Introduction
Evidence has indicated that more than 850,000
patients are diagnosed with liver cancer each year worldwide,
indicating that liver cancer is a major health issue.
Hepatocellular carcinoma (HCC) accounts for approximately 90% of
all primary liver cancer cases and is known as the second leading
cause of cancer-related deaths worldwide (1,2).
Notably, investigations have revealed that the age-standardized
incidence rates (ASIRs) of liver cancer in eastern Asia are higher
than those in other countries worldwide. In Taiwan, liver cancer
was among the top four most common cancers in 2014, and the ASIR
has decreased over the past several years (3). The development of liver cancer has
been linked to a variety of risk factors, including sex, ethnicity,
chronic viral hepatitis, cirrhosis, inherited metabolic disorders,
alcohol abuse, tobacco use, aflatoxins, obesity, and type-2
diabetes (4,5).
The incidence of primary liver cancer is largely
explained by infection with hepatitis B and C viruses, and such
infections account for over 80% of liver cancer cases worldwide
(6). In fact, numerous studies
have demonstrated a strong correlation between chronic viral
hepatitis, particularly hepatitis induced by hepatitis B and C
viruses and liver cancer development (6-9). A
previous study using an algorithm reported that patients who met
the criteria for chronic hepatitis B virus (HBV) infection had a
significantly higher incidence, ranging from 30 to 140 times, of
developing HCC compared with patients without HBV (10). Another cohort study also indicated
that hepatitis C virus (HCV) infection was associated with the
highest incidence of HCC in patients with cirrhosis, particularly
in Japan (11). More than half of
HCC cases worldwide are attributable to HBV infection. Case-control
and cohort studies reported that the relative risk of HCC in
patients with HBV infection ranges from 5 to 49 and from 7 to 98,
respectively (12,13).
Evidence has indicated that chronic HBV or HCV
infection may cause liver cirrhosis, which is also the most
important risk factor for liver cancer. Various studies have
reported that sustained reduction in HBV/HCV replication lowers the
risk of HCC in patients with HBV/HCV-associated cirrhosis (7,11).
Accordingly, the primary strategy for liver cancer prevention is
the elimination of viral infection by antiviral therapy (14,15). However, information concerning the
anti-liver cancer effects of antiviral drugs remains obscure.
Therefore, the present study investigated the effects of two
anti-hepatitis virus drugs, lamivudine and ribavirin, and one
anti-influenza virus drug, oseltamivir, on liver cancer cells to
identify alternative methods for the treatment of liver cancer.
Materials and methods
Cell culture
Normal human liver epithelial cell line THLE-3
[CRL-11233; American Type Culture Collection (ATCC)] and two liver
cancer cell lines, Huh-7 (JCRB0403; JCRB Cell Bank) and C3A
[HepG2/C3A, derivative of HepG2 (ATCC HB-8065)] (CRL-10741; ATCC)
were maintained following the manufacturers' instructions in
bronchial epithelial cell growth medium (BEGM) (Lonza Group, Ltd.)
or Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.),
respectively. The cell lines used in the present study were
subjected to short tandem repeat (STR) profiling through the
National Cheng Kung University (NCKU) Center for Genomic Medicine
to confirm their authenticity. The antiviral drugs, including
lamivudine (Zeffix Tablet 100 mg; GlaxoSmithKline), ribavirin
(Robatrol capsule 200 mg) and oseltamivir (Tamiflu capsule 75 mg;
both from Roche Diagnostics) were obtained from Changhua Christian
Hospital, Taiwan.
Cell viability
To determine the survival of cells, a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT)
assay was performed. A total of 1×105 cells were
cultured overnight at 37°C in each well of a 24-well plate in a
cell incubator. Following incubation with different concentrations
of antiviral drugs (lamivudine and ribavirin: 0, 1,000, 2,000,
3,000, 4,000 and 5,000 µM; oseltamivir: 0, 50, 100, 250, 500
and 1,000 µM), the culture medium was removed and MTT
reagent (0.5 mg/ml) was added to each well and incubated for
another 4 h. A total of 0.3 ml dimethyl sulfoxide (DMSO) was then
added to each well of the plate and the absorbance was measured at
570 nm with a microplate reader (SpectraMax M5; Molecular Devices,
LLC).
Wound healing assay
To verify the effects of oseltamivir on migration of
liver cancer cells, a wound healing assay was performed. Briefly,
Huh-7 and HepG2 cells were cultured in serum-free DMEM medium
overnight in a 6-well plate (5×106 cells/well) until
reaching 90% confluency. A sterilized 200-µl pipette tip was
used to make a wound by scratching across the well surface.
Following washing out the debris with fresh medium, the cells were
incubated at 37°C for 24 and 48 h in the presence of various
concentrations (0, 50, 100, 250, 500 and 1,000 µM) of
oseltamivir and images of the wound gaps were captured at 0, 24 and
48 h using Zeiss AxioVert 200 inverted fluorescence microscope. The
cell-migrated areas were calculated with Motic Images 2.0 software
(Motic Incoporation, Ltd.).
Transwell migration and invasion
assays
To verify the effects of oseltamivir on hepatoma
cell migration and invasion, 24-well Millicell Hanging Cell Culture
inserts (8-µm pore size; EMD Millipore) were used. For the
invasion assay, the upper chambers were precoated with 0.4 mg/ml
Matrigel (BD Biosciences) at 37°C for 24 h. Briefly, the upper
chamber containing serum-free DMEM medium (2×105 cells)
and various concentrations (0, 50, 100, 250, 500 and 1,000
µM) of oseltamivir, and the bottom chamber containing
standard medium (DMEM with 10% FBS) was combined and incubated at
37°C for 24 and 48 h in a cell incubator. Next, the migrating cells
were fixed with neutral-buffered formalin (10%) at 25°C for 2 h and
then stained with 0.05% Giemsa stain at 25°C for 2 h. A total of
six random fields were counted for each experiment under a light
microscope at a magnification of ×200 per filter.
Flow cytometry
For flow cytometric analysis, the cells were
incubated with various concentrations of oseltamivir (0, 50, 100,
250, 500 and 1,000 µM) at 37°C for 24 and 48 h. Following
incubation, the 1×106 cells were harvested, washed with
phosphate-buffered saline (PBS), and fixed with 70% alcohol for 16
h at -20°C. The cells were then washed with PBS and transferred
into 12×75-mm tubes. A total of 10 µl of propidium iodide
(PI) staining solution was added and chilled on ice in the dark.
Following filtration through a 40-µm nylon screen, the cells
were analyzed with a FACSCalibur analyzer (Nippon Becton Dickinson)
and data analysis was performed using WinMDI 2.9 (The Scripps
Research Institute, San Diego, USA).
Protein preparation and
immunoblotting
The cell pellets were collected by centrifugation at
800 × g for 5 min at 4°C and suspended in 600 µl PRO-PREP™
buffer (iNtRON Biotechnology, Inc.) for lysis. The supernatant was
then obtained by centrifugation at 16,600 × g for 5 min at 4°C. The
concentrations of protein were measured by a modified Bradford's
assay using a spectrophotometer (Hitachi U 3000; HITACHI) at 595 nm
with BSA (Sigma-Aldrich; Merck KGaA) as the standard. For
immunoblotting, extracted proteins (25 µg/lane) were
separated by 8-12% SDS-PAGE and electrophoretically transferred to
PVDF membranes (Immobilon-E, 0.45 µM; MilliporeSigma). After
blocking in 5% non-fat dry milk for 1 h at 25°C, the membranes were
incubated with antibodies against Apaf-1 (1:2,000; product code
ab2000; Abcam), cleaved caspase-3 (1:500; product no. AB3623;
Sigma-Aldrich; Merck KGaA), cleaved PARP-1 (1:500; cat. no.
sc-7150; Santa Cruz Biotechnology, Inc.), LC3 (1:5,000; cat. no.
NB100-2220), Beclin-1 (1:10,000; cat. no. NB110-87318), and
p62/SQSTM1 (1:4,000; cat. no. NBP1-48320; all from Novus
Biologicals, LLC) and β-actin (1:5,000; cat. no. MAB1501; EMD
Millipore) were used to detect apoptosis and autophagy. Briefly,
the PVDF membranes (Immobilon-E, 0.45 µM; MilliporeSigma)
were incubated with the antibodies for 3 h at 25°C. Next, the
secondary antibodies conjugated with horse-radish peroxidase (HRP)
(1:5,000; cat. no. sc-2004 or sc-2005; Santa Cruz Biotechnology,
Inc.) were added and incubated for 1 h at 25°C. To detect the
antigen-antibody complexes, Immobilion Western HRP Chemiluminescent
Substrate (EMD Millipore) and a densitometry apparatus LAS-4000
(Image Analysis Software: GE ImageQuant TL 8.1; GE Healthcare Life
Sciences) were used. In addition, an autophagy inhibitor,
chloroquine diphosphate (CQ; cat. no. L10382; LC3B Antibody Kit;
Invitrogen; Thermo Fisher Scientific Inc.), was used to verify the
participation of the autophagic mechanism to oseltamivir-induced
death in Huh-7 and HepG2 cells. Following pre-treatment of Huh-7
and HepG2 cells with CQ (25 µM) for 1 h, the cells were then
incubated with 1 mM oseltamivir for 24 h at 37°C.
Enzyme-linked immunosorbent assay
(ELISA)
An active caspase-3 ELISA Kit (Human active
caspase-3 (Ser29) SimpleStep ELISA kit; product code ab181418;
Abcam) was used to measure active caspase-3 according to the
manufacturer's protocol.
Immunofluorescence staining
An LC3B Antibody kit (cat. no. L10382; Invitrogen;
Thermo Fisher Scientific Inc.) was used for autophagy detection
according to the manufacturer's protocol. Briefly, the
1×106 cells were seeded on Millicell EZ SLIDE 8-well
glass slides and maintained with fresh DMEM with 10% FBS medium
containing various doses of oseltamivir (0, 50, 100, 250, 500 and
1,000 µM) for 24 h. Next, the cells were blocked in 2% BSA
buffer at 25°C for 1 h and then washed with 1X PBS and fixed with
4% paraformaldehyde at 25°C for 15 min. Subsequently, the cells
were permeabilized with 0.3% Triton X-100 at 25°C for 15 min,
followed by reacting with antibodies against LC3-B (0.5
µg/ml). Following incubation with Alexa Fluor®
488 goat anti-rabbit IgG (H+L) antibodies (1:500; cat. no. A21206,
Invitrogen; Thermo Fisher Scientific Inc.) at 25°C for 1 h, one
drop ProLong™ Gold Antifade Mountant with DAPI (Thermo Fisher
Scientific Inc.) was used to mount the coverslips at 25°C for 1
min. The cells were observed under a ZEISS AXioskop2 fluorescence
microscope (Carl Zeiss Microscopy, LLC).
