1. Introduction
The tumor microenvironment (TME) refers to the
surrounding microenvironment of tumor cells, including surrounding
fibroblasts, immune cells, blood vessels, inflammatory cells,
various signal molecules and the extracellular matrix (ECM). The
TME is a complex environment for the survival and development of
tumor cells. The normal functions of non-malignant cells within the
TME can be effectively 'hijacked' by malignant cells to facilitate
tumor progression. The interaction network between cancer cells and
their microenvironment promotes cell proliferation and
angiogenesis, inhibits cell apoptosis and immune detection, and
activates immune cells to support cell invasion and migration
(1). Given the crucial roles of
the TME in tumorigenesis and tumor progression, it is evident that
the pathways involved serve as potential targets for cancer
treatment (2,3).
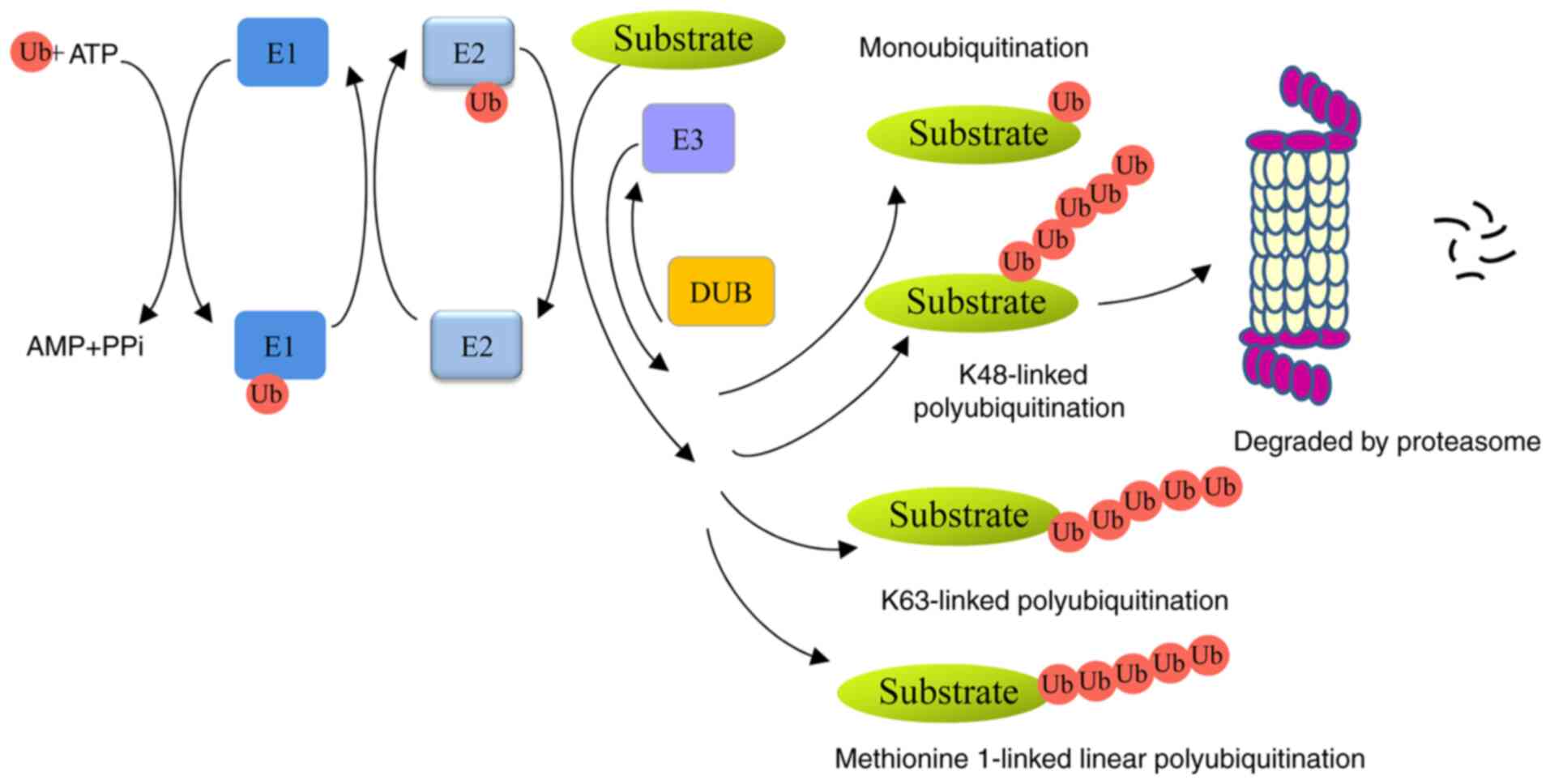
Ubiquitin, a small 76-amino-acid protein, can be
covalently tagged to target proteins by monoubiquitination or
polyubiquitination. A series of enzymes, including
ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes
(E2s) and ubiquitin ligases (E3s), are involved in this process
(4). Polyubiquitin chains, linked
at 48 lysine sites (K48) or K11 sites lead to proteolysis in a 26S
proteasome-dependent manner. Monoubiquitination or K63-linked
polyubiquitin chains are non-proteolytic ubiquitination signals and
participate in autophagy, signal transduction and DNA damage repair
(5,6). In addition, methionine 1-linked
linear ubiquitin chains are assembled by a linear ubiquitin chain
assembly complex (LUBAC), and are involved in inflammation,
immunity and cell death (7,8).
Ubiquitins can be removed from ubiquitinated proteins by
deubiquitinases (DUBs) (Fig. 1).
Members of the family can be divided into six types:
ubiquitin-specific proteases (USPs), ubiquitin carboxyl-terminal
hydrolases, ovarian-tumor proteases, Machado-Joseph disease protein
proteases, JAB1/MPN/MOV34 metalloenzymes, and monocyte chemotactic
protein-induced proteases (9).
Protein ubiquitination is a critical mechanism that
modulates the levels and activities of proteins. This process is
involved in various activities, including protein degradation, DNA
repair activity, gene transcription, and signal transduction
(10). The dysregulation of this
system is closely associated with various human diseases, including
cancers (11). Accumulating
evidence demonstrates that E3 ligases and DUBs participate in
tumorigenesis and progression through various biological processes,
such as the cell cycle, cell proliferation, apoptosis, DNA damage
repair and cell signaling (10,12,13). However, the roles of
ubiquitination in the crosstalk between a tumor and the TME have
rarely been discussed. The present review article comprehensively
discusses the roles of ubiquitination in the interaction between
tumors and the TME, such as the regulation of the hypoxic
environment, angiogenesis and chronic inflammation-mediated tumor
formation, as well as the modulation of the function of
cancer-associated fibroblasts (CAFs) and infiltrating immune cells
(tumor-associated macrophages, T-cells, myeloid-derived suppressor
cells, dendritic cells and natural killer cells). In addition, the
potential targets of the ubiquitination proteasome system (UPS)
within the TME in cancer therapy and their therapeutic effects are
reviewed.
2. Roles of the tumor microenvironment in
tumorigenesis and progression
The TME consists of fibroblasts, endothelial cells,
immune cells and ECM (Fig. 2).
These cells provide support for tumor cells and encourage their
proliferation, invasion and migration.
