Introduction
Breast cancer is one of the most common cancers and
the leading cause of cancer-related deaths among women world-wide.
It is estimated that there were 1.7 million breast cancer cases and
520,000 breast cancer-related deaths in 2012. Breast cancer
accounts for 25% of all cancer cases and for 15% of all cancer
deaths among women (1). In recent
years, significant advances in early diagnosis and effective
treatments for the disease have been achieved resulting in improved
overall survival of breast cancer patients. However, metastasis
represents a large obstacle to reduce the mortality rate in breast
cancer and remains the primary cause of breast cancer-related
mortalities (2). Bone is the most
common site for breast cancer metastasis (3). It has been reported that 50-60% of
metastatic breast cancer patients develop bone metastases in the
advanced stage of breast cancer (4); 5-6% of all breast cancer patients
exhibit bone metastases at diagnosis (5). Bone metastasis remains incurable and
causes skeletal-related events, such as pain, hypercalcemia,
pathological fracture, and spinal cord compression, which lead to
the poor quality of life and/or activities of the patients
(6). It has also been revealed
that breast cancer patients with complications due to bone
metastasis have shorter median survival time than that of patients
without bone complications (7).
Thus, understanding the mechanism underlying bone metastasis is
crucial to treat it in its early stages.
Breast cancer cells themselves are not capable of
carrying out the highly specialized function of bone resorption
(8). The formation of bony
osteolytic lesions by solid tumor metastasis mainly depends on the
activation of osteoclasts (9,10).
Osteoclastogenesis is regulated by exogenous hormones, cytokines,
and various transcriptional factors. It is known that cancer cells
are capable of promoting osteoclast differentiation, function, and
activities (8,11). Breast cancer cells have been
reported to secrete various factors that act on pre-osteoclasts,
osteoblasts, and bone stromal cells, which stimulate the
differentiation of mature osteoclasts (11,12). Hussein et al reported that
the macrophage colony-stimulating factor and the phospholipase C-γ
produced by metastatic breast cancer cells could prolong osteoclast
survival and block the apoptotic effect of bisphosphonates
(13).
MicroRNAs (miRNAs or miRs) are small, non-coding
RNAs that regulate gene expression by translational inhibition and
degradation of mRNAs (14,15).
They have been recognized to play important roles in various
biological processes, including cell differentiation,
proliferation, apoptosis, and tumorigenesis (14,15). Therefore, miRNAs are considered to
have significant clinical potential; for example, they may be used
as new therapeutic targets and disease-specific biomarkers
(16). Previous studies have also
suggested that several miRNAs could play important roles in the
progression of breast cancer, and bone metastasis with osteoclast
differentiation and/or function (17,18). Ell et al reported that
miR-16 levels are higher in the serum of patients with breast
cancer bone metastasis than that in healthy donors (17). In addition, in the case of breast
cancer with bone metastasis, miR-16 levels were increased in
osteoclast differentiation and bone metastasis (18). Conversely, there are several
studies that have reported that miR-133a and miR-223 may act as
suppressors not only in breast cancer progression but also in bone
destruction (14,17-19). It has been reported that miR-133a
acts as a tumor suppressor in breast cancer by targeting LIM and
SH3 domain protein 1 (LASP1) (19), and that the ectopic expression of
miR-133a may inhibit osteoclast differentiation and bone resorption
(17). Other studies indicated
that miR-223 reduces tumor growth in breast cancer (20), and plays essential roles during
osteoclast differentiation (21,22). However, there is no detailed study
in which the roles of miRNAs in osteoclastic bone destruction
caused by breast cancer bone metastasis have been revealed. Based
on this background, it was hypothesized that miR-16, miR-133a and
miR-223 could regulate osteoclast differentiation, function, and/or
activities in breast cancer bone metastasis, thereby controlling
metastatic bone destruction. In the present study, to investigate
the roles of these miRNAs in bone destruction caused by breast
cancer metastasis, their effects on the expression of osteolytic
factors and osteoclast activity were examined in vitro, and
on bone destruction in vivo.
Materials and methods
Cell culture
A human-derived breast cancer cell line, MDA-MB-231
(HTB-26), and a mouse macrophage cell line, RAW264.7 (TIB-71), both
obtained from the American Type Culture Collection (ATCC), were
used in the present study. The RAW 264.7 cells are known as
preosteoclast cells, which readily differentiate into osteoclasts
upon exposure to receptor activator for nuclear factor-κB ligand
(RANKL) (23). Cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100
µg/ml streptomycin (all from Sigma-Aldrich; Merck KGaA), and
maintained in a humidified atmosphere with 5% CO2 at
37°C.
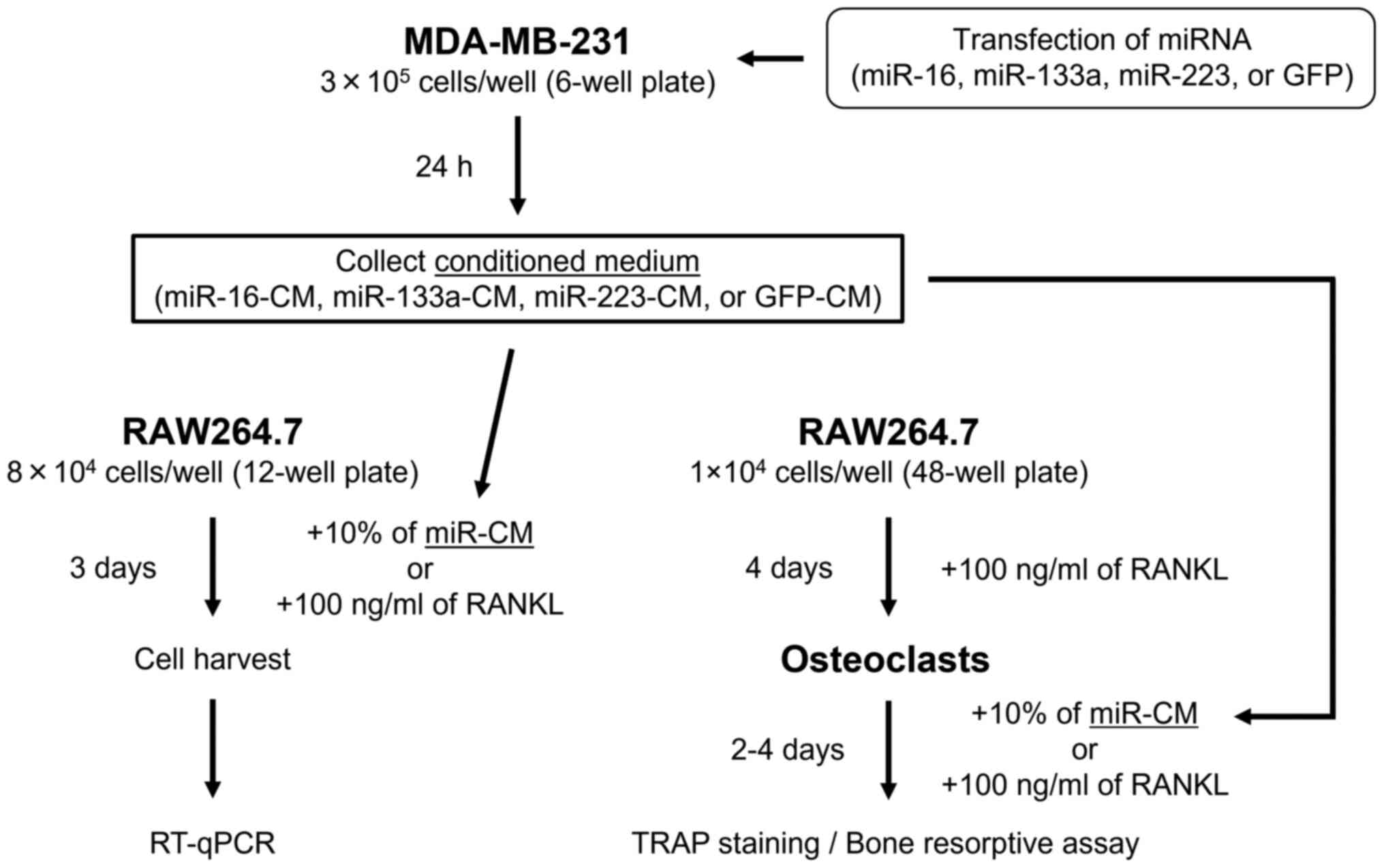
Transfection of MDA-MB-231 cells with
miRNAs and collection of conditioned medium
MDA-MB-231 cells (3×105 cells/well) were
seeded into 6-well plates on the day before transfection.
