Introduction
Drug repositioning is a strategy for repurposing
approved or investigational drugs outside the scope of their
original medical indication. Memantine is a promising agent for
cancer therapy (1–3). Memantine, an antagonist of the
N-methyl-D-aspartate (NMDA) receptor, exerts beneficial
effects and is widely used in the treatment of Alzheimer's disease.
Memantine acts on the glutamatergic system by blocking NMDA
receptors and inhibiting glutamate overstimulation (4). Of note, NMDA receptors have been found
in several types of cancer, such as glioma (5), medulloblastoma, neuroblastoma
(6), oral squamous cell carcinoma
(7), laryngeal cancer (8), gastric cancer (9,10) and
prostate cancer (11). There is
increasing evidence to suggest that memantine regulates tumor
growth, invasion and metastasis in a number of types of cancer,
such as high-grade glioma (5),
neuroblastoma (12), lung cancer
(13), breast cancer (11), prostate cancer (2,11),
colon cancer (11), skin cancer
(14), and leukemia (15). One of its tumor-suppressive effects
is considered to be the blockade of the NMDA receptor followed by
glutamine depletion in cancer cells (16).
Fibroblast growth factors (FGFs) are potent
regulators of cell proliferation and differentiation (17,18).
Accordingly, the FGF receptor (FGFR) pathway plays a major role in
several biological processes during oncogenesis (19,20).
Several aberrations, including gene amplifications, point mutations
and chromosomal translocations, have been reported in various types
of cancer (21,22). Moreover, the upregulation of FGFR
signaling is a common event in a number of tumor types. Thus, the
FGFR pathway is a promising target for cancer treatment (23). Several FGFR inhibitors are currently
used in the clinical setting (20,24).
In gliomas, the expression levels of FGFs and their
receptors (FGFRs) are elevated, serving as autocrine or paracrine
growth accelerators (25–27). In addition, the upregulation of FGFs
and FGFRs in breast cancer has been reported to result in brain
metastasis and treatment-resistant cancer (28,29).
Of note, FGFs bind to three distinct types of molecules: i) FGF
receptor tyrosine kinases (FGFR1-4); ii) heparan sulfate
proteoglycans (HSPGs); and iii) Golgi glycoprotein 1 [GLG1; also
known as MG-160, cysteine-rich FGF receptor and E-selectin-ligand 1
(ESL-1)] (30,31).
In particular, GLG1 is a 160-kDa membrane
sialoglycoprotein, originally isolated from the Golgi apparatus of
rat neurons (32,33). Two homologs, the chicken
cysteine-rich fibroblast growth factor receptor (CFR) (34) and ESL-1 (35–37),
were identified in embryonic chick and murine myeloid cells,
respectively. This FGF-binding protein has 16 cysteine-rich repeats
in the extracellular luminal amino-terminal region, 21 amino acids
in the transmembrane domain, and 13 amino acids in the
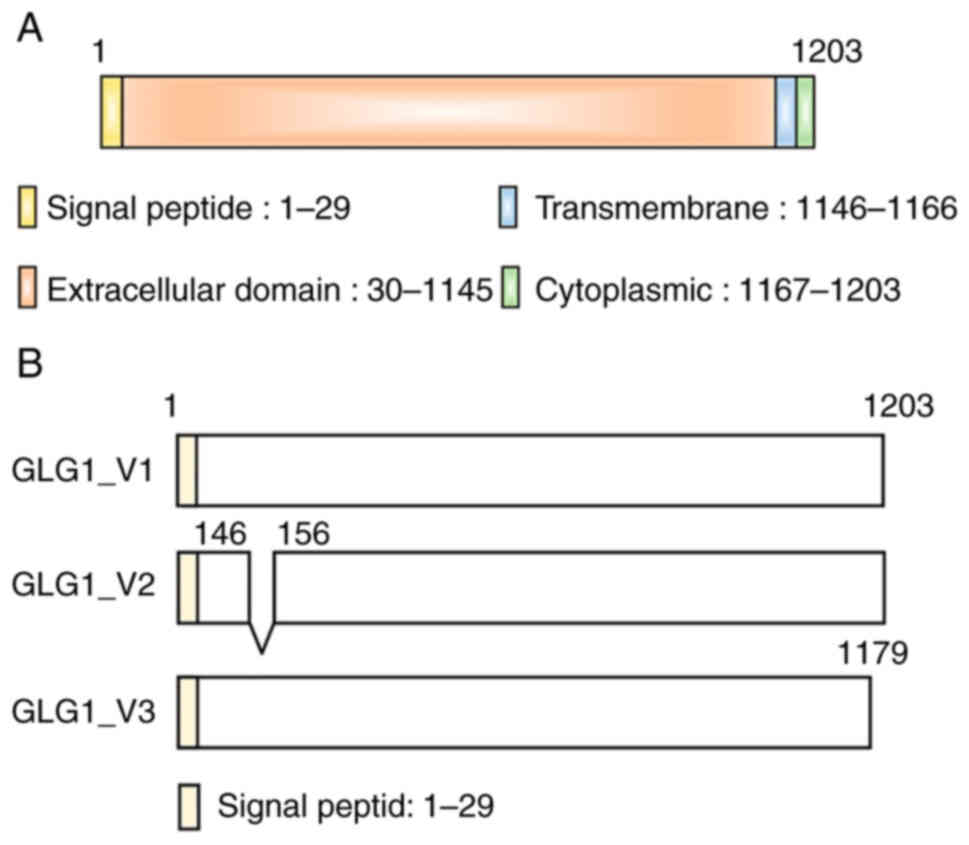
intracellular cytoplasmic carboxy-terminal domain (Fig. 1A) (38). Three major Glg1 splice variants are
known. Variant 1 is a full-length form. Compared with variant 1,
GLG1 variant 2 lacks an in-frame coding exon, resulting in lack of
an internal segment. Compared with variant 1, GLG1 variant 3 has an
additional segment in the 3′ region, resulting in a shorter
C-terminus region. GLG1 (Fig. 1B).
GLG1 is a conserved membrane sialoglycoprotein of the Golgi
apparatus in the majority of cells, displaying >90% amino acid
sequence identity with CFR (30)
and ESL-1 (30,31,33–35,39).
The GLG1 gene was assigned to human chromosome 16q22-23
(40).
FGF1, 2, 3, 4 and 18 are known to bind to this
protein (30,39). Although GLG1 does not have a
tyrosine kinase domain, which plays a main role in a variety of
cellular processes, including growth, motility, differentiation and
metabolism, it has been reported to participate in the
intracellular trafficking of FGF that is integrated into the cell
following ligand-receptor conjugation (40–43).
According to a previous study, the overexpression of GLG1 induces
cell death (42). However, the
pattern of expression of GLG1 and its function in tumors remain
unexplored.
In the present study, in order to gain insight into
the possible involvement of GLG1 in the treatment of neoplasia with
memantine, the changes in its expression were analyzed in several
types of memantine-treated human glioma and breast cancer cells
known to frequently metastasize to the brain. All memantine-treated
tumor cells exhibited an upregulated expression of GLG1. The
induction of the differential expression of GLG1 variants and
changes in its intracellular distribution in memantine-treated
cells were also identified. The results presented herein suggest
the possibility that memantine exerts a suppressive effect on cell
proliferation partly through the modulation of the expression of
GLG1, which has an FGF traffic control function. The aim of the
present study was to elucidate the intracellular behavior of tumor
growth-related factors under treatment with memantine.
Materials and methods
Cancer cell lines
The U87MG cell line was established from
glioblastoma of unknown origin. T98G is a glioblastoma cell line.
The MDA-MB-231 cell line was established from triple-negative
breast adenocarcinoma. These cell lines were purchased from the
American Type Culture Collection (ATCC). The catalog numbers for
these cell lines are HTB-14 for U87MG, CRL-1690 for T98G, and
CRM-HTB-26 for MDA-MB-231.
SNB19 is a glioblastoma cell line. The
characterization of this cell line has been precisely studied by
Welch et al (44). The SNB19
cell line used in the present study was a gift from Professor
Richard S. Morrison, Department of Neurological Surgery, University
of Washington, Seattle, WA, USA, who is one of the authors of the
aforementioned study. In their laboratory, the authors of the
present study used the early passage of dispensed frozen cells. The
authenticity of the SNB-19 cell line was confirmed by STR analysis
(45).
All cell lines were cultured in Dulbecco's modified
Eagle's medium (cat. no. 11885084, DMEM, Nacalai Tesque)
supplemented with 10% fetal bovine serum (FBS; Biosera) in a
humidified atmosphere of 5% CO2 at 37°C and harvested
when a confluency of ~70–80% was achieved.
