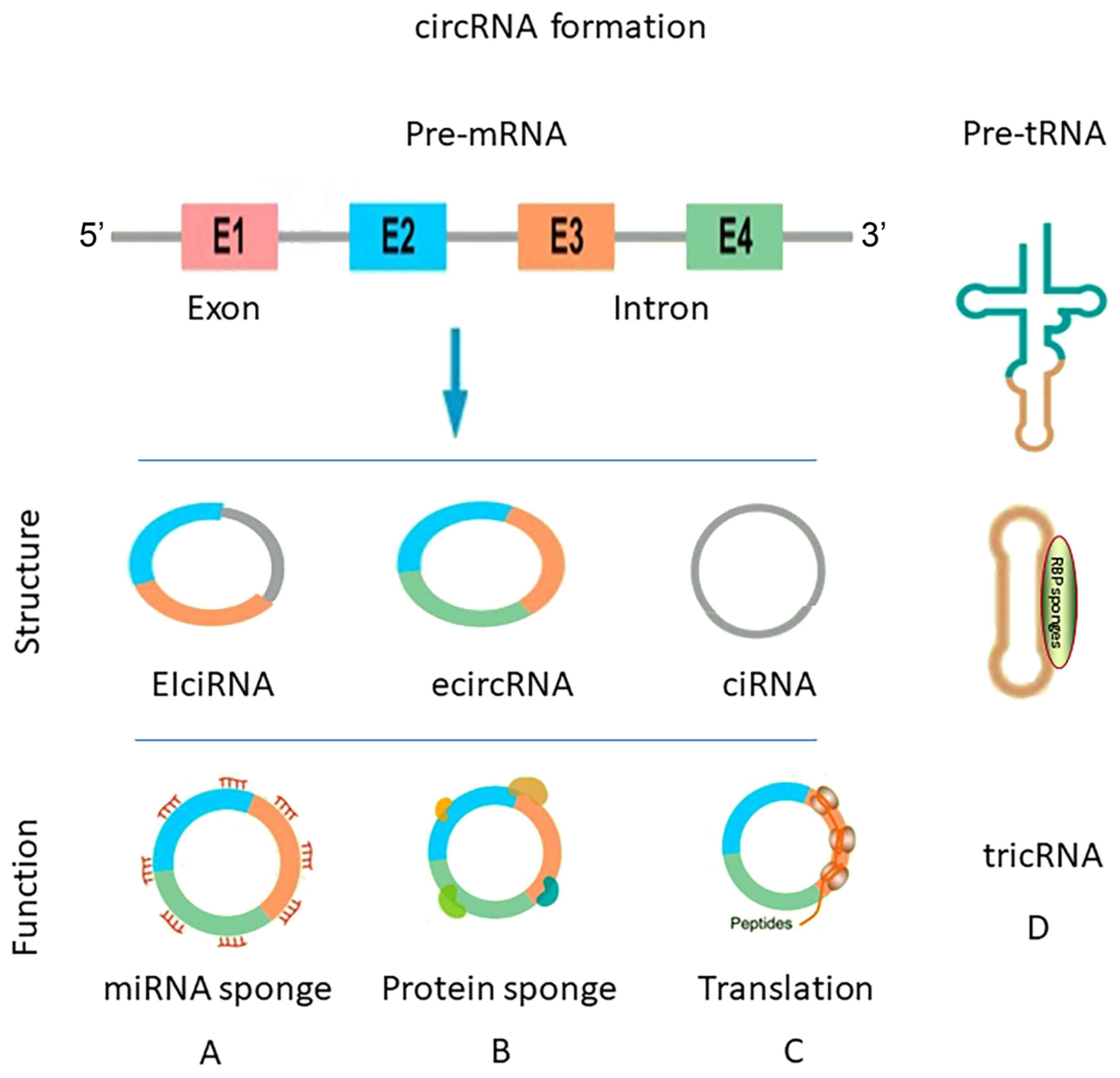
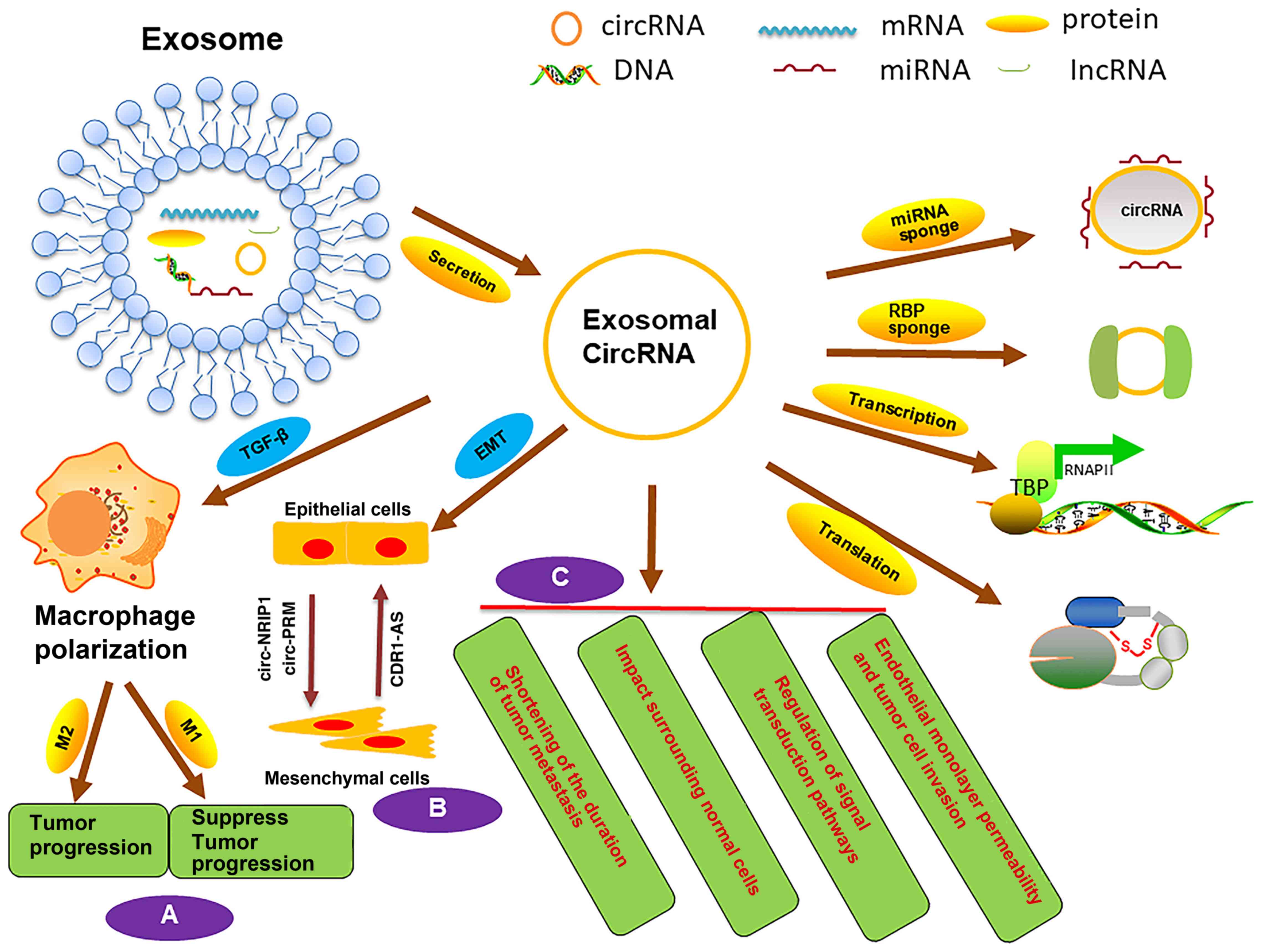
|
1
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar :
|
|
2
|
Lin J, Zhang Y, Zeng X, Xue C and Lin X:
CircRNA CircRIMS Acts as a MicroRNA sponge to promote gastric
cancer metastasis. ACS Omega. 5:23237–23246. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang Y, Mo Y, Gong Z, Yang X, Yang M,
Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, et al: Circular RNAs in
human cancer. Mol Cancer. 16:252017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Martins VR, Dias MS and Hainaut P:
Tumor-cell-derived microvesicles as carriers of molecular
information in cancer. Curr Opin Oncol. 25:66–75. 2013. View Article : Google Scholar
|
|
5
|
Whiteside TL: Tumor-derived exosomes and
their role in cancer progression. Adv Clin Chem. 74:103–141. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
7
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Funes SC, Rios M, Escobar-Vera J and
Kalergis AM: Implications of macrophage polarization in
autoimmunity. Immunology. 154:186–195. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mantovani A, Sozzani S, Locati M, Allavena
P and Sica A: Macrophage polarization: Tumor-associated macrophages
as a paradigm for polarized M2 mononuclear phagocytes. Trends
Immunol. 23:549–555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang JJ, Lei KF and Han F: Tumor
microenvironment: Recent advances in various cancer treatments. Eur
