Introduction
Hepatocellular carcinoma (HCC) was the seventh
leading type of cancer and the fourth leading cause of
cancer-related death worldwide in 2017 (1,2).
Liver cancer has a poor prognosis, with a ratio of mortality to
incidence of 0.93 worldwide (2).
Surgical resection, transplantation and percutaneous ablation are
clinically available for the treatment of early-stage HCC (3,4).
However, resistance to chemotherapy or radiotherapy has resulted in
an absence of effective therapeutic options and a poor prognosis,
especially for patients with advanced HCC (5). Therefore, it is important to develop
novel treatments or to reverse chemoresistance to improve the
outcomes of patients with advanced HCC.
Eukaryotic translation initiation factor 5A (eIF5A
or eIF5A1) is highly conserved throughout eukaryotic evolution and
serves roles in mRNA translation, cellular proliferation,
differentiation and inflammation (6). eIF5A and its homolog eIF5A2, a 17-kDa
protein comprising 153 amino acids, are the only known proteins
that contain a unique amino acid residue, hypusine, which is
crucial for their function to maintain polyamine homeostasis
(6,7). eIF5A2 is of particular interest since
it is rare in the majority of normal tissues but abundant in the
testes, parts of the brain and several types of malignant tissues
including bladder and colorectal cancer as well as hepatocellular
carcinoma (8,9). eIF5A2 has been proposed to be an
oncogene, and overexpression of eIF5A2 has been demonstrated to
promote cell aggressiveness in colorectal carcinoma (10), bladder cancer (11), melanoma (12) and gastric cancer (13). Furthermore, inhibition of eIF5A2 by
N1-guanyl-1, 7-diaminoheptane, an inhibitor of deoxyhypusine
synthase, has been demonstrated to enhance the chemotherapeutic
effects of gemcitabine in pancreatic ductal adenocarcinoma cells
(14) and that of doxorubicin in
bladder cancer cells (11).
Although a number of studies have been performed to understand the
relationship between eIF5A2 and chemosensitivity (11,14),
the mechanism underlying eIF5A2 and chemosensitivity in HCC remains
unclear.
Autophagy, a lysosomal degradation pathway for
providing energy and macromolecular precursors (15), has also been implicated in the
development of chemoresistance. When cells encounter environmental
stressors, such as nutrient starvation and pathogen infection,
autophagy is induced to digest and recycle cellular proteins and
organelles to sustain cellular metabolism, or to eliminate
pathogens and apoptotic cells (16,17).
Autophagy is also coordinated with the ubiquitin-proteasome system
to remove polyubiquitinated and aggregated proteins (18). Therefore, autophagy may promote
cell survival and contribute to chemoresistance. A number of
studies have demonstrated that inhibition of autophagy overcomes
chemoresistance in prostate cancer, leukemia and bladder cancer
(19-21). The induction of autophagy has also
been reported to cause a form of programmed cell death termed
autophagic cell death (22-24).
Furthermore, Segala et al (25) have reported that dendrogenin A, a
cholesterol metabolite, directly controls a nuclear receptor to
trigger lethal autophagy in melanoma. Autophagy has been identified
as a cytoprotective mechanism in gastric carcinoma, leukemia and
squamous cell carcinoma (26-28).
In addition, autophagy serves a cytocidal role in breast and
colorectal cancer (29,30). However, the role of autophagy in
the chemoresistance or chemosensitivity in HCC remains
controversial.
The present study aimed to determine the potential
roles of eIF5A2 in doxorubicin sensitivity and to investigate the
effects of autophagy during this process.
Materials and methods
Ethics statement
The present study was approved by the Research
Ethics Committee of the Second Affiliated Hospital of Zhejiang
University School of Medicine (approval no. 2018-238; Hangzhou,
China). All samples were anonymously coded in accordance with local
ethical guidelines (based on the Declaration of Helsinki), and
written informed consent was obtained from all patients. All
animals used received appropriate care according to the
Institutional Animal Care and Use Committee at the Second
Affiliated Hospital of Zhejiang University School of Medicine
(approval no. 2018-311). All efforts were made to minimize animal
suffering.
Cell lines and culture
The human hepatocellular carcinoma cell lines
SNU449, SNU387 and Huh7 were purchased from the Cell Bank of Type
Culture Collection of Chinese Academy of Sciences, Shanghai
Institute of Cell Biology, Chinese Academy of Sciences. SNU449 and
SNU387 cells were cultured in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc.). Huh7 cells were cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.). All culture media were
supplemented with 10% heat-inactivated fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100
mg/ml streptomycin, and all cells were maintained at 37°C in a
humidified incubator with 5% CO2.
Antibodies and reagents
Anti-LC3B (1:1,000; cat. no. 3868S), SQSTM1/p62
(1:1,000; cat. no. 8025S), Beclin 1 (1:1,000, cat. no. 3495P),
HRP-conjugated anti-mouse IgG (1:2,000; cat. no. 7076S) and
HRP-conjugated anti-rabbit IgG (1:2,000; cat. no. 7074S) antibodies
were purchased from Cell Signaling Technology, Inc. The anti-eIF5A2
(1:1,000; cat. no. ab126735) and anti-KI67 (1:200; cat. no.
ab16667) antibodies were obtained from Abcam, and the anti-β-actin
(1:1,000; cat. no. 66009-1-ig) antibody was from ProteinTech Group,
Inc..
The eIF5A2 small interfering RNA (siRNA) and
negative control siRNA were synthesized by Shanghai GenePharma Co.,
Ltd. The PT3-EF1a-eIF5A2-flag and PT3-EF1a plasmids were purchased
from Wuhan Yuling Biological Technology Co., Ltd. Cell Counting
Kit-8 (CCK-8; cat. no. AD10) was obtained from Dojindo Molecular
Technologies, Inc. The 5-ethynyl-2′-deoxyuridine (EdU) kit (cat.
no. A10044) and Lipofectamine® 2000 Transfection Reagent
(cat. no. 11668019) were purchased from Invitrogen; Thermo Fisher
Scientific, Inc. The autophagy inhibitor chloroquine (CQ; cat. no.
C6628) and the mTOR inhibitor rapamycin (Rapa; cat. no. V900930)
were obtained from Sigma-Aldrich; Merck KGaA. Doxorubicin (cat. no.
S1208) was purchased from Selleck Chemicals. Monomeric red
fluorescent protein (mRFP)-green fluorescent protein (GFP)-LC3
adeno-associated virus (AAV) was obtained from Hanbio Biotechnology
Co., Ltd. 2′-O-methoxyethyl (2′-Ome)- and 5′-cholesterol
(5′-chol)-modified eIF5A2 siRNA was chemically synthesized by
Guangzhou RiboBio Co., Ltd.
Survival analysis
The tissue microarray containing 90 paired HCC and
adjacent tissues was obtained from Shanghai Xinchao Biological
Technology Co. Ltd. (cat. no. HLiv-HCC180Sur-04). The
clinicopathological information about patient age, sex, tumor stage
and survival was provided by Shanghai Xinchao Biological Technology
Co. Ltd. The follow-up period ranged between 1 and 6 years.
Immunohistochemistry (IHC) staining of eIF5A2 was performed as
follows: The tissue microarray was incubated with 3%
H2O2 for 10 min at room temperature, followed
by antigen retrieval in Tris-EDTA buffer containing 0.05% Tween 20
(pH 9.0) in a water bath at 95-100°C for 15 min, incubation with
10% FBS in PBS for 30 min at room temperature, and further
incubation with an anti-eIF5A2 (1:100; cat. no. ab126735; Abcam)
antibody at 4°C overnight. The tissues were subsequently incubated
with an HRP-conjugated goat anti-rabbit IgG secondary antibody
(1:200; cat. no. ab205718; Abcam) at room temperature for 30 min,
followed by incubation with DAB (cat. no. ab64238; Abcam) at room
temperature for 1-2 min according to the manufacturer's
instructions and counterstaining with hematoxylin. The stained
tissues were dehydrated and stabilize with mounting medium (cat.
no. ab64230; Abcam). Images (magnification, ×200) were captured
using AperioImageScope software 11.1.2.752 (Leica Biosystems), and
5-10 fields of view per sample were analyzed. IHC analysis of the
paired HCC and adjacent tissues in the microarray was used to
evaluate eIF5A2 staining intensity and relative expression levels
in tumors and adjacent tissues. Images (magnification, ×200) were
captured using Aperio ImageScope software 11.1.2.752 (Leica
Biosystems), and 5-10 fields of view per sample were analyzed. The
tissue scoring was confirmed by two indepen-dent pathologists who
were blinded to the grouping. eIF5A2 staining score was based on
staining intensity as follows: 0, none; 1,
equivocal/uninterpretable; 2, weak; and 3, intermediate-strong. The
proportion score was based on expression rate as follows: 0, none;
1, 1-25%; 2, 26-50%; 3, 51-75%; and 4, 76-100%. A composite score
was obtained by multiplying the intensity score by the proportion
score. A composite score <4 was considered low expression, and a
score ≥4 was considered high expression. Long-term follow-up was
obtained through tumor registries and telephone interviews. The
survival curve was drawn by Kaplan-Meier method, and the difference
was evaluated by log-rank test.
