Introduction
At present, ~1/3 of esophageal cancer cases are
diagnosed at an advanced stage, and are no longer eligible for
surgical resection (1).
Chemotherapy is the main treatment for advanced esophageal cancer,
and cisplatin-based chemotherapy is considered to be a first-line
therapy (2). However, the majority
of patients will benefit from cisplatin treatment for the initial
4-6 chemotherapy cycles, then subsequently develop cisplatin
resistance (3,4). Cisplatin resistance is the main
reason for treatment failure and mortality in patients with
esophageal cancer (5).
Circular RNAs (circRNAs) are a novel class of
endogenous non-coding RNAs characterized by a covalently closed
loop without 5' caps and 3' tails (6). circRNAs are conserved and stable, and
may therefore serve as suitable markers for disease diagnosis and
treatment; previous studies have shown that circRNAs serve
important roles in various tumors, and the abnormal expression of
circRNAs may be involved in the occurrence and development of
cancer (7,8). Furthermore, circRNAs may act as
microRNA (miRNA/miR) sponges by competitively binding to miRNA
response elements to influence downstream target gene expression,
as well as affecting gene function at a post-translational level
(9).
The present study aimed to investigate the role of
circRNAs in cisplatin-resistant esophageal cancer. A circRNA chip
assay revealed that circRNA_001275 was significantly upregulated in
cisplatin-resistant esophageal cancer. Furthermore, circRNA_001275
was found to serve as an miR-370-3p sponge, upregulating Wnt family
member 7A (Wnt7a) expression and consequently promoting cisplatin
resistance. To the best of our knowledge, the present study was the
first to demonstrate the role of the
circRNA_001275/miR-370-3p/Wnt7a axis in cisplatin-resistant
esophageal cancer.
Materials and methods
Cell lines
The parental cisplatin-sensitive cells lines KYSE30
and ECA109 and the corresponding cisplatin-resistant cell lines
KYSE30/DDP and ECA109/DDP were purchased from The Cell Bank of Type
Culture Collection of the Chinese Academy of Sciences. The cells
after resuscitation were cultured in high-glucose DMEM containing
10% FBS (both Sigma-Aldrich; Merck KGaA) and incubated at 37°C with
5% CO2.
Clinical specimens
Three pairs of pathologically confirmed
cisplatin-resistant and adjacent cisplatin-sensitive tissue were
collected from patients with esophageal cancer at the Department of
Thoracic Surgery of The First Affiliated Hospital of China Medical
University between May 2017 to March 2018. The clinicopathological
characteristics of the three patients are presented in Table SI. The present study was approved
by the Ethics Committee of The First Affiliated Hospital of China
Medical University and performed in accordance with the Declaration
of Helsinki (10). Informed
consent was obtained from all patients.
circRNA chip detection
The circRNA expression profiles of the three pairs
of cisplatin-resistant and corresponding adjacent tissues were
evaluated using a circRNA chip assay (ArrayStar Co., Ltd.). The
assay was performed and analyzed by Shanghai GeneChem Co., Ltd.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and reversed
transcribed into cDNA using a riboSCRIPT™ reverse transcription kit
(Guangzhou RiboBio Co., Ltd.). RT was conducted at 42°C for 60 min
and 70°C for 10 min. The qPCR primers were designed using Primer
Premier software (version 5; Premier Biosoft International) and
synthesized by Shanghai GeneChem Co., Ltd. The following primers
were used: circRNA_001275 forward, 5'-TCT TCT TCT CCA CTC CTG AA-3'
and reverse, 5'-GAG CAA GGG CCC TAG CTC AA-3'; miR-370-3p forward,
5'-TGT AAC CAG AGA GCG GGA TGT-3' and reverse, 5'-TTT TGG CAT AAC
TAA GGC CGA A-3'; Wnt7a forward, 5'-CTC CGG ATC GGT GGC TAT G-3'
and reverse, 5'-CCC ATT TGT GAG CCT TCT CCT-3'; U6 forward, 5'-CTC
GCT TCG GCA GCA CA-3' and reverse 5'-AAC GCT TCA CGA ATT TGC GT-3';
and GAPDH forward, 5'-CGT CAC CAA CTG GGA CGA CA-3' and reverse,
5'-CTT CTC GCG GTT GGC CTT GG-3'. The qPCR thermocycling conditions
were as follows: 94°C for 15 sec, then 55°C for 30 sec and 70°C for
30 sec for 40 cycles. mRNA levels were quantified using the
2-Δ∆Cq method (11);
circRNA_001275 and Wnt7a were normalized to GAPDH, whereas
miR-370-3p was normalized to U6.
Cell transfection
The circRNA_001275 overexpression (OE) vector
(circRNA_001275 OE), pcDNA3.1 empty vector, small interfering
(si)RNA targeting circRNA_001275 (si-circRNA_001275), si-negative
control (NC), miR-370-3p mimic, miR-370-3p inhibitor and miR-370-3p
NC labeled with green fluorescent protein were designed and
synthesized by Shanghai GeneChem Co., Ltd. Sequences were as
follows: si-circRNA_001275, 5'-GTT GAA GGG GGA GCT CCT GTC ATA AAA
GCC AA-3'; si-NC, 5'-GAG CCC CAG CCT TCT CCA TG-3'. miR-370-3p
mimic, 5'-GCC TGC TGG GGT GGA ACC TGG T-3'; miR-370-3p inhibitor,
5'-CAG GUC ACG UCU CUG CAG UUA C-3'; and miR-370-3p NC, 5-GCC TGC
TGG GGT GGA ACC TCC T-3'. Cells (1x105/well) were
transfected with 50 nM RNA or plasmid using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). At 48 h later, the transfection efficiency was
assessed by RT-qPCR. The circRNA_001275 OE vector or pcDNA3.1 empty
vector were transfected into KYSE30/DDP cells, whereas
si-circRNA_001275 or si-NC were transfected into ECA109/DDP
cells.
MTT assay
Transfected cells in the logarithmic growth phase
were digested with trypsin at room temperature for 5 sec and seeded
into 96-well plates at a density of 1x104 cells/well. A
total of 20 µl MTT solution (2.5 mg/ml) was added to the cells at
0, 12, 24, 48 and 72 h. The cells were subsequently incubated for
an additional 4 h at 37°C with 5% CO2. The purple
formazan crystals were dissolved by the addition of 100 µl/well
DMSO (Guangzhou RiboBio Co., Ltd.) and the absorbance was measured
at a wavelength of 490 nm.
Hoechst 33258 staining
Transfected cells in the logarithmic growth phase
were digested with trypsin and seeded into 6-well plates at a
density of 5x105 cells/well. Following incubation for 24
h at 37°C, the cells were fixed with 3% paraformaldehyde for 5 min
at 37°C and washed twice with PBS. The cells were stained with 10
µl Hoechst 33258 for 15 min in the dark at 37°C and subsequently
sealed with neutral balsam. Apoptotic cells in five randomly
selected fields imaged under a fluorescence microscope
(magnification, x400).
Luciferase assay
The 3'-untranslated region (3'-UTR) of
circRNA_001275 and Wnt7a were cloned into the XbaI site of
the pGL3 control vector (Promega Corporation). Cells were
co-transfected with the miR-370-3p mimic or miR-370-3p NC along
with wild-type (wt) or mutated (mut) circRNA_001275-3'-UTR or
Wnt7a-3'-UTR using Lipofectamine 2000 at a final concentration of
50 nM for each RNA/plasmid. Firefly and Renilla luciferase
activity was detected using a dual-luciferase reporter assay system
(Promega Corporation). Firefly luciferase activity was normalized
to Renilla luciferase activity (Promega Corporation). The
luciferase assay was performed by Shanghai GeneChem Co., Ltd.
Prediction of target genes
Target genes were predicted using miRNA target gene
prediction databases. miRanda v5 (http://www.microrna.org/microrna/home.do), TargetScan
v7.1 (http://www.targetscan.org) and miRBase
(http://www.mirbase.org/). Genes which appeared in
>2 databases simultaneously were predicted to be target
genes.
