Introduction
In 2020, the morbidity of lung cancer ranked second
and its mortality ranked first in both females and males worldwide
(1,2). Lung adenocarcinoma (LUAD) is the
major type of lung cancer. Therapeutic options include surgery and
treatment using radiotherapy, adjuvant chemotherapy, targeted drugs
and immunotherapy (3). However,
the 5-year survival rate in patients with LUAD remains low, and the
risk of recurrence is high due to the metastatic nature of the
tumor (4). Therefore, further
progress is required to identify effective prognostic biomarkers in
high-risk patients with LUAD and to elucidate the molecular
mechanisms underlying LUAD development.
Abnormal spindle-like microcephaly (ASPM) protein
was first identified as a microcephaly-associated protein that
controls spindle architecture (5), which is essential for neurogenesis
and brain size determination (6,7).
Additionally, ASPM participates in the process of symmetric stem
cell division by promoting cyclin E ubiquitination (8). Previous studies have suggested that
ASPM regulates microtubule degradation at the mitotic spindle pole
by promoting katanin-mediated cleavage of dynamic microtubules
(9,10). In particular, ASPM exhibits both
cytoplasmic and nuclear localization during interphase, and its
cytoplasmic expression levels vary significantly among different
types of tumor (11-13). This suggests that ASPM may have
diverse biological functions in malignant tissues. Furthermore,
ASPM is an oncofetal protein that is overexpressed in numerous
malignant neoplasms, including pancreatic ductal adenocarcinoma
(14), prostate cancer (13), ovarian cancer (15), hepatocellular carcinoma (16) and bladder cancer (17). The oncogenic function of ASPM has
been demonstrated to be involved in regulation of cell cycle
completeness and stem cell properties (8). However, the role of ASPM in LUAD
remains unclear. Therefore, the present study aimed to explore the
clinical role of ASPM in LUAD.
Materials and methods
Human tissue samples and cell lines
A total of 109 fresh paired normal and
pathologically confirmed LUAD tumor tissues (7 pairs used for
RNA-sequencing, 94 pairs for tissue microarray and 8 pairs for
western blotting) were obtained from Qilu Hospital (Shandong
University, Jinan, China) between September 2014 and June 2020. The
normal tissues (≥5 cm from tumour tissues) were obtained from
dissected lung tissues. The median age of patients was 62 years and
the age range was 30-84 years among all 109 patients, with 59 males
and 50 females in total. Both overall survival (OS) and
progression-free survival (PFS) were analyzed. TNM stages were
established according to the American Joint Committee on Cancer 8th
edition (18). Patients did not
receive any chemotherapy or radiotherapy before surgery. Patients
provided written informed consent for the use of their samples in
the study. All pathological results were confirmed by pathology.
Complete clinicopathological and follow-up data for the 94 paired
LUAD samples were available. All experiments were approved and
supervised by the Medical Ethics Committee of Qilu Hospital of
Shandong University (approval no. KYLL-2016-097).
Normal lung epithelial cells (HBE) and human lung
cancer cell lines (HCC827, A549, H157, H1299 and H1975) were
obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. The H157 cell line was authenticated
by STR profiling. Cells were cultured in RPMI 1640 medium (cat. no.
CF0001; Shandong Sparkjade Biotechnology Co., Ltd.) supplemented
with 10% FBS (cat. no. 04-001-1A; Biological Industries) and 1%
antibiotic-antimycotic agents at 37°C in a 5% CO2 cell
culture incubator.
Cell transfection using small interfering
(si)RNA
siRNA silencing was performed by transient
transfection using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.). Cells were seeded in a 6-well plate at a density
of 2×105 cells/well. Once the cells had adhered, they
were transfected with 5 µl siRNA (20 µM) or negative
control (NC) for 8 h at 37°C. Cells were incubated with the
transfection mixture for 6-8 h, after which the medium was replaced
with fresh RPMI 1640 medium. siRNA molecules against ASPM were
purchased from Shanghai GenePharma Co., Ltd., with a scrambled
siRNA used as the NC. The siRNA sequences were as follows: NC
sense, 5′-UUC UCC GAA CGU GUC ACG UTT-3′ and antisense, 5′-ACG UGA
CAC GUU CGG AGA ATT-3′; siRNA1 sense, 5′-CCC ACC ACC ACU GUA UGA
UTT-3′ and antisense, 5′-AUC AUA CAG UGG UGG UGG GTT-3′; and siRNA2
sense, 5′-GCU UGC AAU ACA GCA AUA ATT-3′ and antisense, 5′-UUA UUG
CUG UAU UGC AAG CTT-3′.
Recombinant vector construction and cell
transfection
The interference vector, pGMLV-SC5 (iGene
Biotechnology Co., Ltd) was linearized using double restriction
digestion and BamHI and EcoRI (Fermentas; Thermo
Fisher Scientific, Inc.), purified using a Gel-Spin DNA Extraction
kit (Tiangen Biotech Co., Ltd.) and then ligated with a short
hairpin (sh) RNA against ASPM (shASPM) or a negative control shRNA
(shNC) (both iGene Biotechnology Co., Ltd) using T4 ligase
(Fermentas; Thermo Fisher Scientific, Inc.). DH5α competent cells
(Tiangen Biotech Co., Ltd.) were mixed with the constructed
vectors, which were then harvested using DNA Gel/PCR Purification
Maxiprep kit (Generay Biotech Co., Ltd.). All the vectors were then
transfected into 293T cells (The Cell Bank of Type Culture
Collection of The Chinese Academy of Sciences) using the HG
transgene reagent (iGene Biotechnology Co., Ltd) according to the
manufacturer's instructions. High titers (>108
transforming units/ml) of the concentrated lentivirus solutions
were harvested from the supernatant. The shRNAs were diluted to 0.1
nmol for the downstream reaction, and the transfected cells were
passaged to at least 5 generations (transfected after 24 h of
culturing). Stable ASPM-knockdown H1299 and A549 cell lines were
infected with lentivirus (MOI, 10 and 20, respectively) for 72 h
and then selected with 1 mg/ml puromycin for ~1 week. After
selection, the cells were used for further experiments.
Cell Counting Kit-8 (CCK-8) and
5-Ethynyl-2′-deoxyuridine (EDU) assay
Cell proliferation was measured using the CCK-8
(Beyotime Institute of Biotechnology) and EDU (Guangzhou RiboBio
Co., Ltd.; cat. no. C10310) assays, according to the manufacturer's
instructions. The cells were seeded in 96-well plates. Cell numbers
were counted every 24 h after cell adhesion lasting for 4 days. The
cells were incubated in CCK-8 reagent for 2 h. Subsequently, the
absorbance was measured at a wavelength of 450 nm using a
microplate reader (Bio-Rad Laboratories, Inc.). The experiment was
repeated three times independently.
For EDU analysis, an EDU assay kit (cat. no.
C10310-1; Guangzhou RiboBio Co., Ltd.) was used following the
manufacturer's protocol. Cells were cultured in RPMI 1640 medium
containing EDU for 4 h at 37°C. Subsequently, cells were fixed with
4% paraformaldehyde at room temperature for 60 min, after removing
EDU-containing medium. After washing by penetrant (0.5% TritonX-100
in PBS) and incubating on a decolorizing shaker for 10 min, add
Apollo staining reaction solution to each well lasting for 30 min
on a decolorizing shaker in the dark at room temperature. Then
Hoechst reaction solution was added to each well. After incubating
for 30 min on a decolorizing shaker in the dark at room
temperature, discard the staining reaction solution. After PBS
washing, images were acquired using a fluorescence microscope
(magnification, ×400).
Wound healing, migration and invasion
assay
For the wound healing assay, transfected and control
cells were separately plated in 6-well plates. After the cells were
completely attached, a sterile tip was used to draw straight lines
on the bottom of the plate. After the scratch was made, the
original medium was replaced by medium without FBS. The cells were
washed with PBS, and the initial wound size was recorded with a
light microscope (Olympus Corporation; magnification, ×50). After
incubation for 36 h at 37°C, the current wound size was recorded.
The wound closure percentage was calculated using the following
formula: [1-(current wound size/initial wound size)] ×100.
Cell migration or invasion were performed using
Transwell chambers coated without or with Matrigel, respectively
(both Corning, Inc.). Matrigel was added to Transwell chambers at
4°C and then incubated at 37°C for 30 min. In brief, cells
(1×105) were seeded into the upper chamber for adhesion
with or without a monolayer of 5% Matrigel and then serum-starved
for 6 h. The chambers were then placed in 24-well plates containing
medium with 10% FBS in the bottom chambers. After 24 h at 37°C, the
cells adhering to the lower surface were fixed with methanol at 4°C
for 1 h and stained with a 0.1% crystal violet solution at room
temperature for 1 h (Sigma-Aldrich; Merck KGaA). The number of
cells on the membrane was counted from five randomly selected
visual fields with a fluorescence microscope (Olympus Corporation;
magnification, ×200).
Transcriptome sequencing and
bioinformatics analysis
Transcriptional sequencing of cells and tissues was
performed by Hangzhou Lianchuan Biotechnology Co., Ltd. (https://www.omicstudio.cn/index). Total RNA was
isolated from cell lines and tissues using the RNeasy Mini kit
(Qiagen GmbH). Related data were uploaded in the Gene Expression
Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) dataset GSE140797
(19). The gene expression levels
and clinical data of patients with LUAD from The Cancer Genome
Atlas (TCGA) were downloaded from UCSC Xena Browser (https://xenabrowser.net/). The gene expression levels
and clinical data of patients with LUAD from the GEO database were
obtained using the GSE19804 (20), GSE116959 (21), GSE31210 (22) and GSE68465 (23) datasets (https://www.ncbi.nlm.nih.gov/gds). The volcano plot
representing differentially expressed genes in LUAD and normal
tissues was performed using R software (v3.6.1; www.r-project.org) according to P<0.05 and
fold-change >2. Kyoto Encyclopedia of Genes and Genomes (KEGG)
and Gene Ontology (GO) enrichment analysis were performed using R
software. P<0.05 was defined as a significant enrichment
analysis result (24,25).