Xenograft study
A total of 15 female athymic nude mice (5-weeks old;
weight 15-17 g) were acquired from National Center for Experimental
Animals of Taiwan and kept in specific-pathogen-free (SPF)
facilities with a 12-h light/dark cycle and a relative humidity in
an airconditioned room of 55%. Animals were allowed free access to
sterilized water and chow (Lab Diet 5001; PMI Nutrition
International Inc.) at Chung Shan Medical University (Taichung,
Taiwan). Study protocols were authorized by the Institutional
Animal Care and Use Committee of Chung Shan Medical University,
Taiwan, R.O.C. (IACUC approval no. 2542). The study was carried out
in compliance with the ARRIVE guidelines (Animal Research:
Reporting of In Vivo Experiments). A total of
5×106 Huh-7 cells in PBS were hypodermically injected
into the flank of mice at the age of 6-weeks old. The doses of
oseltamivir used in the xenograft study were based on a previous
study (16). When the tumor
volumes reached ~100 mm3, the mice were randomly divided
into three groups including control, low dose- and high-dose groups
and were daily intraperitoneally injected with PBS, 15 and 60 mg/kg
oseltamivir, respectively. The tumor diameters and volume were
measured every two days using a caliper. All the mice were
sacrificed (performed on August 3, 2021) when the tumor volumes of
the mice from the control group reached ~2,000 mm3.
Inhalation of carbon dioxide (CO2) was used for mice
euthanasia. The flow rate of CO2 was 50% of the chamber
volume/min. Following visual confirmation of respiratory cessation
of the mice, the CO2 flow was maintained for 1 min to
ensure the death of mice.
Statistical analysis
GraphPad Prism 5 software (GraphPad Software, Inc.)
was used to calculate the significant differences among groups. For
the MTT, wound healing, and Transwell migration assays, as well as
ELISA and immunoblotting, two-way ANOVA with Bonferroni's post hoc
test for multiple comparisons was performed to calculate the
effects of drug treatment. P<0.05 was considered to indicate a
statistically significant difference. All values are presented as
the mean ± SEM.
Results
Effects of lamivudine, ribavirin and
oseltamivir on the proliferation, migration and invasion of liver
cancer cells
To assess the effects of antiviral drugs, including
lamivudine, ribavirin, and oseltamivir, on liver cancer, Huh-7 and
HepG2 cells were first treated with various concentrations of
lamivudine, ribavirin, and oseltamivir. THLE-3 cells were used as
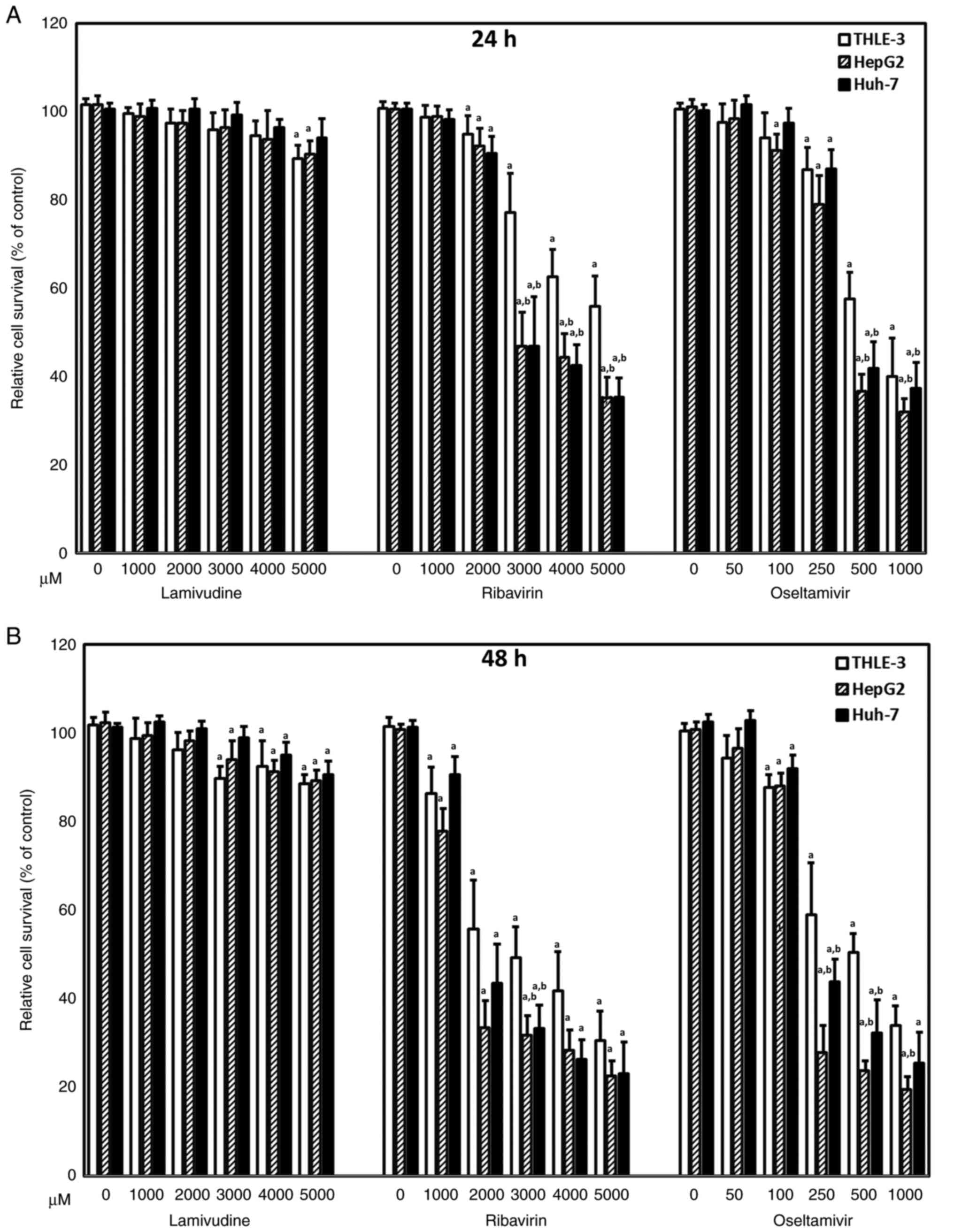
normal control cells. Although lamivudine caused a statistically
significant difference in cell viability compared with the control
treatment (0 µM), lamivudine had little effect on the
survival of THLE-3, Huh-7, and HepG2 cells (Fig. 1). All three cell lines exhibited
significantly decreased viability in the presence of ribavirin and
oseltamivir in a dose-dependent manner at 24 (Fig. 1A) and 48 h (Fig. 1B). Notably, Huh-7 and HepG2 cells
exhibited significantly lower viability in the presence of 250 and
500 µM oseltamivir at 48 h compared with THLE-3 cells
(Fig. 1B). Since more aggravated
clinical adverse effects have been reported with ribavirin than
oseltamivir, the following studies focused on the effects of
oseltamivir on liver cancer cells. To further investigate the
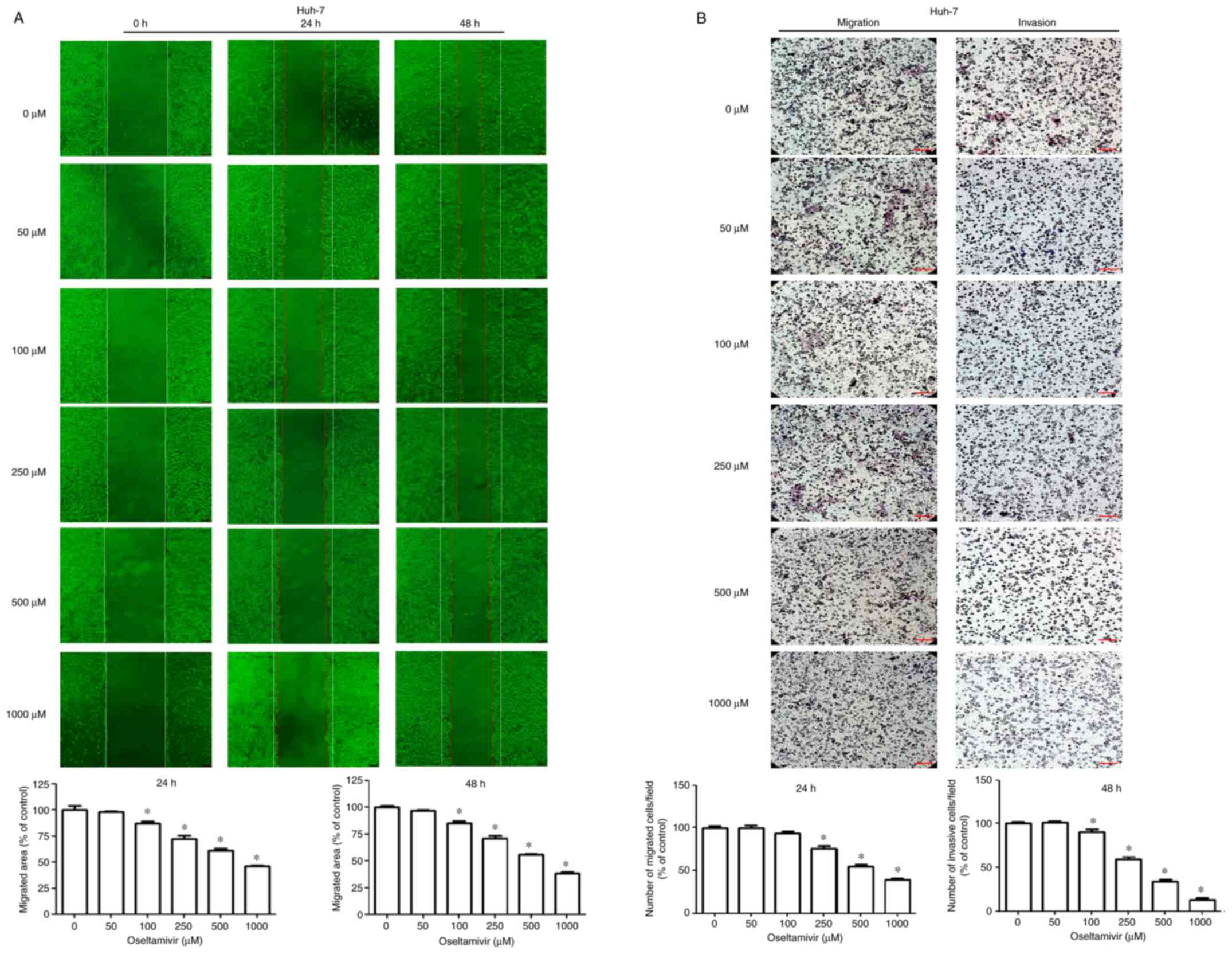
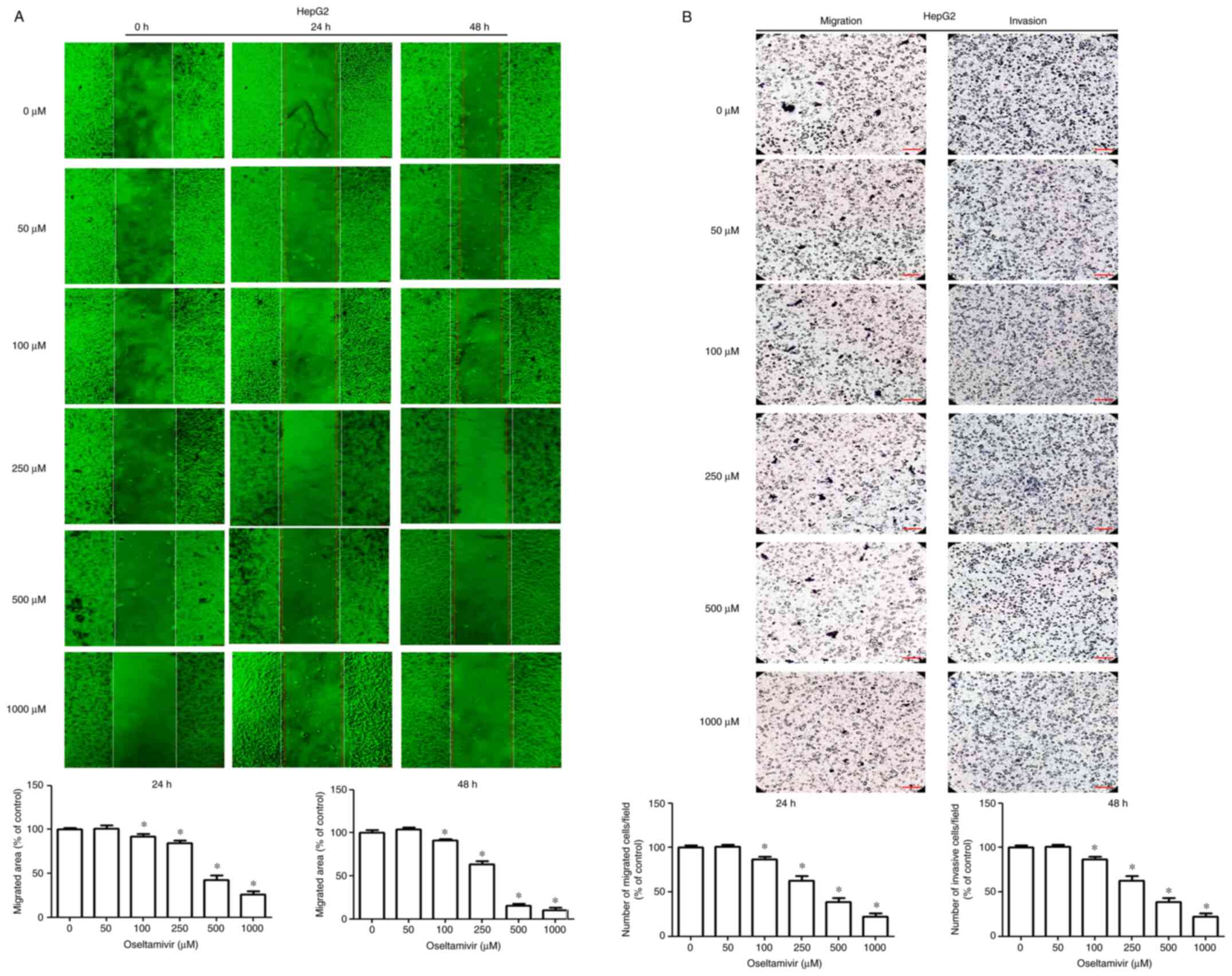
effects of oseltamivir on the migration and invasion of Huh-7 and
HepG2 cells, wound healing and Transwell assays were performed.
