Epigenetic modifications, including DNA methylation
and histone modifications, play an important role in gene
regulation. The dysregulation of these modifications can result in
pathogenicity, including tumorigenicity. Research has indicated an
important influence of the trimethylation modification at lysine 27
on histone H3 (H3K27me3) within chromatin. This methylation is
involved in the repression of multiple genes within the genome by
condensing DNA to reduce access to the transcription start site
(TSS) within gene promoter sequences (1). The recruitment of H1.2, an H1 histone
subtype, by the H3K27me3 modification has been a suggested as a
mechanism for mediating this compaction (1). The present review discusses the
function of the H3K27me3 regulator, polycomb repressive complex 2
(PRC2), and its contribution to cancer, specifically highlighting
alterations in breast cancer.
PRC2 is a multi-subunit protein complex that
mediates the mono-, di and tri-methylation of H3K27 to silence gene
expression (1-6). First established in
Drosophila, the poly-comb group (PcG) genes are well
conserved across species through to human cells (7-10).
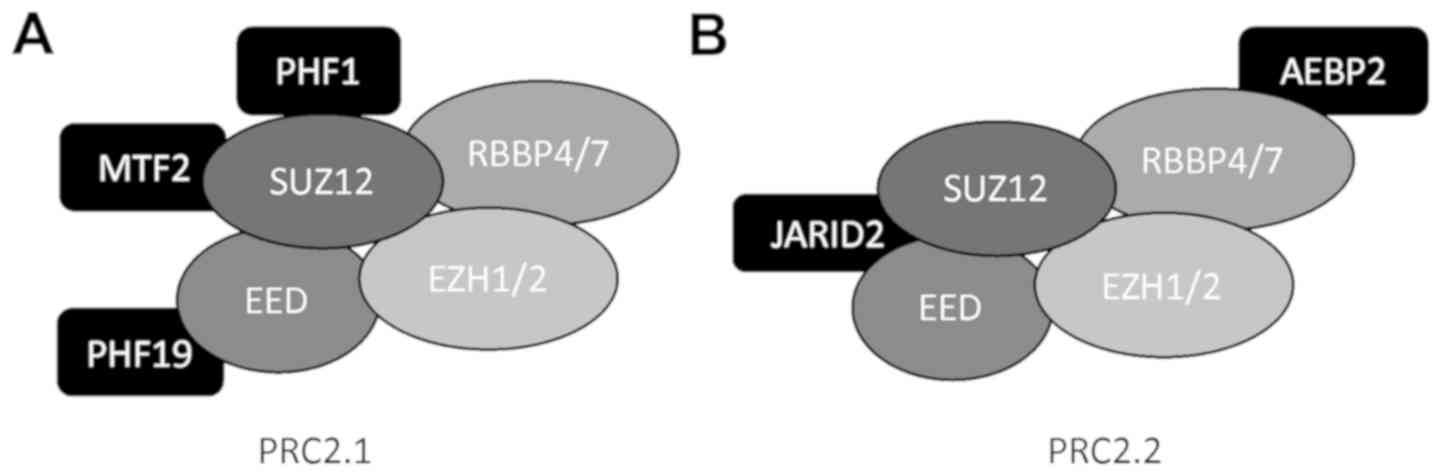
The PRC2 complex contains 4 core subunits, including the enhancer
of zeste homolog 1 or 2 (EZH1/2), embryonic ectoderm development
(EED), suppressor of zeste 12 (SUZ12) and retinoblastoma associated
protein 46/48, (RbAP46/48), also known as RBBP4/7 (Fig. 1) (9,11).
This core complex can additionally interact with accessory proteins
consisting of the jumonji, AT-rich interactive domain 2 (JARID2),
AE binding protein 2 (AEBP2) and polycomb-like protein (PCL)
(11-15). Together, these subunits can form a
holo-PRC2 complex to efficiently methylate histone H3. In general,
approximately 70% of H3 histones can be methylated by PRC2,
indicating the main regulatory role PRC2 has within the genome
(4).
Although there are 4 core subunits of PRC2, only
SUZ12, EED and EZH2 are required for the basic function of this
complex (16-18). Within the core complex, there are
two functional lobes that govern catalytic and regulatory duties.
The catalytic lobe consists of the VEFS domain in SUZ12 and the
SANT1L/2L, CXC motif and suppressor of variegation, enhancer of
zeste, trithorax (SET) domains all within the EZH2 subunit
(19). The methyltransferase
activity of PRC2 has been specifically linked to the SET domain of
EZH2 (2,3,5,20).
By contrast, the regulatory lobe consists of the SR motif and SET
activation loop domain (SAL) of the EED subunit (19). The SAL domain is required for
catalytic function by playing a main role in the coordination of
these two lobes (19,21). The interactions between these
domains allow EED to recognize the presence of the H3K27me3
modification through the WD40 domain and stimulate the
methyltransferase activity produced through the EZH2 subunit
(17,22). Overall, the EED subunit is
important for reading the modifications, SUZ12 is important for the
stabilization of this interaction and EZH2 is the methyltransferase
subunit. When individually isolated, these subunits do not have
PRC2 methyltransferase activity, suggesting that these peptides
have to cooperate to accomplish their functions (18,23).
The interaction between EED and EZH2 is facilitated
through the EED binding domain (EBD) of EZH2 (24). This interaction entails the
formation of a β-propeller through a 7 WD40 repeat sequence on the
EED subunit to establish Van der Waals interactions and hydrogen
bonds with the EBD (24). EZH2
envelops EED through multiple associations, closely linking these
subunits structurally and as a result, functionally. In general,
methyltransferase activity is initiated by the EED WD40 domain
reading the H3K27me3 modification to induce a rotational change of
the PRC2 confirmation and signals to the EZH2 subunit to activate
catalytic function (19). The
knockdown of EED is associated with a global reduction in H3K27me3
levels (25).
The second subunit, SUZ12, is involved in the
stabilization of the PRC2 complex during catalytic activity
(26). This stabilizing action is
considered to be specifically linked to the VEFS domain of SUZ12
(27). More detailed research is
required for the full analysis of processes involved with this
subunit; however, the overall importance of this subunit is upheld
by the observation that the knockdown of SUZ12 is also associated
with a global reduction in H3K27me3 levels (18,26).
The third subunit, EZH1/2, is considered the writer
protein in which the catalytic function of PRC2 is carried out
through. This peptide is a methyltransferase that adds a methyl
group to lysine 27 on histone H3 through activity within the SET
domain (21,28). Both EZH1 and EZH2 have the capacity
to form PRC2 complexes, but may not be completely interchangeable.
EHZ2 has been observed to have a higher methyltransferase activity
compared to EZH1 (29). For
instance, EZH1 has been observed to specialize in only the mono-
and di-methylation of H3K27, while EZH2 catalyzes mono-, di- and
trimethylations (30). In addition
to this complex, EZH2 may have PRC2-independent activity that will
be discussed below in the present review.
Finally, some research considers RBBP4/7 part of the
core subunits of PRC2, while others consider it as an accessory
protein. In general, these peptides play a role in the
stabilization and regulation of PRC2-chromatin interactions by
binding the unmethylated histone H3K4 tail (31). This binding is impaired when
H3K4me3 is present, therefore indicating to the PRC2 complex that
the gene is currently active (31,32).
Although only SUZ12, EED and EZH2 are required for any enzymatic
activity, RBBP4/7 may be considered a core subunit due to the
control it contributes to the complex (18,23,33).
Furthermore, RBBP4 and RBBP7 are part of the WD40 repeat protein
family that acts to stabilize the PRC2-chromatin interaction. This
occurs through the simultaneous binding of PRC2, H3 and H4
(31,34,35).
The binding of H4 by RBBP4/7 causes a structural change to unfold
helix 1 of H4 to support this process (34). Within the PRC2 complex, RBBP7 is
closely associated with SUZ12, but only weakly to EED (16). Moreover, this incorporation into
the PRC2 complex is SUZ12-dependent (16). Functionally, RBBP4 and RBBP7 are
similar and have been studied closely together.
