Background
Precision medicine
The paradigm shift (1) that emphasizes preemptive over
responsive medicine, while also actively involving the individual
patients in their therapy, is coined with the term precision
medicine (PM) (2). PM, in
essence, is a medical approach that uses individual genotypes and
phenotypes for tailoring the right therapeutic strategy to the
right individual at the right time (3). This medical process classifies
patients into subgroups based on their characteristics, which is a
significant divergence from the 'one-size-fits-all' model, rather
than forming strategies unique to each patient (4). Oncology currently stands at the
forefront of the medical fields which has benefited from PM
(5) due to the in silico, in
vitro and in vivo improvements in disease modelling
(6), and also due to the genomic
characterization of thousands of cases (7).
The majority of cancerous malignancies are driven by
genomic alterations that influence key oncogenic pathways leading
to the genesis and progression of the disease (5,8).
Early detection of those drivers brings forth the promise of
genome-driven oncology care (9).
Neither the idea of the characterization of malignancies based on
the genomic profile of a patient, nor the notion that medicine
needs to be more anthropocentric and preemptive, are novel
approaches. For example, the relevance of karyotypes to the
decision as to whether a patient with acute myelogenous leukemia
(AML) should be administered chemotherapy (10), and the mention of the '4Ps'
(predictive, preventive, personalized and participatory) in
medicine (11-14), are both ideas that were proposed
at times when both the expertise and the technological capabilities
were inadequate for addressing them any further.
The framework changed with the completion of the
Human Genome Project (HGP) (15)
followed by the emergence and establishment of next-generation
sequencing (NGS) as the de facto sequencing method. While
triggering fundamental changes in the field of biology and
practically encouraging biological research to adopt more
data-driven approaches (16), the
unique features of NGS also cover the needs of the routine clinical
practice (17), by improving the
clinical outcomes for many patients with cancer. Using NGS (also
known as massively parallel or deep sequencing), an entire human
genome can be sequenced within a single day, whereas with the
previous Sanger sequencing technology, the first assembly of the
human genome required over a decade (18). With the availability of the genome
sequence and the technological advancements in sequencing
technologies, which became progressively more accurate and
affordable, large-scale initiatives such as The Cancer Genome Atlas
(TCGA), and the International Cancer Genome Consortium (ICGC)
(19), have paved the road for
the development of the 'cancer genomics' field (5). In cancer genomics the fundamental
premise is that cancer is caused by somatically acquired mutations
and therefore it is a disease of the genome (18).
Biobanking
The 'fuel' that drives progress in cancer genomics
consists of the samples that individual patients provide to the
research teams worldwide. These samples are stored in
biorepositories that accept, process, store and distribute them
along with their associated data and meta-data (20). These biorepositories are referred
to as 'biobanks', a term that was used for the first time by Loft
and Poulsen in 1996 (21), and
are ranked among the most important research infrastructures in
cancer research (22). The
Organization for Economic Co-Operation and Development (OECD)
defines a biobank, 'as a collection of biological material and the
associated data and information stored in an organized system, for
a population or a large subset of a population' (23,24). While the biobanking taxonomy
incorporates a wide area of biobank types (e.g., commercial,
DNA/RNA, project-driven, virtual and more) (20), a generic distinction can be made
between disease-centric and population-based biobanks (25). An important issue of biobanks is
their heterogeneity and therefore, a proper biobank and sample
classification is of utmost importance (26).
The field of biobanking is constantly evolving.
Individual and small university-based repositories and sample
collections have gradually become institutional, commercial, and
government supported repositories, while the complexity of the data
associated with the samples has also increased exponentially, from
basic information such as the date of the sample's collection, to
the extensive information accompanying the various '-omic'
(genomic, transcriptomic, proteomic) technologies (20). The number of samples and the
available data that accompany them should adequately cover the
broad spectrum of sub-entities emerging in malignant neoplasm
diseases in reference to PM; these are constantly growing and
differentiating, thus offering more and more areas of study for the
application of both the available targeted therapies and those that
are being developed at a rapid pace. Additionally, the operating
regulations of modern biobanks must guarantee both the management
of high-quality samples, as well as the compliance with ethical,
legal, and societal requirements, while allowing transparency and
effective procedures regarding access to samples and data (3).
Pan-European initiatives
While decentralized individual collections can be
well organized and accessible, there is a need for harmonization
between the collections. Accessibility and funding issues, ethical
frameworks and protocols regarding the sample collections and their
storage, must be approached in a way that ensures interoperability
and 'common ground' between biobanks. The difference between
'harmonization' and 'standardization' of biobanks is that the
former is a more flexible approach that facilitates the effective
interchange of valid information and samples, while the latter is
an approach that demands that the same protocols and standard
operating procedures (SOPs) are used by all biobanks (27,28).
To address these issues, the European Commission
developed a Biobanking and BioMolecular Resources Infrastructure
(BBMRI) (http://www.bbmri-eric.eu/), providing
a legal and ethical framework for biobanking across the European
Union. BBMRI includes comprehensive collections of biological
samples from different sub-populations of Europe, linked with
continuously updated data on the health status, lifestyle, and
environmental exposure of the sample donors (29). Another project that has worked in
tandem with BBMRI inside the EU is the Biobank Standardization and
Harmonization for Research Excellence in the European Union project
(BioSHaRE-EU) (http://www.bioshare.eu/). Biobank development has been
stated as one of the key challenges of the last two decades in EU.
Europe is not the only continent that progresses the biobanking
field. In the international landscape, the effort for instigating
the inter-communication in biobanking is driven by the National
Cancer Institute (NCI) (https://biospecimens.cancer.gov/), the International
Cancer Genome Consortium (ICGC) (https://daco.icgc.org/), the Public Population Project
in Genomics and Society (P3G, renamed to
P3G2) (http://p3g2.org/), and the
International Society of Biological and Environmental Repositories
(ISBER) (https://www.isber.org/). As also
indicated by the current SARS-CoV-2 pandemic, the collaboration
between ISBER and BBMRI is expanding so as to cover a broader
spectrum (30).
The Hellenic Network of Precision
Medicine on Cancer
In order to address the needs of oncology patients,
the Hellenic Network of Precision Medicine on Cancer (HNPM) was
founded in Greece on 17/05/2018 at the initiative of the Research
and Innovation Department of the Ministry of Education, Research
and Religion in close collaboration with the Ministry of Health.
The project is supervised by the General Secretariat of Research
and Innovation (GSRI) and funded by the Hellenic Republic-Siemens
Settlement Agreement framework. This multi-stakeholder initiative
has grown rapidly since its inception and aims to establish a
nation-wide infrastructure which provides high-quality healthcare
to Greek citizens, enrich diagnosis knowledge and prediction
outcome and improve the targeted therapeutic treatments of cancer
patients (3).
A partner institution collaborating for the
formation of the HNPM is the 1st Department of Pathology, which
belongs to the Medical School of the National and Kapodistrian
University of Athens (NKUA). In addition to academic activities, it
conducts histologic examinations for 'Laiko' Hospital and a
substantial number of other hospitals all over Greece. More than
200,000 paraffin blocks are stored in the files of the department,
making it the largest tissue collection in Greece. The vast
majority of specimens represent malignant neoplasms, among which,
solid tumors, hematologic malignancies, myoskeletal and nervous
system tumors, figure prominently. This is because the 1st
Department of Pathology is a reference center for these tumors. New
diagnostic techniques include, but are not limited to, the analysis
of biomarkers in the context of molecular targeted therapeutic
approach of malignant neoplasms. During the last decade, the 1st
Department of Pathology has been actively involved in a European
brain tissue bank network named BrainNet Europe II (Network of
European Brain and Tissue Banks for Clinical and Basic
Neuroscience), a project funded by the European Commission's 6th
Framework Program for Research, aiming to define 'gold-standards'
in tissue sampling practice and neuropathological diagnostics. In
the context of the HNPM, the 1st Department of Pathology has
implemented the biobanks sub-project, in close cooperation with the
Biomedical Research Foundation of the Academy of Athens (BRFAA),
which is the central hub of the Greek biobanks, all of which
together form the BBMRI-GR network, an official member of the
pan-European Biobanking and BioMolecular resources Research
Infrastructure, European Research Infrastructure Consortium
(BBMRI-ERIC).
Purpose
This publication focuses on a) delineating the
regulatory framework regarding biobanking development in Greece, b)
analyzing the methodologies that were followed in order to create a
pilot biobank of rare malignant neoplasms, in the context of the
HNPM, and c) presenting the clinical importance of the biobank. In
respect to the multidimensionality of the pilot biobank project,
the basic goals that were set are the following: i) the
organization of sample collections consisting of specimens from
patient cohorts with specific types of malignant neoplasms and with
available clinical information; ii) implementation of a Pilot
Biobank Management System (PBMS) compatible with the Minimum
Information About BIobank data Sharing (MIABIS) model; iii)
harmonization of best practices regarding biobank functions; iv)
ethical and legal framework clarifications aligned with the General
Data Protection Regulation (GDPR); and v) connection of the biobank
of rare malignant neoplasms of the 1st Department of Pathology with
the BBMRI-GR network.
Methodologies and tools
Research and methods
Throughout the course of the HNPM's biobank project,
including this publication, a wide selection of manuals and schemas
were reviewed, and a number of best practices were adopted
(Table I) (27,31-47). The following sections analyze the
methodologies and the tools that were utilized in creating the
infrastructure of the pilot biobank of rare malignant
neoplasms.
 | Table ISelected publications on biobank
operations and best practices. |
Table I
Selected publications on biobank
operations and best practices.
| Name of
publication | (Refs.) |
|---|
| Biobanking for
Epidemiological Research and Public Health | (31) |
| Biobanking for
Interdisciplinary Clinical Research | (32) |
| Biobanks for
Europe: A Challenge for Governance | (27) |
| Common Minimum
Technical Standards and Protocols for Biobanks Dedicated to Cancer
Research | (33) |
| GDPR and
Biobanking: Individual Rights, Public Interest and Research
Regulation across Europe | (34) |
| Human Tissue
Monitoring and Specimen Banking | (35) |
| Innovation in
Scientific Research and Emerging Technologies: A Challenge to
Ethics and Law | (36) |
| ISBER 2005, 2008,
2012 and 2018 Best Practices for Repositories Publications | (37-40) |
| Minimum Information
About BIobank data Sharing (MIABIS) Publications | (41-44) |
| NCI Best Practices
for Biospecimen Resources | (45) |
| Standard
Preanalytical Coding for Biospecimens: Defining the Sample
PREanalytical Code | (46) |
| The Legal
Regulation of Biobanks: National Report: Greece | (47) |
Biological material
The Biobank currently comprises of 553 samples,
specifically 384 samples of hematopoietic and lymphoid tissue
malignancies, 72 samples of pediatric brain tumors and 97 samples
of malignant skin neoplasms. These samples have been collected from
the collaborating groups of scientists at the 1st Department of
Pathology and the Laboratory of Biological Chemistry, Medical
School of the NKUA, and also from the 'Andreas Syggros' Hospital of
Cutaneous and Venereal Diseases, under the supervision of the 1st
department of Pathology (Fig. 1).
