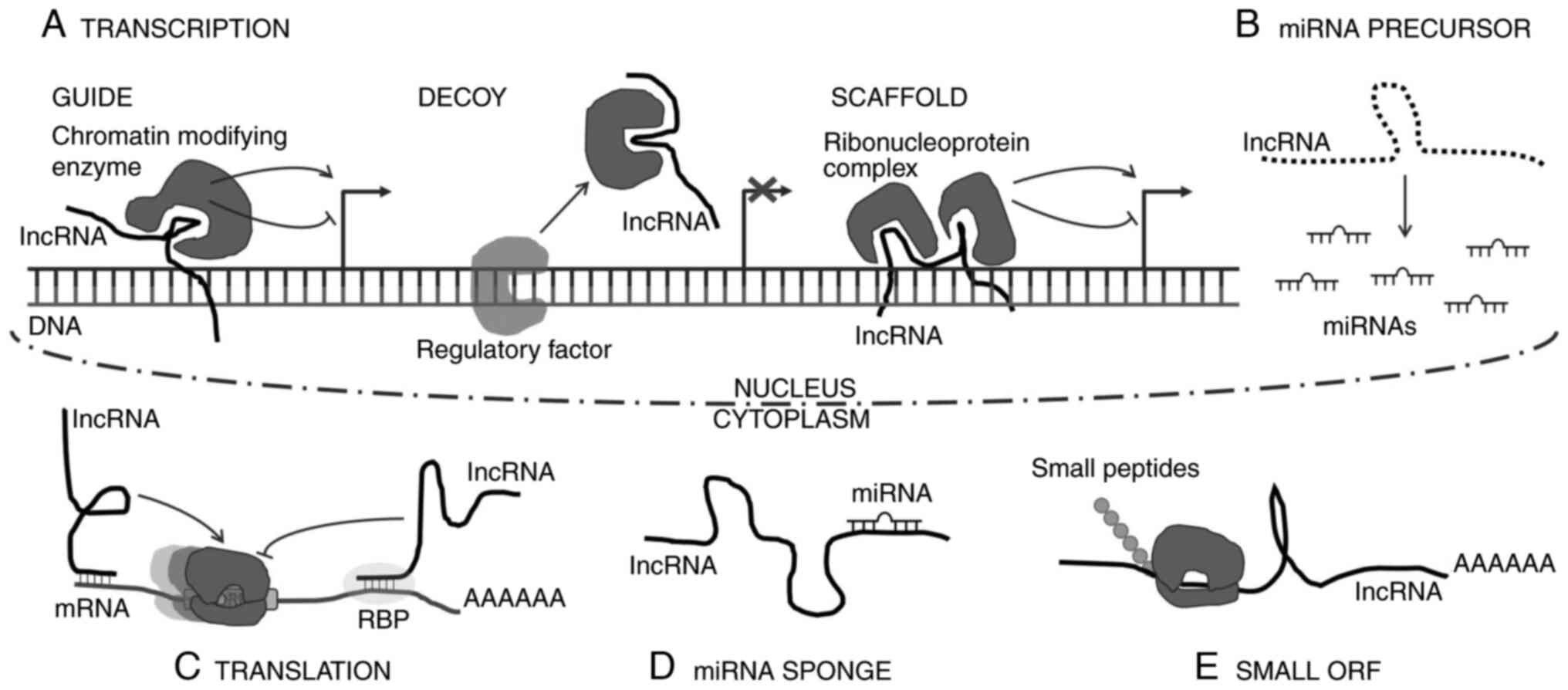
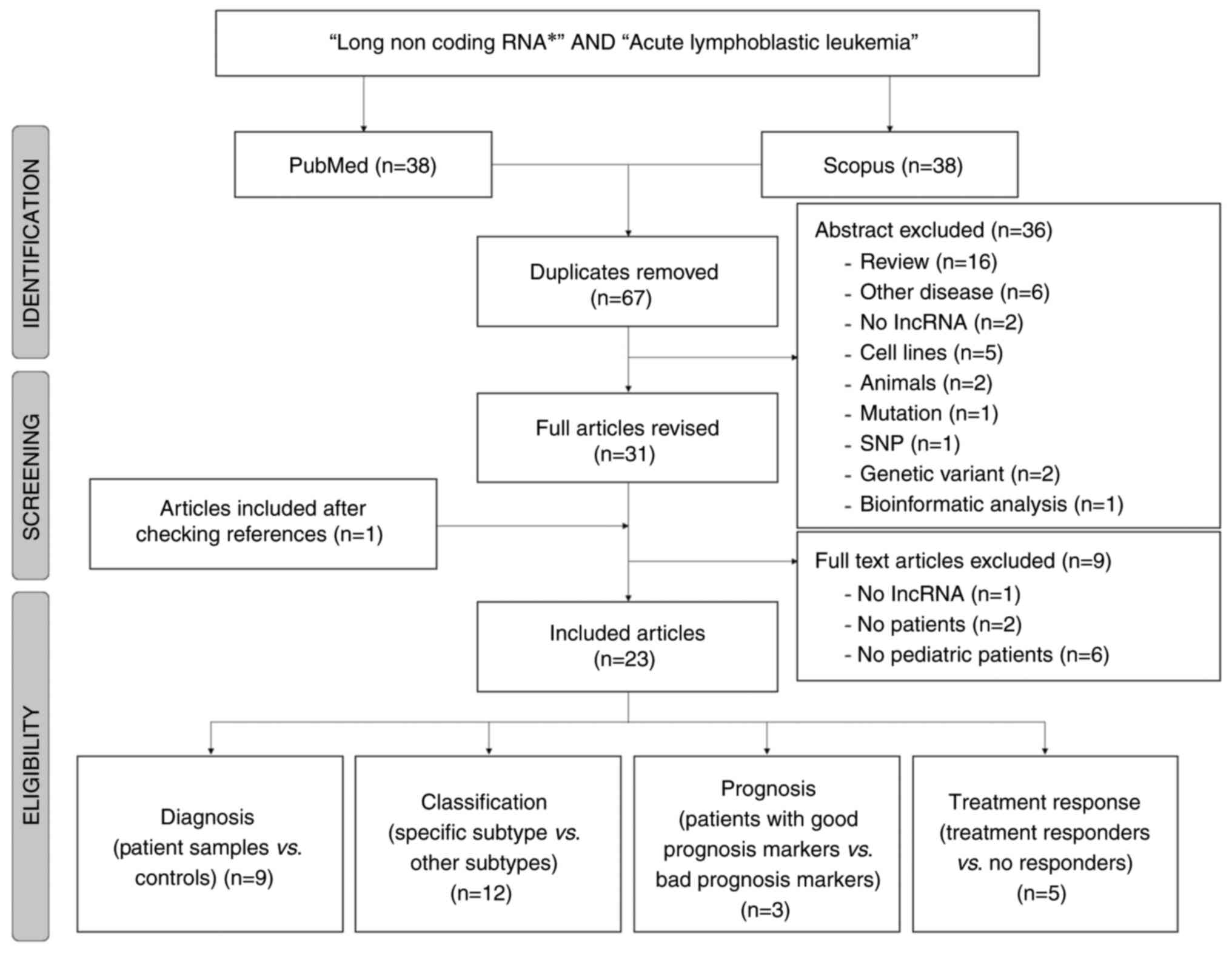
|
1
|
Hunger SP and Mullighan CG: Acute
lymphoblastic leukemia in children. N Engl J Med. 373:1541–1552.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arber DA, Orazi A, Hasserjian R, Thiele J,
Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M and Vardiman JW:
The 2016 revision to the world health organization classification
of myeloid neoplasms and acute leukemia. Blood. 127:2391–2405.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nachman JB, Heerema NA, Sather H, Camitta
B, Forestier E, Harrison CJ, Dastugue N, Schrappe M, Pui CH, Basso
G, et al: Outcome of treatment in children with hypodiploid acute
lymphoblastic leukemia. Blood. 110:1112–1115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lilljebjorn H and Fioretos T: New
oncogenic subtypes in pediatric B-cell precursor acute
lymphoblastic leukemia. Blood. 130:1395–1401. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jain N, Lamb AV, O'Brien S, Ravandi F,
Konopleva M, Jabbour E, Zuo Z, Jorgensen J, Lin P, Pierce S, et al:
Early T-cell precursor acute lymphoblastic leukemia/lymphoma
(ETP-ALL/LBL) in adolescents and adults: A high-risk subtype.
