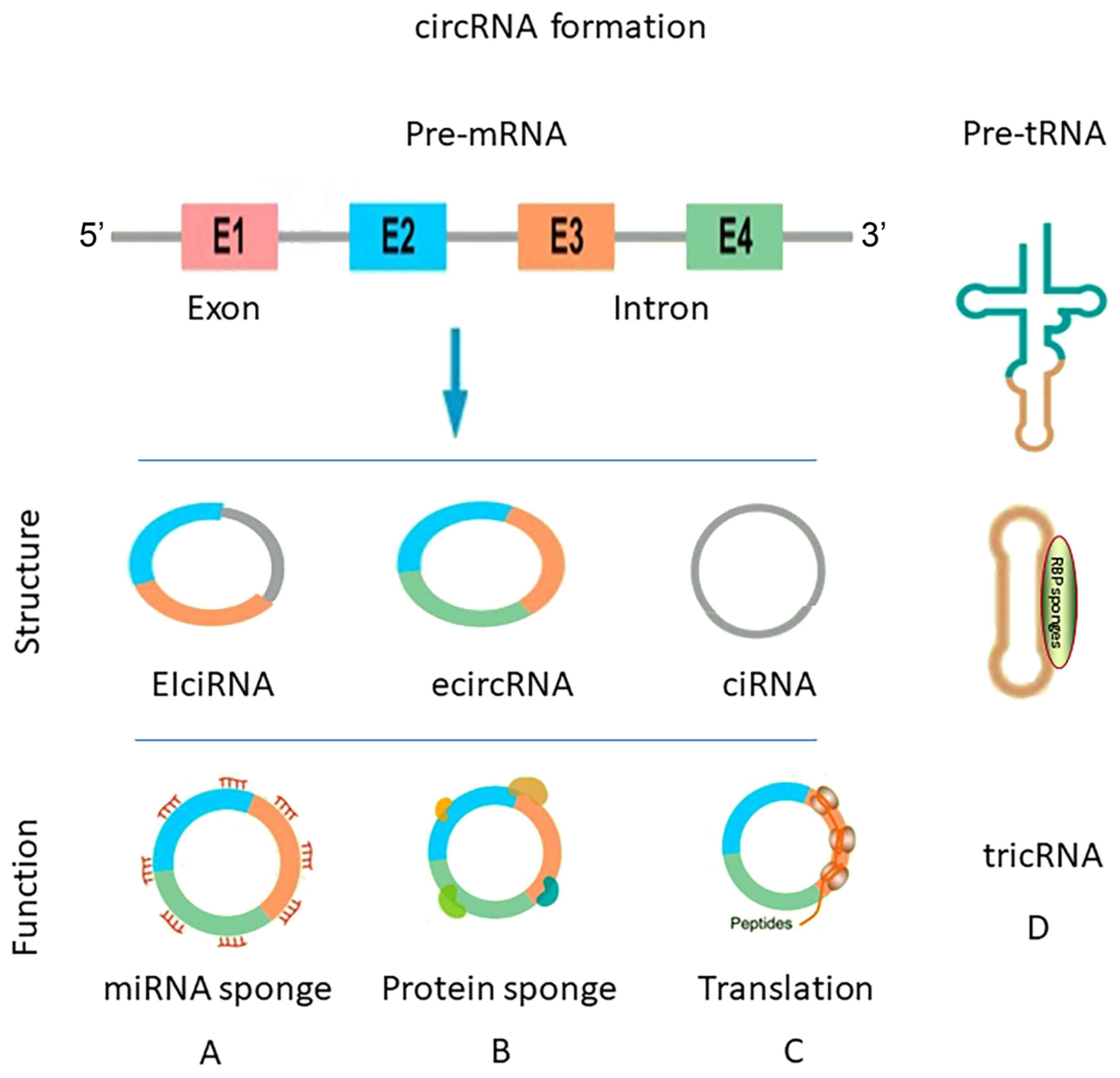
Circular RNA (circRNA) is a type of endogenous
non-coding RNA with diverse biological functions in tumors
(1). circRNA molecules are
involved in the initiation of tumorigenesis and tumor metastasis.
Differential expression of circRNA occurs in diverse stages of
tumor progression (2). circRNA can
sponge microRNA (miRNA/miR) molecules and affect mRNA translation
into proteins (3).
Exosomes contain various cargos, including proteins,
lipids, DNA and RNA. Exosomes mediate intercellular communication
by transferring cargo between cells, including cancer cells. The
exosomal contents are transferred to the recipient cell, inducing
changes in gene expression and alterations to the tumor
microenvironment (TME) (4,5). This process is associated with tumor
metastasis. Moreover, circRNA molecules are abundant and stable in
exosomes (6). Exosomal circRNA has
several extracellular functions and plays critical roles following
exosome uptake by recipient cells (7). Exosomal circRNA may induce
trans-differentiation processes, such as epithelial-to-mesenchymal
transition (EMT), macrophage polarization (8,9) and
changes to the TME (10,11). These processes often impact tumor
metastasis, as remodeling the extracellular microenvironment is a
crucial condition for metastasis. The distant movement of malignant
cells requires changes to the pre-metastatic niche (12). However, the detailed mechanism
underlying the functions of exosomal circRNA in tumor metastasis
are unclear. Thus, deciphering the roles of exosomal circRNA may
provide insight into the mechanism of tumor progression.
In this review, research advances into the
underlying mechanism and clinical application of exosomal circRNA
in tumorigenesis and metastasis are discussed. In particular, the
present review focuses on the relationship between circRNA,
exosomes and the TME. Furthermore, we propose that exosomal circRNA
might indirectly regulate the TME, EMT and macrophage polarization,
which in turn affects tumor progression. In addition, exosomal
circRNA may represent a diagnostic molecular biomarker and clinical
target for cancer treatment.
circRNA has several biological characteristics. It
is predominantly expressed in the cytoplasm and regulates the
expression of target genes by mediating miRNA activity (24). Most circRNA molecules are found at
high levels in extracellular body fluids (liquid biopsy), including
serum, urine, saliva and cerebrospinal fluid (25). They are highly resistant to RNA
exonuclease or RNase R (26).
circRNA may encode a protein under a certain conditions; for
example, in vitro and in vivo evidence suggests that
hsa_circ_0000423 carries an open reading frame that encodes a
functional protein in colon cancer (CC) cells (27).
miRNA are small, non-coding RNA molecules that play
pivotal roles in the regulation of target gene expression by
regulating mRNA degradation or inhibiting translation (28). Previous studies have demonstrated
that miRNA molecules are significantly associated with tumor
growth, metastasis and progression (29,30).
circRNA may also regulate cell signal transduction and RNA
transcription via circRNA-miRNA regulatory networks that control
target genes (31). Thus, circRNA
can represent a switch that controls miRNA. Currently, circRNA is
considered an important factor in the regulation of tumorigenesis
and metastasis. For instance, hsa_circRNA_102958 expression is
significantly increased in colorectal cancer (CRC) tissue and
silencing the expression of this circRNA markedly suppresses CRC
growth, migration and invasion (32). Moreover, hsa_circRNA_102958 acts as
a sponge for miR-585, whereby hsa_circRNA_102958 promotes cell
division cycle 25B expression by inhibiting miR-585 activity in CRC
cells (32). The circRNA
cerebellar degeneration-related protein 1-antisense (CDR1-AS) binds
to miR-7 and acts as its molecular sponge (33). miR-7-5p is the mature form of miR-7
and is closely related to the occurrence and development of tumors
(34). For example, miR-7 inhibits
the ability of gastric cancer cells to divide, migrate and invade,
while promoting their apoptosis (35). circRNA circR-7 acts as a miR-7
sponge that inhibits its biological function (36). Thus, circRNA binding to miRNA
inhibits its functions and since these functions can be associated
with oncogenesis, circRNA could regulate tumor cell
differentiation, growth and metastasis indirectly.
