Introduction
Colon cancer is a malignant tumour, with the second
and fifth highest mortality rates in USA and China, respectively.
Its morbidity and mortality rates are currently increasing in China
(1-3). Of note, <20% of first-time
diagnosed patients with metastatic colon cancer survive for >5
years and 60% of these patients are already in the advanced stage
of the disease at the time of consultation (4). Current colon cancer treatments are
based on surgical resection and supplemented with concurrent
chemotherapy, targeted therapy and biological therapy (5). 5-Fluorouracil (5-FU) drug-based
chemotherapeutic regimens have been reported to effectively improve
the tumour-free survival rate of patients with colorectal cancer
(CRC); however, patients with CRC have exhibited varying degrees of
resistance to 5-FU (6). This
innate or acquired chemoresistance affects the treatment efficacy
(7). Therefore, the
identification of a molecular target that can effectively predict
and combat resistance to 5-FU is essential in improving CRC
treatment efficacy and reducing mortality rates. Visfatin, also
known as nicotinamide phosphoribosyl transferase (NAMPT), has been
reported to decrease the sensitivity of cancer cells to
chemotherapeutic drugs (8).
Visfatin is an adipocytokine secreted by visceral
fat tissue. The human visfatin gene, NAMPT, is located
between chromosomes 7q22.1 and 7q31.33 and has a relative molecular
weight of 52,000, comprising 473 amino acid residues (9). Clinical studies have found that the
extracellular expression of visfatin in T4 stage tumour tissues is
significantly increased compared to its expression in initial stage
II-III tumour tissues, and this elevated expression of visfatin is
a strong risk factor for advanced- and early-stage CRC (10,11). Clinical studies have confirmed the
association of visfatin with cancer staging, lymph node metastasis
and other adverse factors, which affect patient survival, in colon
cancer and gliomas (12,13). It is worth further investigating
whether the upregulation of visfatin is part of the stem cell
phenotype or simply the result of other malignant changes. Visfatin
catalyses the conversion of nicotinamide mononucleotide, which is
the rate-limiting step in the NAD salvage pathway (14), thereby enhancing cancer cell
proliferation. However, decreasing proliferation could decrease
tumour sensitivity to drugs that target proliferating cells.
Therefore, NAMPT has been proposed as a promising anticancer target
(14). For example, NAMPT
inhibitors combined with platinum-based chemotherapy have been
shown to suppress senescence-associated cancer stem-like cells
(CSCs) and reduce tumour recurrence (15). The overexpression of visfatin can
also downregulate the sensitivity of non-small-cell lung cancer
(NSCLC) cells to doxorubicin, and can upregulate the mRNA and
protein expression of ATP binding cassette subfamily C member 1
(ABCC1) (16). Moreover, as
previously demonstrated, NAMPT inhibition by FK866 inhibits cell
viability and aggravates apoptosis in cancer cells treated with
4-hydroxytamoxifen, while NAMPT overexpression promotes xenograft
tumour growth in nude mice (17).
Thus, previous studies prove the suppressive effects of visfatin on
sensitivity to chemotherapeutic drugs. PI3K/Akt is a typical
downstream pathway of visfatin in various types of cancer,
including CRC (18,19), and it has been frequently found to
be associated with stromal cell-derived factor-1 (SDF-1) in cancer
(20-22). Therefore, it was hypothesised that
the role and regulatory mechanisms of visfatin in CRC may be
related to changes in SDF-1/chemokine receptor type 4 (CXCR4)
levels; this hypothesis was preliminarily confirmed in a previously
published study by the authors (23).
The present study aimed to further explore the
effects and mechanisms of action of visfatin on the sensitivity of
colon cancer cells to 5-FU. Visfatin expression in colon cancer
tissues and cells was determined, followed by the verification of
the suppressive effects of visfatin on the sensitivity of CRC to
5-FU chemotherapy in vitro. Furthermore, the downstream
signalling pathway involved in the effects of visfatin on
sensitivity to 5-FU was analysed and verified using correlation
analysis, rescue experiments, as well as other analyses.
Materials and methods
Tissue microarray (TMA)
A TMA was purchased from the Xian Alenabio
Biotechnology Company (China), which included 57 tissue samples
from patients with colon cancer whose clinical characteristics are
presented in Table SI.
Immunohistochemistry was performed on the TMA samples, and each
tumour included three 0.6-mm core biopsies. Briefly, the chips were
placed in a 67°C oven for 2 h. Citrate buffer [pH 6.0, Sangon
Biotech (Shanghai) Co., Ltd.] was added to the microwave box and
microwaved at mid-range for 10 min. Primary antibody dilutions of
visfatin (1:1,000, cat. no. ab236091) or SDF-1 (1:1,000, cat. no.
ab25117) (both from Abcam) were added to the chip and incubated
overnight at 4°C. The secondary antibody (S0001, 1:5,000, Affinity
Biosciences) was then added in a dropwise manner followed by
incubation at 37°C for 2 h. The TMA slides were scanned and the
Quant centre (Panoramic MIDI II, Sysmex Europe GmbH) was used to
analyse the digital images. Each slide was annotated using
automatic TMA de-arrangement tools (Quant Center) and detection
classifiers to distinguish tumours. The immunostaining percentage
and staining intensity (0, negative; 1+, weak; 2+, moderate; and
3+, strong) were recorded. Furthermore, the H-score was used to
score the staining as follows: H-score=(percentage of cells of weak
intensity ×1) + (percentage of cells of moderate-intensity ×2) +
percentage of cells of strong intensity ×3), as previously
described (24). The nuclear
tests were visually inspected and then manually corrected to
eliminate staining artefacts.
Clinical samples
Colon cancer tissues were obtained from 3 patients
with colon cancer and 3 control volunteers at the First People's
Hospital of Yunnan Province from April, 2020 to May, 2020. All
patients provided written informed consent. The present study was
approved and supervised by the Ethics Committee of the First
People's Hospital of Yunnan Province in accordance with the
International Ethical Guidelines for Biomedical Research Involving
Human Subjects (CIOMS).
Cells and cell culture
Colon cancer DLD-1 [CCL-221, American Type Culture
Collection (ATCC)], SW48 (CCL-231, ATCC), HCT116 (CCL-247EMT, ATCC)
and SW620 (CCL-227, ATCC) cells were cultured using Dulbecco
modified Eagle's medium (DMEM, Invitrogen; Thermo Fisher
Scientific, Inc.), containing 10% foetal bovine serum (FBS), at
37°C and 5% CO2. Human colonic epithelial NCM460 cells
were incubated in DMEM containing 10% FBS [Sangon Biotech
(Shanghai) Co., Ltd.], 100 U/ml penicillin [Sangon Biotech
(Shanghai) Co., Ltd.] and 100 µg/ml streptomycin [Sangon
Biotech (Shanghai) Co., Ltd.]. These cell lines were purchased from
the Institute of Biochemistry and Cell Biology, Chinese Academy of
Sciences (Shanghai, China) or the American Type Culture Collection.
Additionally, cells were tested for mycoplasma contamination.
Cell transfection
Full-length visfatin or SDF-1 coding sequences were
amplified using polymerase chain reaction (PCR), according to the
sequence available at the National Center for Biotechnology
Information database (Shanghai, China) and cloned into pcDNA3.1
[Sangon Biotech (Shanghai) Co., Ltd.]. Following the manufacturer's
instructions, Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to transfect the pcDNA-visfatin
or pcDNA-SDF-1 vector (20 nmol/l) into the DLD-1 and SW48 cells
cultured in six-well plates. Using Lipofectamine™ 2000, a short
hairpin RNA [shRNA, Sangon Biotech (Shanghai) Co., Ltd.] that
specifically targets visfatin was transiently transfected into the
DLD-1 and SW48 cells. The transfection group with a scrambled shRNA
sequence was used as the mock group. The sequences were as follows:
Visfatin-shRNA-1 Sense, ccg gCT TGA GGA ATA TGG TCA GGA Tct cga gAT
CCT GAC CAT ATT CCT CAA Gtt ttt g and antisense, AAT Tca aaa aCT
TGA GGA ATA TGG TCA GGA Tct cga gAT CCT GAC CAT ATT CCT CAA G;
Visfatin-shRNA-2 sense, ccg gTT CTC CAT ACT GTC TTC AAG Act cga gTC
TTG AAG ACA GTA TGG AGA Att ttt g and antisense, AAT Tca aaa aTT
CTC CAT ACT GTC TTC AAG Act cga gTC TTG AAG ACA GTA TGG AGA A;
SDF-1 shRNA sense, ccg gCC GTC AGC CTG AGC TAC AGA Tct cga gAT CTG
TAG CTC AGG CTG ACG Gtt ttt g and antisense, AAT Tca aaa aCC GTC
AGC CTG AGC TAC AGA Tct cga gAT CTG TAG CTC AGG CTG ACG G;
scrambled shRNA sense, ccg gTA CTG AAC CGT AAA GCA GGT Act cga gTA
CCT GCT TTA CGG TTC AGT Attt ttg and antisense, AAT Tca aaa aTA CTG
AAC CGT AAA GCA GGT Act cga gTA CCT GCT TTA CGG TTC AGT A. PI3K
inhibitor (LY294002, 10 µmol/l, Selleck Chemicals) was used
to treat the cells for 24 h to verify the association between SDF-1
and the PI3K/AKT axis. Total RNA was isolated using TRIzol reagent
(Takara Biotechnology Co., Ltd.) following 48 h of transfection,
and visfatin and SDF-1 expression levels were detected using
reverse transcription-quantitative PCR (RT-qPCR) and western blot
analysis.
