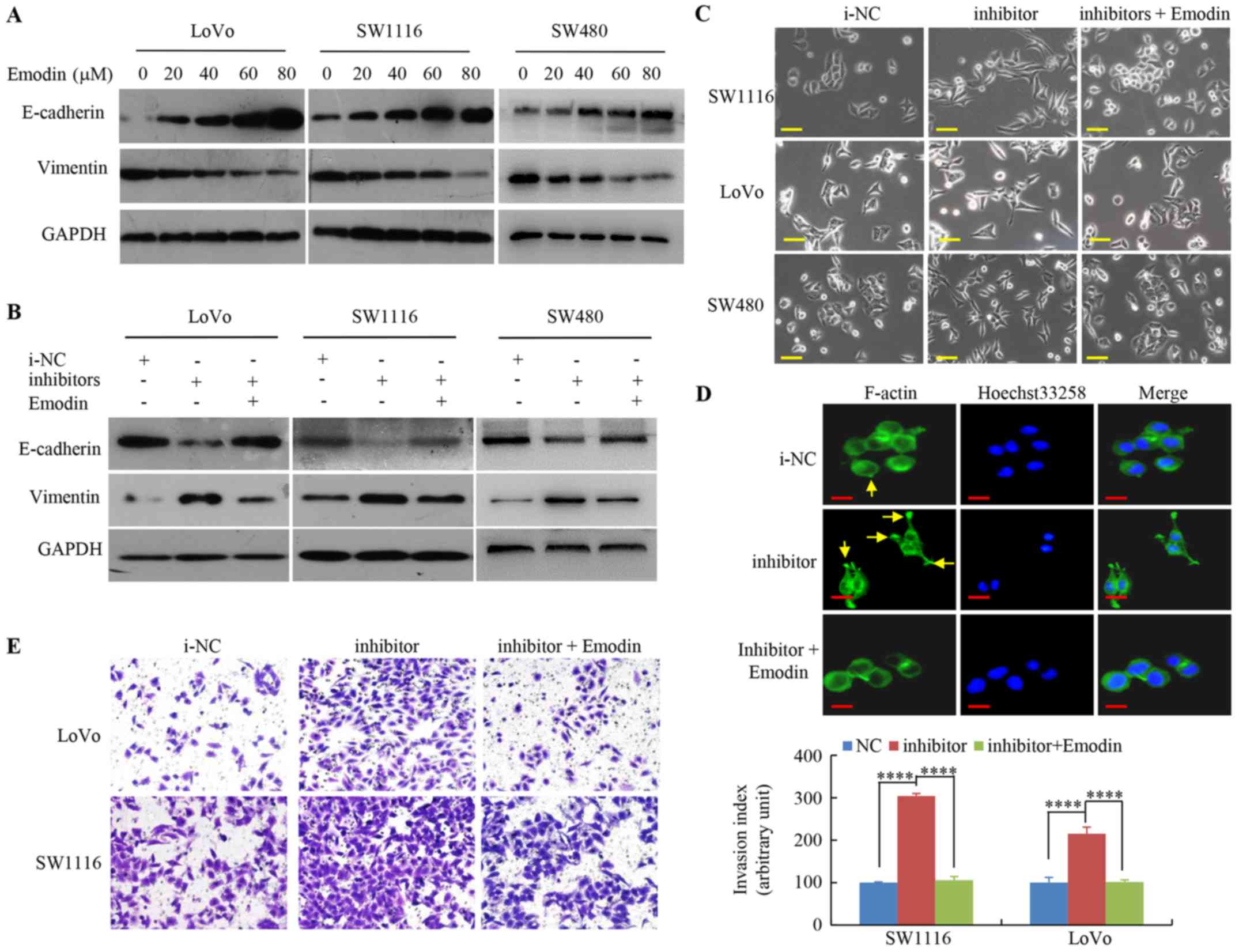
undergoing EMT. The western blot analysis indicated that increasing
emodin concentrations markedly decreased the mesenchymal marker
expression, vimentin, but increased the epithelial marker,
E-cadherin, levels in SW1116, SW480 and LoVo cells (Fig. 3A).
