Introduction
The epithelial membrane protein 3 (EMP3), a member
of the peripheral myelin protein 22-kDa (PMP22) gene family, is a
small hydrophobic membrane glycoprotein, and a myelin-related gene.
The protein encoded by this gene contains two N-linked
glycosylation sites and four transmembrane domains (1). In several studies, EMP3 has been
reported to function as a tumor suppressor gene in solid tumors
(2-4). However, EMP3 has also been shown to
function as an oncogene in several other malignancies, particularly
brain tumors, breast carcinoma and hepatocellular carcinoma
(5-7). Primary glioblastoma multiforme (GBM)
frequently exhibits upregulated expression of EMP3 (5). In addition, EMP3 mRNA upregulation
may be a suitable molecular marker to predict clinical outcomes in
patients with GBM (8). Taken
together, these studies suggest that EMP3 is an important gene that
promotes tumorigenesis in primary GBM. However, the molecular
mechanism of EMP3 in non-small cell lung cancer (NSCLC) has not
been studied previously, to the best of our knowledge, and thus
requires investigation.
Worldwide, lung cancer has a poor prognosis for both
men and women. NSCLC accounts for the majority (≥75%) of lung
cancer cases, and is the leading cause of cancer-related death
worldwide (9,10). Despite the use of conventional
chemotherapy and radiation therapy, NSCLC cannot be effectively
treated, and lung injury may occur as a side effect of these
treatments (11). Although
various studies have been performed on cell proliferation, invasion
and migration of NSCLC, the mechanism underlying the development of
NSCLC in the first place is unclear, and the 5-year survival rate
of patients with NSCLC is <15%. Metastasis is the cause of death
in >90% of patients with solid tumors, including in patients
with NSCLC (12), and cancer stem
cells (CSCs) are involved in recurrence and in metastasis (13). CSCs are involved in tumor
initiation, growth and maintenance and are also called tumor
initiating cells (14). CSCs
exhibit the self-renewal ability of normal stem cells, can
differentiate into cells of various phenotypes, and are resistant
to radiation or chemotherapy (15,16). CSCs can be identified using
specific marker proteins for each tissue. In the case of lung
cancer, cancer stem cells can be selected by CD133, CD44 and ALDH
(17,18). In particular, ALDH is used as a
marker protein in cancer stem cells of various tissues, including
breast cancer, brain cancer, and colorectal cancer, in addition to
lung cancer (19-21).
Lung CSCs (LCSCs) are becoming an increasingly
studied target for treatment of lung cancer (22). Accordingly, several pharmaceutical
companies are attempting to develop anticancer drugs targeting CSCs
specifically, and several drugs are being used in patients
(23-25). Various tumors, including LCSCs,
affect the malignancy and clinical prognosis by activation of the
TGF-β signaling pathway (26,27). The activation of TGF-β signaling
enhances the ability of cells to migrate and promote the
epithelial-mesenchymal transition (EMT), which is associated with
metastasis (28,29). Cell motility is important for
metastasis from the primary site to the secondary site via lymph or
blood vessels (30,31). In the present study, it was shown
for the first time that EMP3 is involved in maintaining the
characteristics of LCSCs via direct binding to TGFBR2.
Materials and methods
Cell culture and sphere-formation
assays
Human A549 and H460 lung cancer cell lines were
purchased from the Korea Cell Line Bank and grown using RPMI-1640
medium (cat. no. SH30027.01; HyClone; Cytiva) supplemented with 10%
(v/v) FBS (cat. no. SH30919.03; HyClone; Cytiva), 1% streptomycin
and 1% penicillin (cat. no. SV30010; HyClone; Cytiva). Cells were
cultured in a humidified incubator with 5% CO2 at 37°C.
During the sphere formation assay, cells were cultured in stem
cell-acceptable DMEM (DMEM-F12; cat. no. 11320-033; Invitrogen;
Thermo Fisher Scientific, Inc.) supplemented with basic fibroblast
growth factor (bFGF; 20 ng/ml; cat. no. 13256-029; Invitrogen;
Thermo Fisher Scientific, Inc.), epidermal growth factor (20 ng/ml;
cat. no E9644; Sigma-Aldrich; Merck KGaA) and B27 Serum-Free
Supplement (cat. no. 17504-044; Invitrogen; Thermo Fisher
Scientific, Inc.). Single cell experiments were set up with
floating cells in an ultra-low adhesion 96-well plate (cat. no.
3474; Corning, Inc.) with 1 or 2 cells distributed per well. The
cells were cultured in a humidified incubator as above. The
following day, only wells with single cells in each well were
selected visually under a light microscope (magnification, ×400),
and after 10-14 days, spheres were quantified based on absolute
count as well as diameter, and photographed using an inverted phase
contrast microscope.
Sorting CSCs from A549 cells
An ALDEFLUOR™ assay (cat. no. 01700; Stemcell
Technologies, Inc.) was used to isolate the CSC population from
A549 cells according to the manufacturer's protocol. A total of
1×106 cells were harvested and resuspended in ALDEFLUOR
assay buffer containing ALDH substrate. As a negative control, an
aliquot of cells exposed to ALDEFLUOR was immediately quenched with
the specific ALDH inhibitor N,N-diethylaminobenzaldehyde. After 30
min of incubation at 37°C, cells were washed and sorted into
ALDH1high and ALDH1low cells using a FACSAria flow cytometer (BD
Biosciences), and analyzed using BD FACSDiva version 6.1.3 (BD
Biosciences).
DNA microarray for gene expression
profiling
The quality of total RNA was measured using
Agilent's 2100 Bioanalyzer System (Agilent Technologies, Inc.).
Amplification and labeling were performed using the Low RNA Input
Linear Amplification kit PLUS (cat. no. 5185-5818; Agilent
Technologies, Inc.). Microarray hybridization was performed using
the Gene Expression Hybridization kit (cat. no. 5188-5242; Agilent
Technologies, Inc.). Microarray washes were performed using the
Gene Expression Wash Buffer kit (cat. no. 5190-0448; Agilent).
Finally, scanning and image analysis was performed using a DNA
microarray scanner (cat. no. G4900DA; Agilent Technologies, Inc.)
with the Feature Extraction Software (cat. no. G4460AA; Agilent
Technologies, Inc.). All procedures were performed according to the
manufacturer's protocol.
Small interfering RNA (siRNA) mediated
knockdown of EMP3 and TGFBR2
A549 cells were transfected with siRNA targeting
EMP3 or TGFBR2 (Bioneer Corporation) the sequences of which are
listed in Table SI. 10 pmol
siRNAs were transfected using Lipofectamine RNAi MAX reagent (cat.
no. 13-778-150; Invitrogen; Thermo Fisher Scientific, Inc.).
Stealth RNAi Negative Control Medium GC (cat. no. 12935-300;
Invitrogen; Thermo Fisher Scientific, Inc.) was used as the
negative control. Cells were incubated at 37°C for 72 h after
transfection.
DNA constructs
cDNA encoding full-length human EMP3 was generated
by reverse transcription-PCR from total RNA extracted from A549
cells. RNA was extracted using TRIzol® (cat no. TR118;
MRC). cDNA was synthesized from the extracted RNA using an RT
PreMix at 60°C for 1 h (cat no. 25264; Intron Biotechnology, Inc.).