Western blotting
Transfected cells in the logarithmic growth phase
were digested, and total protein was collected using RIPA buffer
(Beijing Solarbio Science &Technology Co., Ltd.) with 1 mM
phenylmethylsulfonyl fluoride and subsequently quantified using a
bicinchoninic acid protein assay kit. Total protein (50 µg/lane)
was separated via 10% SDS-PAGE and then transferred onto PVDF
membranes. The membranes were blocked for 1 h in blocking buffer
with 5% non-fat milk at room temperature. Subsequently, the PVDF
membranes were incubated with primary antibodies against Wnt7a
(1:1,000; cat. no. sc-365665; Santa Cruz Biotechnology, Inc.) and
b-actin (1:1,000; cat. no. sc-69879; Santa Cruz Biotechnology,
Inc.) overnight at 4°C. Following primary antibody incubation, the
membranes were incubated with horseradish peroxidase-conjugated
secondary antibody (1:1,500; cat. no. sc-516102; Santa Cruz
Biotechnology, Inc.). The membranes were washed twice with PBS and
protein bands were visualized using a ECL-PLUS kit (GE Healthcare
Life Sciences). Densitometry analysis was performed using ImageJ
software v1.5 (National Institutes of Health) with β-actin as the
loading control.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 17; SPSS, Inc.). Data are presented as the mean ±
SD of three experimental repeats. Student's t-test was used to
analyze the differences between two groups. One-way ANOVA followed
by Tukey's multiple comparison test was used to analyze differences
among three groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
circRNA expression profiles
circRNA expression profiles were evaluated using a
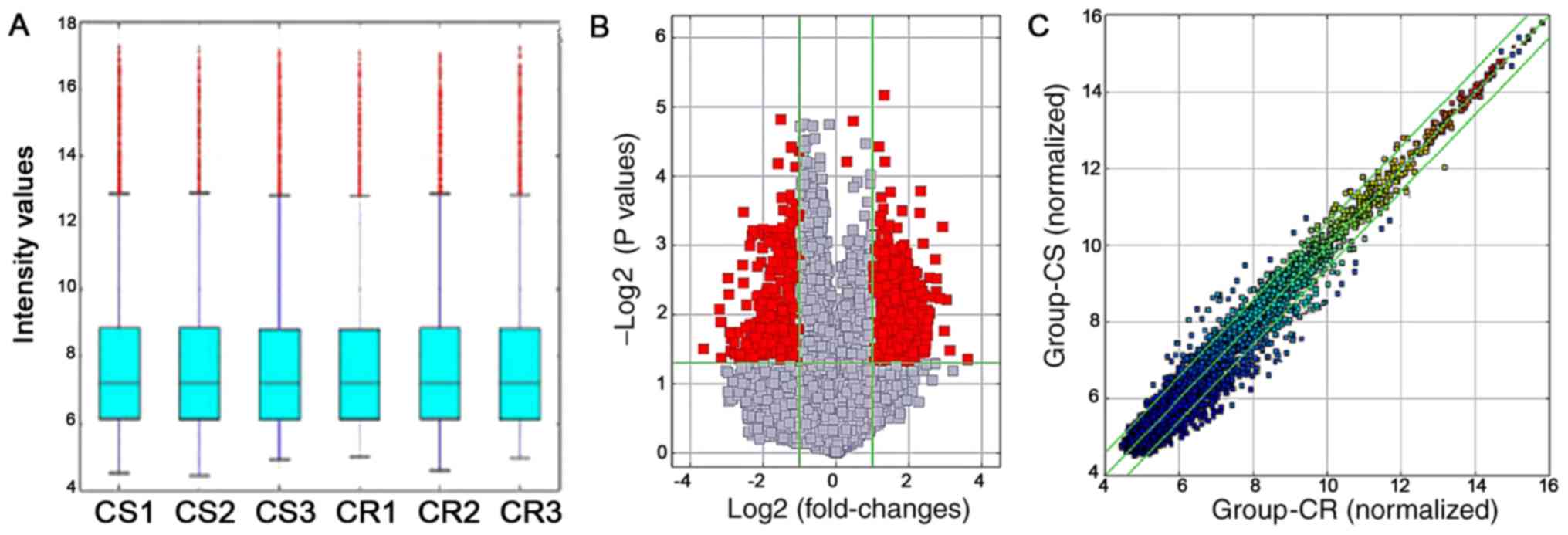
circRNA chip assay. Polyacrylamide gel electrophoresis revealed
that the total RNA obtained from the three pairs of tissues was of
high purity and undegraded (Fig.
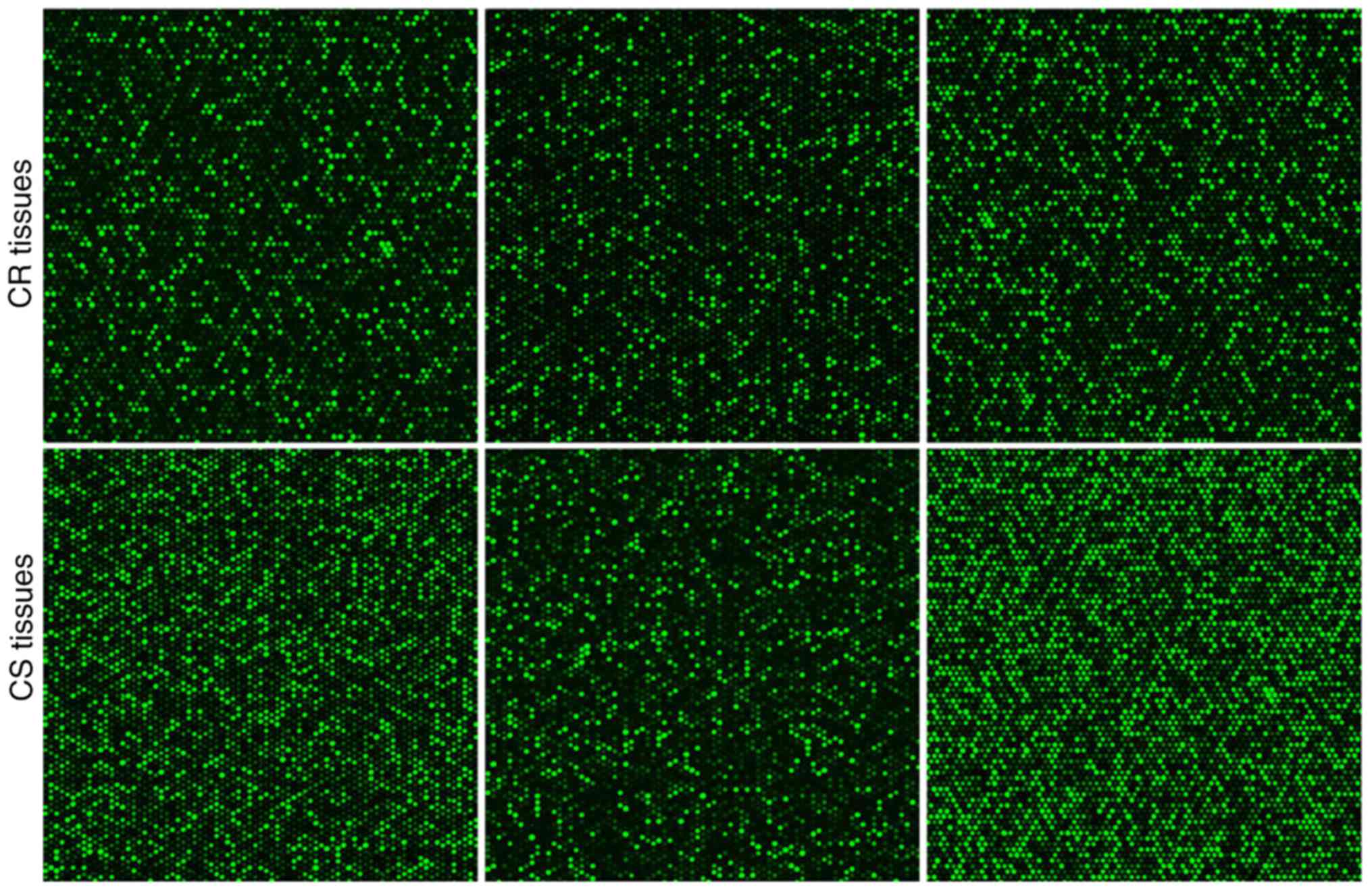
1A). The circRNA microarray hybridization signal diagram of the
three tissue pairs acquired by the circRNA chip scanner is
presented in Fig. 1B. The box plot
(Fig. 1C), volcano plot (Fig. 2A) and scatter plot (Fig. 2B) revealed variable circRNA
expression.