To highlight the emodin regulation of miR-500a-5p in
EMT during CRC cell metastasis, the EMT marker levels were
investigated. The protein expression level of E-cadherin was
significantly decreased, and expression level of vimentin was
increased by miR-500a-5p inhibitor, whereas emodin treatment
partially reversed EMT compared with that in i-NC CRC cells
(Fig. 3B).
Furthermore, miR-500a-5p inhibitor promoted EMT and
lead to mesenchymal changes in CRC cells, which resulted in a long
fusiform shape, whereas cells treated with emodin and miR-500-a-5p
inhibitor did not exhibit scattering as seen in cells transfected
with miR-500a-5p inhibitor only (Fig.
3C).
In addition, in the miR-500a-5p inhibitor group,
F-actin was distributed along the long axis of the cells, which is
a hallmark of the mesenchymal phenotype. However, treatment of the
miR-500a-5p inhibitor-transfected cells with emodin caused
mesenchymal-to-epithelial transition, which is the opposite of EMT
(Fig. 3D).
Lastly, it was observed that cells transfected with
miR-500a-5p inhibitor significantly increased the numbers of
invading cells, whereas the activating effect of the miR-500a-5p
inhibitor on CRC cell invasion was abolished by emodin (Fig. 3E). This result indicates that
emodin suppresses miR-500a-5p inhibitor-induced EMT in CRC
cells.
miR-500a-5p affects tumor metastasis by
regulating EMT in CRC cells in vivo
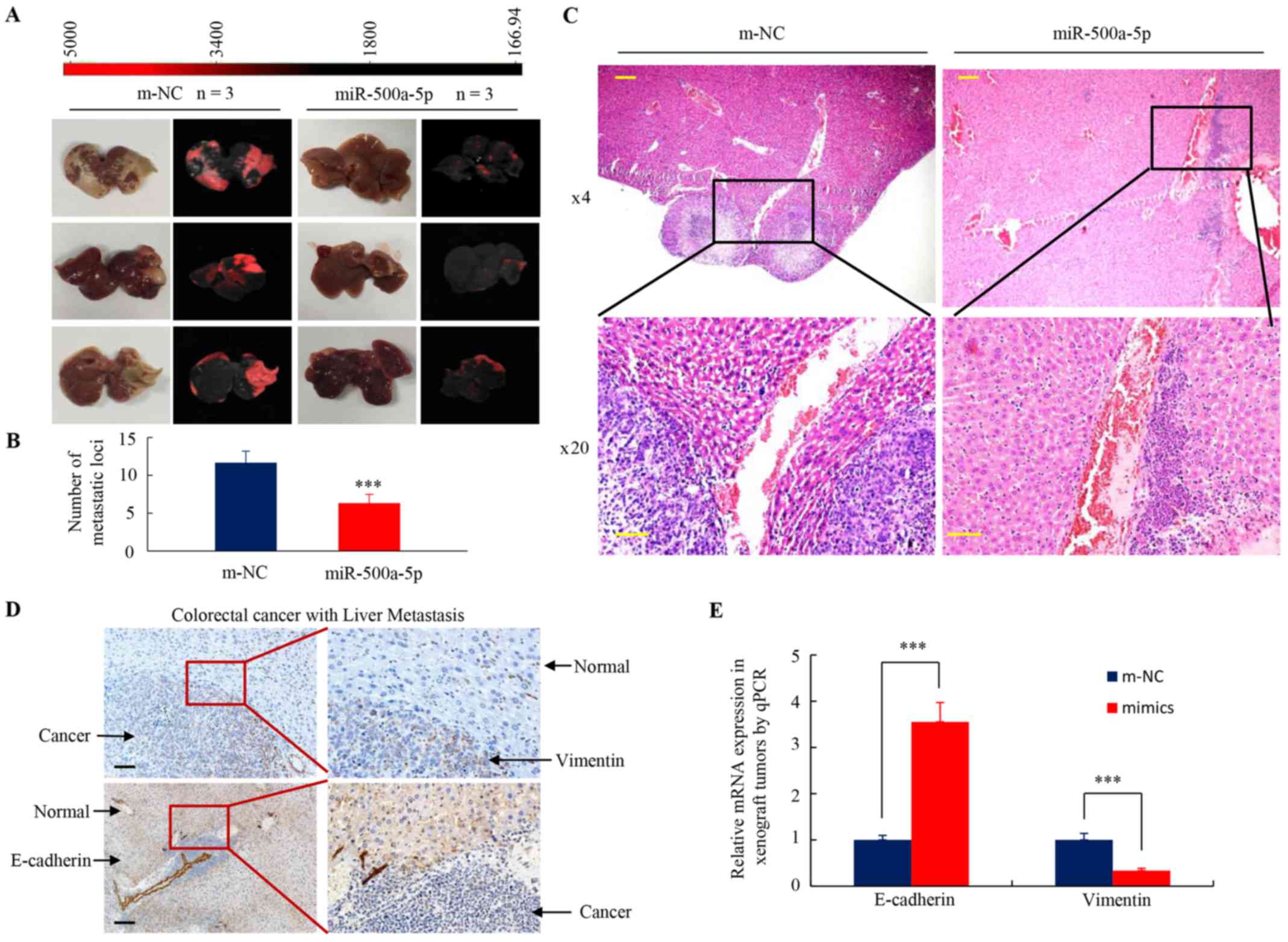
To investigate the association between miR-500a-5p
and tumor metastasis via EMT regulation, miR-500a-5p/LoVo cells and
m-NC/LoVo cells, with RFP expression, were established to visualise
tumor metastasis. A total of one month following orthotopic spleen
injections in nude mice, the animals were euthanised and their
livers were examined. The number of tumor metastatic liver nodules
in the miR-500a-5p/LoVo mouse group was significantly lower
compared with that in the m-NC/LoVo group (Fig. 4A and B). The corresponding HE and
IHC images of the liver are shown in Fig. 4C and D, respectively. Brown-yellow
signals indicate vimentin or E-cadherin staining. The protein
expression level of E-cadherin was downregulated, while that of
vimentin was upregulated in liver cancer tissues compared with that
in adjacent normal liver tissues in miR-500a-5p mimic group
(Fig. 4D).
Moreover, the orthotopic implantation of
miR-500a-5p/LoVo cells led to increased E-cadherin, and decreased
vimentin mRNA expression levels in liver cancer tissues, compared
with that in the mice implanted with m-NC (Fig. 4E). These results indicate that
miR-500a-5p affects tumor metastasis by regulating EMT in CRC cells
in vivo.
Discussion