The sequences of the primers used for PCR were: RMP3 forward, TAT
AAG CTT ATG TCA CTC CTC TTG CTG GTG G and reverse, ATA TGA ATT CTC
ACT CCC GCT TCC GTA GG. For reverse transcription-PCR, PCR PreMix
(cat no. 250256; Intron Biotechnology, Inc.) was used. The
thermocycling conditions were; Initial denaturation at 94°C for 30
sec; followed by 34 cycles of denaturation at 94°C for 30 sec,
annealing at 57.2°C for 40 sec, and extension at 72°C for 1 min.
The results were confirmed using a 2% agarose gel (cat no.
161-3102; Bio-Rad Laboratories, Inc.). The cDNA was cloned into the
mammalian expression vector pcDNA3.1 (Invitrogen; Thermo Fisher
Scientific, Inc.).
Limiting dilution assay
Cells were plated in 100 µl spheroid
formation assay medium in ultra-low adhesion 96-well plates. A
total of 1, 10, 50, 100 or 200 cells/well were plated, with 48
wells for each starting density of cells. Analysis of oncospheres
was performed using alight microscope (magnification, ×400) after
12-20 days of incubation. A well with at least one spheroid with a
diameter ≥100 µm was defined as a positive well, and the
number of positive wells were counted.
Antibodies
Antibodies against EMP3 (cat. no. ab236671; Abcam),
Sox2 (sex determining region Y-box 2; cat. no. 3579; Cell Signaling
Technology, Inc.), phosphorylated (p)-Smad2 (cat. no. 18338; Cell
Signaling Technology, Inc.), p-Smad3 (cat. no. 9520; Cell Signaling
Technology, Inc.), Smad2/3 (cat. no. 5678; Cell Signaling
Technology, Inc.), β-actin (cat. no. sc-7963; Santa Cruz
Biotechnology, Inc.), TGFBR1 (cat. no. sc-518086; Santa Cruz
Biotechnology, Inc.), TGFBR2 (cat. no. sc-17791; Santa Cruz
Biotechnology, Inc.), TGFBR3 (cat. no. sc-74511; Santa Cruz
Biotechnology, Inc.), CD44 (cat. no. 5640; Cell Signaling
Technology, Inc.), β-catenin (cat. no. sc-7963; Santa Cruz
Biotechnology, Inc.), Twist (cat. no. sc-15393; Santa Cruz
Biotechnology, Inc.), Vimentin (MA5-16409; Thermo Fisher
Scientific, Inc.), aldehyde dehydrogenase (ALDH1)A1 (cat. no.
ab52492; Abcam), ALDH1A3 (cat. no. ab129815; Abcam), Snail (cat.
no. sc-10432; Santa Cruz Biotechnology, Inc.), Slug (cat. no.
sc-166476; Santa Cruz Biotechnology, Inc.), ZEB1 (Zinc finger
E-box-binding homeobox 1; cat. no. sc-25388; Santa Cruz
Biotechnology, Inc.), E-cadherin (cat. no ab15148; Abcam),
N-cadherin (cat. no. 610921; BD Biosciences) and Oct4 (cat. no.
2750; Cell Signaling Technology, Inc.) were used. Thsese antibodies
were used for immunofluorescence assays, western blot analysis
and/or immunoprecipitation (IP).
Neutralization assay
The anti-EMP3 antibody (1:100) was used for
the neutralization assay. The normal mouse IgG1 anti-body (cat. no.
sc-3877; Santa Cruz Biotechnology, Inc.) was used as the control
antibody. The experiment was conducted in the same manner as the
cell culture environment, and the subsequent experiments or results
were performed/obtained after 24-48 h.
TGF-β2 treatment
A total of 20 ng/ml TGFB3 (cat. no. 302-B2;
Bio-Techne) was added to cells, and cells were cultured in an
incubator for 24 h.
Western blotting and IP
Cells were mixed with lysis buffer [150 mM NaCl, 1
mM EDTA, 1 mM EGTA, 0.5% Triton X-100 in 20 mM Tris-HCl (pH 7.5)]
and protease inhibitor cocktail (cat. no. P8340; Sigma-Aldrich;
Merck KGaA). The protein concentration was measured using Bradford
reagent (cat. no. 5000006, Bio-Rad Laboratories, Inc.) and
normalized to a standard curve developed using known concentrations
of BSA. For western blot analysis, 20 µg protein sample was
loaded on 8-15% SDS-gels, resolved using SDS-PAGE and transferred
to Hybond nitrocellulose membranes (cat. no. 10-6000-04; Amersham;
Cytiva). Membranes were treated with specific antibodies all at a
dilution of 1:1,000 overnight in a cold chamber at 4°C. After
washing with Tris-buffered saline (cat. no. TR2008-100-00;
Biosesang), the membrane was treated with an horseradish
peroxidase-conjugated secondary antibody (cat. no. anti-rabbit;
cat. no. ab205718; or anti-mouse; cat. no. ab205719; both 1:10,000;
Abcam) for 2 h at room temperature, and a Westzol enhanced
chemiluminescence detection kit (cat. no. sc-2048; Santa Cruz
Biotechnology, Inc.) was used to visualize signals.
IP was performed overnight at 4°C using a
concentration of 1:200 concentration-specific antibody (anti-EMP3;
cat. no. ab236671; Abcam; or anti-TGFBR2; cat. no. sc-17791; Santa
Cruz Biotechnology, Inc.) with 2 mg cell lysate. Then, 0.2
µg protein A/G Ultralink Resin (cat. no. sc-2003; Santa Cruz
Biotechnology, Inc.) was added and incubated for 2 h at 4°C. After
washing with lysis buffer on the reacted samples, the
immunoprecipitate was resuspended in 2× SDS sample buffer, and
analyzed by western blotting using the specific antibodies
(anti-EMP3 using the same antibody as above; and anti-TGFBR1; cat.
no. sc-518086; anti-TGFBR2; cat. no. sc-17791; and anti-TGFBR3;
cat. no. sc-74511; all from Santa Cruz Biotechnology, Inc.).
Invasion and migration assays
Migration assays were performed using an uncoated
chamber (cat. no. 3422; 8-µm pore; Corning, Inc.) and the
ability of cells to migrate was measured. Invasion assays were
performed by coating the chamber with Matrigel®
according to the manufacturer's protocol. The lower chamber of the
Transwell inserts (Cell Biolabs) was filled with 800 µl
RPMI-1640 supplemented with 10% FBS. In the upper chamber, 150
µl serum-free medium (Opti-MEM®; cat. no.
31985-070; Invitrogen; Thermo Fisher Scientific, Inc.) containing
2×105 cells was added. The cells were incubated for 24 h
at 37°C in a humidified incubator with 5% CO2. Cells
that had migrated/invaded to the bottom of the chamber were stained
with crystal violet (cat. no. HT90132-1L; Sigma-Aldrich; Merck
KGaA) and the cells were counted under a light microscope (×400,
magnification).
Wound healing assay
Cells were plated in a 60 mm culture dish and grown
to 80% confluence. A wound was created by scraping the monolayer of
cells with a 200 µl pipette tip in the middle. Floating
cells were removed by washing with PBS and fresh medium containing
10% FBS was added. The doubling time of the A549 cells used was 22
h. Cells were incubated at 37°C for 24 h, and imaged using
phase-contrast microscopy (magnification, ×400). The distance
between the edges of the wounds shown in the image was measured
randomly at three or more places and the mean of the three
measurements were obtained.