The accessory proteins interact with the core
subunits ultimately to enhance PRC2 function by increasing
stability, regulating catalytic function or aiding in the
recruitment of the PRC2 complex to a locus. Accessory proteins that
interact with the PRC2 complex include JARID2, AEBP2 and PCLs.
Finally, PCL is a group of peptides that contain
Tudor domains that can bind H3K36me3 markers, known to increase
PRC2 recruitment (43,44). Recruitment processes will be
discussed in detail in the following section. Overall, there are 3
mammalian PCLs important for PRC2 activity. These include PHF1,
MTF2 and PHF19, also known as PCL1, PCL2 and PCL3, respectively
(12,45). They all contain Tudor domains, 2
plant homeodomain fingers, an extended homologous region N terminal
cluster and C terminal Chromo-like domain (46). These proteins may be particularly
associated with promoting new PcG modifications of genes not
previously silenced. More recently, PHF20L1, another TUDOR domain
containing protein has been identified as a H3K27me2 reader that
can recruit PRC2 (47).
The mapping of the PRC2 complex has revealed that
these accessory proteins form 2 distinct complexes, identified as
PRC2.1 and PRC2.2 (Fig. 1)
(12). PRC2.1 was identified as
containing PCLs as the accessory proteins, while PRC2.2 exclusively
contained JARID2 and AEBP2 (Fig.
1) (12). Further analysis
suggested that PRC2.1 was involved in the de novo
recruitment of PRC2 to CpG islands that lack H3K27me3 (46,48).
By contrast, PRC2.2 has been suggested to be involved in
methyltransferase activity through recruitment to chromatin that
has PcG-dependent modifications, H3K119ub or H3K27me3, already
present (39,41).
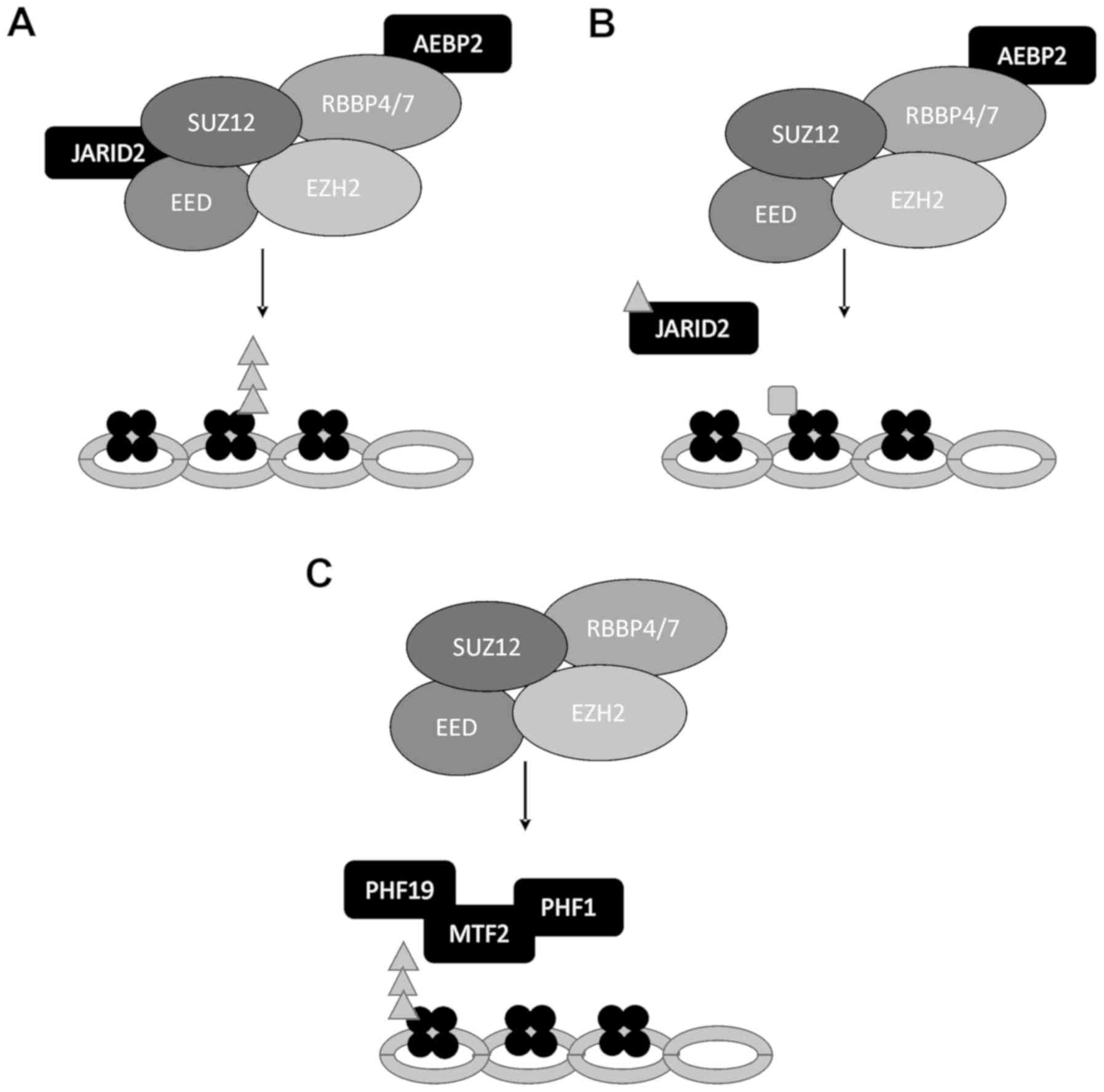
Once H3K27 is trimethylated, the histone must remain
in this state to continue the repression of the gene at hand,
avoiding improper transcription. When an H3K27me3 is already
present, PRC2 can be recruited to the chromatin, allowing EED to
read the marker (Fig. 2A)
(22,49,50).
A positive feedback loop can then be initiated to stimulate PRC2
methyltransferase activity. This activation can propagate the
H3K27me2/3 marker in cis and far cis through
long-range contacts (50). In
general, this method of recruitment is involved in the maintenance
and propagation of H3K27me3, although this is not the sole form of
recruitment.
PRC1, another PcG complex, also plays a significant
role during the recruitment of PRC2. PRC1 and PRC2 have an
interdependent association where they can recruit one another. This
is supported by the co-localization of PRC1 and PRC2 to
approximately 10-15% of all genes (51-54).
PRC1 is an ubiquitin ligase that can be recruited to H3K27me3 by
chromobox subunits (CBX), CBX7 and CBX8 (55,56).
Subsequently, PRC1 recruitment and activation initiate the
ubiquitination of lysine 119 at histone 2A (H2AK119ub1) (40,41).
As a result, the ubiquitination marker can recruit and activate
PRC2, again spreading H3K27 trimethylation (Fig. 2B) (41,57,58).
TRIM37 has also been observed to ubiquitinate H2AK119 (59). This protein binds both PRC1 and
PRC2, specifically targeting genes for silencing (59). Although the mechanism of PRC2
recruitment by TRIM37 has not yet been fully established, the
knockdown of TRIM37 induces the re-expression of silenced genes due
to the dissociation of PRC1 and PRC2 from the target locus
(59).
Overall, recruitment to sites with H3K27me3 or
H2AK119ub already present occurs through JARID2 (39,41).
This is largely a PRC1-dependent process in which JARID2 can dock
on DNA by interacting with the H3K119ub modification (39,41).
JARID2 then catalytically stimulates the PRC2 complex through the
methylation of JARID2 at lysine 116 (Fig. 2B) (60). The function of this modification is
to allow JARID2 to bind EED at the WD40 domain, inducing a
conformational change and leading to the increased catalytic
activity of EZH2 (60). Additional
interactions of other proteins within the holo-protein complex help
stabilize the PRC2 interaction with the chromatin. This includes
JARID2, AEBP2, RBBP4/7 with histone H3 or H4, EED and SUZ12.