Significantly decreased migrating areas were detected in both Huh-7
and HepG2 cells treated with oseltamivir for 24 and 48 h (Figs. 2A and 3A). Significantly decreased migration
and invasion were observed in both Huh-7 and HepG2 cells that were
treated with oseltamivir for 24 h (Figs. 2B and 3B).
Oseltamivir induces autophagy but not
apoptosis in Huh-7 cells
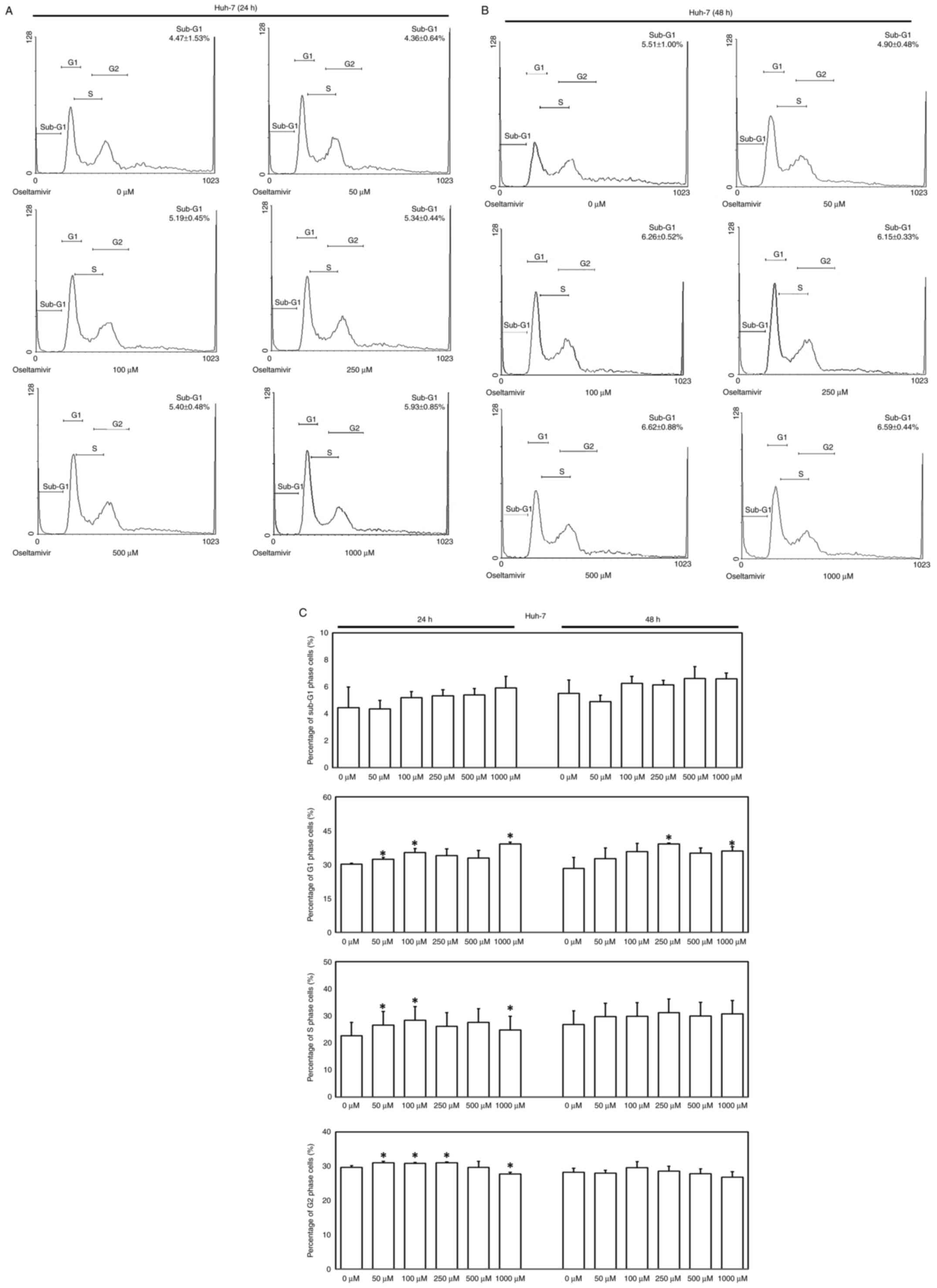
To verify whether apoptosis and autophagy are
involved in oseltamivir-induced Huh-7 cell death, flow cytometry,
immunoblotting, immunofluorescence, and caspase-3 ELISAs were
performed. A slightly increased proportion of Huh-7 cells in the
sub-G1 phase were observed following treatment with different
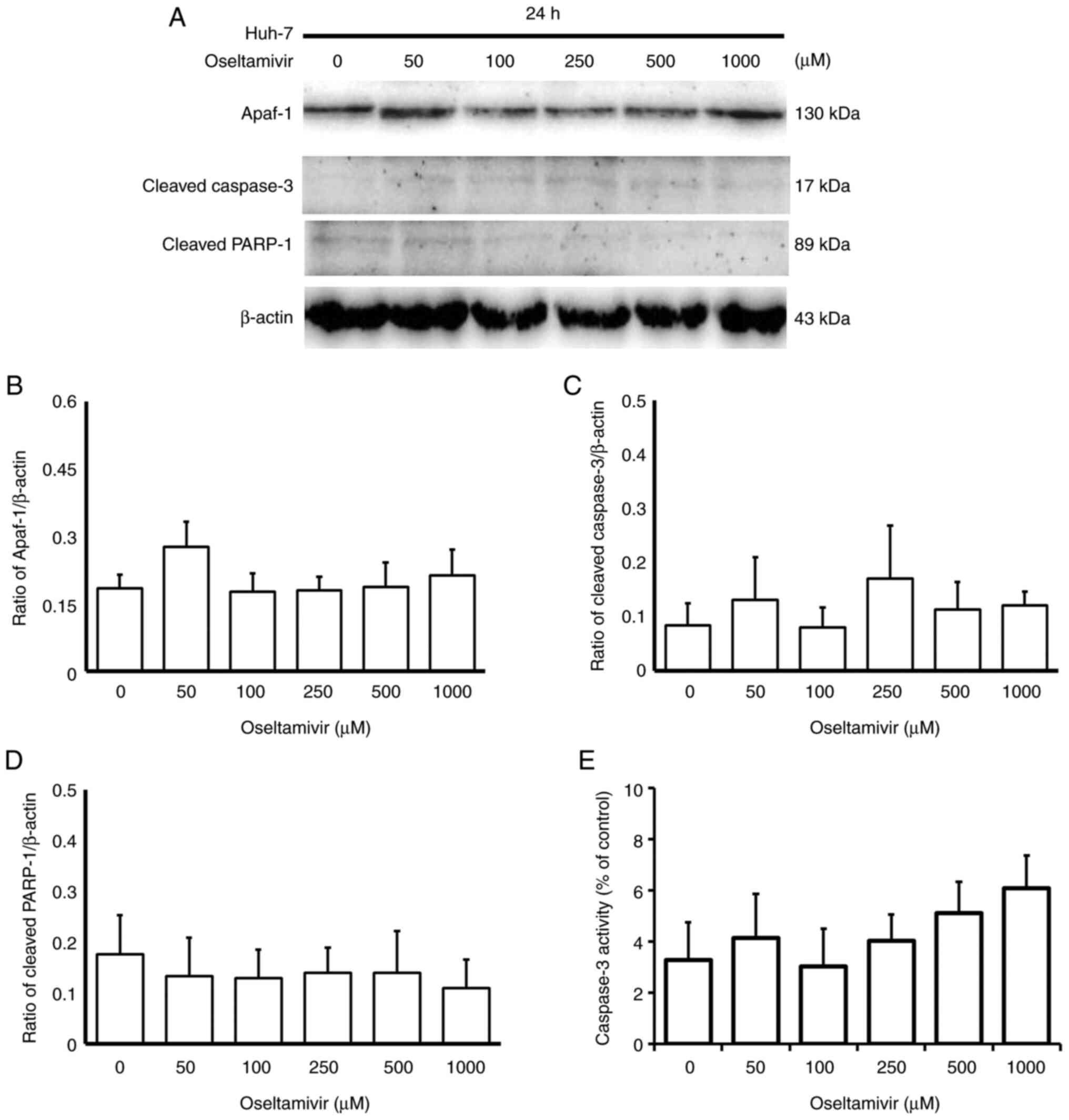
concentrations of oseltamivir for 24 and 48 h (Fig. 4A-C). No statistically significant
differences in the protein expression of Apaf-1, cleaved caspase-3
and cleaved PARP-1 (Fig. 5A-D) or
the activity of caspase-3 (Fig.