All formalin-fixed paraffin-embedded (FFPE) samples were evaluated
by hematoxylin and eosin (H&E) and microscopy regarding the
quality of tissue, histological aspects, and any potential damage
during processing. The samples that deviated from optimal quality
have been excluded from the biobank. A number of these samples have
also been evaluated by PCR and immunohistochemistry (IHC) to assess
the genetic and molecular profile of the tissues. Cryopreserved
samples were evaluated by qPCR, RT-PCR, and western blot analysis
for assessment of RNA and protein quality. The majority of the
samples have also been included in research protocols and published
studies in international peer-reviewed journals, indicating both
their quality as biological materials as well as their reliability
(48-62).
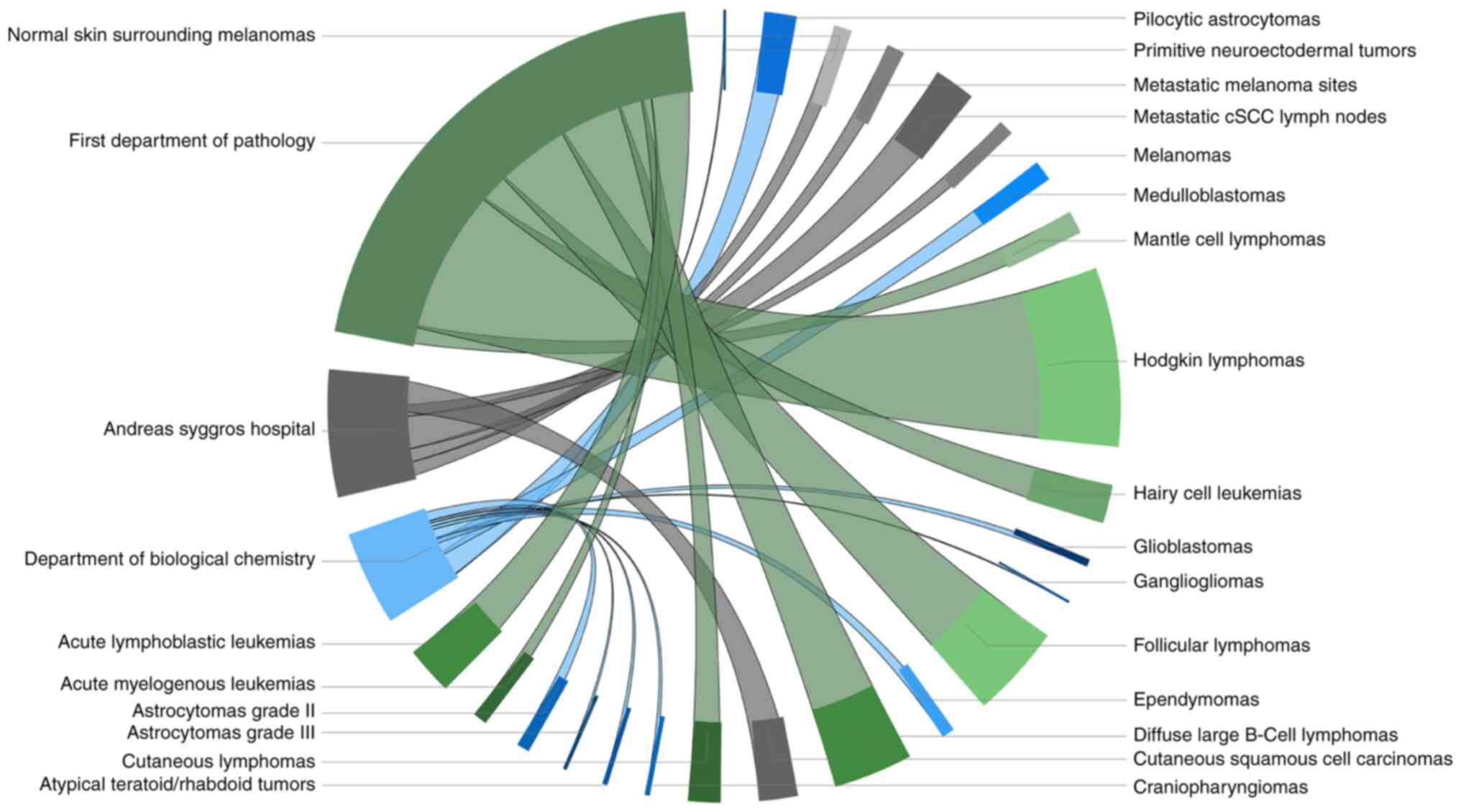 | Figure 1Sample distribution by department and
tumor type. Each disease collection is illustrated by the same
color palette as the department to which it belongs, while the part
of the circle it occupies is proportional to the number of samples
it contains. Specifically, collections of hematopoietic and
lymphoid tissue malignancies (384 samples) from the 1st Department
of Pathology are represented with green (follicular lymphomas,
diffuse large b-cell lymphomas, hodgkin lymphomas, mantle cell
lymphomas, hairy cell leukemias, cutaneous lymphomas, acute
myelogenous leukemias, acute lymphoblastic leukemias), pediatric
brain tumors (72 samples) from the Department of Biological
Chemistry with blue (pilocytic astrocytomas, astrocytomas grade ll,
astrocytomas grade lll, glioblastomas, medulloblastomas,
ependymomas, atypical teratoid/rhabdoid tumors, craniopharyngiomas,
gangliogliomas, primitive neuroectodermal tumors) and lastly
malignant skin neoplasms (97 samples) from the 'Andreas Syggros'
Hospital are rendered in gray (melanomas, normal skin surrounding
melanomas, metastatic melanoma sites, cutaneous squamous cell
carcinomas, metastatic cSCC lymph nodes). This graph was
illustrated with the open-source visual 'Chord', a PowerBI business
analytics service add-on by Microsoft. cSCC, cutaneous squamous
cell carcinoma. |
As far as hematologic malignancies are concerned,
the tumor sample collection and its associated data consisted of a
multistep procedure that required the collaboration of clinicians
from the Hematology Department of General Hospital 'Laiko', NKUA.
First, patients with hematologic malignancies were retrieved
throughout a clinical database according to their initial
histologic diagnosis. The treating physician of each patient was
responsible to decide who was eligible for the biobank formation
depending on the availability of data regarding each case. A
process of matching the selected patients to their respective
biological material, consisting mainly of FFPE tumor samples (lymph
node, bone marrow and spleen biopsies) was the next step. A
specially authorized team collected, assessed, archived, and stored
the samples of each collection. The team consisted of experienced
pathologists who confirmed the diagnosis based on the histological
examination of each sample in respect to the clinical information
collected from the treating physician and biochemists who evaluated
the quality of the samples, and other life scientists who performed
the collection/archiving processes. Finally, a list of FFPE tumor
samples and the associated clinical data for each individual
hematologic disease was obtained. Patient anonymity as well as
confidentiality of the clinical information was ensured at every
step of this procedure.
Regarding the pediatric brain tumors, two different
tissue collections were assembled from the 1st Pediatric
Neurosurgery Department of 'Mitera' Hospital and the Department of
Neurosurgery of 'Aghia Sofia' Children's Hospital along with their
histopathological diagnosis. All cases included were of clinical
interest due to their rarity and their heterogeneous
clinical/pathological characteristics. The executive clinician for
the malignant brain tumors, collected all FFPE samples, archived,
and stored the samples of each collection separately.
Concerning malignant skin tumors and their
metastatic sites, two different collections were created based on
the clinical information from the Plastic Surgery Department of
'Andreas Syggros' Hospital and the histological diagnosis from the
Department of Pathology of 'Andreas Syggros' Hospital. The two
collections concern metastatic (late-stage) cutaneous squamous cell
carcinomas (CSCCs) along with metastatically involved lymph nodes
and primary melanomas with adjacent surrounding tissue and either
regional or distant metastases (lymph nodes, lung, brain). All
cases included were of clinical interest due to their rarity or
distinguishing clinical/pathological characteristics. The executive
clinician for the malignant skin tumors, collected all FFPE
samples, archived, and stored the samples of each collection
separately. All collections were formed guarantying patient
anonymity, each of whom was enrolled with a code number based on
the number of their FFPE tumor sample.
The pilot biobank of rare malignant neoplasms mainly
consists of FFPE samples (535 samples). FFPE blocks are stored in a
controlled low-humidity environment at room temperature (20-25°C),
as per the current best practice for FFPE tissue block and slide
storage (40,63). The biobank also contains 18
cryopreserved astrocytoma samples, stored at −80°C.
Qualification of samples
Downstream analysis largely depends on the existence
of homogenous biobanks. Preanalytical factors may have an impact on
the quality of the collected biospecimens by introducing
uncontrollable variables (64).
At any failure sample testing process, the results of subsequent
analysis may be of little to no value. Inappropriate sample
collection can obstruct the identification of novel biomarkers,
while also having a negative economic impact; e.g. up to 10 million
euros of funding are lost each year in clinical trials due to
preanalytical and analytical problems (63). Therefore, qualification processes,
such as quality assurance (QA) and quality control (QC) processes,
are mandatory in order to assess the quality of biospecimens before
their storage into a biobank. The terms are utilized as they were
defined in the study of Dr Fay Betsou in 'Quality Assurance and
Quality Control in Biobanking'. In the aforementioned work,
detailed definitions of common terms used in biobanking are also
provided (65).