Rev Med Pharmacol Sci. 22:3855–3864. 2018.PubMed/NCBI
|
|
11
|
Sun Y: Tumor microenvironment and cancer
therapy resistance. Cancer Lett. 380:205–215. 2016. View Article : Google Scholar
|
|
12
|
Xie F, Zhou X, Fang M, Li H, Su P, Tu Y,
Zhang L and Zhou F: Extracellular vesicles in cancer immune
microenvironment and cancer immunotherapy. Adv Sci (Weinh).
6:19017792019. View Article : Google Scholar
|
|
13
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J, Mohammed-Elsabagh M, Paczkowski F
and Li Y: Circular nucleic acids: Discovery, functions and
applications. Chembiochem. 21:1547–1566. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Patop IL, Wüst S and Kadener S: Past,
present, and future of circRNAs. EMBO J. 38:e1008362019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meng X, Chen Q, Zhang P and Chen M:
CircPro: An integrated tool for the identification of circRNAs with
protein-coding potential. Bioinformatics. 33:3314–3316. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao X, Cai Y and Xu J: Circular RNAs:
Biogenesis, mechanism, and function in human cancers. Int J Mol
Sci. 20:39262019. View Article : Google Scholar :
|
|
20
|
Wilusz JE: A 360° view of circular RNAs:
From biogenesis to functions. Wiley interdisciplinary reviews. RNA.
9:e14782018.
|
|
21
|
Chen X, Yang T, Wang W, Xi W, Zhang T, Li
Q, Yang A and Wang T: Circular RNAs in immune responses and immune