Patient samples
A total of 30 paired tumor and adjacent tissues (≥3
cm from the tumor margin) were collected from patients with HCC
(mean age, 56 years; range, 38-78 years; 21 male and 9 female
patients) undergoing surgery at the Second Affiliated Hospital of
Zhejiang University School of Medicine between January and December
2018. The histological diagnosis of HCC was confirmed by two
independent pathologists, and the samples were used for reverse
transcription-quantitative PCR (RT-qPCR) and mouse model
establishment.
RNA isolation
A total of 50-100 mg of HCC or adjacent tissue was
homogenized in 1 ml TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) using a power homogenizer. For cells, 1 ml
TRIzol® reagent was added to a culture dish to lyse the
cells directly. The homogenized samples were incubated for 5 min at
room temperature, and 0.2 ml chloroform per 1 ml of TRIzol reagent
was added, followed by centrifugation of the samples at 12,000 × g
for 15 min at 4°C. Subsequently, the upper aqueous phase was
transferred into fresh tubes, and 0.5 ml isopropyl alcohol was
added, followed by centrifugation at 12,000 × g at 4°C for 10 min.
The supernatant was removed, and the RNA pellet was washed with 75%
ethanol. Finally, the RNA was dissolved in DEPC-treated water at
room temperature.
RT-qPCR
Following total RNA extraction, reverse
transcription of mRNA was performed using a PrimeScript RT reagent
kit with gDNA Eraser (Perfect Real Time) (Takara Bio, Inc.)
according to the manufacturer's instructions. The mRNA levels of
eIF5A2 and β-actin were quantified by qPCR using specific primers
and a TB Green qPCR Master Mix (Takara Bio, Inc.) on a
LightCycler® 480 II instrument (Roche Diagnostics). The
thermocycling conditions were as follows: 95°C for 30 sec; 40
cycles of 95°C for 5 sec and 60°C for 30 sec; and 50°C for 30 sec.
The primer sequences were as follows: eIF5A2 forward, 5′-TAT GCA
GTG CTC GGC CTT G-3′ and reverse, 5′-TTG GAA CAT CCA TGT TGT GAG
TAG A-3′; and β-actin forward, 5′-TTC CAG CCT TCC TTC CTG-3′ and
reverse, 5′-CTT TGC GGA TGT CCA CGT-3′. Relative changes in the
mRNA expression levels were analyzed using the 2−ΔΔCq
method (31). All experiments were
carried out in triplicate and independently repeated three
times.
Cell viability assay
Tumor cells were seeded into 96-well plates at 3,000
cells/well. Following adherence, the medium was replaced with
medium containing 0, 0.125, 0.25, 0.5, 1, 2 or 5 µg/ml
doxorubicin for 48 h at 37°C. Subsequently, CCK-8 solution was
added into the wells (1:1,000) for 2 h at 37°C according to the
manufacturer's instructions, and cell viability was determined at
450 nm using an Epoch Microplate Spectrophotometer (BioTek
Instruments, Inc.). For the co-treatment with CQ or rapamycin,
SNU449 cells were pretreated with 10 µM CQ (dissolved in
ddH2O) or 100 nM rapamycin (dissolved in DMSO) for 6 h
and subsequently treated with eIF5A2 siRNA and doxorubicin.
In vivo analysis
A total of 20 male athymic BALB/c mice (age, 4-5
weeks; weight, 15-18 g) were obtained from the Model Animal
Research Center of Nanjing University, Nanjing, China. The mice
were housed in specific pathogen-free barrier facilities in a
temperature-controlled room (24°C) with a 12 h light-dark cycle and
35-40% relative humidity, and were permitted free access to food
and drinking water. The body weight of each mouse was measured
every 2 days. The mice were humanely sacrificed by cervical
dislocation at the study endpoint.
Patient-derived xenograft (PDX) tumor tissues were
used to establish the tumor model. Patient tumor in which eIF5A2
was detected by RT-qPCR was used to establishing the mouse model.
The resected tumor was cut into 1 mm3 fragments and
inoculated subcutaneously into the flanks of the mice (one fragment
per mouse). After 3 weeks, when the tumor volumes reached 50-100
mm3, the mice were randomly divided into the following
groups (n=5 mice/group): i) Control (saline); ii) doxorubicin; iii)
eIF5A2 siRNA; and iv) eIF5A2 siRNA combined with doxorubicin.
Saline and doxorubicin (2 mg/kg) were injected via the tail vein
every 2 days. eIF5A2 siRNA (1 nmol/mouse) was administered via
intratumoral injection every 2 days. To maintain the the stability
of eIF5A2 siRNA in vivo, a 2′-OMe and 5′-chol modified siRNA
was used. The 2′-OMe modification inhibits the ability of RNase H
to cleave the bound sense RNA strand within the heteroduplex formed
between the nucleic acid and the target RNA (32), and the 5′-chol modification alters
protein binding in serum, extending the circulation time, and
facilitates direct cellular uptake (33). The body weights and tumor
dimensions were measured every other day. The tumor volume was
calculated using the modified ellipsoid formula: Volume=1/2 ×
(length × width2). After 14 days, all mice were
sacrificed, and the blood, liver, kidney and tumor tissues were
harvested. The tissues were fixed with 4% formaldehyde for 48 h at
25°C, and the liver, kidney and a part of each tumor were used for
IHC. The remaining tumor tissues used for protein and mRNA
expression analysis of eIF5A2 and autophagy-related genes.
IHC analysis and TUNEL assay
After the mice were sacrificed, the tumor tissues
were dissected and fixed in formaldehyde. Following paraffin
embedding, sectioning (5 µm), deparaffinization, incubation
in 3% H2O2 for 10 min at room temperature,
antigen retrieval by boiling in 0.01 M sodium citrate buffer (pH
6.0) for 15 min, the tissues were blocked with 10% FBS in PBS for
30 min at room temperature and stained with an anti-ki67 antibody
(1:200) at 4°C overnight. Subsequently, the sections were incubated
with an HRP-conjugated goat anti-rabbit IgG secondary antibody
(1:200) for 30 min at room temperature, incubated with DAB at for
1-2 min room temperature, counterstained with hematoxylin,
dehydrated and stabilized with mounting medium (cat. no. ab64230;
Abcam). For TUNEL assay, an in situ Cell Death Detection kit
(Roche Molecular Diagnostics) was used to detect apoptosis
according to the manufacturer′s instructions following
deparaffinization. The sections were counterstained with
hematoxylin for 3 min at room temperature following dehydration and
stabilizing with mounting medium. The slides were visualized under
a Leica DM300 microscope (magnification, ×200; Leica Microsystems
GmbH), and 5-10 fields of view per sample were quantified using
Leica Application Suite X software 3.0.0.15697 (Leica Microsystems
GmbH).
Transfection
HCC cells (2×105) in the logarithmic
phase of growth were seeded into 6-well plates and transfected with
eIF5A2 siRNA (50 pmol), control siRNA (50 pmol),
pT3-EF1a-eIF5A2-flag plasmid (2 µg) or pT3-EF1a plasmid (2
µg) when the cells reached a density of 50-60% using
Lipofectamine® 2000 according to the manufacturer's instructions.
The transfection medium (Opti-MEM; Gibco; Thermo Fisher Scientific,
Inc.) was replaced with complete medium following 6-h incubation at
37°C, and the cells were harvested after 48 h. The sequences of
siRNAs were as follows: Negative control-homo sense, 5′-UUC UCC GAA
CGU GUC ACG UTT-3′ and anti-sense, 5′-ACG UGA CAC GUU CGG AGA
ATT-3′; eIF5A2-homo sense, 5′-GCA GAC GAA AUU GAU UUC ATT-3′ and
anti-sense, 5′-UGA AAU CAA UUU CGU CUG CTT-3′; and Beclin 1-homo
sense, 5′-GUA CCG ACU UGU UCC CUA UTT-3′ and anti-sense, 5′-AUA GGG
AAC AAG UCG GUA CTT-3′.
