Introduction
Colorectal cancer (CRC) was the third most
widespread cancer and the second most deadly cancer in 2018,
worldwide. There were 1,800,977 new cases of CRC and 861,663
related deaths in 2018 (1,2).
In the past few decades, improved treatment options have become
available, including surgery, radiotherapy, chemotherapy and
molecular-targeted therapy for advanced CRC (3,4).
However, the 5-year survival rate of CRC is <65% due to cancer
relapse (2).
MicroRNAs (miRNAs) are short (18-25 nucleotides)
non-coding RNAs that mainly bind to the 3′-untranslated region
(3′UTR) of target mRNAs, contributing to mRNA cleavage or
translational suppression (5,6).
miRNAs play an important role in numerous biological processes,
such as immune response (7),
neurogenesis (8) and insulin
secretion (9). In addition,
miRNAs contribute to various processes in cancer, including tumor
growth, apoptosis, invasion and survival of patients, which are
closely related to oncogenesis and tumor progression (10,11).
Previous studies have demonstrated that the
pathological mechanisms underlying CRC depend on a variety of
signaling pathways, including Wnt/β-catenin, epidermal growth
factor receptor (EGFR), transforming growth factor-β (TGF-β), tumor
protein p53 and epithelial-to-mesenchymal transition (EMT)
(12-16). Furthermore, miRNAs have been
determined to play a pivotal role in regulating these pathways. For
example, miR-4689 exerts an anti-tumor effect on mutant kirsten rat
sarcoma virus (KRAS) in CRC by inhibiting the EGFR and AKT pathway
(17). MIRTX, a byproduct of
miR-29b-1-5p suppresses the NF-κB signaling pathway in KRAS-mutated
CRC cells by directly binding to CXC chemokine receptor 2 and
phosphatidylinositol 3-kinase regulatory subunit alpha mRNA
(18). In addition, miR-34a can
inhibit cell proliferation and increase the expression of
p21WAF1/CIP1 in HCT116 and RKO CRC cells (19). It has been recently reported that
miR-4711-5p regulates cancer stemness and the cell cycle in CRC
cells by targeting Kruppel-like factor 5, mouse double minute 2
homolog and transcription factor Dp-1 (20).
Cancer stem cells (CSCs) have distinct capability
for self-renewal and multi-potential differentiation (21). CSCs confer resistance to
anticancer drugs and radiotherapy, and thus cause distant
metastases and recurrence (22-24). Several CSC markers for CRC have
been identified, including CD133, B cell-specific Moloney murine
leukemia virus integration site 1 (BMI1) and leucine rich repeat
containing G protein-coupled receptor 5 (23,25,26). Previously, doublecortin-like
kinase 1 (DCLK1) was demonstrated to be a CSC marker in CRC
(27,28). DCLK1 belongs to the protein kinase
superfamily and is overexpressed in several human malignancies,
including colorectal, pancreas and kidney cancer (29-31). Our preliminary data also
demonstrated that miR-1291 exhibited a potent growth inhibitory
effect on pancreatic cancer stem-like cells in which the ornithine
decarboxylase (ODC) degron was transduced (32-34).
It has been reported that miR-1291 sensitizes cells
to doxorubicin by suppressing multidrug resistance-associated
protein 1 (35). Additionally,
miR-1291 is considered to be a candidate biomarker for the
diagnosis of acute myocardial infarction (36) and Bullous Pemphigoid (37), and is also a marker for the
prediction of severe symptoms in SARS-CoV-2 infection (38). In relation to human cancers,
miR-1291 has been reportedly upregulated in liver cirrhosis and
hepatocellular carcinoma (39).
In addition, miR-1291 is a biologically relevant regulator of
glypican-3 expression in hepatoma cells and acts by silencing the
endoplasmic reticulum (ER) stress sensor, inositol-requiring
transmembrane kinase/endoribonuclease 1α (40). Several studies have demonstrated
that miR-1291 inhibits cell proliferation and tumorigenesis, and
sensitizes pancreatic cancer cells to chemotherapy (41-43). Furthermore miR-1291 is reportedly
downregulated in kidney, esophagus and prostate carcinoma, serving
anti-tumor effects (44-46). The target molecules for miR-1291
include solute career family 2 member 1/glucose transporter 1 in
renal cancer cells, mucin 1 (MUC1) in human esophagus cancer cells
(44,45) and forkhead box protein A2-anterior
gradient 2 pathway in PANC-1 pancreatic cancer cells (43). Furthermore, miR-1291 has been
demonstrated to inhibit cell growth and tumorigenesis in prostate
cancer by binding to Mediator Complex Subunit 1 (46).
Detailed functional assessments of miR-1291 have not
yet been conducted in CRC. However, Salehi et al (47) reported that miR-1291 levels were
slightly increased in liver metastasis compared with primary CRC by
using the NCBI Gene Expression Omnibus (GEO) database (47). Therefore, the aim of the present
study was to clarify the anti-tumor effect of miR-1291 in CRC
cells, partially including DCLK1 regulation.
Materials and methods
Cell lines and cell culture
Human CRC cell lines (CACO-2, COLO205, DLD-1,
HCT116, LoVo, RKO and SW480), the human pancreatic cancer cell line
Panc-1, and non-tumor cell lines (293, CCD-18Co and MRC5) were
purchased from the American Type Culture Collection. The HT29 cell
line (cat. no. KBN0398_01) was obtained from Japanese Collection of
Research Bioresources Cell Bank. Short tandem repeat (STR)
profiling for authentication indicated that the HT29 cell line was
the same as the one registered with the American Type Culture
Collection (HTB-38 HT-29: human adenocarcinoma; Colorectal). KM12SM
(48) was a gift from Professor
Toshinari Minamoto (Cancer Research Institute, Kanazawa University,
Kanazawa, Japan). These cell lines were authenticated by
morphological inspection, STR profiling and mycoplasma testing.
Cells were cultured either in RPMI 1640 medium or in DMEM (each,
Nissui Pharmaceutical Co., Ltd.) supplemented with 10% FBS (Biowest
SAS), 100 U/ml penicillin and 100 µg/ml streptomycin
(Nacalai Tesque, Inc.). Cells were cultured in a humidified
incubator at 37°C in an atmosphere containing 5%
CO2.
Retroviral transduction of the degron
reporter into pancreatic cancer Panc-1 cells
The degron sequence of ornithine decarboxylase (ODC)
is recognized directly by proteasomes, which leads to the
destruction of the involved protein. The retroviral expression
vector pQCXIN-ZsGreen-cODC, containing green fluorescence
ZsGreen-labeled degron ODC (Gdeg) was kindly provided by Dr Frank
Pajonk (Jonsson Comprehensive Cancer Center, UCLA, CA, USA). The
plasmid was transfected into Platinum retroviral packaging cells
using Lipofectamine® 2000 (Thermo Fisher Scientific,
Inc.). The plasmid and Lipofectamine® 2000 were diluted
with Opti-MEM I reduced serum medium (Thermo Fisher Scientific,
Inc.) for 5 min at room temperature, separately. The diluted
Lipofectamine® 2000 was subsequently mixed with the
diluted plasmid and incubated for 15 min at room temperature. The
mixture was then added to cells immediately at room temperature,
after which the cells were incubated at 37°C. And the retrovirus
collected from the supernatant was used for Panc-1 cell infection
as previously described (32-34). Stable transfectants were selected
with G418 solution (Sigma-Aldrich; Merck KGaA) and maintained in
0.1 mg/ml G418 solution.
Clinical tissue samples
When the tumor diameter >3 cm, paired clinical
tissue specimens (normal mucosa and CRC tissue) were randomly
collected from 20 patients (10 males and 10 females; age range,
18-87 years) who had surgery for colorectal cancer at Osaka
University Hospital (Osaka, Japan) between October 2016 and April
2017. All tissue specimens were stored at -80°C until RNA
extraction. This study was performed in accordance with the
Declaration of Helsinki. All patients provided written informed
consent, in accordance with the guidelines approved (approval no.
08226) by the Institutional Research Board of the institute. The
present study was conducted under the supervision of and approved
by the Ethics Board of Osaka University Hospital.
miRNA, antagomir, siRNA and plasmid
transfection
Mimic-hsa-miR-1291 (miR-1291) sense (5′-UGG CCC UGA
CUG AAG ACC AGC AGU-3′) and antisense (5′-ACU GCU GGU CUU CAG UCA
GGG CCA-3′) sequences, along with the negative control miR (miR-NC)
sense (5′-AUC CGC GCG AUA GUA CGU A-3′) and antisense (5′-UAC GUA
CUA UCG CGC GGA U-3′) sequences were designed and synthesized by
Ajinomoto Bio-Pharma. Hsa-miR-1291 inhibitor S-Tud (antagomir-1291)
sense [5′-GAC GGC GCU AGG AUC AUC AAC ACU GCU GGU CUU CAG UCA GGG
CCA CAA GUA UUC UGG U-3′] and antisense [5′-ACC AGA AUA CAA CAC UGC
UGG UCU UCA GUC AGG GCC ACA AGA UGA UCC UAG CGC CGUC-3′] were
2-OMethyl modified, and were also designed and synthesized by
Ajinomoto Bio-Pharma. Three small interfering RNAs (siRNAs)
targeting DCLK1 were obtained from Thermo Fisher Scientific, Inc.
(Assay IDs: s17584, s17585 and s17586). Cells were transfected with
miRNAs, antagomir, and siRNAs at final concentration of 30-50 nM
and plasmids at concentration of 50 ng per well (96-well plate) or
1 µg per well (six-well plate) using
Lipofectamine® 2000 or Lipofectamine® RNAiMAX
(both from Thermo Fisher Scientific, Inc.) The nucleotide (miRNAs,
antagomir, siRNAs and plasmids) and transfection reagent
(Lipofectamine® 2000 or Lipofectamine®
RNAiMAX) were diluted with serum free medium (RPMI-1640 or DMEM)
for 5 min at room temperature, separately. The diluted transfection
reagent was subsequently mixed with the diluted nucleotide and
incubated for 15 min at room temperature. The mixture was then
added to cells immediately at room temperature, after which the
cells were incubated at 37°C. The subsequent experiments were
performed at 4 and 24 h after transfection for miRNA uptake into
cells and from 24 to 72 h for various anti-tumor feature
assessments.
RNA isolation
Total RNA was collected from each cell line using
TRIzol® Reagent (Thermo Fisher Scientific, Inc.)
followed by phenol-chloroform extraction and ethanol precipitation.
Subsequently, miRNA was collected from tissue specimens and
cultured cells using the miRNeasy kit (Qiagen GmbH) according to
the manufacturer.s protocol. Total RNA concentration and purity
were measured using a NanoDrop one spectrophotometer (Thermo Fisher
Scientific, Inc.) at 260 and 280 nm (A260/280) wavelengths.
Reverse transcription-quantitative (RT-q)
PCR analysis of mRNA
A High Capacity cDNA Reverse Transcription kit
(Thermo Fisher Scientific, Inc.) was used to synthesize
complementary DNA from 2.5 µg of total RNA according to the
manufacturer.s protocol. qPCR for DCLK1, BMI1 and CD133 RNA was
performed using oligonucleotide primers and the LightCycler 480
Real-Time PCR system (Roche Diagnostics). The amplification
products were detected using the THUNDERBIRD SYBR qPCR Mix (Toyobo
Life Science), and the level of target gene expression was
calculated. The qPCR conditions were as follows: 95°C for 30 sec;
followed by 40 cycles of 95°C for 10 sec, 60°C for 10 sec and 72°C
for 30 sec. The expression of the target gene was normalized to
endogenous GAPDH expression. Relative expression was quantified by
the 2−ΔΔCq method (49). The PCR primers are listed in
Table SI.
RT-qPCR analysis of miRNA
The TaqMan MicroRNA Reverse Transcription kit
(Thermo Fisher Scientific, Inc.) was used to synthesize the
complementary DNA from 25 ng of total RNA according to the
manufacturer.s protocol. qPCR of miRNA was then performed using
TaqMan Universal PCR Master Mix, No AmpErase UNG (Thermo Fisher
Scientific, Inc.) with a 7900 HT Sequence Detection System (Thermo
Fisher Scientific, Inc.). RNU6B was used as the endogenous control.
The primers for miR-1291 and RNU6B were designed by Thermo Fisher
Scientific, Inc. (miR-1291, Assay ID: 002838; RNU6B, Assay ID:
001093; cat. no. 4427975). The qPCR conditions were as follows:
95°C for 10 min; followed by 45 cycles of 95°C for 15 sec, and 60°C
for 1 min and 72°C for 1 sec; cooling to 40°C for 30 sec. Relative
expression was quantified with the 2−ΔΔCq method.
Cell viability assay
Cells were seeded in 96-well plates at a density of
4,000-8,000 cells per well and were transfected with miR-NC or
miR-1291 or antagomir-1291 or DCLK1-siRNA at a final concentration
of 30 nM as aforementioned, the second day after seeding. A total
of 24, 48 and 72 h after transfection, 10 µl Cell Counting
Kit-8 (Dojindo Molecular Technologies, Inc.) solution was added to
each well, after which the 96-well plates were kept in dark for 2 h
at 37°C. The absorbance was then detected using a Multiskan Go
plate reader (Thermo Fisher Scientific, Inc.). The differences in
absorption at 630 and 450 nm wavelengths were then subtracted and
used to determine cell viability.
Matrigel invasion assay
Corning BioCoat Matrigel Invasion Chambers (pore
size: 8.0 µm; Corning, Inc.; cat. no. 354480) were used. The
upper chamber, which was pre-coated with Matrigel, was rehydrated
with culture medium (RPMI-1640 or DMEM) for 2 h at 37°C in 5%
CO2 before seeding the cells. The medium (RPMI-1640 or
DMEM) was then removed, and DLD-1, HT29 and HCT116 cells were
seeded into the upper chambers at a density of 1-2×105
cells per chamber with medium (RPMI-1640 or DMEM) containing 0.1%
bovine serum albumin. Medium (RPMI-1640 or DMEM) containing 10% FBS
were put in the lower wells. The cells were transfected with the
miRNAs or antagomir or DCLK1-siRNA at a final concentration of 50
nM and incubated at 37°C after transfection. After incubation for
48, 72 and 96 h, cells passing through the Matrigel were fixed with
10% formalin for 1 h at room temperature and then stained with
hematoxylin for 1 h at room temperature. To count cells passing
through the Matrigel, images were captured using a bright field
light microscope (CKX53; Olympus Corporation) with Visualix camera
(Visualix, Corporation) at a magnification of ×200.