Blood. 127:1863–1869. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yeoh EJ, Ross ME, Shurtleff SA, Williams
WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A,
et al: Classification, subtype discovery, and prediction of outcome
in pediatric acute lymphoblastic leukemia by gene expression
profiling. Cancer Cell. 1:133–143. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ross ME, Zhou X, Song G, Shurtleff SA,
Girtman K, Williams WK, Liu HC, Mahfouz R, Raimondi SC, Lenny N, et
al: Classification of pediatric acute lymphoblastic leukemia by
gene expression profiling. Blood. 102:2951–2959. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schotte D, De Menezes RX, Akbari Moqadam
F, Khankahdani LM, Lange-Turenhout E, Chen C, Pieters R and Den
Boer ML: MicroRNA characterize genetic diversity and drug
resistance in pediatric acute lymphoblastic leukemia.
Haematologica. 96:703–711. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang H, Luo XQ, Zhang P, Huang LB, Zheng
YS, Wu J, Zhou H, Qu LH, Xu L and Chen YQ: MicroRNA patterns
associated with clinical prognostic parameters and CNS relapse
prediction in pediatric acute leukemia. PLoS One. 4:e78262009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Morlando M, Ballarino M and Fatica A: Long
non-coding RNAs: New players in hematopoiesis and leukemia. Front
Med (Lausanne). 2:232015.PubMed/NCBI
|
|
12
|
Nobili L, Lionetti M and Neri A: Long
non-coding RNAs in normal and malignant hematopoiesis. Oncotarget.
7:50666–50681. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sweta S, Dudnakova T, Sudheer S, Baker AH
and Bhushan R: Importance of long non-coding RNAs in the
development and disease of skeletal muscle and cardiovascular
lineages. Front Cell Dev Biol. 7:2282019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ling H, Vincent K, Pichler M, Fodde R,
Berindan-Neagoe I, Slack FJ and Calin GA: Junk DNA and the long
non-coding RNA twist in cancer genetics. Oncogene. 34:5003–5011.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Garitano-Trojaola A, Jose-Eneriz ES,
Ezponda T, Unfried JP, Carrasco-León A, Razquin N, Barriocanal M,
Vilas-Zornoza A, Sangro B, Segura V, et al: Deregulation of
linc-PINT in acute lymphoblastic leukemia is implicated in abnormal
proliferation of leukemic cells. Oncotarget. 9:12842–12852. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bárcenas-López DA, Núñez-Enríquez JC,
Hidalgo-Miranda A, Beltrán-Anaya FO, May-Hau DI, Jiménez-Hernández
E, Bekker-Méndez VC, Flores-Lujano J, Medina-Sansón A, Tamez-Gómez
EL, et al: Transcriptome analysis identifies LINC00152 as a
biomarker of early relapse and mortality in acute lymphoblastic
leukemia. Genes (Basel). 11:3022020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Q, Ma H, Sun X, Liu B, Xiao Y, Pan S,
Zhou H, Dong W and Jia L: Correction to: The regulatory
ZFAS1/miR-150/ST6GAL1 crosstalk modulates sialylation of EGFR via
PI3K/akt pathway in T-cell acute lymphoblastic leukemia. J Exp Clin
Cancer Res. 38:3572019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fernando TR, Contreras JR, Zampini M,
Rodriguez-Malave NI, Alberti MO, Anguiano J, Tran TM, Palanichamy
JK, Gajeton J, Ung NM, et al: The lncRNA CASC15 regulates SOX4
expression in RUNX1-rearranged acute leukemia. Mol Cancer.
16:1262017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li X, Song F and Sun H: Long non-coding
RNA AWPPH interacts with ROCK2 and regulates the proliferation and
apoptosis of cancer cells in pediatric T-cell acute lymphoblastic
leukemia. Oncol Lett. 20:2392020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bramer WM, Rethlefsen ML, Kleijnen J and
Franco OH: Optimal database combinations for literature searches in
systematic reviews: A prospective exploratory study. Syst Rev.
