Introduction
Pancreatic cancer (PC) is one of the most
biologically aggressive solid organ tumors, with >70% of
patients presenting with locally advanced or metastatic disease
(1). Since PC is usually
diagnosed at an advanced stage, only ~20% of patients qualify for
initial resection (2,3). Even if curative resection is
performed, most patients experience recurrence. The 5-year survival
rate in patients undergoing complete resection is only ~25%
(4). The molecular basis of this
aggressive clinical behavior remains only partially clear (5-7).
Surgical resection is the only potentially curative treatment for
PC; however, a considerable proportion of patients undergo
unnecessary laparotomy due to the underestimation of the extent of
the cancer during preoperative radiographic examinations (8). Imaging modalities have poor
sensitivity for identifying small liver or peritoneal metastases
(9,10). A proportion (40%) of patients who
undergo surgical exploration have tumors that are unresectable due
to occult distant metastasis, which cannot be identified during
preoperative examinations and is only discovered intraoperatively
or by infiltration of local structures (8,11-14). For patients with distant occult
metastasis, surgical resection does not prolong survival in the
majority of patients (11,15,16).
Therefore, it is critical to identify patients with PC who are
likely to have occult distant metastasis to avoid unnecessary
surgery and offer tailored treatments in a timely manner. Although
there have been reports on the correlation between clinical factors
such as preoperative tumor marker levels and tumor size and the
presence of occult distant metastasis, these biomarkers have not
yet been recognized as useful biomarkers (1,11,12).
Epigenetic alterations are among the most common
features of human cancers and are associated with cancer
development and progression (17,18). A major epigenetic alteration is
DNA methylation abnormality, which is the post-replicative addition
of a methyl group to the fifth carbon of the cytosine ring in CpG
dinucleotides. DNA methylation alterations of CpG islands in gene
promoter regions lead to transcriptional suppression of tumor
suppressor genes, which are involved in multistep carcinogenesis.
Genome-wide DNA hypomethylation is another type of epigenetic
alteration that is mainly caused by the demethylation of DNA
repetitive sequences that are normally methylated. Repetitive DNA
sequences are widely distributed in the human genome and are ideal
targets for DNA hypomethylation. These sequences are involved in
tumor progression in ovarian epithelial and hepatocellular
carcinomas (19,20). Long interspersed nucleotide
element-1 (LINE-1) is a transposable element in the human genome
that comprises a repetitive sequence and constitutes a substantial
proportion (~17%) of the human genome (21-24). The level of LINE-1 methylation is
associated with genome-wide DNA methylation (24-27) and with poor prognosis in various
human cancers (27,28).
Chromosomal instability (CIN) is a major driver of
tumor evolution and a hallmark of cancer, that results from ongoing
errors in chromosome segregation during mitosis (29,30). Experimental studies using mouse
models have provided evidence that genome-wide DNA hypomethylation
induces CIN (17,31). The development of CIN has been
reported to be correlated with tumor metastasis (32,33). Additionally, LINE-1
hypomethylation has been reported to be significantly associated
with CIN in gastrointestinal stromal tumors (34). Indeed, it was previously
demonstrated that DNA hypomethylation and its connection with DNA
copy number changes is associated with a poor prognosis in colon
and stomach cancers (35,36). Similarly, overexpression of
repetitive sequences induced by retrovirus-expressing vectors leads
to changes in copy number at specific chromosomes (37). These data suggested that
genome-wide DNA hypomethylation drives the cancer phenotype to be
more invasive by affecting the machinery that maintains chromosomal
stability. However, no study has been conducted to elucidate the
impact of genome-wide DNA hypomethylation on the biological
behavior of PC in connection with CIN.
In the present study, it was investigated whether
genome-wide DNA hypomethylation promotes the invasive ability and
metastatic potential via CIN in human PC cell lines. The
significance of genome-wide DNA hypomethylation on the biological
behavior of PC and their correlation with clinical outcomes were
also verified.
Materials and methods
Cell lines and cell culture
The human PC cell lines PANC-1 and Capan-1 were
obtained from the American Type Culture Collection. Cultured PANC-1
cells were maintained in RPMI-1640 (FUJIFILM Wako Pure Chemical
Corporation) supplemented with 10% fetal bovine serum (FBS; Cytiva)
and Capan-1 cell was cultured in IMDM (FUJIFILM Wako Pure Chemical
Corporation) supplemented with 20% FBS at 37°C in a humidified
atmosphere containing 5% CO2.
Clinical samples
A total of 49 clinical samples were collected from
patients with PC who underwent surgery between September 2010 and
July 2017 at Saitama Medical Center of Jichi Medical University
(Saitama, Japan). No metastasis was diagnosed based on preoperative
imaging studies or intended curative surgery. The patients did not
undergo adjuvant chemotherapy before surgery. Clinical data were
collected by a review of patient medical records. The preoperative
variables included sex, age, smoking, alcoholic consumption, serum
carcinoembryonic antigen (CEA) content, serum carbohydrate antigen
19-9 (CA19-9) content, serum duke pancreatic monoclonal antigen
type-2 content, tumor size, tumor location and clinical stage
classified with the Union for International Cancer Control 7th
edition (38). Intraoperative
variables that were recorded included the presence of distant
metastases such as liver or peritoneal metastases, and were defined
as occult distant metastasis. Overall survival (OS) in these
patients was calculated as the time from surgery to the occurrence
of the event. Additional clinical samples from 5 patients with
stage IV were collected to compare their metastatic potential with
that of 49 patients. These 5 patients did not undergo curative
surgery, and one patient had received chemotherapy before clinical
samples were collected. The study protocol was approved (approval
no. 21-09) by the research ethics committee of Jichi Medical
University (Saitama, Japan) and conformed to the ethical guidelines
of the World Medical Association Declaration of Helsinki (seventh
revision).
Treatment with 5-aza-2′-deoxycytidine
(5-Aza-dC)
The demethylating agent 5-Aza-dC was purchased from
FUJIFILM Wako Pure Chemical Corporation. The drug was dissolved in
phosphate-buffered saline as a 1 mM stock solution, passed through
a 0.22-µm filter, and stored at -20°C in aliquots that were
thawed immediately prior to use. The stock was diluted in cell
culture medium at different concentrations and was applied to cells
24 h after cell seeding. Fresh medium supplemented with 5-Aza-dC
was replaced every 24 h. Cultures without 5-Aza-dC were used as the
control group.
MTT cell viability assay
PANC-1 and Capan-1 cells were seeded at a density of
5,000 cells/well in 96-well plates. A total of 24 h after seeding,
cells were treated with 0.1, 1, 5, 10, and 20 µM 5-Aza-dC,
with fresh 5-Aza-dC solutions replaced every 24 h. Assays were
conducted on extracts harvested every 24 h for 1 to 7 days after
the beginning of the 5-Aza-dC treatment. Cell viability was
determined by measuring
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
colorimetric dye reduction using the MTT assay kit (TOX-1;
Sigma-Aldrich; Merck KGaA) according to the manufacturer's
instructions. After the MTT reagent was added, the cells were
incubated for 4 h at 37°C, after which the solubilization solution
(10% Triton X-100, 0.1 N HCl and anhydrous isopropanol) was added
and absorbance was measured using a microplate reader (Varioskan
LUX multimode reader; Thermo Fisher Scientific, Inc.) at a
wavelength of 570 nm. Background absorbance at 690 nm was
subtracted from the 570 nm measurement, and the percentage of
viable cells was determined relative to the control. Each
experiment was performed in triplicate for each treatment
condition.
DNA extraction and bisulfite
modification
Genomic DNA was isolated and purified from the
cultured cells treated or without 5-Aza-dC using an EZ1 Advanced XL
(Qiagen GmbH), and from clinical samples using the QIAamp DNA FFPE
Tissue kit (Qiagen GmbH) according to the manufacturer's
instructions. DNA purity was assessed using a NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.) at absorbances of 260 and 280 nm. In all instances, the
A260/A280 ratio exceeded 1.8. Sodium bisulfite conversion of
genomic DNA was performed using an EpiTect Bisulfite kit (Qiagen
GmbH). DNA quantities of 100 ng in a volume of up to 40 µl
were processed using this standard protocol. The treatment of
genomic DNA with sodium bisulfite converted unmethylated cytosine
to uracil, which was then converted to thymidine during subsequent
PCR steps. This process revealed sequence differences between
methylated and unmethylated DNA.
MethyLight methods
Following bisulfite modification, each sample was
examined using MethyLight technology for the LINE-1 sequence. Two
sets of primers and probes specifically designed to bind to
bisulfite-converted DNA were used in the reaction: one set of
LINE-1 primers and a probe for unmethylated target analyses
(unmethylated reaction) and another set of primers for the
reference locus, ALU-C (normalization control reaction), as
previously described (26,37,39).