Gap closure assay
Ibidi culture inserts (8.4 width ×8.4 length ×5 mm
height; Ibidi GmbH) with two 70 µl wells were put on the
24-well plates before cell seeding. DLD-1, HT29 and HCT116 cell
suspensions (70 µl) were then added into each 70 µl
well at a density of 2×105, 2.5×105 and
8×105 cells per ml, respectively. The inserts were
removed after 24 h to create gap with an area of 6×105
pixel2. After that, the miRNAs or antagomir or
DCLK1-siRNA were transfected at a final concentration of 30 nM. To
enhance gap closure, cells were cultured in the medium (RPMI-1640
or DMEM) supplemented with 10% FBS as previously described
(50,51). At 24 and 48 h after transfection,
images were captured by a bright field light microscope (CKX53;
Olympus Corporation) with Visualix camera (Visualix, Corporation)
at a magnification of ×100. The areas of the gaps were measured
using ImageJ 1.52v software (National Institutes of Health).
Colony formation assay
Cells were seeded in a six-well plate at a density
of 1×105 cells per well, incubated at 37°C overnight and
transfected with miR-NC or miR-1291 at a final concentration of 30
nM for 8 h. Samples were then reseeded in six-well plates at a
density of 500 cells per well. A colony was considered to consist
of ≥50 cells. Cells were incubated for 10 days at 37°C after
transfection, after which cells were fixed with 100% methanol for
30 min at room temperature, and stained by 0.5% crystal violet for
10 min at room temperature for counting. The images of each well
were scanned using an Epson scanner GT-X970 (Seiko Epson
Corporation), and the colonies were counted using ImageJ 1.52v
software (National Institutes of Health).
Cell cycle assay
DLD-1, HT29 and HCT116 cells were seeded to six-well
plates at a density of 3×105, 4×105 and
3.5×105 cells per well, respectively. Cells were starved
in serum-free medium (RPMI-1640 or DMEM) for 48 h. A total of 24 h
before the end of starvation, miR-NC or miR-1291 was transfected at
a final concentration of 30 nM. Cells were collected at the
indicated times (0, 12, 24 and 48 h) and fixed in 70% ethanol for
30 min at 4°C. After fixation, cells were washed twice with PBS and
incubated with RNase (Sigma Aldrich; Merck KGaA) for 20 min at
37°C. Cells were treated with propidium iodide (PI; Dojindo
Molecular Technologies, Inc.) for 20 min on ice and analyzed by
flow cytometry (Spectral Analyzer SA3800; Sony Biotechnology, Inc.)
with SA3800 2.0 software (Sony Biotechnology, Inc.).
Bromodeoxyuridine (BrdU) Proliferation
assay
Cell proliferation assays were performed using
CysLex Cellular BrdU ELISA kit Ver.2. (Medical & Biological
Laboratories Co., Ltd.). Cells were cultured with BrdU labeling
reagent at 10 µM for 2 h at 37°C, then incubated with 50
µl per well of anti-BrdU monoclonal antibody (provided in
the kit) for 1 h at room temperature, followed by 50 µl per
well of secondary antibody reaction with HRP-conjugated anti-mouse
IgG (provided in the kit), for 1 h at room temperature. After the
addition of the substrate reagent (provided in the kit), the
absorbance in each well was measured using a Multiskan Go
microplate spectrophotometer (Thermo Fisher Scientific, Inc.) at
dual wavelengths of 450/540 nm.
Annexin V apoptosis assay
Apoptotic cells were assessed using an Alexa Fluor
488. Annexin V/Dead Cell Apoptosis kit (Thermo Fisher Scientific,
Inc.). A total of 2×105 cells were diluted with 100
µl 1X annexin-binding buffer, after which 5 µl Alexa
Flour 488 Annexin V and 1 µl 100 µg/ml PI was added.
Samples were subsequently incubated for 15 min at room temperature.
A total of 400 µl 1X annexin-binding buffer was added to
each cell suspension and apoptotic cells were counted by flow
cytometry using Spectral Analyzer SA3800 (Sony Biotechnology, Inc.)
with SA3800 2.0 software (Sony Biotechnology, Inc.).
Western blot analysis
Cells were seeded in six-well plates at a density of
1×105-2×105 per well and transfected with
miR-NC, miR-1291 or DCLK1-siRNA at a final concentration of 30-50
nM. After 48 and 72 h, cells were rinsed twice with PBS and lysed
by RIPA buffer (0.05 M Tris-HCl, pH 7.6, 0.15 M NaCl, 1% Nonidet
P40, 0.5% sodium deoxycholate, 0.1% SDS) with 1% proteinase
inhibitor cocktail (Nacalai Tesque, Inc.). The protein samples (30
µg/lane) were electrophoresed by SDS-PAGE using 10, 13 or
15% acrylamide gel and transferred to PVDF transfer membranes
(Bio-Rad Laboratories, Inc.). The membranes were blocked with 5%
non-fat dry milk (Cell Signaling Technology, Inc.) in TBS with
Tween-20 (TBS-T; 50 mM Tris, 158 mM NaCl, 2.7 mM KCl, pH 7.5, 0.1%
Tween-20) for 1 h at room temperature and incubated with the
following primary antibodies overnight at 4°C: Anti-ACTB (1:4,000;
Rabbit mAb, cat. no. 4970; Cell Signaling Technology, Inc.), and
anti-DCLK1 (1:2,000; cat. no. ab31704; Abcam)
anti-p21WAF1/CIP1 (1:1,000; cat. no. ab80633; Abcam),
anti-p27KIP1 (1:1,000; sc-528, Santa Cruz Biotechnology,
Inc.), anti-CD133 (1:1,000; ab216323; Abcam), anti-CDC25A (1:1,000;
cat. no. 3652; Cell Signaling Technology, Inc.), anti-CDC25B
(1:1,000; cat. no. 9525; Cell Signaling Technology, Inc.),
anti-CDC25C Rabbit mAb (1:1,000; cat. no. 4688; Cell Signaling
Technology, Inc.), anti-CDK4 (1:1,000; cat. no. MAB8879;
MilliporeSigma), anti-CDK6 (1:1,000; cat. no. SAB4300596;
Sigma-Aldrich; Merck KGaA), anti-Cyclin D1 (1:1,000; cat. no. 2922;
Cell Signaling Technology, Inc.), anti-Cyclin E1 (1:1,000; cat. no.
sc-247; Santa Cruz Biotechnology, Inc.), anti-retinoblastoma (Rb;
1:1,000; cat. no. ab24; Abcam), anti-cdc2 (1:1,000; cat. no. 77055;
Cell Signaling Technology, Inc.) and anti-phosphorylated (p)-cdc2
[(Tyr15); 1:1,000; cat. no. 9111; Cell Signaling Technology,
Inc.)]. Subsequently, the membranes were incubated with secondary
antibodies, including HRP anti-mouse IgG (1:3,000; cat. no. NA931;
GE Health Care Life Sciences) or anti-rabbit IgG antibodies
(1:3,000; cat. no. NA934; GE Healthcare Life Sciences) for 1 h at
room temperature. The bands were visualized by the ECL Detection
System (GE Healthcare Life Sciences) and analyzed using ImageJ
1.52v software (National Institutes of Health).
pmirGLO plasmid vector construction
The 3′UTR of DCLK1 mRNA was amplified by PCR using
the following primer sequences (amplified product size, 211 bp):
forward, 5′-GCT CGC TAG CCT CGA GCT AGT GTA CTG AGC CTGCG G-3′ and
reverse, 5′-ATG CCT GCA GGT CGA CTG ACT GGT CAC ATT CCA CTG-3′. The
amplified products were subcloned and ligated into the multicloning
site between SalI and XhoI in the pmirGLO
Dual-Luciferase miRNA Target Expression Vector (Promega
Corporation) using the In-Fusion HD Cloning kit (Clontech
Laboratories, Inc.) The vectors with mismatched 3′UTR sequences
were constructed using the QuikChange Site Directed Mutagenesis kit
(Agilent Technologies, Inc.) according to the manufacturer.s
protocol. The PCR primers for mutated type and deleted type
plasmids are listed in Table SI.
The entire sequence (insert and vector) was confirmed by Sanger
sequencing (outsourced to Genome Information Research Center, Osaka
University, Suita, Osaka).
Luciferase reporter assay
Cells were seeded in 96-well plates at a density of
1×104 cells per well and transfected with 50 ng of DCLK1
wild type, 2-nucleotide mutated type (Mut) or 3-nucleotide deleted
type (Del) 3′UTR containing pmirGLO Dual-Luciferase miRNA Target
Expression Vector (aforementioned) using Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.). Additionally, 50 nM of
either miR-NC sense (5′-AUC CGC GCG AUA GUA CGU A-3′) and antisense
(5′-UAC GUA CUA UCG CGC GGA U-3′) sequences or miR-1291 sense
(5′-UGG CCC UGA CUG AAG ACC AGC AGU-3′) and antisense (5′-ACU GCU
GGU CUU CAG UCA GGG CCA-3′) sequences, which were designed and
synthesized by Ajinomoto Bio-Pharma, were transfected using
Lipofectamine® RNAiMAX (Thermo Fisher Scientific, Inc.).
After 24 h of transfection, cells were assayed for both firefly and
Renilla luciferase using the Dual-Luciferase Reporter Assay
System (Promega Corporation).
Transient overexpression of DCLK1 in
HCT116
Total complementary DNA (cDNA) was synthesized using
the High Capacity cDNA Reverse Transcription kit (Thermo Fisher
Scientific, Inc.) from total RNA, which was obtained from the SW480
CRC cell line as aforementioned, and performed in accordance with
the manufacturers protocol. The coding sequence of DCLK1 (NCBI
Reference Sequence: NM_001195416.2) was amplified from total cDNA
via PCR using the KOD FX Neo (Toyobo Life Science). The PCR
conditions were as follows: 94°C for 2 min; followed by 40 cycles
of 60°C for 10 sec, 62°C for 30 sec and 68°C for 60 sec. The
concentration and purity of DCLK1 cDNA were measured using a
NanoDrop one spectrophotometer (Thermo Fisher Scientific, Inc.) at
260 and 280 nm (A260/280) wavelengths. The primer sequences
(amplified product size, 1302 bp) were as follows:
BamHI_DCLK1_forward, 5′-TAC CGA GCT CGG ATC CAT GTT AGA ACT CAT AGA
AGT TA-3′ and reverse, 5′-GAT ATC TGC AGA ATT CTT AAA AGG GCG AGT
TAG GG-3′. The amplified product was subcloned and ligated into the
multicloning site between BamHI and EcoRI in the
pcDNA3.1 plasmid (Thermo Fisher Scientific, Inc.) using the
In-Fusion HD Cloning kit (Clontech Laboratories, Inc.). The
DCLK1-inserted vector or empty vector was transfected to HCT116
cells by Lipofectamine® 2000.
ShDCLK1 HCT116 clones
Sh (short hairpin)-DCLK1 HCT116 clones were
generated as previously described (29). ShDCLK1 #1 (Clone ID:
TRCN0000002145) targeted the sequence 5′-GAA CTG TAT CTT GTC ATG
GAA-3′. ShDCLK1 #2 (Clone ID: TRCN0000002146) targets the sequence
5′-CAG GTA TCT TTG TAG CGG TTT-3′.
Sphere formation assay
HCT116 cells were seeded in six-well plates at a
density of 1×105 cells per well, incubated at 37°C
overnight, and transfected with miR-NC or miR-1291 or DCLK1-siRNA
at a final concentration of 50 nM. After 24 h of transfection,
single cells were reseeded in 96-Well Clear Ultra Low Attachment
Microplates (Corning, Inc.) at a density of 1,000 cells per well.
The cells were cultured in DMEM/F-12 serum-free medium (Thermo
Fisher Scientific, Inc.) supplemented with 20 ng/ml epithelial
growth factor, 10 ng/ml basic fibroblast growth factor-2
(PeproTech, Inc.), 100 U/ml penicillin and 100 µg/ml
streptomycin. Cells were cultured in a humidified incubator at 37°C
and 5% CO2. Images were captured by a bright field light
microscope (CKX53; Olympus Corporation) with Visualix camera
(Visualix, Corporation). The number of spheres was counted manually
on days 4 or 7 after reseeding.
Flow cytometric analysis
HCT116 cells were seeded in six-well plates at a
density of 1×105 cells per well, incubated at 37°C
overnight and transfected with miR-NC or miR-1291 at a final
concentration of 50 nM. For CD133 marker expression, after 48 h of
transfection, cells were resuspended and one million cells were
incubated with antibodies against human CD133 (1:50;
APC-conjugated; cat. no. 130-113-106; Miltenyi Biotec GmbH) on ice
for 20 min in the dark. Samples were then washed twice with PBS
containing 2% FBS. For CD166 marker expression, after 72 and 96 h
of transfection, the cells were resuspended and one million cells
were incubated with PE Mouse Anti-Human CD166 antibody (1:6.7; cat.
no. 559263; BD Biosciences on ice for 20 min in the dark. Samples
were then washed twice with PBS containing 2% FBS. The Spectral
Analyzer SA3800 (Sony Biotechnology, Inc.) with SA3800 2.0 software
(Sony Biotechnology, Inc.) was used for flow cytometric analyses.
Dead cells were excluded by utilizing forward and side scatter.
