Introduction
Prostate cancer (PCa) is one of the most common
types of cancer, and is a leading cause of cancer-associated among
men worldwide (1). Tumor growth in
the early-stages of prostate cancer development is dependent on the
presence of androgens, and androgen deprivation therapy (ADT) is
the primary treatment used for androgen-dependent prostate cancer
(2). However, a notable subset of
patients experience recurrence and progression following the
failure of ADT, which is the leading cause of PCa-associated death
(3). Therefore, understanding the
pathogenesis of PCa and identifying novel treatment choices are
urgently required.
Tetramethylpyrazine (TMP) is one of the active
compounds extracted from the Chinese medicinal plant Ligusticum
chuanxiong (4). TMP has been
widely used in Chinese herbal medicines for various purposes,
including treating neurovascular and cardiovascular diseases, as it
possesses anti‑oxidant and anti‑inflammatory properties (5). TMP has been shown to influence cancer
progression in lung cancer (6),
bladder cancer (7), breast cancer
(8) and gastric cancer (9), beyond its traditional roles. In our
previous study, it was shown that TMP exerted anti-cancer effects
in PCa (10). To the best of our
knowledge, there are no studies examining the molecular mechanisms
underlying the effects of TMP in cancer.
Long-chain non-coding RNA (lncRNA) is a type of
non-coding RNA, 200-100,000 bases in length, which lack an
effective open reading framework and is transcribed by RNA
polymerase II without protein coding function (11). LncRNAs serve as transcription
regulators through interfering with the binding of transcription
factors to promoters, interfering with gene transcription and
chromatin remodeling; or by binding to proteins through chaperones,
regulating subcellular localization of proteins (12-14).
The antisense lncRNAs are a subtype of lncRNA molecules transcribed
from the opposite DNA strand of protein-coding or
non-protein-coding genes (15). At
present, only a few antisense lncRNAs have been extensively
studied, such as AGAP2-AS1 (16)
and ZEB1-AS1 (17). Whether
antisense lncRNAs participate in the progression of PCa and the
TMP-induced anti-cancer effects, and the mechanisms underlying the
regulatory mechanisms of TMP have not been studied.
In the present study, the antisense lncRNA termed
DPP10-AS1, which is located on chr 2q14.1, and contains 744 nt was
examined in PCa. The status of DPP10-AS1 in PCa has not been
investigated to the best of our knowledge, thus, the expression
levels and functional roles of DPP10-AS1 of DPP10-AS1 in PCa, and
its role in TMP-induced anti-cancer effects were determined. It was
shown that DPP10-AS1 expression was downregulated in TMP-treated
PCa cells, and upregulation of DPP10-AS1 reversed the anti-cancer
effects induced by TMP. Mechanistically, DPP10-AS1 increased the
expression of forkhead box M1 (FOXM1) via H3K27 acetylation
modification at the FOXM1 promoter region.
Materials and methods
Clinical specimen
A total of 35 pairs of PCa tissues and adjacent
normal tissues were collected by puncture biopsy between January
2014 and June 2017, and snap-frozen in liquid nitrogen instantly.
The median age of the patients recruited in the present study was
53 years (age range, 42-69 years). Patients who received
radiotherapy and chemotherapy prior to puncture biopsy were
excluded. The enrolled patients provided written informed consent,
and the present study was approved by the Research Ethics Committee
of Peking Union Medical College Hospital.
Cell culture and treatment
The PCa cell lines, PC3 and DU145, and one prostate
epithelial cell line, hPrEC, were all purchased from Shanghai
Institutes for Biological Sciences Cell Resource Center (Shanghai,
China). PC3 and DU145 cells were grown in DMEM (Thermo Fisher
Scientific, Inc.) supplemented with glucose and 10% FBS (Thermo
Fisher Scientific, Inc.). hPrEC cells were cultured in prostate
epithelium basal media (Shanghai Bangjing Industry Co., Ltd.) and
all the cells were cultured in a humidified incubator at 37°C with
5% CO2. The TMP-resistant PC3 sub-line (PC3-R) was
established by continuously treating PC3 cells with TMP (500
µg/l; Mansite Biotechnology, Co., Ltd.) for 3 months.
Cell transfection
Small interfering (si)RNAs targeting DPP10-AS1
(si-DPP10-AS1, GFP-labeled) and FOXM1 (si-FOXM1) plasmids
were synthesized by Shanghai GenePharma Co., Ltd. DPP10-AS1, CREB
binding protein (CBP) and FOXM1 cDNA was generated and
subcloned into a pcDNA3.1 vector, termed p-DPP10-AS1, p-CBP or
p-FOXM1, respectively. p-DPP10-AS1 was further cloned into an
Ad.Max™ adenovirus vector (Shanghai GeneChem Co., Ltd.) for in
vivo studies. Plasmid constructs were transfected into cells at
70‑90% confluency using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) for 6 h with a final concentration
of 100 nM plasmid according to the manufacturer's protocol. The
sequences of the siRNAs are presented in Table I.
 | Table ISequences of the primers and
siRNAs. |
Table I
Sequences of the primers and
siRNAs.
| Gene | Sequence,
5'-3' |
|---|
| DPP10-AS1 | |
| Forward |
AGATTGTGGCCTGAGGTGC |
| Reverse |
TTAGGAGTTCCACCGACGTG |
| FOXM1 | |
| Forward |
GGAGGAAATGCCACACTTAGCG |
| Reverse |
TAGGACTTCTTGGGTCTTGGGGTG |
| CREB binding
protein | |
| Forward |
GTGCTGGCTGAGACCCTAAC |
| Reverse |
GGCTGTCCAAATGGACTTGT |
| GAPDH | |
| Forward |
GCACCGTCAAGGCTGAGAAC |
| Reverse |
ATGGTGGTGAAGACGCCAGT |
| si-DPP10-AS1 |
CCUAAAGGGAUGCCUUCAATT |
| si-FOXM1 |
UAGUAACUCUGGCCAUAGCTT |
| si-negative
control |
CAGGUGGACUCACAAUUCCTT |
RNA extraction and reverse
transcription‑quantitative (RT‑q)PCR
Total RNA was extracted using TRIzol®
reagent (Thermo Fisher Scientific, Inc.). The concentration of
extracted RNA was determined using a Qubit 4.0 spectrophotometer
(Thermo Fisher Scientific, Inc.). For DPP10‑AS1 and FOXM1
expression analysis, RT was performed using a TaqMan High-Capacity
cDNA Reverse Transcription kit and a TaqMan Fast PCR Master mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. The thermocycling conditions for qPCR
were: 95°C for 30 sec; followed by 45 cycles of 95°C for 5 sec and
60°C for 30 sec. At the beginning of the logarithmic phase of PCR
amplification, cycle quantifications (Cq) were appointed and the
repeated Cq values were analyzed using the 2‑∆∆cq method
(18). GAPDH was used to normalize
the relative expression levels of lncRNA and mRNA. The sequences of
the primers are shown in Table
I.
Cell viability assay
PCa cells (1×105 cells per well) were
seeded in 96-well plates and cultured for 24 h prior to analysis of
cell proliferation using a Cell Counting Kit-8 (CCK-8) assay
(Dojindo Molecular Technologies, Inc.). Cells were cultured for a
further 24, 48 or 72 h. After the set incubation times, 10
µl CCK-8 solution was added and cells were further incubated
at 37°C for 4 h. To obtain cell growth curves, the optical density
of the plates were measured at 450 nm using a microplate
spectrophotometer (Thermo Fisher Scientific, Inc.). All experiments
were performed in triplicate.
TUNEL assay
An in‑situ cell death detection kit (Roche
Diagnostics) was used to measure cell apoptosis according to the
manufacturer's protocol. Transfected cells (5×105
cells/well) were cultured in a 24-well plate for 24 h.
Subsequently, cells were fixed with 4% paraformaldehyde for 60 min
at room temperature and subsequently incubated in 0.1% Triton X-100
for 2 min on ice. After permeabilization with Triton X-100, a total
of 50 µl mixed TUNEL solution (prepared according to the
manufacturer's protocol) was added and cells were incubated at 37°C
for 1 h. The nuclei of cells were stained using DAPI for 1 h at
room temperature and images were taken using a fluorescence
microscope (magnification, ×20; Nikon Corporation).
