Introduction
The B7 family of immunoregulatory proteins comprises
several members expressed on antigen-presenting cells (APCs),
T-cells and tumour cells (1).
These are transmembrane glycoproteins, with one or more
amino-terminal extracellular immunoglobulin domains, a single
transmembrane alpha-helical domain, and a short intracellular
region.
B7 proteins are expressed on the surface of APCs and
are considered to interact in trans with specific receptors
on subsets of T-cells as part of the immune synapse that forms
between these two types of cells. In so doing, they provide a
co-signal for T-cell regulation that acts in concert with the
trans interaction between the T-cell receptor and the
antigen-MHC complex from the APC (2,3). The
best characterised example is the protein B7-H1 (PD-L1), which
recognises the programmed death-1 (PD-1) protein expressed on the
surface of T-cells. This interaction can inhibit T-cell activity by
stimulating T-cell apoptosis (4,5). By
contrast, the structurally related protein B7-H4 (B7S1, VTCN1, B7x)
is less well understood. The binding of B7-H4 to CD4+
and CD8+ T-cells inhibits their activation and
proliferation (6). However, the
trans-binding partner for B7-H4 at the T-cell plasma
membrane has not yet been confirmed (7).
Although generally absent from most normal human
tissues, the B7-H4 protein is often expressed on the plasma
membrane of human cancer cells, and a high level of expression is
correlated with poor prognosis (8). Expression of B7-H4 is particularly
common in lung, ovarian, oesophageal and breast cancers (8-11),
and is associated with an enhanced metastatic potential (12,13).
It is believed that B7-H4 promotes tumour survival, at least in
part, by attenuating the immune response of T-cells and other
immune cells that infiltrate the tumour microenvironment (14,15).
There is increasing interest in the potential of
antibody-based cancer therapy as a mechanism for blocking
immune-inhibitory signals from the B7 proteins beyond the
well-established PD-1:PD-L1 interaction (16,17).
However, to realise the potential of this approach, a clearer
understanding of the molecular context of the B7 microenvironment,
both on the plasma membrane of the B7-expressing tumour cells and
the immune cells that recognise them, is required. For example, an
important unresolved question is whether the B7 proteins are
uniformly distributed over the tumour cell plasma membrane, or
whether they form a co-localised cluster of specific proteins. Such
local cis-clustering is common for immunoregulatory
molecules and may facilitate the formation of the immune synapse
with the interacting immune cell (18). Identifying the cis-molecular
neighbours of B7 family proteins using new proximity proteomic
technologies may provide new insights into their function. However,
to the best of our knowledge, there are no reports in the
literature that investigate this issue. In the present study, a
proximity labelling method was used to take a snapshot of the
plasma membrane proteins surrounding B7-H4 and characterise these
proteins using orthogonal approaches.
Cis-interacting proteins need only interact
weakly to maintain their associations on the two-dimensional
surface of the plasma membrane; such low-affinity interactions may
not be maintained once the cell is lysed. Furthermore, proteins
assembled into more extended cis-clusters need not interact
directly to be spatially and functionally associated (19). These characteristics make it
difficult to characterise the composition of such clusters by
simple proteomic analysis of immunoprecip-itated proteins. As an
alternative approach to the investigation and analysis of these
two-dimensional clusters, our group has previously developed a
specific proteomic proximity labelling assay using tyramide
(SPPLAT) (20). In this method,
peroxidase is targeted via a specific antibody to the plasma
membrane protein of interest, and a biotin-tyramide derivative is
briefly added. The peroxidase generates a biotin-tyramide free
radical that covalently biotinylates proteins within a few tens of
nm from the target. These proteins can then be isolated by
streptavidin affinity capture and identified by mass spectrometry
(MS) (19-21).
The SPPLAT method was used to investigate the
molecular neighbours of the B7-H4 immune checkpoint protein on the
surface of the human breast cancer cell line SK-BR-3. This cell
line expresses B7-H4 on its plasma membrane, and has been
extensively used as a model to understand the biochemistry, cell
biology and pathophysiology of breast cancer (22). The aim of the present study was to
determine whether the B7-H4 molecule, as well as regulating T-cell
immune responses, may also play a role in modulating the
cell-matrix interactions when expressed on cancer cells, which is
likely to be important in metastatic survival.
Materials and methods
Experimental workflow
Stable isotope labelling of amino acids in culture
(SILAC) SPPLAT methodology is schematically outlined in Fig. S1.
Cell culture
Human breast adenocarcinoma SK-BR-3 cells (ATCC)
were grown in McCoy's medium or RPMI-1640 containing 2 mM
L-glutamine and supplemented with 10% foetal bovine serum
(Invitrogen; Thermo Fisher Scientific, Inc.). The medium was
changed every 2-3 days. For SILAC experiments, the RPMI-1640 heavy
medium contained 13C6 labelled L-lysine and
L-arginine (K6R6), while the light medium was
RPMI-1640 containing non-labelled L-lysine and L-arginine
(K0R0), both of which were supplemented with
10% dialyzed foetal bovine serum (all from Dundee Cell Products), 2
mM L-glutamine, 100 U/ml penicillin and 100 µg/ml
streptomycin (all from Invitrogen; Thermo Fisher Scientific,
Inc.).
Flow cytometry analysis
The cells were washed with PBS and were detached
from tissue culture flasks by accutase (Gibco; Thermo Fisher
Scientific, Inc.). Cells were incubated with LIVE/DEAD®
fixable violet dead cell stain (20 min, 4°C, Thermo Fisher
Scientific, Inc.), washed with flow cytometry buffer
(eBiosciences), and stained with AF647 (Thermo Fisher Scientific,
Inc.)-conjugated anti-B7-H4 (MedImmune) or isotype control (clone
R347, MedImmune) antibodies for 30 min at 4°C. Cells were washed
with flow cytometry buffer and fixed with 1% paraformaldehyde for
20 min at 4°C before analysis. Compensation was performed using
antibody-stained AbC beads (Invitrogen; Thermo Fisher Scientific,
Inc.) and LIVE/DEAD® violet-stained ArC beads
(Invitrogen; Thermo Fisher Scientific, Inc.). Flow cytometry was
performed on an FACS Canto II with 405 and 633 nm lasers (BD
Biosciences), with data analysis in FlowJo v10.0.6 software (Tree
Star, Inc.).
Immunofluorescence
SK-BR-3 cells were spun onto coverslips pre-coated
with 0.05% poly L-lysine, fixed with 4% parafor-maldehyde at room
temperature for 10 min, washed in PBS and incubated with
anti-B7-H4-488 (MedImmune) for 1 h at room temperature. To
determine all locations of B7-H4, the cells were permeabilised with
1% saponin for 30 min prior to antibody incubation and imaged at
×60 magnification using an Olympus Fluoview IX81 laser scanning
confocal microscope (Olympus Corporation).
Biotinylation of surface B7-H4 and
neighbours
The biotin tyramide reagent was prepared as
previously described (21). A
total of 1 mg each of Human IgG1 anti-B7-H4 monoclonal antibody
(in-house; MedImmune) and anti-B7-H1 (PD-L1) monoclonal antibody
(in-house; MedImmune) were conjugated with HRP using the EZ-link
activated peroxidase kit at pH 9.4 (cat. no. 31497; Thermo Fisher
Scientific, Inc.) and purified by gel filtration. This resulted in
~1:1 stoichiometry. Exponentially growing SK-BR-3 cells were
pelleted and washed in PBS at room temperature. Approximately
1×106 live cells were incubated with end-over-end
rotation for 2 h with either 20 µg/ml human anti-B7-H4-HRP,
20 µg/ml human anti-B7-H4 with 200 nM free HRP (BioRad
Laboratories, Inc.), or 20 µg/ml human anti-B7-H1-HRP in 20
ml PBS with 10% BSA (Sigma-Aldrich; Merck KGaA) at 4°C. For SILAC
SPPLAT ~5×108 heavy (K6R6) cells
were incubated with end-over-end rotation for 2 h with 20
µg/ml HRP-conjugated human anti-B7-H4 and 5×108
light (K0R0) with 20 µg/ml human
anti-B7-H4 and 200 nM free HRP.
Cells were pelleted and re-suspended in 10 ml
tyramide-labelling buffer (50 mM Tris HCl pH7.4, fresh 0.03%
H2O2 and 80 µg/µl tyramide
biotin label (Thermo Fisher Scientific, Inc.) and incubated with
end-over-end rotation at room temperature for 5 min. After
incubation, 100 U/ml cata-lase (Sigma-Aldrich; Merck KGaA) was
added and incubated with the samples for a further 5 min to quench
H2O2. Cells were washed gently with 45 ml
antibody strip buffer [50 mM glycine (pH 3.0), 150 mM NaCl, 0.9 mM
CaCl2, 0.5 mM MgCl2] and left on ice for 5
min. The extent of biotinylation was assessed by immunofluorescence
using non-permeabilised cells stained with Streptavidin-488 (at
1:100) for 1 h at room temperature.