miRNA-expressing plasmid (2 µg) was mixed with 4 µl
of ViaFect (Promega Corporation) in 200 µl of serum-free
DMEM, and incubated at room temperature for 15 min according to the
manufacturer's protocol. Then, the mixture was added to the
MDA-MB-231 cells and incubated at 37°C. Following incubation for 24
h, gene transfection efficiency was confirmed by observing GFP
expression under a fluorescence microscope (BZ-X700 microscope and
BZ-X Viewer; Keyence Corporation) (Fig. S1), and culture medium was
collected, filtered through a 0.22-µm filter, and used as
the miRNA-transfected conditioned medium (miR-16-CM, miR-133a-CM,
miR-223-CM and GFP-CM). The miRNA-expressing plasmids used were:
pcDNA3-pri-miR-16-1 (cat. no. 51382), pcDNA3.2 mir-1-1 reporter
hsa-mir-133a-1 (cat. no. 46676), pcDNA3.2/V5 mmu-mir-223 (cat. no.
26334), and pCDNA3-GFP as the control (cat. no. 74165; all from
Addgene, Inc.).
Expression of osteoclast differentiation
markers and osteolytic factors in miR-CM
To evaluate the effect of miRNA transfection on the
expression of osteoclast differentiation markers and osteolytic
factors in MDA-MB-231 cells, total RNA was extracted from
miRNA-transfected cells and each CM using the Total Exosome RNA and
Protein Isolation Kit (cat. no. 4478545; Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Subsequently, cDNA was synthesized using the MystiCq microRNA cDNA
Synthesis Mix (product no. MIRRT-100RXN; Sigma-Aldrich; Merck KGaA)
according to the manufacturer's protocol. The expression of each
transfected miRNA (miR-16, miR-133a and miR-223) in the cells, and
osteoclast differentiation markers and osteolytic factors including
RANKL, interleukin (IL)-1β, IL-6, parathyroid hormone-related
protein (PTHrP), and tumor necrosis factor (TNF), in each miR-CM
were examined by reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) (Fig.
1).
Effect of miR-CM on osteoclast
differentiation in RAW264.7 preosteoclasts in vitro
RAW264.7 cells (8×104 cells/well) were
seeded in 12-well plates, and after 24 h, the medium was replaced
with medium containing 10% of each miR-CM or 100 ng/ml of
recombinant RANKL (PeproTech EC, Ltd.) as a positive control.
Subsequently, 3 days after miR-CM or RANKL treatment, total RNA was
isolated from RAW264.7 cells using the RNeasy mini kit (cat. no.
74104; Qiagen, Inc.) and cDNA synthesis was performed using the
High-capacity cDNA Transcription kit (cat. no. 4368814; Applied
Biosystems; Thermo Fisher Scientific, Inc.), according to the
manufacturers' protocol. Thereafter, mRNA expression of osteoclast
differentiation markers, including nuclear factor of activated
T-cells, cytoplasmic 1 (NFATc1), osteoclast-associated receptor
(OSCAR), β3-integrin, cathepsin-K, and tartrate-resistant acid
phosphatase (TRAP), in the cells was assessed by RT-qPCR (Fig. 1).
RT-qPCR
RT-qPCR reactions were performed in 20 µl
reaction mixture volumes using the SYBR-Green master mix reagent
(Applied Biosystems; Thermo Fisher Scientific, Inc.) on the ABI
Prism 7500 sequence-detection system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). qPCR conditions were as follows: one
cycle at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec
and 60°C for 1 min. Forward primers for each miRNA (MystiCq
microRNA qPCR Assay Primer) were purchased from Sigma-Aldrich;
Merck KGaA, and MystiCq Universal PCR Primer (product no. MIRUP;
Sigma-Aldrich; Merck KGaA) was used as a reverse primer for the
RT-qPCR reactions. Pre-designed primers specific for RANKL, IL-1β,
IL-6, PTHrP, TNF, NFATc1, OSCAR, β3-integrin, cathepsin-K and TRAP
were obtained from Invitrogen; Thermo Fisher Scientific, Inc.
Primer sequences are listed in Table
I. The obtained values were normalized to those for SNORD43
(product no. MIRCP00004; Sigma-Aldrich; Merck KGaA) for miRNA and
β-actin for other genes, and expression levels were quantified
using the 2−∆∆Cq method (24).
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Primer name | Sequence
(5′→3′) |
|---|
| miR-16
(MIRAP00030) | F: CCA GUA UUA ACU
GUG CUG CUGA |
| miR-133a
(MIRAP00155) | F: UUU GGU CCC CUU
CAA CCA GCUG |
| miR-223
(MIRAP00282) | F: CGU GUA UUU GAC
AAG CUG AGUU |
| SNORD43
(MIRCP00004) | F: CAC AGA UGA UGA
ACU UAU UGA CGG GCG GAC AGA AAC UGU GUG CUG AUU GUC ACG UUC UGA
UU |
| RANKL | F: CCC AGA TCA AGG
TGG TGT CT |
| R: TGC TGA CCA ATG
AGA GCA TC |
| IL-1β | F: GGA CAA GCT GAG
GAA GAT GC |
| R: TCG TTA TCC CAT
GTG TCG AA |
| IL-6 | F: AAA GAG GCA CTG
GCA GAA AA |
| R: TTT CAC CAG GCA
AGT CTC CT |
| PTHrP | F: CAT CAG CTC CTC
CAT GAC AA |
| R: TCA GCT GTG TGG
ATT TCT GC |
| TNF | F: CCT GTG AGG AGG
ACG AAC AT |
| R: GGT TGA GGG TGT
CTG AAG GA |
| NFATc1 | F: GAA GCA AAG ACT
GAC CGG GA |
| R: ATC CTC TGG TTG
CGG AAA GG |
| OSCAR | F: GAG CTC TGC CTT
TGA TGG TC |
| R: CAA GGA TCC CAG
CTT CTC TG |
| β3-integrin | F: GAA AGG CCA GTC
AGA ACT GC |
| R: TGT GGC CTC CCA
GAT TAA AG |
| Cathepsin-K | F: TTC TCC TCT CGT
TGG TGC TT |
| R: AAA AAT GCC CTG
TTG TGT CC |
| TRAP | F: GAT GAC TTT GCC
AGT CAG CA |
| R: AAC TGC TTT TTG
AGC CAG GA |
| β-actin | F: GAT GAG ATT GGC
ATG GCT TT |
| R: CAC CTT CAC CGT
TCC AGT TT |
In vitro effects of miR-CM on osteoclast
activity and function
In vitro effects of miR-CM on osteoclast
activity and function in osteoclasts derived from RAW264.7 cells
were evaluated (Fig. 1). To
generate osteoclasts, RAW264.7 cells were cultured in medium
containing 100 ng/ml of RANKL at 37°C for 4 days, and osteoclast
formation was confirmed by the presence of TRAP-positive
osteoclasts with three or more nuclei under a light microscope.
Following confirmation of osteoclast formation, the medium was
replaced with medium containing 10% of miR-CM or 100 ng/ml RANKL
(positive control). TRAP staining was performed after 2 days of
miR-CM or RANKL treatment using the standard naphthol AS-BI
phosphate post-coupling method (25). Briefly, the cells were fixed in
3.6% PBS-buffered formalin for 5 min at room temperature, then
incubated at 37°C in sodium acetate buffer (pH 5.0) containing
0.01% naphthol AS-BI phosphate, 0.5 M L-(+)-tartaric acid, and 0.05
M pararosaniline chloride for 20 min, and finally counterstained
with hematoxylin at room temperature for 2 min. The number of
TRAP-positive multinucleated cells containing three or more nuclei
were counted as osteoclasts per three random fields under a light
microscope.