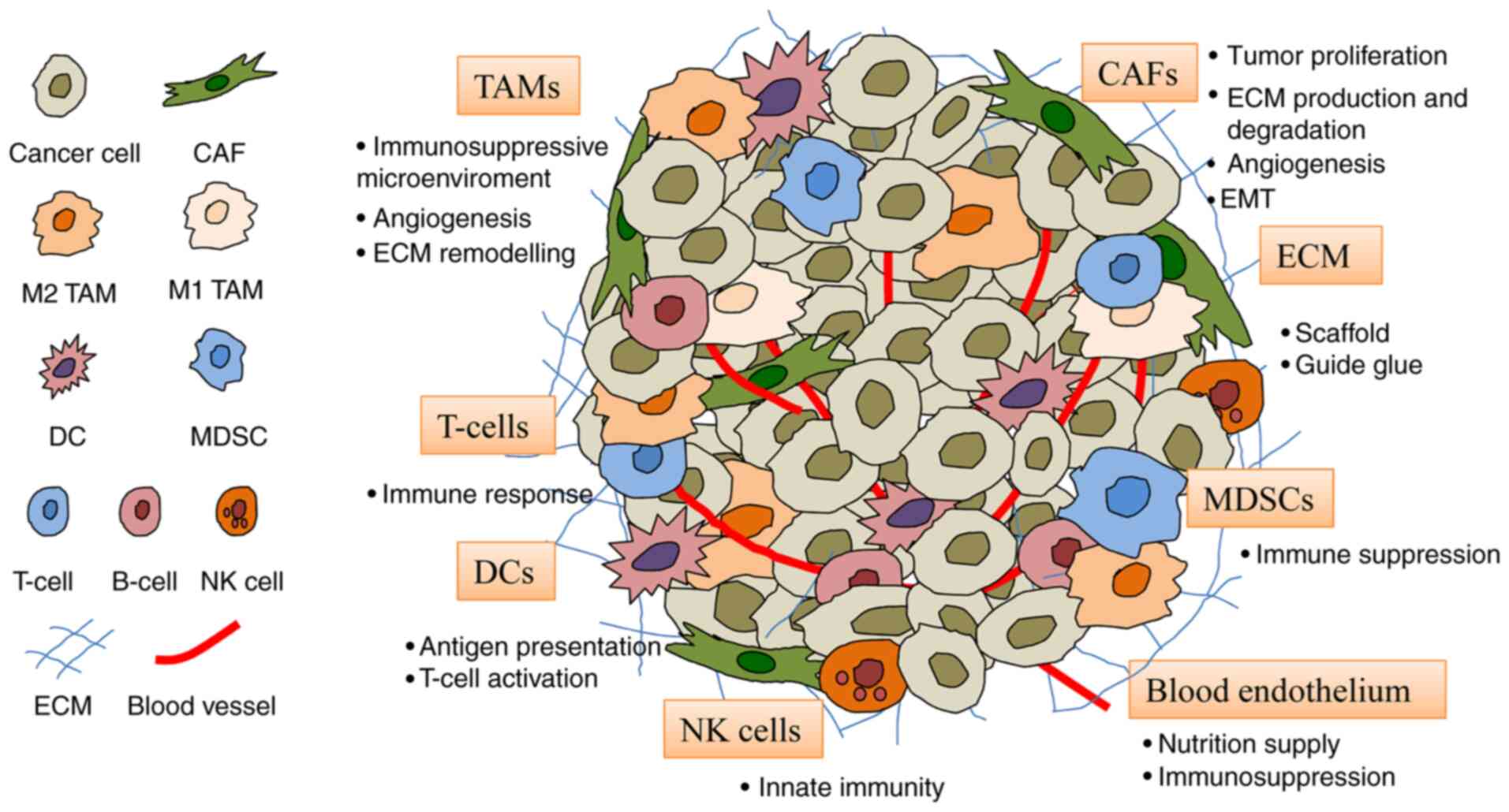 | Figure 2Constituents of the TME. The TME
comprises cells including CAFs, blood endothelial cells, immune
cells (TAMs, DCs, T-cells, MDSCs and NK cells) and the ECM. CAF,
cancer-associated fibroblast; DC, dendritic cell; ECM,
extracellular matrix; MDSC, myeloid-derived suppressor cell; NK,
natural killer; TAM, tumor-associated macrophage; TME, tumor
microenvironment. |
CAFs
CAFs, major components of stromal cells in solid
tumors, play pivotal roles in tumor progression via multiple
mechanisms, including paracrine and direct interactions, immune
response regulation and ECM remodeling (14). CAFs can secrete tumor-promoting
growth factors [e.g., transforming growth factor (TGF)-β,
fibroblast growth factor (FGF), vascular endothelial growth factor
(VEGF) and endothelial growth factor (EGF)], inflammatory cytokines
[e.g., interleukin (IL)-6 and IL-8] and chemokines (e.g., CXCL8 and
CXCL12), subsequently mediating cell proliferation, angiogenesis,
migration, immunogenicity and drug resistance. As the main source
of collagen-producing cells, CAFs synthesize the ECM and directly
communicate with cancer cells, as well as other TME cells, such as
endothelial and inflammatory cells. CAFs can also degrade and
remodel the ECM by producing matrix metalloproteinases (MMPs) and
then contribute to tumor invasion and metastasis in various cancers
(15).