Animal experiments
A total of 15 NOD/SCID mice (Shanghai SLAC
Laboratory Animal Co., Ltd.; 6 weeks old, female), were
accommodated at a constant temperature of 22°C, with a relative
humidity of 30% and a 12-h light/dark cycle, and standard
conditions with free access to food and water. When the confluency
of the cells subjected to visfatin knockdown or overexpression
reached 80-90%, 0.25% trypsin digestion was performed to obtain a
single-cell suspension. The cells were resuspended in
phosphate-buffered saline (PBS) and adjusted to 1×107/ml
density. Each mouse was inoculated with 0.2 ml cell suspension
subcutaneously into the right armpit. Mice were randomly divided
into the nock group, shRNA2 group, and shRNA2 + visfatin group,
with 5 mice in each group. When the tumour tissues of the mice in
the mock group reached a volume of 1,000 mm3, all mice
were sacrificed by an intraperitoneal injection of sodium
pentobarbital (150 mg/kg). The tumour tissue was collected for
volume and weight measurements. The experimental protocol was
approved by the Animal Protection and Use Agency Committee of
Kunming University of Science and Technology and conducted in
accordance with the Laboratory animal Guideline for ethical review
of animal welfare (GB/T 35892-2018).
Cell viability assay
The Cell Counting Kit-8 (CCK-8; Beyotime Institute
of Biotechnology) was used to detect cell viability in each group.
The experimental procedures were performed following the
manufacturer's instructions. Briefly, cells were seeded in a
96-well plate at a density of 5.0×103/well. Following
gradient 5-FU (10, 20, 30, 40, 50, 60 µM; ST1060, Beyotime
Institute of Biotechnology) treatments and vector transfection for
24 h, 10 µl CCK-8 reagent were added to each well and
incubated at 37°C for 2 h. The absorbance was evaluated using a
microplate reader at 450 nm (Tecan Group, Ltd.).
Colony formation assay
The clone formation experiment was used to detect
the passage and proliferative ability of the cells in each group.
The cells were evenly spread into a six-well plate (300 cells/well)
following suspension treatment after 5-FU (40 µM) treatment,
and incubated at 37°C, with 5% CO2 and saturated
humidity for 2 to 3 weeks. When macroscopic clones were observed in
the Petri dish under an inverted light microscope (CKX3-SLP,
Olympus Corporation), the supernatant was discarded and the cells
were fixed with 4% paraformaldehyde (Dalian Meilun Biotechnology
Co., Ltd.) at room temperature for 20 min. The cells were then
stained with crystal violet (Dalian Meilun Biotechnology Co., Ltd.)
at room temperature for 20 min, and the clone formation number was
calculated using ImageJ 1.51J8 software (National Institutes of
Health) after obtaining images under an inverted light microscope
(CKX3-SLP, Olympus Corporation).
Immunofluorescence
Immunofluorescence was used to detect caspase-3
expression in cells. After immunofluorescence processing, the cells
were mounted onto slides. The slides were washed thrice with PBS
for 3 min each time. After fixing the cells with 4%
paraformaldehyde at room temperature for 15 min, they were washed
thrice with PBS for 3 min each time. The cells were infiltrated
with 0.5% Triton X-100 (prepared in PBS) for 20 min and then
blocked using goat serum for at room temperature 30 min. After
absorbing the excess blocking solution with an absorbent paper, a
sufficient diluted cleaved caspase-3 primary antibody (1:1,000,
cat. no. ab32042, Abcam) was added to each slide. The slides were
placed in a wet box and incubated overnight at 4°C. Subsequently,
the corresponding fluorescent secondary antibody [IgG H&L (HRP)
(S0001, 1:500, Affinity Biosciences)] was added to the glass slide,
followed by the addition of DAPI (C1002, Beyotime Institute of
Biotechnology) and incubation at room temperature for 5 min in a
dark room. After the addition of an anti-fluorescence quencher
(P0126, Beyotime Institute of Biotechnology) to the glass slide and
incubated at room temperature, the images were observed under a
fluorescence microscope.
Flow cytometric analysis
The cells were seeded in a 12-well plate
(5×104 cells per well) for apoptosis analysis using flow
cytometry. Following 12 h of transfection, each well was replaced
with a cell culture medium containing 5-FU and further incubated at
37°C for 72 h. An AnnexinV-FITC/PI Apoptosis Kit (Multisciences
(Lianke) Biotech Co., Ltd.) was used to measure cell apoptosis,
following the manufacturer's instructions. Flow cytometric analysis
was performed using the BD FACSCalibur™ (BD Bioscience) flow
cytometer system.
Caspase-3/7 assay
Apoptosis was detected using the caspase-3/7 kit
(AAT Bioquest). The operation steps were as follows: The cells were
seeded into 96-well plates at 20,000 cells/well in a growth medium
(DMEM and 10% FBS). Subsequently, 10 µl per well of 10× the
test compound was added to a 96-well plate for cell processing. For
blank wells, compound buffer (10 µl/well) was added. The
cell plate was incubated at 37°C in 5% CO2. Furthermore,
a caspase-3/7 substrate working solution (100 µl/well) was
added to the plate. The cells were incubated for at least 1 h in a
dark room at room temperature. A fluorescence microplate reader
(Fluoroskan ascent FL, Thermo Fisher Scientific, Inc.) was used to
monitor the fluorescence intensity.
RT-qPCR
According to the manufacturer's instructions, total
RNA was extracted from the DLD-1 and SW-48 cells using TRIzol
reagent and a high-purity total RNA rapid extraction kit (Takara
Biotechnology Co., Ltd.) and reverse transcribed into complementary
DNA (cDNA) using the 5X Prime Script RT Master Mix (Takara Bio,
Inc.) by incubating at 85°C for 5 min. RNase inhibitor
diethylpyrocarbonate (DEPC, ST036, Beyotime Institute of
Biotechnology) were used to inhibit RNA degradation. SYBR-Green and
other reagents (Takara Bio, Inc.) were prepared in accordance with
the manufacturer's instructions for an RT-PCR reaction. The PCR
reaction conditions were 95°C for 5 min, 95°C for 30 cycles, 60°C
for 30 sec, 72°C for 30 sec, and finally 72°C for 7 min. The primer
sequences used were as follows: SDF-1 forward, 5′-CTA CAG ATG CCC
ATG CCG AT-3′ and reverse, 5′-CAG CCG GGC TAC AAT CTG AA-3′; CXCR4
forward, 5′-ATC AGT CTG GAC CGC TAC CT-3′ and reverse, 5′-CCA CCT
TTT CAG CCA ACA GC-3′; β-actin forward, 5′-ACA TGC AGC CAG AAG AGG
AC-3′ and reverse, 5′-ACA CCA TGC CGT GAA TGT CT-3′. The relative
expression was obtained using the 2−ΔΔCq method
(25). β-actin was used as an
internal control.
Western blot analysis
Proteins were extracted from cells after processing
using RIPA lysis buffer (Beyotime Institute of Biotechnology), and
the protein concentration was measured using Bradford analysis
(Bio-Rad Laboratories, Inc.). Proteins (20 µg/lane) were
separated using sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE, 8-12%) and transferred to a PVDF
membrane (MilliporeSigma). The membrane was then sealed with 5%
(w/v) skimmed milk at room temperature for 1 h and subsequently
combined with visfatin (1:1,000, cat. no. ab23609), SDF-1 (1:1,000,
cat. no. ab25117), CXCR4 (1:1,000, cat. no. ab181020), caspase-3
(1:1,000, cat. no. ab32351), caspase-7 (1:1,000, cat. no. ab32522),
proliferating cell nuclear antigen (PCNA; 1:1,000, cat. no.
ab18197), Ki-67 (1:1,000, cat. no. ab92742), cleaved caspase-3
(1:1,000, cat. no. ab32042), survivin (1:1,000, cat. no. ab76424),
Livin (1:1,000, cat. no. ab236500), p53 (1:1,000, cat. no.
ab32389), Bax (1:1,000, cat. no. ab32503), Bcl-2 (1:1,000, cat. no.
ab32124), Akt (1:1,000, cat. no. ab8805), phosphorylated (p-)Akt
(1:1,000, cat. no. ab38449), PI3K (1:1,000, cat. no. ab140307),
p-PI3K (1:1,000, cat. no. ab278545), glycogen synthase kinase
(GSK)3β (1:1,000, cat. no. ab32391), p-GSK3β (1:1,000, cat. no.
ab75814) and β-actin (1:5,000, cat. no. ab8227) primary antibodies
(all from Abcam) at 4°C overnight. Following this, the membrane was
incubated with the secondary antibody (S0001, 1:5,000, Affinity
Biosciences) at 37°C for 2 h. The liquid on the membrane was dried
using a filter paper and reacted in the enhanced chemiluminescence
(Amersham; Cytiva) reaction mixture in a dark room for 2-3 min. For
the stabilisation experiments, cells were collected after being
treated with cycloheximide (CHX, 100 µg/ml DMSO, C8500,
USBiological) for 0, 2, 4, 8 or 24 h, and western blot analysis was
used to examine the stabilisation of SDF-1 and CXCR4. Specific
bands were detected. ImageJ 1.51J8 software (National Institutes of
Health) was used to count the grayscale of the strips.