There are two main approaches for qualification: a)
the careful collection of samples with pre-analytical annotation by
implementing the Standard PREanalytical Code (SPREC) (46), and b1) the
classification of biospecimens, contained in retrospective
collections, in different categories corresponding to in
vivo parameters (quality stratification) or b2) the
examination and validation of a single biospecimen or a collection
on the basis of objective analytical parameters (sample
qualification) (25,66). The quality process used in the
biobank of rare malignant neoplasms, considering that the biobank
mainly consists of FFPE tissues, was quality stratification
(Table II) (66) of archival samples stored in the
collection of the 1st Department of Pathology. All pediatric brain
tumor samples have been evaluated by H&E staining and
microscopy. Astrocytoma samples have also been evaluated by qPCR,
RT-PCR, and western immunoblotting.
 | Table IIQC measurands for quality
stratification of FFPE tissue specimens (66). |
Table II
QC measurands for quality
stratification of FFPE tissue specimens (66).
| Biospecimen
type | Quality
stratification parameter | Quality
stratification parameter category | Measurand | Quality
stratification threshold | Measurement method
and reference |
|---|
| Tumor FFPE | % tumor | Tumor-rich | Tumor | >70% | H&E staining,
digital pathology |
| Fixation time
NBF | >72 h | None to date | TBD | RT-qPCR |
| Fixation
conditions | NBF (no acidic
formalin) | Size range RT
PCR | ~250 bp | RT-PCR |
| Cold ischemia | >12 h | None to date | TBD | RT-qPCR |
| Tumor frozen
tissue | Cold ischemia | >12 h | None to date | TBD | H&E staining,
RT-PCR |
Formalin-fixed paraffin-embedded
tissues
FFPE is a form of biopsy specimen preservation which
prevents tissue degradation and has become the standard
preservation procedure for diagnostic pathology. The biobank of
rare malignant neoplasms largely consists of FFPE blocks. Although
there are other alternatives such as fresh-frozen (FF) tissue, FFPE
blocks remain the most common and cost-effective preservation
method. FFPE samples can sometimes match with FF samples in the
quality of the data produced by NGS experiments (67-71). NGS can be used to study FFPE
specimens in both prospective and retrospective archive-based
studies in which FF specimens are not available (71). Both DNA and RNA can be
successfully extracted from FFPE blocks, but it is a more delicate
procedure than extraction from FF tissues. The long-term storage at
room temperatures, may generate genomic mutations resulting in the
identification of false single nucleotide variations (SNVs); this
is correctable by choosing high sequencing depths (80X at least)
when analyzing FFPE materials (72). Another study suggests that the
routine processing of FFPE samples may have a detectable, albeit
negligible effect on NGS data, therefore not impacting the
reliability of clinical NGS testing (67). The last potential threat to the
quality and amplifiability of the extracted DNA is the method of
deparaffinization. Higher melting temperature can result in the
denaturation of the double-stranded DNA, while lower melting
temperature can result in improper melting of paraffin and
therefore reducing the DNA yield. It is suggested that using a
temperature of 75°C for 5 min, results in no loss of DNA content
(72).
Other sample types
Frozen samples of 18 pediatric astrocytomas are
stored at -80°C. Fresh freezing is currently the preservation
method that is most compatible with subsequent proteomic analyses.
Fixation by formaldehyde crosslinking is incompatible with some
proteomic analyses, necessitating a change in the routines of
histopathological sample handling, such that the tissue must be
received unfixed.
Associated data and clinical
information
All pediatric brain as well as hematologic tumor
samples are accompanied by histopathological records, demographic
data, and informed consent of patients/parents of the patients or
their legal guardians. All samples have additional
clinicopathological data (molecular data, IDH1, R132H, H3K27M, BRAF
mutations, treatment information, response to therapy and patient
follow up). To ensure the anonymity of patients and the
confidentiality of their information, each patient is assigned a
number and only authorized members have access to this data.
Creating the pilot biobank management
system
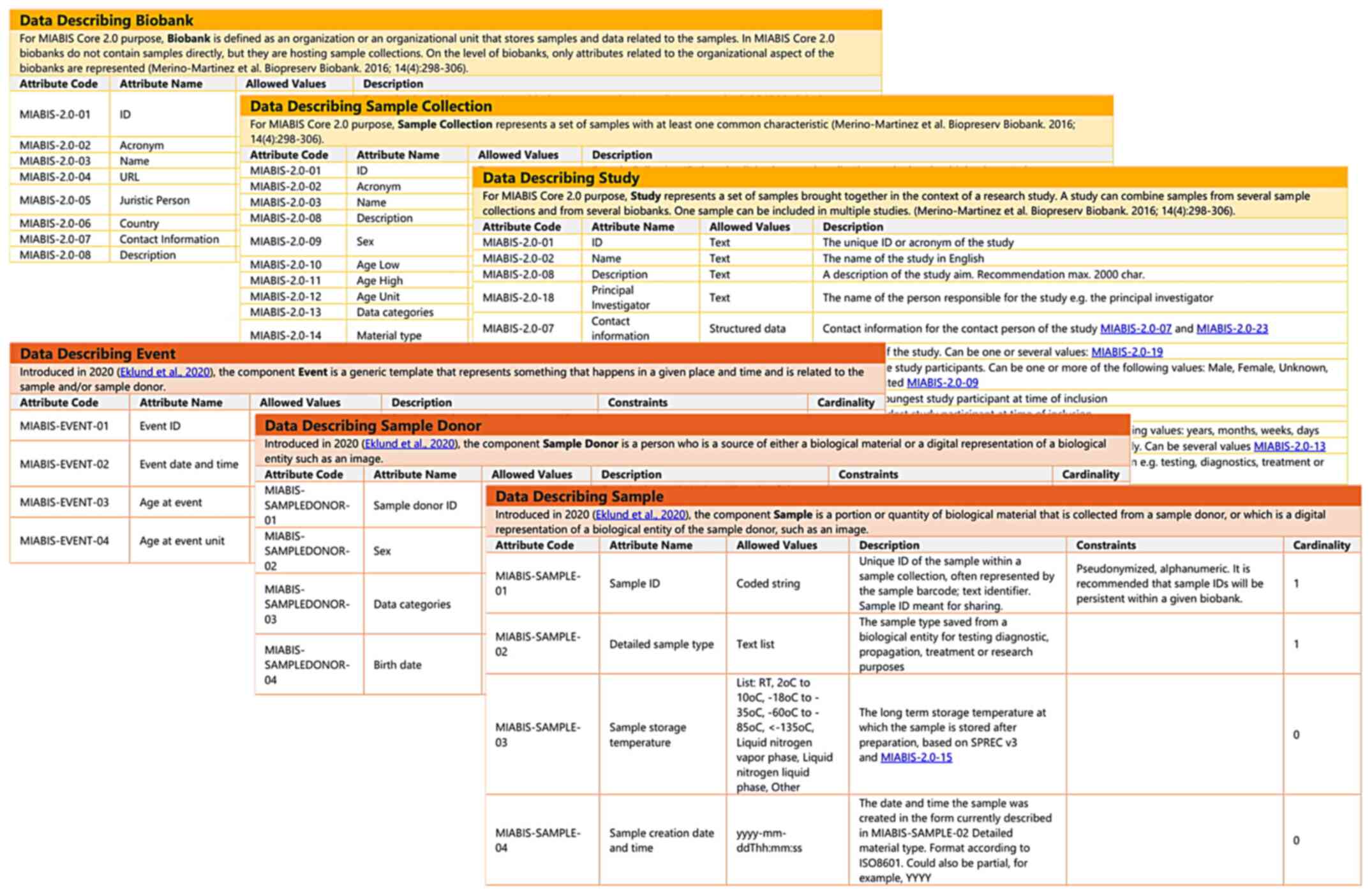
Minimum information about biobank data
sharing model
A crucial tool for effective networking and resource
sharing that was adopted in this present work is the MIABIS model
(Fig. 2). MIABIS 1.0 was launched
in 2012 and aimed to facilitate data discovery through
harmonization of data elements describing a biobank at the
aggregate level (44). It
recommended the minimum data items and the format of the items
required to enable the exchange of biological samples and data
(64). MIABIS 2.0 was released in
2016 and provided the ontology that represents the administrative
entities regarding a biobank, while also defining the core
components needed to describe biobanks (biobanks, sample
collections and studies) (41,43). An extension of MIABIS was released
in 2020 and included an updated terminology to describe samples,
sample donors and events (42).
MIABIS is currently being updated to version 3.0 and will include
updates identified by BBMRI-ERIC Common Service IT (CS-IT) working
group (73,74). Every MIABIS component along with
the structured data and lists [as defined in the MIABIS main
repository (73)] were
implemented in the PBMS.
Pilot biobank management system
The digital representation of the biobank is
implemented as a relational database, using MySQL language. Tables
and relations are structured in accordance with MIABIS. In order to
easily access and modify the biobank's samples and their associated
data, a desktop interface was developed as to enable non-IT
personnel to perform basic CRUD (create, read, update, delete)
operations to the database. The interface was developed via the
open-source graphical subsystem: Windows Presentation Foundation
(WPF), using C# as the coding language. To ensure anonymity,
sensitive information is encrypted as described in the Privacy and
Data Protection subsection. On top of the pseudonymization, log-in
authentication was implemented on the interface and the database
levels as an additional measure.
Framework and processes
The European regulatory framework
Ethical, legal and societal issues (ELSI) and
certain bioethical aspects are a constantly evolving debate topic
among committees, scholars, and policy makers. The ELSI approach is
considered to facilitate the advancement of genomic technology
rather than hinder it (75).
Biobanking regulations within the EU are highly heterogenous.
Biobanks, in general, are governed under the general regulatory
framework for biomedical research. Biobanking development is
inhibited by the lack of EU legal-binding documents specifically
applicable to biobanks, barring legislation at the national level
(27). Several attempts have been
made for the unification of European regulations regarding
research. For example, the 04/04/1997 Oviedo Convention on Human
Rights and Biomedicine (76), is
the basis for safeguarding the rights of human subjects regarding
scientific progress within the EU countries which have signed and
ratified the convention (27). At
the national level, research ethics committees (RECs) determine
whether research should proceed within the national legal
frameworks instead. Various other regulations that are applicable
to biomedical research at the European level have been introduced
(Table III) (77-86).
 | Table IIILegally binding or non-binding
instruments regarding biobank regulation within Europe. |
Table III
Legally binding or non-binding
instruments regarding biobank regulation within Europe.
| Regulation no. | Brief
description | (Refs.) |
|---|
| 2001/20/EC | Clinical Trial
Directive | (77) |
| EU 536/2014 | Clinical Trials
Regulation | (77) |
| ETS No. 5 | Convention for the
Protection of Human Rights and Fundamental Freedoms | (78) |
| ETS No. 108 | Convention for the
Protection of Individuals with regard to Automatic Processing of
Personal Data | (79) |
| ETS No. 164 | Convention on Human
Rights and Biomedicine | (80) |
| 95/48/EC | Data Protection
Directive (Repealed by GDPR) | (81) |
| CM/Rec/2016/6 | Recommendation on
Research on Biological Materials of Human Origin | (82) |
| A/RES/217(III) | The Universal
Declaration of Human Rights | (83) |
| 2004/23/EC | Tissues and Cells
Directive | (84) |
| WMA/1964 | World Medical
Association Declaration of Helsinki: Ethical Principles for Medical
Research Involving Human Subjects | (85) |
| WMA/2002 | World Medical
Association Declaration of Taipei: Ethical Considerations Regarding
Health Databases and Biobanks | (86) |
Regardless, due to the existence of national-level
regulations, further ameliorations need implementation at a
cross-border level in the EU. The complexity that researchers
across the EU face, might place them at risk of operating
unlawfully by sharing data and samples across borders where
different laws are in force (27).
The Greek landscape
Lack of specific biobanking legislation along with
the absence of discrete terminology concerning biobanking (in the
context of the Greek law), heavily obstructs further scientific
research. Biobanking governance in Greece consists of a complex web
of generally applied constitutional provisions, laws, regulations,
codes of practice, guidelines and so forth (Table IV) (34,47,87), each of which can potentially apply
to biobanks (47). The only piece
of legislation wherein collections of tissues are referred to in a
systematic way, is the Presidential Decree (26/2008), that sets
quality and safety standards for the donation, procurement,
testing, processing, preservation, storage, disposal of dangerous
substances, and distribution of human tissues and cells. While not
directly applicable to biobanks, it applies to biobanks which store
stem cells, in which case the Hellenic Transplant Organization
(HTO/EOM) is responsible for authorization (34). Since biobanks contain personal
data along with biological samples, the participants' privacy is
regulated by Law 2472/1997 (Protections of Individuals with regard
to the Processing of Personal Data) and more recently, GDPR.