Tumor-associated endothelial cells
Angiogenesis plays a crucial role in tumor growth
and metastasis, and is regarded as a prognostic marker of cancer
(16). Endothelial cells and
vascular blood vessel cells not only supply nutrition to tumor
tissues, but also promote immune tolerance. For example,
endothelial cells reduce the expression of E-selectin,
intercellular adhesion molecule 1 and 2, and vascular cell adhesion
molecule 1, and also impair the recruitment of cytotoxic T-cells to
tumor lesions (17). In addition,
the endothelial cells also express inhibitory molecules, such as
programmed cell death-ligand1 (PD-L1) and PD-L2, and release
soluble factors (prostaglandin E2, IL-6, TGF-β and VEGF) to
regulate T-cell responses (18-20). Moreover, endothelial cells allow
immunosuppressive myeloid cells to migrate from the blood to the
tumor, thus impairing the antitumor immune response (21).
Immune cells
The immune-associated cells in the TME include
macrophages, T-cells, myeloid-derived suppressor cells (MDSCs),
dendritic cells (DCs) and natural killer (NK) cells (22).
Tumor-associated macrophages (TAMs) are the main
immune cell population of the TME in the majority of cancers.
Macrophages are polarized by various microenvironments to form
heterogeneous populations and can be divided into pro-inflammatory
M1 macrophages or anti-inflammatory M2 macrophages. M1 TAMs are
activated by Toll-like receptors (TLRs) or Th1 cytokines, such as
tumor necrosis factor α (TNF-α), interferon (IFN)-γ, and
colony-stimulating factor 2, and exert antitumor effects. M2 TAMs
are regulated by IL-4, IL-10, IL-13, TGF-β or prostaglandin E2, and
produce anti-inflammatory cytokines, including TGF-β and IL-10,
with anti-inflammatory and tumor-promoting effects (23). In tumor tissues, the majority of
macrophages are polarized into M2 macrophages, and subsequently
promote angiogenesis, inhibit the antitumor immune response,
support tumor growth and secrete different factors to regulate ECM
remodeling, tumor cell motility and intravasation (24).
T-cells play vital roles in the host defense against
cancer. External signals activate immature T-cells and trigger
their immune function. Cytotoxic CD8+ T-cells and
CD4+ T-helper 1 cells are the main factors of antitumor
immunity (25). However, the
immunosuppressive TME can induce T-cell dysfunction. Generally,
T-cells express programmed cell death protein-1 (PD-1) and tumor
cells express PD-L1. Their interaction leads to T-cell suppression
and tumor cell survival, that is, cancer cell immunosuppression. In
addition, Foxp3+ T-regulatory cells (Tregs) are another
T-cell subpopulation involved in the escape of tumor cells.
MDSCs, a heterogeneous population of immature
myeloid cells, are the key mediators of immune suppression in the
TME. MDSCs inhibit the T-cell immune response mainly by expressing
arginase-1, inducible nitric oxide synthase and anti-inflammatory
cytokines, such as IL-10 and cyclooxygenase-2 (COX2) (26). MSDCs also express PD-L1 and
cytotoxic-T-lymphocyte-antigen-4, and possess the ability to
suppress CD8+ T-cell function (27).