Statistical analyses
The experiments requiring statistical analyses were
repeated thrice. The experimental data are expressed as the mean ±
standard deviation. Statistical analyses were performed using SPSS
software (version 13.0, SPSS, Inc.) with one-way or two-way
analysis of variance, followed by Tukey's post hoc test. Pearson's
correlation analysis was used for correlation analysis. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
Close association between visfatin
expression and colon cancer
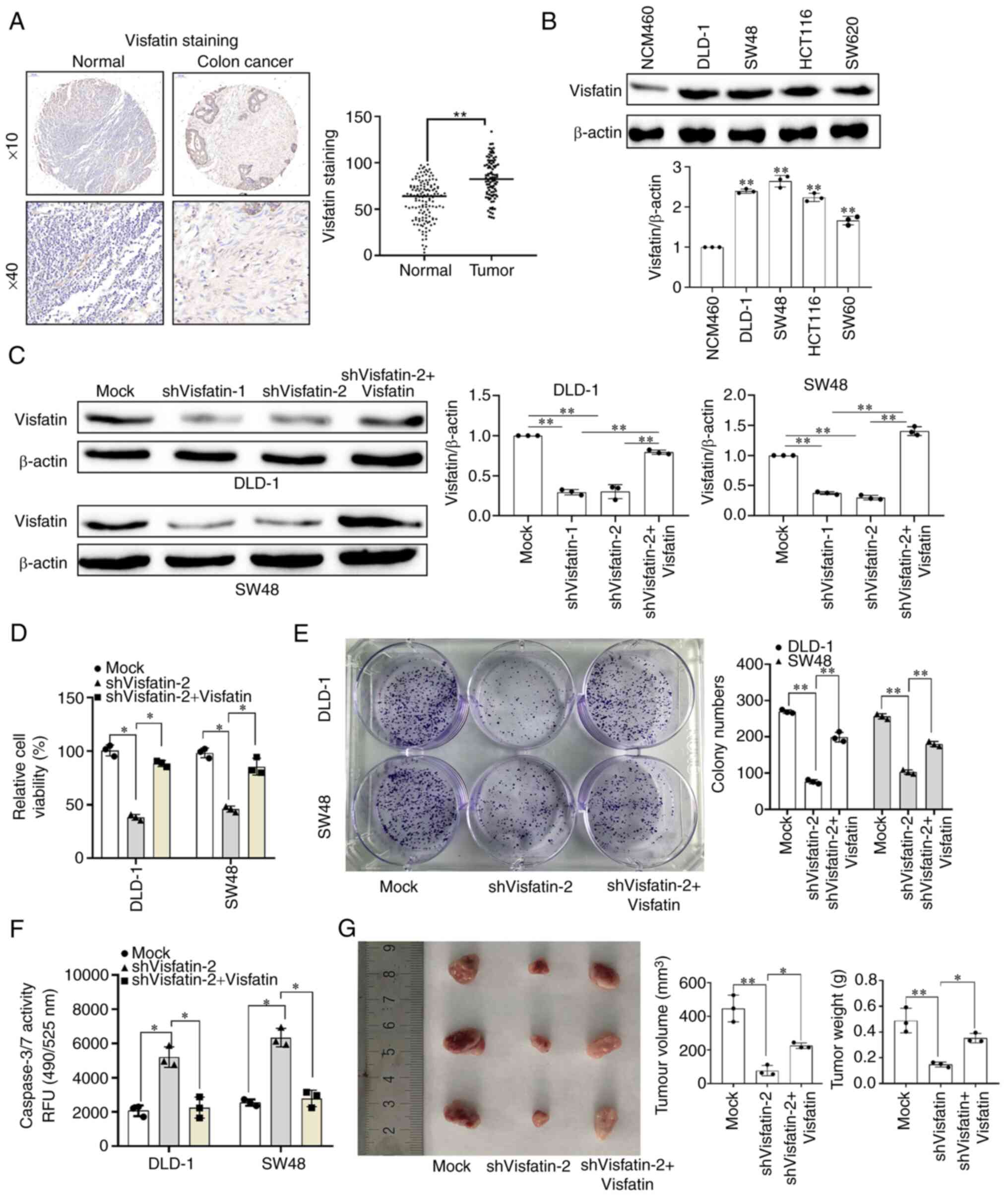
The TMA results revealed that visfatin was highly
expressed in colon cancer tissues (Fig. 1A). The evaluation of visfatin
expression in various colon cancer cell lines in vitro
revealed that visfatin was highly expressed in colon cancer cells
(Fig. 1B). Among these, the DLD-1
and SW48 cells exhibited the highest protein expression levels;
hence, they were selected for use in subsequent experiments.
Following transfection with shRNA1 and shRNA 2, visfatin protein
expression was significantly downregulated; however, upon
transfection with the overexpression vector, its expression was
significantly upregulated (Figs.
1C and S1). shRNA2 was
selected for subsequent studies based on the rotational efficiency
and stability. Following visfatin knockdown, the activity and
proliferative ability of the DLD-1 and SW48 cells were
significantly decreased; however, this decrease was reversed on the
overexpression of visfatin (Fig. 1D
and E). Apoptosis-related protein detection (caspase-3/7) also
revealed that visfatin knockdown promoted colon cancer cell
apoptosis, whereas visfatin overexpression inhibited it (Fig. 1F). Following the construction of a
colon cancer mouse model in which mice were injected with cells in
which visfatin expression was knocked down or overexpressed, the
tumour volume and weight were significantly reduced in the group
injected with the cells in which visfatin expression was knocked
down compared with the mock group. However, following visfatin
overexpression, the tumour volume and weight increased (Fig. 1G).
Visfatin regulates the sensitivity of
DLD-1 and SW48 cells to 5-FU
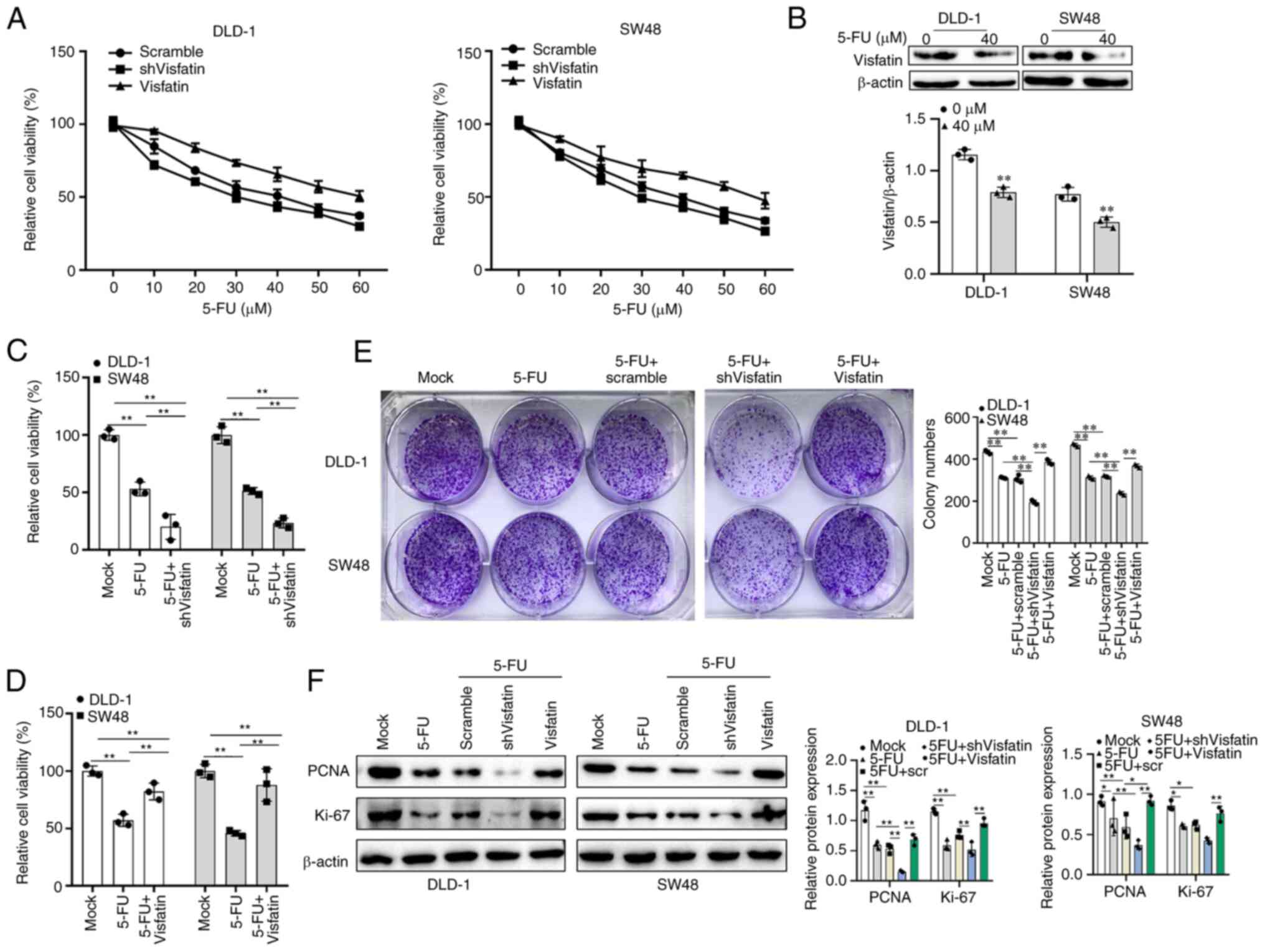
The DLD-1 and SW48 cells were treated with a
gradient of 5-FU and cell viability was detected. The concentration
of 40 µM was selected for subsequent 5-FU treatment
(Fig. 2A). Following treatment
with 40 µM 5-FU, visfatin protein expression in the DLD-1
and SW48 cells was significantly decreased (Fig. 2B). Following the knockdown of
visfatin in colon cancer cells treated with 5-FU, cell viability
was significantly decreased. However, the overexpression of
visfatin, cell viability was significantly increased compared with
that of the cells treated with 5-FU alone (Fig. 2C and D). The colony formation
assay demonstrated the proliferative ability of the colon cancer
cells. Following visfatin knockdown, the staining of the
5-FU-treated cells was significantly reduced, whereas visfatin
overexpression reversed this effect (Fig. 2E). Similar results were observed
for the other two tumour cell proliferation protein markers, PCNA
and Ki-67. Therefore, the knockdown of visfatin decreased the
expression of PCNA and Ki-67, while the overexpression of visfatin
increased their expression (Fig.
