Introduction
Senescence is a cellular stress response triggered
by various stressors, including oncogene activation, telomere
shortening and genotoxic treatments (1). The definitive arrest of
proliferation in most cases is led by the p53/p21 and p16/Rb
signaling pathways in response to DNA damage (2,3),
which then induces irreversible proliferative arrest. Furthermore,
senescent cells present specific hallmarks, such as DNA damage,
that can be revealed by γH2Ax staining, the Senescence-Associated
Secretory Phenotype, and a compaction of proliferative genes termed
Senescence-Associated Heterochromatin Foci (4).
Senescence can be induced in response to
chemotherapy (Chemotherapy-Induced Senescence; CIS) and was
initially considered as a favorable outcome (5). The proliferative arrest observed
during senescence has long been perceived as a definitive end to
proliferation (1,6). However, several studies and
experiments show that cells are able to overcome this state and
proliferate again. The authors and other laboratories have
demonstrated that chemotherapy-induced senescence is incomplete. As
a result, some cells can proliferate and become more aggressive
(1,7,8).
Proteomics is a powerful tool to search for markers
in various pathological processes and particularly in oncology. The
authors' laboratory uses a mass spectrometry approach to identify
and study deregulated proteins, such as Olfactomedin 4 (OLFM4) and
Thrombospondin 1 (TSP1), in tumor samples from patients (9,10).
A proteome study identified Human Anterior Gradient protein 2
(AGR2), which is implicated in cancer development and metastasis
induction, especially in breast cancer (9).
AGR2, a member of the human Protein Disulfide
Isomerase family, is involved in protein folding in the endoplasmic
reticulum (ER) (11,12). A number of studies have shown that
AGR2, which is resident in the ER to regulate peptide maturation,
can be secreted and act on the tumor niche in an auto- and
paracrine manner (13-16). These two forms of the protein seem
to result from a state of equilibrium between the monomeric and
dimeric forms regulating the activities of the molecule (17). Although it was originally
described as a developmental protein (18), a number of studies have shown
variation in its expression in different types of cancer cells
(15,19-23). Its overexpression and pro-tumoral
role were first shown in breast cancer and described in other
tissues such as the esophagus, pancreas, lungs and ovaries
(23).
In breast cancer tumors, AGR2 expression correlates
with the expression of estrogen receptor (24). This receptor regulates AGR2
expression following stimulation by estradiol or tamoxifen
(25). The overexpression of AGR2
in breast cancer tumors can be responsible for the induction of
cell proliferation through the regulation of proliferative proteins
such as cyclin D1, c-Myc and E2F1 (26). AGR2 expression is also associated
with tumor aggressivity by inducing metastasis (27). Finally, it was observed that cells
overexpressing AGR2 show treatment resistance toward tamoxifen and
doxorubicin (28,29).
The author's laboratory studies the expression of
various proteins in colorectal cancer and breast cancer to identify
new biomarkers that allow tumor and metastasis detection. These
proteomic studies are also combined with a CIS escape model to
determine the pathways that lead to senescence escape.
The aim of the present study was to determine a role
of the AGR2 protein in vivo and evaluate its potential role
as a biomarker of prognosis in patients with breast cancer. It also
determined its new role in vitro, specifically in a
chemotherapy-induced senescence model and the impact of its
intracellular and extracellular forms on the activation of
proliferative pathways.
Material and methods
Cell lines and treatments
MCF-7 and LS174T cell lines were obtained from the
American Type Culture Collection. Cell lines were authenticated by
STR profiling and regularly tested to exclude mycoplasma
contamination. To induce senescence, MCF-7 and LS174T cells were
treated in RPMI medium (Dutscher; cat. no. L0500-500) containing 3%
fetal bovine serum (FBS) (Eurobio; cat. no. CVFSVF00 01)
respectively with doxorubicin (Tocris Bioscience; cat. no. 2252)
(25 ng/ml) and sn38 (Tocris Bioscience; cat. no. 2684) (5 ng/ml)
for 96 h. To promote senescence escape, cells were washed with PBS
and stimulated with fresh medium containing 10% FBS for the
indicated time. AKT inhibitor
[iAKT1/2:1,3-Dihydro-1-(1-[(4-[6-phenyl-1H-imidazo(4,5-g)quinoxalin-7-yl]phenyl)methyl]-4-piperidinyl)-2H-benzimidazol-2-one
trifluoroacetate; Sigma-Aldrich; Merck KGaA; cat. no. A6730] was
used at a concentration of 100 µM and Torin (Cell Signaling
Technology, Inc.; cat. no. 14379) at 10 nM.
Emerging cells
MCF-7 and LS174T cells were treated in RPMI medium
containing 3% FBS respectively with doxorubicin (25 ng/ml) and sn38
(5 ng/ml) for 96 h. To obtain an emerging population, comprising
proliferative and senescent cells, senescent cells were washed with
PBS and fresh media was added for 7 or 11 days.
Small interfering (si)RNA
transfection
Cells were transfected with 50 nM of siRNA directed
against AGR2 (5′-CUG AUU AGG UUA UGG UUU ATT-3′) (30) and prevalidated control siRNA
(Dharmacon; cat. no. D-001810-10-20) using DharmaFECT-4
(Dharmacon). The cells were incubated for 24 h at 37°C, then the
media was changed into fresh RPMI 10% FBS for an extra 24 h for
western blot experiments or 9 days for emergence experiments.
Extracellular (e)AGR2 Plasmid
transfection and conditioned media generation
For transfection experiments, MCF-7 and 293 cells
were seeded into 10 cm culture dishes and grown until 80%
confluence. Then, 2.5 µg of the empty vector pcDNA3.0 and
the pcDNA3.0/AGR2 plasmid (Genewiz; Sigma-Aldrich; Merck KGaA) were
transfected using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h at 37°C
according to the manufacturer's instructions. The cells were
starved in non-complemented RPMI media for 24 h. The conditioned
media were harvested and centrifuged at 690 × g at room temperature
to eliminate dead cells. The presence of extracellular AGR2 (eAGR2)
was assessed using western blot analysis of media from
non-transfected cells and transfected cells with pcDNA3.0 or
pcDNA3.0/AGR2. The media were concentrated using Vivaspin 15R 5kDa
following the manufacturer's protocol (Sartorius AG).
Recombinant AGR2
Recombinant AGR2 (RayBiotech, Inc.; cat. no.
230-00596) was reconstituted in 1X PBS at 20 µg/ml and used
at 200 ng/ml to treat cells during senescence escape.
SA-β galactosidase staining
Cells were fixed for 10 min at room temperature in
1% formaldehyde, washed with PBS and incubated at 37°C for 16 h in
the absence of CO2 with freshly made staining solution:
0.3 mg/ml of 5-bromo-4-chloro-3-in dolyl-β-d-galactopyranoside
(X-Gal; Promega Corporation; cat. no. V394A), 40 mM citric acid
(Sigma-Aldrich; Merck KGaA), 40 mM sodium phosphate (Sigma-Aldrich;
Merck KGaA) [stock solution (400 mM citric acid, 400 mM sodium
phosphate) held at pH 6], 5 mM potassium hexacyanoferrate
(Sigma-Aldrich; Merck KGaA), 5 mM potassium ferricyanide
(Sigma-Aldrich; Merck KGaA), 150 mM NaCl (Sigma-Aldrich; Merck
KGaA) and 150 mM MgCl2 (Sigma-Aldrich; Merck KGaA). SA-β
galactosidase staining was observed using a light microscope (Life
Technologies; EVOS XL Core) and images were captured at ×40, ×100
and ×200 magnification in different areas in examples of each
condition (31).
Western blotting
Following cell lysis with FASP Buffer (0.1 M
Tris-HCL, 4% SDS, pH 7.6) containing a cocktail of inhibitors (10
µg/ml aprotinin, 10 µg/ml leupeptin, 10 µg/ml
pepstatin, 1 mM Na3VO4, 50 mM NaF), lysates were sonicated for 20
sec at room temperature and then boiled for 10 min. Proteins were
quantified using BCA kit (Thermo Scientific Inc.; Pierce Protein
Assay kit cat. no. 23225) and 50 µg of each sample was
separated on a SDS polyacrylamide gel (8 and 10% for AGR2, p21,
p53, AKT, pAKT evaluation, 6% for RICTOR, pRICTOR evaluation) and
transferred to a PVDF membrane. Following 1 h incubation in 5% milk
(5% BSA for pRICTOR), Tris-buffered saline (TBS) and 0.1% Tween-20,
the membranes were incubated overnight at 4°C with the following
primary antibodies at 1:1,000: AGR2 (Abnova; cat. no.
0001055-1-M03), Actin (Santa Cruz Biotechnology, Inc.; cat. no.
sc-8432), AKT pan (Cell Signaling Technology, Inc.; cat. no.
2920S), phosphorylated (p)-AKT S473 (Cell Signaling Technology,
Inc.; cat. no. 4058S), HSC70 (Santa Cruz, sc-7298), p21Waf1 (Cell
Signaling Technology, Inc.; cat. no. 2947S), p53 (Santa Cruz,
sc-98), p-p53 S15 (Cell Signaling Technology, Inc.; cat. no. 9286),
RICTOR (Cell Signaling Technology, Inc.; cat. no. 2114T), p-RICTOR
T1135 (Cell Signaling Technology, Inc.; cat. no. 3806S) and p-S6
ribosomal protein (S235/236) (Cell Signaling Technology, Inc.; cat.
no. 2211). Membranes were then washed three times with TBS with
0.1% Tween 20 and incubated for 45 min with the secondary
antibodies at 1:3,000: anti-rabbit IgG, horseradish peroxidase
(HRP)-linked antibody (Cell Signaling Technology, Inc.; cat. no.
7074), anti-mouse IgG and HRP-linked antibody (Cell Signaling
Technology, Inc.; cat. no. 7076). Visualization was performed by
chemiluminescence with Fusion Solo (Vilber Lourmat) and
quantification performed on Evolution Capt Solo (v 6 17.00) (Vilber
Lourmat).
Flow cytometry
Data acquisition and analysis were performed on BD
LSR II flow cytometry device and on Diva 6 software (BD
Bioscience).
γ-H2AX (Ser 139) staining
A total of 250,000 cells (MCF7) were incubated with
4% paraformaldehyde at 37°C for 10 min and permeabilized with cold
90% methanol in ice for 30 min. Cells were then washed and
incubated with 16 ng of A488 mouse anti-γ-H2AX (Ser 139) (BD
Pharmingen; cat. no. 560445) or 16 ng of A488 mouse IgG1K (BD
Pharmingen; cat. no. 557721) in the dark at room temperature for 1
h.