In vivo experiments
DLD-1 cells were mixed with Matrigel (Corning, Inc.)
and RPMI-1640 at a 1:1 ratio (vol:vol). Subsequently,
~2×106 cells in 100 µl RPMI-1640/Matrigel
solution were injected subcutaneously into both sides of the lower
back regions of 17 4-week-old female nude mice (CLEA Japan, Inc.).
The mice were divided randomly into a parent group (n=5), a miR-NC
group (n=6) and a miR-1291 group (n=6) for the evaluation of
anti-tumor growth effects and safety. After tumor volumes reached
80 mm3, miRNA was formulated with super carbonate
apatite (sCA), which was intravenously administered as the vehicle
via the tail vein at a dose of 40 µg per injection as
previously described (17,18,20,52-55).
Mice were treated eight times with formulated miR-NC or miR-1291
over 2 weeks. The tumors were resected on day 14. Tumor volumes
were determined as previously described (17,18). The animal facility was specific
pathogen free and was kept at 20-24°C with 40-60% humidity. The
dark/light cycle was 12/12 h. All animals could access food and
water ad libitum. All animal experiments were performed in
accordance with currently prescribed guidelines and the Animal
(Scientific Procedures) 1986 Act. Regarding the tumor burden, a
marked increase in tumor size (≥10% body weight) was applied as one
a humane endpoints according to Guidelines for Proper Conduct of
Animal Experiments by Science Council of Japan in 2006 (56). The present study was approved
(approval no. 13377-5) by the Ethics Board of Osaka University
(Osaka, Japan). Physical methods of euthanasia were applied. Thus,
mice were anesthetized by isoflurane (4-5%), followed by immediate
incision in the abdomen, and successive cut of diaphragm and the
post caval vein. After that, the death of mice was verified by
assessing the observation of respiratory arrest, cessation of heart
beat (lack of activity for ≥5 min) (56) and pupillary response to light
(57). The weight of the animals
ranged from 15.6 to 20.3 g/mouse on day 0, and at the time of
sacrifice (on day 14) ranged from 16.2 to 21.8 g/mouse.
In silico analysis
TargetScan human version 7.2 (http://www.targetscan.org/vert_72/), miRwalk
(http://mirwalk.umm.uni-heidelberg.de/), miRabel
(http://bioinfo.univ-rouen.fr/mirabel/view/result.php?page=mir),
and miRmap (https://mirmap.ezlab.org/app/) were utilized to search
and crosscheck the target candidates of miR-1291. The tissue atlas
database (https://ccb-web.cs.uni-saarland.de/tissueatlas/patterns)
was used to examine basal expression of miRNAs in normal
tissues.
Statistical analysis
Data are presented as the mean ± SEM. Statistical
analyses were performed using GraphPad Prism 5 (GraphPad Software,
Inc.) and Microsoft Excel (Microsoft Corporation). Statistical
differences between the miR-NC and miR-1291 groups were analyzed by
Student.s t-test (two-tailed, unpaired). The expression levels of
miRNAs in normal and cancer colorectal tissues were analyzed using
the Wilcoxon signed-rank test (two-tailed, paired). In Table SII, Student.s t-test (two-tailed,
unpaired) was used for analyzing the statistical differences of age
and tumor size, and Fisher.s exact test was used for other
clinicopathological characteristics of tumors. P<0.05 was
considered to indicate a statistically significant difference. To
perform statistical evaluation, each experiment, except for western
blot analysis which was performed twice, was performed three
times.
Results
Screening of candidate miRNAs
Firstly, 1,749 miRNAs that targeted DCLK1 were
identified using TargetScan Human. Using the Ingenuity Pathway
Analysis miRNA Target Filter, candidate miRNAs whose target genes
were associated with Notch Signaling, Wnt/β-catenin signaling or
Wnt/Ca2+ signaling pathway were screened. Eventually, 30
candidate miRNAs were selected for cell viability assessment
(Fig. S1A). For the assessment,
a CSC model was constructed by transducing ODC-degron to the
pancreatic cancer Panc-1 cells as previously described (32-34) to test the effects of these miRNAs
on stem [degron (+)] cells and non-stem [degron (-)] cells.
miR-34a, which reached phase I clinical trial as a therapy for
human solid tumors (5), was used
as a therapeutic control in this experiment. Among the 30 miRNAs,
miR-1291 (the 16th miRNA) markedly suppressed both the stem [degron
(+)] and non-stem [degron (-)] viability of Panc-1 cells compared
with either miR-NC or positive control miR-34a (Fig. S1B). This result is consistent
with previous studies reporting that miR-1291 presented anti-tumor
function in pancreatic cancer (42,43). However, functional assessment of
miR-1291 in CRC has not been conducted, and therefore it was
investigated in the present study whether miR-1291 would be a
therapeutic option against CRC.
Expression of miR-1291 in CRC cells and
clinical tissue specimens
The expression of miR-1291 was evaluated in nine CRC
cell lines and three non-tumor cell lines (293, CCD-18Co and MRC5)
by RT-qPCR (Fig. 1A). The
expression of miR-1291 in the nine CRC cell lines was generally
lower when compared with the normal colon tissue cell lines.
According to the tissue atlas database, miR-1291 levels in normal
colon tissues is 1/50 and 1/1,500 of putative anti-oncomiR
miR-34a-5p and oncomiR miR-21-5p, respectively (Fig. S2). The expression levels of
miR-1291 in normal mucosa and CRC tissues from 20 paired clinical
tissue specimens were examined in the present study. The results
revealed that miR-1291 expression was significantly lower in tumor
tissues compared with normal mucosa (Fig. 1B). When the clinicopathological
characteristics of tumors were assessed, there was no significant
difference of the clinicopathological characteristics between high
and low miR-1291 levels (Table
SII).
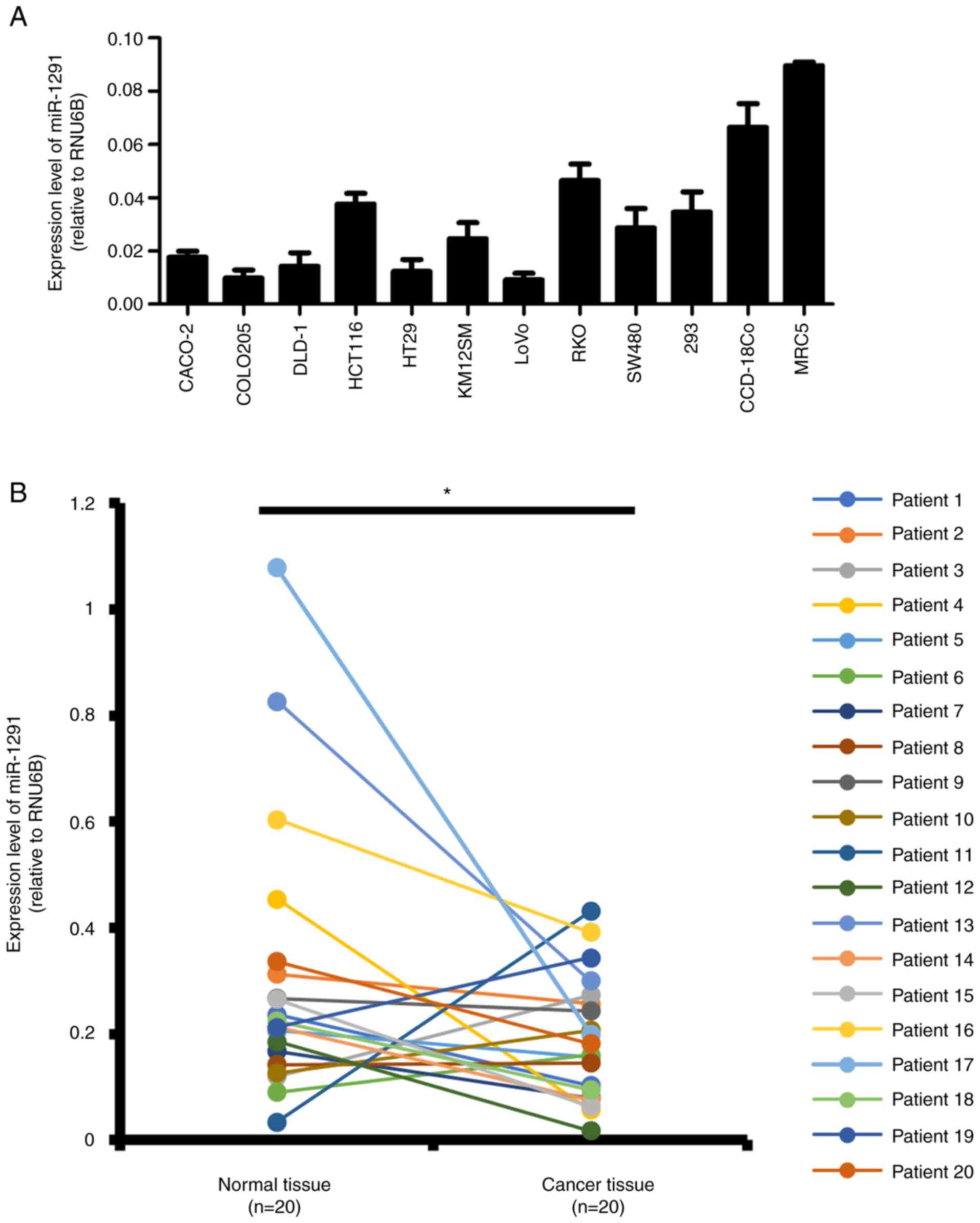 | Figure 1Expression levels of miR-1291. (A)
CRC cell lines and non-tumor cell lines. Reverse
transcription-quantitative PCR was used to detect the expression of
miR-1291 in CRC cell lines (CACO-2, COLO205, DLD-1, HCT116, HT29,
KM12SM, LoVo, RKO and SW480) and human non-tumor cell lines (293,
CCD-18Co and MRC5). RNU6B was used as a loading control. All
experiments were performed in triplicate. All data are presented as
the mean ± SEM. (B) Expression levels of miR-1291 were detected in
CRC tissues and corresponding normal colon mucosa (n=20). Different
colored lines represent the individual patient analyzed.
*P<0.05 as indicated. CRC, colorectal cancer; miR,
microRNA. |
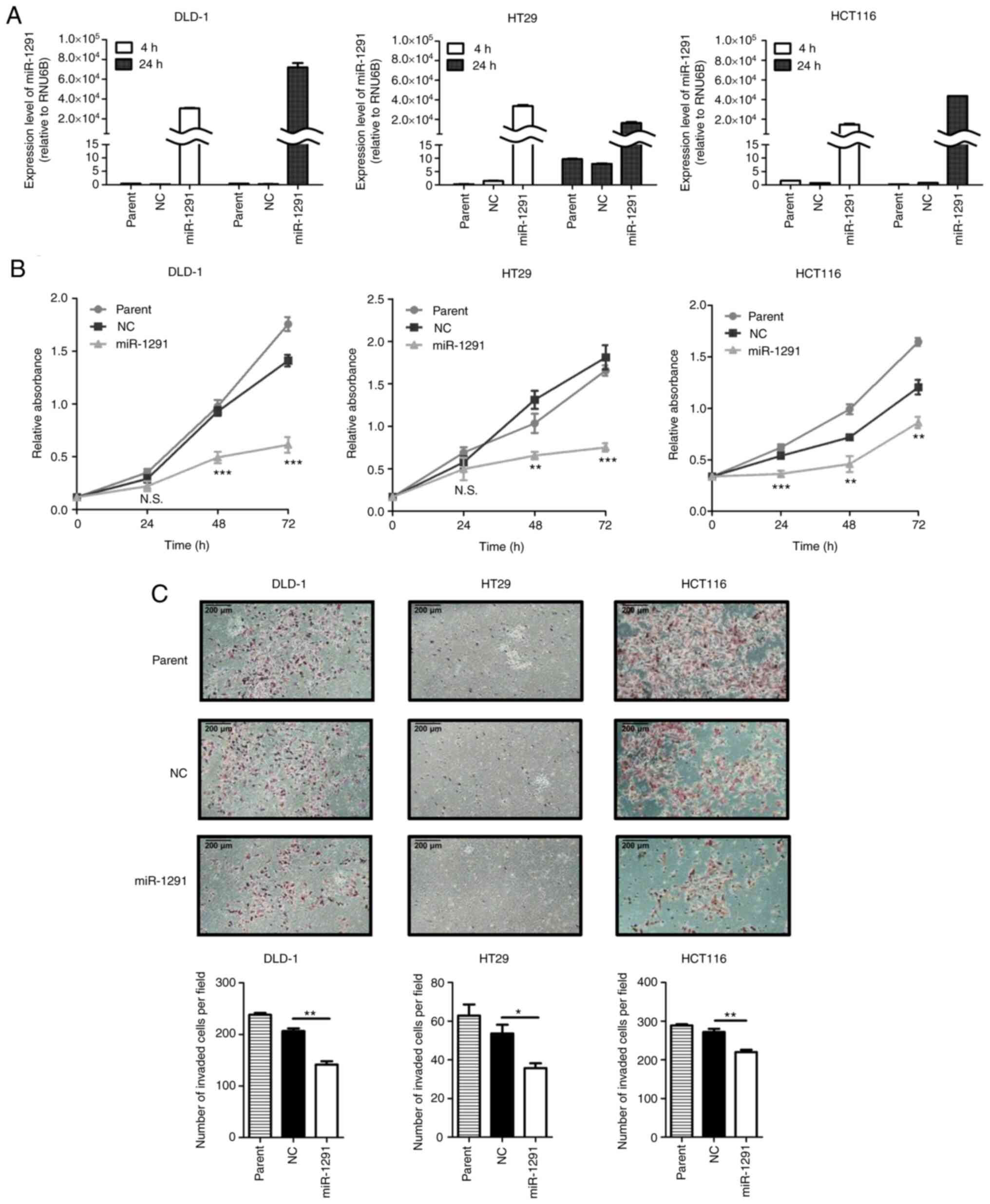
miR-1291 overexpression after
transfection
At 4 or 24 h after miRNA transfection,
miR-1291-transfected cells presented markedly higher miR-1291
expressions compared with the miR-NC transfection group in DLD-1,
HT29 and HCT116 cells (Fig.
