Introduction
Lung cancer, as one of the most common types of
cancer, causes more deaths than breast, prostate, colorectal and
brain cancer combined in 2017 in the United States, with the 5-year
relative survival rate being 5% (1). Non-small cell lung cancer (NSCLC)
accounted for >80% of all lung cancer cases worldwide in 2016,
which fall into several categories, including adenocarcinoma,
squamous cell carcinoma and large cell carcinoma (2). The 5-year survival rate for
metastatic NSCLC is <5%, and that for patients with early-stage
NSCLC is <50% (3). Therefore,
finding novel target biomarkers is urgently required.
Accumulating evidence has revealed that most RNA
transcripts do not encode proteins in mammals (4,5).
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs of
>200 nucleotides in length (6).
lncRNAs used to be considered non-functional (7), but an increasing number of lncRNAs
have now been reported to serve important roles in biological
processes; for example, lncRNAs form extensive networks of
ribonucleoprotein complexes with numerous chromatin regulators and
then target enzymatic activities to appropriate locations in the
genome (8,9). Rigorous evidence has suggested that
certain lncRNAs exert important regulatory effects in
carcinogenesis and the progression of prostate, hepatocellular and
lung cancer, with potential roles in oncogenic or tumor-suppressive
signaling pathways (4,10,11-13).
FEZ family zinc finger 1 antisense RNA 1 (FEZF1-AS1)
is a lncRNA that is highly expressed in colorectal carcinoma (CRC),
and its overexpression can promote the aggressive behavior of CRC
cells, both in vitro and in vivo (14). In gastric cancer, high FEZF1-AS1
expression promotes cell proliferation (14). In addition, the inhibition of
FEZF1-AS1 can suppress the activation of the Wnt/p-catenin
signaling pathway (15).
Additionally, FEZF1-AS1 expression is upregulated in lung
adenocarcinoma and is mediated by FEZF1 (16). However, the mechanism of FEZF1-AS1
in NSCLC remains unclear. Although FEZF1-AS1 expression is
upregulated in multiple types of cancer, there are different views
on the location and mechanism of FEZF1-AS1 in the existing
literature. He et al (17)
reported that FEZF1-AS1 is located in the cytoplasm and nucleus in
lung adenocarcinoma A549 and SPC-A1 cells, while in other studies
FEZF1-AS1 has been reported to be mainly located in the cytoplasm
in osteosarcoma, pancreatic ductal adenocarcinoma and myeloma
(18-20). Therefore, further elucidation of
the mechanism and location of FEZF1-AS1 in NSCLC is required.
In the present study, the association between the
expression levels of FEZF1-AS1 in NSCLC tissues and overall
survival (OS) was explored. In addition, the function of FEZF1-AS1
in NSCLC cells, the N6-methyladenosine (m6A)
modification and the involvement of the FEZF1-AS1/integrin subunit
all (ITGA11)/microRNA (miRNA/miR)-516b-5p axis in NSCLC were
investigated. The present results may provide novel biomarkers for
NSCLC.
Materials and methods
Patient and clinical data collection
The study protocol was approved by the Institutional
Review Board of Hebei Medical University (Shijiazhuang, China).
Frozen surgical tumor tissues and corresponding normal lung tissues
(5 cm from the edge of the tumor) of 45 patients with NSCLC
(including 17 squamous cell carcinoma, 27 adenocarcinoma and 1
atypical carcinoid) were obtained from The Fourth Hospital of Hebei
Medical University (Shijiazhuang, China) between February 2009 and
February 2018. The median age of the patients was 61 years (range,
44-73 years). All patients were diagnosed with NSCLC on
histopathological evaluation, and were reviewed by experienced
pathologists and staged according to the 8th edition lung cancer
classification of the American Joint Committee of Cancer (21). EGFR was detected via PCR, and ALK
was detected via fluorescence in situ hybridization (FISH)
by the Department of Pathology of the Fourth Hospital of Hebei
Medical University. PD-1 and PD-L1 were detected via
immunohistochemistry (IHC) by the Department of Pathology of the
Fourth Hospital of Hebei Medical University. PCR, FISH and IHC were
performed according to routine hospital protocols. No patients
received any other treatment prior to surgery, and all patients
were followed up until February 2020. The smoking index was
calculated as cigarettes/day x smoking time (years). Since a
smoking index >400 indicated a high risk of lung cancer,
patients were divided into never smokers (0, smoking index=0),
light smokers (1, smoking index <400) and heavy smokers (2,
smoking index >400). The study was conducted in accordance with
the Declaration of Helsinki, and approval was obtained from the
Research Ethics Committee of the Fourth Hospital of Hebei Medical
University.
Cell culture
Human NSCLC H358, H1299, A549, H520, SK-MES-1, H1703
and normal human bronchial epithelial BEAS-2B cells were purchased
from the China Infrastructure of Cell Line Resources. SK-MES-1
cells were cultured in Minimum Essential Medium (Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gemini Bio Products),
10,000 U/ml penicillin and 10,000 μg/ml streptomycin (Thermo
Fisher Scientific, Inc.). H520 cells were cultured in DMEM (Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Gemini Bio
Products), 10,000 U/ml penicillin and 10,000 μg/ml
streptomycin (Thermo Fisher Scientific, Inc.). The other cells were
cultured in RPMI-1640 medium (Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS, 10,000 U/ml penicillin and 10,000
μg/ml streptomycin. BEAS-2B cells were cultured in Bronchial
Epithelial Cell Medium (ScienCell Research Laboratories, Inc.) with
1% cell growth supplements (cat. no. 3962; ScienCell Research
Laboratories, Inc.) and 1% penicillin/streptomycin (Thermo Fisher
Scientific, Inc.). All cells were incubated at 37°C with 5%
CO2.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA from frozen tissues or cultured cells was
extracted using TRIzol® reagent (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Total
RNA (500 ng) was reverse transcribed in a 10-μl reaction
volume, using random primers according to the manufacturer's
protocol of the ReverTra Ace™ qPCR RT Master mix with gDNA Remover
kit (Toyobo Life Science). The primer sequences for lncRNA and mRNA
expression are shown in Table I.
qPCR was performed on the obtained cDNA using SYBR Green (Promega
Corporation) following the manufacturer's protocol. PCR was
performed using a Gene Amp PCR System MX3005P (Agilent
Technologies, Inc.) with the following protocol: 94°C for 5 min,
followed by 40 cycles of denaturation at 94°C for 15 sec and
annealing/extension at 60°C for 1 min, and a final extension step
at 72°C for 5 min. p-actin was used as the reference gene. Analysis
of relative gene expression was performed using the
2−ΔΔCq method (22).
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5′-3′) |
|---|
| FEZF1-AS1
forward |
ACCTGCCTTCTTGACTGAATG |
| FEZF1-AS1
reverse |
GCAGTAACCATAGCCAGAAACT |
| YTHDF1 forward |
GTGGACACCCAGAGAACAAA |
| YTHDF1 reverse |
CAGTAAGGTAGGGCTCAAAGTC |
| YTHDF2 forward |
ACAGCCAAGGCCCAATAA |
| YTHDF2 reverse |
GCAGCTTCACCCAAAGAATAG |
| METTL3 forward |
CACTGATGCTGTGTCCATCT |
| METTL3 reverse |
CTTGTAGGAGACCTCGCTTTAC |
| METTL14
forward |
GAAACTGGCATCACTGCTAATG |
| METTL14
reverse |
CCAGAACCACACCAGAGAAA |
| ITGA11 forward |
CCTACAGCACGGTCCTAAATATC |
| ITGA11 reverse |
CTCCTCGTTCACACACTCAAT |
| GAPDH forward |
GCACCGTCAAGGCTGAGAAC |
| GAPDH reverse |
GCCTTCTCCATGGTGGTGAA |
| MALAT1 forward |
GCTCAGTTGCGTAATGGAAAG |
| MALAT1 reverse |
GTGTTCTCTTGAGGGACAGTAG |
| RHOV forward |
ACACCTTCTCTGTGCAAGTC |
| RHOV reverse |
GGGAACGAAGTCGGTCAAA |
| ONECUT2
forward |
ATGTGGAAGTGGCTTCAGG |
| ONECUT2
reverse |
GGGACTTCTTCTGGGAATTGT |
| SOX4 forward |
CAGCGACAAGATCCCTTTCA |
| SOX4 reverse |
GCCGGACTTCACCTTCTTC |
| MARK4 forward |
GGGAGGTTGCCATCAAGATTAT |
| MARK4 reverse |
CAGCGTCTTCTCAGTCTCAATC |
| UBN1 forward |
CCCAGAGCTGGTGAAGAATATC |
| UBN1 reverse |
GGGCCTCTACTTTATGCCTTT |
| COL1A1 forward |
CTAAAGGCGAACCTGGTGAT |
| COL1A1 reverse |
TCCAGGAGCACCAACATTAC |
| PRKAB2 forward |
AGCACCAAGATTCCACTGATTA |
| PRKAB2 reverse |
CCACTGTCCATCCACAAAGA |
| PIP5K1A
forward |
CGGCCCGATGATTACTTGTAT |
| PIP5K1A
reverse |
CGTCGCTGGACACATAGAATAG |
| CDK6 forward |
GGATAAAGTTCCAGAGCCTGGAG |
| CDK6 reverse |
GCGATGCACTACTCGGTGTGAA |
| SKA3 forward |
AATCTGCTCAGAACACCTACAC |
| SKA3 reverse |
TGGGTGGCACTGCTTTAAT |
| XKR9 forward |
AGGCTGCCCACAACTTATTC |
| XKR9 reverse |
AGAAATAGCACAGCAAGAGACC |
| β-actin
forward |
AGCGAGCATCCCCCAAAGTT |
| β-actin
reverse |
GGGCACGAAGGCTCATCATT |
Small interfering RNA (siRNA) silencing
and miRNA mimics
H1299 and H520 cells were transfected with siRNA
(si-NC, si-FEZF1-AS1, si-ITGA11-1, si-ITGA11-2, si-ITGA11-3,
si-METTL3, si-METTL14, si-YTHDF1 and si-YTHDF2; 40 pmol/well;
Shanghai GenePharma Co., Ltd.) or miRNA mimics (NC mimics and
miR-516b-5p mimics, miR-126a mimics, miR-29b mimics, miR-145
mimics, miR-486 mimics, miR-584 mimics; 100 nmol; Guangzhou RiboBio
Co., Ltd.) using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.) and incubated at 37°C with 5% CO2 for
48 h before subsequent experiments. The siRNA and miRNA mimic
sequences are shown in Table
II.
 | Table IIsiRNA and miRNA mimics sequences. |
Table II
siRNA and miRNA mimics sequences.
| siRNA | Sequence
(5′-3′) |
|---|
| si-NC |
UUCUCCGAACGUGUCACGUTT |
| si-FEZF1-AS1 |
GCAAUAGGCCUGGGAAAGUTT |
| si-METTL3 |
CTGCAAGTATGTTCACTATGA |
| si-METTL14 |
AAGGATGAGTTAATAGCTAAA |
| si-YTHDF1 |
CCGCGTCTAGTTGTTCATGAA |
| si-YTHDF2 |
TTGGCTATTGGGAACGTCCTT |
| si-ITGA11-1 |
CCCAGUGGUUCAGAUCAAUTT |
| si-ITGA11-2 |
GACGGCAUUUGGCAUUGAATT |
| si-ITGA11-3 |
GACCUUCUCAGUCGAGUAUTT |
|
Biotinylated-miR-516b-mutated |
AGUCAACAGUAAGAAGCACUUU |
|
Biotinylated-miR-516b-wild-type |
AUCUGGAGGUAAGAAGCACUUU |
|
Biotinylated-miR-NC |
UUGUACUACACAAAAGUACUG |
Plasmid transfection
The pcDNA-3.1-NC and pcDNA3.1-FEZF1-AS1 plasmids for
FEZF1-AS1 overexpression were purchased from GenScript. The
plasmids (1 μg/well) were transfected into H1299 and H520
cells using Lipofectamine 2000 according to the manufacturer's
protocol.