Without H3K27me3, the recruitment of PRC2 occurs
through the presence of different accessory proteins. While PRC2.2,
including JARID2 and AEBP2 can be recruited effectively through
H3K27me3 or H2AK119ub markers, de novo recruitment appears
to be limited to the PRC2.1 complex containing PCLs (46,48).
This recruitment appears to occur at CpG islands (CGI), an area of
DNA that has a high frequency of cytosine and guanine (Fig. 2C) (46,48).
This area may have fewer nucleosomes; however, this observation is
not a direct predictor of recruitment (61). More specifically, polycomb response
elements (PREs) within these CGI are suggested to recruit PRC2 and
PRC1 (62). These have been well
characterized in Drosophila; however, identification and
characterization within mammals have yet to be confirmed (63). More distinguished within mammalian
cells is the de novo recruitment of PRC2 through the
H3K36me3 modification (Fig. 2C).
H3K4me3 in combination with the H3K36me3 modification is associated
with active genes (64). PRC2 may
use the H3K36me3 modification to create balance within the
chromatin, initiating the silencing of genes that were previously
active. PCL proteins are specifically involved in this de
novo recruitment. PHF19 can interact with H3K36me3 through its
Tudor domain to create a contact point to recruit PRC2 (Fig. 2C) (43,65,66).
Additionally, the demethylase, NO66, is recruited through PHF19
interactions with H3K36me3, overall resulting the gain of H3K27me3
and the loss of methylation at H3K36 by NO66 activity (65). MTF2 has also been observed to bind
CGIs in the presence of H3K36me3 and is an essential recruiter for
PRC2 through de novo mechanisms in both mice and in
vitro models (46,48). Both MTF2 and PHF1 are observed to
fold into a winged helix to successfully bind unmethylated CpG
motifs (Fig. 2C) (46). Once PRC2 is successfully recruited,
a positive feedback loop through the EED subunit sustains the
methyltransferase activity at the CGIs to promote and propagate
H3K27me3 (22).
Finally, long non-coding RNAs (lncRNAs) may
additionally contribute to the recruitment of PRC2. A lncRNA termed
HOX transcript antisense RNA (HOTAIR) has been identified to
promote PRC2 binding in trans (67,68).
The HOTAIR-PRC2 mechanism seems to regulate specific genes,
including HOXD10, PCDH10, PCDHBS, APC2
and NLK (69,70). HOTAIR provides scaffolding
for histone modification enzymes including PRC2 and LSD1, a lysine
4 demethylase (68). The binding
of these enzymes occurs at the 5′ and 3′ ends of HOTAIR,
respectively (68). Alterations to
HOTAIR levels have been observed in breast cancers, which
will be addressed in detail below.
The altered regulation of genes is an important
contributor to the pathogenicity of cancer. Since PRC2 alters
epigenetic markers to repress genes, this complex may act to
silence tumor suppressors, advancing tumorigenesis. Due to PRC2
targeting multiple genes, pin-pointing individual genes being
silenced that directly contribute to cancer progression is a
complex task. Some highlighted alterations in breast cancer that
research has discovered are described below; however, it is
unlikely this is the entirety of the effects of this complex.
The proliferation of tumor cells is reduced when
PRC2 subunits are knocked down, suggesting that this complex plays
a role in breast tumor growth (20,71).
Specific genes linked to this process have yet to be identified
within breast cancer cells, but are overall linked to EZH2
overexpression (20,72,73).
The levels of positive regulators of the cell cycle, including
cyclinD1, cyclinE1, cyclinA2 and cyclinB1 are significantly reduced
in cells that lack EZH2 expression (71). EZH2, as well as EED and SUZ12 are
downstream targets of the pRB-E2F pathway, in that E2F can regulate
the expression of these PCR2 subunits (71). Additionally, EED has also been
identified as critical for proliferation, as the inhibition of
EED-EZH2 inter-action impairs tumor cell proliferation, thus
providing further evidence that PRC2 activity facilitates
proliferation (71,74).
Metastatic breast cancer cells within adjacent lymph
nodes have been shown to overexpress PRC2, which could suggest
cells have acquired this alteration to gain the capacity to
successfully metastasize (75).
Within the primary tumor, matrix metallopeptidases (MMPs) play a
key role in the degradation of the extracellular matrix (ECM) in
the tumor microenvironment (TME) (76). The degradation of the ECM results
in a higher frequency of the invasion and metastasis of breast
cancer (77). Elevated EZH2 levels
are associated with the repression of tissue inhibitors
metalloproteinases (TIMPs) in triple-negative breast cancer (TNBC)
(78). Specifically, EZH2
represses TIMP expression through the induction of the
H3K27me3 silencing modification (78). Without TIMP proteins, MMP2 and MMP9
activity is increased, thus potentiating the metastatic capacity of
breast cancer cells (78). This
has also been observed in other types of cancer, such as ovarian
and prostate cancer (79,80).
Another potential target of the PRC2 complex that
affects metastatic potential is the repression of growth
differentiation factor-15 (GDF15) (81). Yes-associated protein (YAP) is an
upstream regulator of the PRC2 complex that promotes trimethylation
of H3K27 at the GDF15 promoter, leading to the suppression
of GDF15 (81). When YAP is
knocked down, the metastatic potential is decreased, suggesting
that this recruitment of PRC2 to GDF15 may affect the
aggressiveness of breast cancer cells (81). YAP expression and its role
in breast cancers have been controversial with some research
indicating a tumor-suppressive role, while others suggesting an
oncogenic role (81-84).
Compounds that suppress PRC2 activity, such as
3-deazaneplanocin A (DZNeP), have been observed to re-induce
apoptosis, leading to targeted cell death (85). F-box protein 32 (FBXO32) has been
identified as a key effector of apoptosis in this process (85). Due to this re-expression, it can be
suggested that during abnormal PRC2 activity, apoptosis is
suppressed by reducing FBXO32 levels. Further support for a role of
PRC2 in apoptosis includes the observation that elevated levels of
EZH2 are associated with reduced apoptosis following DNA damage,
while decreased levels of EZH2 potentially facilitate apoptosis
(86-89). Mechanistically, FBXO32 is repressed
by EZH2 (86). FBXO32 normally
directly binds p21 for protease degradation within p53-proficient
cells and additionally induces apoptosis in p53-deficient cells
through CHK1 activation (86).
When EZH2 is depleted, G1 and G2/M checkpoints of the cell cycle
are inhibited, leading to the initiation of apoptosis with or
without p53 involvement (86).
Another suggested pathway for evading apoptosis
involves the deregulation of the pRB/E2F pathway. When EZH2 is
activated by E2F1, E2F1-mediated apoptosis is suppressed (87). This regulation of EZH2 leads to
methyltransferase activity at the promoter of BIM1 to induce
the trimethylation of H3K27, and furthermore, to suppress
BIM1 expression (87). This
process, induced by the pRB/E2F pathway, suggests that suppressing
BIM1 is a potential alteration that occurs in cancer cells
to escape apoptosis through epigenetic regulations. Within clinical
cases, a high EZH2 expression is associated with a poor
outcome, while BMI1 overexpression is associated with good
outcomes (90).
Although EZH2 is a core PRC2 protein, it may also
function independent of the PRC2 complex (95,96).
It has been suggested that EZH2 forms an additional complex with
foxhead box M1 (FOXM1) under hypoxic conditions (97). Hypoxia-inducible factor 1-alpha
(HIF1A) induces EZH2 expression, but suppresses SUZ12
and EED expression and this scenario of elevated EZH2
protein levels in the absence of SUZ12 and EED promotes EZH2-FOXM1
interaction, resulting in the elevated expression of MMPs and
increases tumor invasion (97).
With this in mind, research that aims to address the effects of the
PRC2 complex on cancer progression cannot solely measure EZH2
activity and increased H3K27me3 levels should be measured to
confirm PRC2 activity.
lncRNAs have been emphasized more recently as
regulators of PRC2 activity. The long non-coding RNA termed
HOTAIR has been observed to promote PRC2 binding by acting
as a scaffold (68). The
dysregulation of this lncRNA has been observed during the
progression of breast cancer and is associated with poorer clinical
outcomes (98,99). Particularly, the increased
expression of HOTAIR is observed in primary breast cancers
and metastases, and predicts the detachment of tumor cells,
potentially leading to metastasis (69). By contrast, the knock-down of
HOTAIR represses migration, invasion and metastasis within
in vitro and in vivo models (69). Another HOX transcript antisense RNA
termed HOTAIRM1 has recently been implicated in promoting
tamoxifen resistance in breast cancer through inhibition of PRC2
activity (100).