Furthermore, researchers and physicians acting within a biobank are
bound by the Code of Medical Ethics/Deontology (Law 3418/2005),
which secures protection to the donors' privacy. Several agencies
are also in charge of monitoring privacy, and subsequently,
biobanking in Greece.
 | Table IVLegally binding or non-binding acts
and experts' opinions regarding biobank regulation in Greece. |
Table IV
Legally binding or non-binding acts
and experts' opinions regarding biobank regulation in Greece.
| Law no. | Brief
description | (Refs.) |
|---|
| Law 2472/1997 | On the Protection
of Individuals with regard to the Processing of Personal Data,
Implementing Directive 95/46/EC | (34,47) |
| Law 3305/2005 | Application of the
Methods of Medically Assisted Reproduction | (47) |
| Law 3418/2005 | Code of Medical
Ethics/Deontology | (34,47) |
| DYG
3/89292/2003 | Ministerial
Decision, Implementing Directive 2001/20/EC | (34) |
| 52/2006 | National Bioethics
Commission Recommendation on Banks of Biological Material of Human
Origin (Biobanks) in Biomedicine Research | (87) |
| Law 2723/1999 | On Transplantations
of Tissues and Organs | (47) |
| 115/2001 | Opinion of the
Hellenic Data Protection Authority on Processing Employees'
Personal Data | (47) |
| Law 4624/2019 | Personal Data
Protection Authority, Implementing Measures for Regulation EU
2016/679 | (34) |
| 26/2008 | Presidential
Decree, Implementing Directive 2004/23/EC | (34) |
| Law 2619/1998 | Ratification of the
Oviedo Convention | (34) |
| Law 4386/2016 | Regulations for
Research, Regulating Administrative Aspects of Research | (34) |
| Law 4521/2018 | Research Ethics and
Deontology Committees | (34) |
The establishment of Research Ethics and Deontology
Committees (REDCs) in all universities and research institutions,
provides additional overseeing by examining whether research
projects respect humans' inherent value as well as participants'
autonomy, private life, and personal data (34). The Hellenic Data Protection
Authority (HDPA) is responsible for assessing data protection
violations and HDPA's prior permission is a precondition for the
collection and processing of sensitive data (47).
Privacy and data protection
Biobanks should always protect the patient's
identity in order to ensure their privacy. A distinction enshrined
in the GDPR, is the one between anonymized and pseudonymized data.
Anonymized data fall outside the jurisdiction of GDPR; even so, the
act of anonymization itself is considered as an act of processing
personal data, which should occur in compliance with the GDPR
(34). However, in the context of
biobanks, the patient must be able to be re-identified to provide
relevant information back to researchers (24), so irreversible anonymization is
not applicable to most types of biobanks. Contrastingly,
pseudonymization of personal data is a reversible act.
Pseudonymized data, as defined in the GDPR, are data that 'can no
longer be attributed to a specific data subject without the use of
additional information, provided that such additional information
is kept separately and is subject to technical and organizational
measures to ensure that the personal data are not attributed to an
identified or identifiable natural person' (34). The cryptographic algorithm that
was implemented for the confidentiality of patient and researcher
data is the 128-bit block cipher: Advance Encryption Standard
(AES-Rijndael) (88). AES was
included in the ISO/IEC 18033-3 standard and is currently effective
as a US federal government standard since 26/11/2001 (FIPS PUB 197)
(89).
Informed consent
The collection of several samples is dated before
the GDPR's implementation in EU; and therefore a number of samples
were obtained by the laboratory's physicians via the 'broad
consent' model, which is the agreement for the utilization of the
patient's sample for an unspecified range of future or current
research subjects to a few content and/or process restrictions.
Broad consent is less specific than consent that changes scope
overtime (e.g., 'dynamic consent'), but more narrow than open-ended
permission without limitations ('blanket consent') (90,91). In the context of HNPM, a special
questionnaire and leaflet were formulated to effectively inform
competent individuals about the collection of their specimens. The
purpose was to reassure the individual patients that their sample
may be utilized, with complete anonymity and strictly for research
purposes only, while underlying that 'no ownership of biological
samples exists' as declared by the International Agency for
Research on Cancer (IARC) (33).
In addition, as previously stated, the majority of samples are part
of studies already published in international peer-reviewed
journals, and ethical approval has been obtained for all tissue
samples from the University Hospitals' Ethics Boards and/or the
Bioethics Committee of the University of Athens Medical School.
Governance model and access processes for
samples and data
As previously mentioned, this is a pilot biobank and
therefore the final governance model has not yet been defined. An
exploratory analysis has identified six types of commonly adopted
biobank governance strategies: communication, compliance, expert
advice, external review, internal procedures, and partnerships
(92). Some strategies that were
adopted in the formation of this biobank are the following.
Communication
The structure and the activities regarding this
biobank have been included in HNPM's technical conferences/online
workshops and have been presented to Greek cancer patient
associations and the public, therefore gathering impactful
feedback.
Compliance
This biobank followed GDPR regulations, and a number
of best practices were adopted.
Internal procedures
All samples and data were collected following
informed consent processes. Moreover, various QC measurands are
used to assess the quality of the final samples, and
pseudonymization processes ensure the privacy of the donors.
Partnerships
This biobank is a result of three collaborating
entities (1st Department of Pathology, Laboratory of Biological
Chemistry and 'Andreas Syggros' Hospital of Cutaneous and Venereal
Diseases) under the context of a Panhellenic initiative currently
comprising more than 13 partner institutions.
While the pilot biobank of rare malignant neoplasms
is currently housed at the 1st Department of Pathology, it is
created in the context of HNPM and will subsequently be part of the
BBMRI-GR network, therefore the governance model is subject to
further change. Besides regulations of the three collaborating
entities, HNPM consists of two governing entities, the Scientific
Committee, and the Technical Committee.
As far as sample and data access is concerned, HNPM
has comprehensive forms that interested parties need to fill, and
they contain instructions for accessing the samples and associated
data regarding the biobank of rare malignant neoplasms. Researchers
can apply to study the samples as well as the information stored in
the biobank of rare malignant neoplasms. This includes researchers
from universities or research institutes, the government, and drug-
or health-related companies. Each application will be reviewed by
the Scientific Committee of HNPM, which evaluates each candidacy
based on the following criteria: a) activity related to the subject
of HNPM, b) available infrastructure, c) professionally trained and
experienced staff, and d) process standardization.
When needed an REC will also be formed. This kind of
review ensures that risks are minimized and that the rights and
welfare of people who participate in research are protected. When a
study is approved by the scientific committee, a part of each
biospecimen and additional clinical information might be
distributed to the researchers. Researchers will not be given any
participants' names and/or any other information that could
potentially reveal patient's identity.
Pilot biobank of rare malignant
neoplasms
Sample collections: 1st Department of
Pathology, Medical School, NKUA
Acute leukemia
Acute leukemias are a group of life threatening
hematologic malignant disorders, affecting both children and
adults. The diagnosis relies on cytomorphology, cytochemistry, flow
cytometry, immunophenotyping in FFPE, cytogenetics and molecular
genetics, including detecting recurrent genetic abnormalities. Each
case requires an integrated, multimodality approach, which is a
prerequisite for optimal diagnosis and management. As leukemias are
characterized by extreme heterogeneity, both at the molecular level
and in regards to the clinical outcome, they represent an excellent
field of research, as molecular genetics has led to the
identification of abnormal gene products and pathways and
delineation of specific entities (93,94).
Cutaneous lymphoma
Primary cutaneous lymphomas are non-Hodgkin
lymphomas affecting the skin as the primary site, without
extracutaneous involvement at the time of diagnosis. The majority
consists of cutaneous T-cell lymphomas (CTCLs), accounting for
65-75% of all cases. Mycosis fungoides is the most common CTCL (50%
of all primary cutaneous lymphomas) (95), characterized by skin infiltrates
of small to medium-sized lymphocytes, which is clinically
characterized by the evolution of patches, plaques and tumors.
Among the cutaneous B-cell lymphomas (CBCLs), primary cutaneous
marginal zone lymphoma and follicle center cell lymphoma follow a
more indolent behavior, whereas primary cutaneous diffuse large
B-cell lymphoma, leg type, accounts of 4% of all cutaneous
lymphomas and is associated with a significant risk of systemic
dissemination (48,49,60,96-99).
Diffuse large B-cell lymphoma
Diffuse large B-cell lymphoma represents
approximately 25-35% of all adult non-Hodgkin lymphomas. It is an
entity mainly affecting the elderly, but it can uncommonly occur in
children and young adults. It may present as nodal or extranodal
disease, the latter representing 40% of the cases, affecting the
gastrointestinal tract, bone, testis, spleen and virtually any body
site. It can arise de novo or as aggressive transformation of an
indolent B-cell lymphoma. Risk factors for development include
underlying immunodeficiency and Epstein-Barr infection. Currently
there is novel data elucidating the mutational land-scape of
diffuse large B-cell lymphoma and its pathogenesis. As a highly
heterogeneous group of neoplasms, it is difficult to distinguish
patients that will benefit from more aggressive treatment strategy.
Therefore, it is an excellent candidate for further research in the
context of PM (50,100).
Follicular lymphoma
Follicular lymphoma is a common, indolent B-cell
lymphoma, clinically characterized by diffuse lymphadenopathy, bone
marrow involvement, and splenomegaly with a median age at diagnosis
of 65 years. It originates from germinal center B cells, and the
neoplastic population is composed of centrocytes and centroblasts
with at least partially follicular pattern. Hallmark genetic
finding is the t(14;18) (q32;q21) translocation, present in 90% of
low-grade cases, between the IGH and BCL2 genes (101,102). Transformation to diffuse large
B-cell lymphoma is not uncommon, with an estimated risk of 2% per
year (103).
Hairy cell leukemia
Hairy cell leukemia is a rare and indolent B-cell
lymphoma, clinically characterized by pancytopenia, weakness,
fatigue, and splenomegaly, due to massive infiltration of the bone
marrow and the spleen by neoplastic cells, and absence of
lymphadenopathy. The neoplastic lymphocytes acquire hairy-like
projections and classically express annexinA1, CD25, TRAP, CD123,
CD11c and cyclinD1 by immunohistochemistry. In over 95% of the
cases the BRAFV600E mutation is detected,
constitutionally activating the MAPK pathway, and therefore these
patients are susceptible to BRAF inhibition. Interferon α,
purine analogues and/or anti-CD20 antibodies remain the first-line
treatment. In case of relapse or resistance, BRAF inhibitors
are an option only in patients carrying the mutation (56,104). As a rare disease, gathering all
eligible patients in a cohort is immensely helpful in future study
design (51).