Rescue assays
Cells were divided into 4 groups for transfection,
which were NC, pcDNA3.1-FEZF1-AS1 only, miR-516b-5p mimics only and
pcDNA3.1-FEZF1-AS1 plus miR-516b-5p mimics co-transfection,
performed as aforementioned. Cells were harvested after 48 h of
transfection.
Cell viability assay
Transfected cells (H1299 and H520) were seeded onto
96-well plates at a density of 1.5×104 cells/well after
8 h of transfection. The cell viability was evaluated every 24 h
using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies,
Inc.) assay, following the manufacturer's protocol.
Cell migration and invasion assay
H1299 and H520 cells were plated in their respective
medium (RPMI-1640 and DMEM, respectively) with 1% FBS in the upper
chamber of Transwell inserts (8.0-μm pores; Corning, Inc.).
For the migration assays, the cells (5×104) were
suspended in 200 μl RPMI-1640 medium or DMEM in the upper
chamber. For the invasion assays, the cells (2×105) were
suspended in 200 μl RPMI-1640 medium or DMEM, and placed
into the upper chamber (precoated with Matrigel at 37°C for 1 h;
Merck KGaA). The bottom chamber contained medium with 10% FBS.
Following incubation at 37°C for 48 h, the cells that had migrated
or invaded through the membrane were fixed with 4% formaldehyde for
30 min at room temperature, stained with 0.5% crystal violet for 8
min at room temperature and imaged using a light microscope at a
magnification of ×400.
Cell cycle analysis
The cells were collected by centrifugation at 160 ×
g at 4°C for 5 min and washed twice with cold PBS. For cell cycle
analysis, the cells were fixed overnight in 70% ethanol at 4°C.
Following centrifugation for 5 min at 160 × g at 4°C, the pellet
was stained with 50 μg/ml propidium iodide (BD Biosciences)
in 0.1% Triton X-100, 100 μg/ml RNase A (Thermo Fisher
Scientific, Inc.) and 0.02 mg/ml EDTA. Cell suspensions were
analyzed via flow cytometry using a FACSCalibur system (BD
Biosciences). The cell cycle distribution was analyzed using Flow
Jo 7.6 software (FlowJo LLC).
Coding potential tool
The coding potential of FEZF1-AS1 was calculated
using the Coding-Potential Assessment Tool (lilab.research.bcm.edu/cpat/).
m6A prediction and
analysis
The sequence-based RNA adenosine methylation site
predictor (SRAMP) software (http://www.cuilab.cn/sramp) was used for
m6A prediction and analysis (23). Treatment with 3-Deazaadenosine can
cause tumor cells to be in an RNA demethylation state (24). Therefore, H1299 cells were
incubated with 10 μM 3-Deazaadenosine at 37°C for 4 or 24
h.
Subcellular fractionation
The Minute Cytoplasmic and Nuclear Extraction kit
(Invent Biotechnologies, Inc.) was used to extract cytoplasmic and
nuclear fractions from NSCLC cell lines, following the
manufacturer's protocol. The RNA was extracted using TRIzol
reagent, as aforementioned. Subsequently, RT-qPCR was performed as
aforementioned to evaluate the expression levels of each fraction
of nuclear control transcript [metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1)], cytoplasmic control
transcript (GAPDH) and FEZF1-AS1.
Western blot analysis
Total protein was extracted from cell lines using
freshly prepared RIPA lysis buffer (150 mM NaCL, 1% NP-40, 0.1%
SDS, 2 μg/ml Aprotinin, 2 μg/ml Leupeptin, 1 mM PMSF,
1.5 mM EDTA, 1 mM NaVanadate) and protein concentration was
determined using a BCA assay. A total of 40 μg protein/lane
was separated via 15% SDS-PAGE and transferred electrophoretically
to nitrocellulose membranes. To evaluate protein expression, the
blots were blocked with 5% skimmed milk in TBS with 0.05% Tween-20
at 37°C for 1.5 h and incubated at 4°C overnight with a primary
rabbit monoclonal antibody against ITGA11 (1:1,000; cat. no.
ab198826; Abcam) and an antibody against p-actin (1:10,000; cat.
no. ac009; ABclonal Biotech Co., Ltd.) used as a control. The blots
were then probed with HRP-conjugated goat anti-rabbit (1:5,000;
cat. no. sc-2004; Santa Cruz Biotechnology, Inc.) or anti-mouse
(1:8,000; cat. no. sc-2005; Santa Cruz Biotechnology, Inc.)
secondary antibodies at 37°C for 1.5 h, visualized using enhanced
chemiluminescence (Thermo Fisher Scientific, Inc.) and scanned
using ImageQuant LAS 4010 Imaging System (GE Healthcare Life
Sciences).
RNA-binding protein immunoprecipitation
(RIP)
RIP experiments were performed using the Magna RIP
RNA-binding protein immunoprecipitation kit (EMD Millipore) and
anti-argonaute 2 (Ago2) antibody (5 μg; cat. no. ab32381;
Abcam), according to the manufacturer's protocol. Purified,
co-precipitated RNAs were subjected to RT-qPCR analysis, as
aforementioned.
RNA pulldown assay with biotinylated
miRNA
H1299 cells were transfected with biotinylated
miRNAs (30 nM; Table II) as
aforementioned, and harvested 48 h after transfection. The cell
lysates were incubated at 4°C for 6 h with M-280 streptavidin
magnetic beads (Thermo Fisher Scientific, Inc.) as previously
described (25). The bound RNAs
were purified using TRIzol reagent for further RT-qPCR analysis as
aforementioned.
Luciferase reporter assay
To create a 3′-untranslated region (UTR) luciferase
reporter construct of ITGA11, the sequences from putative
miR-516b-5p binding sites were synthesized and cloned into the
pmirGLO vector (Promega Corporation). The following wild-type (WT)
and mutated (MUT) primers were used to construct the 3′-UTR of
ITGA11: WT forward,
5′-ACTCGAGcccgagcaatggcgcctgctccctccagaatggaactcaagctg gttAGATC-3′
and WT reverse, 5′-CGCGTGAGCTCgggctcg
ttaccgcggacgagggaggtcttaccttgagttcgaccaaT-3′; MUT forward,
5′-ACTCGAGcccgacaatggcgcctgctccagaactcatggaactcaagctggttAGATC-3′
and MUT reverse, 5′-CGCGT GAGCT Cgggctcg
ttaccgcggacgaggtcttgagtaccttgagttcgaccaaT-3′. The amplified
fragment was cloned into the pmirGLO luciferase reporter vector at
the Mlul and XbaI sites. Briefly, cancer cells
(5×104 per well) were seeded in a 24-well plate the day
before transfection, and then co-transfected using Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.) with the firefly
luciferase-3′-UTR (pmirGLO or pmirGLO-ITGA11; 500 ng), along with
miR-516b-5p or control mimics (Guangzhou RiboBio Co., Ltd.). After
48 h, luciferase activity was measured using the Luc-Pair™
Duo-Luciferase HS assay kit (GeneCopoeia, Inc.) and normalized to
the Renilla luciferase activity. All experiments were
repeated at least three times.
Microarray analysis
Arraystar Human LncRNA Microarray V4.0 (Kangchen
BioTech Co., Ltd.) was used to screen the global profiling of human
lncRNAs and protein-coding transcripts. The samples included eight
frozen tissues of NSCLCs (four samples of squamous cell carcinoma
and four of adenocarcinoma) and paired normal lung tissues
(GSE137445) (26). The total RNA
was extracted from the cells or tissues as aforementioned. Sample
labeling and array hybridization were performed according to the
Agilent One-Color Microarray-Based Gene Expression Analysis
protocol (Agilent Technologies, Inc.) with minor modifications.
Briefly, mRNA was purified from total RNA after removal of
ribosomal RNA (mRNA-ONLY™ Eukaryotic mRNA Isolation kit; Epicentre;
Illumina, Inc.). Subsequently, each sample was amplified and
transcribed into fluorescent cRNA along the entire length of the
transcripts without 3′bias utilizing a random priming method
(Arraystar Flash RNA Labeling kit; Arraystar, Inc.). The labeled
cRNAs were purified using an RNeasy Mini kit (Qiagen, Inc.). The
concentration and specific activity of the labeled cRNAs (pmol
Cy3/μg cRNA) were measured by NanoDrop ND-1000 (Thermo
Fisher Scientific, Inc.). A total of 1 μg of each labeled
cRNA was fragmented by adding 5 μl 10X blocking agent and 1
μl 25X fragmentation buffer; the mixture was heated at 60°C
for 30 min, and finally 25 μl 2X GE hybridization buffer was
added to dilute the labeled cRNA. A total of 50 μl
hybridization solution was dispensed into the gasket slide and
assembled to the LncRNA expression microarray slide. The slides
were incubated for 17 h at 65°C in an Agilent Hybridization Oven
(Agilent Technologies, Inc.). The hybridized arrays were washed,
fixed and scanned using the Agilent DNA Microarray Scanner (part
no. G2505C; Agilent Technologies, Inc.). Quantile normalization and
subsequent data processing were performed using the GeneSpring GX
v11.5.1 software package (Agilent Technologies, Inc.). The
thresholds used to screen the upregulated or downregulated lncRNAs
were fold-change (FC) >2.0 and P<0.05. In addition, the
competing endogenous (ce)RNA network of FEZF1-AS1 was analyzed by
integrating the Arraystar Human mRNA microarray data and miRNA
bioinformatic analysis software (TargetScan v7.2; http://www.microrna.org) by Kangchen BioTech Co.,
Ltd..
Statistical analysis
All data are presented as the mean ± SEM. All
experiments were repeated >3 times. SPSS 22.0 (IBM Corp.) was
used for statistical analysis. The association between the
expression levels of FEZF1-AS1, ITGA11 and miR-516b-5p, and the
clinicopathological variables of patients with NSCLC was determined
using the χ2 test. Paired t-test was used to examine the
expression levels of FEZF1-AS1, ITGA11 and miR-516b-5p in normal
versus tumor tissue samples. Unpaired t-test was utilized to
examine the differences between two groups in vitro.