2F).
Visfatin inhibits the 5-FU-induced
apoptosis of DLD-1 and SW48 cells
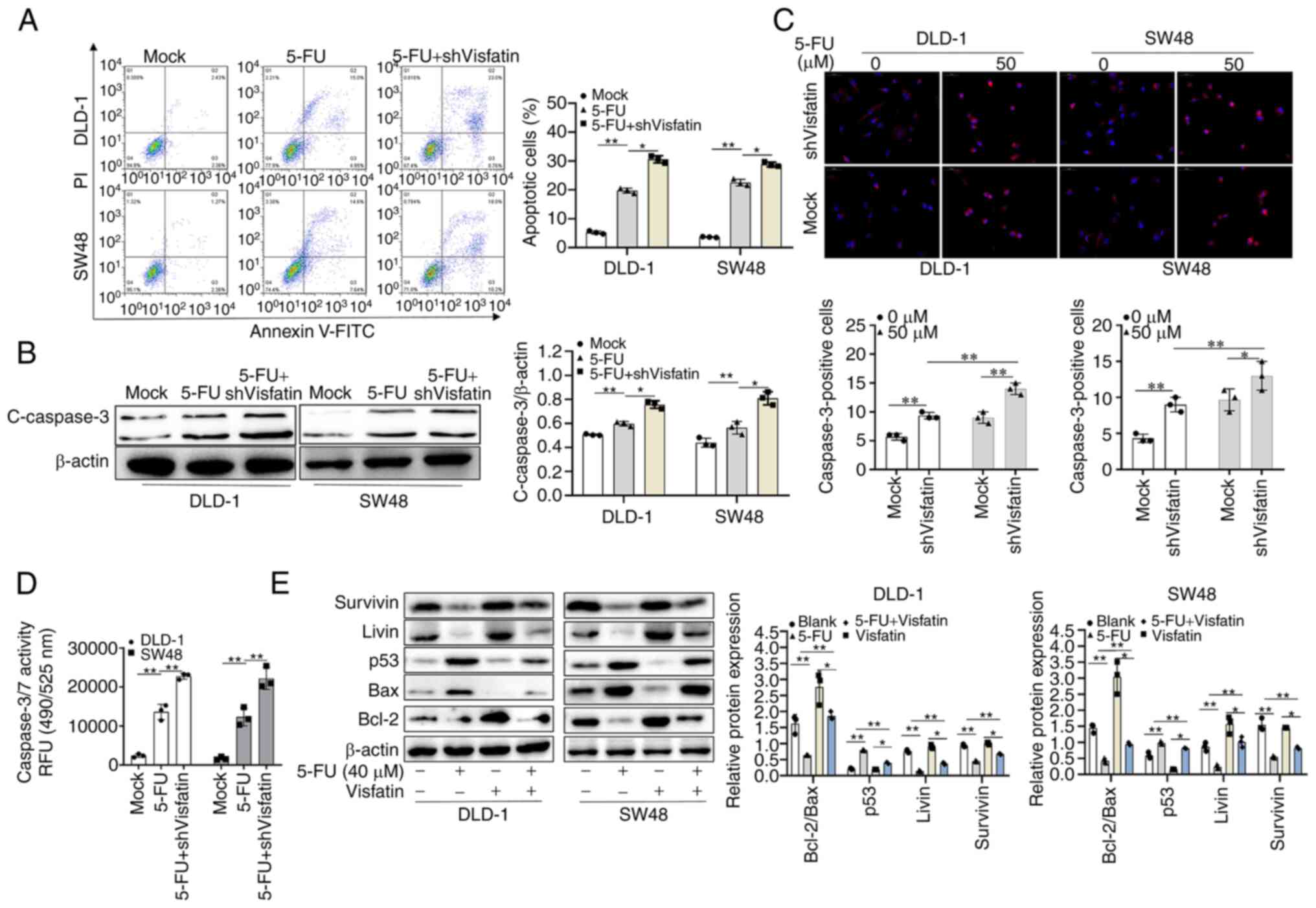
The evaluation of apoptosis using flow cytometry
revealed that visfatin knockdown increased the apoptotic cell
numbers following treatment with 5-FU (Fig. 3A). In addition, the knockdown of
visfatin significantly increased the expression of cleaved
caspase-3 following treatment with 5-FU (Fig. 3B). However, following the
overexpression of visfatin and treatment with 5-FU, cell apoptosis
was decreased and the expression of cleaved caspase-3 did not
increase (Fig. S2A and B). The
expression of cleaved caspase-3 in DLD-1 and SW48 cells treated
with 5-FU was examined using immunofluorescence staining. The
results revealed that following visfatin knockdown, the
fluorescence intensity increased (Fig. 3C); however, this decreased upon
visfatin overexpression (Fig.
S2C). 5-FU treatment increased the activity of caspase-3/7 in
colon cancer cells, with visfatin knockdown significantly
increasing this effect (Fig. 3D).
However, visfatin overexpression suppressed this effect (Fig. S2D). Additionally, the other
apoptotic proteins tested revealed the same results. In the colon
cancer cells treated with 5-FU, the expression levels of Bcl-2/Bax,
Livin and survivin were significantly reduced, whereas those of the
pro-apoptotic p53 protein were significantly increased. Following
the overexpression of visfatin and treatment with 5-FU, the effects
of 5-FU on these pro-apoptotic proteins were attenuated (Fig. 3E).
Positive correlation between visfatin and
SDF-1
The suppressive effects of visfatin on the
sensitivity of the colon cancer cells to 5-FU were confirmed.
However, the underlying mechanisms required further investigation.