Memantine
Memantine hydrochloride
(1-amino-3,5-dimethyladamantane hydrochloride) was purchased from
FUJIFILM Wako Pure Chemical Corporation. A 10-mM stock solution was
prepared in distilled H2O, filtered through a 0.22-µm
PES syringe filter (PES013022; Membrane Solutions LLC), and stored
at −80°C until use.
Memantine treatment
The cells (4×105) grown in a 30-mm
culture dish were exposed to memantine at final concentrations of
0–1,000 µM for 3 days.
Cell viability assay
A cell viability assay was performed by a trypan
blue exclusion assay using 0.4%-Trypan blue solution (15250-061;
Gibco; Thermo Fisher Scientific, Inc.). The cells were suspended in
0.2% trypan blue and counted using a Bio-Rad cell counter TC20
(Bio-Rad Laboratories, Inc.), according to the manufacturer's
protocol.
Microscopic observation
Morphological changes in treated tumor cells were
observed under a microscope (TS100; Nikon Corporation) and recorded
using a Nikon Digital Sight 1000 (Nikon Corporation).
mRNA extraction, cDNA preparation and
reverse transcription
Total RNA was extracted from the tumor cells using a
RNeasy Mini kit according to the manufacturer's instructions (cat.
no. 74134; Qiagen GmbH). Each RNA sample was quantified using a
NanoDrop spectrophotometer (serial no. G188; Thermo Fisher
Scientific, Inc.). cDNA synthesis was performed with 1 µg total RNA
using a PrimeScript RT reagent kit (cat. no. RR037A; Perfect Real
Time, Takara Bio Inc.), in which oligo dT primer and random 6mers
were used according to the manufacturer's instructions, followed by
reverse transcription for 15 min at 37°C and enzyme inactivation
for 5 sec at 85°C.
Quantitative PCR (qPCR)
qPCR was performed using a StepOnePlus™
real-time PCR system (Thermo Fisher Scientific, Inc.) according to
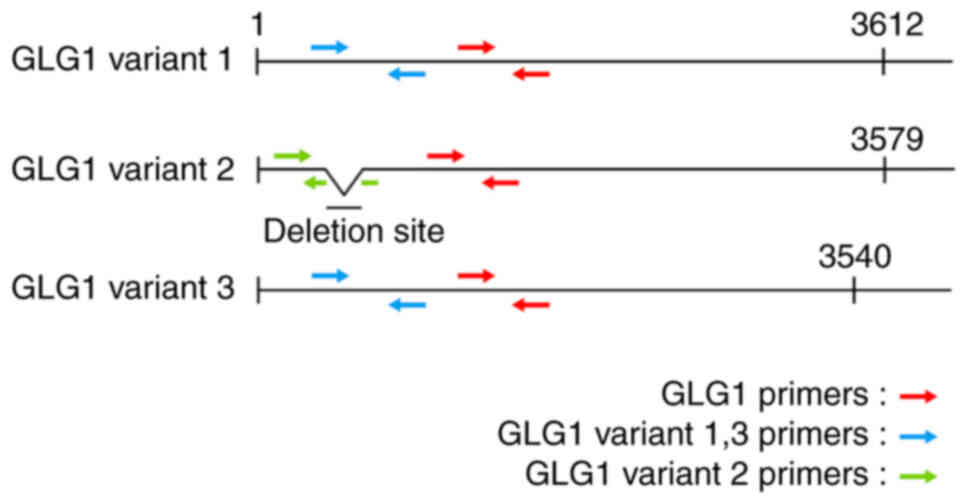
the manufacturer's instructions. qPCR was performed using GLG1
primers (Fig. 2 and Table I). Glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) was used as an mRNA loading and integrity
control (Table I). SYBR-Green-based
qPCR was performed using THUNDERBIRD® SYBR®
qPCR Mix (QSP-201; Toyobo Co., Ltd.), following the manufacturer's
instructions. Each qPCR assay was performed in a 20-µl reaction
volume containing 10 µl 2X one-step SYBR enzyme mix, 0.4 µl ROX
reference dye, 0.4 µl (10 µM) of each primer, 7.8 µl RNase-free
water and 1.0 µl template. Thermal cycling was performed under the
following conditions: 40 cycles at 95°C for 1 sec and 60°C for 20
sec. Each experiment was performed in triplicate. Relative gene
expression was analyzed using the 2−ΔΔCq method
(46). The levels of expression of
target genes were normalized against those of GAPDH using
StepOne™ software (v2.3; Thermo Fisher Scientific,
Inc.). Relative quantity (RQ) values were standardized by setting
the same threshold values.
 | Table I.Sequences of primers used for
RT-qPCR. |
Table I.
Sequences of primers used for
RT-qPCR.
| Gene symbol | Forward primer | Reverse primer |
|---|
| GLG1 |
CCAAGATGACGGCCATCATTT |
AGCCGAATACTGCCACATTTC |
| GLG1 variant
1–3 |
GTGAGGGAGCCTGAAAATGAA |
GGTGATCCACCAAGCAGGAA |
| GLG1 variant
2 |
CCTAAGCACACCTGGAGCAA |
TTCCACAACAACTCCCTCACA |
| GAPDH |
GGAGCGAGATCCCTCCAAAAT |
GGCTGTTGTCATACTTCTCATGG |
Western blot analysis
Samples were prepared using the same number of
treated cells that were homogenized in 100 µl whole-cell lysis
buffer (20 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA and 1% NP40)
supplemented with a complete protease inhibitor cocktail (cat. no.
05056489001; Roche Diagnostics), and then lysed on ice. Equal
amounts of protein samples (30 µg) measured with the Pierce BCA
Protein Assay kit (cat. no. 23227; Thermo Fisher Scientific, Inc.)
were subjected to 7.5% SDS-PAGE Mini-PROTEAN®
TGX™ precast gels (cat. no. 4561024, Bio-Rad
Laboratories, Inc.) and transferred to polyvinylidene difluoride
membranes (cat. no. 1704156, mini PVDF transfer packs; Bio-Rad
Laboratories, Inc.) and blocked using 5% BSA for 1 h at room
temperature. The membranes were then incubated with primary
antibodies against GLG1 (cat. no. MAB78791, monoclonal mouse IgG1,
clone #858238; R&D Systems, Inc.; 1:1,000), GAPDH (cat. no.
ab181602; Abcam; 1:1,000) at 4°C overnight or anti-mouse secondary
antibody (cat. no. 330; Medical and Biological Laboratories Co.,
Ltd.; 1:10,000) for 1 h at room temperature. Signals were
visualized using the ECL™ Prime Western Blotting System
(cat. no. RPN2232, Cytiva), and the FUSION SL-chemiluminescence
imaging system (Vilber-Lourmat, Collégien) was used as the digital
image processor. Quantitative analysis of relative protein
expression and band size were performed with the software provided
with FUSION SL.
Immunofluorescence staining
All cells (2×104 cells per well) were
grown in 4-chamber slides (Watson Co., Ltd.) for 2 days. Following
treatment with memantine for 2 days, the cells were fixed with 4%
PFA (cat. no. 09154-85; Nacalai Tesque, Inc.), permeabilized with
0.1% Triton X-100 (cat. no. 35501-15; Nacalai Tesque, Inc.),
blocked with 2% BSA (Sigma-Aldrich; Merck KGaA) for 1 h at room
temperature, and incubated overnight at 4°C with the following
antibody: anti-GLG1 mouse monoclonal antibody (cat. no. MAB78791;
R&D Systems, Inc.; 1:1,000). Subsequently, the slides were
incubated with Alexa Fluor 488-labeled goat anti-mouse (cat. no.
A-11017; Thermo Fisher Scientific, Inc.; 1:10,000) secondary
antibody for 1 h at room temperature. Nuclei were counterstained
with DAPI (cat. no. D523, Cellstain® DAPI solution;
Dojindo Laboratories, Inc.) for 5 min at room temperature. Samples
were analyzed using the BZ-X800 all-in-one fluorescence microscope
(Keyence Corporation).
Statistical analysis
Experimental data are presented as the mean ± SEM
from three or more independent experiments (unless otherwise
indicated). The levels of significance (one-way ANOVA followed by
Tukey's HSD test) were calculated in each treatment, and the 50%
effective concentration (EC50) value were determined in
four parameter logistic models using GraphPad Prism 9 software
(GraphPad Software, Inc.).