Significant differences among multiple groups were investigated
using one-way ANOVA followed by Bonferroni's post hoc test.
Kaplan-Meier analysis with log-rank test was used to assess the
overall survival rate. The correlation between FEZF1-AS1, ITGA11
and miR-516b-5p expression in NSCLC tissues was analyzed using
Spearman's correlation analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
FEZF1-AS1 is upregulated in NSCLC tissues
and is associated with a poor prognosis
To identify the regulatory networks of mRNA and
lncRNAs in NSCLC, eight pairs of NSCLC and non-tumorous adjacent
tissues, including four pairs of adenocarcinoma and four pairs of
squamous cell carcinoma, were analyzed using the Arraystar Human
lncRNA microarray. A total of 1,157 lncRNAs were found to be
significantly differentially expressed, of which 505 were
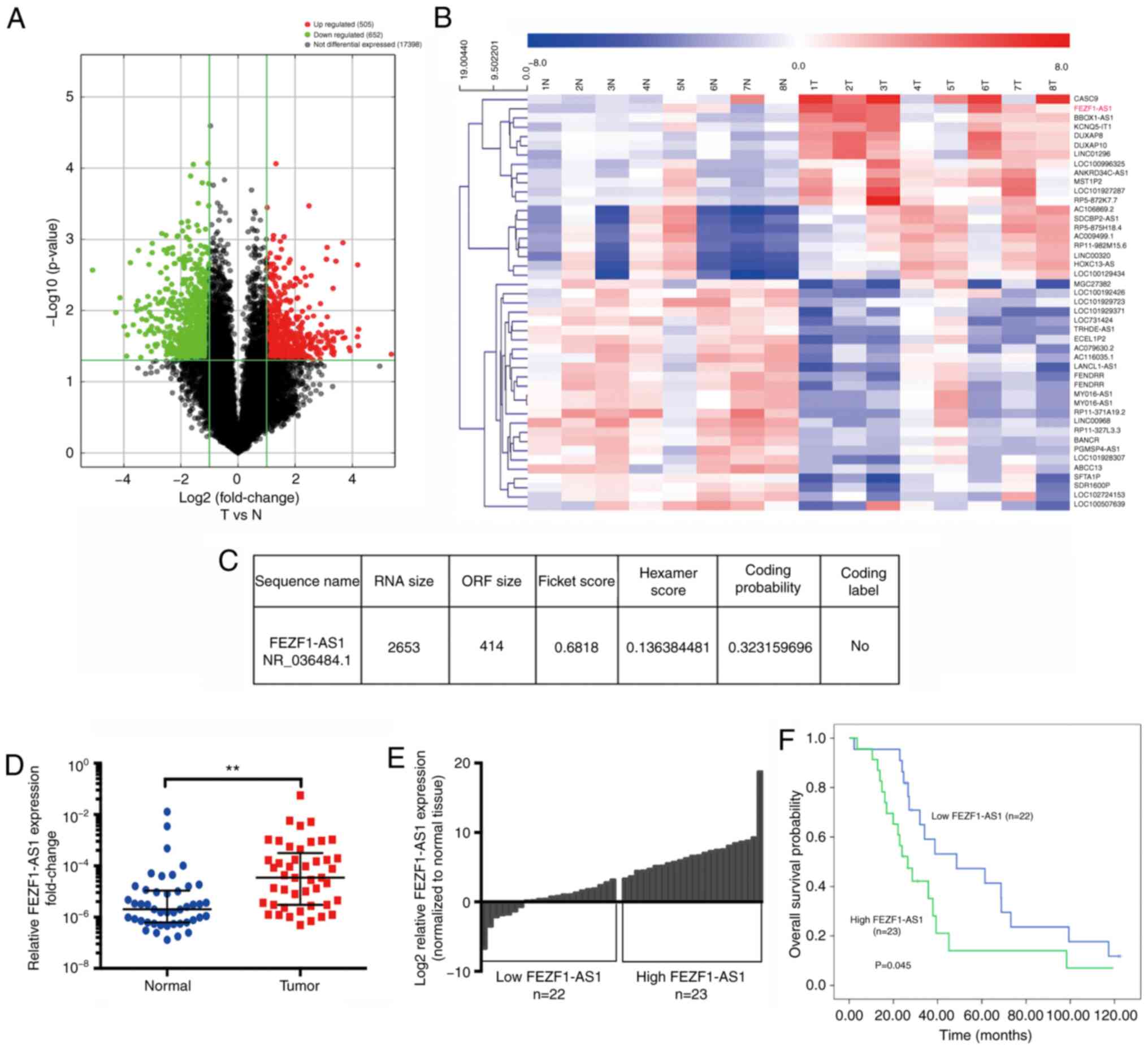
upregulated and 652 were downregulated by >2-fold (Fig. 1A). With the further restriction of
FC≥5 and P<0.05, FEZF1-AS1 was one of the most upregulated
lncRNAs (Fig. 1B), suggesting the
potential crucial role of FEZF1-AS1 in NSCLC tumorigenesis and
development. The coding potential of FEZF1-AS1 was calculated using
the Coding-Potential Assessment Tool (lilab.research.bcm.edu/cpat/), revealing that the
possible open reading frame of FEZF1-AS1 was very short, with a
very low coding probability (Fig.
1C).
To investigate the expression levels of FEZF1-AS1 in
NSCLC, RT-qPCR analysis was performed using the total RNA extracted
from 45 pairs of NSCLC tissues and their matched non-neoplastic
counterparts. The results revealed that FEZF1-AS1 expression was
significantly upregulated in NSCLC samples compared with that in
the corresponding normal tissues (Fig.
1D). To determine the association between FEZF1-AS1 and
clinicopathological features, the 45 patients were divided into the
high (FC>9.00; n=23) or low (FC≤9.00; n=22) FEZF1-AS1 expression
groups according to the median of FEZF1-AS1 expression (Fig. 1E). Kaplan-Meier analysis
demonstrated that high FEZF1-AS1 expression was significantly
associated with a poor OS rate (P=0.045; Fig. 1F). However, there were no
significant associations between FEZF1-AS1 expression and other
important clinicopathological features, such as lymph node
metastasis and clinical staging in patients with NSCLC (all
P>0.05; Tables III and
SI).
 | Table IIIAssociation between low (n=22) and
high (n=23) FEZF1-AS1 expression and clinicopathological features
in patients with non-small cell lung cancer. |
Table III
Association between low (n=22) and
high (n=23) FEZF1-AS1 expression and clinicopathological features
in patients with non-small cell lung cancer.
| Parameter | FEZF1-AS1
expression, n (%)
| P-value |
|---|
| Low | High |
|---|
| Age, years | | | >0.999 |
| <60 | 11 (47.8) | 12 (52.2) | |
| ≥60 | 11 (50.0) | 11 (50.0) | |
| Sex | | | 0.722 |
| Male | 18 (51.4) | 17 (48.6) | |
| Female | 4 (40.0) | 6 (60.0) | |
| Smoking
historya | | | 0.867 |
| Never | 7 (46.7) | 8 (53.3) | |
| Light | 2 (40.0) | 3 (60.0) | |
| Heavy | 13 (52.0) | 12 (48.0) | |
| Family history | | | 0.090 |
| No | 21 (53.8) | 18 (46.2) | |
| Yes | 1 (16.7) | 5 (83.3) | |
| Tumor size, cm | | | 0.542 |
| ≤5 | 13 (44.8) | 16 (55.2) | |
| >5 | 9 (56.3) | 7 (43.8) | |
| Lymph node
metastasis | | | >0.999 |
| Negative | 11 (47.8) | 12 (52.2) | |
| Positive | 11 (50.0) | 11 (50.0) | |
| Stageb | | | >0.999 |
| I-II | 10 (47.6) | 11 (52.4) | |
| III-IV | 12 (50.0) | 12 (50.0) | |
FEZF1-AS1 silencing inhibits lung cancer
cell proliferation, migration and invasion
The function of FEZF1-AS1 was investigated in NSCLC
cell lines. Compared with BEAS-2B cells, FEZF1-AS1 expression was
significantly upregulated in H358, H1299, H520 and SK-MES-1 NSCLC
cells (Fig. 2A). In order to
elucidate the mechanism of FEZF1-AS1 activity in oncogenesis, the
expression levels of FEZF1-AS1 were further examined in H1299 cells
by cytoplasmic and nuclear extraction, revealing that FEZF1-AS1 was
mostly located in the cytoplasmic fraction (Fig. 2B). To investigate the biological
function of FEZF1-AS1, knockdown efficiency of si-FEZF1-AS1 was
measured using RT-qPCR in H1299 and H520 cells (Fig. 2C). The CCK-8 assay revealed that
FEZF1-AS1-knowckdown significantly decreased the proliferative
capacity of NSCLC cells after ≥72 h (Fig. 2D). Additionally, it was observed
that FEZF1-AS1 silencing induced G2/M arrest (Figs. 2E and S1A), and Transwell assays demonstrated
that migration and invasion were significantly inhibited following
FEZF1-AS1 silencing (Fig. 2F and
G).
