Introduction
Cancer was responsible for 9.6 million deaths in
2018 worldwide, according to the World Health Organization Global
Cancer Observatory (1). Breast
cancer is the most common cancer in women with 2.1 million newly
diagnosed cases worldwide in 2018 (1). Breast cancer is a heterogeneous
disease with several subtypes that are classified based upon a
variety of clinical and histopathological features (2-4). The
lack of expression of oestrogen receptor (ER) and progesterone
receptor (PR) and the absence of human epidermal growth factor
Receptor-2 (HER2) overexpression assessed by immunohistochemistry
defines the triple-negative breast cancer (TNBC) subtype. Poor
long-term outcomes are associated with TNBC compared with other
breast cancer subtypes (5-8). Mammography screening accounted for a
decrease in breast cancer mortality in the recent years (9,10).
However, despite the numerous therapeutic options, including
chemotherapy, hormonal therapy and radiotherapy, about 30% of
patients who are treated at early-stages relapse (11). In most cases, treatment is
effective at first, but resistance to therapy and disease
progression eventually occur. Because treatment resistance has
become common (11), it is crucial
to provide novel treatment strategies. Alternative treatments may
emerge from the development of either novel/unconventional
therapeutic compounds or novel pharmacological approaches. The
natural phytochemical compound curcumin exerts some effects on
several biochemical pathways and drug targets. At present, curcumin
is being widely investigated for its potential therapeutic use in
various diseases, including Alzheimer's disease, cystic fibrosis
and several types of cancer (12-19).
Curcumin has been reported to display antioxidant,
anti-inflammatory, antiproliferative, proapoptotic and anticancer
properties (20). The antioxidant
effects of curcumin have been used in the treatment of pancreatitis
(21), spinal cord injury
(22) and complications associated
with diabetes (23). The
anti-inflammatory effects of curcumin have been demonstrated in
diabetes (23) and in the
treatment of cancer therapy-induced oral mucositis (24). Curcumin also displays anticancer
properties that are applicable to the clinic (25-27).
The clinical use of curcumin has been limited by its low aqueous
solubility and stability, resulting in low oral bioavailability
(28,29). Structural modification and
encapsulation of curcumin have been used to improve its
pharmacokinetic profile (30).
Furthermore, reformulations of curcumin have been investigated in
order to improve its delivery directly to the tumour mass within
the breast tissue (31-33). Improved curcumin stability has been
achieved by structural modifications, including replacement of the
β-diketone moiety with metal ions (34,35);
however, the effects of these modifications on the cytotoxic
properties of curcumin have not yet been investigated. In addition,
whether these novel complexes can enter the cell and localize in a
similar manner to curcumin remain unknown. A suitable breast cancer
cell line may be employed to investigate the localization and
movement of curcumin-derived compounds into cells. The MDA-MB-231
cell line (36) is an aggressive
and invasive breast cancer cell line, which lacks ER, PR and HER2
expression. This cell line is therefore referred to as a
triple-negative breast cancer cell line (37). The absence of ER expression results
in MDA-MB-231 cell insensitivity to antihormone-based therapies,
including tamoxifen (38).
Clinically, triple-negative breast cancer has limited treatment
options. The MDA-MB-231 cell line is thus commonly used to
investigate the molecular basis of this type of breast cancer and
for the development of novel therapeutic approaches.
Resistance to apoptosis is a key characteristic of
cancer cells (39). Curcumin has
been reported to inhibit proliferation by inducing apoptosis in
MDA-MB-231 breast cancer cells via increasing the Bax/Bcl-2 ratio
or upregulating p21 expression (40,41).
It was also reported that curcumin can decrease MDA-MB-231 cell
migration and invasion without affecting apoptosis (42). However, the anticancer activity of
curcumin in relation to apoptosis, and the expression of
proapoptotic and antiapoptotic proteins have not been fully
described.
Ion channels directly or indirectly influence most
basic cellular processes. Subsequently, ion channels also affect
most malignant processes in cancer cells, including sustained
proliferation (43-45), tissue invasion (43), metastasis (46,47)
and programmed cell death (48,49).
A previous study demonstrated that molecular, biological and
pharmacological inhibition of voltage-gated potassium channels
(Kv) reduces cancer cell proliferation, whereas
overexpression of certain Kv channels can stimulate cell
proliferation (50). Furthermore,
when cells receive a death stimuli, they decreases in size during a
process called apoptotic volume decrease (AVD). AVD occurs before
the cascade of biochemical events that induces apoptosis (51) and is primarily induced by potassium
efflux across the plasma membrane. Numerous potassium channels are
essential for controlling and regulating the flow of potassium into
and out of the cell. Because of the pivotal role of potassium
channels during AVD, cancer cells may evade apoptosis by
downregulating potassium channel expression. For example,
Kv1.3 and Kv1.5 are expressed at reduced levels in
many types of human cancer cells compared with normal cells
(52). Other potassium channels,
including voltage-gated potassium channels Kv1.1,
Kv1.3, Kv1.5, Kv2.1 and Kv11.1, serve
crucial roles during apoptosis (53). In addition to AVD associated with a
decrease in cytoplasmic potassium concentration, this potassium
movement promotes various cellular events that are critical for
programmed cell death. These events include mitochondrial
depolarization and cytochrome c release from mitochondria as well
as cleavage of procaspase 3 and enhanced endonuclease activity
(53). Furthermore, Bax is known
to inhibit mitochondrial potassium channels downstream of
pro-apoptotic signals. The activation of the mitochondrial pathway
of apoptosis by the pro-apoptotic protein Bax is mediated via the
direct inhibition of mitochondrial Kv1.3 channels by BAX in the
outer mitochondrial membrane (54). It has been reported that curcumin
displays inhibitory effects on potassium channels, mediating the
curcumin-induced anticancer and anti-inflammatory properties in
monocytic leukaemia and effector memory T cells (55,56).
In addition, curcumin decreases potassium channel currents by
inactivating the gating of Kv2.1 in 293 cells (57).
It has been demonstrated that the solubility and
stability of curcumin can be increased by complexation with boron
and iron (34). The aim of this
study was to assess the anti-cancer properties of these curcumin
complexes. The present study examined the effects of soluble
structurally modified curcumin-based compounds on the MDA-MB-231
cell line. Furthermore, the inhibitory effects of curcumin,
boron-curcumin [B(Cur)2] and iron-curcumin
[Fe(Cur)3] on MDA-MB-231 cell proliferation, migration
and invasion were investigated. In addition, the effect of the
curcumin compounds on the apoptosis-proteome and gene expression of
certain ion-channels was assessed.
Materials and methods
Cell lines
The oestrogen negative MDA-MB-231 cell line was
obtained from the American Type Culture Collection. Cells were
cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.; cat. no.
12491015) supplemented with 5% fetal bovine serum (FBS;
Sigma-Aldrich; Merck KGaA; cat. no. F2442), 100 U/ml penicillin,
100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific,
Inc.; cat. no. 15070063), 600 µg/ml L-glutamine
(Sigma-Aldrich; Merck KGaA; cat. no. G7513) and 6/500 ml 100X
non-essential amino acids (Gibco; Thermo Fisher Scientific, Inc.;
cat. no. 11140050) and placed at 37°C in a humidified incubator
containing 5% CO2.
Drugs and reagents
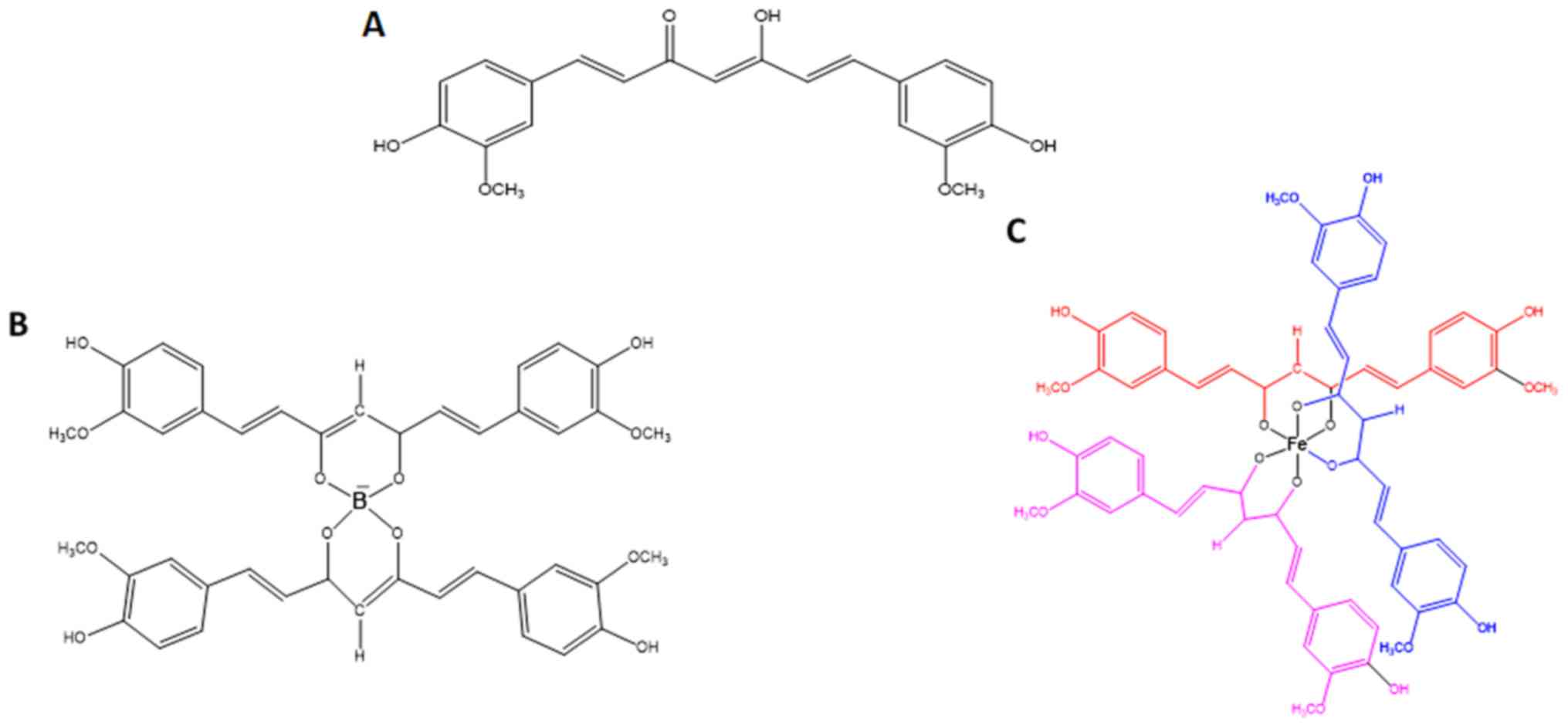
Curcumin was purchased from Sigma-Aldrich: Merck
KGaA (cat. no. C7727). B(Cur)2 and Fe(Cur)3
were synthesized as previously described by Khalil et al
(58). The improved photostability
of B(Cur)2 and Fe(Cur)3 relative to curcumin
has been previously validated (34). Curcumin, B(Cur)2 and
Fe(Cur)3 were initially dissolved in DMSO into a 10 mM
stock solution that was stored at -20°C. Stock solutions were later
diluted in DMEM to the desired final concentrations. The structures
of curcumin, B(Cur)2 and Fe(Cur)3 are
presented in Fig. 1.