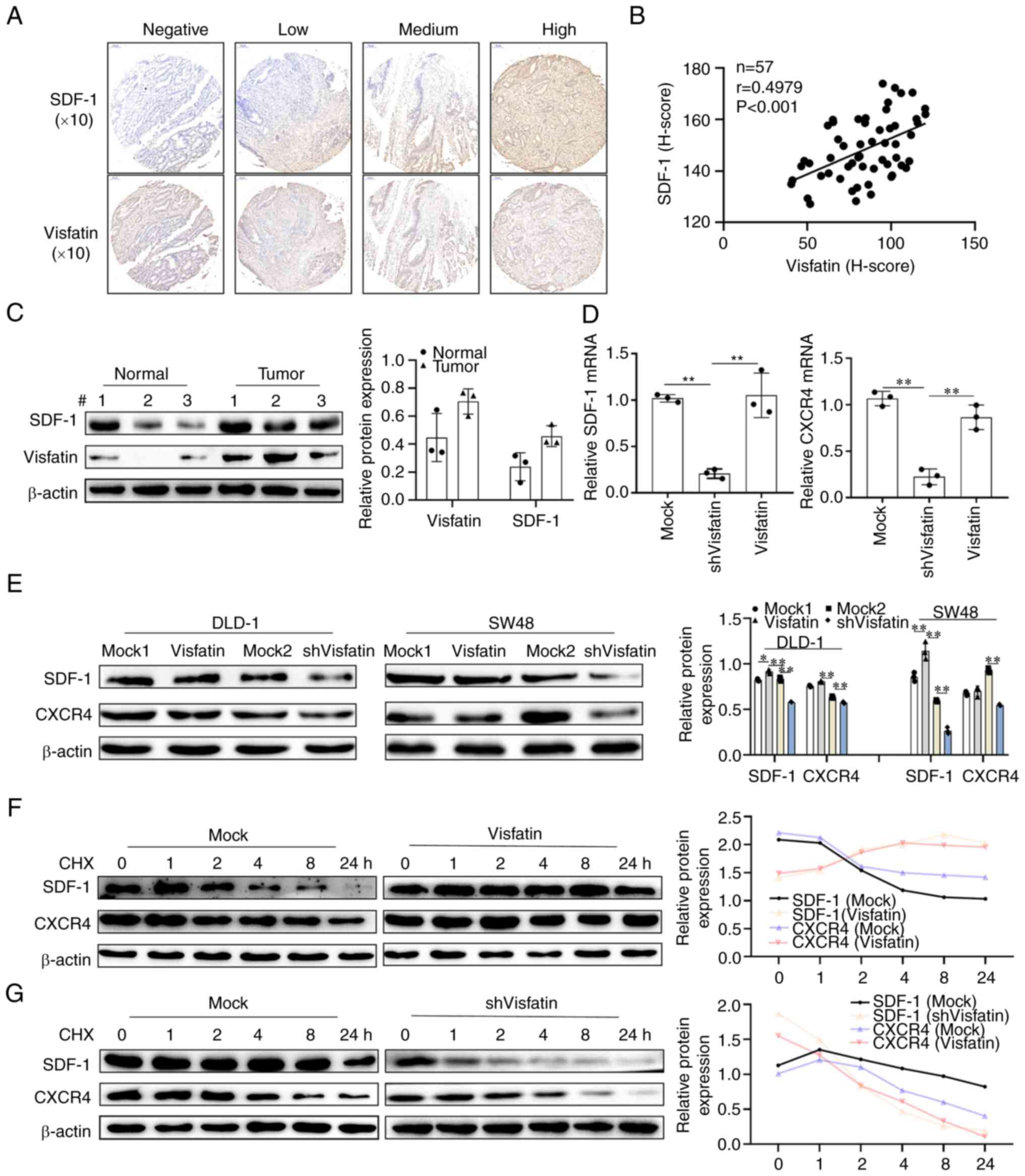
The TMA results revealed that when visfatin exhibited negative
staining, SDF-1 staining was also negative. The positive degree of
SDF-1 staining increased with the increase in the positive degree
of visfatin staining. In addition, the correlation analysis of the
two H-scores revealed that visfatin and SDF-1 positively correlated
(Fig. 4A and B). Both SDF-1 and
visfatin were found to be highly expressed in tumour tissues
compared to normal tissues (Fig.
4C). As the receptor of SDF-1, the mRNA and protein expression
of CXCR4 is the same as that of SDF-1; this was found to decrease
with visfatin knockdown (Fig. 4D and
E). Visfatin also affected the stability of SDF-1 and CXCR4.
Following treatment of the colon cancer cells with CHX for
different periods of time, the SDF-1 and CXCR4 levels gradually
decreased; however, visfatin overexpression reversed the
instability of SDF-1 and CXCR4. Visfatin knockdown further
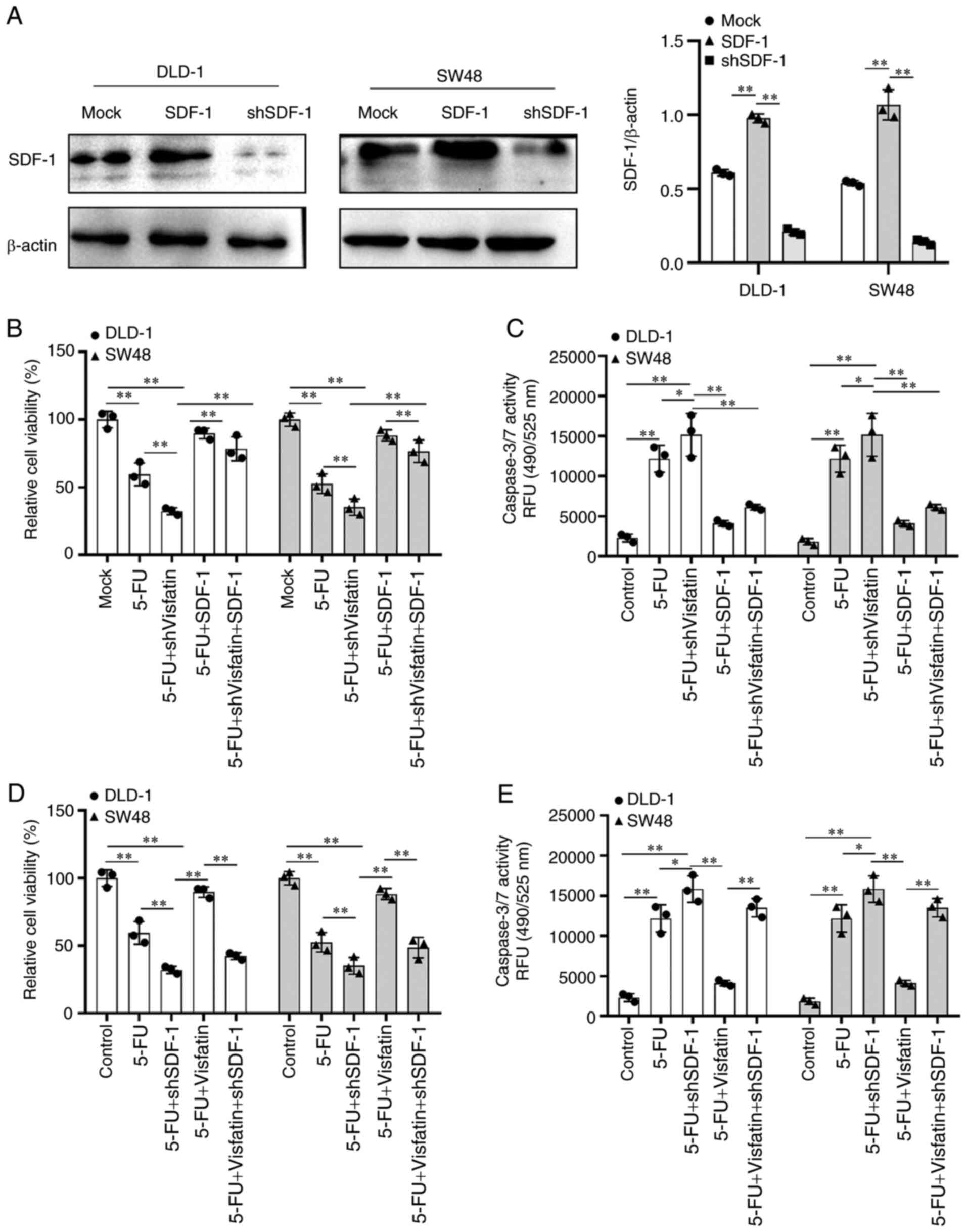
decreased the SDF-1 and CXCR4 levels (Fig. 4F and G).
Visfatin inhibits the sensitivity of
DLD-1 and SW48 cells to 5-FU through SDF-1
Further experiments were conducted to verify the
role of SDF-1 in the regulatory effects of visfatin on the
sensitivity of the colon cancer cells to 5-FU. First, SDF-1 was
overexpressed in the DLD-1 and SW48 cells, and the transfection
efficiency was examined (Fig.
5A). Upon SDF-1 overexpression in colon cancer cells treated
with 5-FU or 5-FU and subjected to visfatin knockdown, cell
viability was significantly increased, indicating that SDF-1
overexpression inhibited the suppressive effects of 5-FU and
visfatin knockdown on cell viability (Fig. 5B). Second, apoptosis-related
protein caspase-3/7 expression was increased following treatment
with 5-FU or 5-FU treatment plus visfatin knockdown, whereas SDF-1
overexpression significantly decreased caspase-3/7 expression
(Fig. 5C). Upon SDF-1 knockdown
in colon cancer cells treated with 5-FU or in those treated with
5-FU and subjected to visfatin overexpression, cell viability was
significantly decreased, indicating that SDF-1 knockdown promoted
the inhibitory effects of 5-FU on cell viability even in the cells
overexpressing visfatin (Fig.
5D). Apoptosis-related protein caspase-3/7 expression was
increased following 5-FU treatment and decreased following 5-FU
treatment and visfatin overexpression, whereas SDF-1 knockdown
significantly increased the apoptosis even of the cells treated
with 5-FU and overexpressing visfatin (Fig. 5E), confirming that SDF-1 is an
intermediate molecule in the regulatory effects of visfatin on
sensitivity to 5-FU.