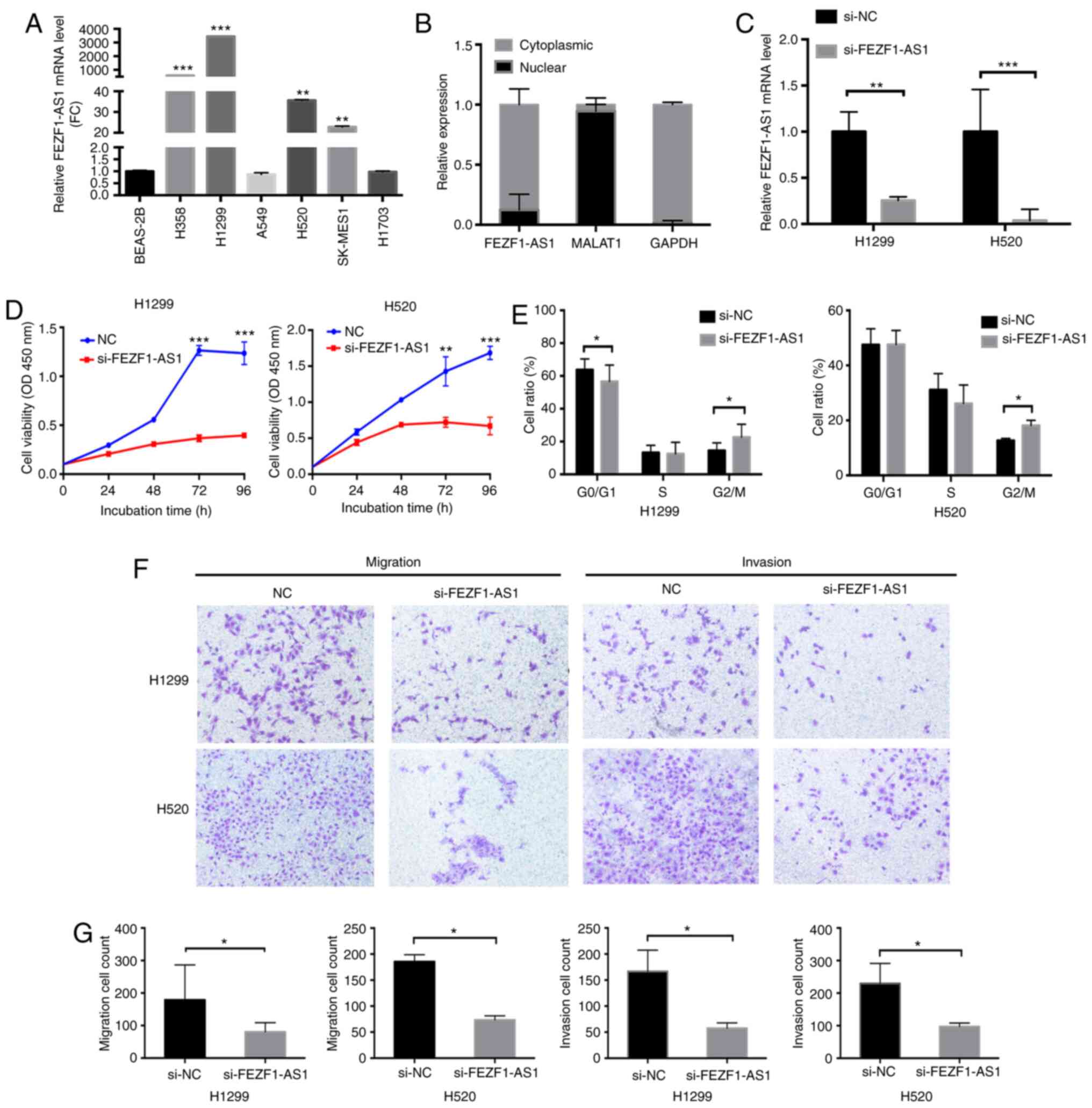 | Figure 2FEZF1-AS1-knockdown inhibits lung
cancer cell proliferation, migration and invasion. (A) Reverse
transcription-quantitative PCR analysis of FEZF1-AS1 expression in
BEAS-2B and six NSCLC cell lines. **P<0.01 and
***P<0.001 vs. BEAS-2B. (B) Subcellular fractionation
revealed that FEZF1-AS1 was mostly located in the cytoplasmic
compartment. (C) Efficacy of FEZF1-AS1-knockdown. (D) Growth curves
of H1299 and H520 cells following transfection with siFEZF1-AS1 or
siNC, as revealed using a Cell Counting Kit-8 assay. (E)
FEZF1-AS1-knockdown induced G2/M arrest in both H1299
and H520 cells. (F and G) FEZF1-AS1-knockdown significantly
decreased the migration and invasion of NSCLC cells in the
Transwell assays (magnification, ×400). Data are presented as the
mean ± SEM. *P<0.05; **P<0.01;
***P<0.001. NSCLC, non-small cell lung cancer; NC,
negative control; si, small interfering; FEZF1-AS1, FEZ family zinc
finger 1 antisense RNA 1; MALAT1, metastasis-associated lung
adenocarcinoma transcript 1; OD, optical density; FC,
fold-change. |
High FEZF1-AS1 expression in NSCLC cells
is regulated by m6A methylation
To reveal the motif of the high expression levels of
FEZF1-AS1 in NSCLC, m6A prediction and analysis software
SRAMP was used to predict the full-length sequence (2,653 bp) of
FEZF1-AS1. As presented in Fig. 3A and
B, seven m6A modified sites were predicted,
suggesting that FEZF1-AS1 expression may be regulated by
m6A modification.
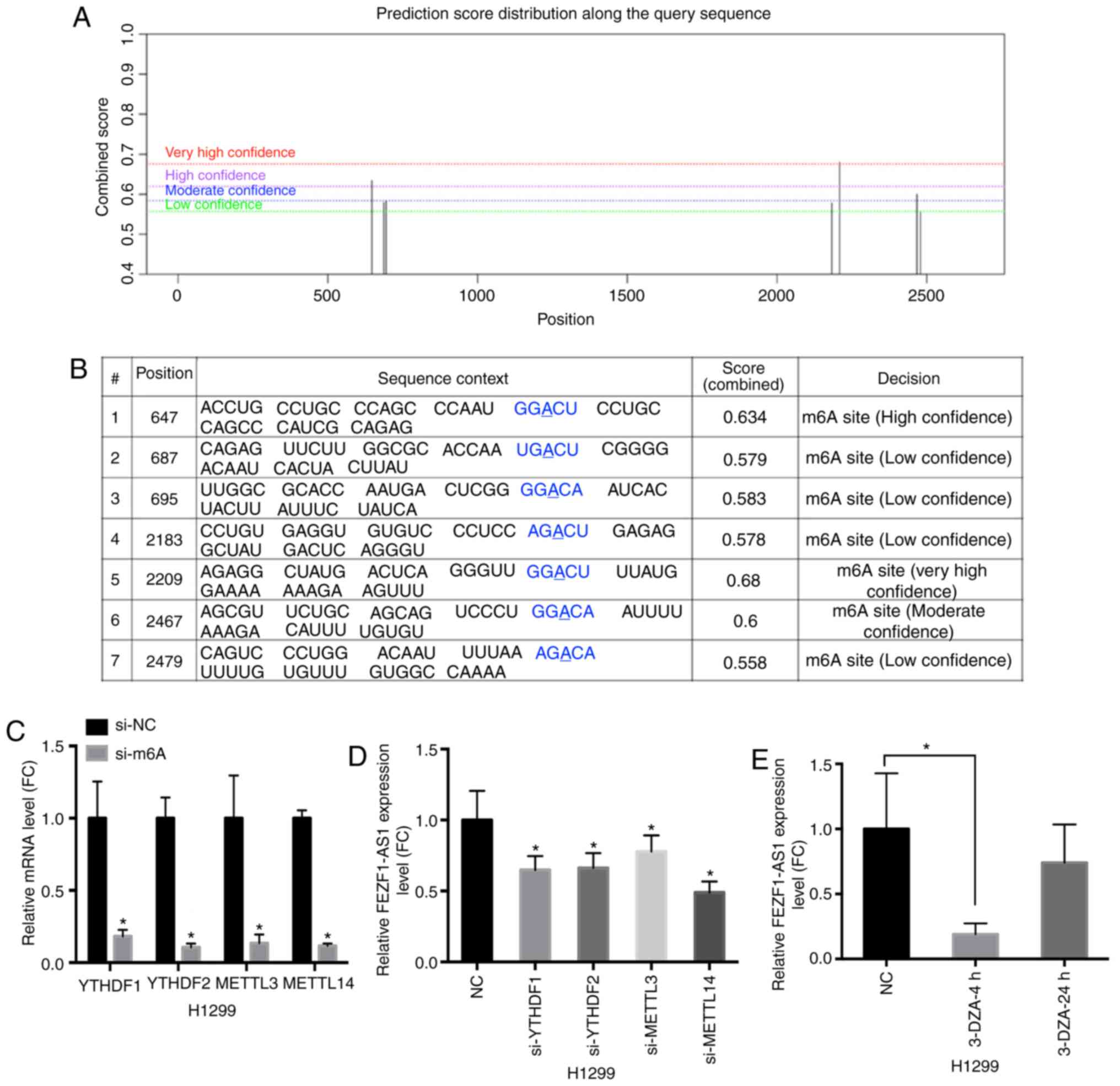 | Figure 3M6A modification is
involved in the high expression levels of FEZF1-AS1. (A) FEZF1-AS1
sequence-based m6A modification site prediction using
the SRAMP software combined with the prediction score. (B)
Positions of m6A modification sites. (C) Knockdown
efficiency of m6A siRNAs specific to YTHDF1, YTHDF2,
METTL3 and METTL14 in H1299 cells, and their respective expression
levels. (D) FEZF1-AS1 expression in H1299 cells following
si-YTHDF1, si-YTHDF2, si-METTL3 and si-METTL14 transfection. (E)
FEZF1-AS1 expression in H1299 cells treated with the demethylation
drug 3-DZA for 4 or 24 h. Data are presented as the mean ± SEM.
*P<0.05 vs. NC. m6A, N6-methyladenosine; NC, negative
control; si, small interfering; FEZF1-AS1, FEZ family zinc finger 1
antisense RNA 1; METTL3/14, methyltransferase-like 3/14; YTHDF1/2,
YTH N6-methyladenosine RNA binding protein 1/2; 3-DZA,
3-Deazaadenosine; FC, fold-change. |
m6A is the most widely used mRNA
modification method in mammals; this modification can be added,
deleted and preferentially combined with m6A through
three regulators, including 'writers' that generate m6A,
'erasers' that exhibit demethylation activity and 'readers' that
decode the m6A modification (27). On the basis of this prediction,
specific siRNAs targeting the 'readers' [methyltransferase-like 3
(METTL3) and METTL14] and 'writers' [YTH N6-methyladenosine RNA
binding protein 1 (YTHDF1) and YTHDF2] were transfected into H1299
cells to examine their effects on FEZF1-AS1 expression. The
efficacy of siRNA interference against METTL3, METTL14, YTHDF1 and
YTHDF2 was analyzed via RT-qPCR (Fig.
3C). Compared with the control group, downregulating these four
m6A modified molecules inhibited the expression levels
of FEZF1-AS1 (Fig. 3D).
Subsequently, H1299 cells were treated with 3-Deazaadenosine, an
RNA demethylation drug, at a concentration of 10 μM.
FEZF1-AS1 expression was detected in the cells collected at 4 and
24 h following administration. Compared with in the control group,
FEZF1-AS1 expression was significantly decreased in the 4-h group
and reversed in the 24-h group (Fig.
3E). The present results suggested that the high expression
levels of FEZF1-AS1 in NSCLC cells was positively regulated by
m6A methylation.
ITGA11 is a potential target of FEZF1-AS1
through miRNAs in NSCLC cells
FEZF1-AS1 was mostly located in the cytoplasm. It
has been demonstrated that lncRNAs in the cytoplasm can interact
with miRNAs as ceRNAs (7). Ago2 is
a crucial factor in miRNA biogenesis (28). An RIP experiment was performed
using an Ago2 antibody to investigate whether FEZF1-AS1 may bind to
miRNAs. As expected, FEZF1-AS1 was detected in Ago2
immunoprecipitates from control cells, indicating that the RNA
could indeed bind to miRNAs (Fig.
4A).