The miR-200 family is a family of small non-coding
RNAs consisting of miR-200a, miR-200b, miR-200c. miR-141 and
miR-429 that are expressed as two clusters, the miR-200c/141 and
miR-200b/200a/429 cluster (105-107). miRNAs are involved in the
post-transcriptional regulation of genes and inhibit the
translation of target genes (108-110). Research from the authors'
laboratory and those of other researchers has indicated that
miR-200s are expressed at high levels in luminal breast cancer and
luminal breast cancer cell lines; however, their expression is
significantly reduced in TNBC and TNBC cell lines (111-117). The miR-200 family negatively
regulates the expression of genes involved in EMT, such as
TWIST1/2, ZEB1/2 and SNAI1/2 (108,118-120). Additionally, SUZ12 may
also be an important target of the miR-200 family. SUZ12
translation is directly inhibited by miR-200b (121,122) and thus modifications resulting in
the reduced expression of miR-200b in breast cancer cells promote
elevated SUZ12 levels and increased H3K27me3 levels by PRC2.
Increased PRC2 activity and the elevated expression of EMT-related
genes, such as ZEB1 ensure that CDH1 transcription is
repressed and maintains breast cancer cells in a mesenchymal
phenotype (92,123). Of note, miR-200s can also be
regulated by PRC2. The miR-200b/a/429 cluster can be silenced
through PRC2-mediated histone H3K27me3 modification (124,125). Therefore, feedback loops between
miR-200 expression and PRC2 activity may exist in cells.
PRC2 has been identified as a potential target for
the treatment of breast cancer. DZNep can effectively reactivate
genes that have been silenced by PRC2 in breast cancer (85). This drug functions to inhibit the
methyl group donor, S-adenosylhomocysteine hydrolase, and results
in the degradation of EZH2, leading to a decrease in H3K27 di and
tri-methylation in the promoters of tumor suppressor genes
(126,127). DZNep also appears to promote
apoptosis by increasing the expression of apoptosis-related genes,
such as FBXO32, TGFBI, IGFBP3 and
PPPIR15A (85,126). It should be noted that DZNep is
not a specific EZH2 inhibitor and thus, other intracellular targets
may also mediate the effects of DZNep (127). More recently, MS1943 (1) has been described as a EZH2 selective
degrader that is cytotoxic to multiple TNBC cell lines in
vitro and in xenograft models (128).
Tazemetostat is an EZH2 inhibitor which functions
competitively to inhibit the co-factor, S-adenosyl-L-methionine
(SAM), an essential component for EZH2 activity (129,130). Tazemetostat recently gained FDA
approval for epithelial sarcomas and clinical trials are currently
underway in other types of cancer (131). Other EZH2 inhibitors that target
SAM are in early-stage clinical trials, including GSK2816126 and
CPI1205 (129) (NCT02082977,
NCT02395601, NCT03525795 and NCT03480646).
In addition to targeting the EZH2 subunit of the
PRC2 complex, other PRC2 components are also being investigated as
therapeutic targets. The EED-EZH2 interaction is critical for EZH2
function and stabilized alpha-helix of EZH2 (SAH-EZH2) peptides
have been developed to disrupt the EED-EZH2 interaction, which
results in decreased H3K27me3 levels and the degradation of EZH2
(132).
As research uncovers the mechanisms that regulate
H3K27me3, additional targets outside of the core PRC2 proteins have
been identified as possible targets. As mentioned previously, the
long non-coding RNA HOTAIR has been observed to recruit PRC2
to target genes (68). The small
molecule AC1Q3QWB (AQB) has been identified to selectively disrupt
the interaction of HOTAIR with EZH2 during PRC2 recruitment,
resulting in elevated levels of HOTAIR target genes such as
APC2 (133). Elevated APC2 levels
result in the degradation of β-catenin and the suppression of
pro-tumorigenic Wnt/β-catenin signaling (133). Other research has aimed to
disrupt HOTAIR-EZH2 interaction using peptide nucleic acids
(PNAs) that bind to single-stranded regions of HOTAIR
(134). Treatment with PNAs has
been shown to result in less invasion, reduced tumor formation and
increased chemosensitivity in breast cancer cells, as well as
ovarian cancer cells (134).
Another long non-coding RNA, named DANCR has also been
targeted. The nanoparticle-mediated RNA inhibition of DANCR
has been shown to knockdown DANCR 80-90% in cells up to 7
days (135). This administration
at a cellular level has been shown to be associated with the
downregulation of PRC2 methyltransferase activity at H3K27me3, as
well as the reduced Wnt/EMT signaling, migration and proliferation
of TNBC cells (135).
Therapies designed to target PRC2 may enhance the
efficacy of existing therapeutic strategies. For example, PRC2
decreases accessibility to regions of the DNA through chromatin
compaction. Chromatin compaction can decrease the sensitivity of
cancer cells to chemotherapeutic agents, such as anthracycline
(136). In addition, the use of
EZH2 inhibitors to decrease global H3K27me3 may enhance the
efficacy of monoclonal antibodies targeting HER2 (137). De novo resistance in
patients has been associated with elevated levels of global
H3K27me3 levels in HER2+ breast cancer and the
administration of EZH2 inhibitor GSK126 in combination with an
anti-ErbB2 monoclonal antibody has been shown to significantly
suppress tumor growth in an orthotopic mouse tumor model (137).
In summary, increased global levels of H3K27me3 can
be associated with breast cancer progression, including
proliferation, invasion, metastasis, evading apoptosis and EMT.
This modification involves the dysregulation of the PRC2
multi-subunit protein which, as stated, may occur through
mechanisms involving altered levels of the miR-200 family or
lncRNAs. Finally, recent studies utilizing the overexpression of
PRC2 as a possible mechanism of pathogenicity have seen some
promise in targeting this protein complex. Advancing the
understanding of this complex and its potential role in
tumorigenicity may provide novel breast cancer therapies.
The present study was funded by a project grant
from the Canadian Institutes of Health Research (CIHR, PJT-162218)
awarded to RAM.
Not applicable.
CJM wrote the manuscript and RAM edited the
manuscript. Both authors have read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Kim JM, Kim K, Punj V, Liang G, Ulmer TS,
Lu W and An W: Linker histone H1. 2 establishes chromatin
compaction and gene silencing through recognition of H3K27me3. Sci
Rep. 5:167142015. View Article : Google Scholar
|
|
2
|
Cao R, Wang L, Wang H, Xia L,
Erdjument-Bromage H, Tempst P, Jones RS and Zhang Y: Role of
histone H3 lysine 27 methylation in Polycomb-group silencing.
Science. 298:1039–1043. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Czermin B, Melfi R, McCabe D, Seitz V,
Imhof A and Pirrotta V: Drosophila enhancer of Zeste/ESC complexes
have a histone H3 methyltransferase activity that marks chromosomal
Polycomb sites. Cell. 111:185–196. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferrari KJ, Scelfo A, Jammula S, Cuomo A,
Barozzi I, Stützer A, Fischle W, Bonaldi T and Pasini D:
Polycomb-dependent H3K27me1 and H3K27me2 regulate active
transcription and enhancer fidelity. Mol Cell. 53:49–62. 2014.
View Article : Google Scholar
|
|
5
|
Kuzmichev A, Nishioka K, Erdjument-Bromage
H, Tempst P and Reinberg D: Histone methyltransferase activity
associated with a human multiprotein complex containing the
Enhancer of Zeste protein. Genes Dev. 16:2893–2905. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Laugesen A, Højfeldt JW and Helin K:
Molecular mechanisms directing PRC2 recruitment and H3K27
methylation. Mol Cell. 74:8–18. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Levine SS, Weiss A, Erdjument-Bromage H,
Shao Z, Tempst P and Kingston RE: The core of the polycomb
repressive complex is compositionally and functionally conserved in
flies and humans. Mol Cell Biol. 22:6070–6078. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lewis EB: A gene complex controlling
segmentation in Drosophila. Genes, development and cancer.