The identification of circRNA functions provided a
novel molecular mechanism for cancer. circRNA exerts several
verified functions. For example, a previous study has demonstrated
that circzinc finger protein 609 (circ-ZNF609), an oncogenic
circRNA, is significantly upregulated in the human glioma cell
lines U87 and U251 (37). In
addition, circ-ZNF292 silencing suppresses tube formation by
inhibiting glioma cell proliferation and cell cycle progression
(38). Based on these findings, it
has been proposed that abnormal miRNA or circRNA expression may
alter cancer growth. Similarly, recent studies have suggested that
circRNA can display tissue-dependent expression in
neurodegenerative diseases (39).
circRNA is differentially expressed in various tumor tissue types,
including breast cancer (40),
hepatocellular carcinoma (HCC) (41,42)
and CRC (32).
Exosomes are nanoscale (30-150 nm) extracellular
vesicles (EVs) of endocytic origin that are shed by most types of
cells and circulate in bodily fluids, such as blood, urine, saliva
and breast milk (43) They contain
a specific cargo of diverse growth factors, proteins, lipids and
nucleic acids, including long non-coding RNA and circRNA, which can
modulate recipient cell behavior and might be used as biomarkers
for diagnosis, such as bladder and breast cancer (44,45).
Different of exosomes can carry these cargos from cell to cell and
mediate intercellular communication. In cancer progression,
exosomal cargos are the link between the disruption of normal
tissues and oncogenesis (46).
Exosomes can facilitate tumor progression by altering their
contents and remodeling the microenvironment, thus accelerating the
progression of oncogenic processes (44).
RNA-seq analysis has revealed that circRNA are
present in exosomes, representing a novel class of RNA species in
exosomes, which might be regulated by the levels of associated
miRNA in cells and result in the transfer of biological cargos into
recipient cells (47). In
addition, a previous study has suggested that the concentration of
circRNA in exosomes is greater in cholangiocarcinoma cells than in
normal cells (48). To date,
>1,000 exosomal circRNA molecules have been identified in human
serum. For example, circkelch domain containing 10 was identified
in a patient with CRC, and it has been demonstrated that this
circRNA could be measured by RNA-seq and further validated
individually with quantitative reverse transcription PCR analysis
in serum from cancer patients (7).
Therefore, exosomal circRNA could be used to screen for tumors
using patient serum, representing a new type of diagnostic
biomarker for cancer (7). Another
study has demonstrated that adipose-derived exosomes mediate the
delivery of circRNA and promote the tumorigenesis of HCC (42).
Exosomal circRNA not only induces tumorigenesis of
HCC by affecting cell signaling, but also facilitates HCC
metastasis. Su et al (49)
reported that circRNA CDR1-AS was directly transferred from HCC
cells into the surrounding normal cells via exosomes, thereby
mediating the proliferative and migratory abilities of surrounding
cells. Moreover, it was observed that CDR1-AS could sponge miR-1270
and facilitate the expression of α-fetoprotein, a specific
biomarker of HCC (49). Another
study by Shen et al (36)
has also provided insight into the association between exosomal
circRNA and tumor metastasis. Therefore, exosomal circRNA may
represent a future therapeutic target for tumor metastasis in
clinical applications (36).
Tumor metastasis is a complex process that involves
genetic mutations and TME changes. Evidence shows that the TME
facilitates tumor initiation and metastasis. Indeed, the TME
comprises multiple categories of substances, such as T cells,
exosomes, tumor-associated macrophages (TAMs), cytokines,
fibroblasts, endothelial cells, mesenchymal stem cells (MSCs) and
neutrophils, which exert a great influence on cancer growth and
generate a pre-metastatic niche favorable to tumor progression
(50-52). The pre-metastatic niche is a
pre-formed microenvironment made possible by exosomes secreted by
the primary tumor site before widespread metastasis (53).
Exosomes are released into the TME and bloodstream,
subsequently acting as messengers that impact distant tissues
(54). Exosomes release various
substances into the extracellular milieu to accelerate tumor cell
migration. For example, cancer stem cell exosomes might target
cancer cells and organs, leading to higher capacity of clear cell
renal cell carcinoma to the metastasize to the lungs (55). Matrix metalloproteinases (MMPs) can
be transferred by exosomes to facilitate cancer cell invasion by
degrading the extracellular stroma (56). Stromal infiltration alters the TME
and facilitates tumor metastasis (57). As a constituent of exosomes,
exosomal circRNA molecules are directly and indirectly involved in
the crosstalk between the TME and tumor cells. The diverse
intercellular exchange of exosomal circRNA might drive tumor
metastasis.
EVs are circular membrane fragments released from
the endosomal compartment as exosomes or shed from the surfaces of
the cell membranes. They comprise a lipid bilayer membrane, forming
a vesicle containing several types of biomolecules, including
proteins, lipids, polysaccharides and nucleic acids. These membrane
fragments can be transferred to a recipient cell. When the
recipient cell absorbs the biomolecules, changes to the function
and gene expression of the recipient cells are elicited (58,59).
Non-coding RNA derived from exosomes might stimulate
the clearance function of macrophages. TAMs are key cells that
create an immunosuppressive TME by inhibiting immune checkpoint
proteins to release T cells (60).
Macrophages can display very different functions depending on the
nature of the microenvironment, such as cancer and inflammatory
environments. Activated macrophages are classified into M1
(classically-activated macrophages) and M2 (alternatively-activated
macrophages) phenotypes (61).
Activated M1 macrophages exhibit anti-tumor activity and elicit
anti-tumor adaptive immunity in the early stages of carcinogenesis.
Activated M2 macrophages tend to display an immunosuppressive
phenotype, suppress tissue repair and promote tumor progression
(62). However, M2 macrophage
differentiation facilitates tumor cell immune escape; however, the
exact ratio of M2/M1 macrophages in response to tumors is unclear
(63).
TAMs play multi-functional roles in tumor
progression, including cancer initiation and promotion, immune
regulation, metastasis and angiogenesis (60). Activated M2 macrophages accumulate
in the TME and facilitate tumor cell metastasis. Increased levels
of exosomes may modulate the expression of proteins and stimulates
M1 macrophages, which can result in the removal of cancer cells.
However, further research is needed to test this hypothesis.
Exosomal circRNA molecules affect target genes in
order to facilitate biological functions through communication
between different cell types. This characteristic distinguishes
exosomal circRNA from classical endocrine circulating RNA. In a
recent study, exosomal circRNA could be transported throughout the
whole body, thus acting as critical mediators of intercellular
communication and inducing neoplasm metastasis (64). A previous study has suggested that
the level of exosomal circRNA might reflect the expression levels
of these circRNA in cells and the physiological activity of the
cells to a certain degree (65).