Colony-formation assay and
irradiation
Cells were seeded at a density of 1×103
cells per 35-mm cell culture dish (cat. no. 430165; Corning, Inc.),
and then allowed to adhere for 24 h in a humidified incubator with
5% CO2 at 37°C. The following day, cells were irradiated
with 3 Gy γ-radiation (KAERI). After 10-14 days, cells were stained
for colonies (defined as clusters of ≥50 cells) with 0.5% crystal
violet for 1 h at room temperature, and stained colonies were
counted. Clonal survival rates are expressed as a percentage of the
non-irradiated control group.
Immunocytochemistry
In 6 well plates (cat. no. 3516; Corning, Inc.),
5×104 cells were grown on glass coverslips and fixed
with 4% paraformaldehyde (cat. no. P2031; Biosesang) for 1 h at
room temperature. After fixing, cells were incubated overnight at
4°C with antibodies in a solution of PBS containing 0.1% Triton
X-100 (cat. no. 161-0781; Bio-Rad Laboratories, Inc.) and 1% BSA
(cat. no. BSAS 0.1; Bovogen). The antibodies used were: Human
anti-EMP3 (mouse polyclonal antibody; 1:200), ALDH1A1 (mouse
polyclonal antibody; 1:200), ALDH1A3 (rabbit polyclonal antibody;
1:200), CD44 (mouse polyclonal antibody; 1:200), E-cadherin (rabbit
polyclonal antibody; 1:200), N-cadherin (rabbit polyclonal
antibody; 1:200), Vimentin (goat polyclonal antibody; 1:200) and
TGFBR2 (rabbit polyclonal antibody; 1:200). Staining was visualized
using an Alexa Fluor 488-conjugated anti-rabbit IgG antibody (cat.
no. A21206; Invitrogen; Thermo Fisher Scientific, Inc.). Nuclei
were stained using DAPI (Sigma-Aldrich; Merck KGaA) for 30 min at
room temperature. The stained cell samples were observed using a
fluorescence microscope (Zeiss LSM510; Carl Zeiss AG; ×400
magnification).
Kaplan-Meier plotter
Using the published genetic information system,
Kaplan-Meier survival values were obtained (kmplot.com/analysis). This was based on results of
mRNA gene chip analysis using tissues from lung cancer patients.
The gene symbol used was EMP3. All conditions were set as total
lung cancer patients.
Statistical analysis
All experiments were performed by repeating at least
three independent experiments, and the results are expressed as the
mean ± standard deviation. Each exact n value is displayed in the
corresponding figure legend. To validate the data, all graphs were
compared using a two-sided paired Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
EMP3 expression is increased in
ALDH+ NSCLC
CSCs that are resistant to cancer treatment
overexpress certain proteins. Amongst these, a protein that is
overexpressed together with a stem cell marker protein is
classified as a CSC marker protein (32). In previous experiments, ALDH1 was
used as a marker protein to classify overexpressed genes in LCSCs
(33). Through classification
using the feature extraction software, information on 4,300 genes
that were expressed >2× higher in ALDH1+ cells than
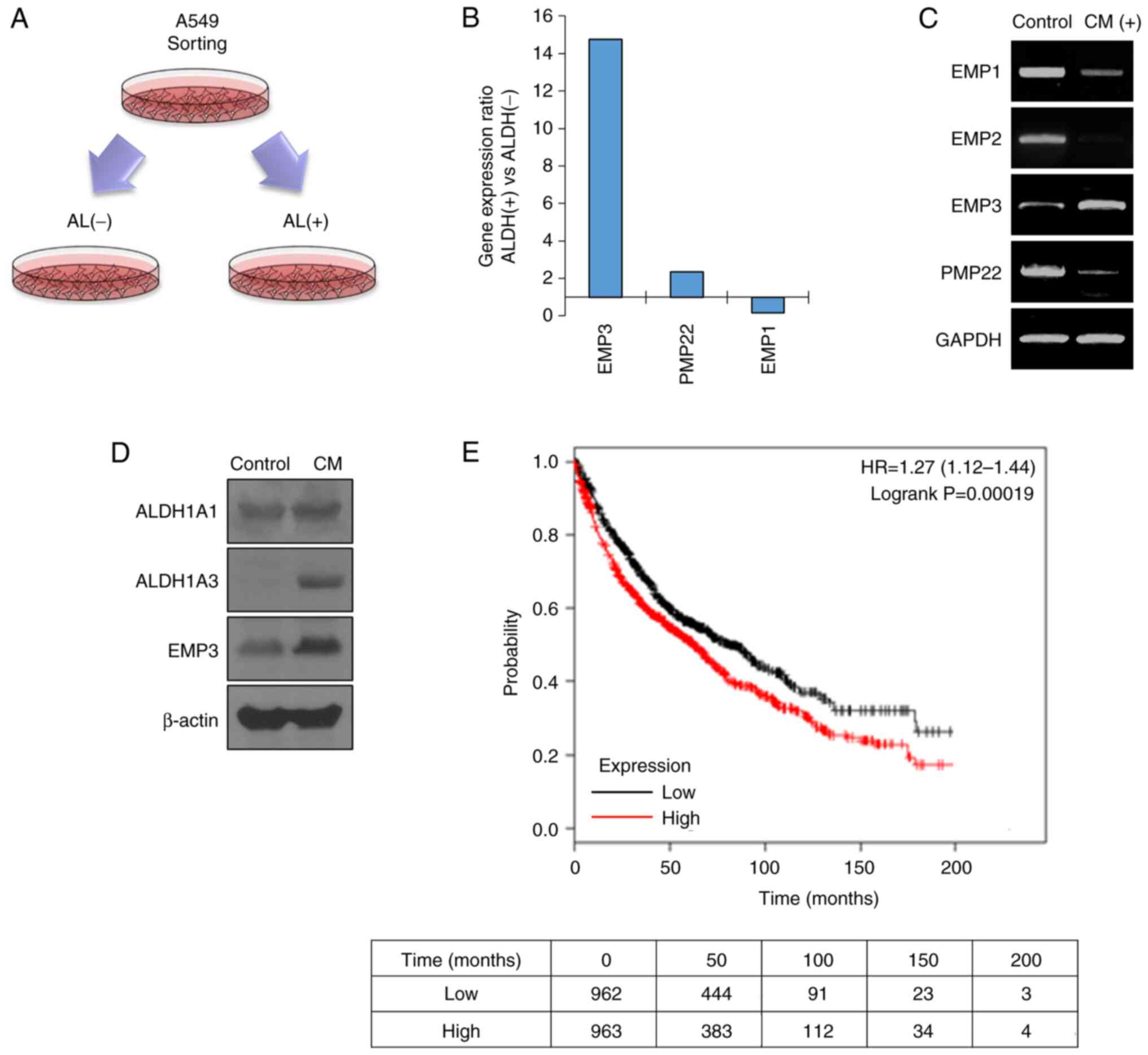
in the control group was obtained (Fig. 1A). Amongst these, it was confirmed
that EMP3 was highly upregulated by selecting genes associated with
cell membrane proteins, and this was selected as they often serve
as initiation points for signal transduction (Fig. 1B). To confirm that EMP3 was
specifically overexpressed in CSCs, LCSCs were prepared by
treatment with the spheroid formation assay medium, and EMP3 gene
expression was increased in the LCSCs (Fig. 1C). In addition, the expression of
EMP3 together with CSC marker proteins was increased in LCSCs
(Fig. 1D). Based on Kaplan-Meier
analysis, the prognosis of patients with high EMP3 expression
amongst patients with lung cancer was significantly worse compared
to those with low EMP3 expression (Fig. 1E). A total of 1,925 patients with
lung cancer were included in the Kaplan-Meier analysis. Patients
with low EMP3 gene expression survived an average of 81.2 months,
and those with high EMP3 gene expression survived an average of
62.3 months after diagnosis. These results showed that EMP3 may be
involved in the prognosis of patients with malignant lung
cancer.
