Introduction
Glioma are aggressive lethal solid brain tumors
derived from astrocytes and oligodendrocytes present in the central
nervous system (CNS). The most prevalent and aggressive type of
adult gliomas is the grade IV astrocytomas, which are also known as
glioblastoma multiforme (GBM) (1). In the present study, the role of
acid-sensing ion channel 1a (ASIC1a) in gliomagenesis and stemness
was explored as the tumor microenvironment is typically acidic due
to increased glycolysis in tumor cells. In general, due to an
oxygen poor environment, tumor cells switch to aerobic glycolysis
to generate considerable amounts of energy to support their rapid
growth and progression. This results in the continuous generation
of metabolic acids. While acidity is harmful to normal cells,
long-time coevolution of tumor cells with the host has enabled them
to be more adaptable to acidic microenvironments (2,3).
Accumulating evidence has indicated that the acidity of the tumor
microenvironment is associated with stemness phenotype, poor
prognosis of tumor patients, and stimulation of a chemo- and
radio-therapy resistant phenotype (4). Ion channels are transmembrane
proteins involved in regulating various physiological and
pathological functions across biological membranes. The precise
role of ion channels during regulation of cell survival and death
is far from being understood, as ion channels may cause cell
proliferation, cancer development, and metastasis in some cell
types, but they may support regulated cell death in other cell
types. The acid-sensing ion channels (ASICs) are extracellular pH
sensors that are acid responsive and can be transiently activated
by extracellular acidosis to be cation permeable (5). ASIC1 has been reported to contribute
to tumorigenesis in breast, prostate, and pancreatic cancers. The
conclusion regarding the role of ASIC1 in glioma is inconsistent
among different groups. Previous studies revealed that the
knockdown of ASIC1 inhibited glioblastoma cell migration (6-10).
However, previous available microarray data from The Cancer Genome
Atlas (TCGA) revealed that glioma patients with high ASIC1
expression had increased survival compared with those with low
ASIC1 expression, which indicates that the preserved susceptibility
to extracellular pH may impair tumor growth (11). In addition, Tian et al
recently revealed that glioblastoma stem cells (GSCs), which mainly
account for the failure of current treatment against malignant
glioma, express functional ASIC1 and ASIC3 channels (11). Glioblastoma is driven by stem
cell-like cells and is characterized by a block of cellular
differentiation. However, the mechanisms that accompany
differentiation remain poorly understood. Any mechanisms identified
in GSCs with regard to astrocytes, oligodendrocytes, and neuron
differentiation will potentially lead to new strategies to treat
glioblastoma (12,13). The scope that GSCs permanently
develop into a non-proliferative and terminally differentiated
state highlights the significance of differentiation therapy. In
the present study, it was revealed that ASIC1a functions as a tumor
suppressor in glioma stemness and tumorigenesis, which may provide
therapeutic applications for GBM patients by directing GSCs toward
differentiation.
Materials and methods
Antibodies and reagents
The following primary antibodies were used in the
present study: Rabbit monoclonal anti-ASIC1a antibody (cat. no.
35-156465) was purchased from American Research Products, Inc.
Rabbit polyclonal anti-Notch4 antibody (cat. no. 07-189) and
anti-β-actin antibody (product no. A3854) were purchased from
Sigma-Aldrich; Merck KGaA. Rabbit polyclonal anti-Notch3 antibody
(product code ab60087) was purchased from Abcam. Mouse monoclonal
anti-Notch1 antibody (product no. N6786) was purchased from
Sigma-Aldrich; Merck KGaA. Rabbit monoclonal anti-Notch2 antibody
(product no. 5732) and rabbit monoclonal anti-survivin antibody
(clone 71G4B7; product no. 2808) were purchased from Cell Signaling
Technology, Inc. Rabbit polyclonal anti-CD133 antibody (cat no.
NB120-16518) was purchased from Novus Biologicals, LLC. Rabbit
polyclonal anti-aldehyde dehydrogenase 1 antibody (ALDH1; cat no.
GTX123973) was purchased from GeneTex, Inc. Mouse monoclonal
anti-p21 antibody (cat no. sc-817), mouse monoclonal anti-Fas
antibody (sc-8009), and mouse monoclonal anti-cyclin D1 antibody
(sc-8396) were purchased from Santa Cruz Biotechnology, Inc. All
secondary antibodies (goat anti-rabbit, peroxidase-conjugated, cat.
no. AP132P; and goat anti-mouse antibody, peroxidase-conjugated,
cat. no. AP124P) used for western blotting were purchased from
Calbiochem; Merck KGaA. Psalmotoxin (PcTx1) was obtained from
Tocris (cat. no. 5042).
Tissue microarray (TMA)
Glioma tissue arrays from Chinese patients were
purchased from BioCoreUSA Corporation (https://biocoreusa.com/default.aspx) and US Biomax,
Inc. (https://www.biomax.us/). Biopsy features
included age, sex, organ or anatomic site involved, grading, and
pathological diagnosis (H&E-stained sections). Slides from
BioCoreUSA (product no. GL1001b) contained 75 cases of glioma:
grade II, n=51 (astrocytoma, n=47; oligodendroglioma, n=2;
oligoastrocytoma, n=2); grade III, n=12 (anaplastic astrocytoma);
grade IV, n=12 (glioblastoma), and 10 cases of normal brain
tissues. Slides from Biomax (product no. GL803c) contained 68 cases
of glioma: grade II, n=27 (astrocytoma, n=14; oligoastrocytoma,
n=13), grade III, n=4 (astrocytoma); grade IV, n=37 (glioblastoma,
n=31; pleomorphic glioblastoma, n=6), and 5 cases of normal brain
tissues.
Immunohistochemistry (IHC)
IHC staining was performed on 5-µm thick
microarray slides. The slides were fixed using 4% paraformaldehyde
for 30 min at room temperature and blocked by 10% normal horse
serum at room temperature for 20 min. The immunohistochemical
staining for ASIC1a was performed using the rabbit monoclonal
anti-ASIC1a antibody, which is specific for ASIC1a, and a
streptavidin-biotin unlabeled immunoperoxidase technique
(ABC-Elite; Vector Laboratories, Inc.) with diaminobenzidine (DAB)
as a chromogen for ASIC1a. The sections were pretreated in citrate
buffer of pH 6 for 10 min at 100°C, and incubated with primary
antibody ASIC1a diluted at 1:100 at 4°C overnight. The secondary
antibody was diluted at 1:200 and incubation was conducted at room
temperature for 60 min. Mayer's hematoxylin was used for nuclear
counterstaining for 2 min. The slides were then visualized under a
light microscope.
HSORE determination
The staining intensity of cells in TMA was evaluated
as negative or positive in three different bright fields (≥100
cells/field). The semi-quantitative HSCORE was calculated for
ASIC1a using the following equation: HSCORE=Σpi (i + 1), where 'i'
is the intensity with a value of 0, 1, 2, or 3 (negative, weak,
moderate or strong, respectively) and 'pi' is the percentage of
stained cells for each intensity (14). Immunohistochemically stained
slides were blindly reviewed and scored by two independent
investigators.
Plasmids
The generation of short hairpin ASIC1a (shASIC1a),
the plasmid pEGFP-ASIC1a, and corresponding controls were
previously described (15).
Briefly, shASIC1a and control shRNA were purchased from SuperArray
Bioscience Corporation. Each vector contained shRNA under the
control of U1 promoter and green fluorescent protein (GFP) gene for
enrichment of transiently transfected cells. In detail,
SureSilencing shRNA plasmid for human ACCN2 (ASIC1a,
amiloride-sensitive cation channel 2, neuronal) was designed to
specifically knockdown the expression of ASIC1a gene by RNA
interference under transient transfection conditions after
performing appropriate enrichment. The vector contained shRNA under
the control of U1 promoter and GFP gene for enrichment of
transiently transfected cells. The RefSeq accession number
(NM_020039) refers to the representative sequence used to design
the enclosed shRNA. The insert sequence is: GCCAAGAAGTTCAACAAATCT.
The sequence of normal control (NC) is GGAATCTCATTCGATGCATAC. The
plasmid overexpressing ASIC1a named pEGFP-ASIC1a was constructed as
previously described (15).
Briefly, the rat ASIC1a cDNA (NM_024153) was fused with a GFP at
the c-terminal and inserted into pcDNA3. The rat ASIC1a cDNA
(NM_024153) tagged with epitope FLAG (YKDDDDK) at the C terminus
was constructed in plasmid pCDNA3.
Cell culture
Human glioblastoma cell lines: A172 (RRID:
CVCL_0131) and U87MG (HTB-14), a glioblastoma of unknown origin
(RRID: CVCL_0022) were obtained from American Type Culture
Collection (ATCC). All cells were cultured in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% fetal bovine serum
(FBS; both from Thermo Fisher Scientific, Inc.), 50 U/ml penicillin
and 50 µg/ml streptomycin at 37°C. All cell lines had been
authenticated using short tandem repeat profiling within the last
three years. Patient-derived xenoline (PDX) lines: Primary tumor
tissue cubes stored at liquid nitrogen [provided by Dr Yancey G.
Gillespie at the University of Alabama at Birmingham (UAB),
Birmingham, USA] were implanted subcutaneously into the flanks of
male or female 6-8 week-old nude mice under anesthesia
(ketamine/xylazine 90/6 mg/kg BW). A total of 4 athymic nude mice
that were 6-8-weeks old and with an average weight of 25 g,
obtained from Charles River Laboratories, Inc., were maintained on
12-h light/dark cycle with access to food and water ad
libitum. The mice were housed at temperature of 18-23°C with
40-60% humidity. Mice with tumors exceeding 1,000 mm3
were euthanized (by cervical dislocation) and the tumors were
removed for further study. The time interval between implantation
and the end of the experiment ranged from 3-4 weeks. Briefly,
cryopreserved tumor tissues were thawed at 37°C and washed with
phosphate-buffered saline (PBS) before subcutaneous implantation.
To prepare single-cell suspension of viable tumor cells, the
xenograft tumor tissues were harvested and minced with scalpel
blades followed by passing through cell strainers. The cells were
then cultured in DMEM/F-12 media plus 10% FBS, 50 U/ml penicillin
and 50 µg/ml streptomycin for future use. All experiments
were performed with mycoplasma-free cells. The study was carried
out in strict accordance with the recommendations in the Guide for
the Care and Use of Laboratory Animal of the National Institutes of
Health. The protocol was approved (approval no. 20-14) by the
Institutional Animal Care and Usage Committee (IACUC) of Morehouse
School of Medicine (Atlanta, USA).
Transfection of shRNA and DNA
constructs
When the glioblastoma cells reached ~50-75%
confluency in 35-mm dishes, 5 µg of ASIC1a shRNA, or
pEGFP-ASIC1a, FLAG-ASIC1a or corresponding controls were
transfected into glioma cells using the Lipofectamine RNAiMAX or
Lipofectamine 3000 transfection reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) at room temperature for 30 min, according to the
manufacturer's instructions. The transiently transfected glioma
cells expressing each specific construct were maintained in DMEM
containing 10% FBS for further growth for 72 h.
MTT assay
All glioma cells were seeded at 2.5×104
cells in 100 µl of medium per well into 96-well plates and
transfected with specific shRNA or DNA constructs or controls using
Lipofectamine RNAiMAX or Lipofectamine 3000 reagents for the
indicated time-points. Another set of glioma cells was treated with
PcTx1 and the corresponding control. A total of 10 µl of
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
reagent (the ratio of MTT reagent to the medium was 1:10;
Sigma-Aldrich; Merck KGaA) was added into each well and incubated
in the dark at 37°C for 2-4 h. Isopropanol was used to dissolve the
formazan. The absorbance was measured at 570 nm using 690 nm as the
reference and the absorbances were obtained using a CytoFluor™ 2300
plate reader.