The primer and probe sequences are summarized in Table I. Whole-genome amplification
provided fully unmethylated DNA obtained from human genomic DNA
(Promega Corporation), which served as the demethylation constant
reference that enabled determination of the relative demethylation
level (RDL). The LINE-1 RDL was defined as (LINE-1 reaction/ALU-C
reaction) sample/(LINE-1 reaction/ALU-C reaction) fully
unmethylated control DNA, as previously described (37,39). In each MethyLight reaction, 1
µl bisulfite-modified DNA solution was used. Thermal cycling
was initiated with a denaturation step at 95°C for 10 sec, followed
by 50 cycles of 95°C for 5 sec and 60°C for 30 sec. Real-time
quantitative PCR (40) was
performed on a QuantStudio 12 K Flex Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with a final reaction
volume of 20 µl containing Premix Ex Taq (Takara Bio, Inc.),
600 nM of each primer and 200 nM probe. Table SI shows the Ct (cycle threshold)
values and their variability in the different reactions in the
present study.
 | Table IPCR primers and TaqMan probes used in
the MethyLight method. |
Table I
PCR primers and TaqMan probes used in
the MethyLight method.
| Alu-C | F:
GGTTAGGTATAGTGGTTTATATTTGTAATTTTAGTA |
| R:
ATTAACTAAACTAATCTTAAACTCCTAACCTCA |
| Probe:
FAM-CCTACCTTAACCTCCC-MGB |
| LINE-1 | F:
TTTATTAGGGAGTGTTAGATAGTGGGTG |
| R:
CCTTACACTTCCCAAATAAAACAATACC |
| Probe:
FAM-CACCCTACTTCAACTCATACACAATACACACACCC-MGB |
Microarray-based comparative genomic
hybridization (array CGH)
Array CGH was performed using a SurePrint G3 Human
CGH Microarray kit, 8×60 K (Agilent Technologies, Inc.). Labeling
and hybridization were performed using a SureTag DNA Labeling kit
and Oligo aCGH/ChiP-on-chip hybridization kit (both from Agilent
Technologies, Inc.) according to the manufacturer's protocol
(Protocol v8.0). In brief, ≥0.2 µg DNA from the samples and
an equal amount of control DNA were digested with AluI and RsaI for
2 h at 37°C. The digested DNA was labeled by random priming and
then the sample and control DNA were labeled with Cy5-dUTP and
Cy3-dUTP, respectively. The labeled products were purified using an
Amicon Ultra-0.5 Centrifugal Filter Unit with an Ultracel-30
membrane (MilliporeSigma) and concentrated to 9.5 µl. Dye
incorporation and DNA yield were measured using a NanoDrop ND-1000
spectrophotometer. Equal amounts of genomic DNA extracted from
samples and control DNA were mixed with human Cot-1 DNA and then
dissolved in hybridization buffer (Agilent Technologies, Inc.),
denatured, and hybridized to the CGH array at 67°C for 24 h.
Following hybridization, the microarrays were washed with Oligo
aCGH/ChIP-on-Chip Wash Buffer (Agilent Technologies, Inc.). After
washing, the slides were scanned using an Agilent Technologies
Microarray Scanner. Microarray images were analyzed using a feature
extraction software program v.12.0.3.1 (Agilent Technologies,
Inc.), and the resulting data were subsequently imported into the
Agilent Cytogenomics software program, v5.1.
Immunocytochemistry
Cells were cultured on cover glass in 24-well
plates. A total of 24 h after seeding, the cells were treated with
400 ng/ml nocodazole (product code ab120630; Abcam) for 18 h to
synchronize the cell cycle with the mitotic phase as previously
described (41-43). A total of 40 min after release
from nocodazole arrest, the prepared cover glass samples were fixed
in 4% paraformaldehyde/PBS at room temperature for 10 min and then
washed three times with PBS. The cells were permeabilized with 0.1%
Triton X-100 (Agilent Technologies, Inc.) at room temperature for
10 min and blocked with 10% normal goat serum (Thermo Fisher
Scientific, Inc.) at room temperature for 30 min. The cells were
washed three times with PBS and incubated with anti-α-tubulin
(1:500; product code ab7291) and anti-γH2AX (1:1,000; product code
ab11174; both from Abcam) at 4°C overnight. Following extensive
washing with PBS, the samples were incubated with
Alexa-594-conjugated anti-mouse IgG secondary antibody (1:200;
product code ab150120) and Alexa-488-conjugated antirabbit IgG
secondary antibody (1:200; product code ab150081; both from Abcam)
for nuclear staining at room temperature for 60 min. The cover
glass samples were washed three times with PBS, mounted in
VECTASHIELD Vibrance Antifade Mounting Medium with DAPI (cat. no.
H-1800; Vector Laboratories, Inc.), and sealed with nail polish.
Images were acquired using a Keyence BZ-X700 fluorescence
microscope (Keyence Corporation).
Invasion assay
The invasion ability of PC cells was assessed using
a Corning BioCoat Matrigel Invasion Chamber assay
(Corning® 354480; Corning, Inc.) according to the
manufacturer's protocol. After PANC-1 and Capan-1 cells were
treated with 1 µM 5-Aza-dC for 3 days, they were seeded at
25,000 cells/well in serum-free medium in the upper chambers of
24-well plates. A culture medium containing 10% FBS was used in the
lower chamber as a chemoattractant. After incubation for 22 h at
37°C, the medium was removed. After the removal of the
non-migratory cells, the cells in the upper chamber were fixed and
permeabilized for 2 min at room temperature, and stained for 4 min
at room temperature using a Diff-Quick® simple and rapid
staining solution (Sysmex Corporation) according to the
manufacturer's protocol. A total of 5 random fields per well in
four independent experiments were observed using a Keyence BZ-X700
fluorescence microscope (Keyence Corporation) to assess cell
migration. Captured images were analyzed using ImageJ software (v.
1.52a; National Institutes of Health). The invasion ability of PC
cells was evaluated as percentage of invasion and was calculated by
the following formula: Percentage of invasion=(average number of
cells invading Matrigel coated membrane)/(average number of cells
migrating through control (no Matrigel coating) membrane) ×100,
according to the manufacturer's instructions.
Statistical analyses
All statistical analyses were performed using EZR
version 1.41 (Saitama Medical Center; Jichi Medical University),
which is a graphical user interface for R version 3.6.1 (The R
Foundation for Statistical Computing, Vienna, Austria) (44). Continuous variables were evaluated
using the Shapiro-Wilk normality test, and the means or medians
were then compared with the paired samples t-test for normally
distributed variables, or the Mann-Whitney U-test and
Kruskal-Wallis test for non-normally distributed variables.
Logistic regression analysis was used to examine the associations
between two categorical variables. The correlation between the two
variables was evaluated using the Spearman's rank correlation
coefficient. The survival time distribution was assessed using the
Kaplan-Meier method with a log-rank test. A Cox proportional
hazards regression model was used to evaluate the association
between overall mortality and other factors in univariate and
multivariate analyses. All reported P-values were two-sided, and
P-values <0.05 were considered to indicate a statistically
significant difference.