diseases. Theranostics. 9:588–607. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Das A, Rout PK, Gorospe M and Panda AC:
Rolling Circle cDNA synthesis uncovers circular RNA splice
variants. Int J Mol Sci. 20:39882019. View Article : Google Scholar :
|
|
24
|
Chan JJ and Tay Y: Noncoding RNA:RNA
Regulatory networks in cancer. Int J Mol Sci. 19:13102018.
View Article : Google Scholar :
|
|
25
|
Pardini B, Sabo AA, Birolo G and Calin GA:
Noncoding RNAs in extracellular fluids as cancer biomarkers: The
New Frontier of Liquid Biopsies. Cancers. 11:11702019. View Article : Google Scholar :
|
|
26
|
Liu K, Zhang Q, Pan F, Wang XD, Wenjing H
and Tong H: Expression of circular RNAs in gynecological tumors: A
systematic review. Medicine. 98:e157362019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z,
Xu B, Wu C, Zhou Q, Hu W, Wu C and Jiang J: A novel protein encoded
by a circular RNA circPPP1R12A promotes tumor pathogenesis and
metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer.
18:472019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Correia de Sousa M, Gjorgjieva M, Dolicka
D, Sobolewski C and Foti M: Deciphering miRNAs' Action through
miRNA Editing. Int J Mol Sci. 20:62492019. View Article : Google Scholar
|
|
29
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol. 9:4022018. View Article : Google Scholar
|
|
30
|
Tan W, Liu B, Qu S, Liang G, Luo W and
Gong C: MicroRNAs and cancer: Key paradigms in molecular therapy.