Fluorescence microscopy
M DA-M B-231 cells (350×103 cell/ml) were
seeded in 8-chamber slides (Eppendorf; cat no. 30742036) and
incubated for 24 h at 37°C with 5% CO2.
Subsequently, cells were treated with curcumin (25 µM),
B(Cur)2 (30 µM) and Fe(Cur)3 (8
µM) or vehicle (control group) for 5 min and immediately
fixed with 3.7% formaldehyde at room temperature for 10 min. Cells
were counterstained with DAPI at room temperature for 5 min to
visualize the nucleus. Stained cells were observed using a BX43
fluorescence microscope (Olympus Corporation; magnification, ×100)
to determine the cellular localization of curcumin and its metal
derivatives. Images were analysed using CellSens Standard software
(version 1.9; Olympus Corporation).
Sulforhodamine B (SRB) assay
The in vitro effect of curcumin,
B(Cur)2 and Fe(Cur)3 on breast cancer cell
proliferation was evaluated by using the SRB assay as previously
described by Skehan et al (59). Briefly, MDA-MB-231 cells were
seeded in triplicate at the density of 1×104 cells/well
into a 96-well plate and incubated overnight at 37°C with 5%
CO2. Cells were treated with vehicle or various
concentrations (5, 10, 20, 30, 40, 50 and 100 µM) of
curcumin, B(Cur)2 and Fe(Cur)3 for 48 h.
Subsequently, cells were fixed overnight with cold 10%
trichloroacetic acid (Sigma-Aldrich; Merck KGaA; cat. no. 91228) at
room temperature, washed with distilled H2O and
air-dried. Cells were stained with 100 µl of 0.4% SRB stain
(Sigma-Aldrich; Merck KGaA; cat. no. S1402) (diluted in 1% acetic
acid) for 30 min at room temperature, washed with acetic acid
(Sigma-Aldrich; Merck KGaA; cat no. 33209) and air-dried. SRB stain
was solubilized in 10 mM unbuffered Tris base solution. Absorbance
was measured at a wavelength of 540 nm with reference wavelength of
650 nm using a micro-plate reader. Cell proliferation was
calculated according to the following formula: Cell
proliferation=100-[(absorbance of treated cells/absorbance of
untreated cells) x100]. IC50 values were calculated
using GraphPad Prism software version 5 (GraphPad Software, Inc.)
to compare the cytotoxicity of the two complexes. For subsequent
cell migration and invasion assays, the IC50 dose, one
dose above the IC50 and one dose below the
IC50 were used. For cell localization, mRNA and protein
expression assays, cells were treated with the IC50
dose.
Cell migration by wound healing
assay
MDA-MB-231 cells were seeded in 24-well plates and
incubated at 37°C with 5% CO2 until they reach 80-90%
confluence. A 100-µl pipette tip was used to make a single
scratch in the cell monolayer and image of the scratch was captured
at 0 h with a light microscope (magnification, ×4). Subsequently,
cells were treated with various concentrations of curcumin,
B(Cur)2 and Fe(Cur)3 or vehicle (control
group) and incubated for 24 h at 37°C with 5% CO2. The
concentrations used were 5, 10, 15, 20, 25 and 30 µM for
curcumin and B(Cur)2, and 2, 4, 5, 6 and 8 µM for
Fe(Cur)3. On day 2, the width of the scratch was
photographed and measured. Wound closure was calculated according
to the following formula: Wound closure=100-[(scratch width at 24
h/scratch width at 0 h) x100]. Cells were not serum-starved during
the assay.
Agarose invasion assay
The under-agarose assay was performed to determine
the random invasion of treated and untreated cells towards serum
components found in the agarose gel. Ultra-pure agarose (0.9%;
Invitrogen; Thermo Fisher Scientific, Inc.) was dissolved in PBS
supplemented with minimum essential medium (Gibco; Thermo Fisher
Scientific, Inc.) containing 5% FBS and incubated at room
temperature in 6-well plates until it solidifies. Subsequently,
2-mm wells were formed in the agarose gel as previously described
(60) and cells (4×104
cells/well) were pre-treated with 5, 10, 15, 20, 25 and 30
µM curcumin and B(Cur)2 and 2, 4, 5, 6 and 8
µM Fe(Cur)3 (or vehicle for control) before being
loaded into the wells. Following 24 h incubation, the number of
cells that had invaded the agarose gel were manually counted under
a light microscope (magnification, ×4).
Profiling of apoptosis-associated
proteins
The effect of curcumin, B(Cur)2 and
Fe(Cur)3 on the expression of apoptosis-associated
proteins was determined using the Proteome Profiler™ Array Human
Apoptosis Array kit (R&D Systems, Inc.; cat no. ARY009)
according to the manufacturer's instructions. Briefly, MDA-MB-231
cells were cultured in a 75 cm2 flask and treated for 48
h with curcumin (25 µM), B(Cur)2 (35 µM),
Fe(Cur)3 (8 µM) or vehicle. Cells (80% confluent)
were lysed and total proteins of treated and untreated cells were
extracted and quantified using a Bradford Assay Protein
Quantitation kit (Abcam). Extraction was performed using the
solution provided in the kit 'Lysis Buffer 17' (Part no. 895943).
The extraction solution contained aprotinin (10 µg/ml),
leupeptin (10 µg/ml) and pepstatin (10 µg/ml).
Protein arrays (nitrocellulose membranes spotted with 35 apoptotic
proteins) were blocked with the array buffer 1 for 1 h at room
temperature. Subsequently, proteins (280 µg) were
transferred onto the arrays and incubated overnight at 4°C. After
being washed, the arrays were incubated at room temperature with
antibody detection cocktail for 1 h followed by the addition of
Streptavidin-HRP for 30 min. The signal was finally obtained by
adding Chemi Reagent Mix on the membrane. Protein spots were
visualized using LI-COR detection system and the pixel density of
each spot was quantified using Image J software (National
Institutes of Health, version 1.46r). The relative expression of
proteins in the treated cells were normalized to those in the
untreated cells. The relative changes were based on differences in
the pixel density of each spot displayed on images of the membrane
array. The assay was performed in duplicate and the mean of the
pixel density value was used for calculations.
RNA extraction
Total RNA was extracted from MDA-MB-231 cells
following treatment with curcumin, B(Cur)2,
Fe(Cur)3 or vehicle by using the RNeasy kit (Qiagen;
cat. no. 74104) according to the manufacturer's protocol. The
concentration of RNA was determined using a NanoDrop 1,000
spectrophotometer (Thermo Fisher Scientific, Inc.).
Ion-channel gene expression analysis by
reverse transcription-quantitative (RT-q) PCR
Total RNA (1 µg) from cells treated with
curcumin, B(Cur)2, Fe(Cur)3 or vehicle was
reverse transcribed into cDNA using the RT2 First Strand
kit (Qiagen; cat. no. 330401) according to the manufacturers'
protocol. cDNA was then subjected to RT-qPCR array analysis for a
panel of 84 neuronal ion channel genes. RT-qPCR was performed on a
96-well Human Neuronal Ion Channels RT2 profiler PCR
array (cat. no. PAHS-036ZA; Qiagen) using RT2 SYBR
Green/ROX PCR Master Mix (Qiagen) and an ABI 7500 real time PCR
machine (Applied Biosystems; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The reaction conditions
were as follows: 10 min at 95°C followed by 40 cycles of 15 sec at
95°C and 1 min at 60°C. Each sample was run in biological
triplicates. The gene expression data were uploaded and analyzed
using an online analysis software package (Qiagen, Inc., processed
via the Qiagen portal at http://www.qiagen.com/geneglobe, July 2018). The array
included five housekeeping genes as internal standards for data
normalization, ACTB, B2M, GAPDH, HPRT1 and RPLP0. Relative gene
expression was represented by fold change which was calculated by
RT2-analysis software using the threshold cycle (Ct) and
based upon the 2−ΔΔCt method (61). Fold change was defined by the
normalised gene expression (2−ΔCt) in the Test Sample
divided by the normalized gene expression (2−ΔCt) in the
Control Sample (vehicle).