Cell migration and invasion assays
The migration and invasion potential were assessed
as previously described (16,17). Briefly, cell culture chambers with
8-µm pore size polycarbonate membrane filters (Corning,
Inc.) were used for cell invasion assays with the filters precoated
with Matrigel at 37°C for 3 h (50 µl; 1.25 mg/ml). Each of
the glioma cell lines/PDX were transfected with or without shASIC1
or ASIC1-GFP for 48 h, and harvested and seeded with
5×105 cells in 200 µl of DMEM supplemented with
1% FBS in the upper chambers. The bottom chambers were filled with
500 µl DMEM supplemented with 10% FBS. Following another 24
h of incubation at 37°C, Matrigel and cells on the upper surface of
the filter were wiped off thoroughly with Q-tips. Cells attached on
the lower surface of the membrane filters were fixed with 4%
paraformaldehyde/PBS for 10 min and stained with 0.5% crystal
violet/methanol for 10 min at room temperature. The cells were then
counted using light microscopy with a magnification of ×10 in 3-4
random fields. Cell numbers under different treatments were
normalized to the appropriate controls. Assays were performed in
triplicate samples and performed in three independent
experiments.
Western blotting
Cells were lysed with lysis buffer (50 mM HEPES, 150
mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, 1%
Nonidet P-40, 100 mM NaF, 10 mM sodium pyrophosphate, 0.2 mM sodium
orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 µg/ml
aprotinin and 10 µg/ml leupeptin). The protein concentration
was determined by BCA assay. Samples (50 µg) were separated
using 4-15% SDS PAGE, and separated proteins were transferred to
nitrocellulose membranes and identified by immunoblotting. The
membranes were blocked using 5% milk for 1 h at room temperature.
Primary antibodies, incubated at 4°C overnight, were obtained from
commercial sources and were diluted at a ratio of 1:1,000 according
to the manufacturer's instructions. Subsequently, the membranes
were incubated with secondary peroxidase-conjugated antibodies (at
a dilution of 1:2,000) at room temperature for 1 h. The blots were
developed with Supersignal Pico or Femto substrate (Pierce; Thermo
Fisher Scientific, Inc.). Densitometric analysis of the bands was
performed with ImageQuant software version 6.1 (Bio-Rad
Laboratories, Inc.).
Flow cytometry
For cell cycle analysis and the apoptosis assay, a
total of 1×106 cells were harvested, fixed in ice-cold
70% ethanol at 4°C for 20 min, and resuspended in PBS for 1 min at
room temperature. Following room temperature centrifugation at 450
× g for 5 min with the brake on low, the cells were resuspended in
200 µl Guava Cell Cycle Reagent (part no. 4500-0220; Luminex
Corporation; containing propidium iodide) and incubated at room
temperature for 30 min while shielded from the light. All samples
were transferred to 96-well microplate plates with a round bottom
and acquired on a Guava easyCyte 8HT Base System (Luminex
Corporation). The percentage of cells in G0/G1, S, and G2/M phases
was determined from the DNA content using guavaSoft 3.1.1 (Luminex
Corporation). The apoptotic glioma cells were detected by flow
cytometry using Annexin V-PE and 7-AAD. The staining procedure was
conducted with a Guava Nexin Reagent kit (part no. 4500-0455;
Luminex Corporation) according to the manufacturer's protocol.
Briefly, after desired treatments (knockdown of ASIC1 by shASIC1a
or overexpression of ASIC1a), cells were collected and resuspended
in 100 µl of 1% FBS (cell concentration was estimated to be
between 2×105 and 1×106 cells/ml) followed by
incubation with 100 µl of Guava Nexin Reagent for 20 min at
room temperature in the dark. The samples were then acquired on a
Guava easyCyte 8HT Base System, which was used to detect apoptotic
cells. Data were analyzed using InCyte software 3.1. To evaluate
CD133 expression by flow cytometry, cells were harvested, washed
with Cell Staining Buffer (cat no. 420201; Biolegend, Inc.), and
then incubated with PE anti-human CD133 antibody (1:200; cat no.
372803; Biolegend, Inc.), for 15-20 min on ice in the dark. Cells
were then washed and suspended in Cell Staining Buffer (at room
temperature for 5 min) for analysis. The data acquired on the Guava
easyCyte 8HT Base System were analyzed using the InCyte software.
ALDH1 enzymatic activity was assessed using an Aldefluor kit (cat
no. 01700; STEMCELL Technologies Inc.), according to the
manufacturer's instructions. Cells suspended in the Aldefluor assay
buffer were incubated with ALDH enzyme substrate,
BODIPY-aminoacetaldehyde (BAAA; 1:200), for 30-60 min at 37°C. As a
control for baseline fluorescence, cells were also treated for
30-60 min at 37°C with the ALDH inhibitor, diethylaminobenzaldehyde
(DEAB; at a 1:100 dilution) contained in the Aldefluor kit.
Fluorescence was detected using the Guava easyCyte 8HT Base System
and analyzed using the InCyte software. Statistical significance
was determined by the paired Student's t-test or one-way ANOVA
test.
Bioinformatics analysis
Kaplan-Meier analysis of the 5-year overall survival
(OS) rates (the cut-off value was the median), according to the
ASIC1 (ACCN2), and the expression of ASIC1 transcript in brain
normal and tumor tissues, were obtained from microarray analysis of
454 glioblastoma patients in the TCGA dataset Affymetrix HT HG
U133A (http://www.betastasis.com/glioma/tcga_gbm/). The
P-value is based on log-rank test or one-way ANOVA test,
respectively.
Statistical analysis
The results obtained in the present study are
expressed as the mean ± SD of at least 3 independent experiments
conducted in triplicate. GraphPad Prism 9 (GraphPad Software, Inc.)
was used for statistical analysis. Paired Student's t-test or
one-way ANOVA followed by Holm-Sidak post hoc tests were performed
for data analysis, and P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of ASIC1a is associated with
improved survival in GBM patients and reduced ASIC1a protein
expression is associated with grade in glioma patients
To determine whether ASIC1 gene expression is
related to patient survival, the TCGA database was analyzed. ASIC1
mRNA expression levels in glioma tissue were lower than those in
normal brain, irrespectively of GBM subtypes (classical,
mesenchymal, neural, and proneural) as detected by Affymetrix HT HG
U133A (P<0.01; Fig. 1A). The
dataset contained information on 454 glioblastoma patients,
classified based on WHO classification as GBM (WHO grade IV
glioma). When all GBM patients were analyzed in a pooled setting,
and the median value was selected as the cut-off point, the 5-year
OS rate, as revealed by the Kaplan-Meier survival curves (Fig. 1B), was significantly higher
(P=0.0184; log-rank test) in those with high ASIC1 transcript
levels (red curve) compared with those with low expression (blue
curve). When glioma patients were grouped into multiple strata, GBM
patients with high ASIC1 levels survived significantly longer than
those with low expression levels (Fig. S1). In GBM patients with classical
(Fig. S1A), neural (Fig. S1B) and proneural (Fig. S1C) subtypes, patients with high
ASIC1 levels were positively associated with longer survivals. GBM
patients in groups with chemotherapy (Fig. S1D), and groups without
chemotherapy (Fig. S1E) or
hormonal therapy demonstrated beneficial survival when ASIC1
expression levels were high. Collectively, the Kaplan-Meier
analyses revealed a significant survival benefit in glioma patients
with elevated ASIC1 expression. The ASIC1 protein expression was
then examined by IHC in glioma brain TMA obtained from Chinese
patients at BioCoreUSA and US Biomax, Inc. ASIC1 protein was
expressed in grade II astrocytoma (Fig. 1C), grade III astrocytoma (Fig. 1D), grade IV GBM (Fig. 1E) and normal brain tissues
(Fig. 1F). Quantification of the
IHC results revealed that positive cytoplasmic staining of ASIC1a
was significantly lower in grade IV GBM (n=49; P<0.001; 95% CI:
0.7674-1.857), grade III gliomas (n=16; P<0.001; 95% CI:
0.5730-1.903), and grade II gliomas (n=78; P<0.001; 95% CI:
0.4293-1.475) compared with that of normal brain tissue (n=15;
Fig. 1G). Similarly, nuclear
staining of ASIC1a in grade IV GBM (P<0.001; 95% CI:
0.5711-1.701) and grade III gliomas (P<0.05; 95% CI:
0.1143-1.494) was significantly lower than that of normal brain
tissue (Fig. 1G). Next, the
expression of ASIC1a was compared in different grades of glioma. It
was revealed that cytoplasmic staining of ASIC1a was significantly
decreased in grade IV GBM compared with that in grade II gliomas
(P<0.05) (Fig. 1H), while
nuclear staining of ASIC1a was significantly decreased in grade IV
GBM compared with that of grade II gliomas (P<0.001) (Fig. 1H). The inverse association between
increased ASIC1a protein expression and glioma grades strongly
indicated the potential of ASIC1a as a prognostic marker in glioma
patients.