Springer; pp. 205–217. 1978, View Article : Google Scholar
|
|
9
|
Margueron R and Reinberg D: The Polycomb
complex PRC2 and its mark in life. Nature. 469:343–349. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rajasekhar VK and Begemann M: Concise
review: Roles of polycomb group proteins in development and
disease: A stem cell perspective. Stem Cells. 25:2498–2510. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kasinath V, Faini M, Poepsel S, Reif D,
Feng XA, Stjepanovic G, Aebersold R and Nogales E: Structures of
human PRC2 with its cofactors AEBP2 and JARID2. Science.
359:940–944. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hauri S, Comoglio F, Seimiya M, Gerstung
M, Glatter T, Hansen K, Aebersold R, Paro R, Gstaiger M and Beisel
C: A high-density map for navigating the human polycomb complexome.
Cell Rep. 17:583–595. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim H, Kang K and Kim J: AEBP2 as a
potential targeting protein for Polycomb Repression Complex PRC2.
Nucleic Acids Res. 37:2940–2950. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li G, Margueron R, Ku M, Chambon P,
Bernstein BE and Reinberg D: Jarid2 and PRC2, partners in
regulating gene expression. Genes Dev. 24:368–380. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peng JC, Valouev A, Swigut T, Zhang J,
Zhao Y, Sidow A and Wysocka J: Jarid2/Jumonji coordinates control
of PRC2 enzymatic activity and target gene occupancy in pluripotent
cells. Cell. 139:1290–1302. 2009. View Article : Google Scholar
|
|
16
|
Cao R and Zhang YI: SUZ12 is required for
both the histone methyltransferase activity and the silencing
function of the EED-EZH2 complex. Mol Cell. 15:57–67. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Denisenko O, Shnyreva M, Suzuki H and
Bomsztyk K: Point mutations in the WD40 domain of Eed block its
interaction with Ezh2. Mol Cell Biol. 18:5634–5642. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pasini D, Bracken AP, Jensen MR, Denchi EL
and Helin K: Suz12 is essential for mouse development and for EZH2
histone methyltransferase activity. EMBO J. 23:4061–4071. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Moritz LE and Trievel RC: Structure,
mechanism, and regulation of polycomb-repressive complex 2. J Biol
Chem. 293:13805–13814. 2018. View Article : Google Scholar :
|
|
20
|
Varambally S, Dhanasekaran SM, Zhou M,
Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt
RG, Otte AP, et al: The polycomb group protein EZH2 is involved in
progression of prostate cancer. Nature. 419:624–629. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiao L and Liu X: Structural basis of
histone H3K27 trimethylation by an active polycomb repressive
complex 2. Science. 350:aac43832015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Margueron R, Justin N, Ohno K, Sharpe ML,
Son J, Drury WJ III, Voigt P, Martin SR, Taylor WR, De Marco V, et
al: Role of the polycomb protein EED in the propagation of
repressive histone marks. Nature. 461:762–767. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nekrasov M, Wild B and Müller J:
Nucleosome binding and histone methyltransferase activity of
Drosophila PRC2. EMBO Rep. 6:348–353. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Han Z, Xing X, Hu M, Zhang Y, Liu P and
Chai J: Structural basis of EZH2 recognition by EED. Structure.
15:1306–1315. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Montgomery ND, Yee D, Chen A, Kalantry S,
Chamberlain SJ, Otte AP and Magnuson T: The murine polycomb group
protein Eed is required for global histone H3 lysine-27
methylation. Curr Biol. 15:942–947. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pasini D, Bracken AP, Hansen JB, Capillo M
and Helin K: The polycomb group protein Suz12 is required for
embryonic stem cell differentiation. Mol Cell Biol. 27:3769–3779.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Højfeldt JW, Laugesen A, Willumsen BM,
Damhofer H, Hedehus L, Tvardovskiy A, Mohammad F, Jensen ON and
Helin K: Accurate H3K27 methylation can be established de novo by
SUZ12-directed PRC2. Nat Struct Mol biol. 25:225–232. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kleer CG, Cao Q, Varambally S, Shen R, Ota
I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al: EZH2
is a marker of aggressive breast cancer and promotes neoplastic
transfor-mation of breast epithelial cells. Proc Natl Acad Sci USA.
100:11606–11611. 2003. View Article : Google Scholar
|
|
29
|
Margueron R, Li G, Sarma K, Blais A,
Zavadil J, Woodcock CL, Dynlacht BD and Reinberg D: Ezh1 and Ezh2
maintain repressive chromatin through different mechanisms. Mol
Cell. 32:503–518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee CH, Holder M, Grau D, Saldaña-Meyer R,
Yu JR, Ganai RA, Zhang J, Wang M, LeRoy G, Dobenecker MW, et al:
Distinct stimulatory mechanisms regulate the catalytic activity of
polycomb repressive complex 2. Mol Cell. 70:435–448.e5. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen S, Jiao L, Shubbar M, Yang X and Liu
X: Unique structural platforms of Suz12 dictate distinct classes of
PRC2 for chromatin binding. Mol Cell. 69:840–852.e5. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schmitges FW, Prusty AB, Faty M, Stützer
A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al:
Histone methylation by PRC2 is inhibited by active chromatin marks.
Mol Cell. 42:330–341. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pasini D, Cloos PAC, Walfridsson J, Olsson
L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J and
Helin K: JARID2 regulates binding of the Polycomb repressive
complex 2 to target genes in ES cells. Nature. 464:306–310. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Murzina NV, Pei XY, Zhang W, Sparkes M,
Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR,
Verreault A, Luisi BF and Laue ED: Structural basis for the
recognition of histone H4 by the histone-chaperone RbAp46.
Structure. 16:1077–1085. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang W, Tyl M, Ward R, Sobott F, Maman J,
Murthy AS, Watson AA, Fedorov O, Bowman A, Owen-Hughes T, et al:
Structural plasticity of histones H3-H4 facilitates their
allosteric exchange between RbAp48 and ASF1. Nat Struct Mol Biol.