Statistical analysis
Data were presented as the means ± standard error of
the mean of at least three independent experiments. Statistical
analyses were performed using SPSS version 25 (IBM Corp.) and
GraphPad Prism version 5 (GraphPad Software, Inc.). Data were
compared using Student's t-test and two-way ANOVA followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Curcumin, B(Cur)2 and
Fe(Cur)3 localize in the cell cyto- plasm and around the
nucleus
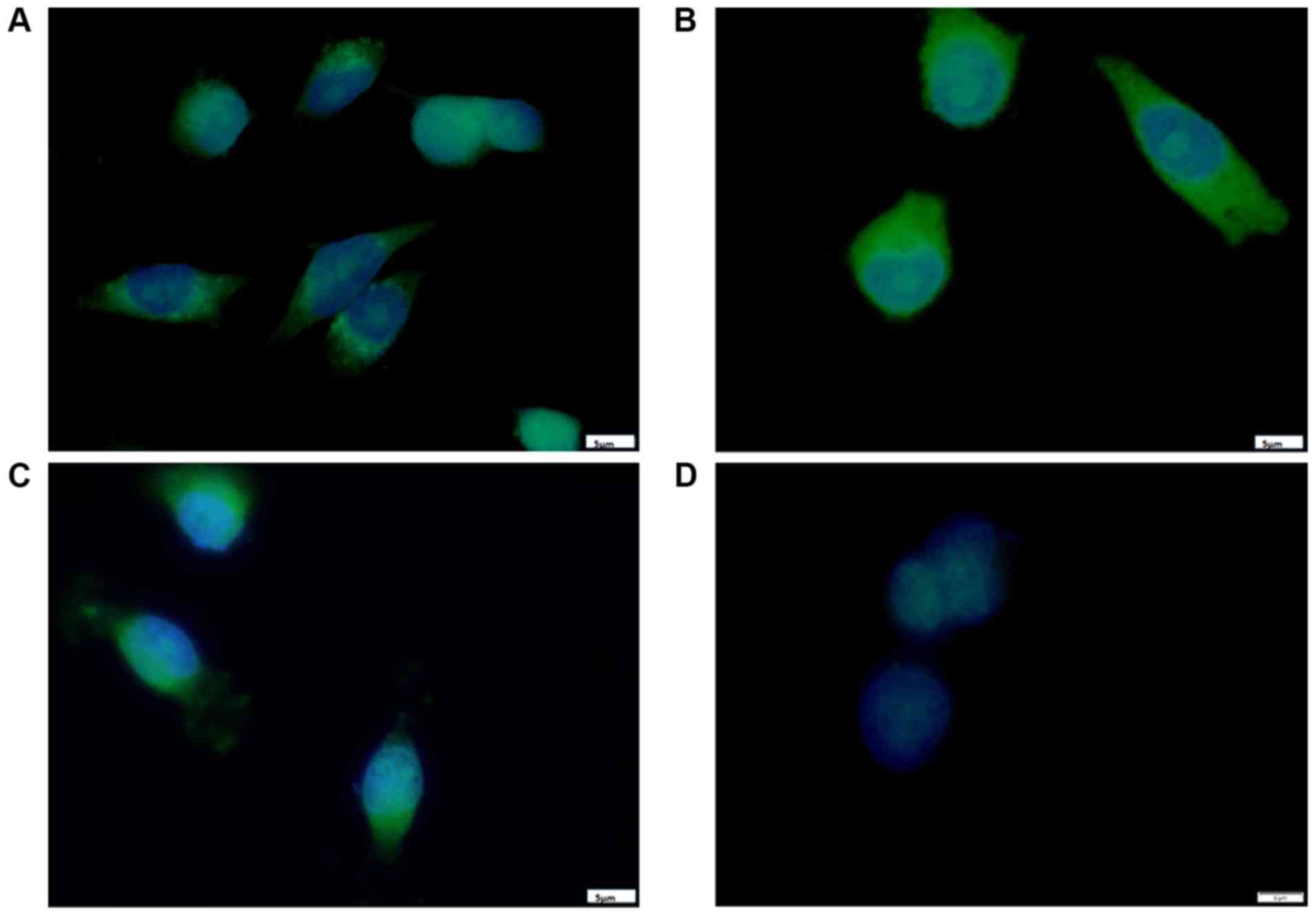
The results from fluorescence microscopy displayed a
cytoplasmic localization of curcumin B(Cur)2 and
Fe(Cur)3 in MDA-MB-231 cells (Fig. 2); however, the three compounds
displayed differences in their exact localization after a short
incubation period. As presented in Fig. 2A, the curcumin-treated cells
displayed fluorescence mainly around the perinuclear region with a
punctate pattern. In Fig. 2B,
cells incubated with B(Cur)2 displayed homogenous
fluorescence in the cytoplasm, and a few cells displayed a distinct
halo around the nucleus, presumably within the nuclear membrane. As
presented in Fig. 2C,
Fe(Cur)3-treated cells displayed a nuclear-centric
pattern of localization with no obvious fluorescence in the nuclear
membrane or nucleus. Furthermore, the three compounds altered the
morphology of MDA-MB-231 cells from the typical spindle-shape to a
more spherical shape.
Curcumin, B(Cur)2 and
Fe(Cur)3 are cytotoxic for MDA-MB-231 cells
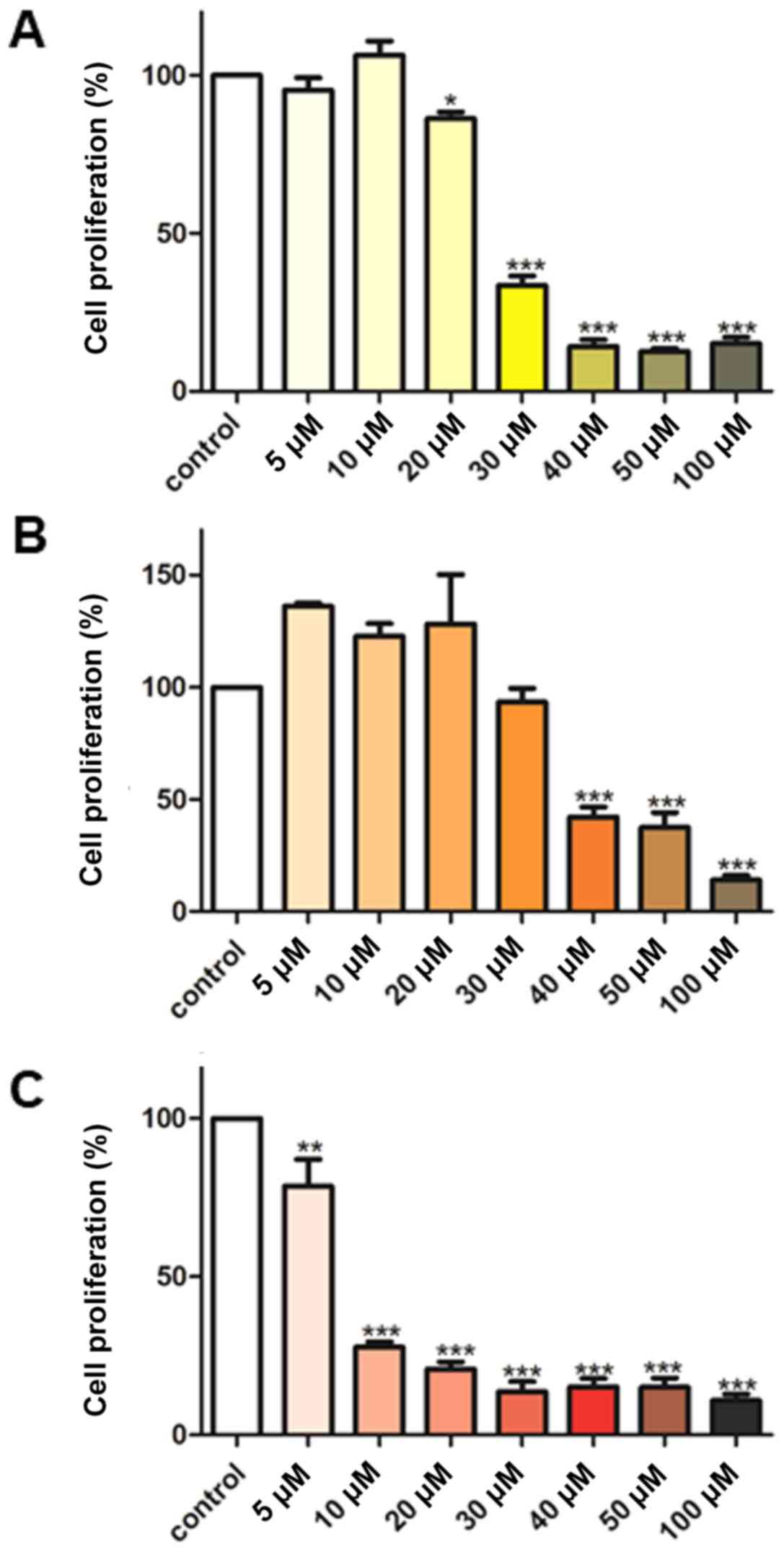
The effect of various concentrations (5-100
µM) of curcumin, B(Cur)2 and Fe(Cur)3
on cell proliferation was assessed using the SRB assay. The results
demonstrated that curcumin displayed an inhibitory effect on
MDA-MB-231 cell proliferation of 66-85%, with an effective dose
starting at 30 µM curcumin (Fig. 3A). By contrast, B(Cur)2
displayed a weaker cytotoxic effect (60%) on MDA-MB-231 cells,
which occurred with a higher dose (40 µM; Fig. 3B). Fe(Cur)3 displayed
the most potent cytotoxic effect on MDA-MB-231 cell proliferation
(20-90%), with a significant decrease in cell proliferation at the
lowest dose (5 µM; Fig.
3C). In addition, the IC50 values of curcumin,
B(Cur)2 and Fe(Cur)3 were 25, 35 and 8
µM, respectively.
