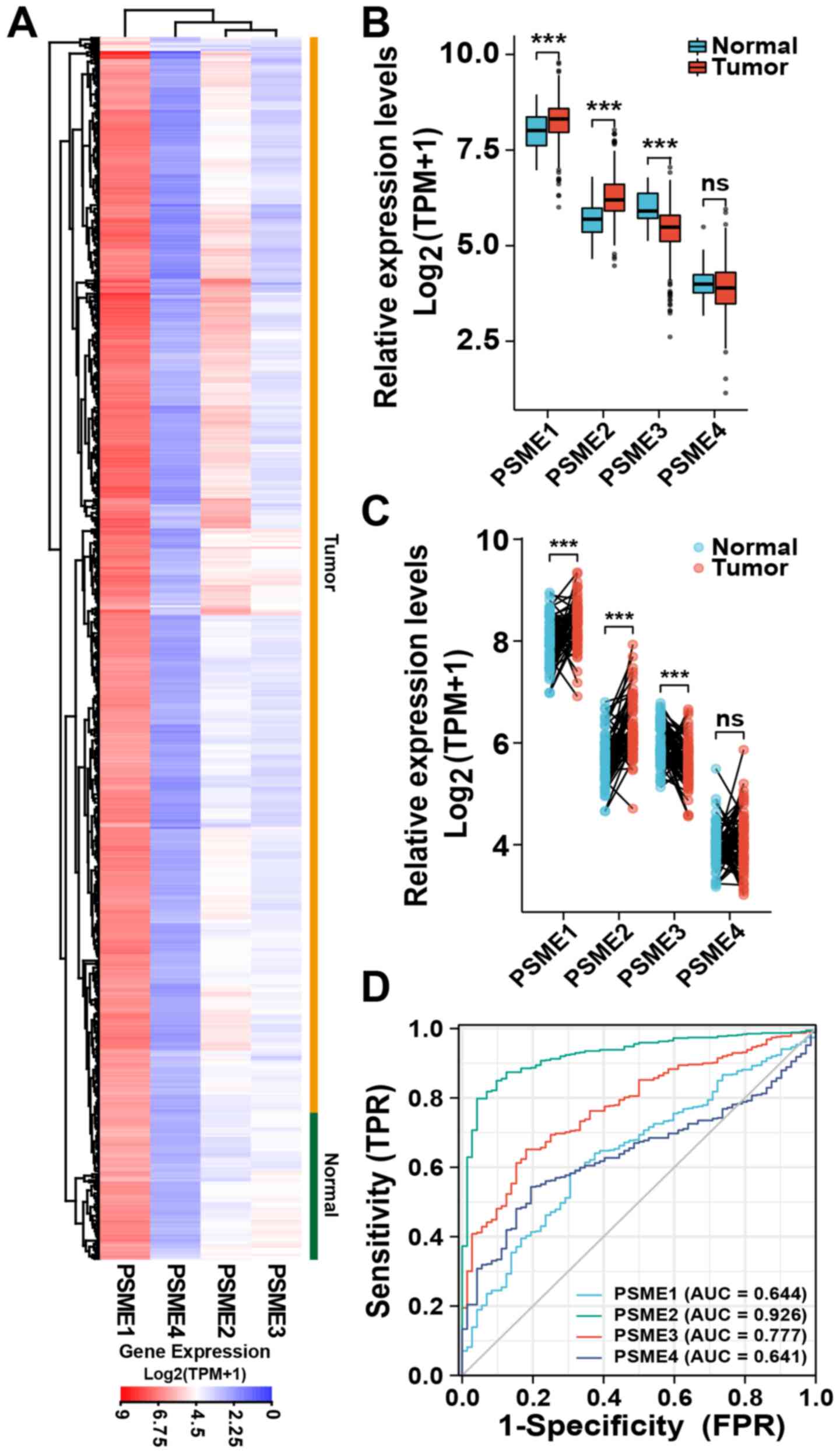
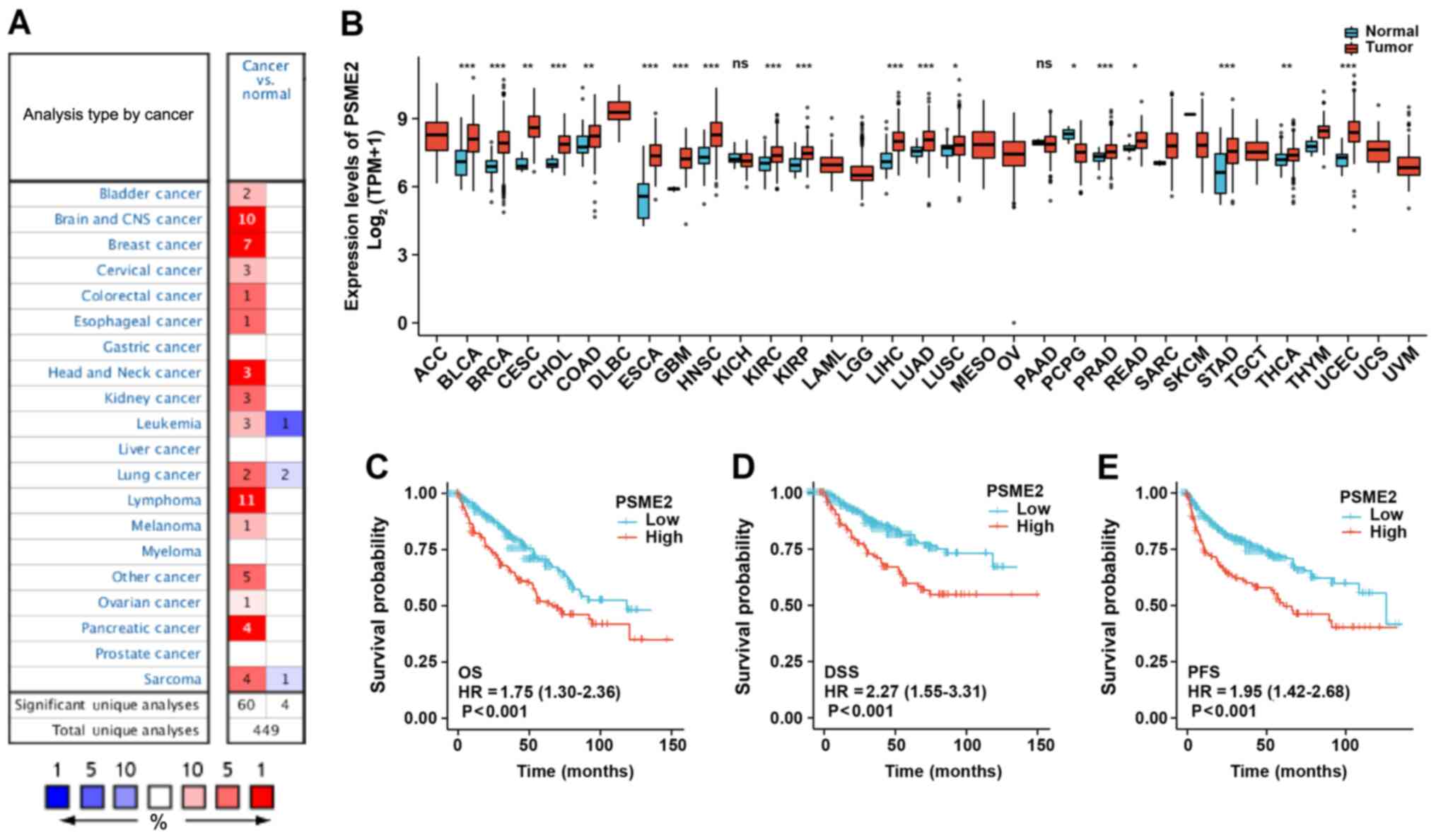
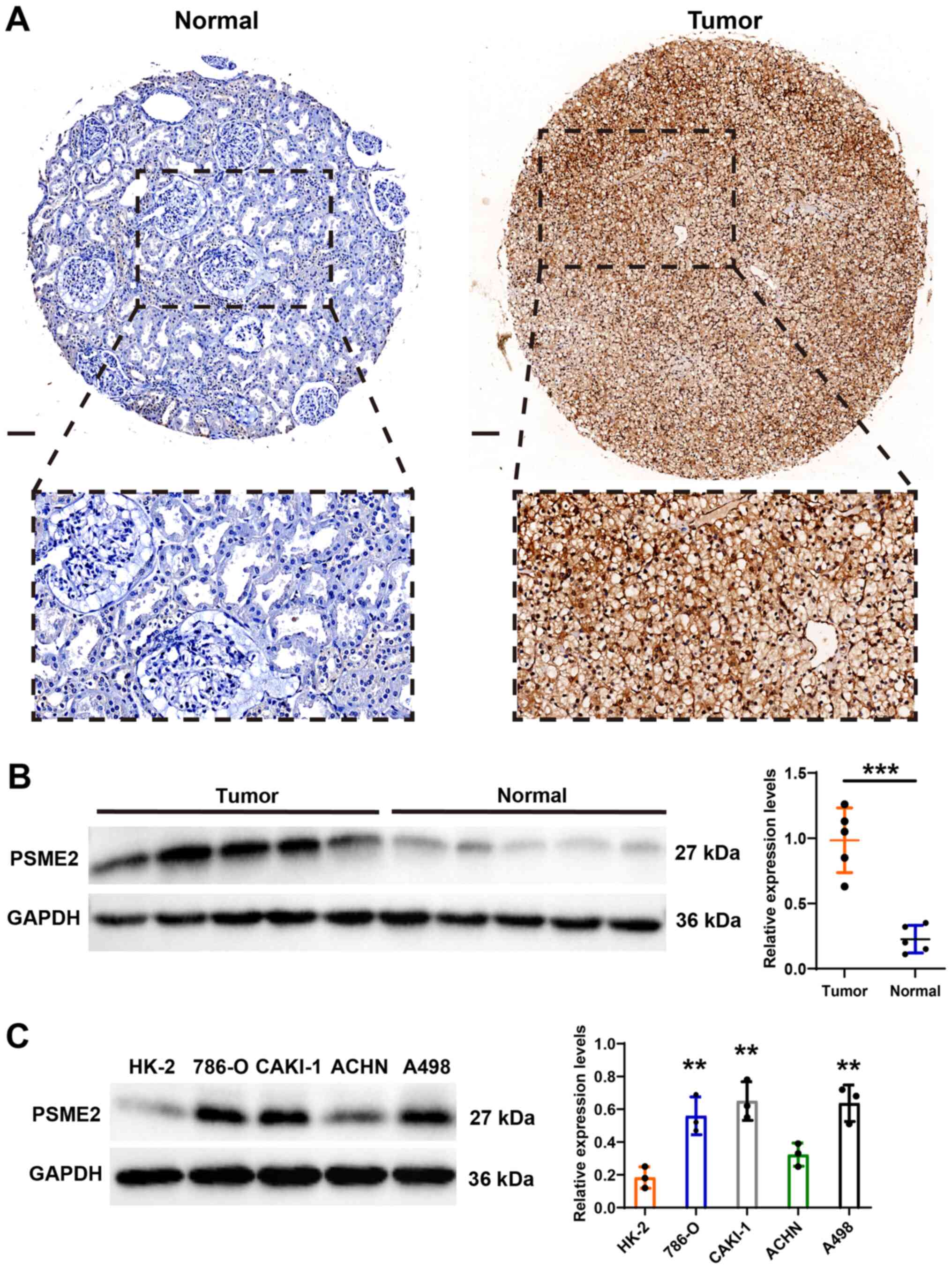
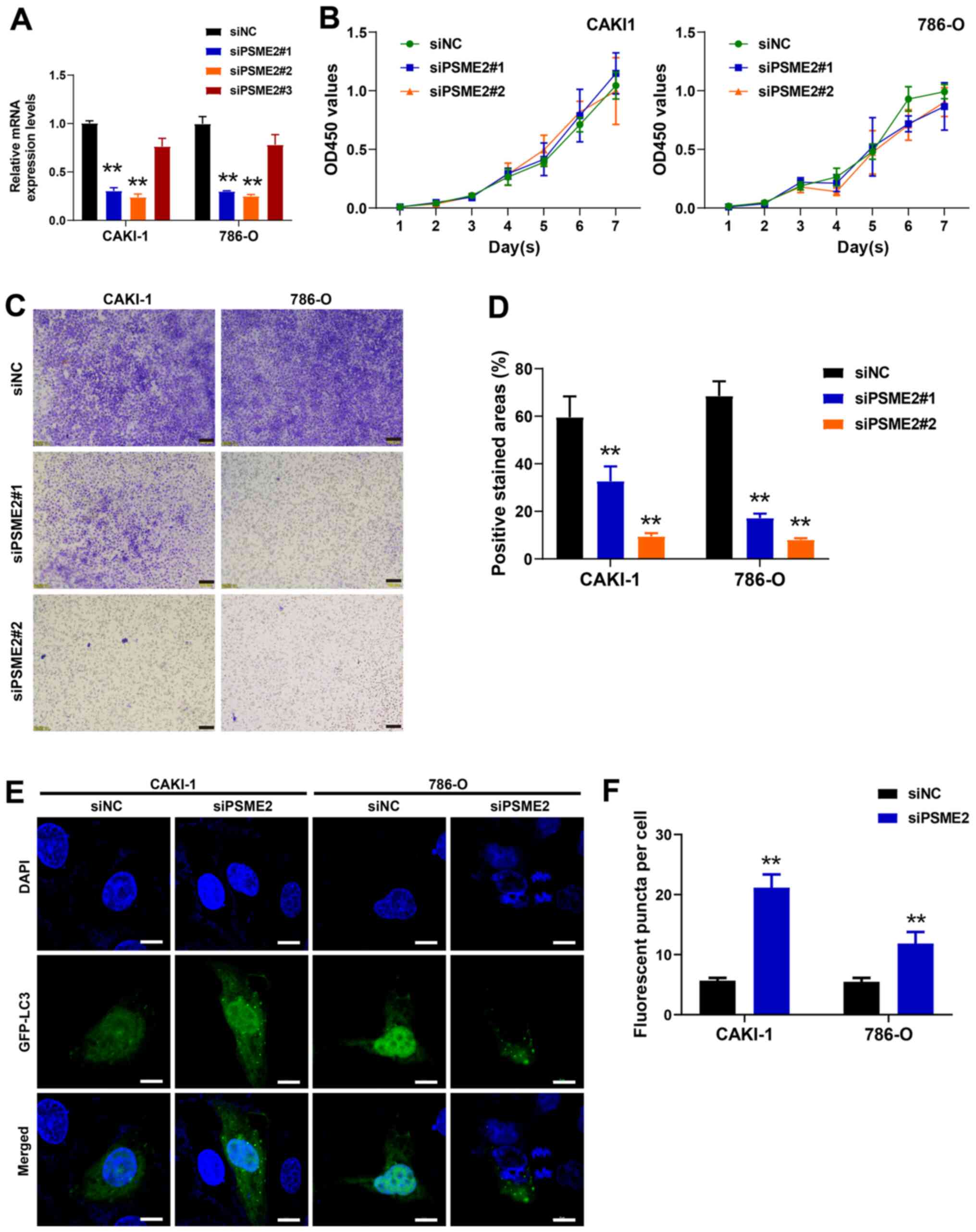
|
1
|
Ahn K, Erlander M, Leturcq D, Peterson PA,
Früh K and Yang Y: In vivo characterization of the proteasome
regulator PA28. J Biol Chem. 271:18237–18242. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Coux O, Tanaka K and Goldberg AL:
Structure and functions of the 20S and 26S proteasomes. Annu Rev
Biochem. 65:801–847. 1996. View Article : Google Scholar
|
|
3
|
Ma CP, Willy PJ, Slaughter CA and
DeMartino GN: PA28, an activator of the 20 S proteasome, is
inactivated by proteolytic modification at its carboxyl terminus. J
Biol Chem. 268:22514–22519. 1993. View Article : Google Scholar
|
|
4
|
Kuroda K and Liu H: The proteasome
inhibitor, bortezomib, induces prostate cancer cell death by
suppressing the expression of prostate-specific membrane antigen,