circRNA_001275 is significantly
upregulated in cisplatin- resistant esophageal cancer
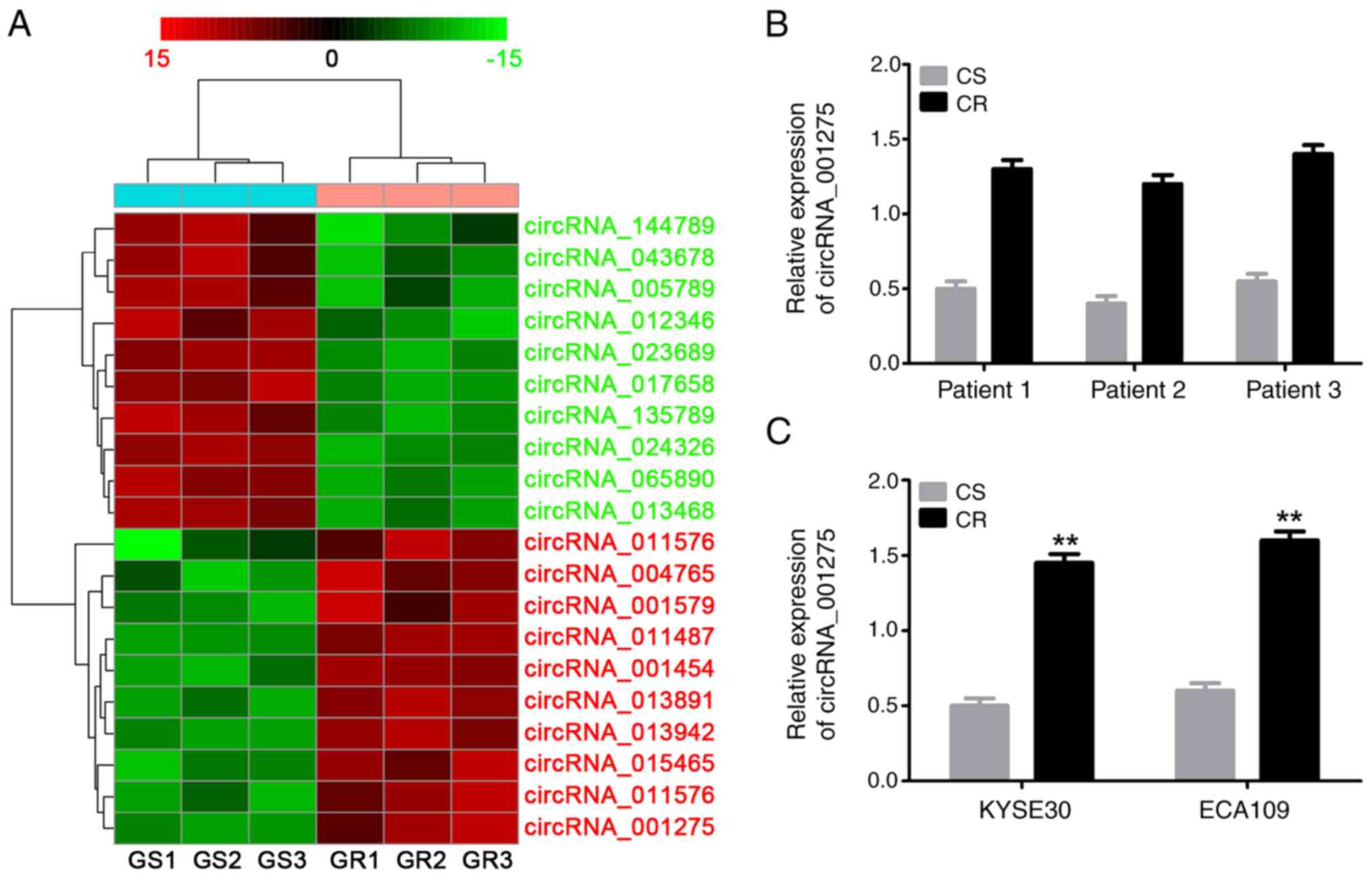
The cluster heat map (Fig. 3A) revealed the top ten most
upregulated and downregulated circRNAs. circRNA_001275 was the most
highly upregulated (14.85-fold) circRNA in cisplatin-resistant
tissues with highest raw intensity. Therefore, circRNA_001275 was
selected as the primary focus of the study. The circRNA chip
results were validated via RT-qPCR; circRNA_001275 was upregulated
in cisplatin-resistant tissues (Fig.
3B), as well as KYSE30/DDP and ECA109/DDP cells (P<0.05;
Fig. 3C), compared with
corresponding adjacent tissues and sensitive cells.
Transfection efficiency
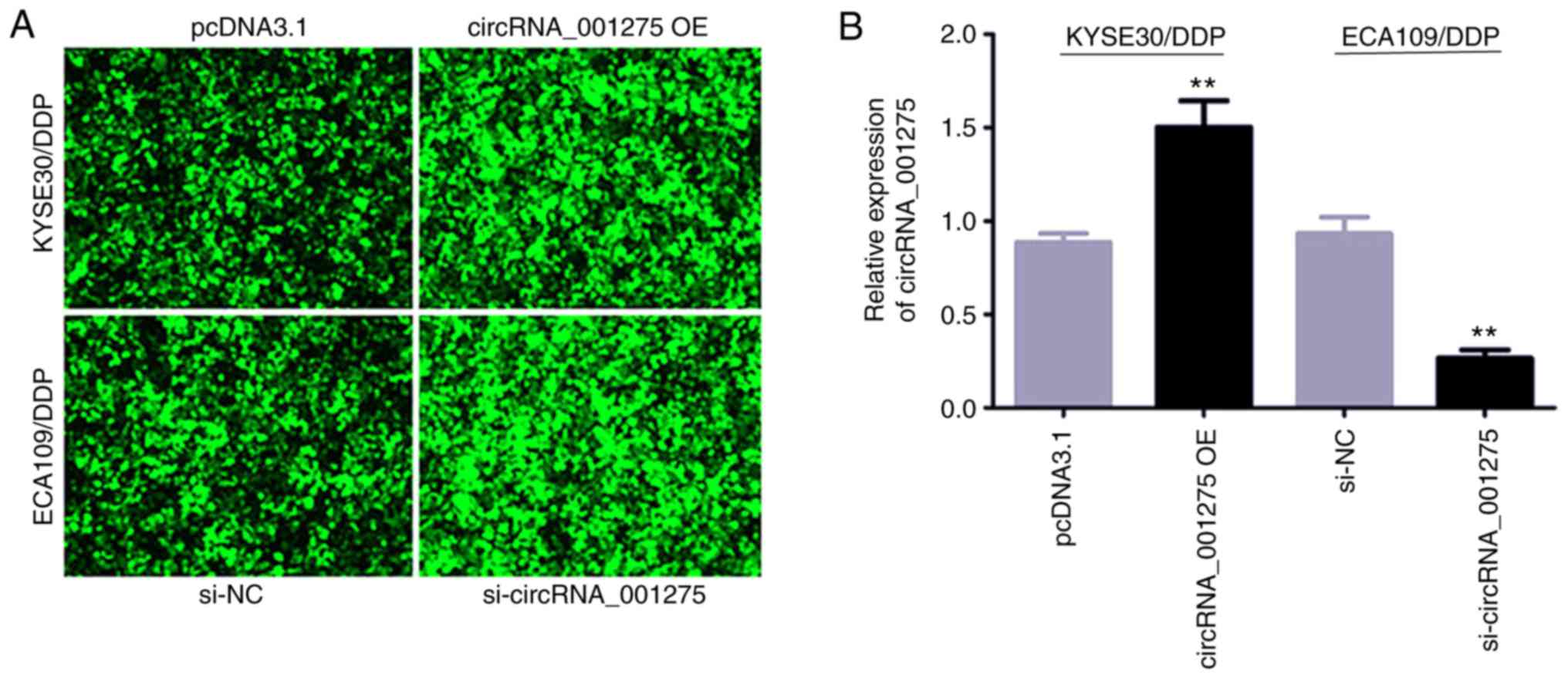
Fluorescence micrographs were captured at 24 and 48
h post-transfection. KYSE30/DDP and ECA109/DDP cells appeared green
under a fluorescence microscope (Fig.