5E) were observed in the presence of oseltamivir for 24 h.
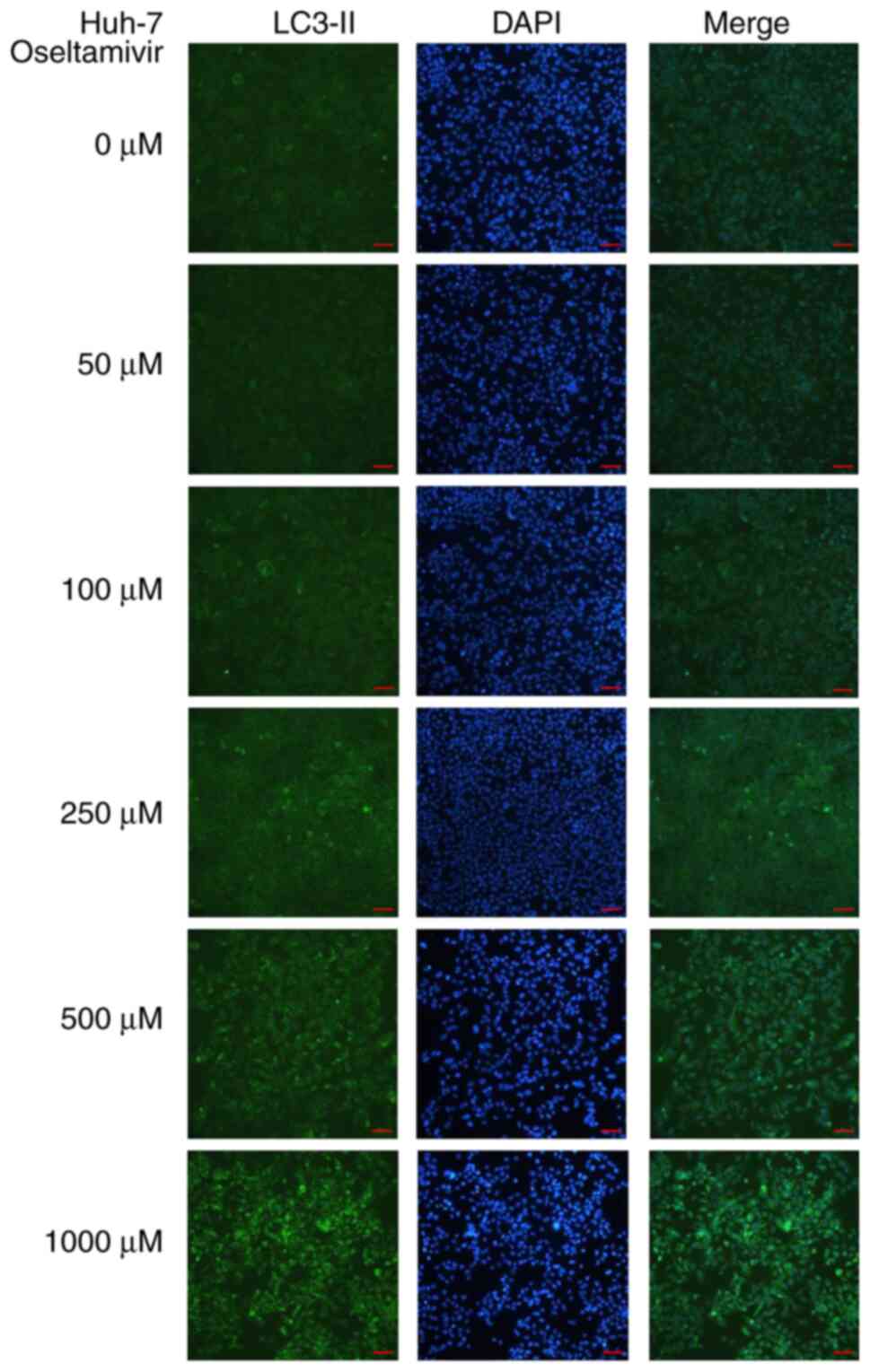
Notable protein expression of LC3-II was detected in Huh-7 cells
treated with 250, 500, and 1,000 µM oseltamivir for 24 h
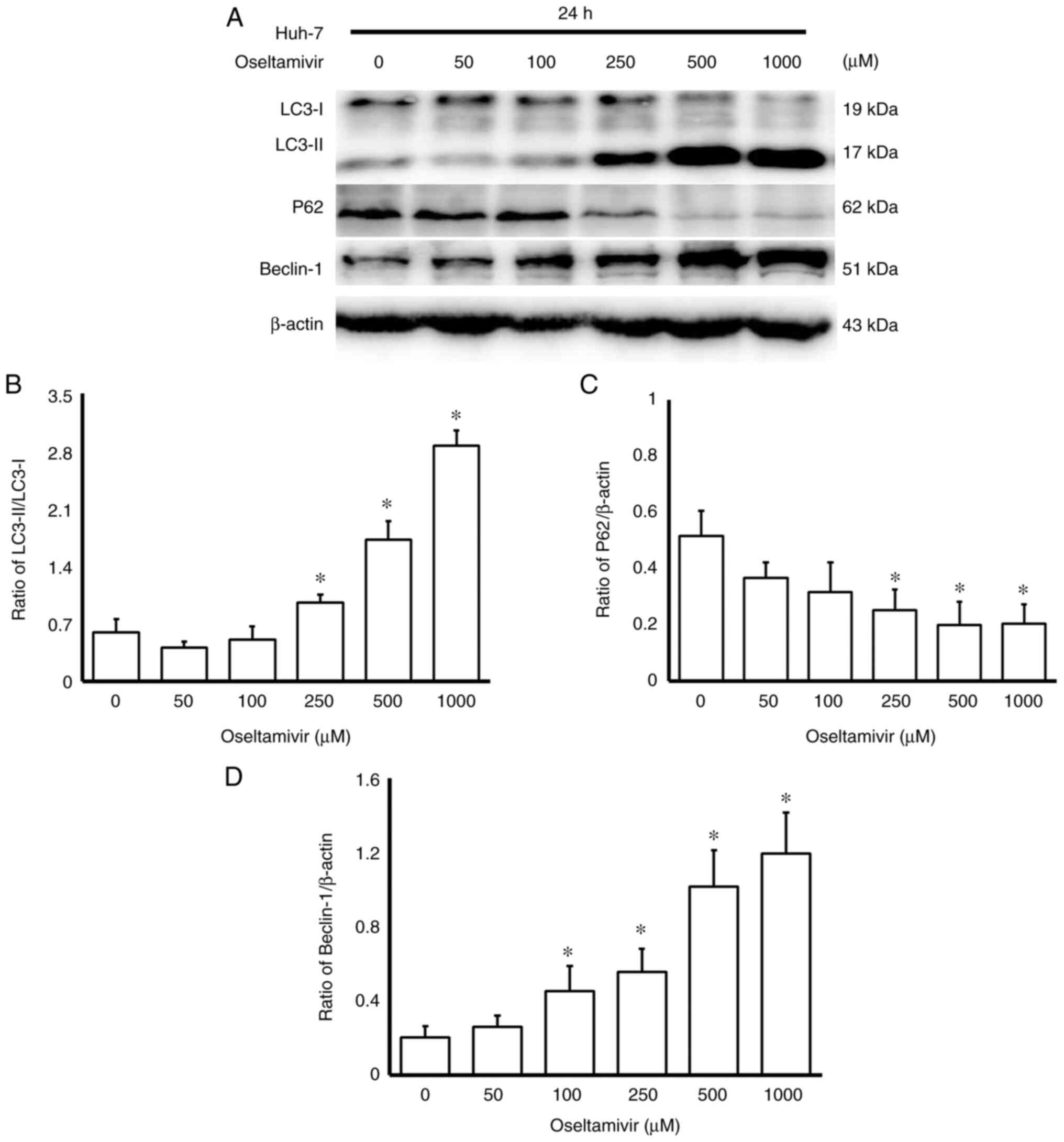
(Fig. 6). In addition, a
significantly higher ratio of LC3-II/LC3-I and increased protein
expression of Beclin-1 was observed in Huh-7 cells in a
dose-dependent manner (Fig. 7A and
B). Conversely, significantly decreased protein expression of
p62 was detected in Huh-7 cells treated with 250, 500, and 1,000
µM oseltamivir for 24 h (Fig.
7C).
Oseltamivir induces both apoptosis and
autophagy in HepG2 cells
To verify whether apoptosis and autophagy are
involved in oseltamivir-induced HepG2 cell death, flow cytometry,
immunoblotting, immunofluorescence, and caspase-3 ELISAs were
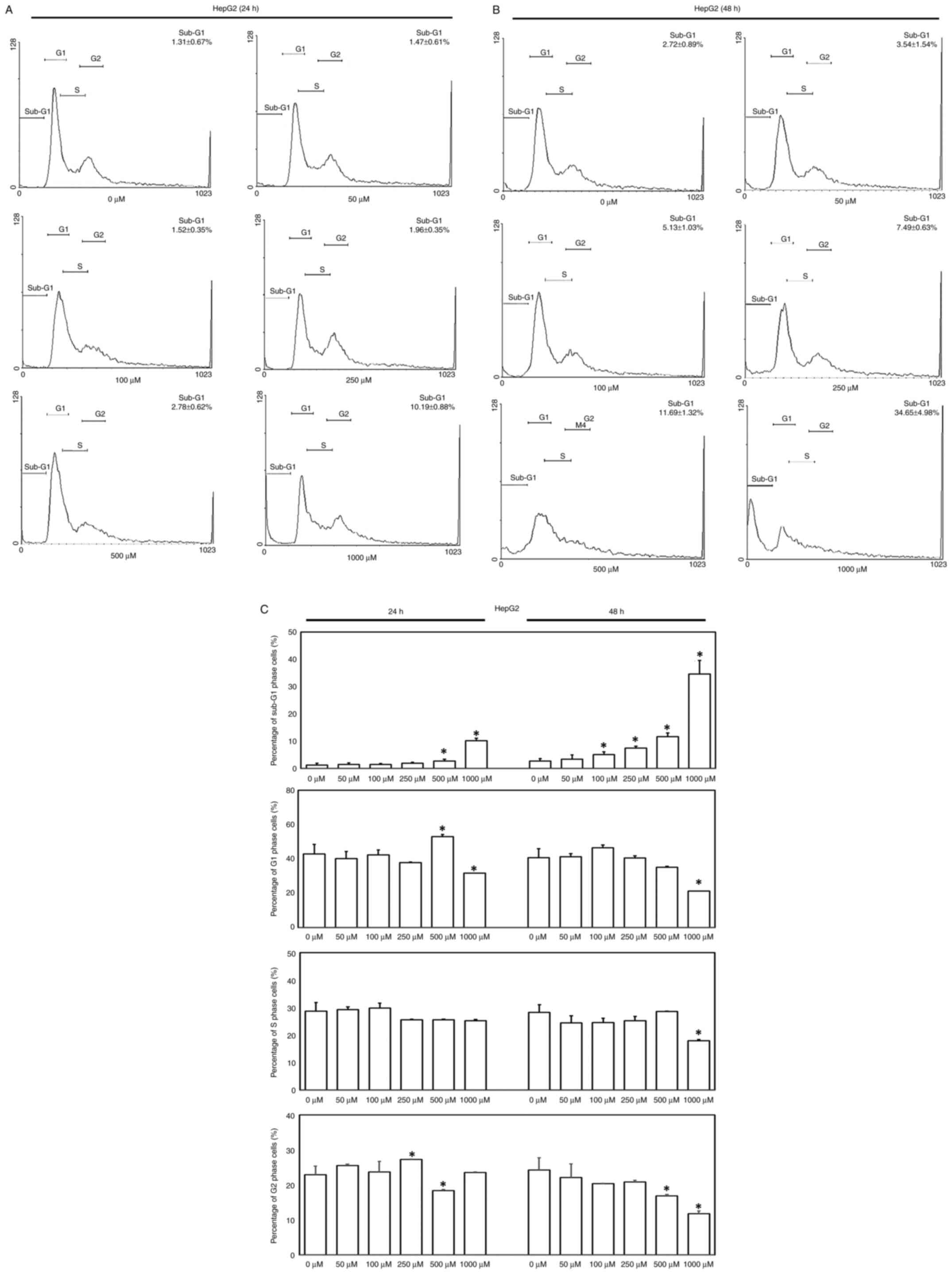
performed. Significantly increased proportion of HepG2 cells in the
sub-G1 phase were observed following treatment with 500 and 1,000
µM oseltamivir for 24 h and with 100, 250, 500, and 1,000
µM for 48 h (Fig. 8A-C).