as well as androgen receptor. Int J Oncol. 54:1357–1366.
2019.PubMed/NCBI
|
|
5
|
Rechsteiner M and Hill CP: Mobilizing the
proteolytic machine: Cell biological roles of proteasome activators
and inhibitors. Trends Cell Biol. 15:27–33. 2005. View Article : Google Scholar : PubMed/NCBI
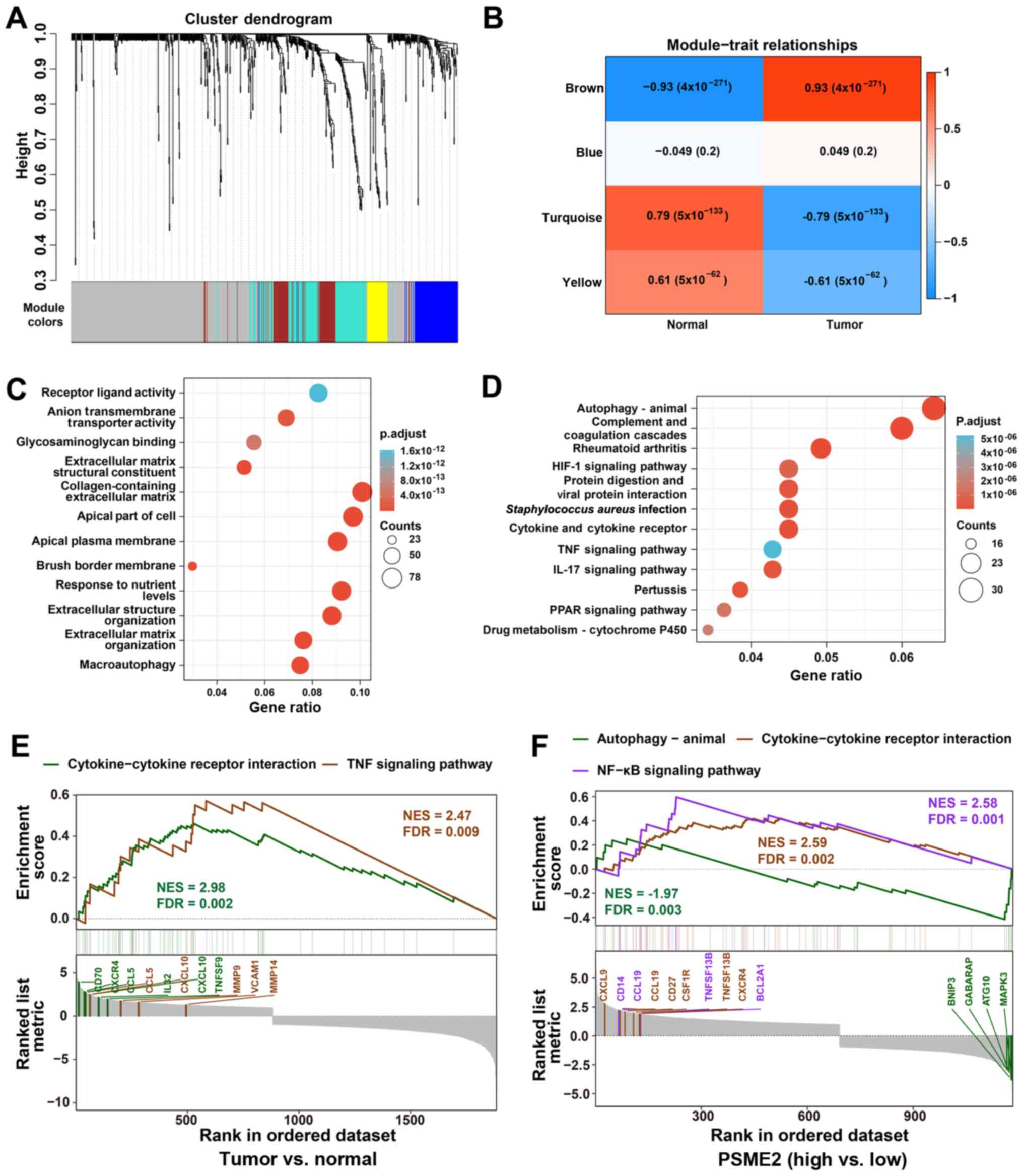
|
|
6
|
Song X, von Kampen J, Slaughter CA and
DeMartino GN: Relative functions of the alpha and beta subunits of
the proteasome activator, PA28. J Biol Chem. 272:27994–28000. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang YF, Yu M, te Pas MFW, Yerle M, Liu B,
Fan B, Xiong TA and Li K: Sequence characterization, polymorphism
and chromosomal localizations of the porcine PSME1 and PSME2 genes.
Anim Genet. 35:361–366. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McCusker D, Wilson M and Trowsdale J:
Organization of the genes encoding the human proteasome activators