Affinity purification of biotinylated
proteins
Cells were re-suspended in 15 ml cell lysis buffer
[20 mM Tris-HCl (pH 7.5), 5 mM EDTA, 1X protease inhibitor cocktail
(Roche Diagnostics), 150 mM NaCl, 1% v/v Triton X-100, 0.1 M sodium
thiocyanate (Sigma-Aldrich; Merck KGaA)] and incubated for 30 min
on ice. Insoluble material was removed by centrifugation at 10,000
× g for 10 min at 4°C, and the protein-containing soluble fraction
was recovered for streptavidin-bead capture.
For the initial SPPLAT, 1 ml lysate from
2×106 SPPLATed cells was added to 100 µl slurry
of high-capacity Neutravidin resin (Thermo Fisher Scientific,
Inc.), incubated with end-over-end rotation for 1 h at 4°C, washed
three times with lysis buffer containing sodium thiocyanate to
reduce non-specific interactions, then biotinylated proteins were
eluted with lysis buffer containing 10 mM biotin. Eluted proteins
were separated by SDS-PAGE (10%) and stained with Simply Blue™
SafeStain Coomassie (Invitrogen; Thermo Fisher Scientific, Inc.)
for 1 h at room temperature prior to dividing and excising equally
into 16 bands. Gel slices were de-stained in ddH2O and
20 mM NH4HCO3, reduced with 2 mM DTT and
alkylated with 10 mM iodoacetamide prior to overnight digestion
with 2 µg sequencing grade trypsin (Promega Corporation).
Peptides were extracted with acetonitrile and 1% formic acid and
re-suspended in water with 1% formic acid after vacuum-drying.
For SILAC SPPLAT, 5 ml cell lysates were quantified
and equal amounts (1 mg) of protein from both heavy and light cell
lysates were added to 0.5 ml slurry of high-capacity Neutravidin
resin and incubated as mentioned above. Reciprocal labelling
(specific-light, control-heavy) and affinity purification was also
performed. Two rounds of 200 µl eluted proteins were
combined and precipitated with 80% ice cold acetone, re-suspended
in 1X SDS sample buffer and separated by SDS-PAGE (10%) as before,
but were separated for only 1 cm; 4 bands per lane were excised for
mass spectrometry (MS) analysis and prepared as before.
Identification of proteins and
biotinylated proteins by MS
All liquid chromatography (LC)-MS/MS SPPLAT
experiments were performed using a NanoAcquity UPLC system (Waters
Corp.) and an LTQ Orbitrap Velos hybrid ion trap mass spectrometer
(Thermo Fisher Scientific, Inc.). Separation of peptides was
performed by reverse-phase chromatography using a Waters
reverse-phase nano column (BEH C18, 75 µm i.d. x250 mm, 1.7
µm particle size) at a flow rate of 300 nl/min. Peptides
were initially loaded onto a pre-column (Waters UPLC Trap Symmetry
C18, 180 µm i.d. x20 mm, 5 µm particle size) from the
NanoAcquity sample manager with 0.1% formic acid for 3 min at a
flow rate of 10 µl/min. After this period, the column valve
was switched to allow the elution of peptides from the pre-column
onto the analytical column, where a linear gradient of increasing
acetonitrile (5-35%) over 60 min was employed. The LC eluant was
sprayed into the mass spectrometer by means of a nanospray source.
All m/z values of eluting ions were measured in the Orbitrap Lumos
Velos mass analyser, set at a resolution of 30,000. Data-dependent
scans (top 10) were employed to automatically isolate and generate
fragment ions by collision-induced dissociation in the linear ion
trap, resulting in the generation of MS/MS spectra. Ions with
charge states of ≥2+ were selected for fragmentation. The raw MS
data files were converted to mgf files and searched against the
Swissprot Human database (accessed in May 2017; 71,567 entries)
using the Mascot search algorithm (version 2.3.02, Matrix Science)
with methionine oxidation (M) as a variable modification and
cysteine carbamidomethylation (C) as a fixed modification, allowing
2 missed cleavages, a peptide mass tolerance of ±1 Da and a
fragment mass tolerance of 0.8 Da. Single-peptide hits were removed
from the lists.
Quantitation was performed using MaxQuant (version
1.6.0.1; https://www.maxquant.org/). Raw data
were searched using Andromeda (http://www.coxdocs.org/doku.php?id=maxquant:andromeda),
with Arg-6 and Lys-6 set as heavy labels, methionine oxidation and
N-acetylation as variable modifications, and cysteine
carbamidomethylation as a fixed modification. The proteins were
identified if there was at least one unique peptide and quantified
if there were at least two unique peptides. Only unique peptides
were used for quantitation.
Identification of the SK-BR-3 proteome by
MS
SK-BR-3 cells (1×105) were grown in
either McCoy's medium or RPMI-1640 and lysed, and the soluble
fraction was separated by 10% SDS-PAGE, Coomassie-stained as
described above, and the entire lane was excised to allow the
analysis of all proteins by MS. The stained bands were reduced and
alkylated, and MS was performed as described above.
Co-localisation of B7-H4 with its
molecular neighbours
To confirm the co-localisation of B7-H4 with
integrins, HLA-E or plexin, SK-BR-3 cells (~1×104) were
spun onto 13 mm coverslips, fixed, but not permeabilised, washed,
blocked and incubated with goat anti-B7-H4 polyclonal IgG Ab
(1:250; cat. no. PA5-47261; Thermo Fisher Scientific, Inc.), and
then either integrin α1 (mouse anti-human IgG1 at 2
µg/ml; cat. no. MAB1973; EMD Millipore), HLA-E (mouse
anti-human IgG1 at 1:50; cat. no. sc-71262; Santa Cruz
Biotechnology, Inc.) or plexin B2 (mouse anti-human IgM at 1:50;
cat. no. sc-373969; Santa Cruz Biotechnology, Inc.) for 1 h at room
temperature. Integrin α1 incubations were also performed at 4 and
37°C. Incubations with anti-goat IgG-488 (1:1,000; cat. no.
A-11078; Thermo Fisher Scientific, Inc.) and anti-mouse IgG-647
(1:1,000; cat. no. A-21239; Thermo Fisher Scientific, Inc.) for 1 h
at room temperature were performed to visualise the cell surface
proteins using confocal microscopy, as mentioned before.
Proximity ligation assay (PLA)
SK-BR-3 cells (~1×104) were spun onto
coverslips pre-coated with 0.05% poly L-lysine. Live cells were
washed and blocked in 10% BSA in PBS and incubated with goat
anti-B7-H4 (2.5 µg/106 cells) in 10% BSA for 30
min at room temperature to induce clustering, then fixed with 4%
paraformaldehyde at room temperature for 10 min and washed in PBS.
For controls, cells were first incubated in antibodies against
integrin α1 (2 µg/ml; cat. no. MAB1973; EMD Millipore),
HLA-E (1:50; cat. no. sc-373969; Santa Cruz Biotechnology, Inc.) or
plexin B2 (1:50; cat. no. sc-373969; Santa Cruz Biotechnology,
Inc.) for 1 h at room temperature. Controls included fixing cells
prior to B7-H4 antibody incubation and omission of the primary
antibodies. Sigma PLA probes (goat plus and mouse minus; Sigma
Aldrich; Merck KGaA) were used and the ligation and amplification
(far red) was performed according to the manufacturer's
instructions. Cells on coverslips were mounted onto slides and
imaged using an Olympus Fluoview IX81 laser scanning confocal
microscope (Olympus Corporation) at x60 magnification.
Real-time integrin binding
SK-BR-3 cells were seeded at 1×104
cells/well in an E-plate (Roche Applied Science) pre-coated with
either 10 µg/ml fibronectin or 10 µg/ml collagen
peptides or 5% albumin. The wells were incubated with 20
µg/ml human anti-B7-H4 (MedImmune), or non-specific human
IgG (1:100 dilution of ascitic fluid; in-house; MedImmune). To
quantitate cell-line binding to collagen peptides, the
impedance-based xCELLigence system (Acea), which allows label-free,
dynamic monitoring of cell adhesion in real time, was used
(23). The assay system expresses
the adhesion-dependent rise in well impedance in units of cell
index (CI), defined as (Rn-Rb)/15, where
Rn is the electrical impedance of each cell-containing
well and Rb is the background impedance of the well with
medium alone. CI was measured for up to 2 h. All experiments were
performed in triplicate in the presence of either 5 mM
Mg2+ or 5 mM EDTA. Data are presented as the mean ±
standard deviation and were analysed for statistical significance
using one-way ANOVA followed by Tukey's post hoc test using the
GraphPad Prism software package (version 7.0d, GraphPad Software,
Inc.).