Cell cycle analysis
A total of 125,000 cells were incubated with 150
µl of solution A (trypsin 30 µg/ml; Sigma-Aldrich;
Merck KGaA) at room temperature in the dark for 10 min. 125
µl of solution B (trypsin inhibitor 0.5 mg/ml, RNAse A 0.1
mg/ml; Sigma-Aldrich; Merck KGaA) was then added in the dark for 10
min. Finally, cells were incubated with 125 µl of solution C
(propidium iodide 0.6 mM, spermine tetrahydrochloride 3.3 mM;
Sigma-Aldrich; Merck KGaA) at 4°C for 10 min. All the solutions
were prepared in a storage buffer pH 7.6 containing 3.4 mM sodium
citrate 2H20 (Sigma-Aldrich; Merck KGaA), 0.1% Igepal
CA-630 (Sigma-Aldrich; Merck KGaA), 3 mM spermine
tetrahydrochloride and 1 mM tris-aminomethane (32).
Patient samples
The research protocol was approved by the Institut
de Cancerologie de l'Ouest Paul Papin (ICO, Angers, France) and
written informed consent was obtained from all participating
patients (approval number NCT02653105). A total of 74 samples were
included in this protocol with patients' age range from 50 to 74
(recruitment date: March 2016). Tumor specimens were embedded in
paraffin as normally performed for routine clinical analysis.
Following histopathological diagnosis, the FFPE (Formalin-Fixed
Paraffin-Embedded) tissues were sectioned at 20 µm, mounted
on glass slides and compared with hematoxylin and eosin-stained (10
sec hematoxylin, 3 sec eosin, at room temperature) slides from the
same block to identify tumor-rich tissue regions.
Sera from female patients with breast cancer were
collected at the ICO in Angers between 2014 and 2017. All sera were
collected following written informed consent. The study protocol
was approved by UNICANCER (approval no. NCT00630032) and 197
patients >18 years old were included in this protocol. The sera
were obtained from blood after centrifugation at 3,700 g at 4°C for
10 min, then stored at −80°C. All samples were obtained prior to
surgery or neoadjuvant treatment.
AGR2 measurement by ELISA
The AGR2 concentration was determined using ELISA
kit from USCN Life Science Inc. (ref. SEC285Hu). Briefly, sera were
collected from healthy donor or patients with breast cancer by
centrifugating blood samples at 4°C at 3,700 × g for 10 min, then
diluted to 1:1,000 using 1X PBS before assay proceeding. Samples
and standards were added into the provided microplates precoated
with AGR2 antibody, before adding a biotin-conjugated antibody
specific to AGR2. In the presence of avidin-conjugated HRP a color
change occurred and the microplates were read in a Tecan microplate
reader (Tecan Group, Ltd.) at 450 nm. The analysis was performed
using Magellan software (version 7.0; Tecan Group, Ltd.;
intra-assay coefficient <10%, inter-assay coefficient
<12%).
Mass spectrometry
Creation of the spectral library
To build the spectral library, peptide solutions of
several protein samples were analyzed by shotgun approach using
micro-LC-MS/MS. A total of 5 pooled samples of breast tissues were
prepared to obtain a good representation of the peptides. Each
sample was fractionated by OFFGEL fractionator (3100 OFFGEL
Fractionator; Agilent Technologies, Inc.) into 24 fractions. Each
fraction was separated into a micro-LC system Ekspert nLC400
(Eksigent Technologies LLC) using a ChromXP C18CL column (0.3 mm x
15 cm, 3 µm, 120 Å; Eksigent Technologies LLC) at a flow
rate of 5 µl/min. Water and acetonitrile, both containing
0.1% formic acid, were used as solvents A and B, respectively. The
following gradient of solvent B was used: 0 to 5 min 5% B, 5 to 125
min 5 to 35% B, then 9 min at 95% B and finally 9 min at 5% B for
column equilibration. As the peptides eluted, they were directly
injected into a hybrid quadrupole-TOF mass spectrometer Triple TOF
5600+ (Sciex) operated with a 'top 30' data-dependent acquisition
system using positive ion mode (pressure at the curtain plate: 60
psi without heating; flow rate 5 µl/min). The acquisition
mode consisted of a 250 msec survey MS scan from 400 to 1,250 m/z,
followed by an MS/MS scan from 200 to 1,500 m/z (75 msec
acquisition time, 350 mDa mass tolerance, rolling collision energy)
of the top 30 precursor ions from the survey scan. The peptide and
protein identifications were performed using Protein Pilot software
(version 5.0; Sciex) with a human Swiss-Prot/TrEMBL concatenated
target-reverse decoy database (https://www.uniprot.org/, downloaded in March 2016)
containing 142,441 human protein sequences, specifying MMTS as Cys
alkylation. The false discovery rate (FDR) was set to 0.01 for both
peptides and proteins. The MS/MS spectra of the identified peptides
were then used to generate the spectral library for SWATH peak
extraction using the add-in for PeakView Software (version 2.2,
Sciex) MS/MSALL with SWATH Acquisition MicroApp (version 2.0,
Sciex). Peptides with a confidence score above 99% were obtained
from Protein Pilot database searches were included in the spectral
library.
Relative quantification by SWATH
acquisition
MCF-7 cells were analyzed using a Data Independent
Acquisition method (33). Each
sample (5 µg) was analyzed using the LC-MS equipment and LC
gradient described in the previous section following a SWATH-MS
acquisition method. The method involved repeating the whole
gradient cycle, which consisted of the acquisition of 35 TOF MS/MS
scans of overlapping sequential precursor isolation windows (25 m/z
isolation width, 1 m/z overlap, high sensitivity mode) covering the
400 to 1,250 m/z mass range, with a previous MS scan for each
cycle. The accumulation time was 50 msec for the MS scan (400-1,250
m/z) and 100 msec for the product ion scan (230-1,500 m/z), making
a 3.5 sec total cycle time.
Data analysis
The targeted data extraction of the SWATH runs was
performed by PeakView using the MS/MSALL with SWATH Acquisition
MicroApp. PeakView processed the data using the spectral library
created from the shotgun data. Up to ten peptides per protein and
seven fragments per peptide were selected, based on signal
intensity; any shared and modified peptides were excluded from the
extraction. The retention times from the peptides selected for each
protein were realigned in each run according to the iRT peptides
(Biognosys AG) spiked in each sample and eluting along the
whole-time axis; the extracted ion chromatograms were generated for
each selected fragment ion. PeakView computed a score and FDR for
each assigned peptide using chromatographic and spectra components;
only peptides with an FDR <5% were used for protein
quantitation. The peak areas for peptides were obtained by summing
the peak areas of the corresponding fragment ions; protein
quantitation was calculated by summing the peak areas of the
corresponding peptides. MarkerView (version 1.2; Sciex) was used
for signal normalization and differential abundance was tested by
applying a t-test at protein level.
GSEA Analysis
GSEA analysis was performed using GSEA software from
Broad Institute (https://www.gsea-msigdb.org/gsea), the raw data tables
were uploaded on the software. The data bases used are Hallmarks
(h.all.v.7.0.symbol), Oncogenic signatures (C6.all.v.7.symbol) and
Senescence signature (C2.CP.REACTOME.REACTOME_OXIDATIVE_
STRESS_INDUCED_SENESCENCE).
Data deposition
The mass spectrometry proteomics data have been
deposited to the ProteomeXchange Consortium via the PRIDE
(http://www.proteomexchange.org/)
(34) partner repository with the
dataset identifier PXD014194 for breast cancer analysis and
PXD028073 for emergent MCF7 analysis.
Statistical analysis
All data were expressed as mean ± standard
deviation. Differences were analyzed using nonparametric tests
(Mann-Whitney, Kolmogorov-Smirnov and one-way ANOVA with Dunnett's
multiple comparison test). Pearson's correlation test was performed
to assess the correlation between protein expression in breast
cancer tumors. P<0.05 was considered to indicate a statistically
significant difference.
Results
AGR2 is significantly overexpressed in
sera of metastasized patients
Previous studies have shown that analysis of the
proteome in colon and breast cancer makes it possible to identify
proteins, such as OLFM4 and TSP1, that are involved in tumor
progression and aggressiveness (9,10).
Proteomic analysis of a cohort of colorectal tumors shows that AGR2
is expressed in colonic adenomas and that its expression is
increased in the invasive stages of adenocarcinomas (stage IV)
(9). AGR2 is involved in various
cancer types, especially breast and prostate and is associated with
poor prognosis (21,35).