PA28alpha and beta. Immunogenetics. 49:438–445. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zaiss DMW and Kloetzel PM: A second gene
encoding the mouse proteasome activator PA28beta subunit is part of
a LINE1 element and is driven by a LINE1 promoter. J Mol Biol.
287:829–835. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kuehn L and Dahlmann B: Structural and
functional properties of proteasome activator PA28. Mol Biol Rep.
24:89–93. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Demartino GN and Gillette TG: Proteasomes:
Machines for all reasons. Cell. 129:659–662. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee J, An S, Jung JH, Kim K, Kim JY, An IS
and Bae S: MUL1 E3 ligase regulates the antitumor effects of
metformin in chemoresistant ovarian cancer cells via AKT
degradation. Int J Oncol. 54:1833–1842. 2019.PubMed/NCBI
|
|
13
|
Kandil E, Kohda K, Ishibashi T, Tanaka K
and Kasahara M: PA28 subunits of the mouse proteasome: Primary
structures and chromosomal localization of the genes.
Immunogenetics. 46:337–344. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Q, Pan F, Li S, Huang R, Wang X, Wang
S, Liao X, Li D and Zhang L: The prognostic value of the proteasome
activator subunit gene family in skin cutaneous melanoma. J Cancer.
10:2205–2219. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Feng J, Xiao T, Lu SS, Hung XP, Yi H, He
QY, Huang W, Tang YY and Xiao ZQ: ANXA1 derived peptides suppress
gastric and colon cancer cell growth by targeting EphA2
degradation. Int J Oncol. 57:1203–1213. 2020.PubMed/NCBI
|
|
16
|
Wójcik C, Tanaka K, Paweletz N, Naab U and
Wilk S: Proteasome activator (PA28) subunits, alpha, beta and gamma
(Ki antigen) in NT2 neuronal precursor cells and HeLa S3 cells. Eur
J Cell Biol. 77:151–160. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Khor B, Bredemeyer AL, Huang CY, Turnbull
IR, Evans R, Maggi LB Jr, White JM, Walker LM, Carnes K, Hess RA,
et al: Proteasome activator PA200 is required for normal
spermatogenesis. Mol Cell Biol. 26:2999–3007. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Noda C, Tanahashi N, Shimbara N, Hendil KB
and Tanaka K: Tissue distribution of constitutive proteasomes,
immunoproteasomes, and PA28 in rats. Biochem Biophys Res Commun.
277:348–354. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang L, Haratake K, Miyahara H and Chiba
T: Proteasome activators, PA28 gamma and PA200, play indispensable
roles in male fertility. Sci Rep. 6:92016.
|
|
20
|
Kaymaz Y, Oduor CI, Yu H, Otieno JA,
Ong'echa JM, Moormann AM and Bailey JA: Comprehensive Transcriptome
and Mutational Profiling of Endemic Burkitt Lymphoma Reveals EBV
Type-Specific Differences. Mol Cancer Res. 15:563–576. 2017.