4A). RT-qPCR validation demonstrated that circRNA_141539
expression was significantly increased in the circRNA_001275 OE
group, and significantly reduced in the si-circRNA_001275 group,
compared with the corresponding control groups (both P<0.05;
Fig. 4B).
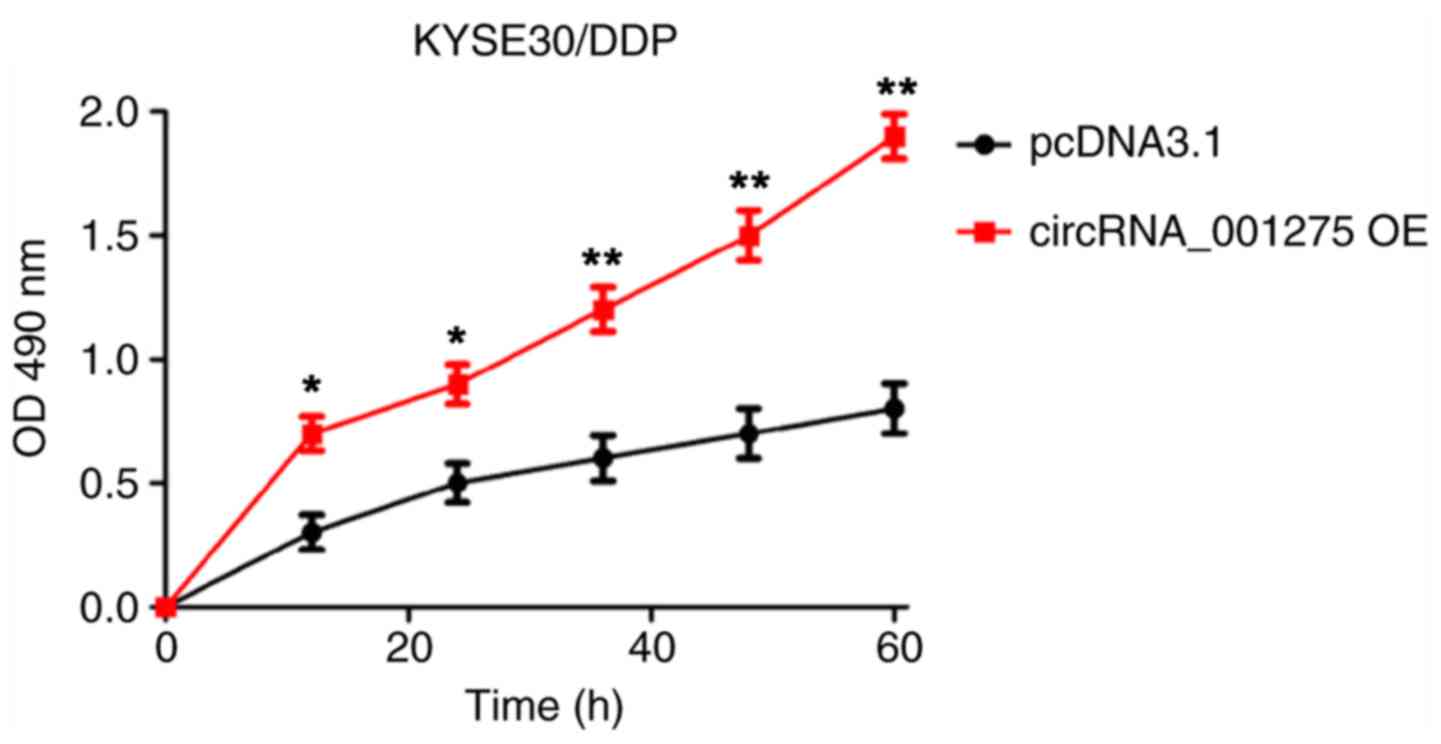
circRNA_001275 promotes cell
proliferation
MTT and EdU assays were used to detect the effects
of circRNA_001275 on cell proliferation. The results showed that
cell viability (P<0.05; Fig.
5A) and the ratio of EdU-positive cells (P<0.05; Fig. 5B) were significantly increased in
the circRNA_001275 OE group, and significantly decreased in the
si-circRNA_001275 group (both P<0.05), compared with the
corresponding control groups.
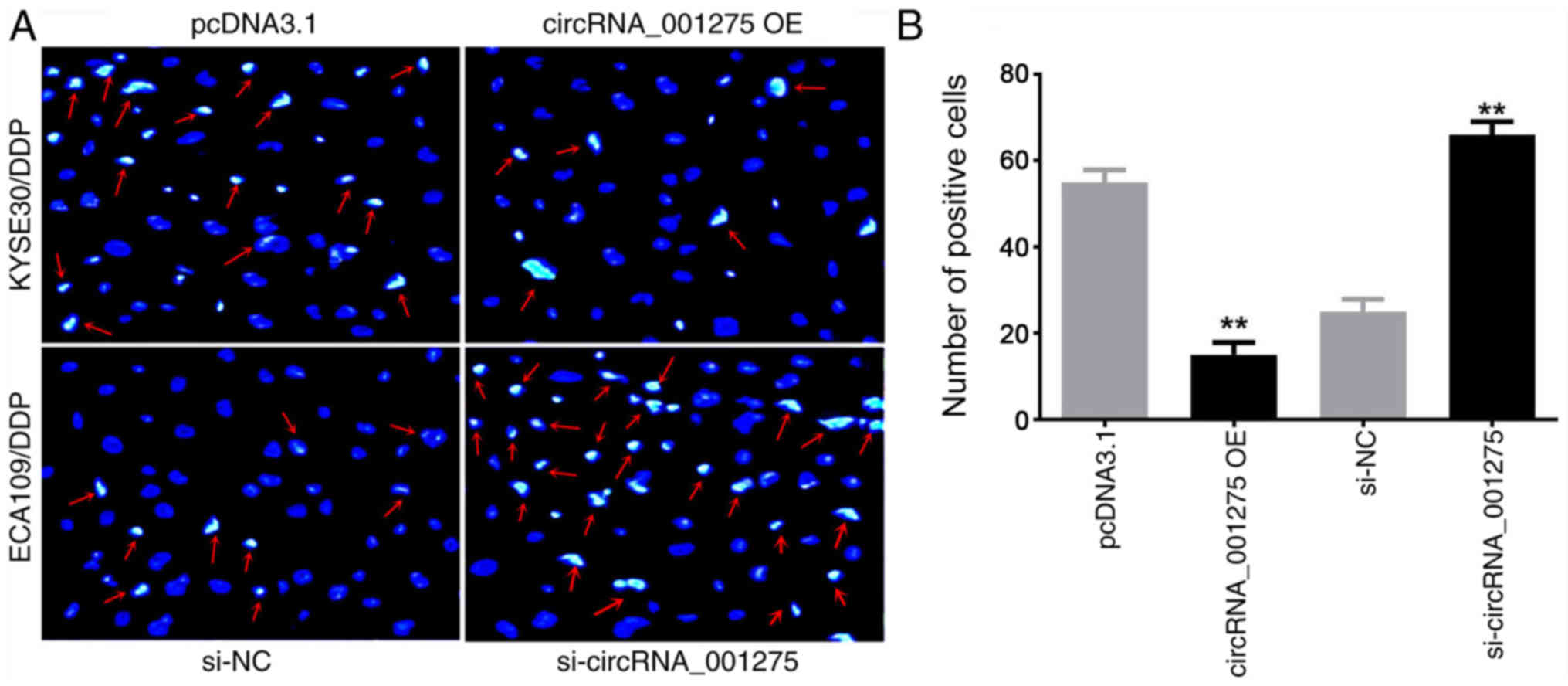
circRNA_001275 promotes cell invasion and
inhibits cell apoptosis
Transwell invasion assays and Hoechst 33258 staining
were used to detect the effects of circRNA_001275 on cell invasion
and apoptosis, respectively. The apoptosis rate was significantly
decreased in the circRNA_001275 OE group, and significantly
increased in the si-circRNA_001275 group, compared with the
corresponding control group (both P<0.05; Fig. 6).
circRNA_001275 serves as an miR-370-3p
sponge
A schematic representation of the potential binding
sites between miR-370-3p and the circRNA_001275-3'-UTR is presented
in Fig. 7A. miR-370-3p NC, mimic
or inhibitor were transfected into KYSE30/DDP and ECA109/DDP cells.