CDR1-AS, also known as CiRS-7, a circRNA sponge for miR-7, was
confirmed to have ~70 conserved binding-sites for miR-7 (66). CDR1-AS expression levels are
increased at metastasis sites compared with primary sites (65). Zou et al (67) have suggested that CDR1-AS plays a
specific role in immune and stromal cell infiltration in tumor
tissue. They observed that high CDR1-AS expression was associated
with a higher proportion of M2 macrophages. Moreover, CDR1-AS
expression correlated negatively with the number of CD8+
T cells, activated natural killer cells, monocytes and neutrophils
and correlated positively with the M2/M1 macrophage ratio.
EMT is essential in tumor metastasis. During EMT,
epithelial cells lose their polarity and obtain invasive properties
to become MSCs (69). Exosomal
circRNA is closely associated with EMT in tumors (70). Chen et al (71) have demonstrated that exosomal
circ-protein arginine methyltransferase 5 (circ-PRMT5) was
upregulated in patients with urinary carcinoma of the bladder (UCB)
and might predict metastatic progress. Indeed, upregulated exosomal
circ-PRMT5 was positively associated with advanced clinical stage
and poor survival. In addition, circ-PRMT5 also promoted EMT in
UCB.
Other studies have also indicated that exosomal
circRNA might induce EMT. In a mouse model, Zhang et al
(72) investigated the role of
exosomal circ-nuclear receptor interacting protein 1 (NRIP1) in
distant metastasis via tail vein injection of gastric cancer cells
co-cultured with exosomes. This study confirmed that exosomal
circ-NRIP1 could promote EMT and metastasis in vivo
(72). However, in another study,
the opposite observation was made (73).
CDR1-AS1 binds to miR-7, which inhibits insulin like
growth factor 1 receptor expression, thereby increasing the
expression of E-cadherin to partially reverse EMT, subsequently
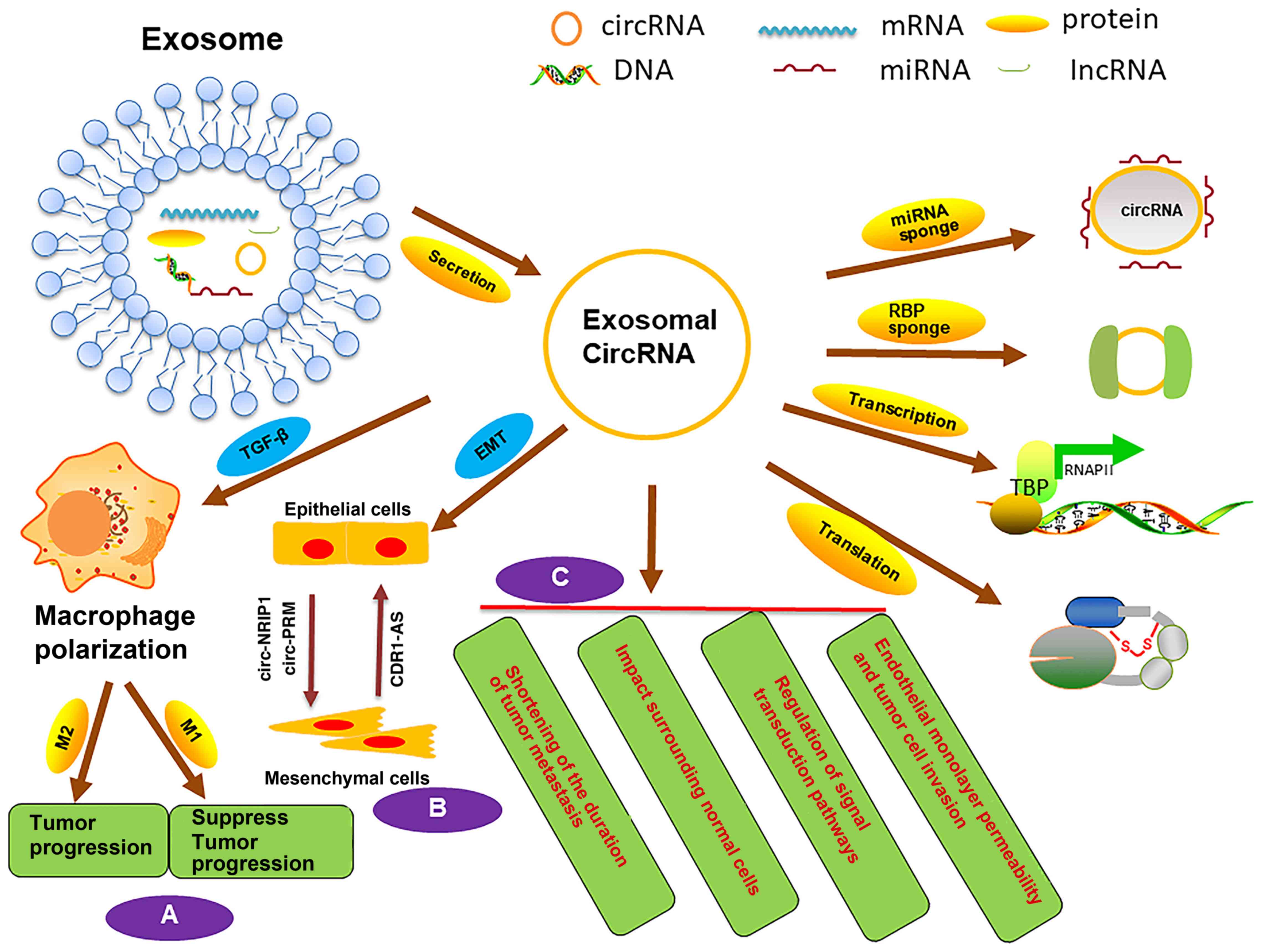
inhibiting tumor cell metastasis (Fig.
2B) (74).
MSCs may migrate to tumor sites and secrete a
variety of factors, such as VEGF and platelet-derived growth factor
(75). Exosomes exert number of
functions that induce and support regenerative processes in
necrotic tissue (76). MSC-derived
exosomes also have antitumor effects. Exosomes derived from
placental MSCs selectively inhibit the growth of prostate cancer
cells through numerous molecular species within their cargo, such
as mRNA, miRNA, lipids and proteins (77). MSC-derived exosomes transport mRNA
and non-coding RNA to target cells and induce endothelial cell
proliferation (78). For example,
the transfer of miR-143 from MSC-derived exosomes to recipient
cells can decrease the migration of osteosarcoma cells in
vitro (79). Moreover, tumor
cells can also reprogram surrounding MSCs to support the growth of
tumors via inter-cellular communication, especially by releasing
EVs. This long-term 'education' by tumor cells favors tumorigenesis
(80). MSCs may secrete exosomes
to promote the migration and invasion of breast cancer cells
(81).
circRNA controls MSC identity and differentiation by
sponging miRNA. For example, circforkhead box P1 (circ-FOXP1) is
also enriched in MSCs. Silencing circ-FOXP1 expression markedly
impairs MSC differentiation, Indeed, a previous study demonstrated
that circ-FOXP1 regulates MSC molecular networks through the Wnt
pathway (82). Tumor-derived
exosomes also regulate MSCs in the distant metastatic
microenvironment to promote metastasis (83). However, further experiments are
required to verify the regulatory relationship between MSCs and
exosomal circRNA.