Hodgkin lymphoma
Hodgkin lymphoma consists of two distinct disease
entities, classical Hodgkin lymphoma, and nodular
lymphocyte-predominant Hodgkin lymphoma. The classical Hodgkin
lymphoma is further subclassified into four subgroups: nodular
sclerosis (the most common one, 90% of all Hodgkin lymphomas),
mixed cellularity, lymphocyte depletion, and lymphocyte-rich
Hodgkin lymphoma. Each subtype has different clinical features and
outcomes. Histologically, the hallmark of classical Hodgkin
lymphoma is the presence of large Hodgkin and Reed-Sternberg cells,
expressing CD30 and CD15 and negative for B-cell markers, admixed
with various mature non-neoplastic inflammatory cells. Nodular
lymphocyte-predominant Hodgkin lymphoma is characterized by
preservation of the B-cell program in the neoplastic cells. Optimal
management consists of chemotherapy and autologous stem cell
transplantation, but in the case of relapse the treatment
approaches are limited. PM may offer new options in this setting
(50,54,55,61,62,105,106).
Mantle cell lymphoma
Mantle cell lymphoma (MCL) is a relatively rare
(3-10% of all non-Hodgkin lymphomas) B-cell lymphoma with poor
prognosis (median survival 3-5 years). It is characterized by the
t(11;14)(q13;q32) translocation, present in >95% of the cases
(107). Commonly it presents
with nodal disease, with the spleen, bone marrow and the
gastrointestinal tract being also infiltrated. Classic presentation
follows an aggressive clinical course, but other variants as
leukemic non-nodal mantle cell lymphoma or in situ mantle
cell lymphoma are considered indolent. Despite the improvement in
response durations with currently available treatment options,
patients will eventually relapse, making the need for novel
therapies urgent (52,53,108).
Clinical importance
Hematologic malignancies are a group of
heterogeneous diseases requiring careful histopathologic evaluation
and molecular characterization that is essential for optimal
therapeutic management. Hematologic tumor biobanks would contribute
to the evolution of the current knowledge, hoping to benefit
thousands of patients around the world. More precisely, integration
of clinical as well as tissue-derived data such as molecular
characteristics, can be used for detection of biomarkers, screening
of patients for clinical trials, offering opportunities to refine
therapeutic decision-making, foster multidisciplinary high-impact
research, and ultimately improve patient outcomes. They can also
promote the study of genomics in oncology through a public data
sharing platform.
Sample collections: Laboratory of
Biological Chemistry, Medical School, NKUA
Pediatric brain tumors
This sample collection contains 72 samples of
pediatric brain tumors, of which 54 are FFPE samples and 18 are
cryopreserved astrocytoma samples, stored at -80°C. Specifically,
it comprises 40 astrocytomas (24 pilocytic astrocytomas, 9
astrocytomas grade ll, 2 astrocytomas grade lll, and 5
glioblastomas), 15 medulloblastomas, 9 ependymomas, 3 atypical
teratoid/rhabdoid tumors, 3 craniopharyngiomas, 1 ganglioglioma and
1 primitive neuroectodermal tumor. These tumors belong to ICD-10-CM
Code C71 (malignant neoplasms of the brain). They have been
molecularly characterized using RT-PCR, western immunoblotting, and
immunohistochemistry for several epigenetic markers, and research
data have been recently published (59,109).
Clinical importance
Due to the sensitive location of tumors within or
adjacent to the brain, biobanking of brain tumor specimens in
neuro-oncology is highly challenging and valuable. Pediatric brain
tumor repositories are important for neuro-oncology progress and
promotion of PM.
Sample collections: 'Andreas Syggros'
Hospital of Cutaneous and Venereal Diseases
The collections of malignant skin tumors contain 97
samples. The collection of metastatic melanomas includes 11 samples
of primary tumor together with 13 samples of adjacent normal skin
surrounding the melanoma and 13 samples of the metastatic sites
(lymph nodes, lung, brain). The second collection of metastatic
cutaneous squamous cell carcinomas (cSCCs) includes 29 samples of
primary tumors and 31 samples of lymph nodes with metastatic SCC
deposits.
Metastatic melanomas
Melanoma is a malignant tumor that arises from
melanocytes and primary involves the skin. Even small tumors may
have a tendency to metastasize, either by the lymphatic or the
hematogenous route, thus leading to a relatively unfavorable
prognosis. The mean time of overall survival for metastatic
melanoma patients ranges from 6 up to 9 months, with a limited
five-year overall survival rate 1-2% (110). The incidence of metastasis seems
to be related to the Breslow thickness, melanoma type, presence of
ulceration, increased mitotic rate and intense regression.
Two-thirds of metastases are originally confined to the drainage
area of regional lymph nodes and can appear as: satellite
metastases (defined as up to 2 cm from the primary tumor);
in-transit metastases (located in the skin between 2 cm from the
site of the primary tumor and the first draining lymph node);
micro-metastases in the regional lymph nodes identified via
sentinel lymph node biopsy (SLNB).
Clinically or radiologically recognizable
regional lymph node metastases
Distant metastases can be clinically defined by the
number of organs involved, presence of brain metastases, and serum
levels of lactate dehydrogenase (LDH) (111). As early as 2002, the depiction
of BRAFV600 mutations in melanoma set the stage
for an explosion of interest for developing oncogene-directed
therapy in this disease (BRAFV600 inhibitor-vemurafenib)
(112). In the last decade,
additional targets have been achieved, creating a new era of
molecular treatment for advanced diseases. Novel agents that impact
on various signaling pathways or modulate the immune system
[(anti-PD-1 drugs (nivolumab, pembrolizumab) and anti-CTLA-4
antibody (ipilimumab)] hold the promise of a whole new therapeutic
landscape for metastatic melanoma patients (57,58,113).
Cutaneous squamous cell carcinomas
Cutaneous squamous cell carcinoma (cSCC) originates
from squamous cells of the epidermis or its appendages. The
majority of cSCCs are low risk, with a 90% 5-year survival rate
after surgical excision. Low-risk patients are unlikely to
experience local recurrence (10%) or lymph node metastasis (5%)
(114). Nevertheless, there is a
subgroup of cSCC patients with an increased risk of local
recurrence (10-47.2%) and metastatic spread (15-38%), which are
characterized as high-risk. In this category of patients, SLNB is
encouraged for nodal staging and detecting micro-metastasis. No
study yet has sufficiently assessed the impact of SLNB on the
survival of these patients. As there are no solid criteria, the
management of these high-risk cSCC patients, is usually in the
clinician's discretion (115,116). Although early cSCC is a common
tumor and totally curable with surgical excision, metastatic or
advanced cSCC is relatively rare, potentially life-threatening and
without established treatment options. In the past, diverse options
have been used with unsatisfactory results including chemotherapy
(cisplatin, fluoropyrimidines, bleomycin, doxorubicin),
13-cis-retinoic acid, and interferon-a2a (117). Other treatments, such as
epidermal growth factor receptor inhibitors have also been used
with moderate success. In recent years, a better understanding of
the biological behavior of cSCC, estimating clinical and
histological risk factors, have led to new promising immunotherapy.
These treatments include PD-1 inhibitors such as cemiplimab and
pembrolizumab whose efficacy is still being tested (118).
Clinical importance
Metastatic melanomas and cSCCs are of great clinical
interest due to their association with the high morbidity and
mortality of the patients. Tissue-derived data concerning
histological characteristics in correlation with clinical data
should be integrated for new biomarker development, and for the
elucidation of molecular tumor profiles resulting in new management
guidelines for these patients, while also presenting novel
treatment opportunities with the very promising immunotherapeutic
approaches. High-risk cSCCs and the detection of possible
micro-metastasis in sentinel lymph nodes are the new challenges in
dermato-oncology. Clinical trials globally are trying to develop
and provide dependable prognostic models for better nodal staging
and reasonable treatment options for high-risk cSCCs.
Despite the immense advances in the field of
oncology, cancer remains one of the main causes of mortality and
morbidity worldwide. Personalized medicine has evolved greatly in
the current era and has significantly altered patient care and
management. Biobanks are a crucial part of personalized medicine
enabling a systematic way of data collection, processing, and
functioning. They provide stable ground for the enhancement of
cancer genomics, transcriptomics, proteomics, metabolomics and
epigenomics, which form the main pillars for biomarker
establishment and future drug development. Moreover, the emergence
of large data collections in cancer research has clearly underlined
the importance of biobanks. Biospecimens are critical fuels for
human disease-oriented research; cancer biobanks that integrate
clinical and tissue derived data, offer extensive opportunities to
refine therapeutic decision making, to foster high impact research
and to improve patient outcomes. Having carried out a thorough
review of the regulations governing the creation and establishment
of biobanks in the Greek landscape, we formed a pilot biobank of
rare malignant neoplasms with a total of 553 samples of patients
with different tumor types. By laying the groundwork with the
establishment of this pilot biobank, while conforming to
international guidelines and recommended governance models, we
aspire that this endeavor shall assist the intricate biobanking
harmonization process. This biobank aspires to further bridge the
gap between translational and routine medical practice by aiding
clinicians and researchers alike. Molecular profiling in precision
oncology can identify the best possible treatments, while also
aiding researchers for investigating potential biomarkers. The
biobank's samples along with their associated data have aim to meet
the rising demand of quality biosamples in cancer research. It is
our belief that the future of medical research is entwined with
accessible, effective, and ethical biobanking and that our project
will facilitate research planning in the '-omic' era by
contributing high-quality samples along with their associated
data.
Availability of data and materials
Not applicable.
Authors' contributions
CP and PK shared supervision of this work. DSK
coordinated the activity of all participants, and carried out the
majority of manuscript writing, reviewing and bibliographic search.
In addition, he provided the ethical background and analysis. AP
carried out sample acquisition and data quality assessment,
analysis, interpretation, and management. DMV carried out all
software and database development and functionality. DSK, AP and
DMV carried out the conceptualization and testing of the interface
development. EAK, DNZ, ED, AK, GK, VT and EC carried out sample and
data acquisition. SS, EL and PK provided their expertise as
pathologists. DKS, AP, DMV, EAK, DNZ, ED, SS, EL, CP and PK carried
out manuscript editing and participated in the bibliographic
research. SS, EL, CP and PK revised and gave final approval of the
manuscript. All authors read and approved the final manuscript for
publication.
Ethics approval and consent to
participate
The study was conducted in accordance with the
Declaration of Helsinki and ethical approval was obtained for all
tissue samples from the University Hospitals' Ethics Boards and/or
the Bioethics Committee of the University of Athens Medical
School.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
This project was supported by the Hellenic Network
of Precision Medicine on Cancer (HNPM). We are especially thankful
to HNPM's Coordinator, Dr Kostas Stamatopoulos, for his support and
for his constructive comments.
Funding
This work was funded by the Hellenic Network of Precision
Medicine on Cancer (HNPM).