Immunohistochemistry (IHC)
Tissue sections fixed with 4% paraformaldehyde at
4°C for 24 h and embedded into paraffin were cut into
4-µm-thick sections and dried at room temperature for 24 h.
Paraffin-embedded LUAD and normal tissues sections were dewaxed
with xylene and ethanol. Antigen retrieval was performed by
incubation with sodium citrate buffer (pH 6.0) at 95°C for 15 min.
Sections were then blocked for endogenous peroxidase using 3%
H2O2 for 15 min at room temperature, washed
by PBS for 5 min at room temperature and blocked in 10% goat serum
(Beijing Solarbio Science & Technology Co., Ltd.) for 1 h at
room temperature. After washing for three times, the tissue
sections were incubated with primary antibodies against ASPM
(1:200; Abcam; cat. no. 238106), IgG isotype control (1:1,000; Cell
Signaling Technology, Inc.; cat. no. 3900), E-cadherin (1:400; Cell
Signaling Technology, Inc.; cat. no. 14472), N-cadherin (1:400;
Cell Signaling Technology, Inc.; cat. no. 13116) and Ki67 (1:400;
Cell Signaling Technology, Inc.; cat. no. 9027) in a wet box at 4°C
overnight. Subsequently, tissue sections were incubated with a
secondary antibody (1:200; Abcam; cat. no. 6721) for 2 h at room
temperature. After staining with DAB and H&E for 10 min at room
temperature, tissue sections were observed and photographed under a
light microscope (magnifications, ×100). The tissue sections were
scanned using a scanner to be scored using the nuclei quant
software (v3.1; 3DHISTECH Ltd.) according to previous studies
(26,27).
Western blotting
Protein was extracted from tissues and cells using
lysis buffer (Thermo Fisher Scientific, Inc.) with protease and
phosphatase inhibitors (Roche Diagnostics). A BCA kit (Shanghai
Yeasen Biotechnology Co., Ltd.; cat. no. B68010) was used to
determine the protein concentration. Protein extracts (20
µg/lane) were separated by 10% SDS-PAGE and transferred to
PVDF membranes (EMD Millipore). After blocking with 5% milk in
TBS-Tween (TBST; 0.1% Tween 20) for 1 h at room temperature,
membranes were incubated with primary antibodies against ASPM
(1:500; cat. no. sc-488883; Santa Cruz Biotechnology, Inc.), GAPDH
(used as a reference protein; 1:1,000; Cell Signaling Technology,
Inc.; cat. no. 5174), E-cadherin (1:1,000; Cell Signaling
Technology, Inc.; cat. no. 14472), N-cadherin (1:1,000; Cell
Signaling Technology, Inc.; cat. no. 13116), Snail (1:1,000; Cell
Signaling Technology, Inc.; cat. no. 3879), PI3K p110α (1:1,000;
Cell Signaling Technology, Inc.; cat. no. 4249), phosphorylated (p)
AKT (1:1,000; Cell Signaling Technology, Inc.; cat. no. 4060) and
AKT (1:1,000; Cell Signaling Technology, Inc.; cat. no. 4691)
overnight at 4°C. Subsequently, the membranes were incubated with
an HRP-conjugated secondary antibody (1:1,000; Cell Signaling
Technology, Inc.; cat. no. 7074) at room temperature for 1 h. After
washing with TBST 3 times, the proteins on the membranes were
visualized by enhanced chemiluminescence (EMD Millipore; cat. no.
WBKLS0010). The bands were visualized using the ChemiDoc™ MP
Imaging System (Bio-Rad Laboratories, Inc.). The gray value was
calculated using ImageJ v2.4.1 (National Institutes of Health). All
of the protein expression levels, except pAKT expression, were
calculated relative to GAPDH protein expression. pAKT expression
was calculated relative to AKT protein expression.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from cell lines using the
RNeasy Mini kit (Qiagen GmbH) and cDNA was synthesized using the
PrimeScript® RT Reagent Perfect Real Time kit (Takara
Biotechnology Co., Ltd.) according to the manufacturer's protocol.
The cDNA was subjected to qPCR using the LightCycler®
480 SYBR Green I Master (Roche Diagnostics) following the
manufacturer's instructions. GAPDH was used to normalize the amount
of cDNA between different samples. The thermocycling conditions
were as previously described (28,29): Initial denaturation at 95°C for 10
min, followed by 40 cycles of denaturation at 95°C for 15 sec,
annealing at 60°C for 15 sec and extension at 70°C for 20 sec. Data
analysis was performed using the 2−∆∆Cq method for
relative quantification (30).
All of the gene expression levels were calculated relative to the
GAPDH gene. Sequences of primers were as follows: ASPM forward,
5′-GGC CCT AGA CAA CCC TAA CGA-3′ and reverse, 5′-AGC TTG GTG TTT
CAG AAC ATC A-3′; and GAPDH forward, 5′-GGA GCG AGA TCC CTC CAA
AAT-3′ and reverse, 5′-GGC TGT TGT CAT ACT TCT CAT GG-3′.
PI3K/AKT pathway activator
740 Y-P was obtained from MedChemExpress. The
transfected LUAD cells were further treated with 740 Y-P (25
µg/ml) for 24 h at 37°C.
Animal experiments
All animal experiments were approved by the Medical
Ethics Committee of Shandong University (approval no.
KYLL-2016-097). A total of 18 male nude BalB/c mice (age, 6-8 weeks
old; weight, 20-22 g) were purchased from Beijing Vital River
Laboratory Animal Technology Co., Ltd. Animals were housed in
polycarbonate standard mouse cages. Housing conditions were kept
constant at a temperature of 18-21°C and humidity of 40-70%, with a
12-h light/dark cycle. Pelleted mouse food and water was provided
ad libitum. For the in vivo proliferation assay,
~1×106 H1299 cells (transfected with shASPM or shNC) in
100 µl PBS were subcutaneously injected into the left oxter
of nude mice (n=5 mice/group). After 7 days, the first measurements
of the length and width of tumors were made by caliper, and tumor
growth was measured for 17 days after this point. After the tumor
emerged, tumor volume was measured every three days, and tumor
growth curves were calculated and compared. For the in vivo
metastasis assay, 3×105 H1299 cells (transfected with
shASPM or control lentivirus) in 100 µl PBS were injected
into the caudal vein of each nude mice (n=4 mice/group). After 7
weeks, the mice were sacrificed by decapitation under anesthesia
with phenobarbital (100 mg/kg) intraperitoneal injection and the
number of nodules in the lungs was confirmed by hematoxylin and
eosin (H&E) staining and counted.
H&E staining
Tumor and lung tissues from mice were fixed in 4%
paraformaldehyde at 4°C for 24 h, embedded in paraffin and cut into
10-µm-thick sections. The sections were deparaffinized in
xylene, rehydrated in serially diluted ethanol and stained with
H&E (Sigma-Aldrich; Merck KGaA) at 4°C for 10 min.
Representative photomicrographs were captured using a light
microscope (Olympus Corporation; magnifications, ×100 and
×400).
Statistical analysis
Statistical analysis was performed using R software
(R version 3.6.1) and GraphPad Prism 6.0 (GraphPad Software, Inc.).
The median cut-off value was used to separate high- and
low-expression groups for Kaplan-Meier analysis. X-title software
(v1.0.4; http://kinzler.com/me/xtitle) was
used to select a suitable cut-off value to avoid crossover issues.
For comparisons, unpaired and paired Student's t-test (two-sided),
Pearson's χ2 test, Kaplan-Meier survival analysis with
log-rank test, and Fisher's exact test were performed as indicated.
One-way ANOVA was used to compare the differences among >2
groups followed by Tukey's multiple comparisons post-hoc test.
Univariate and multivariate Cox proportional-hazards regression
analyses were performed to evaluate factors affecting survival.
Data are presented as the mean ± SD. Each experiment was performed
in triplicate. P<0.05 was considered to indicate a statistically
significant difference.