RT-qPCR results confirmed that miR-370-3p were significantly
increased in the miR-370-3p mimic group, and significantly reduced
in miR-370-3p inhibitor group, compared with the miR-370-3p NC
group (both P<0.05; Fig. S1).
A luciferase assay revealed that the relative luciferase activity
in the circRNA_001275-wt and miR-370-3p mimic co-transfection group
was significantly decreased compared with the circRNA_001275-mut or
miR-370-3p NC groups (both P<0.05; Fig. 7B). RT-qPCR revealed that miR-370-3p
was downregulated in cisplatin-resistant cells compared with
corresponding sensitive cells (P<0.05; Fig. 7C); additionally, miR-370-3p levels
were markedly decreased in cisplatin-resistant tissues compared
with adjacent tissues. Furthermore, the expression of miR-370-3p
was significantly decreased in the circRNA_001275 OE group, and
significantly increased in the si-circRNA_001275 group, compared
with the corresponding control groups (both P<0.05; Fig. 7D). Therefore, the results
demonstrated that circRNA_001275 may serve as a sponge for
miR-370-3p.
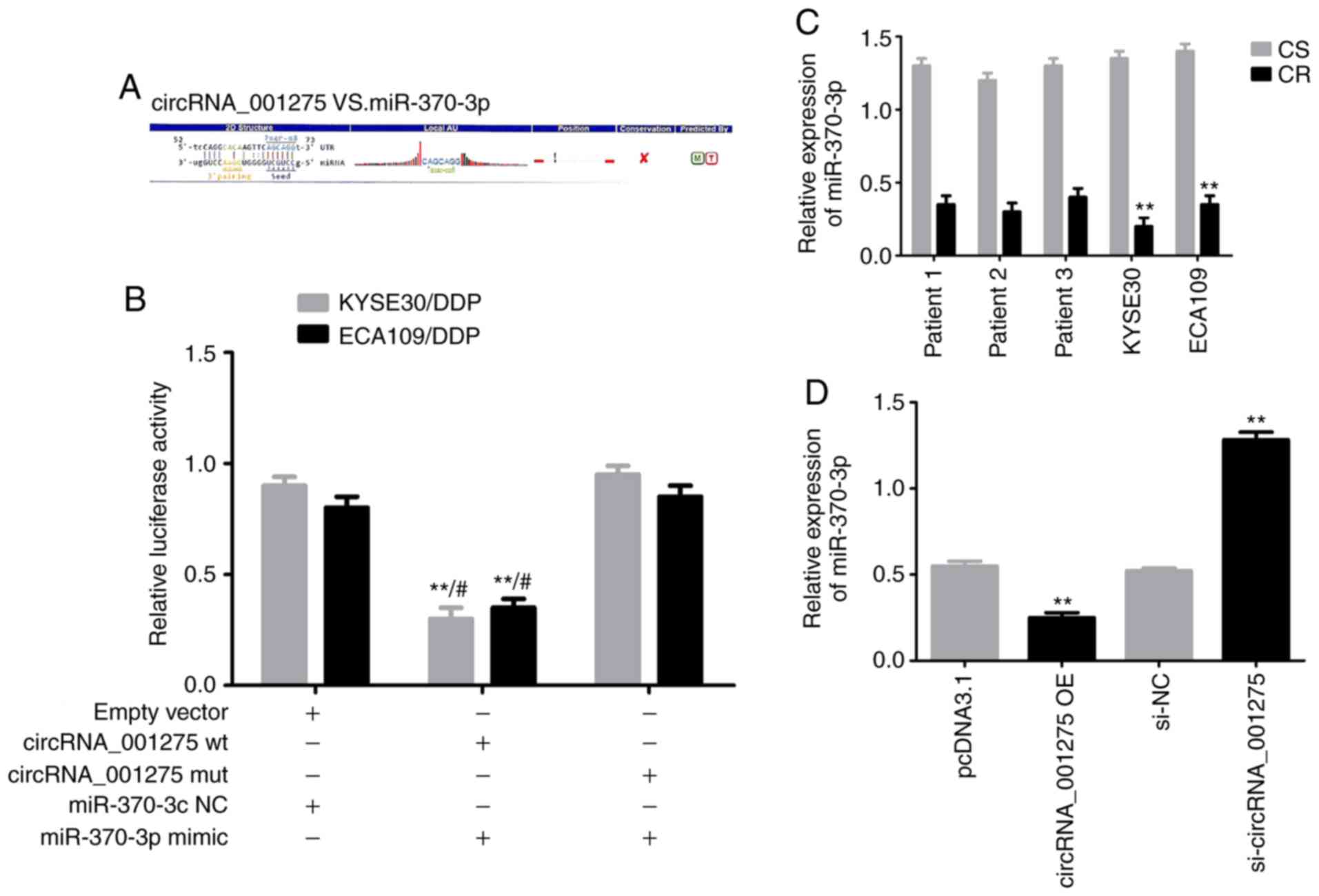 | Figure 7circRNA_001275 serves as an miR-370-3p
sponge. (A) Schematic representation of potential binding sites of
miR-370-3p with 3'-UTR of circRNA_001275. (B) Dual luciferase
assays results. **P<0.01 vs. empty
vector;#P<0.01 vs. mut + mimic. (C) RT-qPCR analysis
of miR-370-3p expression in tissues and
cells.**P<0.01 vs. CS. (D) RT-qPCR analysis
miR-370-3p expression in circRNA_001275 transfection groups.