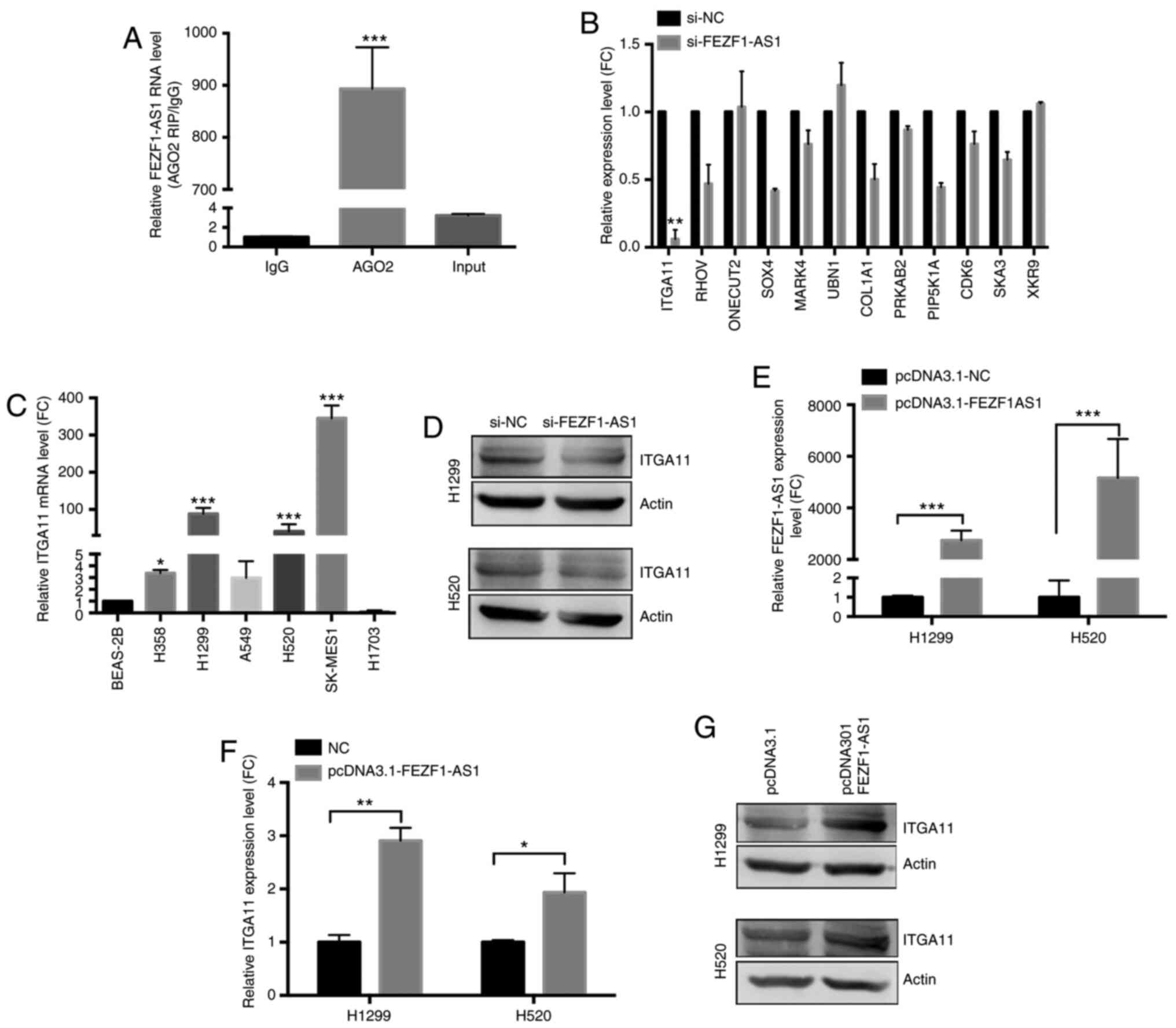 | Figure 4ITGA11 is a potential target of
FEZF1-AS1 in NSCLC cells. (A) FEZF1-AS1 was detected in Ago2
immunoprecipitates in the RNA-binding protein immunoprecipitation
assay. ***P<0.001 vs. IgG. (B) Following
FEZF1-AS1-knockdown, ITGA11 was the most downregulated mRNA of the
12 selected mRNAs, as determined using RT-qPCR.
**P<0.01 vs. si-NC. (C) RT-qPCR analysis of ITGA11
expression in BEAS-2B and NSCLC cell lines. *P<0.05
and ***P<0.001 vs. BEAS-2B. (D) Western blot analysis
verified that ITGA11 expression was regulated by FEZF1-AS1. (E)
Efficacy of FEZF1-AS1 overexpression. FEZF1-AS1 overexpression
upregulated ITGA11 expression, as determined using (F) RT-qPCR and
(G) western blot analysis. Data are presented as the mean ± SEM.
*P<0.05; **P<0.01;
***P<0.001. ITGA11, integrin subunit all; NSCLC,
non-small cell lung cancer; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; si, small interfering; FEZF1-AS1,
FEZ family zinc finger 1 antisense RNA 1; Ago2, argonaute 2; FC,
fold-change. |
lncRNAs can regulate miRNAs to influence the
expression levels of mRNAs as ceRNAs (8). Combined with the lncRNA microarray
and miRNA bioinformatic analysis using TargetScan software programs
results (data not shown), mRNA-ceRNA analysis was performed to
identify target genes. Based on the gene frequency, a total of 12
mRNAs (ITGA11, RHOV, ONECUT2, SOX4, MARK4, UBN1, COL1A1, PRKAB2,
PIP5K1A, CDK6, SKA3 and XKR9) were selected for further validation.
Following the knockdown of FEZF1-AS1, ITGA11 was the most
downregulated mRNA, as revealed using RT-qPCR (Fig. 4B). Compared with BEAS-2B cells, the
expression levels of ITGA11 were significantly increased in H358,
H1299, H520 and SK-MES-1 cells (Fig.
4C). Additionally, western blot analysis confirmed that ITGA11
protein levels were decreased following FEZF1-AS1 knockdown in
H1299 and H520 cells (Fig. 4D).
Subsequently, the role of FEZF1-AS1 overexpression on ITGA11
expression was further investigated. As shown in Fig. 4E-G, FEZF1-AS1 overexpression
significantly upregulated ITGA11 expression both at the mRNA and
protein levels in H1299 and H520 cells. Therefore, it was concluded
that FEZF1-AS1 was able to upregulate ITGA11 expression.
FEZF1-AS1 can compete with miR-516b-5p
for direct binding to ITGA11 in NSCLC cells
Based on the present ceRNA networks results and gene
ontology analysis in the GSE137445 array dataset, three miRNAs
(miR-486, miR-516b-5p and miR-584-3p) binding to both FEZF1-AS1 and
ITGA11 were identified. Furthermore, three other miRNAs (miR-126a,
miR-29b and miR-145) targeting ITGA11 were selected based on
previous reports (29-31). After miRNA overexpression,
miR-516b-5p significantly downregulated FEZF1-AS1 expression
(Fig. 5A). Proof of transfection
was performed for all miRNA mimics (data not shown). Additionally,
it was verified that ITGA11 expression was negatively regulated by
miR-516b-5p at both the transcriptional (Fig. 5B) and translational (Fig. 5D) levels, as measured using RT-qPCR
and western blot analysis, respectively, after verifying that
miR-516b-5p was successfully overexpressed in H1299 and H520 cells
(Fig. S1C). Compared with BEAS-2B
cells, the expression levels of miR-516b-5p were significantly
decreased in H358, H1299, H520, A549 and SK-MES-1 cells (Fig. 5C). Subsequently, it was examined
whether miR-516b-5p-mediated ITGA11 regulation occurred through
direct targeting of miRNA-binding sites in the ITGA11 sequence.
According to the bioinformatics analysis using TargetScan software,
the construction diagram of the miR-516b-5p binding site reporter
gene in the ITGA11 3′-UTR region (WT and MUT) is shown in Fig. 5E. ITGA11 was subcloned into the
pmirGLO dual-luciferase reporter vector, and luciferase assays were
performed in H1299 cells by inducing miR-516b-5p overexpression
using miR-516b-5p mimics. As shown in Fig. 5F, co-transfection with
pmirGLO-ITGA11-WT and miR-516b-5p mimics demonstrated a significant
decrease in luciferase reporter activity compared with the negative
control (NC) group (P<0.05). This repressive effect was
abolished by directed mutagenesis of the miR-516b-5p binding seed
region in ITGA11.
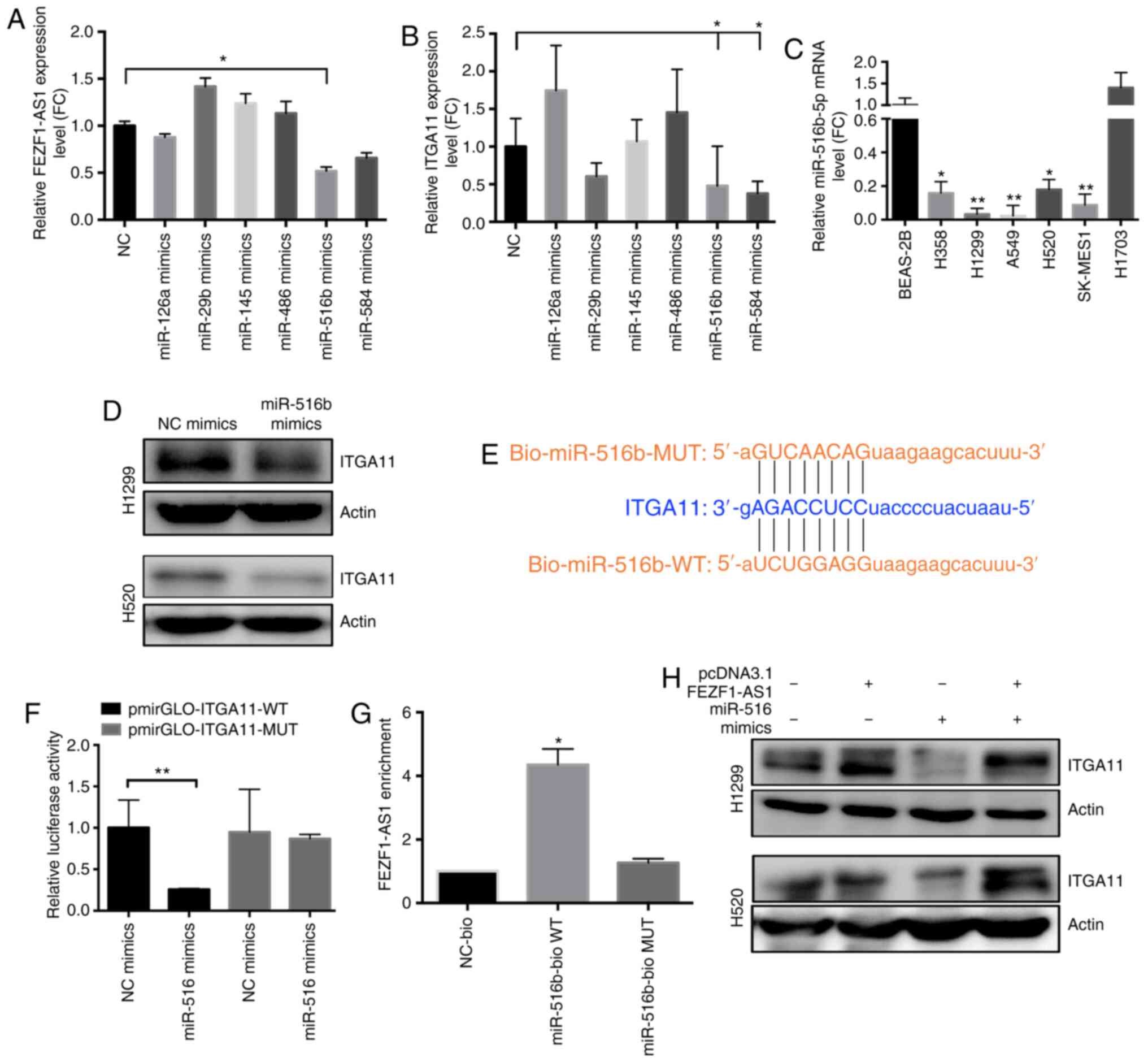 | Figure 5FEZF1-AS1 can compete with
miR-516b-5p for direct binding to ITGA11 in NSCLC cells. After
miRNA overexpression, miR-516b-5p significantly downregulated (A)
FEZF1-AS1 and (B) ITGA11 expression. *P<0.05 vs. NC.