Osteoclast function was assessed using bone
resorptive assays. RAW264.7 cells (1×104 cells/well)
were plated on dentin slices (FUJIFILM Wako Pure Chemical
Corporation) in 48-well plates. Following confirmation of
osteoclast formation after 4 days of 100 ng/ml RANKL treatment, the
medium was replaced with medium containing 10% of miR-CM or 100
ng/ml RANKL and refreshed every 2 days. Subsequently, after 4 days
of incubation with miR-CM or RANKL at 37°C, cells were detached
from the dentine slices by sonication for 5 min in 0.5 M
NH4OH (FUJIFILM Wako Pure Chemical Corporation). The
slices were stained with 1% toluidine blue (Muto Pure Chemicals
Co., Ltd.) at room temperature for 1 min to visualize resorption
pits. The area of the resorption pits was measured in three random
fields using a light microscope. All morphometric studies were
performed by two examiners blinded to treatment conditions.
Animal models
Female 5-week-old BALB/c nude mice (18-20 g) were
purchased from CLEA Japan Inc. Animals were maintained under
pathogen-free conditions in accordance with institutional
guidelines. All animal procedures were performed in accordance with
the Japanese Physiological Society Guidelines for the Care and Use
of Laboratory Animals, and the study protocol was approved by the
Institutional Animal Care and Use Committee (approval no. P191207)
and carried out according to the Kobe University Animal
Experimentation Regulations (Kobe, Japan). Animals were fed
pathogen-free laboratory chow and were permitted free access to
autoclaved water in an air-conditioned room at 25°C and 50-60%
humidity with a 12-h light/dark cycle. To create the in vivo
bone metastasis model of breast cancer, 24 mice were randomly
divided into four groups with six mice per group: miR-16, miR-133a,
miR-223, and GFP (as control). Based on our previous pilot studies,
it was determined that six samples would be required in each group
to detect differences as calculated using G*power 3.1 when α was
set at 0.05 and power was set at 0.9 (26). MDA-MB-231 cells transfected with
miRNA (1×106 cells suspended in 10 µl PBS) were
implanted intramedullary in the proximal epiphysis of the left
tibia of 6-week-old mice (27,28). For surgical procedures, mice were
anesthetized by intraperitoneal injection of 50 mg/kg pentobarbital
sodium (Kyoritsu Seiyaku) for induction and isoflurane inhalation
at a concentration of 2% for maintenance. At 4 weeks following cell
transplantation, all mice were euthanized and their tibiae were
removed. Humane endpoints were determined to be when the xenograft
tumor reached >10% of the animal body weight, the tumor diameter
was >20 mm, body weight loss >20% occurred due to tumor
growth, and the signs of immobility, the inability to eat,
ulceration, infection, or necrosis were observed. All mice reached
the study endpoint and were euthanized by cervical dislocation
under anesthesia by intraperitoneal injection of 100 mg/kg
pentobarbital sodium. Death was verified by the cessation of the
heartbeat of the mice and the dilatation of their pupils.
Micro-computed tomography (µCT)
analysis
Quantitative analysis of the tibia was performed 4
weeks following cell transplantation using a µCT Scanner
(R_mCT; Rigaku Mechatronics Co., Ltd.). The bone samples were
scanned with parameter settings of tube voltage, 90 kV; tube
current, 160 µA; and FOV, 10 mm. For morphometric analyses
of the bone samples, a region of interest (ROI) was set 1 mm below
the growth plate with an offset of 200 µm from a reference
slice. Bone volume/total volume (BV/TV) was assessed using an image
analysis system (TRI/3D-BON; RATOC System Engineering).
Hematoxylin and eosin (H&E) staining
and TRAP staining
Tibia samples were fixed in 10% formalin at room
temperature for 48 h, decalcified in 10% ethylenediaminetetraacetic
acid for 2 weeks, and then embedded in paraffin. Paraffin-embedded
tibia samples were sliced into 6-µm thick sections. After
deparaffinization and rehydration, the sections were stained with
hematoxylin for 5 min followed by rinsing in distilled water. Then,
the sections were stained with eosin for 2 min followed by
dehydration with graded alcohol and clearing in xylene. All
procedures were performed at room temperature. Sections were
evaluated using a light microscope to confirm bone destruction and
the presence of tumor cells.
For TRAP staining, paraffin-embedded tibia sections
were deparaffinized and incubated at 37°C in sodium acetate buffer
(pH 5.0) containing 0.01% naphthol AS-BI phosphate and 0.5 M
L-(+)-tartaric acid for 20 min. The sections were then incubated in
the same buffer containing 0.05 M pararosaniline chloride at 37°C
for 20 min, followed by washing in distilled water. The sections
were counterstained with hematoxylin at room temperature for 30
sec. The number of TRAP-positive multinucleated cells containing
three or more nuclei were counted as osteoclasts in three random
fields under a light microscope.
Immunohistochemical staining
Tibia samples were fixed in 10% formalin for 48 h,
decalcified in 10% ethylenediaminetetraacetic acid for 2 weeks,
then embedded in paraffin and sliced into 6-µm thick
sections. Paraffin-embedded tibia sections were deparaffinized, and
treated with proteinase K (Dako; Agilent Technologies, Inc.) for 10
min and 3% H2O2 for 5 min at room temperature
followed by overnight incubation at 4°C with the following primary
antibodies: anti-RANKL rabbit polyclonal antibody (1:200; cat. no.
23408-1-AP), anti-IL-1β rabbit polyclonal antibody (1:200; cat. no.
16806-1-AP), anti-IL-6 mouse monoclonal antibody (1:200; cat. no.
66146-1-Ig), anti-PTHrP rabbit polyclonal antibody (1:200; cat. no.
10817-1-AP), and anti-TNF-α mouse monoclonal anti-body (1:200; cat.
no. 60291-1-Ig; all from ProteinTech Group, Inc.). Subsequently,
the sections were incubated with horseradish peroxidase-conjugated
labeled anti-mouse antibody (1:200; cat. no. 424131; Histofine
Simplestain Max PO(M); Nichirei Biosciences Inc.) or anti-rabbit
antibody (1:200; cat. no. 424141; Histofine Simplestain Max PO(R);
Nichirei Biosciences Inc.) for 30 min at room temperature, then
treated with 3,3′-diaminobenzidine tetrahydrochloride substrate
chromogen (cat. no. 415171; Simplestain DAB Solution; Nichirei
Biosciences Inc.) for 5 min, and counterstained with hematoxylin
for 1 min at room temperature. Immuno-positive cells were counted
in three random fields under a high-power field. All morphometric
studies were performed by two examiners blinded to treatment
conditions.
Statistical analysis
All experiments were performed independently in
triplicate, and statistically analyzed using a software package
(GraphPad Prism version 5.02; GraphPad Software, Inc.). Values are
presented as the mean ± standard error of the mean (SEM).
Comparisons between two groups were performed using a paired
Student's t-test, and comparisons among multiple groups were
performed using a one-way analysis of variance (ANOVA) followed by
post hoc testing with Tukey's procedure. P<0.05 was considered
to indicate a statistically significant difference.
Results
mRNA expression of osteoclast
differentiation and osteolytic factors is increased in miR-16-CM
and decreased in miR-133a-CM and miR-223-CM
RT-qPCR analyses revealed that the expression level
of miR-16, miR-133a, or miR-223 was significantly increased in the
transfected MDA-MB-231 cells (P<0.05) (Fig. 2A). To examine the effect of miRNA
transfection on bone destruction, the expression of osteoclast
differentiation markers and osteolytic factors (RANKL,
IL-1β, IL-6, PTHrP, and TNF) in the CM
of miRNA-transfected MDA-MB-231 cells was evaluated by RT-qPCR
analysis. The mRNA expression of RANKL, IL-1β,
PTHrP, and TNF was significantly increased in
miR-16-CM compared with that in GFP-CM (P<0.05) (Fig. 2B). The mRNA expression of these
genes was significantly decreased in miR-133a-CM (P<0.05)
compared with that in GFP-CM. The expression was also decreased in
miR-223-CM, and the decrease in RANKL, IL-6, and
PTHrP was significant compared with that in GFP-CM
(P<0.05) (Fig. 2B).