Previous research suggests that miRNAs modulate
tumor-associated gene expression and affect cancer progression
(38-41). For example, miR-300 inhibits cell
invasion and regulates EMT-related marker genes by targeting TWIST
in head and neck squamous cell carcinoma (39). Moreover, overexpression of miR-300,
using miR-330 mimics, suppressed cell invasion in vitro and
experimental metastasis in vivo (39). Several previous studies have also
reported the role of miR-500a-5p in the development of the
malignant tumor, in liver cancer (8), chronic lymphocytic leukemia (9) and breast cancer (10). However, research on the function of
miR-500a-5p in CRC development in the literature is limited. In the
present study, miR-500a-5p was found to inhibit the metastasis and
EMT in CRC cells. Moreover, increased miR-500a-5p expression
attenuated TGF-b1-induced EMT. In addition, miR-500a-5p
inhibitor-induced EMT was suppressed by emodin. These findings
suggest that miR-500a-5p affects CRC progression and may be
beneficial as a therapeutic target.
EMT occurs during embryonic stage, healing of
lesions and cancer progression (13,15,42,43).
During EMT, cyto-skeletal restructuring occurs, resulting in loss
of polarity and mesenchymal phenotype changes, loss of cellular
junctions and increased migratory potential. miRNAs may act as
tumor suppressors or oncogenes (5,6,33,36,39).
It has been suggested that some miRNAs regulate EMT during cancer
progression and metastasis. For example, Liu et al (19) confirmed that miR-1271 modulates EMT
in pancreatic cancer cells metastasis by targeting ZEB1 and TWIST1,
increasing E-cadherin and decreasing vimentin protein expression
levels. In the present study it was revealed that miR-500a-5p is
involved in CRC metastasis through EMT regulation. It was
demonstrated that miR-500a-5p inhibitor induces EMT and the
metastatic phenotype. The results were consistent with the outcomes
of a previous investigation, which revealed that overexpression of
miR-708 inhibits motility, migration and invasion of CRC cells and
suppresses EMT (44). The results
of the present study indicate that miR-500a-5p plays a role in EMT
of CRC cells, and this may be one of the factors affecting the
development of CRC. In addition, miRNAs play key roles by
regulating target gene expression. Therefore, two algorithms that
predict the mRNA targets of a miRNA were used. Zinc finger E-box
binding homeobox ZEB1 and ZEB2 were identified as potential target
genes of miR-500a-5p in cancer cells, including CRC cells using
bioinformatics analysis. ZEB family is known as a transcription
factor, whose family members include ZEB1 and ZEB2, which are
important factors for EMT (45).