Curcumin and B(Cur)2 alter
MDA-MB-231 cell migratory ability
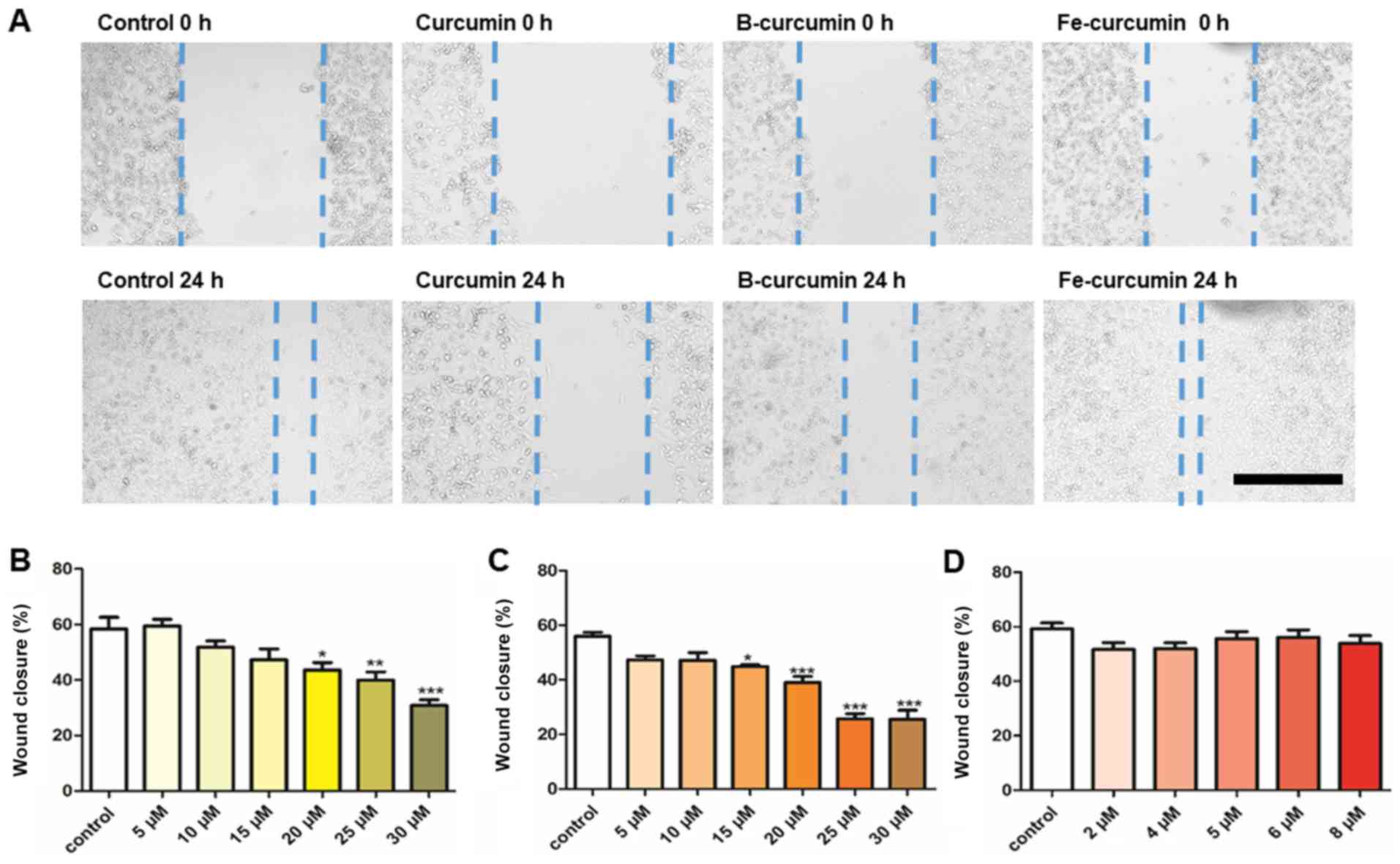
The results from the wound healing assay
demonstrated that curcumin and B(Cur)2 had an inhibitory
effect on MDA-MB-231 cell migratory ability (Fig. 4A). Curcumin and B(Cur)2
inhibited cell migration by 60-70% at 20-30 µM and 15-35
µM, respectively (Fig. 4B and
C). Fe(Cur)3 had no effect on cell migratory ability
(Fig. 4C).
Curcumin, B(Cur)2 and
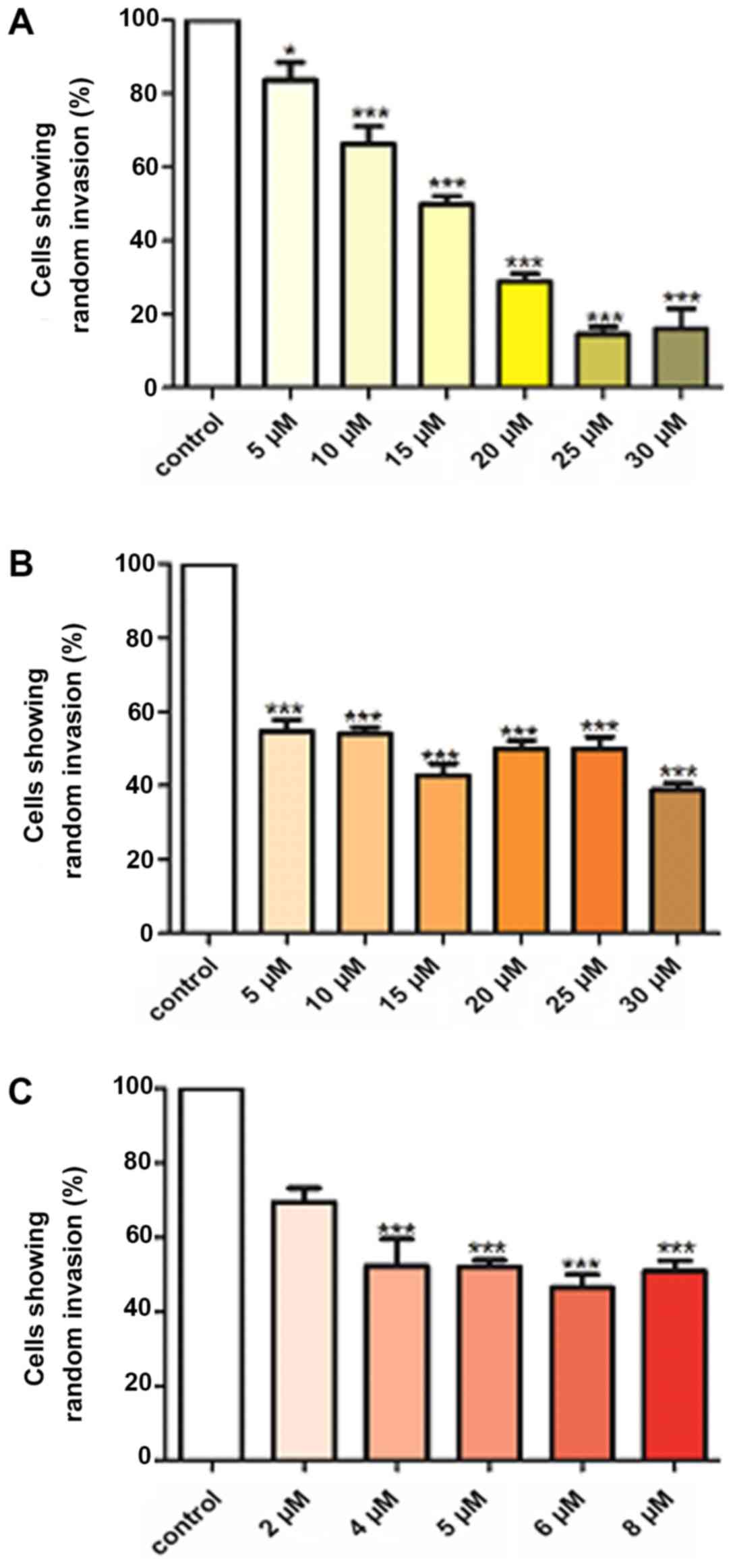
Fe(Cur)3 inhibit MDA-MB-231 cell random invasion
Curcumin, B(Cur)2 and Fe(Cur)3
significantly inhibited cell invasion (Fig. 5). Curcumin was the most effective,
inhibiting cell invasion by 16-84% at concentrations between 5 and
30 µM (Fig. 5A).
B(Cur)2 and Fe(Cur)3 inhibited cell invasion
by 45-56 and 48-53%, respectively (Fig. 5B and C).
Effects of curcumin, B(Cur)2
and Fe(Cur)3 on apoptosis- associated proteins
The relative expression levels of 35
apoptosis-related proteins were assessed. A total of 17 proteins
were detected in the MDA-MB-231 cell extracts according to the
proteome profiler assay (Table I).
The relative expression of the proapoptotic proteins cytochrome c
and cleaved caspase-3 was markedly increased in MDA-MB-231 cells
following treatment with B(Cur)2 and
Fe(Cur)3. Cell treatment with curcumin also increased
the expression of cytochrome c and cleaved caspase-3, but in a more
modest way. Furthermore, cell treatment with curcumin,
B(Cur)2 and Fe(Cur)3 decreased the expression
of the three antiapoptotic proteins cIAP-1, claspin and survivin by
<10-fold. In addition, an increase in the expression of haem
oxygenase-1 (HO-1) was observed in MDA-MB-231 cells following
treatment with curcumin, B(Cur)2 and
Fe(Cur)3, in particular with B(Cur)2 and
Fe(Cur)3 higher compared with curcumin.
 | Table IRegulation of pro-apoptotic and
anti-apoptotic proteins in MDA-MB-231 cell line following treatment
with curcumin, B(Cur)2 and Fe(Cur)3. |
Table I
Regulation of pro-apoptotic and
anti-apoptotic proteins in MDA-MB-231 cell line following treatment
with curcumin, B(Cur)2 and Fe(Cur)3.
