|
1
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dong Y, He D, Peng Z, Peng W, Shi W, Wang
J, Li B, Zhang C and Duan C: Circular RNAs in cancer: An emerging
key player. J Hematol Oncol. 10:22017. View Article : Google Scholar
|
|
4
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak S, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dupont C, Armant DR and Brenner CA:
Epigenetics: Definition, mechanisms and clinical perspective. Semin
Reprod Med. 27:351–357. 2009. View Article : Google Scholar :
|
|
7
|
Bolisetty MT and Graveley BR: Circuitous
route to transcription regulation. Mol Cell. 51:705–706. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suzuki H and Tsukahara T: A view of
pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci.
15:9331–9342. 2014. View Article : Google Scholar :
|
|
9
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar
|
|
10
|
Gruner H, Cortés-López M, Cooper DA, Bauer
M and Miura P: CircRNA accumulation in the aging mouse brain. Sci
Rep. 6:389072016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar
|
|
12
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: CircRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: CircRNA: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16:942017. View Article : Google Scholar
|
|
15
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res. 27:626–641.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou C, Molinie B, Daneshvar K, Pondick
JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC and Mullen
AC: Genome-wide maps of m6A circRNAs identify widespread and
cell-type-specific methylation patterns that are distinct from
mRNAs. Cell Rep. 20:2262–2276. 2017. View Article : Google Scholar
|
|
17
|
Gapp K, Woldemichael BT, Bohacek J and
Mansuy IM: Epigenetic regulation in neurodevelopment and
neurodegenerative diseases. Neuroscience. 264:99–111. 2014.
View Article : Google Scholar
|
|
18
|
Trowbridge JJ, Snow JW, Kim J and Orkin
SH: DNA methyltransferase 1 is essential for and uniquely regulates
hematopoietic stem and progenitor cells. Cell Stem Cell. 5:442–449.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Harman MF and Martín MG: Epigenetic
mechanisms related to cognitive decline during aging. J Neurosci
Res. 98:234–246. 2020. View Article : Google Scholar
|
|
20
|
Feinberg AP and Tycko B: The history of
cancer epigenetics. Nat Rev Cancer. 4:143–153. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jones PA: Functions of DNA methylation:
Islands, start sites, gene bodies and beyond. Nat Rev Genet.
13:484–492. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hájková H, Marková J, Haškovec C, Šárová
I, Fuchs O, Kostečka A, Cetkovský P, Michalová K and Schwarz J:
Decreased DNA methylation in acute myeloid leukemia patients with
DNMT3A mutations and prognostic implications of DNA methylation.
Leuk Res. 36:1128–1133. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bröske AM, Vockentanz L, Kharazi S, Huska
MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et
al: DNA methylation protects hematopoietic stem cell multipotency
from myeloerythroid restriction. Nat Genet. 41:1207–1215. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bock C, Beerman I, Lien WH, Smith ZD, Gu
H, Boyle P, Gnirke A, Fuchs E, Rossi DJ and Meissner A: DNA
methylation dynamics during in vivo differentiation of blood and
skin stem cells. Mol Cell. 47:633–647. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hodges E, Molaro A, Dos Santos CO, Thekkat
P, Song Q, Uren PJ, Park J, Butler J, Rafii S, McCombie WR, et al:
Directional DNA methylation changes and complex intermediate states
accompany lineage specificity in the adult hematopoietic
compartment. Mol Cell. 44:17–28. 2011. View Article : Google Scholar
|
|
26
|
Hogart A, Lichtenberg J, Ajay SS, Anderson
S; NIH Intramural Sequencing Center; Margulies EH and Bodine DM:
Genome-wide DNA methylation profiles in hematopoietic stem and
progenitor cells reveal overrepresentation of ETS transcription
factor binding sites. Genome Res. 22:1407–1418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tadokoro Y, Ema H, Okano M, Li E and
Nakauchi H: De novo DNA methyltransferase is essential for
self-renewal, but not for differentiation, in hematopoietic stem
cells. J Exp Med. 204:715–722. 2007. View Article : Google Scholar :
|
|
28
|
Jiang Y, Dunbar A, Gondek LP, Mohan S,
Rataul M, O'Keefe C, Sekeres M, Saunthararajah Y and Maciejewski
JP: Aberrant DNA methylation is a dominant mechanism in MDS
progression to AML. Blood. 113:1315–1325. 2009. View Article : Google Scholar :
|
|
29
|
Chen J, Odenike O and Rowley JD:
Leukaemogenesis: More than mutant genes. Nat Rev Cancer. 10:23–36.
2010. View Article : Google Scholar
|
|
30
|
Schoofs T, Berdel WE and Müller-Tidow C:
Origins of aberrant DNA methylation in acute myeloid leukemia.
Leukemia. 28:1–14. 2014. View Article : Google Scholar
|
|
31
|
Figueroa ME, Lugthart S, Li Y,
Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J,
van Putten W, Skrabanek L, et al: DNA methylation signatures
identify biologically distinct subtypes in acute myeloid leukemia.