**P<0.01 vs. pcDNA3.1 or si-NC. circRNA, circular
RNA; CR, cisplatin-resistant; CS, cisplatin-sensitive; DDP,
cisplatin; miR/miRNA, microRNA; mut, mutated; NC, negative control;
OE, overexpression vector; RT-qPCR, reverse
transcription-quantitative PCR; si, small interfering RNA; UTR,
untranslated region; wt, wild-type. |
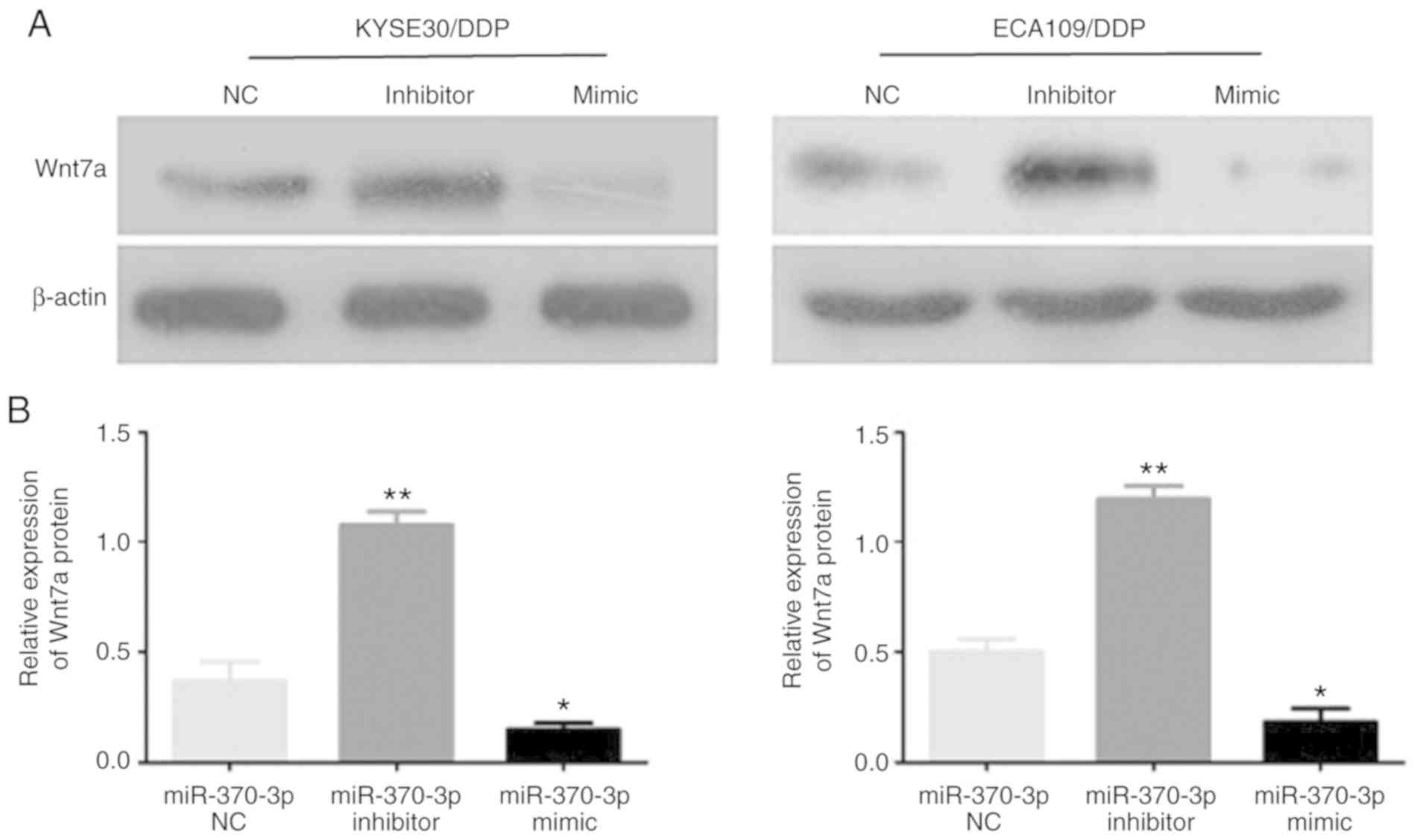
Wnt7a is a direct target gene of
miR-370-3p
The potential binding sites of miR-370-3p with
Wnt7a-wt and Wnt7a-mut were predicted. The results indicated a
potential binding site for miR-370-3p in the Wnt7a 3'-UTR (Fig. 8A). A luciferase assay revealed that
the relative luciferase activity in the Wnt7a-3'-UTR-wt and
miR-370-3p mimic co-transfection group was significantly decreased
compared with the miR-370-3p NC or Wnt7a-3'-UTR-mut groups (both
P<0.05; Fig. 8B). RT-qPCR
revealed that Wnt7a mRNA was significantly upregulated in
cisplatin-resistant cells compared with corresponding sensitive
cells (both P<0.05; Fig. 8C);
additionally, Wnt7a levels were markedly increased in
cisplatin-resistant tissues compared with adjacent tissues.
Furthermore, Wnt7a mRNA was significantly decreased in the
miR-370-3p mimic group, and significantly increased in the
miR-370-3p inhibitor group, compared with the miR-370-3p NC group
(both P<0.05; Fig. 8D).
Additionally, the Wnt7a protein level was significantly decreased
in the miR-370-3p mimic group, and significantly increased in the
miR-370-3p inhibitor group, compared with the miR-370-3p NC group
(both P<0.05; Fig. 9A and B).
Therefore, the results revealed that Wnt7a is a direct target gene
of miR-370-3p.
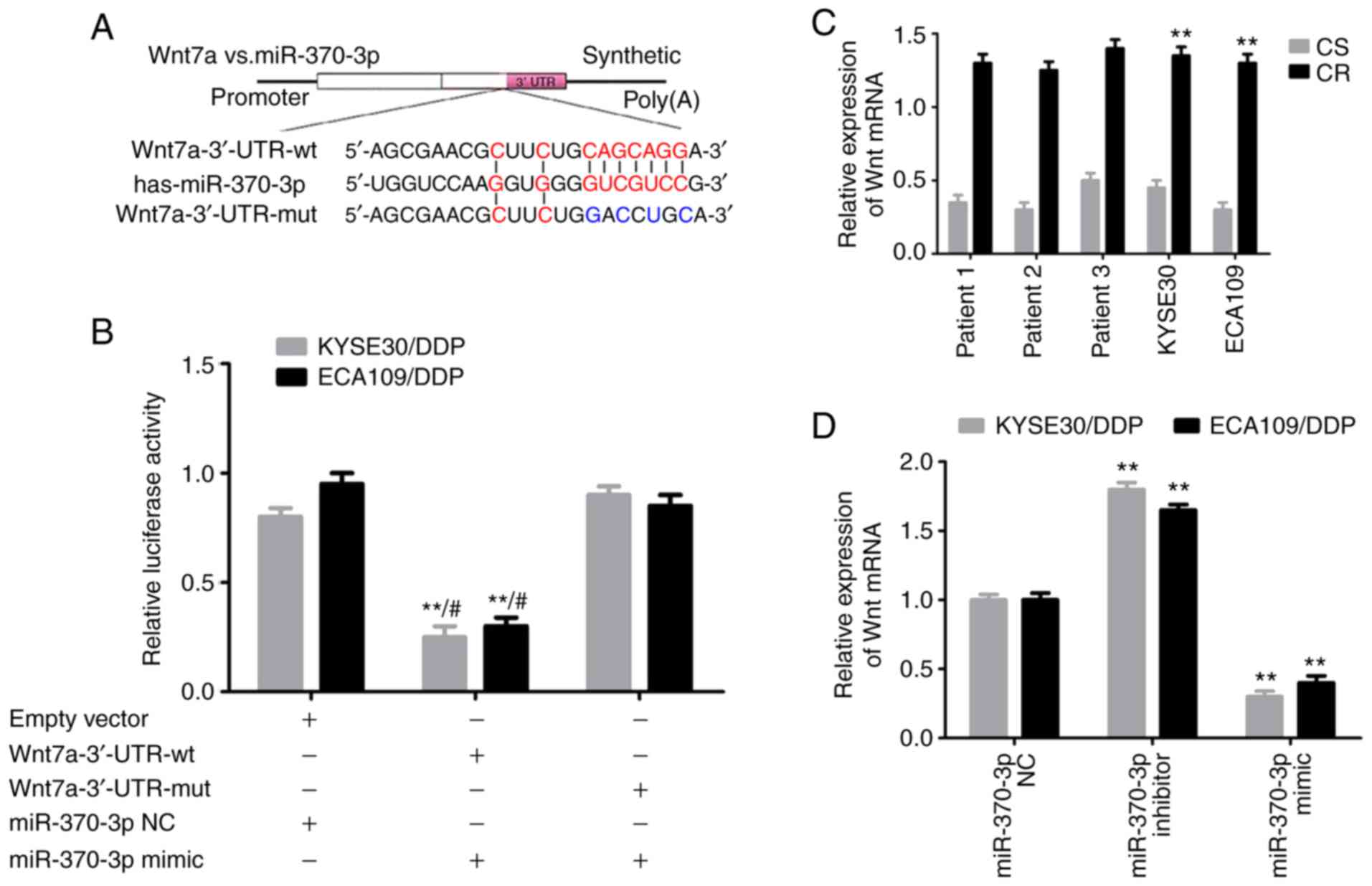 | Figure 8Wnt7a is a direct target of
miR-370-3p. (A) Schematic representation showing that miR-370-3p
specifically binds to the wt 3'-UTR sequence of Wnt7a. (B) Dual
luciferase assays results. **P<0.01 vs. empty vector;
#P<0.01 vs. mut + mimic. (C) RT-qPCR analysis of
Wnt7a mRNA expression in tissues and cells. **P<0.01
vs. CS. (D) RT-qPCR analysis of Wnt7a mRNA expression in
circRNA_001275 transfection groups. **P<0.01 vs.