EMP3 promotes self-renewal and
tumorigenic capacity of NSCLC
Although several studies have confirmed the
functionality of the PMP22 family of proteins, little is known
regarding the functionality of EMP3 in NSCLC. Therefore, in the
present study, to investigate whether EMP3 was involved in the
enrichment of NSCLC stem cells, A549 cells were used, an
adenocarcinoma cell line that exhibits a high level of resistance
to radiation and high ALDH1 expression (22). Knockdown of EMP3 expression via
transfection siRNA reduced the expression of the CSC marker
proteins CD133, ALDH1 and CD44. EMP3 was also overexpressed in the
lung cancer cells to confirm the function of EMP3 more accurately.
The cells used for overexpression were the H460 lung cancer cells,
which are known to be relatively less malignant than the A549
(22). The expression levels of
EMP3 in the two cells was compared (Fig. S1A). The expression of EMP3 in
H460 cells was significantly lower than in A549 cells. EMP3
overexpression was thus performed using the H460 cells. The
expression of CSC marker proteins was increased in the
EMP3-overexpressing cells (Fig. S1B
and C). Overexpression of EMP3 also reduced the expression of
the CSC regulator proteins Sox2, Oct-4 and Nanog (Fig. 2A). In Fig. 2B, the difference in the expression
of marker proteins according to the knockdown of EMP3 expression
was confirmed using immunocytochemistry. To confirm the effect of
EMP3 on CSCs, A549 cells were cultured in a serum-free medium
containing epidermal growth factor and bFGF to generate spheroids.
When the expression of EMP3 was suppressed using siRNA, the size
and number of CSC spheres decreased significantly (Fig. 2C). Conversely, when the expression
of EMP3 was overexpressed, the size and number of CSC spheres
increased significantly (Fig.
S1D). In addition, EMP3 is a cell membrane protein, and direct
inhibitory action using an antibody was confirmed. Treatment with
the anti-EMP3 antibody resulted in similar outcomes as those
obtained from treatment with siRNA (Fig. 2D). A single cell assay and a
limited dilution assay were performed to confirm the effects of
EMP3 on self-renewal, a characteristic of CSCs. The self-renewal
ability was decreased in the si-EMP3 treated group (Fig. 2E and G). As shown above, EMP3 can
be directly inhibited by treatment with the anti-EMP3 antibodies.
Treatment with the anti-EMP3 antibody resulted in similar outcomes
as those obtained from treatment with siRNA (Fig. 2F). To assess whether EMP3 was
involved in the characterization of tumor resistance to ionizing
radiation, the effect of EMP3 on the clonal formation of A549 cells
was investigated. Colony formation was suppressed in the EMP3
knockdown cells. These results indicate that the sensitivity to
radiation was modulated by EMP3. As above, antibody treatment has a
similar effect to that of EMP3 knockdown (Fig. 2H and I). CSCs have been shown to
possess more than one aberration in various signaling pathways
(13). Amongst them, Notch,
Hedgehog (HH) and Wnt signals control stem cell self-renewal and
play an important role in embryonic development and
differentiation, but within CSCs, they contribute to a signaling
system that promotes recurrence and metastasis of cancer by
engaging in abnormal signaling. siRNA was used to confirm whether
EMP3 was involved in the regulation of these important signaling
systems in CSCs. The results showed that Notch, HH and Wnt signals
were all regulated by EMP3 (Fig.
2J).
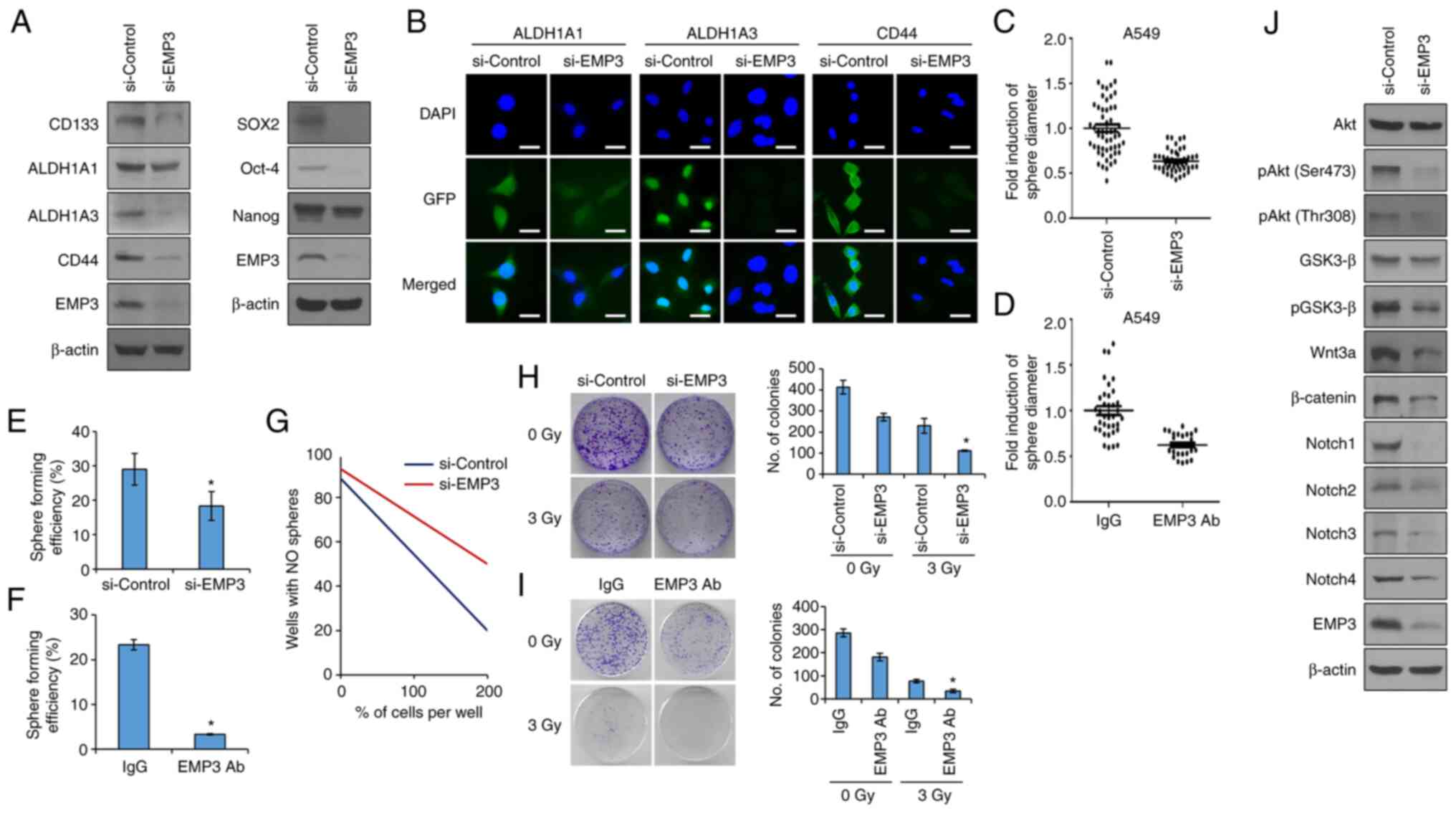 | Figure 2EMP3 regulates CSC properties in lung
CSC. (A) Western blot analysis of the CSC marker proteins CD133,
ALDH1A1, ALDH1A3 and CD44. The CSC regulatory proteins Sox2, Oct-4
and Nanog were also analyzed by western blotting. A549 cells were
transfected with siRNA targeting EMP3. (B) Immunocytochemistry
analysis of CSC marker proteins using siRNA treated A549 cells. The
green signal is produced by the GFP tag on the secondary antibody.