Cancer Cell. 17:13–27. 2010. View Article : Google Scholar :
|
|
32
|
Cole CB, Verdoni AM, Ketkar S, Leight ER,
Russler-Germain DA, Lamprecht TL, Demeter RT, Magrini V and Ley TJ:
PML-RARA requires DNA methyltransferase 3A to initiate acute
promyelocytic leukemia. J Clin Invest. 126:85–98. 2016. View Article : Google Scholar :
|
|
33
|
Ley TJ, Miller C, Ding L, Raphael BJ,
Mungall AJ, Robertson AG, Hoadley K, Triche TJ Jr, Laird PW, Batty
JD, et al: Genomic and epigenomic landscapes of adult de novo acute
myeloid leukemia. N Engl J Med. 368:2059–2074. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Thol F, Damm F, Lüdeking A, Winschel C,
Wagner K, Morgan M, Yun H, Göhring G, Schlegelberger B, Hoelzer D,
et al: Incidence and prognostic influence of DNMT3A mutations in
acute myeloid leukemia. J Clin Oncol. 29:2889–2896. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Marková J, Michková P, Burčková K,
Březinová J, Michalová K, Dohnalová A, Maaloufová JS, Soukup P,
Vítek A, Cetkovský P and Schwarz J: Prognostic impact of DNMT3A
mutations in patients with intermediate cytogenetic risk profile
acute myeloid leukemia. Eur J Haematol. 88:128–135. 2012.
View Article : Google Scholar
|
|
36
|
Alvarez S, Suela J, Valencia A, Fernández
A, Wunderlich M, Agirre X, Prósper F, Martín-Subero JI, Maiques A,
Acquadro F, et al: DNA methylation profiles and their relationship
with cytogenetic status in adult acute myeloid leukemia. PLoS One.
5:e121972010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Akalin A, Garrett-Bakelman FE, Kormaksson
M, Busuttil J, Zhang L, Khrebtukova I, Milne TA, Huang Y, Biswas D,
Hess JL, et al: Base-pair resolution DNA methylation sequencing
reveals profoundly divergent epigenetic landscapes in acute myeloid
leukemia. PLoS Genet. 8:e10027812012. View Article : Google Scholar :
|
|
38
|
Cimmino L, Dawlaty MM, Ndiaye-Lobry D, Yap
YS, Bakogianni S, Yu Y, Bhattacharyya S, Shaknovich R, Geng H,
Lobry C, et al: Erratum: TET1 is a tumor suppressor of
hematopoietic malignancy. Nat Immunol. 16:8892015. View Article : Google Scholar
|
|
39
|
Moran-Crusio K, Reavie L, Shih A,
Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar
A, Patel J, Zhao X, et al: Tet2 loss leads to increased
hematopoietic stem cell self-renewal and myeloid transformation.
Cancer Cell. 20:11–24. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Z, Cai X, Cai CL, Wang J, Zhang W,
Petersen BE, Yang FC and Xu M: Deletion of Tet2 in mice leads to
dysregulated hematopoietic stem cells and subsequent development of
myeloid malignancies. Blood. 118:4509–4518. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Abdel-Wahab O, Mullally A, Hedvat C,
Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara
O, Bhat R, et al: Genetic characterization of TET1, TET2, and TET3
alterations in myeloid malignancies. Blood. 114:144–147. 2009.
View Article : Google Scholar :
|
|
42
|
Tefferi A, Lim KH, Abdel-Wahab O, Lasho
TL, Patel J, Patnaik MM, Hanson CA, Pardanani A, Gilliland DG and
Levine RL: Detection of mutant TET2 in myeloid malignancies other
than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML.
Leukemia. 23:1343–1345. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bacher U, Haferlach C, Schnittger S,
Kohlmann A, Kern W and Haferlach T: Mutations of the TET2 and CBL
genes: Novel molecular markers in myeloid malignancies. Ann
Hematol. 89:643–652. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sato H, Wheat JC, Steidl U and Ito K:
DNMT3A and TET2 in the pre-leukemic phase of hematopoietic
disorders. Front Oncol. 6:1872016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chan SM and Majeti R: Role of DNMT3A,
TET2, and IDH1/2 mutations in pre-leukemic stem cells in acute
myeloid leukemia. Int J Hematol. 98:648–657. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Weissmann S, Alpermann T, Grossmann V,
Kowarsch A, Nadarajah N, Eder C, Dicker F, Fasan A, Haferlach C,
Haferlach T, et al: Landscape of TET2 mutations in acute myeloid
leukemia. Leukemia. 26:934–942. 2012. View Article : Google Scholar
|
|
47
|
Shih AH, Jiang Y, Meydan C, Shank K,
Pandey S, Barreyro L, Antony-Debre I, Viale A, Socci N, Sun Y, et
al: Mutational cooperativity linked to combinatorial epigenetic
gain of function in acute myeloid leukemia. Cancer Cell.