PCR of integrin isoforms
Total RNA was extracted from ~5×106
SK-BR-3 cells using an RNA extraction kit (Qiagen GmbH). RT-PCR was
carried out with 1 mg of RNA, using the One-Step RT-PCR kit (Qiagen
GmbH), following the manufacturer's instructions. The amplification
primers used were as follows: Human integrin α1, forward 5′-GGT GAA
TCA TTA CCT TGC GT-3′ and reverse 5′-AGC ACA TCT CCA GAA GAA GC-3′;
human integrin β1, forward 5′-AGG AAC AGC AGA GAA GCT CA-3′ and
reverse 5′-CAT TTT CTT CAA TTT TCC CC-3′.
Co-immunoprecipitation (co-IP)
To confirm direct binding of α1 integrin and surface
B7-H4, antibodies to α1 integrin (EMD Millipore) and B7-H4
(MedImmune) were coupled to separate preparations of Protein G
beads (Thermo Fisher Scientific, Inc.) overnight at 4°C with gentle
agitation. To assess only surface interactions of α1 integrin, IP
was performed with live cells, as lysates would contain
predominantly cytosolic B7-H4. Live cells (1×106) were
washed in PBS and allowed to bind antibody-bound Protein G beads.
Control cells were first incubated in goat serum with a
non-specific antibody, and then Protein G beads were added. Unbound
beads were washed off and cells were then lysed in lysis buffer as
before. Soluble lysates were separated by SDS-PAGE (10%),
transferred to a nitrocellulose membrane and probed with either
antibodies to α1 integrin (1:3,000; EMD Millipore) or B7-H4
(1:2,000; Thermo Fisher Scientific, Inc.) for the B7-H4 and α1
integrin co-IP, respectively.
Results
Analysing the SK-BR-3 proteome
A partial SK-BR-3 proteome was obtained, which
enables estimation of the abundance of common proteins that may
also reside in the footprint of the SPPLAT biotin label and thus
specifically bind affinity resins. For a stringent list,
single-peptide hit protein assignments were removed, enabling
positive identification of 1,220 proteins that were ranked by
empirical abundance using the protein EMPAI score (Table SI). These equated to ~0.77%
coverage of the expected total human proteome (currently 159,552
protein entries) and were interpreted with caution if they appeared
in SPPLAT downstream purifications. Gene Ontology (GO) annotation
analysis identified no over- or under-representation of any 'cell
components' compared with the human reference proteome, indicating
successful solubilisation and extraction of all protein classes.
These abundant proteins included keratins, histones, metabolic
enzymes, structural and heat shock proteins. The target protein,
B7-H4, was not identified as a high-abundance protein, as expected,
thereby highlighting the fact that a method for specifically
labelling these B7 cell surface membrane proteins is required to
identify proximal proteins.
This analysis also compared SK-BR-3 cells grown in
McCoy's medium and the RPMI-1640 medium required for SILAC
labelling. There was a large (86%) overlap in identified peptides
for the most abundant proteins from each condition, indicating that
the culture of SK-BR-3 cells in RPMI-1640 medium for quantitative
analysis would not result in a significant change in the
proteome.
The plasma membrane B7-H4
microenvironment
The localisation of B7-H4 in SK-BR-3 cells was first
characterised and its weak plasma membrane expression compared with
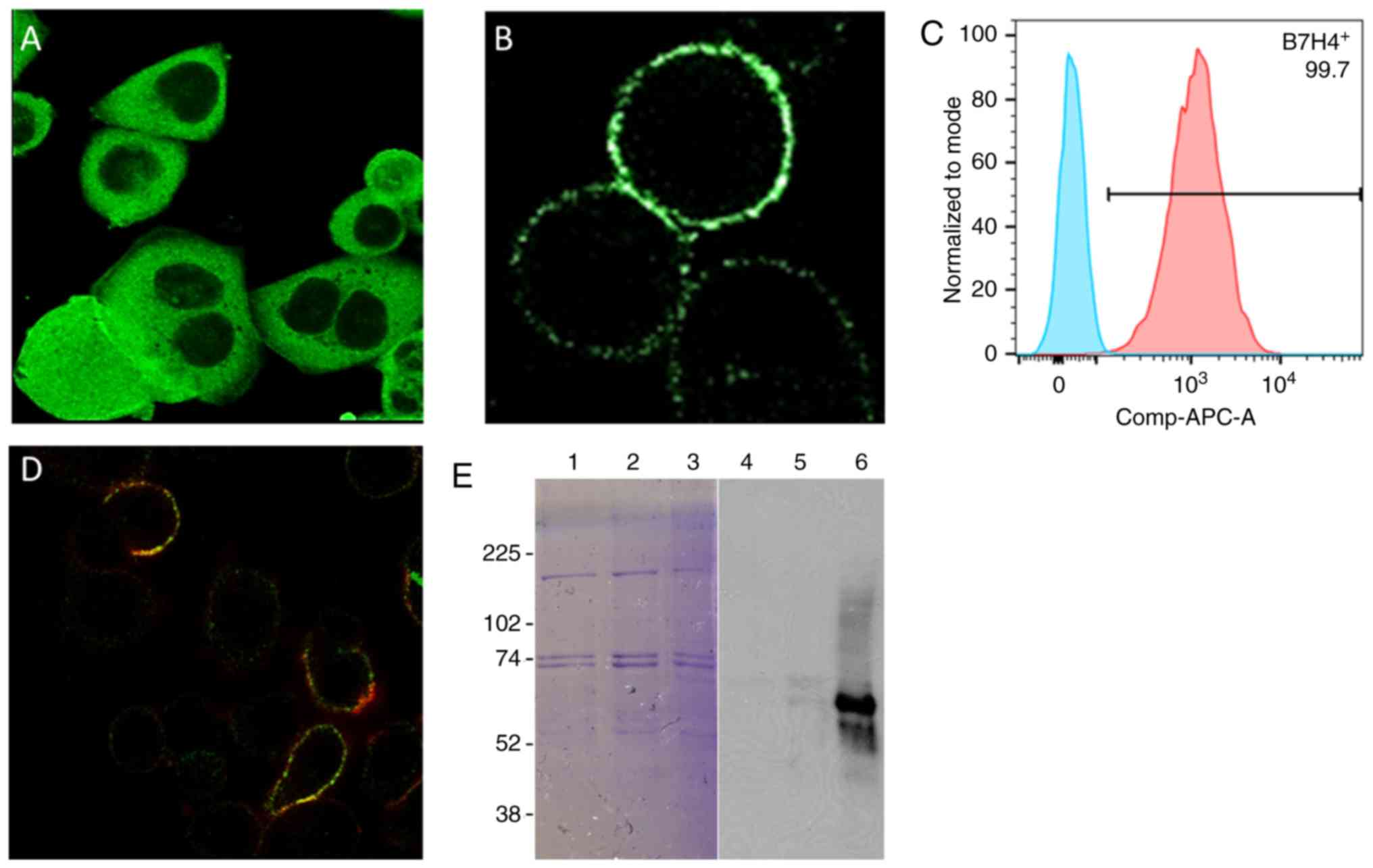
abundant intracellular expression was confirmed (Fig. 1A-C), suggesting its suitability for
investigation via the SPPLAT method. Analysing B7-H4 3D structures
using PDB structures identified B7-H4 only having two surface
exposed tyrosine residues that are required for SPPLAT labelling.
Confocal imaging and western blotting confirmed that B7-H4 and its
neighbours were biotinylated (Fig. 1D
and E). As a comparison, targeting B7-H1 resulted in
biotinylation of a different, and smaller, set of proteins to
B7-H4, as determined by western blotting (Fig. 1E). The negative control, using an
HRP-tagged non-specific IgG, did not lead to biotinylation of any
proteins.
This non-quantitative B7-H4 SPPLAT experiment
followed by affinity purification (AP)-MS resulted in the
identification of 2,004 biotinylated proteins (Table SII). The B7-H4 protein and its
proximal proteins were enriched and identified, a number of which
were not identified in the initial SK-BR-3 proteome screen. GO
annotation revealed an enrichment of cell surface and peripheral
membrane proteins compared with the control, as expected. The top
80 cell surface-associated proteins from the anti-B7-H4 SPPLAT
experiment that include the target, VTCN1 (B7-H4), ICOS ligand
(B7-H2), a number of integrins, plexins, cell adhesion molecules
and proteins from the Ig superfamily are shown in Table I. A number of cytosolic proteins
were also identified, suggesting that the biotin-tyramide can
permeate the plasma membrane in the 5-min labelling time and/or
that cytosolic proteins can be recruited to the plasma membrane
upon B7-H4 activation by antibody. As a further comparison, B7-H1
was also labelled (Fig. 1E), and
several immune signalling molecules were identified, a number of
which were also associated with B7-H4 (Table SI), although the protein
microenvironments differed. The negative control did not
non-specifically label signalling molecules.
 | Table IProteins from replicate SPPLAT
experiments. |
Table I
Proteins from replicate SPPLAT
experiments.
| Uniprot
identifier | Gene symbol | Protein
description | EMPAI score | Fold
enrichment | No. Peptides | % sequence
coverage |
|---|
| A8K6Q8 | TFRC | cDNA FLJ75881. highly similar to H.