Results
Suppressive effects of memantine on
cell proliferation
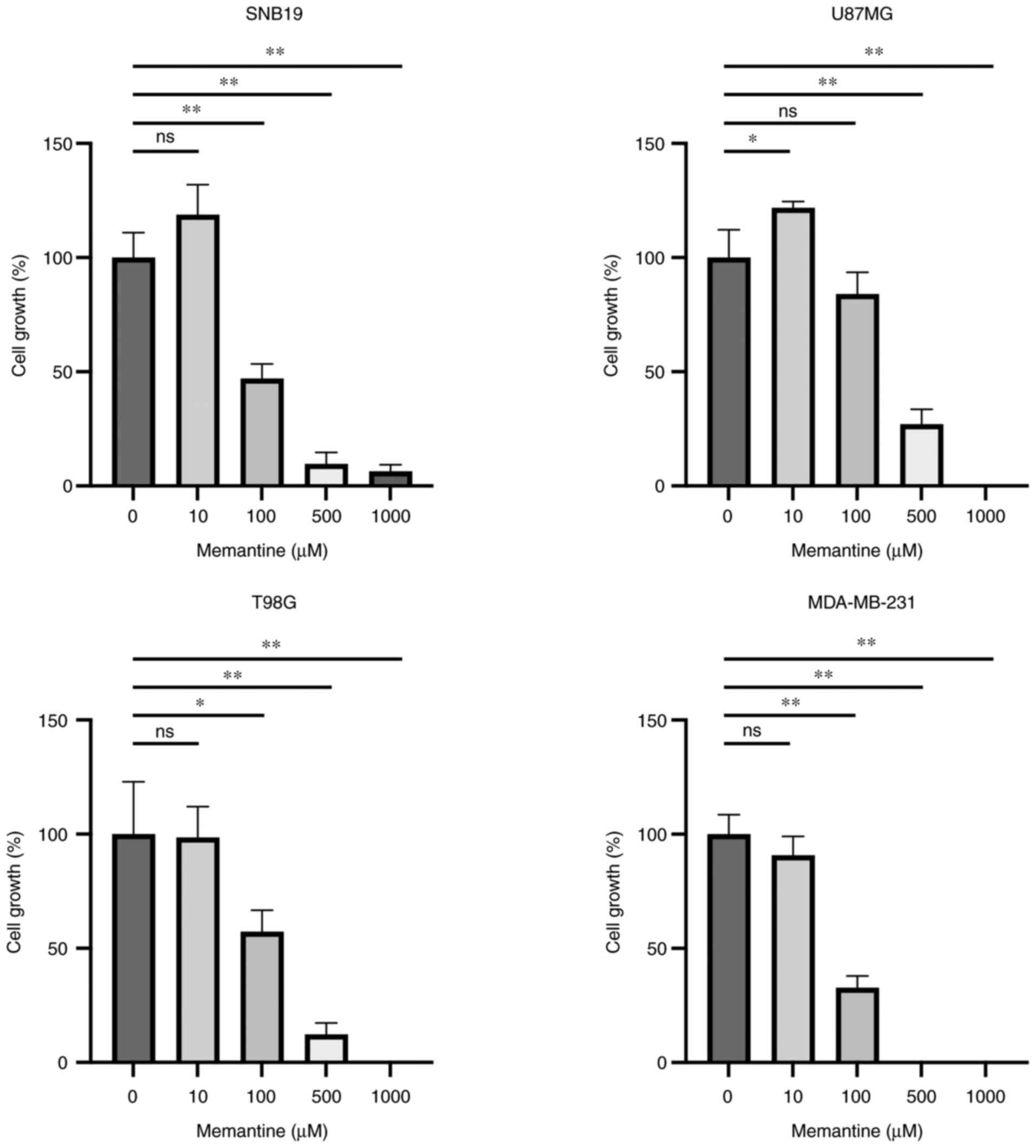
To confirm the suppressive effects of memantine on
the proliferation of cancer cells, three glioma cell lines and one
breast cancer cell line were cultured in the presence of memantine
at various concentrations (0–1,000 µM) for 3 days and the number of
live cells was counted. It was found that memantine suppressed the
growth of malignant glioma and breast cancer cells in a
concentration-dependent manner (Fig.
3). As regards the SNB19 glioma cells, it was observed that
treatment with 100 µM memantine suppressed cell growth to 53.0%.
This phenomenon was more apparent in the MDA-MB-231 cells, which
exhibited an 67.1% decrease in proliferation (Fig. 3). It was also noted that the
MDA-MB-231 breast cancer cells were more prominently affected by
memantine than the glioma cells. Exogenously applied memantine
suppressed the growth of all cell lines, exhibiting half maximal
effective concentration (EC50) values of 87.54, 131.2,
201.6 and 98.26 µM in the SNB19, U87MG, T98G and MDA-MB-231 cells,
respectively (Fig. S1).
Memantine leads to an increased mRNA
expression of GLG1
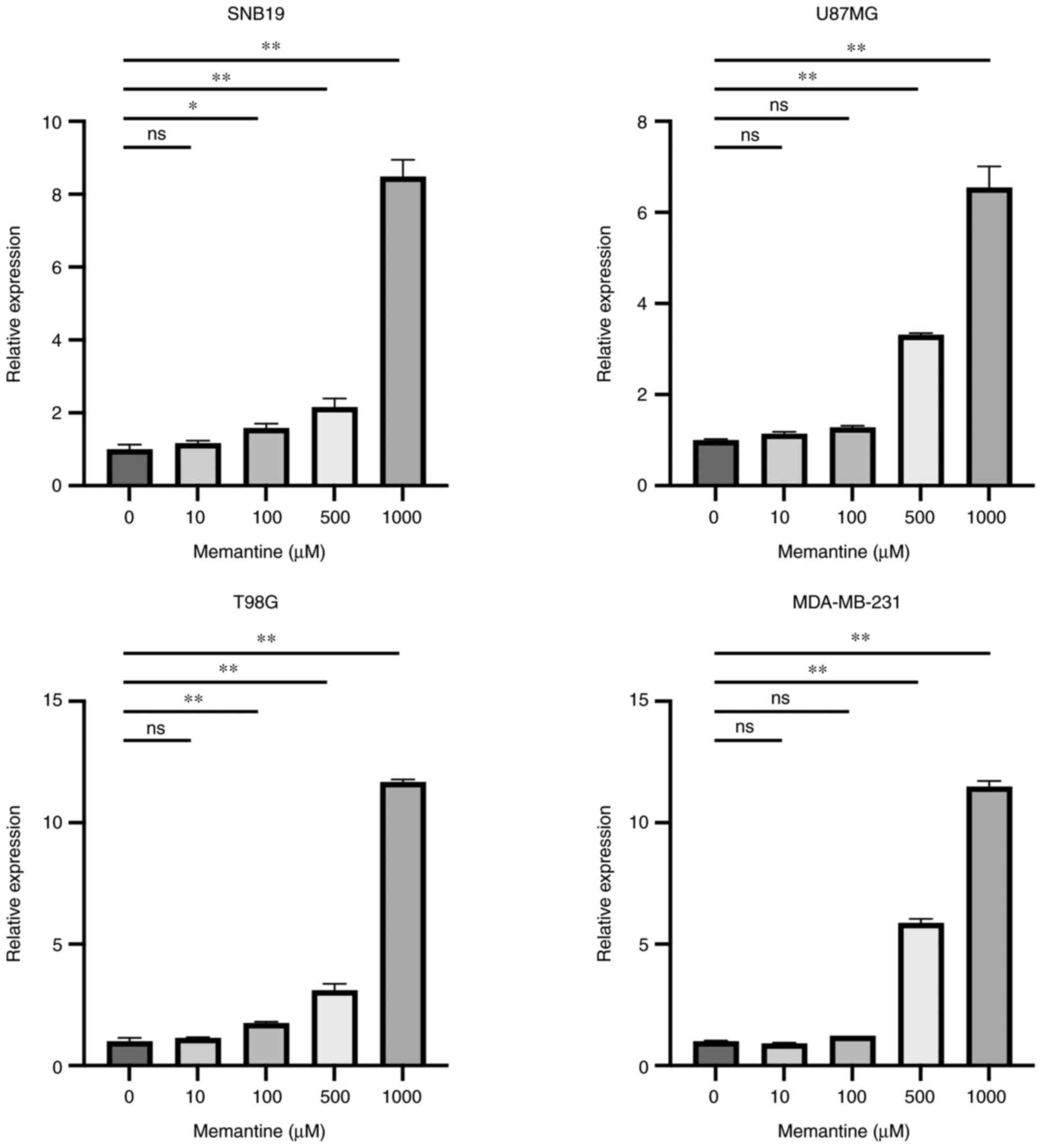
To determine the effects of memantine on the
expression of GLG1 in cancer cells, RT-qPCR was performed using
primers that detect all three GLG1 mRNA variants. The results
revealed that the mRNA expression of GLG1 was increased in a
concentration-dependent manner in all cell lines (Fig. 4). Of note, all these changes were
apparent in treatments using ≥100 µM memantine. Of note, the
highest increase in the mRNA level GLG1 was observed in the T98G
glioma cells treated with 1,000 µM memantine, whereas the
MDA-MB-231 breast cancer cells exhibited a significant increase in
GLG1 expression when treated with ≥500 µM memantine.