27:502–515. 2015. View Article : Google Scholar
|
|
48
|
Rasmussen KD, Jia G, Johansen JV, Pedersen
MT, Rapin N, Bagger F, Porse BT, Bernard OA, Christensen J, Helin
K, et al: Loss of TET2 in hematopoietic cells leads to DNA
hypermethylation of active enhancers and induction of
leukemogenesis. Genes Dev. 29:910–922. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Berger SL: The complex language of
chromatin regulation during transcription. Nature. 447:407–412.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Podobinska M, Szablowska-Gadomska I,
Augustyniak J, Sandvig I, Sandvig A and Buzanska L: Epigenetic
modulation of stem cells in neurodevelopment: The role of
methylation and acetylation. Front Cell Neurosci. 11:232017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang Y, Gilquin B, Khochbin S and
Matthias P: Two catalytic domains are required for protein
deacetylation. J Biol Chem. 281:2401–2404. 2006. View Article : Google Scholar
|
|
52
|
Uchida T, Kinoshita T, Nagai H, Nakahara
Y, Saito H, Hotta T and Murate T: Hypermethylation of the p15INK4B
gene in myelodysplastic syndromes. Blood. 90:1403–1409. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Melki JR, Vincent PC and Clark SJ:
Concurrent DNA hyper-methylation of multiple genes in acute myeloid
leukemia. Cancer Res. 59:3730–3740. 1999.PubMed/NCBI
|
|
54
|
Herman JG, Jen J, Merlo A and Baylin SB:
Hypermethylation-associated inactivation indicates a tumor
suppressor role for p15INK4B. Cancer Res. 56:722–727.
1996.PubMed/NCBI
|
|
55
|
Jenuwein T: Translating the histone code.
Science. 293:1074–1080. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
van Dijk AD, Hu CW, de Bont ESJM, Qiu Y,
Hoff FW, Yoo SY, Coombes KR, Qutub AA and Kornblau SM: Histone
modification patterns using RPPA-based profiling predict outcome in
acute myeloid leukemia patients. Proteomics. 18:17003792018.
View Article : Google Scholar
|
|
57
|
Zaghlool A, Halvardson J, Zhao JJ,
Etemadikhah M, Kalushkova A, Konska K, Jernberg-Wiklund H,
Thuresson AC and Feuk L: A role for the chromatin-remodeling factor
BAZ1A in neurodevelopment. Hum Mutat. 37:964–975. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Olave IA, Reck-Peterson SL and Crabtree
GR: Nuclear actin and actin-related proteins in chromatin
remodeling. Annu Rev Biochem. 71:755–781. 2002. View Article : Google Scholar
|
|
59
|
Choi KY, Yoo M and Han JH: Toward
understanding the role of the neuron-specific BAF chromatin
remodeling complex in memory formation. Exp Mol Med. 47:e1552015.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Redner RL, Wang J and Liu JM: Chromatin
remodeling and leukemia: New therapeutic paradigms. Blood.
94:417–428. 1999. View Article : Google Scholar
|
|
61
|
Sperlazza J, Rahmani M, Beckta J, Aust M,
Hawkins E, Wang SZ, Zu Zhu S, Podder S, Dumur C, Archer K, et al:
Depletion of the chromatin remodeler CHD4 sensitizes AML blasts to
genotoxic agents and reduces tumor formation. Blood. 126:1462–1472.
2015. View Article : Google Scholar
|
|
62
|
Denslow SA and Wade PA: The human
Mi-2/NuRD complex and gene regulation. Oncogene. 26:5433–5438.
2007. View Article : Google Scholar
|
|
63
|
D'Alesio C, Punzi S, Cicalese A, Fornasari
L, Furia L, Riva L, Carugo A, Curigliano G, Criscitiello C, Pruneri
G, et al: RNAi screens identify CHD4 as an essential gene in breast
cancer growth. Oncotarget. 7:80901–80915. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
O'Shaughnessy A and Hendrich B: CHD4 in
the DNA-damage response and cell cycle progression: Not so NuRDy
now. Biochem Soc Trans. 41:777–782. 2013. View Article : Google Scholar :
|
|
65
|
Polo SE, Kaidi A, Baskcomb L, Galanty Y
and Jackson SP: Regulation of DNA-damage responses and cell-cycle
progression by the chromatin remodelling factor CHD4. EMBO J.
29:3130–3139. 2010. View Article : Google Scholar :
|
|
66
|
Xia L, Huang W, Bellani M, Seidman MM, Wu
K, Fan D, Nie Y, Cai Y, Zhang YW, Yu LR, et al: CHD4 has oncogenic
functions in initiating and maintaining epigenetic suppression of
multiple tumor suppressor genes. Cancer Cell. 31:653–668.e7. 2017.
View Article : Google Scholar :
|
|
67
|
Heshmati Y, Türköz G, Harisankar A,
Kharazi S, Boström J, Dolatabadi EK, Krstic A, Chang D, Månsson R,
Altun M, et al: The chromatin-remodeling factor CHD4 is required
for maintenance of childhood acute myeloid leukemia. Haematologica.
103:1169–1181. 2018. View Article : Google Scholar :
|
|
68
|
Zhen T, Kwon EM, Zhao L, Hsu J, Hyde RK,
Lu Y, Alemu L, Speck NA and Liu PP: Chd7 deficiency delays
leukemogenesis in mice induced by Cbfb-MYH11. Blood. 130:2431–2442.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Fazi F, Racanicchi S, Zardo G, Starnes LM,
Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco
F, et al: Epigenetic silencing of the myelopoiesis regulator
microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 12:457–466.
2007. View Article : Google Scholar
|
|
70
|
Li Y, Gao L, Luo X, Wang L, Gao X, Wang W,
Sun J, Dou L, Li J, Xu C, et al: Epigenetic silencing of
microRNA-193a contributes to leukemogenesis in t(8;21) acute
myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood.