pcDNA3.1 or si-NC. circRNA, circular RNA; CR, cisplatin-resistant;
CS, cisplatin-sensitive; DDP, cisplatin; miR, microRNA; mut,
mutated; NC, negative control; OE, overexpression vector; RT-qPCR,
reverse transcription-quantitative PCR; siRNA, small interfering
RNA; UTR, untranslated region; Wnt7a, Wnt family member 7A; wt,
wild-type. |
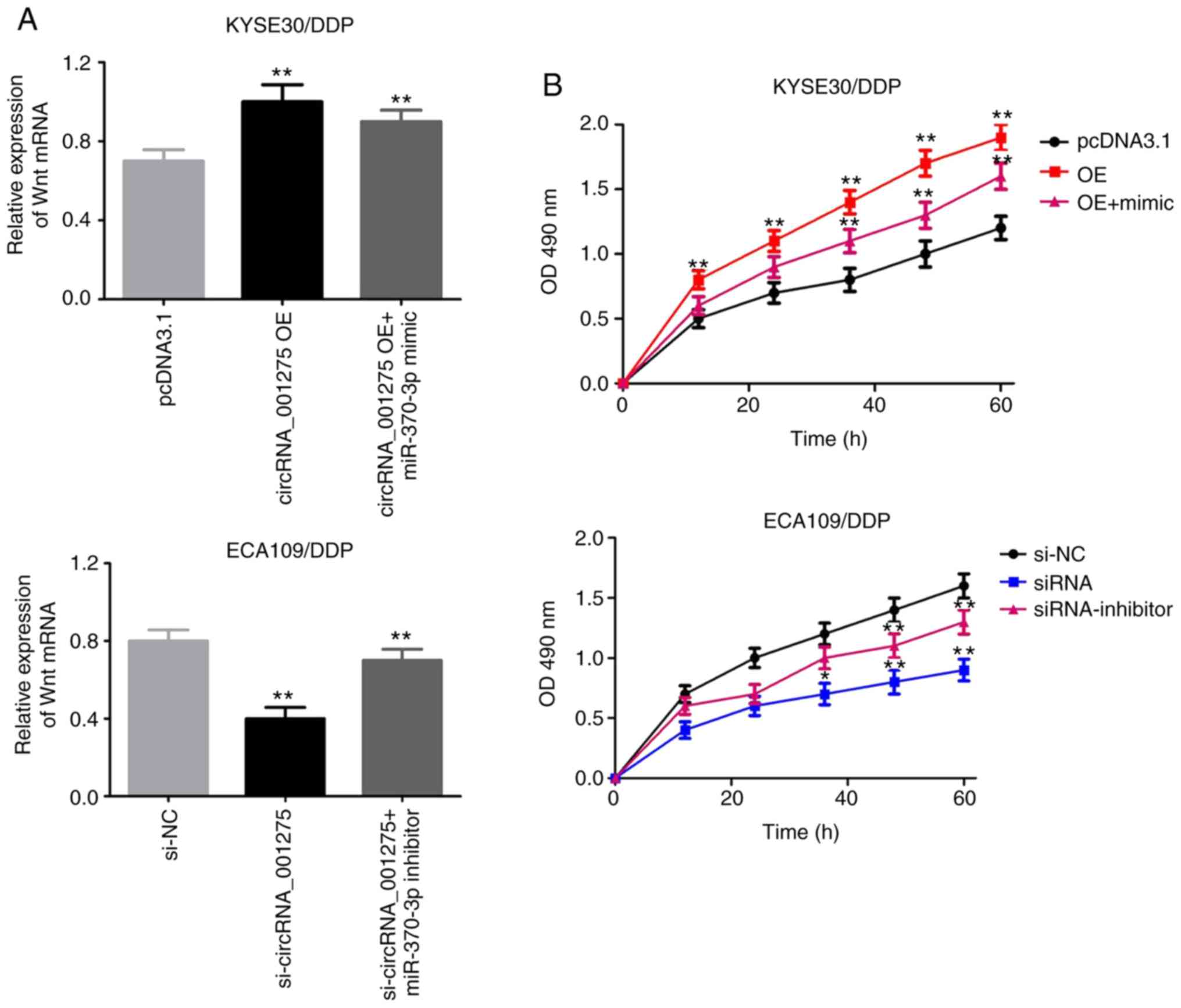
Wnt7a is activated by the
circRNA_001275/miR-370-3p axis
The effects of the circRNA_001275/miR-370-3p axis on
Wnt7a expression were investigated. RT-qPCR revealed that Wnt7a
mRNA expression was significantly increased in the circRNA_001275
OE group and significantly decreased in the si-circRNA_001275
group; however, these effects were attenuated by miR-370-3p mimic
and inhibitor, respectively (P<0.05; Fig. 10A). Furthermore, MTT results
showed that overexpression of circRNA_141539 increased cell
viability, and that circRNA_001275 silencing decreased cell
viability; these effects were abrogated by miR-370-3p mimic and
inhibitor, respectively (P<0.05; Fig. 10B).
Discussion
Previous studies have reported that
circRNAs-miRNA-mRNA axes play important roles in tumorigenesis and
development. For example, Xie et al (12) revealed that circ_001569 is
upregulated in colorectal cancer. Similarly, Zhong et al
(13) demonstrated that circRNA
transcription factor 25 (circTCF25) is upregulated in bladder
carcinoma, and that circTCF25 down-regulates miR-103a-3p, increases
cyclin-dependent kinase 6 expression and promotes proliferation. Li
et al (14) found that
circRNA RNA-binding motif, single-stranded-interacting protein 3
(circRBMS3) is upregulated in gastric cancer, and that increased
circRBMS3 expression is associated with advanced
tumor-node-metastasis stage. Gao et al (15) reported that the
circ_0006528/miR-7-5p/Raf1 axis serves a key role in
adriamycin-resistant breast cancer. Kun-Peng et al (16) reported that circRNA PVT1 promotes
doxorubicin and cisplatin resistance in osteosarcoma by regulating
ATP-binding cassette subfamily B member 1 expression. Dai et
al (17) reported that
circRNAs may be involved in epidermal growth factor
receptor-tyrosine kinase inhibitor resistance. However, the roles
of circRNAs in cisplatin-resistant esophageal cancer remain
unclear.
The present study revealed that circRNA_001275 was
upregulated in cisplatin-resistant esophageal cancer tissues and
cell lines. Furthermore, circRNA_001275 was found to promote the
proliferation of cisplatin-resistant cells. To the best of the
authors' knowledge, circRNA_001275 has only been investigated in a
study by Zhao (18), who reported
that circRNA_001275 was significantly upregulated in
post-menopausal osteoporosis and may serve as a novel potential
diagnostic biomarker. The present study revealed that
circRNA_001275 may function as an oncogenic factor and serve as a
promising biomarker for patients with cisplatin-resistant
esophageal cancer.
The present study revealed that circRNA_001275 may
act as an miR-370-3p sponge to upregulate Wnt7a expression.
miR-370-3p is located at the ∆-like homolog 1/D103 region of
chromosome 14. Several reports have shown that miR-370-3p may
promote the progression of bladder cancer (19) and glioma (20), and affect temozolomide sensitivity
in glioblastoma (21).
Furthermore, the Wnt signaling pathway plays an important role in
embryogenesis and cancer development by regulating the expression
of genes involved in cell proliferation, differentiation and
survival (22). Wnt7a, one of Wnt
ligands, activates the canonical Wnt/β-catenin signaling pathway to
promote cell proliferation (23).