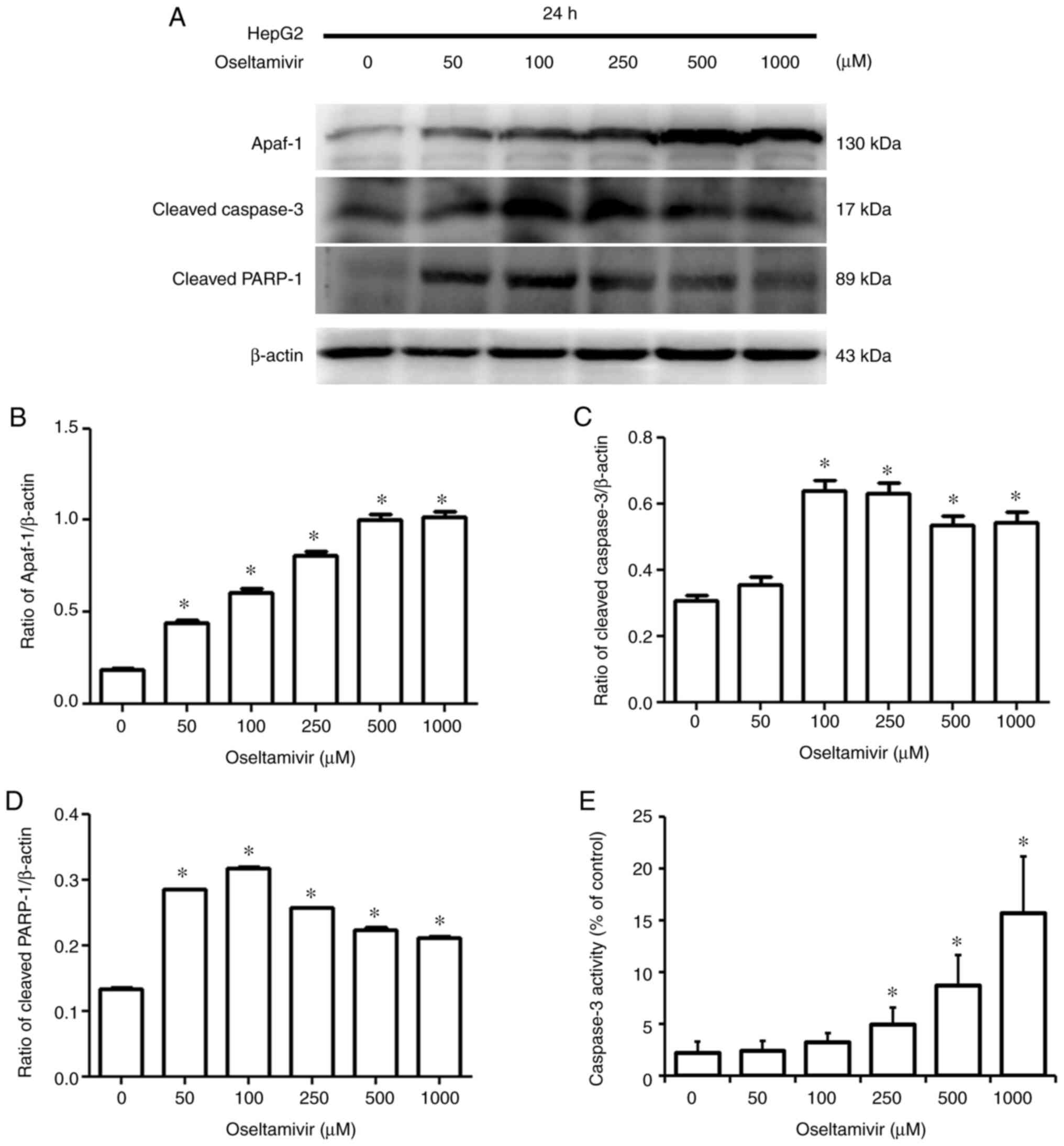
Accordingly, significantly increased protein expression of Apaf-1,
cleaved caspase-3, and cleaved PARP-1 was detected in HepG2 cells
treated with oseltamivir for 24 h (Fig. 9A-D). Additionally, significantly
increased caspase-3 activity was observed following treatment with
250, 500, and 1,000 µM oseltamivir for 24 h (Fig. 9E). A notably higher amount of
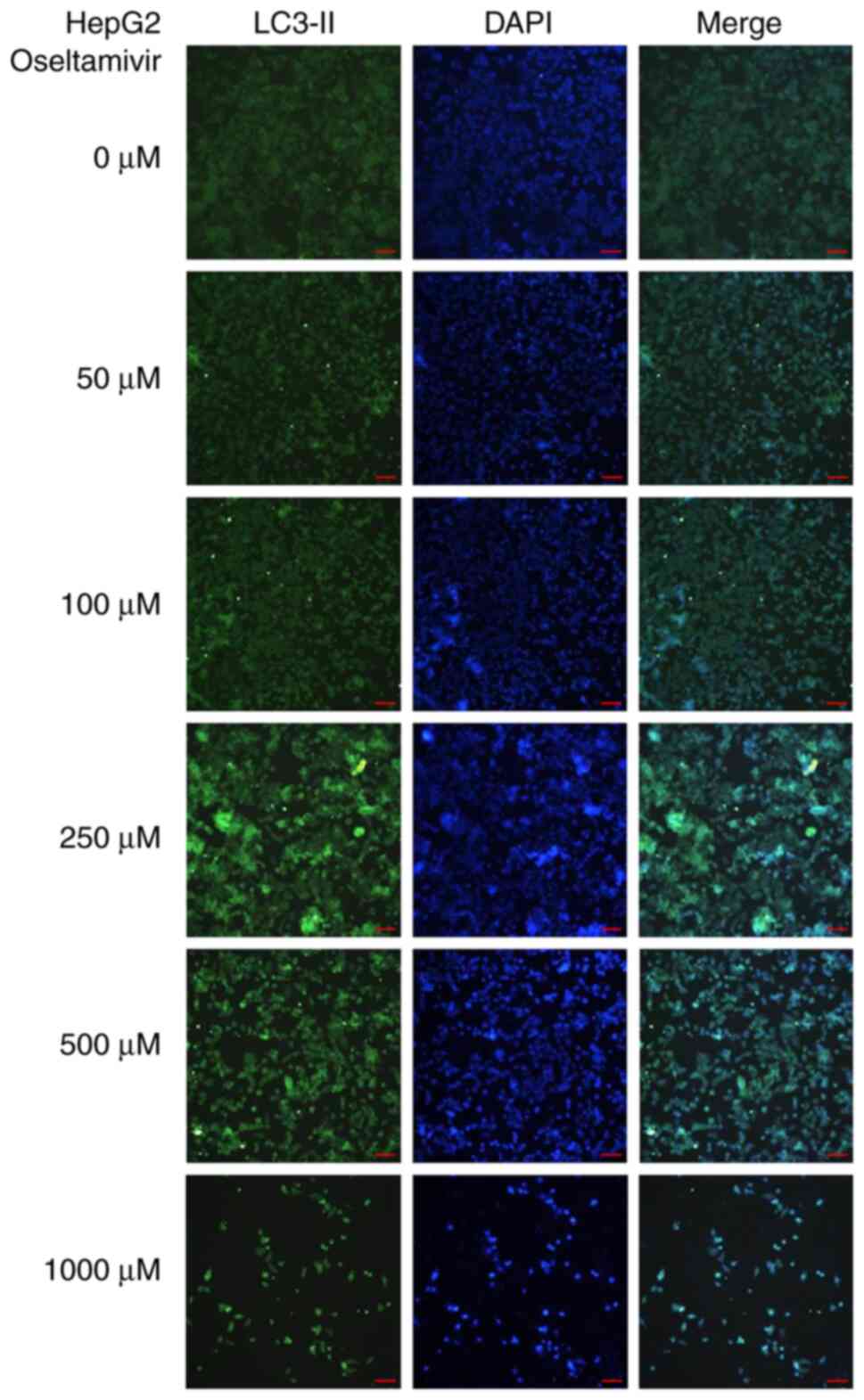
LC3-II protein was observed in HepG2 cells treated with 250, 500,
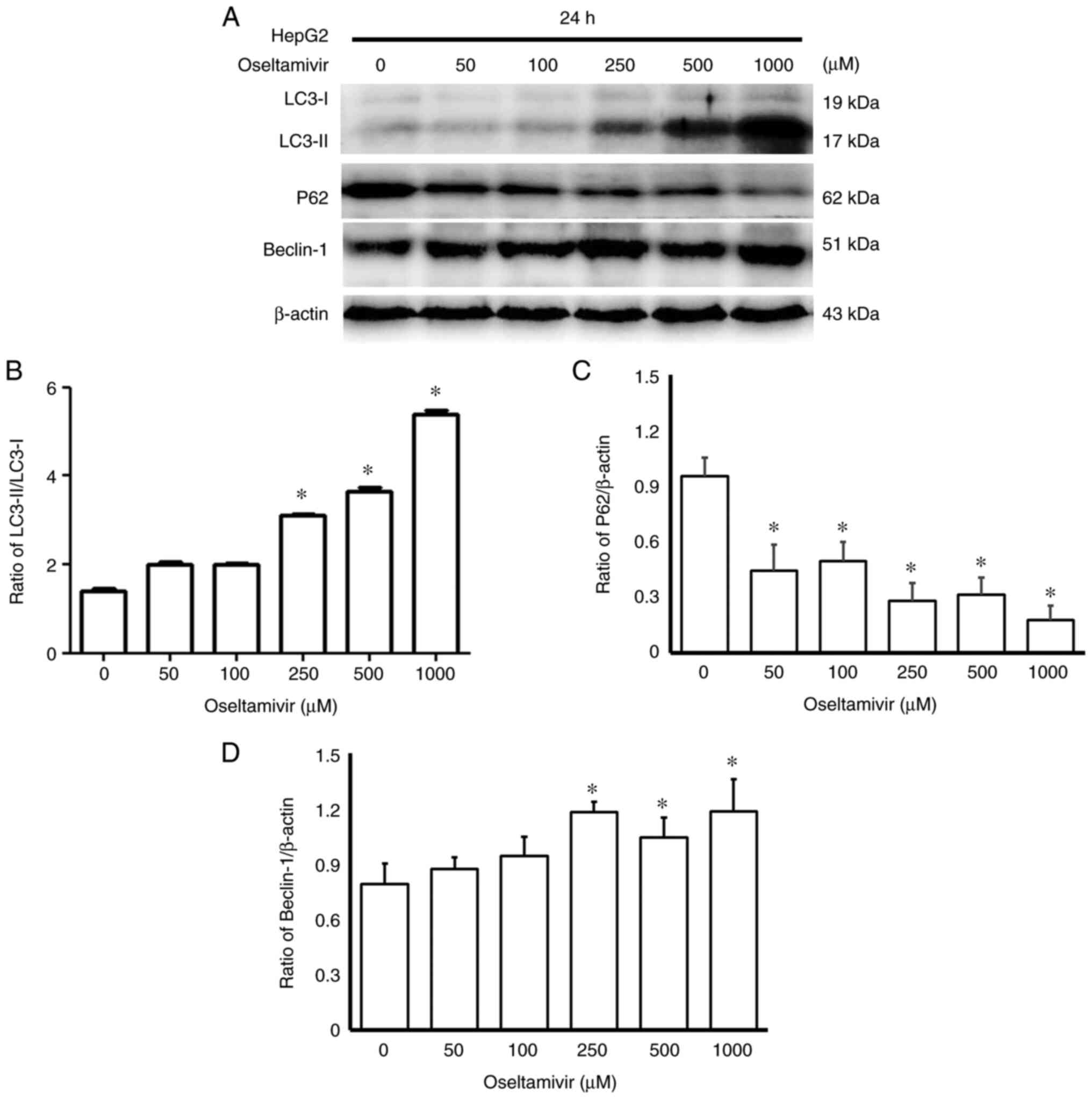
and 1,000 µM oseltamivir for 24 h (Fig. 10). A significantly higher ratio
of LC3-II/LC3-I and increased protein expression of Beclin-1 were
detected in Huh-7 cells in a dose-dependent manner, whereas
significantly decreased protein expression of p62 was detected
following treatment with 50, 100, 250, 500 and 1,000 µM
oseltamivir for 24 h (Fig. 11).