| Protein | Effect on
apoptosis | Curcumin |
B(Cur)2 |
Fe(Cur)3 |
|---|
| Bad | Pro | ↓ | | ↓ |
| Pro-caspase 3 | Pro | ↓ | ↓ | ↓ |
| Cleaved caspase
3 | Pro | ↓ | ↑↑↑ | ↑↑↑ |
| Cytochrome c | Pro | ↓ | ↑↑ | ↑↑ |
| TRAIL R2/DR5 | Pro | ↓ | | ↓ |
| FADD | Pro | ↓ | ↓ | ↓ |
| HIF-1α | Pro | ↓ | | |
| HSP70 | Pro | ↓ | ↓ | ↓ |
| HTRA2/Omi | Pro | ↓ | ↓ | ↓ |
| Phospho-p53
(S15) | Pro | ↓ | ↓ | ↓ |
| Phospho-p53
(S46) | Pro | ↓ | ↓ | ↓ |
| Phospho-p53
(S392) | Pro | ↓ | ↓ | ↓ |
| SMAC/Diablo | Pro | ↓ | | ↓ |
| cIAP-1 | Anti | ↓ | ↓ | ↓ |
| Claspin | Anti | ↓ | ↓ | ↓ |
| HO-1 | Anti | ↑↑↑↑ | ↑↑↑↑↑ | ↑↑↑↑↑ |
| Survivin | Anti | ↓ | ↓ | ↓ |
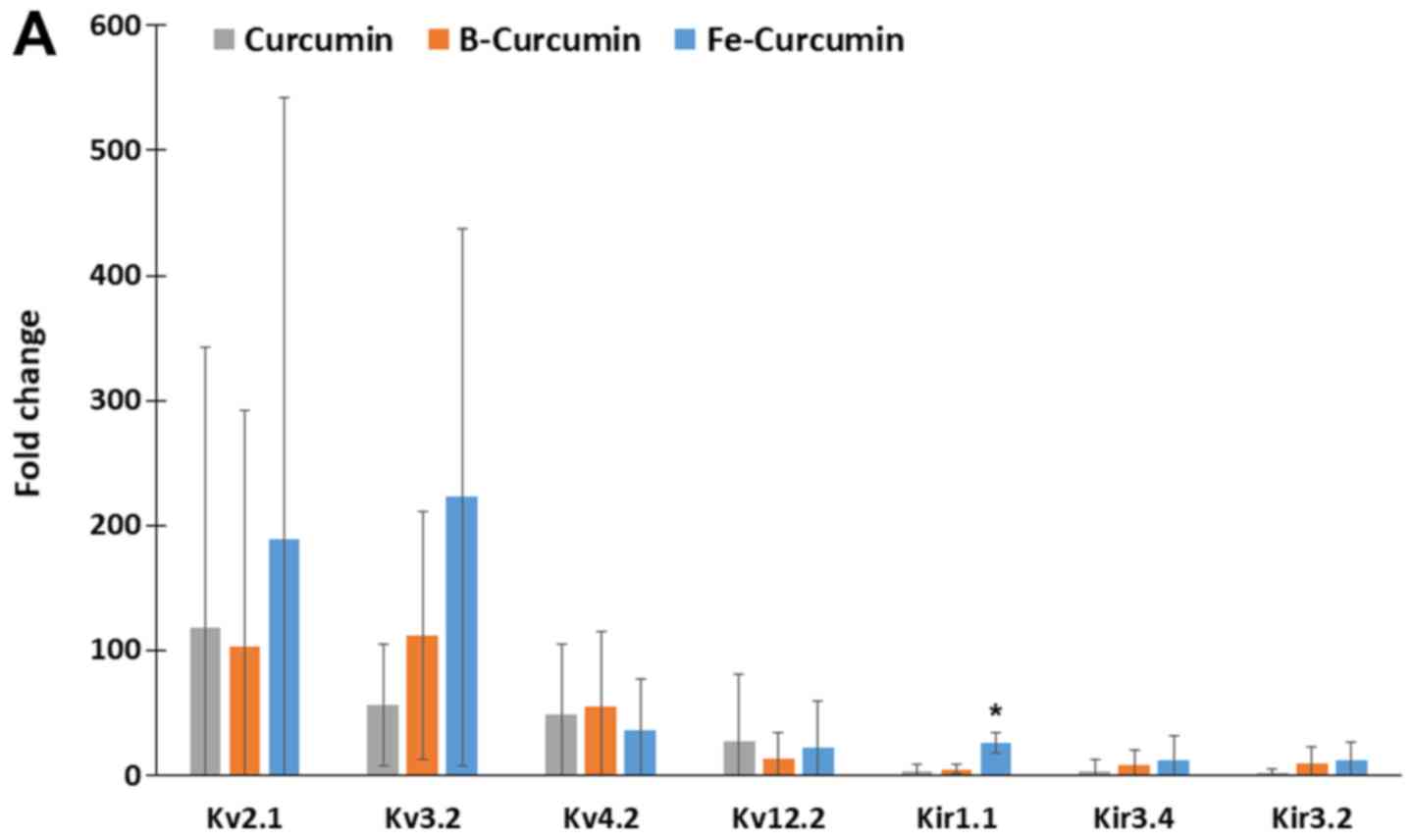
Ion channel gene expression
The relative expression of 84 ion-channel genes was
analysed using the ion channel array profiler. The relative
expression levels of 26 ion-channel genes were statistically
significantly different in cells treated with curcumin,
B(Cur)2 or Fe(Cur)3 compared with the
untreated control cells. Furthermore, MDA-MB-231 cells treated with
curcumin, B(Cur)2 and Fe(Cur)3 displayed
expression levels >10-fold higher for 16 ion channel genes,
including seven potassium channels and nine non-potassium channels,
compared with the untreated control cells (vehicle; Fig. 6A and B). The expression levels of
Kv2.1 and Kv3.2 were increased by >50-fold in
MDA-MB-cells treated with curcumin, B(Cur)2 or
Fe(Cur)3. Furthermore, cells treated with
B(Cur)2 displayed a 50-fold increase in Kv4.2
expression level compared with the control cells. In addition, the
relative expression levels of bestrophin-1 (BEST1) and calcium
voltage-gated channel auxiliary subunit γ4 (CACNG4) were increased
by >50-fold in cells treated with Fe(Cur)3 compared
with control cells.
Discussion
The present study demonstrated that curcumin,
B(Cur)2 and Fe(Cur)3 may be cytotoxic for
MDA-MB-231 cells. In addition to inhibiting cell proliferation, the
three compounds inhibited cell invasive ability; however, only
curcumin and B(Cur)2 reduced cell migratory ability.
As B(Cur)2 and Fe(Cur)3 are
new compounds, their effect on breast cancer cells requires further
investigation. It has been reported that curcumin localized
primarily in the perinuclear region of the cell (62). Despite structural differences,
B(Cur)2 and Fe(Cur)3 displayed a similar
pattern of cellular localization to curcumin in the present study.
Kunwar et al (63) reported
that curcumin localization was mainly at the plasma membrane,
followed by the cytoplasm and the nucleus in MCF-7 breast cancer
cells (63). Cytoplasmic
localization of iron-containing curcumin derivative has also been
reported in MCF-7 breast cancer cells (64). In the present study, curcumin,
B(Cur)2 and Fe(Cur)3 induced morphological
changes in MDA-MB-231 cells from a typical spindle shape to a
rounded structure with no visible blebs near the plasma membrane.
Ganguly et al (65) also
described this feature, suggesting that curcumin may modify the
cell shape and reduce cell attachment by downregulating focal
adhesion kinase expression.
To further investigate the cell death mechanism,
reverse transcription-quantitative PCR was performed to assess the
expression of ion channels, and apoptosis proteome profiling was
also conducted. The expression levels of the proapoptotic proteins
cleaved caspase 3 and cytochrome c were increased in cells treated
with curcumin, B(Cur)2 or Fe(Cur)3. The
mitochondrial release of cytochrome c induces the activation of the
caspase cascade associated with the intrinsic pathway of apoptosis
(66). The increased expression of
cytochrome c and cleaved caspase-3 reported in the present study
suggested that the intrinsic pathway of apoptosis was associated
with curcumin exposure in MDA-MB-231 cells. Furthermore,
phosphorylated p53 expression levels were slightly decreased
following treatment with B(Cur)2 and
Fe(Cur)3. Chiu and Su (14,40)
reported similar effect of curcumin on MDA-MB-231 cells, and
suggested that curcumin may induce apoptosis via a p53 independent
pathway or a p53-dependent Bax pathway.
In the present study, HO-1 expression level was
increased in MDA-MB-231 cells in response to treatment with
curcumin, B(Cur)2 and Fe(Cur)3. Previous
studies also reported that curcumin treatment induces HO-1
overexpression in vitro and in vivo (67-71).
In hepatoma epithelial cells, the induction of HO-1 expression
occurs via a mechanism involving increased oxidative stress and p38
activation (72), whereas in renal
epithelial cells, the mechanism involves activation of the
protein-1 activator transcription factor (73). Although the protein expression of
the antiapoptotic and proproliferative HO-1 was induced by curcumin
in the present study, previous studies indicated that curcumin
displays anticancer effects. It has been reported that high HO-1
expression favours cancer cell proliferation, poor prognosis and
resistance to therapy (74-82).
Furthermore, it was demonstrated that HO-1 is associated with
antiapoptotic (83,84), proangiogenic (85) and prometastatic (86) activities. HO-1 also promotes cancer
cell proliferation via a mechanism that is independent of its
catalytic activity, the HO-1 nuclear translocation-induced
alterations of gene transcription (87,88).
At present, HO-1 is considered as a potential therapeutic target
for various cancers (74,89). However, an increase in HO-1 enzyme
activity has also been reported to display anticancer effects
(90,91), including via the promotion of
apoptosis (92,93). Furthermore, it was reported that
HO-1 overexpression in breast cancer cell lines can inhibit cell
proliferation and invasive ability and induce apoptosis (94-96).
Similarly, Lee et al (48,69)
demonstrated that curcumin can induce HO-1 protein overexpression
in MDA-MB-231 cells, leading to decreased cell proliferation and
invasive ability.
In the present study, curcumin, B(Cur)2
and Fe(Cur)3 had variable effects on the expression of
certain ion channel genes. High gene expression levels were
observed for the Kv2.1, Kv3.2 and Kv4.2
potassium channels following treatment with curcumin,
B(Cur)2 or Fe(Cur)3. Furthermore, high
expression levels of CACNG4, which is a calcium channel coding
gene, and BEST1, which is a calcium-activated chloride channel
coding gene, were observed in MDA-MB-231 cells following treatment
with curcumin, B(Cur)2 and Fe(Cur)3.
Ion channel overexpression may result in a
proapoptotic and antiproliferative response by increasing the
movement of ions that favours a decrease in cell volume, which may
be involved in initiating apoptosis (43-45,48-51).
The cellular potassium efflux induces AVD and subsequent apoptotic
events. By downregulating potassium channel expression, cancer
cells can therefore evade apoptosis (44,53,97).
Numerous voltage-gated potassium channels are known to influence
AVD in cancer cells, including Kv1.1, Kv1.3,
Kv1.5, Kv2.1 and Kv11.1 (53), with several studies focusing on
Kv1.3 and Kv1.5 (51,52,54,98-101). In the present study, no
significant alteration in Kv1.3 and Kv1.5 expression
levels were observed; however, the expression level of
Kv2.1, which is a potassium channel associated with
apoptosis, was increased. Previous studies reported that neuronal
apoptosis requires a potassium efflux via the Kv2.1 channel
(97,102). Curcumin has also been reported to
reduce Kv2.1 potassium currents by modulating the
inactivation gating of this channel (57). Kv2.1 overexpression, which
was observed in the present study, may be a cellular response to
the inhibition of the function of this voltage-gates potassium
channel.
Cancer cell survival is partly attributed to the
evasion of apoptotic processes (39). In the present study, the results
from the proteome profile and ion channel expression analyses
identified two potential mechanisms underlying cancer cell
apoptosis evasion. The findings suggested that curcumin may
increase the expression level of HO-1 and Kv2.1 and that
HO-1 may counterbalance the effects of Kv2.1. HO-1 catalyses
the breakdown of haem to generate biliverdin, iron and carbon
monoxide (CO), which inhibit apoptosis. Subsequently, HO-1 and its
products may display some antiapoptotic effects in certain cells.