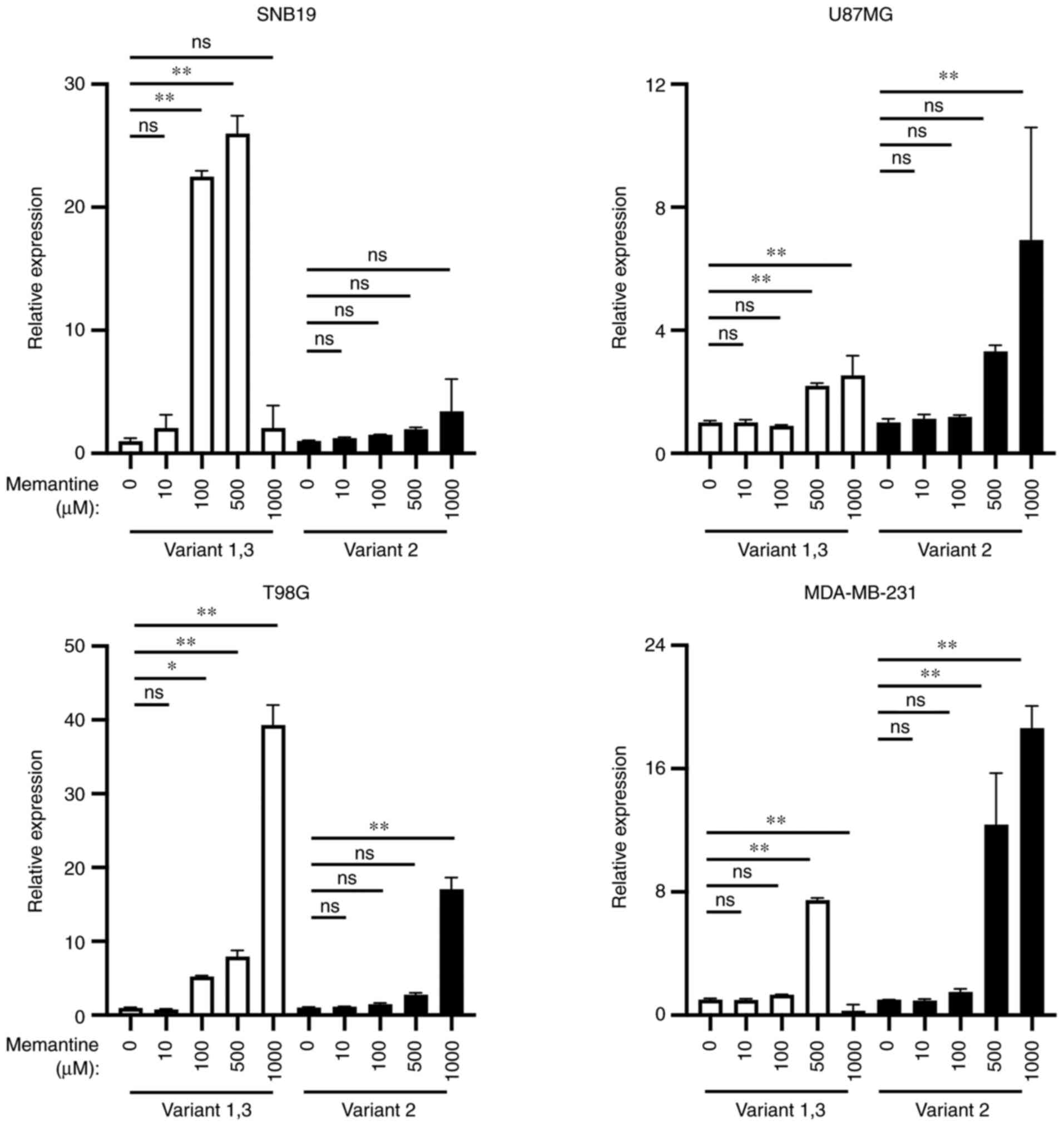
Differential mRNA expression of
GLG1
The present study also examined the changes in the
expression of GLG1 variants following treatment with memantine. It
was detected that the expression level of variants 1 and 3 was
increased to a greater extent than that of variant 2 in the SNB19
and T98G cells, whereas the expression level of variant 2 was
higher than that of variants 1 or 3 in the U87MG and MDA-MB-231
cells (Fig. 5).
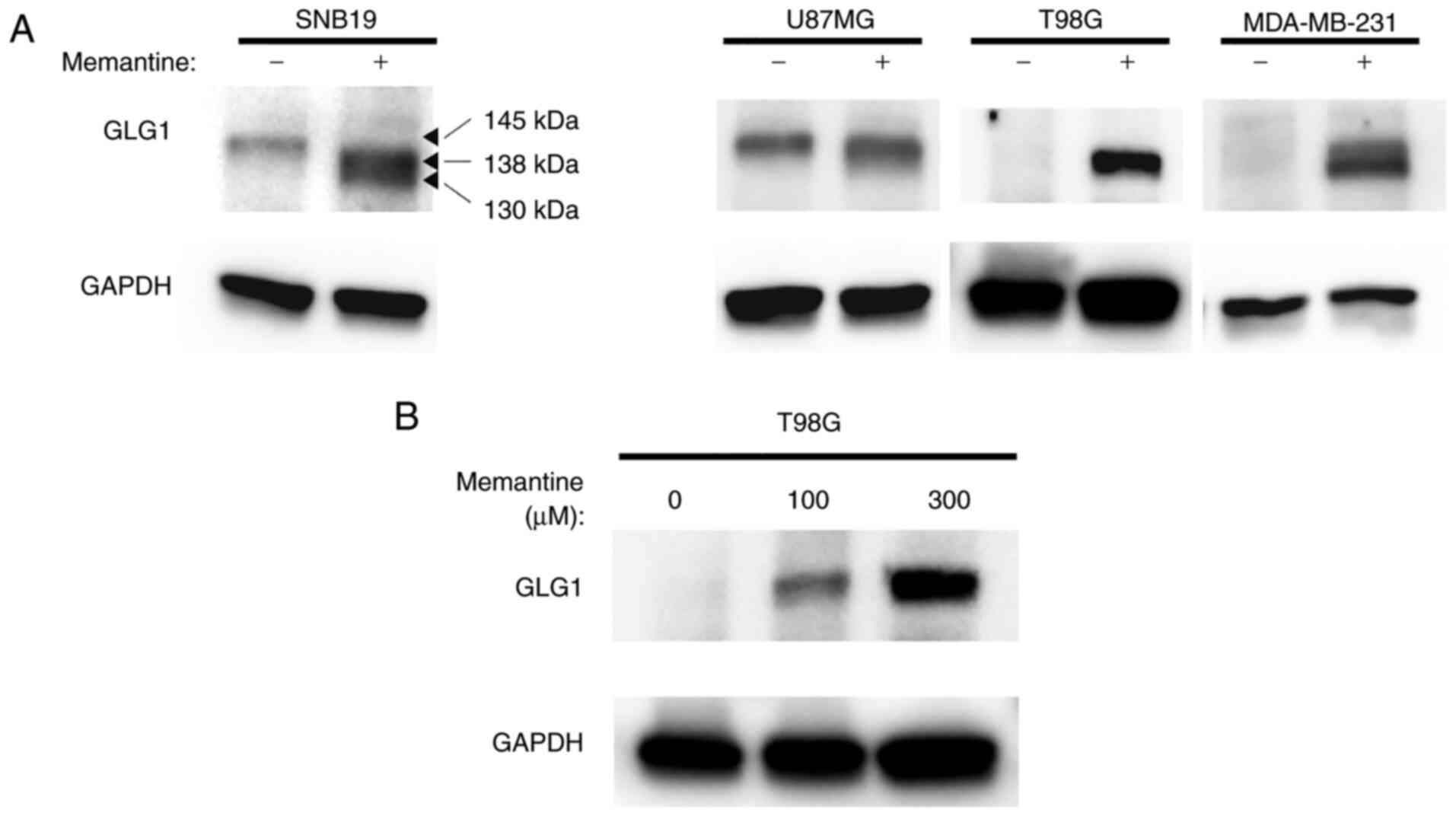
GLG1 protein expression
Western blot analysis was then performed to
determine whether the protein expression of GLG1 in cancer cells
was altered following treatment with memantine. Of note, despite
differences in its expression levels among cell lines, GLG1 was
expressed in all control cell lines. It was found that the U87MG
glioma cells exhibited a relatively high expression under normal
(untreated) conditions. Following treatment with memantine, the
expression of GLG1 was induced in all cell lines (Fig. 6A). Notably, it was found that the
protein expression level of GLG1 increased in a
concentration-dependent manner in the T98G glioma cells (Fig. 6B). It was also observed that the
size of the GLG1 protein detected by the GLG1 antibody in the
untreated tumor cells was ~145 kDa. However, it was found that the
memantine-treated cells expressed two smaller-size GLG1 proteins.