2A).
In vitro tumor inhibitory effects of
miR-1291
miR-1291 significantly suppressed cell viability
compared with the miR-NC transfection group in DLD-1, HT29 and
HCT116 cells after 48 and 72 h of transfection (Fig. 2B).
The invasion of cells was subsequently assessed at
different time points since the time required for each cell type to
pass through Matrigel is different, as previously reported
(53,58). These included incubations for 48,
72 and 96 h for DLD-1, HCT116 and HT29 cells, respectively. The
invasion of miR-1291-transfected cells was significantly inhibited
compared with miR-NC-transfected cells in the three cell lines
examined (Fig. 2C). The effect of
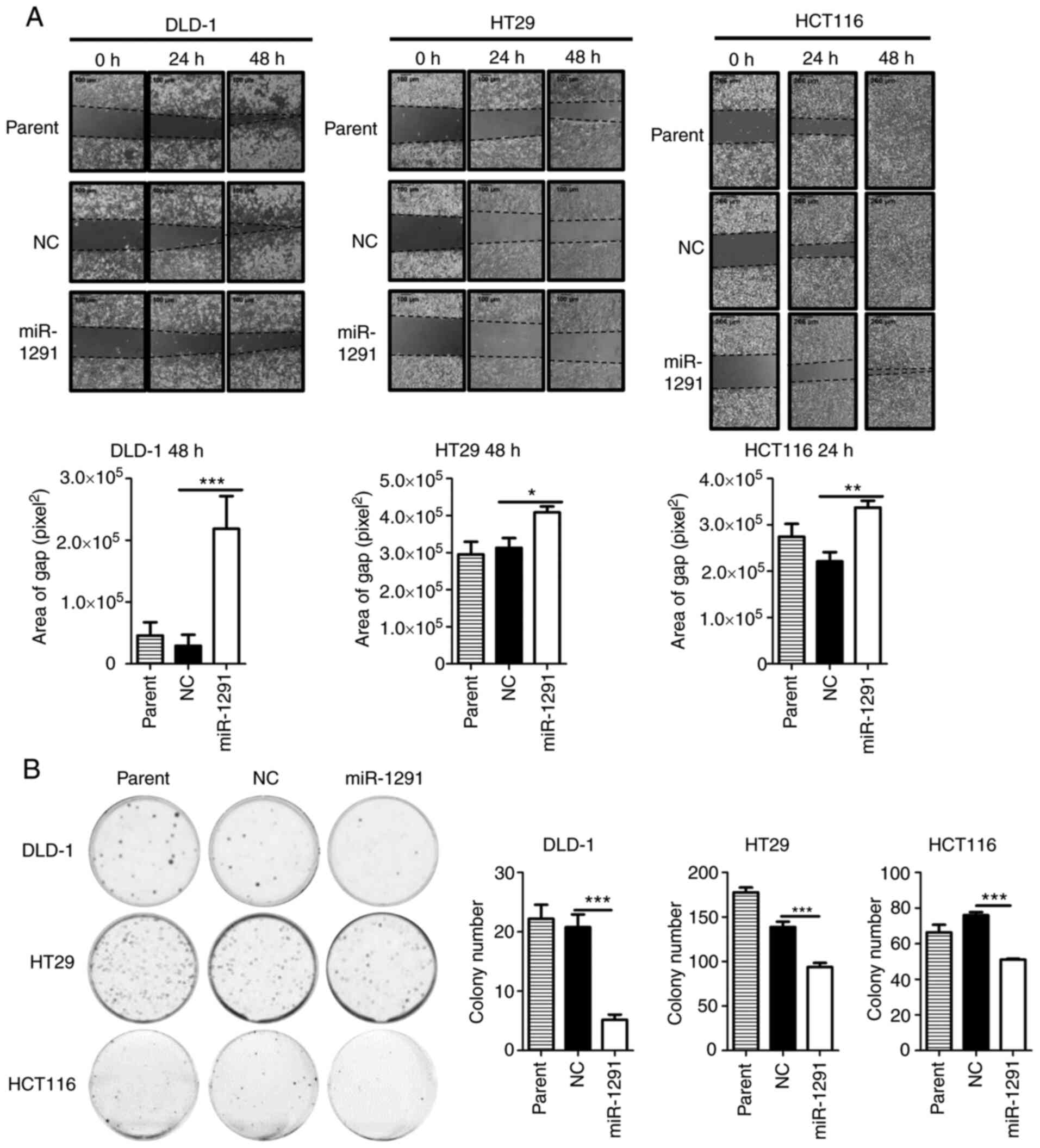
miR-1291 on the gap closure ability of CRC cells was then
evaluated. The migration of cells was significantly suppressed in
the miR-1291 group compared with the miR-NC group in 10% FBS
supplemented conditions (Fig.
3A). Furthermore, miR-1291 significantly inhibited the colony
formation of cells compared with miR-NC (Fig. 3B). In repeat experiments the miR
inhibitor (antagomir-1291) was additionally included as previously
reported (59-61). It was revealed that the effects of
miR-1291 on cell viability, invasion and migration were not
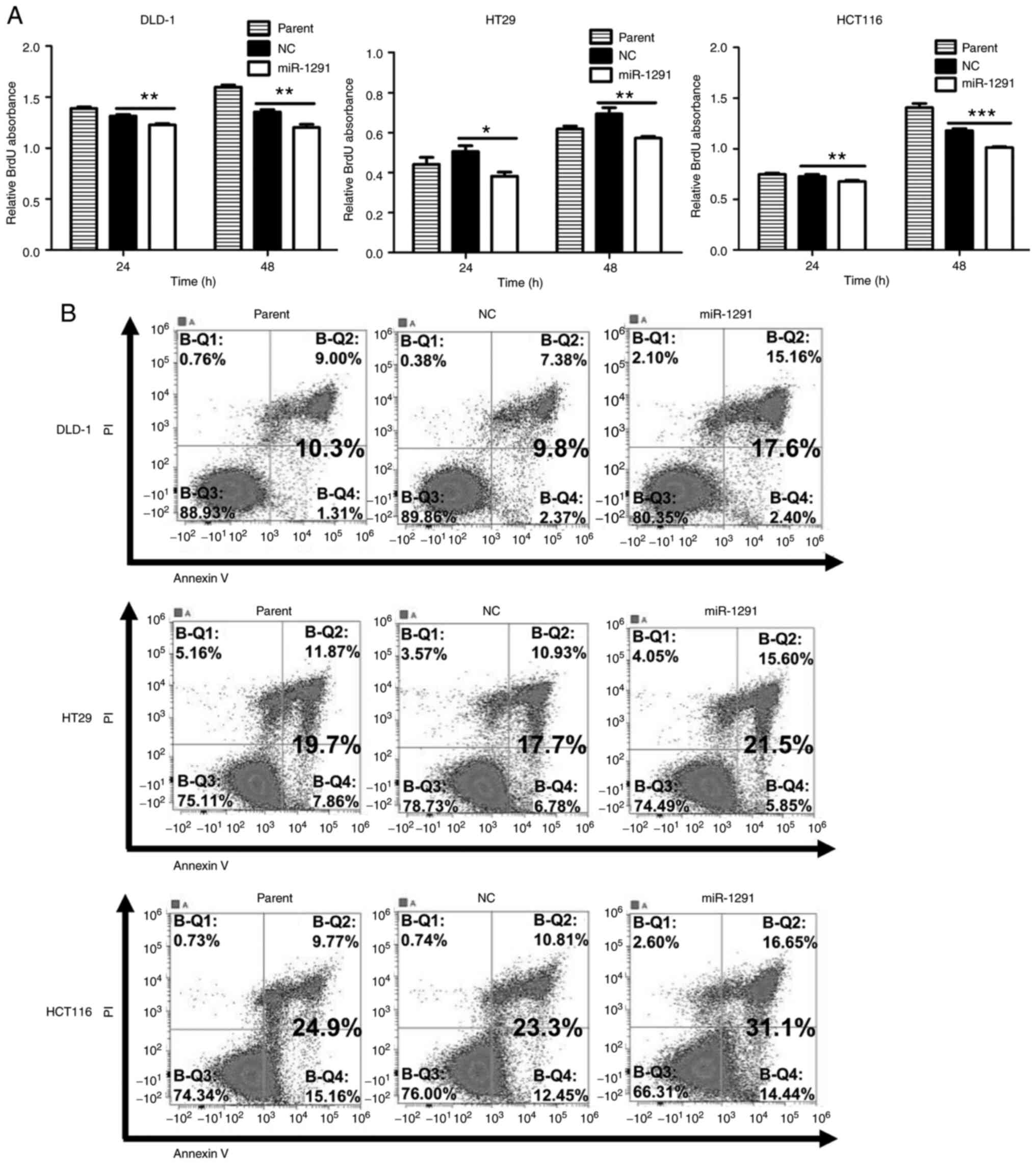
observed when CRC cells were treated by antagomir-1291 (Fig. S3A-C). To further investigate the
decrease in cell viability by miR-1291, BrdU proliferation assays
and Annexin V apoptosis assays were performed. It was demonstrated
that miR-1291 significantly suppressed the proliferation of DLD-1,
HT29 and HCT116 cells at 24 and 48 h (Fig. 4A) and induced higher rates of
apoptosis, especially in DLD-1 cells (Fig. 4B).
miR-1291 directly targets DCLK1
In silico analysis using TargetScan Human
indicated that miR-1291 may directly bind to the 3′UTR of DCLK1
mRNA (Fig. 5A). Other miR
databases, including miRwalk, miRabel and miRmap also supported
that DCLK1 is a potential target of miR-1291. Therefore, luciferase
reporter assays were performed to confirm whether miR-1291 could
directly bind to DCLK1 mRNA. Co-transfection with miRNA-1291 at 50
nM significantly inhibited the luciferase activity of wild type of
the DCLK1-3′UTR reporter vector compared with miR-NC. However, no
significant difference in luciferase activity was observed between
miR-1291 and miR-NC groups following 2-nucleotide Mut or
3-nucleotide Del DCLK1-3′UTR reporter vector transfection (Fig. 5B). Our previous study on the role
of DCLK1 in CRC revealed that HCT116 expressed the DCLK1 protein,
but DLD-1 and HT29 did not (29).
This result was confirmed in the present study (Fig. 5C). Transfection with miR-1291
significantly suppressed the expression of DCLK1 mRNA compared with
miR-NC, and a repeat assay including antagomir-1291 demonstrated
that antagomir-1291 had no inhibitory effects on DCLK1 mRNA
expression (Fig. 5D and E).
Transfection with miR-1291 also significantly decreased the
expression of the short form of DCLK1 protein compared with miR-NC
(Fig. 5F).
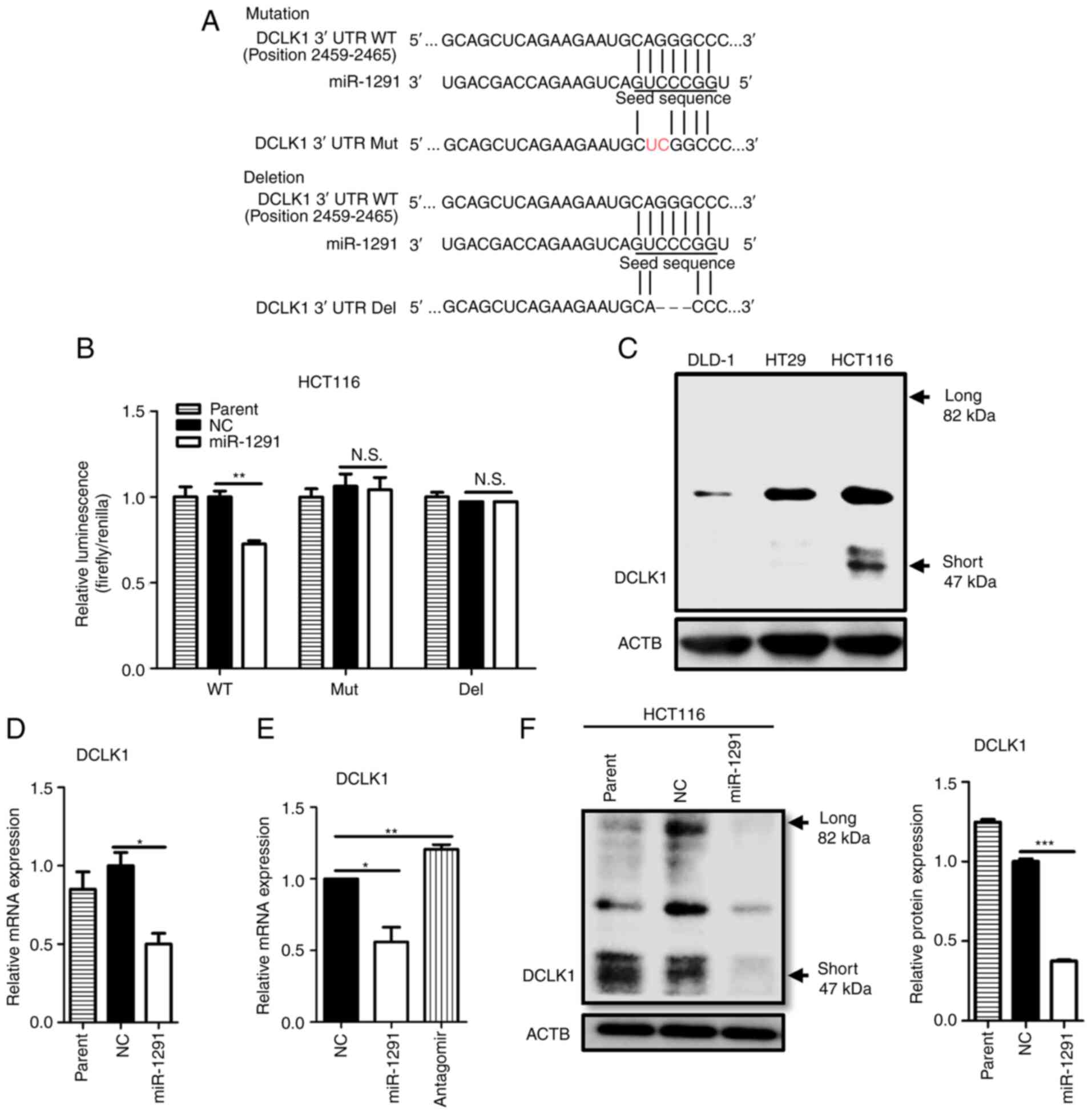 | Figure 5miR-1291 directly targets the 3′UTR
of DCLK1 in HCT116 cells. (A) TargetScan (http://www.targetscan.org/vert_72/) was used to
identify a binding site at position 2,459-2,465 of the DCLK1 mRNA
3′UTR that was complementary to the seed sequence of miR-1291 (WT).