Tumor cells affect surrounding cells through direct
contact, paracrine secretion and autocrine secretion (84). Intercellular communication can
occur via exosomes, which has an important role in tumor metastasis
and invasion (85). Exosomes
containing non-coding RNA molecules are detectable in body fluids,
particularly the blood, and are frequently released by tumor cells,
implying that exosomes can act as messengers between cancer cells
and immune cells to promote cancer cell escape from immune
surveillance, thus contributing to tumor formation (86). As aforementioned, a previous study
has indicated that circRNA is abundant and stable in exosomes and
can continue to exert its role following uptake by recipient cells
(7).
Metastasis is a phenomenon that can occur in the
majority of cancer types. Tumor cells and normal cells can
communicate with each other through exosomes, which secrete
substances that affect tumor metastasis (87). The first metastasis mechanism
occurs when exosomal circRNA induce the progression of a tumor by
regulating cell signal transduction pathways (88). In ovarian cancer cells,
circ-Wolf-Hirschhorn syndrome candidate 1 binds to miR-145 and
miR-1182, which upregulates the expression of downstream targets
mucin 1 and telomerase reverse transcriptase, thus promoting
proliferation and invasion (89).
It can also facilitate peritoneal dissemination via exosomes
(89). Li et al (90) also found that high expression
levels of exosomal circ-phosphodiesterase 8A (circ-PDE8A) in
pancreatic ductal adenocarcinoma tissue was positively associated
with invasion. Further research revealed that circ-PED8A promoted
tumor cell growth by upregulating the MET proto-oncogene (91). Exosomal circRNA also regulates the
progress of tumor metastasis via cell signal transduction.
Moreover, circ-protein phosphatase 1 regulatory subunit 12A
(circ-PPP1R12A) encodes a conserved 73-amino-acid peptide,
PPP1R12A-C, which promotes the proliferation, migration and
invasion of CC cells by activating the Hippo/YES-activating protein
signaling pathway (27). Another
study suggested that exosomal circ-NRIP1 promoted energy production
by activating the protein kinase B/AKT/mTOR signaling pathway to
facilitate gastric cancer tumor growth (72). Moreover, exosomal circ-NRIP1
promoted gastric cancer metastasis via EMT (72).
The second mechanism of tumor metastasis occurs
through endothelial monolayer permeability and tumor cell invasion.
In pancreatic cancer cells, exosomal circisoleucyl-tRNA synthetase
1 (circ-IARS) increases the expression levels of RhoA and F-actin
and downregulates zona occludens-1, which increases the
permeability of endothelial monolayer cells (92). Animal experiments have confirmed
that circ-IARS can increase the number of tumor cells that can pass
through the endothelial monolayer, which promotes metastasis
(92).
Other evidence has suggested that exosomal circRNA
modulates tumor cell migration by sponging miRNA, thus affecting
metastasis. Exosomal circRNA may shorten the duration of tumor
metastasis by sponging miRNA and regulating mRNA expression. For
example, circ-PTGR1 is detectable in three HCC cell lines, namely
the non-metastatic HepG2, the low-metastatic 97L and the highly
metastatic LM3 cell lines (91).
Exosomal circ-prostaglandin reductase 1 (circ-PTGR1) derived from
highly metastatic HCC cells can promote metastasis in cells that
normally have low metastatic potential (93). In particular, LM3 exosome-derived
circ-PTGR1 promotes the progression of HepG2 and 97L cells in
vitro and in vivo, which results from circ-PTGR1
competing with MET to target miR-449a. Thus, circ-PTGR1 can promote
migration and metastasis in HCC through the circ-PTGR1/miR-449a/MET
pathway (93).
In tumor tissues, exosomal circRNA mediates the
progression of cancer by modulating surrounding normal cells to
accelerate invasion and metastasis. Su et al (49) found that exosomal circRNA CDR1-AS
from HCC cells accelerated the proliferative and migratory
abilities of surrounding normal cells. Exosomal circRNA CDR1-AS
serves as a competing endogenous RNA (ceRNA) to promote the
progression of HCC (94). It is
directly transferred from HCC cells to surrounding normal cells via
exosomes to further mediate its biological functions (49). In nasopharyngeal carcinoma (NPC)
tissues, CDR1-AS upregulates E2F transcription factor 3 expression
by binding to miR-7-5p, which promotes the growth of NPC cells.
Moreover, CDR1-AS could promote NPC progression by negatively
regulating miR-7-5p in a nasopharyngeal carcinoma xenograft tumor
model (95).
In prostate cancer, exosomal circRNA acts as miRNA
sponge to induce EMT and facilitate tumor cell migration and
invasion. Li et al (96)
found that exosome circ-0044516 was significantly upregulated in
patients with prostate cancer and in cell lines. Exosomal
circ-0044516 downregulates miR-29a-3p expression, which inhibits
the progression of prostate cancer cells (96). High expression levels of exosomal
circ-0000284 enhance the migration, invasion and proliferation
abilities of cholangiocarcinoma cells by acting as a ceRNA that
binds to miR-637, further regulating lymphocyte antigen 6 family
member E expression in vivo and in vitro (97). In bladder cancer, circhomeodomain
interacting protein kinase 33 suppresses bladder cell migration,
invasion and angiogenesis by sponging miR-558, which subsequently
inhibits heparanase, MMP-9 and VEGF expression in vitro (98) (Fig.
2C).
Currently, clinical therapeutic progress includes
improvements in early diagnosis, surgery, radiotherapy,
chemotherapy and other types of therapeutic methods. Early
effective diagnosis biomarkers are insufficient. Final diagnosis is
often established by performing a biopsy, which may delay early
diagnosis of the majority cancer patients. Biopsies can be painful,
invasive for the patients and can lead to associated complications.
Nevertheless, non-invasive early diagnostic biomarkers are lacking
for clinical use and many types of carcinoma lack an authoritative
indicator. Thus, exosomal circRNA could be developed as
authoritative indicators. Interestingly, a previous study has shown
circRNA are abundant and stable in exosomes, which can be detected
in the circulation and urine (99). The most effective approach is
RNA-seq analyses detection of circRNA molecules in exosomes
(100,101).
There is an urgent need to identify novel diagnostic
biomarkers and develop more efficient therapeutic molecular targets
in cancer. A recent study has revealed that the exosomes secreted
by tumor cells are more abundant compared with those from normal
cells (102). In addition, the
contents of exosomes are different under normal physiological
conditions compared with pathological conditions, even for the same
tissue or body fluid (103).
Exosomal circRNA is resistant to degradation and its secretion into
the extracellular environment could be exploited in many biological
applications; for example, as novel diagnostic biomarkers. For
example, exosomal circRNA has been proposed as a potential
diagnostic biomarker of idiopathic membranous nephropathy (104). A previous study has shown that
both esophageal epithelial cells and esopha-geal squamous-cell
carcinoma (ESCC) cells secrete exosomal circRNA and suggested that
circRNA from patient plasma could be used for early diagnosis
(105). hsa-circ-0001946 and
hsa-circ-0043603 are found in exosomes of cell-conditioned culture
conditioned media and may be potential diagnostic biomarkers for
ESCC (106).