6:2452017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Larrabeiti-Etxebarria A, Lopez-Santillan
M, Santos-Zorrozua B, Lopez-Lopez E and Garcia-Orad A: Systematic
review of the potential of MicroRNAs in diffuse large B cell
lymphoma. Cancers (Basel). 11:1442019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gutierrez-Camino A, Garcia-Obregon S,
Lopez-Lopez E, Astigarraga I and Garcia-Orad A: MiRNA deregulation
in childhood acute lymphoblastic leukemia: A systematic review.
Epigenomics. 12:69–80. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Page MJ, McKenzie JE, Bossuyt PM, Boutron
I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan
SE, et al: The PRISMA 2020 statement: An updated guideline for
reporting systematic reviews. BMJ. 372:n712021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Affinito O, Pane K, Smaldone G, Orlandella
FM, Mirabelli P, Beneduce G, Parasole R, Ripaldi M, Salvatore M and
Franzese M: lncRNAs-mRNAs Co-Expression network underlying
childhood B-Cell acute lymphoblastic leukaemia: A pilot study.
Cancers (Basel). 12:24892020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
das Chagas PF, de Sousa GR, Kodama MH, de
Biagi Junior CAO, Yunes JA, Brandalise SR, Calin GA, Tone LG,
Scrideli CA and de Oliveira JC: Ultraconserved long non-coding RNA
uc.112 is highly expressed in childhood T versus B-cell acute
lymphoblastic leukemia. Hematol Transfus Cell Ther. 43:28–34. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen L, Shi Y, Li J, Yang X, Li R, Zhou X
and Zhu L: LncRNA CDKN2B-AS1 contributes to tumorigenesis and
chemoresistance in pediatric T-cell acute lymphoblastic leukemia
through miR-335-3p/TRAF5 axis. Anticancer Drugs. Sep 25–2020.(Epub
ahead of print). View Article : Google Scholar
|
|
30
|
Cuadros M, Andrades A, Coira IF, Baliñas
C, Rodríguez MI, Álvarez-Pérez JC, Peinado P, Arenas AM, García DJ,
Jiménez P, et al: Expression of the long non-coding RNA TCL6 is
associated with clinical outcome in pediatric B-cell acute
lymphoblastic leukemia. Blood Cancer J. 9:932019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Durinck K, Wallaert A, Van de Walle I, Van
Loocke W, Volders PJ, Vanhauwaert S, Geerdens E, Benoit Y, Van Roy
N, Poppe B, et al: The notch driven long non-coding RNA repertoire
in T-cell acute lymphoblastic leukemia. Haematologica.
99:1808–1816. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fang K, Han BW, Chen ZH, Lin KY, Zeng CW,
Li XJ, Li JH, Luo XQ and Chen YQ: A distinct set of long non-coding
RNAs in childhood MLL-rearranged acute lymphoblastic leukemia:
Biology and epigenetic target. Hum Mol Genet. 23:3278–3288. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fernando TR, Rodriguez-Malave NI, Waters
EV, Yan W, Casero D, Basso G, Pigazzi M and Rao DS: lncRNA
expression discriminates karyotype and predicts survival in
B-lymphoblastic leukemia. Mol Cancer Res. 13:839–851. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gasic V, Stankovic B, Zukic B, Janic D,
Dokmanovic L, Krstovski N, Lazic J, Milosevic G, Lucafò M, Stocco
G, et al: Expression pattern of long non-coding RNA growth
arrest-specific 5 in the remission induction therapy in childhood
acute lymphoblastic leukemia. J Med Biochem. 38:292–298. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ghazavi F, De Moerloose B, Van Loocke W,
Wallaert A, Helsmoortel HH, Ferster A, Bakkus M, Plat G, Delabesse
E, Uyttebroeck A, et al: Unique long non-coding RNA expression
signature in ETV6/RUNX1-driven B-cell precursor acute lymphoblastic
leukemia. Oncotarget. 7:73769–73780. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
James AR, Schroeder MP, Neumann M, Bastian
L, Eckert C, Gökbuget N, Tanchez JO, Schlee C, Isaakidis K,
Schwartz S, et al: Long non-coding RNAs defining major subtypes of
B cell precursor acute lymphoblastic leukemia. J Hematol Oncol.