Western blotting analysis
Cells were harvested in cell lysis buffer (Cell
Signaling Technology, Inc.) containing protease inhibitors
(Sigma-Aldrich; Merck KGaA) following treatment with eIF5A2 siRNA
and doxorubicin for 48 h. The protein concentration was determined
by BCA assay (Applygen Technologies, Inc.). A total of 10 µg
protein was loaded per lane, separated by 15% SDS-PAGE and
transferred to 0.45-µm polyvinylidene fluoride membranes
(EMD Millipore). The membranes were blocked with TBS-Tween-20 (0.1%
Tween-20) containing 0.5% BSA (cat. no. 4240GR100; NeoFroxx GmbH)
and incubated with the aforementioned primary antibodies at 4°C
overnight. The membranes were washed with TBS-0.1% Tween-20 three
times. Following incubation with anti-mouse IgG or anti-rabbit IgG
secondary antibodies at 4°C for 2 h, protein expression was
detected by chemiluminescence with an EZ-ECL Chemiluminescence
Detection Kit (Biological Industries).
EdU assay
Tumor cells were seeded in a 96-well plate at 3,000
cells/well and incubated with medium (RPMI-1640 medium for SNU449
and DMEM for Huh7) containing doxorubicin at the IC50
concentration for 48 h. Prior to fixation with 4% paraformaldehyde
at 25°C for 15 min, the cells were incubated with 10 µM EdU
at 37°C for 2 h. Next, membrane permeabilization for 15 min, EdU
incubation for 30 min and Hoechst 33342 (Invitrogen; Thermo Fisher
Scientific, Inc.) staining for 15 min were performed to stain EdU
and the cell nuclei at 25°C, respectively. Images were captured
under na Olympus IX73 fluorescence microscope (magnification, ×200;
Olympus Corporation), and 5-10 fields of view per sample were
analyzed.
Transmission electron microscopy (TEM)
analysis
Following transfection with eIF5A2 siRNA for 48 h,
SNU449 cells were fixed with glutaraldehyde at 4°C for 16 h,
followed by treatment with 1% osmic acid at 25°C for >2 h, then
rinsed with 0.1 M phosphate buffer, dehydrated sequentially in 50,
70 and 90% ethanol, 90% ethanol plus 90% acetone, 90 and 100%
acetone for 15 min per step, and then embedded in Epon 812
embedding medium (Structure Probe, Inc.) according to the
manufacturer's instructions. The samples were sectioned at 50-60
nm, stained with 3% uranyl acetate plus lead citrate, and images
were captured using a transmission electron microscope
(magnification, ×25,000), and five fields of view per sample were
analyzed.
Confocal microscopy
SNU449 cells were seeded into confocal dishes at
3×105 cells/well. Cells were treated with RPMI-1640
medium containing the mRFP-GFP-LC3 AAV at 37°C for 6 h, incubated
with eIF5A2 siRNA and doxorubicin at 37°C for 48 h and fixed with
4% paraformaldehyde for 15 min at 25°C. Images were captured using
an Olympus IX81-FV1000 confocal microscope (magnification, ×1,600;
Olympus Corporation), and 5-10 fields of view of per sample were
analyzed.
Apoptosis assay
Apoptosis was detected using Annexin V, FITC
Apoptosis Detection kit (Dojindo Molecular Technologies, Inc.)
After eIF5A2 siRNA and doxorubicin treatment for 48 h,
1×106 SNU449 cells were harvested by trypsinization and
collected by centrifugation (300 × g at 4° for 3 min), resuspended
in 100 µl binding buffer, stained with 5 µl annexin
V-FITC and 5 µl propidium iodide (PI) at room temperature in
the dark for 15 min according to the manufacturer's instructions,
and then analyzed using a BD FACScanto II flow cytometer with BD
FACSDiva software v8.0.1 (both from BD Biosciences).
Lactate dehydrogenase (LDH) release
assay
HCC cells were seeded into a 96-well plate at 3,000
cells/well and administered eIF5A2 siRNA, eIF5A2 plasmid, CQ and
rapamycin, alone or in combination at 37°C for 48 h. The culture
medium was collected by centrifugation at 300 × g for 3 min (4°C)
and transferred to a new 96-well plate, and the release of LDH was
measured using a Cytotoxicity LDH Assay kit at 25°C (Dojindo
Molecular Technologies, Inc.) according to the manufacturer's
instructions. LDH release was detected at 490 nm using an Epoch
Microplate Spectrophotometer.
Cell death assay
SNU449 cells (2×105) in the logarithmic
phase of growth were seeded in 6-well plates. After cells were
administered eIF5A2 siRNA and Beclin 1 siRNA, alone or in
combination at 37°C for 48 h, cells were harvested by
trypsinization and collected by centrifugation at 300 × g for 3
min, resuspended in trypan blue solution (T10282, Thermo Fisher
Scientific, Inc.) and counted using a Countess II Automated Cell
Counter with Countess II/II FL software v1.0.247 (both from Thermo
Fisher Scientific, Inc.). The cell counter automatically quantified
the trypan blue-positive dead cells and the trypan blue-negative
living cells.
Calcein-AM/PI staining
SNU449 cells were seeded in a 96-well plate at 3,000
cells/well and administered eIF5A2 siRNA, eIF5A2 plasmid, CQ and
rapamycin, alone or in combination at 37°C for 48 h. The cells were
then cultured with 1 µM calcein-AM and 3 µM PI (Cell
Signaling Technology, Inc.) at 25°C for 30 min. Subsequently,
images were captured using an Olympus IX73 fluorescence microscope
(magnification, ×100), and five fields of view per were
analyzed.
Statistical analysis
Data are presented as the mean ± SD from ≥3
independent replicates. Statistical analyses were performed using
GraphPad Prism 7.0 (GraphPad Software, Inc.). Unpaired two-tailed
Student's t-test was used to analyze differences between two
groups, and one-way ANOVA with Tukey's post hoc test was used for
multiple comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results
eIF5A2 expression is negatively
associated with the prognosis of patients with HCC
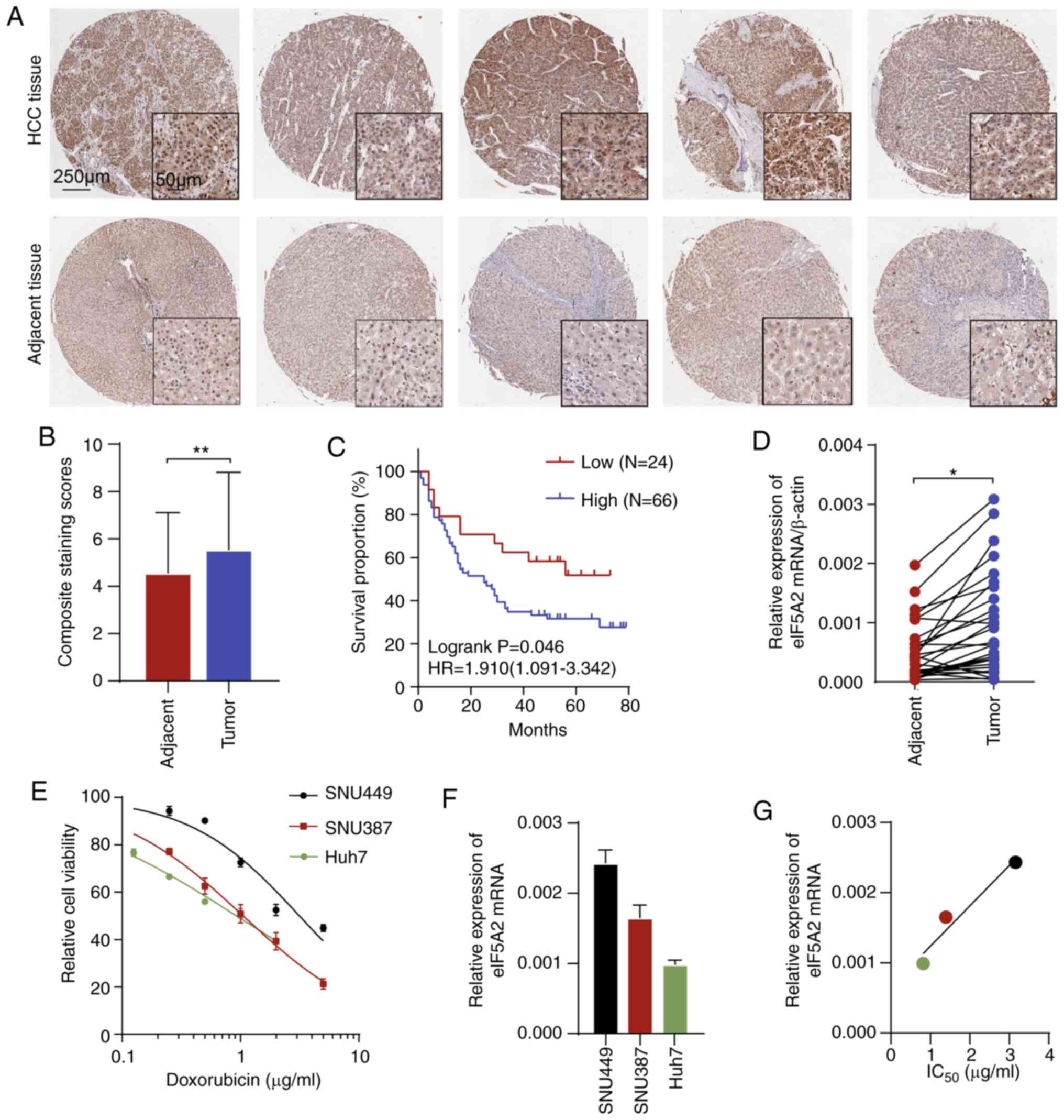
To investigate the role of eIF5A2 in HCC, a tissue
microarray was used. Immunohistochemical staining of eIF5A2 protein
in paired tumor and adjacent tissues from 90 patients with HCC
revealed that the staining intensity of eIF5A2 was higher in tumor
tissues compared with that in normal tissues (Fig. 1A and B). Based on the aberrant
eIF5A2 expression, survival analysis revealed that high expression
levels of eIF5A2 were associated with a shorter overall survival
time of patients with HCC (Fig.