sapiens transferrin receptor (p90. CD71) (TFRC). mRNA | 6.11 | 2.01 | 48 | 51.7 |
| A5YM53 | ITGAV | Integrin
a-5 | 2.27 | 7.09 | 44 | 37.2 |
| A0A087WXM8 | BCAM | Basal cell adhesion
molecule | 1.99 | | 20 | 43.0 |
| B7Z9S8 | ATP1B1 |
Sodium/potassium-transporting ATPase
subunit β | 1.97 | | 8 | 37.2 |
| O15031 | PLXNB2 |
Plexin-B2 | 1.84 | 4.97 | 60 | 35.2 |
| D3DVF0 | F11R | F11 receptor. isoform CRA
a | 1.64 | 3.49 | 10 | 29.3 |
| P05026 | ATP1B1 | Sodium/potassium - transporting ATPase
subunit β-1 | 1.45 | 2.59 | 8 | 28.0 |
| O75054 | IGSF3 | Immunoglobulin superfamily member
3 | 1.19 | 3.31 | 34 | 31.4 |
| P14384 | CPM | Carboxypeptidase M | 1.12 | 4.00 | 12 | 28.6 |
| Q969P0-3 | IGSF8 | Isoform 3 of
immunoglobulin superfamily member 8 | 1.09 | | 12 | 28.3 |
| P01860 | IGHG3 | Immunoglobulin heavy constant gamma 3
(G3m marker) | 0.97 | 12.13 | 9 | 21.4 |
| B4DDZ4 | ANXA4 | Annexin | 0.92 | | 8 | 27.0 |
| H0YIC4 | CS | Citrate
synthase | 0.91 | | 3 | 24.3 |
| B2R6C4 | REEP5 | Receptor expression-enhancing
protein | 0.8 | 2.35 | 3 | 10.2 |
| O43570-2 | CA12 | Isoform 2 of
carbonic anhydrase 12 | 0.78 | | 7 | 24.1 |
| Q5U0H8 | MPZL1 | Myelin protein
zero-like 1 | 0.71 | | 4 | 15.9 |
| Q2TTR7 | EGFR | Receptor
protein-tyrosine kinase | 0.67 | | 23 | 21.3 |
|
A0A0A8LFF7 |
HLA-E | MHC class I
antigen |
0.66 | |
2 |
20.1 |
| Q5T2L0 | VTCN1 | V-set
domain-containing T-cell activation inhibitor 1 | 0.65 | | 5 | 18.5 |
| Q6N093 | IGHG2 | Putative
uncharacterized protein DKFZp686104196 | 0.61 | | 7 | 20.1 |
| Q16625-3 | OCLN | Isoform 3 of
occludin | 0.6 | | 9 | 25.0 |
| Q8WTV0-2 | SCARB1 | Isoform 1 of scavenger receptor class
B member 1 | 0.56 | 4.67 | 7 | 15.9 |
| A4D1S0 | KLRG2 | Killer cell
lectin-like receptor subfamily G member 2 | 0.55 | | 6 | 16.8 |
| P18084 | ITGB5 | Integrin β -5 | 0.53 | | 12 | 16.0 |
| Q14126 | DSG2 | Desmoglein-2 | 0.52 | 3.06 | 16 | 17.6 |
| P10586-2 | PTPRF | Isoform 2 of receptor-type
tyrosine-protein phosphatase F | 0.51 | 6.38 | 31 | 21.8 |
| Q7Z7H5-2 | TMED4 | Isoform 2 of
transmembrane emp24 domain-containing protein 4 | 0.47 | | 3 | 13.2 |
|
P05556 |
ITGB1 | Integrin
β-1 |
0.42 |
3.82 |
11 |
16.2 |
| Q7Z3Z9 | L1CAM | L1 cell adhesion
molecule | 0.41 | 3.42 | 19 | 16.4 |
| P17301 | ITGA2 | Integrin a-2 | 0.41 | 2.16 | 16 | 1 4. 9 |
| F8VY02 | ERP29 | Endoplasmic
reticulum resident protein 29 | 0.41 | | 2 | 10.6 |
| Q9UEI6 | PVRL2 | Polio virus-related
protein 2. a isoform | 0.39 | 2.79 | 5 | 12.9 |
| B4DW34 | SMPDL3B | cDNA FLJ56798,
highly similar to acid sphingomyelinase-like phosphodies terase
3b | 0.37 | 2.85 | 5 | 12.0 |
| B4DL19 | ANXA1 | Annexin | 0.37 | | 2 | 9.8 |
| A0A087WUV8 | BSG | Basigin | 0.35 | | 2 | 14.8 |
| A0A0A0MSA9 | PVR | Poliovirus receptor | 0.34 | 4.25 | 4 | 14.2 |
| O00622 | CYR61 | Protein CYR61 | 0.34 | | 4 | 10.8 |
|
O75144 |
ICOSLG | ICOS
ligand |
0.32 | |
3 |
10.2 |
| B4DU18 | CDH5 | cDNA FLJ51093, highly similar to
Cadherin-5 | 0.31 | 7.75 | 8 | 12.5 |
| F5GXJ9 | ALCAM | CD166 antigen | 0.31 | 6.20 | 6 | 13.3 |
| Q9BS26 | ERP44 | Endoplasmic reticulum resident protein
44 | 0.31 | 2.21 | 4 | 11.0 |
| Q969N2-5 | PIGT | Isoform 5 of GPI
transamidase component PIG-T | 0.31 | | 5 | 8.4 |
| B1AP13 | CD55 | Complement
decay-accelerating factor | 0.29 | | 4 | 7.4 |
| A8K6K4 | IL1RAP | cDNA FLJ77565,
highly similar to H. sapiens interleukin 1 receptor accessory
protein (IL1RAP) | 0.27 | | 5 | 7.5 |
| B1AMW1 | CD58 | CD58 antigen,
(lymphocyte function- associated antigen 3), isoform CRA_c | 0.26 | | 2 | 7.5 |
| Q9UNN8 | PROCR | Endothelial protein
C receptor | 0.26 | | 2 | 9.2 |
| B4DPN0 | APOH | cDNA FLJ51265,
moderately similar to β-2-glycoprotein 1 (β-2-glycoprotein I) | 0.22 | | 2 | 5.4 |
| G3V124 | TMPRSS4 | Transmembrane
protease serine 4 | 0.18 | | 2 | 8.9 |
| A0A024R798 | SLC44A2 | Choline transporter-like protein 2
isoform 2 | 0.17 | 4.25 | 6 | 7.8 |
| B3KP89 | GNAO1 | cDNA FLJ31446 fis,
highly similar to Guanine nucleotide-binding protein G(o) subunit α
1 | 0.17 | | 2 | 6.4 |
| Q5TG12 | PTPRK | Receptor-type
tyrosine-protein phosphatase ĸ | 0.17 | | 8 | 6.7 |
| D6RBJ7 | GC | Vitamin D-binding
protein | 0.17 | | 2 | 8.6 |
| B4DDE5 | SLC5A6 | cDNA FLJ56614,
highly similar to sodium-dependent multivitamin transporter | 0.15 | 2.14 | 3 | 10.0 |
| P06727 | APOA4 | Apolipoprotein
A-IV | 0.15 | | 2 | 4.5 |
| P36955 | SERPINF1 | Pigment
epithelium-derived factor | 0.15 | | 2 | 6.4 |
| Q13641 | TPBG | Trophoblast
glycoprotein | 0.15 | | 2 | 5.0 |
| B2RAF9 | ST14 | Suppressor of
tumorigenicity 14 protein homolog | 0.14 | | 4 | 5.6 |
| Q6EMK4 | VASN | Vasorin | 0.14 | | 4 | 7.8 |
| Q96GQ5 | C16orf58 | RUS1 family protein
C16orf58 | 0.13 | 2.17 | 2 | 7.6 |
| Q9Y5L3-2 | ENTPD2 | Isoform short of