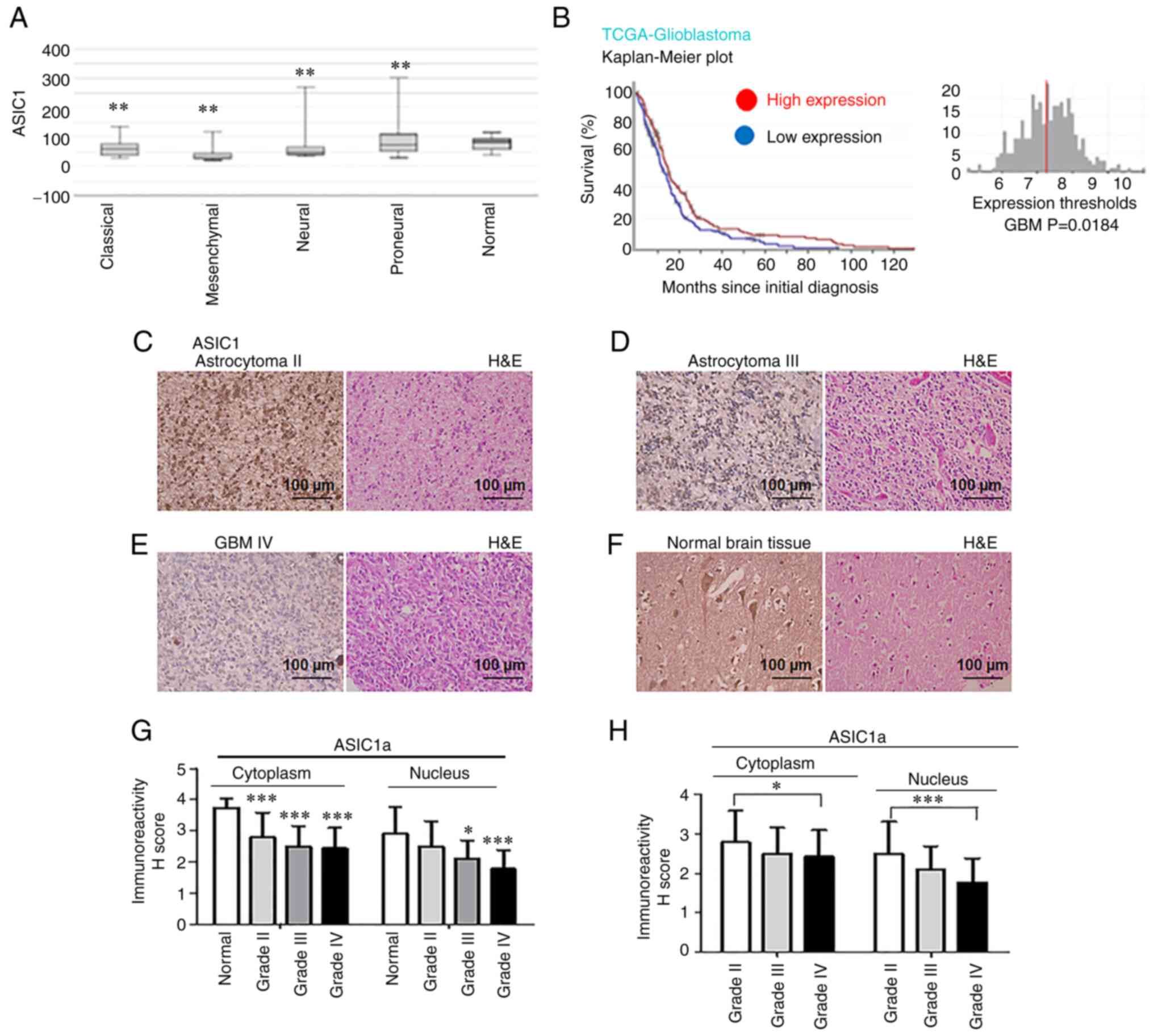 | Figure 1Expression of ASIC1 is associated
with improved survival in glioblastoma patients, and reduced ASIC1a
protein expression is associated with grade progression in glioma
patients. (A) ASIC1 mRNA expression levels in brain tissues of
different molecular subtypes of glioma patients were analyzed by
Affymetrix HT HG U133ATCGA data. (B) Kaplan-Meier survival curve
for the 5-year OS rate of glioma patients. The cut-off was set at
the median. (C-F) ASIC1a protein was expressed in (C) grade II
gliomas, (D) grade III gliomas, (E) grade IV GBM and (F) normal
brain tissue. Images were captured at a magnification of ×40. (G)
Quantification of IHC results in grade IV GBM, grade III, and II
gliomas compared with that of normal brain tissue. (H)
Quantification of IHC results compared among different grades (IV
GBM, grade III, and II gliomas). *P<0.05,
**P<0.01 and ***P<0.001. ASIC1a,
acid-sensing ion channel 1a; GBM, glioblastoma multiforme; IHC,
immunohistochemistry; H&E, hematoxylin and eosin. |
Downregulation of ASIC1a promotes glioma
cell proliferation, while overexpression of ASIC1a inhibits its
growth
To evaluate the function of ASIC1a on glioma cell
growth, glioma A172 and U87MG cells were cultured at ~70%
confluence followed by transfection with either a shASIC1a or
ASIC1a-expressing construct (ASIC1-pEGFP) for 24, 48, 72, and 96 h
to decrease or increase ASIC1a expression levels, respectively, as
previously described (14). The
effects of ASIC1a on glioma cell proliferation were then detected
at indicated time-points using MTT assays. As revealed in Fig. 2, reduced ASIC1a expression by
transient transfection of shASIC1a significantly increased the
growth rate of A172 (Fig. 2A,
left panel) and U87MG cells (Fig.
2C, left panel), where cell proliferation was increased in A172
and U87MG cells by a maximum of 32.8 and 30% at 96 h, respectively.
Fig. 2B (upper panel) and D
(upper panel) revealed the high transfection efficiency in A172 and
U87MG cells, respectively, by transient transfection of shASIC1a
vectors. Conversely, elevated ASIC1a expression by ASIC1a-GFP
impeded the growth of the two aforementioned cell lines, where cell
proliferation was decreased in A172 (Fig. 2A, middle panel) and U87MG cells
(Fig. 2C, middle panel) by a
maximum of 42.3 and 30.3% at 96 h, respectively. Fig. 2B (lower panel) and D (lower panel)
revealed the high transfection efficiency in A172 and U87MG cells
by transient transfection of ASIC1a-GFP vectors. Collectively,
these studies of loss- and gain-of-function indicated that ASIC1a
may act as a tumor suppressor in glioma tumorigenesis. To provide
additional evidence, the effects of PcTx1 (10 nM), a potent and
selective inhibitor to the ASIC1 channels either by homomeric
ASIC1a channels (18) or
heteromeric ASIC1a/2b channels (19), on glioma cell proliferation were
examined at room temperature for 3 days. Similar to shASIC1a, the
growth rate of A172 (Fig. 2A,
right panel) and U87MG cells (Fig.
2C, right panel) was significantly increased by incubation with
10 nM PcTx1 at a maximum of 49.7 and 39.5% at 96 h, respectively.
Similar results were obtained in PDX, which represented individual
patient tumors in an improved way. PDX-L12 cells, a PDX with neural
subtype, have wild-type genes of EGFR, PTEN, CDKN2A, NF-κB, and
amplified genes of CDK4/MDM2 and CSNK2A with a deleted TP53
(20). Silencing ASIC1a by
shASIC1a or PcTx1 treatment increased the percentage of growth of
PDX-L12 cells at a maximum of 32.5 (Fig. 2E, left panel) and 40.6% (Fig. 2E, right panel), respectively, at
96 h. The anticipated results were observed when PDX-L12 cells
overexpressed the ASIC1a gene and their growth capacity decreased
gradually and reached a maximal level of ~40.6% at 96 h (Fig. 2E, middle panel). Fig. 2F revealed the high transfection
efficiency in PDX-L12 cells by transient transfection of shASIC1a
(Fig. 2F, upper panel) and
ASIC1a-GFP (Fig. 2F, lower
panel). Collectively, these results strongly indicated that ASIC1a
decreased glioma cell proliferation and ASIC1a may act in a tumor
suppressor-like role in glioma growth (P<0.05; one-way ANOVA).
All data were from triplicate samples performed in three different
independent experiments.
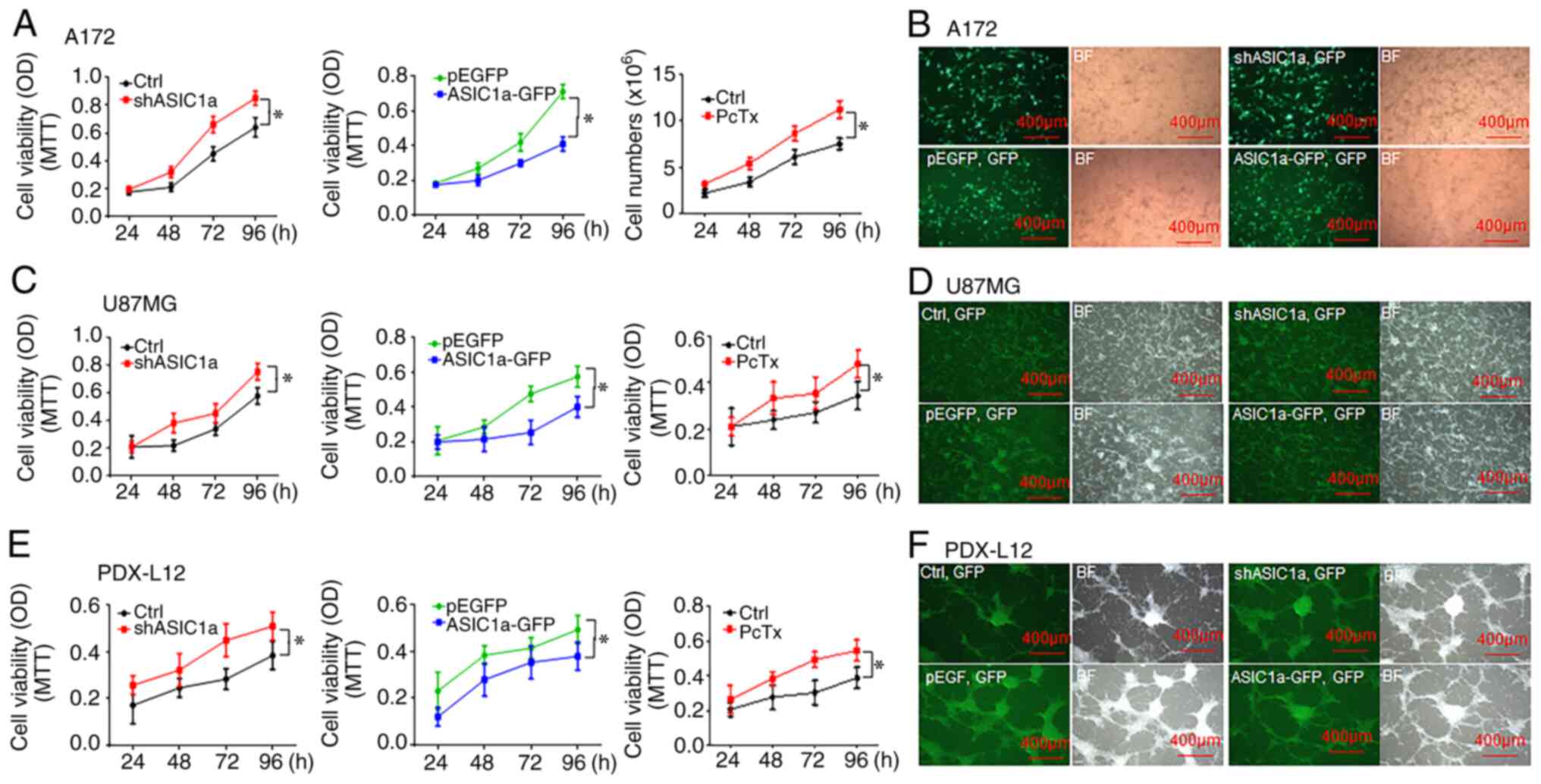 | Figure 2Downregulation of ASIC1a promotes
glioma cell proliferation, while overexpression of ASIC1a inhibits
its growth. (A) A172 cells were transfected with shASIC1a (left),
treated with ASIC1a selective inhibitor PcTx1 (right), and
transfected with ASIC1a-GFP (middle) for 24 to 96 h followed by an
MTT assay. (B) Transfection efficiency of A172 cells was revealed
by the expression of GFP marker. (C) U87MG cells were transfected
with shASIC1a (left), treated with ASIC1a selective inhibitor PcTx1
(right), and transfected with ASIC1a-GFP (middle) for 24 to 96 h
followed by an MTT assay. (D) Transfection efficiency of U87MG