High-throughput sequencing analysis has demonstrated
that the expression levels of three types of circRNA
(hsa-circ-007293, hsa-circ-031752 and hsa-circ-020135) are
upregulated in the serum of patients with papillary thyroid
carcinoma. Thus, these exosome circRNA might represent potential
diagnostic molecular biomarkers (107).
In cholangiocarcinoma, exosome-transmitted
circ-0000284 stimulates the migration and proliferation of
surrounding normal cells and promotes the progression of the tumor;
thus, exosomal circ-0000284 may serve as a potential metastatic
diagnostic biomarker (97).
Exosomal circ-PTGR1 is abundant and aberrantly expressed in
malignant cells of patients with metastatic HCC. Thus, circ-PTGR1
could act as a prognostic biomarker and therapeutic target
(93). Circulating exosomal
hsa-circ-0004771 is significantly upregulated in patients with CRC
and may serve as a novel potential diagnostic biomarker of CRC
(108). A previous study also
suggested that hsa_circ_0001492 and hsa_circ_0001346 might be novel
potential early diagnosis candidate markers in lung adenocarcinoma
(109). Altogether, these studies
indicated the possibility of using circRNA as tumor diagnostic
markers. However, further research remains to be done to in
different diseases and tumor subtypes.
Drug resistance (chemotherapy insensitivity) is
common during cancer therapy. Exosomal miRNA associated with drug
resistance have been revealed in certain types of cancer, such as
ovarian cancer (110). However,
the detailed molecular mechanism underlying the relationship
between exosomal circRNA and drug resistance is unclear and should
be investigated to develop new target drugs. A study has
highlighted epigenetic changes, such as resistance to cytotoxic
drugs and angiogenic properties conferred by exosomes in the
recipient cells through this 'cargo delivery' process (111). Zhao et al (112) have proposed that exosomal circRNA
affects drug sensitivity. CDR1-AS is 1,500 nucleotides in length
and is transcribed in the antisense orientation with respect to the
CDR1 gene. Upregulation of exosomal CDR1-AS could increase the
cisplatin sensitivity of ovarian cancer cells (112). CDR1-AS functions as a molecular
sponge for miR-1270, which weakens the inhibitory effect of the
miRNA on the downstream target gene suppressor of cancer cell
invasion (113). CDR1-AS might
regulate the sensitivity of ovarian cancer cells to cisplatin and
the progression of ovarian cancer (112,114). Modulating the release of exosomal
circRNA might be a potential therapeutic strategy to improve drug
sensitivity in tumors. However, further trials are required to test
this hypothesis.
Exosomal circRNA has stable biological function and
structure. Importantly, circRNA is resistant to degradation by RNA
exonuclease or RNase R and easily obtained from body fluid or serum
(115). Recently, exosomal
circRNA molecules have been proposed as novel cancer diagnostic
biomarkers that can be extracted non-invasively. They have been
used to study cancer progression and to evaluate prognosis. Several
studies have demonstrated that exosomes are closely related to
tumor development. The function of exosomes as biological delivery
vehicles is of considerable interest in cancer research because
modulating the TME using variety of RNA species may help increase
the sensitivity of chemotherapeutics. Exosomal circRNA might
promote the generation of the TME. Upregulation of exosomal circRNA
expression can aggravate tumor progression. By contrast,
downregulation of exosomal circRNA expression might inhibit tumor
metastasis. The exosomal circRNA functions that impact on tumor
progression can be divided into three pathways: i) sponging miRNA
and regulating its function; ii) regulating transcription and
translation; and iii) interacting with proteins to modulate gene
expression. Thus, an improved understanding of exosome circRNA
characteristics and functional roles in tumor progression is
required. Although exosomal circRNA are known to display various
functions, controlled studies in the majority of disease are
lacking. In conclusion, it is essential to understand the detailed
molecular mechanisms underlying the functions of exosomal circRNA,
as this may lead to the development of novel diagnostic tools and
treatment targets for tumors. Thus, further investigation of
exosomal circRNA candidates and tumor metastasis may contribute to
the control of cancer.
This work was supported by The Department of Science
and Technology of Liaoning province (grant no. 2017225038) and 345
Talent Project funding from Sheng Jing Hospital of China Medical
University (grant no. 2019345).
Not applicable.
XW wrote and edited the manuscript. BY was involved
in acquisition of the data and provided direction and guidance
throughout the preparation of this manuscript. XM generated the
figures and revised the manuscript. YS, ZD and PW conceived the
review and revised the manuscript. All authors read and approved
the final manuscript. YS and ZD confirm the authenticity of the
data.
Not applicable.
Not applicable.
Not applicable.
|
1
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar :
|
|
2
|
Lin J, Zhang Y, Zeng X, Xue C and Lin X:
CircRNA CircRIMS Acts as a MicroRNA sponge to promote gastric
cancer metastasis. ACS Omega. 5:23237–23246. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang Y, Mo Y, Gong Z, Yang X, Yang M,
Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, et al: Circular RNAs in
human cancer. Mol Cancer. 16:252017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Martins VR, Dias MS and Hainaut P:
Tumor-cell-derived microvesicles as carriers of molecular
information in cancer. Curr Opin Oncol. 25:66–75. 2013. View Article : Google Scholar
|
|
5
|
Whiteside TL: Tumor-derived exosomes and
their role in cancer progression. Adv Clin Chem. 74:103–141. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
7
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Funes SC, Rios M, Escobar-Vera J and
Kalergis AM: Implications of macrophage polarization in
autoimmunity. Immunology. 154:186–195. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mantovani A, Sozzani S, Locati M, Allavena
P and Sica A: Macrophage polarization: Tumor-associated macrophages
as a paradigm for polarized M2 mononuclear phagocytes. Trends
Immunol. 23:549–555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang JJ, Lei KF and Han F: Tumor
microenvironment: Recent advances in various cancer treatments. Eur
Rev Med Pharmacol Sci. 22:3855–3864. 2018.PubMed/NCBI
|
|
11
|
Sun Y: Tumor microenvironment and cancer
therapy resistance. Cancer Lett. 380:205–215. 2016. View Article : Google Scholar
|
|
12
|
Xie F, Zhou X, Fang M, Li H, Su P, Tu Y,
Zhang L and Zhou F: Extracellular vesicles in cancer immune
microenvironment and cancer immunotherapy. Adv Sci (Weinh).
6:19017792019. View Article : Google Scholar
|
|
13
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J, Mohammed-Elsabagh M, Paczkowski F
and Li Y: Circular nucleic acids: Discovery, functions and
applications. Chembiochem. 21:1547–1566. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Patop IL, Wüst S and Kadener S: Past,
present, and future of circRNAs. EMBO J. 38:e1008362019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meng X, Chen Q, Zhang P and Chen M:
CircPro: An integrated tool for the identification of circRNAs with
protein-coding potential. Bioinformatics. 33:3314–3316. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao X, Cai Y and Xu J: Circular RNAs:
Biogenesis, mechanism, and function in human cancers. Int J Mol
Sci. 20:39262019. View Article : Google Scholar :
|
|
20
|
Wilusz JE: A 360° view of circular RNAs:
From biogenesis to functions. Wiley interdisciplinary reviews. RNA.