To confirm these observations, the present study
first evaluated the concentration of AGR2 in human serum samples
from patients with breast cancer with or without metastasis and
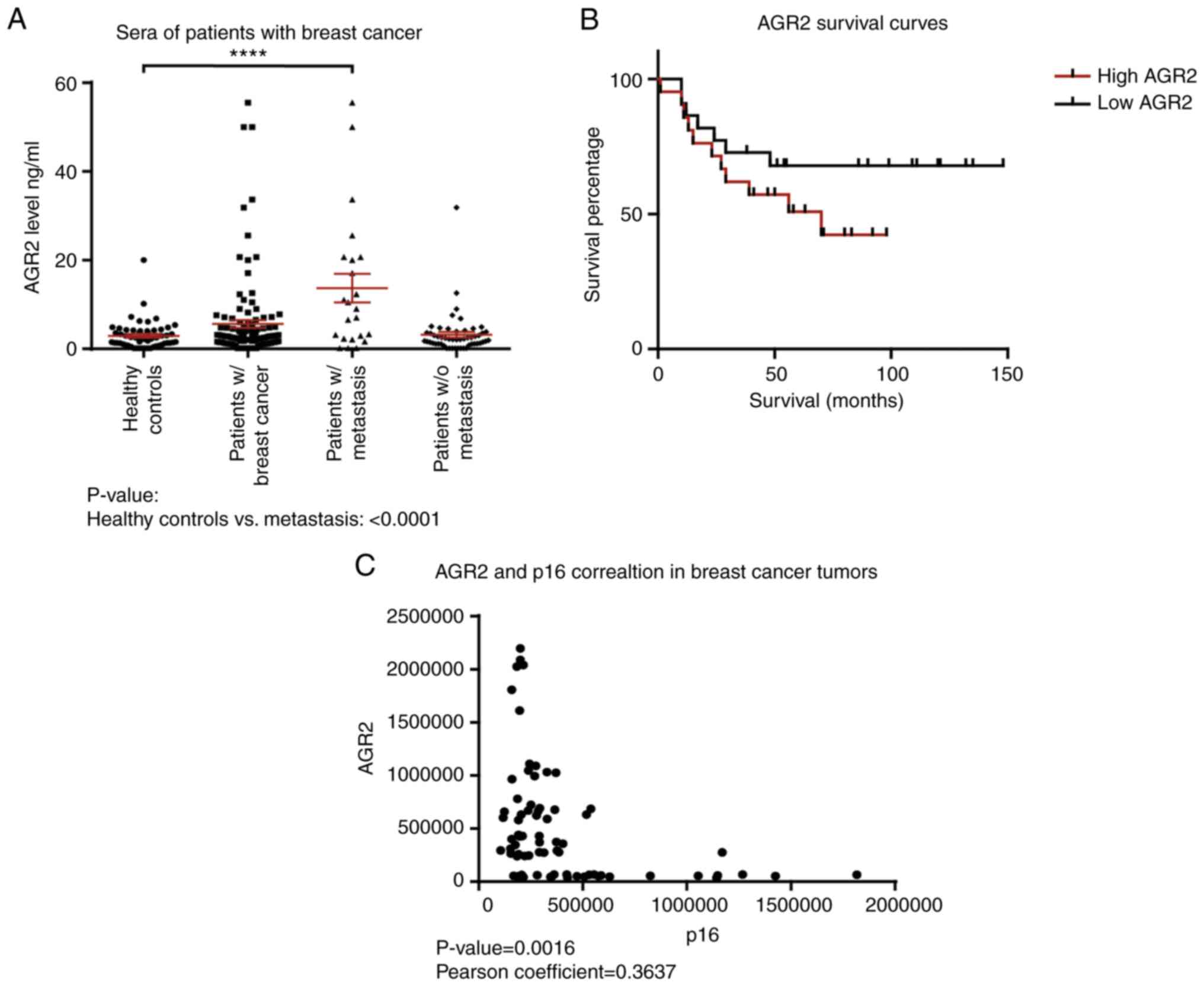
healthy controls using an ELISA approach. The results presented in
Table I show that the mean value
for healthy donors is 2.93±0.42 ng/ml (n=56), 5.62±0.87 ng/ml
(n=118) in the breast cancer group and 13.7±3.2 ng/ml (n=23) in the
breast cancer with metastasis group. AGR2 is significantly higher
in patients with breast cancer with metastasis than healthy
controls (P-value<0.0001; Fig.
1A; Table I).
 | Table IAGR2 concentrations in sera from
healthy donors, patients with breast cancer and patients with
breast cancer with metastasis. |
Table I
AGR2 concentrations in sera from
healthy donors, patients with breast cancer and patients with
breast cancer with metastasis.
| Cases | Number | AGR2 mean
concentration ng/ml | Standard
deviation |
|---|
| Healthy donors | 56 | 2.93 | 0.42 |
| Patients with
breast cancer | 118 | 5.62 | 0.87 |
| Patients with
breast cancer and with metastasis | 23 | 13.7 | 3.2 |
Data were examined for the correlation between AGR2
and overall survival. Kaplan-Meier survival curves based on AGR2
expression were derived from Tang et al (36). This study compared the proteome of
proteins extracted from breast tumors and adjacent noncancerous
tissues by mass spectrometry. The cohort contained samples from the
three molecular subtypes of breast cancer: Luminal A, HER2-positive
and triple negative (36). The
present study used the data on AGR2 expression from this breast
cancer study to assess if overall survival can be related to AGR2.
The results show that patients harboring high AGR2 expression
present low survival compared to patients with low AGR2 expression
(Fig. 1B; P-value=0.07).
To understand AGR2's implication in breast cancer
progression, its correlation with tumor suppressors was
investigated using the SWATH-MS approach on 119 breast tumors. The
results presented in Fig. 1C show
that AGR2 expression is inversely correlated with p16, a major
senescence regulator (P-value=0.0016). They suggest that AGR2 could
induce tumor progression through the regulation of suppressive
mechanisms such as senescence.
All these results showed that AGR2 was a protein
detected in breast cancer. In addition, high concentration in the
serum of patients was associated with a metastatic state and its
expression in breast tumors is associated with poor prognosis.
AGR2 is overexpressed during senescence
escape
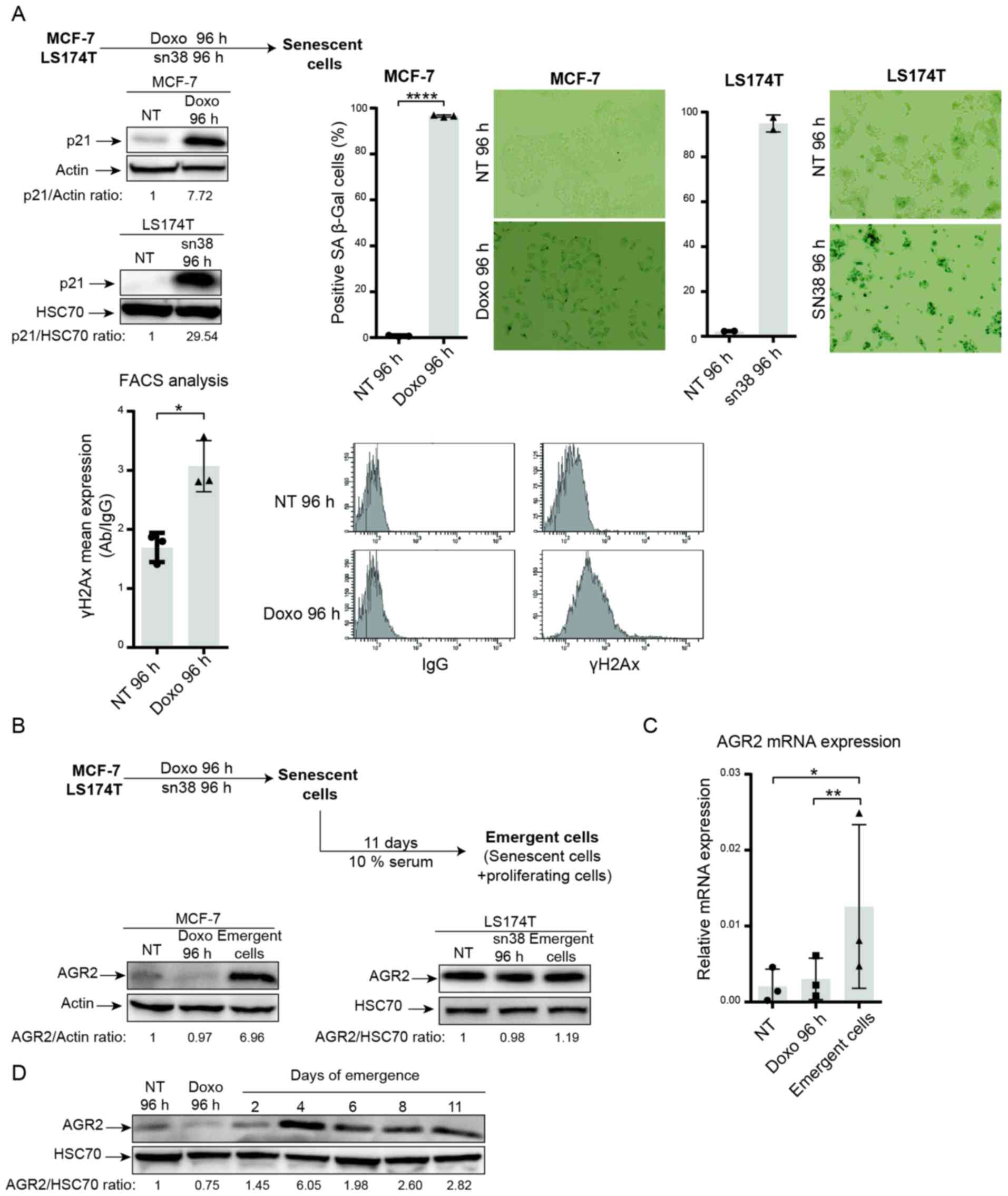
In breast and colorectal cell lines, it was shown
that genotoxic treatment induced senescence. As depicted in
Fig. 2A, doxorubicin (25 ng/ml)
and sn38 (5 ng/ml) were used to treat respectively the MCF-7 and
LS174T cell lines for 96 h. CIS was confirmed using p21WAF1
expression (Fig. 2A, upper left)
and SA-β-galactosidase (Fig. 2A,
upper right.). These experiments revealed that both MCF-7 and
LS174T cells showed upregulation of the cell cycle inhibitor p21
and high SA-β-galactosidase staining. The MCF-7 cell line also
showed an elevation in γ-H2AX staining, which reflected an increase
in DNA damage (Fig. 2A, lower
part).
The present authors recently reported that breast
and colorectal cells can adapt to CIS and resume proliferation. CIS
escape generates heterogeneous populations called emerging cells
and comprising senescent cells and dividing cells (37). Taking into account the results
obtained from proteomic analysis and the anticorrelation between
AGR2 and p16, AGR2 expression was analyzed during CIS induction and
CIS escape in emerging cells (Fig.
2B, upper part). AGR2 expression was first analyzed in the two
CIS escape models. Analysis of the MCF-7 model shows that
expression of the protein AGR2 (Fig.
2B, left) and its mRNA (Fig.
2C) was induced in the emerging population but not in senescent
cells. Whereas in the LS174T model, western blot analysis shows no
significant variation in AGR2 expression between senescent and
emergent cells. To analyze AGR2 expression more accurately in the
LS174T model, mass spectrometry analysis was performed to compare
senescent and emergent cells. The result showed a slight increase
in AGR2 expression in emergent LS174T compared to senescent LS174T
(data not shown). Finally, AGR2 expression during CIS escape was
assessed with two days' interval. The result presented in Fig. 2D showed that AGR2 expression
during CIS escape increased after four days of emergence.
These initial results led to study AGR2's potential
role in CIS escape, primarily by inhibiting its expression or
adding its extracellular form.