Results
Viability of PANC-1 and Capan-1 cells
treated with 5-Aza-dC
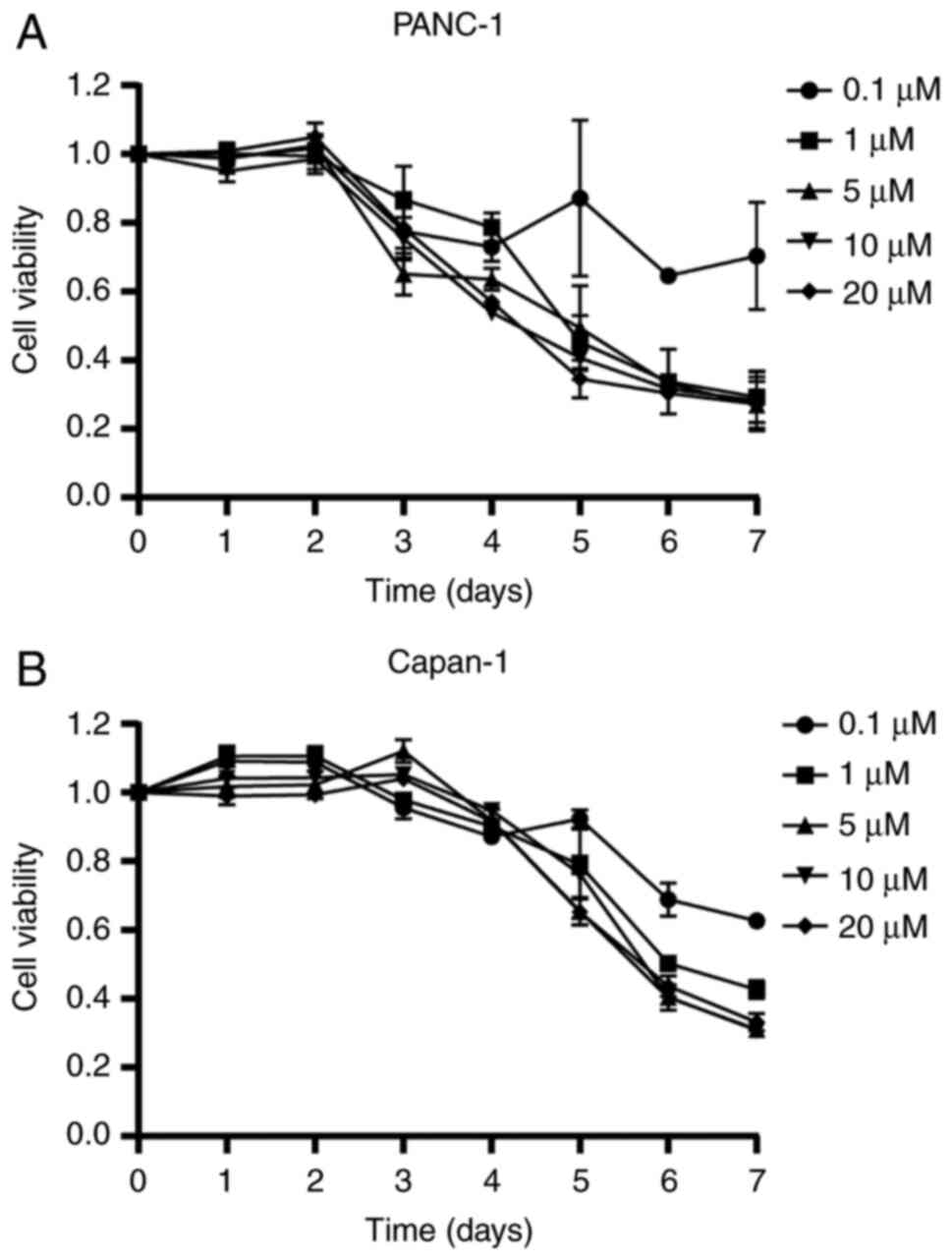
First, the optimal concentration of 5-Aza-dC and the
duration of treatment for the PC cell lines were determined. The
MTT cell viability assays showed that cell viability gradually
decreased in a dose-dependent manner after treatment with 1
µM 5-Aza-dC for 3 days; however, doses of 5-Aza-dC greater
than 1 µM or treatment periods longer than 3 days led to
significant cytotoxicity (Fig. 1A and
B and Table SII). Therefore,
5-Aza-dC was used at a concentration of 1 µM for 3 days in
the present study.
5-Aza-dC enhancement of DNA
hypomethylation
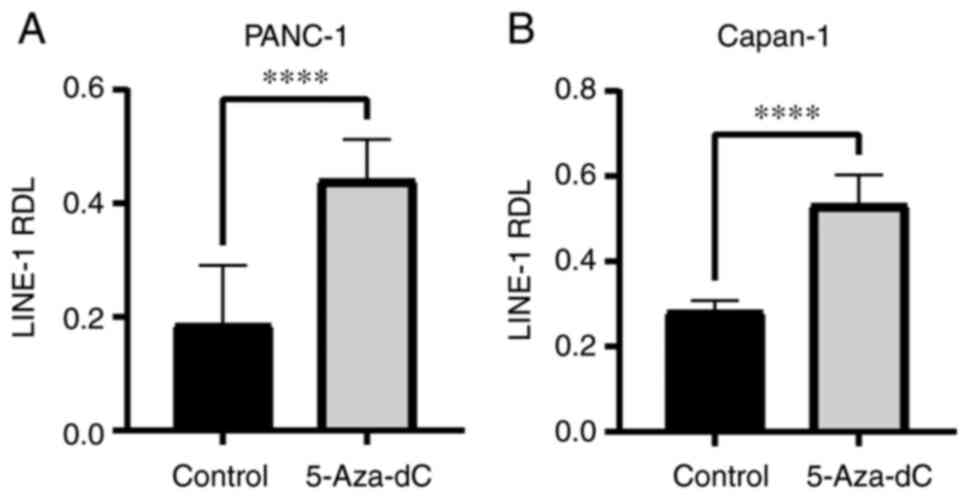
It was next verified whether 5-Aza-dC induced DNA
hypomethylation in PC cell lines. PANC-1 and Capan-1 cells were
cultured with or without 1 µM 5-Aza-dC for 3 days before the
RDL of the LINE-1 repetitive sequence was used to determine the
level of genome-wide DNA hypomethylation (24-27). LINE-1 RDLs in PANC-1 and Capan-1
cells were significantly elevated after treatment with 5-Aza-dC
(PANC-1, 0.19±0.097 to 0.44±0.067, P=0.00002; Capan-1, 0.28±0.024
to 0.53±0.066, P=0.000005; Fig. 2A
and B). These results indicated that the 5-Aza-dC treatment
could induce DNA hypomethylation in PC cells.
Abnormal segregation of chromosomes, DNA
damage and chromosomal instability in response to 5-Aza-dC
treatment
Immunocytochemistry was performed to detect abnormal
segregation of chromosomes and DNA damage, which could lead to CIN.
DNA damage response was evaluated using the number of anti-γH2AX
positive cells. The number of cells with abnormal segregation of
chromosomes including micronuclei, lagging, anaphase bridging, and
multiple nuclei significantly increased (3.9±2.7 to 12.5±3.6%,
P=0.0005; Fig. 3A-E), with
elevated numbers of anti-γH2AX positive cells (15.1±3.2 to
55.6±7.9%, P=0.028; Fig. 3F) in
PANC-1 cells treated with 5-Aza-dC when compared with the untreated
control. In Capan-1 cells, the nuclei were fused with each other,
making it difficult to identify abnormal segregations; however, the
number of anti-γH2AX positive cells significantly increased
(47.8±8.4 to 58.9±8.8%, P=0.0032; Fig. 3G). Fig. 3H demonstrates representative
images of anti-γH2AX positive cells. The incidences of abnormal
segregation and anti-γH2AX positive cells were investigated in more
than 100 mitotic cells per sample and were representative of three
distinct experiments. Changes in copy number were analyzed using
CGH array analysis. Successful induction of DNA hypomethylation led
to copy number changes in specific regions of the chromosomes in PC
cells after treatment with 5-Aza-dC (Fig. 3I and J), with copy number changes
being more likely to occur in PANC-1 cells than in Capan-1 cells.
These results suggested that 5-Aza-dC induced DNA hypomethylation
that led to CIN in human PC cells.
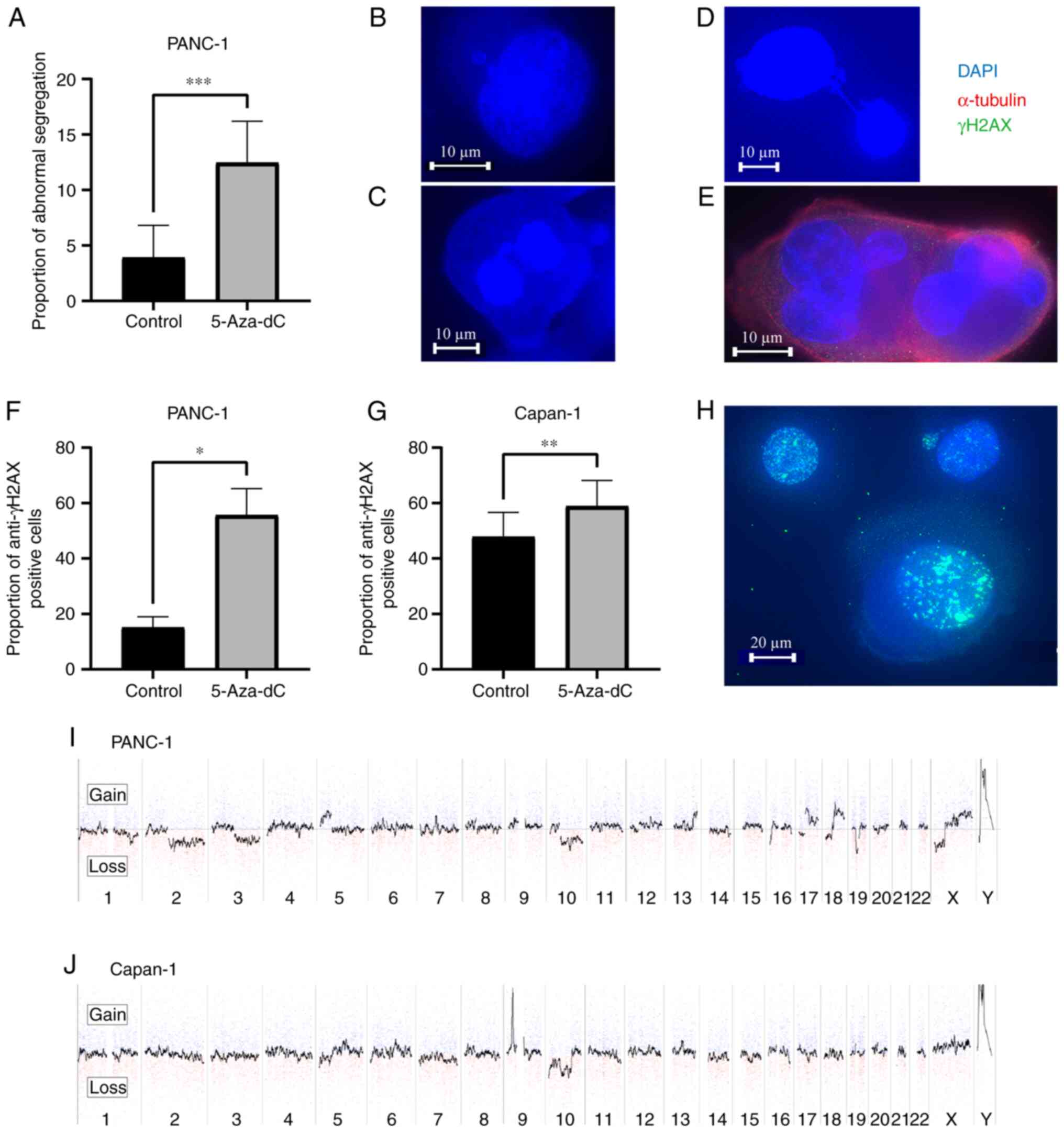 | Figure 3Abnormal segregation of chromosomes,
DNA damage and chromosomal instability in response to 5-Aza-dC
treatment in pancreatic cancer cell lines. (A) The number of cells
with abnormal segregation in PANC-1 cells increased after treatment
with 1 µM 5-Aza-dC for 3 days when compared with untreated
controls. (B-E) Representative images of abnormal segregation
including (B) micronuclei, (C) lagging, (D) anaphase bridge and (E)
multiple nuclei in PANC-1 cells treated with 5-Aza-dC (Scale bar,
10 µm). (F and G) Increased number of cells with DNA damage
in (F) PANC-1 and (G) Capan-1 cell lines treated with 1 µM
5-Aza-dC for 3 days compared with the untreated controls. DNA
damage was evaluated by the number of anti-γH2AX positive cells.