DCs are natural immune cells and effective
antigen-presenting cells, and participate in immune-mediated cancer
elimination via antigen presentation and T-cell activation. In
addition, DCs also provide antigens, co-stimulatory molecules, and
cytokines for T-cell activation and differentiation, thereby
inducing the immune response (28).
NK cells are cytotoxic effectors against cancer in
innate immunity that are able to recognize and eliminate tumor
cells without antigen presentation. NK cells express a variety of
receptors with activated or inhibitory function and NK cell
function is determined by the balance of signal input from these
receptors. Cytokines (IL-12, IL-15 and IL-18) and transcription
factors (e.g., T-bet) promote the activation of NK cells and
produce IFN-γ, which plays a critical role in innate and adaptive
immune responses (29).
ECM
The ECM is a protein network around tumor cells and
TME cells. The main components of the ECM include collagen,
proteoglycans, laminin and fibronectin. The ECM not only serves as
a scaffold for tissue, but also provides key biochemical and
biomechanical cues to guide cell growth, survival, migration and
differentiation. In addition, the ECM also plays a role in the
regulation of vascular development and immune function (30). Therefore, the ECM also plays a
vital role in tumor proliferation, invasion, metastasis and
angiogenesis.
3. Roles of ubiquitination in modulating the
crosstalk between tumors and the TME
A number of studies have indicated that the UPS
regulates tumorigenesis and progression by modulating the crosstalk
between a tumor and the TME (Fig.
3). Some of the E3 ligases and their substrates involved are