cells was revealed by the expression of GFP marker. (E) PDX-L12
cells were transfected with shASIC1a (left), treated with ASIC1a
selective inhibitor PcTx1 (right), and transfected with ASIC1a-GFP
(middle) for 24 to 96 h followed by an MTT assay. (F) Transfection
efficiency of PDX-L12 cells was revealed by the expression of GFP
marker. *P<0.05. ASIC1a, acid-sensing ion channel 1a;
sh, short hairpin; PcTx1, psalmotoxin; GFP, green fluorescent
protein. |
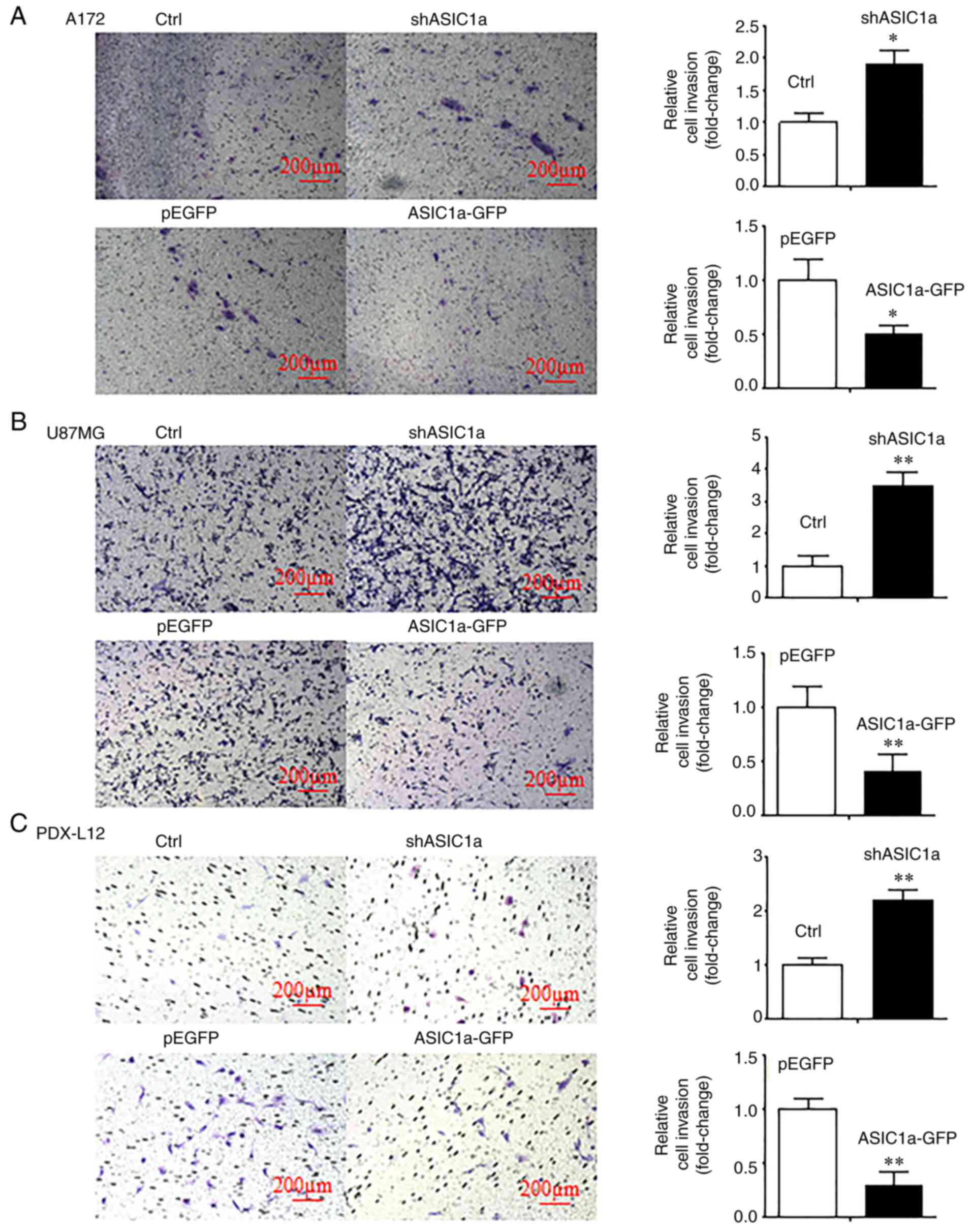
Downregulation of ASIC1a increases glioma
cell invasion, while overexpression of ASIC1a decreases its
invasion
Next, the possible roles of ASIC1a in glioma cell
invasion were determined. Transwell invasion assays were conducted
on A172, U87MG and PDX-L12 cells that were transiently transfected
with either shASIC1a or ASIC1a-GFP and corresponding controls. The
results revealed that the silencing of ASIC1a enhanced the number
of invasive cells by 1.9 (Fig.
3A, upper panels), 3.50 (Fig.
3B, upper panels), and 2.2 fold (Fig. 3C, upper panels), respectively, as
compared with those of the controls. Conversely, the ectopic
expression of ASIC1a inhibited the number of invasive cells by 50
(Fig. 3A, lower panels), 70
(Fig. 3B, lower panels) and 83%
(Fig. 3C, lower panels),
respectively. These results indicated that ASIC1a decreases glioma
cell invasion, and ASIC1a may act in a tumor suppressor-like role
in glioma metastasis (P<0.05 and P<0.01; paired Student's
t-test). All data were from triplicate samples performed in three
different independent experiments.
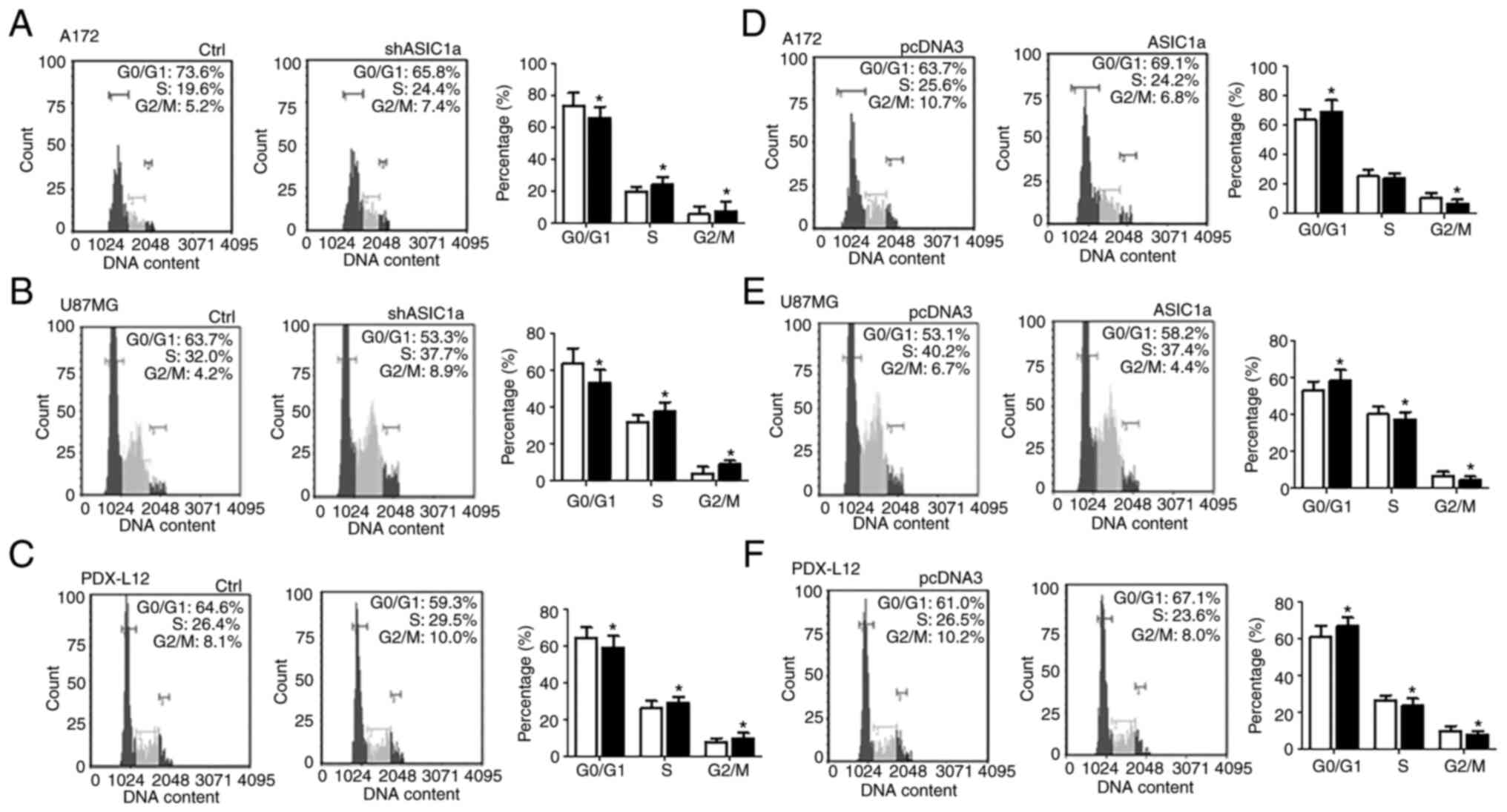
ASIC1a suppresses the growth and
proliferation of glioma cells through G1/S arrest and the induction
of apoptosis
To gain further insight into the role of ASIC1a in
inhibiting glioma cell growth, the contribution of ASIC1a to cell
proliferation vs. cell death was investigated. Cell cycle
distribution was firstly assessed using propidium iodide flow
cytometric analysis (Fig. 4).
Treatment with shASIC1a increased the percentage of A172 (Fig. 4A) glioma cells in the S phase
(19.6 to 24.4%) and G2/M phase (5.2 to 7.4%), and concomitantly
decreased cells in the G0/G1 phase (73.6 to 65.8%). Similar results
were observed in U87MG and PDX-L12 cells. Treatment with shASIC1a
increased the percentage of U87MG cells (Fig. 4B) in the S phase from 32.0 to
37.7% and the G2/M phase from 4.2 to 8.9%, while it decreased the
G0/G1-phase cells from 63.7 to 53.3%. Concurrently, shASIC1a
treatment increased the percentage of PDX-L12 cells (Fig. 4C) in the S phase from 26.4 to
29.5% and the G2/M phase from 8.1 to 10.0%, while it decreased the
G0/G1-phase cells from 64.6 to 59.3%. As anticipated, increased
expression of ASIC1a levels by introduction of ectopic ASIC1
resulted in a decrease in the S and G2/M phases, and an increase in
the G0/G1 phase to varying degrees, depending on the cell lines. In
detail, ASIC1a-FLAG-transfected A172 cells revealed a decrease in
the percentage of cells from 25.6 to 24.2% in the S phase, from
10.7 to 6.8% in the G2/M phase, while the percentage of cells in
the G0/G1 phase was increased from 63.7 to 69.1% (Fig. 4D). Similarly, for U87MG cells,
increased ASIC1a expression reduced cells in the S phase (40.2 to
37.4%) and the G2/M phase (6.7 to 4.4%) and increased the
G0/G1-phase cells (53.1 to 58.2%) (Fig. 4E). The data from PDX-L12 cells
further consolidated the findings obtained from the two glioma cell
lines A172 and U87MG. ASIC1a-FLAG-transfected PDX-L12 cells
exhibited decreased cell numbers in the S phase from 26.5 to 23.6%
and decreased cell numbers in the G2/M phase from 10.2 to 8.0%, as
well as increased cell numbers in the G0/G1 phase from 61.0 to
67.1% (Fig. 4F).
Next, it was determined whether shASIC1a decreased
apoptosis by flow cytometry with Annexin V (detects phosphatidyl
serine in the outer leaflet of the plasma membrane) and 7-AAD
(detects cells with disrupted membrane integrity). Early apoptotic
cells were detected by Annexin V positivity, while late apoptotic
cells were positive for both markers. It was revealed that shASIC1a
decreased the rates of early (16.90 to 12.44% for A172, Fig. 5A, upper panels; and 8.85 to 1.99%
for U87MG, Fig. 5B, upper panels)
and late (1.64 to 1.50% for A172, Fig. 5A, upper panels; and 5.38 to 0.99%
for U87MG, Fig. 5B, upper panels)
apoptotic cells in glioma cell lines. As anticipated, PDX-L12
behaved in the same way as the two aforementioned glioma cell lines
with decreased early (28.32 to 12.26%) and late apoptotic cells
(12.65 to 8.00%) (Fig. 5C, upper
panels) upon ASIC1a silencing. Conversely, the elevated levels of
ASIC1a in ASIC1a-FLAG-transfected glioma cells revealed an increase
in early and late apoptosis to varying degrees, depending on cell
lines. Upon transfection of ASIC1a-FLAG, the early and late
apoptotic cells were increased i) in A172 cells: from 16.4 to
17.91% and 15.46 to 22.18% (Fig.