The binding sequence of DCLK1 was mutated by changing two
nucleotides (Mut) or deleting three nucleotides (Del). (B) A
luciferase reporter assay was performed in HCT116 cells following
transfection with miR-1291. (C) DCLK1 expression in HCT116 cells
was assessed using an anti-human rabbit polyclonal antibody against
DCAMKL1. (D) The effect of miR-1291 on the expression of DCLK1 was
assessed by RT-qPCR. (E) A repeat RT-qPCR experiment was performed
using antagomir-1291. GAPDH was utilized as an endogenous control.
All experiments were performed in triplicate. All data are
presented as the mean ± SEM. (F) Western blotting was performed to
determine the expression of DCLK1 protein after 48 h of
transfection in HCT116 cells. Anti-human rabbit polyclonal
antibodies against DCAMKL1 were used. ACTB was used as a loading
control and two independent experiments were performed.
*P<0.05, **P<0.01 and
***P<0.001 as indicated. Del, deletion; DCLK1,
doublecortin-like kinase 1; miR, microRNA; Mut, mutant; NC,
negative control; N.S., not significant; RT-qPCR, reverse
transcription-quantitative PCR; UTR, untranslated region; WT, wild
type. |
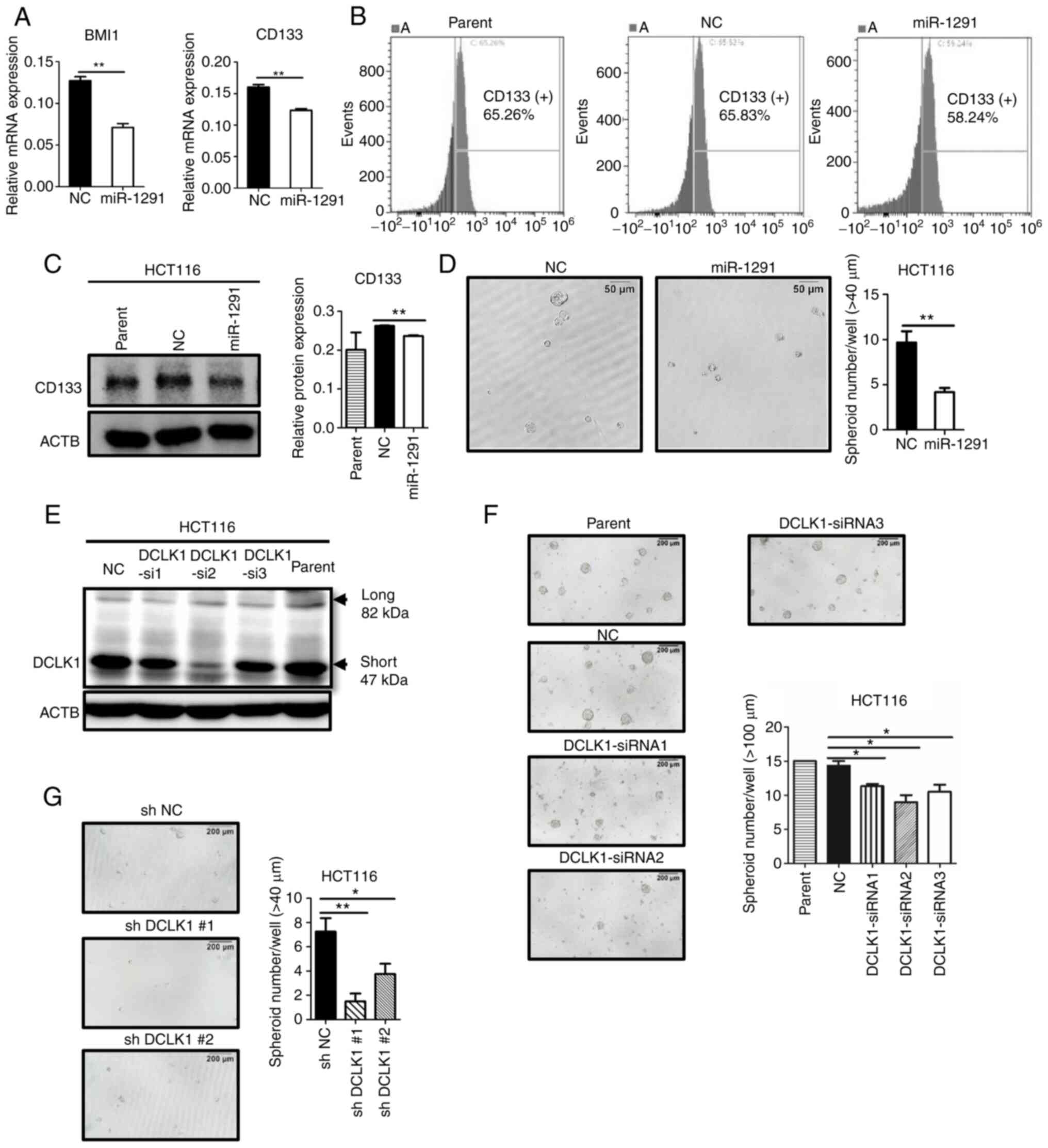
miR-1291 suppresses stem-like properties
in HCT116 cells
It was then demonstrated that other CSC markers in
addition to DCLK1, including BMI1 and CD133, were significantly
downregulated by miR-1291 overexpression at the mRNA level
(Fig. 6A). In addition, the ratio
of CD133, but not CD166, positive cells decreased following
miR-1291 transfection, as revealed by flow cytometric analysis
(Figs. 6B and S4). Western blotting also indicated
that CD133 protein expression was significantly decreased by
miR-1291 treatment compared with miR-NC transfection (Fig. 6C). Furthermore, miR-1291 treatment
significantly decreased the sphere formation of HCT116 cells, which
is a hallmark of CSC (24,33)
(Fig. 6D). To verify the
functional role of DCLK1 in sphere formation of HCT116, siRNAs
(siRNA1, siRNA2 and siRNA3) were used to knock down DCLK1. Western
blot analyses revealed that the protein expression of DCLK1
decreased by these three siRNAs (Fig.
6E), and that siRNA transfection significantly downregulated
sphere formation in HCT116 cells (Fig. 6F) compared with miR-NC-treated
group. Furthermore, it was verified that shDCLK1 clones, which were
produced in our previous study (29), exhibited decreased sphere
formation in HCT116 cells compared with sh-NC cells (Fig. 6G).
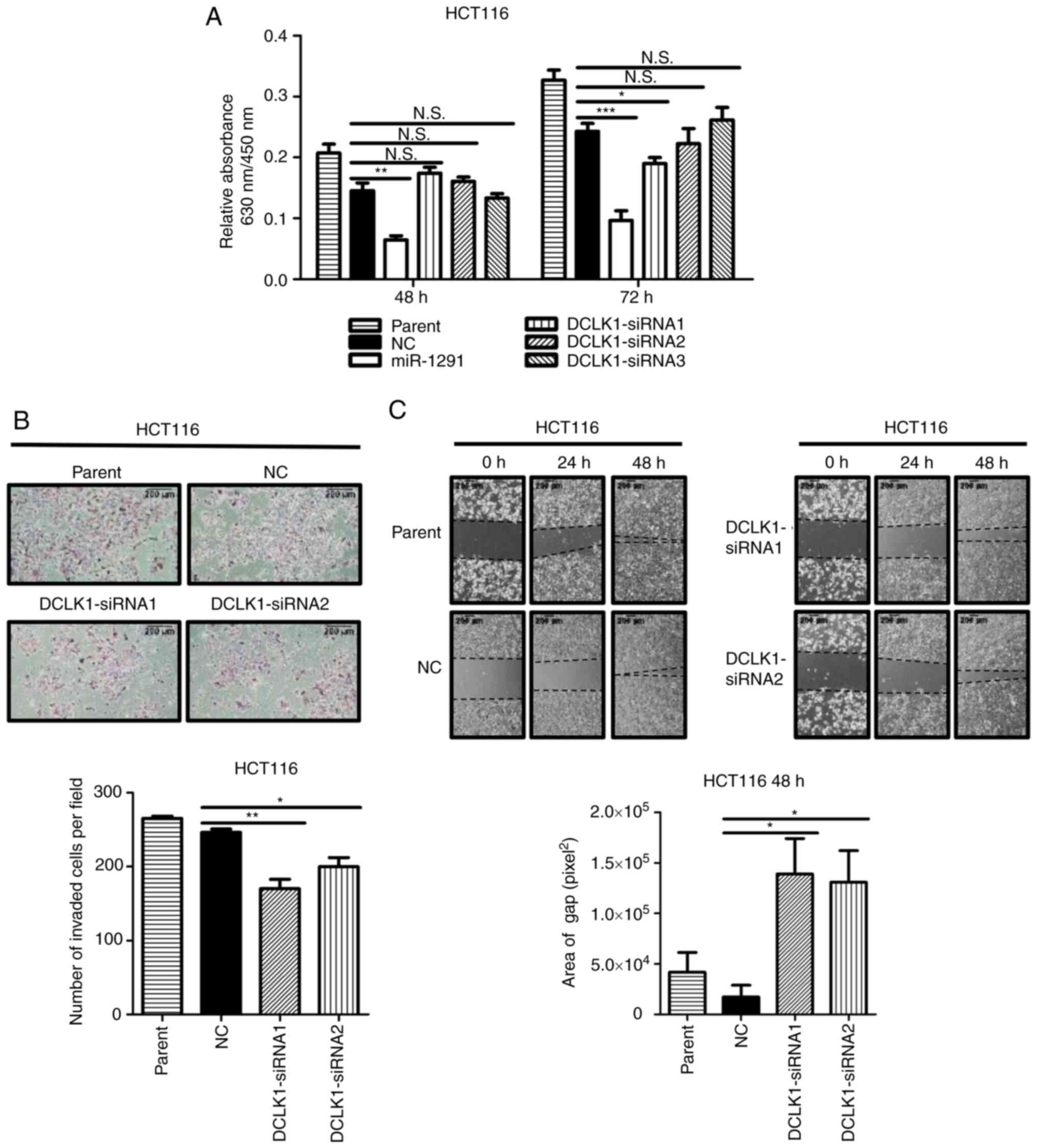
In vitro tumor inhibitory effects of
DCLK1 siRNAs
The effects of DCLK1 siRNAs on cell viability,
invasion and gap closure were examined. DCLK1 siRNAs significantly
suppressed the migration and invasion of HCT116 cells. For the
effects on cell viability, although miR-1291 significantly
decreased the viability of HCT116 cells at 48 and 72 h (Fig. 7A), only one of the DCLK1 siRNAs
(siRNA1) significantly suppressed the cell viability at 72 h after
transfection compared with the miR-NC (Fig. 7A). However, DCLK1-siRNA
significantly suppressed the invasion and gap closure of HCT116
cells compared with miR-NC groups (Fig. 7B and C).
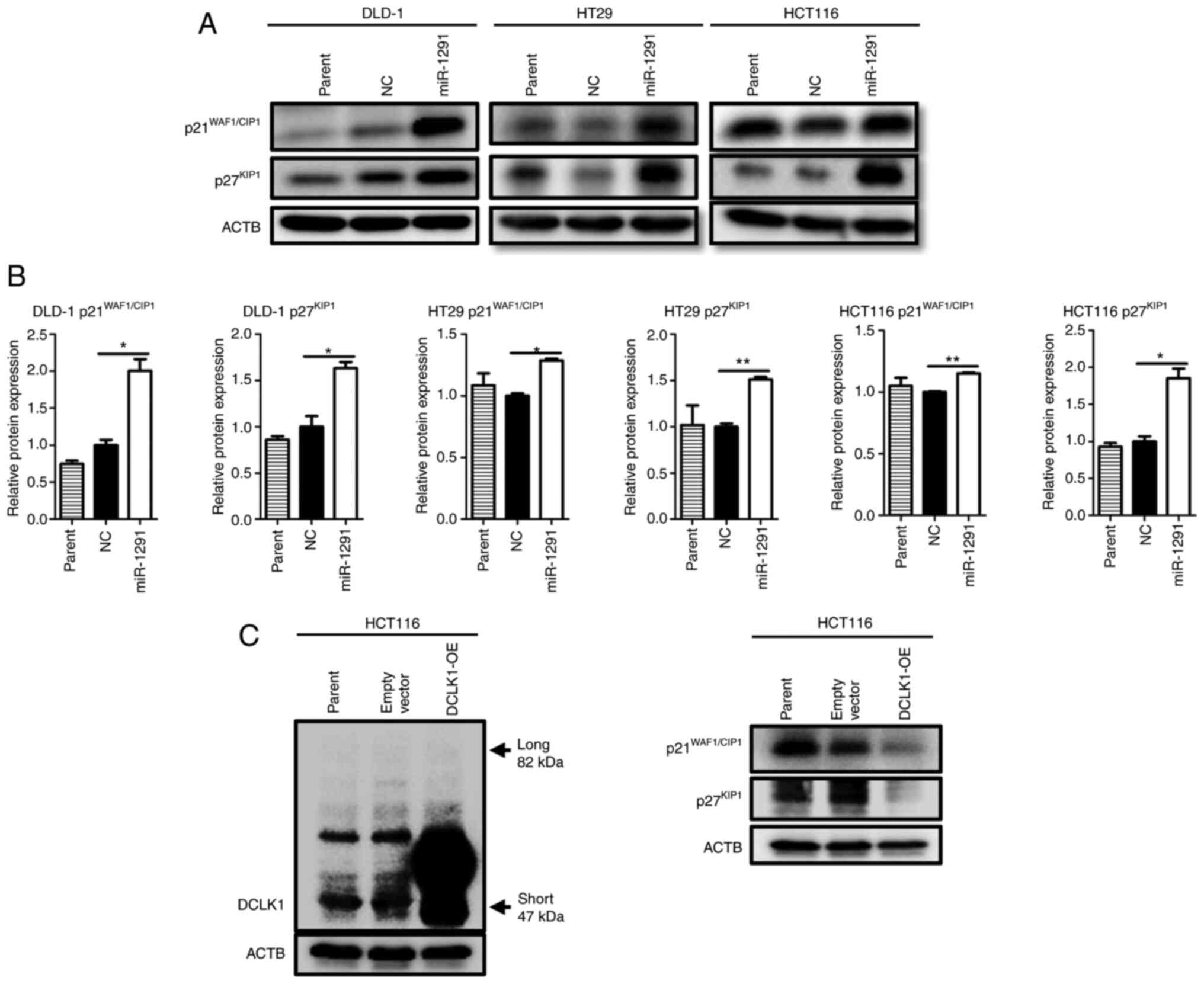
Altered expression of cell cycle
components by treatment of miR-1291
Since DLD-1 and HT29 cells had no or scarce DCLK1
expression (Fig. 5C), another
mechanism should exist for miR-1291-mediated anti-tumor effect.