However, further studies are required to elucidate how miR-500a-5p
modulates ZEB1 and/or ZEB2 to promote EMT in CRC cells.
Rearrangement of the actin cytoskeleton is the major
mechanism driving cell migration and invasion during the process of
EMT, and promoting invasive behaviour of cancer cells (46,47).
Previous studies indicated that miRNAs modulate actin cyto-skeleton
organization in cancer cells. For example, Liu et al
(48) revealed that miR-144-3p
significantly inhibited proliferation, migration and invasion in GC
cells. Moreover, overexpression of miR-144-3p by its mimics
disrupted the cytoskeleton of GC cells by decreasing f-actin
expression, thus may affect GC metastasis and invasion by
regulating EMT. Jurmeister et al (49) reported that miR-200c also inhibits
breast cancer cell migration via the same cytoskeletal
reorganization, which markedly reduces the formation of
lamellipodia, and reverses EMT. This is consistent with the results
from the present study as it was found that the miR-500a-5p
inhibitor promoted rearrangements in the actin cytoskeleton and
resulted in an increase of F-actin microfilaments. Thus, it was
demonstrated that cytoskeletal organization was also regulated by
miR-500a-5p in CRC cells.
TGF-β signaling can suppress or promote the
development of tumors, which may vary according to cancer stage
(14,18,50).
TGF-β signaling prevents tumor progression by initiating programmed
cell death, particularly during the early stages of cancer,
including CRC (50). However,
TGF-β signaling promotes EMT-mediated cancer cell invasion and
metastasis in the later stages. For example, upregulated TGF-β can
induce EMT through Smad2 and Smad3 activation, thus, promoting
cancer cell invasion and metastasis to the progressive stage
(51). Numerous studies have
reported the involvement of miRNAs in TGF-β-induced EMT (5,6,36).
For example, Chen et al (36) demonstrated that upregulation of
miR-141 suppresses EMT and lymph node metastasis in laryngeal
cancer through suppressing HOXC6-dependent TGF-β signaling pathway.
In the present study, miR-500a-5p expression decreased in a dose
and time-dependent manner in the CRC cells treated with TGF-β.
miR-500a-5p reversed the EMT phenotype of CRC cells. Moreover,
incubation of cells transfected with miR-500a-5p decreased
TGF-β-mediated cell invasion, which was similar to the effect of
miR-630 on hepatocellular carcinoma cells (52). The findings from the present study
strongly suggest that miR-500a-5p may be a downstream inhibitor of
the TGF-β signaling pathway, reducing the effects of TGF-β on CRC
cells.
Emodin exerts a direct inhibitory effect on the
growth and metastasis of a variety of malignant tumor cells. For
example, emodin causes inhibition of growth of K562 leukaemia cells
and improves survival (53).
Emodin inhibits CRC cell invasion and migration (37) and also inhibits EMT (54), as demonstrated by increased
E-cadherin and decreased fibronectin, a-smooth muscle actin,
N-cadherin and vimentin protein expression levels, together with
reduced expression of the transcriptional repressors Slug, Twist
and Snail. Multiple previous studies have demonstrated that
treatment with emodin inhibits the EMT process and metastasis by
regulating miRNAs (28,29). For example, emodin inhibits
pancreatic cancer EMT and invasion by upregulating miR-1271
(29). The results from the
present study are consistent with previous studies, which indicates
that the miR-500a-5p inhibitor initiates EMT in CRC cells, whereas
emodin can prevent this process. Treatment with emodin blocked
miR-500a-5p inhibitor-initiated cell invasion and caused
alterations in cellular morphology. Moreover, emodin inhibited
miR-500a-5p inhibitor-induced cell invasion. Thus, emodin
suppressed miR-500a-5p inhibitor-induced EMT, thereby exerting
therapeutic and preventive effects by suppressing the metastasis of
CRC cells.