(C) miR-516b-5P expression in BEAS-2B and NSCLC cell lines using
RT-qPCR. *P<0.05 and **P<0.01 vs.
BEAS-2B. (D) miR-516b-5p overexpression downregulated ITGA11
protein expression, as determined via western blot analysis. (E)
Predicted binding site of miR-516b-5p and ITGA11 sequence. (F)
Luciferase activity of H1299 cells co-transfected with
pmirGLO-ITGA11-WT or pmirGLO-ITGA11-MUT reporter plasmids and
miR-516b-5p mimic. (G) Using a biotin-avidin pulldown system,
miR-516b-5p precipitated FEZF1-AS1, as determined via RT-qPCR.
*P<0.05 vs. NC. (H) Rescue assay supporting the
FEZF1-AS1-miR-516b-5p-ITGA11 axis. Data are presented as the mean ±
SEM. NSCLC, non-small cell lung cancer; ITGA11, integrin subunit
all; WT, wild-type; MUT, mutated; NC, negative control; bio,
biotinylated; RT-qPCR, reverse transcription-quantitative PCR;
miR/miRNA, microRNA; FEZF1-AS1, FEZ family zinc finger 1 antisense
RNA 1; FC, fold-change. |
To assess whether miR-516b-5p interacted with
FEZF1-AS1, the biotin-avidin pulldown system was used, revealing
that FEZF1-AS1 co-precipitated miR-516b-5p (Fig. 5G). For the rescue assay, FEZF1-AS1
and miR-516b-5p were overexpressed either alone or together using
transfection. Compared with the NC group, the results revealed that
overexpression of miR-516b markedly suppressed ITGA11 expression,
whereas overexpression of FEZF1-AS1 partly abolished the silencing
effect of miR-516b-5p on ITGA11, suggesting that ITGA11 was
regulated by miR-516b-5p and FEZF1-AS1 (Fig. 5H). Therefore, FEZF1-AS1 may
function as a ceRNA, regulating miR-516b-5p and its target gene
ITGA11 in NSCLC.
Inhibiting ITGA11 or overexpressing
miR-516b-5p inhibits cell proliferation and invasion in NSCLC
ITGA11 expression was examined in 45 pairs of NSCLC
tissues and their matched non-neoplastic counterparts. It was
identified that ITGA11 expression was significantly upregulated in
the NSCLC samples compared with that in the corresponding normal
tissues (Fig. 6A). To determine
the association between ITGA11 and clinicopathological features,
the 45 patients were divided into high (FC>6.96; n=23) or low
(FC<6.96; n=22) ITGA11 expression groups according to the median
of ITGA11 expression. However, there were no significant
associations between ITGA11 expression and clinicopathological
features, such as lymph node metastasis and clinical staging in
patients with NSCLC (all P>0.05; Table IV).
 | Table IVAssociation between low (n=22) and
high (n=23) ITGA11 or miR-516b-5p expression and
clinicopathological features in patients with non-small cell lung
cancer. |
Table IV
Association between low (n=22) and
high (n=23) ITGA11 or miR-516b-5p expression and
clinicopathological features in patients with non-small cell lung
cancer.
| Parameter | ITGA11 expression,
n (%)
| P-value | miR-516b-5p
expression, n (%)
| P-value |
|---|
| Low | High | Low | High |
|---|
| Age, years | | | 0.652 | | | 0.884 |
| <60 | 12 (52.2) | 11 (47.8) | | 11 (47.8) | 12 (52.2) | |
| ≥60 | 10 (45.5) | 12 (54.5) | | 11 (50.0) | 11 (50.0) | |
| Sex | | | 0.936 | | | 0.936 |
| Male | 17 (48.6) | 18 (51.4) | | 17 (48.6) | 18 (51.4) | |
| Female | 5 (50.0) | 5 (50.0) | | 5 (50.0) | 5 (50.0) | |
| Smoking
historya | | | 0.739 | | | 0.014 |
| Never | 8 (53.3) | 7 (46.7) | | 11 (73.3) | 4 (26.7) | |
| Light | 3 (60.0) | 2 (40.0) | | 0 (0) | 5 (100.0) | |
| Heavy | 11 (44.0) | 14 (56.0) | | 11 (44.0) | 14 (56.0) | |
| Family history | | | 0.413 | | | 0.349 |
| No | 20 (51.3) | 19 (48.7) | | 18 (46.2) | 21 (53.8) | |
| Yes | 2 (33.3) | 4 (66.7) | | 4 (66.7) | 2 (33.3) | |
| Tumor size, cm | | | 0.912 | | | 0.009 |
| ≤5 | 14 (48.3) | 15 (51.7) | | 10 (34.5) | 19 (65.5) | |
| >5 | 8 (50.0) | 8 (50.0) | | 12 (75.0) | 4 (25.0) | |
| Lymph node
metastasis | | | 0.181 | | | 0.652 |
| Negative | 9 (39.1) | 14 (60.9) | | 12 (52.2) | 11 (47.8) | |
| Positive | 13 (59.1) | 9 (40.9) | | 10 (45.5) | 12 (54.5) | |
| Stageb | | | 0.051 | | | 0.449 |
| I-II | 7 (33.3) | 14 (66.7) | | 9 (42.9) | 12 (57.1) | |
| III-IV | 15 (62.5) | 9 (37.5) | | 13 (54.2) | 11 (45.8) | |
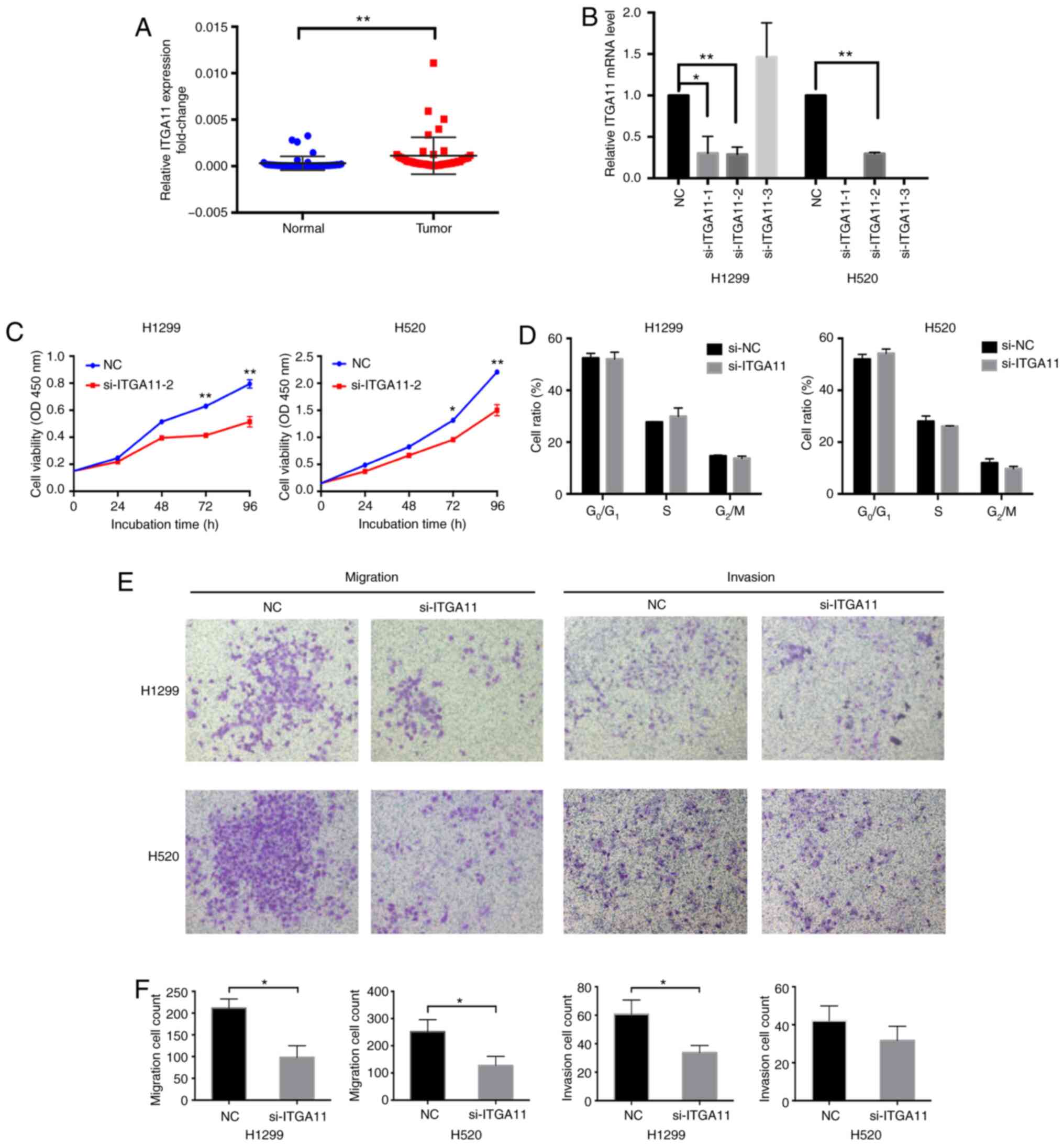
To examine the role of ITGA11 in cell proliferation,
ITGA11 expression was knocked down in H1299 and H520 cells
(Fig. 6B), revealing that the
proliferative capacities of these cells were significantly
decreased after >72 h (Fig.
6C). Transwell assays revealed that the knockdown of ITGA11
significantly decreased the migratory and invasive abilities of
NSCLC cells (Fig. 6E and F).
However, ITGA11-knockdown did not influence the cell cycle
(Figs. 6D and S1B).
Compared with surrounding healthy tissues,
miR-516b-5p expression was significantly inhibited in tumor tissues
compared with in normal tissues (Fig.