Oncol Lett. 15:2735–2742. 2018.PubMed/NCBI
|
|
31
|
Xue D, Wang H, Chen Y, Shen D, Lu J, Wang
M, Zebibula A, Xu L, Wu H, Li G and Xia L: Circ-AKT3 inhibits clear
cell renal cell carcinoma metastasis via altering
miR-296-3p/E-cadherin signals. Mol Cancer. 18:1512019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li R, Wu B, Xia J, Ye L and Yang X:
Circular RNA hsa_ circRNA_102958 promotes tumorigenesis of
colorectal cancer via miR-585/CDC25B axis. Cancer Manag Res.
11:6887–6893. 2019. View Article : Google Scholar :
|
|
33
|
Xu H, Guo S, Li W and Yu P: The circular
RNA Cdr1as, via miR-7 and its targets, regulates insulin
transcription and secretion in islet cells. Sci Rep. 5:124532015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu W, Wang Y, Zhang D, Yu X and Leng X:
MiR-75p functions as a tumor suppressor by targeting SOX18 in
pancreatic ductal adenocarcinoma. Biochem Biophys Res Commun.
497:963–970. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin J, Liu Z, Liao S, Li E, Wu X and Zeng
W: Elevated microRNA-7 inhibits proliferation and tumor
angiogenesis and promotes apoptosis of gastric cancer cells via
repression of Raf-1. Cell Cycle. 19:2496–2508. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shen B, Wang Z, Li Z, Song H and Ding X:
Circular RNAs: An emerging landscape in tumor metastasis. Am J
Cancer Res. 9:630–643. 2019.PubMed/NCBI
|
|
37
|
Tong H, Zhao K, Wang J, Xu H and Xiao J:
CircZNF609/miR-134-5p/BTG-2 axis regulates proliferation and
migration of glioma cell. J Pharm Pharmacol. 72:68–75. 2020.
View Article : Google Scholar
|
|
38
|
Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu
C, Li G and Zhu Y: Silencing of cZNF292 circular RNA suppresses
human glioma tube formation via the Wnt/β-catenin signaling
pathway. Oncotarget. 7:63449–63455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Z, Chen Z, Hu G and Jiang Y: Roles of
circular RNA in breast cancer: Present and future. Am J Transl Res.
11:3945–3954. 2019.PubMed/NCBI
|
|
41
|
Sun S, Wang W, Luo X, Li Y, Liu B and Li
X, Zhang B, Han S and Li X: Circular RNA circ-ADD3 inhibits
hepatocellular carcinoma metastasis through facilitating EZH2
degradation via CDK1-mediated ubiquitination. Am J Cancer Res.
9:1695–1707. 2019.PubMed/NCBI
|
|
42
|
Liu Z, Yu Y, Huang Z, Kong Y, Hu X, Xiao
W, Quan J and Fan X: CircRNA-5692 inhibits the progression of
hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP
expression. Cell Death Dis. 10:9002019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Boriachek K, Islam MN, Möller A, Salomon
C, Nguyen NT, Hossain MSA, Yamauchi Y and Shiddiky MJA: Biological
functions and current advances in isolation and detection
strategies for exosome nanovesicles. Small. 14:17021532018.
View Article : Google Scholar
|
|
44
|
Braicu C, Tomuleasa C, Monroig P, Cucuianu
A, Berindan-Neagoe I and Calin GA: Exosomes as divine messengers:
Are they the Hermes of modern molecular oncology? Cell Death
Differ. 22:34–45. 2015. View Article : Google Scholar
|
|
45
|
Pant S, Hilton H and Burczynski ME: The
multifaceted exosome: Biogenesis, role in normal and aberrant
cellular function, and frontiers for pharmacological and biomarker
opportunities. Biochem Pharmacol. 83:1484–1494. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ngalame NNO, Luz AL, Makia N and Tokar EJ:
Arsenic alters exosome quantity and cargo to mediate stem cell
recruitment into a cancer stem cell-like phenotype. Toxicol Sci.