(H) Images of anti-γH2AX positive cells (Scale bar, 20 µm).
(I and J) Copy number changes in chromosomes detected by CGH array
in (I) PANC-1 and (J) Capan-1 cells treated with 1 µM
5-Aza-dC for 3 days. Copy number changes were observed more in
PANC-1 cells when compared with Capan-1 cells. Gains are shown in
the upper area and losses in the lower area. Numbers represent the
chromosome number. In each chromosome area, the short arm is
located on the left side, and the long arm is on the right side.
*P<0.05, **P<0.01 and
***P<0.001. 5-Aza-dC, 5-aza-2′-deoxycytidine; CGH,
comparative genomic hybridization. |
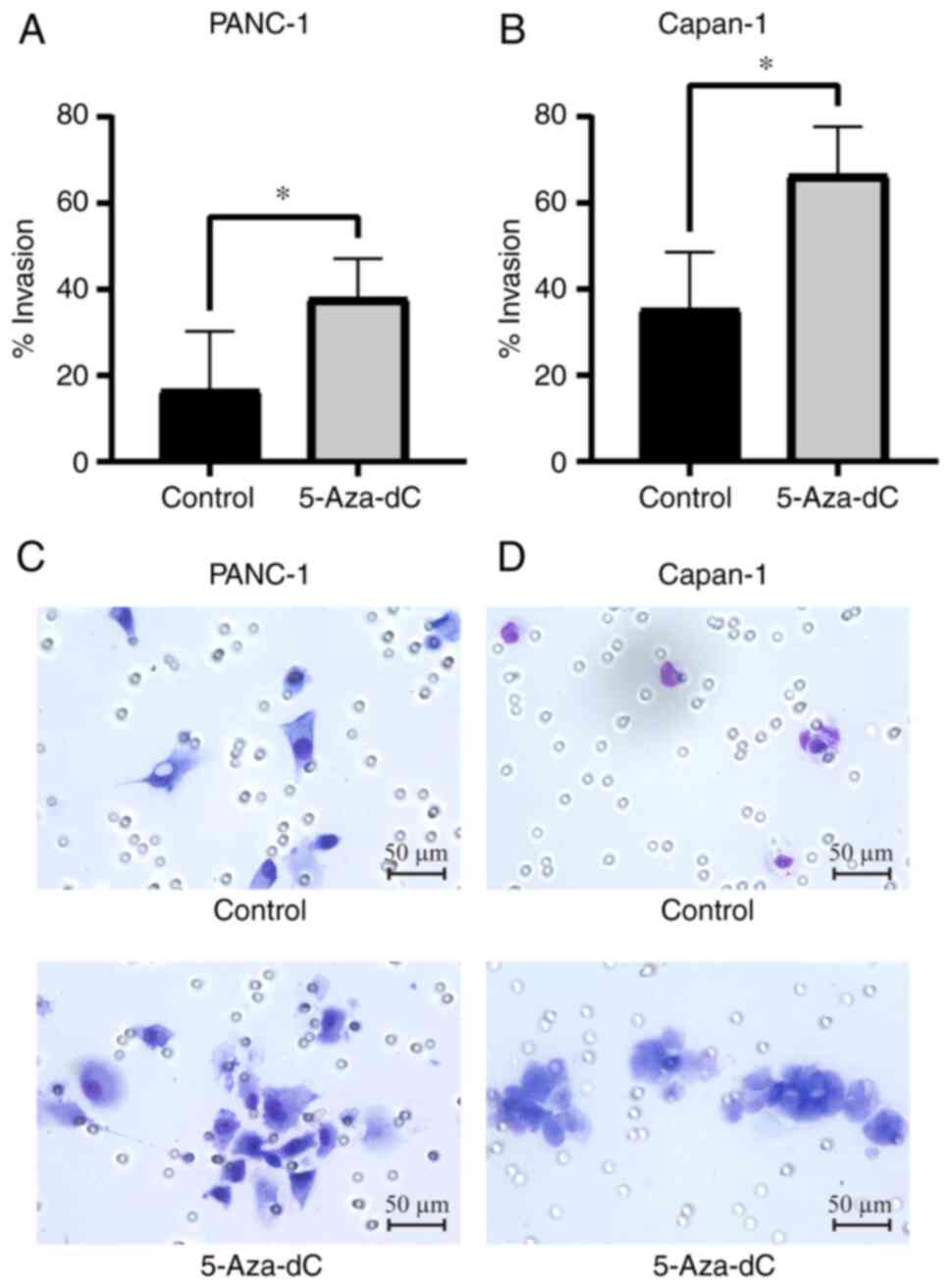
Invasion ability of PC cells after
5-Aza-dC treatment
The invasion ability of PANC-1 and Capan-1 PC cells
was assessed after treatment with 5-Aza-dC using the Matrigel
invasion assay. The number of cells that migrated through the
membrane represented the invasive and metastatic potential of the
cancer cells. Before 5-Aza-dC treatment, Capan-1 cells migrated
through the membrane more than twice as much as PANC-1 (PANC-1,
16.6±11.9%; Capan-1, 35.4±11.4%; Fig.
4A and B), indicating that Capan-1 cells harbored more invasive
potential than PANC-1 cells. After 5-Aza-dC treatment, the number
of PANC-1 and Capan-1 cells that migrated increased significantly
compared with the untreated cells, respectively (PANC-1, 16.6±11.9
to 37.9±8.0%, P=0.042; Capan-1, 35.4±11.4 to 66.5±9.7%; P=0.011;
Fig. 4A and B). Representative
images of cells invading through the Matrigel-coated membrane of
PANC-1 and Capan-1 cell lines with and without 5-Aza-dC treatment
are presented in Fig. 4C and D.
Cells treated with 5-Aza-dC appeared to be more invasive than
untreated cells. Notably, the increased number of migratory PANC-1
cells after 5-Aza-dC treatment (37.9±8.0%) was the same as that of
naïve Capan-1 before treatment (35.4±11.4%), which indicated that
PANC-1 cells acquired an invasive potential similar to Capan-1
cells, further suggesting that genome-wide DNA
hypomethylation-induced CIN drives the PC phenotype to be more
invasive.