Results
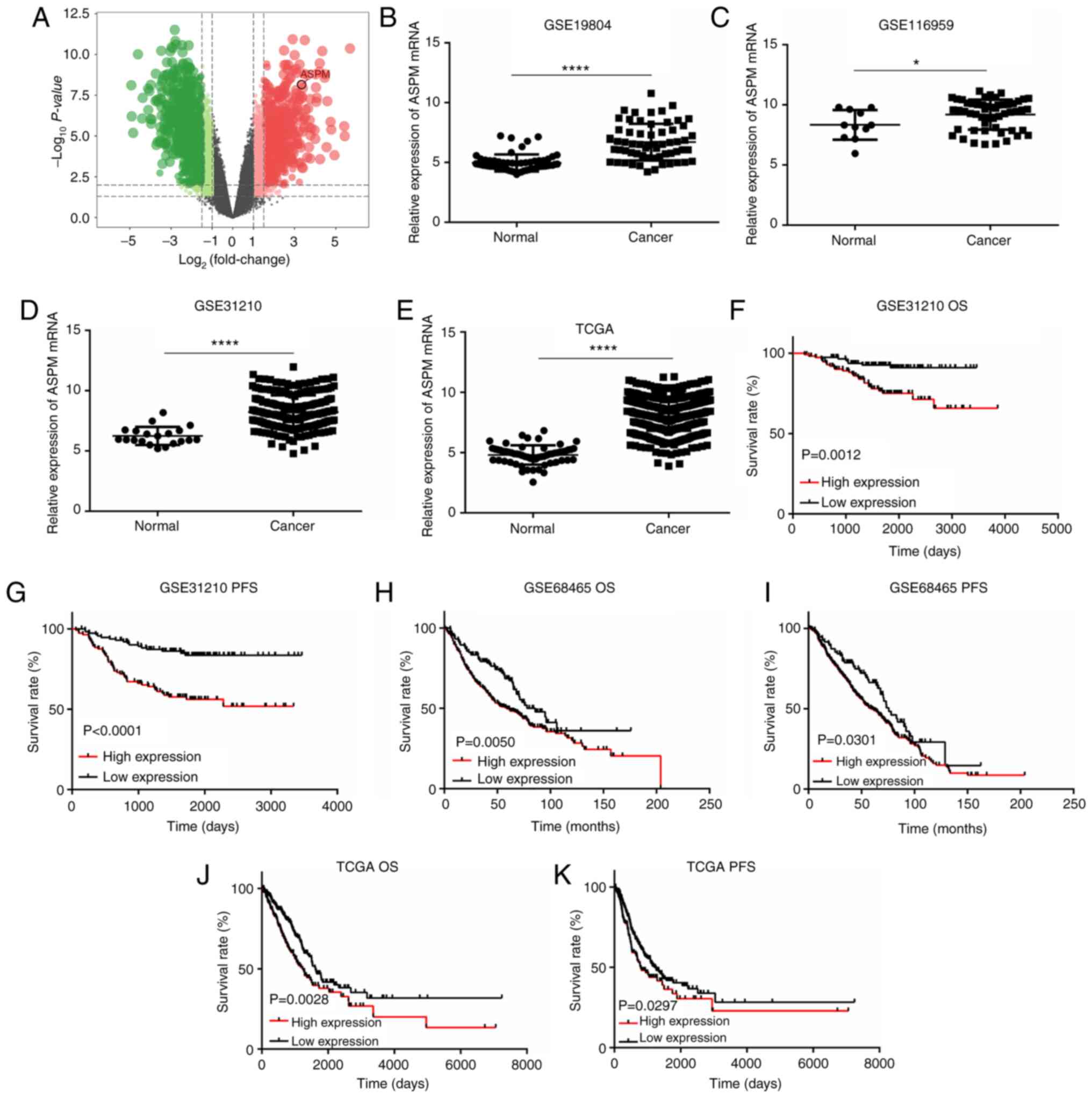
Bioinformatics analysis reveals
upregulated ASPM expression and predicts a poor prognosis in
patients with LUAD
First, seven pairs of frozen LUAD and adjacent
normal tissues were collected to perform transcriptome analysis to
screen for potential biomarkers (GSE140797). According to this
analysis, it was revealed that ASPM mRNA expression was
significantly increased in LUAD tissues compared with that in
adjacent normal tissues (Fig.
1A). To confirm this result, the GEO LUAD datasets (GSE19804,
GSE116959 and GSE31210) and TCGA LUAD dataset were analyzed via
bioinformatics analysis. Similarly, ASPM mRNA expression was
significantly upregulated in LUAD tissues compared with that in
normal lung tissues (Fig.
1B-E).
Subsequently, the association between
clinicopathological parameters and ASPM mRNA expression in patients
with LUAD was analyzed. The GSE68465 dataset was used as the
discovery cohort (Table I) and
TCGA dataset as the validation cohort (Table II). ASPM expression was
significantly associated with pathological tumor (pT) status
(discovery cohort, P<0.0001; validation cohort, P=0.0135), TNM
stage (discovery cohort, P=0.0001; validation cohort, P=0.0056) and
survival state (discovery cohort, P=0.0004; validation cohort,
P=0.0110) in both cohorts (Tables
I and II). Additionally,
ASPM expression was significantly associated with lymph node status
(P=0.0129) and differentiation status (P<0.0001) in the
discovery cohort, but not in the validation cohort, and with pM
status in the validation cohort (P=0.0221) (Tables I and II).
 | Table IAssociation between ASPM expression
and clinicopathological features in patients with lung
adenocarcinoma from the GSE68465 dataset (n=436). |
Table I
Association between ASPM expression
and clinicopathological features in patients with lung
adenocarcinoma from the GSE68465 dataset (n=436).
|
Characteristics | All cases, n
(%) | ASPM expression, n
| P-valuea |
|---|
| Low expression
(n=219) | High expression
(n=217) |
|---|
| Sex | | | | 0.0699 |
| Male | 220 (50.5) | 101 | 119 | |
| Female | 216 (49.5) | 118 | 98 | |
| Ageb | | | | 0.7746 |
| <65 | 208 (47.7) | 106 | 102 | |
| ≥65 | 228 (52.3) | 113 | 115 | |
| pT status | | | | <0.0001 |
| T1 | 148 (33.9) | 94 | 54 | |
| T2 | 248 (56.9) | 116 | 132 | |
| T3 | 28 (6.4) | 7 | 21 | |
| T4 | 12 (2.8) | 2 | 10 | |
| pN status | | | | 0.0129 |
| N0 | 296 (67.9) | 156 | 140 | |
| N1 | 79 (18.1) | 43 | 36 | |
| N2+N3 | 61 (14.0) | 20 | 41 | |
| TNM stage | | | | 0.0001 |
| I | 112 (25.7) | 73 | 39 | |
| II | 255 (58.5) | 123 | 132 | |
| III+IV | 69 (15.8) | 23 | 46 | |
|
Differentiation | | | | <0.0001 |
| Well | 60 (13.8) | 58 | 2 | |
| Medium | 209 (47.9) | 112 | 97 | |
| Poor | 167 (38.3) | 49 | 118 | |
| Event | | | | 0.0004 |
| Alive | 202 (46.3) | 120 | 82 | |
| Dead | 234 (53.7) | 99 | 135 | |
 | Table IIAssociation between ASPM expression
and clinicopathological features in patients with lung
adenocarcinoma in The Cancer Genome Atlas database (n=337). |
Table II
Association between ASPM expression
and clinicopathological features in patients with lung
adenocarcinoma in The Cancer Genome Atlas database (n=337).
|
Characteristics | All cases, n
(%) | ASPM expression, n
| P-valuea |
|---|
| Low expression
(n=168) | High expression
(n=169) |
|---|
| Sex | | | | 0.3841 |
| Male | 166 (49.3) | 87 | 79 | |
| Female | 171 (50.7) | 81 | 90 | |
| Ageb | | | | 0.5852 |
| <66 | 162 (48.1) | 78 | 84 | |
| ≥66 | 175 (51.9) | 90 | 85 | |
| pT status | | | | 0.0135 |
| T1 | 102 (30.3) | 64 | 38 | |
| T2 | 189 (56.1) | 82 | 107 | |
| T3 | 28 (8.3) | 15 | 13 | |
| T4 | 18 (5.3) | 7 | 11 | |
| pN status | | | | 0.0630 |
| N0 | 212 (62.9) | 116 | 96 | |
| N1 | 72 (21.4) | 29 | 43 | |
| N2+N3 | 53 (15.7) | 23 | 30 | |
| pM status | | | | 0.0221 |
| M0 | 316 (93.8) | 163 | 153 | |
| M1 | 21 (6.2) | 5 | 16 | |
| TNM stage | | | | 0.0056 |
| I | 172 (51.1) | 100 | 72 | |
| II | 83 (24.6) | 36 | 47 | |
| III | 61 (18.1) | 27 | 34 | |
| IV | 21 (6.2) | 5 | 16 | |
| Event | | | | 0.0110 |
| Alive | 202 (59.9) | 113 | 89 | |
| Dead | 137 (40.1) | 57 | 80 | |
Furthermore, Kaplan-Meier survival analysis
indicated that patients with higher expression levels of ASPM
exhibited a poorer survival rate both in OS and PFS (Fig. 1F-K; P<0.05) than those who had
lower expression levels of ASPM both in the GEO and TCGA datasets.
To further explore the prognostic significance of the mRNA
expression levels of ASPM in LUAD, univariate and multivariate Cox
proportional-hazards regression analyses were performed. Univariate
Cox regression analysis revealed that ASPM expression [hazard ratio
(HR), 1.49; 95% CI, 1.147-1.936; P=0.003 in discovery cohort; HR,
1.926; 95% CI, 1.364-2.718; P<0.001 in the validation cohort)
was a significant negative prognostic factor for OS (Tables III and IV). Multivariate analysis revealed that
ASPM expression was an independent prognostic factor of OS (HR,
1.323; 95% CI, 1.007-1.739; P=0.045 in the discovery cohort; HR,
1.725; 95% CI, 1.219-2.441; P=0.002 in the validation cohort)
(Tables III and IV). Both univariate and multivariate
Cox regression analyses revealed that age, pT stage, pN stage and
ASPM expression were significant prognostic factors for OS
(Table III). In TCGA cohort,
only pT stage and pN stage were significant prognostic factors for
OS, in addition to ASPM expression (Table IV).
 | Table IIIUnivariate and multivariate Cox
regression analysis of different prognostic factors in patients
with lung adenocarcinoma from the GSE68465 dataset. |
Table III
Univariate and multivariate Cox
regression analysis of different prognostic factors in patients
with lung adenocarcinoma from the GSE68465 dataset.
|
Characteristics | Sample, n (%) | Univariate
analysisa
| Multivariate
analysisa
|
|---|
| HR | 95% CI | P-valueb | HR | 95% CI | P-valueb |
|---|
| Agec | | 1.544 | 1.191-2.002 | 0.001 | 1.571 | 1.210-2.040 | <0.001 |
| ≤65 | 224 (51.6) | | | | | | |
| >65 | 210 (48.4) | | | | | | |
| Sex | | 1.436 | 1.107-1.863 | 0.006 | 1.231 | 0.943-1.607 | 0.127 |
| Female | 215 (49.5) | | | | | | |
| Male | 219 (50.5) | | | | | | |
| ASPM
expression | | 1.490 | 1.147-1.936 | 0.003 | 1.323 | 1.007-1.739 | 0.045 |
| Low
expression | 217 (50.0) | | | | | | |
| High
expression | 217 (50.0) | | | | | | |
| T stage | | 1.652 | 1.376-1.983 | <0.001 | 1.374 | 1.130-1.671 | 0.001 |
| T1 | 147 (33.9) | | | | | | |
| T2 | 248 (57.1) | | | | | | |
| T3 | 28 (6.5) | | | | | | |
| T4 | 11 (2.5) | | | | | | |
| N stage | | 2.012 | 1.710-2.368 | <0.001 | 1.987 | 1.687-2.34 | <0.001 |
| N1 | 296 (68.2) | | | | | | |
| N2 | 86 (19.8) | | | | | | |
| N3 | 52 (12.0) | | | | | | |
| Histological
stage | | 1.135 | 0.934-1.379 | 0.204 | | | |
| Well | 60 (13.8) | | | | | | |
| Moderate | 208 (47.9) | | | | | | |
| Poor | 166 (38.2) | | | | | | |
 | Table IVUnivariate and multivariate Cox
regression analysis of different prognostic factors in patients
with lung adenocarcinoma from The Cancer Genome Atlas database. |
Table IV
Univariate and multivariate Cox
regression analysis of different prognostic factors in patients
with lung adenocarcinoma from The Cancer Genome Atlas database.