Therefore, alterations of the cell cycle and cell cycle-related
protein expressions were investigated. By performing cell cycle
distribution analysis after cells were serum-starved, the results
revealed that the G1-S population increased in DLD-1 cells at 12 h
(60.81 vs. 67.89%) and that the G2-M fraction increased in HT29
cells at 48 h (12.01 vs. 17.21%) following treatment with miR-1291
when compared with the NC group (Fig. S5A). Western blot analyses
indicated that the expression of CDK inhibitors
p21WAF1/CIP1 and p27KIP1 were significantly
upregulated in DLD-1, HT29 and HCT116 cells (Fig. 8A and B). Moreover, G1-S
accelerators CDK4 (at 48 h) and Rb (at 72 h) in DLD-1 cells, CDK4
(at 48 and 72 h) in HT29 cells and CDK4 (at 72 h) in HCT116 cells
were downregulated. G2-M accelerators CDC25B (at 72 h), along with
CDC25C (at 48 and 72 h), and phosphorylated-cdc2/cdc2 ratio (at 48
h) also decreased in HT29 cells (Fig. S5B). In addition, it was
identified that the DCLK1-overexpressing HCT116 clone expressed
considerably higher levels of short form DCLK1 protein at 48 h
after transfection compared with parental cells or empty vector
transfected control cells. The expression of
p21WAF1/CIP1 and p27KIP1 protein decreased in
the DCLK1-OE clone compared with parental or control cells
(Fig. 8C).
Anti-tumor effects of miR-1291 in
vivo
A DLD-1 tumor xenograft mouse model was used to
verify the anti-tumor effect of miR-1291 in vivo. Compared
with miR-NC group or parent group, the systemic administration of
miR-1291 on sCA significantly inhibited the growth of tumors
(Fig. 9A). No obvious body weight
loss of mice was observed among the three groups (Fig. 9B). Western blotting using the
resultant tumors indicated that the systemic administration of
miR-1291 led to significant upregulation of p21WAF1/CIP1
and p27KIP1 compared with miR-NC treatment (Fig. 9C).
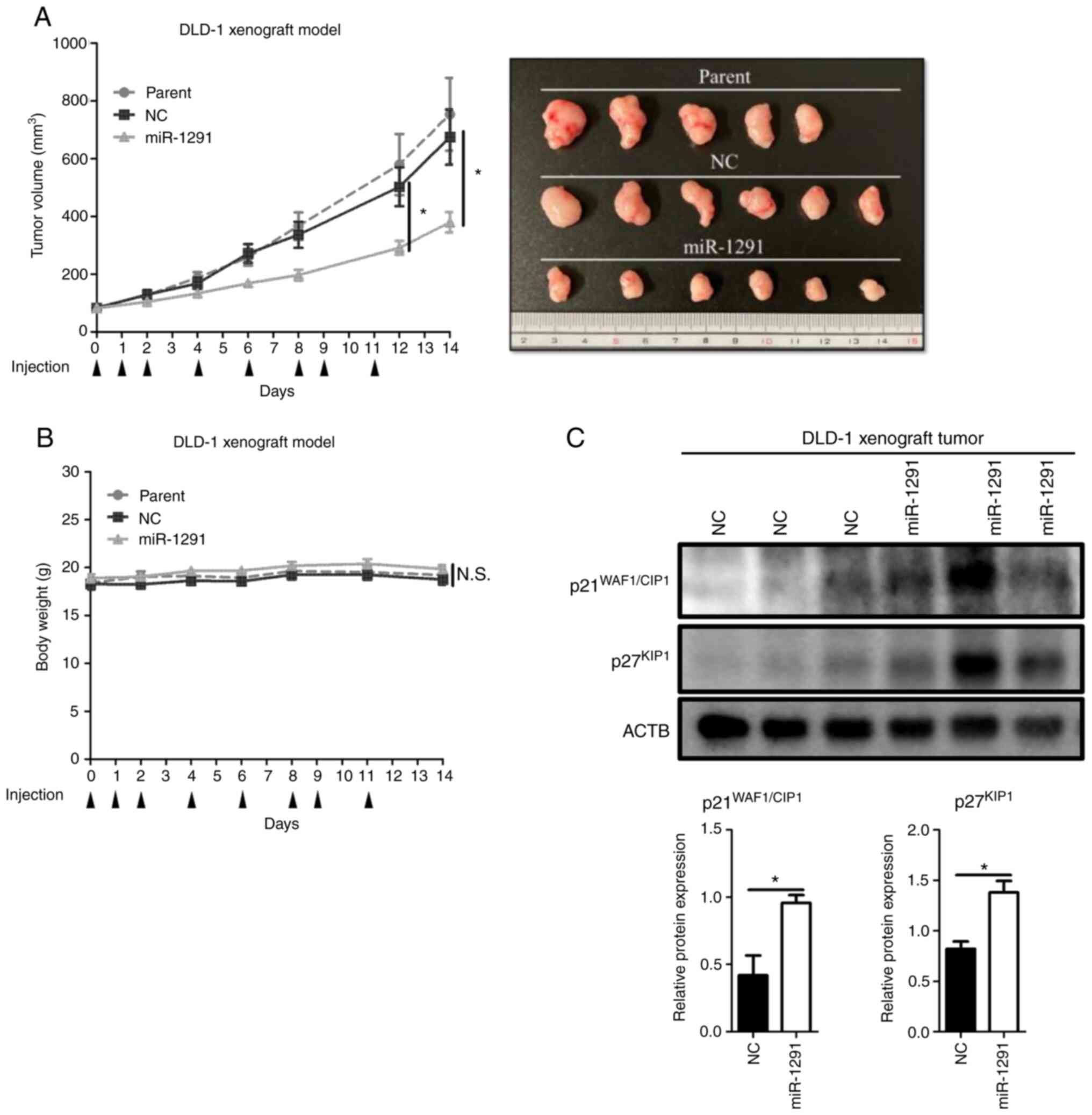 | Figure 9Systemic administration of formulated
sCA-miR-1291 in vivo. (A) Tumor volume and images of
resected tumors. When the mean tumor volume reached 80
mm3 (day 0), tumor xenograft mouse models received
intravenous administrations of miR-1291 or miR-NC using sCA as a
vehicle. This was administered on days 0, 1, 2, 4, 6, 8, 9 and 11
via a tail vein injection (arrow heads indicate days of
injections). Each injection contained 40 µg of formulated
oligo. Data are presented as the mean ± SEM. (B) Body weight did
not significantly differ between the groups. (C) Protein expression
of cell cycle components in the tumors resected on day 14. ACTB was
used as a loading control. *P<0.05. sCA, super
carbonate apatite; miR, microRNA; NC, negative control; N.S., not
significant. |
Discussion
miRNA is emerging as a next generation cancer
treatment (62-65). Previous studies have revealed that
miR-1291 has anti-tumor effects in multiple types of cancer,
including hepatocellular carcinoma, renal, esophagus, pancreatic
and prostate cancer (39-46). The present study revealed that
miR-1291 inhibited the viability of cancer stem-like pancreatic
cancer cells. As for CRC, which is one of the most widespread
cancers in the world, Salehi et al (47) analyzed 14 paired primary CRC
samples and liver metastasis samples using NCBI GEO database. It
was reported that miR-1291 only demonstrated a 1.065-fold increase
(logFC 0.0895) in liver metastasis compared with primary CRC
samples among 58 upregulated miRNAs. By using TargetScan, it was
also suggested that DCLK1 may be a potential target of miR-1291. In
the present study, the direct binding of miR-1291 to DCLK1 mRNA was
verified and functional in vitro and in vivo
assessments of miR-1291 were performed in CRC cells.
The tissue atlas database has indicated that
miR-1291 levels in colon tissue are relatively low, at ~1/50 and
1/1,500 of the putative anti-oncogenic miR-34a and the oncogenic
miR-21 (5), respectively. In the
present study, it was revealed that miR-1291 expression in CRC cell
lines was generally lower compared with the colon tissue cell line
CCD-18Co. However, the absence of the other normal human colon cell
lines, such as CCD-112CoN and CCD-33Co is a limitation of the
present study. When comparing miR-1291 levels in normal and cancer
colorectal tissues, it was found that cancer tissues expressed
significantly lower levels of miR-1291 compared with normal mucosa.
This result is in line with those of other cancer types, such as
renal cell cancer, esophageal squamous cell cancer and prostate
cancer (44-46), and emphasizes the tumor
suppressive abilities of miR-1291. Furthermore, this situation is
advantageous as miR-1291 is endogenously expressed in normal
colorectal tissue, thus the restoration of tumor suppressive
miR-1291 would be a relatively safe strategy for CRC treatment
(17). When analyzing the
relationship between with miR-1291 expression and various
clinicopathological parameters, it was determined that miR-1291
levels were not associated with any clinicopathological factors in
the present study. Further clinical study assessing a larger number
of patients with CRC is therefore essential.
Each CRC cell line in the current study was used
based on the representative characteristics of CRC, and included:
i) Cancer stem-like HCT116 cells (66-68); ii) differentiated carcinoma HT29
cells exhibiting goblet-like features (66); iii) aggressive phenotype DLD-1
cells obtained from a murine model displaying moderate to poorly
differentiated adenocarcinoma (69). CRC is mostly composed of
differentiated adenocarcinoma and <5% of CRC is poorly
differentiated carcinoma, which is extremely aggressive (1-3).
CSC exists as a portion of the tumor population and is considered
to be a cause of recurrence (22-24). HCT116 cells are considered to
contain mainly CSCs since they do not differentiate or express
transcription factor caudal-type homeobox 1 and since they have
high colonosphere forming capacities (66-68). Additionally,
CD133+CD44+ stem-like cancer cells are highly
enriched in HCT116 (68).
In the present study, it was revealed that miR-1291
was successfully transfected into three CRC cell lines between 4
and 24 h. miR-1291 levels reached >1~5×104 fold
higher than those of NC-treated cells. This result is in line with
that of a previous study, which revealed that miR-522 levels were
increased by 1~6×104 fold in ovarian cancer cell lines
(70). In a previous time-course
study, it was found that 18mer oligo DNA was incorporated into 293
or HCT116 cell lines within 1 h, with its peak effect being
observed at 6 h and a gradual decline following thereafter
(71). Small RNA sequences were
also highly incorporated into various cell types at 4 h (52).
The present study demonstrated that miR-1291, but
not antagomir-1291, exhibited anti-tumor effects in HCT116, DLD-1
and HT29 CRC cells with regard to cell proliferation, invasion,
mobility, colony-forming and cell cycle regulation. The effect of
miR-1291 should be compared with NC, but not with parental cells,
because the transfection process itself occasionally causes stress
or damage to the cells. This NC sequence was utilized and
established in our previous studies (17,18,20,52-54). During fundamental in vitro
assays, the negative control displayed data compatible with
parental cells in most experiments.
Of the three CRC cell lines, DLD-1 was selected for
in vivo experiments since miR-1291 exhibited the best
efficacy in DLD-1 during in vitro experiments. DLD-1 is
derived from a patient with colon cancer and the cells produce
aggressive tumors in nude mice, as previously described (69), thus DLD-1 is also an important CRC
cell line and has been used in our previous in vivo studies
(17,18,20). Previous studies have demonstrated
that the intravenous administration of miR-1291 loaded on an sCA
delivery system (17,18,20,52-55) significantly inhibits the in
vivo tumor growth of DLD-1 cells. It is of note that
transfection efficiency when using an sCA system is markedly higher
than when using a liposome system (52). Additionally, the sCA delivery
system produces high uptake (~500-1,500 fold higher miRNA levels)
in tumor tissues (18,20,54,55).
In the current study, miR-1291 inhibited the sphere
formation of HCT116 cells, which is a hallmark of cancer stemness
(24,33). It has been reported that excision
of DCLK1-positive CSC cells results in the regression of the
intestinal tumor without apparent impairment of normal tissue,
indicating that DCLK1 may be a novel target for CSC-targeted
therapy (27). It was recently
revealed that shDCLK1 clones, in which the expression of short form
DCLK1 protein was silenced by shDCLK1 RNA, exhibited decreased cell
growth, invasion, migration and EMT in HCT116 cells (29). The short form of DCLK1 is
considered to play a crucial role in CRC. It has been demonstrated
that the short form of DCLK1, but not the long form, is an
important target for the suppression of CRC (72,73). Furthermore, patients with CRC
exhibiting high expressions of short form, but not long form, DCLK1
tend to demonstrate a worse overall survival (72,73).