20:29–35. 2013. View Article : Google Scholar :
|
|
36
|
Kouznetsova VL, Tchekanov A, Li X, Yan X
and Tsigelny IF: Polycomb repressive 2 complex-molecular mechanisms
of function. Protein Sci. 28:1387–1399. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Landeira D, Sauer S, Poot R, Dvorkina M,
Mazzarella L, Jørgensen HF, Pereira CF, Leleu M, Piccolo FM,
Spivakov M, et al: Jarid2 is a PRC2 component in embryonic stem
cells required for multi-lineage differentiation and recruitment of
PRC1 and RNA Polymerase II to developmental regulators. Nat Cell
Biol. 12:618–624. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Son J, Shen SS, Margueron R and Reinberg
D: Nucleosome- binding activities within JARID2 and EZH1 regulate
the function of PRC2 on chromatin. Genes Dev. 27:2663–2677. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cooper S, Grijzenhout A, Underwood E,
Ancelin K, Zhang T, Nesterova TB, Anil-Kirmizitas B, Bassett A,
Kooistra SM, Agger K, et al: Jarid2 binds mono-ubiquitylated H2A
lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and
PRC2. Nat Commun. 7:136612016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Endoh M, Endo TA, Endoh T, Isono K, Sharif
J, Ohara O, Toyoda T, Ito T, Eskeland R, Bickmore WA, et al:
Histone H2A mono-ubiquitination is a crucial step to mediate
PRC1-dependent repression of developmental genes to maintain ES
cell identity. PLoS Genet. 8:e10027742012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kalb R, Latwiel S, Baymaz HI, Jansen PW,
Müller CW, Vermeulen M and Müller J: Histone H2A monoubiquitination
promotes histone H3 methylation in Polycomb repression. Nat Struct
Mol Biol. 21:569–571. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shen X, Kim W, Fujiwara Y, Simon MD, Liu
Y, Mysliwiec MR, Yuan GC, Lee Y and Orkin SH: Jumonji modulates
polycomb activity and self-renewal versus differentiation of stem
cells. Cell. 139:1303–1314. 2009. View Article : Google Scholar
|
|
43
|
Ballaré C, Lange M, Lapinaite A, Martin
GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, et al:
Phf19 links methylated Lys36 of histone H3 to regulation of
Polycomb activity. Nat Struct Mol Biol. 19:1257–1265. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Musselman CA, Avvakumov N, Watanabe R,
Abraham CG, Lalonde ME, Hong Z, Allen C, Roy S, Nuñez JK, Nickoloff
J, et al: Molecular basis for H3K36me3 recognition by the Tudor
domain of PHF1. Nat Struct Mol Biol. 19:1266–1272. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Boulay G, Rosnoblet C, Guérardel C,
Angrand PO and Leprince D: Functional characterization of human
Polycomb-like 3 isoforms identifies them as components of distinct
EZH2 protein complexes. Biochem J. 434:333–342. 2011. View Article : Google Scholar
|
|
46
|
Li H, Liefke R, Jiang J, Kurland JV, Tian
W, Deng P, Zhang W, He Q, Patel DJ, Bulyk ML, et al: Polycomb-like
proteins link the PRC2 complex to CpG islands. Nature. 549:287–291.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hou Y, Liu W, Yi X, Yang Y, Su D, Huang W,
Yu H, Teng X, Yang Y, Feng W, et al: PHF20L1 as a H3K27me2 reader
coordinates with transcriptional repressors to promote breast
tumorigenesis. Sci Adv. 6:eaaz03562020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Perino M, van Mierlo G, Karemaker ID, van
Genesen S, Vermeulen M, Marks H, van Heeringen SJ and Veenstra GJC:
MTF2 recruits polycomb repressive Complex 2 by
helical-shape-selective DNA binding. Nat Genet. 50:1002–1010. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hansen KH, Bracken AP, Pasini D, Dietrich
N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M and Helin K: A
model for transmission of the H3K27me3 epigenetic mark. Nat Cell
Biol. 10:1291–1300. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Oksuz O, Narendra V, Lee CH, Descostes N,
LeRoy G, Raviram R, Blumenberg L, Karch K, Rocha PP, Garcia BA, et
al: Capturing the onset of PRC2-mediated repressive domain
formation. Mol Cell. 70:1149–1162.e5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Boyer LA, Plath K, Zeitlinger J, Brambrink
T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et
al: Polycomb complexes repress developmental regulators in murine
embryonic stem cells. Nature. 441:349–353. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bracken AP, Dietrich N, Pasini D, Hansen
KH and Helin K: Genome-wide mapping of Polycomb target genes
unravels their roles in cell fate transitions. Genes Dev.
20:1123–1136. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lee TI, Jenner RG, Boyer LA, Guenther MG,
Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K,
et al: Control of developmental regulators by Polycomb in human
embryonic stem cells. Cell. 125:301–313. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Schwartz YB, Kahn TG, Nix DA, Li XY,
Bourgon R, Biggin M and Pirrotta V: Genome-wide analysis of
polycomb targets in drosophila melanogaster. Nat Genet. 38:700–705.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ren C, Morohashi K, Plotnikov AN, Jakoncic
J, Smith SG, Li J, Zeng L, Rodriguez Y, Stojanoff V, Walsh M and
Zhou MM: Small-molecule modulators of methyl-lysine binding for the
CBX7 chromodomain. Chem Biol. 22:161–168. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhen CY, Tatavosian R, Huynh TN, Duc HN,
Das R, Kokotovic M, Grimm JB, Lavis LD, Lee J, Mejia FJ, et al:
Live-cell single-molecule tracking reveals co-recognition of
H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin. Elife.
5:e176672016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Blackledge NP, Farcas AM, Kondo T, King
HW, McGouran JF, Hanssen LLP, Ito S, Cooper S, Kondo K, Koseki Y,
et al: Variant PRC1 complex-dependent H2A ubiquitylation drives
PRC2 recruitment and polycomb domain formation. Cell.
157:1445–1459. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Cooper S, Dienstbier M, Hassan R,
Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S,
Koseki H, et al: Targeting polycomb to pericentric heterochromatin
in embryonic stem cells reveals a role for H2AK119u1 in PRC2
recruitment. Cell Rep. 7:1456–1470. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bhatnagar S, Gazin C, Chamberlain L, Ou J,
Zhu X, Tushir JS, Virbasius CM, Lin L, Zhu LJ, Wajapeyee N and
Green MR: TRIM37 is a new histone H2A ubiquitin ligase and breast
cancer oncoprotein. Nature. 516:116–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sanulli S, Justin N, Teissandier A,
Ancelin K, Portoso M, Caron M, Michaud A, Lombard B, da Rocha ST,
Offer J, et al: Jarid2 methylation via the PRC2 complex regulates
H3K27me3 deposition during cell differentiation. Mol Cell.
57:769–783. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Riising EM, Comet I, Leblanc B, Wu X,
Johansen JV and Helin K: Gene silencing triggers polycomb
repressive complex 2 recruitment to CpG islands genome wide. Mol
Cell. 55:347–360. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kahn TG, Dorafshan E, Schultheis D, Zare
A, Stenberg P, Reim I, Pirrotta V and Schwartz YB: Interdependence
of PRC1 and PRC2 for recruitment to Polycomb Response Elements.
Nucleic Acids Res. 44:10132–10149. 2016.PubMed/NCBI
|
|
63
|
Bauer M, Trupke J and Ringrose L: The
quest for mammalian Polycomb response elements: Are we there yet?
Chromosoma. 125:471–496. 2016. View Article : Google Scholar :
|
|
64
|
Mikkelsen TS, Ku M, Jaffe DB, Issac B,
Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP,
et al: Genome-wide maps of chromatin state in pluripotent and
lineage-committed cells. Nature. 448:553–560. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Brien GL, Gambero G, O'connell DJ, Jerman
E, Turner SA, Egan CM, Dunne EJ, Jurgens MC, Wynne K, Piao L, et
al: Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase
NO66 to embryonic stem cell genes during differentiation. Nat
Struct Mol Biol. 19:1273–1281. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Cai L, Rothbart SB, Lu R, Xu B, Chen WY,
Tripathy A, Rockowitz S, Zheng D, Patel DJ, Allis CD, et al: An
H3K36 methylation-engaging Tudor motif of polycomb-like proteins
mediates PRC2 complex targeting. Mol Cell. 49:571–582. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhou X, Ren Y, Zhang J, Zhang C, Zhang K,
Han L, Kong L, Wei J, Chen L, Yang J, et al: HOTAIR is a
therapeutic target in glioblastoma. Oncotarget. 6:8353–8365. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Bracken AP, Pasini D, Capra M, Prosperini
E, Colli E and Helin K: EZH2 is downstream of the pRB-E2F pathway,
essential for proliferation and amplified in cancer. EMBO J.
22:5323–5335. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bachmann IM, Halvorsen OJ, Collett K,
Stefansson IM, Straume O, Haukaas SA, Salvesen HB, Otte AP and
Akslen LA: EZH2 expression is associated with high proliferation
rate and aggressive tumor subgroups in cutaneous melanoma and
cancers of the endometrium, prostate, and breast. J Clin Oncol.
24:268–273. 2006. View Article : Google Scholar
|
|
73
|
Derfoul A, Juan AH, Difilippantonio MJ,
Palanisamy N, Ried T and Sartorelli V: Decreased microRNA-214
levels in breast cancer cells coincides with increased cell
proliferation, invasion and accumulation of the Polycomb Ezh2
methyltransferase. Carcinogenesis. 32:1607–1614. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kong X, Chen L, Jiao L, Jiang X, Lian F,
Lu J, Zhu K, Du D, Liu J, Ding H, et al: Astemizole arrests the
proliferation of cancer cells by disrupting the EZH2-EED
interaction of poly-comb repressive complex 2. J Med Chem.