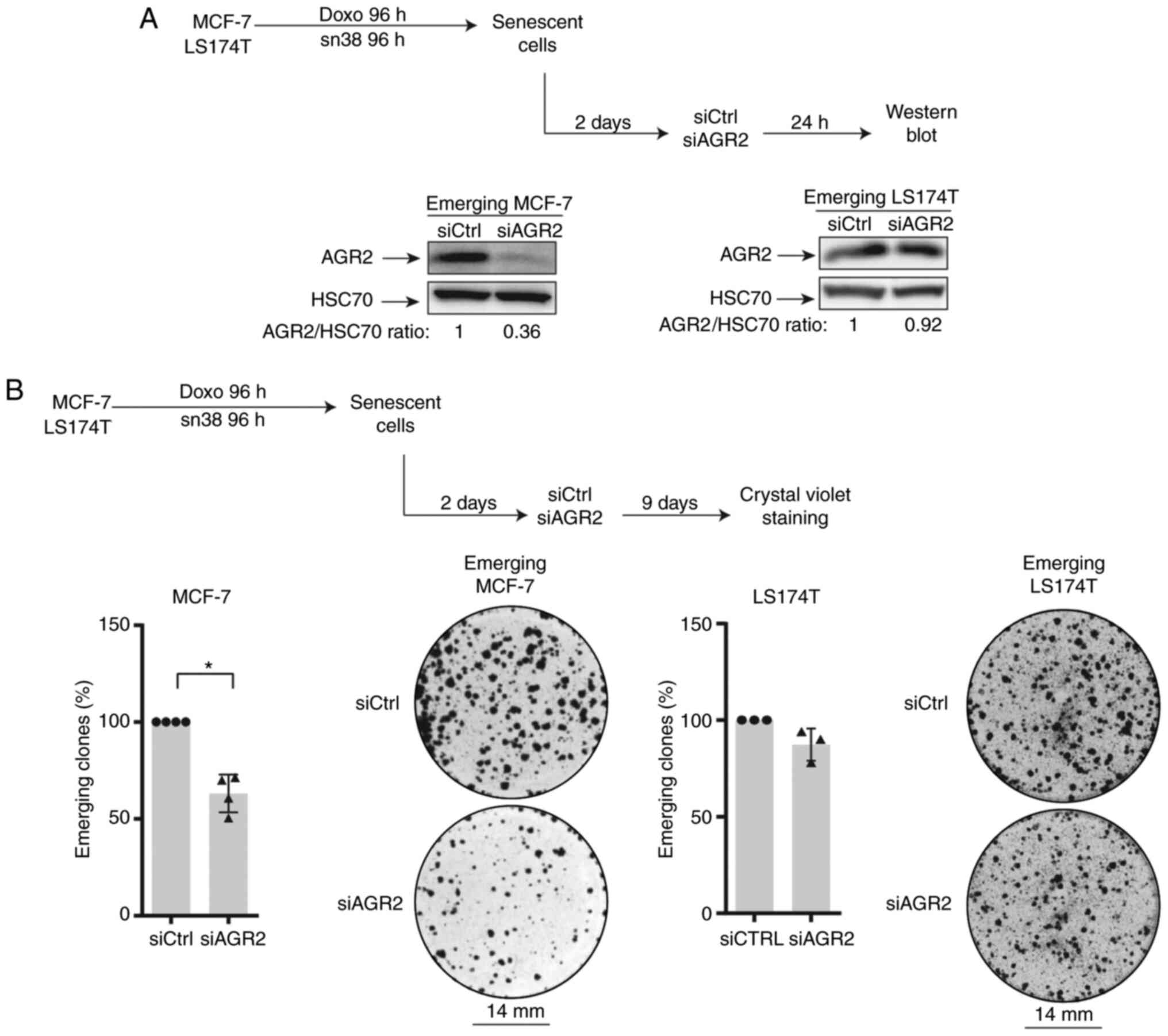
AGR2 inhibition prevents CIS escape
To evaluate AGR2's involvement in CIS escape, its
expression was inhibited in senescent cells. For this, siRNA was
transfected two days after the end of treatment (in accordance with
the kinetic of expression) as specified in Fig. 3A (top). Western blot analysis
confirmed the protein's downregulation (Fig. 3A, lower left). The impact of AGR2
inhibition on the number of emerging clones was then evaluated
(Fig. 3B, upper part). As shown
in Fig. 3B (left lower part),
AGR2 downregulation significantly decreased the percentage of
emerging clones.
The present study also transduced the siRNA against
AGR2 into emerging colorectal cell line. It was not possible to
inhibit AGR2 expression efficiently due to the high amount of
protein (Fig. 3A, lower right).
However, a slight decrease in the number of proliferating clones
was observed following siRNA transduction during the emergence of
LS174T cell line (Fig. 3B,
right).
Taken together these results indicated that AGR2
expression favors cell emergence after CIS. This effect is
heterogenous and seems to depend on AGR2 expression on cell lines.
Although a slight decrease in the number of emerging clones was
observed after siRNA transfection in colorectal cells, the present
study chose to investigate the CIS escape mechanism only in the
breast cancer model.
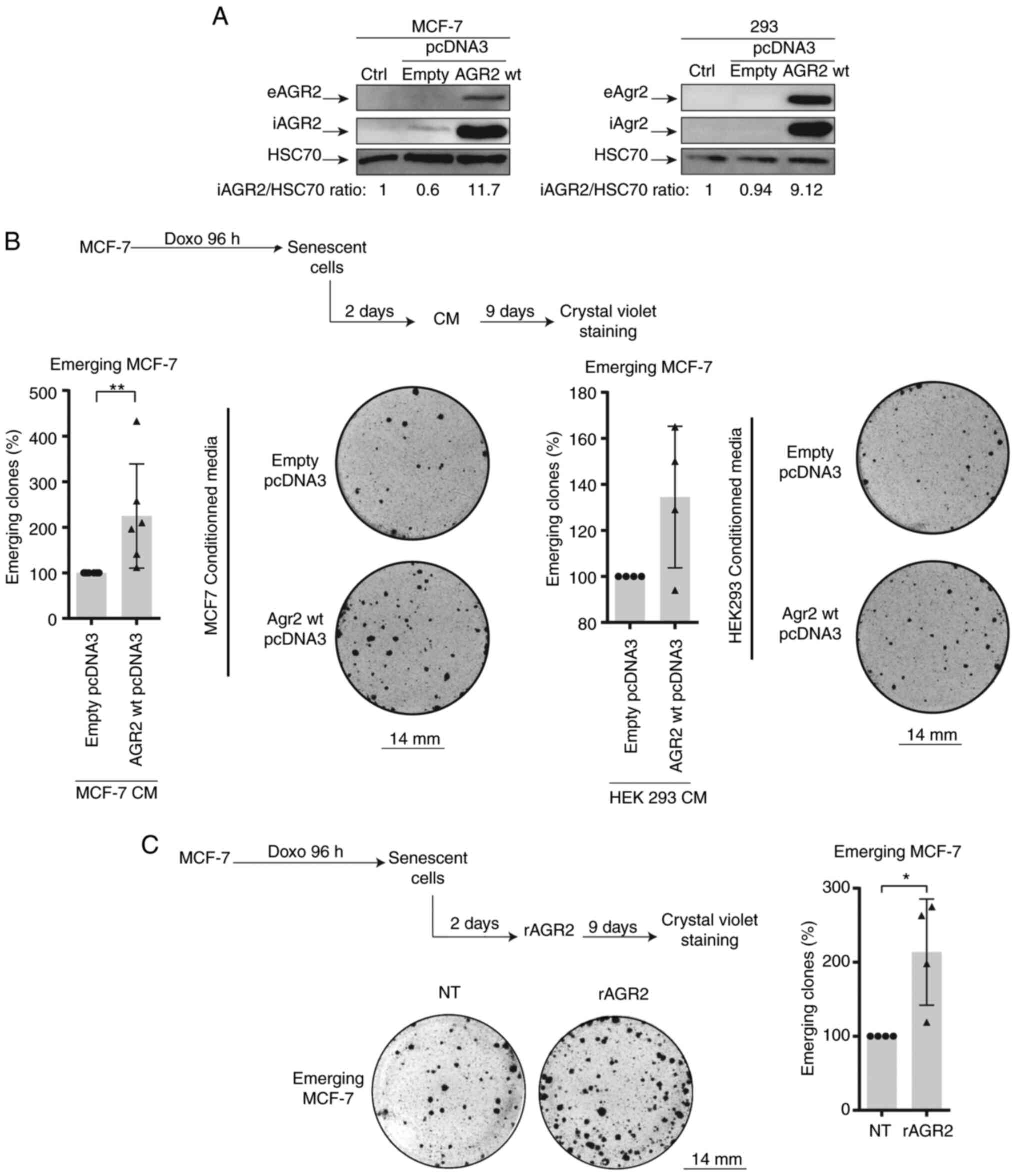
eAGR2 enhances CIS escape in MCF-7
cells
To confirm the role of AGR2 in CIS escape, a plasmid
construct coding for AGR2 protein was transduced into emerging
MCF-7. This experiment could not be of a use as the transduction
lead to cell death. Therefore, the present study chose to assess
the role of extracellular AGR2 on CIS escape.
Previous studies have shown that the extracellular
form of AGR2 (eAGR2) is implicated in the proliferation and
aggressiveness of tumor cells (15,17). Therefore the effect of eAGR2 on
CIS escape was studied. For this, the growing MCF-7 cells and 293
cells were transfected with a plasmid-encoding AGR2 or with a
control plasmid. After two days of transfection, the culture media
was collected and added to MCF-7 senescent cells. eAGR2 production
was monitored by western blotting (Fig. 4A). eAGR2 significantly increases
the number of emergent clones, as shown in Fig. 4B. Recombinant eAGR2 (200 ng/ml)
was also added during CIS escape (Fig. 4C, upper left). The count of
emerging clones showed that the recombinant molecule significantly
increased CIS escape (Fig. 4C,
right and lower part).
The effect of eAGR2 on CIS escape after inhibition
AGR2 expression was also assessed using siRNA. This experiment
showed no influence of eAGR2 on cell emergence (data not shown),
thus eAGR2 alone is not sufficient to favor CIS escape and a
cooperation is needed between its two forms.