|
Characteristics | Sample, n (%) | Univariate
analysisa
| Multivariate
analysisa
|
|---|
| HR | 95% CI | P-valueb | HR | 95% CI | P-valueb |
|---|
| Aged | | 1.086 | 0.774-1.523 | 0.635 | | | |
| <66 | 162 (48.1) | | | | | | |
| ≥66 | 175 (51.9) | | | | | | |
| Sex | | 0.923 | 0.660-1.292 | 0.641 | | | |
| Male | 166 (49.3) | | | | | | |
| Female | 171 (50.7) | | | | | | |
| ASPM
expression | | 1.926 | 1.364-2.718 | <0.001 | 1.725 | 1.219-2.441 | 0.002 |
| Low
expression | 168 (49.9) | | | | | | |
| High
expression | 169 (50.1) | | | | | | |
| TNM stage | | 1.590 | 1.360-1.860 | <0.001 | 1.235 | 0.969-1.574 | 0.088 |
| I | 172 (51) | | | | | | |
| II | 83 (24.6) | | | | | | |
| III | 61 (18.1) | | | | | | |
| IV | 21 (6.2) | | | | | | |
| T stage | | 1.617 | 1.330-1.967 | <0.001 | 1.310 | 1.05-1.635 | 0.017 |
| T1 | 102 (30.3) | | | | | | |
| T2 | 189 (56.1) | | | | | | |
| T3 | 28 (8.3) | | | | | | |
| T4 | 18 (5.3) | | | | | | |
| N stage | | 1.804 | 1.473-2.208 | <0.001 | 1.346 | 1.024-1.769 | 0.033 |
| N0 | 212 (62.9) | | | | | | |
| N1 | 72 (21.4) | | | | | | |
| N2+N3 | 53 (15.7) | | | | | | |
| M stage | | 1.849 | 1.042-3.283 | 0.036 | | | NAc |
| M0 | 316 (93.8) | | | | | | |
| M1 | 21 (6.2) | | | | | | |
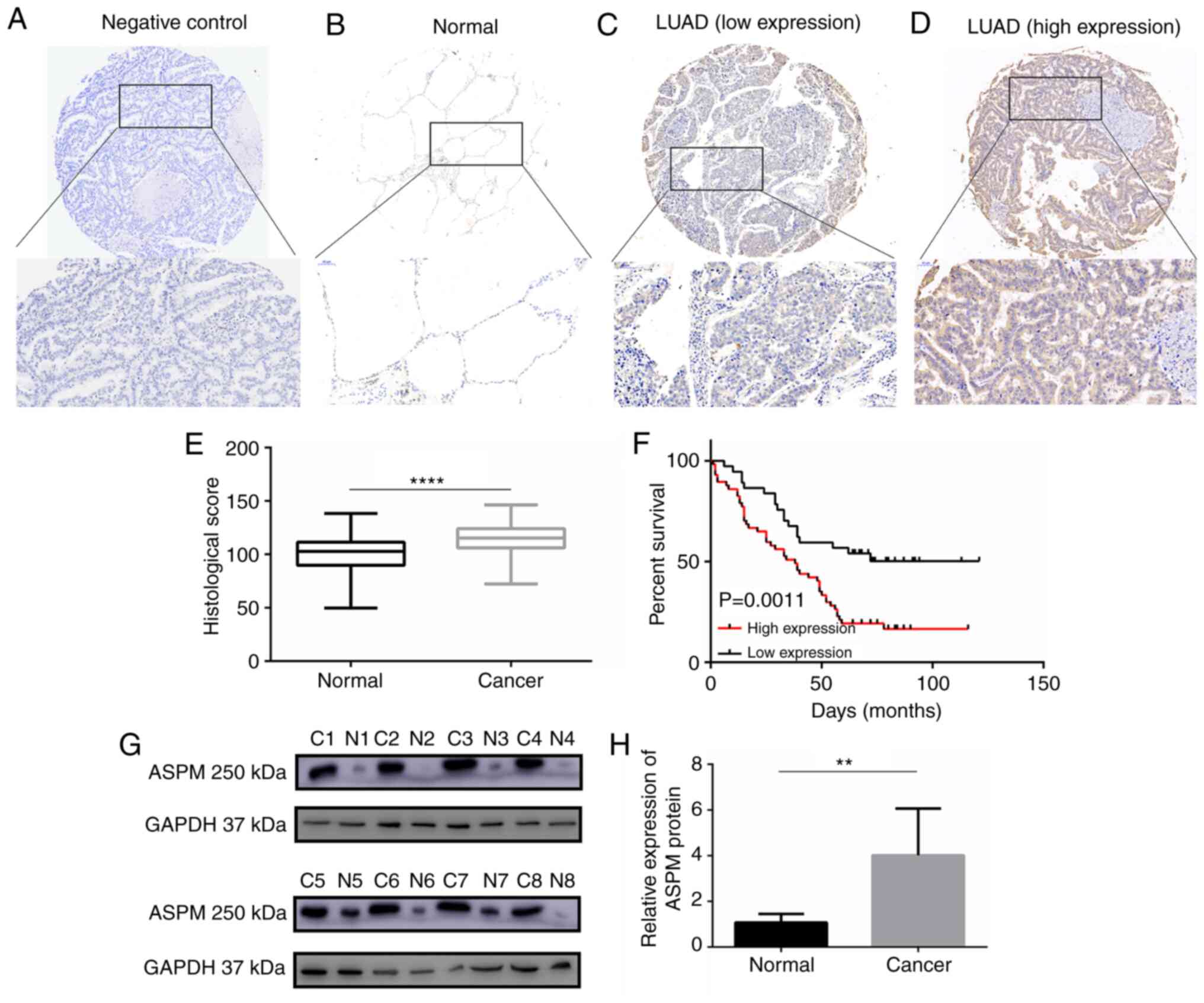
Clinical specimens confirm that high ASPM
expression is associated with a poor prognosis in patients with
LUAD
Subsequently, to investigate the expression pattern
of ASPM protein in LUAD, IHC staining we performed to analyze ASPM
protein expression in 94 paired tissues of LUAD collected from
patients at Qilu Hospital (Fig.
2A-D). IHC staining revealed that ASPM protein was localized in
the cytoplasm. A scoring system indicated that ASPM protein
expression was significantly higher in LUAD tissues than in
adjacent normal tissues (Fig.
2E). Similarly, western blotting confirmed that ASPM protein
expression was increased in eight paired LUAD tissues compared with
in their adjacent non-tumor tissues (Fig. 2G and H). The associations between
ASPM expression and clinicopathological parameters are presented in
Table V, indicating that ASPM
expression was significantly associated with pathological lymph
node (pN) status (P=0.041), TNM stage (P=0.027) and survival status
(P=0.001). However, ASPM expression was not significantly
associated with sex, age, pT status, pathological metastasis (pM)
status and pathological stage (Table
V). Additionally, Kaplan-Meier curve analysis demonstrated a
significant negative association between ASPM expression and OS
(P=0.0011; Fig. 2F).
 | Table VAssociation between ASPM expression
and clinicopathological characteristics in patients with lung
adenocarcinoma (n=94). |
Table V
Association between ASPM expression
and clinicopathological characteristics in patients with lung
adenocarcinoma (n=94).
|
Characteristics | All cases, n
(%) | ASPM expression
| P-valuea |
|---|
| Low expression
(n=37) | High expression
(n=57) |
|---|
| Sex | | | | 0.786 |
| Male | 53 (56.4) | 22 | 31 | |
| Female | 41 (43.6) | 15 | 26 | |
| Ageb | | | | 0.304 |
| <60 | 43 (45.7) | 14 | 29 | |
| ≥60 | 51 (54.3) | 23 | 28 | |
| pT status | | | | 0.762 |
| T1 | 20 (21.3) | 8 | 12 | |
| T2 | 50 (53.2) | 21 | 29 | |
| T3 | 18 (19.1) | 7 | 11 | |
| T4 | 6 (6.4) | 1 | 5 | |
| pN status | | | | 0.041 |
| N0 | 42 (44.7) | 21 | 21 | |
| N1 | 17 (18.1) | 8 | 9 | |
| N2+N3 | 35 (37.2) | 8 | 27 | |
| pM status | | | | >0.999 |
| M0 | 93 (98.9) | 37 | 56 | |
| M1 | 1 (1.1) | 0 | 1 | |
| TNM stage | | | | 0.027 |
| I | 30 (31.9) | 16 | 14 | |
| II | 20 (21.3) | 10 | 19 | |
| III+IV | 44 (46.8) | 11 | 33 | |
| Pathological
stage | | | | 0.280 |
| I | 11 (11.7) | 6 | 5 | |
| II | 52 (55.3) | 17 | 35 | |
| III | 31 (33.0) | 14 | 17 | |
| Event | | | | 0.001 |
| Alive | 29 (30.9) | 19 | 10 | |
| Dead | 65 (69.1) | 18 | 47 | |
ASPM promotes carcinogenesis of LUAD in
vitro and in vivo
Considering that high ASPM expression was associated
with poor survival outcomes in patients with LUAD, the function of
high ASPM expression in the biological behavior of LUAD cells was
further investigated. The baseline expression levels of endogenous
ASPM were evaluated in a panel of six cell lines (HBE, HCC827,
A549, H157, H1299 and H1975) using RT-qPCR and western blotting
(Fig. S1A and B). The H1299 cell
line demonstrated the highest endogenous ASPM expression and was
thus selected as the cell model for ASPM-knockdown. The ASPM gene
was knocked down using RNA interference (RNAi) by introducing two
targeted siRNAs (siRNA1 and siRNA2) in H1299 cells. The knockdown
efficiency was verified using RT-qPCR, revealing that both siRNAs
significantly knocked down ASPM expression (Fig. 3A). CCK-8 and EDU assays were used
to monitor cell proliferation, and the results indicated that LUAD
cells with silenced ASPM exhibited significantly impaired
proliferation (Fig. 3B and C).