View Article : Google Scholar :
|
|
21
|
Tanahashi N, Yokota K, Ahn JY, Chung CH,
Fujiwara T, Takahashi E, DeMartino GN, Slaughter CA, Toyonaga T,
Yamamura K, et al: Molecular properties of the proteasome activator
PA28 family proteins and gamma-interferon regulation. Genes Cells.
2:195–211. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Minor MM, Hollinger FB, McNees AL, Jung
SY, Jain A, Hyser JM, Bissig KD and Slagle BL: Hepatitis B Virus
HBx Protein Mediates the Degradation of Host Restriction Factors
through the Cullin 4 DDB1 E3 Ubiquitin Ligase Complex. Cells.
9:92020. View Article : Google Scholar
|
|
23
|
Li J, Powell SR and Wang X: Enhancement of
proteasome function by PA28α overexpression protects against
oxidative stress. FASEB J. 25:883–893. 2011. View Article : Google Scholar :
|
|
24
|
Grune T, Catalgol B, Licht A, Ermak G,
Pickering AM, Ngo JK and Davies KJ: HSP70 mediates dissociation and
reassociation of the 26S proteasome during adaptation to oxidative
stress. Free Radic Biol Med. 51:1355–1364. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Adelöf J, Andersson M, Porritt M, Petersen
A, Zetterberg M, Wiseman J and Hernebring M: PA28αβ overexpression
enhances learning and memory of female mice without inducing 20S
proteasome activity. BMC Neurosci. 19:702018. View Article : Google Scholar
|
|
26
|
Miyagi T, Tatsumi T, Takehara T, Kanto T,
Kuzushita N, Sugimoto Y, Jinushi M, Kasahara A, Sasaki Y, Hori M,
et al: Impaired expression of proteasome subunits and human
leukocyte antigens class I in human colon cancer cells. J
Gastroenterol Hepatol. 18:32–40. 2003. View Article : Google Scholar
|
|
27
|
Cerruti F, Martano M, Petterino C, Bollo
E, Morello E, Bruno R, Buracco P and Cascio P: Enhanced expression
of interferon-gamma-induced antigen-processing machinery components
in a spontaneously occurring cancer. Neoplasia. 9:960–969. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ebert MPA, Krüger S, Fogeron ML, Lamer S,
Chen J, Pross M, Schulz HU, Lage H, Heim S, Roessner A, et al:
Overexpression of cathepsin B in gastric cancer identified by
proteome analysis. Proteomics. 5:1693–1704. 2005. View Article : Google Scholar
|
|
29
|
Perroud B, Lee J, Valkova N, Dhirapong A,
Lin PY, Fiehn O, Kültz D and Weiss RH: Pathway analysis of kidney
cancer using proteomics and metabolic profiling. Mol Cancer.
5:642006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Milioli HH, Santos Sousa K, Kaviski R, Dos
Santos Oliveira NC, De Andrade Urban C, De Lima RS, Cavalli IJ and
De Souza Fonseca Ribeiro EM: Comparative proteomics of primary
breast carcinomas and lymph node metastases outlining markers of
tumor invasion. Cancer Genomics Proteomics. 12:89–101. 2015.
|
|
31
|
Huang Q, Huang Q, Chen W, Wang L, Lin W,
Lin J and Lin X: Identification of transgelin as a potential novel
biomarker for gastric adenocarcinoma based on proteomics
technology. J Cancer Res Clin Oncol. 134:1219–1227. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang Q, Huang Q, Lin W, Lin J and Lin X:
Potential roles for PA28beta in gastric adenocarcinoma development
and diagnosis. J Cancer Res Clin Oncol. 136:1275–1282. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zheng D-L, Huang Q-L, Zhou F, Huang Q-J,
Lin J-Y and Lin X: PA28β regulates cell invasion of gastric cancer
via modulating the expression of chloride intracellular channel 1.
J Cell Biochem. 113:1537–1546. 2012.
|
|
34
|
Kim JE, Koo KH, Kim YH, Sohn J and Park
YG: Identification of potential lung cancer biomarkers using an in
vitro carcinogenesis model. Exp Mol Med. 40:709–720. 2008.
View Article : Google Scholar
|
|
35
|
Chen JY, Xu L, Fang WM, Han JY, Wang K and
Zhu KS: Identification of PA28β as a potential novel biomarker in
human esophageal squamous cell carcinoma. Tumour Biol.
39:10104283177197802017.
|
|
36
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Langfelder P and Horvath S: Eigengene
networks for studying the relationships between co-expression
modules. BMC Syst Biol. 1:542007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
40
|
Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar :
|
|
41
|
Azhar RA, de Castro Abreu AL, Broxham E,
Sherrod A, Ma Y, Cai J, Gill TS, Desai M and Gill IS: Histological
analysis of the kidney tumor-parenchyma interface. J Urol.