ectonucleoside triphosphate diphosphohydrolase 2 | 0.13 | | 2 | 4.6 |
| Q53G72 | BCAP31 | B-cell
receptor-associated protein 31 variant | 0.12 | | 2 | 8.1 |
| B4DTS6 | CD97 | cDNA FLJ54117,
highly similar to CD97 antigen | 0.12 | | 3 | 5.1 |
| Q9Y490 | TLN1 | Talin-1 | 0.1 | 2.00 | 11 | 4.0 |
| A0A087WVP1 | FAT1 | Protocadherin Fat
1 | 0.1 | | 20 | 4.5 |
| S4R3V8 | LSR |
Lipolysis-stimulated lipoprotein
receptor | 0.1 | | 2 | 4.3 |
| P29317 | EPHA2 | Ephrin type-A
receptor 2 | 0.09 | | 3 | 2.4 |
| B4E0H8 | ITGA3 | cDNA FLJ60385,
highly similar to integrin α-3 | 0.09 | | 3 | 2.7 |
|
B4DTY8 |
ITGA1 | cDNA
FLJ61587, highly similar to Integrin α-1 |
0.08 | |
3 |
1.8 |
| Q5R3F8 | ELFN2 | Protein phosphatase
1 regulatory subunit 29 | 0.07 | | 2 | 1.7 |
| B2RBY8 | ENPP1 | FLJ95771, highly
similar to H. sapiens ectonucleotide
pyrophosphatase/phosphodiesterase 1 (ENPP1) | 0.07 | | 2 | 1.9 |
| P05543 | SERPINA7 | thyroxine-binding
globulin | 0.07 | | 2 | 4.0 |
| H0Y858 | n/a | Uncharacterized
protein IL1 r binding | 0.07 | | 3 | 4.3 |
| Q08345-5 | DDR1 | Isoform 4 of
epithelial discoidin domain-containing receptor 1 | 0.06 | | 3 | 3.1 |
| O14678 | ABCD4 | ATP-binding
cassette sub-family d member 4 | 0.05 | | 3 | 2.1 |
| Q9UIW2 | PLXNA1 | Plexin-A1 | 0.05 | | 3 | 1.5 |
| A0A024R9Q1 | THBS1 | Thrombospondin 1,
isoform CRA_a | 0.05 | | 2 | 1.9 |
| Q04721 | NOTCH2 | Neurogenic locus
notch homolog protein 2 | 0.04 | | 3 | 1.4 |
| B2R7F8 | PLG | Plasminogen | 0.04 | | 2 | 1.8 |
| Q9UHN6 | TMEM2 | Transmembrane
protein 2 | 0.04 | | 3 | 1.3 |
| Q9Y4D7 | PLXND1 | Plexin-D1 | 0.03 | | 2 | 0.9 |
In order to quantify the enrichment of proteins in
an anti-B7-H4-SPPLAT experiment compared with a negative control
IgG, a SILAC SPPLAT analysis was performed, where cells cultured in
isotope-labelled media were labelled via SPPLAT for 2 min to
minimise membrane diffusion and cytosolic labelling. In performing
two biological replicates and two reciprocal labellings, 938
proteins were identified, of which 452 were present in all four
datasets (Table SIII). By
performing control experiments with a non-specific IgG, a
significant proportion (~90%) of proteins that demonstrated minimal
expression changes between the anti-B7-H4 and non-specific IgG
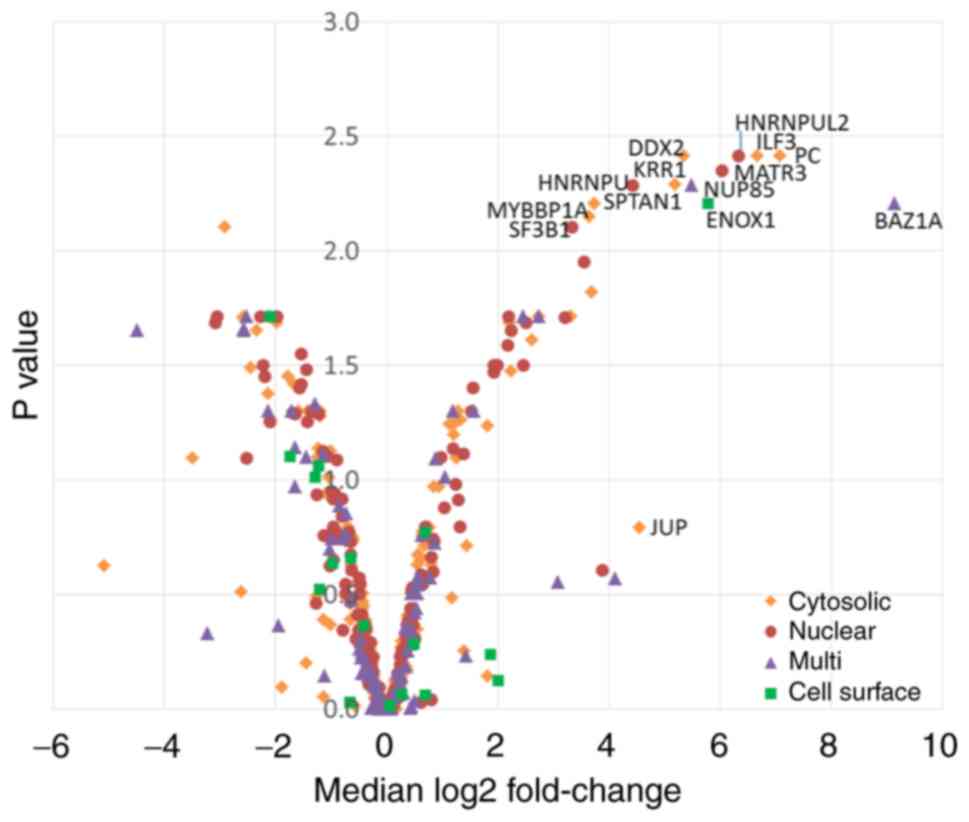
control were eliminated. Subsequently, the 13 proteins that
demonstrated a log2 fold-change of >2 and significance of
P>0.01 (log10 adjusted P-value of 2) between the test and
control experiments were analysed; in addition to plasma
membrane-localised proteins, there were some cytoplasmic and
nuclear proteins, despite a short labelling time (Fig. 2). One identified plasma membrane
protein, Enox1, an Ecto-NOX disulphidethiol exchanger, is involved
in electron transport to the cell surface and has not previously
been reported to be associated with B7-H family members. A total of
13 predicted binding partners of Enox1 were not found in our list,
notably Protein GREB1, which is considered to play a role in
oestrogen-stimulated cell proliferation by acting as a regulator of
hormone-dependent cancer growth in breast and prostate cancers. A
cytosolic candidate, interleukin enhancer-binding factor 3,
interacts with and inhibits viral mRNAs (24); therefore, it was not further
investigated. The remainder of increased fold changed proteins were
metabolic or nucleus-localised and were also not further
investigated. As B7-H4 was exclusively SPPLATed and not detected in
the SILAC SPPLAT, the present study focused on the proteins that
were present only in the B7-H4 labellings and absent in controls
with relevant GO annotation, good peptide coverage and abundance,
as indicated by EmPAI scoring or peak intensity from MaxQuant
analysis (listed in Table I). Of
particular interest were several integrins, HLA-E and plexins.
These were further investigated using PLA, CoIP and binding
studies.
Validation of B7-H4 proximal proteins by
PLA
The SPPLAT method can biotinylate proteins within a
100-nm footprint of the target HRP-coupled antibody-ligand
interaction. PLA can validate our identified proteins and further
positions within 40 nm. PLA was performed on integrin α1, HLA-E and
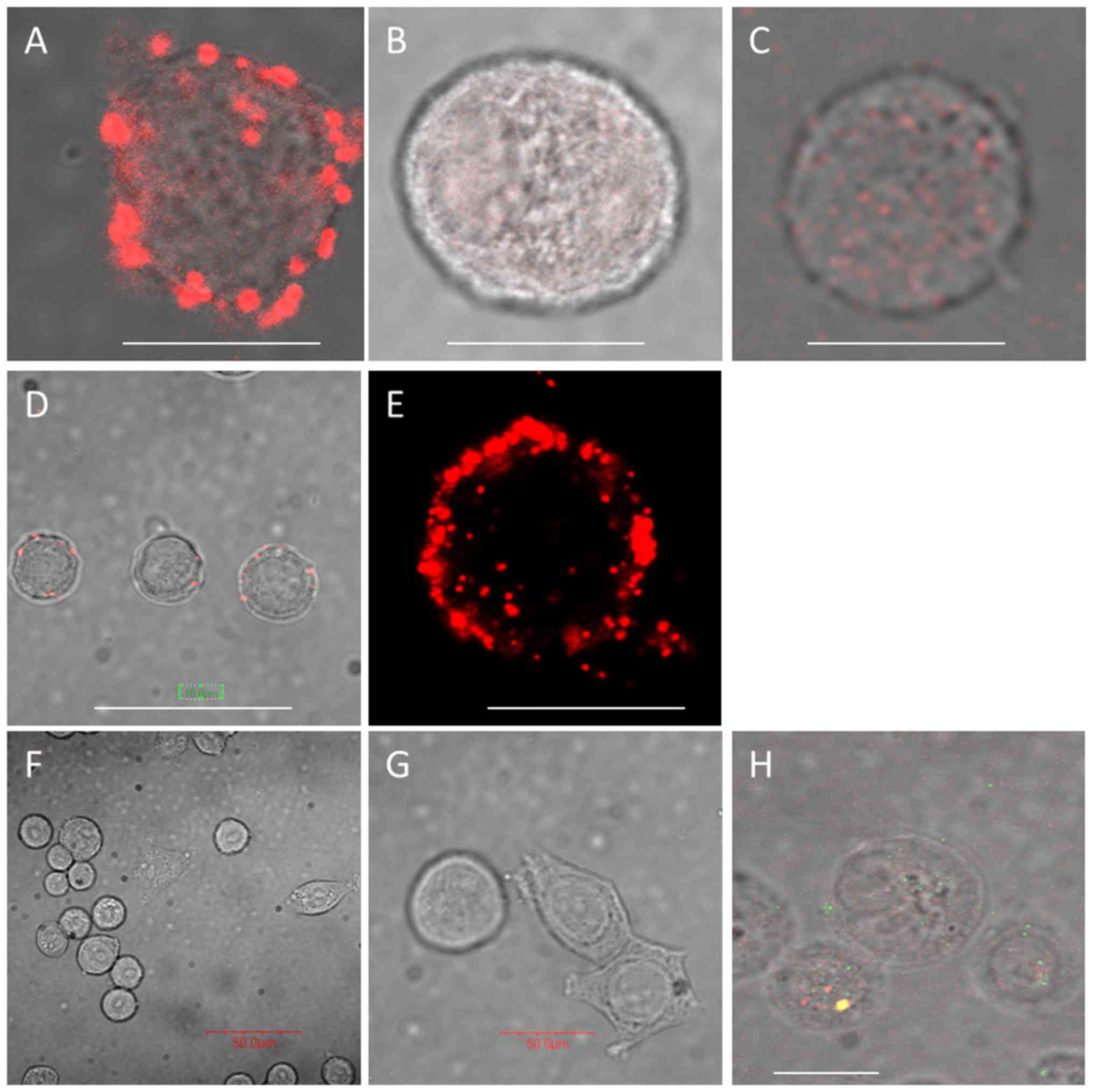
plexin B2. As shown in Fig. 3,
when SK-BR-3 cells are pre-incubated with B7-H4 antibody, this
induces B7-H4 clustering at the cell surface, and both integrin α1
and HLA-E co-localise with B7-H4 forming distinct clusters compared
to when no B7-H4 antibody incubation occurs (Fig. 3A-E). Plexin B2 was not found to
co-localise with B7-H4 at the cell surface (Fig. 3F and G); however, standard
immunofluores-cence demonstrated co-localisation just beneath the
plasma membrane (Fig. 3H).