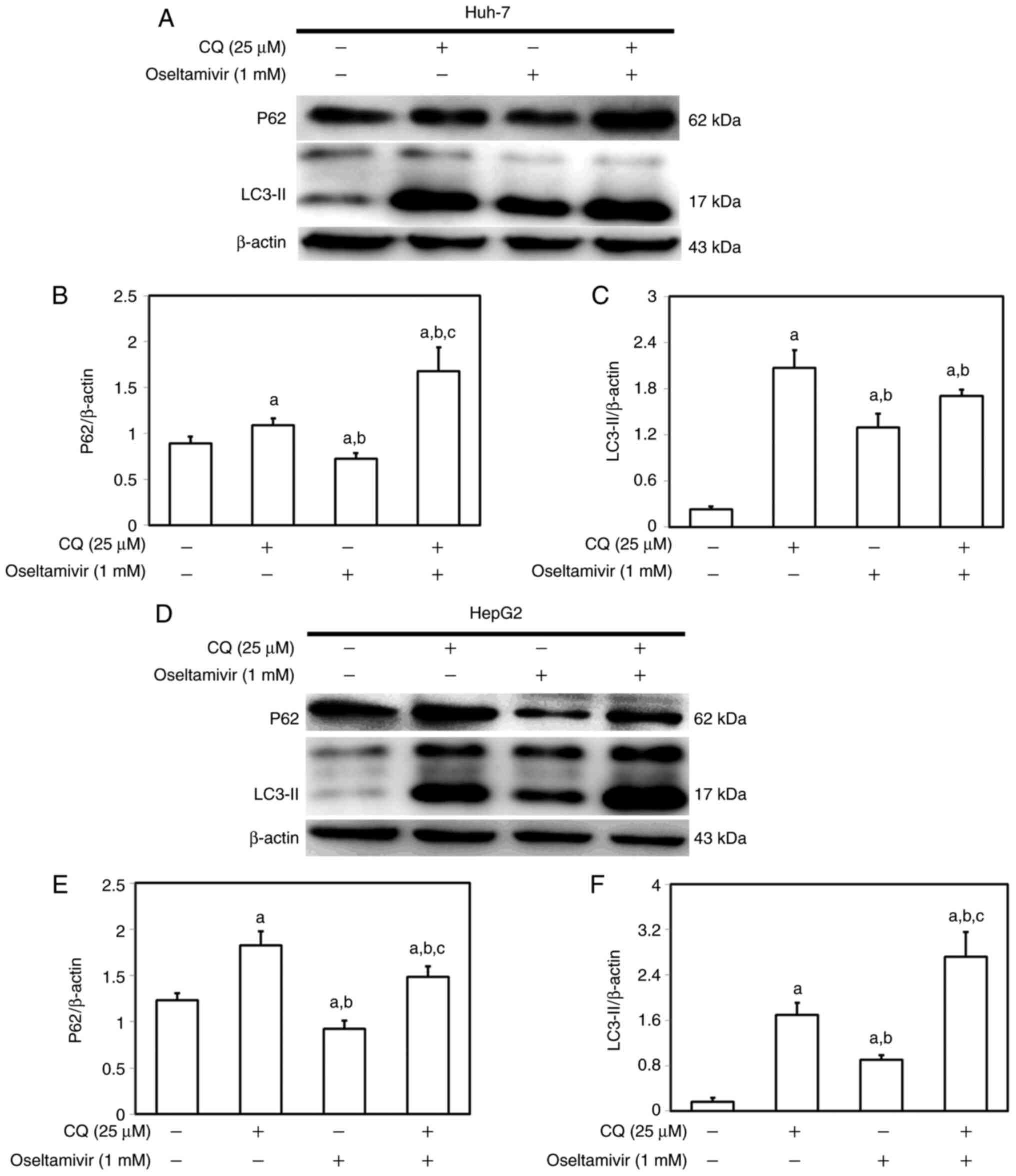
In addition, an autophagy inhibitor, CQ, was used to verify the
participation of the autophagic mechanism to oseltamivir-induced
death in Huh-7 and HepG2 cells (Fig.
12). Following pre-treatment of Huh-7 and HepG2 cells with CQ
(25 µM) for 1 h, the cells were then incubated with 1 mM
oseltamivir for 24 h. As anticipated, reduced p62 and increased
LC3-II protein levels were observed in both Huh-7 and HepG2 cells
that were treated with 1 mM oseltamivir compared with the cells
cultured in absence of both CQ and oseltamivir. Inhibition of
lysosomal degradation by pre-treatment with CQ prevented the
decomposition of LC3-II and p62 and resulted in accumulation of
LC3-II and p62 (Fig. 12).
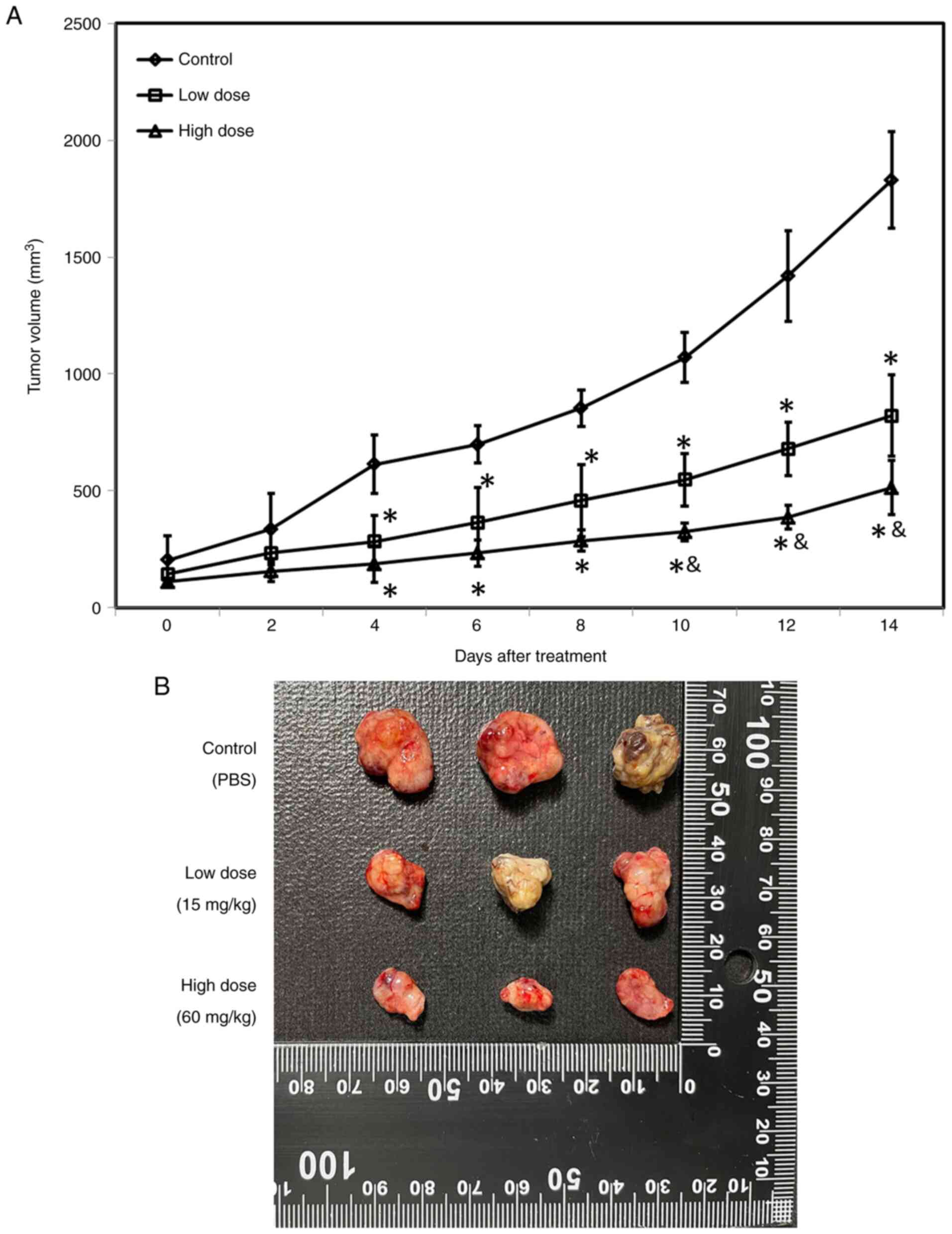
Oseltamivir inhibits the growth of
xenografted Huh-7 cells in nude mice
To assess the effects of oseltamivir in vivo,
xenografted tumors were generated by hypodermic injection of
5×106 Huh-7 cells into athymic nude mice. When the tumor
volume was ~100 mm3, the animals were treated daily with
PBS (control group), 15 mg/kg oseltamivir (low-dose group) and 60
mg/kg oseltamivir (high-dose group), respectively. A significantly
smaller mean tumor volume was detected in the mice treated with 15
mg/kg or 60 mg/kg oseltamivir as compared with the mice treated
with PBS from day-4 (Fig. 13A).