CO inhibits the potassium currents generated by the proapoptotic
Kv2.1 channel (49,50,103,104). The increase in Kv2.1
expression level observed in the present study may be a cellular
response to the HO-1-mediated inhibition of the Kv2.1
channel activity. However, the association between HO-1 and
Kv2.1 in breast cancer cells requires further
investigation.
Conversely with Kv2.1, only a few studies
reported an association between Kv3.2 and apoptosis. Lan
et al (105) reported high
relative expression of Kv1.3, Kv1.5, Kv1.6,
Kv2.1 and Kv2.2 in numerous gastric cancer cell
lines; however, Kv3.2 expression level was only just
detectable. The increased expression level of Kv3.2 observed
in the present study may therefore be specific to breast cancer
cells. Some Kv channels are considered oxygen sensors, and
Song et al (106)
demonstrated that Kv3.1 and Kv3.4 are tumour
hypoxia-related channels involved in cancer cell migratory and
invasive abilities. Kv3.1 and Kv3.4 protein
expression in A549 and MDA-MB-231 cells increase in a cell
density-dependent manner. When Kv3.1 and Kv3.4 were
inhibited using a Kv3 subfamily-specific blocker, both cell
migration and invasion were inhibited (106). In the present study, cell
treatment with Fe(Cur)3 induced the largest increase in
Kv3.2 expression; however, Fe(Cur)3 was the only
compound that failed to inhibit the breast cancer cell migratory
ability in the present study. The overexpression of Kv3.2 may be
linked to an attenuation in MDA-MB-231 cell ability to migrate.
In the present study, BEST1 was the only anion
channel that was significantly upregulated in response to curcumin
treatment. BEST1 is responsible for the production of an outward
flow of chloride ions, which counterbalances transient membrane
potentials induced by potassium efflux for cellular
electroneutrality (51,107). Several family members of the
calcium-activated chloride channels, including BEST1, have been
reported to promote cell proliferation and tumorigenesis (52,53,108,109). Furthermore, it was reported that
both potassium voltage-gated channel subfamily H member 1 and BEST1
are involved in the transformation of slow-growing T84
colonic carcinoma cells into fast-growing T84 cells
(109).
In the present study, the effects of curcumin,
B(Cur)2 and Fe(Cur)3 were evaluated in one
breast cancer cell line only. Although the MDA-MB-231 cell line
(36), has proven useful in
laboratory investigations of genetics, molecular biology and
biology of TNBC (37), this
represents a limitation to this study. Further investigation in
additional breast cancer cell lines, including MCF-7, is therefore
required. Both MCF-7 and MDA-MB-231 cell lines are invasive
ductal/breast carcinoma cells; however, these cells display
significant phenotypic and genotypic differences. MCF-7 is a
hormone dependent cell line due to its expression of ER and PR,
whereas the MDA-MB-231 cell line lacks ER and PR. Since MDA-MB-231
cells are highly metastatic and more aggressive compared with MCF-7
cells, the MDA-MB-231 cell line was the most appropriate cell line
for investigating the anticancer properties of curcumin compounds
in the present study.
Previous studies reported that B(Cur)2
and Fe(Cur)3 display improved photostability and high
fluorescence efficiency compared with curcumin (9,10,34,35).
The present study investigated whether curcumin complexes possessed
similar biological properties as curcumin. The results demonstrated
that Fe(Cur)3 was more effective at inhibiting cell
proliferation and random invasion at low doses compared with
curcumin. Conversely, B(Cur)2 was only effective at
inhibiting the cell migratory ability. B(Cur)2 and
Fe(Cur)3 displayed similar anticancer properties to
curcumin. Curcumin may serve as an adjuvant for chemotherapy. The
increase in Kv2.1, Kv3.2 and HO-1 highlight important
changes in molecular responses to curcumin induced inhibition of
proliferation and invasion of breast cancer cells. In conclusion,
the present study demonstrated that HO-1 and potassium channels may
be considered as important therapeutic targets in breast
cancer.
Funding
This study was supported by the RCSI (Bahrain)
internal research grants (grant no. BR00061) and the Dr Ali
Al-Khalifa Research Fund (Bahrain) (grant no. AK-2017).
Availability of data and material
The datasets used and/or analysed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
FM designed the study, performed cell and molecular
experiments and analysed data. FR had the original research idea,
developed the experimental design, conducted data analysis and
reading and revised the manuscript. ST performed cell and molecular
experiments and analysed data. SC synthesised the curcumin
complexes. SF was responsible for funding acquisition, data
curation and manuscript writing. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Blows FM, Driver KE, Schmidt MK, Broeks A,
van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO,
Blomqvist C, et al: Subtyping of breast cancer by
immunohistochemistry to investigate a relationship between subtype
and short and long term survival: A collaborative analysis of data
for 10,159 cases from 12 studies. PLoS Med. 7:pp. e10002792010,
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hugh J, Hanson J, Cheang MC, Nielsen TO,
Perou CM, Dumontet C, Reed J, Krajewska M, Treilleux I, Rupin M, et
al: Breast cancer subtypes and response to docetaxel in
node-positive breast cancer: Use of an immunohistochemical
definition in the BCIRG 001 trial. J Clin Oncol. 27:1168–1176.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nielsen TO, Hsu FD, Jensen K, Cheang M,
Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler
L, et al: Immunohistochemical and clinical characterization of the
basal-like subtype of invasive breast carcinoma. Clin Cancer Res.
10:5367–5374. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carey LA, Dees EC, Sawyer L, Gatti L,
Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML and Perou
CM: The triple negative paradox: Primary tumor chemosensitivity of
breast cancer subtypes. Clin Cancer Res. 13:2329–2334. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li X, Yang J, Peng L, Sahin AA, Huo L,
Ward KC, O'Regan R, Torres MA and Meisel JL: Triple-negative breast
cancer has worse overall survival and cause-specific survival than
non-triple-negative breast cancer. Breast Cancer Res Treat.
161:279–287. 2017. View Article : Google Scholar
|
|
8
|
Liedtke C, Mazouni C, Hess KR, André F,
Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B,
Green M, et al: Response to neoadjuvant therapy and long-term
survival in patients with triple-negative breast cancer. J Clin
Oncol. 26:1275–1281. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Massat NJ, Dibden A, Parmar D, Cuzick J,
Sasieni PD and Duffy SW: Impact of screening on breast cancer
mortality: The UK program 20 years on. Cancer Epidemiol Biomarkers
Prev. 25:455–462. 2016. View Article : Google Scholar
|
|
10
|
Kalager M, Zelen M, Langmark F and Adami
HO: Effect of screening mammography on breast-cancer mortality in
Norway. N Engl J Med. 363:1203–1210. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gonzalez-Angulo AM, Morales-Vasquez F and
Hortobagyi GN: Overview of resistance to systemic therapy in
patients with breast cancer. Adv Exp Med Biol. 608:1–22. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Choudhuri T, Pal S, Das T and Sa G:
Curcumin selectively induces apoptosis in deregulated cyclin
D1-expressed cells at G2 phase of cell cycle in a p53-dependent
manner. J Biol Chem. 280:20059–20068. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pal S, Bhattacharyya S, Choudhuri T, Datta
GK, Das T and Sa G: Amelioration of immune cell number depletion
and potentiation of depressed detoxification system of
tumor-bearing mice by curcumin. Cancer Detect Prev. 29:470–478.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shim JS, Kim JH, Cho HY, Yum YN, Kim SH,
Park HJ, Shim BS, Choi SH and Kwon HJ: Irreversible inhibition of
CD13/aminopeptidase N by the antiangiogenic agent curcumin. Chem
Biol. 10:695–704. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang X, Thomas DP, Zhang X, Culver BW,
Alexander BM, Murdoch WJ, Rao MN, Tulis DA, Ren J and Sreejayan N:
Curcumin inhibits platelet-derived growth factor-stimulated
vascular smooth muscle cell function and injury-induced neointima
formation. Arterioscler Thromb Vasc Biol. 26:85–90. 2006.
View Article : Google Scholar
|
|
16
|
Zeitlin P: Can curcumin cure cystic
fibrosis? N Engl J Med. 351:606–608. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Carroll RE, Benya RV, Turgeon DK, Vareed
S, Neuman M, Rodriguez L, Kakarala M, Carpenter PM, McLaren C,
Meyskens FL Jr and Brenner DE: Phase IIa clinical trial of curcumin
for the prevention of colorectal neoplasia. Cancer Prev Res
(Phila). 4:354–364. 2011. View Article : Google Scholar
|
|
18
|
He ZY, Shi CB, Wen H, Li FL, Wang BL and
Wang J: Upregulation of p53 expression in patients with colorectal
cancer by administration of curcumin. Cancer Invest. 29:208–213.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hejazi J, Rastmanesh R, Taleban FA, Molana
SH, Hejazi E, Ehtejab G and Hara N: Effect of curcumin
supplementation during radiotherapy on oxidative status of patients
with prostate cancer: A double blinded, randomized,
placebo-controlled study. Nutr Cancer. 68:77–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Epstein J, Sanderson IR and Macdonald TT:
Curcumin as a therapeutic agent: The evidence from in vitro, animal
and human studies. Br J Nutr. 103:1545–1557. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Durgaprasad S, Pai CG, Vasanthkumar,
Alvres JF and Namitha S: A pilot study of the antioxidant effect of
curcumin in tropical pancreatitis. Indian J Med Res. 122:315–318.
2005.
|
|
22
|
Sahin Kavaklı H, Koca C and Alıcı O:
Antioxidant effects of curcumin in spinal cord injury in rats. Ulus
Travma Acil Cerrahi Derg. 17:14–18. 2011. View Article : Google Scholar
|
|
23
|
Meng B, Li J and Cao H: Antioxidant and
antiinflammatory activities of curcumin on diabetes mellitus and
its complications. Curr Pharm Des. 19:2101–2113. 2013.
|
|
24
|
Lüer S, Troller R and Aebi C:
Antibacterial and antiinflammatory kinetics of curcumin as a
potential antimucositis agent in cancer patients. Nutr Cancer.
64:975–981. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Beevers CS and Huang S: Pharmacological
and clinical properties of curcumin. Botanics Targets Ther. 1:5–18.