121:499–509. 2013. View Article : Google Scholar
|
|
71
|
Berger SL, Kouzarides T, Shiekhattar R and
Shilatifard A: An operational definition of epigenetics. Genes Dev.
23:781–783. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Sun WJ, Li JH, Liu S, Wu J, Zhou H, Qu LH
and Yang JH: RMBase: A resource for decoding the landscape of RNA
modifications from high-throughput sequencing data. Nucleic Acids
Res. 44:D259–D265. 2016. View Article : Google Scholar :
|
|
73
|
Lee M, Kim B and Kim VN: Emerging roles of
RNA modification: m6A and U-tail. Cell. 158:980–987. 2014.
View Article : Google Scholar
|
|
74
|
Flamand MN and Meyer KD: The
epitranscriptome and synaptic plasticity. Curr Opin Neurobiol.
59:41–48. 2019. View Article : Google Scholar
|
|
75
|
Maden BE: The numerous modified
nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol
Biol. 39:241–303. 1990. View Article : Google Scholar
|
|
76
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar
|
|
77
|
Zhang X and Jia GF: RNA epigenetic
modification: N6-methyladenosine. Yi Chuan. 38:275–288. 2016.
|
|
78
|
Wei CM, Gershowitz A and Moss B:
Methylated nucleotides block 5′ terminus of HeLa cell messenger
RNA. Cell. 4:379–386. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Niu Y, Zhao X, Wu YS, Li MM, Wang XJ and
Yang YG: N6-methyl-adenosine (m6A) in RNA: An old modification with
a novel epigenetic function. Genomics Proteomics Bioinformatics.
11:8–17. 2013. View Article : Google Scholar
|
|
80
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Dominissini D, Moshitch-Moshkovitz S,
Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K,
Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human
and mouse m6A RNA methylomes revealed by m6A-seq. Nature.
485:201–206. 2012. View Article : Google Scholar
|
|
82
|
Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N,
Han D, Dominissini D, Dai Q, Pan T and He C: High-resolution
N(6)-methyladenosine (m(6) A) map using photo-crosslinking-assisted
m(6) A sequencing. Angew Chemie Int Ed. 54:1587–1590. 2015.
View Article : Google Scholar
|
|
83
|
Linder B, Grozhik AV, Olarerin-George AO,
Meydan C, Mason CE and Jaffrey SR: Single-nucleotide-resolution
mapping of m6A and m6Am throughout the transcriptome. Nat Methods.
12:767–772. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Roundtree IA and He C: RNA
epigenetics-chemical messages for posttranscriptional gene
regulation. Curr Opin Chem Biol. 30:46–51. 2016. View Article : Google Scholar
|
|
85
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2014. View Article : Google Scholar :
|
|
87
|
Bokar JA, Rath-Shambaugh ME, Ludwiczak R,
Narayan P and Rottman F: Characterization and partial purification
of mRNA N6-adenosine methyltransferase from HeLa cell nuclei.
Internal mRNA methylation requires a multisubunit complex. J Biol
Chem. 269:17697–17704. 1994. View Article : Google Scholar
|
|
88
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mrna methylation at internal and 5′ sites. Cell
Rep. 8:284–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Ke S, Alemu EA, Mertens C, Gantman EC, Fak
JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al:
A majority of m 6 A residues are in the last exons, allowing the
potential for 3′ UTR regulation. Genes Dev. 29:2037–2053. 2015.
View Article : Google Scholar
|
|
90
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m6A promotes cap-independent translation. Cell. 163:999–1010. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR
and Qian SB: Dynamic m6A mRNA methylation directs translational
control of heat shock response. Nature. 526:591–594. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Pendleton KE, Chen B, Liu K, Hunter OV,
Xie Y, Tu BP and Conrad NK: The U6 snRNA m 6 A methyltransferase
METTL16 regulates SAM synthetase intron retention. Cell.
169:824–835.e14. 2017. View Article : Google Scholar
|
|
93
|
Dina C, Meyre D, Gallina S, Durand E,
Körner A, Jacobson P, Carlsson LMS, Kiess W, Vatin V, Lecoeur C, et
al: Variation in FTO contributes to childhood obesity and severe
adult obesity. Nat Genet. 39:724–726. 2007. View Article : Google Scholar
|
|
94
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-Methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zheng G, Dahl JA, Niu Y, Fedorcsak P,
Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al: ALKBH5
is a mammalian RNA demethylase that impacts RNA metabolism and
mouse fertility. Mol Cell. 49:18–29. 2013. View Article : Google Scholar :
|
|
96
|
Fu Y, Jia G, Pang X, Wang RN, Wang X, Li
CJ, Smemo S, Dai Q, Bailey KA, Nobrega MA, et al: FTO-mediated
formation of N6-hydroxymethyladenosine and N6-formyladenosine in
mammalian RNA. Nat Commun. 4:17982013. View Article : Google Scholar
|
|
97
|
Zhao X, Yang Y, Sun BF, Shi Y, Yang X,
Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al: FTO-dependent
demethylation of N6-methyladenosine regulates mRNA splicing and is
required for adipogenesis. Cell Res. 24:1403–1419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Hess ME, Hess S, Meyer KD, Verhagen LAW,
Koch L, Brönneke HS, Dietrich MO, Jordan SD, Saletore Y, Elemento
O, et al: The fat mass and obesity associated gene (Fto) regulates
activity of the dopaminergic midbrain circuitry. Nat Neurosci.