The current study aimed to determine whether
miR-1291 binds to 3′UTR of DCLK1 mRNA according to an in
silico search using TargetScan and other databases. As a
result, it was verified that miR-1291 directly bound to the 3′UTR
of the DCLK1 mRNA sequence and that miR-1291, but not
antagomir-1291, decreased DCLK1 mRNA expression in HCT116 cells
in vitro. In silico analysis and in vitro selection
revealed that miR-1291 was an upstream modulator of DCLK1. miR-1291
also decreased the expression of CSC markers BMI1 and CD133, but
scarcely decreased CD166 expression (25,26,68,74). Since these molecules are not
likely to be direct targets of miR-1291 according to TargetScan
predictions, it is postulated that the decrease in BMI1 and CD133
may reflect certain diminution of the stem-like properties of
HCT116 cells, which could be an indirect effect of miR-1291 through
DCLK1.
DCLK1 siRNAs displayed relatively weak inhibition
of cell viability, but exhibited potent anti-tumor effects in
invasion and cell mobility assays. Additionally, DCLK1 siRNAs
inhibited sphere formation, which was subsequently confirmed by
shDCLK1 clones in HCT116 cells. The results regarding sphere
formation are consistent with other studies, which demonstrated
that DCLK1 levels were closely associated with spheroid formation
in HCT116 cells (75,76). Taken together, it was suggested
that miR-1291 may be involved in the regulation of stem cell
properties through the inhibition of DCLK1 in HCT116 cells.
DLD-1 and HT29 cells, which express no or scarce
DCLK1, exerted potent tumor inhibitory effects, suggesting that an
additional mechanism may be operating in these CRC cells. In this
regard, it is reported that miR-1291 regulates MUC1 in human
esophagus cancer EC9706 and EC-1 cells (45). However, when MUC1 expression was
examined with treatment of miR-1291 in CRC cells, its level was not
decreased (unpublished data), suggesting that the target molecule
may differ between tumor types. Instead, one of the major effects
observed following miR-1291 treatment was the drastic increase in
the cell cycle components p21WAF1/CIP1 and
p27KIP1. These CDK inhibitors bind to and block the G1-S
accelerators Cyclin D1-CDK4/6 complex and Cyclin E1-CDK2 complex,
leading to arrest from the G1 to S phase (77-79). In addition,
p21WAF1/CIP1 has an ability to inhibit G2-M transition
by inhibiting Cyclin B and cdc2 (80). Under conditions supplemented with
FBS, no obvious change could be observed in the cell cycle
distribution between miR-NC and miR-1291 treatment (unpublished
data). However, following detailed cell cycle distribution analysis
after cells were serum-starved, it was found that the G1-S
population increased in DLD-1 cells at 12 h and that the G2-M
fraction increased in HT29 cells at 48 h with treatment of miR-1291
compared with NC treatment. Accordingly, western blotting indicated
downregulation of G1-S accelerators CDK4 in DLD-1 cells, HT29 cells
and HCT116 cells. G2-M accelerators CDC25B, along with CDC25C, and
phosphorylated-cdc2/cdc2 ratio decreased in HT29 cells. In
vivo experiments also revealed that systemic administrations of
miR-1291 led to an upregulation of p21WAF1/CIP1 and
p27KIP1 in DLD-1 derived tumors. Although
p27KIP1 protein levels differed among the treated
tumors, this is not unique, as the cell cycle during in vivo
treatment could be affected by several factors such as tumor size,
tumor vascularization and stromal development. Collectively, these
findings suggested that miR-1291 could cause dysregulation of cell
cycle control, which may partially account for mechanism underlying
the anti-tumor effect of DLD-1 and HT29 cells. Although the precise
mechanism is yet to be fully elucidated, DCLK1 in HCT116 cells may
also be involved in the regulation of the CDK inhibitors, since it
was determined that the overexpression of DCLK1 decreased
p21WAF1/CIP1 and p27KIP1 protein
expression.
In conclusion, miR-1291 presented a potent
anti-tumor effect in CRC cells. Furthermore, miR-1291 could
suppress cancer stemness in HCT116 cells. To the best of our
knowledge, this is the first study that conducted detailed
functional assessments of miR-1291 in CRC. Considering the
anti-tumor effect of miR-1291 in the broad range of cancer types
(39-46), this miRNA may be a candidate for
next generation nucleic acid medicine. Furthermore, it is also
expected to serve as a CSC-targeted therapeutic strategy for
DCLK1-expressing CRC when the practical drug delivery systems are
equipped (62-65,81,82).
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors. contributions
HY and MM designed the study. HY, MM, YY and MU
supervised the study. HT and YM are responsible for methodology. SS
and SKou analyzed and interpreted of data and also confirmed the
authenticity of all the raw data. JW, SB, YS, HH, KM and SuT
performed the experiments. SKob and HI are responsible for
statistical analysis. MU collected and provided the normal and
colorectal cancer tissue samples and their clinical data. JW wrote
the original draft. HY and MU reviewed and edited the manuscript.
ShT and XW performed ODC degron-related experiments. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was performed in accordance with
the Declaration of Helsinki and all animal procedures (approval no.
13377-5) and patient-derived tissue experiments were approved by
the Ethics Board of Osaka University (Osaka, Japan). All patients
provided written informed consent, in accordance with the
guidelines approved (approval no. 08226) by the Institutional
Research Board of the institute.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Dr Masaaki Miyo
(Department of Surgery, Gastroenterological Surgery, Graduate
School of Medicine, Osaka University, Osaka, Japan.) for editing
this manuscript, Dr Koki Takeda (Department of Surgery,
Gastroenterological Surgery, Graduate School of Medicine, Osaka
University, Osaka, Japan.) for technical advice and Professor
Toshinari Minamoto (Cancer Research Institute, Kanazawa University,
Kanazawa, Japan) for the generous gift of the KM12SM cell line. The
authors would also like to thank Dr Frank Pajonk (Jonsson
Comprehensive Cancer Center, UCLA, CA, USA) for providing the
retroviral expression vector pQCXIN-ZsGreen-cODC containing green
fluorescence ZsGreen-labeled degron ODC (Gdeg).
Funding
The present study was supported by a grant from Kagoshima
Shinsangyo Sousei Investment Limited Partnership (its general
partner is Kagoshima Development Co., Ltd.) and by Grant-in-Aid for
Young Scientists (B), JSPS KAKENHI (grant no. 18K16361).
Abbreviations:
|
Antagomir-1291
|
Hsa-miR-1291 inhibitor S-Tud
|
|
BMI1
|
B cell-specific Moloney murine
leukemia virus integration site 1
|
|
BrdU
|
Bromodeoxyuridine
|
|
CDK
|
cyclin dependent kinase
|
|
CSC
|
cancer stem cell
|
|
CRC
|
colorectal cancer
|
|
Del
|
deleted type
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
EGFR
|
epidermal growth factor receptor
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
ER
|
endoplasmic reticulum
|
|
KRAS
|
kirsten rat sarcoma virus
|
|
miRNA
|
microRNA
|
|
miR-1291
|
mimic-hsa-miR-1291
|
|
miR-NC
|
negative control miR
|
|
MUC1
|
mucin 1
|
|
Mut
|
mutated type
|
|
ODC
|
ornithine decarboxylase
|
|
RT-qPCR
|
quantitative real-time PCR
|
|
sCA
|
super carbonate apatite
|
|
sh
|
short hairpin
|
|
siRNA
|
small interfering RNA
|
|
STR
|
short tandem repeat
|
|
TGF-β
|
transforming growth factor-β
|
|
3′UTR
|
3′-untranslated region
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rawla P, Sunkara T and Barsouk A:
Epidemiology of colorectal cancer: Incidence, mortality, survival,
and risk factors. Prz Gastroenterol. 14:89–103. 2019.PubMed/NCBI
|
|
3
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar
|
|
4
|
Graham JS and Cassidy J: Adjuvant therapy
in colon cancer. Expert Rev Anticancer Ther. 12:99–109. 2012.
View Article : Google Scholar
|
|
5
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fischer SE: RNA Interference and
MicroRNA-mediated silencing. Curr Protoc Mol Biol. Oct 1. pp.
201Epub ahead of print 0.1002/0471142727.mb2601s112.
|
|
7
|
Mehta A and Baltimore D: MicroRNAs as
regulatory elements in immune system logic. Nat Rev Immunol.
16:279–294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pons-Espinal M, de Luca E, Marzi MJ,
Beckervordersandforth R, Armirotti A, Nicassio F, Fabel K,
Kempermann G and De Pietri Tonelli D: Synergic functions of miRNAs
determine neuronal fate of adult neural stem cells. Stem Cell
Reports. 8:1046–1061. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aghaei M, Khodadadian A, Elham KN, Nazari
M and Babakhanzadeh E: Major miRNA involved in insulin secretion
and production in beta-cells. Int J Gen Med. 13:89–97. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ali Syeda Z, Langden SSS, Munkhzul C, Lee
M and Song SJ: Regulatory mechanism of MicroRNA expression in
cancer. Int J Mol Sci. 21:17232020. View Article : Google Scholar :
|
|
11
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar :
|
|
12
|
Li VS, Ng SS, Boersema PJ, Low TY,
Karthaus WR, Gerlach JP, Mohammed S, Heck AJ, Maurice MM, Mahmoudi
T and Clevers H: Wnt signaling through inhibition of β-catenin
degradation in an intact Axin1 complex. Cell. 149:1245–1256. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Spano JP, Lagorce C, Atlan D, Milano G,
Domont J, Benamouzig R, Attar A, Benichou J, Martin A, Morere JF,
et al: Impact of EGFR expression on colorectal cancer patient
prognosis and survival. Ann Oncol. 16:102–108. 2005. View Article : Google Scholar
|
|
14
|
Markowitz S, Wang J, Myeroff L, Parsons R,
Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein
B, et al: Inactivation of the type II TGF-beta receptor in colon
cancer cells with microsatellite instability. Science.
268:1336–1338. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baker SJ, Fearon ER, Nigro JM, Hamilton
SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF,
Nakamura Y, et al: Chromosome 17 deletions and p53 gene mutations
in colorectal carcinomas. Science. 244:217–221. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelialmesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hiraki M, Nishimura J, Takahashi H, Wu X,
Takahashi Y, Miyo M, Nishida N, Uemura M, Hata T, Takemasa I, et
al: Concurrent Targeting of KRAS and AKT by MiR-4689 is a novel
treatment against mutant KRAS Colorectal cancer. Mol Ther Nucleic
Acids. 4:e2312015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Inoue A, Mizushima T, Wu X, Okuzaki D,
Kambara N, Ishikawa S, Wang J, Qian Y, Hirose H, Yokoyama Y, et al:
miR-29b byproduct sequence exhibits potent tumour-suppressive
activities via inhibition of NF-κB signaling in KRAS-mutant colon
cancer cells. Mol Cancer Ther. 17:977–987. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tazawa H, Tsuchiya N, Izumiya M and
Nakagama H: Tumor-suppressive miR-34a induces senescence-like
growth arrest through modulation of the E2F pathway in human colon
cancer cells. Proc Natl Acad Sci USA. 104:15472–15477. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morimoto Y, Mizushima T, Wu X, Okuzaki D,
Yokoyama Y, Inoue A, Hata T, Hirose H, Qian Y, Wang J, et al:
miR-4711-5p regulates cancer stemness and cell cycle progression
via KLF5, MDM2 and TFDP1 in colon cancer cells. Br J Cancer.
122:1037–1049. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ayob AZ and Ramasamy TS: Cancer stem cells
as key drivers of tumour progression. J Biomed Sci. 25:202018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao J: Cancer stem cells and
chemoresistance: The smartest survives the raid. Pharmacol Ther.
160:145–58. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
de Sousa e Melo F, Kurtova AV, Harnoss JM,
Kljavin N, Hoeck JD, Hung J, Anderson JE, Storm EE, Modrusan Z,
Koeppen H, et al: A distinct role for Lgr5+ stem cells
in primary and metastatic colon cancer. Nature. 543:676–680. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vermeulen L, Todaro M, de Sousa Mello F,
Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G and Medema
JP: Single-cell cloning of colon cancer stem cells reveals a
multi-lineage differentiation capacity. Proc Natl Acad Sci USA.
105:13427–1332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ren F, Sheng WQ and Du X: CD133: A cancer
stem cells Marker, is used in colorectal cancers. World J
Gastroenterol. 19:2603–2611. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Soheilifar MH, Moshtaghian A, Maadi H,
Izadi F and Saidijam M: BMI1 roles in cancer stem cells and its
association with MicroRNAs dysregulation in cancer: Emphasis on
colorectal cancer. Int J Cancer Manag. 11:92018.
|
|
27
|
Nakanishi Y, Seno H, Fukuoka A, Ueo T,
Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M, et
al: Dclk1 distinguishes between tumor and normal stem cells in the
intestine. Nat Genet. 45:98–103. 2013. View Article : Google Scholar
|
|
28
|
Chandrakesan P, Weygant N, May R, Qu D,
Chinthalapally HR, Sureban SM, Ali N, Lightfoot SA, Umar S and
Houchen CW: DCLK1 facilitates intestinal tumor growth via enhancing
pluripotency and epithelial mesenchymal transition. Oncotarget.
5:9269–9280. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Makino S, Takahashi H, Okuzaki D, Miyoshi
N, Haraguchi N, Hata T, Matsuda C, Yamamoto H, Mizushima T, Mori M,
et al: DCLK1 integrates induction of TRIB3, EMT, drug resistance
and poor prognosis in colorectal cancer. Carcinogenesis.