5A, lower panels); ii) in U87MG cells: from 4.3 to 7.00% and
2.15 to 2.82% (Fig. 5B, lower
panels); and iii) in PDX-L12 cells: from 20.25 to 26.00% and 13.45
to 16.04% (Fig. 5C, lower
panels). Collectively, our data indicated that ASIC1a suppresses
the growth and proliferation of glioma cells through G1/S arrest
and the induction of apoptosis.
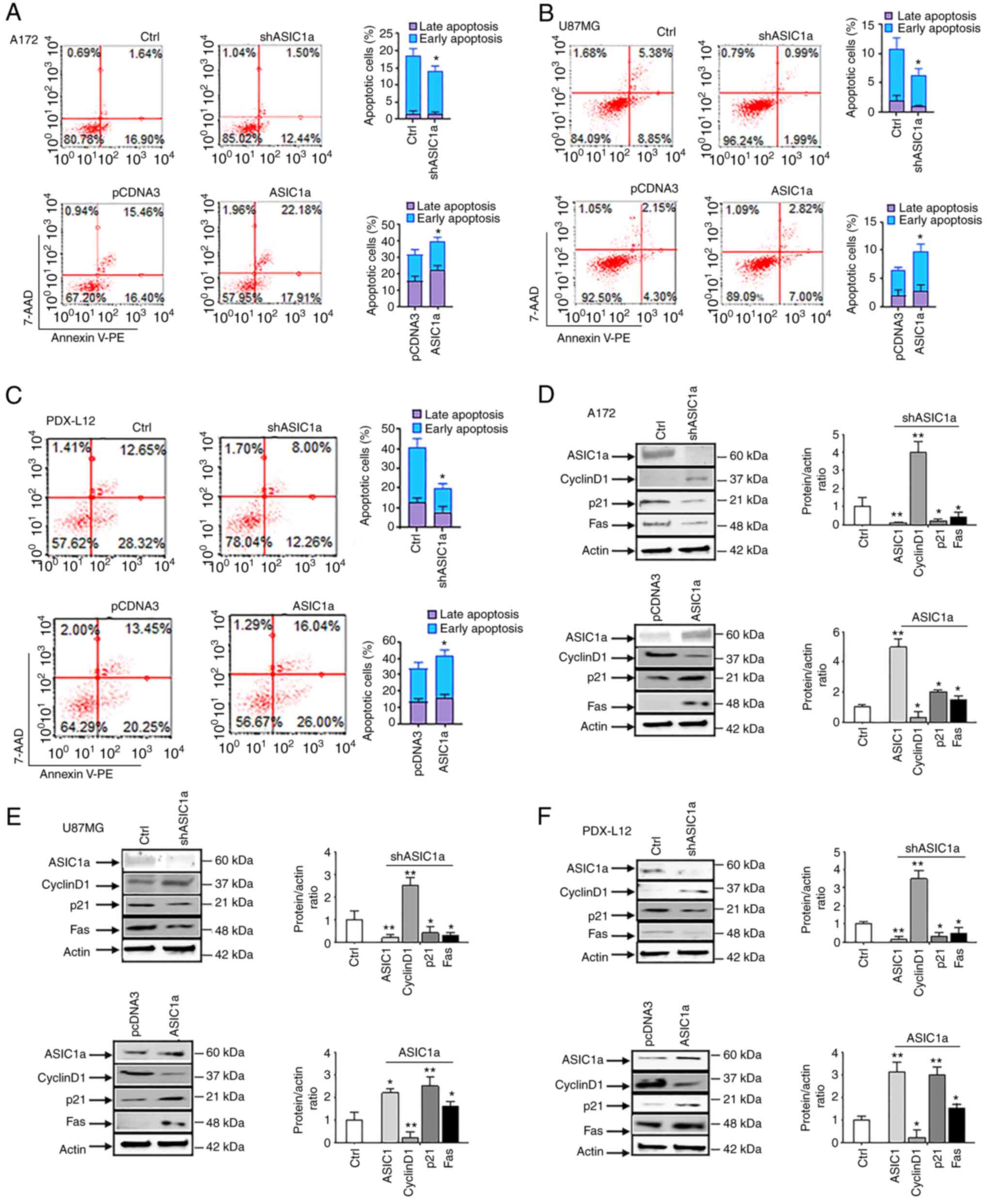 | Figure 5ASIC1a suppresses the growth and
proliferation of glioma cells through the induction of apoptosis.
(A-C) Apoptosis was analyzed by flow cytometric analysis of Annexin
V-PE staining after shASIC1 treatment in (A, upper panel) glioma
cells A172, (B, upper panel) U87MG and (C, upper panel) PDX-L12
cells. Apoptosis was also examined by overexpression of ASIC1a in
(A, lower panel) A172, (B, lower panel) U87MG and (C, lower panel)
PDX-L12 cells. (D-F) The cell cycle regulatory proteins cyclin D1
and p21, as well as apoptotic-related protein Fas were examined
when the ASIC1 gene was silenced by shASIC1a in (D, upper panel)
A172, (E, upper panel) U87MG, and (F, upper panel) PDX-L12 cells.
Cyclin D1, p21, and Fas were also detected when the ASIC1a gene was
overexpressed in (D, lower panel) A172, (E, lower panel) U87MG and
(F, lower panel) PDX-L12 cells. The bar graphs indicate the mean ±
SD of three independent experiments. All data represent a
representative experiment from three independent experiments.
*P<0.05 and **P<0.01. ASIC1a,
acid-sensing ion channel 1a; sh, short hairpin. |
ASIC1a modulates the expression of cell
cycle regulatory and apoptosis-related proteins in glioblastoma
cells
Cyclin D1 plays crucial roles in the progression of
cells through the S and G2/M phases (21), and it is amplified and
overexpressed in numerous cancers (22). Results from western blot analysis
revealed that ASIC1a negatively regulated cyclin D1. In detail,
shASIC1a-transfected glioma cells exhibited elevated levels of
cyclin D1 expression (Fig. 5D-F,
upper panels) whereas ASIC1a-FLAG-transfected glioma cells had
decreased cyclin D1 expression (Fig.
5D-F, lower panels). These findings were in agreement with the
previous data obtained by the flow cytometric analyses (Fig. 4) which indicated that ASIC1a
induces a G1 arrest at the expense of S and G2/M phases. The
protein p21, a cyclin/CDK inhibitor which accompanies increased
levels of cyclin D1, is induced by cyclin D1 through an E2F
mechanism (22). Our results
corroborated these findings that ASIC1a positively regulated p21
protein (Fig. 5D-F).
Interestingly, A172 and U87MG cells have wild-type TP53 (p53)
genes, while PDX-L12 has a deleted p53 gene which suggests that in
PDX-L12, ASIC1a upregulates p21 in a p53-independent manner. It was
further confirmed that apoptosis occurred at a molecular level with
changes in levels of Fas protein (Fig. 5D-F). Apoptotic-inducing protein
Fas was decreased by silencing of ASIC1a (5D-F, upper panels) while
Fas protein was elevated by overexpression of ASIC1a (5D-F, lower
panels). The bar graphs indicate the mean ± SD of three independent
experiments. All data represent a representative experiment from
three independent experiments.
GSC Markers CD133, ALDH1 and Notch
signaling are negatively associated with ASIC1a in GBM
The changes in the number of CD133+ cells
in response to changes in ASIC1a expression in glioma cells were
firstly examined by flow cytometry. It was revealed that once
ASIC1a was knocked down by shASIC1a, the number of
CD133+ cells were increased in A172 (Fig. 6A, left two panels), U87MG
(Fig. 6B, left two panels) and
PDX-L12 cells (Fig. 6C, left two
panels). These results indicated that the downregulation of ASIC1a
resulted in an enlarged GSC population. When ASIC1a expression
levels were elevated by introducing ASIC1a-FLAG into glioma cells,
the number of CD133+ cells revealed a decrease in A172
(Fig. 6A, right two panels),
U87MG (Fig. 6B, right two panels)
and PDX-L12 cells (Fig. 6C, right
two panels). To further verify our findings, the effects of PcTx1
on the expression of CD133 on PDX-L12 glioma cells were examined.
As anticipated, PcTx1 effectively increased expression of CD133
compared with that of the control (Fig. S2). To address the question of
whether ASIC1a would affect ALDH1, another GSC marker, the
ALDEFLUOR assay was performed on an identical model of glioma cells
aforementioned, in which ASIC1a was either underexpressed or
overexpressed by shASIC1- or ASIC1-FLAG-related constructs. ASIC1a
silencing increased the number of ALDH1-positive cells from 3.45 to
13.54% in A172 (Fig. 6D, left
five panels), from 4.01 to 7.03% in U87MG (Fig. 6E, left five panels), and from 8.49
to 14.91% in PDX-L12 cells (Fig.
6F, left five panels). In accordance with the relationship of
ASIC1a and CD133, here, our results produced further evidence that
ASIC1a knockdown resulted in an increased GSC population as defined
by the ALDH1+ population. Similarly, overexpression of
ASIC1a decreased the ALDH1+ cells in all of the three
glioma cells examined. In detail, compared with corresponding
controls, the ALDH1+ cells decreased from 3.61 to 1.77%
in A172-transfected ASIC1a-FLAG cells (Fig. 6D, right five panels), from 4.65 to
2.25% in U87MG-transfected ASIC1a-FLAG cells (Fig. 6E, right five panels), and from
8.71 to 2.58% in PDX-L12-transfected ASIC1a-FLAG cells (Fig. 6F, right five panels).