Abbreviations:
|
AML
|
acute myelogenous leukemia
|
|
AES
|
Advanced Encryption Standard
|
|
BioSHaRE-EU
|
Biobank Standardization and
Harmonization for Research Excellence in the European Union
|
|
BBMRI
|
Biobanking and BioMolecular Resources
Research Infrastructure
|
|
BRFAA
|
Biomedical Research Foundation of the
Academy of Athens
|
|
CS-IT
|
Common Service IT
|
|
CRUD
|
Create, Read, Update, Delete
|
|
CBCLs
|
cutaneous B-cell lymphomas
|
|
cSCC
|
cutaneous squamous cell carcinoma
|
|
CTCLs
|
cutaneous T-cell lymphomas
|
|
ELSI
|
ethical, legal and societal
issues
|
|
ERIC
|
European Research Infrastructure
Consortium
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
FF
|
fresh-frozen
|
|
GDPR
|
General Data Protection
Regulation
|
|
GSRI
|
General Secretariat of Research and
Innovation
|
|
HDPA
|
Hellenic Data Protection
Authority
|
|
HNPM
|
Hellenic Network of Precision
Medicine
|
|
HTO/EMO
|
Hellenic Transplant Organization
|
|
HGP
|
Human Genome Project
|
|
IARC
|
International Agency for Research on
Cancer
|
|
IHC
|
immunohistochemistry
|
|
ICGC
|
International Cancer Genome
Consortium
|
|
ISBER
|
International Society of Biological
and Environmental Repositories
|
|
LDH
|
lactate dehydrogenase
|
|
MCL
|
mantle cell lymphoma
|
|
MIABIS
|
minimum information about BIobank
data sharing
|
|
NKUA
|
National and Kapodistrian University
of Athens
|
|
NCI
|
National Cancer Institute
|
|
NGS
|
next-generation sequencing
|
|
OECD
|
Organization for Economic
Co-Operation and Development
|
|
PBMS
|
Pilot Biobank Management System
|
|
PM
|
precision medicine
|
|
P3G
|
Public Population Project in Genomics
and Society
|
|
QA
|
quality assurance
|
|
QC
|
quality control
|
|
REDCs
|
Research Ethics and Deontology
Committees
|
|
RECs
|
Research Ethics Committees
|
|
SLNB
|
sentinel lymph node biopsy
|
|
SNVs
|
single nucleotide variants
|
|
SOPs
|
standard operating procedures
|
|
SPREC
|
Standard PREanalytical Code
|
|
TCGA
|
The Cancer Genome Atlas
|
|
UDHR
|
The Universal Declaration of Human
Rights
|
|
WPF
|
Windows Presentation Foundation
|
References
|
1
|
Kuhn TS: The structure of scientific
revolutions. 2nd edition. University of Chicago Press; Chicago, IL:
1970
|
|
2
|
Toward precision medicine: Building a
knowledge network for biomedical research and a new taxonomy of
disease. National Academies Press (US); Washington, DC: 2011
|
|
3
|
Hellenic Network of Precision Medicine on
Cancer: What does precision medicine mean? https://oncopmnet.gr/?page_id=2784&lang=en.
Accessed July 15, 2021.
|
|
4
|
Kohler S: Precision medicine-moving away
from one-size-fits-all. Quest Science for South Africa. 14. Academy
of Science of South Africa; Pretoria: pp. 12–15. 2018
|
|
5
|
Garraway LA and Lander ES: Lessons from
the cancer genome. Cell. 153:17–37. 2013. View Article : Google Scholar
|
|
6
|
Re A, Nardella C, Quattrone A and Lunardi
A: Editorial: Precision medicine in oncology. Front Oncol.
8:4792018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garraway LA, Verweij J and Ballman KV:
Precision oncology: An overview. J Clin Oncol. 31:1803–1805. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garraway LA: Genomics-driven oncology:
Framework for an emerging paradigm. J Clin Oncol. 31:1806–1814.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hyman DM, Taylor BS and Baselga J:
Implementing genome-driven oncology. Cell. 168:584–599. 2017.
View Article : Google Scholar :
|
|
10
|
Sakurai M and Sandberg AA: Chromosomes and
causation of human cancer and leukemia. XI. Correlation of
karyotypes with clinical features of acute myeloblastic leukemia.
Cancer. 37:285–299. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Flores M, Glusman G, Brogaard K, Price ND
and Hood L: P4 medicine: How systems medicine will transform the
healthcare sector and society. Per Med. 10:565–576. 2013.
View Article : Google Scholar
|
|
12
|
Hood L, Balling R and Auffray C:
Revolutionizing medicine in the 21st century through systems
approaches. Biotechnol J. 7:992–1001. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hood L, Heath JR, Phelps ME and Lin B:
Systems biology and new technologies enable predictive and
preventative medicine. Science. 306:640–643. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weston AD and Hood L: Systems biology,
proteomics, and the future of health care: Toward predictive,
preventative, and personalized medicine. J Proteome Res. 3:179–196.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
International Human Genome and Sequencing
Consortium: Finishing the euchromatic sequence of the human genome.
Nature. 431:931–945. 2004. View Article : Google Scholar
|
|
16
|
Harmston N, Filsell W and Stumpf MP: What
the papers say: Text mining for genomics and systems biology. Hum
Genomics. 5:17–29. 2010. View Article : Google Scholar
|
|
17
|
Dong L, Wang W, Li A, Kansal R, Chen Y,
Chen H and Li X: Clinical next generation sequencing for precision
medicine in cancer. Curr Genomics. 16:253–263. 2015. View Article : Google Scholar
|
|
18
|
Behjati S and Tarpey PS: What is next
generation sequencing? Arch Dis Child Educ Pract Ed. 98:236–238.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
International Cancer Genome Consortium;
Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabé RR, Bhan
MK, Calvo F, Eerola I, et al International network of cancer genome
projects. Nature. 464:993–998. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
De Souza YG and Greenspan JS: Biobanking
past, present and future: Responsibilities and benefits. AIDS.
27:303–312. 2013. View Article : Google Scholar
|
|
21
|
Loft S and Poulsen HE: Cancer risk and
oxidative DNA damage in man. J Mol Med (Berl). 74:297–312. 1996.
View Article : Google Scholar
|
|
22
|
Castillo-Pelayo T, Babinszky S, LeBlanc J
and Watson PH: The importance of biobanking in cancer research.
Biopreserv Biobank. 13:172–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kinkorová J: Biobanks in the era of
personalized medicine: Objectives, challenges, and innovation:
Overview. EPMA J. 7:42016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Patil S, Majumdar B, Awan KH, Sarode GS,
Sarode SC, Gadbail AR and Gondivkar S: Cancer oriented biobanks: A
comprehensive review. Oncol Rev. 12:3572018.PubMed/NCBI
|
|
25
|
Coppola L, Cianflone A, Grimaldi AM,
Incoronato M, Bevilacqua P, Messina F, Baselice S, Soricelli A,
Mirabelli P and Salvatore M: Biobanking in health care: Evolution
and future directions. J Transl Med. 17:1722019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Watson PH and Barnes RO: A proposed schema
for classifying human research biobanks. Biopreserv Biobank.
9:327–333. 2011. View Article : Google Scholar
|
|
27
|
European Commission: Biobanks for Europe:
A Challenge for Governance. Directorate-General for Research and
Innovation Science in society, EUR 25302 EN; 2012
|
|
28
|
Harris JR, Burton P, Knoppers BM,
Lindpaintner K, Bledsoe M, Brookes AJ, Budin-Ljøsne I, Chisholm R,
Cox D, Deschênes M, et al: Toward a roadmap in global biobanking
for health. Eur J Hum Genet. 20:1105–1111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuille M, van Ommen GJ, Bréchot C,
Cambon-Thomsen A, Dagher G, Landegren U, Litton JE, Pasterk M,
Peltonen L, Taussig M, et al: Biobanking for Europe. Brief
Bioinform. 9:14–24. 2008. View Article : Google Scholar
|
|
30
|
Catchpoole DR, Florindi F, Ahern C, Garcia
DL, Mullins P, Van Enckevort E, Zaayenga A, Mayrhofer MT and Holub
P: Expanding the BBMRI-ERIC directory into a global catalogue of
COVID-19-ready collections: A joint initiative of BBMRI-ERIC and
ISBER. Biopreserv Biobank. 18:479–480. 2020. View Article : Google Scholar
|
|
31
|
Brand AM and Probst-Hensch NM: Biobanking
for epidemiological research and public health. Pathobiology.
74:227–238. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Riegman PH, Dinjens WN and Oosterhuis JW:
Biobanking for interdisciplinary clinical research. Pathobiology.
74:239–244. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mendy M, Caboux E, Lawlor RT, Wright J and
Wild CP: Common minimum technical standards and protocols for
biobanks dedicated to cancer research. (IARC Technical Publication
No. 44). International Agency for Research on Cancer; Lyon:
2017
|
|
34
|
Slokenberga S, Tzortzatou O and Reichel J:
GDPR and biobanking: Individual rights, public interest and
research regulation across Europe. Springer International
Publishing; Cham: pp. 291–307. 2021
|
|
35
|
Lee LW, Griffith J, Zenick H and Hulka BS:
Human tissue monitoring and specimen banking: Opportunities for
exposure assessment, risk assessment, and epidemiologic research.
Environ Health Perspect. 103(Suppl 3): S3–S8. 1995.
|
|
36
|
Palazzani L: Innovation in Scientific
Research and Emerging Technologies: A Challenge to Ethics and Law.
Springer Nature; 2019, View Article : Google Scholar
|
|
37
|
Pitt KE, Campbell L, Skubitz APN, Aamodt
RL, Anouna A, Baird P, Beck JC, Bledsoe M, De Souza Y, Grizzle W,
et al: Best practices for repositories I: Collection, storage, and
retrieval of human biological materials for research. Cell Preserv
Technol. 3:5–48. 2005. View Article : Google Scholar
|
|
38
|
ISBER: Best practices for repositories:
Collection storage, retrieval and distribution of biological
materials for research. Cell Preserv Technol. 6:3–58. 2008.
View Article : Google Scholar
|
|
39
|
Campbell LD, Betsou F, Garcia DL, Giri JG,
Pitt KE, Pugh RS, Sexton KC, Skubitz AP and Somiari SB: Development
of the ISBER best practices for repositories: Collection, storage,
retrieval and distribution of biological materials for research.
Biopreserv Biobank. 10:232–233. 2012. View Article : Google Scholar
|
|
40
|
Campbell LD, Astrin JJ, DeSouza Y, Giri J,
Patel AA, Rawley-Payne M, Rush A and Sieffert N: The 2018 revision
of the ISBER best practices: Summary of changes and the editorial
team's development process. Biopreserv Biobank. 16:3–6. 2018.
View Article : Google Scholar
|
|
41
|
Brochhausen M, Fransson MN, Kanaskar NV,
Eriksson M, Merino-Martinez R, Hall RA, Norlin L, Kjellqvist S,
Hortlund M, Topaloglu U, et al: Developing a semantically rich
ontology for the biobank-administration domain. J Biomed Semantics.