Additionally, ASPM deficiency significantly attenuated wound
healing ability compared with that in control cells (Fig. 3D). Moreover, ASPM silencing
significantly suppressed migration and invasion of H1299 cells
(Fig. 3D). To validate whether
ASPM silencing affected epithelial-mesenchymal transition (EMT) in
LUAD cells, western blotting was performed. The current results
revealed that ASPM-silenced cells exhibited increased protein
expression levels of the epithelial marker E-cadherin, while the
expression levels of the mesenchymal marker proteins N-cadherin and
Snail were decreased compared with those in the control group
(Figs. 3E and S2). Overall, the present results
suggested that ASPM may exert a crucial role in the proliferation,
migration and invasion of LUAD cells.
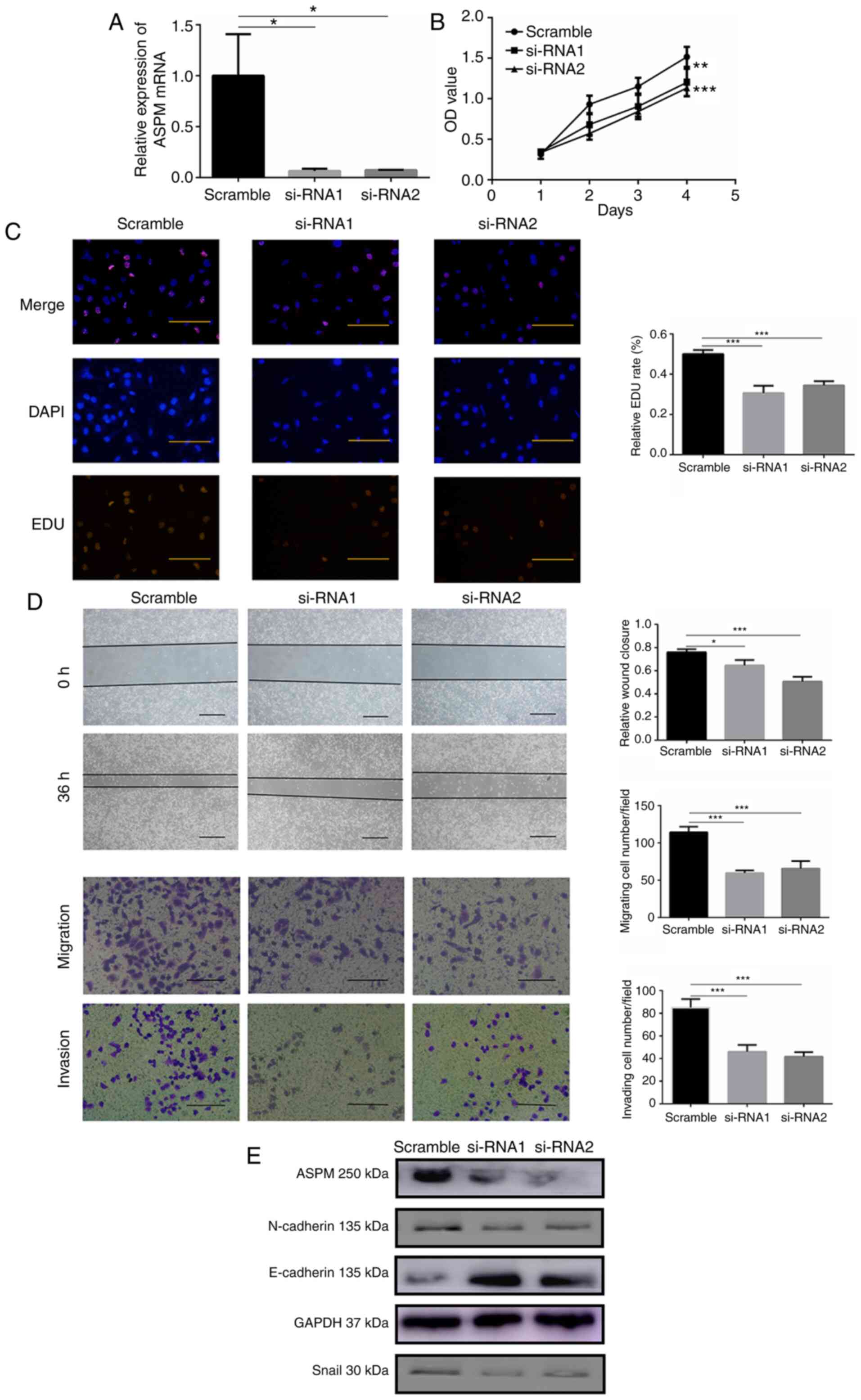 | Figure 3ASPM promotes lung adenocarcinoma
cell proliferation, migration and invasion in vitro. (A)
Reverse transcription-quantitative PCR was used to confirm
ASPM-knockdown using two siRNAs in H1299 cells. (B) Cell Counting
Kit-8 and (C) EDU assays were performed to identify proliferation
after ASPM-knockdown in H1299 cells (scale bar, 100 µm). (D)
Wound healing (scale bar, 100 µm), migration and invasion
(scale bar, 50 µm) assays were performed to identify the
migratory and invasive abilities after ASPM-knockdown in H1299
cells. (E) Changes in the expression levels of the
epithelial-mesenchymal transition biomarkers E-cadherin, N-cadherin
and Snail after ASPM-knockdown were detected by western blotting.
Data are presented as the mean ± SD (n=3). *P<0.05,
**P<0.01 and ***P<0.001 vs. scramble.
ASPM, abnormal spindle-like microcephaly; si-RNA, small interfering
RNA; OD, optical density; EDU, 5-Ethynyl-2′-deoxyuridine. |
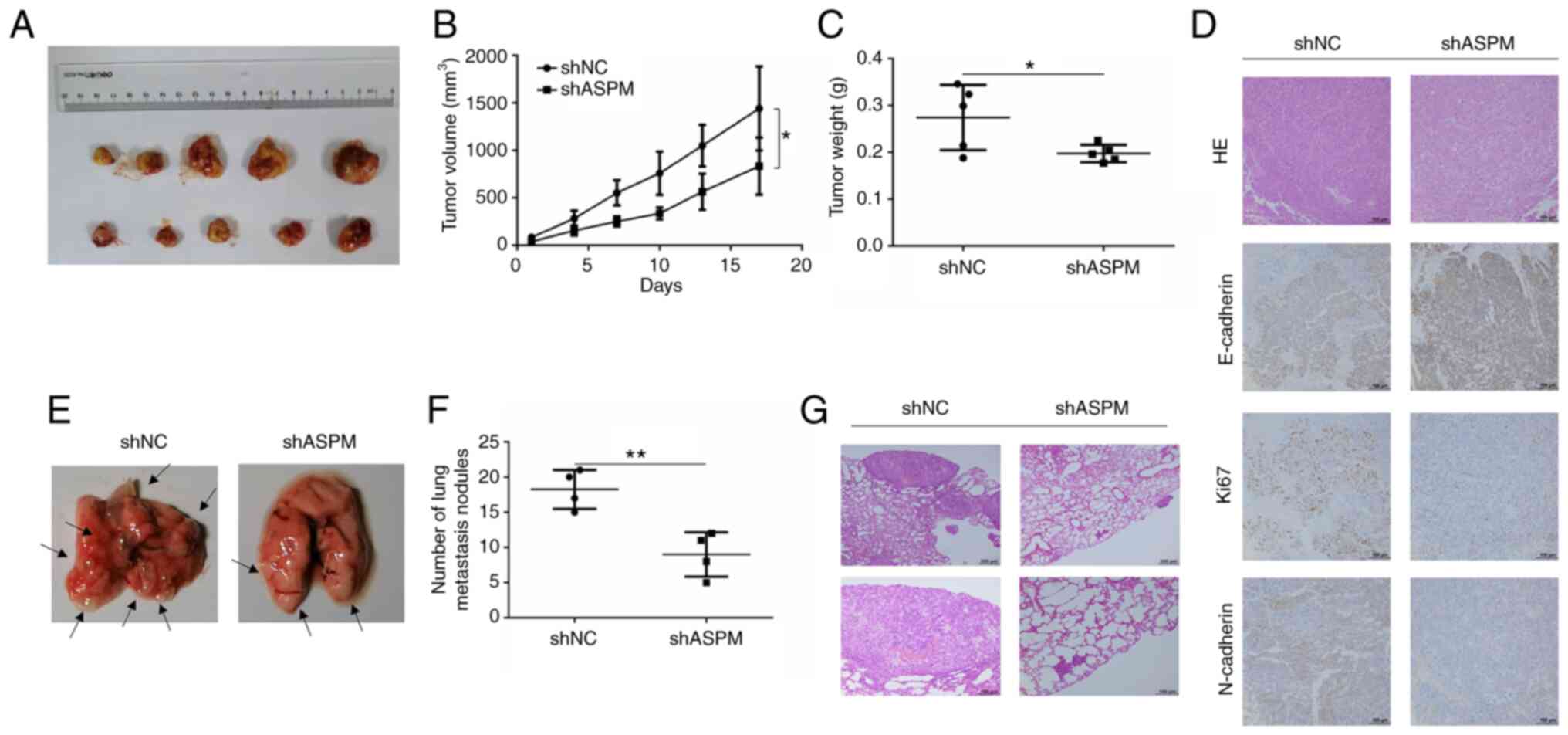
Subsequently, animal experiments were performed to
evaluate the oncogenic effect of ASPM. Stable ASPM-knockdown cells
(H1299 and A549) were established via lentiviral transfection using
a shRNA against ASPM, and RT-qPCR results indicated that shASPM
significantly inhibited ASPM expression in both cell lines
(Fig. S1C and D). H1299 cells
were injected subcutaneously to establish the xenograft tumor model
in nude mice (Fig. 4A). The mice
that were transplanted with ASPM-knockdown H1299 cells exhibited a
significant inhibition of tumor growth and reduction in tumor
weight (Fig. 4B and C).