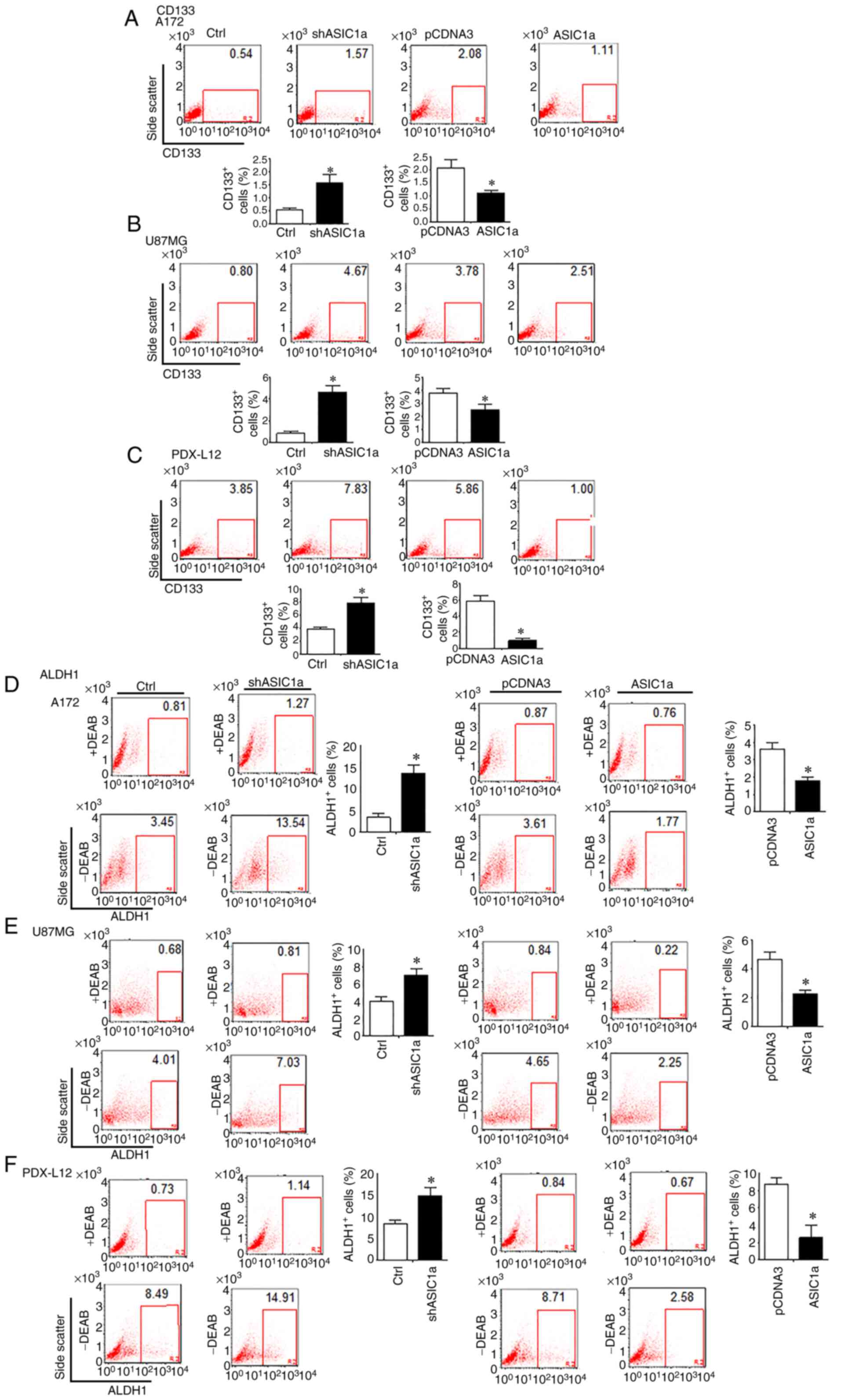 | Figure 6GSC markers CD133 and ALDH1 are
negatively associated with ASIC1a in glioblastoma multiforme. (A-C)
Flow cytometric analysis to assess CD133 expression in (A, left two
panels) A172, (B, left two panels) U87MG, and (C, left two panels)
PDX-L12 glioma cells by ASIC1a silencing. CD133 expression was
determined when (A, right two panels) A172, (B, right two panels),
U87MG, and (C, right two panels) PDX-L12 cells overexpressed
ASIC1a. (D-F) ALDH1 enzymatic activities were determined by the
ALDEFLUOR assay which was performed in ASIC1a-knockdown (D, left
five panels) A172, (E, left five panels), U87MG and (F, left five
panels) PDX-L12 cells. ALDH1 enzymatic activities were also
determined in (D, right five panels) A172, (E, right five panels)
U87MG and (F, right five panels) PDX-L12 cells overexpressing
ASIC1a. Bars on the right of each image represent the mean ± SD
after normalization to control. All results are representative of
three separate experiments. *P<0.05. GSC,
glioblastoma stem cells; ALDH1, aldehyde dehydrogenase 1; ASIC1a,
acid-sensing ion channel 1a; sh, short hairpin. |
It was previously reported that ASIC1 promotes
differentiation of neuroblastoma by negatively regulating the Notch
signaling pathway (14). The
Notch signaling pathways plays critical roles in the maintenance
and differentiation of neural stem cells (NSC) (23) and can maintain GSCs in an
undifferentiated state (24). It
was therefore determined whether ASIC1a is a critical regulator of
Notch1 gene expression during gliomagenesis. Upon ASIC1a
downregulation by shASIC1a, in A172 cells, the active form of
Notch, intracellular domains of Notch1 (Notch1/NICD), Notch2/NICD,
Notch3/NICD, Notch4/NICD, along with the Notch target survivin
expression were increased. The GSC markers CD133 and ALDH1 were
increased as well (Fig. 7A).
U87MG (Fig. 7B) and PDX-L12 cells
(Fig. 7C) exhibited similar
patterns based on ASIC1a silencing, with enhanced expression of
Notch active forms of Notch1/NICD, Notch2/NICD, Notch3/NICD,
Notch4/NICD, Notch target survivin, and GSC markers CD133 and ALDH1
in response to ASIC1a knockdown. To further detect the association
between ASIC1a and Notch receptors, A172, U87MG and PDX-L12 cells
were transfected with ASIC1-FLAG to overexpress ASIC1a protein
along with the control vector pCDNA3. The anticipated results were
observed in that ASIC1a-overexpressing A172 (Fig. 7D), U87MG (Fig. 7E) and PDX-L12 (Fig. 7F) cells exhibited decreased Notch
active forms of Notch1/NICD, Notch2/NICD, Notch3/NICD, Notch4/NICD,
Notch target survivin and GSC makers CD133 and ALDH1. These results
indicated that ASIC1a causatively induced the inactivation of
Notch, reduced the expression of GSC markers CD133 and ALDH1, and
played a critical role in glioma stemness.
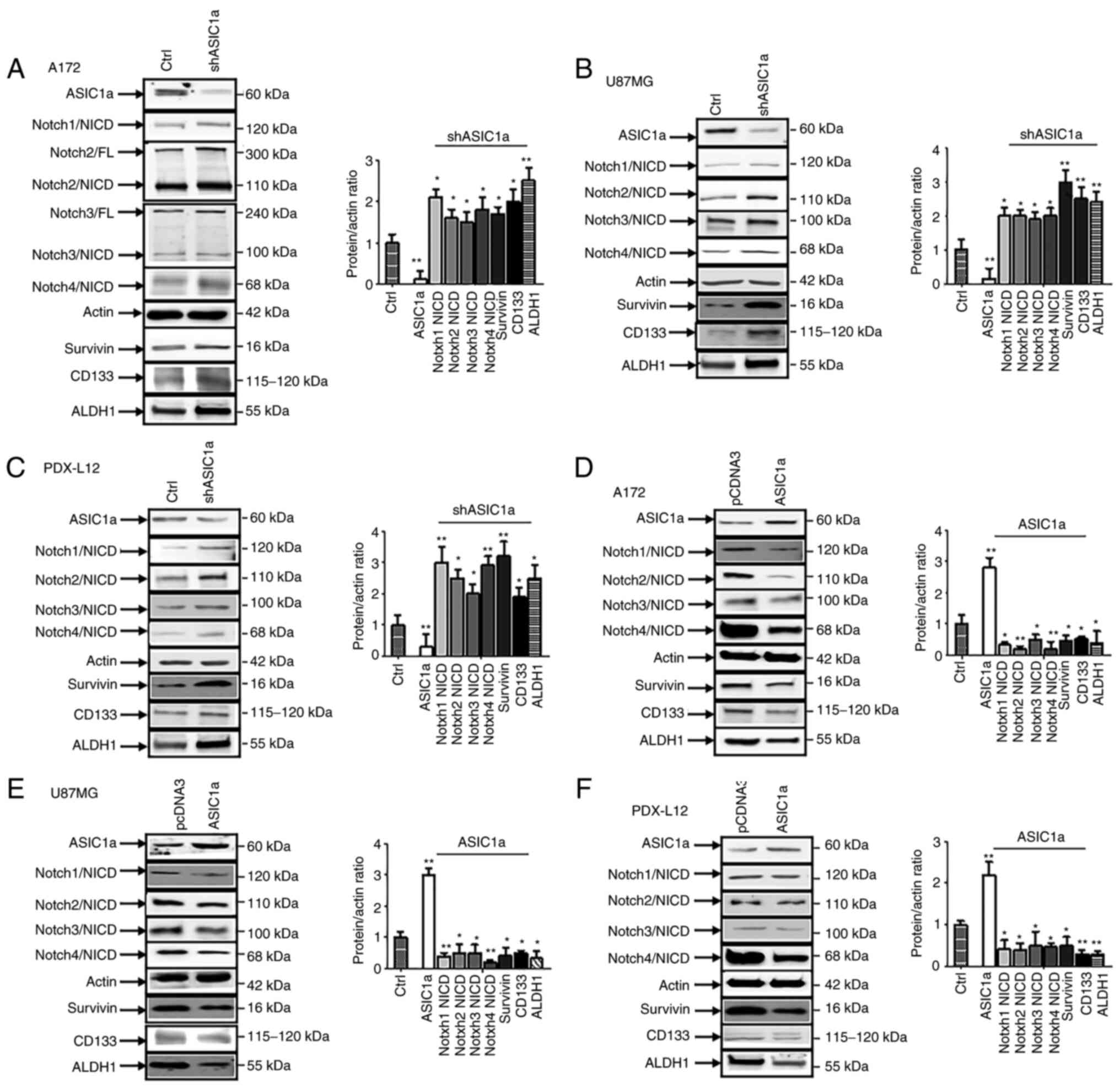 | Figure 7Notch signaling is negatively
associated with ASIC1a in glioblastoma multiforme. (A-C) The glioma
cells were transfected with shASIC1a and control followed by
assaying protein expression of ASIC1a, active form of Notch1,
Notch2, Notch3, Notch4, and Notch target survivin, as well as GSC
markers CD133 and ALDH1 by western blotting in (A) A172, (B) U87MG,
and (C) PDX-L12 cells. (D-F) Subsequently, the glioma cells (D)
A172, (E) U87MG and (F) PDX-L12 were transfected with ASIC1a-FLAG
followed by assaying protein expression of ASIC1a, active form of
Notch1, Notch2, Notch3, Notch4 and Notch target survivin, as well
as GSC markers CD133 and ALDH1 by western blotting.
*P<0.05 and **P<0.01. ASIC1a,
acid-sensing ion channel 1a; sh, short hairpin; GSC, glioblastoma
stem cells; ALDH1, aldehyde dehydrogenase 1. |
Discussion
In the present study, two glioma cell lines, A172
and U87MG and one PDX line were utilized based on their different
molecular characteristics. A172 and U87MG both have wild-type
TP53, PTEN mutations, and CDKN2A
(p14ARF/p16INK4a) deletion (25). However, U87MG cells have another
CDKN2C (p18INK4c) mutation, express high
levels of VEGF as compared with A172 expressing high levels of bFGF
(26). PDX-L12 cells, a PDX with
neural subtype, have wild-type genes of EGFR, PTEN,
CDKN2A, NF-κB, and amplified genes of CDK4/MDM
and CSNK2A with a deleted TP53. Major findings from
the present study include: i) Expression of ASIC1 was associated
with improved survival in glioblastoma patients and reduced ASIC1a
protein expression was associated with grade progression in glioma
patients; ii) downregulation of ASIC1a increased glioma cell
proliferation and invasion, while upregulation of ASIC1a decreased
their proliferation and invasion; iii) ASIC1a suppressed the growth
and proliferation of glioma cells through G1/S arrest and induced
apoptosis; and iv) ASIC1a causatively induced the inactivation of
Notch, reduced expression of GSC markers CD133 and ALDH1, and
played an important role in glioma stemness.
A total of 4 ASIC genes (ASIC1, ASIC2, ASIC3 and
ASIC4) and splice variants for ASIC1 (ASIC1a, ASIC1b, and ASIC1b2)
and ASIC2 (ASIC2a and ASIC2b) have been identified and found to be
expressed in a variety of cell types (27,28). The functional ASICs are trimeric
assemblies with each subunit consisting of two transmembrane
domains (2). ASICs are
voltage-independent ion channels and have the highest expression in
the brain, mainly in the central nervous system (28,29); in addition, they are also
expressed in the retina (27,30,31), lung epithelia (31), bone and cartilage (32), pituitary gland (33), and testis (34). As extracellular acidosis is
typically concomitant with brain injury, ASICs, the main neuronal
H+ receptor in neurons, play an important role in
neuronal injury under various injurious conditions in the brain
(35). In ischemia-induced brain
injury, multiple endogenous factors (lactate, sperimine, and
dynorphins) potentiate ASIC1a channel-mediated ischemic injury.