4:232013. View Article : Google Scholar
|
|
42
|
Eklund N, Andrianarisoa NH, van Enckevort
E, Anton G, Debucquoy A, Müller H, Zaharenko L, Engels C, Ebert L,
Neumann M, et al: Extending the minimum information about biobank
data sharing terminology to describe samples, sample donors, and
events. Biopreserv Biobank. 18:155–164. 2020. View Article : Google Scholar
|
|
43
|
Merino-Martinez R, Norlin L, van Enckevort
D, Anton G, Schuffenhauer S, Silander K, Mook L, Holub P, Bild R,
Swertz M and Litton JE: Toward global biobank integration by
implementation of the minimum information about biobank data
sharing (MIABIS 2.0 Core). Biopreserv Biobank. 14:298–306. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Norlin L, Fransson MN, Eriksson M,
Merino-Martinez R, Anderberg M, Kurtovic S and Litton JE: A minimum
data set for sharing biobank samples, information, and data:
MIABIS. Biopreserv Biobank. 10:343–348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
National Cancer Institute (NCI): NCI Best
Practices for Biospecimen Resources. NCI; Bethesda, MD: 2021,
https://biospecimens.cancer.gov/.
Accessed January 13, 2021.
|
|
46
|
Betsou F, Lehmann S, Ashton G, Barnes M,
Benson EE, Coppola D, DeSouza Y, Eliason J, Glazer B, Guadagni F,
et al: Standard preanalytical coding for biospecimens: Defining the
sample PREanalytical code. Cancer Epidemiol Biomarkers Prev.
19:1004–1011. 2010. View Article : Google Scholar
|
|
47
|
Sándor J, Drakopoulou A and Bárd P: The
legal regulation of biobanks: National report: Greece. (CELEB Paper
Series, No 2). 2009. View Article : Google Scholar
|
|
48
|
Papadavid E, Korkolopoulou P, Levidou G,
Saetta AA, Papadaki T, Siakantaris M, Nikolaou V, Oikonomidi A,
Chatziandreou I, Marinos L, et al: In situ assessment of PI3K and
PTEN alterations in mycosis fungoides: Correlation with
clinicopathological features. 23:931–933. 2014.PubMed/NCBI
|
|
49
|
Argyropoulos KV, Pulitzer M, Perez S,
Korkolopoulou P, Angelopoulou M, Baxevanis C, Palomba ML and
Siakantaris M: Tumor-infiltrating and circulating granulocytic
myeloid-derived suppressor cells correlate with disease activity
and adverse clinical outcomes in mycosis fungoides. Clin Transl
Oncol. 22:1059–1066. 2020. View Article : Google Scholar
|
|
50
|
Mansouri L, Noerenberg D, Young E, Mylonas
E, Abdulla M, Frick M, Asmar F, Ljungström V, Schneider M, Yoshida
K, et al: Frequent NFKBIE deletions are associated with poor
outcome in primary mediastinal B-cell lymphoma. Blood.
128:2666–2670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lakiotaki E, Levidou G, Angelopoulou MK,
Adamopoulos C, Pangalis G, Rassidakis G, Vassilakopoulos T, Gainaru
G, Flevari P, Sachanas S, et al: Potential role of AKT/mTOR
signalling proteins in hairy cell leukaemia: Association with
BRAF/ERK activation and clinical outcome. Sci Rep. 6:212522016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sachanas S, Pangalis GA, Vassilakopoulos
TP, Korkolopoulou P, Kontopidou FN, Athanasoulia M, Yiakoumis X,
Kalpadakis C, Georgiou G, Masouridis S, et al: Combination of
rituximab with chlorambucil as first line treatment in patients
with mantle cell lymphoma: A highly effective regimen. Leuk
Lymphoma. 52:387–393. 2011. View Article : Google Scholar
|
|
53
|
Pouliou E, Xochelli A, Kanellis G, Stalika
E, Sutton LA, Nava r ro A, Agathangelidis A, Dimosthenous K,
Anagnostopoulos A, Patsouris E, et al: Numerous ontogenetic roads
to mantle cell lymphoma: Immunogenetic and immunohistochemical
evidence. Am J Pathol. 187:1454–1458. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Vassilakopoulos P: Levidou T, Milionis G,
Hartmann V, Lakiotaki S, Sepsa E, Thymara A, Ntailiani I, Spanou P,
Angelopoulou K K M, et al Thioredoxin-1, chemokine (C-X-C motif)
ligand-9 and interferon-γ expression in the neoplastic cells and
macrophages of Hodgkin lymphoma: Clinicopathologic correlations and
potential prognostic implications. Leuk Lymphoma. 58:1–13. 2017.
View Article : Google Scholar
|
|
55
|
Dimtsas GS, Georgiadi EC, Karakitsos P,
Vassilakopoulos TP, Thymara I, Korkolopoulou P, Patsouris E, Kittas
C and Doussis-Anagnostopoulou IA: Prognostic significance of
immunohistochemical expression of the angiogenic molecules vascular
endothelial growth factor-A, vascular endothelial growth factor
receptor-1 and vascular endothelial growth factor receptor-2 in
patients with classical Hodgkin lymphoma. Leuk Lymphoma.
55:558–564. 2014. View Article : Google Scholar
|
|
56
|
Lakiotaki E, Giaginis C, Tolia M,
Alexandrou P, Delladetsima I, Giannopoulou I, Kyrgias G, Patsouris
E and Theocharis S: Clinical significance of cannabinoid receptors
CB1 and CB2 expression in human malignant and benign thyroid
lesions. Biomed Res Int. 2015:8394032015. View Article : Google Scholar
|
|
57
|
Dimonitsas E, Liakea A, Sakellariou S,
Thymara I, Giannopoulos A, Stratigos A, Soura E, Saetta A and
Korkolopoulou P: An update on molecular alterations in melanocytic
tumors with emphasis on Spitzoid lesions. Ann Transl Med.
6:2492018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kostaki M, Manona AD, Stavraka I,
Korkolopoulou P, Levidou G, Trigka EA, Christofidou E, Champsas G,
Stratigos AJ, Katsambas A, et al: High-frequency p16(INK) (4A)
promoter methylation is associated with histone methyltransferase
SETDB1 expression in sporadic cutaneous melanoma. Exp Dermatol.
23:332–338. 2014. View Article : Google Scholar
|
|
59
|
Davies NP, Wilson M, Harris LM, Natarajan
K, Lateef S, Macpherson L, Sgouros S, Grundy RG, Arvanitis TN and
Peet AC: Identification and characterisation of childhood
cerebellar tumours by in vivo proton MRS. NMR Biomed. 21:908–918.
2008. View Article : Google Scholar
|
|
60
|
Levidou G, Siakantaris M, Papadaki T,
Papadavid E, Vassilakopoulos TP, Angelopoulou MK, Marinos L,
Nikolaou V, Economidi A, Antoniou C, et al: A comprehensive
immunohistochemical approach of AKT/mTOR pathway and p-STAT3 in
mycosis fungoides. J Am Acad Dermatol. 69:375–384. 2013. View Article : Google Scholar
|
|
61
|
Rassidakis GZ, Medeiros LJ,
Vassilakopoulos TP, Viviani S, Bonfante V, Nadali G, Herling M,
Angelopoulou MK, Giardini R, Chilosi M, et al: BCL-2 expression in
Hodgkin and Reed-Sternberg cells of classical Hodgkin disease
predicts a poorer prognosis in patients treated with ABVD or
equivalent regimens. Blood. 100:3935–3941. 2002. View Article : Google Scholar
|
|
62
|
Georgiadi EC, Sachinis N, Dimtsas G,
Vassilakopoulos TP, Kittas C and Doussis-Anagnostopoulou IA:
Evaluation of apoptosis in classical Hodgkin's lymphoma comparing
different methods. J BUON. 17:746–752. 2012.
|
|
63
|
Lippi G, Simundic AM, Rodriguez-Manas L,
Bossuyt P and Banfi G: Standardizing in vitro diagnostics tasks in
clinical trials: A call for action. Ann Transl Med. 4:1812016.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Mendy M, Caboux E, Lawlor RT, Wright J and
Wild CP: Common minimum technical standards and protocols for
biobanks dedicated to cancer research. World Health Organization,
International Agency for Research on Cancer; Lyon: 2017
|
|
65
|
Betsou F: Quality assurance and quality
control in biobanking. Biobanking of Human Biospecimens: Principles
and Practice. Hainaut P, Vaught J, Zatloukal K and Pasterk M:
Springer International Publishing; Cham: pp. 23–49. 2017,
View Article : Google Scholar
|
|
66
|
Betsou F, Bulla A, Cho SY, Clements J,
Chuaqui R, Coppola D, De Souza Y, De Wilde A, Grizzle W, Guadagni
F, et al: Assays for qualification and quality stratification of
clinical biospecimens used in research: A technical report from the
isber biospecimen science working group. Biopreserv Biobank.
14:398–409. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Spencer DH, Sehn JK, Abel HJ, Watson MA,
Pfeifer JD and Duncavage EJ: Comparison of clinical targeted
next-generation sequence data from formalin-fixed and fresh-frozen
tissue specimens. J Mol Diagn. 15:623–633. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Graw S, Meier R, Minn K, Bloomer C, Godwin
AK, Fridley B, Vlad A, Beyerlein P and Chien J: Robust gene
expression and mutation analyses of RNA-sequencing of
formalin-fixed diagnostic tumor samples. Sci Rep. 5:123352015.
View Article : Google Scholar
|
|
69
|
Norton N, Sun Z, Asmann YW, Serie DJ,
Necela BM, Bhagwate A, Jen J, Eckloff BW, Kalari KR, Thompson KJ,
et al: Gene expression, single nucleotide variant and fusion
transcript discovery in archival material from breast tumors. PLoS
One. 8:e819252013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kokkat TJ, Patel MS, McGarvey D, LiVolsi
VA and Baloch ZW: Archived formalin-fixed paraffin-embedded (FFPE)
blocks: A valuable underexploited resource for extraction of DNA,
RNA, and protein. Biopreserv Biobank. 11:101–106. 2013. View Article : Google Scholar
|
|
71
|
Hedegaard J, Thorsen K, Lund MK, Hein AM,
Hamilton-Dutoit SJ, Vang S, Nordentoft I, Birkenkamp-Demtröder K,
Kruhøffer M, Hager H, et al: Next-generation sequencing of RNA and
DNA isolated from paired fresh-frozen and formalin-fixed
paraffin-embedded samples of human cancer and normal tissue. PLoS
One. 9:e981872014. View Article : Google Scholar
|
|
72
|
Kerick M, Isau M, Timmermann B, Sültmann
H, Herwig R, Krobitsch S, Schaefer G, Verdorfer I, Bartsch G,
Klocker H, et al: Targeted high throughput sequencing in clinical
cancer settings: Formaldehyde fixed-paraffin embedded (FFPE) tumor
tissues, input amount and tumor heterogeneity. BMC Med Genomics.