More specifically, it was detected that the molecular weight of
each band was ~8 and 15 kDa smaller than that of the full-length
GLG1 protein (Fig. 6A). It was thus
hypothesized that treatment with memantine induced the expression
of truncated proteins, such as GLG1 variants 2 and 3 (Fig. 1B).
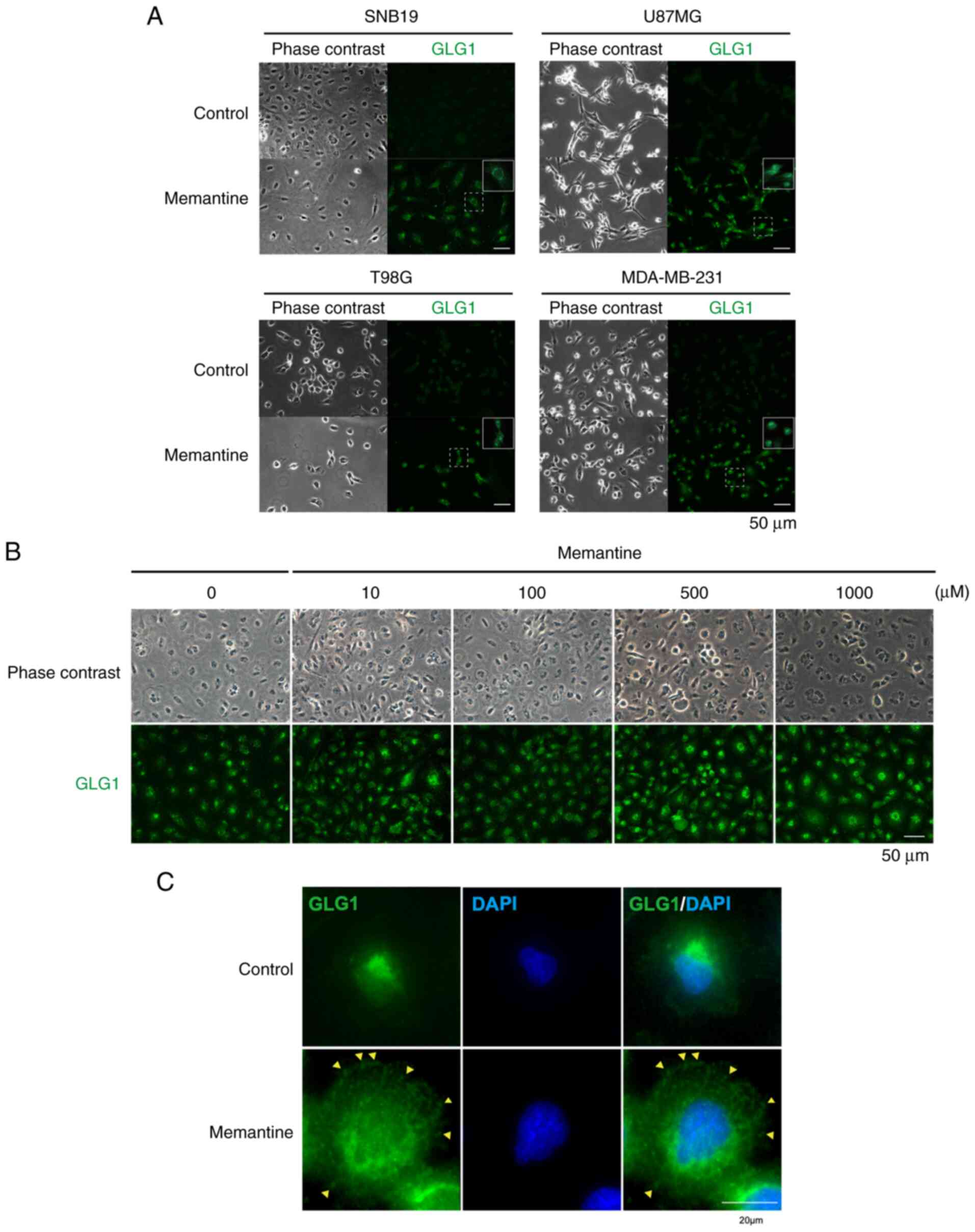
Subcellular localization of GLG1
protein
Both GLG1 variants 2 and 3 are widely expressed in
cells (41). Therefore, it was
hypothesized that treatment with memantine may result in
alterations in the cellular localization of GLG1, which is
typically the Golgi apparatus. The immunohistochemical staining of
GLG1 demonstrated that GLG1 was localized in the Golgi apparatus in
untreated cells; however, following treatment with memantine, the
localization of GLG1 was altered in all cell lines. In particular,
it was found that its expression was spread into the cytosol in
memantine-treated cells (Fig. 7A).
It was further noted that the fluorescence intensity of GLG1
increased following treatment with memantine in a
concentration-dependent manner, suggesting an increase in its
expression at the protein level (Fig.
7B). In addition, immunofluorescence staining demonstrated that
GLG1 was present throughout the cell in memantine-treated SNB19
cells, whereas it was localized near the nucleus in the control
cells. GLG1 was observed on the cell surface (Fig. 7C, yellow arrowheads). GLG1 protein
detected on the cell surface was considered to be a putative
truncated form, consistent with the results of western blot
analysis. These results suggest that the memantine-induced increase
in the expression of GLG1 and variations in the intracellular
distribution of GLG1 play a crucial role in the suppression of
tumor growth (Fig. 8).
Discussion
The accumulation of several genetic alterations,
such as the loss of tumor suppressor functions and the induction of
oncogene functions, results in the transformation of normal cells
into cells with highly malignant features (47,48).
The deregulation of FGF signaling through the genetic modification
or overexpression of FGFs and FGFRs has been observed in numerous
tumors, with FGFs playing a key role in tumorigenesis and
angiogenesis during tumor growth (28,49,50).
There is evidence to indicate that inhibition of FGFR signaling
results in anti-proliferative or pro-apoptotic effects (51); thus, an increasing number of drugs
against FGF pathways are currently in clinical use (18,23,28).
The high-affinity cell surface FGF receptors belong
to a family of receptor tyrosine kinases. Their intracellular
tyrosine kinase domain is activated upon ligand binding and induces
various intracellular downstream signaling pathways, leading to the
positive regulation of cell proliferation. In addition to FGFRs,
HSPGs serve as low-affinity receptors for FGFs. It has been
suggested that the low-affinity HSPG receptor provides easier
access of FGFs to FGFRs, inducing the dimerization of FGFR and the
activation of tyrosine kinase inhibitors (52).
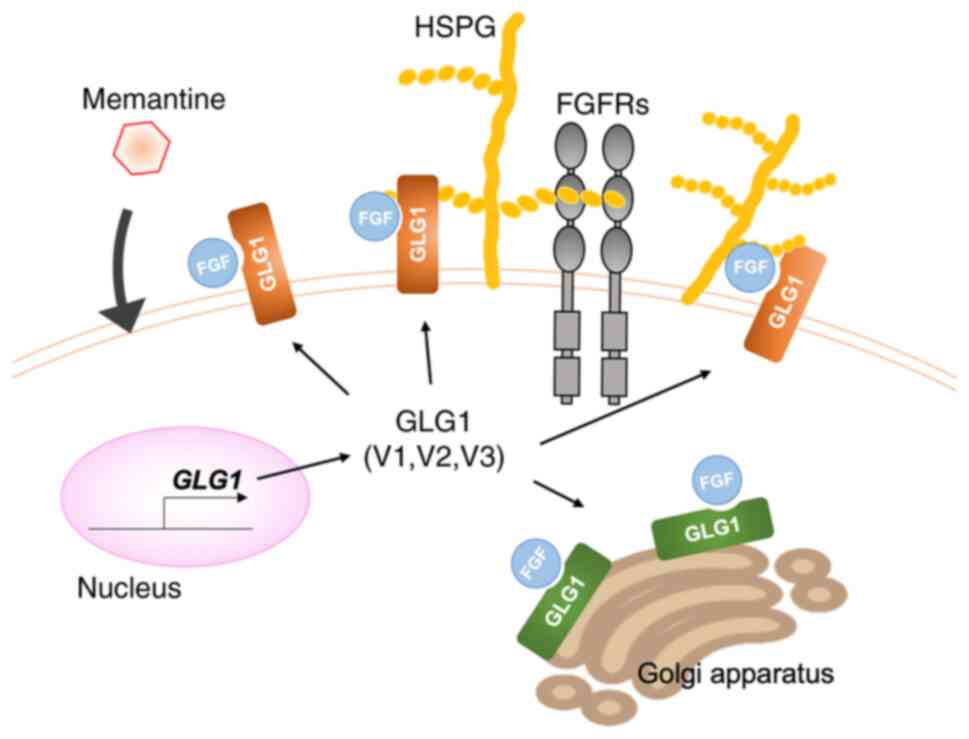
Apart from signal-transducing FGFRs, GLG1 is known
to bind FGFs. GLG1 was originally identified as an FGF2 receptor,
found in the Golgi complex (31).