Previous studies have revealed that Wnt7a expression is
dysregulated in several tumors, including gastric, lung, liver,
pancreatic, colorectal, cervical and breast cancers (24,25).
Wnt7a is a target gene of miR-150 (26), miR-34a (27) and miR-29b (28) in several types of cancer. The
present study revealed that Wnt7a was significantly upregulated in
cisplatin-resistant esophageal cancer.
Therefore, miR-370-3p-induced Wnt7a upregulation may
be one of the mechanisms underlying cisplatin resistance. However,
whether the circRNA_001275/miR-370-3p/Wnt7a axis serves as an
independent or auxiliary factor driving cisplatin resistance, as
well as the associated mechanisms, require further
investigation.
To the best of our knowledge, the present study was
the first to determine that circRNA_001275 was upregulated in
cispl-atin-resistant esophageal cancer, and that circRNA_001275 may
promote cisplatin resistance by sponging miR-370-3p to upregulate
Wnt7a expression. However, future studies investigating a larger
number of samples are required to validate these results.
Collectively, the results of the present study suggested that the
circRNA_001275/miR-370-3p/Wnt7a axis may serve as a novel potential
diagnostic biomarker and therapeutic target for patients with
cisplatin-resistant esophageal cancer.
Supplementary Data
Funding
This work was supported by Chinese Postdoctoral
Science Foundation (grant no. 2018M640266) and Postgraduate
Independent Exploration and Innovation Project of Central South
University (grant no. 2019zzts356).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Author contributions
ZHL, SX, and CHH designed the study and supervised
the experiments. FWZ conducted the majority of the molecular and
cellular experiments, data analysis, interpretation of data and
preparation of the manuscript. SZY and WYL were responsible for
bioinformatics and statistical analysis of the data. CYL and XHL
participated in the interpretation of the data and preparation of
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of China Medical
University and conducted in accordance with the Declaration of
Helsinki. Informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
We thank Shanghai GeneChem Co., Ltd. for conducting
and analyzing the circRNA chip assay and luciferase assay.
References
|
1
|
Hall KA, Spicer MT, Ilich JZ and Levenson
CW: Nutritional care for patients with esophageal cancer. Topics
Clin Nutr. 34:2–13. 2019. View Article : Google Scholar
|
|
2
|
Su LL, Chang XJ, Zhou HD, Hou LB and Xue
XY: Exosomes in esophageal cancer: A review on tumorigenesis,
diagnosis and therapeutic potential. World J Clin Cases. 7:908–916.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi H, Mao Y, Ju Q, Wu Y, Bai W, Wang P,
Zhang Y and Jiang M: C-terminal binding protein-2 mediates
cisplatin chemoresistance in esophageal cancer cells via the
inhibition of apoptosis. Int J Oncol. 53:167–176. 2018.PubMed/NCBI
|
|
4
|
Huang XP, Li X, Situ MY, Huang LY, Wang
JY, He TC, Yan QH, Xie XY, Zhang YJ, Gao YH, et al: Entinostat
reverses cisplatin resistance in esophageal squamous cell carcinoma
via down-regulation of multidrug resistance gene 1. Cancer Lett.
414:294–300. 2018. View Article : Google Scholar
|
|
5
|
Sun Y, Zhai L, Ma S, Zhang C, Zhao L, Li
N, Xu Y, Zhang T, Guo Z, Zhang H, et al: Down-regulation of RIP3
potentiates cisplatin chemoresistance by triggering HSP90-ERK
pathway mediated DNA repair in esophageal squamous cell carcinoma.
Cancer Lett. 418:97–108. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meng X, Li X, Zhang P, Wang J, Zhou Y and
Chen M: Circular RNA: An emerging key player in RNA world. Brief
Bioinform. 18:547–557. 2017.
|
|
7
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang C and Shan G: What happens at or
after transcription: Insights into circRNA biogenesis and function.
Transcription. 6:61–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Panda AC: Circular RNAs Act as miRNA
sponges. Adv Exp Med Biol. 1087:67–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
World Medical Association Inc: Declaration
of Helsinki. Ethical principles for medical research involving
human subjects. J Indian Med Assoc. 107:403–405. 2009.PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
12
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li G, Xue M, Yang F, Jin Y, Fan Y and Li
W: CircRBMS3 promotes gastric cancer tumorigenesis by regulating
miR-153-SNAI1 axis. J Cell Physiol. 234:3020–3028. 2019. View Article : Google Scholar
|
|
15
|
Gao D, Zhang X, Liu B, Meng D, Fang K, Guo
Z and Li L: Screening circular RNA related to chemotherapeutic
resistance in breast cancer. Epigenomics. 9:1175–1188. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kun-Peng Z, Xiao-Long M and Chun-Lin Z:
Overexpressed circPVT1, a potential new circular RNA biomarker,
contributes to doxorubicin and cisplatin resistance of osteosarcoma
cells by regulating ABCB1. Int J Biol Sci. 14:321–330. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dai Y, Huang Y, Huang J, Wen C, Zhou H and
Wu L: Differential expression of circular RNAs in
gefitinib-acquired resistant non-small cell lung cancer cells.
Tumor. 37:1128–1135. 2017.
|
|
18
|
Zhao K: Study on the expression profile of
cyclic RNA in postmenopausal osteoporosis and screening of
potential molecular markers (unpublished PhD thesis). Southern
Medical University; 2018
|
|
19
|
Huang X, Zhu H, Gao Z, Li J, Zhuang J,
Dong Y, Shen B, Li M, Zhou H, Guo H, et al: Wnt7a activates
canonical Wnt signaling, promotes bladder cancer cell invasion, and
is suppressed by miR-370-3p. J Biol Chem. 293:6693–6706. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peng Z, Wu T, Li Y, Xu Z, Zhang S, Liu B,
Chen Q and Tian D: MicroRNA-370-3p inhibits human glioma cell
proliferation and induces cell cycle arrest by directly targeting
β-catenin. Brain Res. 1644:53–61. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao YT, Chen XB and Liu HL: Up-regulation
of miR-370-3p restores glioblastoma multiforme sensitivity to
temozolomide by influencing MGMT expression. Sci Rep. 6:329722016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Anastas JN and Moon RT: WNT signalling
pathways as therapeutic targets in cancer. Nat Rev Cancer.
13:11–26. 2013. View
Article : Google Scholar
|
|
23
|
Tríbulo P, Jumatayeva G, Lehloenya K, Moss
JI, Negrón-Pérez VM and Hansen PJ: Effects of sex on response of
the bovine preimplantation embryo to insulin-like growth factor 1,
activin A, and WNT7A. BMC Dev Biol. 18:162018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang W, Yan X, Lin Y, Ge H and Tan Q:
Wnt7a promotes wound healing by regulation of angiogenesis and
inflammation: Issues on diabetes and obesity. J Dermatol Sci. Feb
15–2018.Epub ahead of print. View Article : Google Scholar
|
|
25
|
Liu Y, Dong Y, Zhao L, Su L and Luo J:
Circular RNA-MTO1 suppresses breast cancer cell viability and
reverses monastrol resistance through regulating the TRAF4/Eg5
axis. Int J Oncol. 53:1752–1762. 2018.PubMed/NCBI
|
|
26
|
MacLean JA II, King ML, Okuda H and
Hayashi K: WNT7A regulation by miR-15b in ovarian cancer. PLoS One.
11:e01561092016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Feng G, Fan Y, Li L, et al: MicroRNA-34a
sensitizes breast cancer cells to paclitaxel treatment by targeting
WNT7A. Bachu Med J. 15:6–10. 2018.In Chinese.
|
|
28
|
Avasarala S, Van Scoyk M, Wang J, Sechler
M, Vandervest K, Brzezinski C, Weekes C, Edwards MG, Arcaroli J,
Davis RE, et al: hsa-miR29b, a critical downstream target of
non-canonical Wnt signaling, plays an anti-proliferative role in
non-small cell lung cancer cells via targeting MDM2 expression.
Biol Open. 2:675–685. 2013. View Article : Google Scholar : PubMed/NCBI
|