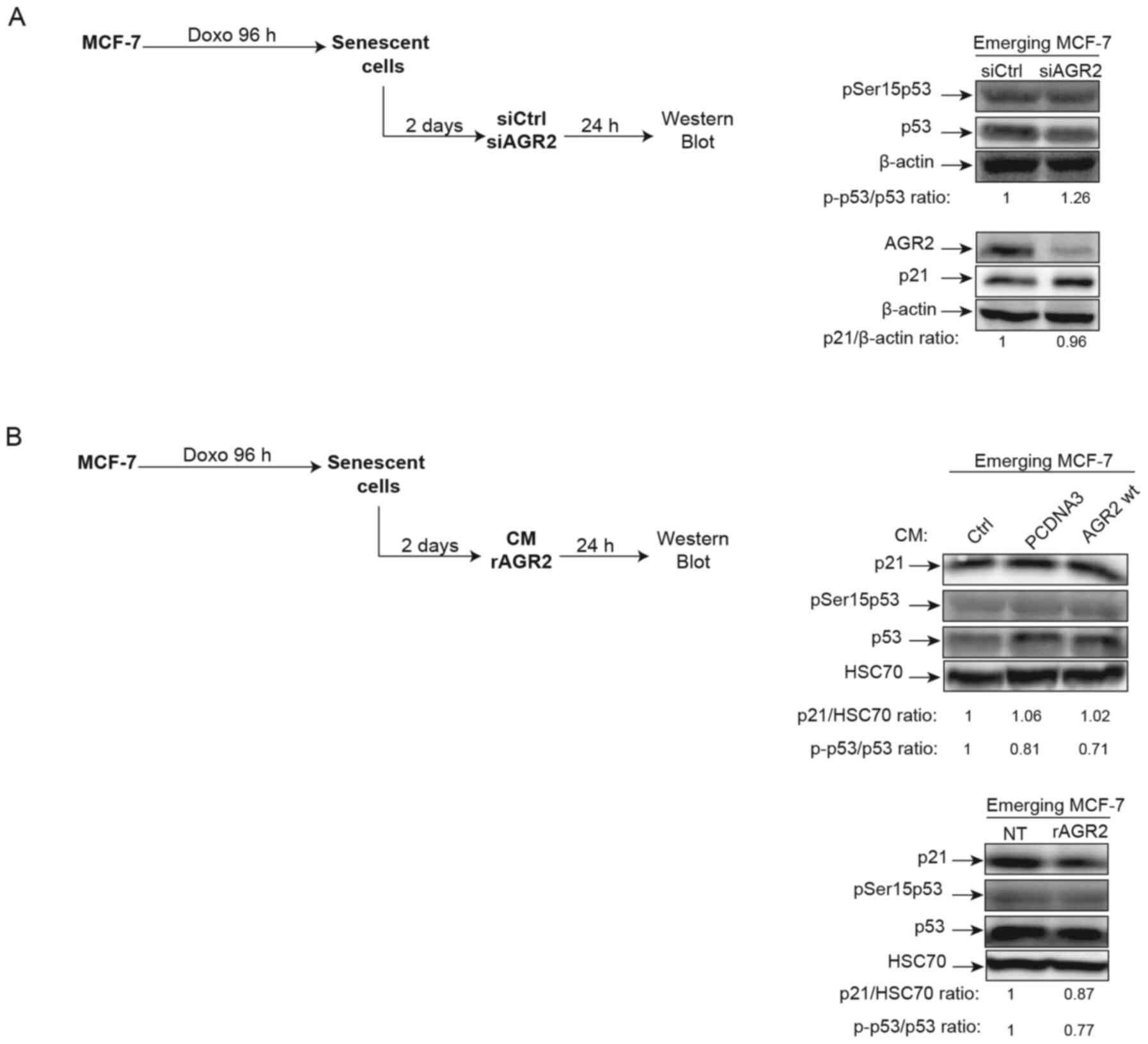
AGR2 does not regulate p21/p53 during CIS
escape
In light of AGR2's observed effects on senescence
escape, whether this effect is regulated or not by the p21/p53
signaling pathway was investigated. Indeed, several studies have
shown that AGR2 may regulate p53 activation following DNA damage by
UV or genotoxic treatments. In these studies, suppression of AGR2
leads to higher p53 activation by allowing phosphorylation of
serine 15 (38,39). Based on these observations, the
effect of AGR2 on the phosphorylation of p53 on serine 15 and the
expression of its target gene p21 was assessed during senescence
escape. To this end, senescent MCF-7 cells were transfected with a
siRNA directed against AGR2, the cells recovered after 24 h
(Fig. 5A, left) and activation of
the p53/p21 pathway analyzed. However, neither the phosphorylation
level of p53 serine 15 nor the level of p21 expression were
modified by the inhibition of AGR2 expression (Fig. 5A, right). The same results were
obtained when senescent MCF-7 cells were stimulated by
extracellular AGR2 (Fig. 5B,
left) CM (Fig. 5B, upper right)
or recombinant (r)AGR2 (Fig. 5B,
lower right). These results show that AGR2 does not modify the
p53/p21 pathway during emergence.
AGR2 facilitates senescence escape by
regulating the mTOR/AKT pathway
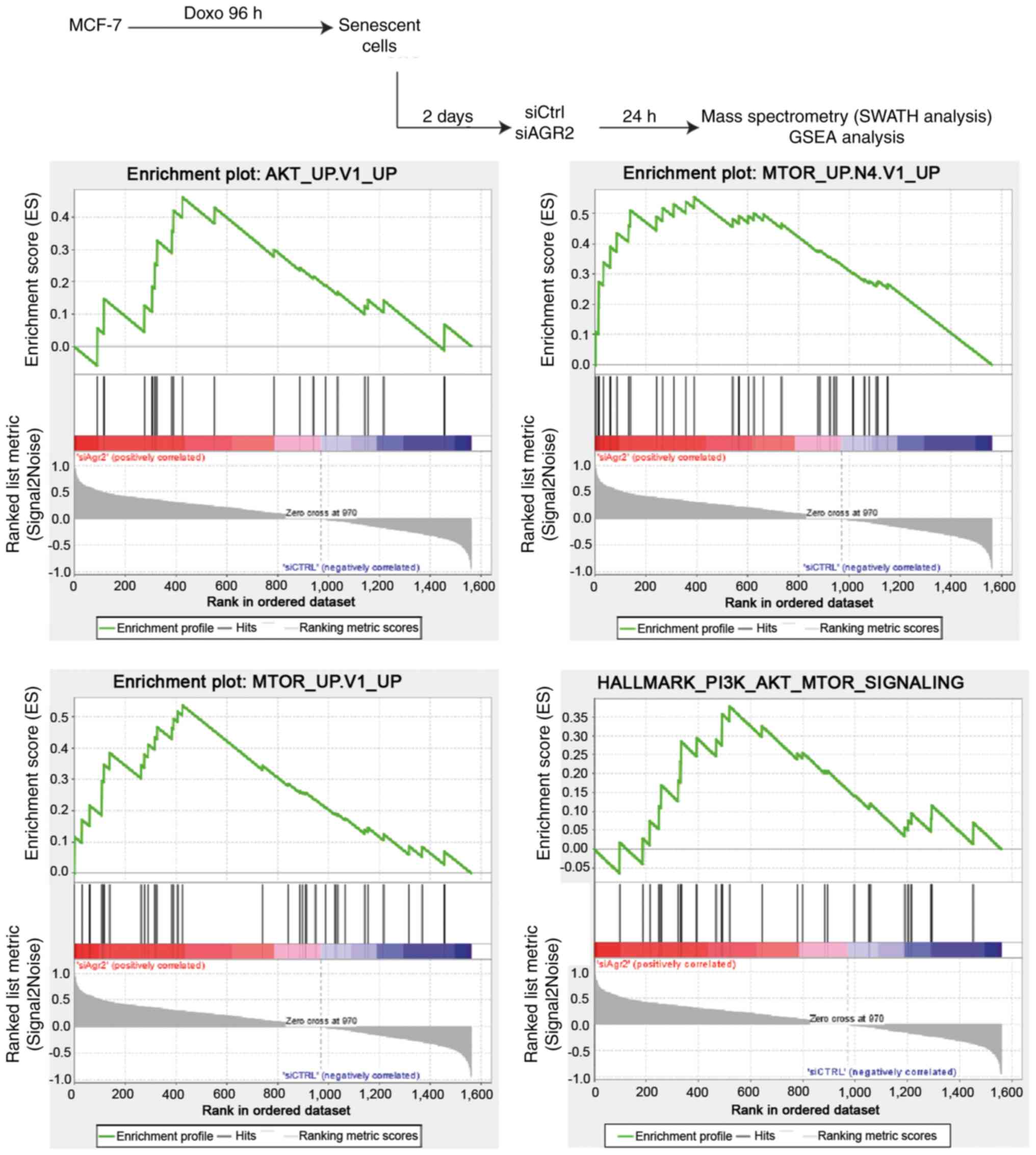
To investigate new pathways by which AGR2 could
control the emergence of MCF-7 cells, a proteomic analysis was
performed using SWATH-MS as previously described (40). Gene Set Enrichment Analysis (GSEA)
of proteomes from emergent MCF-7 transduced with siRNA directed
against AGR2 or control siRNA allowed the identification of
pathways that are significantly correlated with the loss of AGR2
during emergence (Fig. 6, upper
part). This analysis revealed only two gene sets from the Hallmarks
data-base with a nominal P-value <0.05 and a FDR >25%.
However, assessing the oncogenic signature database revealed seven
significantly enriched gene sets (P-value <0.05), including five
with a FDR <25% (Table II).
The enriched gene sets in the results were related to AKT and mTOR
signaling pathways, suggesting that AGR2 could regulate these
pathways during emergence (Fig.
6, lower part).
 | Table IIMolecular signatures that are
significantly deregulated following AGR2 suppression. |
Table II
Molecular signatures that are
significantly deregulated following AGR2 suppression.