In conclusion, EMT and CRC cell invasion are
inhibited by miR-500a-5p. In addition, overexpression of
miR-500a-5p attenuates TGF-β1-induced EMT, whereas emodin
suppresses miR-500a-5p inhibitor-induced EMT in CRC cells. These
results uncovered the EMT-suppressive role of miR-500a-5p, which
may be used for the development of novel therapeutic strategies for
CRC.
Supplementary Data
Abbreviations:
|
EMT
|
epithelial-mesenchymal transition
|
|
CRC
|
colorectal cancer
|
|
TGF-β1
|
recombinant human transforming growth
factor β-1
|
Acknowledgments
Not applicable.
Funding
This project was funded by the National Natural
Science Funds of China (grant nos. 81672875, 81772964 and
81974448); Guangdong Gastrointestinal Disease Research Center
(grant no. 2017B02029003); Special Scientific Research Fund of
Public Welfare Profession of National Health and Family Planning
Commission (grant no. 201502026); Guangdong Medical Science and
Technology Research Foundation (grant no. B2019126); Science and
technology innovation foundation of Shenzhen (grant no.
JCYJ20180306170328854).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author upon
reasonable request.
Authors' contributions
WT and WD performed the in vitro experiments.
JLi, HZ, JLin and YW contributed to acquisition, analysis and
interpretation of the data. QY, ML and YX performed the in
vivo experiments. ZL, YZ, XW and JinW revised the manuscript
critically for important intellectual content. YC, HH and SL made
substantial contributions to conception and design of the study.
JidW and LX drafted the manuscript. LH revised the manuscript. The
final manuscript for publication was read and approved by all
authors.
Ethics approval and consent to
participate
The study was ethically approved by Southern Medical
University Experimental Animal Ethics Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Finlay IG and McArdle CS: Effect of occult
hepatic metastases on survival after curative resection for
colorectal carcinoma. Gastroenterology. 85:596–599. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kemp Z, Thirlwell C, Sieber O, Silver A
and Tomlinson I: An update on the genetics of colorectal cancer.
Hum Mol Genet. 13(Spec No 2): R177–185. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Konishi M, Kikuchi-Yanoshita R, Tanaka K,
Muraoka M, Onda A, Okumura Y, Kishi N, Iwama T, Mori T, Koike M, et
al: Molecular nature of colon tumors in hereditary nonpolyposis
colon cancer, familial polyposis, and sporadic colon cancer.
Gastroenterology. 111:307–317. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Erning FN, Crolla RM, Rutten HJ,
Beerepoot LV, van Krieken JH and Lemmens VE: No change in lymph
node positivity rate despite increased lymph node yield and
improved survival in colon cancer. Eur J Cancer. 50:3221–3229.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang F, Luo Y, Shao Z, Xu L, Liu X, Niu
Y, Shi J, Sun X, Liu Y, Ding Y and Zhao L: MicroRNA-187, a
downstream effector of TGFβ pathway, suppresses Smad-mediated
epithelial-mesen-chymal transition in colorectal cancer. Cancer
Lett. 373:203–213. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang P, Tang WM, Zhang H, Li YQ, Peng Y,
Wang J, Liu GN, Huang XT, Zhao JJ, Li G, et al: MiR-646 inhibited
cell proliferation and EMT-induced metastasis by targeting FOXK1 in
gastric cancer. Br J Cancer. 117:525–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu X, Li J, Yu Z, Li J, Sun R and Kan Q:
miR-935 promotes liver cancer cell proliferation and migration by
targeting SOX7. Onco Res. 25:427–435. 2017. View Article : Google Scholar
|
|
8
|
Guo Y, Chen L, Sun C and Yu C:
MicroRNA-500a promotes migration and invasion in hepatocellular
carcinoma by activating the Wnt/β-catenin signaling pathway. Biomed
Pharmacother. 91:13–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ruiz-Lafuente N, Alcaraz-Garcia MJ,
Sebastian-Ruiz S, Garcia-Serna AM, Gomez-Espuch J, Moraleda JM,
Minguela A, Garcia-Alonso AM and Parrado A: IL-4 Up-regulates
MiR-21 and the MiRNAs hosted in the CLCN5 gene in chronic
lymphocytic leukemia. PLoS One. 10:e01249362015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Degli Esposti D, Aushev VN, Lee E, Cros
MP, Zhu J, Herceg Z, Chen J and Hernandez-Vargas H: miR-500a-5p
regulates oxidative stress response genes in breast cancer and
predicts cancer survival. Sci Rep. 7:159662017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang M, Zhou LY, Xu N and An Q:
Down-regulation of miR-500 and miR-628 suppress non-small cell lung
cancer proliferation, migration and invasion by targeting ING1.