Functional studies show that integrin α1
associates with B7-H4
Several integrin subunits (α1, α2, α3, αV, β1 and
β5) were identified in the B7-H4 SPPLAT AP-MS screen. Integrins
function as αβ heterodimers in various combinations, and several,
such as αV and β5, have been implicated in tumour growth, so were
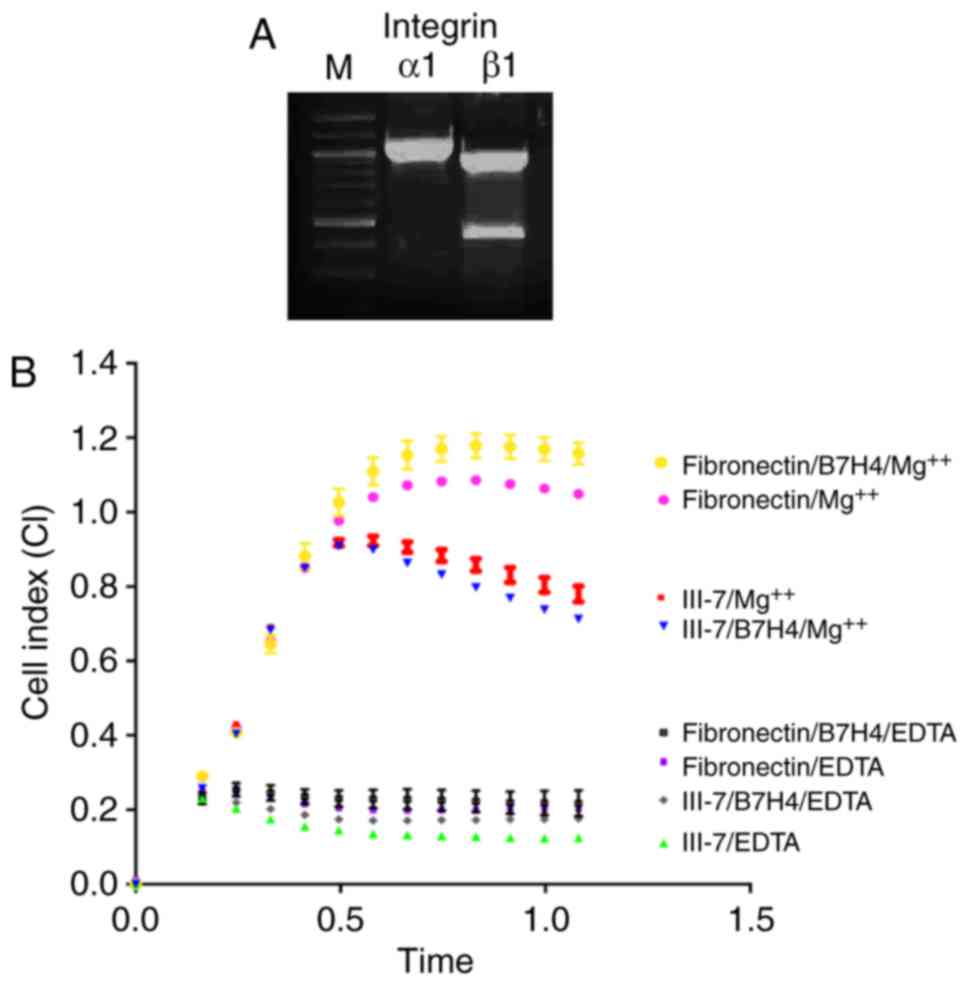
used as positive controls in validation experiments. Integrin α1
operates in partnership with β1 and β5. The PLA experiment
demonstrated that integrin α1 was indeed within 40 nm of B7-H4 on
the plasma membrane (Fig. 3A) and,
in parallel, it was confirmed by RT-PCR that α1 and β1 are
expressed in SK-BR-3 cells (Fig.
4A). To assess whether α1 and β1 interact with B7-H4 on the
cell surface, integrin real-time binding assays were performed with
known substrates of integrin heterodimers α1β1 (collagen receptor)
and α5β1 (fibronectin receptor) in the presence and absence of
anti-B7-H4 (blocking) antibody. The presence of B7-H4 in close
proximity to integrins α1 and β1 could modulate integrin-substrate
binding and, furthermore, the presence of a blocking antibody for
B7-H4 may alter binding. Indeed, the presence of anti-B7-H4 reduced
binding of SK-BR-3 cells, via α1, to collagen substrates at 0.5 h
(Fig. 4B), whereas the cell
binding response to fibronectin was significantly higher
(P<0.0001) in the presence of an anti-B7-H4.
To ensure that integrins were not being
non-specifically activated due to the PLA being performed at 37°C,
antibody incubation at 4°C was also performed prior to fixation; no
differences in the expression or internalisation were observed
(Fig. S2A-D). To assess whether
integrin α1 is a direct binding partner of B7-H4, co-IP was
performed in SK-BR-3 cells using an antibody to B7-H4 to pull down
B7-H4-bound proteins, and subsequent detection of integrin α1 by
western blotting (Fig. S2E). The
anti-B7-H4 pulled down integrins, whilst a control anti-B7-H4 Fab
alone did not pull down B7-H4 with integrins α1 and β1, thus
confirming that integrin α1 is in a complex with B7-H4 (Fig. S2E). Thus, multiple lines of
evidence show that integrin α1 is found in a complex with B7-H4 on
the cell surface.
HLA-E was another interesting find, as its role is
typically in negative regulation of the immune system. Its close
proximity to B7-H4 confirmed by SPPLAT and PLA (Fig. 3D and E) was further validated by
co-IP and found to be in complex with B7-H4 at the cell surface
(Fig. S2F).
Only a limited number of SPPLAT hits that had both
high and low scoring EMPAI values were investigated. There were
also several Ig superfamily members and antigens, such as
CD58/LFA-3, ALCAM and L1CAM, EGFR and ST14 in the B7-H4
microenvironment that may play a role that would warrant further
validation.
Discussion
The role of B7-H4 in tumorigenesis is unclear. In
order to gain an insight into the function of B7-H4, the human
breast cancer cell-line SK-BR-3 was used to identify proteins
closely located in the cell membrane. SPPLAT is a useful tool for
surveying protein environments on the cell surface, especially low
abundance proteins such as the B7 family. The specific targeting of
membrane proteins by SPPLAT, using antibodies in this case,
facilitates their identification and characterisation. However,
understanding highly abundant proteins is crucial for carefully
designing such targeted experiments in order to minimise false
positives especially as upon cell lysis, all proteins mix and may
vary in concentration of 100 orders of magnitude. These include
cytosolic carboxylases, which bind streptavidin affinity resins in
our downstream purification steps. Therefore, a partial SK-BR-3
proteome was obtained, which enables estimation of the abundance of
common proteins that may also reside in the footprint of the biotin
label and thus specifically bind affinity resins. These may often
dominate low-abundance cell surface proteins that are more
difficult to extract and characterise, which are the focus of the
present study. Thus far, few proteomic analyses of SK-BR-3 cells
have been reported comparing proteins to a normal human mammary
epithelial cell line (25), both
of which used 2D-DIGE, which is inefficient at resolving membrane
proteins. Thus, a more comprehensive list of the more abundant
proteins was also presented in the present study. The aim was not
to characterise the entire proteome, but rather to be aware of the
top 1% of abundant proteins, as these may bind non-specifically to
proteins that have been labelled for identifying surface
microenvironments. No over- or under-representation of Protein
Groups was observed, and abundant proteins were mostly structural
proteins.