Bioinformatics analysis
The putative binding site of H3K27ac at the promoter
region of DPP10-AS1 gene was predicted using UCSC Genome Browser
(19). The full sequence of
DPP10-AS1 gene can be accessed with the NCBI Reference Sequence:
NC_00002.12.
RNA immunoprecipitation (RIP) and
chromatin immune-precipitation (ChIP)
RIP analysis was performed in PCa cells using a
Magna RIP RNA-binding protein immunoprecipitation kit (EMD
Millipore) according to manufacturer's protocol. Briefly, cells
were collected after washing with cold PBS and RIP lysis buffer was
added. The suspension was then centrifuged and 100 µl from
each cell lysate was transferred to the RIP immunoprecipitation
buffer, which contained CBP antibody (1:100; Abcam; cat. no.
ab2832) or IgG as the negative control (1:200; EMD Millipore; cat.
no. 12-371). The magnetic beads were washed with RIP wash buffer
and incubated with proteinase K at 55°C for 30 min. Subsequently,
RNA was extracted for RT-qPCR analysis.
ChIP assay was performed using an EZ-Magna ChIP kit
(EMD Millipore). To generate the chromatin fragments (200-300 bp in
length) cells were treated with formaldehyde for 10 min followed by
sonication at 4°C. Subsequently, ChIP‑specific antibodies against
H3K27ac (1:100; Abcam; cat. no. ab4729), CBP (1:100; Abcam; cat.
no. ab2832) and normal mouse IgG polyclonal antibody (1:100; EMD
Millipore; cat. no. 12-371) were used for immunoprecipitation. RNA
was recovered using an EpiTect ChIP qPCR assays (cat. no. 334001;
Qiagen, Inc.), according to the manufacturer's protocol and
analyzed by qPCR.
Nucleocytoplasmic separation
Nuclear and cytosolic fractions were separated using
a Nuclear/Cytosol Fractionation kit (cat. no. K266-25; BioVision,
Inc.) according to the manufacturer's protocol. Subsequently,
RT-qPCR was used to determine the expression of DPP10-AS1, GAPDH
and U1 in the nucleus and cytoplasm of PC3 and PC3-R cells.
RNA florescent in situ hybridization
(FISH)
Hybridization was performed using a FISH kit
(Guangzhou RiboBio Co., Ltd.) according to the manufacturer's
protocol. Briefly, 4% paraformal-dehyde was used to fix the PCa
cells (15 min at room temperature) followed by permeabilization
with 0.5% Triton X-100 for 15 min at room temperature.
Subsequently, the cells were cultured with specific GFP‑labeled
DPP10‑AS1 probes (Guangzhou RiboBio Co., Ltd.) overnight at 4°C.
All fluorescent images were captured using Nikon A1Si Laser
Scanning Confocal Microscope (magnification ×100; Nikon
Corporation).
Immunohistochemistry (IHC) analysis and
scoring methods
Tumor tissues from nude mice were rehydrated using a
graded sequence of ethanol solutions (70, 85, 95 and 100%) followed
by deionized water. Subsequently, tissues were immersed in citrate
buffer (0.01 mol/l) and heated to 95°C for 30 min. Slides were
washed with PBS solution followed by treatment with 1% Triton X-100
solution for 30 min at room temperature, and stained using a
biotin-streptavidin CytoScan™ horseradish peroxidase Detection
system according to the manufacturer's protocol (EMD Millipore),
followed by incubation with a primary antibody targeting FOXM1
(1:200; Abcam; cat. no. ab184637) overnight at 4°C. The presence of
brown chromogen in the cytoplasm indicated positive
immunoreactivity.
The immunostaining intensity of each sample was
graded as follows: Negative, 0; weak, 1; moderate, 2; or strong, 3.
The proportion of positively stained cells is represented as a
percentage of the total number of cells. The final score was then
calculated as the intensity score multiplied by the percentage of
cells stained. Images were visualized using a confocal Nikon
ECLIPSE Ti (magnification ×40; Nikon Corporation) and processed
using Nikon NIS-Elements software (version 1.0; Nikon
Corporation).
In vivo nude mouse model
Tumor xenografts were established using male BALB/c
nude mice (4-6 weeks old), which were purchased from the Model
Animal Research Center of Nanjing University. PC3 cells were stably
infected with p-DPP10-AS1 or p-NC, and 3×106 cells were
subcutaneously injected into nude mice followed by treatment with
TMP or PBS (50 mg/kg) once every two days for 6 weeks. The mice
were divided into 3 groups (5 mice in each group): i) Group I, PBS
treatment + p-NC; ii) group II, TMP treatment + p-NC; and iii)
group III, TMP treatment + p-DPP10-AS1. Subsequently the mice were
sacrificed after 30 days, the grafted tumors were removed, and the
weights of the neoplasms were measured immediately after resection.
After weighing, the tissues were fixed for sectioning with 4%
paraformaldehyde followed by IHC staining for FOXM1, as described
above. Animal experiments were approved by the Institutional Review
Board of Peking Union Medical College Hospital.
Western blotting
RIPA lysis buffer consisting of 150 mM NaCl, 1%
Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris (pH 8)
and protease inhibitors cocktail (Promega Corporation) was used to
lyse the cells and exosome samples. Protein concentration was
measured using a bicinchoninic acid assay (cat. no. BCA1-1KT, Sigma
Aldrich; Merck KGaA). Proteins (25 mg per lane) were loaded on a
10% SDS gel, resolved using SDS-PAGE and transferred to PVDF
membranes using a Trans-Blot system (Bio-Rad Laboratories, Inc.).
Subsequently, the membranes were incubated with specific primary
antibodies against FOXM1 (1:1,000; Abcam; cat. no. ab184637), CBP
(1:1,000; Abcam; cat. no. ab2832), and GAPDH (1:5,000; Abcam; cat.
no. ab9485) at 4°C overnight and subsequently incubated with the
goat anti-rabbit polyclonal horseradish peroxidase-conjugated
secondary antibody (1:5,000; Abcam; cat. no. ab7090) for 1 h at
room temperature. Membranes were developed using an enhanced
chemiluminescent reagent and visualized using a ChemiDoc XRS
Imaging system and analyzed using the accompanying software Image
Lab version 3.0 (Bio-Rad Laboratories, Inc.).
Statistical analysis
GraphPad Prism Software version 5.0.1 (GraphPad
Software, Inc.) was used to perform statistical analysis. A
two-tailed Student's t-test, one-way ANOVA with a Tukey's post-hoc
test or Pearson's correlation analysis were used to compare
differences. Data are presented as the mean ± standard error of the
mean. P<0.05 was considered to indicate a statistically
significant difference.