1C). In addition, eIF5A2 mRNA expression was detected in 30
pairs of tumors and adjacent tissues from patients with HCC. The
tumor tissues exhibited higher expression levels of eIF5A2 compared
with those in the adjacent normal tissues (Fig. 1D). These results suggested that
eIF5A2 mRNA and protein expression levels were upregulated in HCC
compared with those in normal tissues, and were associated with a
poor prognosis.
To characterize the relationship between eIF5A2 and
doxorubicin sensitivity, three HCC cell lines (SNU449, SNU387 and
Huh7) were treated with 0.125-5 µg/ml doxorubicin.
Sensitivity to doxorubicin was measured after 48-h treatment by
CCK-8 assay. As demonstrated in Fig.
1E, among the tested cell lines, SNU449 was the most resistant
to doxorubicin, followed by SNU387 and Huh7. Basal levels of eIF5A2
mRNA in the three cell lines were also assessed by RT-PCR (Fig. 1F). SNU449 had the highest
expression of eIF5A2 while Huh7 had the lowest. Notably, the
IC50 values for doxorubicin appeared to be associated
with eIF5A2 expression (Fig. 1G).
Collectively, these results suggested that eIF5A2 may serve as a
biomarker of HCC and may be involved in doxorubicin
sensitivity.
eIF5A2 silencing improves doxorubicin
chemotherapy in vivo
To investigate the role of eIF5A2 in
chemosensitivity, three specific siRNAs were designed to silence
the expression of eIF5A2 (Fig.
S1), and the most efficient one (sieIF5A2-3, further referred
to as eIF5A2 siRNA) was selected for further experiments.
To further determine the effects of eIF5A2 on
doxorubicin sensitivity, a patient-derived xenograft (PDX) mouse
model was established. An HCC tumor with high expression levels of
eIF5A2 (Fig. S2) was
subcutaneously injected into mice. Following 14-day treatment, both
doxorubicin administration and eIF5A2 siRNA injection were observed
to inhibit tumor growth (Fig.
2A-C). Notably, co-administration of eIF5A2 siRNA markedly
enhanced the therapeutic effects of doxorubicin on tumor
suppression (Fig. 2A-C).
Doxorubicin and eIF5A2 siRNA exhibited no significant effects on
the mouse body weight (Fig. 2C).
In addition, the combination of doxorubicin and eIF5A2 siRNA
inhibited the proliferation of tumor cells (as demonstrated by IHC
staining of ki67) and increased the apoptotic rate (analyzed by
TUNEL assay) compared with those in the other three groups
(Fig. 2D), suggesting that
co-treatment with of eIF5A2 siRNA may improve the therapeutic
efficiency of doxorubicin in vivo.
eIF5A2 is involved in regulating
autophagy and chemosensitivity of HCC cells
To further determine the tumor-suppressing effects
of eIF5A2 siRNA observed in the in vivo experiment, SNU449
and Huh7 cells were transfected with eIF5A2 siRNA or an
eIF5A2-overexpressing plasmid, respectively, and doxorubicin
sensitivity was evaluated using the CCK-8 assay. The results
demonstrated that silencing of eIF5A2 enhanced the effects of
doxorubicin on cell viability compared with those in the negative
control group, whereas eIF5A2 upregulation impaired the cytotoxic
effects of doxorubicin (Fig. 3A and
B). In addition, doxorubicin treatment alone notably reduced
the proliferation rate in SNU449 and Huh7 cells according to the
results of the EdU incorporation assay, and transfection with
eIF5A2 siRNA and doxorubicin treatment exhibited additive
antiproliferative effects (Fig.
3C). By contrast, eIF5A2 overexpression promoted HCC cell
proliferation of doxorubicin-treated cells. These results
demonstrated that eIF5A2 silencing enhanced doxorubicin efficacy
in vitro.
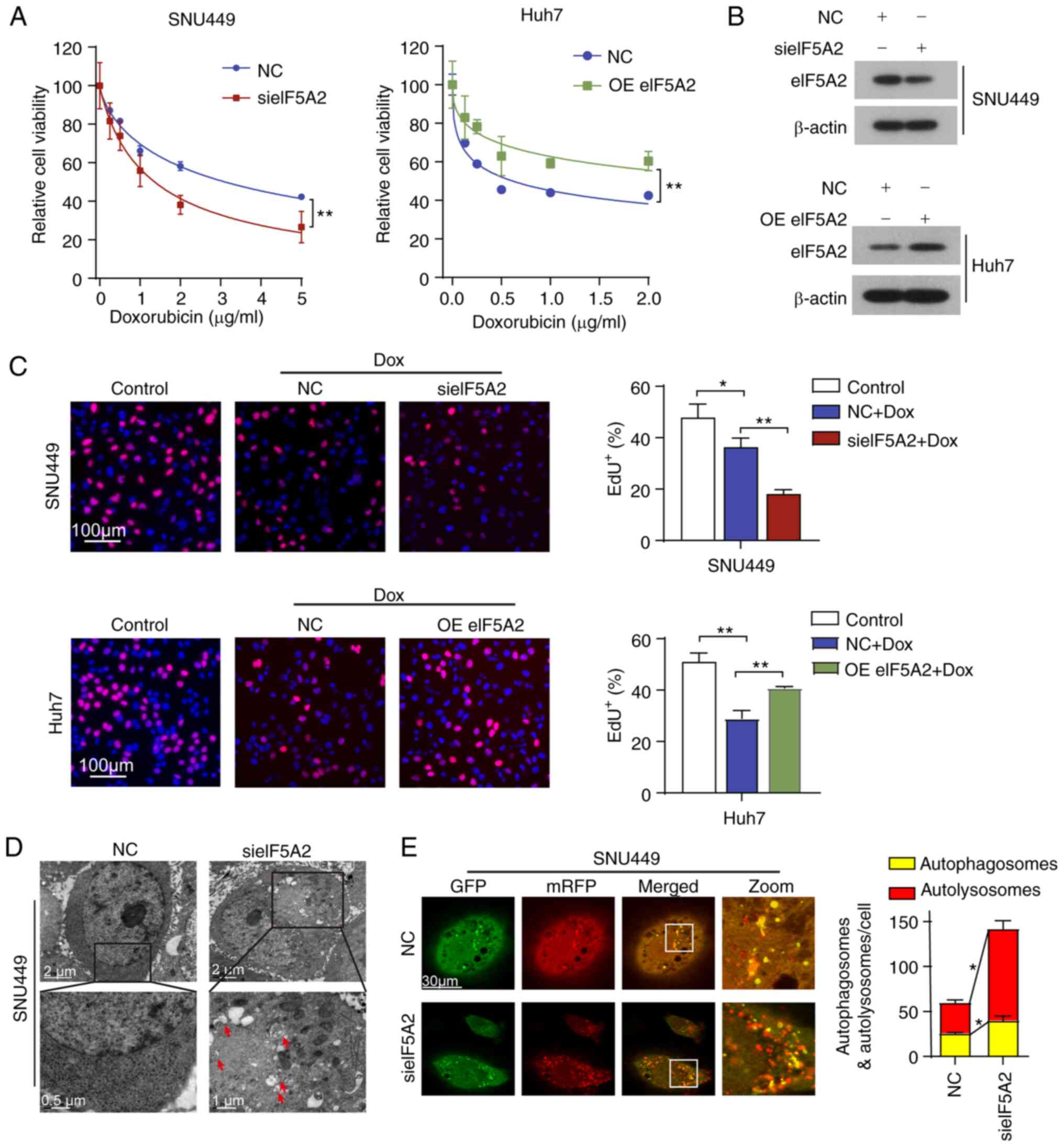 | Figure 3eIF5A2 is involved in the regulation
of autophagy and chemosensitivity in HCC cells. (A) SNU449 cells
were treated with indicated concentrations of Dox for 48 h
following transfection with NC siRNA or sieIF5A2. Huh7 cells were
treated with indicated concentrations of Dox for 48 h following
transfection with an empty plasmid or OE eIF5A2. Cell viability was
measured by the Cell Counting Kit-8 assay. (B) Western blotting
analysis of eIF5A2 protein expression levels in HCC cells
transfected with sieIF5A2 or OE eIF5A2. (C) HCC cells were treated
with or without Dox for 48 h in the presence or absence of sieIF5A2
or OE eIF5A2. The cell proliferation rate was evaluated by EdU
incorporation assay, and the frequency of EdU positive cells was
quantified. (D) Autophagosomes were visualized in SNU449 cells
treated with NC siRNA or sieIF5A2 by transmission electron
microscopy. (E) SNU449 cells were infected with the mRFP-GFP-LC3
adenovirus and subsequently transfected with NC siRNA or sieIF5A2
and visualized by confocal microscopy. The numbers of
GFP+/mRFP+-LC3 (yellow, autophagosomes) and GFP-/mRFP+-LC3 (red,
autolysosomes) dots were counted and analyzed.