GLG1 binds FGF1, 2, 3, 4 and 18 (30,40);
however, it does not have a tyrosine kinase domain, which plays a
crucial role in a variety of cellular processes, including growth,
motility, differentiation and metabolism.
Therefore, it functions as a so-called ‘decoy
receptor’ that is able to recognize and bind FGFs efficiently;
however, it is not structurally able to signal or activate the
intended receptor complex. This mechanism is known to regulate the
intracellular levels of FGFs (42).
In the present study, it was demonstrated that GLG1
expression was upregulated in a concentration-dependent manner in
memantine-treated glioma and breast cancer cells. The behavior of
each cell line differed, and this was presumably due to the
different reactions in other pathways, such as NMDA-receptor
blocking. This phenomenon was observed in three malignant glioma
cell lines and one breast cancer cell line. However, other breast
cancer cell lines need to be analyzed in the future in order to
confirm whether a change in GLG1 expression can be universally
detected.
As GLG1 functions as a decoy to interfere with the
tyrosine kinase FGF receptor, this may downregulate the
intracellular levels of FGFs, resulting in the loss of
proliferative effects, in accordance with the findings of a
previous study by the authors reporting that high-grade glioma
expressed lower levels of GLG1, whereas low-grade glioma higher
levels of GLG1 (53).
Although GLG1 is a 150-kDa integral membrane
glycoprotein that is primarily located in the cis-medial Golgi
complex, a substantial proportion of GLG1 is secreted (40). Structurally, compared with variant
1, variant 2 lacks an in-frame coding exon, resulting in the lack
of the internal segment of aa 147–157 (Fig. 1B). Conversely, compared with variant
1, variant 3 has an additional segment in the 3′ region, resulting
in a shorter isoform at the C-terminus, which contains a 14 aa
substitution for aa 685–1179 (Fig.
1B). This shorter cytoplasmic segment allows for presentation
at the cell membrane, whereas full-length GLG1 is localized in the
medial cisternae. These truncated variants are widely distributed
in the cell, suggesting that the intraluminal juxtamembrane domain
is important for the targeting and retention of GLG1 to the medial
Golgis (40).
The present study also found alterations in RNA
alternative splicing following the of cancer cells with memantine.
RNA splicing is a post-transcriptional process that is estimated to
affect the regulation of as many as 60% of all human genes
(54). The regulation of
alternative splicing is a complex process involving numerous
interacting components (55). Of
note, alternative splicing has been reported to be associated with
principal biological and pathological processes. FGF receptors are
subjected to this process and by incorporating different exons into
the extracellular binding domain, they are hence affected by both
ligand specificity and binding affinity (56–62).
GLG1, a non-tyrosine kinase FGF receptor, is also
regulated by RNA splicing (39,41,63).
The data of the present study demonstrated that the expression of
variants 2 and 3 increased in memantine-treated tumor cells at the
mRNA level and putatively at the protein level, in addition to the
expression of the GLG1 variant 1. Secreted GLG1 binds to HSPG and
traps FGFs, thereby directly competing with tyrosine kinase
receptors for FGF binding (40,64).
This event inhibits the following dimerization of FGF receptors,
thus potentially diminishing the biological availability of FGFs
(Fig. 8). The mechanism through
which memantine leads to these alterations in alternative splicing
remains unknown, and further studies are thus warranted to
elucidate this mechanism. To achieve this, factors such as enhancer
elements (exonic splicing enhancers and intronic splicing
enhancers), activator proteins (SR protein family), silencer
elements (exonic splicing silencers and intronic splicing
silencers), and repressor proteins (heterogeneous nuclear
ribonucleoproteins protein family) need to be analyzed (55).
To date, the mechanism of the tumor growth
suppressive effect of memantine has been considered to be
attributed to the blockage of the NMDA-receptor activating
glutamate. Malignant gliomas and breast cancers are known to
exhibit high levels of glutamate, which accelerate cell
proliferation following NMDA-receptor stimulation. Therefore,
memantine affects tumor cell growth by blocking the NMDA receptor,
as reported in a number of studies (2,8,11,13–15,65–67).
Albayrak and Demirtas Korkmaz (68)
reported that memantine triggered G0/G1 cell cycle arrest; they
also examined Caspase-3, Bcl-2 and Bax expression, revealing a
change in apoptotic gene expression (69). The present study suggested another
mechanism through which the induction of the expression of GLG1 and
the generation of additional truncated variants may suppress the
FGF and FGFR pathways. The present study revealed the GLG1
expression was altered by memantine. However, the association of
this change with cancer proliferation remains unknown.
The safety of memantine has been demonstrated in a
randomized, placebo-controlled clinical trial in patients with
mild-to-moderate vascular dementia (70). As a result, memantine has been
approved for the treatment of moderate-to-severe Alzheimer's
disease (71). By contrast, there
have been reports on the risk of somnolence, weight gain,
confusion, hypertension, nervous system disorders and falling due
to the administration of memantine, although it has been shown to
be beneficial for patients with Alzheimer's disease as regards the
improvement of cognition (72).
Therefore, further studies are required to evaluate the clinical
use of this medicine for cancer treatment from a safety standpoint.
Two clinical trials for glioblastoma are registered in the USA: ‘A
phase II study of memantine in the treatment of recurrent
glioblastoma’ (NCT01260467) and ‘Temozolomide, memantine
hydrochloride, mefloquine, and metformin hydrochloride in treating
patients with glioblastoma multiforme after radiation therapy’
(NCT01430351). The former trial was terminated, and no adverse
effect was reported. The latter phase I trial is currently ongoing,
and the results are not available at this moment.
The present study observed a change in the protein
and mRNA expression of GLG1. The function of GLG1 and its different
variants has been reported in several studies (35,40–42,45,53,63,73).
The aim of the present study was to report these phenomena.
Determining the extent of the effects of GLG1 on cell proliferation
may be the following step in examining its effects under memantine
exposure.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms. Sachiko Kurihara
from the Division of Reproductive Medicine, Perinatology and
Gynecologic Oncology, Graduate School of Medicine, Nippon Medical
School, Tokyo, Japan and Dr Hidefumi Wakashin from Goi Hospital,
Chiba, Japan for providing technical advice.
Funding
The present study was supported in part by a grant from the
Strategic Research Foundation Grant-aided Project for Private
Universities from the Ministry of Education, Culture, Sport,
Science and Technology, Japan (MEXT) (no. S0801035).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FY designed the experiments. FY, SH and SK performed
the experiments. TA contributed to sample preparation. FY, SH and
YO performed data analyses. SH helped interpret the results and was
involved in the preparation of the manuscript. FY wrote the
manuscript in consultation with SH and YO. All authors critically
revised the manuscript, commented on drafts of the manuscript, and
have read and approved the final draft. FY and SH were equal
contributors to the study and confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GLG1
|
Golgi glycoprotein 1
|
|
FGF
|
fibroblast growth factor
|
|
FGFR
|
fibroblast growth factor receptor
|
|
HSPG
|
heparan sulfate proteoglycan
|
|
NMDA
|
N-methyl-D-aspartate
|
|
CFR
|
chicken cysteine-rich fibroblast
growth factor receptor
|
|
ESL-1
|
E-selectin-ligand 1
|
References
|
1
|
Abbruzzese C, Matteoni S, Signore M,
Cardone L, Nath K, Glickson JD and Paggi MG: Drug repurposing for
the treatment of glioblastoma multiforme. J Exp Clin Cancer Res.
36:1692017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Albayrak G, Konac E, Dikmen AU and Bilen
CY: Memantine induces apoptosis and inhibits cell cycle progression
in LNCaP prostate cancer cells. Hum Exp Toxicol. 37:953–958. 2018.
View Article : Google Scholar
|
|
3
|
Albayrak G and Korkmaz FD: Alzheimer's
drug memantine inhibits metastasis and p-Erk protein expression on
4T1 breast cancer cells. Bratisl Lek Listy. 121:499–503.
2020.PubMed/NCBI
|
|
4
|
Lipton SA: Paradigm shift in
neuroprotection by NMDA receptor blockade: Memantine and beyond.
Nat Rev Drug Discov. 5:160–170. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Müller-Längle A, Lutz H, Hehlgans S, Rödel
F, Rau K and Laube B: NMDA receptor-mediated signaling pathways
enhance radiation resistance, survival and migration in
glioblastoma cells-a potential target for adjuvant radiotherapy.