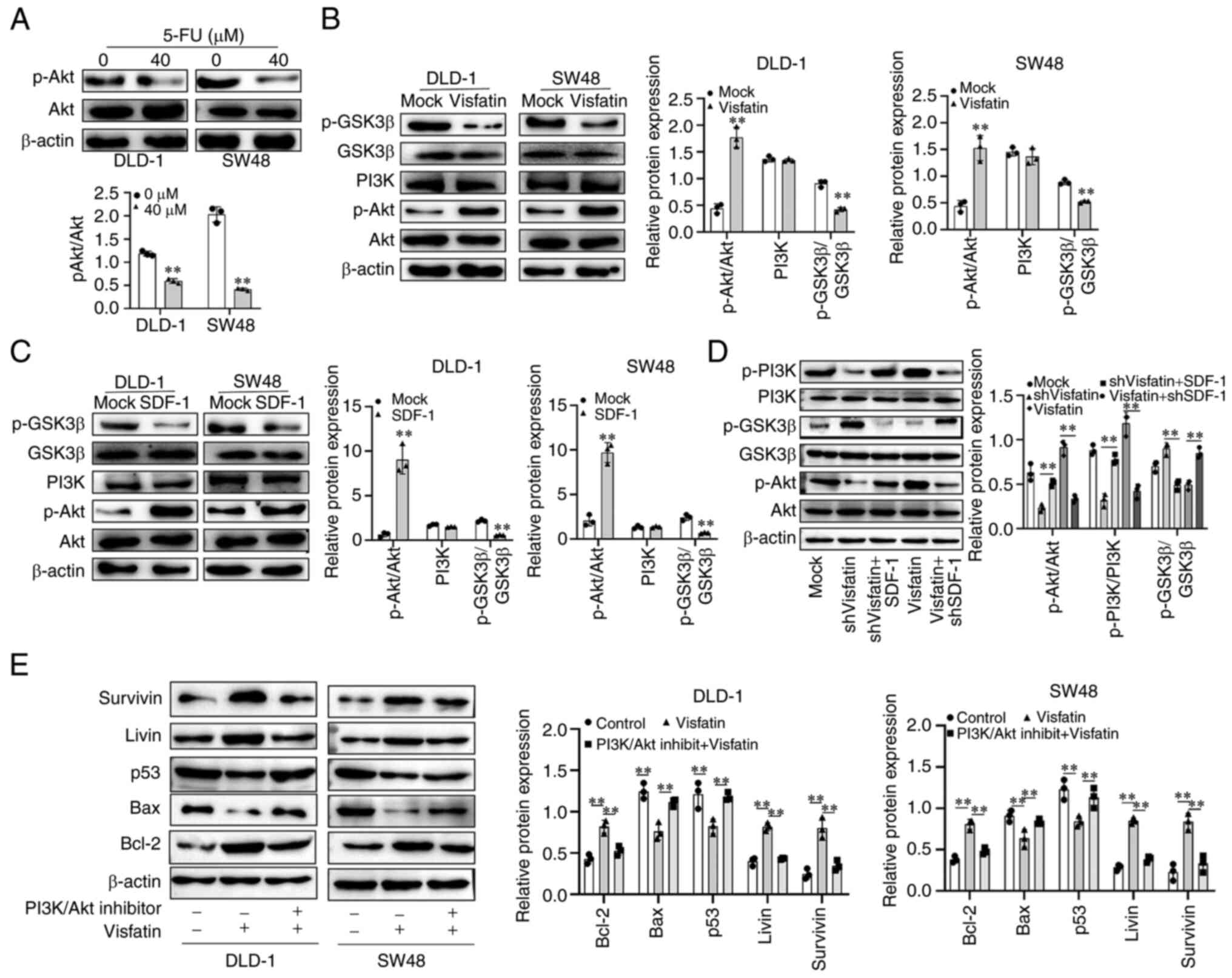
Akt functions as a common target of
visfatin and SDF-1
Furthermore, the regulatory mechanisms of the
SDF-1/CXCR4 axis in the effects of visfatin on sensitivity to 5-FU
were examined. Following treatment of the DLD-1 and SW48 cells with
5-FU, the activity of Akt decreased (Fig. 6A), with was consistent with the
decrease in the expression of visfatin (Fig. 2B). Upon visfatin overexpression in
the colon cancer cells, Akt phosphorylation increased and GSK3β
phosphorylation decreased (Fig.
6B). The results observed following the overexpression of SDF-1
were similar to those observed following the overexpression of
visfatin, wherein the Akt pathway was activated (Fig. 6C). Following the knockdown of
visfatin, the phosphorylation of Akt and PI3K decreased, whereas
that of GSK3β increased. However, upon SDF-1 overexpression, this
effect was attenuated. The overexpression of visfatin increased the
phosphorylation of Akt and PI3K, and decreased the phosphorylation
of GSK3β, and this effect was attenuated by the knockdown of SDF-1
(Fig. 6D). The overexpression of
visfatin increased the expression of Bcl-2, Livin and survivin,
whereas it decreased the expression of the pro-apoptotic proteins,
Bax and p53, in the DLD-1 and SW48 cells. Upon visfatin
overexpression and simultaneous treatment with PI3K/Akt pathway
inhibitor (LY294002, 10 µmol/l), this effect was reversed,
and an increase in cell apoptosis was observed (Fig. 6E).
Discussion
In the present study, the knockdown of visfatin
inhibited colon cancer cell viability and proliferation, promoted
apoptosis and inhibited tumour formation in vivo. The
sensitivity of colon cancer cells to 5-FU was promoted by visfatin
knockdown and inhibited by its overexpression. Furthermore, these
effects were attenuated by the overexpression of SDF-1 and PI3K/Akt
inhibitors. Thus, the present study confirms the modulating effects
of the visfatin/SDF-1/Akt axis on sensitivity to 5-FU.
Globally, the incidence and mortality rates
associated with CRC are high compared to other cancer types
(26). Tumour recurrence or
distant metastasis following surgery, with various other consequent
severe complications, is the main cause of mortality of patients
with CRC. Therefore, radical surgery combined with chemotherapy is
the conventional form of treatment for colon cancer. 5-FU is among
the most important and commonly used drugs for treating patients
with advanced colon cancer. However, the effectiveness of 5-FU in
the treatment of advanced colon cancer varies among individuals,
with resistance to 5-FU chemotherapy contributing to treatment
failure (27,28). The mechanisms of resistance to
5-FU chemotherapy remain unclear (29); however, two main theories exist:
i) Genetic and epigenetic alterations, such as antiapoptotic
protein Bcl-2 and Bcl-xL overexpression; and ii) ATP-binding
cassette transporter overexpression, leading to drug efflux that
decreases intracellular drug accumulation (30). The mechanism of resistance to 5-FU
is also related to the increased expression of nucleoside
metabolizing enzymes, p53 gene mutation or loss of function and the
imbalance of the mismatch repair system (31-33). Strategies against resistance to
5-FU, including gene therapy, drug combination and drug
repurposing, are currently being studied in-depth. miR-122, which
is significantly downregulated in 5-FU-resistant CRC cells, has the
potential to restore chemosensitivity by targeting pyruvate kinase
type M2 (34). Additionally,
metformin has been reported to downregulate the DNA replication
machinery proteins (MCM2 and PCNA) selectively in 5-FU-resistant
colon cancer cell lines, leading to a synergistic effect with
chemotherapy (35). The
combination of celecoxib and 5-FU chemotherapy has been shown to
induce significant therapeutic responses in chemo-refractory CRC
cells and overcome resistance in both in vivo and in
vitro colorectal cancer cells (36). Visfatin promotes resistance to
5-FU by affecting the proliferation and apoptosis of tumour cells
via the SDF-1/CXCR4/Akt axis. As demonstrated herein, after the
blocking of PI3K/AKT, p53 expression increased and 5-FU resistance
was weakened. The present study confirmed that visfatin has the
potential to enhance the sensitivity to chemotherapeutic drugs,
providing a novel foundation to combat drug resistance using drug
combinations.
Chemokines affect tumour and endothelial cells in
the tumour microenvironment, regulating tumour cell growth,
proliferation and invasion, significantly impacting cancer
treatment outcomes (37). SDF-1
is a member of the CXC subfamily of chemokines, and high levels of
SDF-1 have been shown to be associated with CRC tumour progression;
therefore, SDF-1 is considered a key negative prognostic marker in
CRC (38,39). Visfatin is involved in tumour
progression, similar to other adipokines. It has been confirmed
that visfatin promotes the downstream regulator, silent information
regulator 1 (SIRT1), by affecting the activity of nicotinamide
adenine dinucleotide and the proliferation and apoptosis of
prostate cancer cells (40).
Furthermore, visfatin activates STAT3 by inducing the secretion of
the inflammatory factor IL-6, thereby promoting the proliferation
and metastasis of breast cancer (41). In colon cancer, visfatin mRNA and
protein are highly expressed in colon cancer tissues and are
related to distant metastasis, anaemia and poor prognosis (42). Visfatin has also been found to
downregulate E-cadherin and upregulate N-cadherin and transcription
factor 1 (Snail-1) expression in a time- and
concentration-dependent manner, while Snail-1 inhibitors have been
shown to weaken the effect of visfatin on epithelial-mesenchymal
transition markers (43).
Additionally, visfatin participates in mediating cancer stem
pathways in CRC tumours through the downstream effectors, SIRT1 and
PARP1. Moreover, NAMPT inhibition increases the sensitivity to
apoptosis in both NAMPT-expressing cells and tumour spheres
(8). NAMPT affects the
proliferation of colon cancer cells by inhibiting Axin and
activating the Wnt pathway (44).