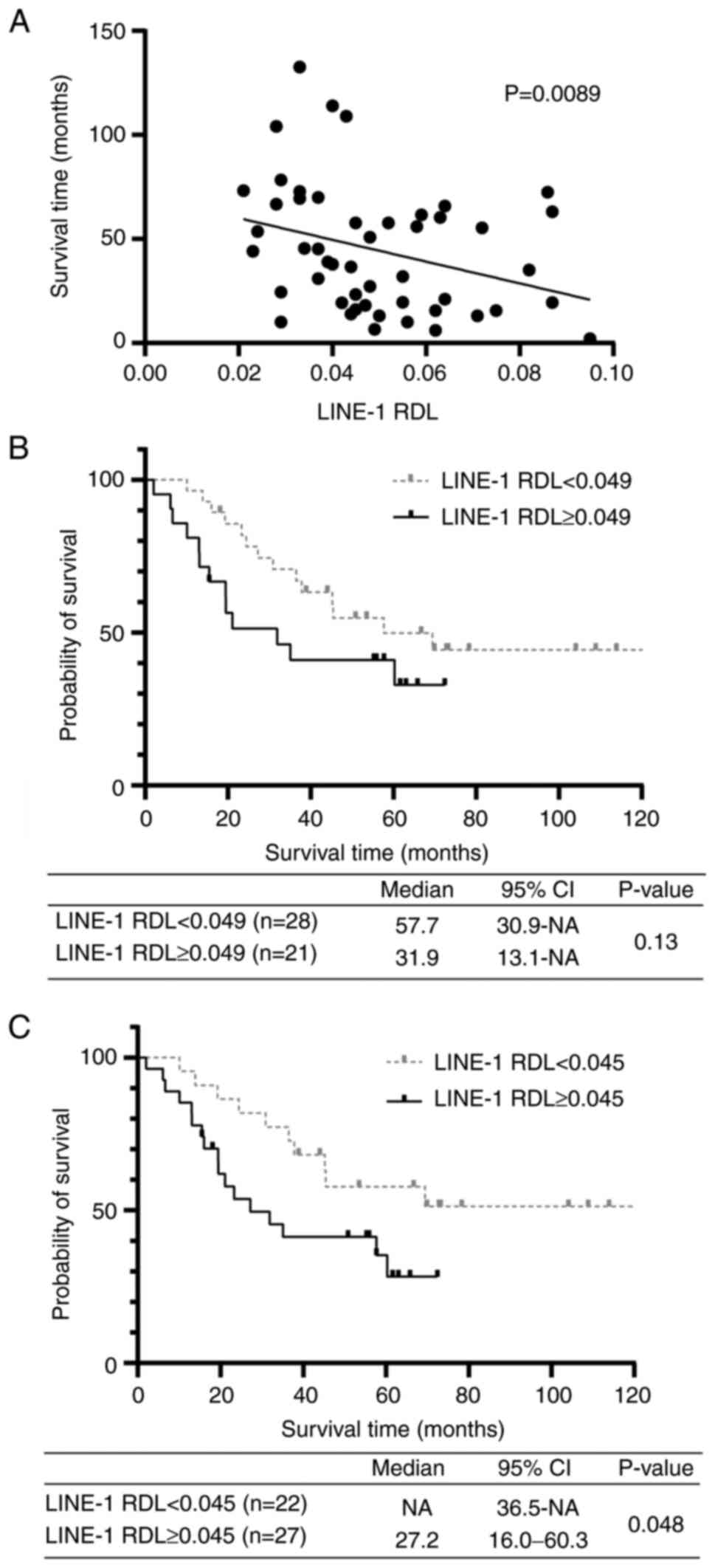
Clinical outcomes and LINE-1 RDL in
patients with PC undergoing curative surgery
The significance of genome-wide DNA hypomethylation
in the biological behavior of PC in 49 patients who underwent
curative surgery was investigated. The clinical features of the 49
patients and their relation to the LINE-1 RDL of the tumor
specimens are shown in Table II.
The mean age of all patients was 68 years, and the median age was
70 (range, 40-83) years. This median age was used as the boundary
for analysis in the present study. Occult distant metastasis was
found in 7 of the 49 patients (14.3%), although all patients were
diagnosed without metastasis by imaging studies performed before
surgery. Significant correlation of the LINE-1 RDL was observed
with tumor size (P=0.0012), clinical stage (P=0.045) and the
presence of occult distant metastasis (P=0.034). The presence of
occult distant metastasis and its relationship to the LINE-1 RDL
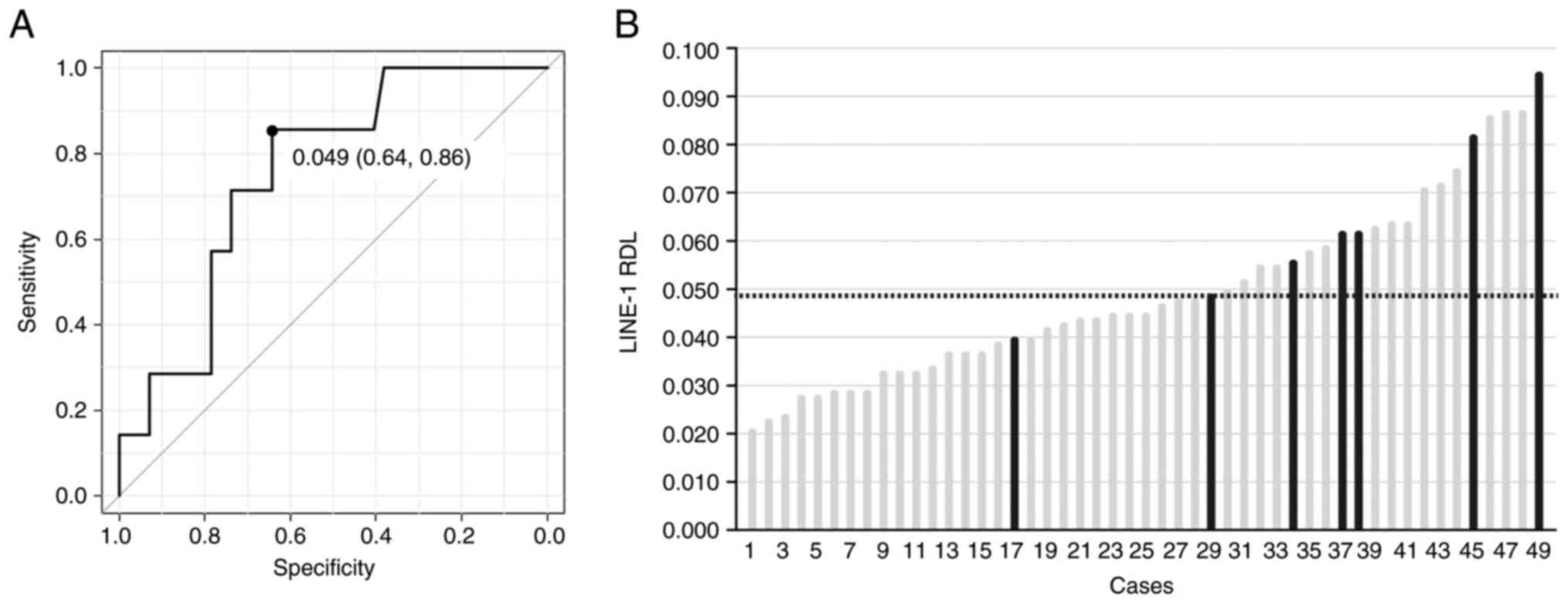
were then addressed by performing receiver operating characteristic
analysis to determine the cut-off value of the LINE-1 RDL to
identify patients with occult distant metastasis (Fig. 5A). The area under the curve was
0.76 (95% confidence interval 0.58-0.93) and the derived cut-off
value of the LINE-1 RDL was 0.049 with a sensitivity and
specificity of 0.86 and 0.64, respectively. The distribution of the
LINE-1 RDL in 49 tumor specimens in increasing order of LINE-1 RDL
is presented in Fig. 5B. A total
of 6 out of 21 patients with high LINE-1 RDL ≥0.049 had occult
distant metastasis, whereas only one in 28 patients with low LINE-1
RDL <0.049 displayed occult distant metastasis. Univariate and
multivariate analyses revealed that LINE-1 RDL was a significant
independent predictor of occult distant metastasis (Table III). In addition, the impact of
LINE-1 RDL on patient prognosis was verified. Correlation
coefficient analysis revealed a significant correlation between
LINE-1 RDL and OS in 49 patients (rho=-0.37, P=0.0089; Fig. 6A), while multivariate analysis
showed that LINE-1 RDL was a significant independent prognostic
factor (Table IV). However, it
was not possible to demonstrate a significant difference in OS
between patients with high and low LINE-1 RDL using Kaplan-Meier
analysis, although patients with high LINE-1 RDL tended to have
worse OS than those with low LINE-1 RDL (median OS, 31.9 M vs. 57.7
M; P=0.13; Fig. 6B). The modified
LINE-1 RDL of 0.045 was then applied as a median value to
Kaplan-Meier analysis, which demonstrated that patients with high
LINE-1 RDL had a significantly worse OS than those with low LINE-1
RDL (median OS, 27.2 M vs. not reached; P=0.048; Fig. 6C). In addition, LINE-1 RDL was
determined in five additional patients diagnosed with clinical
stage IV and significantly higher LINE-1 RDL at 0.052, 0.082,
0.113, 0.230 and 0.331 was confirmed, respectively (data not
shown).