165:40–49. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shi X, Wang B, Feng X, Xu Y, Lu K and Sun
M: circRNAs and exosomes: A mysterious frontier for human cancer.
Mol Ther Nucleic Acids. 19:384–392. 2020. View Article : Google Scholar :
|
|
48
|
Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y,
Zheng Q, Li Y, Wang P, He X and Huang S: exoRBase: A database of
circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids
Res. 46:D106–D112. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A
and Qi F: CircRNA Cdr1as functions as a competitive endogenous RNA
to promote hepatocellular carcinoma progression. Aging (Albany NY).
11:8182–8203. 2019.
|
|
50
|
Binnewies M, Roberts EW, Kersten K, Chan
V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI,
Ostrand-Rosenberg S, Hedrick CC, et al: Understanding the tumor
immune microenvironment (TIME) for effective therapy. Nat Med.
24:541–550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ridge SM, Sullivan FJ and Glynn SA:
Mesenchymal stem cells: Key players in cancer progression. Mol
Cancer. 16:312017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kalluri R: The biology and function of
fibroblasts in cancer. Nat Rev Cancer. 16:582–598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li K, Chen Y, Li A, Tan C and Liu X:
Exosomes play roles in sequential processes of tumor metastasis.
Int J Cancer. 144:1486–1495. 2019. View Article : Google Scholar
|
|
54
|
Matei I, Kim HS and Lyden D: Unshielding
exosomal RNA unleashes tumor growth and metastasis. Cell.
170:223–225. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang L, Yang G, Zhao D, Wang J, Bai Y,
Peng Q, Wang H, Fang R, Chen G, Wang Z, et al: CD103-positive CSC
exosome promotes EMT of clear cell renal cell carcinoma: Role of
remote MiR-19b-3p. Mol Cancer. 18:862019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hakulinen J, Sankkila L, Sugiyama N, Lehti
K and Keski-Oja J: Secretion of active membrane type 1 matrix
metalloproteinase (MMP-14) into extracellular space in
microvesicular exosomes. J Cell Biochem. 105:1211–1218. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Turley SJ, Cremasco V and Astarita JL:
Immunological hall-marks of stromal cells in the tumour
microenvironment. Nat Rev Immunol. 15:669–682. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lai CP, Kim EY, Badr CE, Weissleder R,
Mempel TR, Tannous BA and Breakefield XO: Visualization and
tracking of tumour extracellular vesicle delivery and RNA
translation using multiplexed reporters. Nat Commun. 6:70292015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yoon YJ, Kim OY and Gho YS: Extracellular
vesicles as emerging intercellular communicasomes. BMB Rep.
47:531–539. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lin Y, Xu J and Lan H: Tumor-associated
macrophages in tumor metastasis: Biological roles and clinical
therapeutic applications. J Hematol Oncol. 12:762019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Biswas SK and Mantovani A: Macrophage
plasticity and inter-action with lymphocyte subsets: Cancer as a
paradigm. Nat Immunol. 11:889–896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Najafi M, Hashemi Goradel N, Farhood B,
Salehi E, Nashtaei MS, Khanlarkhani N, Khezri Z, Majidpoor J,
Abouzaripour M, Habibi M, et al: Macrophage polarity in cancer: A
review. J Cell Biochem. 120:2756–2765. 2019. View Article : Google Scholar
|
|
63
|
Allavena P, Sica A, Garlanda C and
Mantovani A: The Yin-Yang of tumor-associated macrophages in
neoplastic progression and immune surveillance. Immunol Rev.
222:155–161. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Preußer C, Hung LH, Schneider T, Schreiner
S, Hardt M, Moebus A, Santoso S and Bindereif A: Selective release
of circRNAs in platelet-derived extracellular vesicles. J Extracell
Vesicles. 7:14244732018. View Article : Google Scholar
|
|
65
|
Hou J, Jiang W, Zhu L, Zhong S, Zhang H,
Li J, Zhou S, Yang S, He Y, Wang D, et al: Circular RNAs and
exosomes in cancer: A mysterious connection. Clin Transl Oncol.