12:82019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lajoie M, Drouin S, Caron M, St-Onge P,
Ouimet M, Gioia R, Lafond MH, Vidal R, Richer C, Oualkacha K, et
al: Specific expression of novel long non-coding RNAs in
high-hyperdiploid childhood acute lymphoblastic leukemia. PLoS One.
12:e01741242017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ngoc PCT, Tan SH, Tan TK, Chan MM, Li Z,
Yeoh AEJ, Tenen DG and Sanda T: Identification of novel lncRNAs
regulated by the TAL1 complex in T-cell acute lymphoblastic
leukemia. Leukemia. 32:2138–2151. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ouimet M, Drouin S, Lajoie M, Caron M,
St-Onge P, Gioia R, Richer C and Sinnett D: A childhood acute
lymphoblastic leukemia-specific lncRNA implicated in prednisolone
resistance, cell proliferation, and migration. Oncotarget.
8:7477–7488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pouyanrad S, Rahgozar S and Ghodousi ES:
Dysregulation of miR-335-3p, targeted by NEAT1 and MALAT1 long
non-coding RNAs, is associated with poor prognosis in childhood
acute lymphoblastic leukemia. Gene. 692:35–43. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Verboom K, Van Loocke W, Volders PJ,
Decaesteker B, Cobos FA, Bornschein S, de Bock CE, Atak ZK,
Clappier E, Aerts S, et al: A comprehensive inventory of TLX1
controlled long non-coding RNAs in T-cell acute lymphoblastic
leukemia through polyA + and total RNA sequencing. Haematologica.
103:e585–e589. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang Y, Wu P, Lin R, Rong L, Xue Y and
Fang Y: LncRNA NALT interaction with NOTCH1 promoted cell
proliferation in pediatric T cell acute lymphoblastic leukemia. Sci
Rep. 5:137492015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang T, Jin X, Lan J and Wang W: Long
non-coding RNA SNHG16 has tumor suppressing effect in acute
lymphoblastic leukemia by inverse interaction on hsa-miR-124-3p.
IUBMB life. 71:134–142. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang L, Xu H and Lu C: A novel long
non-coding RNA T-ALL-R-LncR1 knockdown and par-4 cooperate to
induce cellular apoptosis in T-cell acute lymphoblastic leukemia
cells. Leuk Lymphoma. 55:1373–1382. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu C, Han B, Xin J and Yang C:
LncRNA-AWPPH activates TGF-β1 in colorectal adenocarcinoma. Oncol
Lett. 18:4719–4725. 2019.PubMed/NCBI
|
|
46
|
Zhao X, Liu Y and Yu S: Long noncoding RNA
AWPPH promotes hepatocellular carcinoma progression through YBX1
and serves as a prognostic biomarker. Biochim Biophys Acta Mol
Basis Dis. 1863:1805–1816. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zheng J, Song Y, Li Z, Tang A, Fei Y and
He W: The implication of lncRNA expression pattern and potential
function of lncRNA RP4-576H24.2 in acute myeloid leukemia. Cancer
Med. 8:7143–7160. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ju H, Zhang L, Mao L, Wu Y, Liu S, Ruan M,
Hu J and Ren G: A comprehensive genome-wide analysis of the long
noncoding RNA expression profile in metastatic lymph nodes of oral
mucosal melanoma. Gene. 675:44–53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang B, Tang D, Zhang Z and Wang Z:
Identification of aberrantly expressed lncRNA and the associated
TF-mRNA network in hepatocellular carcinoma. J Cell Biochem.
121:1491–1503. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zuo X, Chen Z, Gao W, Zhang Y, Wang J,
Wang J, Cao M, Cai J, Wu J and Wang X: M6A-mediated upregulation of
LINC00958 increases lipogenesis and acts as a nanotherapeutic
target in hepatocellular carcinoma. J Hematol Oncol. 13:52020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cui Y, Xie M and Zhang Z: LINC00958
involves in bladder cancer through sponging miR-378a-3p to elevate
IGF1R. Cancer Biother Radiopharm. 35:776–788. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang L, Zhong Y, Yang B, Zhu Y, Zhu X, Xia
Z, Xu J and Xu L: LINC00958 facilitates cervical cancer cell
proliferation and metastasis by sponging miR-625-5p to upregulate
LRRC8E expression. J Cell Biochem. 121:2500–2509. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rong D, Dong Q, Qu H, Deng X, Gao F, Li Q
and Sun P: M6A-induced LINC00958 promotes breast cancer
tumorigenesis via the miR-378a-3p/YY1 axis. Cell Death Discov.