Biomed Pharmacother. 108:1628–1639. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tang W, Zhou W, Xiang L, Wu X, Zhang P,
Wang J, Liu G, Zhang W, Peng Y, Huang X, et al: The
p300/YY1/miR-500a-5p/HDAC2 signalling axis regulates cell
proliferation in human colorectal cancer. Nat Commun. 10:6632019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang MH, Wu MZ, Chiou SH, Chen PM, Chang
SY, Liu CJ, Teng SC and Wu KJ: Direct regulation of TWIST by
HIF-1alpha promotes metastasis. Nat Cell Biol. 10:295–305. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang W, Jiang B, Guo Z, Sardet C, Zou B,
Lam CS, Li J, He M, Lan HY, Pang R, et al: Four-and-a-half LIM
protein 2 promotes invasive potential and epithelial-mesenchymal
transition in colon cancer. Carcinogenesis. 31:1220–1229. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yan Q, Zhang W, Wu Y, Wu M, Zhang M, Shi
X, Zhao J, Nan Q, Chen Y, Wang L, et al: KLF8 promotes
tumorigenesis, invasion and metastasis of colorectal cancer cells
by transcriptional activation of FHL2. Oncotarget. 6:25402–25417.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang X, Xiang L, Li Y, Zhao Y, Zhu H,
Xiao Y, Liu M, Wu X, Wang Z, Jiang P, et al: Snail/FOXK1/Cyr61
signaling axis regulates the epithelial-mesenchymal transition and
metastasis in colorectal cancer. Cell Physiol Biochem. 47:590–603.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nature reviews Mol
Cell Biol. 15:178–196. 2014. View Article : Google Scholar
|
|
18
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: TGF-β-induced upregulation of malat1 promotes bladder
cancer metastasis by associating with suz12. Clin Cancer Res.
20:1531–1541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu H, Wang H, Liu X and Yu T: miR-1271
inhibits migration, invasion and epithelial-mesenchymal transition
by targeting ZEB1 and TWIST1 in pancreatic cancer cells. Biochem
Biophys Res Commun. 472:346–352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Demirezer LO, Kuruuzum-Uz A, Bergere I,
Schiewe HJ and Zeeck A: The structures of antioxidant and cytotoxic
agents from natural source: Anthraquinones and tannins from roots
of Rumex patientia. Phytochemistry. 58:1213–1217. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang Q, Lu G, Shen HM, Chung MC and Ong
CN: Anti-cancer properties of anthraquinones from rhubarb. Med Res
Rev. 27:609–630. 2007. View Article : Google Scholar
|
|
22
|
Li J, Liu P, Mao H, Wanga A and Zhang X:
Emodin sensitizes paclitaxel-resistant human ovarian cancer cells
to paclitaxel-induced apoptosis in vitro. Oncol Rep. 21:1605–1610.
2009.PubMed/NCBI
|
|
23
|
Dong X, Fu J, Yin X, Cao S, Li X, Lin L,
Huyiligeqi and Ni J: Emodin: A review of its pharmacology, toxicity
and pharmaco-kinetics. Phytother Res. 30:1207–1218. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ko JC, Su YJ, Lin ST, Jhan JY, Ciou SC,
Cheng CM and Lin YW: Suppression of ERCC1 and Rad51 expression
through ERK1/2 inactivation is essential in emodin-mediated
cytotoxicity in human non-small cell lung cancer cells. Biochem
Pharmacol. 79:655–664. 2010. View Article : Google Scholar
|
|
25
|
He Y, Huang J, Wang P, Shen X, Li S, Yang
L, Liu W, Suksamrarn A, Zhang G and Wang F: Emodin potentiates the
antiproliferative effect of interferon α/β by activation of
JAK/STAT pathway signaling through inhibition of the 26S
proteasome. Oncotarget. 7:4664–4679. 2016.
|
|
26
|
Cai J, Niu X, Chen Y, Hu Q, Shi G, Wu H,
Wang J and Yi J: Emodin-induced generation of reactive oxygen
species inhibits RhoA activation to sensitize gastric carcinoma
cells to anoikis. Neoplasia. 10:41–51. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
van Gorkom BA, Timmer-Bosscha H, de Jong
S, van der Kolk DM, Kleibeuker JH and de Vries EG: Cytotoxicity of
rhein, the active metabolite of sennoside laxatives, is reduced by
multidrug resistance-associated protein 1. Br J Cancer.