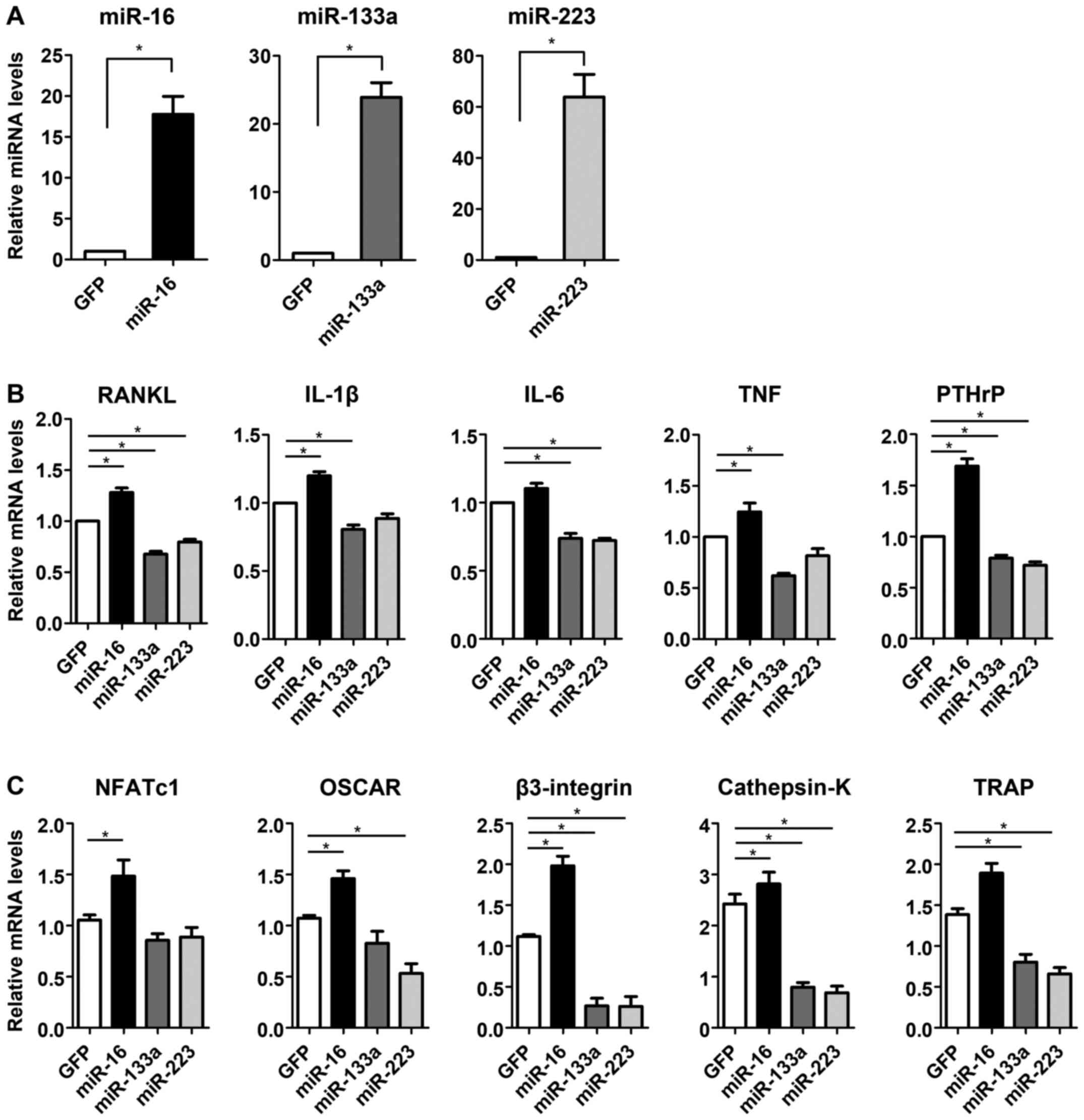 | Figure 2Transfection of miR-16, miR-133a and
miR-223 in MDA-MB-231 cells and gene expression in the conditioned
media of miRNA-transfected cells (miR-CM). (A) RT-qPCR analyses of
the expression levels of miR-16, miR-133a or miR-223 in the cells
of each miRNA-transfected group. (B) RT-qPCR analyses for mRNA
expression of osteoclast differentiation and osteolytic factors in
miR-CM. (C) RT-qPCR analyses of mRNA expression of osteoclast
differentiation markers in RAW264.7 cells treated with miR-CM.
*P<0.05. miR, microRNA; CM, conditioned medium;
RT-qPCR, reverse transcription-quantitative PCR; RANKL, receptor
activator for nuclear factor-κB ligand; NFATc1, nuclear factor of
activated T-cells cytoplasmic 1; OSCAR, osteoclast-associated
receptor; TRAP, tartrate-resistant acid phosphatase; IL,
interleukin; PTHrP, parathyroid hormone-related protein; TNF, tumor
necrosis factor. |
mRNA expression of osteoclast
differentiation markers in RAW264.7 pre-osteoclasts is increased by
miR-16-CM treatment, but decreased by both miR-133a-CM and
miR-223-CM treatments
To investigate the effects of miR-CM on the
osteoclast differentiation process, the expression of osteoclast
differentiation markers was evaluated in miR-CM-treated RAW264.7
cells. The mRNA expression levels of NFATc1, OSCAR,
β3-integrin, and cathepsin-K were significantly
increased by miR-16-CM treatment compared with that in the control
(P<0.05) (Fig. 2C). The
expression of β3-integrin, cathepsin-K, and
TRAP in miR-133a-CM treated-cells, and that of OSCAR,
β3-integrin, cathepsin-K, and TRAP in
miR-223-CM-treated cells was significantly decreased compared with
the respective levels in the control (P<0.05) (Fig. 2C).