 | Table IIAssociation between clinical features
and LINE-1 RDLs of patients with pancreatic cancer. |
Table II
Association between clinical features
and LINE-1 RDLs of patients with pancreatic cancer.
| Clinical
features | Total n (%) | LINE-1 RDL
(median) | P-value |
|---|
| All cases | 49 (100) | 0.045 | |
| Age, years | | | 0.21 |
| <70 | 24 (41.0) | 0.047 | |
| ≥70 | 25 (49.0) | 0.045 | |
| Sex | | | 0.49 |
| Male | 25 (49.0) | 0.046 | |
| Female | 24 (41.0) | 0.045 | |
| Smoking | | | 0.41 |
| No | 24 (41.0) | 0.045 | |
| Yes | 25 (49.0) | 0.049 | |
| Alcohol
consumption | | | 0.24 |
| No | 41 (83.7) | 0.045 | |
| Yesa | 8 (16.3) | 0.045 | |
| CEA (ng/ml) | | | 0.058 |
| ≤5 | 39 (79.6) | 0.044 | |
| >5 | 10 (20.4) | 0.063 | |
| CA19-9 (U/ml) | | | 0.72 |
| ≤37 | 11 (22.4) | 0.044 | |
| >37 | 38 (77.6) | 0.046 | |
| DUPAN-2 (U/ml) | | | 0.12 |
| ≤150 | 28 (57.1)b | 0.044 | |
| >150 | 19 (38.8) | 0.055 | |
| Tumor size
(cm) | | | 0.0011 |
| ≤2 | 18 (36.7) | 0.037 | |
| >2 | 31 (63.3) | 0.052 | |
| Clinical
stagec | | | 0.045 |
| I | 15 (30.6) | 0.040 | |
| II | 33 (67.3) | 0.052 | |
| III | 1 (2.0) | 0.043 | |
| IV | 0 | NA | |
| Tumor location | | | 0.42 |
| Head | 28 (57.1) | 0.048 | |
| Body | 11 (22.4) | 0.040 | |
| Tail | 10 (20.4) | 0.045 | |
| Occult distant
metastasis | | | 0.034 |
| Absent | 42 (86) | 0.045 | |
| Present | 7 (14) | 0.062 | |
 | Table IIIUnivariate and multivariate analyses
of presenting occult distant metastasis in patients with pancreatic
cancer. |
Table III
Univariate and multivariate analyses
of presenting occult distant metastasis in patients with pancreatic
cancer.
| Clinical
features | Univariate analysis
| Multivariate
analysis
|
|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value |
|---|
| Age (<70 vs.
≥70) | 0.68
(0.14-3.43) | 0.64 | | |
| Sex (male vs.
female) | 0.68
(0.14-3.43) | 0.64 | | |
| Smoking (no vs.
yes) | 1.33
(0.27-6.70) | 0.73 | | |
| Alcohol consumption
(no vs. yes) | 2.40
(0.38-15.3) | 0.36 | | |
| CEA (ng/ml) (≤5 vs.
>5) | 1.70
(0.28-10.4) | 0.57 | 0.39
(0.033-4.59) | 0.46 |
| CA19-9 (U/ml) (≤37
vs. >37) | 2.61e+07
(0-Inf) | 0.99 | 2.33e+08
(0-Inf) | 0.99 |
| DUPAN-2 (U/ml)
(≤150 vs. >150) | 4.64
(0.8-27.1) | 0.088 | 5.07
(0.43-59.8) | 0.20 |
| Tumor size (cm) (≤2
vs. >2) | 4.08
(0.45-37.0) | 0.21 | 5.71
(0.16-208) | 0.34 |
| Clinical
stagea (I vs. II, III) | 3.0
(0.33-27.4) | 0.33 | 0.057
(0.00063-5.14) | 0.21 |
| Tumor location
(head vs. body, tail) | 0.48
(0.084-2.78) | 0.42 | 0.62
(0.069-5.51) | 0.66 |
| LINE-1 RDLb (<0.049 vs. ≥0.049) | 10.8
(1.19-98.4) | 0.035 | 22.2
(1.06-464) | 0.046 |
 | Table IVUnivariate and multivariate analyses
of overall survival in patients with pancreatic cancer. |
Table IV
Univariate and multivariate analyses
of overall survival in patients with pancreatic cancer.
| Prognostic
factors | Univariate analysis
| Multivariate
analysis
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (<70 vs.
≥70) | 1.13
(0.53-2.40) | 0.76 | | |
| Sex (male vs.
female) | 0.90
(0.42-1.94) | 0.79 | | |
| Smoking (no vs.
yes) | 1.00
(0.47-2.14) | 1.00 | | |
| Alcohol consumption
(no vs. yes) | 0.83
(0.25-2.77) | 0.77 | | |
| CEA (ng/ml) (≤5 vs.
>5) | 0.60
(0.21-1.76) | 0.53 | 0.33
(0.076-1.39) | 0.13 |
| CA19-9 (U/ml) (≤37
vs. >37) | 1.45
(0.55-3.84) | 0.45 | 2.03
(0.66-6.26) | 0.22 |
| DUPAN-2 (U/ml)
(≤150 vs. >150) | 1.03
(0.47-2.28) | 0.93 | 1.04
(0.39-2.77) | 0.93 |
| Tumor size (cm) (≤2
vs. >2) | 0.79
(0.37-1.68) | 0.54 | 0.88
(0.32-2.39) | 0.80 |
| Clinical
stagea (I vs. II, III) | 0.85
(0.38-1.90) | 0.69 | 0.48
(0.15-1.59) | 0.23 |
| Tumor location
(head vs. body, tail) | 0.68
(0.32-1.48) | 0.33 | 0.49
(0.21-1.15) | 0.099 |
| LINE-1 RDLb (<0.049 vs. ≥0.049) | 1.80
(0.84-3.85) | 0.13 | 3.40
(1.23-9.38) | 0.018 |
Discussion
To the best of our knowledge, the present study is
the first to show that genome-wide DNA hypomethylation induced by
treatment with 5-Aza-dC can drive the invasive phenotype of PC via
CIN in PC cells. Furthermore, by using LINE-1 RDL, the extent of
genome-wide DNA hypomethylation that is involved in the aggressive
behavior of PC, such as occult distant metastasis and poor
prognosis, was determined. These findings suggested that LINE-1 RDL
is a potent epigenetic biomarker for the selection of patients with
PC with occult distant metastases who are candidates for curative
surgery.
In the present study, genome-wide DNA
hypomethylation was induced in human PC cell lines with 5-Aza-dC
treatment. A significant increase in DNA hypomethylation was
observed in both cell lines regardless of their invasive potential.
In a previous study, DNA hypomethylation was successfully induced
in colon cancer cell lines with 5-Aza-dC treatment (45). Induction of DNA hypomethylation
was verified by methylation-sensitive amplified fragment-length
polymorphism, which is a fingerprinting technique that
simultaneously analyzes DNA methylation in hundreds of loci
(45,46). Genome-wide DNA hypomethylation was
reported to be induced in several tumors after treatment with
5-Aza-dC and is widely assessed by LINE-1 (18,47-50). Consistent with these findings, it
was reported that genome-wide DNA hypomethylation was induced in PC
cell lines.
Since the experimental mouse model has been used to
show that genome-wide DNA hypomethylation induces CIN (17,31), it has also been used to
investigate aneuploidy of chromosomes. The successful induction of
genome-wide DNA hypomethylation in the present study led to copy
number changes in specific regions of chromosomes that was
concomitant with mitotic chromosomal errors and DNA damage in PC
cell lines, which was consistent with the report that aneuploidy of
chromosomes increased when genome-wide DNA hypomethylation was
induced in colon cancer cells treated with 5-Aza-dC (18). However, no other studies have
clarified the relationship between genome-wide DNA hypomethylation
and CIN in PC cells after 5-Aza-dC treatment. Furthermore, the
present study demonstrated that changes in copy number were more
likely to be observed in PANC-1 than in Capan-1 cells, suggesting
that cells that do not have invasive potential are more susceptible
to CIN induced by genome-wide DNA hypomethylation.
CIN is an evolutionary process of PC metastasis
(32-34) and poor patient prognosis. Bakhoum
et al (51) showed that
CIN promotes metastasis by sustaining a tumor-cell autonomous
response to cytosolic DNA in breast cancer. In the present study,
the number of migratory cells in PANC-1 cells treated with 5-Aza-dC
(37.9±8.0%) was the same as that in naïve Capan-1 cells before
treatment (35.4±11.4%), indicating that PANC-1 acquired invasive
potential similar to Capan-1 (Fig.
4) and that genome-wide DNA hypomethylation affects the
invasive nature of the PC phenotype through the induction of
CIN.
Metastasis is the most common cause of
cancer-related death in patients with cancer in clinical practice,
particularly in patients with PC that have a poor prognosis.