| Database
(https://www.gsea-msigdb.org/gsea) | Signatures | Number of
proteins | P-value | FDR q-value |
|---|
| Hallmarks
(h.all.v7.0. symbols) |
HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 41 | 0.01988 | 0.25301 |
|
HALLMARK_PI3K_AKT_MTOR_SIGN ALING | 27 | 0.04490 | 0.53485 |
| Oncogenic
signatures (c6.all.v7.0.symbols) |
MTOR_UP.N4.V1_UP | 30 | <0.001 | 0.00820 |
| MTOR_UP.V1_UP | 33 | <0.001 | 0.03366 |
| AKT_UP.V1_UP | 20 | 0.01362 | 0.15083 |
|
GCNP_SHH_UP_LATE.V1_UP | 37 | 0.03137 | 0.14104 |
|
CYCLIN_D1_UP.V1_UP | 22 | 0.01183 | 0.16396 |
| NRL_DN.V1_DN | 17 | 0.02236 | 0.27494 |
| Senescence
signature |
REACTOME_OXIDATIVE_STRESS_INDUCED_SENESCENCE | 7 | 0.03400 | 0.03600 |
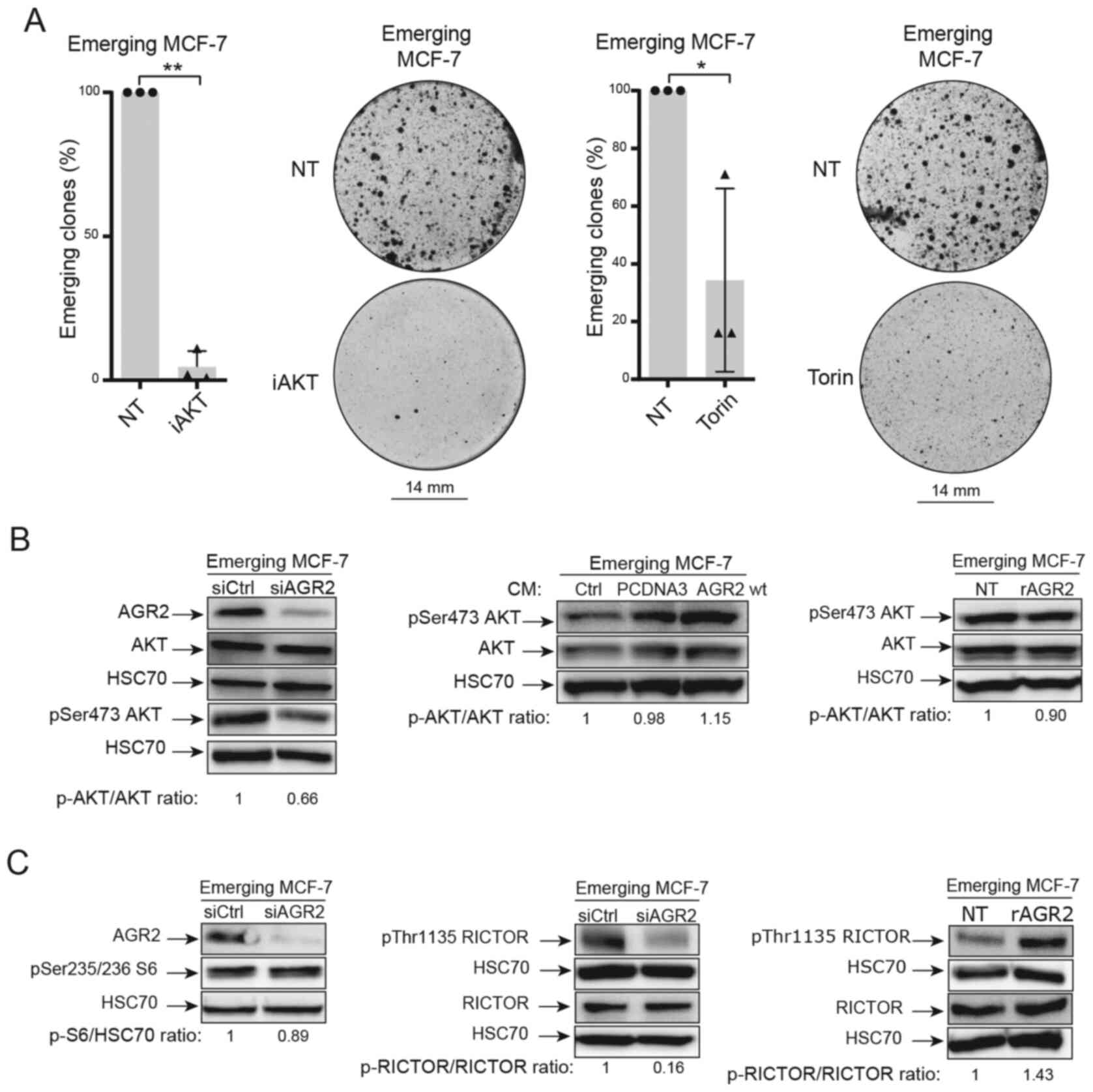
Previous studies in the authors' laboratory have
shown that these two pathways are involved in CIS escape (7). Therefore, AKT and mTOR pathways were
inhibited using two inhibitors (iAKT1/2 and Torin). The results
presented in Fig. 7A show that
these inhibitors prevented CIS escape.
The present study first focused on the AKT pathway.
The importance of AKT phosphorylation (Ser 473) during CIS escape
has been previously shown (7).
The results led to the study of this phosphorylation when AGR2
expression was modulated. When AGR2 expression was inhibited by
siRNA in senescent MCF-7 cells, a decrease in AKT phosphorylation
on its serine 473 residue was observed (Fig. 7B, left). However, AKT
phosphorylation was not increased by extracellular forms of AGR2
[conditioned media, Fig. 7B
(middle) and rAGR2, Fig. 7B
(right)].
Several studies show that AKT phosphorylation is
regulated by the mTOR pathway (41-43). The proteomic study identified the
mTOR signaling pathway as being regulated by AGR2 during CIS
escape. It was therefore determined if mammalian target of
rapamycin complex mTORC1 and mTORC2 were regulated by AGR2 during
CIS escape. First, one of the main targets of mTORC1, the ribosomal
protein S6 was studied. The results presented in Fig. 7C (left) show that siRNA-mediated
inhibition of AGR2 in senescent cells did not change S6
phosphorylation. The total form of S6 was not analyzed as variation
of its phosphorylated form was not observed.
Given these results, the regulation of other mTOR
signaling pathways by AGR2 were studied. The activation of mTORC2
complex is attested by the phosphorylation of RICTOR on threonine
1135. The western blotting results presented in Fig. 7C (middle) show a decrease in
RICTOR phosphorylation when AGR2 was inhibited by siRNA.
Conversely, RICTOR phosphorylation on Thr1135 level is higher when
MCF-7 cells are stimulated with eAGR2 (Fig. 7C, right). Taken together, the
results showed that AGR2's effect on CIS escape could be mediated
by activation of the mTORC2/AKT signaling pathway.
Discussion
Chemotherapy-induced senescence is a complex
mechanism which has been described as a first step in tumor cell
proliferation arrest and elimination by the immune system (44). Long considered irreversible, this
mechanism is being called into question (1,6).
It has been shown in different cell types, notably in a breast
cancer model, that cells treated with doxorubicin go into
senescence. Some of these cells are able to emerge after
chemotherapy and reproliferate (7,8).
This observation suggests that the phenomenon can be considered as
therapeutic failure or treatment resistance. The present study
aimed to identify and establish new proteins and pathways involved
in the induction of CIS escape. Given the proteomic studies in
patients with breast cancer and the proteins identified the present
study focused on the protein AGR2.
In breast cancer tumors, AGR2 is overexpressed and
serves a major role in cell proliferation and aggressiveness. It
has been shown that AGR2 expression in breast cancer cell lines
induces proliferation through the regulation of a number of
proliferative proteins (26).
AGR2 is also known as a tumor aggressiveness marker and its
overexpression leads to metastasis induction (27). However, among the number of
studies published on the role of AGR2 in tumor progression
(15,21,26,45,46), none refers to its contribution to
chemotherapy-induced senescence and particularly the escape leading
to tumor cell proliferation.
The present study showed for the first time that the
presence of AGR2 in the sera of patients with breast cancer is a
marker of metastasis and that its expression in breast tumors is
inversely correlated to p16 expression. It also highlighted that
AGR2 is involved in the emergence of cells after senescence
induction by doxorubicin. The present study underlined that AGR2
was detectable in the serum of untreated patients with breast
cancer and that its level was significantly higher in patients
compared to healthy donors. In addition, the amounts of AGR2 were
significantly higher in the metastatic patient group. From these
observations the present study demonstrated that AGR2 is
anticorrelated with p16, a marker of cellular senescence as has
been shown in ovarian cancer (47) and therefore it studied the
potential role of AGR2 during CIS and implication in the appearance
of emerging clones.
AGR2's role in senescence is not known. Although one
study showed that its loss induces the senescence of prostate tumor
cells (48), there is no research
on AGR2's potential role in the different forms of senescence,
especially during CIS. The present study proposed in its CIS escape
model that AGR2 serves an important role and that this action is
mediated by the cellular and secreted forms of the molecule.
Indeed, it showed that the loss of AGR2 in senescent cells induced
a loss of the number of emergent clones, but that the contribution
of the extracellular form (eAGR2) made it possible to increase the
number of reproliferate clones. Mass spectrometry proteomic
approaches demonstrated that the role of AGR2 is linked to the
mTORC and AKT signaling pathways. Indeed, the AKT signaling pathway
is involved in CIS escape (7).
Furthermore, AKT is implicated in AGR2 regulation following
tamoxifen treatment, leading to cell invasion (49). It is also shown that AGR2
knockdown reduces chemotherapy resistance by negatively regulating
AKT/ERK signaling pathways and promoting apoptosis (50,51). The present study then evaluated
this pathway's activation in the model and the results showed that
AGR2 suppression during CIS escape led to a decrease in AKT
phosphorylation on Ser473. Moreover, it has been shown that mTORC1
regulates AGR2 expression by regulating the length of its mRNA
(52). The CIS escape model of
the present study investigated the potential feedback regulation
between mTORC1 and AGR2 by assessing the protein S6
phosphorylation. The results showed no modification in the
activation of mTORC1 signaling pathway following AGR2 suppression
during cell emergence. It has been shown that mTORC2 is regulated
by AGR2 through the phosphorylation of RICTOR (53). The present study then also
analyzed this pathway's activation in its model. The results showed
that AGR2 suppression during CIS escape led to a reduction in
RICTOR phosphorylation on Threonine 1135. By contrast, the
stimulation of emerging cells by recombinant AGR2 induced RICTOR
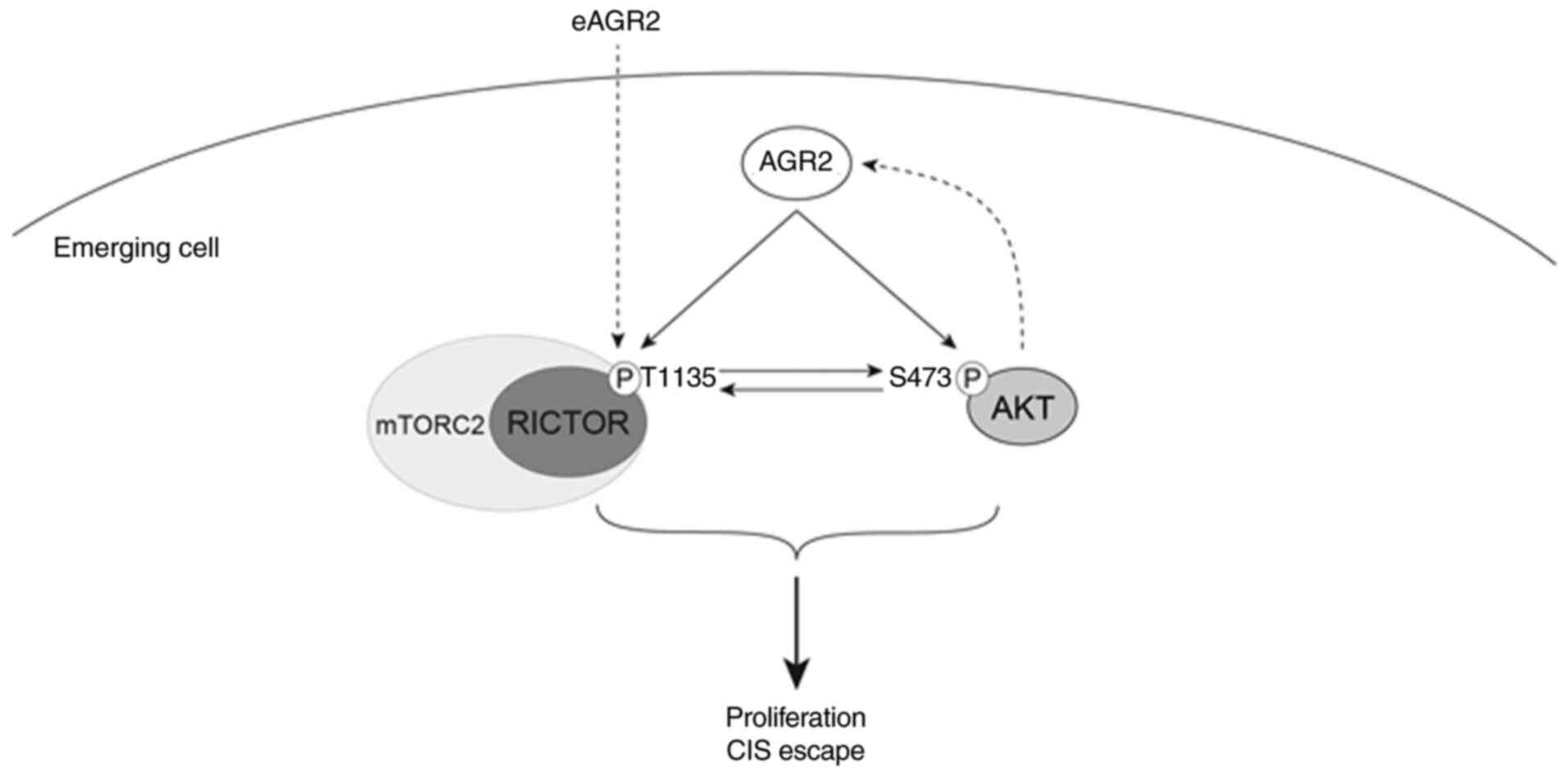
phosphorylation. These results suggested that AGR2 induced the
proliferation of senescent cells by activating AKT and mTORC2
signaling, independently of the p53/p21 pathway (Fig. 8). However, the role of AGR2 in CIS
escape was only confirmed in a breast cancer cell line and only a
slight effect was observed in a colorectal cell line. This
limitation needs more investigations using other breast and
colorectal cell lines expressing AGR2 moderately, to inhibit
efficiently its expression.