In 5-FU-resistant colon cancer cell lines, the Wnt pathway is
activated, thereby inhibiting the p53 pathway and enhancing cell
survival. Consistent with the findings of previous studies, the
present study confirmed the importance of NAMPT the resistance of
CRC to 5-FU (45). The present
study further demonstrated that visfatin promoted tumorigenesis and
inhibited sensitivity to 5-FU via the SDF-1/CXCR4 axis.
SDF-1/CXCR4 signal transduction is a complex process
influenced by a number of factors (such as signal network system).
It activates various signalling pathways to affect tumour
proliferation, migration and prognosis. PI3K/Akt and extracellular
signalling-regulated protein kinases 1 and 2 (ERK 1/2) are two key
signalling pathways involved in cell signal transduction,
colorectal cancer cell survival and 5-FU resistance (45,46). CSCs initiate heterogeneous tumours
and repopulate metastatic outgrowths (47). A previous clinical study found
that: SDF-1 and CXCR4 expression was significantly elevated in CRC
tissues and patients with CXCR4 nuclear-type expression exhibited
more frequent lymph node metastasis. The data of the present study
suggest that there is a significant association between SDF-1/CXCR4
to enhance the liver metastases causing poor prognosis in CRC
(48). The high expression of
SDF-1/CXCR4 in CD44+/CD133+ prostate CSCs
indicates that a potential function of the CXCR12/CXCL4 axis that
promotes prostate cancer metastasis could be to maintain the
stemness of CSCs (49). This axis
promotes tumour metastasis mediated by actin polymerisation and
prosthetic foot formation, playing a key role in CSC's self-renewal
in drug-resistant NSCLC (50).
The present study further suggests that visfatin is closely related
to SDF-1 and is involved in affecting the sensitivity of colon
cancer cells to 5-FU. In the present study, SDF-1 exhibited a
positive correlation with visfatin in colon cancer tissue samples.
Furthermore, the present study confirmed that the SDF-1/CXCR4 axis
functioned as a downstream module of visfatin and affected the
PI3K/Akt signalling pathway, which is involved in regulating the
sensitivity of colon cancer cells to 5-FU.
Owing to time and funding constraints, a complete
in vivo experimental verification could not be conducted.
Another limitation of the present study is that only the effects of
visfatin on the sensitivity of colon cancer cells to 5-FU were
investigated, disregarding other chemotherapeutic drugs. Therefore,
further research is required to elucidate the mechanisms involved
in the effects of visfatin on sensitivity to chemotherapeutics.
In conclusion, the present study demonstrated that
visfatin inhibited the chemotherapeutic effects of 5-FU on colon
cancer cells via the visfatin/SDF-1/Akt axis. Therefore, the
application of 5-FU combined with visfatin inhibition may provide a
novel strategy which may be used to reduce the resistance of colon
cancer to chemotherapeutic drugs.
Supplementary Data
Availability of data and materials
All data generated or analysed during this study are
included in this published article or in the accompanying
supplementary material.
Authors' contributions
KG and JZ contributed to the study conception and
design. Material preparation, data collection and analysis were
performed by QZ and YaxinL, who contributed equally to the study.
The initial draft of the manuscript was written by QZ and YaxinL.
WC, YH, JL, YuejinL, XL, XG, YuL and GL were responsible for data
visualisation and the literature review. KG and JZ contributed to
the critical revision of the manuscript. QZ and JZ confirmed the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First People's Hospital of Yunnan Province in
accordance with the International Ethical Guidelines for Biomedical
Research Involving Human Subjects (CIOMS). The experimental
procedure was approved by the Animal Protection and Use Agency
Committee of the Kunming University of Science and Technology and
conducted in accordance with the Laboratory animal Guideline for
ethical review of animal welfare (GB/T 35892-2018).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Health and Science
Foundation of Yunnan Province (grant no. 2017NS208), the Open
Project of Vascular Disease Clinical Medical Center of Yunnan First
People's Hospital in 2021 (grant no. 2021LCZXXF-XG01), and the
Special Fund for Clinical Research of Wu Jieping Medical Foundation
(grant no. 320.6750.18403).
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar
|
|
3
|
Shi Q, Paul J and Grothey A: Duration of
adjuvant chemotherapy for stage III colon cancer. N Engl J Med.
379:396–397. 2018.
|
|
4
|
Dienstmann R, Vermeulen L, Guinney J,
Kopetz S, Tejpar S and Tabernero J: Consensus molecular subtypes
and the evolution of precision medicine in colorectal cancer. Nat
Rev Cancer. 17:79–92. 2017. View Article : Google Scholar
|
|
5
|
Labianca R, Beretta GD, Kildani B, Milesi
L, Merlin F, Mosconi S, Pessi MA, Prochilo T, Quadri A, Gatta G, et
al: Colon cancer. Crit Rev Oncol Hematol. 74:106–133. 2010.
View Article : Google Scholar
|
|
6
|
Vodenkova S, Buchler T, Cervena K,
Veskrnova V, Vodicka P and Vymetalkova V: 5-fluorouracil and other
fluoropyrimidines in colorectal cancer: Past, present and future.
Pharmacol Ther. 206:1074472020. View Article : Google Scholar
|
|
7
|
Marin JJG, Macias RIR, Monte MJ, Her raez
E, Peleteiro-Vigil A, Blas BS, Sanchon-Sanchez P, Temprano AG,
Espinosa-Escudero RA, Lozano E, et al: Cellular mechanisms
accounting for the refractoriness of colorectal carcinoma to
pharmacological treatment. Cancers (Basel). 12:26052020. View Article : Google Scholar
|
|
8
|
Lucena-Cacace A, Otero-Albiol D,
Jiménez-García MP, Muñoz-Galvan S and Carnero A: NAMPT is a potent
oncogene in colon cancer progression that modulates cancer stem
cell properties and resistance to therapy through Sirt1 and PARP.
Clin Cancer Res. 24:1202–1215. 2018. View Article : Google Scholar
|
|
9
|
Fukuhara A, Matsuda M, Nishizawa M, Segawa
K, Tanaka M, Kishimoto K, Matsuki Y, Murakami M, Ichisaka T,
Murakami H, et al: Visfatin: A protein secreted by visceral fat
that mimics the effects of insulin. Science. 307:426–430. 2005.
View Article : Google Scholar
|
|
10
|
Hufton SE, Moerkerk PT, Brandwijk R, de
Bruïne AP, Arends JW and Hoogenboom HR: A profile of differentially
expressed genes in primary colorectal cancer using suppression
subtractive hybridization. FEBS Lett. 463:77–82. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nakajima TE, Yamada Y, Hamano T, Furuta K,
Matsuda T, Fujita S, Kato K, Hamaguchi T and Shimada Y:
Adipocytokines as new promising markers of colorectal tumors:
Adiponectin for colorectal adenoma, and resistin and visfatin for
colorectal cancer. Cancer Sci. 101:1286–1291. 2010. View Article : Google Scholar
|
|
12
|
Lucena-Cacace A, Otero-Albiol D,
Jiménez-García MP, Peinado-Serrano J and Carnero A: NAMPT
overexpression induces cancer stemness and defines a novel tumor
signature for glioma prognosis. Oncotarget. 8:99514–99530. 2017.
View Article : Google Scholar
|
|
13
|
Chen M, Wang Y, Li Y, Zhao L, Ye S, Wang
S, Yu C and Xie H: Association of plasma visfatin with risk of
colorectal cancer: An observational study of Chinese patients. Asia
Pac J Clin Oncol. 12:e65–74. 2016. View Article : Google Scholar
|
|
14
|
Lucas S, Soave C, Nabil G, Ahmed ZSO, Chen
G, El-Banna HA, Dou QP and Wang J: Pharmacological inhibitors of
NAD biosynthesis as potential an ticancer agents. Recent Pat
Anticancer Drug Discov. 12:190–207. 2017. View Article : Google Scholar
|
|
15
|
Nacarelli T, Fukumoto T, Zundell JA,
Fatkhutdinov N, Jean S, Cadungog MG, Borowsky ME and Zhang R: NAMPT
inhibition suppresses cancer stem-like cells associated with
therapy-induced senescence in ovarian cancer. Cancer Res.
80:890–900. 2020. View Article : Google Scholar
|
|
16
|
Cao Z, Liang N, Yang H and Li S: Visfatin
mediates doxorubicin resistance in human non-small-cell lung cancer
via Akt-mediated up-regulation of ABCC1. Cell Prolif.
50:e123662017. View Article : Google Scholar
|
|
17
|
Ge X, Zhao Y, Dong L, Seng J, Zhang X and
Dou D: NAMPT regulates PKM2 nuclear location through 14-3-3ζ:
Conferring resistance to tamoxifen in breast cancer. J Cell
Physiol. 234:23409–23420. 2019. View Article : Google Scholar
|
|
18
|
Bi TQ and Che XM: Nampt/PBEF/visfatin and
cancer. Cancer Biol Ther. 10:119–125. 2010. View Article : Google Scholar
|
|
19
|
Li XQ, Lei J, Mao LH, Wang QL, Xu F, Ran
T, Zhou ZH and He S: NAMPT and NAPRT, key enzymes in NAD salvage
synthesis pathway, are of negative prognostic value in colorectal
cancer. Front Oncol. 9:7362019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma J, Sun X and Wang Y, Chen B, Qian L and
Wang Y: Fibroblast-derived CXCL12 regulates PTEN expression and is
associated with the proliferation and invasion of colon cancer
cells via PI3k/Akt signaling. Cell Commun Signal. 17:1192019.