86:1494–1500. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Niu Y, Zhang J, Tong Y, Li J and Liu B:
Physcion 8-O-β-glucopyranoside induced ferroptosis via regulating
miR-103a-3p/GLS2 axis in gastric cancer. Life Sci. 237:1168932019.
View Article : Google Scholar
|
|
29
|
Li N, Wang C, Zhang P and You S: Emodin
inhibits pancreatic cancer EMT and invasion by upregulating
microRNA1271. Mol Med Rep. 18:3366–3374. 2018.PubMed/NCBI
|
|
30
|
Wang J, Yang Y, Xia HH, Gu Q, Lin MC,
Jiang B, Peng Y, Li G, An X, Zhang Y, et al: Suppression of FHL2
expression induces cell differentiation and inhibits gastric and
colon carcinogenesis. Gastroenterology. 132:1066–1076. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
32
|
Liu B, Li X, Li C, Xu R and Sun X: miR-25
mediates metastasis and epithelial-mesenchymal-transition in human
esophageal squamous cell carcinoma via regulation of E-cadherin
signaling. Bioengineered. 10:679–688. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li S, Hou X, Wu C, Han L, Li Q, Wang J and
Luo S: MiR-645 promotes invasiveness, metastasis and tumor growth
in colorectal cancer by targeting EFNA5. Biomed Pharmacother.
125:1098892020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang HC, Hu CH, Tang MC, Wang WS, Chen PM
and Su Y: Thymosin beta4 triggers an epithelial-mesenchymal
transition in colorectal carcinoma by upregulating integrin-linked
kinase. Oncogene. 26:2781–2790. 2007. View Article : Google Scholar
|
|
35
|
Schelch K, Wagner C, Hager S, Pirker C,
Siess K, Lang E, Lin R, Kirschner MB, Mohr T, Brcic L, et al: FGF2
and EGF induce epithelial-mesenchymal transition in malignant
pleural mesothelioma cells via a MAPKinase/MMP1 signal.
Carcinogenesis. 39:534–545. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen L, Sun DZ, Fu YG, Yang PZ, Lv HQ, Gao
Y and Zhang XY: Upregulation of microRNA-141 suppresses
epithelial-mesenchymal transition and lymph node metastasis in
laryngeal cancer through HOXC6-dependent TGF-β signaling pathway.
Cell Signal. 66:1094442020. View Article : Google Scholar
|
|
37
|
Gu J, Cui CF, Yang L, Wang L and Jiang XH:
Emodin inhibits colon cancer cell invasion and migration by
suppressing epithelial-mesenchymal transition via the Wnt/β-catenin
pathway. Oncol Res. 27:193–202. 2019. View Article : Google Scholar
|
|
38
|
Larsson O, Li S, Issaenko OA, Avdulov S,
Peterson M, Smith K, Bitterman PB and Polunovsky VA: Eukaryotic
translation initiation factor 4E induced progression of primary
human mammary epithelial cells along the cancer pathway is
associated with targeted translational deregulation of oncogenic
drivers and inhibitors. Cancer Res. 67:6814–6824. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yu J, Xie F, Bao X, Chen W and Xu Q:
miR-300 inhibits epithelial to mesenchymal transition and
metastasis by targeting Twist in human epithelial cancer. Mol
Cancer. 13:1212014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh
T, Kojima K, Nakashima R, Kitade Y and Naoe T: Role of
anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer
Gene Ther. 17:398–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu F, Cai Y, Rong X, Chen J, Zheng D,
Chen L, Zhang J, Luo R, Zhao P and Ruan J: MiR-661 promotes tumor
invasion and metastasis by directly inhibiting RB1 in non small
cell lung cancer. Mol Cancer. 16:1222017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Huang C, Xie D, Cui J, Li Q, Gao Y and Xie
K: FOXM1c promotes pancreatic cancer epithelial-to-mesenchymal
transition and metastasis via upregulation of expression of the
urokinase plas-minogen activator system. Clin Cancer Res.