Identification of markers that can successfully predict occult
distant metastasis is critical for improving patient prognosis and
identifying appropriate candidates for curative surgery. Liu et
al (11) have reported that
younger age, male sex, larger tumor size, low serum ALT level, and
high serum CA19-9 level are independent predictors of unexpected
distant metastasis on exploration. Although certain studies have
suggested a role for tumor location, serum CEA level, and clinical
findings such as weight loss and jaundice, currently insufficient
evidence exists for the applicability of these variables in
predicting resectability (1,12).
The present study demonstrated that there was a significant
correlation before the LINE-1 RDL and clinical variables such as
tumor size, clinical stage and the presence of occult distant
metastasis. The significance of genome-wide DNA hypomethylation was
then elucidated using LINE-1 RDL as a predictor of occult distant
metastasis. Occult distant metastasis was recognized in six out of
21 patients with high LINE-1 RDL ≥0.049, although these patients
were diagnosed to not show metastasis by imaging studies performed
before surgery. The analysis of LINE-1 RDL facilitates the
identification of patients likely to have occult distant
metastasis, who require further exploration for unresectability
before surgery. De Rosa et al (12) have reported that serum CA19-9
level and tumor size are potential predictors of unresectability in
selecting patients for staging laparoscopy. By contrast, only one
in 28 patients with low LINE-1 RDL <0.049 displayed occult
distant metastasis. These patients with a low risk of occult
distant metastasis, as assessed by LINE-1 RDL, would be appropriate
candidates for curative surgery. Total assessment of LINE-1 RDL in
combination with serum CA19-9 level and tumor size would be
reliable surrogate markers for selecting patients for curative
surgery without staging laparoscopy before operation.
Previous studies have reported that LINE-1
hypomethylation is associated with clinical outcome in patients
with several types of cancer (27,28). In the present study, a significant
correlation was also observed between LINE-1 RDL and OS in patients
with PC, with multivariate analysis indicating that LINE-1 RDL is a
significant independent predictor of prognosis (Table IV). Yamamura et al
(28) reported no significant
association between LINE-1 hypomethylation and prognosis in
patients with PC using a cohort that included 10% of patients with
stage IV PC, which may have affected prognosis. Patients with stage
IV disease were excluded and focus was addressed on patients who
underwent curative surgery without extensive invasion or distant
metastasis. Although no significant difference in OS was identified
between patients with high and low LINE-1 RDL in the present study,
patients with high LINE-1 RDL tended to have a worse OS than those
with low LINE-1 RDL. A modified LINE-1 RDL of 0.045 revealed a
significant difference in OS between patients with high and low
LINE-1 RDL. The difference in patient selection using LINE-1
hypomethylation may affect its association of clinical outcomes
between the present study and that of Yamamura et al
(28).
The present study had certain limitations. The
optimal concentration of 5-Aza-dC to prevent cell death in PC cell
lines was determined, but the possibility that the cytotoxicity
induced by 5-Aza-dC was associated with the induction of CIN could
not be excluded. Furthermore, 5-Aza-dC may alter the transcription
of multiple genes that could potentially affect chromosomal
segregation fidelity and invasiveness. In addition, CIN was induced
by genome-wide DNA hypomethylation following treatment with
5-Aza-dC, but it is unknown which was more responsible for
enhancing the invasive potential of the treated cells. Further
studies with a larger and more inclusive patient cohort are also
required to confirm the significance of LINE-1 hypomethylation as a
potent epigenetic biomarker in the prognosis of patients with stage
IV cancer in clinical practice.
In conclusion, the present findings indicated that
successful induction of CIN by genome-wide DNA hypomethylation
induces the phenotype of PC to become more invasive in
vitro, and that genome-wide DNA hypomethylation is involved in
the biological behavior of PC, such as occult distant metastasis
and poor patient prognosis in PC. LINE-1 RDL could be a potent
epigenetic biomarker for the selection of patients who could
benefit from staging laparoscopy to avoid unnecessary laparotomy
and develop bespoke treatments in a timely manner. It is considered
that the present data shed light on new strategies for treating
patients with PC who could benefit from curative surgery.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YE designed the study and wrote the initial draft of
the manuscript. KS contributed to the analysis and interpretation
of the data and assisted in the preparation of the manuscript. YK,
ST, HA, IA, FW, TK, MS, KF, HN, FK and TR contributed to data
collection and interpretation, and critically reviewed the
manuscript. YE and KS confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of the Jichi Medical University (Saitama, Japan).
Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by a Grant-in-Aid for Scientific
Research (grant no. JP 16K10514) from the Ministry of Education,
Culture, Sports, Science and Technology and the JKA Foundation
through its promotion funds from the Keirin Race [grant no.
27-1-068 (2)].
Abbreviations:
|
PC
|
pancreatic cancer
|
|
CIN
|
chromosomal instability
|
|
LINE-1
|
long interspersed nucleotide
element-1
|
|
RDL
|
relative demethylation level
|
|
5-Aza-dC
|
5-aza-2′-deoxycytidine
|
|
CGH
|
comparative genomic hybridization
|
|
CEA
|
carcinoembryonic antigen
|
|
CA19-9
|
carbohydrate antigen 19-9
|
|
DUPAN-2
|
duke pancreatic monoclonal antigen
type 2
|
|
OS
|
overall survival
|
References
|
1
|
Fong ZV, Alvino DML, Fernández-Del
Castillo C, Mehtsun WT, Pergolini I, Warshaw AL, Chang DC, Lillemoe
KD and Ferrone CR: Reappraisal of staging laparoscopy for patients
with pancreatic adenocarcinoma: A contemporary analysis of 1001
patients. Ann Surg Oncol. 24:3203–3211. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gillen S, Schuster T, Meyer Zum
Büschenfelde C, Friess H and Kleeff J: Preoperative/neoadjuvant
therapy in pancreatic cancer: A systematic review and meta-analysis
of response and resection percentages. PLoS Med. 7:e10002672010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kamisawa T, Wood LD, Itoi T and Takaori K:
Pancreatic cancer. Lancet. 388:73–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mihaljevic AL, Michalski CW, Friess H and
Kleeff J: Molecular mechanism of pancreatic cancer-understanding
proliferation, invasion, and metastasis. Langenbecks Arch Surg.
395:295–308. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Du YX, Liu ZW, You L, Wu WM and Zhao YP:
Advances in understanding the molecular mechanism of pancreatic
cancer metastasis. Hepatobiliary Pancreat Dis Int. 15:361–370.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Missiaglia E, Donadelli M, Palmieri M,
Crnogorac-Jurcevic T, Scarpa A and Lemoine NR: Growth delay of
human pancreatic cancer cells by methylase inhibitor
5-aza-2′-deoxycytidine treatment is associated with activation of
the interferon signalling pathway. Oncogene. 24:199–211. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Allen VB, Gurusamy KS, Takwoingi Y, Kalia
A and Davidson BR: Diagnostic accuracy of laparoscopy following
computed tomography (CT) scanning for assessing the resectability
with curative intent in pancreatic and periampullary cancer.
Cochrane Database Syst Rev. 7:CD0093232016.PubMed/NCBI
|
|
9
|
Vargas R, Nino-Murcia M, Trueblood W and
Jeffrey RB Jr: MDCT in Pancreatic adenocarcinoma: Prediction of
vascular invasion and resectability using a multiphasic technique
with curved planar reformations. AJR Am J Roentgenol. 182:419–425.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pietryga JA and Morgan DE: Imaging
preoperatively for pancreatic adenocarcinoma. J Gastrointest Oncol.
6:343–357. 2015.PubMed/NCBI
|
|
11
|
Liu X, Fu Y, Chen Q, Wu J, Gao W, Jiang K,
Miao Y and Wei J: Predictors of distant metastasis on exploration
in patients with potentially resectable pancreatic cancer. BMC
Gastroenterol. 18:1682018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
De Rosa A, Cameron IC and Gomez D:
Indications for staging laparoscopy in pancreatic cancer. HPB
(Oxford). 18:13–20. 2016. View Article : Google Scholar
|
|
13
|
Durczynski A, Kumor A, Hogendorf P,
Szymanski D, Grzelak P and Strzelczyk J: Preoperative high level of
D-dimers predicts unresectability of pancreatic head cancer. World
J Gastroenterol. 20:13167–13171. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schlieman MG, Ho HS and Bold RJ: Utility
of tumor markers in determining resectability of pancreatic cancer.
Arch Surg. 138:951–956. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gleisner AL, Assumpcao L, Cameron JL,
Wolfgang CL, Choti MA, Herman JM, Schulick RD and Pawlik TM: Is
resection of periampullary or pancreatic adenocarcinoma with
synchronous hepatic metastasis justified? Cancer. 110:2484–2492.