Results
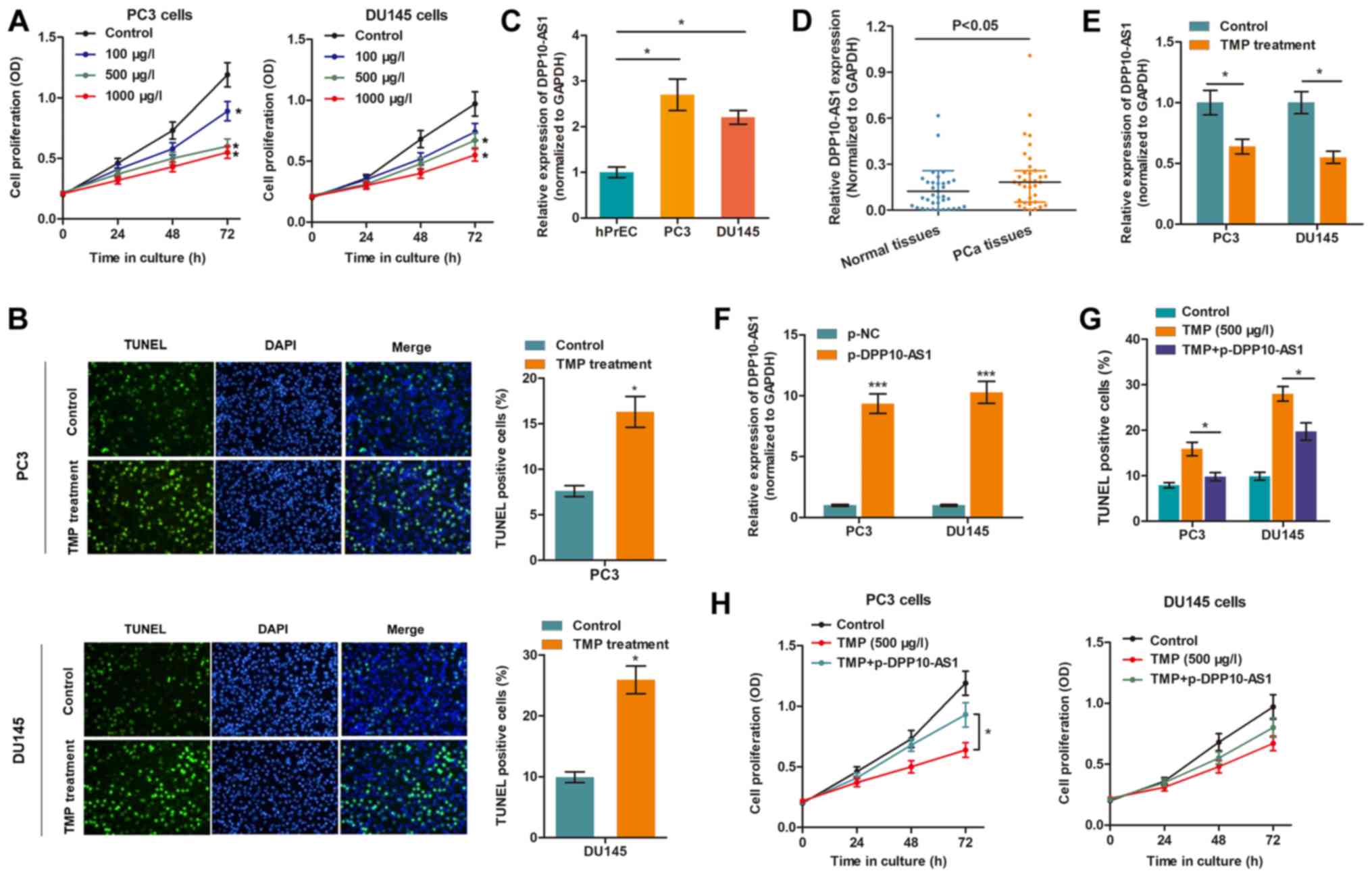
TMP reduces the proliferation and
promotes apoptosis of PCa cells by targeting DPP10‑AS1
Our previous study showed that PC3 and DU145 are
appropriate cell models for assessing the effects of TMP in PCa
(10). To confirm the anti-cancer
effects of TMP in PCa, the cell viability of cells treated with
different concentrations of TMP was determined. The CCK-8 assay
showed that cell viability was suppressed in a
concentration-dependent manner (Fig.
1A), whereas the TUNEL assay showed that TMP increased cell
death (Fig. 1B), suggesting that
TMP reduced progression of PCa. To determine whether DPP10-AS1 was
involved in the TMP-induced anti-cancer effects, the expression
levels of DPP10-AS1 in PCa were measured. Fig. 1C showed that DPP10-AS1 expression
was upregulated in PC3 and DU145 cells compared with prostate
epithelial cells. In addition, DPP10-AS1 levels were also increased
in PCa tissues compared with normal tissues (Fig. 1D). Importantly, TMP treatment
significantly reduced DPP10-AS1 levels in PCa cells compared with
the control cells (Fig. 1E). To
verify the functional role of DPP10-AS1, DPP10-AS1 was
overexpressed in PCa cells treated with TMP (Fig. 1F), and it was shown that DPP10‑AS1
significantly reversed the anti‑cancer effects of TMP in PCa cells,
as evidenced by increase in apoptosis and proliferation (Fig. 1G and H).
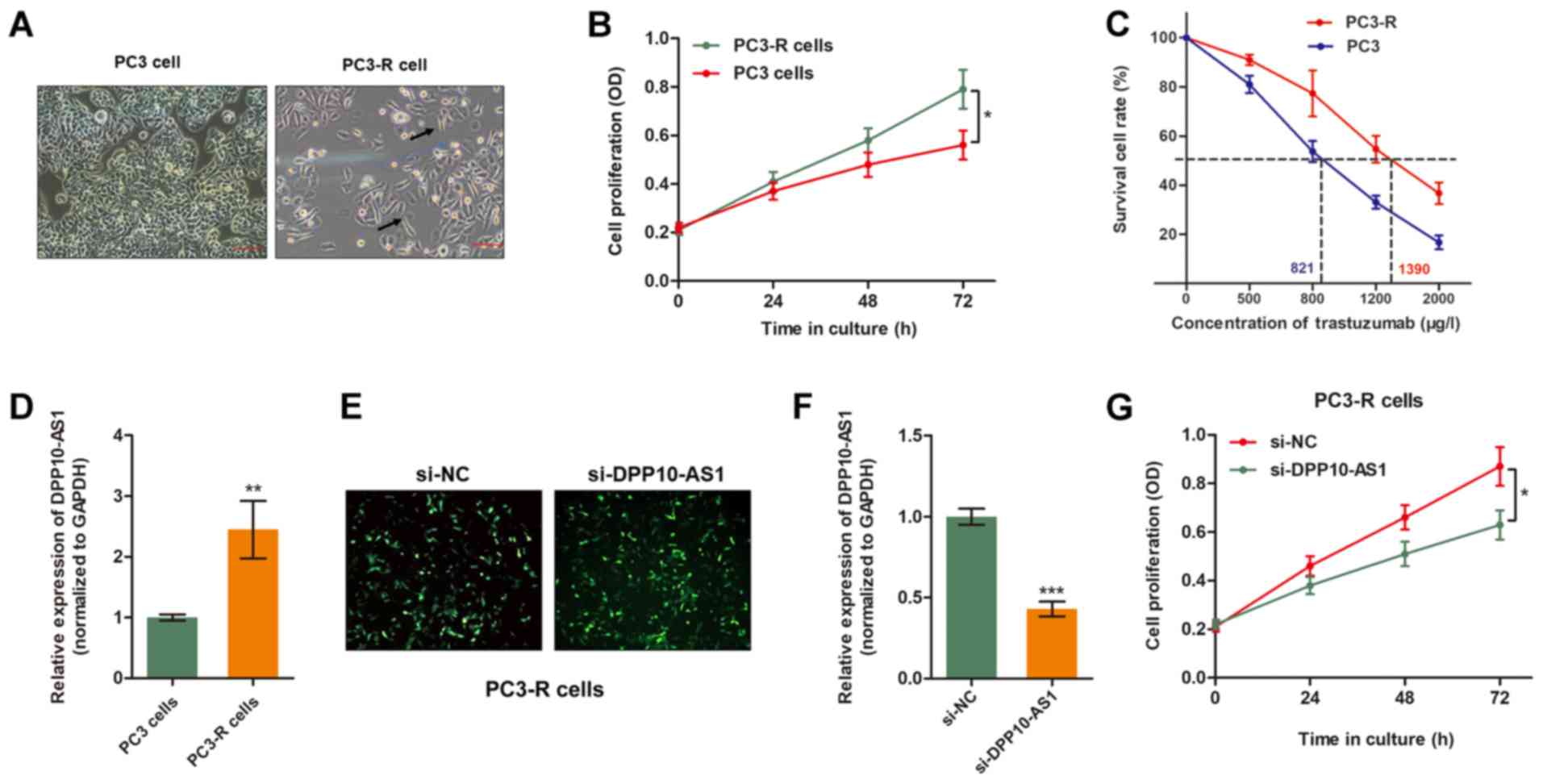
Overexpression of DPP10‑AS1 reverses TMP
resistance in PCa cells
To further verify the role of DPP10-AS1 in
TMP-induced anti-tumor effects, a TMP-resistant PC3 sub-line
(PC3-R) was established by continuously treating PCa cells with TMP
(500 µg/l) for 3 months. As shown in Fig. 2A, PC3‑R cells showed specific
morphological changes in contrast to the sensitive parental cell
lines, such as reduced cell polarity, increased number of
pseudopodia and enlarged inter-cellular separation. Furthermore,
the established PC3-R cells showed increased cell viability
compared with the parental cells when treated with 500 mg/l TMP
(Fig. 2B and C). RT-qPCR analysis
showed expression of DPP10-AS1 was significantly higher in the
PC3‑R cells compared with the PC3 cells (Fig. 2D). Therefore, DPP10-AS1 expression
was down-regulated in PC3‑R cells by transient transfection of
specific siRNA (Fig. 2E and F). As
shown in Fig. 2F, knockdown of
DPP10-AS1 partially reversed TMP resistance of PC3-R cells as
evidenced by increased cell cytotoxicity caused by TMP treatment.