7:272021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gioia R, Drouin S, Ouimet M, Caron M,
St-Onge P, Richer C and Sinnett D: LncRNAs downregulated in
childhood acute lymphoblastic leukemia modulate apoptosis, cell
migration, and DNA damage response. Oncotarget. 8:80645–80650.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Mousavi Z, Ghorbian S, Rezamand A,
Roshangar L and Jafari B: Expression profile of LncRNAs in
childhood acute lymphoblastic leukemia: A pilot study. Pharm Sci.
27:385–392. 2021. View Article : Google Scholar
|
|
56
|
Hou J, Xiu B, Ji W, Zhang Q, Guo R, Chi Y
and Wu J: Abstract P1-05-08: LINC00926 interacts with HNRNPC and
suppress metastasis in breast cancer. Cancer Res. 80 (Suppl
4):Abstract nr P1-05-08. 2020.
|
|
57
|
Wang F, Tian X, Zhou J, Wang G, Yu W, Li
Z, Fan Z, Zhang W and Liang A: A threelncRNA signature for
prognosis prediction of acute myeloid leukemia in patients. Mol Med
Rep. 18:1473–1484. 2018.PubMed/NCBI
|
|
58
|
Liang Y, Zhu H, Chen J, Lin W, Li B and
Guo Y: Construction of relapse-related lncRNA-mediated ceRNA
networks in hodgkin lymphoma. Arch Med Sci. 16:1411–1418. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhou Y, Huan L, Wu Y, Bao C, Chen B, Wang
L, Huang S, Liang L and He X: LncRNA ID2-AS1 suppresses tumor
metastasis by activating the HDAC8/ID2 pathway in hepatocellular
carcinoma. Cancer Lett. 469:399–409. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yao K, Wang Q, Jia J and Zhao H: A
competing endogenous RNA network identifies novel mRNA, miRNA and
lncRNA markers for the prognosis of diabetic pancreatic cancer.
Tumour Biol. Jun 22–2017.(Epub ahead of print). View Article : Google Scholar
|
|
61
|
Li S, Chen S, Wang B, Zhang L, Su Y and
Zhang X: A robust 6-lncRNA prognostic signature for predicting the
prognosis of patients with colorectal cancer metastasis. Front Med
(Lausanne). 7:562020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Stewart G: Alteration of non-coding RNAs
as a mechanism of lung cancer gene deregulation. In: The Faculty Of
Graduate And Postdoctoral Studies. University of British Columbia;
2020
|
|
63
|
Lai C, Wu Z, Shi J, Li K, Zhu J, Chen Z,
Liu C and Xu K: Autophagy-related long noncoding RNAs can predict
prognosis in patients with bladder cancer. Aging (Albany NY).
12:21582–21596. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang Z, Liu Y, Zhang J, Zhao R, Zhou X and
Wang H: An immune-related long noncoding RNA signature as a
prognostic biomarker for human endometrial cancer. J Oncol.
2021:99724542021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hou J and Yao C: Potential prognostic
biomarkers of lung adenocarcinoma based on bioinformatic analysis.
Biomed Res Int. 2021:88599962021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chi H, Yang R, Zheng X, Zhang L, Jiang R
and Chen J: LncRNA RP11-79H23.3 functions as a competing endogenous
RNA to regulate PTEN expression through sponging hsa-miR-107 in the
development of bladder cancer. Int J Mol Sci. 19:25312018.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Neri LM, Cani A, Martelli AM, Simioni C,
Junghanss C, Tabellini G, Ricci F, Tazzari PL, Pagliaro P, McCubrey
JA and Capitani S: Targeting the PI3K/akt/mTOR signaling pathway in
B-precursor acute lymphoblastic leukemia and its therapeutic
potential. Leukemia. 28:739–748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chen C, Wang P, Mo W, Zhang Y, Zhou W,
Deng T, Zhou M, Chen X, Wang S and Wang C: lncRNA-CCDC26, as a
novel biomarker, predicts prognosis in acute myeloid leukemia.