(C) Sphere-forming capacity analysis of A549 cells in siRNA
transfected EMP3 cells and (D) the ability to of an anti-EMP3
antibody to abrogate sphere formation was assessed. (E) Single-cell
assay of si-EMP3 transfected cells and (F) the ability of the
anti-EMP3 antibody to abrogate this. (G) Limiting dilution assays
were performed in 96 well plates. Wells were plated with 1, 50,
100, 150 or 200 cells/well, and conditioned media was added.
Results were confirmed 10 days after seeding of cells.
Colony-formation assays to observe the clonogenicity of the (H)
EMP3-knockdown A549 and (I) anti-EMP3 antibody treated A549 cells.
Cells were irradiated with 3 Gy radiation. After 10 days of
incubation, the colonies were stained with crystal violet. (J)
Western blot analysis of members of the Sonic hedgehog,
Wnt/β-catenin and Notch signaling pathways in CSCs. Data are
presented as the mean ± standard deviation of three repeats. Scale
bar, 50 µm. *P<0.05 vs. control. EMP3,
epithelial membrane protein 3; ALDH1, aldehyde dehydrogenase 1;
CSC, cancer stem cell; Sox2, sex determining region Y-box 2; Oct-4,
octamer-binding transcription factor 4; siRNA, small interfering
RNA; p, phosphorylated; GSK3-β, glycogen synthase kinase 3-β. |
EMT is regulated by EMP3
Previously, a direct link has been reported between
EMT progression and acquisition of stem cell properties (25,34). In general, CSC and EMT exhibit
similar signaling mechanisms (35). Amongst the several features of
EMT, the most distinctive is related to cancer metastasis caused by
increased mobility and invasiveness of cancer cells. Cancer
metastasis may be preventable by studying the signaling mechanisms
associated with EMT and establishing effective control methods.
However, whether EMP3 participates in EMT remains unknown, and is
of interest as EMT is closely associated with CSCs. In the present
study, the intracellular expression levels of E-cadherin,
N-cadherin and Vimentin, which are EMT marker proteins, and Snail,
slug, Twist and ZEB1, which are EMT-related transcription factors,
were investigated by western blotting. When EMP3 expression was
knocked down using siRNA, expression of EMT markers and EMT-related
transcription factors was decreased (Fig. 3A). Immunofluorescence staining
confirmed that inhibition of EMP3 expression upregulated the levels
of the epithelial marker protein E-cadherin and downregulated the
levels of the mesenchymal marker proteins N-cadherin and Vimentin
(Fig. 3B). To confirm these
results, the A549 cell line was used to evaluate the effect of EMP3
on cell migration and invasion. First, an in vitro wound
healing assay was performed. The migration of A549 cells into the
wound was significantly reduced when EMP3 gene expression was
knocked down when compared with the control group (Fig. 3C). Likewise, when the A549 cell
line was treated with an anti-EMP3 antibody, the migration of the
cells to the wound site was significantly reduced (Fig. 3D). Conversely, when the EMP3 gene
was overexpressed in the H460 cell line, cell migration into the
wound was increased (Fig. S1E).
Additionally, the effect of EMP3 on cell migration and cell
invasion were assessed using Transwell assays. EMP3 notably
regulated the invasion and migration of A549 cells (Fig. 3E and F). In the EMP3
overexpressing H460 cells, it was confirmed that cell migration and
invasion was increased (Fig.
S1F).
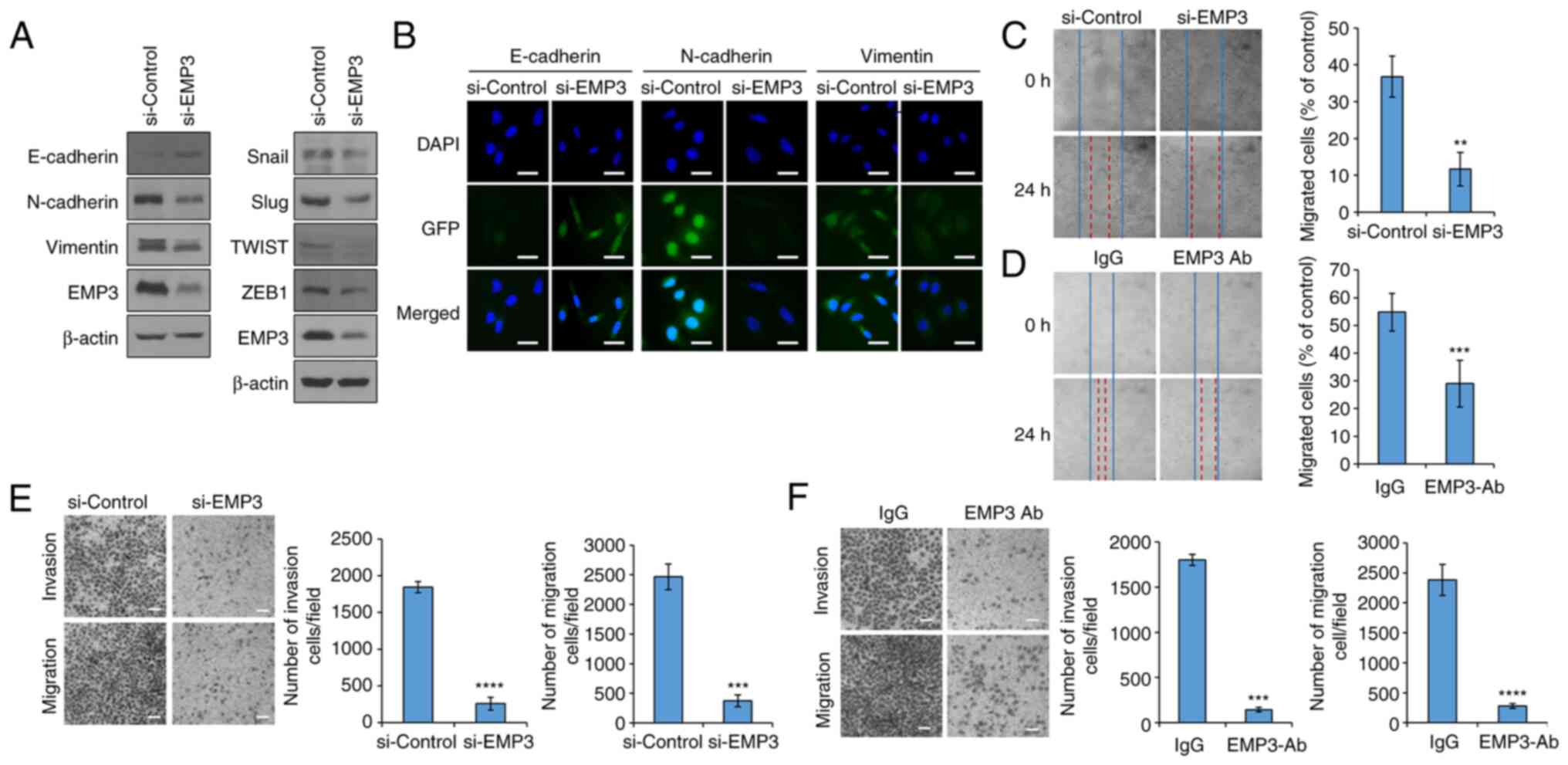 | Figure 3EMP3 is involved in EMT in lung
cancer stem cells. (A) Western blot analysis of EMT marker proteins
E-cadherin, N-cadherin and Vimentin. EMT regulatory proteins Snail,
Slug, TWIST and ZEB1 were also analyzed by western blot. A549 cells
were transfected with siRNA targeting EMP3. (B) Immunocytochemistry
analysis of EMT marker proteins after transfection with siRNA
targeting EMP3 in A549 cells. A primary antibody targeting each
marker protein was used, and the secondary antibody used was tagged
with GFP. Wound healing assays of the (C) EMP3-knockdown A549 cells
and (D) anti-EMP3 antibody treated A549 cells. Migration and
invasion assays of (E) EMP3-knockdown A549 cells and (F) anti-EMP3
antibody treated A549 cells. Data are presented as the mean ±
standard deviation of three repeats. Scale bar, 50 µm.