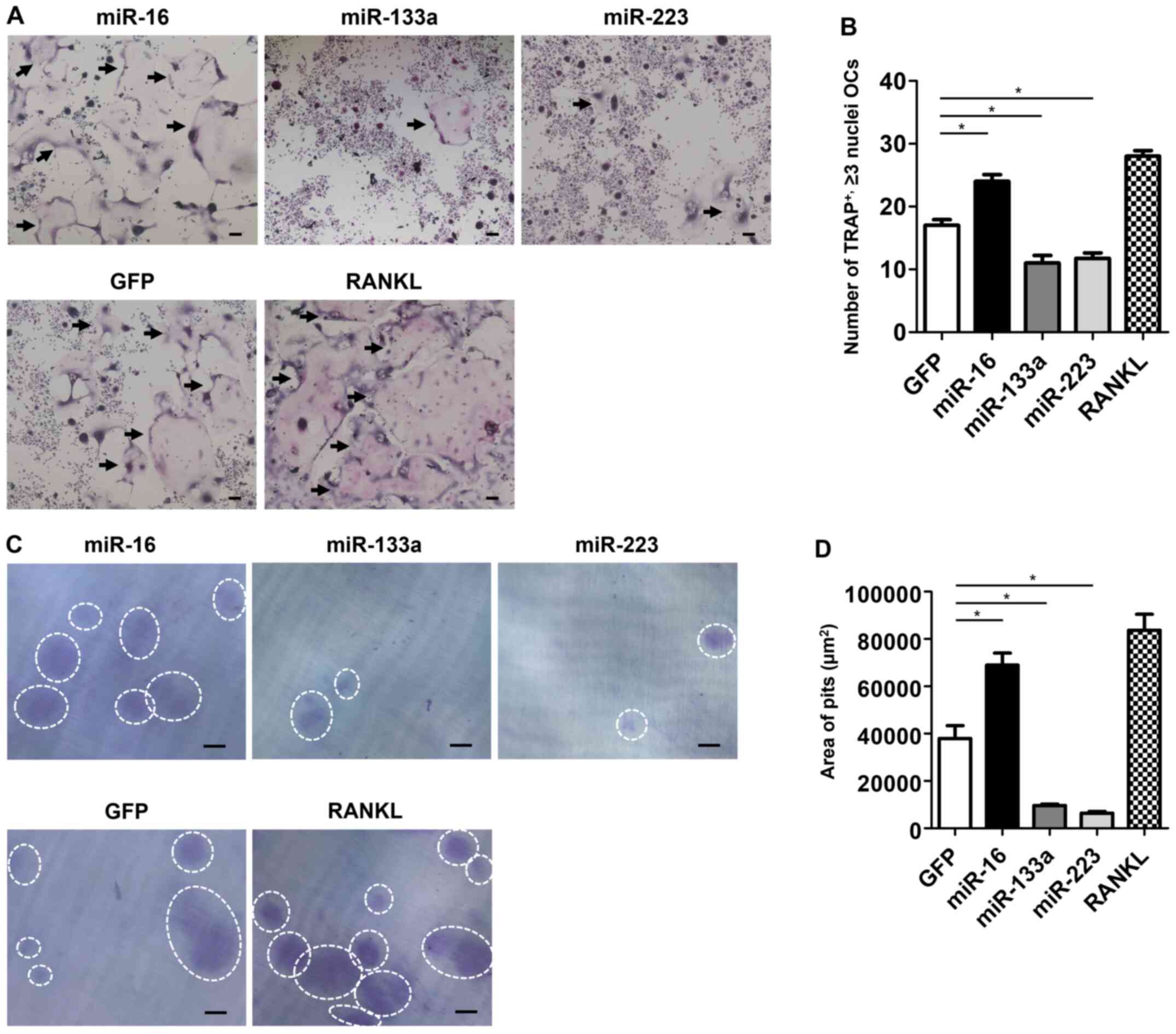
Osteoclast formation and activities are
promoted by miR-16-CM treatment, but suppressed by both miR-133a-CM
and miR-223-CM treatments
The effect of miR-CM treatment on osteoclast
formation was evaluated by TRAP staining of miR-CM-treated
osteoclasts. The number of TRAP-positive multinucleated osteoclasts
was significantly higher in miR-16-CM-treated cells, but lower in
both miR-133a-CM- and miR-223-CM-treated cells, compared with that
of control cells (P<0.05) (Fig. 3A
and B).
Considering that bone resorption is a characteristic
feature of osteoclasts, the effect of miR-CM treatment on
osteoclastic resorptive function in dentin slices was evaluated.
Significantly more resorption pits on dentin slices were observed
in miR-16-CM-treated osteoclasts, whereas the pits were lesser in
miR-133a- or miR-223-treated osteoclasts compared with those in the
control (Fig. 3C). The area of
resorption pits in miR-16-CM-treated osteoclasts was as large as
that in the RANKL-treated osteoclasts. In contrast, in both
miR-133a- and miR-223-treated osteoclasts, the area of pits was
significantly smaller than that in the control (P<0.05)
(Fig. 3D).
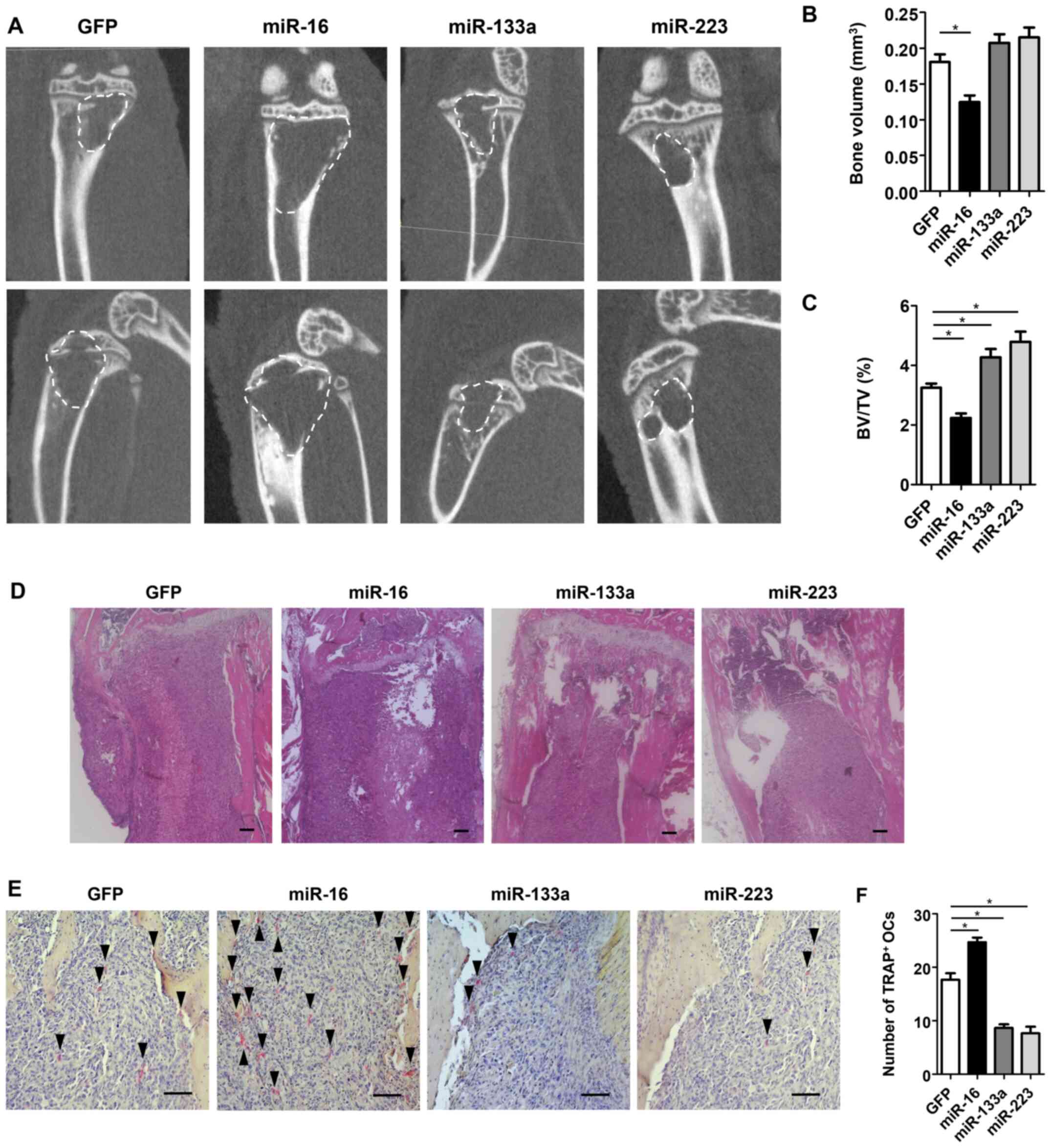
Bone destruction by breast cancer cells
is enhanced by miR-16 transfection, but suppressed by either
miR-133a or miR-223 transfection along with osteoclast
activities
To evaluate the effects of each miRNA on osteoclast
formation and bone destruction at the site of bone metastasis,
miRNA-transfected MDA-MB-231 cells were transplanted into the
tibiae of nude mice, and histological and morphometric evaluation
were performed. µCT analyses revealed that the bone volume
of the tibia in the miR-16 group was significantly lower with
enhanced bone destruction compared with that in the control group,
whereas the bone volume in both the miR-133a and miR-223 groups was
significantly higher compared with that in the control group
(P<0.05) (Fig. 4A-C).