Furthermore, the depletion of ASPM decreased the expression levels
of Ki67 and N-cadherin, and increased those of E-cadherin compared
with the control group (Fig. 4D).
Additionally, metastasis experiments in BALB/c nude mice were
performed using shASPM-transfected H1299 cells. H1299 cells were
injected into BALB/c nude mice through the caudal vein, and
metastasis lesions were evaluated 7 weeks after injection (Fig. 4E). The results revealed that
ASPM-knockdown significantly decreased the number of metastatic
lesions in the lung (Fig. 4F).
Tumor metastasis was confirmed using hematoxylin and eosin staining
(Fig. 4G). Therefore, both in
vitro and in vivo data supported the proliferative and
metastatic effects of ASPM in LUAD.
ASPM-knockdown suppresses the PI3K/AKT
signaling pathway
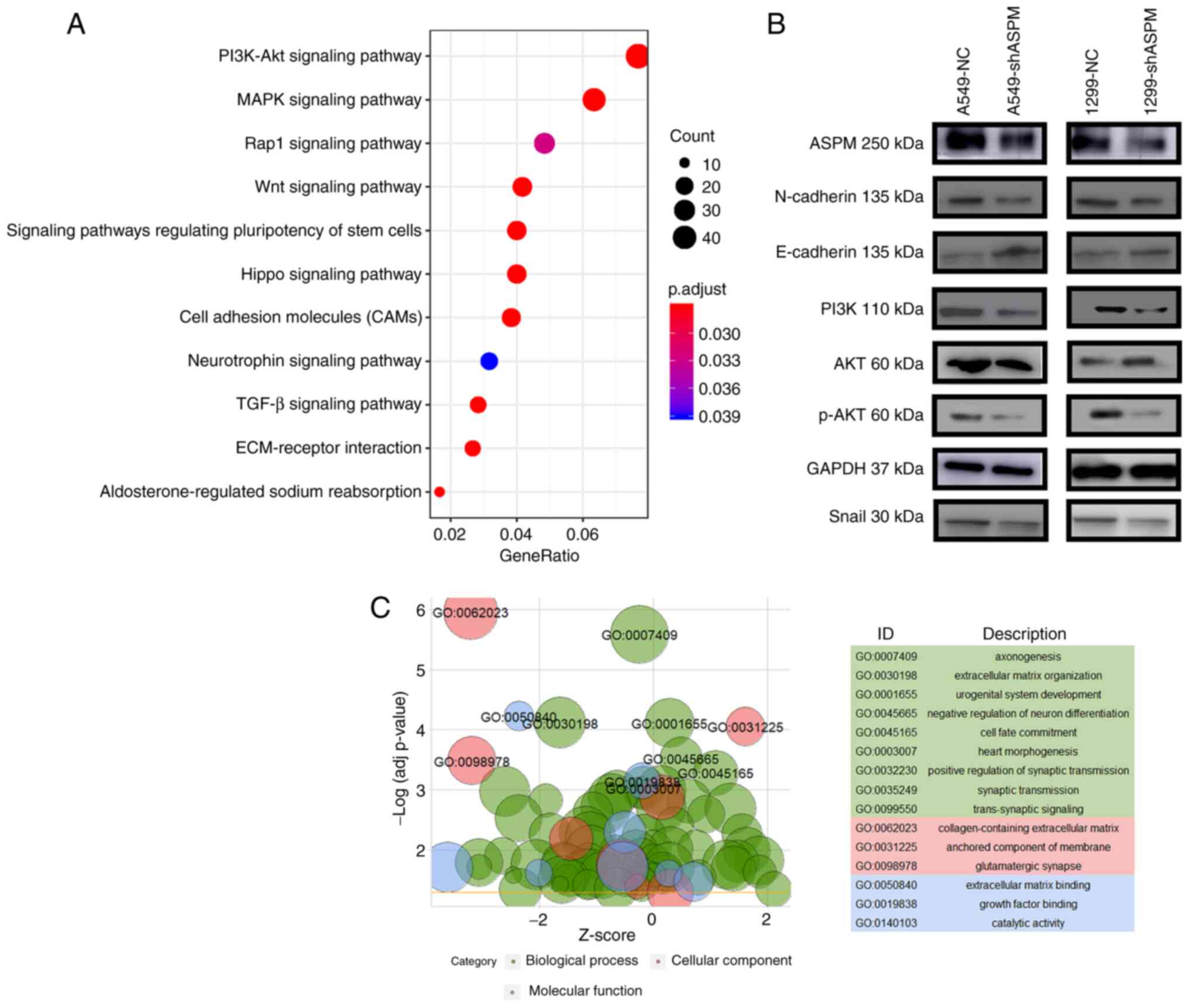
In order to explore the molecular mechanism of ASPM
in LUAD, transcriptome sequencing was performed in
shASPM-transfected H1299 and control cells. KEGG analysis indicated
that ASPM-knockdown had important effects on genes that were mainly
associated with the PI3K/AKT, mitogen-activated protein kinase
(MAPK), Ras-proximate-1 (Rap1), Wnt, Hippo and stem cell signaling
pathways, as well as with cell adhesion molecules (CAMs) and
extracellular matrix (ECM)-receptor interaction (Fig. 5A). GO revealed genes involved in
'extracellular matrix organization' (GO:0030198),
'collagen-containing extracellular matrix' (GO:0062023), 'anchored
component of membrane' (GO:0031225) and 'extracellular matrix
binding' (GO:0050840) (Fig. 5C).
It is well known that activation of the PI3K/AKT signaling pathway
can lead to EMT in cancer and that the extracellular matrix
organization is altered to promote metastasis during EMT (31,32). Therefore, it was hypothesized that
ASPM promoted LUAD metastasis by activating EMT via the PI3K/AKT
signaling pathway. Western blotting results further proved that
ASPM-knockdown led to increased protein expression levels of the
epithelial marker E-cadherin and decreased protein expression
levels of the mesenchymal marker proteins N-cadherin and Snail, and
molecules in PI3K signaling pathway (PI3K and pAKT) in both H1299
and A549 cells (Figs. 5B and
S3).
Activation of the PI3K/AKT signaling
pathway can restore the migration and invasion of
shASPM-transfected LUAD cells
The role of the PI3K/AKT signaling pathway in LUAD
metastasis caused by ASPM was further investigated. A specific
activator of the PI3K/AKT signaling pathway, 740 Y-P, was used to
activate PI3K activity after ASPM-knockdown. Consequently,
ASPM-knockdown inhibited EMT (leading to decreased N-cadherin and
Snail expression, and increased E-cadherin expression) in H1299 and
A549 cells, whereas 740 Y-P treatment partially restored these
changes (Figs. 6A and S4). Moreover, 740 Y-P treatment
significantly reversed the inhibition of H1299 and A546 cell
invasion and migration induced by ASPM-knockdown (Fig. 6B and C), suggesting that the
PI3K/AKT signaling pathway was the key effector of the
ASPM-mediated invasion and migration.
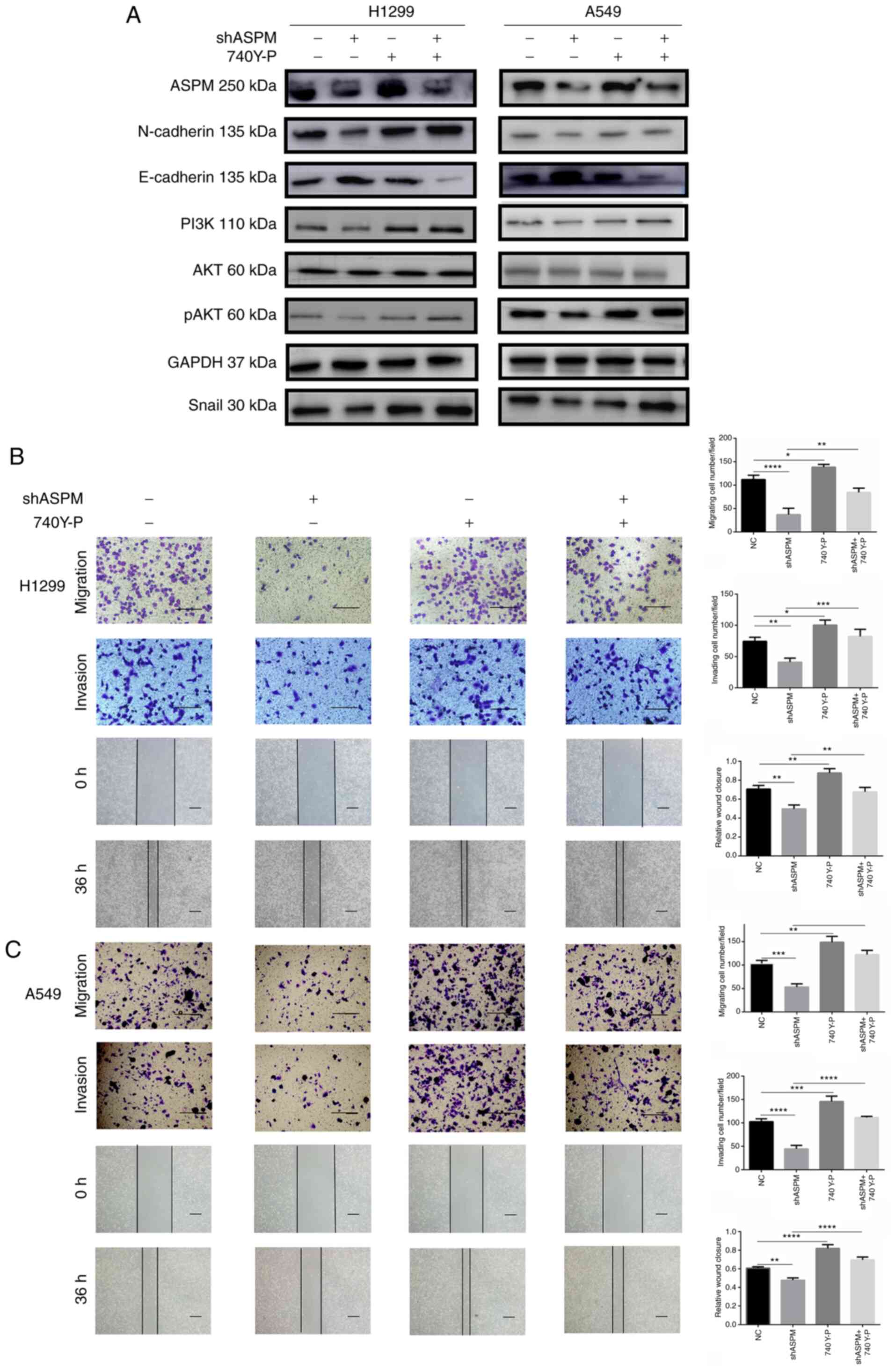 | Figure 6Activation of the PI3K/AKT signaling
pathway restores the migratory and invasive abilities of
shASPM-transfected lung adenocarcinoma cells. (A) Changes in
epithelial-mesenchymal transition biomarker expression, including
N-cadherin, E-cadherin and Snail, were detected in ASPM-knockdown
H1299 and A549 cells with or without 740 Y-P treatment for 24 h.