7A). To determine the association between miR-516b-5p
expression and clinicopathological features, the 45 patients were
divided into high (FC>0.29; n=23) or low miR-516b-5p expression
groups (FC <0.29; n=22) according to the median of miR-51b-5p
expression. Smoking history (P=0.014) and tumor size (P=0.009) were
associated with miR-516b-5p expression using a χ2 test
in patients with NSCLC (Table
IV). However, due to the small sample size of NSCLC tissues, a
statistical association between miR-516b-5p expression and OS was
not identified (data not shown), nor a correlation between
FEZF1-AS1, ITGA11 and miR516b-5p expression (data not shown). The
CCK-8 assay revealed that overexpression of miR-516b-5p
significantly inhibited NSCLC cell proliferation after >72 h
(Fig. 7B). Additionally, in the
Transwell assay, the migratory and invasive abilities of tumor
cells were inhibited in the miR-516b-5p mimic group (Fig. 7C and D) compared with in the
control group. The current data indicated that inhibiting ITGA11 or
overexpressing miR-516b-5p inhibited cell proliferation and
invasion in NSCLC.
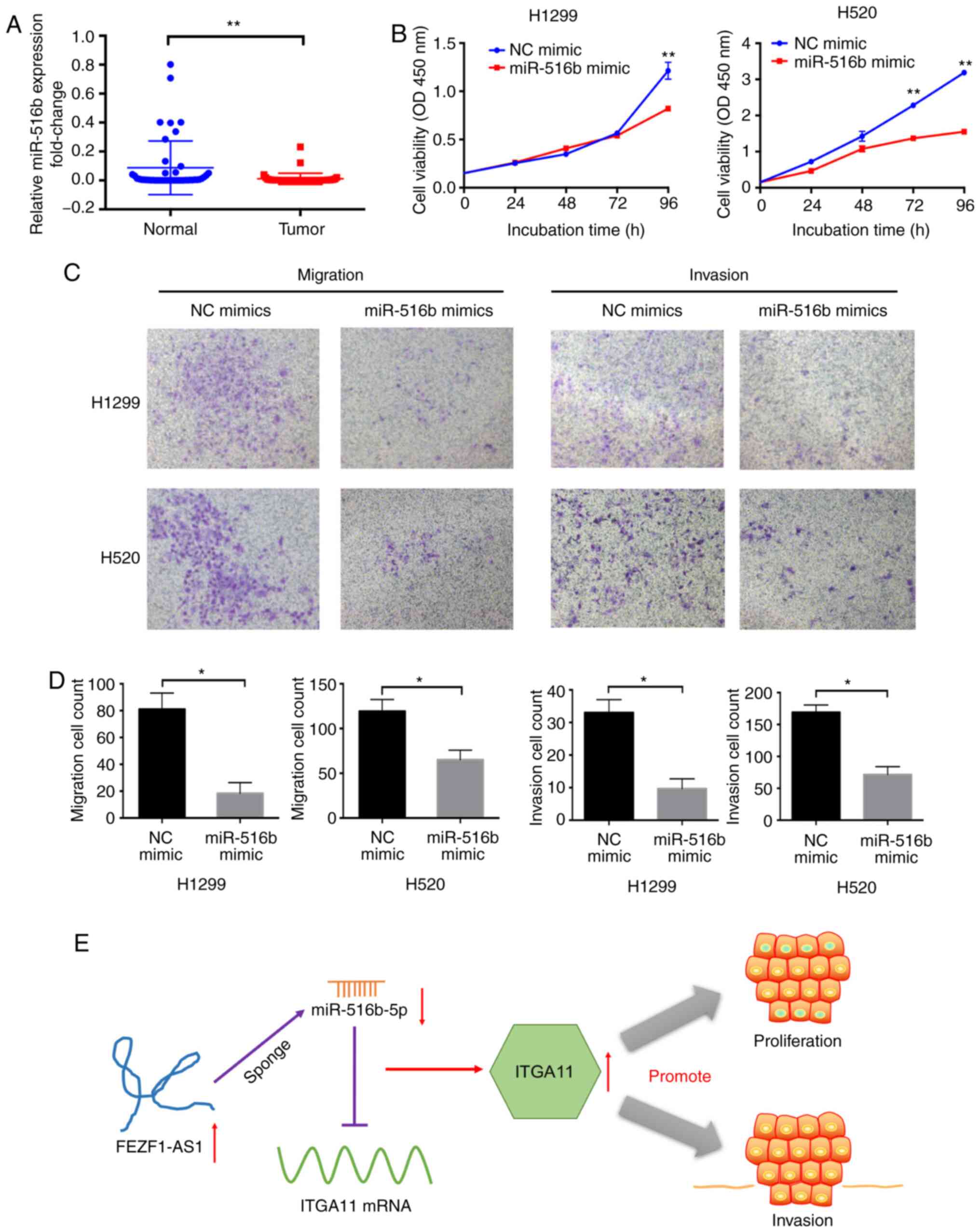 | Figure 7miR-516b-5p overexpression inhibits
cell proliferation and invasion in NSCLC. (A) Reverse
transcription-quantitative PCR analysis of miR-516b-5p expression
in 45 pairs of NSCLC tissues and corresponding non-tumor lung
tissues analyzing using a paired Student's t-test. (B) Growth
curves of H1299 and H520 cells following miR-516b-5p or NC
overexpression were determined using a Cell Counting Kit-8 assay.
(C and D) miR-516b-5p upregulation significantly decreased
migration and invasion in NSCLC cells in the Transwell assays
(magnification, ×400). (E) Graph of the potential mechanism of the
regulatory network of FEZF1-AS1, miR-516b-5p and ITGA11 in NSCLC
cells. FEZF1-AS1 may upregulate ITGA11 via sponging miR-516b-5p,
thus promoting the proliferation and invasion of NSCLC cells. Data
are presented as the mean ± SEM. *P<0.05 and
**P<0.01 vs. NC or indicated groups. NSCLC, non-small
cell lung cancer; ITGA11, integrin subunit α11; NC, negative
control; si, small interfering; FEZF1-AS1, FEZ family zinc finger 1
antisense RNA 1; OD, optical density; miR, microRNA. |
Discussion
NSCLC accounts for the vast majority of lung cancer
cases (2). In past years, numerous
studies regarding non-coding RNAs, including lncRNAs, have been
conducted (4,6-9).
lncRNA FEZF1-AS1 is the antisense RNA of FEZF1. In various types of
cancer, FEZF1-AS1 expression in cancer tissues is significantly
higher compared with that in normal tissues, and high FEZF1-AS1
expression indicates a poor survival in patients with colon,
gastric and cervical cancer (15,16,32).
The present data revealed that FEZF1-AS1 was highly expressed in
NSCLC tissues, compared with normal tissues, and was associated
with a poor prognosis in patients with NSCLC. FEZF1-AS1
downregulation in NSCLC cells inhibited cell proliferation,
migration and invasion, and arrested the cell cycle at the
G2/M phase, in accordance with findings previously
reported on the function of FEZF1-AS1 in NSCLC and breast cancer
cell lines (18,33).
Subsequently, the mechanism underlying the high
FEZF1-AS1 expression in NSCLC was explored, and m6A
modification was identified. m6A is the most widely used
mRNA modification method in mammals (27). The modification can be added,
deleted and preferentially combined with m6A through
three regulators of 'writers', 'erasers' and 'readers' (27). The m6A modification is
involved in a variety of mRNA processes, including mRNA translation
and decay (34). In addition to
mRNA, non-coding RNAs are also regulated by m6A, and
miRNAs and lncRNAs are important classes of non-coding RNAs
(35). It has been reported in
several studies that the expression levels of lncRNAs are regulated
by m6A modification. For example, the change of lncRNA
MALAT1 cut by m6A 'reading' molecules is associated with
the progression of cancer (36,37).
The downregulation of METTL3 in the 'writing' module can damage the
lncRNA Xist-mediated gene silencing (38). However, to the best of our
knowledge, whether FEZF1-AS1 expression is regulated by
m6A modification has not been reported. In the present
study, knockdown of 'readers' (METTL3 and METTL14) and 'writers'
(YTHDF1 and YTHDF2) resulted in the decrease of FEZF1-AS1
expression. Treatment with 3-Deazaadenosine causes tumor cells to
be in an RNA-demethylation state (24). Following treatment with
3-Deazaadenosine in the present study, FEZF1-AS1 expression was
significantly decreased. The current results demonstrated that
m6A modification may have an important regulatory effect
on FEZF1-AS1 expression.
To investigate the mechanism of FEZF1-AS1 in NSCLC,
mRNAs that could be regulated by FEZF1-AS1 were screened, and
ITGA11, a member of the integrin family, was selected. Integrins
are transmembrane receptors that mediate the connection between
cells and their external environment (39). These proteins are heterodimers
formed by two subunits, the α (120-185 kD) and β (90-110 kD)
subunits (40). In cells, ITGA11
is involved in collagen-mediated biological processes, such as cell
migration, collagen deposition and collagen recombination (41). ITGA11 is highly expressed in
esophageal squamous cell carcinoma, head and neck squamous
carcinoma, prostate cancer and breast cancer, and is closely
associated with the migration and invasion of tumor cells (42-44).
In lung adenocarcinoma A549, H460 and H520 cell lines, tumor growth
was significantly higher in A549+WT, compared with A549+Knockout
(KO) tumors (45). Re-expression
of human a11 cDNA in KO cells rescued the tumor growth rate to a
rate that was comparable with that of the A549+WT tumors (45). In patients with NSCLC, ITGA11
expression is significantly upregulated and is associated with a
poor OS (46). In the present
study, both the migration and invasion of NSCLC cells were
inhibited following ITGA11-knockdown. This was likely due to the
function of integrin in connecting cells to collagen in the
extracellular matrix, but this hypothesis needs to be further
confirmed.
miR-516b-5p was selected and validated in the
present study as a possible binding miRNA to both FEZF1-AS1 and
ITGA11. Low miR-516 expression can significantly improve OS in
patients with lung squamous cell carcinoma (47). In the present study, miR-516b-5p
expression in NSCLC tissues was lower compared with in
non-neoplastic tissues, and the proliferation, migration and
invasion of cells was inhibited following miR-516b-5p upregulation.
In the present study, both ITGA11 and miR-516b-5p were involved in
cell proliferation, migration and invasion, which is consistent
with the biological role in tumor progression of FEZF1-AS1.
Following FEZF1-AS1-knockdown, ITGA11 expression was decreased at
both the RNA and protein levels. Additionally, ITGA11 expression
was decreased following miR-516b overexpression. Therefore, both
FEZF1-AS1 and miR-516b may affect ITGA11 expression. Through RIP
and RNA pull-down assays, together with the effects of miR-516b-5p
upregulation on FEZF1-AS1, it was concluded that miR-516b-5p and
FEZF1-AS1 may share a binding site. The rescue assay further
confirmed this regulatory axis (Fig.
7E).
Future studies should explore the effects of
silencing both ITGA11 and miR-516b-5p, or of overexpressing
FEZF1-AS1 and silencing ITGA11, and to observe phenotypic changes
in proliferation and invasion. In addition to the involvement of
miR-516b-5p and ITGA11, other FEZF1-AS1-regulated genes may also
contribute to the pro-tumorigenic function of FEZF1-AS1. Future
studies are warranted to comprehensively explore the role of
FEZF1-AS1 in NSCLC pathogenesis.