16:1042–1048. 2013. View Article : Google Scholar
|
|
99
|
Geula S, Moshitch-Moshkovitz S,
Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V,
Peer E, Mor N, Manor YS, et al: Stem cells. m6A mRNA methylation
facilitates resolution of naïve pluripotency toward
differentiation. Science. 347:1002–1006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Li Z, Weng H, Su R, Weng X, Zuo Z, Li C,
Huang H, Nachtergaele S, Dong L, Hu C, et al: FTO plays an
oncogenic role in acute myeloid leukemia as a 6-methyladenosine RNA
demethylase. Cancer Cell. 31:127–141. 2017. View Article : Google Scholar
|
|
101
|
Jaffrey SR and Kharas MG: Emerging links
between m6A and misregulated mRNA methylation in cancer. Genome
Med. 9:22017. View Article : Google Scholar :
|
|
102
|
Zhang Z, Theler D, Kaminska KH, Hiller M,
de la Grange P, Pudimat R, Rafalska I, Heinrich B, Bujnicki JM,
Allain FHT and Stamm S: The YTH domain is a novel RNA binding
domain. J Biol Chem. 285:14701–14710. 2010. View Article : Google Scholar :
|
|
103
|
Xu C, Wang X, Liu K, Roundtree IA, Tempel
W, Li Y, Lu Z, He C and Min J: Structural basis for selective
binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol.
10:927–929. 2014. View Article : Google Scholar
|
|
104
|
Luo S and Tong L: Molecular basis for the
recognition of methylated adenines in RNA by the eukaryotic YTH
domain. Proc Natl Acad Sci USA. 111:13834–13839. 2014. View Article : Google Scholar
|
|
105
|
Zhu T, Roundtree IA, Wang P, Wang X, Wang
L, Sun C, Tian Y, Li J, He C and Xu Y: Crystal structure of the YTH
domain of YTHDF2 reveals mechanism for recognition of
N6-methyladenosine. Cell Res. 24:1493–1496. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N6-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar
|
|
107
|
Wang Y, Li Y, Toth JI, Petroski MD, Zhang
Z and Zhao JC: N6-methyladenosine modification destabilizes
developmental regulators in embryonic stem cells. Nat Cell Biol.
16:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Fustin JM, Doi M, Yamaguchi Y, Hida H,
Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I
and Okamura H: RNA-methylation-dependent rna processing controls
the speed of the circadian clock. Cell. 155:793–806. 2013.
View Article : Google Scholar
|
|
109
|
Alarcón CR, Lee H, Goodarzi H, Halberg N
and Tavazoie SF: N6-methyladenosine marks primary microRNAs for
processing. Nature. 519:482–485. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Chen T, Hao YJ, Zhang Y, Li MM, Wang M,
Han W, Wu Y, Lv Y, Hao J, Wang L, et al: m6A RNA methylation is
regulated by MicroRNAs and promotes reprogramming to pluripotency.
Cell Stem Cell. 16:289–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Liu N, Dai Q, Zheng G, He C, Parisien M
and Pan T: N6-methyladenosine-dependent RNA structural switches
regulate RNA-protein interactions. Nature. 518:560–564. 2015.
View Article : Google Scholar
|
|
112
|
Klungland A and Dahl JA: Dynamic RNA
modifications in disease. Curr Opin Genet Dev. 26:47–52. 2014.
View Article : Google Scholar
|
|
113
|
Kwok CT, Marshall AD, Rasko JEJ and Wong
JJL: Erratum to: Genetic alterations of m6A regulators predict
poorer survival in acute myeloid leukemia. J Hematol Oncol.
10:492017. View Article : Google Scholar :
|
|
114
|
Su R, Dong L, Li C, Nachtergaele S,
Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al: R-2HG
exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA
signaling. Cell. 172:90–105.e23. 2018. View Article : Google Scholar
|
|
115
|
Vu LP, Pickering BF, Cheng Y, Zaccara S,
Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al:
The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid
differentiation of normal hematopoietic and leukemia cells. Nat
Med. 23:1369–1376. 2017. View Article : Google Scholar
|
|
116
|
Weng H, Huang H, Wu H, Qin X, Zhao BS,
Dong L, Shi H, Skibbe J, Shen C, Hu C, et al: METTL14 inhibits
hematopoietic stem/progenitor differentiation and promotes
leukemogenesis via mRNA m6A modification. Cell Stem Cell.
22:191–205.e9. 2018. View Article : Google Scholar
|
|
117
|
Chhabra R: miRNA and methylation: A
multifaceted liaison. Chembiochem. 16:195–203. 2015. View Article : Google Scholar
|
|
118
|
Hall RH: Isolation of 3-methyluridine and
3-methylcytidine from soluble ribonucleic acid. Biochem Biophys Res
Commun. 12:361–364. 1963. View Article : Google Scholar
|
|
119
|
Xu L, Liu X, Sheng N, Oo KS, Liang J,
Chionh YH, Xu J, Ye F, Gao YG, Dedon PC and Fu XY: Three distinct
3-methylcytidine (m3C) methyltransferases modify tRNA and mRNA in
mice and humans. J Biol Chem. 292:14695–14703. 2017. View Article : Google Scholar :
|
|
120
|
Glasner H, Riml C, Micura R and Breuker K:
Label-free, direct localization and relative quantitation of the
RNA nucleobase methylations m6A, m5C, m3U, and m5U by top-down mass
spectrometry. Nucleic Acids Res. 45:8014–8025. 2017. View Article : Google Scholar :
|
|
121
|
Li X, Zhu P, Ma S, Song J, Bai J, Sun F
and Yi C: Chemical pulldown reveals dynamic pseudouridylation of
the mammalian transcriptome. Nat Chem Biol. 11:592–597. 2015.