Consistent with these findings, H&E staining of the tibia
revealed that a larger area of breast cancer cells was observed
with normal bone marrow destruction in the miR-16 group, whereas
the same area was smaller with less bone destruction in both the
miR-133a and miR-223 groups compared with that in the control
(Fig. 4D). Additionally, TRAP
staining revealed that the number of TRAP-positive multinucleated
osteoclasts was significantly increased in the miR-16 group, but
significantly decreased in both miR-133a and miR-223 groups
compared with that in the control group (P<0.05) (Fig. 4E and F). Immunohistochemical
staining for osteoclast differentiation markers and osteolytic
factors, such as RANKL, IL-1β, IL-6, TNF and PTHrP, revealed that
the number of immunopositive cells was significantly increased in
the miR-16 group, but significantly decreased in both the miR-133a
and miR-223 groups compared with that in the control group
(P<0.05) (Fig. 5A and B).
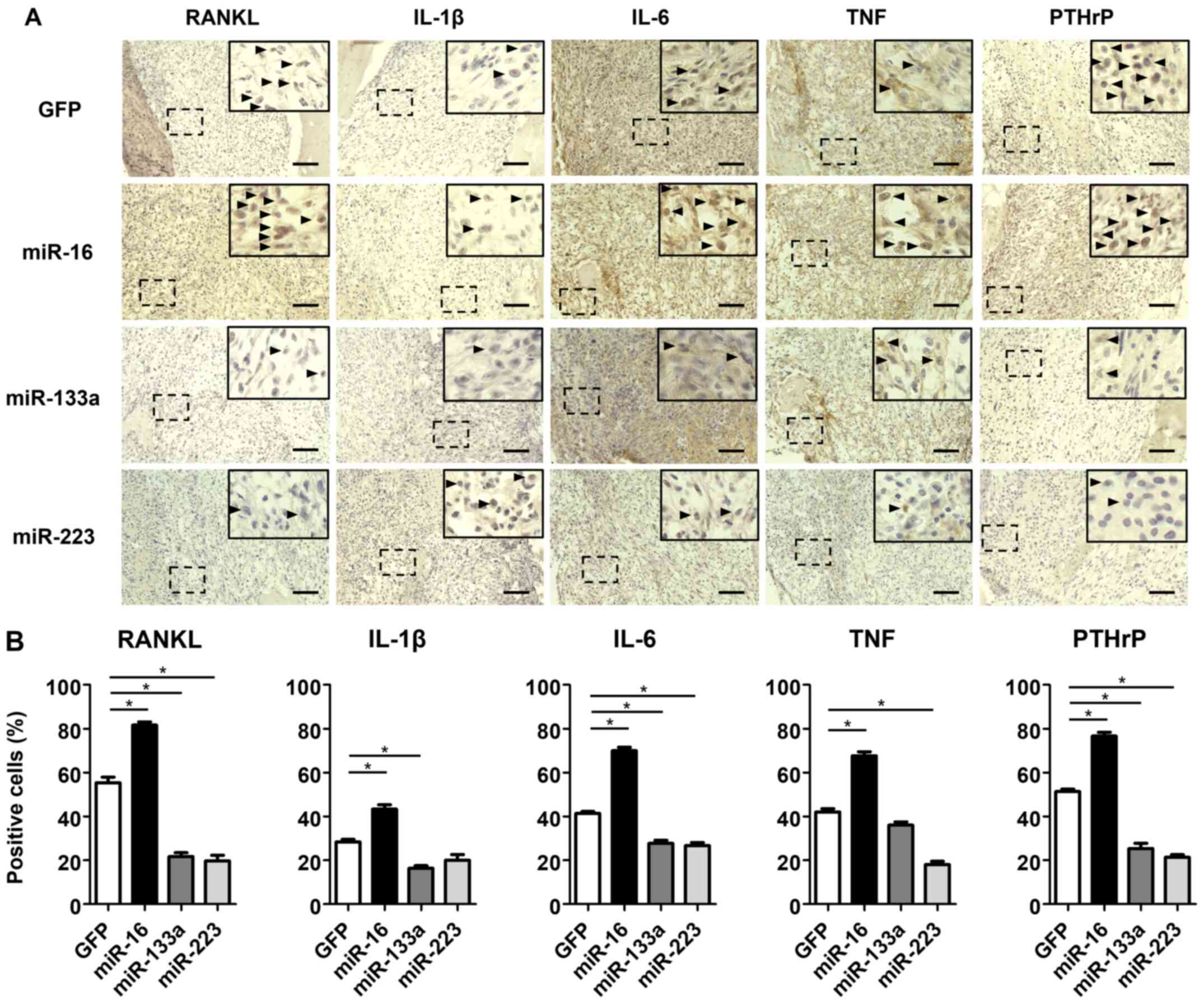 | Figure 5Effects of miRNA on the expression of
osteoclast differentiation markers and osteolytic factors in
vivo. (A) Immunohistochemical staining for RANKL, IL-1β, IL-6,
TNF and PTHrP of proximal tibiae of mice following 4 weeks of cell
transplantation. Right upper panels in each image indicate high
magnification image. Arrow heads indicate immunopositive cells. (B)
The number of cells immunopositive for RANKL, IL-1β, IL-6, TNF, and
PTHrP. Scale bar, 100 µm. *P<0.05. miR,
microRNA; RANKL, receptor activator for nuclear factor-κB ligand;
IL, interleukin; PTHrP, parathyroid hormone-related protein; TNF,
tumor necrosis factor. |
Discussion
In the present study, the roles of miR-16, miR-133a,
and miR-223 on osteoclastic bone destruction by breast cancer
metastasis were investigated using the MDA-MB-231 breast cancer
cell line in vitro and breast cancer bone metastasis mouse
model in vivo. It was demonstrated that the expression of
osteolytic factors, such as RANKL, IL-1β, IL-6, PTHrP and TNF, was
enhanced in the culture medium of breast cancer cells that were
transfected with miR-16, but decreased in that of cells transfected
with miR-133a or 223. In RAW264.7 preosteoclasts, the stimulation
of osteoclast differentiation and function by the breast cancer
culture medium was promoted by miR-16 overexpression, but was
suppressed by miR-133a or miR-223 overexpression. These findings
indicated that osteolytic factors derived from breast cancer cells
stimulated the osteoclast differentiation and function, and that
the effects could be regulated positively by miR-16, but negatively
by miR-133a or miR-223. Consistent with these in vitro
results, in vivo experiments using a breast cancer bone
metastasis animal model demonstrated that miR-16 enhanced bone
destruction with increased osteoclast activities; by contrast,
miR-133a and miR-223 prevented the destruction with decreased
activities.
Bone lesions by cancer metastasis are caused as a
result of an imbalance between bone formation and bone resorption
in the bone microenvironment (10,29,30). Both osteolytic and osteoblastic
lesions could be caused by cancer bone metastases (31), and the majority of the lesions
caused by breast cancer metastasis have been reported as osteolytic
lesions due to the stimulation of osteoclast activities (32,33). Breast cancer cells themselves are
unable to perform bone resorption but may stimulate the bone
resorptive activity of osteoclasts by secreting RANKL (8,9).
Breast cancer cells also secrete factors that promote RANKL
production in osteoblasts, immune cells, and bone stromal cells
(8,11,12), such as PTHrP, one of the most
common tumor-derived factors (34,35). In addition, it has been reported
that breast cancer cells could secrete or induce bone stromal cells
to release other factors enhancing osteoclast differentiation and
bone resorption, such as IL-1β, IL-6, IL-8, IL-11, and TNF
(11,36-39). In addition, it has been reported
that the conditioned medium of breast cancer cells could prolong
the survival of osteoclasts, which die primarily by apoptosis
within several days of formation (13). Our findings in the present study
support the previous studies, and indicate that breast cancer
cell-derived factors, such as RANKL, IL-1β, IL-6, PTHrP, and TNF,
promote bone resorption by affecting osteoclast differentiation and
activity.
miRNAs have been recognized to be important in the
regulation of various biological processes of cancer cells
(40,41), and several miRNAs, such as miR-16,
miR-133a, and miR-223, have been reported to be involved in the
progression of breast cancer, bone metastasis, and osteoclast
differentiation or function (14,17-29). Previous studies have suggested
that miR-16 could be a promoter of breast cancer bone metastasis
(17,18,42). Elevated expression of miR-16 was
reported in clinical samples of bone lesions of breast cancer
metastasis, compared with that in primary tumor samples (17). Microarray analysis of RAW264.7
preosteoclast cells treated with the culture medium of a highly
metastatic breast cancer cell line revealed a significant
upregulation of miR-16 (17). In
addition, miR-16 upregulation has also been reported in osteoclast
differentiation associated with cancer bone metastasis (18,42). By contrast, miR-133a and miR-223
have been widely reported as tumor suppressors in various cancers
including breast cancer (43).