2011.
|
|
26
|
Shishodia S, Sethi G and Aggarwal BB:
Curcumin: Getting back to the roots. Ann N Y Acad Sci.
1056:206–217. 2005. View Article : Google Scholar
|
|
27
|
Aggarwal BB, Kumar A and Bharti AC:
Anticancer potential of curcumin: Preclinical and clinical studies.
Anticancer Res. 23:363–398. 2003.PubMed/NCBI
|
|
28
|
Tønnesen HH, Karlsen J and van Henegouwen
GB: Studies on curcumin and curcuminoids. VIII. Photochemical
stability of curcumin. Z Lebensm Unters Forsch. 183:116–122. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang YJ, Pan MH, Cheng AL, Lin LI, Ho YS,
Hsieh CY and Lin JK: Stability of curcumin in buffer solutions and
characterization of its degradation products. J Pharm Biomed Anal.
15:1867–1876. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cartiera MS, Ferreira EC, Caputo C, Egan
ME, Caplan MJ and Saltzman WM: Partial correction of cystic
fibrosis defects with PLGA nanoparticles encapsulating curcumin.
Mol Pharm. 7:86–93. 2010. View Article : Google Scholar :
|
|
31
|
Liang G, Shao L, Wang Y, Zhao C, Chu Y,
Xiao J, Zhao Y, Li X and Yang S: Exploration and synthesis of
curcumin analogues with improved structural stability both in vitro
and in vivo as cytotoxic agents. Bioorg Med Chem. 17:2623–2631.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wanninger S, Lorenz V, Subhan A and
Edelmann FT: Metal complexes of curcumin-synthetic strategies,
structures and medicinal applications. Chem Soc Rev. 44:4986–5002.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Greish K, Pittalà V, Taurin S, Taha S,
Bahman F, Mathur A, Jasim A, Mohammed F, El-Deeb IM, Fredericks S
and Rashid-Doubell F: Curcumin-copper complex nanoparticles for the
management of triple-negative breast cancer. Nanomaterials (Basel).
8. pp. E8842018, View Article : Google Scholar
|
|
34
|
Mohammed F, Rashid-Doubell F, Cassidy S
and Henari F: A comparative study of the spectral, fluorometric
properties and photostability of natural curcumin, iron- and
boron-complexed curcumin. Spectrochim Acta A Mol Biomol Spectrosc.
183:439–450. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pröhl M, Schubert US, Weigand W and
Gottschaldt M: Metal complexes of curcumin and curcumin derivatives
for molecular imaging and anticancer therapy. Coord Chem Rev.
307:32–41. 2016. View Article : Google Scholar
|
|
36
|
Cailleau R, Olivé M and Cruciger QV:
Long-term human breast carcinoma cell lines of metastatic origin:
Preliminary characterization. In Vitro. 14:911–915. 1978.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chavez KJ, Garimella SV and Lipkowitz S:
Triple negative breast cancer cell lines: One tool in the search
for better treatment of triple negative breast cancer. Breast Dis.
32:35–48. 2010. View Article : Google Scholar
|
|
38
|
Osborne CK: Tamoxifen in the treatment of
breast cancer. N Engl J Med. 339:1609–1618. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chiu TL and Su CC: Curcumin inhibits
proliferation and migration by increasing the Bax to Bcl-2 ratio
and decreasing NF-kappaBp65 expression in breast cancer MDA-MB-231
cells. Int J Mol Med. 23:469–475. 2009.PubMed/NCBI
|
|
41
|
Lv ZD, Liu XP, Zhao WJ, Dong Q, Li FN,
Wang HB and Kong B: Curcumin induces apoptosis in breast cancer
cells and inhibits tumor growth in vitro and in vivo. Int J Clin
Exp Pathol. 7:2818–2824. 2014.PubMed/NCBI
|
|
42
|
Kim HI, Huang H, Cheepala S, Huang S and
Chung J: Curcumin inhibition of integrin (alpha6beta4)-dependent
breast cancer cell motility and invasion. Cancer Prev Res (Phila).
1:pp. 385–391. 2008, View Article : Google Scholar
|
|
43
|
Yee NS: Roles of TRPM8 Ion channels in
cancer: Proliferation, survival, and invasion. Cancers (Basel).
7:2134–2146. 2015. View Article : Google Scholar
|
|
44
|
Pardo LA and Stühmer W: The roles of K(+)
channels in cancer. Nat Rev Cancer. 14:39–48. 2014. View Article : Google Scholar
|
|
45
|
Urrego D, Tomczak AP, Zahed F, Stühmer W
and Pardo LA: Potassium channels in cell cycle and cell
proliferation. Philos Trans R Soc Lond B Biol Sci.
369:201300942014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Litan A and Langhans SA: Cancer as a
channelopathy: Ion channels and pumps in tumor development and
progression. Front Cell Neurosci. 9:862015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Djamgoz MB and Onkal R: Persistent current
blockers of voltage-gated sodium channels: A clinical opportunity
for controlling metastatic disease. Recent Pat Anticancer Drug
Discov. 8:66–84. 2013. View Article : Google Scholar
|
|
48
|
Wang Y, Yang Z, Meng Z, Cao H, Zhu G, Liu
T and Wang X: Knockdown of TRPM8 suppresses cancer malignancy and
enhances epirubicin-induced apoptosis in human osteosarcoma cells.
Int J Biol Sci. 10:90–102. 2013. View Article : Google Scholar
|
|
49
|
Hoffmann EK and Lambert IH: Ion channels
and transporters in the development of drug resistance in cancer
cells. Philos Trans R Soc Lond B Biol Sci. 369:201301092014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang M and Brackenbury WJ: Membrane
potential and cancer progression. Front Physiol. 4:1852013.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bortner CD and Cidlowski JA: Ion channels
and apoptosis in cancer. Philos Trans R Soc Lond B Biol Sci.
369:201301042014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Felipe A, Bielanska J, Comes N, Vallejo A
and Roig S: Targeting the voltage-dependent K(+) channels Kv1.3 and
Kv1.5 as tumor biomarkers for cancer detection and prevention. Curr
Med Chem. 19:661–674. 2012. View Article : Google Scholar
|
|
53
|
Szabò I, Zoratti M and Gulbins E:
Contribution of voltage-gated potassium channels to the regulation
of apoptosis. FEBS Lett. 584:2049–2056. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Szabó I, Bock J, Grassmé H, Soddemann M,
Wilker B, Lang F, Zoratti M and Gulbins E: Mitochondrial potassium
channel Kv1.3 mediates Bax-induced apoptosis in lymphocytes. Proc
Natl Acad Sci USA. 105:14861–14866. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Banderali U, Belke D, Singh A, Jayanthan
A, Giles WR and Narendran A: Curcumin blocks Kv11.1 (erg) potassium
current and slows proliferation in the infant acute monocytic
leukemia cell line THP-1. Cell Physiol Biochem. 28:1169–1180. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lian YT, Yang XF, Wang ZH, Yang Y, Yang Y,
Shu YW, Cheng LX and Liu K: Curcumin serves as a human kv1.3
blocker to inhibit effector memory T lymphocyte activities.
Phytother Res. 27:1321–1327. 2013. View Article : Google Scholar
|
|
57
|
Aréchiga-Figueroa IA, Delgado-Ramirez M,
Morán-Zendejas R and Rodriguez-Menchaca AA: Modulation of Kv2.1
channels inactivation by curcumin. Pharmacol Rep. 67:1273–1279.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Khalil MI, AL-Zahem AM and Qunaibit MM:
Synthesis, characterization, and antitumor activity of binuclear
curcumin-metal(II) hydroxo complexes. Med Chem Res. 23:1683–1689.
2014. View Article : Google Scholar
|
|
59
|
Skehan P, Storeng R, Scudiero D, Monks A,
McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S and Boyd MR:
New colorimetric cytotoxicity assay for anticancer-drug screening.
J Natl Cancer Inst. 82:1107–1112. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Heit B and Kubes P: Measuring chemotaxis
and chemokinesis: The under-agarose cell migration assay. Sci STKE.
2003:PL52003.PubMed/NCBI
|
|
61
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
62
|
Hope-Roberts M and Horobin RW: A review of
curcumin as a biological stain and as a self-visualizing
pharmaceutical agent. Biotech Histochem. 92:315–323. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kunwar A, Barik A, Mishra B, Rathinasamy
K, Pandey R and Priyadarsini KI: Quantitative cellular uptake,
localization and cytotoxicity of curcumin in normal and tumor
cells. Biochim Biophys Acta. 1780:673–679. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sarkar T, Butcher RJ, Banerjee S,
Mukherjee S and Hussain A: Visible light-induced cytotoxicity of a
dinuclear iron(III) complex of curcumin with low-micromolar IC50
value in cancer cells. Inorganica Chimica Acta. 439:8–17. 2016.
View Article : Google Scholar
|
|
65
|
Ganguly KK, Sen T, Pal S, Biswas J and
Chatterjee A: Studies on focal adhesion kinase in human breast
cancer cell MDA-MB-231. Adv Biol Chem. 2:29–42. 2012. View Article : Google Scholar
|
|
66
|
Garrido C, Galluzzi L, Brunet M, Puig PE,
Didelot C and Kroemer G: Mechanisms of cytochrome c release from
mitochondria. Cell Death Differ. 13:1423–1433. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xiao Y, Xia J, Wu S, Lv Z, Huang S, Huang
H, Su X, Cheng J and Ke Y: Curcumin inhibits acute vascular
inflammation through the activation of heme oxygenase-1. Oxid Med
Cell Longev. 2018:32958072018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wu SY, Lee YR, Huang CC, Li YZ, Chang YS,
Yang CY, Wu JD and Liu YW: Curcumin-induced heme oxygenase-1
expression plays a negative role for its anti-cancer effect in
bladder cancers. Food Chem Toxicol. 50:3530–3536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lee WY, Chen YC, Shih CM, Lin CM, Cheng
CH, Chen KC and Lin CW: The induction of heme oxygenase-1
suppresses heat shock protein 90 and the proliferation of human
breast cancer cells through its byproduct carbon monoxide. Toxicol
Appl Pharmacol. 274:55–62. 2014. View Article : Google Scholar
|
|
70
|
Park J and Conteas CN: Anti-carcinogenic
properties of curcumin on colorectal cancer. World J Gastrointest
Oncol. 2:169–175. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Balogun E, Hoque M, Gong P, Killeen E,
Green CJ, Foresti R, Alam J and Motterlini R: Curcumin activates
the haem oxygenase-1 gene via regulation of Nrf2 and the
antioxidant-responsive element. Biochem J. 371:887–895. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
McNally SJ, Harrison EM, Ross JA, Garden
OJ and Wigmore SJ: Curcumin induces heme oxygenase 1 through
generation of reactive oxygen species, p38 activation and
phosphatase inhibition. Int J Mol Med. 19:165–172. 2007.
|
|
73
|
Mimche PN, Taramelli D and Vivas L: The
plant-based immunomodulator curcumin as a potential candidate for
the development of an adjunctive therapy for cerebral malaria.