193:415–422. 2015. View Article : Google Scholar
|
|
42
|
Moch H, Artibani W, Delahunt B, Ficarra V,
Knuechel R, Montorsi F, Patard JJ, Stief CG, Sulser T and Wild PJ:
Reassessing the current UICC/AJCC TNM staging for renal cell
carcinoma. Eur Urol. 56:636–643. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Margulis V, McDonald M, Tamboli P, Swanson
DA and Wood CG: Predictors of oncological outcome after resection
of locally recurrent renal cell carcinoma. J Urol. 181:2044–2051.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kuang P, Deng H, Liu H, Cui H, Fang J, Zuo
Z, Deng J, Li Y, Wang X and Zhao L: Sodium fluoride induces
splenocyte autophagy via the mammalian targets of rapamycin (mTOR)
signaling pathway in growing mice. Aging (Albany NY). 10:1649–1665.
2018. View Article : Google Scholar
|
|
45
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
46
|
Jones J, Otu H, Spentzos D, Kolia S, Inan
M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al: Gene
signatures of progression and metastasis in renal cell cancer. Clin
Cancer Res. 11:5730–5739. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yusenko MV, Kuiper RP, Boethe T, Ljungberg
B, van Kessel AG and Kovacs G: High-resolution DNA copy number and
gene expression analyses distinguish chromophobe renal cell
carcinomas and renal oncocytomas. BMC Cancer. 9:1522009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yusenko MV, Zubakov D and Kovacs G: Gene
expression profiling of chromophobe renal cell carcinomas and renal
oncocytomas by Affymetrix GeneChip using pooled and individual
tumours. Int J Biol Sci. 5:517–527. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhu WK, Xu WH, Wang J, Huang YQ,
Abudurexiti M, Qu YY, Zhu YP, Zhang HL and Ye DW: Decreased SPTLC1
expression predicts worse outcomes in ccRCC patients. J Cell
Biochem. 121:1552–1562. 2020. View Article : Google Scholar
|
|
50
|
Hu J, Chen Z, Bao L, Zhou L, Hou Y, Liu L,
Xiong M, Zhang Y, Wang B, Tao Z, et al: Single-Cell Transcriptome
Analysis Reveals Intratumoral Heterogeneity in ccRCC, which Results
in Different Clinical Outcomes. Mol Ther. 28:1658–1672. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Reustle A, Di Marco M, Meyerhoff C, Nelde
A, Walz JS, Winter S, Kandabarau S, Büttner F, Haag M, Backert L,
et al: Integrative -omics and HLA-ligandomics analysis to identify
novel drug targets for ccRCC immunotherapy. Genome Med. 12:322020.
View Article : Google Scholar :
|
|
52
|
Wan B, Liu B, Huang Y and Lv C:
Identification of genes of prognostic value in the ccRCC
microenvironment from TCGA database. Mol Genet Genomic Med.
8:e11592020. View Article : Google Scholar :
|
|
53
|
Dikic I and Elazar Z: Mechanism and
medical implications of mammalian autophagy. Nat Rev Mol Cell Biol.
19:349–364. 2018. View Article : Google Scholar
|
|
54
|
Tanida I, Ueno T and Kominami E: LC3
conjugation system in mammalian autophagy. Int J Biochem Cell Biol.
36:2503–2518. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tanida I, Ueno T and Kominami E: LC3 and
Autophagy. Methods Mol Biol. 445:77–88. 2008. View Article : Google Scholar
|
|
56
|
Kabeya Y, Mizushima N, Ueno T, Yamamoto A,
Kirisako T, Noda T, Kominami E, Ohsumi Y and Yoshimori T: LC3, a
mammalian homologue of yeast Apg8p, is localized in autophagosome
membranes after processing. EMBO J. 19:5720–5728. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li L, Tan J, Miao Y, Lei P and Zhang Q:
ROS and Autophagy: Interactions and Molecular Regulatory
Mechanisms. Cell Mol Neurobiol. 35:615–621. 2015. View Article : Google Scholar
|
|
58
|
Zhang J, Zhang C, Jiang X, Li L, Zhang D,
Tang D, Yan T, Zhang Q, Yuan H, Jia J, et al: Involvement of
autophagy in hypoxia-BNIP3 signaling to promote epidermal
keratinocyte migration. Cell Death Dis. 10:2342019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Christian F, Krause E, Houslay MD and
Baillie GS: PKA phosphorylation of p62/SQSTM1 regulates PB1 domain
interaction partner binding. Biochim Biophys Acta. 1843:2765–2774.
2014. View Article : Google Scholar
|
|
60
|
Katsuragi Y, Ichimura Y and Komatsu M:
p62/SQSTM1 functions as a signaling hub and an autophagy adaptor.