**P<0.01, ***P<0.001,
****P<0.0001. EMP3, epithelial membrane protein 3;
EMT, epithelial-mesenchymal transition; ZEB1, Zinc finger
E-box-binding homeobox 1; siRNA, small interfering RNA. |
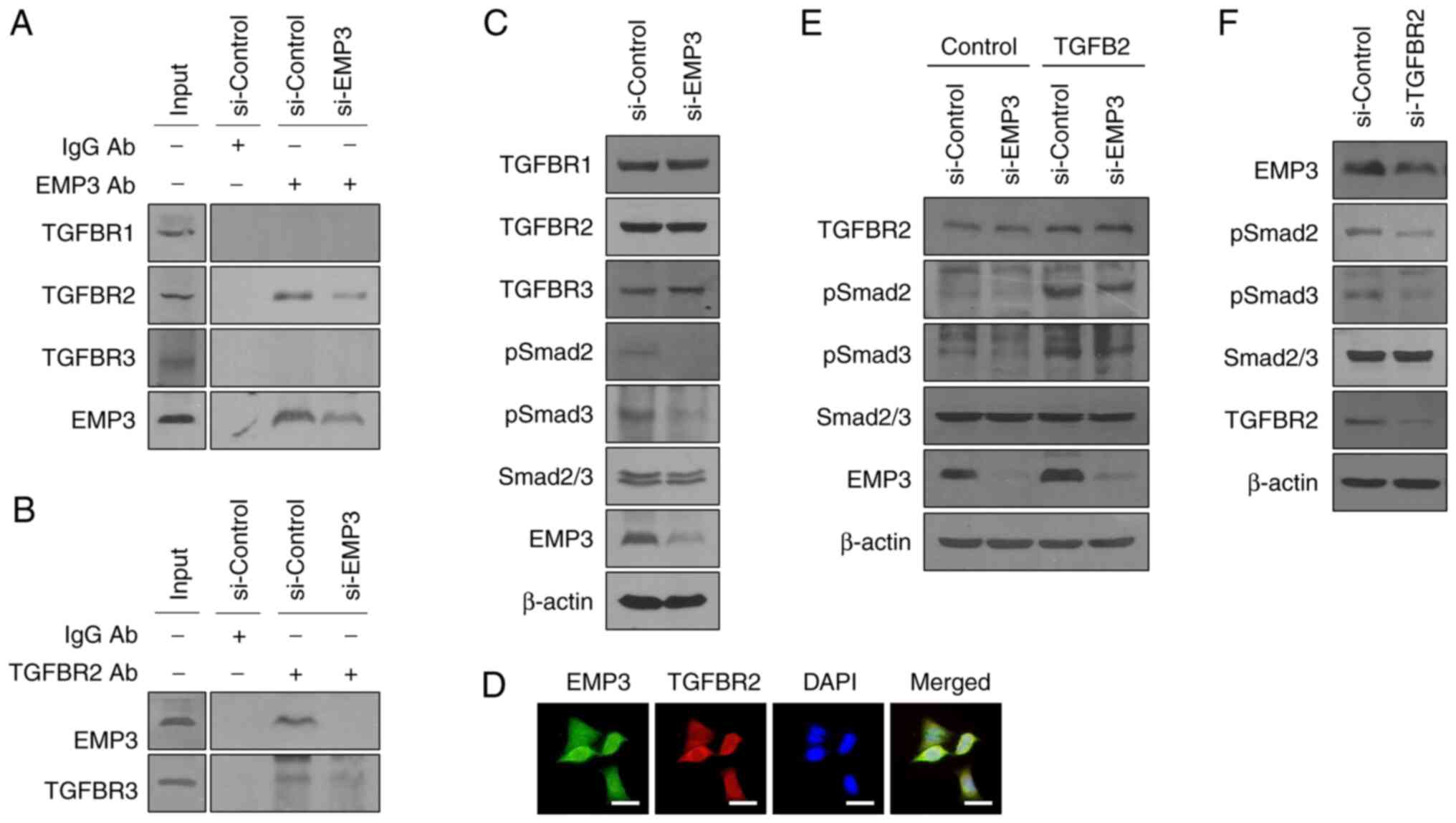
Direct binding to TGFBR2 of EMP3
activates the down-stream TGF-β/Smad signaling pathway
There are three main types of TGF-β receptors: The
transmembrane serine/threonine kinase receptor, type 1 TGF-β
receptor (TGFBR1), type II TGF-β receptor (TGFBR2) and β-glycan,
and type III TGF-β receptor (TGFBR3) (29). IP analysis was performed to
confirm the binding of three types of TGF-β receptors (TGFBR) to
EMP3. EMP3 bound to TGFBR2, and this was further confirmed by IP
analysis using a TGFBR2 antibody (Fig. 4A and B). In addition, western blot
analysis was used to confirm the association of EMP3 with Smad
downstream of TGFBR. The phosphorylation of Smad2 and Smad3 was
lower in the si-EMP3 treated group, suggesting that the TGFBR-Smad
signaling axis was regulated by EMP3. (Fig. 4C). Since the binding between the
two proteins is related to the intracellular location, ICC analysis
was used to confirm the subcellular location of EMP3 and TGFBR2,
and the results showed that EMP3 and TGFBR2 were colocalized at the
membrane, and that EMP3 regulated the TGFBR2-Smad signaling pathway
(Fig. 4D). Additionally,
following TGF-β2 (TGFB2) treatment, the sub-signaling mechanism of
TGFBR was confirmed (Fig. 4E). In
the cells treated with si-EMP3, it was observed that p-Smad2 and
p-Smad3 levels were decreased even when treated with TGFB2. This
confirmed that EMP3 was involved in the downregulation of TGFBR2
signaling through TGF-β. In addition, it was confirmed that the
expression of EMP3 was lower in cells treated with si-TGFBR2
(Fig. 4F). This suggests that
another mechanism linked EMP3 and TGF-β signaling.