20:1477–1488. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qi H, Fu X, Li Y, Pang X, Chen S, Zhu X,
Li F and Tan W: SATB1 promotes epithelial-mesenchymal transition
and metastasis in prostate cancer. Oncol Lett. 13:2577–2582. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sun S, Hang T, Zhang B, Zhu L, Wu Y, Lv X,
Huang Q and Yao H: miRNA-708 functions as a tumor suppressor in
colorectal cancer by targeting ZEB1 through Akt/mTOR signaling
pathway. Am J Transl Res. 11:5338–5356. 2019.PubMed/NCBI
|
|
45
|
Guan T, Dominguez CX, Amezquita RA,
Laidlaw BJ, Cheng J, Henao-Mejia J, Williams A, Flavell RA, Lu J
and Kaech SM: ZEB1, ZEB2, and the miR-200 family form a
counterregulatory network to regulate CD8+ T cell fates.
J Exp Med. 215:1153–1168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sousa-Squiavinato ACM, Rocha MR,
Barcellos-de-Souza P, de Souza WF and Morgado-Diaz JA: Cofilin-1
signaling mediates epithelial-mesenchymal transition by promoting
actin cytoskeleton reorganization and cell-cell adhesion regulation
in colorectal cancer cells. Biochim Biophys Acta Mol Cell Res.
1866:418–429. 2019. View Article : Google Scholar
|
|
47
|
Islam SU, Ahmed MB, Lee SJ, Shehzad A,
Sonn JK, Kwon OS and Lee YS: PRP4 kinase induces actin
rearrangement and epithelial-mesenchymal transition through
modulation of the actin-binding protein cofilin. Exp Cell Res.
369:158–165. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu F, Chen N, Xiao R, Wang W and Pan Z:
miR-144-3p serves as a tumor suppressor for renal cell carcinoma
and inhibits its invasion and metastasis by targeting MAP3K8.
Biochem Biophys Res Commun. 480:87–93. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jurmeister S, Baumann M, Balwierz A,
Keklikoglou I, Ward A, Uhlmann S, Zhang JD, Wiemann S and Sahin O:
MicroRNA-200c represses migration and invasion of breast cancer
cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol
Cell Biol. 32:633–651. 2012. View Article : Google Scholar :
|
|
50
|
Shen W, Tao GQ, Zhang Y, Cai B, Sun J and
Tian ZQ: TGF-β in pancreatic cancer initiation and progression: Two
sides of the same coin. Cell Biosci. 7:392017. View Article : Google Scholar
|
|
51
|
Zhou Q, Zheng X, Chen L, Xu B, Yang X,
Jiang J and Wu C: Smad2/3/4 pathway contributes to TGF-β-Induced
MiRNA-181b expression to promote gastric cancer metastasis by
targeting Timp3. Cell Physiol Biochem. 39:453–466. 2016. View Article : Google Scholar
|
|
52
|
Chen WX, Zhang ZG, Ding ZY, Liang HF, Song
J, Tan XL, Wu JJ, Li GZ, Zeng Z, Zhang BX and Chen XP: MicroRNA-630
suppresses tumor metastasis through the TGF-beta- miR-630-Slug
signaling pathway and correlates inversely with poor prognosis in
hepatocellular carcinoma. Oncotarget. 7:22674–22686.
2016.PubMed/NCBI
|
|
53
|
Chun-Guang W, Jun-Qing Y, Bei-Zhong L,
Dan-Ting J, Chong W, Liang Z, Dan Z and Yan W: Anti-tumor activity
of emodin against human chronic myelocytic leukemia K562 cell lines
in vitro and in vivo. Eur J Pharmacol. 627:33–41. 2010. View Article : Google Scholar
|
|
54
|
Han YT, Chen XH, Gao H, Ye JL and Wang CB:
Physcion inhibits the metastatic potential of human colorectal
cancer SW620 cells in vitro by suppressing the transcription factor
SOX2. Acta Pharmacol Sin. 37:264–275. 2016. View Article : Google Scholar :
|