Multiple sclerosis (MS) is a demyelinating disease in CNS and is
associated with prolonged inflammation and acidification. A recent
clinical study conducted by Arun et al revealed that
amiloride, an ASIC1 inhibitor, has a promising neuroprotective
effect by reducing brain atrophy of MS patients, axonal damage, and
myelin loss (36). Brain pH is
reduced in traumatic brain injury (TBI) patients due to massive
disruption of metabolism; and in TBI patients whose ASIC1a
expression was increased in the brain, amiloride or PcTx1 (ASIC1
selective inhibitor) attenuated the severity of brain injury
(37). ASIC-mediated responses
result in loss of dopaminergic neurons in Parkinson's disease (PD).
In patients with PD, ASIC1 inhibition with amiloride or PcTx1
alleviates the reduction of immunoreactivity of tyrosine
hydroxylase and dopamine transporter, and consequently prevents the
loss of dopaminergic neurons, and therefore impedes PD progression
(38).
Adaptions to the highly acidic microenvironment are
crucial steps in the development of invasive cancer (3). As a result, proton (H+)
concentration increases within the lumen and causes the interior of
the lumen to become highly acidic. Cancer cells have a lower
extracellular pH (pHe) and higher intracellular pH (pHi) than
normal cells in acute acidosis conditions (2). Solid tumors are characterized by a
highly acidic microenvironment that may compromise the
effectiveness of antitumor immunity (39). The potential for future clinical
translation lies with the neutralization of tumor acidity with
bicarbonate therapy to inhibit the growth of some cancer types and
improve antitumor responses to immunotherapy (39). Acidic conditions that are
independent of restricted oxygen promote the expression of GSC
markers, self-renewal, and tumor growth (40). Activity of ion channels is closely
related to malignant features of tumor cells such as the lack of
differentiation, increased proliferation, increased migratory and
invasive phenotypes, and elevated chemoresistance (41-43). It has been revealed that glioma
cells express higher levels of potassium, sodium, and chloride
channels compared with normal astrocytes (29,44,45), indicating that these ion channels
may contribute to glioma progression. The epithelial sodium
channel/degenerin (ENaC/DEG) superfamily includes ENaCs and ASICs
(6,7,46).
ASIC channels are extracellular pH sensors that are acid-responsive
and can be transiently activated by extracellular acidosis and
become permeable to cations (2,28,47). The cells of high-grade gliomas
express RNA for numerous different subunits of the ASIC and ENaC
families (6). Unlike ENaC, ASICs
are proton-gated cation-selective channels most permeable to
Na+ ions (29,47,48). ASIC1a and heteromeric ASIC1a/2b
channels are permeable to Ca2+ and can cause an
accumulation of intracellular Ca2+ in neurons (49,50). The studies on the role of ASIC1 in
gliomagenesis are controversial. Sun et al reported that
ASIC1 and CaMKII form a functional complex at the plasma membrane
in GBM cells, which promotes GBM migration. However, their results
were only based on experimentation in the U251-MG cell line, which
may represent a selection bias (51). Previous studies performed in 2003
and 2009, that support the mitogenic role of ASIC1 reported that
silencing of ASIC1 inhibits glioblastoma cell migration (6,7).
The mechanical studies from this group demonstrated that ASIC
interacted with several biochemical molecules such as integrin-β
and α-actinin (9), ENaC subunits
(8), Hsc70 (52) or cleaved by serine protease
matriptase (53) to accomplish
its functions. The apparent limitation of their studies is lack of
prognostic information drawn from big data bioinformatics, which is
critical to identify the difference between tumor suppressor genes
and oncogenes.
The study supporting a tumor suppressor role of
ASIC1 originated from previous research from Tian et al. In
rat C6 glioma cells, functional activation of ASIC1 induced a short
depolarization or a transient calcium influx even with persistent
acidic stimulation. Notably, GSC expresses functional ASIC1a and
ASIC3. Microarray data from their study revealed that the
expression of ASIC1 and ASIC3 was associated with improved survival
of glioma patients, which indicated that the preserved
susceptibility to extracellular pH may impair tumor growth
(11). In 2017, our group first
revealed that ASIC1 induces neuroblastoma differentiation (14). Later, Zhang et al revealed
that both human-induced pluripotent stem cell-(hiPSC)-derived
neural progenitor cell (hiPSC-NPC) and hiPSC-NPC-derived neurons
express abundant ASIC1 mRNA (54). These findings provided an
indication of the important relationship between stem cells and
ASIC1. As acidic stress maintains (55) or promotes (40) glioblastoma stem cell-like
phenotype, the acid-sensor ASIC1a regulation of GSC markers was
therefore evaluated. In the present study, different glioma cell
lines and PDX were utilized to reveal that ASIC1a serves as a tumor
suppressor in glioma development and progression, which is
consistent with the research of Tian et al (11). It was also revealed that ASIC1a
expression is inversely associated with glioma grade progression by
using human glioma tumor tissues. The role for ASIC1a as a tumor
suppressor is further strengthened by the bioinformatic data from
TCGA, which demonstrated that GBM patients with high expression
ASIC1 have improved OS, indicating that ASIC1 is a promising
prognostic biomarker for GBM patients. The antitumor function of
ASIC1 was also supported by our previous work (14), which revealed that ASIC1a promotes
neurite growth and differentiation by negatively regulating Notch
signaling. In summary, our data strongly indicated that ASIC1a
functions as a tumor suppressor in glioma stemness and
tumorigenesis. Stimulation of ASIC1 activity may inhibit GSC
self-renewal and glioma progression.
All the major findings from the present study were
drawn from in vitro cell cultures from established glioma
cell lines or glioma PDX. PDX has its advantages by recapitulating
the actual disease more closely than the established glioma cell
lines, which may not resemble the original tumor, by adapting to
the environment and acquiring mutations. In the future, the
conclusion from this study, especially from PDX, should be tested
in immunocompromised animals.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SG and ML designed the study protocol. PK, JW, AAG,
and YJ performed experiments based on glioma cell cultures and
evaluated the data with the help of ML. ML performed the
biostatistical evaluation of the data. AAG and ML wrote the
manuscript with contributions and final approval by all authors. SG
and YJ contributed to the critical reading and revision of the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was carried out in strict accordance with
the recommendations in the Guide for the Care and Use of Laboratory
Animals of the National Institutes of Health. The protocol was
approved (approval no. 20-14) by the Institutional Animal Care and
Usage Committee (IACUC) of Morehouse School of Medicine (Atlanta,
USA).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
We are grateful to Dr Yancey G. Gillespie at the
University of Alabama at Birmingham (UAB) (Birmingham, USA) for
providing us with the PDX lines.
Abbreviations:
|
ASIC1a
|
acid-sensing ion channel 1a
|
|
ASICs
|
acid-sensing ion channels
|
|
BAAA
|
BODIPY-aminoacetaldehyde
|
|
CNS
|
central nervous system
|
|
DEAB
|
diethylaminobenzaldehyde
|
|
GBM
|
glioblastoma multiforme
|
|
GSCs
|
glioblastoma stem cells
|
|
IHC
|
immunohistochemistry
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
|
|
OS
|
overall survival
|
References
|
1
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella- Branger D, Cavenee WK, Ohgaki H, Wiestler
OD, Kleihues P and Ellison DW: The 2016 World Health Organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar
|
|
2
|
Damaghi M, Wojtkowiak JW and Gillies RJ:
pH sensing and regulation in cancer. Front Physiol. 4:3702013.
View Article : Google Scholar
|
|
3
|
Lee WY, Huang SC, Hsu KF, Tzeng CC and
Shen WL: Roles for hypoxia-regulated genes during cervical
carcinogenesis: Somatic evolution during the
hypoxia-glycolysis-acidosis sequence. Gynecol Oncol. 108:377–384.
2008. View Article : Google Scholar
|
|
4
|
Ward G, Meehan J, Gray ME, Murray AF,
Argyle DJ, Kunkler IH and Langdon SP: The impact of tumor pH on
cancer progression: Strategies for clinical intervention. Explor
Target Antitumor Ther. 1:71–100. 2020. View Article : Google Scholar
|
|
5
|
Xiong ZG, Chu XP and Simon RP: Acid
sensing ion channels-novel therapeutic targets for ischemic brain
injury. Front Biosci. 12:1376–1386. 2007. View Article : Google Scholar
|
|
6
|
Berdiev BK, Xia J, McLean LA, Markert JM,
Gillespie GY, Mapstone TB, Naren AP, Jovov B, Bubien JK, Ji HL, et
al: Acid-sensing ion channels in malignant gliomas. J Biol Chem.
278:15023–15034. 2003. View Article : Google Scholar
|
|
7
|
Kapoor N, Bartoszewski R, Qadri YJ, Bebok
Z, Bubien JK, Fuller CM and Benos DJ: Knockdown of ASIC1 and
epithelial sodium channel subunits inhibits glioblastoma whole cell
current and cell migration. J Biol Chem. 284:24526–24541. 2009.
View Article : Google Scholar
|
|
8
|
Kapoor N, Lee W, Clark E, Bartoszewski R,
McNicholas CM, Latham CB, Bebok Z, Parpura V, Fuller CM, Palmer CA
and Benos DJ: Interaction of ASIC1 and ENaC subunits in human
glioma cells and rat astrocytes. Am J Physiol Cell Physiol.
300:C1246–C1259. 2011. View Article : Google Scholar
|
|
9
|
Rooj AK, Liu Z, McNicholas CM and Fuller
CM: Physical and functional interactions between a glioma cation
channel and integrin-β1 require α-actinin. Am J Physiol Cell
Physiol. 309:C308–C319. 2015. View Article : Google Scholar
|
|
10
|
Rooj AK, McNicholas CM, Bartoszewski R,
Bebok Z, Benos DJ and Fuller CM: Glioma-specific cation conductance
regulates migration and cell cycle progression. J Biol Chem.
287:4053–4065. 2012. View Article : Google Scholar
|
|
11
|
Tian Y, Bresenitz P, Reska A, El Moussaoui
L, Beier CP and Gründer S: Glioblastoma cancer stem cell lines
express functional acid sensing ion channels ASIC1a and ASIC3. Sci
Rep. 7:136742017. View Article : Google Scholar
|
|
12
|
Carén H, Stricker SH, Bulstrode H, Gagrica
S, Johnstone E, Bartlett TE, Feber A, Wilson G, Teschendorff AE,
Bertone P, et al: Glioblastoma stem cells respond to
differentiation cues but fail to undergo commitment and terminal
cell-cycle arrest. Stem Cell Reports. 5:829–842. 2015. View Article : Google Scholar
|
|
13
|
Park NI, Guilhamon P, Desai K, McAdam RF,
Langille E, O'Connor M, Lan X, Whetstone H, Coutinho FJ, Vanner RJ,
et al: ASCL1 reorganizes chromatin to direct neuronal fate and
suppress tumorigenicity of glioblastoma stem cells. Cell Stem Cell.