Discussion
In the present study, it was confirmed that EMP3
expression is upregulated in LCSCs. However, according to a recent
paper, EMP2, one of the EMP family proteins, has been reported to
prevent cancer malignancy (36).
Contrary to the increase in expression of the EMP3 gene observed in
the present study, all other EMP family genes showed a decreasing
trend. In LCSCs, it is necessary to confirm the change in
expression and function of the other EMP family members, to compare
with EMP3, and to obtain a more holistic picture of EMPs in lung
cancer.
Numerous studies on the signaling process of TGFBR
have been reported. Changes in TGF-β have been linked to TGFBR, and
both have been associated with a variety of diseases, including
cancer and inflammation (37-39). It can be confirmed that the
disruption of TGF-β homeostasis occurred in samples of various
several cancers (26-29). However, the specifics of the
signaling processes regulated EMP3 and TGFBR2 remained to be
determined, and will form the subject of future studies, and may be
considered an important part of the EMP3 signaling process. In
addition, TGFBR signaling has a profound relationship on the
inflammatory response (37,39), and this may serve as an indication
of the relationship with immune cells. CSCs inhibit the activation
of immune cells and allow for evasion of immune cells, which allows
for the formation of malignant cancers and metastases, and
EMP3-TGFBR axis identified in the present may be involved in this
process (13,40,41).
Taken together, the results confirmed that EMP3
regulates the CSC population, EMT and the phosphorylation of Smad
through binding with TGFBR2. EMP3 is a protein present in the cell
membrane, and can bind to various proteins present in the cell
membrane, such as RTK, and integrins, as well as TGFBR2 as
identified herein (42,43). In addition, it is predicted to
modulate or be involved in various signaling mechanisms through
binding to proteins present in cells. Furthermore, since EMP3 is
present at the cell membrane, it may allow for easier targeting by
drugs, a possible advantage of this protein as a potential
druggable target. In general, targets present at cell membranes are
easier targets for development of anticancer agents, and play an
important role in regulating signals inside and outside of cells
(44-46). Thus, EMP3, which is present at the
cell membrane, may be an important target in the development of
anticancer drugs. Furthermore, EMP3 affects sensitivity to
radiation, and CSCs are characterized by resistance to ionizing
radiation (13,15,22). Therefore, inhibition of EMP3 may
reduce tumor resistance to ionizing radiation and maximize
therapeutic efficacy. Considering these results together, it is
hypothesized that EMP3 may be targeted to improve the efficacy of
radiotherapy.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
IGK and RKK conceived and designed and the study.
YJK, UJ and RKK performed the experiments. UJ and RKK performed the
radiation experiments. IGK and RKK analyzed the data. IGK, YJK and
RKK wrote and revised the manuscript. IGK and RKK confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Ben-Porath I and Benvenisty N:
Characterization of a tumor-associated gene, a member of a novel
family of genes encoding membrane glycoproteins. Gene. 183:69–75.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fumoto S, Hiyama K, Tanimoto K, Noguchi T,
Hihara J, Hiyama E, Noguchi T and Nishiyama M: EMP3 as a tumor
suppressor gene for esophageal squamous cell carcinoma. Cancer
Lett. 274:25–32. 2009. View Article : Google Scholar
|
|
3
|
Alaminos M, Dávalos V, Ropero S, Setién F,
Paz MF, Herranz M, Fraga MF, Mora J, Cheung NKV, Gerald WL and
Esteller M: EMP3, a myelin-related gene located in the critical
19q13.3 region, is epigenetically silenced and exhibits features of
a candidate tumor suppressor in glioma and neuroblastoma. Cancer
Res. 65:2565–2571. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xue Q, Zhou Y, Wan C, Lv L, Chen B, Cao X,
Ju G, Huang Y, Ni R and Mao G: Epithelial membrane protein 3 is
frequently shown as promoter methylation and functions as a tumor
suppressor gene in non-small cell lung cancer. Exp Mol Pathol.
95:313–318. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jun F, Hong J, Liu Q, Guo Y, Liao Y, Huang
J, Wen S and Shen L: Epithelial membrane protein 3 regulates TGF-β
signaling activation in CD44-high glioblastoma. Oncotarget.
8:14343–14358. 2017. View Article : Google Scholar
|
|
6
|
Hsieh YH, Hsieh SC, Lee CH, Yang SF, Cheng
CW, Tang MJ, Lin CL, Lin CL and Chou RH: Targeting EMP3 suppresses
proliferation and invasion of hepatocellular carcinoma cells
through inactivation of PI3K/Akt pathway. Oncotarget.
6:34859–34874. 2015. View Article : Google Scholar
|
|
7
|
Hong XC, Fen YJ, Yan GC, Hong H, Yan CH,
Bing LW and Zhong YH: Epithelial membrane protein 3 functions as an
oncogene and is regulated by microRNA-765 in primary breast
carcinoma. Mol Med Rep. 12:6445–6450. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ernst A, Hofmann S, Ahmadi R, Becker N,
Korshunov A, Engel F, Hartmann C, Felsberg J, Sabel M, Peterziel H,
et al: Genomic and expression profiling of glioblastoma stem
cell-like spheroid cultures identifies novel tumor-relevant genes
associated with survival. Clin Cancer Res. 21:6541–6550. 2009.
View Article : Google Scholar
|
|
9
|
Li JJ, Li R, Wang W, Zhang B, Song X,
Zhang C, Gao Y, Liao Q, He Y, You S, et al: IDH2 is a novel
diagnostic and prognostic serum biomarker for non-small-cell lung
cancer. Mol Oncol. 12:602–610. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Alidousty C, Baar T, Heydt C,
Wagener-Ryczek S, Kron A, Wolf J, Buettner R and Schultheis AM:
Advance of theragnosis biomarkers in lung cancer: From clinical to
molecular pathology and biology. J Thorac Dis. 11(Suppl 1): S3–S8.
2019. View Article : Google Scholar :
|
|
11
|
Zappa C and Mousa SA: Non-small cell lung
cancer: Current treatment and future advances. Transl Lung Cancer
Res. 5:288–300. 2016. View Article : Google Scholar :
|
|
12
|
Brognard J, Clark AS, Ni Y and Dennis PA:
Akt/protein kinase B is constitutively active in non-small cell
lung cancer cells and promotes cellular survival and resistance to
chemotherapy and radiation. Cancer Res. 61:3986–3997. 2001.
|
|
13
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen SY, Huang YC, Liu SP, Tsai FJ, Shyu
WC and Lin SZ: An overview of concepts for cancer stem cells. Cell
Transplant. 20:113–120. 2011. View Article : Google Scholar
|
|
15
|
Rycaj K and Tang DG: Cancer stem cells and
radioresistance. Int J Radiat Biol. 90:615–621. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Suresh R, Ali S, Ahmad A, Philip PA and
Sarkar FH: The role of cancer stem cells in recurrent and
drug-resistant lung cancer. Adv Exp Med Biol. 890:57–74. 2016.
View Article : Google Scholar
|
|
17
|
Zhao W, Luo Y, Li B and Zhang T:
Tumorigenic lung tumorospheres exhibit stem-like features with
significantly increased expression of CD133 and ABCG2. Mol Med Rep.