B7-H4 has previously been reported to interact with
only a handful of proteins, mainly involved in adaptive immunity.
Using SPPLAT, another family member of B7-H4, B7-H2 (ICOSL), and a
number of novel interacting and proximal proteins within a 100-nm
radius on the surface of SK-BR-3 cells were identified. The only
experimentally reported interactor of B7-H4 is a trans
interactor, BTLA, a B- and T-lymphocyte attenuator, a lymphocyte
inhibitory receptor that inhibits lymphocytes during immune
response and unlikely to reside on APCs. Our SPPLAT method did not
identify any of the three reported B7-H4 co-expressing interacting
proteins described by STRING, namely IL6 (an activator of B7-H4),
CD80 or CTLA4 found on T-cells. Other putative interactors
predicted by literature text mining, such as PDCD1LG2, a programmed
cell death 1 ligand 2, involved in the costimulatory signal and
essential for T-cell proliferation and IFNG production in a
PDCD1-independent manner, IL4, CD28 and CD86, were not identified.
These were all low-scoring STRING predictions. However, other
programmed cell death proteins, PDCD5 and PDCD6, were identified in
its interacting protein PDCD6IP. By contrast, a whole new family of
proteins were identified in the B7-H4 microenvironment, namely
integrins. These candidates were further narrowed down to within 40
nm. and co-IP was used to confirm direct interaction. Previous
reports have suggested roles for integrin subunits in tumour
development. The present study investigated α1β1, as there has been
no previous report of this heterodimer being associated with B7-H4.
Its substrate is collagen, and its function is to mediate collagen
synthesis. It has been reported than integrin α1β1 mediates a
unique collagen-dependent proliferation pathway in vivo in
pancreatic cancer cells (26), and
this may also be the case in breast cancer cells. Our confirmation
of the presence of integrin α1β1 by RT-PCR, immunofluorescence and
proximity to B7-H4 by PLA and direct binding by xCEL-Ligence cell
binding assays and co-IP suggest that these two integrin subunits
function as a α1β1 heterodimer and mediate a collagen-dependent
proliferation pathway in vivo in breast cancer cells akin to
pancreatic cells mentioned previously.
Several integrin subunits were found in our SPPLAT
screen, a number of which form functional heterodimers: α1β1, α2β1,
α3β1, α5β1, αVβ1 and α1β5. Integrins are trans-membrane adhesion
receptors that provide the physical link between the actin
cytoskeleton and the extracellular matrix. It has been well
established that integrins play a major role in various cancer
stages, such as tumour growth, progression, invasion, metastasis
and angiogenesis. In breast cancer, integrin αV-β3 has been
associated with high malignant potential in cancer cells,
signalling the onset of widespread metastasis. Expression of αV
integrins and identification of the vitronectin receptor have been
reported in breast cancer cells (27). Other reports have confirmed α5β1
integrin as a pertinent therapeutic target (28). Integrins α3 and α2, both
independently dimerising with β1, were also identified in the
SPPLAT experiment along with their interacting partners, basigin
and talin, confirming the presence of multiple integrin subunits
and macromolecular complexes at the cell surface. Whilst reports
suggest overexpression of combinations of different integrin
subunits in a variety of cancers, it was not identified as an
abundant family of proteins from SK-BR-3 lysates in our partial
proteome study.
The present study also identified other interacting
proteins, including HLA-E, a key regulator of both the innate and
adaptive immune response through positive regulation of several
interleukins, such as IL4 and IL13, and interferon signalling
(29). In addition, the present
study identified several plexins, some of which have been
implicated in cancer. One of these, PLXNA4 in complex with SemA3
and neuropilin-1 has been reported to be the receptor of B7-H4 on
regulatory CD4 T-cells (30).
Another plexin identified as being in close proximity to B7-H4 on
the cell surface, PLXNB2, is required for the physiological and
pathological functions of angiogenin (ANG) and has significant
therapeutic potential in solid and hematopoietic cancers and
neurodegenerative diseases (31).
In addition, quantitative SILAC demonstrated increased expression
of Enox1 that has previously been implicated in immune regulation:
As a candidate gene for the autoimmune disease myasthenia gravis
(32), and as a biomarker of
response to an anti-IL6 mAb in rheumatoid arthritis (33).
It appears the B7-H4 surface microenvironment in
SK-BR-3 cells is complex upon binding anti-B7-H4, whereby an
assortment of proteins are recruited to the cell surface upon
induced clustering by the HRP-labelled antibody. The data were
screened using the STRING programme, which compares our data with
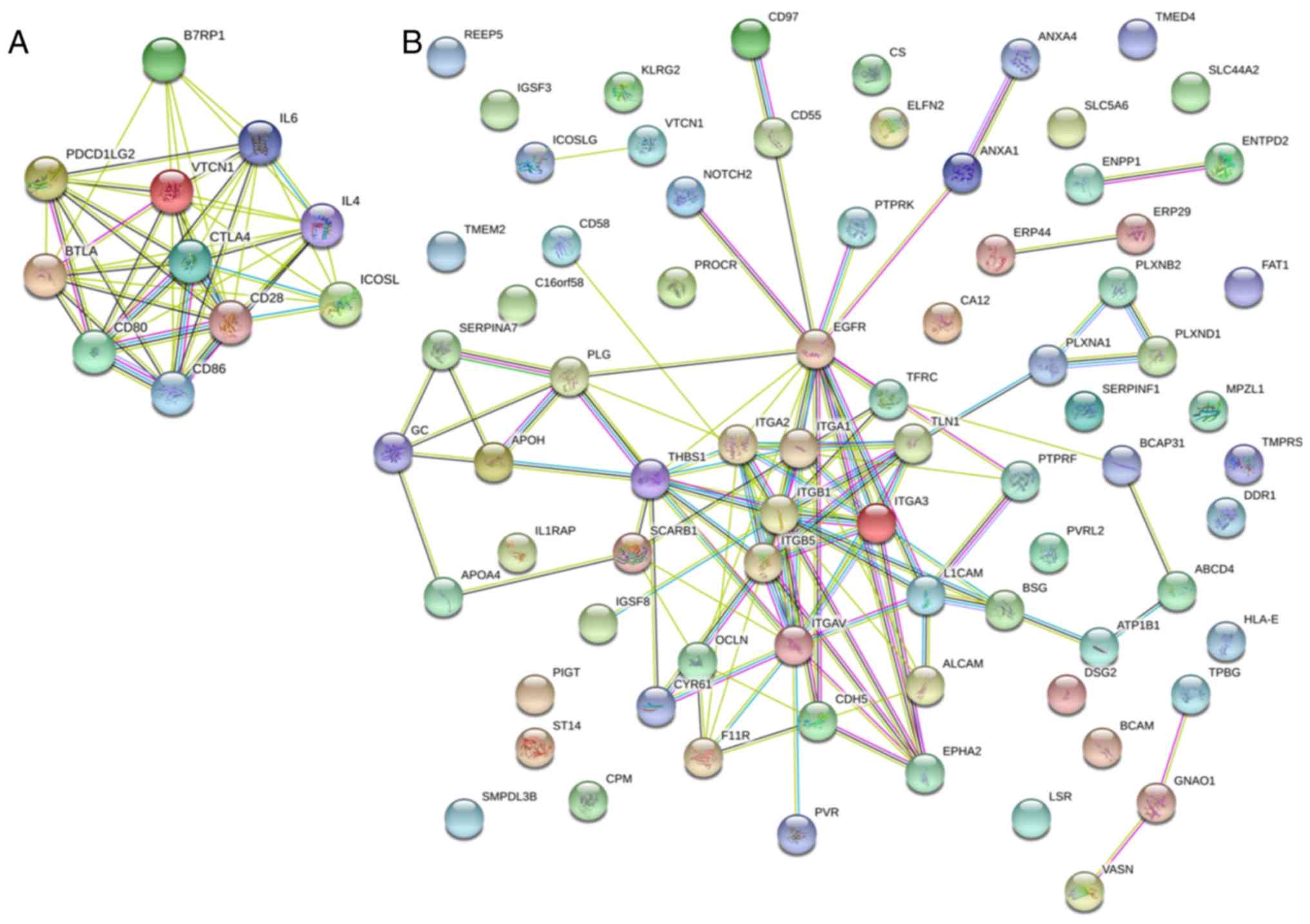
experimental and computationally predicted interactors (Fig. 5A and B) and a model was proposed
based on known and newly identified associate proteins (Fig. 5C). Little overlap of the predicted
(Fig. 5A) and our findings
(Fig. 5B) was observed. Clearly,
integrins are highly connected and, the plexin cluster is
associated with the integrins via talin. In fact, 54% of the
proteins were connected, suggesting a complex process in
signalling. HLA-E appears to be unconnected along with the B7
family, so this is a novel finding.