View Article : Google Scholar
|
|
122
|
Charette M and Gray MW: Pseudouridine in
RNA: What, where, how, and why. IUBMB Life. 49:341–351. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ofengand J: Ribosomal RNA pseudouridines
and pseudouridine synthases. FEBS Lett. 514:17–25. 2002. View Article : Google Scholar
|
|
124
|
Jack K, Bellodi C, Landry DM, Niederer RO,
Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM,
Thompson SR, et al: rRNA pseudouridylation defects affect ribosomal
ligand binding and translational fidelity from yeast to human
cells. Mol Cell. 44:660–666. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Kiss T, Fayet-Lebaron E and Jády BE: Box
H/ACA small ribonucleoproteins. Mol Cell. 37:597–606. 2010.
View Article : Google Scholar
|
|
126
|
Yu AT, Ge J and Yu YT: Pseudouridines in
spliceosomal snRNAs. Protein Cell. 2:712–725. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Karijolich J and Yu YT: Converting
nonsense codons into sense codons by targeted pseudouridylation.
Nature. 474:395–398. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Rosselló-Tortella M, Ferrer G and Esteller
M: Epitranscriptomics in hematopoiesis and hematologic
malignancies. Blood Cancer Discov. 1:26–31. 2020. View Article : Google Scholar
|
|
129
|
Alseth I, Dalhus B and Bjørås M: Inosine
in DNA and RNA. Curr Opin Genet Dev. 26:116–123. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Bass BL, Nishikura K, Keller W, Seeburg
PH, Emeson RB, O'Connell MA, Samuel CE and Herbert A: A
standardized nomenclature for adenosine deaminases that act on RNA.
RNA. 3:947–949. 1997.
|
|
131
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar
|
|
132
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular rna that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar :
|
|
134
|
Haimov O, Sinvani H and Dikstein R:
Cap-dependent, scanning-free translation initiation mechanisms.
Biochim Biophys Acta. 1849:1313–1318. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
Circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar :
|
|
136
|
Zhang M, Huang N, Yang X, Luo J, Yan S,
Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al: A novel protein
encoded by the circular form of the SHPRH gene suppresses glioma
tumorigenesis. Oncogene. 37:1805–1814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Liang WC, Wong CW, Liang PP, Shi M, Cao Y,
Rao ST, Tsui SKW, Waye MMY, Zhang Q, Fu WM and Zhang JF:
Translation of the circular RNA circβ-catenin promotes liver cancer
cell growth through activation of the wnt pathway. Genome Biol.
20:842019. View Article : Google Scholar
|
|
138
|
Huang X, He M, Huang S, Lin R, Zhan M,
Yang D, Shen H, Xu S, Cheng W, Yu J, et al: Circular RNA circERBB2
promotes gallbladder cancer progression by regulating
PA2G4-dependent rDNA transcription. Mol Cancer. 18:1662019.
View Article : Google Scholar
|
|
139
|
Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ,
Ma XD, Han K, Chen JW, Judde JG, Deas O, et al: 6-methyladenosine
modification of circNSUN2 facilitates cytoplasmic export and
stabilizes HMGA2 to promote colorectal liver metastasis. Nat
Commun. 10:46952019. View Article : Google Scholar
|
|
140
|
Wu P, Fang X, Liu Y, Tang Y, Wang W, Li X
and Fan Y: N6-methyladenosine modification of circCUX1 confers
radio-resistance of hypopharyngeal squamous cell carcinoma through
caspase1 pathway. Cell Death Dis. 12:2982021. View Article : Google Scholar
|
|
141
|
Park OH, Ha H, Lee Y, Boo SH, Kwon DH,
Song HK and Kim YK: Endoribonucleolytic cleavage of m6A-containing
RNAs by RNase P/MRP complex. Mol Cell. 74:494–507.e8. 2019.
View Article : Google Scholar
|
|
142
|
Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin
D, Liu J and Sun Z: The role of N6-methyladenosine (m6A)
modification in the regulation of circRNAs. Mol Cancer. 19:1052020.
View Article : Google Scholar
|
|
143
|
Chen YG, Chen R, Ahmad S, Verma R, Kasturi
SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, et al:
N6-methyladenosine modification controls circular RNA immunity. Mol
Cell. 76:96–109.e9. 2019. View Article : Google Scholar
|
|
144
|
Lux S, Blätte TJ, Gillissen B, Richter A,
Cocciardi S, Skambraks S, Schwarz K, Schrezenmeier H, Döhner H,
Döhner K, et al: Deregulated expression of circular RNAs in acute
myeloid leukemia. Blood Adv. 5:1490–1503. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Bell CC, Fennell KA, Chan YC, Rambow F,
Yeung MM, Vassiliadis D, Lara L, Yeh P, Martelotto LG, Rogiers A,
et al: Targeting enhancer switching overcomes non-genetic drug
resistance in acute myeloid leukaemia. Nat Commun. 10:27232019.