57:9512–9521. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yu H, Simons DL, Segall I, Carcamo-Cavazos
V, Schwartz EJ, Yan N, Zuckerman NS, Dirbas FM, Johnson DL, Holmes
SP and Lee PP: PRC2/EED-EZH2 complex is up-regulated in breast
cancer lymph node metastasis compared to primary tumor and
correlates with tumor proliferation in situ. PLoS One.
7:e512392012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Curran S and Murray GI: Matrix
metalloproteinases in tumour invasion and metastasis. J Pathol.
189:300–308. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Merdad A, Karim S, Schulten HJ, Dallol A,
Buhmeida A, Al-Thubaity F, Gari MA, Chaudhary AG, Abuzenadah AM and
Al-Qahtani MH: Expression of matrix metalloproteinases (MMPs) in
primary human breast cancer: MMP-9 as a potential biomarker for
cancer invasion and metastasis. Anticancer Res. 34:1355–1366.
2014.PubMed/NCBI
|
|
78
|
Chien YC, Liu LC, Ye HY, Wu JY and Yu YL:
EZH2 promotes migration and invasion of triple-negative breast
cancer cells via regulating TIMP2-MMP-2/-9 pathway. Am J Cancer
Res. 8:422–434. 2018.PubMed/NCBI
|
|
79
|
Shin YJ and Kim JH: The role of EZH2 in
the regulation of the activity of matrix metalloproteinases in
prostate cancer cells. PLoS One. 7:e303932012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yi X, Guo J, Guo J, Sun S, Yang P, Wang J,
Li Y, Xie L, Cai J and Wang Z: EZH2-mediated epigenetic silencing
of TIMP2 promotes ovarian cancer migration and invasion. Sci Rep.
7:35682017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wang T, Mao B, Cheng C, Zou Z, Gao J, Yang
Y, Lei T, Qi X, Yuan Z, Xu W and Lu Z: YAP promotes breast cancer
metastasis by repressing growth differentiation factor-15. Biochim
Biophys Acta Mol Basis Dis. 1864:1744–1753. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Overholtzer M, Zhang J, Smolen GA, Muir B,
Li W, Sgroi DC, Deng CX, Brugge JS and Haber DA: Transforming
properties of YAP, a candidate oncogene on the chromosome 11q22
amplicon. Proc Natl Acad Sci USA. 103:12405–12410. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Song Q, Mao B, Cheng J, Gao Y, Jiang K,
Chen J, Yuan Z and Meng S: YAP enhances autophagic flux to promote
breast cancer cell survival in response to nutrient deprivation.
PLoS One. 10:e01207902015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yuan M, Tomlinson V, Lara R, Holliday D,
Chelala C, Harada T, Gangeswaran R, Manson-Bishop C, Smith P,
Danovi SA, et al: Yes-associated protein (YAP) functions as a tumor
suppressor in breast. Cell Death Differ. 15:1752–1759. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Tan J, Yang X, Zhuang L, Jiang X, Chen W,
Lee PL, Karuturi RK, Tan PB, Liu ET and Yu Q: Pharmacologic
disruption of Polycomb-repressive complex 2-mediated gene
repression selectively induces apoptosis in cancer cells. Genes
Dev. 21:1050–1063. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wu Z, Lee S, Qiao Y, Li Z, Lee PL, Lee YJ,
Jiang X, Tan J, Aau M, Lim CZ and Yu Q: Polycomb protein EZH2
regulates cancer cell fate decision in response to DNA damage. Cell
Death Differ. 18:1771–1779. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wu ZL, Zheng SS, Li ZM, Qiao YY, Aau MY
and Yu Q: Polycomb protein EZH2 regulates E2F1-dependent apoptosis
through epigenetically modulating Bim expression. Cell Death
Differ. 17:801–810. 2010. View Article : Google Scholar
|
|
88
|
Zhang B, Liu XX, He JR, Zhou CX, Guo M, He
M, Li MF, Chen GQ and Zhao Q: Pathologically decreased miR-26a
antagonizes apoptosis and facilitates carcinogenesis by targeting
MTDH and EZH2 in breast cancer. Carcinogenesis. 32:2–9. 2011.
View Article : Google Scholar
|
|
89
|
Zhang Q, Padi SKR, Tindall DJ and Guo B:
Polycomb protein EZH2 suppresses apoptosis by silencing the
proapoptotic miR-31. Cell Death Dis. 5:e14862014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Pietersen AM, Horlings HM, Hauptmann M,
Langerød A, Ajouaou A, Cornelissen-Steijger P, Wessels LF, Jonkers
J, van de Vijver MJ and van Lohuizen M: EZH2 and BMI1 inversely
correlate with prognosis and TP53 mutation in breast cancer. Breast
Cancer Res. 10:R1092008. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Onder TT, Gupta PB, Mani SA, Yang J,
Lander ES and Weinberg RA: Loss of E-cadherin promotes metastasis
via multiple downstream transcriptional pathways. Cancer Res.
68:3645–3654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Lester RD, Jo M, Montel V, Takimoto S and
Gonias SL: uPAR induces epithelial-mesenchymal transition in
hypoxic breast cancer cells. J Cell Biol. 178:425–436. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Schmalhofer O, Brabletz S and Brabletz T:
E-cadherin, β-catenin, and ZEB1 in malignant progression of cancer.
Cancer and Metastasis Rev. 28:151–166. 2009. View Article : Google Scholar
|
|
94
|
Herranz N, Pasini D, Díaz VM, Francí C,
Gutierrez A, Dave N, Escrivà M, Hernandez-Muñoz I, Di Croce L,
Helin K, et al: Polycomb complex 2 is required for E-cadherin
repression by the Snail1 transcription factor. Mol Cell Biol.
28:4772–4781. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Anwar T, Arellano-Garcia C, Ropa J, Chen
YC, Kim HS, Yoon E, Grigsby S, Basrur V, Nesvizhskii AI, Muntean A,
et al: p38-mediated phosphorylation at T367 induces EZH2
cytoplasmic localization to promote breast cancer metastasis. Nat
Commun. 9:28012018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li J, Xi Y, Li W, McCarthy RL, Stratton
SA, Zou W, Li W, Dent SY, Jain AK and Barton MC: TRIM28 interacts
with EZH2 and SWI/SNF to activate genes that promote mammosphere
formation. Oncogene. 36:2991–3001. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Mahara S, Lee PL, Feng M, Tergaonkar V,
Chng WJ and Yu Q: HIFI-α activation underlies a functional switch
in the paradoxical role of Ezh2/PRC2 in breast cancer. Proc Natl
Acad Sci USA. 113:E3735–E3744. 2016. View Article : Google Scholar
|
|
98
|
Chisholm KM, Wan Y, Li R, Montgomery KD,
Chang HY and West RB: Detection of long non-coding RNA in archival
tissue: Correlation with polycomb protein expression in primary and
metastatic breast carcinoma. PLoS One. 7:e479982012. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Sørensen KP, Thomassen M, Tan Q, Bak M,
Cold S, Burton M, Larsen MJ and Kruse TA: Long non-coding RNA
HOTAIR is an independent prognostic marker of metastasis in
estrogen receptor-positive primary breast cancer. Br Cancer Res
Treat. 142:529–536. 2013. View Article : Google Scholar
|
|
100
|
Kim CY, Oh JH, Lee JY and Kim MH: The
LncRNA HOTAIRM1 promotes tamoxifen resistance by mediating HOXA1
expression in ER+ Breast Cancer Cells. J Cancer. 11:3416–3423.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Hu P, Chu J, Wu Y, Sun L, Lv X, Zhu Y, Li
J, Guo Q, Gong C, Liu B and Su S: NBAT1 suppresses breast cancer
metastasis by regulating DKK1 via PRC2. Oncotarget. 6:32410–32425.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhang J, Sui S, Wu H, Zhang J, Zhang X, Xu
S and Pang D: The transcriptional landscape of lncRNAs reveals the
oncogenic function of LINC00511 in ER-negative breast cancer. Cell
Death Dis. 10:5992019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Sha S, Yuan D, Liu Y, Han B and Zhong N:
Targeting long non-coding RNA DANCR inhibits triple negative breast
cancer progression. Biol Open. 6:1310–1316. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Jia J, Li F, Tang XS, Xu S, Gao Y, Shi Q,
Guo W, Wang X, He D and Guo P: Long noncoding RNA DANCR promotes
invasion of prostate cancer through epigenetically silencing
expression of TIMP2/3. Oncotarget. 7:37868–37881. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Saini HK, Enright AJ and Griffiths-Jones
S: Annotation of mammalian primary microRNAs. BMC Genomics.