4:682011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
GitHub: MIABIS: Minimum information about
biobank data sharing repository. https://github.com/BBMRI-ERIC/miabis. Accessed
January 20, 2021.
|
|
74
|
BBMRI-ERIC: Common Services IT: What are
we aiming to achieve? BBMRI-ERIC, Graz. 2021, https://www.bbmri-eric.eu/bbmri-eric/common-service-it/.
Accessed February 29, 2021.
|
|
75
|
Yesley MS: What's ELSI got to do with it?
Bioethics and the human genome project. New Genet Soc. 27:1–6.
2008. View Article : Google Scholar
|
|
76
|
Council of Europe: Convention for the
protection of human rights and dignity of the human being with
regard to the application of biology and medicine: Convention on
Human Rights and Biomedicine (ETS No. 164). https://www.coe.int/en/web/conven-tions/full-list?module=treaty-detail&treatynum=164.
Accessed January 17, 2021.
|
|
77
|
European Commission: Clinical trials -
Regulation EU 536/2014. https://ec.europa.eu/health/human-use/clinical-trials/regulation_en.
Accessed January 17, 2021.
|
|
78
|
Council of Europe: Convention for the
Protection of Human Rights and Fundamental Freedoms (ETS No. 005).
https://www.coe.int/en/web/conventions/full-list/-/conventions/treaty/005.
Accessed January 16, 2021.
|
|
79
|
Council of Europe: Convention for the
Protection of Individuals with regard to Automatic Processing of
Personal Data (ETS No. 108). https://www.coe.int/en/web/conventions/full-list/-/conventions/treaty/108.
Accessed January 16, 2021.
|
|
80
|
Council of Europe: Convention for the
protection of human rights and dignity of the Human Being with
regard to the application of biology and medicine: Convention on
Human Rights and Biomedicine (ETS No. 164). https://www.coe.int/en/web/conventions/full-list/-/conventions/treaty/164.
Accessed January 15, 2021.
|
|
81
|
Council of Europe: Data Protection
Directive. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A31995L0046.
Accessed January 16, 2021.
|
|
82
|
Council of Europe: Recommendation CM/Rec
(2016) 6 of the Committee of Ministers to member States on research
on biological materials of human origin. https://search.coe.int/cm/Pages/result_details.aspx?ObjectID=090000168064e8ff#globalcontainer.
Accessed January 15, 2021.
|
|
83
|
United Nations: Universal declaration of
human rights. https://www.un.org/en/universal-declaration-human-rights/.
Accessed January 15, 2021.
|
|
84
|
Council of Europe: Setting standards of
quality and safety for the donation, procurement, testing,
processing, preservation, storage and distribution of human tissues
and cells. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32004L0023.
Accessed January 16, 2021.
|
|
85
|
World Medical Association: The declaration
of Helsinki - Ethical principles for medical research involving
human subjects (Last amended 2013). https://www.wma.net/policies-post/wma-decla-ration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/.
Accessed June 13, 2021.
|
|
86
|
World Medical Association: The declaration
of Taipei on Ethical considerations regarding health databases and
biobanks (Last revised 2016). https://www.wma.net/policies-post/wma-declaration-of-taipei-on-ethical-consid-erations-regarding-health-databases-and-biobanks/.
Accessed June 13, 2021.
|
|
87
|
National Bioethics Commission: On banks of
biological material of human origin (biobanks) in biomedical
research. http://www.bioethics.gr/images/pdf/ENGLISH/OPINIONS_REPORTS/biobanks_recom_eng.pdf.
Accessed December 27, 2020.
|
|
88
|
Daemen J and Rijmen V: The block cipher
rijndael. Lecture Notes in Computer Science. 1820. Springer-Verlag;
Berlin: pp. 277–284. 2000, View Article : Google Scholar
|
|
89
|
Nechvatal J, Barker E, Bassham L, Burr W,
Dworkin M, Foti J and Roback E: Report on the development of the
advanced encryption standard (AES). J Res Natl Inst Stand Technol.
106:511–577. 2001. View Article : Google Scholar
|
|
90
|
Wendler D: Broad versus blanket consent
for research with human biological samples. Hastings Cent Rep.
43:3–4. 2013. View Article : Google Scholar
|
|
91
|
Grady C, Eckstein L, Berkman B, Brock D,
Cook-Deegan R, Fullerton SM, Greely H, Hansson MG, Hull S, Kim S,
et al: Broad consent for research with biological samples: Workshop
conclusions. Am J Bioeth. 15:34–42. 2015. View Article : Google Scholar
|
|
92
|
Gille F, Vayena E and Blasimme A:
Future-proofing biobanks' governance. Eur J Hum Genet. 28:989–996.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Juliusson G and Hough R: Leukemia. Prog
Tumor Res. 43:87–100. 2016. View Article : Google Scholar
|
|
94
|
Haferlach T and Schmidts I: The power and
potential of integrated diagnostics in acute myeloid leukaemia. Br
J Haematol. 188:36–48. 2020. View Article : Google Scholar :
|
|
95
|
Willemze R, Jaffe ES, Burg G, Cerroni L,
Berti E, Swerdlow SH, Ralfkiaer E, Chimenti S, Diaz-Perez JL,
Duncan LM, et al: WHO-EORTC classification for cutaneous lymphomas.
Blood. 105:3768–3785. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kempf W, Zimmermann AK and Mitteldorf C:
Cutaneous lymphomas-an update 2019. Hematol Oncol. 37(Suppl 1):
S43–S47. 2019. View Article : Google Scholar
|
|
97
|
Willemze R, Cerroni L, Kempf W, Berti E,
Facchetti F, Swerdlow SH and Jaffe ES: The 2018 update of the
WHO-EORTC classification for primary cutaneous lymphomas. Blood.
133:1703–1714. 2019. View Article : Google Scholar :
|
|
98
|
Hernández JM, del Cañizo MC, Cuneo A,
García JL, Gutiérrez NC, González M, Castoldi G and San Miguel JF:
Clinical, hematological and cytogenetic characteristics of atypical
chronic myeloid leukemia. Ann Oncol. 11:441–444. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Hernández L, Beà S, Pinyol M, Ott G,
Katzenberger T, Rosenwald A, Bosch F, López-Guillermo A, Delabie J,
Colomer D, et al: CDK4 and MDM2 gene alterations mainly occur in
highly proliferative and aggressive mantle cell lymphomas with
wild-type INK4a/ARF locus. Cancer Res. 65:2199–2206. 2005.
View Article : Google Scholar
|
|
100
|
Li S, Young KH and Medeiros LJ: Diffuse
large B-cell lymphoma. Pathology. 50:74–87. 2018. View Article : Google Scholar
|
|
101
|
Horsman DE, Gascoyne RD, Coupland RW,
Coldman AJ and Adomat SA: Comparison of cytogenetic analysis,
southern analysis, and polymerase chain reaction for the detection
of t(14; 18) in follicular lymphoma. Am J Clin Pathol. 103:472–478.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Rowley JD: Chromosome studies in the
non-Hodgkin's lymphomas: The role of the 14;18 translocation. J
Clin Oncol. 6:919–925. 1988. View Article : Google Scholar
|
|
103
|
Freedman A and Jacobsen E: Follicular
lymphoma: 2020 Update on diagnosis and management. Am J Hematol.
95:316–327. 2020. View Article : Google Scholar
|
|
104
|
Maitre E, Wiber M, Cornet E and Troussard
X: Hairy cell leukemia. Presse Med. 48:842–849. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Mathas S, Hartmann S and Küppers R:
Hodgkin lymphoma: Pathology and biology. Semin Hematol. 53:139–147.
2016. View Article : Google Scholar
|
|
106
|
Wang HW, Balakrishna JP, Pittaluga S and
Jaffe ES: Diagnosis of Hodgkin lymphoma in the modern era. Br J
Haematol. 184:45–59. 2019. View Article : Google Scholar
|
|
107
|
Swerdlow SH, Campo E, Pileri SA, Harris
NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz
AD and Jaffe ES: The 2016 revision of the World Health Organization
classification of lymphoid neoplasms. Blood. 127:2375–2390. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Jain P and Wang M: Mantle cell lymphoma:
2019 Update on the diagnosis, pathogenesis, prognostication, and
management. Am J Hematol. 94:710–725. 2019. View Article : Google Scholar
|
|
109
|
Klonou A, Korkolopoulou P, Gargalionis AN,
Kanakoglou DS, Katifelis H, Gazouli M, Chlamydas S, Mitsios A,
Kalamatianos T, Stranjalis G, et al: Histone mark profiling in
pediatric astrocytomas reveals prognostic significance of H3K9
trimethylation and histone methyltransferase SUV39H1.
Neurotherapeutics. 18:2073–2090. 2021. View Article : Google Scholar
|
|
110
|
Treisman J and Garlie N: Systemic therapy
for cutaneous melanoma. Clin Plast Surg. 37:127–146. 2010.
View Article : Google Scholar
|
|
111
|
Garbe C, Amaral T, Peris K, Hauschild A,
Arenberger P, Bastholt L, Bataille V, Del Marmol V, Dréno B,
Fargnoli MC, et al: European consensus-based interdisciplinary
guideline for melanoma. Part 2: Treatment-update 2019. Eur J
Cancer. 126:159–177. 2020. View Article : Google Scholar
|
|
112
|
Nikolaou VA, Stratigos AJ, Flaherty KT and
Tsao H: Melanoma: New insights and new therapies. J Invest
Dermatol. 132:854–863. 2012. View Article : Google Scholar
|
|
113
|
Lugowska I, Teterycz P and Rutkowski P:
Immunotherapy of melanoma. Contemp Oncol (Pozn). 22:61–67.
2018.
|
|
114
|
Karia PS, Han J and Schmults CD: Cutaneous
squamous cell carcinoma: Estimated incidence of disease, nodal
metastasis, and deaths from disease in the United States. 2012.J Am
Acad Dermatol. 68:957–966. 2013. View Article : Google Scholar
|
|
115
|
Stratigos AJ, Garbe C, Dessinioti C, Lebbe
C, Bataille V, Bastholt L, Dreno B, Fargnoli MC, Forsea AM, Frenard
C, et al: European interdisciplinary guideline on invasive squamous
cell carcinoma of the skin: Part 1. Epidemiology, diagnostics and
prevention. Eur J Cancer. 128:60–82. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Parikh SA, Patel VA and Ratner D: Advances
in the management of cutaneous squamous cell carcinoma. F1000Prime
Rep. 6:702014. View
Article : Google Scholar : PubMed/NCBI
|
|
117
|
Cranmer LD, Engelhardt C and Morgan SS:
Treatment of unresectable and metastatic cutaneous squamous cell
carcinoma. Oncologist. 15:1320–1328. 2010. View Article : Google Scholar
|
|
118
|
Soura E, Gagari E and Stratigos A:
Advanced cutaneous squamous cell carcinoma: How is it defined and
what new therapeutic approaches are available? Curr Opin Oncol.
31:461–468. 2019. View Article : Google Scholar
|