Altogether, the findings of the present study
demonstrated that AGR2 could be used as a blood marker of
metastasis and is a poor prognostic biomarker in patients with
breast cancer. It also showed that AGR2 is implicated, by its
secreted and intra-cellular form, in senescence escape via the
activation of new signaling pathways. From these conclusions,
measuring AGR2 concentration in patients could help predict tumor
progression and prevent potential relapse following chemotherapy
treatment.
AGR2 is known to interact with several proteins,
which allows it to participate in different mechanisms (13). This feature should be explored in
the model of the present study, using co-immunoprecipitation
combined with mass spectrometry, to determine the AGR2 partners
responsible for CIS escape induction. Furthermore, it would be
useful to examine whether AGR2 interacts directly with AKT and
RICTOR to induce their phosphorylation. Finally, AGR2 is also known
to be implicated in endoplasmic reticulum stress and UPR (Unfolded
Protein Response) pathways (54).
AGR2 is overexpressed during endoplasmic reticulum stress in
pancreatic cells and contributes to removing that stress by
activating UPR proteins such as binding immunoglobulin protein and
X antigen binding protein 1 (55). Given these observations, AGR2
might be a key protein to link UPR pathways to CIS escape.
Therefore, AGR2 expressed during CIS escape can accelerate
endoplasmic stress repair, allowing good protein folding and more
cell proliferation. Conversely, AGR2 expressed during CIS escape
can be a consequence of UPR pathways activation, which can lead to
the activation of proliferative pathways such as AKT and mTORC2 and
so promote cell proliferation. Moreover, it has been shown that
AGR2 can be found in the cell in monomeric and dimeric forms, with
this balanced status giving AGR2 new functions (17). In the model of the present study,
the dimeric form should be assessed to determine whether during CIS
escape AGR2 promotes proliferation as a monomeric or dimeric
form.
The present study performed proteomic analysis
(ELISA and SWATH-MS) on patient samples to study AGR2 expression
and its outcome in breast cancer. On the other side, a CIS escape
model was used to study the role of AGR2 in senescence escape and
the pathways it regulates. Through the results obtained in
vivo and in vitro it is possible to understand how cells
are able to escape tumors suppression and which pathways are
upregulated to permit cell proliferation.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AM and EL conceived and designed the experiments
with assistance from OC. AM performed and interpreted the
experiments. AB and CH performed ELISA assay on sera samples and
prepared breast cancer, and MCF7 cells samples for mass
spectrometry analysis. CG performed proteomic analysis on mass
spectrometer and analyzed raw data and performed statistical
analysis of ELISA assay data. AM performed GSEA analysis with the
assistance of CG. AM and EL wrote, reviewed, and revised the
manuscript with assistance of OC and CG. AM, EL and GL performed
experiments for reviewing process. AM, EL, and CG confirm the
authenticity of all data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Written consent was obtained from patients upon
registration in the experimental protocol by Institut de
Cancérologie de l'Ouest.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors thank Dr Eric Chevet (INSERM U1242,
Chemistry, Oncogenesis Stress Signaling, University of Rennes,
Rennes, France) for providing AGR2 and control vectors.
Funding
The present study work was supported by donation from Comité
Féminin 49 pour la Prévention et le Dépistage du Cancer Octobre
Rose.
Abbreviations:
|
CIS
|
chemotherapy-induced senescence
|
|
AGR2
|
anterior gradient 2
|
References
|
1
|
Lee S and Schmitt CA: The dynamic nature
of senescence in cancer. Nat Cell Biol. 21:94–101. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rodier F, Coppé J, Patil CK, Hoeijmakers
WA, Muñoz DP, Raza SR, Freund A, Campeau E, Davalos AR and Campisi
J: Persistent DNA damage signalling triggers senescence-associated
inflammatory cytokine secretion. Nat Cell Biol. 11:973–979. 2009.
View Article : Google Scholar :
|
|
3
|
Beauséjour CM, Krtolica A, Galimi F,
Narita M, Lowe SW, Yaswen P and Campisi J: Reversal of human
cellular senescence: Roles of the p53 and p16 pathways. EMBO J.
22:4212–4222. 2003. View Article : Google Scholar :
|
|
4
|
Kuilman T and Peeper DS:
Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev
Cancer. 9:81–94. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
te Poele RH, Okorokov AL, Jardine L,
Cummings J and Joel SP: DNA damage is able to induce senescence in
tumor cells in vitro and in vivo. Cancer Res. 62:1876–1883.
2002.PubMed/NCBI
|
|
6
|
Narita M, Nũnez S, Heard E, Narita M, Lin
AW, Hearn SA, Spector DL, Hannon GJ and Lowe SW: Rb-mediated
hetero-chromatin formation and silencing of E2F target genes during
cellular senescence. Cell. 113:703–716. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vétillard A, Jonchère B, Moreau M, Toutain
B, Henry C, Fontanel S, Bernard AC, Campone M, Guette C and
Coqueret O: Akt inhibition improves irinotecan treatment and
prevents cell emergence by switching the senescence response to
apoptosis. Oncotarget. 6:43342–43362. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jonchère B, Vétillard A, Toutain B, Lam D,
Bernard AC, Henry C, De Carné Trécesson S, Gamelin E, Juin P,
Guette C and Coqueret O: Irinotecan treatment and senescence
failure promote the emergence of more transformed and invasive
cells that depend on anti-apoptotic Mcl-1. Oncotarget. 6:409–426.
2015. View Article : Google Scholar
|
|
9
|
Besson D, Pavageau AH, Valo I, Bourreau A,
Bélanger A, Eymerit-Morin C, Moulière A, Chassevent A,
Boisdron-Celle M, Morel A, et al: A quantitative proteomic approach
of the different stages of colorectal cancer establishes OLFM4 as a
new nonmetastatic tumor marker. Mol Cell Proteomics.
10:M111.0097122011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Campone M, Valo I, Jézéquel P, Moreau M,
Boissard A, Campion L, Loussouarn D, Verriele V, Coqueret O and
Guette C: Prediction of recurrence and survival for triple-negative
breast cancer (TNBC) by a protein signature in tissue samples. Mol
Cell Proteomics. 14:2936–2946. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Persson S, Rosenquist M, Knoblach B,
Khosravi-Far R, Sommarin M and Michalak M: Diversity of the protein
disulfide isomerase family: Identification of breast tumor induced
Hag2 and Hag3 as novel members of the protein family. Mol
Phylogenet Evol. 36:734–740. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Park SW, Zhen G, Verhaeghe C, Nakagami Y,
Nguyenvu LT, Barczak AJ, Killeen N and Erle DJ: The protein
disulfide isomerase AGR2 is essential for production of intestinal
mucus. Proc Natl Acad Sci. 106:6950–6955. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Delom F, Mohtar MA, Hupp T and Fessart D:
The anterior gradient-2 interactome. Am J Physiol Cell Physiol.
318:C40–C47. 2020. View Article : Google Scholar
|
|
14
|
Delom F, Nazaraliyev A and Fessart D: The
role of protein disulphide isomerase AGR2 in the tumour niche. Biol
Cell. 110:271–282. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fessart D, Domblides C, Avril T, Eriksson
LA, Begueret H, Pineau R, Malrieux C, Dugot-Senant N, Lucchesi C,
Chevet E and Delom F: Secretion of protein disulphide isomerase
AGR2 confers tumorigenic properties. Elife. 5:e138872016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fessart D, de Barbeyrac C, Boutin I,
Grenier T, Richard E, Begueret H, Bernard D, Chevet E, Robert J and
Delom F: Extracellular AGR2 triggers lung tumour cell proliferation
through repression of p21CIP1. Biochim Biophys Acta Mol Cell Res.
1868:1189202021. View Article : Google Scholar
|
|
17
|
Maurel M, Obacz J, Avril T, Ding YP,
Papadodima O, Treton X, Daniel F, Pilalis E, Hörberg J, Hou W, et
al: Control of anterior GRadient 2 (AGR2) dimerization links
endoplasmic reticulum proteostasis to inflammation. EMBO Mol Med.