Wound healing (scale bar, 100 µm), migration and invasion
assays (scale bar, 50 µm) in ASPM-knockdown (B) H1299 and
(C) A549 cells with or without 740 Y-P treatment for 24 h. Data are
presented as the mean ± SD (n=3). *P<0.05;
**P<0.01; ***P<0.001;
****P<0.0001. ASPM, abnormal spindle-like
microcephaly; sh, short hairpin RNA; NC, negative control; p,
phosphorylated. |
Discussion
In the present study, the clinical and oncogenic
role of ASPM in LUAD was explored. LUAD is one of the deadliest
tumors worldwide (33). Due to
the lack of effective diagnostic and prognostic evaluation
biomarkers, the morbidity and mortality rates in patients with LUAD
are on the rise (33,34). Identifying new molecular markers
that can detect recurrence and metastasis, guide prognosis and
improve survival rates in patients with LUAD are urgently required.
Although some studies reported a polygene model that included the
ASPM-predicted prognosis in LUAD (35,36), to the best of our knowledge the
present study was the first to employ the GEO database as a
discovery cohort and TCGA as a validation cohort to address the
role of ASPM in LUAD. The current study revealed that ASPM mRNA
expression was upregulated in LUAD tissues, and the upregulation of
ASPM expression independently predicted a poor prognosis in
patients with LUAD. Furthermore, higher ASPM protein expression in
LUAD was associated with disease progression, indicating that ASPM
expression may be used as a potential biomarker. Similarly, ASPM
expression is elevated in multiple types of cancer, including
bladder cancer, pancreatic cancer (37), ovarian cancer (15), breast cancer (38), endometrial cancer (39), glioblastoma (12,40), hepatocellular carcinoma (16) and lung squamous carcinoma
(41), where it confers a poor
prognosis.
A series of in vitro and in vivo
assays were performed in the present study to illustrate the
biological function of ASPM in regulating LUAD cell proliferation
and migration. ASPM-knockdown markedly attenuates the proliferation
and migration of cells in prostate cancer (13), endometrial cancer (39), pancreatic cancer (37), glioma (40) and lung squamous carcinoma
(41). Overall, the findings of
the current study indicated that ASPM served a critical role in
LUAD cell proliferation and migration.
Accumulating evidence has revealed that EMT
participates in the invasion and early metastasis in numerous types
of tumor by inducing the expression of mesenchymal markers and
decreasing that of epithelial markers (42,43). A number of key transcription
factors, including Twist, Snail and Slug, have been considered as
major drivers of the EMT program (31). In the present study,
ASPM-knockdown had a significant negative influence on EMT by
suppressing the expression levels of mesenchymal markers
(N-cadherin and Snail) and inducing the expression levels of the
epithelial marker E-cadherin. Additionally, in vivo assays
indicated upregulation of E-cadherin expression and downregulation
of N-cadherin expression in the ASPM-knockdown group. Thus,
ASPM-knockdown may affect LUAD cell migration and invasion by
interfering with the EMT process.
The activation of EMT depends on a number of
oncogenic signaling pathways, including the intracellular PI3K/AKT
signaling pathway (44). The
current results demonstrated that the PI3K/AKT signaling pathway
was associated with ASPM-induced EMT. A previous study suggested
that the activation of the PI3K/AKT signaling pathway led to LUAD
tumorigenesis, including increased cell proliferation, metastasis
and drug resistance (45).
Phosphorylated AKT can regulate pivotal EMT genes by inducing key
transcription factors, including Snail, that affect the progression
of cancer (44). The present
results indicated that ASPM-knockdown affected the expression
levels of PI3K and phosphorylated AKT. Moreover, the attenuation
effect of ASPM-knockdown on LUAD cell migration and invasion, and
the ASPM-induced EMT was markedly restored when the PI3K/AKT
signaling pathway was activated using 740 Y-P. Therefore, the
mechanism of ASPM-induced promotion of EMT may be mediated through
the activation of the PI3K/AKT signaling pathway, ultimately
driving LUAD cell migration and invasion. However, the specific
mechanisms by which ASPM activates PI3K/AKT should be further
explored. As initial suggestions for possible mechanisms, the
current RNA-sequencing results revealed that ASPM-knockdown had
important effects on genes that are mainly associated with the
MAPK, Rap1, Wnt, Hippo and stem cell signaling pathways, as well as
CAMs and ECM-receptor interaction. This is in agreement with
previous studies indicating that ASPM is involved in the regulation
of cancer stem cells and promotes Wnt pathway activity by
stabilizing the Dvl-3 protein (8,46,47). To the best of our knowledge, the
present study was the first to demonstrate that ASPM promoted
activation of the PI3K/AKT signaling pathway. However, a direct
association between ASPM and the PI3K/AKT signaling pathway was not
observed. Future studies should focus on the mechanism by which
ASPM activates the PI3K/AKT signaling pathway and the association
between ASPM and other signaling pathways, including MAPK, Rap1 and
Hippo.
In conclusion, the current study first demonstrated
that ASPM was highly expressed in LUAD tissues and was an
independent biomarker for predicting prognosis in patients with
LUAD. ASPM-knockdown in LUAD inhibited proliferation, migration and
invasion in LUAD cells. Furthermore, functional and mechanistic
studies revealed that ASPM regulated EMT to promote LUAD metastasis
through the activation of the PI3K/AKT signaling pathway.
Therefore, the current findings provided a theoretical basis for
the biological function of ASPM in LUAD and indicated that ASPM may
serve as a clinically relevant biomarker and a potential
therapeutic target in LUAD.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The datasets generated and/or analyzed during the current
study (GSE140797, GSE19804, GSE116959, GSE31210 and GSE68465) are
available in the GEO repository (GSE140797, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140797;
GSE19804, https://www.ncbi.nlm.nih.gov/gds/?term=GSE19804;
GSE116959, https://www.ncbi.nlm.nih.gov/gds/?term=GSE116959;
GSE31210, https://www.ncbi.nlm.nih.gov/gds/?term=GSE31210; and
GSE68465, https://www.ncbi.nlm.nih.gov/gds/?term=GSE68465)
Authors' contributions
JW and HT designed the study. JW, JL, HL, JH, JJ,
YL, ZF and RZ performed the experiments. JW and JL performed the
statistical analysis. JW wrote the manuscript and HT revised the
manuscript. JW and JL are responsible for confirming the
authenticity of the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All experiments were approved and supervised by the
Medical Ethics Committee of Qilu Hospital of Shandong University
(Jinan, China; approval no. KYLL-2016-097). Patients provided
written informed consent for the use of their samples in the study.
All animal experiments were approved by the Medical Ethics
Committee of Shandong University (approval no. KYLL-2016-097).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 81802397 and 81672292) and the
Taishan Scholar Program of Shandong Province (grant no.
ts201712087).
Abbreviations:
|
LUAD
|
lung adenocarcinoma
|
|
ASPM
|
abnormal spindle-like microcephaly
|
|
EMT
|
epithelial-mesenchymal transition
|
|
CCK-8
|
Cell Counting Kit-8
|
|
EDU
|
5-Ethynyl-2′-deoxyuridine
|
|
TCGA
|
The Cancer Genome Atlas
|
|
IHC
|
immunohistochemistry
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
OS
|
overall survival
|
|
MAPK
|
mitogen-activated protein kinase
|
|
RAP1
|
Ras-proximate-1
|
|
CAM
|
cell adhesion molecule
|
|
GO
|
Gene Ontology
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Genome Atlas and Research Network:
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar
|
|
3
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar
|
|
4
|
Borczuk AC: Prognostic considerations of
the new world health organization classification of lung
adenocarcinoma. Eur Respir Rev. 25:364–371. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhong X, Liu L, Zhao A, Pfeifer GP and Xu
X: The abnormal spindle-like, microcephaly-associated (ASPM) gene
encodes a centrosomal protein. Cell Cycle. 4:1227–1229. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kouprina N, Pavlicek A, Mochida GH,
Solomon G, Gersch W, Yoon YH, Collura R, Ruvolo M, Barrett JC,
Woods CG, et al: Accelerated evolution of the ASPM gene controlling
brain size begins prior to human brain expansion. PLoS Biol.