9:5642008. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Hill L, Browne G and Tulchinsky E:
ZEB/miR-200 feedback loop: At the crossroads of signal transduction
in cancer. Int J Cancer. 132:745–754. 2013. View Article : Google Scholar
|
|
108
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Mei J, Hao L, Wang H, Xu R, Liu Y, Zhu Y
and Liu C: Systematic characterization of non-coding RNAs in
triple-negative breast cancer. Cell Prolif. 53:e128012020.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Jones R, Watson K, Bruce A, Nersesian S,
Kitz J and Moorehead R: Re-expression of miR-200c suppresses
proliferation, colony formation and in vivo tumor growth of murine
Claudin-low mammary tumor cells. Oncotarget. 8:23727–23749. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Watson KL, Jones RA, Bruce A and Moorehead
RA: The miR-200b/200a/429 cluster prevents metastasis and induces
dormancy in a murine claudin-low mammary tumor cell line. Exp Cell
Res. 369:17–26. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Bockmeyer CL, Christgen M, Müller M,
Fischer S, Ahrens P, Länger F, Kreipe H and Lehmann U: MicroRNA
profiles of healthy basal and luminal mammary epithelial cells are
distinct and reflected in different breast cancer subtypes. Breast
Cancer ResTreat. 130:735–745. 2011. View Article : Google Scholar
|
|
114
|
Cochrane DR, Cittelly DM, Howe EN,
Spoelstra NS, McKinsey EL, LaPara K, Elias A, Yee D and Richer JK:
MicroRNAs link estrogen receptor alpha status and Dicer levels in
breast cancer. Horm Cancer. 1:306–319. 2010. View Article : Google Scholar
|
|
115
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Mekala JR, Naushad SM, Ponnusamy L,
Arivazhagan G, Sakthiprasad V and Pal-Bhadra M: Epigenetic
regulation of miR-200 as the potential strategy for the therapy
against triple-negative breast cancer. Gene. 641:248–258. 2018.
View Article : Google Scholar
|
|
117
|
Humphries B, Wang Z, Oom AL, Fisher T, Tan
D, Cui Y, Jiang Y and Yang C: MicroRNA-200b targets protein kinase
Calpha and suppresses triple-negative breast cancer metastasis.
Carcinogenesis. 35:2254–2263. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Bracken CP, Gregory PA, Kolesnikoff N,
Bert AG, Wang J, Shannon MF and Goodall GJ: A double-negative
feedback loop between ZEB1-SIP1 and the microRNA-200 family
regulates epithelial-mesenchymal transition. Cancer Res.
68:7846–7854. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Howe EN, Cochrane DR and Richer JK: The
miR-200 and miR-221/222 microRNA families: Opposing effects on
epithelial identity. J Mammary Gland Biol Neoplasia. 17:65–77.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Radisky DC: miR-200c at the nexus of
epithelial-mesenchymal transition, resistance to apoptosis, and the
breast cancer stem cell phenotype. Breast Cancer Res. 13:1102011.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Iliopoulos D, Lindahl-Allen M, Polytarchou
C, Hirsch HA, Tsichlis PN and Struhl K: Loss of miR-200 inhibition
of Suz12 leads to polycomb-mediated repression required for the
formation and maintenance of cancer stem cells. Mol Cell.
39:761–772. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Peng F, Jiang J, Yu Y, Tian R, Guo X, Li
X, Shen M, Xu M, Zhu F, Shi C, et al: Direct targeting of
SUZ12/ROCK2 by miR-200b/c inhibits cholangiocarcinoma
tumourigenesis and metastasis. Br J Cancer. 109:3092–3104. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Siitonen SM, Kononen JT, Helin HJ, Rantala
IS, Holli KA and Isola JJ: Reduced E-cadherin expression is
associated with invasiveness and unfavorable prognosis in breast
cancer. Am J Clin Pathol. 105:394–402. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Lim YY, Wright JA, Attema JL, Gregory PA,
Bert AG, Smith E, Thomas D, Lopez AF, Drew PA, Khew-Goodall Y and
Goodall GJ: Epigenetic modulation of the miR-200 family is
associated with transition to a breast cancer stem-cell-like state.
J Cell Sci. 126:2256–2266. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Ning X, Shi Z, Liu X, Zhang A, Han L,
Jiang K, Kang C and Zhang Q: DNMT1 and EZH2 mediated methylation
silences the microRNA-200b/a/429 gene and promotes tumor
progression. Cancer Lett. 359:198–205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Fiskus W, Wang Y, Sreekumar A, Buckley KM,
Shi H, Jillella A, Ustun C, Rao R, Fernandez P, Chen J, et al:
Combined epigenetic therapy with the histone methyltransferase EZH2
inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor
panobinostat against human AML cells. Blood. 114:2733–2743. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Miranda TB, Cortez CC, Yoo CB, Liang G,
Abe M, Kelly TK, Marquez VE and Jones PA: DZNep is a global histone
methylation inhibitor that reactivates developmental genes not
silenced by DNA methylation. Mol Cancer Ther. 8:1579–1588. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Ma A, Stratikopoulos E, Park KS, Wei J,
Martin TC, Yang X, Schwarz M, Leshchenko V, Rialdi A, Dale B, et
al: Discovery of a first-in-class EZH2 selective degrader. Nat Chem
Biol. 16:214–222. 2020. View Article : Google Scholar :
|
|
129
|
Gulati N, Béguelin W and Giulino-Roth L:
Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk Lymphoma.
59:1574–1585. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Richart L and Margueron R: Drugging
histone methyltransferases in cancer. Curr Opin Chem Biol.
56:51–62. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Hoy SM: Tazemetostat: First approval.
Drugs. 80:513–521. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Kim W, Bird GH, Neff T, Guo G, Kerenyi MA,
Walensky LD and Orkin SH: Targeted disruption of the EZH2-EED
complex inhibits EZH2-dependent cancer. Nat Chem Biol. 9:643–650.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Li Y, Ren Y, Wang Y, Tan Y, Wang Q, Cai J,
Zhou J, Yang C, Zhao K, Yi K, et al: A compound AC1Q3QWB
selectively disrupts HOTAIR-mediated recruitment of PRC2 and
enhances cancer therapy of DZNep. Theranostics. 9:4608–4623. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Özeş AR, Wang Y, Zong X, Fang F, Pilrose J
and Nephew KP: Therapeutic targeting using tumor specific peptides
inhibits long non-coding RNA HOTAIR activity in ovarian and breast
cancer. Sci Rep. 7:8942017. View Article : Google Scholar
|
|
135
|
Vaidya AM, Sun Z, Ayat N, Schilb A, Liu X,
Jiang H, Sun D, Scheidt J, Qian V, He S, et al: Systemic delivery
of tumor-targeting siRNA nanoparticles against an oncogenic LncRNA
facilitates effective triple-negative breast cancer therapy.
Bioconjug Chem. 30:907–919. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Seoane JA, Kirkland JG, Caswell-Jin JL,
Crabtree GR and Curtis C: Chromatin regulators mediate
anthracycline sensitivity in breast cancer. Nat Med. 25:1721–1727.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Hirukawa A, Singh S, Wang J, Rennhack JP,
Swiatnicki M, Sanguin-Gendreau V, Zuo D, Daldoul K, Lavoie C, Park
M, et al: Reduction of Global H3K27me3 Enhances
HER2/ErbB2 targeted therapy. Cell Rep. 29:249–257. 2019. View Article : Google Scholar
|