20:1109–1116. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sekar S, Cuyugan L, Adkins J, Geiger P and
Liang WS: Circular RNA expression and regulatory network prediction
in posterior cingulate astrocytes in elderly subjects. BMC
Genomics. 19:3402018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zou Y, Zheng S, Deng X, Yang A and Xie X,
Tang H and Xie X: The role of circular RNA CDR1as/ciRS-7 in
regulating tumor micro-environment: A pan-cancer analysis.
Biomolecules. 9:4292019. View Article : Google Scholar
|
|
68
|
Pickup M, Novitskiy S and Moses HL: The
roles of TGFβ in the tumour microenvironment. Nature Revi Cancer.
13:788–799. 2013. View Article : Google Scholar
|
|
69
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Shang BQ, Li ML, Quan HY, Hou PF, Li ZW,
Chu SF, Zheng JN and Bai J: Functional roles of circular RNAs
during epithelial-to-mesenchymal transition. Mol Cancer.
18:1382019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chen X, Chen RX, Wei WS, Li YH, Feng ZH,
Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al: PRMT5 circular RNA
promotes metastasis of urothelial carcinoma of the bladder through
sponging miR-30c to induce epithelial-mesenchymal transition. Clin
Cancer Res. 24:6319–6330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ma C, Shi T, Qu Z, Zhang A, Wu Z, Zhao H,
Zhao H and Chen H: CircRNA_ACAP2 suppresses EMT in head and neck
squamous cell carcinoma by targeting the miR-21-5p/STAT3 signaling
axis. Front Oncol. 10:5836822020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhao X, Dou W, He L, Liang S, Tie J, Liu
C, Li T, Lu Y, Mo P, Shi Y, et al: MicroRNA-7 functions as an
anti-metastatic microRNA in gastric cancer by targeting
insulin-like growth factor-1 receptor. Oncogene. 32:1363–1372.
2013. View Article : Google Scholar
|
|
75
|
Ball SG, Shuttleworth CA and Kielty CM:
Mesenchymal stem cells and neovascularization: Role of
platelet-derived growth factor receptors. J Cell Mol Med.
11:1012–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Altaner C, Altanerova U and Jakubechova J:
Intracellular acting tumor cell-targeted chemotherapy by
MSC-suicide gene exosomes. Oncotarget. 10:5573–5575. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Brossa A, Fonsato V and Bussolati B:
Anti-tumor activity of stem cell-derived extracellular vesicles.
Oncotarget. 10:1872–1873. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ferguson SW, Wang J, Lee CJ, Liu M,
Neelamegham S, Canty JM and Nguyen J: The microRNA regulatory
landscape of MSC-derived exosomes: A systems view. Sci Rep.
8:14192018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Shimbo K, Miyaki S, Ishitobi H, Kato Y,
Kubo T, Shimose S and Ochi M: Exosome-formed synthetic microRNA-143
is transferred to osteosarcoma cells and inhibits their migration.
Biochem Biophys Res Commun. 445:381–387. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li X, Wang S, Zhu R, Li H, Han Q and Zhao
RC: Lung tumor exosomes induce a pro-inflammatory phenotype in
mesenchymal stem cells via NFκB-TLR signaling pathway. J Hematol
Oncol. 9:422016. View Article : Google Scholar
|
|
81
|
Menck K, Klemm F, Gross JC, Pukrop T,
Wenzel D and Binder C: Induction and transport of Wnt 5a during
macrophage-induced malignant invasion is mediated by two types of
extracellular vesicles. Oncotarget. 4:2057–2066. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Cherubini A, Barilani M, Rossi RL, Jalal
MMK, Rusconi F, Buono G, Ragni E, Cantarella G, Simpson HARW,
Péault B and Lazzari L: FOXP1 circular RNA sustains mesenchymal
stem cell identity via microRNA inhibition. Nucleic Acids Res.