2007. View Article : Google Scholar
|
|
16
|
Hackert T, Niesen W, Hinz U, Tjaden C,
Strobel O, Ulrich A, Michalski CW and Büchler MW: Radical surgery
of oligometastatic pancreatic cancer. Eur J Surg Oncol. 43:358–363.
2017. View Article : Google Scholar
|
|
17
|
Gaudet F, Hodgson JG, Eden A,
Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H and Jaenisch R:
Induction of tumors in mice by genomic hypomethylation. Science.
300:489–492. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Costa G, Barra V, Lentini L, Cilluffo D
and Di Leonardo A: DNA demethylation caused by
5-Aza-2′-deoxycytidine induces mitotic alterations and aneuploidy.
Oncotarget. 7:3726–3739. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Itano O, Ueda M, Kikuchi K, Hashimoto O,
Hayatsu S, Kawaguchi M, Seki H, Aiura K and Kitajima M: Correlation
of postoperative recurrence in hepatocellular carcinoma with
demethylation of repetitive sequences. Oncogene. 21:789–797. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Widschwendter M, Jiang G, Woods C, Müller
HM, Fiegl H, Goebel G, Marth C, Müller-Holzner E, Zeimet AG, Laird
PW and Ehrlich M: DNA hypomethylation and ovarian cancer biology.
Cancer Res. 64:4472–4480. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deininger PL, Moran JV, Batzer MA and
Kazazian HH Jr: Mobile elements and mammalian genome evolution.
Curr Opin Genet Dev. 13:651–658. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ehrlich M: DNA methylation in cancer: Too
much, but also too little. Oncogene. 21:5400–5413. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cordaux R and Batzer MA: The impact of
retrotransposons on human genome evolution. Nat Rev Genet.
10:691–703. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li J, Huang Q, Zeng F, Li W, He Z, Chen W,
Zhu W and Zhang B: The prognostic value of global DNA
hypomethylation in cancer: A meta-analysis. PLoS One.
9:e1062902014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang AS, Estécio MR, Doshi K, Kondo Y,
Tajara EH and Issa JP: A simple method for estimating global DNA
methylation using bisulfite PCR of repetitive DNA elements. Nucleic
Acids Res. 32:e382004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Weisenberger DJ, Campan M, Long TI, Kim M,
Woods C, Fiala E, Ehrlich M and Laird PW: Analysis of repetitive
element DNA methylation by MethyLight. Nucleic Acids Res.
33:6823–6836. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Baba Y, Yagi T, Sawayama H, Hiyoshi Y,
Ishimoto T, Iwatsuki M, Miyamoto Y, Yoshida N and Baba H: Long
interspersed element-1 methylation level as a prognostic biomarker
in gastrointestinal cancers. Digestion. 97:26–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yamamura K, Kosumi K, Baba Y, Harada K,
Gao F, Zhang X, Zhou L, Kitano Y, Arima K, Kaida T, et al: LINE-1
methylation level and prognosis in pancreas cancer: Pyrosequencing
technology and literature review. Surg Today. 47:1450–1459. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thompson SL and Compton DA: Examining the
link between chromosomal instability and aneuploidy in human cells.
J Cell Biol. 180:665–672. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cimini D, Howell B, Maddox P, Khodjakov A,
Degrassi F and Salmon ED: Merotelic kinetochore orientation is a
major mechanism of aneuploidy in mitotic mammalian tissue cells. J
Cell Biol. 153:517–527. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Eden A, Gaudet F, Waghmare A and Jaenisch
R: Chromosomal instability and tumors promoted by DNA
hypomethylation. Science. 300:4552003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jamal-Hanjani M, Wilson GA, Mcgranahan N,
Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al: Tracking the evolution of non-small-cell lung
cancer. N Engl J Med. 376:2109–2121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Turajlic S and Swanton C: Metastasis as an
evolutionary process. Science. 352:169–175. 2016. View Article : Google Scholar
|
|
34
|
Igarashi S, Suzuki H, Niinuma T, Shimizu
H, Nojima M, Iwaki H, Nobuoka T, Nishida T, Miyazaki Y, Takamaru H,
et al: A novel correlation between LINE-1 hypomethylation and the
malignancy of gastrointestinal stromal tumors. Clin Cancer Res.
16:5114–5123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Suzuki K, Ohnami S, Tanabe C, Sasaki H,
Yasuda J, Katai H, Yoshimura K, Terada M, Perucho M and Yoshida T:
The genomic damage estimated by arbitrarily primed PCR DNA
fingerprinting is useful for the prognosis of gastric cancer.
Gastroenterology. 125:1330–1340. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Suzuki K, Suzuki I, Leodolter A, Alonso S,
Horiuchi S, Yamashita K and Perucho M: Global DNA demethylation in
gastrointestinal cancer is age dependent and precedes genomic
damage. Cancer Cell. 9:199–207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ichida K, Suzuki K, Fukui T, Takayama Y,
Kakizawa N, Watanabe F, Ishikawa H, Muto Y, Kato T, Saito M, et al:
Overexpression of satellite alpha transcripts leads to chromosomal
instability via segregation errors at specific chromosomes. Int J
Oncol. 52:1685–1693. 2018.PubMed/NCBI
|
|
38
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM classification of malignant tumours. 7th edition.
Chichester, West Sussex; Wiley-Blackwell; Hoboken, NJ: 2010
|
|
39
|
Saito M, Suzuki K, Maeda T, Kato T,
Kamiyama H, Koizumi K, Miyaki Y, Okada S, Kiyozaki H and Konishi F:
The accumulation of DNA demethylation in Sat α in normal gastric
tissues with Helicobacter pylori infection renders susceptibility
to gastric cancer in some individuals. Oncol Rep. 27:1717–1725.
2012.PubMed/NCBI
|
|
40
|
Heid CA, Stevens J, Livak KJ and Williams
PM: Real time quantitative PCR. Genome Res. 6:986–994. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Matsui Y, Nakayama Y, Okamoto M, Fukumoto
Y and Yamaguchi N: Enrichment of cell populations in metaphase,
anaphase, and telophase by synchronization using nocodazole and
blebbistatin: A novel method suitable for examining dynamic changes
in proteins during mitotic progression. Eur J Cell Biol.
91:413–419. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Caravaca JM, Donahue G, Becker JS, He X,
Vinson C and Zaret KS: Bookmarking by specific and nonspecific
binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev.
27:251–260. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Basit A, Cho MG, Kim EY, Kwon D, Kang SJ
and Lee JH: The cGAS/STING/TBK1/IRF3 innate immunity pathway
maintains chromosomal stability through regulation of p21 levels.
Exp Mol Med. 52:643–657. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kanda Y: Investigation of the freely
available easy-to-use software 'EZR' for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar
|
|
45
|
Miyaki Y, Suzuki K, Koizumi K, Kato T,
Saito M, Kamiyama H, Maeda T, Shibata K, Shiya N and Konishi F:
Identification of a potent epigenetic biomarker for resistance to
camptothecin and poor outcome to irinotecan-based chemotherapy in
colon cancer. Int J Oncol. 40:217–226. 2012.
|
|
46
|
Alonso S, Suzuki K, Yamamoto F and Perucho
M: Methylation-sensitive amplification length polymorphism
(MS-AFLP) microarrays for epigenetic analysis of human genomes.
Methods Mol Biol. 1766:137–156. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Daskalos A, Nikolaidis G, Xinarianos G,
Savvari P, Cassidy A, Zakopoulou R, Kotsinas A, Gorgoulis V, Field
JK and Liloglou T: Hypomethylation of retrotransposable elements
correlates with genomic instability in non-small cell lung cancer.
Int J Cancer. 124:81–87. 2009. View Article : Google Scholar
|
|
48
|
Hamm CA, Xie H, Costa FF, Vanin EF, Seftor
EA, Sredni ST, Bischof J, Wang D, Bonaldo MF, Hendrix MJ and Soares
MB: Global demethylation of rat chondrosarcoma cells after
treatment with 5-aza-2′-deoxycytidine results in increased
tumorigenicity. PLoS One. 4:e83402009. View Article : Google Scholar
|
|
49
|
Wang X, Gao H, Ren L, Gu J and Zhang Y and
Zhang Y: Demethylation of the miR-146a promoter by
5-Aza-2′-deoxycytidine correlates with delayed progression of
castration-resistant prostate cancer. BMC Cancer. 14:3082014.
View Article : Google Scholar
|
|
50
|
Zhang C, Fan L, Fan T, Wu D, Gao L, Ling
Y, Zhu J, Li R and Wei L: Decreased PADI4 mRNA association with
global hypomethylation in hepatocellular carcinoma during HBV
exposure. Cell Biochem Biophys. 65:187–195. 2013. View Article : Google Scholar
|
|
51
|
Bakhoum SF, Ngo B, Laughney AM, Cavallo
JA, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, et
al: Chromosomal instability drives metastasis through a cytosolic
DNA response. Nature. 553:467–472. 2018. View Article : Google Scholar : PubMed/NCBI
|