Together, these data suggest that DPP10-AS1 is essential for TMP
resistance in PCa cells.
LncRNA DPP10‑AS1 mediates TMP‑induced
cell cytotoxicity by directly targeting FOXM1
To identify the target genes of DPP10-AS1, StarBase
v2.0 was used (20), and 7
candidate genes associated with DPP10‑AS1 were identified (Table II). In addition, in our previous
study it was shown that TMP inhibited PCa progression by
downregulating the expression of FOXM1 (10). Thus, the expression levels of these
8 potential target genes was assessed. The results showed that the
expression of FOXM1 was upregulated by DPP10-AS1
over-expression and downregulated when DPP10-AS1 was silenced
(Fig. 3A). Furthermore,
FOXM1 expression levels were higher in the TMP-resistant
PC3-R cells compared with the PC3 cells (Fig. 3B). The other 7 genes were not
regulated by DPP10-AS1 (data not shown). To determine whether
FOXM1 was a functional target of DPP10-AS1, FOXM1
knockdown and overexpression vectors were transfected into the PC3
and PC3-R cells, respectively (Fig.
3C). By performing a CCK8 assay, it was shown that silencing of
FOXM1 abrogated the influence of DPP10-AS1 in PC3 cells
treated with TMP (Fig. 3D).
Similarly, overexpression of FOXM1 partially reversed
DPP10-AS1 knockdown-induced effects in PC3-R cells (Fig. 3E).
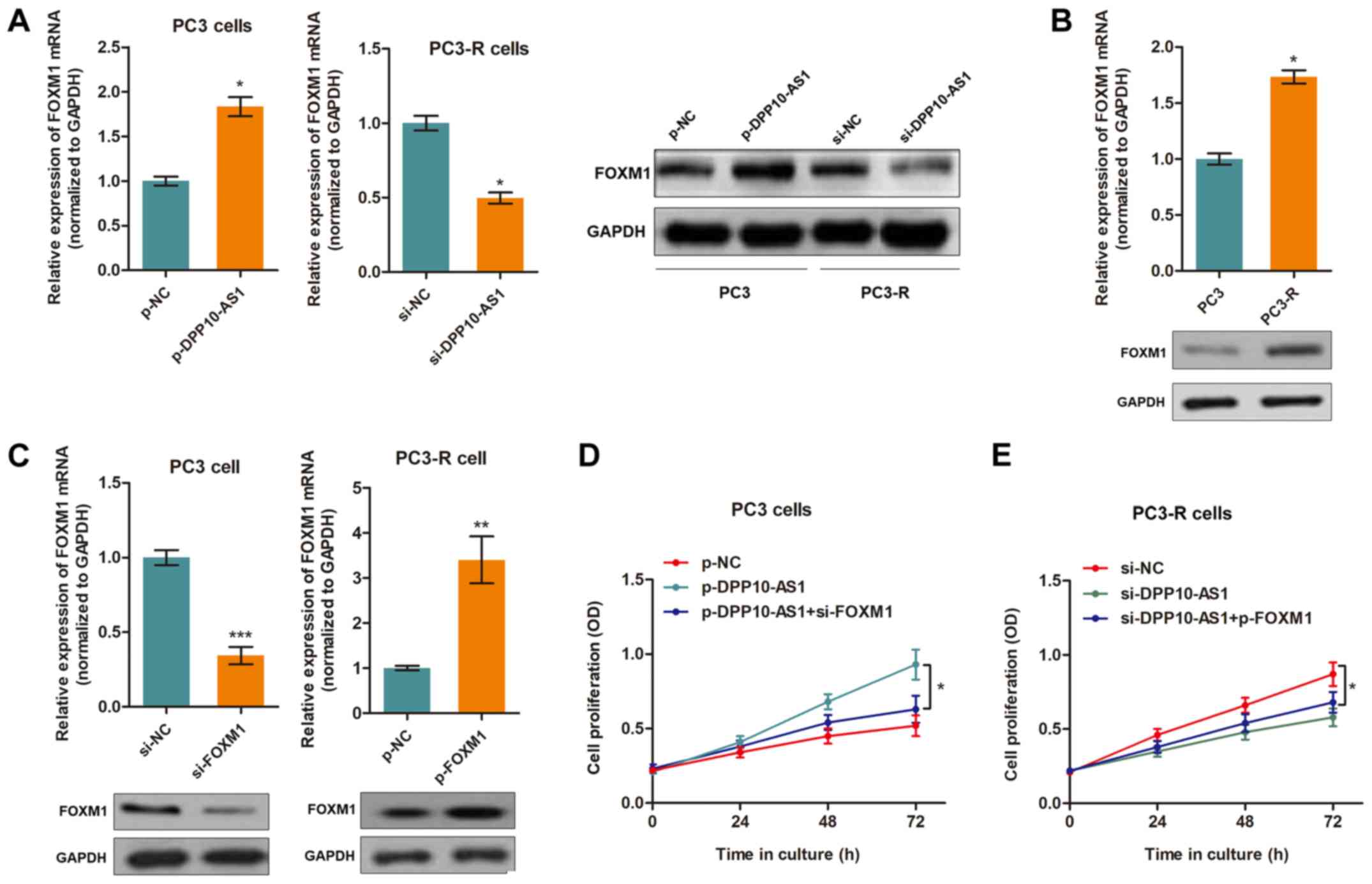 | Figure 3DPP10-AS1 targets FOXM1 in PCa
cells. (A) FOXM1 expression was upregulated in the DPP10-AS1
overexpressing PC3 cells. Silencing of DPP10-AS1 resulted in
downregulation of PC3-R cells at both the mRNA and protein
expression levels. *P<0.05. (B) The mRNA and protein
expression levels of FOXM1 were upregulated in PC3-R cells
compared with PC3 cells. *P<0.05. (C) Successful
knockdown or overexpression of FOXM1 in cells transfected with a
specific siRNA or overexpression vector, respectively.
**P<0.01,***P<0.001. (D) A CCK8 assay
showed that knockdown of FOXM1 partially reversed the
DPP10-AS1-induced increase in cell proliferation in PC3 cells,
*P<0.05. (E) A CCK8 assay showed that overexpression
of FOXM1 abrogated the effects induced by si-DPP10-AS1 in
PC3-R cells, *P<0.05. FOXM1, forkhead box M1;
PCa, prostate cancer; CCK-8, Cell Counting Kit-8; si, small
interfering; OD, optical density; NC, negative control; p-,
pcDNA3.1 overexpression vector. |
 | Table IIPotential targets of
DPP10-AS1a predicted by StarBase
version 2.0. |
Table II
Potential targets of
DPP10-AS1a predicted by StarBase
version 2.0.