9:e14782018.
|
|
21
|
Chen X, Yang T, Wang W, Xi W, Zhang T, Li
Q, Yang A and Wang T: Circular RNAs in immune responses and immune
diseases. Theranostics. 9:588–607. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Das A, Rout PK, Gorospe M and Panda AC:
Rolling Circle cDNA synthesis uncovers circular RNA splice
variants. Int J Mol Sci. 20:39882019. View Article : Google Scholar :
|
|
24
|
Chan JJ and Tay Y: Noncoding RNA:RNA
Regulatory networks in cancer. Int J Mol Sci. 19:13102018.
View Article : Google Scholar :
|
|
25
|
Pardini B, Sabo AA, Birolo G and Calin GA:
Noncoding RNAs in extracellular fluids as cancer biomarkers: The
New Frontier of Liquid Biopsies. Cancers. 11:11702019. View Article : Google Scholar :
|
|
26
|
Liu K, Zhang Q, Pan F, Wang XD, Wenjing H
and Tong H: Expression of circular RNAs in gynecological tumors: A
systematic review. Medicine. 98:e157362019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z,
Xu B, Wu C, Zhou Q, Hu W, Wu C and Jiang J: A novel protein encoded
by a circular RNA circPPP1R12A promotes tumor pathogenesis and
metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer.
18:472019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Correia de Sousa M, Gjorgjieva M, Dolicka
D, Sobolewski C and Foti M: Deciphering miRNAs' Action through
miRNA Editing. Int J Mol Sci. 20:62492019. View Article : Google Scholar
|
|
29
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol. 9:4022018. View Article : Google Scholar
|
|
30
|
Tan W, Liu B, Qu S, Liang G, Luo W and
Gong C: MicroRNAs and cancer: Key paradigms in molecular therapy.
Oncol Lett. 15:2735–2742. 2018.PubMed/NCBI
|
|
31
|
Xue D, Wang H, Chen Y, Shen D, Lu J, Wang
M, Zebibula A, Xu L, Wu H, Li G and Xia L: Circ-AKT3 inhibits clear
cell renal cell carcinoma metastasis via altering
miR-296-3p/E-cadherin signals. Mol Cancer. 18:1512019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li R, Wu B, Xia J, Ye L and Yang X:
Circular RNA hsa_ circRNA_102958 promotes tumorigenesis of
colorectal cancer via miR-585/CDC25B axis. Cancer Manag Res.
11:6887–6893. 2019. View Article : Google Scholar :
|
|
33
|
Xu H, Guo S, Li W and Yu P: The circular
RNA Cdr1as, via miR-7 and its targets, regulates insulin
transcription and secretion in islet cells. Sci Rep. 5:124532015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu W, Wang Y, Zhang D, Yu X and Leng X:
MiR-75p functions as a tumor suppressor by targeting SOX18 in
pancreatic ductal adenocarcinoma. Biochem Biophys Res Commun.
497:963–970. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin J, Liu Z, Liao S, Li E, Wu X and Zeng
W: Elevated microRNA-7 inhibits proliferation and tumor
angiogenesis and promotes apoptosis of gastric cancer cells via
repression of Raf-1. Cell Cycle. 19:2496–2508. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shen B, Wang Z, Li Z, Song H and Ding X:
Circular RNAs: An emerging landscape in tumor metastasis. Am J
Cancer Res. 9:630–643. 2019.PubMed/NCBI
|
|
37
|
Tong H, Zhao K, Wang J, Xu H and Xiao J:
CircZNF609/miR-134-5p/BTG-2 axis regulates proliferation and
migration of glioma cell. J Pharm Pharmacol. 72:68–75. 2020.
View Article : Google Scholar
|
|
38
|
Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu
C, Li G and Zhu Y: Silencing of cZNF292 circular RNA suppresses
human glioma tube formation via the Wnt/β-catenin signaling
pathway. Oncotarget. 7:63449–63455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Z, Chen Z, Hu G and Jiang Y: Roles of
circular RNA in breast cancer: Present and future. Am J Transl Res.
11:3945–3954. 2019.PubMed/NCBI
|
|
41
|
Sun S, Wang W, Luo X, Li Y, Liu B and Li
X, Zhang B, Han S and Li X: Circular RNA circ-ADD3 inhibits
hepatocellular carcinoma metastasis through facilitating EZH2
degradation via CDK1-mediated ubiquitination. Am J Cancer Res.
9:1695–1707. 2019.PubMed/NCBI
|
|
42
|
Liu Z, Yu Y, Huang Z, Kong Y, Hu X, Xiao
W, Quan J and Fan X: CircRNA-5692 inhibits the progression of
hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP
expression. Cell Death Dis. 10:9002019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Boriachek K, Islam MN, Möller A, Salomon
C, Nguyen NT, Hossain MSA, Yamauchi Y and Shiddiky MJA: Biological
functions and current advances in isolation and detection
strategies for exosome nanovesicles. Small. 14:17021532018.
View Article : Google Scholar
|
|
44
|
Braicu C, Tomuleasa C, Monroig P, Cucuianu
A, Berindan-Neagoe I and Calin GA: Exosomes as divine messengers:
Are they the Hermes of modern molecular oncology? Cell Death
Differ. 22:34–45. 2015. View Article : Google Scholar
|
|
45
|
Pant S, Hilton H and Burczynski ME: The
multifaceted exosome: Biogenesis, role in normal and aberrant
cellular function, and frontiers for pharmacological and biomarker
opportunities. Biochem Pharmacol. 83:1484–1494. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ngalame NNO, Luz AL, Makia N and Tokar EJ:
Arsenic alters exosome quantity and cargo to mediate stem cell
recruitment into a cancer stem cell-like phenotype. Toxicol Sci.
165:40–49. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shi X, Wang B, Feng X, Xu Y, Lu K and Sun
M: circRNAs and exosomes: A mysterious frontier for human cancer.
Mol Ther Nucleic Acids. 19:384–392. 2020. View Article : Google Scholar :
|
|
48
|
Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y,
Zheng Q, Li Y, Wang P, He X and Huang S: exoRBase: A database of
circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids
Res. 46:D106–D112. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A
and Qi F: CircRNA Cdr1as functions as a competitive endogenous RNA
to promote hepatocellular carcinoma progression. Aging (Albany NY).
11:8182–8203. 2019.
|
|
50
|
Binnewies M, Roberts EW, Kersten K, Chan
V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI,
Ostrand-Rosenberg S, Hedrick CC, et al: Understanding the tumor
immune microenvironment (TIME) for effective therapy. Nat Med.
24:541–550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ridge SM, Sullivan FJ and Glynn SA:
Mesenchymal stem cells: Key players in cancer progression. Mol
Cancer. 16:312017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kalluri R: The biology and function of
fibroblasts in cancer. Nat Rev Cancer. 16:582–598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li K, Chen Y, Li A, Tan C and Liu X:
Exosomes play roles in sequential processes of tumor metastasis.
Int J Cancer. 144:1486–1495. 2019. View Article : Google Scholar
|
|
54
|
Matei I, Kim HS and Lyden D: Unshielding
exosomal RNA unleashes tumor growth and metastasis. Cell.