FEBS J. 282:4672–4678. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wooten MW, Geetha T, Seibenhener ML, Babu
JR, Diaz-Meco MT and Moscat J: The p62 scaffold regulates nerve
growth factor-induced NF-kappaB activation by influencing TRAF6
polyubiquitination. J Biol Chem. 280:35625–35629. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lamark T, Perander M, Outzen H,
Kristiansen K, Øvervatn A, Michaelsen E, Bjørkøy G and Johansen T:
Interaction codes within the family of mammalian Phox and Bem1p
domain-containing proteins. J Biol Chem. 278:34568–34581. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lin X, Li S, Zhao Y, Ma X, Zhang K, He X
and Wang Z: Interaction domains of p62: A bridge between p62 and
selective autophagy. DNA Cell Biol. 32:220–227. 2013. View Article : Google Scholar
|
|
64
|
Islam MA, Sooro MA and Zhang P: Autophagic
Regulation of p62 is Critical for Cancer Therapy. Int J Mol Sci.
19:152018. View Article : Google Scholar
|
|
65
|
Johansen T and Lamark T: Selective
autophagy mediated by autophagic adapter proteins. Autophagy.
7:279–296. 2011. View Article : Google Scholar
|
|
66
|
Zhang Y, Mun SR, Linares JF, Towers CG,
Thorburn A, Diaz-Meco MT, Kwon YT and Kutateladze TG: Mechanistic
insight into the regulation of SQSTM1/p62. Autophagy. 15:735–737.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kays JK, Koniaris LG, Cooper CA, Pili R,
Jiang G, Liu Y and Zimmers TA: The Combination of Low Skeletal
Muscle Mass and High Tumor Interleukin-6 Associates with Decreased
Survival in Clear Cell Renal Cell Carcinoma. Cancers (Basel).
12:122020. View Article : Google Scholar
|
|
68
|
Liu T, Xia Q, Zhang H, Wang Z, Yang W, Gu
X, Hou T, Chen Y, Pei X, Zhu G, et al: CCL5-dependent mast cell
infiltration into the tumor microenvironment in clear cell renal
cell carcinoma patients. Aging (Albany NY). 12:21809–21836. 2020.
View Article : Google Scholar
|
|
69
|
Xu WH, Shi SN, Xu Y, Wang J, Wang HK, Cao
DL, Shi GH, Qu YY, Zhang HL and Ye DW: Prognostic implications of
Aquaporin 9 expression in clear cell renal cell carcinoma. J Transl
Med. 17:3632019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Mikami S, Mizuno R, Kosaka T, Saya H, Oya
M and Okada Y: Expression of TNF-α and CD44 is implicated in poor
prognosis, cancer cell invasion, metastasis and resistance to the
sunitinib treatment in clear cell renal cell carcinomas. Int J
Cancer. 136:1504–1514. 2015. View Article : Google Scholar
|
|
71
|
Leibovich SJ, Polverini PJ, Shepard HM,
Wiseman DM, Shively V and Nuseir N: Macrophage-induced angiogenesis
is mediated by tumour necrosis factor-alpha. Nature. 329:630–632.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Michalaki V, Syrigos K, Charles P and
Waxman J: Serum levels of IL-6 and TNF-alpha correlate with
clinicopathological features and patient survival in patients with
prostate cancer. Br J Cancer. 90:2312–2316. 2004. View Article : Google Scholar
|
|
73
|
Balkwill F: Tumour necrosis factor and
cancer. Nat Rev Cancer. 9:361–371. 2009. View Article : Google Scholar
|
|
74
|
Liu W, Liu Y, Fu Q, Zhou L, Chang Y, Xu L,
Zhang W and Xu J: Elevated expression of IFN-inducible CXCR3
ligands predicts poor prognosis in patients with non-metastatic
clear-cell renal cell carcinoma. Oncotarget. 7:13976–13983. 2016.
View Article : Google Scholar
|
|
75
|
Choueiri TK, Atkins MB, Rose TL, Alter RS,
Ju Y, Niland K, Wang Y, Arbeit R, Parasuraman S, Gan L, et al: A
phase 1b trial of the CXCR4 inhibitor mavorixafor and nivolumab in
advanced renal cell carcinoma patients with no prior response to
nivolumab monotherapy. Invest New Drugs. 39:1019–1027. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Jiang P, Yueguo W, Huiming H, Hongxiang Y,
Mei W and Ju S: B-Lymphocyte stimulator: A new biomarker for
multiple myeloma. Eur J Haematol. 82:267–276. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Nardelli B, Moore PA, Li Y and Hilbert DM:
B lymphocyte stimulator (BLyS): A therapeutic trichotomy for the
treatment of B lymphocyte diseases. Leuk Lymphoma. 43:1367–1373.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Shivakumar L and Ansell S: Targeting
B-lymphocyte stimulator/B-cell activating factor and a
proliferation-inducing ligand in hematologic malignancies. Clin
Lymphoma Myeloma. 7:106–108. 2006. View Article : Google Scholar : PubMed/NCBI
|