SPPLAT is a useful tool for identifying membrane
interacting proteins. This technique may work in several ways.
First, one can identify proteins proximal to a selected target by
using an antibody, drug or toxin coupled to HRP that can aid
delivery of biotin-tyramide to a region of ~100 nm. Second, one can
identify new receptors or ligands on the cell surface by similarly
HRP-labelling a selected ligand or receptor and SPPLATing the cell
surface to identify the unknown receptor or ligand,
respectively.
Supplementary Data
Funding
JSR and APJ were funded by BBSRC H024085/1 and
AstraZeneca. LC and SWH were funded by the Department of
Biochemistry, University of Cambridge.
Availability of data and material
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JSR performed the majority of the experiments. LCCC
performed the experiments presented in Fig. 1. SWH performed the reverse
transcription-PCR and xCELLigence assay. GD performed flow
cytometry experiments. JSR and APJ wrote the manuscript with
helpful comments from AS, GD and NT. NT, AS, GD, JSR, KSL and APJ
conceived and designed the project and helped analyse and interpret
the results.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
JSR and APJ were funded by AstraZeneca; GD, AS and
NT are employees of AstraZeneca.
Acknowledgments
We would like to thank Richard Farndale for his
helpful discussions.
References
|
1
|
Huppa JB and Davis MM: The
interdisciplinary science of T-cell recognition. Adv Immunol.
119:1–50. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schildberg FA, Klein SR, Freeman GJ and
Sharpe AH: Coinhibitory pathways in the B7-CD28 ligand-receptor
family. Immunity. 44:955–972. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen L and Flies DB: Molecular mechanisms
of T cell co- stimulation and co-inhibition. Nat Rev Immunol.
13:227–242. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Keir ME, Butte MJ, Freeman GJ and Sharpe
AH: PD-1 and its ligands in tolerance and immunity. Annu Rev
Immunol. 26:677–704. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Salomon B and Bluestone JA: Complexities
of CD28/B7: CTLA-4 costimulatory pathways in autoimmunity and
transplantation. Annu Rev Immunol. 19:225–252. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sica GL, Choi IH, Zhu G, Tamada K, Wang
SD, Tamura H, Chapoval AI, Flies DB, Bajorath J and Chen L: B7-H4 a
molecule of the B7 family, negatively regulates T cell immunity.
Immunity. 18:849–861. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prasad DV, Richards S, Mai XM and Dong C:
B7S1, a novel B7 family member that negatively regulates T cell
activation. Immunity. 18:863–873. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Salceda S, Tang T, Kmet M, Munteanu A,
Ghosh M, Macina R, Liu W, Pilkington G and Papkoff J: The
immunomodulatory protein B7-H4 is overexpressed in breast and
ovarian cancers and promotes epithelial cell transformation. Exp
Cell Res. 306:128–141. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Afreen S and Dermime S: The
immunoinhibitory B7-H1 molecule as a potential target in cancer:
Killing many birds with one stone. Hematol Oncol Stem Cell Ther.
7:1–17. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tringler B, Zhuo S, Pilkington G, Torkko
KC, Singh M, Lucia MS, Heinz DE, Papkoff J and Shroyer KR: B7-h4 is
highly expressed in ductal and lobular breast cancer. Clin Cancer
Res. 11:1842–1848. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie N, Cai JB, Zhang L, Zhang PF, Shen YH,
Yang X, Lu JC, Gao DM, Kang Q, Liu LX, et al: Upregulation of B7-H4
promotes tumor progression of intrahepatic cholangiocarcinoma. Cell
Death Dis. 8:32052017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abadi YM, Jeon H, Ohaegbulam KC,
Scandiuzzi L, Ghosh K, Hofmeyer KA, Lee JS, Ray A, Gravekamp C and
Zang X: Host b7x promotes pulmonary metastasis of breast cancer. J
Immunol. 190:3806–3814. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang H, Li C and Ren G: Clinical
significance of the B7-H4 as a novel prognostic marker in breast
cancer. Gene. 623:24–28. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Franchina DG, He F and Brenner D: Survival
of the fittest: Cancer challenges T cell metabolism. Cancer Lett.
412:216–223. 2018. View Article : Google Scholar
|
|
15
|
Ni L and Dong C: New B7 family checkpoints
in human cancers. Mol Cancer Ther. 16:1203–1211. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pardoll DM: The blockade of immune
checkpoints in cancer immunotherapy. Nat Rev Cancer. 12:252–264.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Podojil JR and Miller SD: Potential
targeting of B7-H4 for the treatment of cancer. Immunol Rev.
276:40–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vogt AB, Spindeldreher S and Kropshofer H:
Clustering of MHC-peptide complexes prior to their engagement in
the immunological synapse: Lipid raft and tetraspan microdomains.
Immunol Rev. 189:136–151. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rees JS, Li XW, Perrett S, Lilley KS and
Jackson AP: Protein neighbors and proximity proteomics. Mol Cell
Proteomics. 14:2848–2856. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li XW, Rees JS, Xue P, Zhang H, Hamaia SW,
Sanderson B, Funk PE, Farndale RW, Lilley KS, Perrett S and Jackson
AP: New insights into the DT40 B cell receptor cluster using a
proteomic proximity labeling assay. J Biol Chem. 289:14434–14447.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rees JS, Li XW, Perrett S, Lilley KS and
Jackson AP: Selective proteomic proximity labeling assay using
tyramide (SPPLAT): A quantitative method for the proteomic analysis
of localized membrane-bound protein clusters. Curr Protoc Protein
Sci. 88:19.27.1–19.27.18. 2017. View Article : Google Scholar
|
|
22
|
Neve RM, Chin K, Fridlyand J, Yeh J,
Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al: A
collection of breast cancer cell lines for the study of
functionally distinct cancer subtypes. Cancer Cell. 10:515–527.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hamaia SW, Pugh N, Raynal N, Némoz B,
Stone R, Gullberg D, Bihan D and Farndale RW: Mapping of potent and
specific binding motifs, GLOGEN and GVOGEA, for integrin
alpha-1beta1 using collagen toolkits II and III. J Biol Chem.
287:26019–26028. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin
QF, Wei J, Yao RW, Yang L and Chen LL: Coordinated circRNA
biogenesis and function with NF90/NF110 in viral infection. Mol
Cell. 67:214–227.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ziegler YS, Moresco JJ, Tu PG, Yates JR
III and Nardulli AM: Plasma membrane proteomics of human breast
cancer cell lines identifies potential targets for breast cancer
diagnosis and treatment. PLoS One. 9:e1023412014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pozzi A, Wary KK, Giancotti FG and Gardner
HA: Integrin alpha1beta1 mediates a unique collagen-dependent
proliferation pathway in vivo. J Cell Biol. 142:587–594. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Meyer T, Marshall JF and Hart IR:
Expression of alphav integrins and vitronectin receptor identity in
breast cancer cells. Br J Cancer. 77:530–536. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schaffner F, Ray AM and Dontenwill M:
Integrin alpha5beta1, the fibronectin receptor, as a pertinent
therapeutic target in solid tumors. Cancers (Basel). 5:27–47. 2013.
View Article : Google Scholar
|
|
29
|
Rieger L, Hofmeister V, Probe C, Dietl J,
Weiss EH, Steck T and Kämmerer U: Th1- and Th2-like cytokine
production by first trimester decidual large granular lymphocytes
is influenced by HLA-G and HLA-E. Mol Hum Reprod. 8:255–261. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Podojil JR, Chiang MY, Ifergan I, Copeland
R, Liu LN, Maloveste S, Langermann S, Liebenson D, Balabanov R, Chi
H, et al: B7-H4 modulates regulatory CD4+ T cell
induction and function via ligation of a semaphorin 3a/Plexin
A4/Neuropilin-1 complex. J Immunol. 201:897–907. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu W, Goncalves KA, Li S, Kishikawa H, Sun
G, Yang H, Vanli N, Wu Y, Jiang Y, Hu MG, et al: Plexin-B2 mediates
physiologic and pathologic functions of angiogenin. Cell.
171:849–864.e25. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Landouré G, Knight MA, Stanescu H, Taye
AA, Shi Y, Diallo O, Johnson JO, Hernandez D, Traynor BJ, Biesecker
LG, et al: A candidate gene for autoimmune myasthenia gravis.
Neurology. 79:342–347. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maldonado-Montoro M, Cañadas-Garre M,
González-Utrilla A, Plaza-Plaza JC and Calleja-Hernández MŸ:
Genetic and clinical biomarkers of tocilizumab response in patients
with rheumatoid arthritis. Pharmacol Res. 111:264–271. 2016.
View Article : Google Scholar : PubMed/NCBI
|