| Target gene ID | Pair gene name | Total reads
number | Free energy | Align score |
|---|
|
ENSG00000089123 | TASP1 | 2 | −39.5 | 28 |
|
ENSG00000105700 | KXD1 | 1 | −44.8 | 25 |
|
ENSG00000113163 | COL4A3BP | 1 | −39 | 16 |
|
ENSG00000133316 | WDR74 | 1 | −12.2 | 16 |
|
ENSG00000222328 | RNU2-2P | 1 | −12.2 | 16 |
|
ENSG00000233876 | GAPDHP68 | 1 | −34 | 21.5 |
|
Entrez100008588 | RNA18N5 | 1 | −25.7 | 27 |
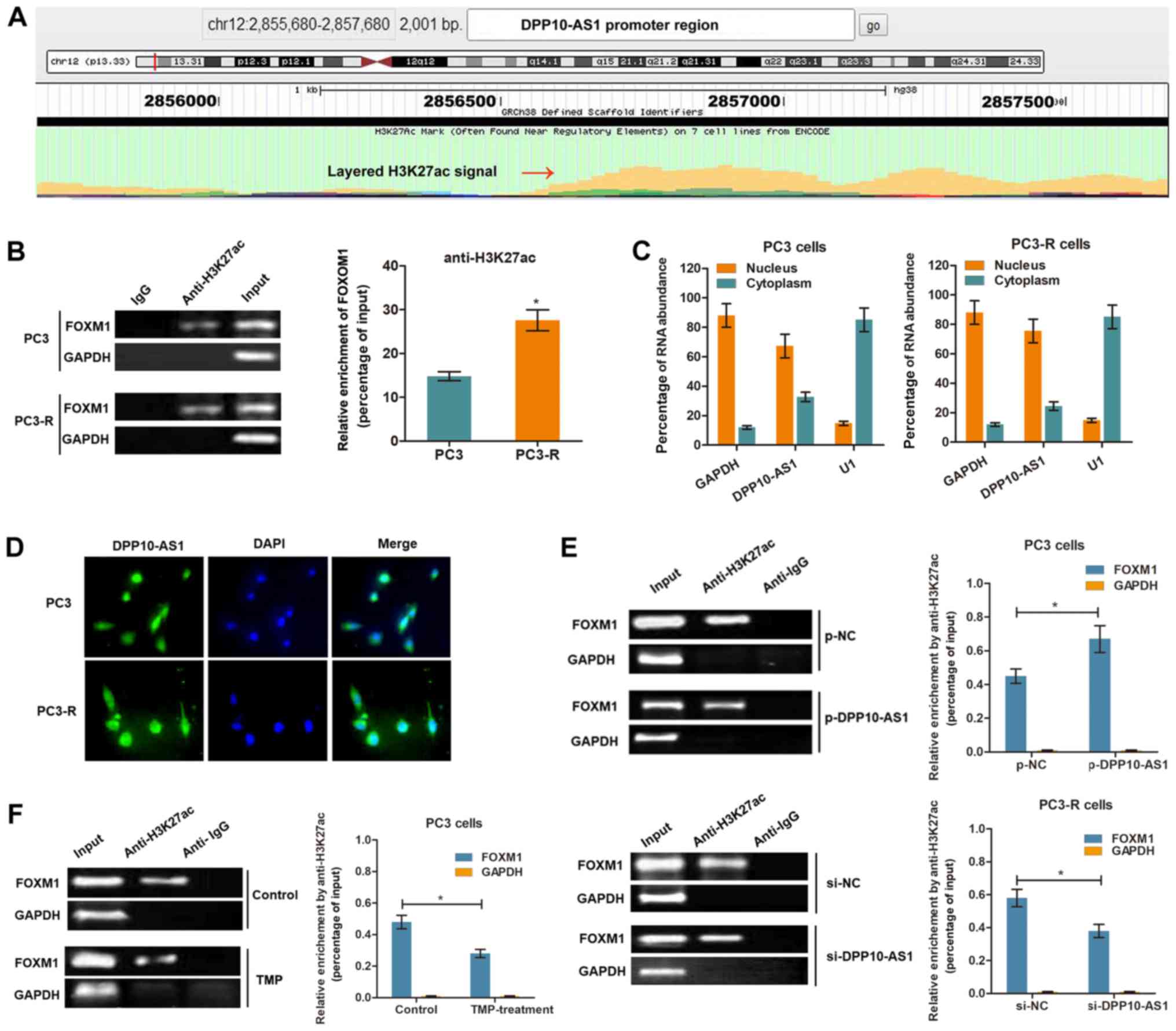
LncRNA DPP10‑AS1 increases FOXM1
expression by inducing H3K27 acetylation at the promoter
region
It has been shown that the CBP, an
acetyltransferase, is recruited to the C-terminal region of FOXM1
and enhances its transcriptional activity at specific stages of the
cell cycle (21). To verify
whether DPP10-AS1 upregulates FOXM1 by histone acetylation,
mediated by H3K27ac modification at the C‑terminal region, the
transcriptional modification regions were analyzed using UCSC
Genome Browser. Fig. 4A showed the
presence of a potential region which may be enriched in H3K27ac
upstream of the FOXM1 promoter region. Subsequently, ChIP
was performed using an anti-H3K27ac antibody in PCa cells. The
results showed that H3K27ac was enriched at the FOXM1
promoter region in both PC3 and PC3-R cells. In addition, the
enrichment level of H3K27ac was significantly increased in PC3-R
cells compared with PC3 cells (Fig.
4B). To further determine whether DPP10-AS1 regulated H3K27ac
enrichment, the subcellular localization of DPP10-AS1 was
determined in PCa cells. RT-qPCR analysis of DPP10-AS1 in the
nucleus and cytoplasm showed that DPP10-AS1 was primarily
distributed in the nucleus (Fig.
4C). The results of a FISH assay with a DPP10‑AS1 specific
probe showed results consistent with the RT-qPCR results (Fig. 4D), suggesting that DPP10-AS1 was
primarily located in the nucleus of PCa cells. Overexpression of
DPP10-AS1 significantly increased the enrichment of H3K27ac,
whereas knockdown of DPP10-AS1 decreased the enrichment levels
(Fig. 4E). Furthermore, TMP
treatment (500 µg/l) decreased H3K27ac enrichment at the
FOXM1 promoter region in PC3 cells (Fig. 4F). Together, these results show
that DPP10-AS1 may increase FOXM1 expression by increasing
H3K27ac modifications at the promoter region of FOXM1.