In conclusion, the present study identified a novel
lncRNA, FEZF1-AS1, associated with a poor prognosis in patients
with NSCLC. The current results suggested that FEZF1-AS1 was an
oncogenic regulator that promoted cell proliferation and invasion.
It induced competitive binding with miR-516b-5p, resulting in the
upregulation of ITGA11 expression. Therefore, the
FEZF1-AS1/miR-516b-5p/ITGA11 axis may be a valuable target for
NSCLC prognosis and treatment.
Supplementary Data
Acknowledgments
Not applicable.
Funding
The present study was supported by Hebei
Postgraduate Innovation Funding Project (grant no.
CXZZBS2017111).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LX designed the study. HeS, HL, XD and ML
contributed to cell transfection, RT-qPCR and western blotting. HeS
contributed to RIP, RNA pull-down and luciferase assays. HL
contributed to data analysis. YL and XZ collected the clinical data
and follow-up data. HaS performed the flow cytometric and data
analysis. HeS and LX participated in writing and revising the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The research protocol conformed to the principles
outlined in the Declaration of Helsinki. All patients provided
written informed consent and the protocol of the study was approved
by the Research Ethics Committee of the Fourth Hospital of Hebei
Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bade BC and Dela Cruz CS: Lung cancer
2020: Epidemiology, etiology, and prevention. Clin Chest Med.
41:1–24. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Velcheti V, Schalper KA, Carvajal DE,
Anagnostou VK, Syrigos KN, Sznol M, Herbst RS, Gettinger SN, Chen L
and Rimm DL: Programmed death ligand-1 expression in non-small cell
lung cancer. Lab Invest. 94:107–116. 2014. View Article : Google Scholar
|
|
4
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Du Z, Sun T, Hacisuleyman E, Fei T, Wang
X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW and Liu XS:
Integrative analyses reveal a long noncoding RNA-mediated sponge
regulatory network in prostate cancer. Nat Commun. 7:109822016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Y, Chen L, Gu J, Zhang H, Yuan J,
Lian Q, Lv G, Wang S, Wu Y, Yang YT, et al: Recurrently deregulated
lncRNAs in hepatocellular carcinoma. Nat Commun. 8:144212017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Seiler J, Breinig M, Caudron-Herger M,
Polycarpou-Schwarz M, Boutros M and Diederichs S: The lncRNA veluct
strongly regulates viability of lung cancer cells despite its
extremely low abundance. Nucleic Acids Res. 45:5458–5469. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Park SM, Choi EY, Bae DH, Sohn HA, Kim SY
and Kim YJ: The LncRNA EPEL promotes lung cancer cell proliferation
through E2F target activation. Cell Physiol Biochem. 45:1270–1283.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen N, Guo D, Xu Q, Yang M, Wang D, Peng
M, Ding Y, Wang S and Zhou J: Long non-coding RNA FEZF1-AS1
facilitates cell proliferation and migration in colorectal
carcinoma. Oncotarget. 7:11271–11283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu X, Zhang P, Zhu H, Li S, Chen X and Shi
L: Long noncoding RNA FEZF1-AS1 indicates a poor prognosis of
gastric cancer and promotes tumorigenesis via activation of Wnt
signaling pathway. Biomed Pharmacother. 96:1103–1108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Z, Zhao P, Han Y and Lu S: LincRNA
FEZF1-AS1 is associated with prognosis in lung adenocarcinoma and
promotes cell proliferation, migration and invasion. Oncol Res.
27:39–45. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He R, Zhang FH and Shen N: LncRNA
FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through
suppressing E-cadherin and regulating WNT pathway in non-small cell
lung cancer (NSCLC). Biomed Pharmacother. 95:331–338. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou C, Xu J, Lin J, Lin R, Chen K, Kong J
and Shui X: Long noncoding RNA FEZF1-AS1 promotes osteosarcoma
progression by regulating the miR-4443/NUPR1 axis. Oncol Res.
26:1335–1343. 2018.PubMed/NCBI
|
|
19
|
Ye H, Zhou Q, Zheng S, Li G, Lin Q, Ye L,
Wang Y, Wei L, Zhao X, Li W, et al: FEZF1-AS1/miR-107/ZNF312B axis
facilitates progression and Warburg effect in pancreatic ductal
adenocarcinoma. Cell Death Dis. 9:342018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li QY, Chen L, Hu N and Zhao H: Long
non-coding RNA FEZF1-AS1 promotes cell growth in multiple myeloma
via miR-610/Akt3 axis. Biomed Pharmacother. 103:1727–1732. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goldstraw P, Chansky K, Crowley J,
Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P,
Mitchell A, Bolejack V, et al: The IASLC lung cancer staging
project: Proposals for revision of the TNM stage groupings in the
forthcoming (Eighth) Edition of the TNM classification for lung
cancer. J Thorac Oncol. 11:39–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Zhou Y, Zeng P, Li YH, Zhang Z and Cui Q:
SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based
on sequence-derived features. Nucleic Acids Res. 44:e912016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Backlund PS Jr, Carotti D and Cantoni GL:
Effects of the S-adenosylhomocysteine hydrolase inhibitors
3-deazaadenosine and 3-deazaaristeromycin on RNA methylation and
synthesis. Eur J Biochem. 160:245–251. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang K, Long B, Zhou LY, Liu F, Zhou QY,
Liu CY, Fan YY and Li PF: CARL lncRNA inhibits anoxia-induced
mitochondrial fission and apoptosis in cardiomyocytes by impairing
miR-539-dependent PHB2 downregulation. Nat Commun. 5:35962014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Ding X, Liu B, Li M, Chang Y, Shen
H, Xie SM, Xing L and Li Y: ETV4 overexpression promotes
progression of non-small cell lung cancer by upregulating PXN and
MMP1 transcriptionally. Mol Carcinog. 59:73–86. 2020. View Article : Google Scholar
|
|
27
|
Fu Y, Dominissini D, Rechavi G and He C:
Gene expression regulation mediated through reversible
m6A RNA methylation. Nat Rev Genet. 15:293–306. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheloufi S, Dos Santos CO, Chong MM and
Hannon GJ: A dicer-independent miRNA biogenesis pathway that
requires Ago catalysis. Nature. 465:584–589. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mao Z, Wu F and Shan Y: Identification of
key genes and miRNAs associated with carotid atherosclerosis based
on mRNA-seq data. Medicine (Baltimore). 97:e98322018. View Article : Google Scholar
|
|
30
|
Li Z, Jia J, Gou J, Tong A, Liu X, Zhao X
and Yi T: Mmu-miR-126a-3p plays a role in murine embryo
implantation by regulating Itga11. Reprod Biomed Online.
31:384–393. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang TC, Renuse S, Pinto S, Kumar P, Yang
Y, Chaerkady R, Godsey B, Mendell JT, Halushka MK, Civin CI, et al:
Identification of miR-145 targets through an integrated omics
analysis. Mol Biosyst. 11:197–207. 2015. View Article : Google Scholar :
|
|
32
|
Zhang HH and Li AH: Long non-coding RNA
FEZF1-AS1 is up-regulated and associated with poor prognosis in
patients with cervical cancer. Eur Rev Med Pharmacol Sci.
22:3357–3362. 2018.PubMed/NCBI
|
|
33
|
Zhang Z, Sun L, Zhang Y, Lu G, Li Y and
Wei Z: Long non-coding RNA FEZF1-AS1 promotes breast cancer
stemness and tumorigenesis via targeting miR-30a/Nanog axis. J Cell
Physiol. 233:8630–8638. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao BS, Roundtree IA and He C:
Post-transcriptional gene regulation by mRNA modifications. Nat Rev
Mol Cell Biol. 18:31–42. 2017. View Article : Google Scholar
|
|
35
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Alarcon CR, Goodarzi H, Lee H, Liu X,
Tavazoie S and Tavazoie SF: HNRNPA2B1 is a mediator of
m(6)A-dependent nuclear RNA processing events. Cell. 162:1299–1308.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo F, Li Y, Liu Y, Wang J, Li Y and Li G:
Inhibition of metastasis-associated lung adenocarcinoma transcript
1 in CaSki human cervical cancer cells suppresses cell
proliferation and invasion. Acta Biochim Biophys Sin (Shanghai).
42:224–229. 2010. View Article : Google Scholar
|
|
38
|
Patil DP, Chen CK, Pickering BF, Chow A,
Jackson C, Guttman M and Jaffrey SR: m(6)A RNA methylation promotes
XIST-mediated transcriptional repression. Nature. 537:369–373.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Takada Y, Ye X and Simon S: The integrins.
Genome Biol. 8:2152007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hegde S and Raghavan S: A skin-depth
analysis of integrins: Role of the integrin network in health and
disease. Cell Commun Adhes. 20:155–169. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tiger CF, Fougerousse F, Grundström G,
Velling T and Gullberg D: alpha11beta1 integrin is a receptor for
interstitial collagens involved in cell migration and collagen
reorganization on mesenchymal nonmuscle cells. Dev Biol.
237:116–129. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chai J, Modak C, Ouyang Y, Wu SY and Jamal
MM: CCN1 induces ß-catenin translocation in esophageal squamous
cell carcinoma through integrin a11. ISRN Gastroenterol.
2012:2072352012. View Article : Google Scholar
|
|
43
|
Parajuli H, Teh MT, Abrahamsen S,
Christoffersen I, Neppelberg E, Lybak S, Osman T, Johannessen AC,
Gullberg D, Skarstein K and Costea DE: Integrin a11 is
overexpressed by tumour stroma of head and neck squamous cell
carcinoma and correlates positively with alpha smooth muscle actin
expression. J Oral Pathol Med. 46:267–275. 2017. View Article : Google Scholar
|
|
44
|
Reigstad I, Smeland HY, Skogstrand T,
Sortland K, Schmid MC, Reed RK and Stuhr L: Stromal integrin a11ß1
affects RM11 prostate and 4T1 breast xenograft tumors differently.
PLoS One. 11:e01516632016. View Article : Google Scholar
|
|
45
|
Zhu CQ, Popova SN, Brown ER,
Barsyte-Lovejoy D, Navab R, Shih W, Li M, Lu M, Jurisica I, Penn
LZ, et al: Integrin alpha 11 regulates IGF2 expression in
fibroblasts to enhance tumorigenicity of human non-small-cell lung
cancer cells. Proc Natl Acad Sci USA. 104:11754–11759. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu P, Wang Y, Wu Y, Jia Z, Song Y and
Liang N: Expression and prognostic analyses of ITGA11, ITGB4 and
ITGB8 in human non-small cell lung cancer. PeerJ. 7:e82992019.
View Article : Google Scholar :
|
|
47
|
Qi L, Gao C, Feng F, Zhang T, Yao Y, Wang
X, Liu C, Li J, Li J and Sun C: MicroRNAs associated with lung
squamous cell carcinoma: New prognostic biomarkers and therapeutic
targets. J Cell Biochem. 120:18956–18966. 2019. View Article : Google Scholar : PubMed/NCBI
|