Malar J. 10(Suppl 1): pp. S102011, View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Nitti M, Piras S, Marinari UM, Moretta L,
Pronzato MA and Furfaro AL: HO-1 induction in cancer progression: A
matter of cell adaptation. Antioxidants(Basel). 6. pp. E292017,
View Article : Google Scholar
|
|
75
|
Jeon WK, Hong HY, Seo WC, Lim KH, Lee HY,
Kim WJ, Song SY and Kim BC: Smad7 sensitizes A549 lung cancer cells
to cisplatin-induced apoptosis through heme oxygenase-1 inhibition.
Biochem Biophys Res Commun. 420:288–292. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kongpetch S, Kukongviriyapan V, Prawan A,
Senggunprai L, Kukongviriyapan U and Buranrat B: Crucial role of
heme oxygenase-1 on the sensitivity of cholangiocarcinoma cells to
chemotherapeutic agents. PLoS One. 7:pp. e349942012, View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Lv X, Song DM, Niu YH and Wang BS:
Inhibition of heme oxygenase-1 enhances the chemosensitivity of
laryngeal squamous cell cancer Hep-2 cells to cisplatin. Apoptosis.
21:489–501. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tracey N, Creedon H, Kemp AJ, Culley J,
Muir M, Klinowska T and Brunton VG: HO-1 drives autophagy as a
mechanism of resistance against HER2-targeted therapies. Breast
Cancer Res Treat. 179:543–555. 2020. View Article : Google Scholar :
|
|
79
|
Zhao Z, Xu Y, Lu J, Xue J and Liu P: High
expression of HO-1 predicts poor prognosis of ovarian cancer
patients and promotes proliferation and aggressiveness of ovarian
cancer cells. Clin Transl Oncol. 20:491–499. 2018. View Article : Google Scholar
|
|
80
|
Becker JC, Fukui H, Imai Y, Sekikawa A,
Kimura T, Yamagishi H, Yoshitake N, Pohle T, Domschke W and
Fujimori T: Colonic expression of heme oxygenase-1 is associated
with a better long-term survival in patients with colorectal
cancer. Scand J Gastroenterol. 42:852–858. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Loboda A, Jozkowicz A and Dulak J: HO-1/CO
system in tumor growth, angiogenesis and metabolism-targeting HO-1
as an anti-tumor therapy. Vascul Pharmacol. 74:11–22. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Maines MD and Abrahamsson PA: Expression
of heme oxygenase-1 (HSP32) in human prostate: Normal,
hyperplastic, and tumor tissue distribution. Urology. 47:727–733.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Busserolles J, Megías J, Terencio MC and
Alcaraz MJ: Heme oxygenase-1 inhibits apoptosis in Caco-2 cells via
activation of Akt pathway. Int J Biochem Cell Biol. 38:1510–1517.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Tanaka S, Akaike T, Fang J, Beppu T, Ogawa
M, Tamura F, Miyamoto Y and Maeda H: Antiapoptotic effect of haem
oxygenase-1 induced by nitric oxide in experimental solid tumour.
Br J Cancer. 88:902–909. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Cherrington JM, Strawn LM and Shawver LK:
New paradigms for the treatment of cancer: The role of
anti-angiogenesis agents. Adv Cancer Res. 79:1–38. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Price JT and Thompson EW: Mechanisms of
tumour invasion and metastasis: Emerging targets for therapy.
Expert Opin Ther Targets. 6:217–233. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Lin Q, Weis S, Yang G, Weng YH, Helston R,
Rish K, Smith A, Bordner J, Polte T, Gaunitz F and Dennery PA: Heme
oxygenase-1 protein localizes to the nucleus and activates
transcription factors important in oxidative stress. J Biol Chem.
282:20621–20633. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Biswas C, Shah N, Muthu M, La P, Fernando
AP, Sengupta S, Yang G and Dennery PA: Nuclear heme oxygenase-1
(HO-1) modulates subcellular distribution and activation of Nrf2,
impacting metabolic and anti-oxidant defenses. J Biol Chem.
289:26882–26894. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Was H, Dulak J and Jozkowicz A: Heme
oxygenase-1 in tumor biology and therapy. Curr Drug Targets.
11:1551–1570. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Duckers HJ, Boehm M, True AL, Yet SF, San
H, Park JL, Clinton Webb R, Lee ME, Nabel GJ and Nabel EG: Heme
oxygenase-1 protects against vascular constriction and
proliferation. Nat Med. 7:693–698. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Taillé C, Almolki A, Benhamed M, Zedda C,
Mégret J, Berger P, Lesèche G, Fadel E, Yamaguchi T, Marthan R, et
al: Heme oxygenase inhibits human airway smooth muscle
proliferation via a bilirubin-dependent modulation of ERK1/2
phosphorylation. J Biol Chem. 278:27160–27168. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ozawa N, Goda N, Makino N, Yamaguchi T,
Yoshimura Y and Suematsu M: Leydig cell-derived heme oxygenase-1
regulates apoptosis of premeiotic germ cells in response to stress.
J Clin Invest. 109:457–467. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Liu XM, Chapman GB, Wang H and Durante W:
Adenovirus-mediated heme oxygenase-1 gene expression stimulates
apoptosis in vascular smooth muscle cells. Circulation. 105:79–84.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Hill M, Pereira V, Chauveau C, Zagani R,
Remy S, Tesson L, Mazal D, Ubillos L, Brion R, Asghar K, et al:
Heme oxygenase-1 inhibits rat and human breast cancer cell
proliferation: Mutual cross inhibition with indoleamine
2,3-dioxygenase. FASEB J. 19:1957–1968. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lin CW, Shen SC, Hou WC, Yang LY and Chen
YC: Heme oxygenase-1 inhibits breast cancer invasion via
suppressing the expression of matrix metalloproteinase-9. Mol
Cancer Ther. 7:1195–1206. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Andreadi CK, Howells LM, Atherfold PA and
Manson MM: Involvement of Nrf2, p38, B-Raf, and nuclear
factor-kappaB, but not phosphatidylinositol 3-kinase, in induction
of hemeoxy-genase-1 by dietary polyphenols. Mol Pharmacol.
69:1033–1040. 2006. View Article : Google Scholar
|
|
97
|
Pal SK, Takimoto K, Aizenman E and Levitan
ES: Apoptotic surface delivery of K+ channels. Cell Death Differ.
13:661–667. 2006. View Article : Google Scholar :
|
|
98
|
Comes N, Bielanska J, Vallejo-Gracia A,
Serrano-Albarrás A, Marruecos L, Gómez D, Soler C, Condom E, Ramón
Y, Cajal S, et al: The voltage-dependent K(+) channels Kv1.3 and
Kv1.5 in human cancer. Front Physiol. 4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lowinus T, Heidel FH, Bose T, Nimmagadda
SC, Schnöder T, Cammann C, Schmitz I, Seifert U, Fischer T,
Schraven B and Bommhardt U: Memantine potentiates
cytarabine-induced cell death of acute leukemia correlating with
inhibition of Kv1.3 potassium channels, AKT and ERK1/2 signalin.
Cell Commun Signal. 17:52019. View Article : Google Scholar
|
|
100
|
Teisseyre A, Palko-Labuz A, Sroda-Pomianek
K and Michalak K: Voltage-gated potassium channel Kv1.3 as a target
in therapy of cancer. Front Oncol. 9:9332019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wu J, Chen Z, Liu Q, Zeng W, Wu X and Lin
B: Silencing of Kv1.5 gene inhibits proliferation and induces
apoptosis of osteo-sarcoma cells. Int J Mol Sci. 16:26914–26926.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Pal S, Hartnett KA, Nerbonne JM, Levitan
ES and Aizenman E: Mediation of neuronal apoptosis by Kv2.1-encoded
potassium channels. J Neurosci. 23:4798–4802. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Al-Owais MM, Dallas ML, Boyle JP, Scragg
JL and Peers C: Heme oxygenase-1 influences apoptosis via
CO-mediated inhibition of K+ channels. Adv Exp Med Biol.
860:343–351. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Al-Owais MM, Scragg JL, Dallas ML, Boycott
HE, Warburton P, Chakrabarty A, Boyle JP and Peers C: Carbon
monoxide mediates the anti-apoptotic effects of heme oxygenase-1 in
medulloblastoma DAOY cells via K+ channel inhibition. J Biol Chem.
287:24754–24764. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Lan M, Shi Y, Han Z, Hao Z, Pan Y, Liu N,
Guo C, Hong L, Wang J, Qiao T and Fan D: Expression of delayed
rectifier potassium channels and their possible roles in
proliferation of human gastric cancer cells. Cancer Biol Ther.
4:1342–1347. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Song MS, Park SM, Park JS, Byun JH, Jin
HJ, Seo SH, Ryu PD and Lee SY: Kv3.1 and Kv3.4, are involved in
cancer cell migration and invasion. Int J Mol Sci. 19:pp.
E10612018, View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Schwab A, Fabian A, Hanley PJ and Stock C:
Role of ion channels and transporters in cell migration. Physiol
Rev. 92:1865–1913. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Duvvuri U, Shiwarski DJ, Xiao D, Bertrand
C, Huang X, Edinger RS, Rock JR, Harfe BD, Henson BJ, Kunzelmann K,
et al: TMEM16A induces MAPK and contributes directly to
tumorigenesis and cancer progression. Cancer Res. 72:3270–3281.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Spitzner M, Martins JR, Soria RB,
Ousingsawat J, Scheidt K, Schreiber R and Kunzelmann K: Eag1 and
Bestrophin 1 are up-regulated in fast-growing colonic cancer cells.
J Biol Chem. 283:7421–7428. 2008. View Article : Google Scholar : PubMed/NCBI
|