14:2598–2606. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Okudela K, Woo T, Mitsui H, Tajiri M,
Masuda M and Ohashi K: Expression of the potential cancer stem cell
markers, CD133, CD44, ALDH1, and β-catenin, in primary lung
adenocarcinoma-their prognostic significance. Pathol Int.
62:762–801. 2012. View Article : Google Scholar
|
|
19
|
Marcato P, Dean CA, Pan D, Araslanova R,
Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S, et al:
Aldehyde dehydrogenase activity of breast cancer stem cells is
primarily due to isoform ALDH1A3 and its expression is predictive
of metastasis. Stem Cells. 29:32–45. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu SL, Liu S, Cui W, Shi Y, Liu Q, Duan
JJ, Yu SC, Zhang X, Cui YH, Kung HF and Bian XW: Aldehyde
dehydrogenase 1A1 circumscribes high invasive glioma cells and
predicts poor prognosis. Am J Cancer Res. 5:1471–1483. 2015.
|
|
21
|
Vishnubalaji R, Manikandan M, Fahad M,
Hamam R, Alfayez M, Kassem M, Aldahmash A and Alajez NM: Molecular
profiling of ALDH1+ colorectal cancer stem cells reveals
preferential activation of MAPK, FAK, and oxidative stress
pro-survival signalling pathways. Oncotarget. 9:13551–13564. 2018.
View Article : Google Scholar
|
|
22
|
Kim IG, Kim SY, Choi SI, Lee JH, Kim KC
and Cho EW: Fibulin-3-mediated inhibition of
epithelial-to-mesenchymal transition and self-renewal of
ALDH+ lung cancer stem cells through IGF1R signaling.
Oncogene. 33:3908–3917. 2014. View Article : Google Scholar
|
|
23
|
Buck E, Eyzaguirre A, Barr S, Thompson S,
Sennello R, Young D, Iwata KK, Gibson NW, Cagnoni P and Haley JD:
Loss of homotypic cell adhesion by epithelial-mesenchymal
transition or mutation limits sensitivity to epidermal growth
factor receptor inhibition. Mol Cancer Ther. 6:532–541. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Creighton CJ, Li X, Landis M, Dixon JM,
Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A,
Herschkowitz JI, et al: Residual breast cancers after conventional
therapy display mesenchymal as well as tumor-initiating features.
Proc Natl Acad Sci USA. 106:13820–13825. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Du S, Bouquet S, Lo CH, Pellicciotta I,
Bolourchi S, Parry R and Barcellos-Hoff MH: Attenuation of the DNA
damage response by transforming growth factor-beta inhibitors
enhances radiation sensitivity of non-small-cell lung cancer cells
in vitro and in vivo. Int J Radiat Oncol Biol Phys. 91:91–99. 2015.
View Article : Google Scholar
|
|
27
|
Murai F, Koinuma D, Shinozaki-Ushiku A,
Fukayama M, Miyaozono K and Ehata S: EZH2 promotes progression of
small cell lung cancer by suppressing the TGF-β-Smad-ASCL1 pathway.
Cell Discov. 1:150262015. View Article : Google Scholar
|
|
28
|
Xu J, Lamouille S and Derynck R:
TGF-beta-induced epithelial to mesenchymal transition. Cell Res.
19:156–172. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hao Y, Baker D and Dijke PT:
TGF-β-mediated epithelial-mesenchymal transition and cancer
metastasis. Int J Mol Sci. 20:27672019. View Article : Google Scholar
|
|
30
|
Wang Y and Shang Y: Epigenetic control of
epithelial-to-mesenchymal transition and cancer metastasis. Exp
Cell Res. 319:160–169. 2013. View Article : Google Scholar
|
|
31
|
Palma Cde S, Grassi ML, Thomé CH, Ferreira
GA, Albuquerque D, Pinto MT, Melo FU, Kashima S, Covas DT, Pitteri
SJ and Faça VM: Proteomic analysis of epithelial to mesenchymal
transition (EMT) reveals cross-talk between SNAIL and HDAC1
proteins in breast cancer cells. Mol Cell Proteomics. 15:906–917.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar
|
|
33
|
Choi SI, Lee JH, Kim RK, Jung U, Kahm YJ,
Cho EW and Kim IG: HSPA1L enhances cancer stem cell-like properties
by activating IGF1Rβ and regulating β-catenin transcription. Int J
Mol Sci. 21:69572020. View Article : Google Scholar
|
|
34
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Morel AP, Lièvre M, Thomas C, Hinkal G,
Ansieau S and Puisieux A: Generation of breast cancer stem cells
through epithelial-mesenchymal transition. PLoS One. 3:e28882008.
View Article : Google Scholar
|
|
36
|
Ma Y, Schröder DC, Nenkov M, Rizwan MN,
Abubrig M, Sonnemann J, Murrieta-Coxca JM, Morales-Prieto DM,
Westermann M, Gassler N and Chen Y: Epithelial membrane protein 2
suppresses non-small cell lung cancer cell growth by inhibition of
MAPK pathway. Int J Mol Sci. 22:29442021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Blobe GC, Schiemann WP and Lodish HF: Role
of transforming growth factor beta in human disease. N Engl J Med.
342:1350–1358. 2000. View Article : Google Scholar
|
|
38
|
de Caestecker MP, Piek E and Roberts AB:
Role of transforming growth factor-beta signaling in cancer. J Natl
Cancer Inst. 92:1388–1402. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoshinaga K, Obata H, Jurukovski V,
Mazzieri R, Chen Y, Zilberberg L, Huso D, Melamed J, Prijatelj P,
Todorovic V, et al: Perturbation of transforming growth factor
(TGF)-beta1 association with latent TGF-beta binding protein yields
inflammation and tumors. Proc Natl Acad Sci USA. 105:18758–18763.
2008. View Article : Google Scholar
|
|
40
|
Korkaya H, Liu S and Wicha MS: Regulation
of cancer stem cells by cytokine networks: Attacking cancer's
inflammatory roots. Clin Cancer Res. 17:6125–6129. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou C, Liu J, Tang Y and Liang X:
Inflammation linking EMT and cancer stem cells. Oral Oncol.
48:1068–1075. 2012. View Article : Google Scholar
|
|
42
|
Christians A, Poisel E, Hartmann C,
Deimling AV and Pusch S: Characterization of the epithelial
membrane protein 3 interaction network reveals a potential
functional link to mitogenic signal transduction regulation. Int J
Cancer. 145:461–473. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang YW, Li WM, Wu WJ, Chai CY, Liu HS,
Lai MD and Chow NH: Potential significance of EMP3 in patients with
upper urinary tract urothelial carcinoma: Crosstalk with
ErbB2-PI3K-Akt pathway. J Urol. 192:242–251. 2014. View Article : Google Scholar
|
|
44
|
Lin CY, Lee CH, Chuang YH, Lee JY, Chiu
YY, Lee YH, Jong YJ, Hwang JK, Huang SH, Chen LC, et al: Membrane
protein-regulated networks across human cancers. Nat Commun.
10:31312019. View Article : Google Scholar :
|
|
45
|
Pasto A, Giordano F, Evangelopoulos M,
Amadori A and Tasiotti E: Cell membrane protein functionalization
of nanoparticles as a new tumor-targeting strategy. Clin Transl
Med. 8:82019. View Article : Google Scholar :
|
|
46
|
Schmit K and Michiels C: TMEM proteins in
cancer: A review. Front Pharmacol. 9:13452018. View Article : Google Scholar :
|