View Article : Google Scholar :
|
|
146
|
Arteaga CL and Engelman JA: ERBB
receptors: From oncogene discovery to basic science to
mechanism-based cancer therapeutics. Cancer Cell. 25:282–303. 2014.
View Article : Google Scholar :
|
|
147
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar
|
|
148
|
L'Abbate A, Tolomeo D, Cifola I,
Severgnini M, Turchiano A, Augello B, Squeo G, D'Addabbo P,
Traversa D, Daniele G, et al: MYC-containing amplicons in acute
myeloid leukemia: Genomic structures, evolution, and
transcriptional consequences. Leukemia. 32:2152–2166. 2018.
View Article : Google Scholar
|
|
149
|
Guarnerio J, Bezzi M, Jeong JC, Paffenholz
SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH and Pandolfi PP:
Oncogenic role of fusion-circRNAs derived from cancer-associated
chromosomal translocations. Cell. 165:289–302. 2016. View Article : Google Scholar
|
|
150
|
Wu DM, Wen X, Han XR, Wang S, Wang YJ,
Shen M, Fan SH, Zhang ZF, Shan Q, Li MQ, et al: Role of circular
RNA DLEU2 in human acute myeloid leukemia. Mol Cell Biol.
38:e00259–e00218. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L
and Ming Z: Silencing of circ_0009910 inhibits acute myeloid
leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol
Dis. 75:41–47. 2019. View Article : Google Scholar
|
|
152
|
Fan H, Li Y, Liu C, Liu Y, Bai J and Li W:
Circular RNA-100290 promotes cell proliferation and inhibits
apoptosis in acute myeloid leukemia cells via sponging miR-203.
Biochem Biophys Res Commun. 507:178–184. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Chen H, Liu T, Liu J, Feng Y, Wang B, Wang
J, Bai J, Zhao W, Shen Y, Wang X, et al: Circ-ANAPC7 is upregulated
in acute myeloid leukemia and appears to target the miR-181 family.
Cell Physiol Biochem. 47:1998–2007. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Li W, Zhong C, Jiao J, Li P, Cui B, Ji C
and Ma D: Characterization of hsa_circ_0004277 as a new biomarker
for acute myeloid leukemia via circular RNA profile and
bioinformatics analysis. Int J Mol Sci. 18:5972017. View Article : Google Scholar :
|
|
155
|
Shang J, Chen WM, Wang ZH, Wei TN, Chen ZZ
and Wu WB: CircPAN3 mediates drug resistance in acute myeloid
leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp Hematol.
70:42–54.e3. 2019. View Article : Google Scholar
|
|
156
|
Hirsch S, Blätte TJ, Grasedieck S,
Cocciardi S, Rouhi A, Jongen-Lavrencic M, Paschka P, Krönke J,
Gaidzik VI, Döhner H, et al: Circular RNAs of the nucleophosmin
(NPM1) gene in acute myeloid leukemia. Haematologica.
102:2039–2047. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar
|
|
158
|
Okcanoğlu TB and Gündüz C: Circular RNAs
in leukemia (Review). Biomed Rep. 10:87–91. 2019.
|
|
159
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar
|
|
161
|
Wang E, Lu SX, Pastore A, Chen X, Imig J,
Lee SC, Hockemeyer K, Ghebrechristos YE, Yoshimi A, Inoue D, et al:
Targeting an RNA-binding protein network in acute myeloid leukemia.
Cancer Cell. 35:369–384.e7. 2019. View Article : Google Scholar
|
|
162
|
Yoshida K, Sanada M, Shiraishi Y, Nowak D,
Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et
al: Frequent pathway mutations of splicing machinery in
myelodysplasia. Nature. 478:64–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Sun YM, Wang WT, Zeng ZC, Chen TQ, Han C,
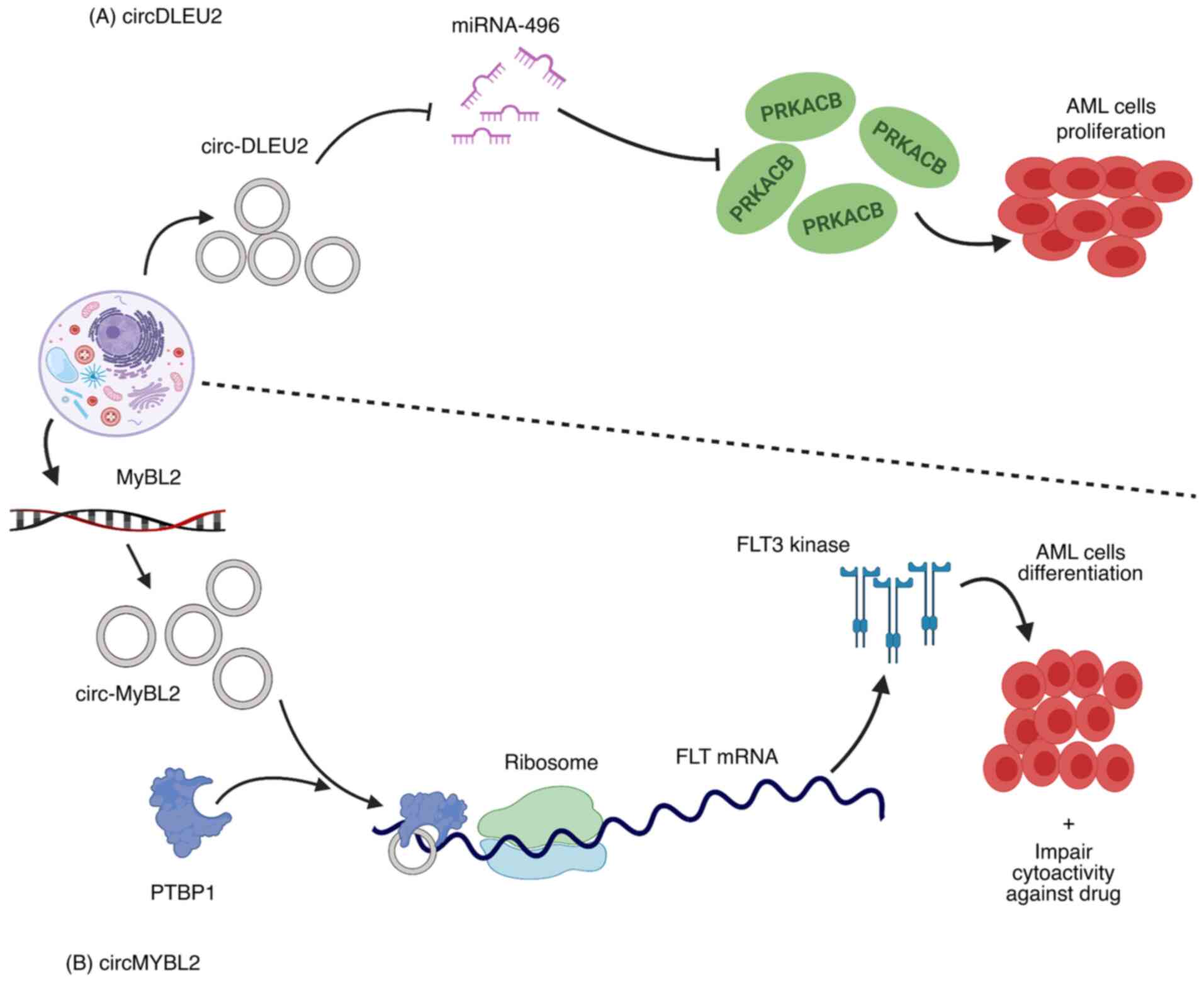
Pan Q, Huang W, Fang K, Sun LY, Zhou YF, et al: circMYBL2, a
circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1
to promote FLT3-ITD AML progression. Blood. 134:1533–1546. 2019.
View Article : Google Scholar
|
|
164
|
Guil S and Esteller M: Cis-acting
noncoding RNAs: Friends and foes. Nat Struct Mol Biol.
19:1068–1075. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar
|
|
166
|
Conn VM, Hugouvieux V, Nayak A, Conos SA,
Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta
C and Conn SJ: A circRNA from SEPALLATA3 regulates splicing of its
cognate mRNA through R-loop formation. Nat Plants. 3:170532017.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Schmitz KM, Mayer C, Postepska A and
Grummt I: Interaction of noncoding RNA with the rDNA promoter
mediates recruitment of DNMT3b and silencing of rRNA genes. Genes
Dev. 24:2264–2269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Starke S, Jost I, Rossbach O, Schneider T,
Schreiner S, Hung LH and Bindereif A: Exon circularization requires
canonical splice signals. Cell Rep. 10:103–111. 2015. View Article : Google Scholar
|
|
169
|
van Rossum D, Verheijen BM and Pasterkamp
RJ: Circular RNAs: Novel regulators of neuronal development. Front
Mol Neurosci. 9:742016. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Chen C, Yuan W, Zhou Q, Shao B, Guo Y,
Wang W, Yang S, Guo Y, Zhao L, Dang Q, et al:
N6-methyladenosine-induced circ1662 promotes metastasis of
colorectal cancer by accelerating YAP1 nuclear localization.
Theranostics. 11:4298–4315. 2021. View Article : Google Scholar
|
|
171
|
Dai F, Wu Y, Lu Y, An C, Zheng X, Dai L,
Guo Y, Zhang L, Li H, Xu W and Gao W: Crosstalk between RNA m6A
modification and non-coding RNA contributes to cancer growth and
progression. Mol Ther Nucleic Acids. 22:62–71. 2020. View Article : Google Scholar
|
|
172
|
Harding CV, Heuser JE and Stahl PD:
Exosomes: Looking back three decades and into the future. J Cell
Biol. 200:367–371. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Melo SA, Sugimoto H, O'Connell JT, Kato N,
Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, et
al: Cancer exosomes perform cell-independent MicroRNA biogenesis
and promote tumorigenesis. Cancer Cell. 26:707–721. 2014.
View Article : Google Scholar :
|
|
174
|
Boyiadzis M and Whiteside TL: Exosomes in
acute myeloid leukemia inhibit hematopoiesis. Curr Opin Hematol.
25:279–284. 2018. View Article : Google Scholar
|