Oncol Lett. 18:2203–2211. 2019.PubMed/NCBI
|
|
69
|
Yan J, Chen D, Chen X, Sun X, Dong Q, Hu
C, Zhou F and Chen W: Downregulation of lncRNA CCDC26 contributes
to imatinib resistance in human gastrointestinal stromal tumors
through IGF-1R upregulation. Braz J Med Biol Res. 52:e83992019.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Li C, Mu J, Shi Y and Xin H: LncRNA CCDC26
interacts with CELF2 protein to enhance myeloid leukemia cell
proliferation and invasion via the circRNA_ANKIB1/miR-195-5p/PRR11
axis. Cell Transplant. Jan 13–2021.(Epub ahead of print).
|
|
71
|
Shen S, Chen X, Cai J, Yu J, Gao J, Hu S,
Zhai X, Liang C, Ju X, Jiang H, et al: Effect of dasatinib vs
imatinib in the treatment of pediatric philadelphia
chromosome-positive acute lymphoblastic leukemia: A randomized
clinical trial. JAMA Oncol. 6:358–366. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang W, Wu F, Ma P, Gan S, Li X, Chen L,
Sun L, Sun H, Jiang Z and Guo F: LncRNA CRNDE promotes the
progression of B-cell precursor acute lymphoblastic leukemia by
targeting the miR-345-5p/CREB axis. Mol Cells. 43:718–727.
2020.PubMed/NCBI
|
|
73
|
Ma X, Zhang W, Zhao M, Li S, Jin W and
Wang K: Oncogenic role of lncRNA CRNDE in acute promyelocytic
leukemia and NPM1-mutant acute myeloid leukemia. Cell Death Discov.
6:1212020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kang Y, Zhang S, Cao W, Wan D and Sun L:
Knockdown of LncRNA CRNDE suppresses proliferation and
P-glycoprotein-mediated multidrug resistance in acute myelocytic
leukemia through the Wnt/β-catenin pathway. Biosci Rep. Jun
26–2020.(Epub ahead of print). View Article : Google Scholar
|
|
75
|
Wang Y, Zhou Q and Ma JJ: High expression
of lnc-CRNDE presents as a biomarker for acute myeloid leukemia and
promotes the malignant progression in acute myeloid leukemia cell
line U937. Eur Rev Med Pharmacol Sci. 22:763–770. 2018.PubMed/NCBI
|
|
76
|
Zhang J, Yin M, Peng G and Zhao Y: CRNDE:
An important oncogenic long non-coding RNA in human cancers. Cell
Prolif. 51:e124402018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Cuadros M, Garcia DJ, Andrades A, Arenas
AM, Coira IF, Baliñas-Gavira C, Peinado P, Rodríguez MI,
Álvarez-Pérez JC, Ruiz-Cabello F, et al: LncRNA-mRNA co-expression
analysis identifies AL133346.1/CCN2 as biomarkers in pediatric
B-cell acute lymphoblastic leukemia. Cancers (Basel). 12:38032020.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Chen D, Li Y, Wang Y and Xu J: LncRNA
HOTAIRM1 knockdown inhibits cell glycolysis metabolism and tumor
progression by miR-498/ABCE1 axis in non-small cell lung cancer.
Genes Genomics. 43:183–194. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Li Q, Dong C, Cui J, Wang Y and Hong X:
Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion
through up-regulating HOXA1 and sequestering G9a/EZH2/dnmts away
from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer
Res. 37:2652018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Hu N, Chen L, Li Q and Zhao H: LncRNA
HOTAIRM1 is involved in the progression of acute myeloid leukemia
through targeting miR-148b. RSC Adv. 9:10352–10359. 2019.
View Article : Google Scholar
|
|
81
|
Inaba H and Pui CH: Glucocorticoid use in
acute lymphoblastic leukaemia. Lancet Oncol. 11:1096–1106. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Volders PJ, Anckaert J, Verheggen K,
Nuytens J, Martens L, Mestdagh P and Vandesompele J: LNCipedia 5:
Towards a reference set of human long non-coding RNAs. Nucleic
Acids Res. 47:D135–D139. 2019. View Article : Google Scholar : PubMed/NCBI
|