170:223–225. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang L, Yang G, Zhao D, Wang J, Bai Y,
Peng Q, Wang H, Fang R, Chen G, Wang Z, et al: CD103-positive CSC
exosome promotes EMT of clear cell renal cell carcinoma: Role of
remote MiR-19b-3p. Mol Cancer. 18:862019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hakulinen J, Sankkila L, Sugiyama N, Lehti
K and Keski-Oja J: Secretion of active membrane type 1 matrix
metalloproteinase (MMP-14) into extracellular space in
microvesicular exosomes. J Cell Biochem. 105:1211–1218. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Turley SJ, Cremasco V and Astarita JL:
Immunological hall-marks of stromal cells in the tumour
microenvironment. Nat Rev Immunol. 15:669–682. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lai CP, Kim EY, Badr CE, Weissleder R,
Mempel TR, Tannous BA and Breakefield XO: Visualization and
tracking of tumour extracellular vesicle delivery and RNA
translation using multiplexed reporters. Nat Commun. 6:70292015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yoon YJ, Kim OY and Gho YS: Extracellular
vesicles as emerging intercellular communicasomes. BMB Rep.
47:531–539. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lin Y, Xu J and Lan H: Tumor-associated
macrophages in tumor metastasis: Biological roles and clinical
therapeutic applications. J Hematol Oncol. 12:762019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Biswas SK and Mantovani A: Macrophage
plasticity and inter-action with lymphocyte subsets: Cancer as a
paradigm. Nat Immunol. 11:889–896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Najafi M, Hashemi Goradel N, Farhood B,
Salehi E, Nashtaei MS, Khanlarkhani N, Khezri Z, Majidpoor J,
Abouzaripour M, Habibi M, et al: Macrophage polarity in cancer: A
review. J Cell Biochem. 120:2756–2765. 2019. View Article : Google Scholar
|
|
63
|
Allavena P, Sica A, Garlanda C and
Mantovani A: The Yin-Yang of tumor-associated macrophages in
neoplastic progression and immune surveillance. Immunol Rev.
222:155–161. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Preußer C, Hung LH, Schneider T, Schreiner
S, Hardt M, Moebus A, Santoso S and Bindereif A: Selective release
of circRNAs in platelet-derived extracellular vesicles. J Extracell
Vesicles. 7:14244732018. View Article : Google Scholar
|
|
65
|
Hou J, Jiang W, Zhu L, Zhong S, Zhang H,
Li J, Zhou S, Yang S, He Y, Wang D, et al: Circular RNAs and
exosomes in cancer: A mysterious connection. Clin Transl Oncol.
20:1109–1116. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sekar S, Cuyugan L, Adkins J, Geiger P and
Liang WS: Circular RNA expression and regulatory network prediction
in posterior cingulate astrocytes in elderly subjects. BMC
Genomics. 19:3402018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zou Y, Zheng S, Deng X, Yang A and Xie X,
Tang H and Xie X: The role of circular RNA CDR1as/ciRS-7 in
regulating tumor micro-environment: A pan-cancer analysis.
Biomolecules. 9:4292019. View Article : Google Scholar
|
|
68
|
Pickup M, Novitskiy S and Moses HL: The
roles of TGFβ in the tumour microenvironment. Nature Revi Cancer.
13:788–799. 2013. View Article : Google Scholar
|
|
69
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Shang BQ, Li ML, Quan HY, Hou PF, Li ZW,
Chu SF, Zheng JN and Bai J: Functional roles of circular RNAs
during epithelial-to-mesenchymal transition. Mol Cancer.
18:1382019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chen X, Chen RX, Wei WS, Li YH, Feng ZH,
Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al: PRMT5 circular RNA
promotes metastasis of urothelial carcinoma of the bladder through
sponging miR-30c to induce epithelial-mesenchymal transition. Clin
Cancer Res. 24:6319–6330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ma C, Shi T, Qu Z, Zhang A, Wu Z, Zhao H,
Zhao H and Chen H: CircRNA_ACAP2 suppresses EMT in head and neck
squamous cell carcinoma by targeting the miR-21-5p/STAT3 signaling
axis. Front Oncol. 10:5836822020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhao X, Dou W, He L, Liang S, Tie J, Liu
C, Li T, Lu Y, Mo P, Shi Y, et al: MicroRNA-7 functions as an
anti-metastatic microRNA in gastric cancer by targeting
insulin-like growth factor-1 receptor. Oncogene. 32:1363–1372.
2013. View Article : Google Scholar
|
|
75
|
Ball SG, Shuttleworth CA and Kielty CM:
Mesenchymal stem cells and neovascularization: Role of
platelet-derived growth factor receptors. J Cell Mol Med.
11:1012–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Altaner C, Altanerova U and Jakubechova J:
Intracellular acting tumor cell-targeted chemotherapy by
MSC-suicide gene exosomes. Oncotarget. 10:5573–5575. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Brossa A, Fonsato V and Bussolati B:
Anti-tumor activity of stem cell-derived extracellular vesicles.
Oncotarget. 10:1872–1873. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ferguson SW, Wang J, Lee CJ, Liu M,
Neelamegham S, Canty JM and Nguyen J: The microRNA regulatory
landscape of MSC-derived exosomes: A systems view. Sci Rep.
8:14192018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Shimbo K, Miyaki S, Ishitobi H, Kato Y,
Kubo T, Shimose S and Ochi M: Exosome-formed synthetic microRNA-143
is transferred to osteosarcoma cells and inhibits their migration.
Biochem Biophys Res Commun. 445:381–387. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li X, Wang S, Zhu R, Li H, Han Q and Zhao
RC: Lung tumor exosomes induce a pro-inflammatory phenotype in
mesenchymal stem cells via NFκB-TLR signaling pathway. J Hematol
Oncol. 9:422016. View Article : Google Scholar
|
|
81
|
Menck K, Klemm F, Gross JC, Pukrop T,
Wenzel D and Binder C: Induction and transport of Wnt 5a during
macrophage-induced malignant invasion is mediated by two types of
extracellular vesicles. Oncotarget. 4:2057–2066. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Cherubini A, Barilani M, Rossi RL, Jalal
MMK, Rusconi F, Buono G, Ragni E, Cantarella G, Simpson HARW,
Péault B and Lazzari L: FOXP1 circular RNA sustains mesenchymal
stem cell identity via microRNA inhibition. Nucleic Acids Res.
47:5325–5340. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Costa-Silva B, Aiello NM, Ocean AJ, Singh
S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, et
al: Pancreatic cancer exosomes initiate pre-metastatic niche
formation in the liver. Nature Cell Biol. 17:816–826. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ahmed N, Escalona R, Leung D, Chan E and
Kannourakis G: Tumour microenvironment and metabolic plasticity in
cancer and cancer stem cells: Perspectives on metabolic and immune
regulatory signatures in chemoresistant ovarian cancer stem cells.
Semin Cancer Biol. 53:265–281. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Steinbichler TB, Dudás J, Riechelmann H
and Skvortsova II: The role of exosomes in cancer metastasis. Semin
Cancer Biol. 44:170–181. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Raposo G and Stoorvogel W: Extracellular
vesicles: Exosomes, microvesicles, and friends. J Cell Biol.