47:5325–5340. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Costa-Silva B, Aiello NM, Ocean AJ, Singh
S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, et
al: Pancreatic cancer exosomes initiate pre-metastatic niche
formation in the liver. Nature Cell Biol. 17:816–826. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ahmed N, Escalona R, Leung D, Chan E and
Kannourakis G: Tumour microenvironment and metabolic plasticity in
cancer and cancer stem cells: Perspectives on metabolic and immune
regulatory signatures in chemoresistant ovarian cancer stem cells.
Semin Cancer Biol. 53:265–281. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Steinbichler TB, Dudás J, Riechelmann H
and Skvortsova II: The role of exosomes in cancer metastasis. Semin
Cancer Biol. 44:170–181. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Raposo G and Stoorvogel W: Extracellular
vesicles: Exosomes, microvesicles, and friends. J Cell Biol.
200:373–383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wortzel I, Dror S, Kenific CM and Lyden D:
Exosome-mediated metastasis: Communication from a distance. Dev
Cell. 49:347–360. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng
T, Yang H, Sun W, Wang X, Zhu K, et al: Exosomal circRNA derived
from gastric tumor promotes white adipose browning by targeting the
miR-133/PRDM16 pathway. Int J Cancer. 144:2501–2515. 2019.
View Article : Google Scholar
|
|
89
|
Zong ZH, Du YP, Guan X, Chen S and Zhao Y:
CircWHSC1 promotes ovarian cancer progression by regulating MUC1
and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer
Res. 38:4372019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li Z, Yanfang W, Li J, Jiang P, Peng T,
Chen K, Zhao X, Zhang Y, Zhen P, Zhu J and Li X: Tumor-released
exosomal circular RNA PDE8A promotes invasive growth via the
miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett.
432:237–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Luna J, Boni J, Cuatrecasas M, Bofill-De
Ros X, Núñez-Manchón E, Gironella M, Vaquero EC, Arbones ML, de la
Luna S and Fillat C: DYRK1A modulates c-MET in pancreatic ductal
adenocarcinoma to drive tumour growth. Gut. 68:1465–1476. 2019.
View Article : Google Scholar
|
|
92
|
Li J, Li Z, Jiang P, Peng M, Zhang X, Chen
K, Liu H, Bi H, Liu X and Li X: Circular RNA IARS (circ-IARS)
secreted by pancreatic cancer cells and located within exosomes
regulates endothelial monolayer permeability to promote tumor
metastasis. J Exp Clin Cancer Res. 37:1772018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo
J, Zhang Y, Li H, Zhang Q, Yang Y and Chen G: Three isoforms of
exosomal circPTGR1 promote hepatocellular carcinoma metastasis via
the miR449a-MET pathway. EBioMedicine. 40:432–445. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yang X, Xiong Q, Wu Y, Li S and Ge F:
Quantitative proteomics reveals the regulatory networks of circular
RNA CDR1as in hepatocellular carcinoma cells. J Proteome Res.
16:3891–3902. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhong Q, Huang J, Wei J and Wu R: Circular
RNA CDR1as sponges miR-7-5p to enhance E2F3 stability and promote
the growth of nasopharyngeal carcinoma. Cancer Cell Int.
19:2522019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li T, Sun X and Chen L: Exosome
circ_0044516 promotes prostate cancer cell proliferation and
metastasis as a potential biomarker. J Cell Biochem. 121:2118–2126.
2020. View Article : Google Scholar
|
|
97
|
Wang S, Hu Y, Lv X, Li B, Gu D, Li Y, Sun
Y and Su Y: Circ-0000284 arouses malignant phenotype of
cholangiocarcinoma cells and regulates the biological functions of
peripheral cells through cellular communication. Clin Sci (Lond).
133:1935–1953. 2019. View Article : Google Scholar
|
|
98
|
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang
C, Liu D, Wang M, Wang L, Zeng F and Jiang G: CircHIPK3 sponges
miR-558 to suppress heparanase expression in bladder cancer cells.