Cancers (Basel). 11:5032019. View Article : Google Scholar
|
|
6
|
Yoshioka A, Ikegaki N, Williams M and
Pleasure D: Expression of N-methyl-D-aspartate (NMDA) and non-NMDA
glutamate receptor genes in neuroblastoma, medulloblastoma, and
other cell lines. J Neurosci Res. 46:164–178. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Choi SW, Park SY, Hong SP, Pai H, Choi JY
and Kim SG: The expression of NMDA receptor 1 is associated with
clinicopathological parameters and prognosis in the oral squamous
cell carcinoma. J Oral Pathol Med. 33:533–537. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stepulak A, Luksch H, Uckermann O,
Sifringer M, Rzeski W, Polberg K, Kupisz K, Klatka J, Kielbus M,
Grabarska A, et al: Glutamate receptors in laryngeal cancer cells.
Anticancer Res. 31:565–573. 2011.PubMed/NCBI
|
|
9
|
Liu JW, Myoung SK, Nagpal J, Yamashita K,
Poeta L, Chang X, Lee J, Park HL, Jeronimo C, Westra WH, et al:
Quantitative hypermethylation of NMDAR2B in human gastric cancer.
Int J Cancer. 121:1994–2000. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Watanabe K, Kanno T, Oshima T, Miwa H,
Tashiro C and Nishizaki T: The NMDA receptor NR2A subunit regulates
proliferation of MKN45 human gastric cancer cells. Biochem Biophys
Res Commun. 367:487–490. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Abdul M and Hoosein N:
N-methyl-D-aspartate receptor in human prostate cancer. J Membr
Biol. 205:125–128. 2005. View Article : Google Scholar
|
|
12
|
Sekiguchi K, Sato M, Yokoyama MK, Sato T,
Tsutiya A, Omoteyama K, Arito M, Suematsu N, Kato T and Kurokawa M:
Effects of memantine on the growth and protein profiles of
neuroblastoma cells. Integr Mol Med. 5:1–8. 2018. View Article : Google Scholar
|
|
13
|
North WG, Liu F, Dragnev KH and Demidenko
E: Small-cell lung cancer growth inhibition: Synergism between NMDA
receptor blockade and chemotherapy. Clin Pharmacol. 11:15–23.
2019.
|
|
14
|
Du S, Sung YS, Wey M, Wang Y, Alatrash N,
Berthod A, MacDonnell FM and Armstrong DW: Roles of
N-methyl-D-aspartate receptors and D-amino acids in cancer cell
viability. Mol Biol Rep. 47:6749–6758. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kamal T, Green TN, Morel-Kopp MC, Ward CM,
McGregor AL, McGlashan SR, Bohlander SK, Browett PJ, Teague L,
During MJ, et al: Inhibition of glutamate regulated calcium entry
into leukemic megakaryoblasts reduces cell proliferation and
supports differentiation. Cell Signal. 27:1860–1872. 2015.
View Article : Google Scholar
|
|
16
|
Rzeski W, Turski L and Ikonomidou C:
Glutamate antagonists limit tumor growth. Proc Natl Acad Sci USA.
98:6372–6377. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Turner N and Grose R: Fibroblast growth
factor signalling: From development to cancer. Nat Rev Cancer.
10:116–129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Katoh M: Therapeutics targeting FGF
signaling network in human diseases. Trends Pharmacol Sci.
37:1081–1096. 2016. View Article : Google Scholar
|
|
19
|
Tiong KH, Mah LY and Leong CO: Functional
roles of fibroblast growth factor receptors (FGFRs) signaling in
human cancers. Apoptosis. 18:1447–1468. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Presta M, Chiodelli P, Giacomini A,
Rusnati M and Ronca R: Fibroblast growth factors (FGFs) in cancer:
FGF traps as a new therapeutic approach. Pharmacol Ther.
179:171–187. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
De Luca A, Esposito Abate R, Rachiglio AM,
Maiello MR, Esposito C, Schettino C, Izzo F, Nasti G and Normanno
N: FGFR fusions in cancer: From diagnostic approaches to
therapeutic intervention. Int J Mol Sci. 21:68562020. View Article : Google Scholar
|
|
22
|
Tomlinson DC and Knowles MA: Altered
splicing of FGFR1 is associated with high tumor grade and stage and
leads to increased sensitivity to FGF1 in bladder cancer. Am J
Pathol. 177:2379–2386. 2010. View Article : Google Scholar
|
|
23
|
Zhu DL, Tuo XM, Rong Y, Zhang K and Guo Y:
Fibroblast growth factor receptor signaling as therapeutic targets
in female reproductive system cancers. J Cancer. 11:7264–7275.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hierro C, Rodon J and Tabernero J:
Fibroblast growth factor (FGF) receptor/FGF inhibitors: Novel
targets and strategies for optimization of response of solid
tumors. Semin Oncol. 42:801–819. 2015. View Article : Google Scholar
|
|
25
|
Gross JL, Morrison RS, Eidsvoog K, Herblin
WF, Kornblith PL and Dexter DL: Basic fibroblast growth factor: A
potential autocrine regulator of human glioma cell growth. J
Neurosci Res. 27:689–696. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takahashi JA, Mori H, Fukumoto M, Igarashi
K, Jaye M, Oda Y, Kikuchi H and Hatanaka M: Gene expression of
fibroblast growth factors in human gliomas and meningiomas:
Demonstration of cellular source of basic fibroblast growth factor
mRNA and peptide in tumor tissues. Proc Natl Acad Sci USA.
87:5710–5714. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Morrison RS, Yamaguchi F, Saya H, Bruner
JM, Yahanda AM, Donehower LA and Berger M: Basic fibroblast growth
factor and fibroblast growth factor receptor I are implicated in
the growth of human astrocytomas. J Neurooncol. 18:207–216. 1994.
View Article : Google Scholar
|
|
28
|
Hynes NE and Dey JH: Potential for
targeting the fibroblast growth factor receptors in breast cancer.
Cancer Res. 70:5199–5202. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Santolla MF and Maggiolini M: The FGF/FGFR
system in breast cancer: Oncogenic features and therapeutic
perspectives. Cancers (Basel). 12:30292020. View Article : Google Scholar
|
|
30
|
Burrus LW, Zuber ME, Lueddecke BA and
Olwin BB: Identification of a cysteine-rich receptor for fibroblast
growth factors. Mol Cell Biol. 12:5600–5609. 1992. View Article : Google Scholar
|
|
31
|
Steegmaier M, Levinovitz A, Isenmann S,
Borges E, Lenter M, Kocher HP, Kleuser B and Vestweber D: The
E-selectin-ligand ESL-1 is a variant of a receptor for fibroblast
growth factor. Nature. 373:615–620. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Croul S, Mezitis SG, Stieber A, Chen YJ,
Gonatas JO, Goud B and Gonatas NK: Immunocytochemical visualization
of the Golgi apparatus in several species, including human, and
tissues with an antiserum against MG-160, a sialoglycoprotein of
rat Golgi apparatus. J Histochem Cytochem. 38:957–963. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gonatas JO, Mezitis SG, Stieber A,
Fleischer B and Gonatas NK: MG-160. A novel sialoglycoprotein of
the medial cisternae of the Golgi apparatus [published eeratum
appears in J Biol Chem 1989 Mar 5;264(7):4264]. J Biol Chem.
264:646–653. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Burrus LW and Olwin BB: Isolation of a
receptor for acidic and basic fibroblast growth factor from
embryonic chick. J Biol Chem. 264:18647–18653. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Steegmaier M, Borges E, Berger J, Schwarz
H and Vestweber D: The E-selectin-ligand ESL-1 is located in the
Golgi as well as on microvilli on the cell surface. J Cell Sci.
110:687–694. 1997. View Article : Google Scholar
|
|
36
|
Levinovitz A, Mühlhoff J, Isenmann S and
Vestweber D: Identification of a glycoprotein ligand for E-selectin
on mouse myeloid cells. J Cell Biol. 121:449–459. 1993. View Article : Google Scholar
|
|
37
|
Lenter M, Levinovitz A, Isenmann S and
Vestweber D: Monospecific and common glycoprotein ligands for E-
and P-selectin on myeloid cells. J Cell Biol. 125:471–481. 1994.
View Article : Google Scholar
|
|
38
|
Mourelatos Z, Gonatas JO, Cinato E and
Gonatas NK: Cloning and sequence analysis of the human MG160, a
fibroblast growth factor and E-selectin binding membrane
sialoglycoprotein of the Golgi apparatus. DNA Cell Biol.