41:303–312. 2020. View Article : Google Scholar
|
|
30
|
Westphalen CB, Takemoto Y, Tanaka T,
Macchini M, Jiang Z, Renz BW, Chen X, Ormanns S, Nagar K, Tailor Y,
et al: Dclk1 defines quiescent pancreatic progenitors that promote
injury-induced regeneration and tumorigenesis. Cell Stem Cell.
18:441–455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Weygant N, Qu D, May R, Tierney RM, Berry
WL, Zhao L, Agarwal S, Chandrakesan P, Chinthalapally HR, Murphy
NT, et al: DCLK1 is a broadly dysregulated target against
epithelial-mesenchymal transition, focal adhesion, and stemness in
clear cell renal carcinoma. Oncotarget. 6:2193–2205. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Adikrisna R, Tanaka S, Muramatsu S, Aihara
A, Ban D, Ochiai T, Irie T, Kudo A, Nakamura N, Yamaoka S and Arii
S: Identification of pancreatic cancer stem cells and selective
toxicity of chemotherapeutic agents. Gastroenterology.
143:234–245.e7. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Munakata K, Uemura M, Tanaka S, Kawai K,
Kitahara T, Miyo M, Kano Y, Nishikawa S, Fukusumi T, Takahashi Y,
et al: Cancer stem-like properties in colorectal cancer cells with
low proteasome activity. Clin Cancer Res. 22:5277–5286. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qian Y, Wu X, Yokoyama Y, Okuzaki D,
Taguchi M, Hirose H, Wang J, Hata T, Inoue A, Hiraki M, et al:
E-cadherin-Fc chimera protein matrix enhances cancer stem-like
properties and induces mesenchymal features in colon cancer cells.
Cancer Sci. 110:3520–3532. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pan YZ, Zhou A, Hu Z and Yu AM: Small
nucleolar RNA-derived microRNA hsa-miR-1291 modulates cellular drug
disposition through direct targeting of ABC transporter ABCC1. Drug
Metab Dispos. 41:1744–1751. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Peng L, Chun-Guang Q, Bei-Fang L, Xue-Zhi
D, Zi-Hao W, Yun-Fu L, Yan-Ping D, Yang-Gui L, Wei-Guo L, Tian-Yong
H and Zhen-Wen H: Clinical impact of circulating miR-133, miR-1291
and miR-663b in plasma of patients with acute myocardial
infarction. Diagn Pathol. 9:892014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qiu L, Zhang L, Qi R, Gao X, Chen H and
Xiao T: miR-1291 Functions as a potential serum biomarker for
bullous pemphigoid. Dis Markers. 2020:95053122020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen Z, Wang X, Li L, Han M, Wang M, Li Z,
Xie X, Du H, Xie Z and Zhang H: Construction of an autophagy
interaction network based on competitive endogenous RNA reveals the
key pathways and central genes of SARS-CoV-2 infection in vivo.
Microb Pathog. 158:1050512021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hagag NA, Ali YB, Elsharawy AA and Talaat
RM: Clinical impact of circulated miR-1291 in plasma of patients
with liver cirrhosis (LC) and Hepatocellular Carcinoma (HCC):
Implication on Glypican-3 Expression. J Gastrointest Cancer.
51:234–241. 2020. View Article : Google Scholar
|
|
40
|
Maurel M, Dejeans N, Taouji S, Chevet E
and Grosset CF: MicroRNA-1291-mediated silencing of IRE1alpha
enhances Glypican-3 expression. RNA. 19:778–788. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tu MJ, Duan Z, Liu Z, Zhang C, Bold RJ,
Gonzalez FJ, Kim EJ and Yu AM: MicroRNA-1291-5p sensitizes
pancreatic carcinoma cells to arginine deprivation and chemotherapy
through the regulation of arginolysis and glycolysis. Mol
Pharmacol. 98:686–694. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tu MJ, Ho PY, Zhang QY, Jian C, Qiu JX,
Kim EJ, Bold RJ, Gonzalez FJ, Bi H and Yu AM: Bioengineered
miRNA-1291 prodrug therapy in pancreatic cancer cells and
patient-derived xenograft mouse models. Cancer Lett. 442:82–90.
2019. View Article : Google Scholar
|
|
43
|
Tu MJ, Pan YZ, Qiu JX, Kim EJ and Yu AM:
MicroRNA-1291 targets the FOXA2-AGR2 pathway to suppress pancreatic
cancer cell proliferation and tumorigenesis. Oncotarget.
7:45547–45561. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yamasaki T, Seki N, Yoshino H, Itesako T,
Yamada Y, Tatarano S, Hidaka H, Yonezawa T, Nakagawa M and Enokida
H: Tumor-suppressive microRNA-1291 directly regulates glucose
transporter 1 in renal cell carcinoma. Cancer Sci. 104:1411–149.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Luo H, Guo W, Wang F, You Y, Wang J, Chen
X, Wang J, Wang Y, Du Y, Chen X, et al: miR-1291 targets mucin 1
inhibiting cell proliferation and invasion to promote cell
apoptosis in esophageal squamous cell carcinoma. Oncol Rep.
34:2665–2673. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cai Q, Zhao A, Ren L, Chen J, Liao K, Wang
Z and Zhang W: MicroRNA-1291 mediates cell proliferation and
tumorigenesis by downregulating MED1 in prostate cancer. Oncol
Lett. 17:3253–3260. 2019.PubMed/NCBI
|
|
47
|
Salehi Z, Hadadi P and Tavallaei O:
Prediction of biomarker miRNAs signature in colorectal cancer
metastasis to liver cancer. Electron J Gen Med. 16:em1002019.
|
|
48
|
Morikawa K, Walker SM, Nakajima M, Pathak
S, Jessup JM and Fidler IJ: Influence of organ environment on the
growth, selection, and metastasis of human colon carcinoma cells in
nude mice. Cancer Res. 48:6863–6871. 1988.PubMed/NCBI
|
|
49
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
50
|
Jin H, Li XJ, Park MH and Kim SM:
FOXM1-mediated downregulation of uPA and MMP9 by
3,3′-diindolylmethane inhibits migration and invasion of human
colorectal cancer cells. Oncol Rep. 33:3171–3177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jung H, Kim HS, Lee JH, Lee JJ and Park
HS: Wound healing promoting activity of tonsil-derived stem cells
on 5-Fluorouracil-Induced oral mucositis model. Tissue Eng Regen
Med. 17:105–119. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wu X, Yamamoto H, Nakanishi H, Yamamoto Y,
Inoue A, Tei M, Hirose H, Uemura M, Nishimura J, Hata T, et al:
Innovative delivery of siRNA to solid tumors by super carbonate
apatite. PLoS One. 10:e01160222015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ogawa H, Wu X, Kawamoto K, Nishida N,
Konno M, Koseki J, Matsui H, Noguchi K, Gotoh N, Yamamoto T, et al:
MicroRNAs induce epigenetic reprogramming and suppress malignant
phenotypes of human colon cancer cells. PLoS One. 10:e01271192015.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Takeyama H, Yamamoto H, Yamashita S, Wu X,
Takahashi H, Nishimura J, Haraguchi N, Miyake Y, Suzuki R, Murata
K, et al: Decreased miR-340 expression in bone marrow is associated
with liver metastasis of colorectal cancer. Mol Cancer Ther.
13:976–985. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fukata T, Mizushima T, Nishimura J,
Okuzaki D, Wu X, Hirose H, Yokoyama Y, Kubota Y, Nagata K,
Tsujimura N, et al: The Supercarbonate Apatite-MicroRNA complex
inhibits dextran sodium sulfate-induced colitis. Mol Ther Nucleic
Acids. 12:658–671. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
SCJ (Science Council of Japan): Guidelines
for Proper Conduct of Animal Experiments. 2006.
|
|
57
|
UCI Office of Research: Euthanasia of
Research Animals. Assessment Criteria for Confirmation of Death.
2014.
|
|
58
|
Mogavero A, Maiorana MV, Zanutto S,
Varinelli L, Bozzi F, Belfiore A, Volpi CC, Gloghini A, Pierotti MA
and Gariboldi M: Metformin transiently inhibits colorectal cancer
cell proliferation as a result of either AMPK activation or
increased ROS production. Sci Rep. 7:159922017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Gharib E, Nasri Nasrabadi P and Reza Zali
M: miR-497-5p mediates starvation-induced death in colon cancer
cells by targeting acyl-CoA synthetase-5 and modulation of lipid
metabolism. J Cell Physiol. 235:5570–5589. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zou P, Zhu M, Lian C, Wang J, Chen Z,
Zhang X, Yang Y, Chen X, Cui X and Liu J: miR-192-5p suppresses the
progression of lung cancer bone metastasis by targeting TRIM44. Sci
Rep. 9:196192019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Weng J, Zhang H, Wang C, Liang J, Chen G,
Li W, Tang H and Hou J: miR-373-3p targets DKK1 to promote
EMT-Induced metastasis via the Wnt/β-Catenin pathway in tongue
squamous cell carcinoma. Biomed Res Int. 2017:60109262017.
View Article : Google Scholar
|
|
62
|
Forterre A, Komuro H, Aminova S and Harada
M: A Comprehensive review of cancer MicroRNA therapeutic delivery
strategies. Cancers (Basel). 12:18522020. View Article : Google Scholar
|
|
63
|
Abd-Aziz N, Kamaruzman NI and Poh CL:
Development of MicroRNAs as Potential Therapeutics against Cancer.
J Oncol. 2020:80297212020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Merhautova J, Demlova R and Slaby O:
MicroRNA-Based therapy in animal models of selected
gastrointestinal cancers. Front Pharmacol. 7:3292016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Takahashi RU, Prieto-Vila M, Kohama I and
Ochiya T: Development of miRNA-based therapeutic approaches for
cancer patients. Cancer Sci. 110:1140–1147. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sadanandam A, Lyssiotis CA, Homicsko K,
Collisson EA, Gibb WJ, Wullschleger S, Ostos LC, Lannon WA,
Grotzinger C, Del Rio M, et al: A colorectal cancer classification
system that associates cellular phenotype and responses to therapy.
Nat Med. 19:619–625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yeung TM, Gandhi SC, Wilding JL, Muschel R
and Bodmer WF: Cancer stem cells from colorectal cancer-derived
cell lines. Proc Natl Acad Sci USA. 107:3722–3727. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chen KL, Pan F, Jiang H, Chen JF, Pei L,
Xie FW and Liang HJ: Highly enriched CD133(+)CD44(+) stem-like
cells with CD133(+) CD44(high) metastatic subset in HCT116 colon
cancer cells. Clin Exp Metastasis. 28:751–763. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Dexter DL, Barbosa JA and Calabresi P: N,
N-dimethylformamide-induced alteration of cell culture
characteristics and loss of tumorigenicity in cultured human colon
carcinoma cells. Cancer Res. 39:1020–1025. 1979.PubMed/NCBI
|
|
70
|
Miyamoto M, Sawada K, Nakamura K,
Yoshimura A, Ishida K, Kobayashi M, Shimizu A, Yamamoto M, Kodama
M, Hashimoto K and Kimura T: Paclitaxel exposure downregulates
miR-522 expression and its downregulation induces paclitaxel
resistance in ovarian cancer cells. Sci Rep. 10:167552020.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Seki Y, Yamamoto H, Ngan CY, Yasui M,
Tomita N, Kitani K, Takemasa I, Ikeda M, Sekimoto M, Matsuura N, et
al: Construction of a novel DNA decoy that inhibits the oncogenic
beta-catenin/T-cell factor pathway. Mol Cancer Ther. 5:985–994.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
O'Connell MR, Sarkar S, Luthra GK, Okugawa
Y, Toiyama Y, Gajjar AH, Qiu S, Goel A and Singh P: Epigenetic
changes and alternate promoter usage by human colon cancers for
expressing DCLK1-isoforms: Clinical implications. Sci Rep.
5:149832015. View Article : Google Scholar
|
|
73
|
Sarkar S, O'Connell MR, Okugawa Y, Lee BS,
Toiyama Y, Kusunoki M, Daboval RD, Goel A and Singh P: FOXD3
Regulates CSC Marker, DCLK1-S, and invasive potential: Prognostic
implications in colon cancer. Mol Cancer Res. 15:1678–1691. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Manhas J, Bhattacharya A, Agrawal SK,
Gupta B, Das P, Deo SV, Pal S and Sen S: Characterization of cancer
stem cells from different grades of human colorectal cancer. Tumour
Biol. 37:14069–14081. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kantara C, O'Connell M, Sarkar S, Moya S,
Ullrich R and Singh P: Curcumin promotes autophagic survival of a
subset of colon cancer stem cells, which are ablated by
DCLK1-siRNA. Cancer Res. 74:2487–2498. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ji D, Zhan T, Li M, Yao Y, Jia J, Yi H,
Qiao M, Xia J, Zhang Z, Ding H, et al: Enhancement of sensitivity
to Chemo/Radiation therapy by using miR-15b against DCLK1 in
colorectal cancer. Stem Cell Reports. 11:1506–1522. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Abukhdeir AM and Park BH: P21 and p27:
Roles in carcinogenesis and drug resistance. Expert Rev Mol Med.
10:e192008. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Slingerland J and Pagano M: Regulation of
the Cdk Inhibitor p27 and Its Deregulation in Cancer. J Cell
Physiol. 183:10–17. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
LaBaer J, Garrett MD, Stevenson LF,
Slingerland JM, Sandhu C, Chou HS, Fattaey A and Harlow E: New
functional activities for the p21 family of CDK inhibitors. Genes
Dev. 11:847–862. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wang Y, Ji P, Liu J, Broaddus RR, Xue F
and Zhang W: Centrosome-associated regulators of the G(2)/M
checkpoint as targets for cancer therapy. Mol Cancer. 8:82009.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
To KK, Tong CW, Wu M and Cho WC: MicroRNAs
in the prognosis and therapy of colorectal cancer: From bench to
bedside. World J Gastroenterol. 24:2949–2973. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Chi Y and Zhou D: MicroRNAs in colorectal
carcinoma-from pathogenesis to therapy. J Exp Clin Cancer Res.
35:432016. View Article : Google Scholar
|