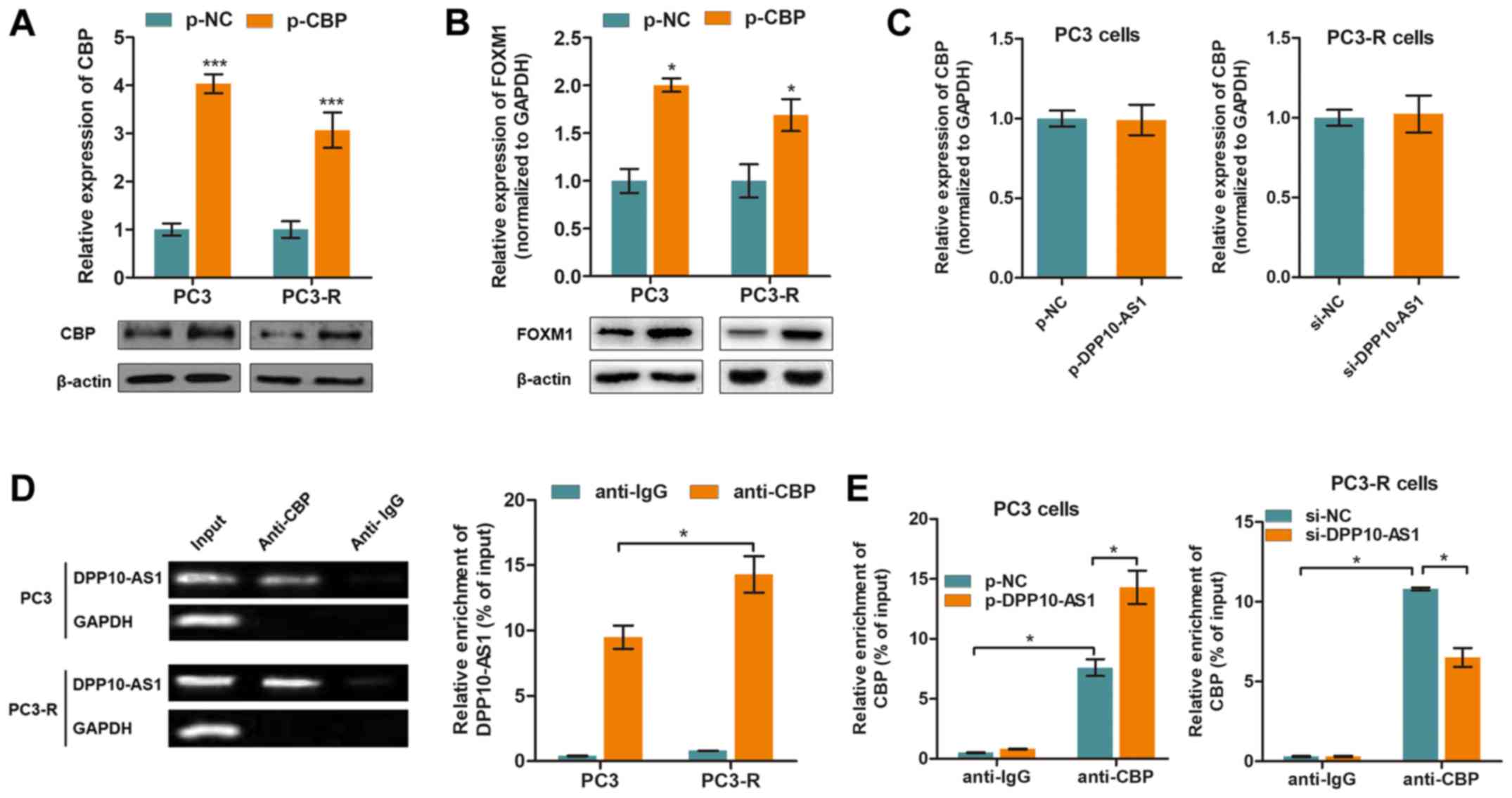
CBP is essential for DPP10‑AS1‑mediated
H3K27ac modifications
CBP is a histone acetyltransferase which serves an
important role in histone acetylation (22). To determine whether CBP
participated in DPP10-AS1-induced histone modification, CBP
overexpression plasmids were transfected into the PC3 and PC3-R
cells (Fig. 5A). As shown in
Fig. 5B, overexpression of CBP
promoted FOXM1 expression at both the mRNA and protein expression
levels. Thus whether DPP10-AS1 directly interacted with CBP was
determined and the results showed that CBP expression was not
altered by downregulation or upregulation of DPP10-AS1 (Fig. 5C). Thus it was hypothesized that
DPP10-AS1 may recruit CBP rather than modulate its expression. By
performing RIP assays with an anti-CBP antibody, it was shown that
DPP10-AS1 was associated with CBP, and enrichment was higher in the
PC3-R cells compared with the PC3 cells (Fig. 5E). In addition, CBP was enriched at
the FOXM1 promoter region, and dysregulated expression of
DPP10-AS1 affected the enrichment of CBP at the FOXM1
promoter (Fig. 5F). Together,
these results showed that DPP10-AS1 interacted with CBP, thereby
inducing H3K27ac modification in the FOXM1 promoter
region.
TMP suppresses tumor growth of PCa in
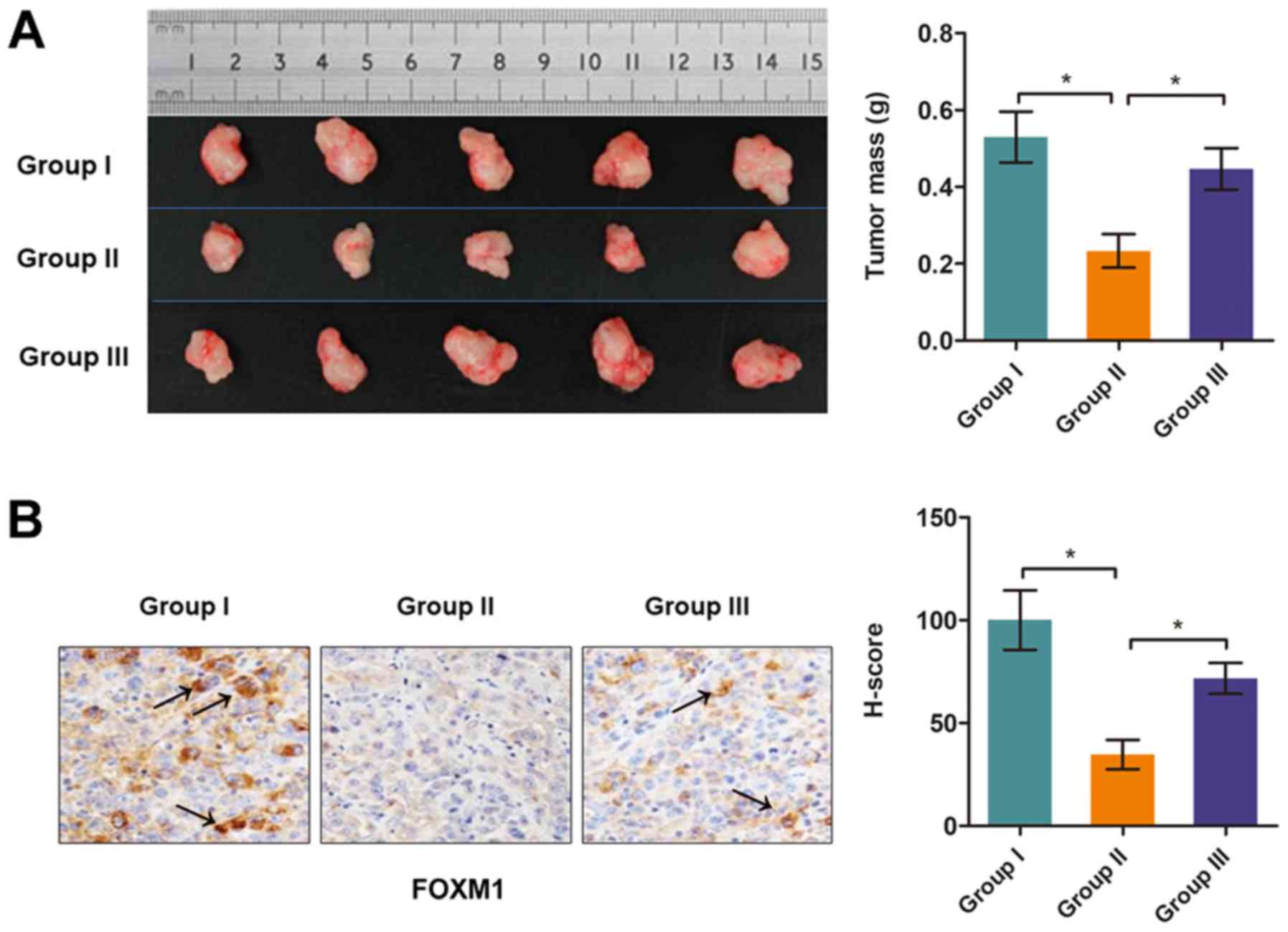
vivo by targeting DPP10‑AS1
To identify the suppressive function of TMP during
PCa progression, an in vivo nude mouse model bearing a PC3
xenograft was established. PC3 cells stably overexpressing
DPP10-AS1 or a negative control vector were injected into the
underarm area of mice. Quantitative data showed that TMP treatment
significantly reduced tumor growth in Group II vs. I; however, this
effect was significantly reversed by co‑expression of DPP10-AS1
(Group III vs. II; Fig. 6A).
The expression levels of FOXM1 was determined in
tumor tissues using an IHC assay. As shown in Fig. 6B, FOXM1 expression levels were
significantly reduced in tumor tissues from mice treated with TMP
(Group II vs. I), and this alteration was partially reversed in
tissues co-transfected with DPP10-AS1 (Group III vs. II).
Discussion
Patients with advanced PCa who develop resistance to
hormone therapy have limited therapeutic options in the clinic at
present, and therefore, several patients turn to alternative
treatments (23). In recent years,
the interest in herbal remedies has grown rapidly in the
industrialized world, since these drugs are increasingly considered
as effective and safe alternatives to synthetic drugs (24-26).