11:e101202019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aberger F, Weidinger G, Grunz H and
Richter K: Anterior specification of embryonic ectoderm: The role
of the Xenopus cement gland-specific gene XAG-2. Mech Dev.
72:115–130. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang JS, Gong A, Cheville JC, Smith DI
and Young CYF: AGR2, an androgen-inducible secretory protein
overexpressed in prostate cancer. Genes Chromosomes Cancer.
43:249–259. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Thompson DA and Weigel RJ: hAG-2, the
human homologue of thexenopus laeviscement gland gene XAG-2, is
coexpressed with estrogen receptor in breast cancer cell lines.
Biochem Biophys Res Commun. 251:111–116. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ramachandran V, Arumugam T, Wang H and
Logsdon CD: Anterior gradient 2 is expressed and secreted during
the development of pancreatic cancer and promotes cancer cell
survival. Cancer Res. 68:7811–7818. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pizzi M, Fassan M, Balistreri M,
Galligioni A, Rea F and Rugge M: Anterior gradient 2 overexpression
in lung adenocarcinoma. Appl Immunohistochem Mol Morphol. 20:31–36.
2012. View Article : Google Scholar
|
|
23
|
Chevet E, Fessart D, Delom F, Mulot A,
Vojtesek B, Hrstka R, Murray E, Gray T and Hupp T: Emerging roles
for the pro-oncogenic anterior gradient-2 in cancer development.
Oncogene. 32:2499–2509. 2013. View Article : Google Scholar
|
|
24
|
Salmans ML, Zhao F and Andersen B: The
estrogen-regulated anterior gradient 2 (AGR2) protein in breast
cancer: A potential drug target and biomarker. Breast Cancer Res.
15:2042013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hrstka R, Nenutil R, Fourtouna A, Maslon
MM, Naughton C, Langdon S, Murray E, Larionov A, Petrakova K,
Muller P, et al: The pro-metastatic protein anterior gradient-2
predicts poor prognosis in tamoxifen-treated breast cancers.
Oncogene. 29:4838–4847. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vanderlaag KE, Hudak S, Bald L,
Fayadat-Dilman L, Sathe M, Grein J and Janatpour MJ: Anterior
gradient-2 plays a critical role in breast cancer cell growth and
survival by modulating cyclin D1, estrogen receptor-alpha and
survivin. Breast Cancer Res. 12:R322010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu D, Rudland PS, Sibson DR,
Platt-Higgins A and Barraclough R: Human homologue of cement gland
protein, a novel metastasis inducer associated with breast
carcinomas. Cancer Res. 65:3796–3805. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hrstka R, Brychtova V, Fabian P, Vojtesek
B and Svoboda M: AGR2 predicts tamoxifen resistance in
postmenopausal breast cancer patients. Dis Markers. 35:207–212.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Z, Zhu Q, Hu L, Chen H, Wu Z and Li D:
Anterior gradient 2 is a binding stabilizer of hypoxia inducible
factor-1a that enhances CoCl 2-induced doxorubicin resistance in
breast cancer cells. Cancer Sci. 106:1041–1049. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Higa A, Mulot A, Delom F, Bouchecareilh M,
Nguyên DT, Boismenu D, Wise MJ and Chevet E: Role of pro-oncogenic
protein disulfide isomerase (PDI) family member anterior gradient 2
(AGR2) in the control of endoplasmic reticulum homeostasis. J Biol
Chem. 286:44855–44868. 2011. View Article : Google Scholar
|
|
31
|
Dimri GP, Lee X, Basile G, Acosta M, Scott
G, Roskelley C, Medrano EE, Linskens M, Rubelj I and Pereira-Smith
O: A biomarker that identifies senescent human cells in culture and
in aging skin in vivo. Proc Natl Acad Sci USA. 92:9363–9367. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vindeløv LL, Christensen IJ and Nissen NI:
A Detergent-trypsin method for the preparation of nuclei for flow
cytometric DNA analysis. Cytometry. 3:323–327. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gillet LC, Navarro P, Tate S, Röst H,
Selevsek N, Reiter L, Bonner R and Aebersold R: Targeted data
extraction of the MS/MS spectra generated by data-independent
acquisition: A new concept for consistent and accurate proteome
analysis. Mol Cell Proteomics. 11:0111.0167172012. View Article : Google Scholar
|
|
34
|
Perez-Riverol Y, Csordas A, Bai J,
Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J,
Mayer G, Eisenacher M, et al: The PRIDE database and related tools
and resources in 2019: Improving support for quantification data.
Nucleic Acids Res. 47:D442–D450. 2019. View Article : Google Scholar :
|
|
35
|
Innes HE, Liu D, Barraclough R, Davies
MPA, O'Neill PA, Platt-Higgins A, de Silva Rudland S, Sibson DR and
Rudland PS: Significance of the metastasis-inducing protein AGR2
for outcome in hormonally treated breast cancer patients. Br J
Cancer. 94:1057–1065. 2006. View Article : Google Scholar
|
|
36
|
Tang W, Zhou M, Dorsey TH, Prieto DA, Wang
XW, Ruppin E, Veenstra TD and Ambs S: Integrated
proteotranscriptomics of breast cancer reveals globally increased
protein-mRNA concordance associated with subtypes and survival.
Genome Med. 10:942018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guillon J, Petit C, Moreau M, Toutain B,
Henry C, Roché H, Bonichon-Lamichhane N, Salmon JP, Lemonnier J,
Campone M, et al: Regulation of senescence escape by TSP1 and CD47
following chemotherapy treatment. Cell Death Dis. 10:1992019.
View Article : Google Scholar :
|
|
38
|
Pohler E, Craig AL, Cotton J, Lawrie L,
Dillon JF, Ross P, Kernohan N and Hupp TR: The barrett's antigen
anterior gradient-2 silences the p53 transcriptional response to
DNA damage. Mol Cell Proteomics. 3:534–547. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hrstka R, Bouchalova P, Michalova E,
Matoulkova E, Muller P, Coates PJ and Vojtesek B: AGR2 oncoprotein
inhibits p38 MAPK and p53 activation through a DUSP10-mediated
regulatory pathway. Mol Oncol. 10:652–662. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Valo I, Raro P, Boissard A, Maarouf A,
Jézéquel P, Verriele V, Campone M, Coqueret O and Guette C: OLFM4
expression in ductal carcinoma in situ and in invasive breast
cancer cohorts by a SWATH-based proteomic approach. Proteomics.
19:e18004462019. View Article : Google Scholar
|
|
41
|
Laplante M and Sabatini DM: MTOR signaling
in growth control and disease. Cell. 149:274–293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang G, Murashige DS, Humphrey SJ and
James DE: A positive feedback loop between akt and mTORC2 via SIN1
phosphorylation. Cell Rep. 12:937–943. 2015. View Article : Google Scholar
|
|
43
|
Sarbassov DD, Guertin DA, Ali SM and
Sabatini DM: Phosphorylation and regulation of Akt/PKB by the
rictor-mTOR complex. Science. 307:1098–1101. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Iannello A, Thompson TW, Ardolino M, Lowe
SW and Raulet DH: p53-dependent chemokine production by senescent
tumor cells supports NKG2D-dependent tumor elimination by natural
killer cells. J Exp Med. 210:2057–2069. 2013. View Article : Google Scholar :
|
|
45
|
Dahal Lamichane B, Jung SY, Yun J, Kang S,
Kim DY, Lamichane S, Kim YJ, Park JH, Jang WB, Ji ST, et al: AGR2
is a target of canonical Wnt/β-catenin signaling and is important
for stemness maintenance in colorectal cancer stem cells. Biochem
Biophys Res Commun. 515:600–606. 2019. View Article : Google Scholar
|
|
46
|
Arumugam T, Deng D, Bover L, Wang H,
Logsdon CD and Ramachandran V: New blocking antibodies against
novel AGR2-C4 4A pathway reduce growth and metastasis of pancreatic
tumors and increase survival in mice. Mol Cancer Ther. 14:941–951.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Armes JE, Davies CM, Wallace S, Taheri T,
Perrin LC and Autelitano DJ: AGR2 expression in ovarian tumours: A
potential biomarker for endometrioid and mucinous differentiation.
Pathology. 45:49–54. 2013. View Article : Google Scholar
|
|
48
|
Hu Z, Gu Y, Han B, Zhang J, Li Z, Tian K,
Young CYF and Yuan H: Knockdown of AGR2 induces cellular senescence
in prostate cancer cells. Carcinogenesis. 33:1178–1186. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hrstka R, Murray E, Brychtova V, Fabian P,
Hupp TR and Vojtesek B: Identification of an AKT-dependent
signalling pathway that mediates tamoxifen-dependent induction of
the pro-metastatic protein anterior. Cancer Lett. 333:187–193.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu Q, Li Y and Yao L: Knockdown of AGR2
induces cell apoptosis and reduces chemotherapy resistance of
pancreatic cancer cells with the involvement of ERK/AKT axis.
Pancreatology. 18:678–688. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dong A, Gupta A, Pai RK, Tun M and Lowe
AW: The human adenocarcinoma-associated gene, AGR2, induces
expression of amphiregulin through hippo pathway co-activator YAP1
activation. J Biol Chem. 286:18301–18310. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Matoulkova E, Sommerova L, Pastorek M,
Vojtesek B and Hrstka R: Regulation of AGR2 expression via 3′UTR
shortening. Exp Cell Res. 356:40–47. 2017.PubMed/NCBI
|
|
53
|
Tiemann K, Garri C, Lee SB, Malihi PD,
Park M, Alvarez RM, Yap LP, Mallick P, Katz JE, Gross ME and Kani
K: Loss of ER retention motif of AGR2 can impact mTORC signaling
and promote cancer metastasis. Oncogene. 38:3003–3018. 2019.
View Article : Google Scholar
|
|
54
|
Dumartin L, Alrawashdeh W, Trabulo SM,
Radon TP, Steiger K, Feakins RM, di Magliano MP, Heeschen C,
Esposito I, Lemoine NR and Crnogorac-Jurcevic T: ER stress protein
AGR2 precedes and is involved in the regulation of pancreatic
cancer initiation. Oncogene. 36:3094–3103. 2017. View Article : Google Scholar :
|
|
55
|
Zhao F, Edwards R, Dizon D, Afrasiabi K,
Mastroianni JR, Geyfman M, Ouellette AJ, Andersen B and Lipkin SM:
Disruption of Paneth and goblet cell homeostasis and increased
endoplasmic reticulum stress in Agr2-/-mice. Dev Biol. 338:270–279.
2010. View Article : Google Scholar
|