200:373–383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wortzel I, Dror S, Kenific CM and Lyden D:
Exosome-mediated metastasis: Communication from a distance. Dev
Cell. 49:347–360. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng
T, Yang H, Sun W, Wang X, Zhu K, et al: Exosomal circRNA derived
from gastric tumor promotes white adipose browning by targeting the
miR-133/PRDM16 pathway. Int J Cancer. 144:2501–2515. 2019.
View Article : Google Scholar
|
|
89
|
Zong ZH, Du YP, Guan X, Chen S and Zhao Y:
CircWHSC1 promotes ovarian cancer progression by regulating MUC1
and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer
Res. 38:4372019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li Z, Yanfang W, Li J, Jiang P, Peng T,
Chen K, Zhao X, Zhang Y, Zhen P, Zhu J and Li X: Tumor-released
exosomal circular RNA PDE8A promotes invasive growth via the
miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett.
432:237–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Luna J, Boni J, Cuatrecasas M, Bofill-De
Ros X, Núñez-Manchón E, Gironella M, Vaquero EC, Arbones ML, de la
Luna S and Fillat C: DYRK1A modulates c-MET in pancreatic ductal
adenocarcinoma to drive tumour growth. Gut. 68:1465–1476. 2019.
View Article : Google Scholar
|
|
92
|
Li J, Li Z, Jiang P, Peng M, Zhang X, Chen
K, Liu H, Bi H, Liu X and Li X: Circular RNA IARS (circ-IARS)
secreted by pancreatic cancer cells and located within exosomes
regulates endothelial monolayer permeability to promote tumor
metastasis. J Exp Clin Cancer Res. 37:1772018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo
J, Zhang Y, Li H, Zhang Q, Yang Y and Chen G: Three isoforms of
exosomal circPTGR1 promote hepatocellular carcinoma metastasis via
the miR449a-MET pathway. EBioMedicine. 40:432–445. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yang X, Xiong Q, Wu Y, Li S and Ge F:
Quantitative proteomics reveals the regulatory networks of circular
RNA CDR1as in hepatocellular carcinoma cells. J Proteome Res.
16:3891–3902. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhong Q, Huang J, Wei J and Wu R: Circular
RNA CDR1as sponges miR-7-5p to enhance E2F3 stability and promote
the growth of nasopharyngeal carcinoma. Cancer Cell Int.
19:2522019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li T, Sun X and Chen L: Exosome
circ_0044516 promotes prostate cancer cell proliferation and
metastasis as a potential biomarker. J Cell Biochem. 121:2118–2126.
2020. View Article : Google Scholar
|
|
97
|
Wang S, Hu Y, Lv X, Li B, Gu D, Li Y, Sun
Y and Su Y: Circ-0000284 arouses malignant phenotype of
cholangiocarcinoma cells and regulates the biological functions of
peripheral cells through cellular communication. Clin Sci (Lond).
133:1935–1953. 2019. View Article : Google Scholar
|
|
98
|
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang
C, Liu D, Wang M, Wang L, Zeng F and Jiang G: CircHIPK3 sponges
miR-558 to suppress heparanase expression in bladder cancer cells.
EMBO Rep. 18:1646–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Ren GL, Zhu J, Li J and Meng XM: Noncoding
RNAs in acute kidney injury. J Cell Physiol. 234:2266–2276. 2019.
View Article : Google Scholar
|
|
100
|
Xu Y, Ku X, Wu C, Cai C, Tang J and Yan W:
Exosomal proteome analysis of human plasma to monitor sepsis
progression. Biochem Biophys Res Commun. 499:856–861. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Fanale D, Taverna S, Russo A and Bazan V:
Circular RNA in Exosomes. Adv Exp Med Biol. 1087:109–117. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Chen R, Xu X, Qian Z, Zhang C, Niu Y, Wang
Z, Sun J, Zhang X and Yu Y: The biological functions and clinical
applications of exosomes in lung cancer. Cell Mol Life Sci.
76:4613–4633. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Lakhal S and Wood MJ: Exosome
nanotechnology: An emerging paradigm shift in drug delivery:
Exploitation of exosome nanovesicles for systemic in vivo delivery
of RNAi heralds new horizons for drug delivery across biological
barriers. Bioessays. 33:737–741. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Ma H, Xu Y, Zhang R, Guo B, Zhang S and
Zhang X: Differential expression study of circular RNAs in exosomes
from serum and urine in patients with idiopathic membranous
nephropathy. Arch Med Sci. 15:738–753. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Liu S, Lin Z, Rao W, Zheng J, Xie Q, Lin
Y, Lin X, Chen H, Chen Y and Hu Z: Upregulated expression of serum
exosomal hsa_circ_0026611 is associated with lymph node metastasis
and poor prognosis of esophageal squamous cell carcinoma. J Cancer.
12:918–926. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Fan L, Cao Q, Liu J, Zhang J and Li B:
Circular RNA profiling and its potential for esophageal squamous
cell cancer diagnosis and prognosis. Mol Cancer. 18:162019.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yang C, Wei Y, Yu L and Xiao Y:
Identification of altered circular RNA expression in serum exosomes
from patients with papillary thyroid carcinoma by high-throughput
sequencing. Med Sci Monit. 25:2785–2791. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Pan B, Qin J, Liu X, He B, Wang X, Pan Y,
Sun H, Xu T, Xu M, Chen X, et al: Identification of serum exosomal
hsa-circ-0004771 as a novel diagnostic biomarker of colorectal
cancer. Front Genet. 10:10962019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Chen F, Huang C, Wu Q, Jiang L, Chen S and
Chen L: Circular RNAs expression profiles in plasma exosomes from
early-stage lung adenocarcinoma and the potential biomarkers. J
Cell Biochem. 121:2525–2533. 2020. View Article : Google Scholar
|
|
110
|
Feng Y, Hang W, Sang Z, Li S, Xu W, Miao
Y, Xi X and Huang Q: Identification of exosomal and non-exosomal
microRNAs associated with the drug resistance of ovarian cancer.
Mol Med Rep. 19:3376–3392. 2019.PubMed/NCBI
|
|
111
|
Sousa D, Lima RT and Vasconcelos MH:
Intercellular transfer of cancer drug resistance traits by
extracellular vesicles. Trends Mol Med. 21:595–608. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhao Z, Ji M, Wang Q, He N and Li Y:
Circular RNA Cdr1as upregulates SCAI to suppress cisplatin
resistance in ovarian cancer via miR-1270 suppression. Mol Ther
Nucleic Acids. 18:24–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Tang W, Ji M, He G, Yang L, Niu Z, Jian M,
Wei Y, Ren L and Xu J: Silencing CDR1as inhibits colorectal cancer
progression through regulating microRNA-7. OncoTargets Ther.
10:2045–2056. 2017. View Article : Google Scholar
|
|
114
|
Sang M, Meng L, Sang Y, Liu S, Ding P, Ju
Y, Liu F, Gu L, Lian Y, Li J, et al: Circular RNA ciRS-7
accelerates ESCC progression through acting as a miR-876-5p sponge
to enhance MAGE-A family expression. Cancer Lett. 426:37–46. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|