Notably, a significantly smaller mean tumor volume was detected in
the mice treated with 60 mg/kg oseltamivir as compared with the
mice treated with 15 mg/kg from day-10 (Fig. 13A). Fig. 13B reveals the representative
xenografted tumors retrieved at the end of the experiments.
Discussion
Since hepatitis virus infection has been strongly
associated with the occurrence of liver cirrhosis and HCC, the
traditional therapeutic strategy for liver cancer is to eliminate
the hepatitis virus (7,17). In addition to drugs that target
hepatitis viruses, various drugs against other viruses have been
used for the treatment of liver cancer. However, information
concerning the effects and related mechanisms of these antiviral
drugs remains limited. In the present study, it was revealed that
oseltamivir, an anti-influenza virus drug, significantly inhibited
the growth and migration of Huh-7 and HepG2 cells. Oseltamivir also
exerted differential effects on these liver cancer cell lines by
inducing apoptosis and autophagy alone or in combination.
Evidence has demonstrated dual roles of autophagy,
namely, both tumor inhibitory and tumor promoting roles, in
cancers. A variety of studies have revealed that autophagy promotes
cancer cell death in early phases and survival in later phases
(18-20). Autophagy may provide energy for
excessive cancer cell proliferation or lead to the insufficient
availability of nutrients for cancer cells by disrupting energy
homeostasis, thus causing cell death (21). A previous study reported that
quinacrine-induced autophagy in ovarian cancer cells leads to
excessive autophagic flux and promotes cell death (22). Another study indicated that
dihydroartemisinin (DHA)-37, an analog of DHA, exhibits significant
anticancer activity against A549 cells by triggering excessive
autophagic cell death (23). A
recent study also revealed that Dendrobium officinale
polysaccharide significantly inhibits CT26 cell proliferation by
inducing mitochondrial dysfunction and excessive autophagy
(24). These findings suggested
that excessive amounts of autophagy could be a potential strategy
for causing cancer cell death. Accordingly, the present study
reported, for the first time to the best of our knowledge, that
oseltamivir induces excessive autophagic cell death in both Huh-7
cells and HepG2 cells, providing an alternative method for the
treatment of liver cancer.
An alternative approach based on the use of
non-oncological drugs for cancer treatment has attracted
considerable attention in recent decades (25,26). Various types of non-oncological
medicines, including antiviral drugs, have yielded promising in
vitro results and are already being assessed in clinical trials
(27). In the present study, it
was revealed that antiviral drugs, including lamivudine, ribavirin,
and oseltamivir, exerted significant inhibitory effects on the
proliferation of Huh-7 and HepG2 cells. However, the concentrations
of lamivudine required to inhibit the proliferation of these liver
cancer cell lines were markedly higher than those of both ribavirin
and oseltamivir. In addition, ribavirin usually needs to be
combined with interferon in clinical treatment and requires a
higher dose and longer treatment time, which lead to more serious
side effects (28). Notably, the
dose of oseltamivir that was significantly effective in inhibiting
proliferation was markedly lower than that of ribavirin. Thus, the
dose of this medication could be reduced while still achieving
therapeutic efficacy, which could prevent adverse side effects,
drug diversion, poisoning, and waste treatment (29). In addition to the significant
inhibitory effects of oseltamivir on Huh-7 and HepG2 cells, these
findings suggested additional potential of oseltamivir in the
treatment of liver cancer in the clinic.
Therapeutic strategies targeting apoptosis (30,31) and autophagy (32,33) have long been used for cancer
treatment. An interesting finding in the present study was the
differential effects of oseltamivir on Huh-7 and HepG2 cells.
Oseltamivir induced both autophagy and apoptosis in HepG2 cells but
induced only autophagy in Huh-7 cells. This phenomenon may be
attributed to the different characteristics of the two cell lines,
particularly the mutation of p53 in Huh-7 cells (33). A previous genetic study reported
that HepG2 cells have a N-ras mutation at codon 61, and Huh-7 cells
have a missense mutation in the p53 gene at codon 220, resulting in
an amino acid change of cysteine for tyrosine (34). p53 is one of the most well
investigated and most frequently mutated genes in various human
cancers (35). A previous study
has indicated that the p53 gene plays an essential role in limiting
cancer formation by modulating metabolism, reactive oxygen species
production, and noncoding RNA expression and by enhancing autophagy
or ferroptosis (36). Indeed,
mutations in the p53 gene have been demonstrated to be involved in
cancer formation and progression and are present in ~50% of
aggressive tumors (35). Notably,
two clinical studies indicated that the presence of missense p53
mutations is significantly associated with breast cancer
specificity and overall mortality (37,38). Interestingly, p53 was also found
to play a key role on controlling the switch between autophagy and
apoptosis. An in vitro model reported that sodium selenite
switched protective autophagy to apoptosis in both a p53-wild type
(NB4 cells) and p53-mutant cell model (Jurkat cells) (39,40). These findings may provide possible
explanations of the difference in oseltamivir-induced death between
Huh-7 and HepG2 cells. However, further investigations such as
xenografted HepG2 experiments and underlying signaling analysis are
required to identify the precise mechanism of oseltamivir-induced
cell apoptosis and/or autophagy in Huh-7 and HepG2 cells.
In addition, the role of p62 in autophagy and
apoptosis has received a great amount of attention in recent years.
The scaffold protein p62, namely sequestosome 1 (SQSTM1), is
well-known as a critical regulator in the autophagic process by
directly binding LC3 for autophagosome generation (41,42). Evidence has indicated that
p62/SQSTM1 mediated a variety of essential cellular processes,
including autophagy and apoptosis, through its different domains
(42,43). In fact, studies have demonstrated
that p62 binds LC3 by the LC3-interacting region (LIR) within p62
and promotes the formation of autophagosomes (44,45). Additionally, p62 has also been
reported to induce apoptosis through the caspase-8 activation at
the autophagosomal membrane (46). In the presence of culin3,
caspase-8 was modified and interacted with p62 and TRAF6, which
leads to the activation of caspase-8 downstream caspase and
apoptosis (47,48). These findings indicated a dual
role of p62 in autophagy and apoptosis and may provide another
possible rationale for explaining the difference of
oseltamivir-induced autophagy/apoptosis in Huh-7 and HepG2 cells.
Definitely, further experiments are merited to investigate the
precise role of p62 on oseltamivir-induced cell death in Huh-7 and
HepG2 cells.
Neuraminidase (NEU) is the enzyme expressed on the
surface of influenza viruses, and it facilitates the release and
trafficking of influenza viruses within the respiratory tract
(39). Oseltamivir (Tamiflu) is
an FDA-approved NEU inhibitor for the prevention and treatment of
influenza A and B infections (49,50). Certain investigations have been
conducted on the alternative use of oseltamivir in cancer
treatment. A previous study on alternative treatments for
pancreatic cancer indicated that oseltamivir inhibits the activity
of NEU-1 (Sialidase) and suppresses intrinsic signaling that
promotes human pancreatic cancer (PANC1) cell survival (51). In addition, oseltamivir also
overcame the chemoresistance of PANC1 cells to cisplatin and
gemcitabine alone or in combination by reversing changes in
E-cadherin and N-cadherin expression (41). A recent study indicated an
association between NEU-1 and HCC (51). Notably increased mRNA and protein
expression of NEU-1 was observed in HBV-related HCC tissues, and
this increased expression was caused by the binding of the HBV core
protein to NF-KB on the NEU-1 promoter, which led to downstream
oncogenic signaling and epithelial-mesenchymal transition (EMT) in
HCC cells, including Huh-7 and HepG2 cells (52). These findings indicated the
involvement of NEU in the carcinogenesis of HCC. Consistently, the
present study was the first to report the anti-liver cancer
activity of oseltamivir by inducing apoptosis in Huh-7 cells and
inducing both apoptosis and autophagy in HepG2 cells. However, the
roles of NEU and intrinsic signaling in oseltamivir-induced Huh-7
and HepG2 cell death remain unknown and warrant further
investigation in order to elucidate the precise mechanism
underlying the oseltamivir-induced cell death of Huh-7 and HepG2
cells. In summary, the present study provided novel findings about
the use of non-oncological drugs for the treatment of liver cancer
and suggests the use of oseltamivir as an alternative approach for
liver cancer treatment.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
PJH and CCC were involved in the study conception
and design. MHH and JLY performed experiments and analysis of data.
TCH was involved in the study conception and design, drafting and
revising of the manuscript, performing experiments, analysis of
data, and study supervision. BST was involved in the study
conception and design, drafting and revising of the manuscript, and
analysis of data. BST and TCH confirm the authenticity of all the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Animal experiments were performed in accordance with
the principles of replacement, refinement and reduction and were
approved (approval no. 2542) by the Institutional Animal Care and
Use Committee (IACUC) of Chung Shan Medical University (Taichung,
Taiwan, R.O.C.).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was funded by Chung Shan Medical University
and Changhua Christian Hospital (grant no. CSMU-CCH-109-05), and
the experimental supplies in the cell study were partially
supported by the Ministry of Science and Technology, Taiwan, R.O.C.
(grant no. MOST-109-2314-B040-021). The funders had no role in the
study design, data collection and analysis, decision to publish, or
preparation of the manuscript.
References
|
1
|
Llovet JM, Zucman-Rossi J, Pikarsky E,
Sangro B, Schwartz M, Sherman M and Gores G: Hepatocellular
carcinoma. Nat Rev Dis Primers. 14:160182016. View Article : Google Scholar
|
|
2
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. The Lancet. 391:1301–1314. 2018.
View Article : Google Scholar
|
|
3
|
Kuo CN, Liao YM, Kuo LN, Tsai HJ, Chang WC
and Yen Y: Cancers in Taiwan: Practical insight from epidemiology,
treatments, biomarkers, and cost. J Formos Med Assoc.
119:1731–1741. 2020. View Article : Google Scholar
|
|
4
|
Polesel J, Zucchetto A, Montella M, Dal
Maso L, Crispo A, La Vecchia C, Serraino D, Franceschi S and
Talamini R: The impact of obesity and diabetes mellitus on the risk
of hepatocellular carcinoma. Ann Oncol. 20:353–357. 2009.
View Article : Google Scholar
|
|
5
|
Li S, Saviano A, Erstad DJ, Hoshida Y,
Fuchs BC, Baumert T and Tanabe KK: Risk factors, pathogenesis, and
strategies for hepatocellular carcinoma prevention: Emphasis on
secondary prevention and its translational challenges. J Clin Med.
9:38172020. View Article : Google Scholar :
|
|
6
|
Bosch FX, Ribes J, Cléries R and Díaz M:
Epidemiology of hepatocellular carcinoma. Clin Liver Dis.
9:191–211. 2005. View Article : Google Scholar
|
|
7
|
Dash S, Aydin Y, Widmer KE and Nayak L:
Hepatocellular carcinoma mechanisms associated with chronic HCV
infection and the impact of direct-acting antiviral treatment. J
Hepatocell Carcinoma. 7:45–76. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sung PS and Shin EC: Immunological
mechanisms for hepatocellular carcinoma risk after direct-acting
antiviral treatment of hepatitis C virus infection. J Clin Med.