The downregulation of miR-133a was reported in both breast cancer
tissues and cell lines, wherein miR-133a inhibited proliferation,
migration, and invasion of breast cancer cells by suppressing LASP1
(19). Both in vitro and
in vivo experiments revealed negative regulation by miR-133a
on breast cancer metastasis via targeting mastermind-like
transcriptional coactivator 1 (44). Ell et al also reported that
miR-133a inhibited osteoclast differentiation and reduced
osteoclast activities (17). In
the case of miR-223, Fabris et al reported that the
overexpression of miR-223 could prevent the recurrence of breast
cancer by mediating the epidermal growth factor signaling pathway
(45). Other studies revealed
that miR-223 inhibited the invasion and migration of breast cancer
cells, and promoted cell apoptosis by suppressing the expression of
epithelial cell transforming sequence 2 oncogene (46), and that miR-223 overexpression in
RAW264.7 osteoclast precursors inhibited osteoclast formation in
vitro (21). Our results are
consistent with previous studies (17-19,21,42,44), indicating that miR-16 positively
affects bone metastasis and osteoclast activity in breast cancer,
whereas miR-133a and miR-223 regulate them negatively. In the
present study, the effect of the miRNAs on the bone resorptive
ability of osteoclasts was additionally evaluated using dentin
slices in vitro and the same effects were examined in the
bone microenvironment using a bone metastasis model in vivo.
The majority of the previous studies have assessed osteoclast
function only by TRAP staining in vitro, and there are few
studies that have evaluated the effect of the miRNAs on the actual
bone resorptive ability of osteoclasts using dentin slices
(3,21). Several miRNAs have been proposed
as diagnostic or predictive biomarkers for breast cancer, but none
have been validated for practical use in a clinical setting
(47,48). In recent years, various studies on
targeting miRNAs for therapeutic purposes have been conducted, and
it has been demonstrated that miRNAs perform several functions. For
example, miR-133a not only acts as a tumor suppressor, but is also
involved in muscle development or function, and cardiac remodeling
(49,50). Similarly, miR-223 is known to be
engaged in the regulation of immune responses in respiratory and
liver diseases (51,52). However, the functions of miRNAs
have not been fully elucidated (17,19). Therefore, it is important to
recognize that miRNA targeting for therapeutic purposes may have
unexpected effects on gene expression and/or cause untoward side
effects.
There are several limitations to the present study.
Firstly, the effects of miRNAs on osteoblasts or stromal cells have
not been investigated. As previously reported, bone resorption
should be orchestrated by the interaction of various factors from
not only osteoclasts or tumor cells but also osteoblasts or stromal
cells (8,11). Secondly, the present study was
conducted only in the bone microenvironment. Cancer bone metastasis
involves multiple processes, including cancer cell dissemination
from the primary site and invasion into the vascular system,
circulation and adhesion to the target site, and invasion and
colonization in the bone microenvironment (53,54). Thirdly, it has been revealed that
the miRNAs could affect osteoclast activity and bone destruction
using MDA-MB-231 human breast cancer cells; however, only one cell
line was used in the present study. Last, only three miRNAs were
employed and other miRNAs and/or their interactions were not
examined. Therefore, further investigations are required to clarify
the effects of miRNAs on breast cancer bone metastasis.
In conclusion, it was demonstrated that miR-16 may
enhance bone destruction caused by breast cancer bone metastasis by
promoting osteoclast function via increased expression of
osteoclast differentiation markers and osteolytic factors, whereas
miR-133a and miR-223 may suppress this process. Although further
studies are required to further elucidate the effect of miRNAs on
cancer bone metastasis, the three miRNAs could serve as biomarkers
and/or therapeutic targets for metastatic bone destruction in
breast cancer.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KKi and YK wrote the manuscript, collected and/or
assembled data and performed data analysis and interpretation. HH,
TT, TMi, YM, YH, KKa, TMatsum, TMatsus and TN collected and/or
assembled data. KKi, TK and YK confirmed the authenticity of all
the raw data. SF and SY performed data analysis and interpretation.
TK, RK and TA conceived/designed, collected and/or assembled data,
performed data analysis and interpretation, and gave final approval
of the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All animal procedures were performed in accordance
with the Japanese Physiological Society Guidelines for the Care and
Use of Laboratory Animals. This study was approved by the
Institutional Animal Care and Use Committee (approval no. P191207)
and carried out according to the Kobe University Animal
Experimentation Regulations (Kobe, Japan).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Ms Minako Nagata, Ms
Maya Yasuda and Ms Kyoko Tanaka (Department of Orthopaedic Surgery,
Kobe University Graduate School of Medicine, Kobe, Japan) for their
expert technical assistance.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li Z and Kang Y: Emerging therapeutic
targets in metastatic progression: A focus on breast cancer.
Pharmacol Ther. 161:79–96. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cai WL, Huang WD, Li B, Chen TR, Li ZX,
Zhao CL, Li HY, Wu YM, Yan WJ and Xiao JR: microRNA-124 inhibits
bone metastasis of breast cancer by repressing Interleukin-11. Mol
Cancer. 17:92018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Coleman RE: Bisphosphonates: Clinical
experience. Oncologist. 9(Suppl 4): 14–27. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
D'Oronzo S, Gregory W, Nicholson S, Chong
YK, Brown J and Coleman R: Natural history of stage II/III breast
cancer, bone metastasis and the impact of adjuvant zoledronate on
distribution of recurrences. J Bone Oncol. 28:1003672021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tanaka R, Yonemori K, Hirakawa A,
Kinoshita F, Takahashi N, Hashimoto J, Kodaira M, Yamamoto H,
Yunokawa M, Shimizu C, et al: Risk factors for developing
skeletal-related events in breast cancer patients with bone
metastases undergoing treatment with bone-modifying agents.
Oncologist. 21:508–513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Coleman RE and Rubens RD: The clinical
course of bone metastases from breast cancer. Br J Cancer.
55:61–66. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu X and Kang Y: Organotropism of breast
cancer metastasis. J Mammary Gland Biol Neoplasia. 12:153–162.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taube T, Elomaa I, Blomqvist C, Beneton MN
and Kanis JA: Histomorphometric evidence for osteoclast-mediated
bone resorption in metastatic breast cancer. Bone. 15:161–166.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Weidle UH, Birzele F, Kollmorgen G and
Rüger R: Molecular mechanisms of bone metastasis. Cancer Genomics
Proteomics. 13:1–12. 2016.
|
|
11
|
Siclari VA, Guise TA and Chirgwin JM:
Molecular interactions between breast cancer cells and the bone
microenvironment drive skeletal metastases. Cancer Metastasis Rev.
25:621–633. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sutherland A, Forsyth A, Cong Y, Grant L,
Juan TH, Lee JK, Klimowicz A, Petrillo SK, Hu J, Chan A, et al: The
role of prolactin in bone metastasis and breast cancer
cell-mediated osteoclast differentiation. J Natl Cancer Inst.