*P<0.05 and **P<0.01. eIF5A2,
eukaryotic translation initiation factor 5A2; EdU,
5-ethynyl-2′-deoxyuridine; HCC, hepatocellular carcinoma; siRNA,
small interfering RNA; NC, negative control; sieIF5A2, siRNA
against eIF5A2; OE, overexpression vector; mRFP, monomeric red
fluorescent protein; GFP, green fluorescent protein; Dox,
doxorubicin. |
eIF5A2 silencing also resulted in increased numbers
of autophagosomes observed by TEM compared with those in the
negative control group (Fig. 3D).
In order to validate the appearance of autophagic characteristics,
the cells were infected with an AAV expressing an mRFP-GFP-LC3
reporter protein for monitoring the autophagic flux (Fig. 3E). The results demonstrated that
eIF5A2 knockdown induced an increase in the numbers of in
autophagosomes and autoly-sosomes compared with those in the
negative control group, suggesting that eIF5A2 siRNA-induced
autophagy may be involved in doxorubicin sensitivity.
Silencing of eIF5A2 induces sensitivity
to doxorubicin in HCC cells by triggering lethal autophagy
To determine the role of autophagy triggered by
eIF5A2 siRNA in doxorubicin treatment, the autophagic flux of cells
treated with doxorubicin and eIF5A2 siRNA alone or in combination
was visualized. Knockdown efficiency of eIF5A2 siRNA was validated
by western blotting (Fig. 4A).
Doxorubicin treatment induced autophagic flux in SNU499 cells
compared with that in the control group (Fig. 4B and C). In addition, combined
eIF5A2 siRNA and doxorubicin treatment further reinforced the
autophagic activity, with an increased number of autophagosomes
compared with that observed in cells treated with doxorubicin alone
(Fig. 4B and C). The protein
expression levels of LC-3 I/II, Beclin 1 and p62 were evaluated by
western blotting. The results demonstrated that eIF5A2 silencing
markedly promoted the conversion of LC3I to LC3II, the formation of
Beclin1 and the degradation of p62 compared with those in cells
treated with doxorubicin alone (Fig.
4C), confirming that eIF5A2 siRNA promoted autophagy induced by
doxorubicin. In addition, an increase of LDH release was observed
in cells treated with eIF5A2 siRNA and doxorubicin compared with
that in cells treated with doxorubicin alone, and eIF5A2 silencing
accelerated doxorubicin-induced cell death (Fig. 4D). Notably, doxorubicin treatment
alone activated apoptosis, but the apoptotic rate (early and late
apoptosis) was only modestly further increased in the presence of
eIF5A2 siRNA (Fig. 4E). These
results suggested a limited additive effect between doxorubicin and
eIF5A2 on apoptotic cell death.
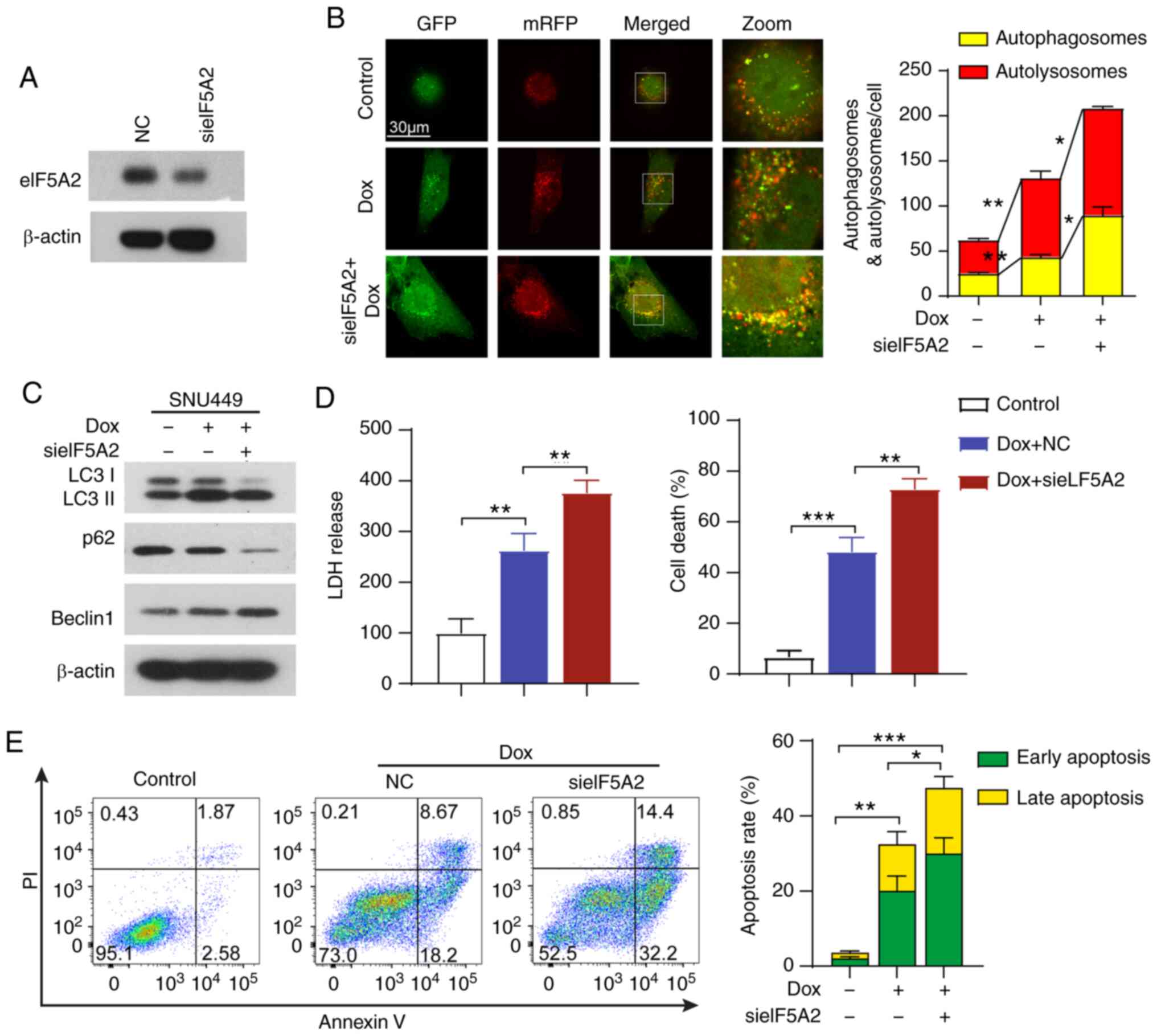 | Figure 4Silencing of eIF5A2 induces
sensitivity to doxorubicin in HCC cells by triggering lethal
autophagy. (A) Transfection efficiency of sieIF5A2 in SNU449 cells.
(B) SNU449 cells were infected with the mRFP-GFP-LC3 adenovirus,
treated with Dox and sieIF5A2 alone or in combination, and
visualized by confocal microscopy. The numbers of
GFP+/mRFP+-LC3 (yellow, autophagosomes) and
GFP−/mRFP+-LC3 (red, autolysosomes) dots were
counted and analyzed. (C) Western blotting analysis of LC3, p62 and
Beclin 1 proteins in SNU449 cells treated with Dox and sieIF5A2
alone or in combination. (D) LDH release was detected in SNU449
cells following the indicated treatments. Cell death of SNU449
cells was detected using trypan blue staining. (E) Apoptotic rates
in SNU449 cells were assessed by annexin V-VITC/PI apoptosis assay
using flow cytometry following the indicated treatments. Early and
late apoptotic rates were analyzed. *P<0.05,
**P<0.01 and ***P<0.001. eIF5A2,
eukaryotic translation initiation factor 5A2; si, small interfering
RNA; NC, negative control; mRFP, monomeric red fluorescent protein;
GFP, green fluorescent protein; Dox, doxorubicin; LDH, lactate
dehydrogenase. |
eIF5A2 enhances HCC cell chemoresistance
by suppressing autophagic cell death via a Beclin 1-dependent
pathway
Since eIF5A2 siRNA and doxorubicin-induced autophagy
was accompanied by cell death, we speculated that eIF5A2 knock-down
may enhance doxorubicin-triggered autophagic cell death. To
determine this, CQ, a pharmacological lysosomal inhibitor, was used
to inhibit autophagy in SNU449 cells. Notably, treatment with CQ
partially reversed doxorubicin sensitivity and diminished eIF5A2
siRNA-mediated doxorubicin sensitivity (Fig. 5A and B). In addition, CQ attenuated
the doxorubicin-induced increase in LDH release and cell death
(Fig. 5C and D). CQ also inhibited
the LDH release and cell death induced by the combination of eIF5A2
siRNA and doxorubicin. By contrast, Rapa, which is an autophagy
activator, significantly decreased doxorubicin-induced cell death
and reversed the cytoprotective effects of the overexpression of
eIF5A2 (Fig. 5E-H). These results
suggested that eIF5A2 siRNA may enhance doxorubicin sensitivity by
inducing autophagic cell death in HCC cells.
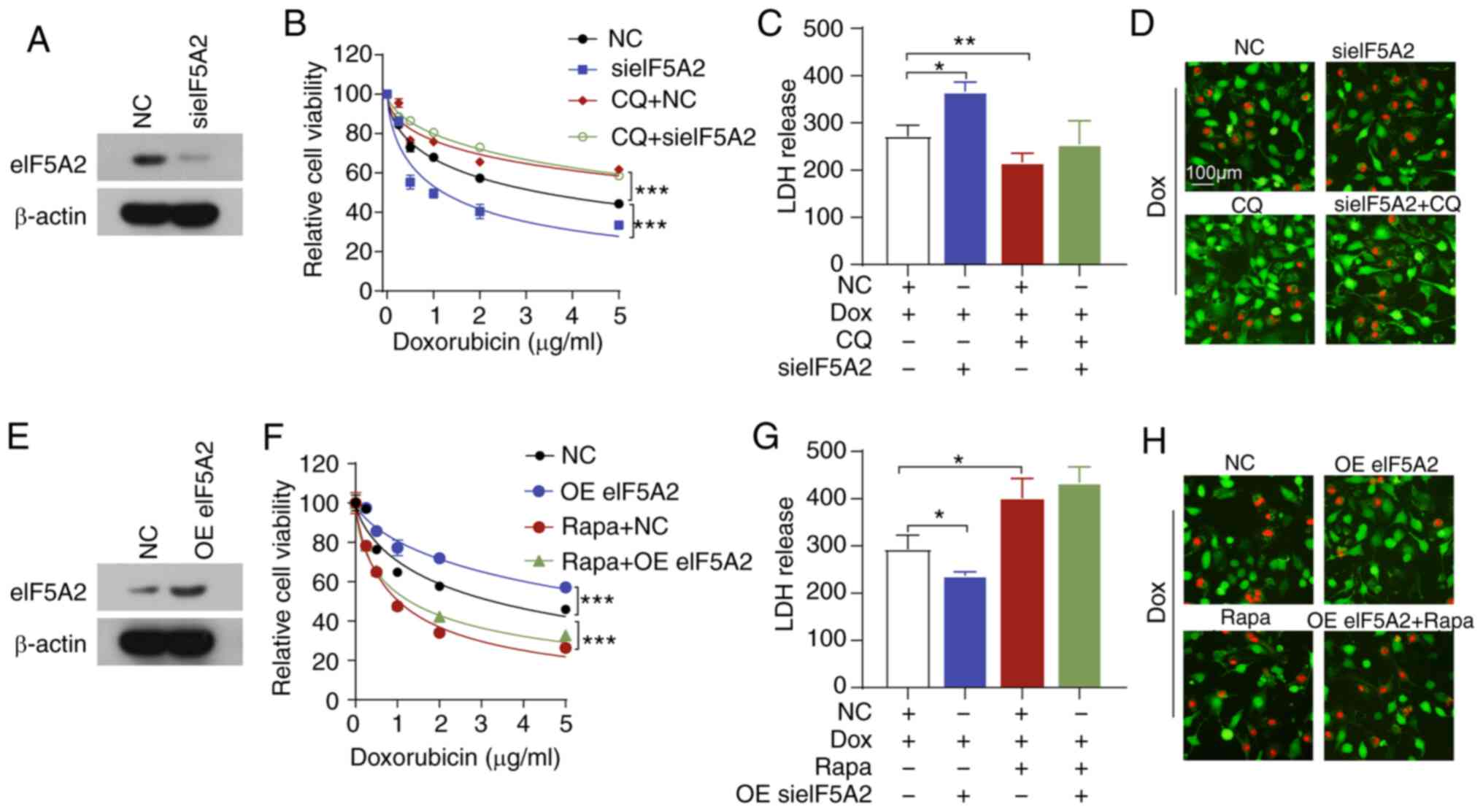 | Figure 5Combination of sieIF5A2 and Dox
induces lethal autophagy. (A-D) SNU449 cells were treated with Dox,
CQ and sieIF5A2 alone or in combination. (A) Transfection
efficiency was assessed by western blotting. (B) Cell viability was
assessed by CCK-8 assay. (C) LDH release was measured using an LDH
assay. (D) Cell death was detected by Calcein-AM/PI staining. (E-H)
SNU449 cells were treated with Dox, Rapa and OE eIF5A2 alone or in
combination. (E) Transfection efficiency was assessed by western
blotting. (F) Cell viability was assessed by CCK-8 assay. (G) LDH
release was measured using an LDH assay. (H) Cell death was
detected by Calcein-AM/PI staining. *P<0.05,
**P<0.01 and ***P<0.001. eIF5A2,
eukaryotic translation initiation factor 5A2; si, small interfering
RNA; NC, negative control; CCK-8, Cell Counting Kit-8; LDH, lactate
dehydrogenase; CQ, chloroquine; Rapa, rapamycin; OE, overexpression
vector; Dox, doxorubicin. |
To further investigate the underlying mechanisms,
the protein levels of eIF5A2, LC-3 I/II, Beclin 1 and p62 in
xeno-graft tumors were detected. Consistently with the results of
the in vitro experiments, doxorubicin and eIF5A2 siRNA
treatment increased the protein expression levels of LC3II and
Beclin 1, but reduced the levels of p62 compared with those in the
control group (Fig. 6A).
Combination of doxorubicin and eIF5A2 siRNA induced higher LC3II
and Beclin 1 and lower p62 protein expression levels compared with
those observed following either therapy alone (Fig. 6A), indicating high levels
autophagic activity in xenograft tumors under combined treatment.
In addition, knockdown of Beclin 1 impaired the sensitivity of HCC
cells to doxorubicin (Fig. 6B and
C), and co-transfection with eIF5A2 siRNA did not restore the
doxorubicin sensitivity of HCC cells following Beclin 1 knockdown
(Fig. 6C). Similarly, knockdown of
Beclin1 inhibited the combination treatment-induced LDH release and
cell death (Fig. 6D and E). Taken
together, these results demonstrated that the effects of eIF5A2
inhibition on autophagic cell death were mediated by Beclin 1.
Discussion
HCC is the third leading cause of cancer-related
death world-wide (2). Multi-drug
resistance diminishes the efficiency of therapeutic agents in HCC;
therefore, developing new strategies to enhance chemotherapy
efficiency is urgent. The results of the present study demonstrated
that the expression levels of eIF5A2 were significantly upregulated
in tumor tissues compared with those in adjacent normal tissues and
were negatively associated with the overall survival of patients
with HCC. In addition, eIF5A2 levels were positively associated
with doxorubicin resistance in HCC cells, and eIF5A2 silencing
enhanced the antitumor effects of doxorubicin in vitro and
in vivo. Further experiments revealed that eIF5A2 siRNA
improved the therapeutic efficiency of doxorubicin in HCC by
triggering autophagic cell death through a Beclin 1-dependent
pathway.
eIF5A2 is an oncogene highly expressed in several
types of human cancer, including melanoma, gastric, breast and
gall-bladder cancer (11,12,34-36).
Accumulating evidence suggests that eIF5A2 is involved in tumor
initiation, growth, metastasis and chemoresistance (37-39).
In particular, previous studies have demonstrated that inhibition
or knockdown of eIF5A2 reverses chemoresistance in gastric and
colorectal cancer, as well as in HCC (34,35,39-41).
In addition, Yang et al (42) have reported that eIF5A2
overexpression significantly enhanced the resistance of esophageal
squamous cell carcinoma cells to 5-fluorouracil by inhibiting
apoptosis. Consistent with these, the present study demonstrated
that eIF5A2 was upregulated in HCC, and eIF5A2 silencing enhanced
the tumor-suppressing effects of doxorubicin both in vivo
and in vitro. The results of the present study also
demonstrated that overexpression of eIF5A2 induced resistance to
doxorubicin. These results suggested a crucial role of eIF5A2 in
the development of chemoresistance in HCC.
Autophagy is a double-edged sword, as the modulation
(either activation or inhibition) of autophagy has been
demonstrated to at least partly overcome or reverse doxorubicin
resistance in breast cancer, hepatocellular carcinoma and
osteosarcoma (43). Previous
studies have suggested that autophagy serves as a pro-survival
role, resulting in acquired resistance to chemotherapeutic agents
in cancer (16,44). However, the cell death-promoting
effects of autophagy have also been reported (25,45).
For example, Tai et al (46) have demonstrated that sorafenib
inhibits tumor growth by inducing autophagic cell death in HCC.
Similarly, Wu et al (47)
have reported that blocking β2 adrenoreceptor signaling enhanced
sorafenib-induced autophagy and improved the antitumor effects of
sorafenib in HCC. The results of the present study demonstrated
that doxorubicin treatment induced autophagy, and that blocking
autophagy with CQ impaired doxorubicin efficiency, suggesting that
doxorubicin caused autophagic cell death. Notably, the results of
further experiments revealed that knockdown of eIF5A2 enhanced cell
death by triggering sustained lethal autophagy induced by
doxorubicin. Beclin 1 was the first autophagy-related protein
identified in mammals and serves a central role in autophagy
(48). Beclin 1 constitutes a
molecular platform for the regulation of autophagosome formation
and maturation by forming distinct PI3K complexes together with the
core lipid kinase VPS34 and the regulatory component VPS15
(49). Therefore, Beclin 1 can be
used as a marker to monitor autophagy (17). The results of the present study
demonstrated that knockdown of Beclin 1 reversed the eIF5A2
siRNA-mediated increase in doxorubicin sensitivity, revealing a
potential mechanism of eIF5A2 siRNA-enhanced doxorubicin
lethality.
In conclusion, the present study described the roles
of eIF5A2 and autophagy in doxorubicin treatment of HCC. The
results of the present study suggested that knockdown of eIF5A2
restored the sensitivity of HCC cells to doxorubicin by triggering
lethal autophagy via a Beclin1-dependent signaling pathway. These
results provide an potential target of HCC chemoresistance.
Supplementary Data
Acknowledgments
Not applicable.
Funding
This study was supported by the Zhejiang Provincial
Ten Thousand Plan for Young Top Talents (2018), the Innovative
Talents Training Project of Zhejiang Health Bureau (2018), the Key
Project Co-constructed by the Zhejiang Province and Ministry (grant
no. WKJ-ZJ-1916), the National High Technology Research and
Development Program of China (grant no. SS2014AA020533), the
Zhejiang Provincial Natural Science Foundation of China (grant no.
LQ13H160006) and the Natural Science Foundation of China (grant
nos. 81302071 and 81673809).
Availability of data and materials
The datasets generated and/or analyzed in the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
YXT, WC and XNZ designed the study. KC and XRL
performed the in vitro experiments and analyzed the data.
JYZ, RRL, XXZ and SZX performed the in vivo experiments. HPK
provided the experimental tools and analyzed the in vivo
results. YXT, KC and XRL wrote the manuscript. WC and XNZ revised
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Second Affiliated Hospital of Zhejiang University
School of Medicine (Hangzhou, China), and written informed consent
was obtained from all participants. All experimental procedures on
animals were approved by the Institutional Animal Care and Use
Committee at Zhejiang University School of Medicine.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Global Burden of Disease Cancer
Collaboration; Fitzmaurice C, Abate D, Abbasi N, Abbastabar H,
Abd-Allah F, Abdel-Rahman O, Abdelalim A, Abdoli A, Abdollahpour I,
et al: Global, regional, and national cancer incidence, mortality,
years of life lost, years lived with disability, and
disability-adjusted life-years for 29 cancer groups, 1990 to 2017:
A systematic analysis for the global burden of disease study. JAMA
Oncol. 5:1749–1768. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma MC, Chen YY, Li SH, Cheng YF, Wang CC,
Chiu TJ, Pei SN, Liu CT, Huang TL, Huang CH, et al: Intra-arterial
chemotherapy with doxorubicin and cisplatin is effective for
advanced hepatocellular cell carcinoma. ScientificWorldJournal.
2014:1601382014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruix J, Reig M and Sherman M:
Evidence-based diagnosis, staging, and treatment of patients with
hepatocellular carcinoma. Gastroenterology. 150:835–853. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma S, Lee TK, Zheng BJ, Chan KW and Guan
XY: CD133+ HCC cancer stem cells confer chemoresistance by
preferential expression of the Akt/PKB survival pathway. Oncogene.
27:1749–1758. 2008. View Article : Google Scholar
|
|
6
|
Schuller AP, Wu CC, Dever TE, Buskirk AR
and Green R: eIF5A functions globally in translation elongation and
termination. Mol Cell. 66:194–205.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taylor CA, Zheng Q, Liu Z and Thompson JE:
Role of p38 and JNK MAPK signaling pathways and tumor suppressor
p53 on induction of apoptosis in response to Ad-eIF5A1 in A549 lung
cancer cells. Mol Cancer. 12:352013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mathews MB and Hershey JW: The translation
factor eIF5A and human cancer. Biochim Biophys Acta. 1849:836–844.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Caraglia M, Park MH, Wolff EC, Marra M and
Abbruzzese A: eIF5A isoforms and cancer: Two brothers for two
functions? Amino Acids. 44:103–109. 2013. View Article : Google Scholar :
|
|
10
|
Zhu W, Cai MY, Tong ZT, Dong SS, Mai SJ,
Liao YJ, Bian XW, Lin MC, Kung HF, Zeng YX, et al: Overexpression
of EIF5A2 promotes colorectal carcinoma cell aggres-siveness by
upregulating MTA1 through C-myc to induce
epithelial-mesenchymaltransition. Gut. 61:562–575. 2012. View Article : Google Scholar
|
|
11
|
Yang J, Yu H, Shen M, Wei W, Xia L and
Zhao P: N1-guanyl-1,7-diaminoheptane sensitizes bladder cancer
cells to doxorubicin by preventing epithelial-mesenchymal
transition through inhibition of eukaryotic translation initiation
factor 5A2 activation. Cancer Sci. 105:219–227. 2014. View Article : Google Scholar
|
|
12
|
Khosravi S, Wong RP, Ardekani GS, Zhang G,
Martinka M, Ong CJ and Li G: Role of EIF5A2, a downstream target of
Akt, in promoting melanoma cell invasion. Br J Cancer. 110:399–408.
2014. View Article : Google Scholar :
|
|
13
|
Meng QB, Kang WM, Yu JC, Liu YQ, Ma ZQ,
Zhou L, Cui QC and Zhou WX: Overexpression of eukaryotic
translation initiation factor 5A2 (EIF5A2) correlates with cell
aggressiveness and poor survival in gastric cancer. PLoS One.
10:e01192292015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yao M, Hong Y, Liu Y, Chen W and Wang W:
N1-guanyl-1, 7-diaminoheptane enhances the sensitivity of
pancreatic ductal adenocarcinoma cells to gemcitabine via the
inhibition of eukaryotic translation initiation factor 5A2. Exp
Ther Med. 14:2101–2107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Levy JMM, Towers CG and Thorburn A:
Targeting autophagy in cancer. Nat Rev Cancer. 17:528–542. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ding ZB, Hui B, Shi YH, Zhou J, Peng YF,
Gu CY, Yang H, Shi GM, Ke AW, Wang XY, et al: Autophagy activation
in hepatocellular carcinoma contributes to the tolerance of
oxaliplatin via reactive oxygen species modulation. Clin Cancer
Res. 17:6229–6238. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hayat MA: Autophagy: Cancer, other
pathologies, inflammation, immunity, infection, and aging. 1. 1st
edition. Elsevier; Academic Press; 2016
|
|
18
|
Pohl C and Dikic I: Cellular quality
control by the ubiquitin-proteasome system and autophagy. Science.
366:818–822. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu F, Zhao Y, Yu Y, Fang JM, Cui R, Liu
ZQ, Guo XL and Xu Q: Docetaxel-mediated autophagy promotes
chemoresistance in castration-resistant prostate cancer cells by
inhibiting STAT3. Cancer Lett. 416:24–30. 2018. View Article : Google Scholar
|
|
20
|
Piya S, Andreeff M and Borthakur G:
Targeting autophagy to overcome chemoresistance in acute
myleogenous leukemia. Autophagy. 13:214–215. 2017. View Article : Google Scholar :
|
|
21
|
Zeng Q, Liu J, Cao P, Li J, Liu X, Fan X,
Liu L, Cheng Y, Xiong W, Li J, et al: Inhibition of REDD1
sensitizes bladder urothelial carcinoma to paclitaxel by inhibiting
autophagy. Clin Cancer Res. 24:445–459. 2018. View Article : Google Scholar
|
|
22
|
Fulda S and Kogel D: Cell death by
autophagy: Emerging molecular mechanisms and implications for
cancer therapy. Oncogene. 34:5105–5113. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang H, Lu Q, Cheng S, Wang X and Zhang H:
Autophagy activity contributes to programmed cell death in
Caenorhabditis elegans. Autophagy. 9:1975–1982. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang X, Wei S, Zhao Y, Shi C, Liu P, Zhang
C, Lei Y, Zhang B, Bai B, Huang Y and Zhang H: Anti-proliferation
of breast cancer cells with itraconazole: Hedgehog pathway
inhibition induces apoptosis and autophagic cell death. Cancer
Lett. 385:128–136. 2017. View Article : Google Scholar
|
|
25
|
Segala G, David M, de Medina P, Poirot MC,
Serhan N, Vergez F, Mougel A, Saland E, Carayon K, Leignadier J, et
al: Dendrogenin A drives LXR to trigger lethal autophagy in
cancers. Nat Commun. 8:19032017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Masui A, Hamada M, Kameyama H, Wakabayashi
K, Takasu A, Imai T, Iwai S and Yura Y: Autophagy as a survival
mechanism for squamous cell carcinoma cells in endonuclease
G-mediated apoptosis. PLoS One. 11:e01627862016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kun Z, Hanqing G, Hailing T, Yuan Y, Jun
Z, Lingxia Z, Kun H and Xin Z: Gastrin enhances autophagy and
promotes gastric carcinoma proliferation via inducing AMPKα. Oncol
Res. 25:1399–1407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Altman JK, Szilard A, Goussetis DJ,
Sassano A, Colamonici M, Gounaris E, Frankfurt O, Giles FJ, Eklund
EA, Beauchamp EM and Platanias LC: Autophagy is a survival
mechanism of acute myelogenous leukemia precursors during dual
mTORC2/mTORC1 targeting. Clin Cancer Res. 20:2400–2409. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu Q, Deng J, Fan D, Duan Z, Zhu C, Fu R
and Wang S: Ginsenoside Rh4 induces apoptosis and autophagic cell
death through activation of the ROS/JNK/p53 pathway in colorectal
cancer cells. Biochem Pharmacol. 148:64–74. 2018. View Article : Google Scholar
|
|
30
|
Sun D, Zhu L, Zhao Y, Jiang Y, Chen L, Yu
Y and Ouyang L: Fluoxetine induces autophagic cell death via
eEF2K-AMPK-mTOR-ULK complex axis in triple negative breast cancer.
Cell Prolif. 51:e124022018. View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
32
|
Yamakawa K, Nakano-Narusawa Y, Hashimoto
N, Yokohira M and Matsuda Y: Development and clinical trials of
nucleic acid medicines for pancreatic cancer treatment. Int J Mol
Sci. 20:42242019. View Article : Google Scholar :
|
|
33
|
Behlke MA: Chemical modification of siRNAs
for in vivo use. Oligonucleotides. 18:305–319. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Y, Du F, Chen W, Yao M, Lv K and Fu P:
EIF5A2 is a novel chemoresistance gene in breast cancer. Breast
Cancer. 22:602–607. 2015. View Article : Google Scholar
|
|
35
|
Sun J, Xu Z, Lv H, Wang Y, Wang L, Ni Y,
Wang X, Hu C, Chen S, Teng F, et al: eIF5A2 regulates the
resistance of gastric cancer cells to cisplatin via induction of
EMT. Am J Transl Res. 10:4269–4279. 2018.
|
|
36
|
Zheng X, Gao L, Wang BT, Shen P, Yuan XF,
Zhang LQ, Yang L, Zhang DP, Zhang Q and Wang XM: Overexpression of
EIF5A2 is associated with poor survival and aggressive tumor
biology in gallbladder cancer. Histol Histopathol. 35:579–587.
2020.
|
|
37
|
Huang PY, Zeng TT, Ban X, Li MQ, Zhang BZ,
Zhu YH, Hua WF, Mai HQ, Zhang L, Guan XY and Li Y: Expression of
EIF5A2 associates with poor survival of nasopharyngeal carcinoma
patients treated with induction chemotherapy. BMC Cancer.
16:6692016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Fu L, Li JB, Qin Y, Zeng TT, Zhou J,
Zeng ZL, Chen J, Cao TT, Ban X, et al: Increased expression of
EIF5A2, via hypoxia or gene amplification, contributes to
metastasis and angiogenesis of esophageal squamous cell carcinoma.
Gastroenterology. 146:1701–1713.e9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xue F, Liu Y, Chu H, Wen Y, Yan L, Tang Q,
Xiao E, Zhang D and Zhang H: eIF5A2 is an alternative pathway for
cell proliferation in cetuximab-treated epithelial hepatocellular
carcinoma. Am J Transl Res. 8:4670–4681. 2016.PubMed/NCBI
|
|
40
|
Fang L, Gao L, Xie L and Xiao G:
Eukaryotic translation initiation factor 5A-2 involves in
doxorubicin-induced epithelial-mesenchymal transition in oral
squamous cell carcinoma cells. J Cancer. 9:3479–3488. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bao Y, Lu Y, Wang X, Feng W, Sun X, Guo H,
Tang C, Zhang X, Shi Q and Yu H: Eukaryotic translation initiation
factor 5A2 (eIF5A2) regulates chemoresistance in colorectal cancer
through epithelial mesenchymal transition. Cancer Cell Int.
15:1092015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang H, Li XD, Zhou Y, Ban X, Zeng TT, Li
L, Zhang BZ, Yun J, Xie D, Guan XY and Li Y: Stemness and
chemotherapeutic drug resistance induced by EIF5A2 overexpression
in esophageal squamous cell carcinoma. Oncotarget. 6:26079–26089.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen C, Lu L, Yan S, Yi H, Yao H, Wu D, He
G, Tao X and Deng X: Autophagy and doxorubicin resistance in
cancer. Anticancer Drugs. 29:1–9. 2018. View Article : Google Scholar
|
|
44
|
Li L, Wang Y, Jiao L, Lin C, Lu C, Zhang
K, Hu C, Ye J, Zhang D, Wu H, et al: Protective autophagy decreases
osimertinib cytotoxicity through regulation of stem cell-like
properties in lung cancer. Cancer Lett. 452:191–202. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dai CH, Shu Y, Chen P, Wu JN, Zhu LH, Yuan
RX, Long WG, Zhu YM and Li J: YM155 sensitizes non-small cell lung
cancer cells to EGFR-tyrosine kinase inhibitors through the
mechanism of autophagy induction. Biochim Biophys Acta Mol Basis
Dis. 1864:3786–3798. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tai WT, Shiau CW, Chen HL, Liu CY, Lin CS,
Cheng AL, Chen PJ and Chen KF: Mcl-1-dependent activation of Beclin
1 mediates autophagic cell death induced by sorafenib and SC-59 in
hepatocellular carcinoma cells. Cell Death Dis. 4:e4852013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wu FQ, Fang T, Yu LX, Lv GS, Lv HW, Liang
D, Li T, Wang CZ, Tan YX, Ding J, et al: ADRB2 signaling promotes
HCC progression and sorafenib resistance by inhibiting autophagic
degradation of HIF1alpha. J Hepatol. 65:314–324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liang XH, Kleeman LK, Jiang HH, Gordon G,
Goldman JE, Berry G, Herman B and Levine B: Protection against
fatal Sindbis virus encephalitis by beclin, a novel
Bcl-2-interacting protein. J Virol. 72:8586–8596. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Guo QQ, Wang SS, Zhang SS, Xu HD, Li XM,
Guan Y, Yi F, Zhou TT, Jiang B, Bai N, et al: ATM-CHK2-Beclin 1
axis promotes autophagy to maintain ROS homeostasis under oxidative
stress. EMBO J. 39:e1031112020. View Article : Google Scholar : PubMed/NCBI
|