In the present study, the effects of TMP on PCa, and the underlying
mechanisms were determined. TMP is the standardized unique extract
from Ligusticum chuanxiong, and is one of the few
well-established plant products. TMP was shown to exhibit
anti-cancer effects in PCa by downregulating the expression levels
of lncRNA DPP10-AS1. Mechanistically, DPP10-AS1 suppressed
FOXM1 expression via histone modification at the
FOXM1 promoter region. These data suggest that TMP may serve
as a useful therapeutic option for treatment of patients with PCa,
and lncRNA DPP10-AS1 may serve as a potential therapeutic
target.
TMP is a major bioa ct ive comp onent of
Ligusticum chuanxiong which has been shown to reduce the
initiation and progression of cardiovascular diseases (27), and exhibits anti‑inflammatory
properties in several pathological processes (28). Several studies have also shown that
TMP exhibits anti-cancer properties. Cao et al (29) showed that TMP inhibited progression
of hepatocellular carcinoma by inducing apoptosis and autophagy.
Jia et al (30)
demonstrated that TMP treatment suppressed lung cancer growth
through disruption of angiogenesis via regulation of the
BMP/Smad/Id-1 signaling pathway. Consistently, in the present
study, the anti-cancer properties of TMP in PCa were demonstrated.
PC3 and DU145 cells exhibit differing sensitivities to TMP,
possibly due to the difference of cell membrane permeability and
material transport (10). In our
previous study, the anti-tumor role of TMP in PCa was shown to be
primarily mediated through downregulation of FOXM1. However,
the detailed regulatory mechanism were unclear. In the present
study, the role of lncRNA DPP10-AS1 in TMP-mediated effects was
demonstrated.
Antisense lncRNAs are a group of noncoding genes
oriented from protein coding or noncoding loci in the opposite
respective direction, and widely participate in the regulation of
multiple biological and pathological processes (31). These noncoding antisense
transcripts, consistent with other types of lncRNAs, may serve as
oncogenes or tumor suppressor genes through upregulation of
transcription of specific genes (32). For example, lncRNA ZEB1-AS1
functions as an oncogene in prostate cancer through epigenetic
activation of ZEB1 and indirectly regulating downstream molecules
(33). LncRNA DPP10-AS1 is
localized in the antisense DNA stand of the DPP10 gene,
which is strongly associated with asthma susceptibility, possibly
through regulating the activities of chemokines and cytokines
(34). However, the function of
DPP10-AS1 in cancer occurrence and other diseases have not been
reported, to the best of our knowledge, and thus the present study
is the first to investigate the role of DPP10‑AS1 in PCa
progression and therapy.
As the essential role of FOXM1 in TMP-induced
anti-tumor effects were demonstrated in our previous study, the
presence of a functional link between DPP10-AS1 and FOXM1
was assessed in the present study. By overexpressing or
knocking-down FOXM1 expression, FOXM1 was verified as
a direct target of DPP10-AS1, and was responsible for the
functional effects of DPP10-AS1. To determine how DPP10-AS1
affected FOXM1 expression, the epigenetic modifications of FOXM1 in
PCa cells were determined, as described previously (35,36).
DNA methylation and acetylation are two common epigenetic
modifications that are essential in the maintenance of
transcription, and closely associated with progression and
prognosis of various diseases (37-39).
A previous study showed that epigenetic modifications may influence
protein coding loci as well as noncoding loci based on a
genome-wide sequence analysis (40). Histone acetylation is a common
epigenetic modification at gene promoter regions which activate
transcription, may induce upregulation of associated transcripts,
and thus may be associated with malignant progression (41). For example, Myd88 is activated due
to H3K27ac modification at the promoter region in hepatocellular
carcinoma, and this results in tumor growth and metastasis
(42); SNHG14 noncoding RNA
activated PABPC1 transcription via the H3K27ac modification at its
promoter region, further regulating chemoresistance in breast
cancer (43).
Histone acetylation is required for regulating gene
transcription by promoting or repressing DNA replication activity
(44,45). The role of CBP in
DPP10-AS1-mediated histone acetylation was also shown in the
present study. CBP/p300 was the first discovered mammalian histone
acetyltransferase, and belongs to the GCN5-related
N-acetyltransferase family (46).
CBP-mediated acetylation of H3K27 facilitates the transcription of
downstream genes (47). CBP has
been reported to be involved in the progression of several types of
cancer, such as gastric cancer and lung adenocarcinoma; however,
the underlying mechanism by which CBP regulates the pathogenesis of
various tumors is still unknown. The present provides an alternate
perspective, where CBP interaction with noncoding RNAs may serve as
the mechanism underlying cancer progression.
In conclusion, the present study verified the
anti‑tumor effects of TMP in PCa, and further identified the
essential role of the DPP10-AS1/CBP/FOXM1 regulatory pathway in
TMP-induced suppression of PCa progression. These data suggest the
potential significance of DPP10‑AS1 as a promising therapeutic
target and predictive indicator of PCa progression.
Funding
The present study was supported by The Science
Foundation of Beijing.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ, ZJ and WY designed the study and performed the
experiments. ZZ, HL and XU analyzed the data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Research
Ethics Committee of Peking union Medical College Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Schatten H: Brief overview of prostate
cancer statistics, grading, diagnosis and treatment strategies. Adv
Exp Med Biol. 1095:1–14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Horiguchi M, Uno H and Wei LJ: Evaluating
noninferiority with clinically interpretable statistics for the
PROSELICA study to assess treatment efficacy of a reduced dose of
cabazitaxel for treating metastatic prostate cancer. J Clin Oncol.
36:825–826. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lunardi A, Ala U, Epping MT, Salmena L,
Clohessy JG, Webster KA, Wang G, Mazzucchelli R, Bianconi M, Stack
EC, et al: A co‑clinical approach identifies mechanisms and
potential therapies for androgen deprivation resistance in prostate
cancer. Nat Genet. 45:747–755. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu N, Zhang Z, Chen P, Zhong Y, Cai X, Hu
H, Yang Y, Zhang J, Li K, Ge J, et al: Tetramethylpyrazine (TMP),
an active ingredient of Chinese herb medicine chuanxiong,
attenuates the degeneration of trabecular meshwork through
SDF-1/CXCR4 axis. PLoS One. 10:e01330552015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zeng Z, Zhu W, Zhou X, Jin Z, Liu H, Chen
X, Pan J, Demura H, Naruse M and Shi Y: Tetramethylpyrazine, a
Chinese drug, blocks coronary vasoconstriction by endothelin-1 and
decreases plasma endothelin-1 levels in experimental animals. J
Cardiovasc Pharmacol. 31(Suppl 1): S313–S316. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang HH, Liu FB, Ruan Z, Zheng J, Su YJ
and Wang J: Tetramethylpyrazine (TMPZ) triggers S-phase arrest and
mitochondria-dependent apoptosis in lung cancer cells. Neoplasma.
65:367–375. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang S, Lei T and Zhang M: The reversal
effect and its mechanisms of tetramethylpyrazine on multidrug
resistance in human bladder cancer. PLoS One. 11:e01577592016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Y, Liu X, Zuo T, Liu Y and Zhang JH:
Tetramethylpyrazine reverses multidrug resistance in breast cancer
cells through regulating the expression and function of
P-glycoprotein. Med Oncol. 29:534–538. 2012. View Article : Google Scholar
|
|
9
|
Yi B, Liu D, He M, Li Q, Liu T and Shao J:
Role of the ROS/AMPK signaling pathway in
tetramethylpyrazine-induced apoptosis in gastric cancer cells.
Oncol Lett. 6:583–589. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou Y, Ji Z, Yan W, Zhou Z, Li H and Xiao
Y: Tetramethylpyrazine inhibits prostate cancer progression by
downregulation of forkhead box M1. Oncol Rep. 38:837–842. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rafiee A, Riazi‑Rad F, Havaskary M and
Nuri F: Long noncoding RNAs: Regulation, function and cancer.