108:1082015.
|
|
13
|
Hussein O, Tiedemann K and Komarova SV:
Breast cancer cells inhibit spontaneous and bisphosphonate-induced
osteoclast apoptosis. Bone. 48:202–211. 2011. View Article : Google Scholar
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Browne G, Taipaleenmäki H, Stein GS, Stein
JL and Lian JB: MicroRNAs in the control of metastatic bone
disease. Trends Endocrinol Metab. 25:320–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ell B, Mercatali L, Ibrahim T, Campbell N,
Schwarzenbach H, Pantel K, Amadori D and Kang Y: Tumor-induced
osteoclast miRNA changes as regulators and biomarkers of osteolytic
bone metastasis. Cancer Cell. 24:542–556. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Usmani A, Shoro AA, Shirazi B, Memon Z and
Hussain M: MiR-16: A novel hereditary marker in breast cancer and
their offspring. J Pak Med Assoc. 67:446–450. 2017.PubMed/NCBI
|
|
19
|
Sui Y, Zhang X, Yang H, Wei W and Wang M:
MicroRNA-133a acts as a tumour suppressor in breast cancer through
targeting LASP1. Oncol Rep. 39:473–482. 2018.
|
|
20
|
Zhang L, Li H, Zang Y and Wang F: NLRP3
inflammasome inactivation driven by miR 223 3p reduces tumor growth
and increases anticancer immunity in breast cancer. Mol Med Rep.
19:2180–2188. 2019.PubMed/NCBI
|
|
21
|
Sugatani T and Hruska KA: MicroRNA-223 is
a key factor in osteoclast differentiation. J Cell Biochem.
101:996–999. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sugatani T and Hruska KA: Impaired
micro-RNA pathways diminish osteoclast differentiation and
function. J Biol Chem. 284:4667–4678. 2009. View Article : Google Scholar :
|
|
23
|
Xu XY, Guo C, Yan YX, Guo Y, Li RX, Song M
and Zhang XZ: Differential effects of mechanical strain on
osteoclastogenesis and osteoclast-related gene expression in
RAW264.7 cells. Mol Med Rep. 6:409–415. 2012.PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Modderman WE, Tuinenburg-Bol Raap AC and
Nijweide PJ: Tartrate-resistant acid phosphatase is not an
exclusive marker for mouse osteoclasts in cell culture. Bone.
12:81–87. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Faul F, Erdfelder E, Lang AG and Buchner
A: G*Power 3: A flexible statistical power analysis program for the
social, behavioral, and biomedical sciences. Behav Res Methods.
39:175–191. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Okada Y, Ueno H, Katagiri M, Oneyama T,
Shimomura K, Sakurai S, Mataga I, Moride M and Hasegawa H:
Experimental study of antiangiogenic gene therapy targeting VEGF in
oral cancer. Odontology. 98:52–59. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mishra S, Tang Y, Wang L, deGraffenried L,
Yeh IT, Werner S, Troyer D, Copland JA and Sun LZ: Blockade of
transforming growth factor-beta (TGFβ) signaling inhibits
osteoblastic tumorigenesis by a novel human prostate cancer cell
line. Prostate. 71:1441–1454. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Papachristou DJ, Basdra EK and
Papavassiliou AG: Bone metastases: Molecular mechanisms and novel
therapeutic interventions. Med Res Rev. 32:611–636. 2012.
View Article : Google Scholar
|
|
30
|
Kingsley LA, Fournier PG, Chirgwin JM and
Guise TA: Molecular biology of bone metastasis. Mol Cancer Ther.
6:2609–2617. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hofbauer LC, Rachner TD, Coleman RE and
Jakob F: Endocrine aspects of bone metastases. Lancet Diabetes
Endocrinol. 2:500–512. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mundy GR: Metastasis to bone: Causes,
consequences and therapeutic opportunities. Nat Rev Cancer.
2:584–593. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Roodman GD: Biology of osteoclast
activation in cancer. J Clin Oncol. 19:3562–3571. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thomas RJ, Guise TA, Yin JJ, Elliott J,
Horwood NJ, Martin TJ and Gillespie MT: Breast cancer cells
interact with osteoblasts to support osteoclast formation.
Endocrinology. 140:4451–4458. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tulotta C and Ottewell P: The role of
IL-1B in breast cancer bone metastasis. Endocr Relat Cancer.
25:R421–R434. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fili S, Karalaki M and Schaller B:
Mechanism of bone metastasis: The role of osteoprotegerin and of
the host-tissue microenvironment-related survival factors. Cancer
Lett. 283:10–19. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Palmqvist P, Persson E, Conaway HH and
Lerner UH: IL-6, leukemia inhibitory factor, and oncostatin M
stimulate bone resorption and regulate the expression of receptor
activator of NF-kappa B ligand, osteoprotegerin, and receptor
activator of NF-kappa B in mouse calvariae. J Immunol.
169:3353–3362. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sunyer T, Lewis J, Collin-Osdoby P and
Osdoby P: Estrogen's bone-protective effects may involve
differential IL-1 receptor regulation in human osteoclast-like
cells. J Clin Invest. 103:1409–1418. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu X, Zeng R, Wu S, Zhong J, Yang L and Xu
J: Comprehensive expression analysis of miRNA in breast cancer at
the miRNA and isomiR levels. Gene. 557:195–200. 2015. View Article : Google Scholar
|
|
41
|
Calin GA and Croce CM: MicroRNA-cancer
connection: The beginning of a new tale. Cancer Res. 66:7390–7394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schwarzenbach H, Hoon DS and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen WS, Leung CM, Pan HW, Hu LY, Li SC,
Ho MR and Tsai KW: Silencing of miR-11 and miR-133a2 cluster
expression by DNA hypermethylation in colorectal cancer. Oncol Rep.
28:1069–1076. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shi W, Tang T, Li X, Deng S, Li R, Wang Y,
Wang Y, Xia T, Zhang Y, Zen K, et al: Methylation-mediated
silencing of miR-133a-3p promotes breast cancer cell migration and
stemness via miR-133a-3p/MAML1/DNMT3A positive feedback loop. J Exp
Clin Cancer Res. 38:4292019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fabris L, Berton S, Citron F, D'Andrea S,
Segatto I, Nicoloso MS, Massarut S, Armenia J, Zafarana G, Rossi S,
et al: Radiotherapy-induced miR-223 prevents relapse of breast
cancer by targeting the EGF pathway. Oncogene. 35:4914–4926. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang X, Tong Z and Liu H: MiR-223-3p
targeting epithelial cell transforming sequence 2 oncogene inhibits
the activity, apoptosis, invasion and migration of MDA-MB-468
breast cancer cells. OncoTargets Ther. 12:7675–7684. 2019.
View Article : Google Scholar
|
|
47
|
McGuire A, Brown JA and Kerin MJ:
Metastatic breast cancer: The potential of miRNA for diagnosis and
treatment monitoring. Cancer Metastasis Rev. 34:145–155. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hamam R, Hamam D, Alsaleh KA, Kassem M,
Zaher W, Alfayez M, Aldahmash A and Alajez NM: Circulating
microRNAs in breast cancer: Novel diagnostic and prognostic
biomarkers. Cell Death Dis. 8:e30452017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Di Mauro V, Crasto S, Colombo FS, Di
Pasquale E and Catalucci D: Wnt signalling mediates miR-133a
nuclear re-localization for the transcriptional control of Dnmt3b
in cardiac cells. Sci Rep. 9:93202019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu N, Bezprozvannaya S, Williams AH, Qi
X, Richardson JA, Bassel-Duby R and Olson EN: microRNA-133a
regulates cardiomyocyte proliferation and suppresses smooth muscle
gene expression in the heart. Genes Dev. 22:3242–3254. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Roffel MP, Bracke KR, Heijink IH and Maes
T: miR-223: A key regulator in the innate immune response in asthma
and COPD. Front Med (Lausanne). 7:1962020. View Article : Google Scholar
|
|
52
|
Ye D, Zhang T, Lou G and Liu Y: Role of
miR-223 in the pathophysiology of liver diseases. Exp Mol Med.
50:1–12. 2018. View Article : Google Scholar
|
|
53
|
Wang M, Xia F, Wei Y and Wei X: Molecular
mechanisms and clinical management of cancer bone metastasis. Bone
Res. 8:302020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fares J, Fares MY, Khachfe HH, Salhab HA
and Fares Y: Molecular principles of metastasis: A hallmark of
cancer revisited. Signal Transduct Target Ther. 5:282020.
View Article : Google Scholar : PubMed/NCBI
|