10:2212021. View Article : Google Scholar :
|
|
9
|
Andrisani O: Epigenetic mechanisms in
hepatitis B virus-associated hepatocellular carcinoma. Hepatoma
Res. 7:122021.PubMed/NCBI
|
|
10
|
Ulcickas Yood M, Quesenberry CP Jr, Guo D,
Wells K, Shan J, Sanders L, Skovron ML, Iloeje U, Caldwell C and
Manos MM: Incidence of hepatocellular carcinoma among individuals
with heaptitis B virus infection identified using an automated data
algorithm. J Viral Hepat. 15:28–36. 2008.
|
|
11
|
Fattovich G, Stroffolini T, Zagni I and
Donato F: Hepatocellular carcinoma in cirrhosis: Incidence and risk
factors. Gastroenterology. 127(5 Suppl 1): S35–S50. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nguyen VT, Law MG and Dore GJ: Hepatitis
B-related hepatocellular carcinoma: Epidemiological characteristics
and disease burden. J Viral Hepat. 16:453–463. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim GA, Lim YS, Han S, Choi J, Shim JH,
Kim KM, Lee HC and Lee YS: High risk of hepatocellular carcinoma
and death in patients with immune-tolerant-phase chronic hepatitis
B. Gut. 67:945–952. 2018. View Article : Google Scholar
|
|
14
|
Pons F, Varela M and Llovet JM: Staging
systems in hepatocellular carcinoma. HPB (Oxford). 7:35–41. 2005.
View Article : Google Scholar
|
|
15
|
Mak LY, Cruz-Ramó V, Chinchilla-López P,
Torres HA, LoConte NK, Rice JP, Foxhall LE, Sturgis EM, Merrill JK,
Bailey HH, et al: Global epidemiology, prevention, and management
of hepatocellular carcinoma. Am Soc Clin Oncol Educ Book.
38:262–279. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Haxho F, Allison S, Alghamdi F, Brodhagen
L, Kuta VE, Abdulkhalek S, Neufeld RJ and Szewczuk MR: Oseltamivir
phosphate monotherapy ablates tumor neovascularization, growth, and
metastasis in mouse model of human triple-negative breast
adenocarcinoma. Breast Cancer (Dove Med Press). 6:191–203.
2014.
|
|
17
|
Xu HZ, Liu YP, Guleng B and Ren JL:
Hepatitis B virus-related hepatocellular carcinoma: Pathogenic
mechanisms and novel therapeutic interventions. Gastrointest
Tumors. 1:135–145. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Levy JM and Thorburn A: Targeting
autophagy during cancer therapy to improve clinical outcomes.
Pharmacol Ther. 131:130–141. 2011. View Article : Google Scholar
|
|
19
|
Chen C, Gao H and Su X: Autophagy-related
signaling pathways are involved in cancer (Review). Exp Ther Med.
22:7102021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee XC, Werner E and Falasca M: Molecular
mechanism of autophagy and its regulation by cannabinoids in
cancer. Cancers (Basel). 13:12112021. View Article : Google Scholar
|
|
21
|
Islam Khan MZ and Law HK: Cancer
Susceptibility Candidate 9 (CASC9) promotes colorectal cancer
carcinogenesis via mTOR-dependent autophagy and
epithelial-mesenchymal transition pathways. Front Mol Biosci.
8:6270222021. View Article : Google Scholar
|
|
22
|
Khurana A, Roy D, Kalogera E, Mondal S,
Wen X, He X, Dowdy S and Shridhar V: Quinacrine promotes autophagic
cell death and chemosensitivity in ovarian cancer and attenuates
tumor growth. Oncotarget. 6:36354–36369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu X, Wu J, Fan M, Shen C, Dai W, Bao Y,
Liu JH and Yu BY: Novel dihydroartemisinin derivative DHA-37
induces autophagic cell death through upregulation of HMGB1 in A549
cells. Cell Death Dis. 9:10482018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang K, Zhou X, Wang J, Zhou Y, Qi W,
Chen H, Nie S and Xie M: Dendrobium officinale polysaccharide
triggers mitochondrial disorder to induce colon cancer cell death
via ROS-AMPK-autophagy pathway. Carbohydr Polym. 264:1180182021.
View Article : Google Scholar
|
|
25
|
Armando RG, Mengual Gómez DL and Gomez DE:
New drugs are not enough-drug repositioning in oncology: An update.
Int J Oncol. 56:651–684. 2020.PubMed/NCBI
|
|
26
|
Nunes M, Henriques Abreu M, Bartosch C and
Ricardo S: Recycling the purpose of old drugs to treat ovarian
cancer. Int J Mol Sci. 21:77682020. View Article : Google Scholar :
|
|
27
|
Hampson L, Maranga IO, Masinde MS, Oliver
AW, Batman G, He X, Desai M, Okemwa PM, Stringfellow H,
Martin-Hirsch P, et al: A single-arm, proof-of-concept trial of
Lopimune (Lopinavir/Ritonavir) as a treatment for HPV-related
pre-invasive cervical disease. PLoS One. 11:e01479172016.
View Article : Google Scholar :
|
|
28
|
Beaucourt S and Vignuzzi M: Ribavirin: A
drug active against many viruses with multiple effects on virus
replication and propagation. Molecular basis of ribavirin
resistance. Curr Opin Virol. 8:10–15. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Daughton CG and Ruhoy IS: Lower-dose
prescribing: Minimizing 'side effects' of pharmaceuticals on
society and the environment. Sci Total Environ. 443:324–337. 2013.
View Article : Google Scholar
|
|
30
|
von Karstedt S, Montinaro A and Walczak H:
Exploring the TRAILs less travelled: TRAIL in cancer biology and
therapy. Nat Rev Cancer. 17:352–366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tan S, Liu X, Chen L, Wu X, Tao L, Pan X,
Tan S, Liu H, Jiang J and Wu B: Fas/FasL mediates
NF-kappaBp65/PUMA-modulated hepatocytes apoptosis via autophagy to
drive liver fibrosis. Cell Death Dis. 12:4742021. View Article : Google Scholar
|
|
32
|
Alvarez-Meythaler JG, Garcia-Mayea Y, Mir
C, Kondoh H and LLeonart ME: Autophagy takes center stage as a
possible cancer hallmark. Front Oncol. 10:5860692020. View Article : Google Scholar
|
|
33
|
Buzun K, Gornowicz A, Lesyk R, Bielawski K
and Bielawska A: Autophagy modulators in cancer therapy. Int J Mol
Sci. 22:58042021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hsu IC, Tokiwa T, Bennett W, Metcalf RA,
Welsh JA, Sun T and Harris CC: p53 gene mutation and integrated
hepatitis B viral DNA sequences in human liver cancer cell lines.
Carcinogenesis. 14:987–992. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Duffy MJ, Synnott NC and Crown J: Mutant
p53 as a target for cancer treatment. Eur J Cancer. 83:258–265.
2017. View Article : Google Scholar
|
|
36
|
Braithwaite AW, Royds JA and Jackson P:
The p53 story: Layers of complexity. Carcinogenesis. 26:1161–1169.
2005. View Article : Google Scholar
|
|
37
|
Berns EM, van Staveren IL, Look MP, Smid
M, Klijn JG and Foekens JA: Mutations in residues of TP53 that
directly contact DNA predict poor outcome in human primary breast
cancer. Br J Cancer. 77:1130–1136. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rossner P Jr, Gammon MD, Zhang YJ, Terry
MB, Hibshoosh H, Memeo L, Mansukhani M, Long CM, Garbowski G,
Agrawal M, et al: Mutations in p53, p53 protein overexpression and
breast cancer survival. J Cell Mol Med. 13:3847–3857. 2009.
View Article : Google Scholar
|
|
39
|
Guo JY and White E: Autophagy, metabolism,
and cancer. Cold Spring Harb Symp Quant Biol. 81:73–78. 2016.
View Article : Google Scholar
|
|
40
|
Shi K, An J, Qian K, Zhao X, Li F, Ma X,
Wang Y and Zhang Y: p53 controls the switch between autophagy and
apoptosis through regulation of PLSCR1 in sodium selenite-treated
leukemia cells. Exp Cell Res. 389:1118792020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Itakura E and Mizushima N: p62 Targeting
to the autophagosome formation site requires self-oligomerization
but not LC3 binding. J Cell Biol. 192:17–27. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Islam MA, Sooro MA and Zhang P: Autophagic
Regulation of p62 is critical for cancer therapy. Int J Mol Sci.
19:14052018. View Article : Google Scholar :
|
|
43
|
Moscat J, Diaz-Meco MT and Wooten MW:
Signal integration and diversification through the p62 scaffold
protein. Trends Biochem Sci. 32:95–100. 2007. View Article : Google Scholar
|
|
44
|
Ichimura Y, Kominami E, Tanaka K and
Komatsu M: Selective turnover of p62/A170/SQSTM1 by autophagy.
Autophagy. 4:1063–1066. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lin X, Li S, Zhao Y, Ma X, Zhang K, He X
and Wang Z: Interaction domains of p62: A bridge between p62 and
selective autophagy. DNA Cell Biol. 32:220–227. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Young MM, Takahashi Y, Khan O, Park S,
Hori T, Yun J, Sharma AK, Amin S, Hu CD and Zhang J: Autophagosomal
membrane serves as platform for intracellular death-inducing
signaling complex (iDISC)-mediated caspase-8 activation and
apoptosis. J Biol Chem. 287:12455–12468. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sanz L, Diaz-Meco MT, Nakano H and Moscat
J: The atypical PKC-interacting protein p62 channels NF-kappaB
activation by the IL-1TRAF6 pathway. EMBO J. 19:1576–1586. 2000.
View Article : Google Scholar
|
|
48
|
Jin Z, Li Y, Pitti R, Lawrence D, Pham VC,
Lill JR and Ashkenazi A: Cullin3-based polyubiquitination and
p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis
signaling. Cell. 137:721–735. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cooper NJ, Sutton AJ, Abrams KR, Wailoo A,
Turner D and Nicholson KG: Effectiveness of neuraminidase
inhibitors in treatment and prevention of influenza A and B:
Systematic review and meta-analyses of randomised controlled
trials. BMJ. 326:12352003. View Article : Google Scholar :
|
|
50
|
Hayden FG, Treanor JJ, Fritz RS, Lobo M,
Betts RF, Miller M, Kinnersley N, Mills RG, Ward P and Straus SE:
Use of the oral neuraminidase inhibitor oseltamivir in experimental
human influenza: Randomized controlled trials for prevention and
treatment. JAMA. 282:1240–1246. 1999. View Article : Google Scholar
|
|
51
|
O'Shea LK, Abdulkhalek S, Allison S,
Neufeld RJ and Szewczuk MR: Therapeutic targeting of Neu1 sialidase
with oseltamivir phosphate (Tamiflu) disables cancer cell survival
in human pancreatic cancer with acquired chemoresistance. Onco
Targets Ther. 7:117–134. 2014.
|
|
52
|
Kong F, Li N, Tu T, Tao Y, Bi Y, Yuan D,
Zhang N, Yang X, Kong D, You H, et al: Hepatitis B virus core
protein promotes the expression of neuraminidase 1 to facilitate
hepatocarcinogenesis. Lab Invest. 100:1602–1617. 2020. View Article : Google Scholar : PubMed/NCBI
|