Biotechnol Genet Eng Rev. 34:153–180. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Josipovic I, Pflüger B, Fork C, Vasconez
AE, Oo JA, Hitzel J, Seredinski S, Gamen E, Heringdorf DMZ, Chen W,
et al: Long noncoding RNA LISPR1 is required for S1P signaling and
endo-thelial cell function. J Mol Cell Cardiol. 116:57–68. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Magistri M, Faghihi MA, St Laurent G III
and Wahlestedt C: Regulation of chromatin structure by long
noncoding RNAs: Focus on natural antisense transcripts. Trends
Genet. 28:389–396. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li W, Sun M, Zang C, Ma P, He J, Zhang M,
Huang Z, Ding Y and Shu Y: Upregulated long non-coding RNA
AGAP2-AS1 represses LATS2 and KLF2 expression through interacting
with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death
Dis. 7:e22252016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen C, Feng Y and Wang X: LncRNA ZEB1-AS1
expression in cancer prognosis: Review and meta-analysis. Clin Chim
Acta. 484:265–271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar
|
|
21
|
Lv C, Zhao G, Sun X, Wang P, Xie N, Luo J
and Tong T: Acetylation of FOXM1 is essential for its
transactivation and tumor growth stimulation. Oncotarget.
7:60366–60382. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang L, Tang Y, Cole PA and Marmorstein R:
Structure and chemistry of the p300/CBP and Rtt109 histone
acetyltrans-ferases: Implications for histone acetyltransferase
evolution and function. Curr Opin Struct Biol. 18:741–747. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Curry EA III and Sweeney CJ: Resistance to
luteinizing hormone releasing hormone agonist therapy for
metastatic prostate cancer. J Urol. 168:1932002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McCarty MF: Current prospects for
controlling cancer growth with non-cytotoxic agents-nutrients,
phytochemicals, herbal extracts, and available drugs. Med
Hypotheses. 56:137–154. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Muhamad N, Plengsuriyakarn T and
Na-Bangchang K: Application of active targeting nanoparticle
delivery system for chemotherapeutic drugs and traditional/herbal
medicines in cancer therapy: A systematic review. Int J
Nanomedicine. 13:3921–3935. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vinayak M: Molecular action of herbal
antioxidants in regulation of cancer growth: Scope for novel
anticancer drugs. Nutr Cancer. 70:1199–1209. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo M, Liu Y and Shi D: Cardiovascular
actions and therapeutic potential of tetramethylpyrazine (Active
Component Isolated from Rhizoma Chuanxiong): Roles and mechanisms.
Biomed Res Int. 2016:24303292016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu JZ, Huang JH, Xiao ZM, Li JH, Li XM and
Lu HB: Tetramethylpyrazine accelerates the function recovery of
traumatic spinal cord in rat model by attenuating inflammation. J
Neurol Sci. 324:94–99. 2013. View Article : Google Scholar
|
|
29
|
Cao J, Miao Q, Miao S, Bi L, Zhang S, Yang
Q, Zhou X, Zhang M, Xie Y, Zhang J and Wang S: Tetramethylpyrazine
(TMP) exerts antitumor effects by inducing apoptosis and autophagy
in hepa-tocellular carcinoma. Int Immunopharmacol. 26:212–220.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jia Y, Wang Z, Zang A, Jiao S, Chen S and
Fu Y: Tetramethylpyrazine inhibits tumor growth of lung cancer
through disrupting angiogenesis via BMP/Smad/Id-1 signaling. Int J
Oncol. 48:2079–2086. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jadaliha M, Gholamalamdari O, Tang W,
Zhang Y, Petracovici A, Hao Q, Tariq A, Kim TG, Holton SE, Singh
DK, et al: A natural antisense lncRNA controls breast cancer
progression by promoting tumor suppressor gene mRNA stability. PLoS
Genet. 14:e10078022018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pian L, Wen X, Kang L, Li Z, Nie Y, Du Z,
Yu D, Zhou L, Jia L, Chen N, et al: Targeting the IGF1R pathway in
breast cancer using antisense lncRNA-mediated promoter cis
competition. Mol Ther Nucleic Acids. 12:105–117. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Su W, Xu M, Chen X, Chen N, Gong J, Nie L,
Li L, Li X, Zhang M and Zhou Q: Long noncoding RNA ZEB1-AS1
epigenetically regulates the expressions of ZEB1 and downstream
molecules in prostate cancer. Mol Cancer. 16:1422017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Belau F, Metzner K, Christ T, Ravens U,
Schaefer M, Künzel S, Li W, Wettwer E, Dobrev D, El-Armouche A and
Kämmerer S: DPP10 is a new regulator of Nav1.5 channels in human
heart. Int J Cardiol. 284:68–73. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pérez‑Peña J, Győrffy B, Amir E, Pandiella
A and Ocaña A: Epigenetic modulation of FOXM1-gene interacting
network by BET inhibitors in breast cancer. Breast Cancer Res
Treat. 172:725–732. 2018. View Article : Google Scholar
|
|
36
|
Zhou Z, Chen H, Xie R, Wang H, Li S, Xu Q,
Xu N, Cheng Q, Qian Y, Huang R, et al: Epigenetically modulated
FOXM1 suppresses dendritic cell maturation in pancreatic cancer and
colon cancer. Mol Oncol. 13:873–893. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shi H, Wei SH, Leu YW, Rahmatpanah F, Liu
JC, Yan PS, Nephew KP and Huang TH: Triple analysis of the cancer
epigenome: An integrated microarray system for assessing gene
expression, DNA methylation, and histone acetylation. Cancer Res.
63:2164–2171. 2003.PubMed/NCBI
|
|
38
|
Tang RZ, Zhu JJ, Yang FF, Zhang YP, Xie
SA, Liu YF, Yao WJ, Pang W, Han LL, Kong W, et al: DNA
methyltransferase 1 and Krüppel‑like factor 4 axis regulates
macrophage inflammation and atherosclerosis. J Mol Cell Cardiol.
128:11–24. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu L, He X, Zhao M, Yang S, Wang S, Yu X,
Liu J and Zang W: Regulation of DNA methylation and 2-OG/TET
signaling by choline alleviated cardiac hypertrophy in
spontaneously hypertensive rats. J Mol Cell Cardiol. 128:26–37.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wan G, Hu X, Liu Y, Han C, Sood AK, Calin
GA, Zhang X and Lu X: A novel non-coding RNA lncRNA-JADE connects
DNA damage signalling to histone H4 acetylation. EMBO J.
32:2833–2847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li D, Bi FF, Cao JM, Cao C, Liu B and Yang
Q: Regulation of DNA methyltransferase 1 transcription in
BRCA1-mutated breast cancer: A novel crosstalk between E2F1 motif
hypermethylation and loss of histone H3 lysine 9 acetylation. Mol
Cancer. 13:262014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xu X, Yin Y, Tang J, Xie Y, Han Z, Zhang
X, Liu Q, Qin X, Huang X and Sun B: Long non-coding RNA Myd88
promotes growth and metastasis in hepatocellular carcinoma via
regulating Myd88 expression through H3K27 modification. Cell Death
Dis. 8:e31242017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dong H, Wang W, Mo S, Liu Q, Chen X, Chen
R, Zhang Y, Zou K, Ye M, He X, et al: Long non-coding RNA SNHG14
induces trastuzumab resistance of breast cancer via regulating
PABPC1 expression through H3K27 acetylation. J Cell Mol Med.
22:4935–4947. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Das C, Lucia MS, Hansen KC and Tyler JK:
CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature.
459:113–117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ghosh TK, Aparicio-Sánchez JJ, Buxton S,
Ketley A, Mohamed T, Rutland CS, Loughna S and Brook JD:
Acetylation of TBX5 by KAT2B and KAT2A regulates heart and limb
development. J Mol Cell Cardiol. 114:185–198. 2018. View Article : Google Scholar
|
|
46
|
Wang YM, Gu ML, Meng FS, Jiao WR, Zhou XX,
Yao HP and Ji F: Histone acetyltransferase p300/CBP inhibitor C646
blocks the survival and invasion pathways of gastric cancer cell
lines. Int J Oncol. 51:1860–1868. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kouzarides T: SnapShot: Histone-modifying
enzymes. Cell. 131:8222007. View Article : Google Scholar : PubMed/NCBI
|