presented in Table I; DUBs are
presented in Table II.
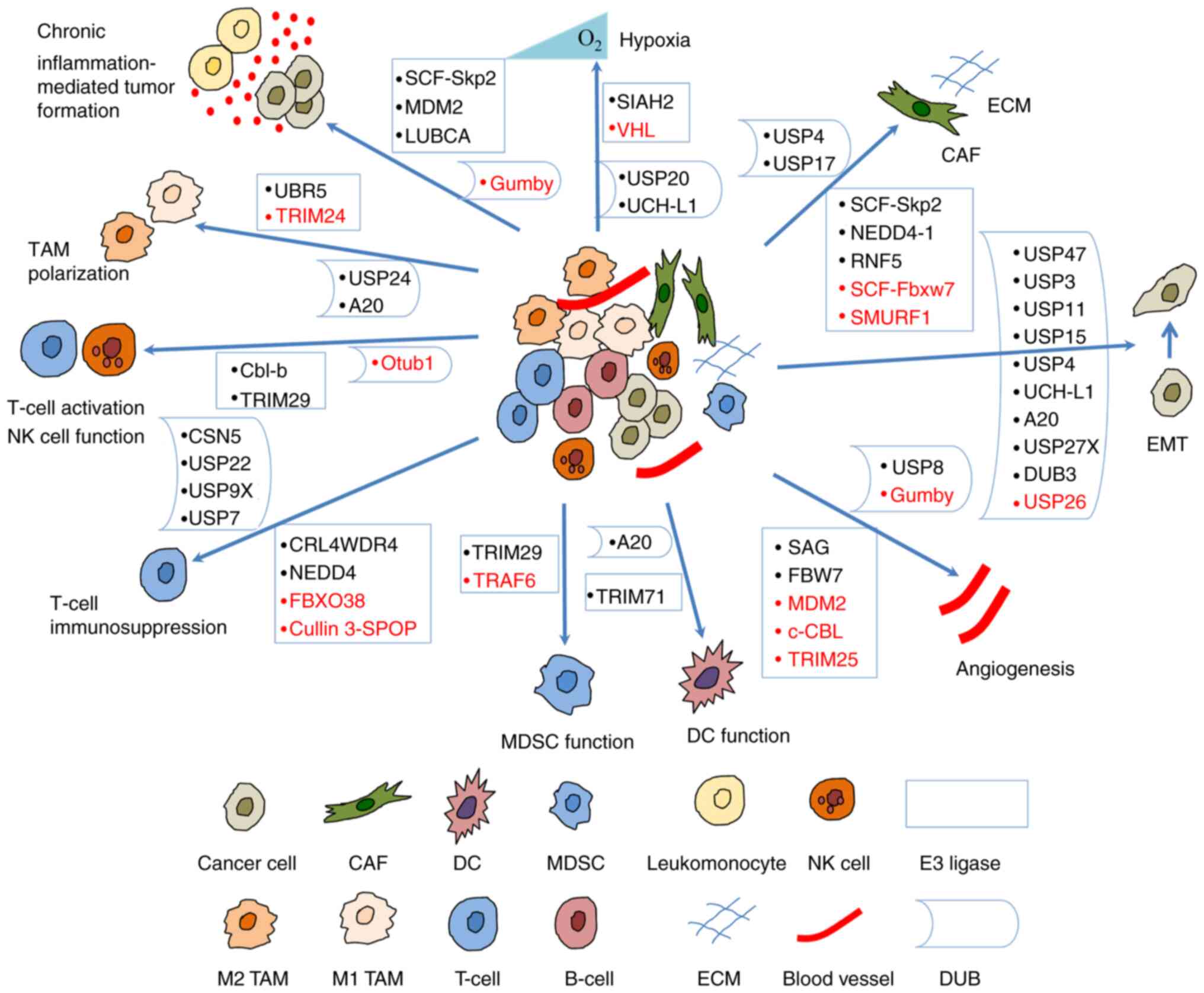 | Figure 3Ubiquitination regulates the
crosstalk between tumors and the tumor microenvironment. E3 ligases
and DUBs regulate hypoxic microenvironment, angiogenesis, EMT,
chronic inflammation-mediated tumor formation, and the crosstalk
between CAFs, TAMs, T-cells, MDSCs, DCs, NK cells and tumor cells.
Some of them suppress tumorigenesis (shown in red) and others
promote tumorigenesis (shown in black). CAF, cancer-associated
fibroblast; DC, dendritic cell; DUB, deubiquitinase; EMT,
epithelial-mesenchymal transition; MDSC, myeloid-derived suppressor
cell; NK, natural killer; TAM, tumor-associated macrophage; UBR5,
ubiquitin protein ligase E3 component n-recognin 5; TRIM,
tripartite motif containing; USP, ubiquitin-specific protease;
Cbl-b, Casitas B-lineage lymphoma proto-oncogene-b; CSN5, COP9
signalosome 5; Otub1, OTU domain-containing ubiquitin
aldehyde-binding protein 1; CRL4WDR4, WD repeat 4-containing
CUL-RING ubiquitin ligase 4; FBXO38, F-box protein 38; cullin
3-SPOP, cullin 3-speckle-type pox virus and zinc finger protein;
TRAF, TNF receptor associated factor; SIAH2, Siah E3 ubiquitin
protein ligase 2; VHL, von Hippel Lindau; UCH-L1, ubiquitin
C-terminal hydrolase-L1; SCF, Skp-Cul1-F-box; Skp2, S-phase
kinase-associated protein-2; RNF5, ring finger protein 5; Fbxw7,
F-box and WD repeat domain containing 7; SMURF1, SMAD Specific E3
ubiquitin protein ligase 1. |
 | Table ISome E3 ligases involved in the
crosstalk between tumors and the TME. |
Table I
Some E3 ligases involved in the
crosstalk between tumors and the TME.