View Article : Google Scholar
|
|
21
|
Ma J, Su H, Yu B, Guo T, Gong Z, Qi J,
Zhao X and Du J: CXCL12 gene silencing down-regulates metastatic
potential via blockage of MAPK/PI3K/AP-1 signaling pathway in colon
cancer. Clin Transl Oncol. 20:1035–1045. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Conley-LaComb MK, Saliganan A, Kandagatla
P, Chen YQ, Cher ML and Chinni SR: PTEN loss mediated Akt
activation promotes prostate tumor growth and metastasis via
CXCL12/CXCR4 signaling. Mol Cancer. 12:852013. View Article : Google Scholar
|
|
23
|
Zhao Q, Li JY, Zhang J, Long YX, Li YJ,
Guo XD, Wei MN and Liu WJ: Role of visfatin in promoting
proliferation and invasion of colorectal cancer cells by
downregulating SDF-1/CXCR4-mediated miR-140-3p expression. Eur Rev
Med Pharmacol Sci. 24:5367–5377. 2020.
|
|
24
|
Azim HA Jr, Peccatori FA, Brohée S,
Branstetter D, Loi S, Viale G, Piccart M, Dougall WC, Pruneri G and
Sotiriou C: RANK-ligand (RANKL) expression in young breast cancer
patients and during pregnancy. Breast Cancer Res. 17:242015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar
|
|
27
|
Wang W, Guo W, Li L, Fu Z, Liu W, Gao J,
Shu Y, Xu Q, Sun Y and Gu Y: Andrographolide reversed 5-FU
resistance in human colorectal cancer by elevating BAX expression.
Biochem Pharmacol. 121:8–17. 2016. View Article : Google Scholar
|
|
28
|
Marjaneh RM, Khazaei M, Ferns GA, Avan A
and Aghaee-Bakhtiari SH: The role of microRNAs in 5-FU resistance
of colorectal cancer: Possible mechanisms. J Cell Physiol.
234:2306–2316. 2019. View Article : Google Scholar
|
|
29
|
Wei L, Wang X, Lv L, Zheng Y, Zhang N and
Yang M: The emerging role of noncoding RNAs in colorectal cancer
chemoresistance. Cell Oncol (Dordr). 42:757–768. 2019. View Article : Google Scholar
|
|
30
|
Hu T, Li Z, Gao CY and Cho CH: Mechanisms
of drug resistance in colon cancer and its therapeutic strategies.
World J Gastroenterol. 22:6876–6889. 2016. View Article : Google Scholar
|
|
31
|
van Staveren MC, Opdam F, Guchelaar HJ,
van Kuilenburg AB, Maring JG and Gelderblom H: Influence of
metastatic disease on the usefulness of uracil pharmacokinetics as
a screening tool for DPD activity in colorectal cancer patients.
Cancer Chemother Pharmacol. 76:47–52. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang W, Cassidy J, O'Brien V, Ryan KM and
Collie-Duguid E: Mechanistic and predictive profiling of
5-Fluorouracil resistance in human cancer cells. Cancer Res.
64:8167–8176. 2004. View Article : Google Scholar
|
|
33
|
Leckband S, Kelsoe J, Dunnenberger H,
George AL Jr, Tran E, Berger R, Müller DJ, Whirl-Carrillo M, Caudle
KE and Pirmohamed M; Clinical Pharmacogenetics Implementation
Consortiu: Clinical pharmacogenetics implementation consortium
guidelines for HLA-B genotype and carbamazepine dosing. Clin
Pharmacol Ther. 94:324–328. 2013. View Article : Google Scholar
|
|
34
|
van Niekerk G and Engelbrecht AM: Role of
PKM2 in directing the metabolic fate of glucose in cancer: A
potential therapeutic target. Cell Oncol (Dordr). 41:343–351. 2018.
View Article : Google Scholar
|
|
35
|
Kim SH, Kim SC and Ku JL: Metformin
increases chemo-sensitivity via gene downregulation encoding DNA
replication proteins in 5-Fu resistant colorectal cancer cells.
Oncotarget. 8:56546–56557. 2017. View Article : Google Scholar :
|
|
36
|
Fong W and To KKW: Drug repurposing to
overcome resistance to various therapies for colorectal cancer.
Cell Mol Life Sci. 76:3383–3406. 2019. View Article : Google Scholar
|
|
37
|
Nagarsheth N, Wicha MS and Zou W:
Chemokines in the cancer microenvironment and their relevance in
cancer immunotherapy. Nat Rev Immunol. 17:559–572. 2017. View Article : Google Scholar
|
|
38
|
Yoshitake N, Fukui H, Yamagishi H,
Sekikawa A, Fujii S, Tomita S, Ichikawa K, Imura J, Hiraishi H and
Fujimori T: Expression of SDF-1 alpha and nuclear CXCR4 predicts
lymph node metastasis in colorectal cancer. Br J Cancer.
98:1682–1689. 2008. View Article : Google Scholar
|
|
39
|
Akishima-Fukasawa Y, Nakanishi Y, Ino Y,
Moriya Y, Kanai Y and Hirohashi S: Prognostic significance of
CXCL12 expression in patients with colorectal carcinoma. Am J Clin
Pathol. 132:202–210; quiz 307. 2009. View Article : Google Scholar
|
|
40
|
Wang B, Hasan MK, Alvarado E, Yuan H, Wu H
and Chen WY: NAMPT overexpression in prostate cancer and its
contribution to tumor cell survival and stress response. Oncogene.
30:907–921. 2011. View Article : Google Scholar
|
|
41
|
Hung AC, Lo S, Hou MF, Lee YC, Tsai CH,
Chen YY, Liu W, Su YH, Lo YH, Wang CH, et al: Extracellular
visfatin-promoted malignant behavior in breast cancer is mediated
through c-Abl and STAT3 activation. Clin Cancer Res. 22:4478–4490.
2016. View Article : Google Scholar
|
|
42
|
Yang J, Zhang K, Song H, Wu M, Li J, Yong
Z, Jiang S, Kuang X and Zhang T: Visfatin is involved in promotion
of colorectal carcinoma malignancy through an inducing EMT
mechanism. Oncotarget. 7:32306–32317. 2016. View Article : Google Scholar
|
|
43
|
Cheng G, Liu C, Sun X, Zhang L, Liu L,
Ouyang J and Li B: Visfatin promotes osteosarcoma cell migration
and invasion via induction of epithelial-mesenchymal transition.
Oncol Rep. 34:987–994. 2015. View Article : Google Scholar
|
|
44
|
Ye C, Qi L, Li X, Wang J, Yu J, Zhou B,
Guo C, Chen J and Zheng S: Targeting the NAD+ salvage
pathway suppresses APC mutation-driven colorectal cancer growth and
Wnt/β-catenin signaling via increasing Axin level. Cell Commun
Signal. 18:162020. View Article : Google Scholar
|
|
45
|
Ding D, Zhang P, Liu Y and Wang Y, Sun W,
Yu Z, Cheng Z and Wang Y: Runx2 was correlated with neurite
outgrowth and schwann cell differentiation, migration after sciatic
nerve crush. Neurochem Res. 43:2423–2434. 2018. View Article : Google Scholar
|
|
46
|
Yu JS and Cui W: Proliferation, survival
and metabolism: The role of PI3K/AKT/mTOR signalling in
pluripotency and cell fate determination. Development.
143:3050–3060. 2016. View Article : Google Scholar
|
|
47
|
Celià-Terrassa T and Jolly MK: Cancer stem
cells and epithelial-to-mesenchymal transition in cancer
metastasis. Cold Spring Harb Perspect Med. 10:a0369052020.
View Article : Google Scholar
|
|
48
|
Amara S, Chaar I, Khiari M, Ounissi D,
Weslati M, Boughriba R, Hmida AB and Bouraoui S: Stromal cell
derived factor-1 and CXCR4 expression in colorectal cancer promote
liver metastasis. Cancer Biomark. 15:869–879. 2015. View Article : Google Scholar
|
|
49
|
Jung Y, Cackowski FC, Yumoto K, Decker AM,
Wang J, Kim JK, Lee E, Wang Y, Chung JS, Gursky AM, et al: CXCL12γ
promotes metastatic castration-resistant prostate cancer by
inducing cancer stem cell and neuroendocrine phenotypes. Cancer
Res. 78:2026–2039. 2018. View Article : Google Scholar
|
|
50
|
Jung MJ, Rho JK, Kim YM, Jung JE, Jin YB,
Ko YG, Lee JS, Lee SJ, Lee JC and Park MJ: Upregulation of CXCR4 is
functionally crucial for maintenance of stemness in drug-resistant
non-small cell lung cancer cells. Oncogene. 32:209–221. 2013.
View Article : Google Scholar
|