EMBO Rep. 18:1646–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Ren GL, Zhu J, Li J and Meng XM: Noncoding
RNAs in acute kidney injury. J Cell Physiol. 234:2266–2276. 2019.
View Article : Google Scholar
|
|
100
|
Xu Y, Ku X, Wu C, Cai C, Tang J and Yan W:
Exosomal proteome analysis of human plasma to monitor sepsis
progression. Biochem Biophys Res Commun. 499:856–861. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Fanale D, Taverna S, Russo A and Bazan V:
Circular RNA in Exosomes. Adv Exp Med Biol. 1087:109–117. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Chen R, Xu X, Qian Z, Zhang C, Niu Y, Wang
Z, Sun J, Zhang X and Yu Y: The biological functions and clinical
applications of exosomes in lung cancer. Cell Mol Life Sci.
76:4613–4633. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Lakhal S and Wood MJ: Exosome
nanotechnology: An emerging paradigm shift in drug delivery:
Exploitation of exosome nanovesicles for systemic in vivo delivery
of RNAi heralds new horizons for drug delivery across biological
barriers. Bioessays. 33:737–741. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Ma H, Xu Y, Zhang R, Guo B, Zhang S and
Zhang X: Differential expression study of circular RNAs in exosomes
from serum and urine in patients with idiopathic membranous
nephropathy. Arch Med Sci. 15:738–753. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Liu S, Lin Z, Rao W, Zheng J, Xie Q, Lin
Y, Lin X, Chen H, Chen Y and Hu Z: Upregulated expression of serum
exosomal hsa_circ_0026611 is associated with lymph node metastasis
and poor prognosis of esophageal squamous cell carcinoma. J Cancer.
12:918–926. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Fan L, Cao Q, Liu J, Zhang J and Li B:
Circular RNA profiling and its potential for esophageal squamous
cell cancer diagnosis and prognosis. Mol Cancer. 18:162019.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yang C, Wei Y, Yu L and Xiao Y:
Identification of altered circular RNA expression in serum exosomes
from patients with papillary thyroid carcinoma by high-throughput
sequencing. Med Sci Monit. 25:2785–2791. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Pan B, Qin J, Liu X, He B, Wang X, Pan Y,
Sun H, Xu T, Xu M, Chen X, et al: Identification of serum exosomal
hsa-circ-0004771 as a novel diagnostic biomarker of colorectal
cancer. Front Genet. 10:10962019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Chen F, Huang C, Wu Q, Jiang L, Chen S and
Chen L: Circular RNAs expression profiles in plasma exosomes from
early-stage lung adenocarcinoma and the potential biomarkers. J
Cell Biochem. 121:2525–2533. 2020. View Article : Google Scholar
|
|
110
|
Feng Y, Hang W, Sang Z, Li S, Xu W, Miao
Y, Xi X and Huang Q: Identification of exosomal and non-exosomal
microRNAs associated with the drug resistance of ovarian cancer.
Mol Med Rep. 19:3376–3392. 2019.PubMed/NCBI
|
|
111
|
Sousa D, Lima RT and Vasconcelos MH:
Intercellular transfer of cancer drug resistance traits by
extracellular vesicles. Trends Mol Med. 21:595–608. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhao Z, Ji M, Wang Q, He N and Li Y:
Circular RNA Cdr1as upregulates SCAI to suppress cisplatin
resistance in ovarian cancer via miR-1270 suppression. Mol Ther
Nucleic Acids. 18:24–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Tang W, Ji M, He G, Yang L, Niu Z, Jian M,
Wei Y, Ren L and Xu J: Silencing CDR1as inhibits colorectal cancer
progression through regulating microRNA-7. OncoTargets Ther.
10:2045–2056. 2017. View Article : Google Scholar
|
|
114
|
Sang M, Meng L, Sang Y, Liu S, Ding P, Ju
Y, Liu F, Gu L, Lian Y, Li J, et al: Circular RNA ciRS-7
accelerates ESCC progression through acting as a miR-876-5p sponge
to enhance MAGE-A family expression. Cancer Lett. 426:37–46. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|