| Biological
process | E3 ligase | Target | Category | Cancer/cell
type | (Refs.) |
|---|
| Hypoxia | VHL | HIF-α | Tumor
suppressor | | (31) |
| SIAH2 | NRF1 | Oncogene | Breast cancer | (36) |
| Angiogenesis | MDM2 | HIF-α | Tumor
suppressor | | (39) |
| SAG | | Oncogene | B16F10
melanoma | (42,110) |
| FBW7 | Notch4 | Oncogene | | (45) |
| c-CBL | β-catenin | Tumor
suppressor | | (48) |
| TRIM25 | SP1 | Tumor
suppressor | Gastric cancer | (52) |
| Chronic
inflammation-mediated tumor formation | SCF-Skp2 | FOXO1 | Oncogene | Hepatoma, prostate
cancer, lymphoma | (58) |
| MDM2 | FOXOs | Oncogene | | (59) |
| SCF-Skp2 | E2F1 | | HepG2 cells | (65) |
| LUBAC | NEMO | Oncogene | B-cell leukemia,
breast cancer | (71,72) |
| CAF | SCF-Fbxw7 | Notch1 | Tumor
suppressor | BMSCs, melanoma
cells | (90) |
| RNF5 | PTEN | Oncogene | Pancreatic ductal
adenocarcinoma cells | (84) |
| ECM | SCF-Skp2 | SP1 | Oncogene | Oral squamous cell
carcinoma, lung cancer cells | (96,97) |
| NEDD4-1 | MT1-MMP | Oncogene | breast cancer cell
MCF-7 | (103) |
| TAM
polarization | TRIM24 | CBP | Tumor
suppressor | Macrophages | (105) |
| UBR5 | | Oncogene | Ovarian cancer | (111) |
| T-cell
immunosuppression | CRL4WDR4 | PML | Oncogene | Lung cancer | (117) |
| FBXO38 | PD-1 | Tumor
suppressor | Colorectal
carcinoma, hepatocellular carcinoma | (118) |
| Cullin 3-SPOP | PD-L1 | Tumor
suppressor | Prostate cancer,
kidney cancer | (120,121) |
| NEDD4 | GITR | Oncogene | Melanoma | (132) |
| MDSC
immunosuppression | TRAF6 | STAT3 | Tumor
suppressor | Lung cancer | (134) |
| MDSC
differentiation | TRIM29 | STING | Oncogene | Nasopharyngeal
carcinoma | (137) |
| Myofibroblastic
activation | SMURF1 | TGF-βRII | Tumor
suppressor | Hepatic stellate
cells | (164) |
| Antigen
presentation | TRIM71 | Rb, p53 | Oncogene | Breast cancer | (141) |
| NK cell
activation | Cbl-b | Tyro3, Mer,
Axl | Oncogene | Melanoma, breast
cancer | (143) |
| NK cell
function | TRIM29 | TAB2 | Oncogene | NK cells | (146) |
 | Table IISome DUBs involved in the crosstalk
between tumors and the TME. |
Table II
Some DUBs involved in the crosstalk
between tumors and the TME.
| Biological
process | DUB | Target | Category | Cancer/cell
type | (Refs.) |
|---|
| Hypoxia | USP20 | HIF-1α | Oncogene | | (33) |
| UCH-L1 | HIF-1α | Oncogene | Breast cancer, lung
cancer | (34) |
| Chronic
inflammation mediated tumor formation | OUTLIN/gumby | NEMO | Tumor
suppressor | Hepatocellular
carcinoma | (73) |
| Angiogenesis | USP8 | VEGFR2 | Oncogene | Endothelial
cells | (37) |
| OUTLIN/gumby | dishevelled2,
β-catenin | Tumor
suppressor | Endothelial
cells | (49,50) |
| EMT | USP47 | Snail | Oncogene | Colorectal cancer
cells | (35) |
| USP3 | SUZ12 | Oncogene | Gastric cancer | (76) |
| USP11 | Snail 1 | Oncogene | Ovarian cancer | (77) |
| USP11 | TGF-β RII | Oncogene | Breast cancer | (78) |
| USP26 | SMAD7 | Tumor
suppressor | Glioblastoma | (79) |
| USP15 | TGF-β RI | Oncogene | Glioblastoma | (80) |
| USP4 | TGF-β RI | Oncogene | Breast cancer | (81) |
| UCH-L1 | | Oncogene | DU145 prostate
cancer cells | (82) |
| A20 | Snail 1 | Oncogene | