Introduction
Glioblastoma multiforme (GBM) is a highly
heterogeneous and incurable intracranial tumor (1). The occurrence of GBM is associated
with various genetic alterations, such as PTEN loss and isocitrate
dehydrogenase (IDH) mutations, as well as with multiple oncogene
signaling pathways, including PI3K/AKT/mTOR and wnt/β-catenin
(1,2). Recently, non-coding RNAs, including
microRNAs (miRNAs/miRs), have been reported to be involved in
glioma tumorigenesis (3,4). Although several novel target
candidates and their complex network regulatory relationships have
been comprehensively investigated using high throughput data and
advanced experimental strategies, such as miR-4528/ETS
transcription factor ELK4, nuclear paraspeckle assembly transcript
1/miR-107/CDK6 axis (3,4), the detailed molecular mechanisms
remain poorly understood. Therefore, it is important to identify
novel molecules and regulatory loops in order to provide additional
insights for the combination targeted therapy of GBM.
In the last few decades, along with the extensive
identification and study of novel non-coding RNAs, such as long
non-coding RNAs and circular RNAs, miRNA-mediated network
dysregulations have been found to serve significant roles in the
development and progression of malignancies, including gliomas
(5,6). Previous studies have reported that
miR-301a is markedly upregulated in a variety of cancer types,
including melanoma, cervical, breast, pancreatic and colorectal
cancer (CRC), indicating that it has an oncogene potential
(7-11). For instance, miR-301a mediates
breast cancer proliferation and invasion by directly targeting
fork-head box F2, BCL2 binding component 3, PTEN and collagen type
II α 1 chain (9). In pancreatic
cancer, miR-301a promotes in vitro migration and in
vivo tumorigenicity by negatively regulating its target SMAD4
(12). Moreover, Yue et al
(13) revealed that miR-301a was
upregulated in gliomas and that it enhanced the malignnat phenotype
via its target gene, septin 7; however, the exact molecular
mechanism of miR-301a on the glioma progression is yet to be fully
elucidated.
Increasing evidence has confirmed that the
wnt/β-catenin signaling is one of the most crucial mediators in the
progression of GBM and forms a network crosstalk with other
signaling pathways, including PI3KAKT/mTOR and Ras/MAPK (14,15).
Zinc and ring finger 3 (ZNRF3) belongs to the E3 ubiquitin ligase
family and represses wnt/β-catenin signaling by promoting the
turnover of the Frizzled and LDL receptor related protein 6 (LRP6)
receptors (16,17). Previous studies have reported that
multiple miRNAs, such as miR-106b-3p and miR-93, directly target
ZNRF3 and subsequently activate the wnt/β-catenin signal pathway
and epithelial-mesenchymal transition (EMT) program (18,19).
However, whether ZNRF3 serves a crucial role in miR-301a-mediated
regulation of the wnt/β-catenin signaling pathway in gliomas
remains unknown.
In the present study, a series of in vitro
and in vivo experiments were performed to confirm whether
mi-301a may function as oncogene via a ZNRF3-mediated wnt/β-catenin
signaling pathway, based on the verification of ZNRF3 as a
functional target of miR-301a, as indicated by luciferase reporter
assay and western blotting. Additionally, the fact that ZNRF3, in
turn, could inhibit miR-301a expression was identified via
luciferase reporter assay and reverse transcription-quantitative
(RT-q)PCR. Overall, the present study identified a new
miR-301a/ZNRF3/wnt/β-catenin feedback loop in gliomas, which
represents a novel therapeutic target.
Materials and methods
Patients and samples
Gene expression data and relevant clinicopathologic
variable information were downloaded from TCGA database (http://cancergenome.nih.gov) (n=489). Additionally,
miRNAs expression data and metadata containing survival information
from the CGGA database (http://www.cgcg.org.cn) (n=198) were analyzed.
Moreover, the ENCORI database (http://starbase.sysu.edu.cn/) was used to analyze the
relationship between miR-301a and ZNRF3 expression in low-grade
gliomas (LGG; n=525).
Furthermore, 39 clinical glioma specimens from 15
female and 24 male patients (age range, 19-76 years; mean age,
42.9±15.4 years) were obtained from the nerve tumor tissues bank at
the Tianjin Institute of Neurosurgery, including 10 low grade (WHO
I and II) and 29 GBMs (WHO IV), who were diagnosed by surgeons and
pathologists at Tianjin Huan Hu Hospital (Tianjin, China) between
January 2010 and July 2018. None of the patients had received any
radiotherapy, chemotherapy or any other anticancer treatments prior
to surgery. In total, five normal adult brain tissue samples were
collected from 4 male and 1 female patient (age range, 58-74 years;
mean age, 66±6.3 years) undergoing post-trauma surgery for severe
traumatic brain injury. All the collected tissues were frozen
immediately in liquid nitrogen and stored at −80°C. This study was
approved by the Institutional Review Board of Tianjin Huan Hu
Hospital, and written informed consent was obtained from all
participants.
Cell culture and transfection
The human glioblastoma cell lines U87, U251 and A172
and the normal astrocyte cell line were obtained from the Peking
Union Medical College Cell Library. Human LN308, LN229 and T98G
glioblastoma cell lines were obtained from the China Academia
Sinica Cell Repository. The U87 cell line was authenticated using
short tandem repeat profiling analysis; the U87 cell line
(glioblastoma of unknown origin) used in the present study was of
the American Type Culture Collection type. Cells were maintained in
DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.). All cell lines were incubated at 37°C with 5%
CO2.
The oligonucleotide sequencess of human miR-301a
inhibitor (AS-miR; 5′- GCUUUGACAAUACUAUUGCACUG-3′), miR-301a mimics
(5′-CAGUGCAAUAGUAUUGUCAA AGC-3′), miRNA-negative control (miR-NC;
5′-UUGUACU ACACAAAAGUACUG-3′), ZNRF3 small interfering (si)RNA (5′-
CACUGGGCCUAUGUAAUAATT-3′) and transcription factor 4 (TCF4)
overexpression plasmid were purchased from Shanghai GenePharma Co.,
Ltd. In brief, LN229 and U87 cells were incubated in a 6-well plate
at a density of 2×105 cells/well and then transfected
with miRNA (2 µg) or siRNA (2 µg), or co-transfected
both of the transfectants at room temperature using X-tremeGENE
transfection reagent (Roche Diagnostics GmbH) according to the
manufacturer's protocol. Subsequent experiments were performed
after 72 h.
Lentivirus-transducing units carrying the ZNRF3
variant 3 (LV-ZNRF3) and Lentivirus vector control (LV-NC) were
purchased from Shanghai Genechem Co., Ltd. For stable transfection,
after seeding in 6-well plates (2×105 cells/well) for 24
h, cells were infected with LV-ZNRF3 or LV-NC at a multiplicity of
infection of 10 in the Opti-DMEM (Gibco; Thermo Fisher Scientific,
Inc.) and incubated at 37°C. In order to establish stable
overexpressing cell lines, infected cells were treated with
puromycin (2 µg/ml for the LN229 and U87 cells) for 7 days.
The overexpression efficiency was evaluated via western blot
analysis.
Total RNA extraction and RT-qPCR
Total RNA was extracted using TRIzol®
reagent (Thermo Fisher Scientific, Inc.) and was reverse
transcribed to cDNA to evaluate the mRNA expression levels of
ZNRF3, c-myc, cyclin D1 and TCF4 using the PrimeScript RT Master
mix (Takara Bio, Inc.) according to the manufacturer's
instructions. The protocol of RT was 37°C for 15 min, 85°C for 5
sec and held at 4°C. The PCR protocol was: Initital denaturation at
95°C for 5 sec, followed by 40 cycles at 60°C for 30 sec and by
annealing and extension at 50°C for 30 sec. To evaluate miR-301a,
the protocol of miRNA reverse transcription was 37°C for 60 min,
95°C for 5 min and held at 4°C, and stem-loop RT-PCR was performed
with an RNA PCR kit (Qiagen GmbH) according to the manufacturer's
instructions. qPCR was performed using the miScript SYBR Premix
Green PCR kit (Qiagen GmbH) and the Roche LC480 quantitative
Real-Time PCR system (Roche Diagnostics). The amplification
conditions were: Initital denaturation at 95°C for 15 min, followed
by 40 cycles at 94°C for 15 sec and by annealing and extension at
55°C for 30 sec. GAPDH or U6 levels were selected as internal
controls, and fold changes were calculated using the relative
quantification (2-ΔΔCq) method (20). The primer sequences are presented
in Table SI. Data were analyzed
from three independent experiments and are presented as the mean ±
SD.
Dual-luciferase reporter assay
StarBaseV2 (http://starbase.sysu.edu.cn/), TargetScan (http://www.targetscan.org/vert_72/) and Pictar
(http://www.pictar.org/) algorithms were applied
to identify targets of miR-301a, and the seed sequence of miR-301a
was identified to match the 3′ untranslated region (3′UTR) of the
ZNRF3 gene. The 3′UTR of ZNRF3 containing the miR-301a conserved
binding sites and the corresponding mutant (MT) sites were inserted
into the pMIR-REPORT vector (Promega Corporation). LN229 and U87
cells were cultured in 96-well plates, and co-transfected with the
wild-type (WT) or MT luciferase reporters (0.15 µg) and the
AS-miR-301a (0.15 µg) using the X-tremeGENE transfection
reagent (Roche Diagnostics GmbH) at room temperature.
To analyze the transcriptional activity of
β-catenin/TCF4, TOP-FLASH and FOP-FLASH luciferase reporter
constructs were constructed. The TOP-FLASH (with repeats of the
Tcf-binding site) or FOP-FLASH (with repeats of the MT Tcf-binding
site) plasmids (EMD Millipore) (0.15 µg) were transfected
into LN229 and U87 cells treated with the miR-301a inhibitor (0.15
µg) in 96-well plates at room temperature. After 72 h
incubation, luciferase activity was measured using the
Dual-Luciferase Reporter Assay system (Promega Corporation), and
Renilla luciferase activity was used as an internal
control.
The PGL3-WT-miR-301a-promoter-Luc reporter or
pGL3-MT-miR-301-promoter-Luc reporter constructs (0.15 µg;
Promega Corporation) were transfected into LN229 and U87 cells or
stable ZNRF3 overexpressing cells, following the co-transfection of
the TCF4 overexpression plasmid or siTCF4 using the X-tremeGENE
transfection reagent (Roche Diagnostics) at 37°C for 72 h. The
luciferase activity was detected after 72 h as aforementioned.
Western blotting and immunohistochemistry
(IHC) analysis
The ExKine Total Protein Extraction kit was
purchased from Abbkine Scientific Co., Ltd. and used to extract
proteins. Protein concentrations were determined using a BCA
Protein assay kit (Thermo Fisher Scientific, Inc.). Then, 40
µg protein lysate from each sample was resolved via 10%
SDS-PAGE and subsequently transferred to PVDF membranes (EMD
Millipore). After blocking in 5% skimmed milk for 1 h at room
temperature, the membranes were incubated with diluted primary
antibodies at 4°C overnight. After washing with TBS-Tween-20 (1%
Tween-20), membranes were then incubated with horseradish
peroxidase-conjugated secondary antibodies (1:5,000; cat. nos.
ab222772 and ab222759; Abcam) for 1 h at room temperature. The
immune-reactive bands were visualized using ECL Western Blot
Detection reagents (EMD Millipore) and semi-quantified using ImageJ
software (version 1.47v; National Institutes of Health). The
primary antibodies used were as follows: ZNRF3 (1:1,000; cat. no.
A16026; ABclonal Biotech Co., Ltd.), cyclin D1 (1:10,000; cat. no.
ab134175; Abcam), c-myc (1:1,000; cat. no. ab39688; Abcam), TCF4
(1:10,000; cat. no. ab76151; Abcam) and β-actin (1:5,000; cat. no.
ab6276; Abcam), which was used as the control.
IHC assays were performed using antibodies against
ZNRF3 (1:200; cat. no. A16026, ABclonal Biotech Co., Ltd.),
β-catenin (1:500; cat. no. ab32572; Abcam), c-myc (1:100; cat. no.
ab39688; Abcam), TCF4 (1:500; cat. no. ab76151; Abcam) and
caspase-3 (1:100; cat. no. 9662; Cell Signaling Technology, Inc.).
The protocols used were performed as described previously (21). Briefly, the animal tumor samples
were fixed with 10% formalin for 48 h at room temperature, embedded
with paraffin and sliced into 4-µm sections, followed by
dewaxing. Subsequently, these sections were incubated with 3%
hydrogen peroxide for 20 min at room temperature to block
endogenous peroxidase. Following antigen retrieval, the slides were
blocked with 10% normal goat serum (Beijing Solarbio Science &
Technology Co., Ltd.) for 30 min at room temperature and then
incubated at 4°C overnight with appropriate primary antibody. After
washing with PBS, the slides were incubated with biotinylated
secondary antibody (cat. no. Sp-9001; 1:1,000; OriGene
Technologies, Inc.) for 1 h at room temperature. A DAB Horseradish
Peroxidase Color Development kit (Beyotime Institute of
Biotechnology) was used for color development, and neutral gum
(Sinopharm Chemical Reagent Co., Ltd.) was used for sealing.
Finally, five randomly selected fields were imaged using the
Olympus X71 inverted microscope (Olympus Corporation;
magnification, x200), and semi-quantitatively analyzed in the form
of mean optical density using Image-Pro Plus 6.0 (Media
Cybernetics, Inc.). The signal density of the tissue areas from
five randomly selected fields were counted in a blinded manner. The
significance of the differences between two groups was determined
using Student's t-test.
Immunofluorescence staining
To examine the regulatory role of miR-301a on
β-catenin/TCF4 activity, LN229 and U87 cells were transfected with
AS-miR-301a or miR-scramble (miR-NC) or were co-transfected with
AS-miR-301a and siZNRF3, followed by immunofluorescence analysis.
The procedures were conducted as previously described (21). In brief, cells were fixed with 0.1%
paraformaldehyde for 20 min at room temperature and were blocked
with 5% BSA (Sigma-Aldrich; Merck KGaA) at room temperature for 2
h. Cells were incubated with anti-β-catenin primary antibody
(1:200; cat. no. ab32572; Abcam) overnight at 4°C. After washing
with PBS at room temperature, Goat anti-rabbit fluorescence
conjugated second antibody (1:10,000; cat. no. A21245; Invitrogen;
Thermo Fisher Scientific, Inc.) was added for 1 h at room
temperature and imaged using a confocal microscope (magnification,
×50).
Cell viability assay
Cell viability was analyzed using a Cell-Counting
Kit 8 (CCK-8; Dojindo Molecular Technologies, Inc.) according to
the manufacturer's instructions. Following transfection with
AS-miR-301a or co-transfection with AS-miR-301a and siZNRF3, LN229
and U87 cells were incubated at 37°C for 24, 48, 72 and 96 h. CCK-8
solution (10 µl) was added to each well, and the absorbance
at 490 nm was measured after incubation at 37°C for 2 h to estimate
the number of viable cells.
Apoptosis assay
Apoptosis was quantified after transfection, using
Annexin V labelling and caspase-3/7 activity. For the Annexin V
assay, the Annexin V-FITC-labelled Apoptosis Detection kit (Abcam)
was used according to the manufacturer's protocol. In brief, cells
were harvested, washed and resuspended, and then 5 µl
Annexin V-FITC and 5 µl PI were added and maintained for 15
min at room temperature in the dark. Within 1 h, the stained cells
were analyzed with a flow cytometry system (BD FACSCalibur™ flow
cytometer; BD Biosciences) equipped with CellQuest software
(version-5.1; BD Biosciences). The apoptotic rate was calculated as
the percentage of early + late apoptotic cells.
Caspase-3/7 activity was analyzed using the
Caspase-Glo-3/7 reagent (cat. no. G8091; Promega Corporation).
Caspase-Glo-3/7 reagent (100 µl) was added to a white-walled
96-well plate, which was gently mixed using a plate shaker at 50 ×
g for 30 sec at room temperature. Following incubation at room
temperature for 1-2 h, the luminescence of each sample at 510 nm
was measured in a plate-reading luminometer.
Xenograft model with nude mice
All animal protocols were performed following the
approval of the Animal Care and Used Committee of Tianjin Huanhu
Hospital. A total of 30 female BALB/c-A nude mice (Age, 5 weeks;
weight,18-21 g) were purchased from the Animal Center of the Cancer
Institute, Chinese Academy of Medical Science. Animal welfare was
considered and all the mice were housed with filtered air, a 12-h
light/dark cycle, regulated temperature (25±2°C) and humidity
(50±10%), and with ad libitum access to sterilized food and
water. Then, ~5×106 U87 GBM cells in 100 µl PBS
were subcutaneously injected into the left flank region of nude
mice. When the tumor volume reached 100 mm3, the mice
were randomly divided into two groups, namely NC group and
AS-miR-301a group (5 mice per group). Each group was treated with
oligonucleotides (10 µg AS-miR-301a or miR-NC) with a
mixture of 10 µl Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) via local injection into the xenograft
tumor at multiple sites on all sides. The treatment was performed
once every 3 days for 27 days, and the tumor volume was monitored
with a caliper, using the formula: Volume = length ×
width2/2.
To evaluate the miR-301a-mediated role of ZNRF3 in
tumor inhibition in vivo, 5×106 U87 cells stably
expressing LV-ZNRF3 or LV-NC in 150 µl PBS were injected
into the left flank region of nude mice. When all the groups formed
a tumor at ~4 weeks, the LV-NC and LV-ZNRF3 injected mice were
separated into two groups (5 mice per group), respectively. One of
the LV-NC groups was treated with miR-scramble (miR-NC), while one
of LV-ZNRF3 groups was treated with miR-301a mimics and another one
was treated with miR-NC every 3 days for 30 days. The tumor
injection of oligonucleotides (10 µg AS-miR-301a or miR-NC)
with a mixture of 10 µl Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) was also performed at multiple
sites. At the end of the experiment, the mice were anesthetized by
injecting 1% pentobarbital sodium (100 mg/kg body weight) and
sacrificed via cervical vertebra dislocation. Finally, tumor weight
was calculated. The removed tumor tissues were subjected to RNA
extraction and immunohistochemistry assays.
Statistical analysis
All statistical analyses were performed using
GraphPad software version 6.0 (GraphPad Software, Inc.) or IBM SPSS
Statistics 23.0 (IBM Corp.). Data are presented as the mean ± SD of
≥3 independent experiments. The unpaired Student's t-test, and
one-way ANOVA were performed to analyze statistically significant
differences between two groups or multiple groups, respectively.
Additionally, a Tukey's test was used for multiple comparisons
following ANOVA. The Kaplan-Meier method was used to evaluate the
differences in survival rates, which were analyzed using the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-301a is significantly upregulated in
glioma specimens and cell lines
To evaluate the potential role of miR-301a in
gliomagenesis, miR-301a expression was analyzed in gliomas with
different grades, including 10 LGG and 29 GBM cases, and seven
malignant glioma cell lines. miR-301a was significantly upregulated
in the clinical samples and GBM cell lines (Fig. 1A and B), and its expression level
was positively associated with the WHO grade (Fig. 1A). The clinicopathological
characteristics of the 39 glioma specimens are summarized in
Table I. There was a statistically
significant statistically correlation between miR-301a expression
and histology, but not for sex and age. Additionally, analysis of
the data from the CGGA database indicated that miR-301a expression
was positively associated with the WHO grade, which was in
accordance with the data from the clinical specimens (Fig. 1C), and a higher level of miR-301a
predicted a poor prognosis (Fig.
1D). Furthermore, the relationship between the miR-301a
expression and prognosis from TCGA database was analyzed, and the
results were consistent with the analysis of the CGGA data
(Fig. S1). These findings
indicated that miR-301a may act as a critical oncogene in
gliomas.
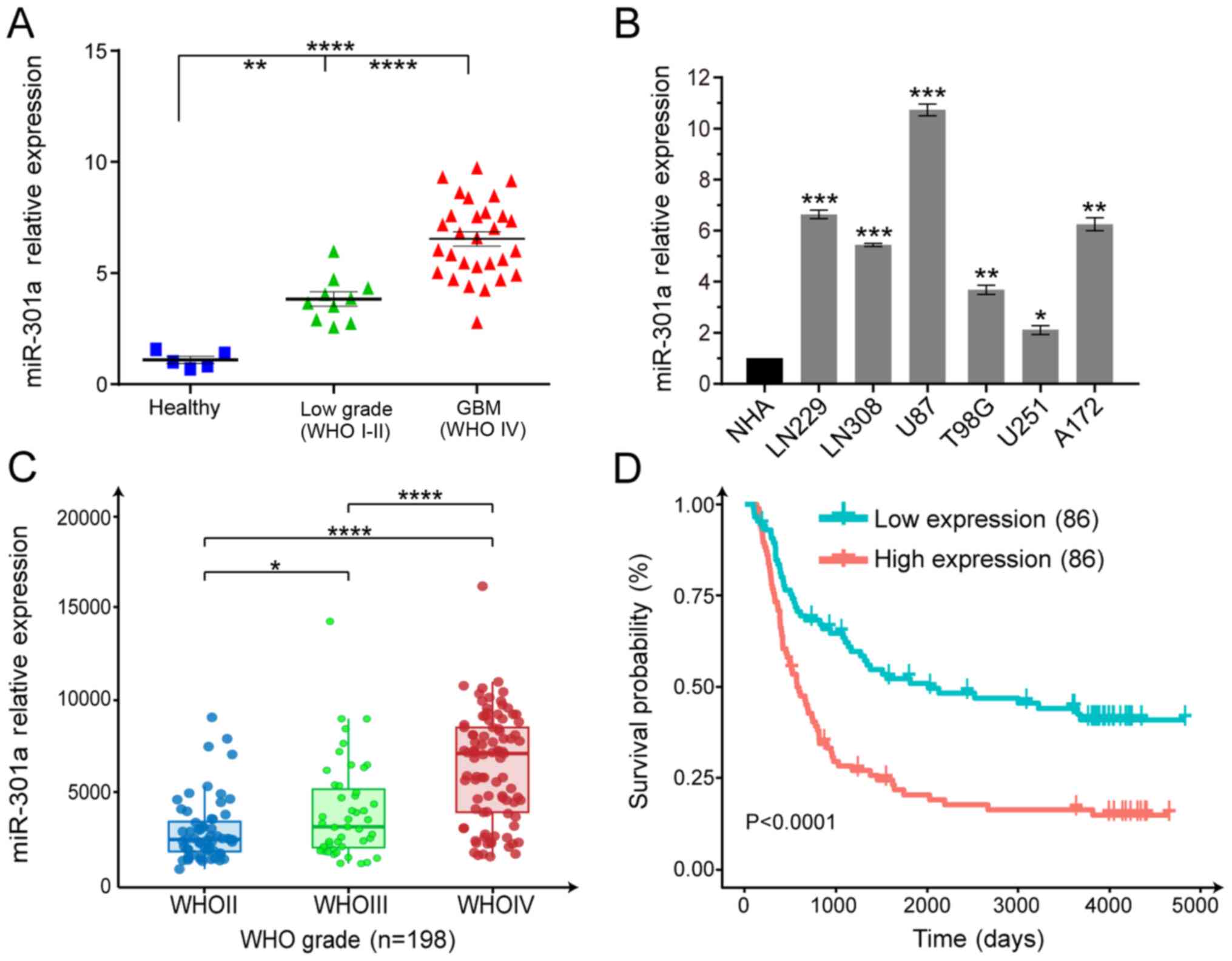 | Figure 1miR-301a is significantly upregulated
in glioma samples and cell lines, and is negatively correlated with
patient survival. (A) RT-qPCR was used to evaluate the miR-301a
expression in clinical specimens of different grades compared with
five healthy brain tissues. **P<0.01,
****P<0.0001. (B) RT-qPCR analysis of GBM cell lines
(LN229, LN308, U87, T98G, U251 and A172) compared with the normal
astrocytes. *P<0.05, **P<0.01,
***P<0.001 vs. NHA. (C) miR-301a expression from
WHOII, WHOIII and WHOIV cases as detected via RT-qPCR from the CGGA
database. *P<0.05, ****P<0.0001. (D)
Kaplan-Meier analysis of survival of the miR-301a low-expressing
(n=86) and high-expressing glioma cases (n=86) from the CGGA
database. miR-301a, microRNA-301a; GBM, glio-blastoma multiforme;
WHO, World Health Organization; CGGA, Chinese Glioma Genome Atlas;
RT-qPCR, reverse transcription-quantitative PCR. |
 | Table IRelationship between miR-301a and
clinicopathological characteristics expression of patients with
glioma. |
Table I
Relationship between miR-301a and
clinicopathological characteristics expression of patients with
glioma.
| Variable | No. of cases | miR-301a expression
| P-value |
|---|
| Low | High |
|---|
| Sex | | | | 0.3332 |
| Female | 15 | 6 | 9 | |
| Male | 24 | 14 | 10 | |
| Age, years | | | | 0.7524 |
| <42 | 20 | 9 | 11 | |
| ≥42 | 19 | 10 | 9 | |
| Histology | | | | 0.0084 |
| Astrocytoma | 10 | 9 | 1 | |
| Glioblastoma | 29 | 11 | 18 | |
ZNRF3 is a direct target of miR-301a
miRNAs serve as oncogenes or tumor-suppressors by
regulating gene expression by either mRNA degradation or
translational inhibition (22).
Thus, StarBaseV2, TargetScan and Pictar algorithms were used to
verify the downstream effectors of miR-301a. Based on the database
evaluation, the seed sequence of miR-301a that matched the 3′UTR of
the ZNRF3 was identified (Fig.
2A).
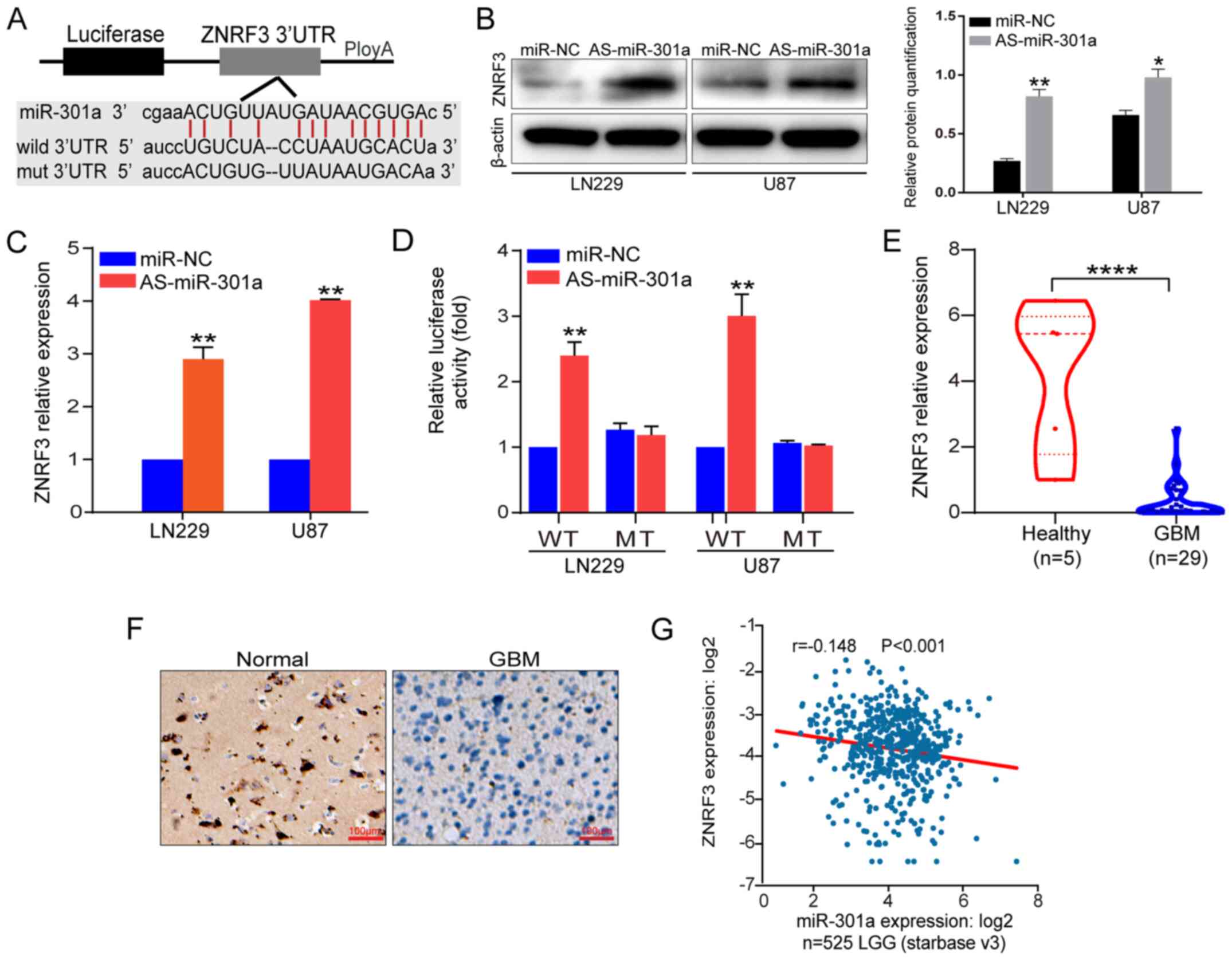 | Figure 2ZNRF3 is a direct functional target
of miR-301a. (A) A schematic diagram of the seed sequence of
miR-301a matching the ZNRF3 3′UTR, and the design of the WT or MT
ZNRF3 3′UTR including reporter constructs. (B) ZNRF3 protein
expression in LN229 and U87 cells transfected with miR-NC or
miR-301a inhibitor was analyzed via western blotting. β-actin was
used as a loading control. The relative protein semi-quantification
was performed with ImageJ software. *P<0.05,
**P<0.01 vs. miR-NC. (C) ZNRF3 mRNA expression in
LN229 and U87 cells transfected with miR-NC or AS-miR-301a was
analyzed using RT-qPCR. (D) Luciferase reporter assays were
performed in LN229 and U87 cells following co-transfection with the
WT or MT 3′UTR of ZNRF3 and AS-miR-301a or miR-NC. The data present
the fold changes as the mean ± SD of three independent experiments.
**P<0.01 vs. miR-NC. (E) ZNRF3 expression was
examined via RT-qPCR in GBM cases compared with the healthy
controls. ****P<0.0001. (F) ZNRF3 expression in
patients with GBM was determined via immunohistochemistry. Scale
bar, 100 µm. (G) Negative correlation analysis from 525 LGG
cases from the public database ENCORI. ZNRF3, Zinc and ring finger
3; miR-301a, microRNA-301a; 3′UTR, 3′untranslated region; NC,
negative control; WT, wild-type; MT, mutant; LGG, low-grade glioma;
RT-qPCR, reverse transcription-quantitative PCR; AS-miR-301a,
miR-301a inhibtor; GBM, glioblastoma multiforme. |
To verify whether ZNRF3 expression was downregulated
by miR-301a, the changes in ZNRF3 mRNA and protein expression
levels were detected in LN229 and U87 cells after knocking down
miR-301a expression. The results demonstrated that ZNRF3 expression
was significantly increased at both the mRNA and protein levels
after miR-301a knockdown (Fig. 2B and
C). Additionally, to assess whether ZNRF3 was the direct
functional target of miR-301a, the p-MIR-WT-ZNRF3-3′UTR and
p-MIR-MT-ZNRF3-3′UTR reporter plasmids were constructed. Reporter
assay results indicated that AS-miR-301a triggered a significantly
increase of p-MIR-WT-ZNRF3-3′UTR luciferase activity, but no change
in luciferase activity was observed for the MT reporter plasmid
(Fig. 2D). Moreover, ZNRF3
expression was significantly downregulated in GBM tissues compared
with healthy tissues (Fig. 2E and
F).
The public database ENCORI was analyzed, and it was
identified that miR-301a expression was weakly negatively
associated with ZNRF3 expression in 525 LGG (r=-0.148; P<0.001;
Fig. 2G). However, the correlation
coefficient was very small, which may due to the LGG or the sample
size. These data suggested that miR-301a directly modulated ZNRF3
expression at both the transcriptional and post-transcriptional
levels via binding to the 3′UTR.
ZNRF3 is required for the biological
effects of miR-301a in the GBM malignant phenotype
To further identify the functional relationship
between miR-301a and its target ZNRF3, the ZNRF3-mediated role on
proliferation and apoptosis exerted by miR-301a was investigated.
The verification of miR-301a knockdown is presented in Fig. 3A. The successful establishment of
lentivirus-mediated ZNRF3 overexpression is presented in Fig. 3B, and the successful establishment
of ZNRF3 knockdown in LN2229 and U87 cells singly transfected with
siZNRF3 is presented in Fig.
S2.
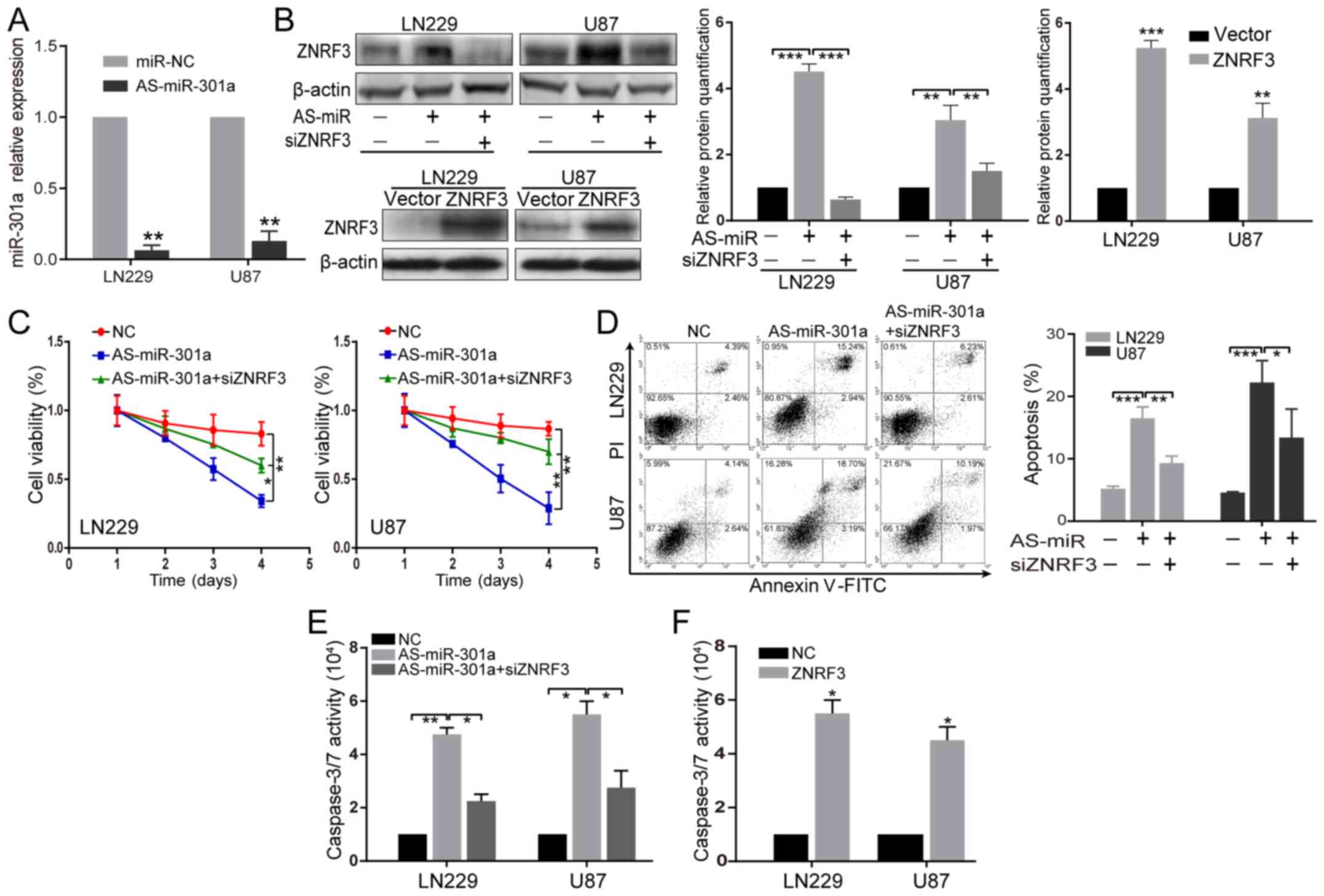 | Figure 3AS-miR-301a inhibits the GBM
malignant phenotype, partially via ZNRF3. (A) Verification of
miR-301a knockdown in LN229 and U87 cells transfected with miR-NC
or miR-301a inhibitor. **P<0.01 vs. miR-NC. (B) ZNRF3
protein expression was examined in LN229 and U87 cells transfected
with AS-miR-301a or co-transfected with AS-miR-301a and siZNRF3
(upper), and in stably ZNRF3-overexpressing LN229 and U87 cells
(below) via western blot analysis. β-actin was used as an internal
loading control. The relative protein semi-quantification was
performed with ImageJ software. **P<0.01,
***P<0.001. (C) LN229 and U87 cells were transfected
with AS-miR-301a or co-transfected with both AS-miR-301a and
siZNRF3, and 72 h later a Cell Counting Kit-8 assay was used to
detect the ZNRF3-mediated effect on proliferation by AS-miR-301a.
*P<0.05, **P<0.01. (D) Annexin V-PI and
(E) caspase-3/7 activity assays were utilized to detect the
ZNRF3-mediated effect on apoptosis by AS-miR-301a in LN229 and U87
cells at 72 h after transection of AS-miR-301a or co-transfection
of both AS-miR-301a and siZNRF3. *P<0.05,
**P<0.01, ***P<0.001. (F) Caspase-3/7
activity assays were performed in LN229 and U87 cells stably
overexpressing ZNRF3. *P<0.05 vs. NC. ZNRF3, Zinc and
ring finger 3; miR-301a, microRNA-301a; NC, negative control;
siRNA, small interfering RNA; AS-miR-301a, miR-301a inhibtor. |
When ZNRF3 expression was knocked down by siZNRF3 in
LN229 and U87 cell lines co-transfected with AS-miR-301a, the ZNRF3
protein expression was significantly down-regulated compared with
cells transfected with AS-miR-301a alone (Fig. 3B), indicating that siZNRF3
transfection could effectively abrogate the effect of
AS-miR-301a-mediated ZNRF3 upregulation. CCK-8, Annexin V and
caspase-3/7 activity analyses demonstrated that the inhibition of
proliferation and the promotion of apoptosis induced by AS-miR-301a
were partially abolished by siZNRF3 (Fig. 3C-E). Furthermore, ZNRF3
overexpression could significantly enhance caspase-3/7 activity
compared with its NC (Fig. 3F),
consistent with the effect mediated by AS-miR-301a. These results
suggested that AS-miR-301a inhibited the GBM malignant phenotype,
partially via ZNRF3.
Knockdown of miR-301a inhibits the
wnt/β-catenin signaling pathway, at least partially via ZNRF3
Our previous studies have reported that ZNRF3 can
repress the wnt/β-catenin signaling pathway (unpublished), and
combined with the fact that ZNRF3 is a direct target of miR-301a,
it was further examined whether miR-301a also regulates the
wnt/β-catenin signaling pathway via ZNRF3. Firstly, a TOP/FOP
luciferase assay was used to analyze the β-catenin/TCF4
transcriptional activity mediated by miR-301a, and the results
indicated that AS-miR-301a suppressed TOP luciferase activity with
no apparent change in FOP activity (Fig. 4A). Additionally, AS-miR-301a could
decrease the mRNA and protein expression levels of TCF4, c-myc and
cyclin D1, as demonstrated by RT-qPCR and western blotting,
respectively (Fig. 4B and C). The
immunofluorescence results indicated that AS-miR-301a repressed the
expression of β-catenin both in the nucleus and cytoplasm (Fig. 4D). It was found that knockdown of
ZNRF3 via siRNA following the transfection with AS-miR-301a could
significantly abrogate the effects mediated by AS-miR-301a
(Fig. 4A-D). Taken together, these
data indicated that AS-miR-301a could attenuate the wnt/β-catenin
signaling pathway, at least partially via ZNRF3, in LN229 and U87
cells.
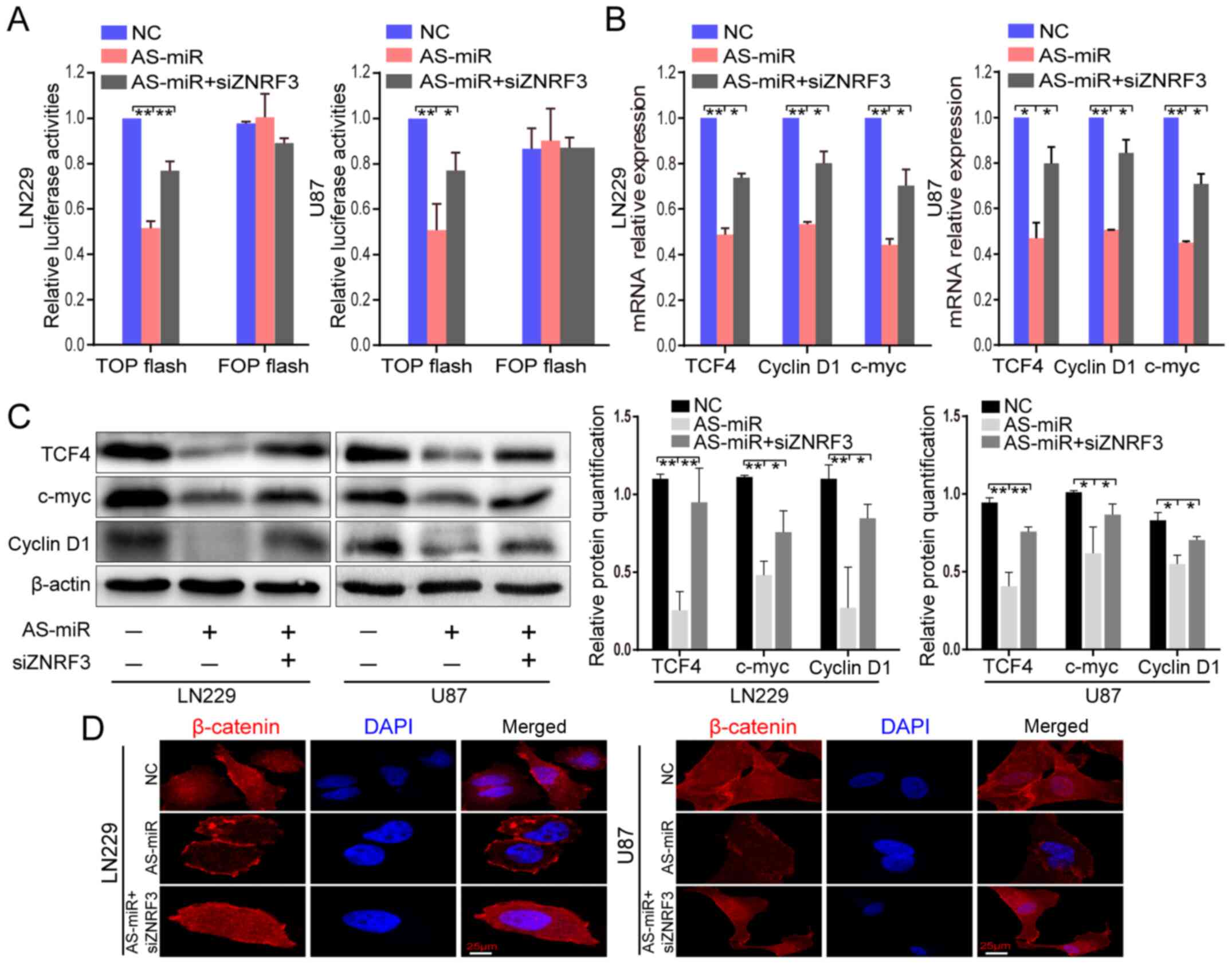 | Figure 4AS-miR-301a suppresses β-catenin/TCF4
transcriptional activity partially via ZNRF3. (A) LN229 and U87
cells were co-transfected with TOP/FOP FLASH luciferase plasmids,
AS-miR-301a or siZNRF3. After 72 h, the luciferase reporter assays
were conducted. *P<0.05, **P<0.01. (B)
Reverse transcription-quantitative PCR and (C) western blotting
were performed to detect TCF4, c-myc and cyclin D1 expression
levels in LN229 and U87 cells transfected with AS-miR-301a or
co-transfected with AS-miR-301a and siZNRF3. *P<0.05,
**P<0.01. (D) Immunofluorescence detection of
β-catenin in the nucleus after transfection with AS-miR-301a or
co-transfection with AS-miR-301a and siZNRF3 (magnification, ×50).
ZNRF3, Zinc and ring finger 3; miR-301a, microRNA-301a; NC,
negative control; siRNA, small interfering RNA; AS-miR-301a,
miR-301a inhibtor; TCF4, transcription factor 4. |
AS-miR-301a represses tumor growth and
wnt signaling in vivo
Based on the in vitro experimental findings,
the effects of AS-miR-301a on tumor growth and wnt/β-catenin
signaling were assessed in vivo. U87 cells were implanted
into the left flanks of nude mice via subcutaneous injection.
AS-miR-301a and miR-NC were treated in a multisite manner every 3
days. After 27 days of treatment, tumors were excised and subjected
to further analysis. The verification of miR-301a knockdown in
tumor tissues is presented in Fig.
5A. The tumor growth curve indicated that AS-miR-301a
significantly inhibited the tumorigenicity of GBM cells, as
observed in AS-miR-301a treated mice (Fig. 5B). Additionally, a significant
decrease in tumor weight was identified in AS-miR-301a-treated
tumors (Fig. 5C). The IHC assay
demonstrated that AS-miR-301a significantly increased ZNRF3
expression, and decreased β-catenin, c-myc and cyclin D1 expression
levels (Fig. 5D), which was
consistent with the in vitro results.
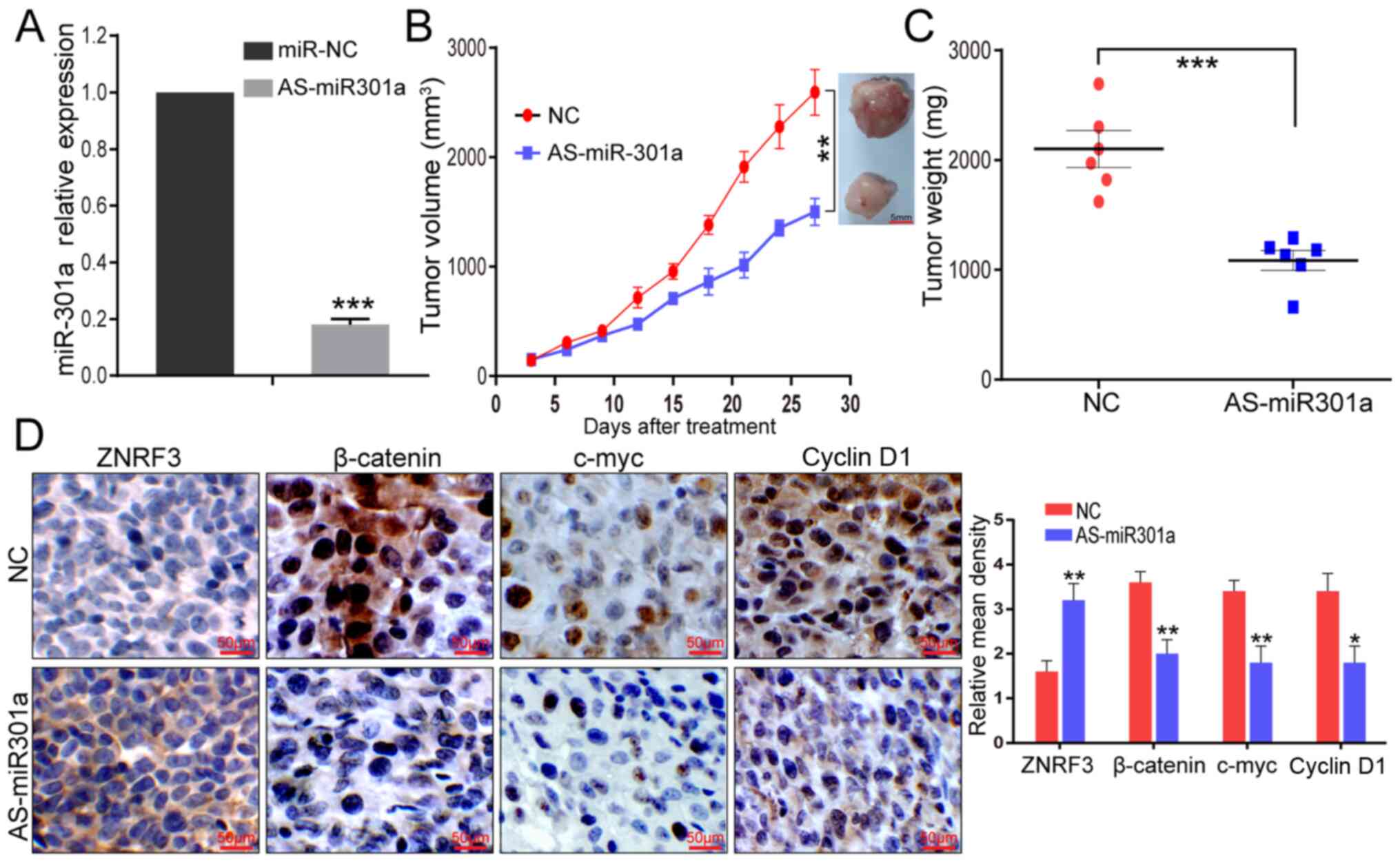 | Figure 5AS-miR-301a inhibits the GBM
malignant phenotype and the wnt signal pathway in vivo. (A)
miR-301a knockdown identification from resected tumor tissues was
detected via reverse transcription-quantitative PCR.
***P<0.001 vs. miR-NC. (B) U87 cells were
subcutaneously injected into nude mice. When tumors were
established uniformly in the two groups, AS-miR-301a was injected
in a multisite injection every 3 days. Tumor volume was evaluated
every 3 days during treatment. **P<0.01. (C) At the
end of the experiment, tumor weight was measured.
***P<0.001. (D) Immunohistochemistry assays were
performed to evaluate ZNRF3, β-catenin, c-myc, and cyclin D1
expression levels from xenograft tumor sections. Scale bar, 50
µm. *P<0.05, **P<0.01. miR-301a,
microRNA-301a; NC, negative control; AS-miR-301a, miR-301a
inhibtor. |
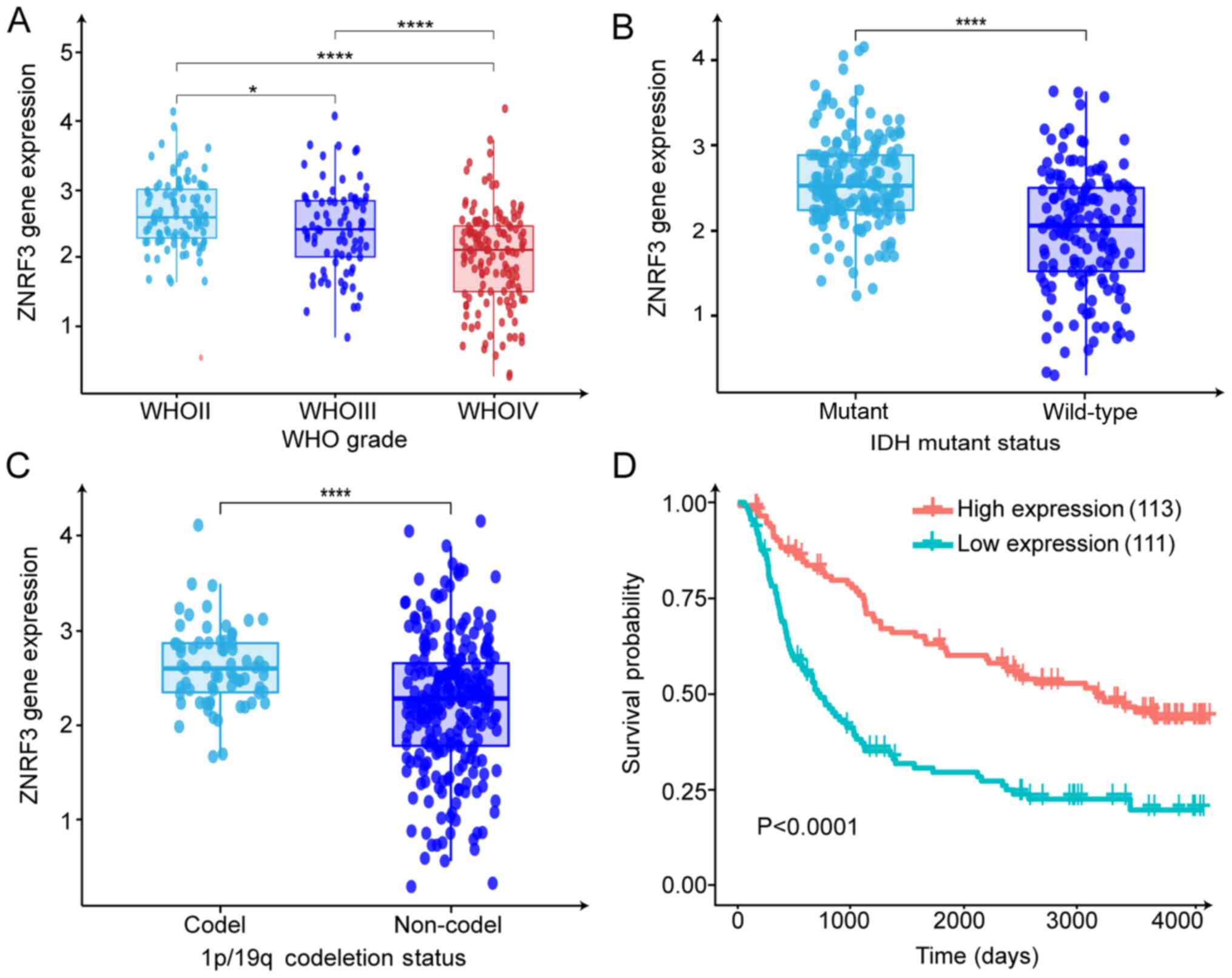
ZNRF3 is downregulated in glioma and is
associated with patient prognosis based on the CGGA dataset
Based on the miR-301a-mediated regulatory effect on
ZNRF3, ZNRF3 expression and its relationship with patient prognosis
was evaluated in the CGGA dataset (CGGA Mseq325). The mRNA
expression of ZNRF3 was significantly down-regulated in high-grade
gliomas compared with the LGG, and ZNRF3 expression was negatively
associated with the WHO grade (Fig.
6A). Analysis of ZNRF3 expression and its relevant clinical
characteristics indicated that ZNRF3 expression was associated with
IDH1 gene mutation status, 1p/19q codeletion status (Fig. 6B and C) and age (Fig. S3B). No statistical significance
was observed for ZNRF3 expression and sex or progression status
(Fig. S3A and C). Furthermore,
Kaplan-Meier survival analysis identified that ZNRF3 upregulation
conferred prolonged overall survival (Fig. 6D). In combination with the ZNRF3
expression data of the clinical specimens, it was concluded that
ZNRF3 may serve as a key tumor suppressor and is involved in GBM
progression.
ZNRF3 can attenuate the miR-301a
expression level dependent on wnt/β-catenin activity
A previous study revealed that miR-301a is activated
by the wnt/β-catenin pathway by direct binding of TCF4 to the
miR-301a promoter region (13).
Thus, it was hypothesized that ZNRF3 restoration could, in turn,
affect miR-301a expression via TCF4. Firstly, ZNRF3 could inhibit
the wnt/β-catenin transcriptional activity, as verified by the
TOP/FOP assay, and the ectopic restoration of miR-301a in stable
ZNRF3-overexpressing LN229 and U87 cells could partially abrogate
this effect mediated by ZNRF3 (Fig.
7A). The successful transfection efficiency of miR-301a
overexpression via RT-qPCR is presented in Fig. S4A. ZNRF3 overexpression could
significantly inhibit miR-301a expression, and the knockdown of
TCF4 (siTCF4) decreased the miR-301a expression. Moreover, the TCF4
overexpression plasmid in stable ZNRF3-overexpressing LN229 and U87
cells could reverse the ZNRF3-mediated inhibitory effect (Fig. 7B). The successful transfection
efficiency of TCF4 overexpression and siTCF4 detected via RT-qPCR
is illustrated in Fig. S4B and C.
Furthermore, the luciferase assay results with the constructed
miR-301a promotor reporter plasmid indicated that ZNRF3
overexpression or the knockdown of TCF4 alone could significantly
decreased the luciferase activities, while the TCF4 restoration in
ZNRF3 stably overexpressing cells could abrogate this inhibitory
effect mediated by ZNRF3 in WT groups as compared with the MT
groups (Fig. 7C), indicating that
TCF4 transcriptionally regulated miR-301a expression by binding to
the promotor of miR-301a. These data demonstrated that ZNRF3 could
repress TCF4-depedent miR-301a expression.
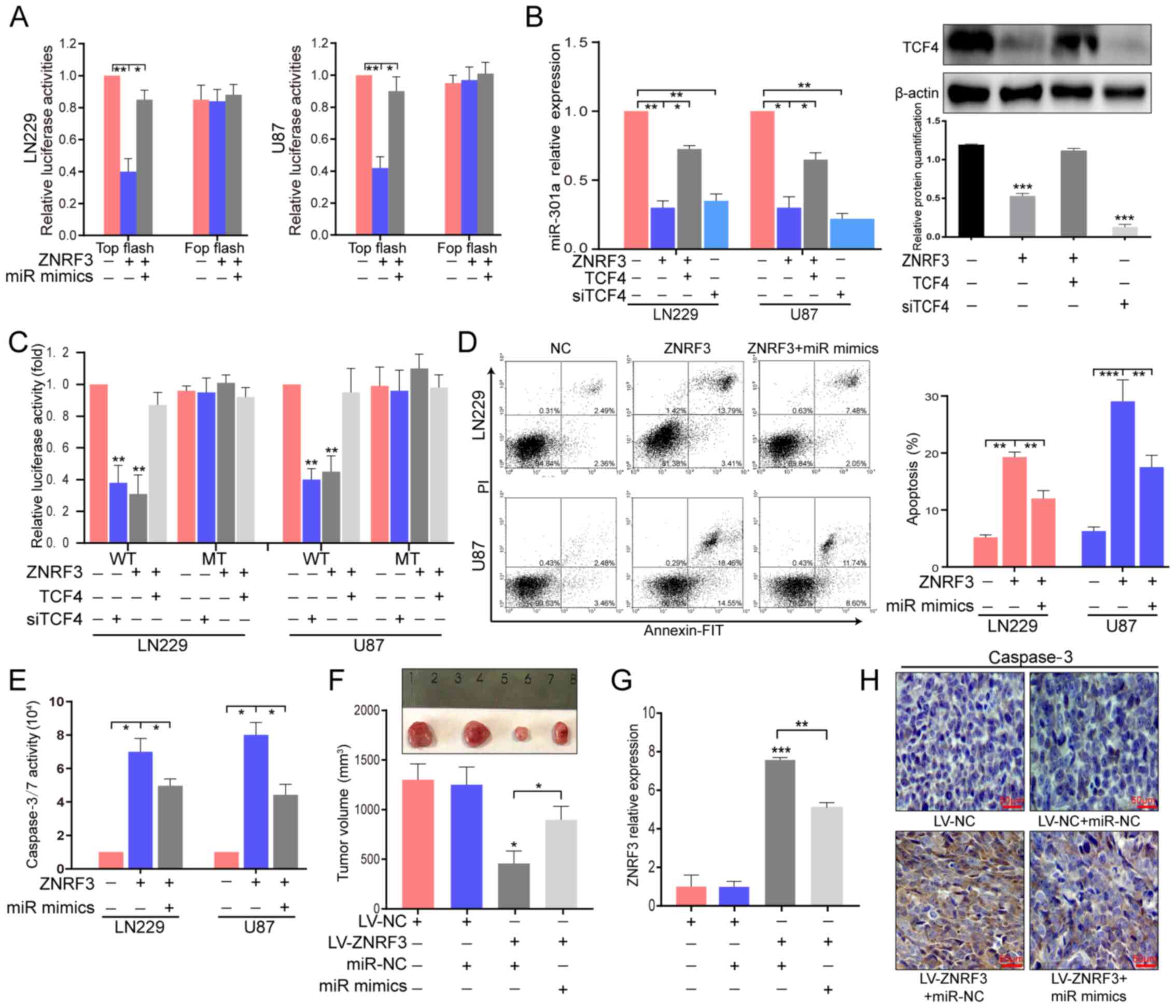 | Figure 7ZNRF3 can inhibit miR-301a expression
that is partially dependent on TCF4. (A) TOP/FOP flash was used to
detect wnt/β-catenin transcriptional activity in ZNRF3 stably
overexpressing LN229 and U87 cells with or without co-transfection
of miR-301a mimics. *P<0.05, **P<0.01.
(B) RT-qPCR was performed to analyze miR-301a expression in L229
and U87 cells transfected with siTCF4, ZNRF3 stable overexpressing
LN229 and U87 cells or those co-transfected with TCF4
overexpression plasmid compared with the cells co-transfected with
NC-siRNA and empty vector (left). Western blotting was used to
identify TCF4 expression after different treatments (right). The
relative protein semi-quantification was performed with ImageJ
software. *P<0.05, **P<0.01,
***P<0.001. (C) pGL3-WT-miR-301a-promotor-luc
reporter and pGL3-MT-miR-301a-promotor-luc reporter were
constructed and co-transfected into LN229 and U87 cells (LV-ZNRF3
or not) with TCF4 overexpression plasmid or siTCF4. Luciferase
activity was further analyzed after 72 h. **P<0.01
compared with NC. (D) Annexin V-PI via flow cytometry and (E)
caspase-3/7 activity assays were used to detect apoptosis in LN229
and U87 cells (LV-ZNRF3 or not) co-transfected with miR-301a mimics
or miR-NC. *P<0.05, **P<0.01,
***P<0.001. In vivo assays investigating the
miR-301a mimic-mediated role on the tumor-suppressive function of
ZNRF3 were performed. (F) Tumor volume and representative images of
excised tumors from different treatment group at the end of the
experiment. *P<0.05. (G) ZNRF3 expression was
determined via RT-qPCR in resected tumor tissues of different
treatment groups. **P<0.01, ***P<0.001.
(H) Representative photomicrographs of immunohistochemistry of
tumor sections for caspase-3 in different groups. Scale bar, 50
µm. ZNRF3, Zinc and ring finger 3; siRNA, small interfering
RNA; miR, microRNA; WT, wild-type; MT, mutant; NC, negative
control; TCF4, transcription factor 4; RT-qPCR, reverse
transcription-quantitative PCR. |
To further evaluate the miR-301a-mediated role of
ZNRF3 on apoptosis, Annexin V-PI and caspase-3/7 activity assays
were performed and the results demonstrated that the transfection
of miR-301a mimics in stable ZNRF3-overexpressing LN229 and U87
cells reversed the ZNRF3-induced effect on apoptosis (Fig. 7D and E). Furthermore, in
vivo experiments indicated that ZNRF3 overexpression
significantly inhibited tumor growth and that the miR-301a mimics
treatment partially reversed this effect mediated by ZNRF3
(Fig. 7F). The ZNRF3 expression
from different treatment groups is presented in Fig. 7G, it was found that ZNRF3
expression in the ZNRF3 + miR-301a group was decreased compared
with the ZNRF3 + miR-NC group (P<0.01; Fig. 7G). The IHC assay results for
caspase 3 expression were in line with the in vitro results
(Fig. 7H). In summary, the results
indicated that the miR-301a/ZNRF3/wnt signal feedback loop may be
involved in gliomagenesis.
Discussion
Previous studies have reported that miR-301a acts as
a crucial oncogene for diagnosis and prognosis evaluation in
various malignant cancer types. For example, Karimi et al
(23) suggested that upregulation
of exosomes miR-301a and miR-23a in serum samples could
discriminate patients with CRC from healthy controls, highlighting
their role as non-invasive biomarkers for the early detection of
CRC. Moreover, Zheng et al (24) revealed that the expression of
miR-301a was associated with decreased overall survival in breast
cancer. It was also observed that the miR-301a upregulation could
predict a poor outcome in pancreatic cancer and hepatocellular
carcinomas (25,26). In addition, a study on glioma
demonstrated that a higher serum exosome miR-301a level was closely
correlated with the WHO grade and lower Karnofsky scores (27). Results from both univariate and
multivariate Cox regression analysis have revealed that miR-301a
could act as a biomarker for glioma diagnosis and prognosis,
especially in advanced grades (27). By analyzing the public datasets,
the present results also verified the prognostic potential of
miR-301a. Moreover, miR-301a expression was measured in glioma
clinical specimens and it was found that miR-301a expression was
positively associated with the WHO grade, which was consistent with
previous reports (13,27). The present study demonstrated that
the knockdown of miR-301a inhibited cell proliferation, as detected
via CCK-8, and promoted apoptosis as shown via Annexin V-FITC assay
and caspase-3/7 activity detection. However, a limitation of the
current study was that some significant apoptosis-related proteins
were not been detected, including Bcl-2 and poly(ADP-ribose)
polymerase 1.
miRNAs are short non-coding RNAs that modulate gene
expression by either mRNA degradation or translational inhibition
(22). miR-301a participates in
the cancer progression by directly targeting a large number of
tumor suppressor genes, such as PTEN, RUNX family transcription
factor 3 (RUNX3) and septin 7 (7,11,13).
Accumulating evidence has demonstrated that miR-301a is innately
associated with cancer cell proliferation, invasion and drug
resistance by targeting PTEN in cervical cancer, malignant melanoma
and pancreatic cancer (7,8,10).
In the present study, according to bioinformatic predictions and
experimental assays, it was identified that ZNRF3 was a functional
target of miR-301a in GBM cell lines. Moreover, the results
suggested a key function of ZNRF3 in miR-301a-mediated promotion of
the glioma malignant phenotype, indicating a novel molecular
mechanism for ZNRF3 inactivation.
Mechanistically, the present data indicated that
miR-301a could activate the wnt/β-catenin signaling pathway, at
least partially via ZNRF3, which is a negative regulator of the wnt
pathway that acts by promoting the ubiquitination, internalization
and degradation of the Wnt receptors Frizzled and LRP5/6 (28,29).
This finding was also reported in various previous cancer studies.
For example, Wang et al (30) revealed that ZNRF3 inhibited the
invasion and EMT by inactivating the wnt/β-catenin pathway in
nasopharyngeal carcinoma. Our previous data also suggested that
ZNRF3 could repress the glioma proliferation and invasion, as well
as promote apoptosis via the inactivation of the wnt/β-catenin
signaling pathway (unpublished data), which supports the
theoretical feasibility of the miR-301a/ZNRF3/wnt pathway.
Additionally, Yue et al (31) reported that exo-miR-301a secreted
by hypoxic glioma cells could suppressed radiation sensitivity by
activating the wnt/β-catenin pathway via targeting transcription
elongation factor A like 7. It has also shown been that RUNX3 is a
direct target of miR-301a in CRC and lung cancer (11,32),
and our previous study demonstrated that RUNX3 repressed the
β-catenin/TCF4 transcriptional activity in gliomas (33), indicating that miR-301a may
regulated the wnt pathway via RUNX3. However, this hypothesis
requires further investigation.
In addition to the wnt/β-catenin signaling, other
important oncogene pathways are also regulated by miR-301a. For
instance, Hu et al (34)
indicated that miR-301a could decrease the expression of suppressor
of cytokine signaling 5 and subsequently result in JAK/STAT3 signal
activation. Lu et al (35)
reported that miR-301a reduced NKRF repressing factor expression,
leading to NF-κB activation in pancreatic cancer. Yin et al
(36) also revealed that miR-301a
could activate ERK/cAMP-response element binding protein signaling
in triple-negative breast cancer. Moreover, based on PTEN target
gene identification, miR-301a participates in biological processes
of cancer via AKT/mTOR signaling (7,9).
Hence, the knockdown of miR-301a may disrupt multiple significant
carcinogenic signaling pathways, and it represents a key candidate
target for cancer treatment.
It is well known that various transcriptional
factors may, to some extent, contribute to miRNA deregulation
(37). Currently, several
transcriptional factors that affect the miR-301a expression have
been identified, including NF-κB, forkhead box M1 and E2F
transcription factor 1, which have been proven to be key oncogene
molecules in the majority of malignancies (34,36,38).
Additionally, Yue et al (13) found that miR-301a was activated by
the wnt/β-catenin pathway, and that TCF4 enhanced miR-301a
expression by directly binding to its promoter region, as
demonstrated by Chromatin immunoprecipitation and luciferase
reporter assays, indicating that the miR-301a-mediated wnt pathway
feedback loop is implicated in glioma tumorigenesis. The present
study investigated whether ZNRF3 overexpression could attenuate the
expression of miR-301a. The results demonstrated that ZNRF3
overexpression significantly inhibited miR-301a expression and that
overexpression of TCF4 in GBM cells stably expressing ZNRF3 could
abrogate the inhibitory effect mediated by ZNRF3. Therefore, these
findings also suggested that the miR-301a/ZNRF3/wnt/β-catenin
signaling feedback loop exists. However, the miR-301a-mediated
regulatory network is complicated and should be further
investigated.
In conclusion, to the best of our knowledge, the
present study demonstrated for the first time that miR-301a
promoted glioma progression via the ZNRF3-mediated wnt/β-catenin
signaling pathway. Additionally, ZNRF3 could repress miR-301a
expression that is dependent on TCF4 transcriptional activity. The
present study identified a novel miR-301a/ZNRF3/wnt/β-catenin
signaling feedback loop that serves critical roles in glioma
tumorigenesis, and it may provide further insights for
understanding the molecular mechanism of glioma.
Supplementary Data
Acknowledgments
Not applicable.
Funding
The study was supported by the Foundation of Tianjin
Science and Technology Committee (grant nos. 14JCZDJC35600 and
12ZCDZSY17700) and the National Key Technology Support Program
(grant nos. 2014BAI04B00 and 2015BAI03B05).
Availability of data and materials
All the datasets generated and analyzed in the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
JS, JWa and CS conceived and designed the
experiments. JS, JX, BL and QM performed the in vitro
experiments. QM, JLi, JLiu and JWu collected the clinical
specimens. JS, JX, JLiu and JWu performed the in vivo
experiment. CS, SZ and JLi helped with the bioinformatics analysis
and data analysis. JS and QM interpreted the data and co-wrote the
manuscript. JS, SZ and JWa reviewed and revised the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was reviewed and approved by
Institutional Review Board of Tianjin Huanhu Hospital (Tianjin,
China; approval no. CK19-190318), and written informed consent was
obtained from patients in all cases. Animal studies were approved
by Animal Ethical and Welfare Committee of Tianjin Huanhu Hospital
of Nankai University ((Tianjin, China; approval no.
SYXK2019-002).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
GBM
|
Glioblastoma multiforme
|
|
ZNRF3
|
Zinc and ring finger 3
|
|
AS-miR-301a
|
miR-301a inhibtor
|
|
TCGA
|
The Cancer Genome Atlas Program
|
|
CGGA
|
Chinese Glioma Genome Atlas
|
|
LGG
|
low-grade gliomas
|
References
|
1
|
Aldape K, Zadeh G, Mansouri S,
Reifenberger G and von Deimling A: Glioblastoma: Pathology,
molecular mechanisms and markers. Acta Neuropathol. 129:829–848.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kanu OO, Hughes B, Di C, Lin N, Fu J,
Bigner DD, Yan H and Adamson C: Glioblastoma multiforme
oncogenomics and signaling pathways. Clin Med Oncol. 3:39–52.
2009.PubMed/NCBI
|
|
3
|
Chan JJ and Tay Y: Noncoding RNA:RNA
regulatory networks in cancer. Int J Mol Sci. 19:192018. View Article : Google Scholar
|
|
4
|
Wang H, Zhang H, Zeng J and Tan Y: ceRNA
network analysis reveals prognostic markers for glioblastoma. Oncol
Lett. 17:5545–5557. 2019.PubMed/NCBI
|
|
5
|
Dragomir M, Mafra ACP, Dias SMG, Vasilescu
C and Calin GA: Using microRNA networks to understand cancer. Int J
Mol Sci. 19:192018. View Article : Google Scholar
|
|
6
|
Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer
A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J,
et al: An extensive microRNA-mediated network of RNA-RNA
interactions regulates established oncogenic pathways in
glioblastoma. Cell. 147:370–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cui L, Li Y, Lv X, Li J, Wang X, Lei Z and
Li X: Expression of MicroRNA-301a and its functional roles in
malignant melanoma. Cell Physiol Biochem. 40:230–244. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peng LN, Shi WT, Feng HR, Wei CY and Yin
QN: Effect of miR-301a/PTEN pathway on the proliferation and
apoptosis of cervical cancer. Innate Immun. 25:217–223. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi W, Gerster K, Alajez NM, Tsang J,
Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, et al:
MicroRNA-301 mediates proliferation and invasion in human breast
cancer. Cancer Res. 71:2926–2937. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia X, Zhang K, Luo G, Cen G, Cao J, Huang
K and Qiu Z: Downregulation of miR-301a-3p sensitizes pancreatic
cancer cells to gemcitabine treatment via PTEN. Am J Transl Res.
9:1886–1895. 2017.PubMed/NCBI
|
|
11
|
Zhang L, Zhang Y, Zhu H, Sun X, Wang X, Wu
P and Xu X: Overexpression of miR-301a-3p promotes colorectal
cancer cell proliferation and metastasis by targeting deleted in
liver cancer-1 and runt-related transcription factor 3. J Cell
Biochem. 120:6078–6089. 2019. View Article : Google Scholar
|
|
12
|
Xia X, Zhang K, Cen G, Jiang T, Cao J,
Huang K, Huang C, Zhao Q and Qiu Z: MicroRNA-301a-3p promotes
pancreatic cancer progression via negative regulation of SMAD4.
Oncotarget. 6:21046–21063. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yue X, Cao D, Lan F, Pan Q, Xia T and Yu
H: MiR-301a is activated by the Wnt/β-catenin pathway and promotes
glioma cell invasion by suppressing SEPT7. Neuro Oncol.
18:1288–1296. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He L, Zhou H, Zeng Z, Yao H, Jiang W and
Qu H: Wnt/β-catenin signaling cascade: A promising target for
glioma therapy. J Cell Physiol. 234:2217–2228. 2019. View Article : Google Scholar
|
|
15
|
Zhang K, Zhang J, Han L, Pu P and Kang C:
Wnt/beta-catenin signaling in glioma. J Neuroimmune Pharmacol.
7:740–749. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nusse R and Clevers H: Wnt/β-catenin
signaling, disease, and emerging therapeutic Modalities. Cell.
169:985–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zebisch M, Xu Y, Krastev C, MacDonald BT,
Chen M, Gilbert RJ, He X and Jones EY: Structural and molecular
basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by
the Wnt agonist R-spondin. Nat Commun. 4:27872013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiao G, Dai C, He Y, Shi J and Xu C:
Effects of miR 106b 3p on cell proliferation and epithelial
mesenchymal transition, and targeting of ZNRF3 in esophageal
squamous cell carcinoma. Int J Mol Med. 43:1817–1829.
2019.PubMed/NCBI
|
|
19
|
Shi J, Jiang X, Yu Z, He G, Ning H, Wu Z,
Cai Y, Yu H and Chen A: ZNRF3 contributes to the growth of lung
carcinoma via inhibiting Wnt/β-catenin pathway and is regulated by
miR-93. Tumour Biol. 37:3051–3057. 2016. View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Sun J, Jia Z, Li B, Zhang A, Wang G, Pu P,
Chen Z, Wang Z and Yang W: MiR-19 regulates the proliferation and
invasion of glioma by RUNX3 via β-catenin/Tcf-4 signaling.
Oncotarget. 8:110785–110796. 2017. View Article : Google Scholar
|
|
22
|
Gebert LFR and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar :
|
|
23
|
Karimi N, Ali Hosseinpour Feizi M,
Safaralizadeh R, Hashemzadeh S, Baradaran B, Shokouhi B and
Teimourian S: Serum overexpression of miR-301a and miR-23a in
patients with colorectal cancer. J Chin Med Assoc. 82:215–220.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zheng JZ, Huang YN, Yao L, Liu YR, Liu S,
Hu X, Liu ZB and Shao ZM: Elevated miR-301a expression indicates a
poor prognosis for breast cancer patients. Sci Rep. 8:22252018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang J, Gusev Y, Aderca I, Mettler TA,
Nagorney DM, Brackett DJ, Roberts LR and Schmittgen TD: Association
of MicroRNA expression in hepatocellular carcinomas with hepatitis
infection, cirrhosis, and patient survival. Clin Cancer Res.
14:419–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner
MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ and Schmittgen
TD: Expression profiling identifies microRNA signature in
pancreatic cancer. Int J Cancer. 120:1046–1054. 2007. View Article : Google Scholar
|
|
27
|
Lan F, Qing Q, Pan Q, Hu M, Yu H and Yue
X: Serum exosomal miR-301a as a potential diagnostic and prognostic
biomarker for human glioma. Cell Oncol (Dordr). 41:25–33. 2018.
View Article : Google Scholar
|
|
28
|
Hao HX, Xie Y, Zhang Y, Charlat O, Oster
E, Avello M, Lei H, Mickanin C, Liu D, Ruffner H, et al: ZNRF3
promotes Wnt receptor turnover in an R-spondin-sensitive manner.
Nature. 485:195–200. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Madan B and Virshup DM: Targeting Wnts at
the source - new mechanisms, new biomarkers, new drugs. Mol Cancer
Ther. 14:1087–1094. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Z, Wang Y, Ren H, Jin Y and Guo Y:
ZNRF3 inhibits the invasion and tumorigenesis in nasopharyngeal
carcinoma cells by inactivating the Wnt/β-catenin pathway. Oncol
Res. 25:571–577. 2017. View Article : Google Scholar
|
|
31
|
Yue X, Lan F and Xia T: Hypoxic glioma
cell-secreted exosomal miR-301a activates Wnt/beta-catenin
signaling and promotes radiation resistance by targeting TCEAL7.
Mol Ther. 27:1939–1949. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li X, Zhong M, Wang J, Wang L, Lin Z, Cao
Z, Huang Z, Zhang F, Li Y, Liu M, et al: miR-301a promotes lung
tumorigenesis by suppressing Runx3. Mol Cancer. 18:992019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun J, Li B, Jia Z, Zhang A, Wang G, Chen
Z, Shang Z, Zhang C, Cui J and Yang W: RUNX3 inhibits glioma
survival and invasion via suppression of the β-catenin/TCF-4
signaling pathway. J Neurooncol. 140:15–26. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu H, Zhang Q, Chen W, Wu T, Liu S, Li X,
Luo B, Zhang T, Yan G, Lu H, et al: MicoRNA-301a promotes
pancreatic cancer invasion and netastasis through the JAK/STAT3
signaling pathway by targeting SOCS5. Carcinogenesis. 41:502–514.
2020. View Article : Google Scholar
|
|
35
|
Lu Z and Li Y, Takwi A, Li B, Zhang J,
Conklin DJ, Young KH, Martin R and Li Y: miR-301a as an NF-κB
activator in pancreatic cancer cells. EMBO J. 30:57–67. 2011.
View Article : Google Scholar
|
|
36
|
Yin J, Chen D, Luo K, Lu M, Gu Y, Zeng S,
Chen X, Song Y, Zhang Z, Zheng G, et al: Cip2a/miR-301a feedback
loop promotes cell proliferation and invasion of triple-negative
breast cancer. J Cancer. 10:5964–5974. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bracken CP, Scott HS and Goodall GJ: A
network-biology perspective of microRNA function and dysfunction in
cancer. Nat Rev Genet. 17:719–732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Besharat ZM, Abballe L, Cicconardi F,
Bhutkar A, Grassi L, Le Pera L, Moretti M, Chinappi M, D'Andrea D,
Mastronuzzi A, et al: Foxm1 controls a pro-stemness microRNA
network in neural stem cells. Sci Rep. 8:35232018. View Article : Google Scholar : PubMed/NCBI
|