15:1121–1128. 1996. View Article : Google Scholar
|
|
39
|
Gonatas JO, Mourelatos Z, Stieber A, Lane
WS, Brosius J and Gonatas NK: MG-160, a membrane sialoglycoprotein
of the medial cisternae of the rat Golgi apparatus, binds basic
fibroblast growth factor and exhibits a high level of sequence
identity to a chicken fibroblast growth factor receptor. J Cell
Sci. 108:457–467. 1995. View Article : Google Scholar
|
|
40
|
Köhl R, Antoine M, Olwin BB, Dickson C and
Kiefer P: Cysteine-rich fibroblast growth factor receptor alters
secretion and intracellular routing of fibroblast growth factor 3.
J Biol Chem. 275:15741–15748. 2000. View Article : Google Scholar
|
|
41
|
Ahn J, Febbraio M and Silverstein RL: A
novel isoform of human Golgi complex-localized glycoprotein-1 (also
known as E-selectin ligand-1, MG-160 and cysteine-rich fibroblast
growth factor receptor) targets differential subcellular
localization. J Cell Sci. 118:1725–1731. 2005. View Article : Google Scholar
|
|
42
|
Zuber ME, Zhou Z, Burrus LW and Olwin BB:
Cysteine-rich FGF receptor regulates intracellular FGF-1 and FGF-2
levels. J Cell Physiol. 170:217–227. 1997. View Article : Google Scholar
|
|
43
|
Szebenyi G and Fallon JF: Fibroblast
growth factors as multifunctional signaling factors. Int Rev Cytol.
185:45–106. 1999. View Article : Google Scholar
|
|
44
|
Welch WC, Morrison RS, Gross JL, Gollin
SM, Kitson RB, Goldfarb RH, Giuliano KA, Bradley MK and Kornblith
PL: Morphologic, immunologic, biochemical, and cytogenetic
characteristics of the human glioblastoma-derived cell line,
SNB-19. In Vitro Cell Dev Biol Anim. 31:610–616. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
An Q, Fillmore HL, Vouri M and Pilkington
GJ: Brain tumor cell line authentication, an efficient alternative
to capillary electrophoresis by using a microfluidics-based system.
Neuro Oncol. 16:265–273. 2014. View Article : Google Scholar
|
|
46
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Solomon E, Borrow J and Goddard AD:
Chromosome aberrations and cancer. Science. 254:1153–1160. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar
|
|
49
|
Yamaguchi F, Saya H, Bruner JM and
Morrison RS: Differential expression of two fibroblast growth
factor-receptor genes is associated with malignant progression in
human astrocytomas. Proc Natl Acad Sci USA. 91:484–488. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jimenez-Pascual A and Siebzehnrubl FA:
Fibroblast growth factor receptor functions in glioblastoma. Cells.
8:7152019. View Article : Google Scholar
|
|
51
|
Yamada SM, Yamaguchi F, Brown R, Berger MS
and Morrison RS: Suppression of glioblastoma cell growth following
antisense oligonucleotide-mediated inhibition of fibroblast growth
factor receptor expression. Glia. 28:66–76. 1999. View Article : Google Scholar
|
|
52
|
Schlessinger J, Lax I and Lemmon M:
Regulation of growth factor activation by proteoglycans: What is
the role of the low affinity receptors? Cell. 83:357–360. 1995.
View Article : Google Scholar
|
|
53
|
Yamaguchi F, Morrison RS, Gonatas NK,
Takahashi H, Sugisaki Y and Teramoto A: Identification of MG-160, a
FGF binding medial Golgi sialoglycoprotein, in brain tumors: An
index of malignancy in astrocytomas. Int J Oncol. 22:1045–1049.
2003.
|
|
54
|
Song SW, Cote GJ, Wu C and Zhang W:
Alternative splicing: Genetic complexity in cancer. Computational
and Statistical Approaches to Genomics. Zhang W and Shmulevich I:
Kluwer Academic Publishers; Bostan: pp. 277–297. 2002
|
|
55
|
Wang Y, Liu J, Huang BO, Xu YM, Li J,
Huang LF, Lin J, Zhang J, Min QH, Yang WM and Wang XZ: Mechanism of
alternative splicing and its regulation. Biomed Rep. 3:152–158.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hou JZ, Kan MK, McKeehan K, McBride G,
Adams P and McKeehan WL: Fibroblast growth factor receptors from
liver vary in three structural domains. Science. 251:665–668. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Werner S, Duan DS, de Vries C, Peters KG,
Johnson DE and Williams LT: Differential splicing in the
extracellular region of fibroblast growth factor receptor 1
generates receptor variants with different ligand-binding
specificities. Mol Cell Biol. 12:82–88. 1992. View Article : Google Scholar
|
|
58
|
Crumley G, Bellot F, Kaplow JM,
Schlessinger J, Jaye M and Dionne CA: High-affinity binding and
activation of a truncated FGF receptor by both aFGF and bFGF.
Oncogene. 6:2255–2262. 1991.PubMed/NCBI
|
|
59
|
Dell KR and Williams LT: A novel form of
fibroblast growth factor receptor 2. Alternative splicing of the
third immunoglobulin-like domain confers ligand binding
specificity. J Biol Chem. 267:21225–21229. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Miki T, Bottaro DP, Fleming TP, Smith CL,
Burgess WH, Chan AML and Aaronson SA: Determination of
ligand-binding specificity by alternative splicing: Two distinct
growth factor receptors encoded by a single gene. Proc Natl Acad
Sci USA. 89:246–250. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yayon A, Zimmer Y, Shen GH, Avivi A,
Yarden Y and Givol D: A confined variable region confers ligand
specificity on fibroblast growth factor receptors: Implications for
the origin of the immunoglobulin fold. EMBO J. 11:1885–1890. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Morrison RS, Yamaguchi F, Bruner JM, Tang
M, McKeehan W and Berger MS: Fibroblast growth factor receptor gene
expression and immunoreactivity are elevated in human glioblastoma
multiforme. Cancer Res. 54:2794–2799. 1994.PubMed/NCBI
|
|
63
|
Yamamoto-Hino M, Abe M, Shibano T,
Setoguchi Y, Awano W, Ueda R, Okano H and Goto S: Cisterna-specific
localization of glycosylation-related proteins to the Golgi
apparatus. Cell Struct Funct. 37:55–63. 2012. View Article : Google Scholar
|
|
64
|
Antoine M, Köhl R, Tag CG, Gressner AM,
Hellerbrand C and Kiefer P: Secreted cysteine-rich FGF receptor
derives from posttranslational processing by furin-like prohormone
convertases. Biochem Biophys Res Commun. 382:359–364. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen R, Xu X, Tao Y, Qian Z and Yu Y:
Exosomes in hepatocellular carcinoma: A new horizon. Cell Commun
Signal. 17:12019. View Article : Google Scholar
|
|
66
|
Yohay K, Tyler B, Weaver KD, Pardo AC,
Gincel D, Blakeley J, Brem H and Rothstein JD: Efficacy of local
polymer-based and systemic delivery of the anti-glutamatergic
agents riluzole and memantine in rat glioma models. J Neurosurg.
120:854–863. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ribeiro MPC, Nunes-Correia I, Santos AE
and Custódio JBA: The combination of glutamate receptor antagonist
MK-801 with tamoxifen and its active metabolites potentiates their
antiproliferative activity in mouse melanoma K1735-M2 cells. Exp
Cell Res. 321:288–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Albayrak G and Demirtas Korkmaz F:
Memantine shifts cancer cell metabolism via AMPK1/2 mediated
energetic switch in A549 lung cancer cells. EXCLI J. 20:223–231.
2021.PubMed/NCBI
|
|
69
|
Albayrak G, Konac E, Dere UA and Emmez H:
Targeting cancer cell metabolism with metformin, dichloroacetate
and memantine in glioblastoma (GBM). Turk Neurosurg. 31:233–237.
2021.PubMed/NCBI
|
|
70
|
Orgogozo JM, Rigaud AS, Stöffler A, Möbius
HJ and Forette F: Efficacy and safety of memantine in patients with
mild to moderate vascular dementia: A randomized,
placebo-controlled trial (MMM 300). Stroke. 33:1834–1839. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sonkusare SK, Kaul CL and Ramarao P:
Dementia of Alzheimer's disease and other neurodegenerative
disorders-memantine, a new hope. Pharmacol Res. 51:1–17. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yang Z, Zhou X and Zhang Q: Effectiveness
and safety of memantine treatment for Alzheimer's disease. J
Alzheimers Dis. 36:445–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fayein NA, Head MW, Jeanny JC, Courtois Y
and Fuhrmann G: Expression of the chicken cysteine-rich fibroblast
growth factor receptor (CFR) during embryogenesis and retina
development. J Neurosci Res. 43:602–612. 1996. View Article : Google Scholar : PubMed/NCBI
|