2:E1262004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Passemard S, Titomanlio L, Elmaleh M,
Afenjar A, Alessandri JL, Andria G, Billette de Villemeur T,
Boespflug-Tanguy O, Burglen L, Del Giudice E, et al: Expanding the
clinical and neuroradiologic phenotype of primary microcephaly due
to ASPM mutations. Neurology. 73:962–969. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Capecchi MR and Pozner A: ASPM regulates
symmetric stem cell division by tuning Cyclin E ubiquitination. Nat
Commun. 6:87632015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gai M, Bianchi FT, Vagnoni C, Verni F,
Bonaccorsi S, Pasquero S, Berto GE, Sgrò F, Chiotto AA, Annaratone
L, et al: ASPM and CITK regulate spindle orientation by affecting
the dynamics of astral microtubules. EMBO Rep. 18:18702017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang K, Rezabkova L, Hua S, Liu Q,
Capitani G, Altelaar AFM, Heck AJR, Kammerer RA, Steinmetz MO and
Akhmanova A: Microtubule minus-end regulation at spindle poles by
an ASPM-katanin complex. Nat Cell Biol. 19:480–492. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
An X, Huang Y and Zhao P: Expression of
ASPM in colonic adenocarcinoma and its clinicopathologic
significance. Int J Clin Exp Pathol. 10:8968–8973. 2017.PubMed/NCBI
|
|
12
|
Chen X, Huang L, Yang Y, Chen S, Sun J, Ma
C, Xie J, Song Y and Yang J: ASPM promotes glioblastoma growth by
regulating G1 restriction point progression and Wnt-β-catenin
signaling. Aging (Albany NY). 12:224–241. 2020. View Article : Google Scholar
|
|
13
|
Xie JJ, Zhuo YJ, Zheng Y, Mo RJ, Liu ZZ,
Li BW, Cai ZD, Zhu XJ, Liang YX, He HC and de Zhong W: High
expression of ASPM correlates with tumor progression and predicts
poor outcome in patients with prostate cancer. Int Urol Nephrol.
49:817–823. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Timaner M and Shaked Y: Elucidating the
roles of ASPM isoforms reveals a novel prognostic marker for
pancreatic cancer. J Pathol. 250:123–125. 2020. View Article : Google Scholar
|
|
15
|
Alsiary R, Bruning-Richardson A, Bond J,
Morrison EE, Wilkinson N and Bell SM: Deregulation of microcephalin
and ASPM expression are correlated with epithelial ovarian cancer
progression. PLoS One. 9:e970592014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin SY, Pan HW, Liu SH, Jeng YM, Hu FC,
Peng SY, Lai PL and Hsu HC: ASPM is a novel marker for vascular
invasion, early recurrence, and poor prognosis of hepatocellular
carcinoma. Clin Cancer Res. 14:4814–4820. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu Z, Zhang Q, Luh F, Jin B and Liu X:
Overexpression of the ASPM gene is associated with aggressiveness
and poor outcome in bladder cancer. Oncol Lett. 17:1865–1876.
2019.PubMed/NCBI
|
|
18
|
Chansky K, Detterbeck FC, Nicholson AG,
Rusch VW, Vallieres E, Groome P, Kennedy C, Krasnik M, Peake M,
Shemanski L, et al: The IASLC lung cancer staging project: External
validation of the revision of the TNM stage groupings in the eighth
edition of the TNM classification of lung cancer. J Thorac Oncol.
12:1109–1121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang J, Li H, Han J, Jiang J, Wang J, Li
Y, Feng Z, Zhao R, Sun Z, Lv B and Tian H: Mex3a interacts with
LAMA2 to promote lung adenocarcinoma metastasis via PI3K/AKT
pathway. Cell Death Dis. 11:6142020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC,
Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC and Chuang EY:
Identification of a novel biomarker, SEMA5A, for non-small cell
lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers
Prev. 19:2590–2597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moreno Leon L, Gautier M, Allan R, Ilie M,
Nottet N, Pons N, Paquet A, Lebrigand K, Truchi M, Fassy J, et al:
The nuclear hypoxia-regulated NLUCAT1 long non-coding RNA
contributes to an aggressive phenotype in lung adenocarcinoma
through regulation of oxidative stress. Oncogene. 38:7146–7165.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar
|
|
23
|
Director's Challenge Consortium for the
Molecular Classification of Lung Adenocarcinoma; Shedden K, Taylor
JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S,
Jurisica I, Giordano TJ, et al: Gene expression-based survival
prediction in lung adenocarcinoma: A multi-site, blinded validation
study. Nat Med. 14:822–827. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar
|
|
26
|
Yeo W, Chan SL, Mo FK, Chu CM, Hui JW,
Tong JH, Chan AWH, Koh J, Hui EP, Loong H, et al: Phase I/II study
of temsirolimus for patients with unresectable hepatocellular
carcinoma (HCC)-a correlative study to explore potential biomarkers
for response. BMC Cancer. 15:3952015. View Article : Google Scholar
|
|
27
|
Azim HA Jr, Peccatori FA, Brohee S,
Branstetter D, Loi S, Viale G, Piccart M, Dougall WC, Pruneri G and
Sotiriou C: RANK-ligand (RANKL) expression in young breast cancer
patients and during pregnancy. Breast Cancer Res. 17:242015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Su D, Liao Z, Feng B, Wang T, Shan B, Zeng
Q, Song J and Song Y: Pulsatilla chinensis saponins cause liver
injury through interfering ceramide/sphingomyelin balance that
promotes lipid metabolism dysregulation and apoptosis.
Phytomedicine. 76:1532652020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shan B, Ai Z, Zeng S, Song Y, Song J, Zeng
Q, Liao Z, Wang T, Huang C and Su D: Gut microbiome-derived lactate
promotes to anxiety-like behaviors through GPR81 receptor-mediated
lipid metabolism pathway. Psychoneuroendocrinology. 117:1046992020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
31
|
Larue L and Bellacosa A:
Epithelial-mesenchymal transition in development and cancer: Role
of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene.
24:7443–7454. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Paolillo M and Schinelli S: Extracellular
matrix alterations in metastatic processes. Int J Mol Sci.
20:49472019. View Article : Google Scholar :
|
|
33
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Oudkerk M, Liu S, Heuvelmans MA, Walter JE
and Field JK: Lung cancer LDCT screening and mortality
reduction-evidence, pitfalls and future perspectives. Nat Rev Clin
Oncol. 18:135–151. 2021. View Article : Google Scholar
|
|
35
|
Li Z, Qi F and Li F: Establishment of a
gene signature to predict prognosis for patients with lung
adenocarcinoma. Int J Mol Sci. 21:84792020. View Article : Google Scholar :
|
|
36
|
Li L, Peng M, Xue W, Fan Z, Wang T, Lian
J, Zhai Y, Lian W, Qin D and Zhao J: Integrated analysis of
dysregulated long non-coding RNAs/microRNAs/mRNAs in metastasis of
lung adenocarcinoma. J Transl Med. 16:3722018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hsu CC, Liao WY, Chan TS, Chen WY, Lee CT,
Shan YS, Huang PJ, Hou YC, Li CR and Tsai KK: The differential
distributions of ASPM isoforms and their roles in Wnt signaling,
cell cycle progression, and pancreatic cancer prognosis. J Pathol.
249:498–508. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tang J, Lu M, Cui Q, Zhang D, Kong D, Liao
X, Ren J, Gong Y and Wu G: Overexpression of ASPM, CDC20, and TTK
confer a poorer prognosis in breast cancer identified by gene
co-expression network analysis. Front Oncol. 9:3102019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou JW, Wang H, Sun W, Han NN and Chen L:
ASPM is a predictor of overall survival and has therapeutic
potential in endometrial cancer. Am J Transl Res. 12:1942–1953.
2020.PubMed/NCBI
|
|
40
|
Bikeye SN, Colin C, Marie Y, Vampouille R,
Ravassard P, Rousseau A, Boisselier B, Idbaih A, Calvo CF, Leuraud
P, et al: ASPM-associated stem cell proliferation is involved in
malignant progression of gliomas and constitutes an attractive
therapeutic target. Cancer Cell Int. 10:12010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yuan YJ, Sun Y, Gao R, Yin ZZ, Yuan ZY and
Xu LM: Abnormal spindle-like microcephaly-associated protein (ASPM)
contributes to the progression of lung squamous cell carcinoma
(LSCC) by regulating CDK4. J Cancer. 11:5413–5423. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Santamaria PG, Moreno-Bueno G, Portillo F
and Cano A: EMT: Present and future in clinical oncology. Mol
Oncol. 11:718–738. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Aiello NM and Kang Y: Context-dependent
EMT programs in cancer metastasis. J Exp Med. 216:1016–1026. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Karimi Roshan M, Soltani A, Soleimani A,
Rezaie Kahkhaie K, Afshari AR and Soukhtanloo M: Role of AKT and
mTOR signaling pathways in the induction of epithelial-mesenchymal
transition (EMT) process. Biochimie. 165:229–234. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Papadimitrakopoulou V: Development of
PI3K/AKT/mTOR pathway inhibitors and their application in
personalized therapy for non-small-cell lung cancer. J Thorac
Oncol. 7:1315–1326. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang F, Li J, Liu J and Zhao Q:
Controversial role of the possible oxyntic stem cell marker ASPM in
gastric cancer. J Pathol. 241:559–561. 2017. View Article : Google Scholar
|
|
47
|
Pai VC, Hsu CC, Chan TS, Liao WY, Chuu CP,
Chen WY, Li CR, Lin CY, Huang SP, Chen LT and Tsai KK: ASPM
promotes prostate cancer stemness and progression by augmenting
Wnt-Dvl-3-β-catenin signaling. Oncogene. 38:1340–1353. 2019.
View Article : Google Scholar
|