21:209–224.e7. 2017. View Article : Google Scholar
|
|
14
|
Lopes C, Madureira TV, Gonçalves JF and
Rocha E: Disruption of classical estrogenic targets in brown trout
primary hepatocytes by the model androgens testosterone and
dihydrotestosterone. Aquat Toxicol. 227:1055862020. View Article : Google Scholar
|
|
15
|
Liu M, Inoue K, Leng T, Zhou A, Guo S and
Xiong ZG: ASIC1 promotes differentiation of neuroblastoma by
negatively regulating Notch signaling pathway. Oncotarget.
8:8283–8293. 2017. View Article : Google Scholar
|
|
16
|
Larco DO, Semsarzadeh NN, Cho-Clark M,
Mani SK and Wu TJ: β-Arrestin 2 is a mediator of GnRH-(1-5)
signaling in immortalized GnRH neurons. Endocrinology.
154:4726–4736. 2013. View Article : Google Scholar
|
|
17
|
Liu M, Inoue K, Leng T, Guo S and Xiong
ZG: TRPM7 channels regulate glioma stem cell through STAT3 and
Notch signaling pathways. Cell Signal. 26:2773–2781. 2014.
View Article : Google Scholar
|
|
18
|
Salinas M, Rash LD, Baron A, Lambeau G,
Escoubas P and Lazdunski M: The receptor site of the spider toxin
PcTx1 on the proton-gated cation channel ASIC1a. J Physiol.
570:339–354. 2006. View Article : Google Scholar
|
|
19
|
Sherwood TW, Lee KG, Gormley MG and
Askwith CC: Heteromeric acid-sensing ion channels (ASICs) composed
of ASIC2b and ASIC1a display novel channel properties and
contribute to acidosis-induced neuronal death. J Neurosci.
31:9723–9734. 2011. View Article : Google Scholar
|
|
20
|
Wan J, Guo AA, King P, Guo S, Saafir T,
Jiang Y and Liu M: TRPM7 induces tumorigenesis and stemness through
notch activation in glioma. Front Pharmacol. 11:5907232020.
View Article : Google Scholar
|
|
21
|
Omoruyi SI, Ekpo OE, Semenya DM, Jardine A
and Prince S: Exploitation of a novel phenothiazine derivative for
its anti-cancer activities in malignant glioblastoma. Apoptosis.
25:261–274. 2020. View Article : Google Scholar
|
|
22
|
Hiyama H, Iavarone A, LaBaer J and Reeves
SA: Regulated ectopic expression of cyclin D1 induces
transcriptional activation of the cdk inhibitor p21 gene without
altering cell cycle progression. Oncogene. 14:2533–2542. 1997.
View Article : Google Scholar
|
|
23
|
Hitoshi S, Alexson T, Tropepe V, Donoviel
D, Elia AJ, Nye JS, Conlon RA, Mak TW, Bernstein A and van der Kooy
D: Notch pathway molecules are essential for the maintenance, but
not the generation, of mammalian neural stem cells. Genes Dev.
16:846–858. 2002. View Article : Google Scholar
|
|
24
|
Purow BW, Haque RM, Noel MW, Su Q, Burdick
MJ, Lee J, Sundaresan T, Pastorino S, Park JK, Mikolaenko I, et al:
Expression of notch-1 and its ligands, delta-like-1 and jagged-1,
is critical for glioma cell survival and proliferation. Cancer Res.
65:2353–2363. 2005. View Article : Google Scholar
|
|
25
|
Ishii N, Maier D, Merlo A, Tada M,
Sawamura Y, Diserens AC and Van Meir EG: Frequent co-alterations of
TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human
glioma cell lines. Brain Pathol. 9:469–479. 1999. View Article : Google Scholar
|
|
26
|
Ke LD, Shi YX, Im SA, Chen X and Yung WK:
The relevance of cell proliferation, vascular endothelial growth
factor, and basic fibroblast growth factor production to
angiogenesis and tumorigenicity in human glioma cell lines. Clin
Cancer Res. 6:2562–2572. 2000.
|
|
27
|
Ettaiche M, Guy N, Hofman P, Lazdunski M
and Waldmann R: Acid-sensing ion channel 2 is important for retinal
function and protects against light-induced retinal degeneration. J
Neurosci. 24:1005–1012. 2004. View Article : Google Scholar
|
|
28
|
Krishtal O: The ASICs: Signaling
molecules? Modulators? Trends Neurosci. 26:477–483. 2003.
View Article : Google Scholar
|
|
29
|
Wemmie JA, Price MP and Welsh MJ:
Acid-sensing ion channels: Advances, questions and therapeutic
opportunities. Trends Neurosci. 29:578–586. 2006. View Article : Google Scholar
|
|
30
|
Brockway LM, Zhou ZH, Bubien JK, Jovov B,
Benos DJ and Keyser KT: Rabbit retinal neurons and glia express a
variety of ENaC/DEG subunits. Am J Physiol Cell Physiol.
283:C126–C134. 2002. View Article : Google Scholar
|
|
31
|
Lingueglia E: Acid-sensing ion channels in
sensory perception. J Biol Chem. 282:17325–17329. 2007. View Article : Google Scholar
|
|
32
|
Jahr H, van Driel M, van Osch GJ, Weinans
H and van Leeuwen JP: Identification of acid-sensing ion channels
in bone. Biochem Biophys Res Commun. 337:349–354. 2005. View Article : Google Scholar
|
|
33
|
Grunder S, Geissler HS, Bässler EL and
Ruppersberg JP: A new member of acid-sensing ion channels from
pituitary gland. Neuroreport. 11:1607–1611. 2000. View Article : Google Scholar
|
|
34
|
Ishibashi K and Marumo F: Molecular
cloning of a DEG/ENaC sodium channel cDNA from human testis.
Biochem Biophys Res Commun. 245:589–593. 1998. View Article : Google Scholar
|
|
35
|
Huang Y, Jiang N, Li J, Ji YH, Xiong ZG
and Zha XM: Two aspects of ASIC function: Synaptic plasticity and
neuronal injury. Neuropharmacology. 94:42–48. 2015. View Article : Google Scholar
|
|
36
|
Arun T, Tomassini V, Sbardella E, de
Ruiter MB, Matthews L, Leite MI, Gelineau-Morel R, Cavey A, Vergo
S, Craner M, et al: Targeting ASIC1 in primary progressive multiple
sclerosis: Evidence of neuroprotection with amiloride. Brain.
136:106–115. 2013. View Article : Google Scholar
|
|
37
|
Yin T, Lindley TE, Albert GW, Ahmed R,
Schmeiser PB, Grady MS, Howard MA and Welsh MJ: Loss of acid
sensing ion channel-1a and bicarbonate administration attenuate the
severity of traumatic brain injury. PLoS One. 8:e723792013.
View Article : Google Scholar
|
|
38
|
Chu XP and Xiong ZG: Physiological and
pathological functions of acid-sensing ion channels in the central
nervous system. Curr Drug Targets. 13:263–271. 2012. View Article : Google Scholar
|
|
39
|
Pilon-Thomas S, Kodumudi KN, El-Kenawi AE,
Russell S, Weber AM, Luddy K, Damaghi M, Wojtkowiak JW, Mulé JJ,
Ibrahim-Hashim A and Gillies RJ: Neutralization of tumor acidity
improves antitumor responses to immunotherapy. Cancer Res.
76:1381–1390. 2016. View Article : Google Scholar
|
|
40
|
Hjelmeland AB, Wu Q, Heddleston JM,
Choudhary GS, MacSwords J, Lathia JD, McLendon R, Lindner D, Sloan
A and Rich JN: Acidic stress promotes a glioma stem cell phenotype.
Cell Death Differ. 18:829–840. 2011. View Article : Google Scholar
|
|
41
|
Arcangeli A, Pillozzi S and Becchetti A:
Targeting ion channels in leukemias: A new challenge for treatment.
Curr Med Chem. 19:683–696. 2012. View Article : Google Scholar
|
|
42
|
Lehen'kyi V, Shapovalov G, Skryma R and
Prevarskaya N: Ion channnels and transporters in cancer. 5. Ion
channels in control of cancer and cell apoptosis. Am J Physiol Cell
Physiol. 301:C1281–C1289. 2011. View Article : Google Scholar
|
|
43
|
Li M and Xiong ZG: Ion channels as targets
for cancer therapy. Int J Physiol Pathophysiol Pharmacol.
3:156–166. 2011.
|
|
44
|
Bubien JK, Keeton DA, Fuller CM, Gillespie
GY, Reddy AT, Mapstone TB and Benos DJ: Malignant human gliomas
express an amiloride-sensitive Na+ conductance. Am J
Physiol. 276:C1405–C1410. 1999. View Article : Google Scholar
|
|
45
|
Olsen ML, Schade S, Lyons SA, Amaral MD
and Sontheimer H: Expression of voltage-gated chloride channels in
human glioma cells. J Neurosci. 23:5572–5582. 2003. View Article : Google Scholar
|
|
46
|
Kellenberger S and Schild L: International
union of basic and clinical pharmacology. XCI. Structure, function,
and pharmacology of acid-sensing ion channels and the epithelial
Na+ channel. Pharmacol Rev. 67:1–35. 2015. View Article : Google Scholar
|
|
47
|
Wemmie JA, Taugher RJ and Kreple CJ:
Acid-sensing ion channels in pain and disease. Nat Rev Neurosci.
14:461–471. 2013. View Article : Google Scholar
|
|
48
|
Jasti J, Furukawa H, Gonzales EB and
Gouaux E: Structure of acid-sensing ion channel 1 at 1.9 a
resolution and low pH. Nature. 449:316–323. 2007. View Article : Google Scholar
|
|
49
|
Sherwood TW and Askwith CC: Endogenous
arginine-phenylalanine-amide-related peptides alter steady-state
desensitization of ASIC1a. J Biol Chem. 283:1818–1830. 2008.
View Article : Google Scholar
|
|
50
|
Sherwood TW, Frey EN and Askwith CC:
Structure and activity of the acid-sensing ion channels. Am J
Physiol Cell Physiol. 303:C699–C710. 2012. View Article : Google Scholar
|
|
51
|
Sun X, Zhao D, Li YL, Sun Y, Lei XH, Zhang
JN, Wu MM, Li RY, Zhao ZF, Zhang ZR and Jiang CL: Regulation of
ASIC1 by Ca2+/calmodulin-dependent protein kinase II in
human glioblastoma multiforme. Oncol Rep. 30:2852–2858. 2013.
View Article : Google Scholar
|
|
52
|
Vila-Carriles WH, Kovacs GG, Jovov B, Zhou
ZH, Pahwa AK, Colby G, Esimai O, Gillespie GY, Mapstone TB, Markert
JM, et al: Surface expression of ASIC2 inhibits the
amiloride-sensitive current and migration of glioma cells. J Biol
Chem. 281:19220–19232. 2006. View Article : Google Scholar
|
|
53
|
Clark EB, Jovov B, Rooj AK, Fuller CM and
Benos DJ: Proteolytic cleavage of human acid-sensing ion channel 1
by the serine protease matriptase. J Biol Chem. 285:27130–27143.
2010. View Article : Google Scholar
|
|
54
|
Zhang XH, Šarić T, Mehrjardi NZ, Hamad S
and Morad M: Acid-sensitive ion channels are expressed in human
induced pluripotent stem cell-derived cardiomyocytes. Stem Cells
Dev. 28:920–932. 2019. View Article : Google Scholar
|
|
55
|
Haley EM, Tilson SG, Triantafillu UL,
Magrath JW and Kim Y: Acidic pH with coordinated reduction of basic
fibroblast growth factor maintains the glioblastoma stem cell-like
phenotype in vitro. J Biosci Bioeng. 123:634–641. 2017. View Article : Google Scholar
|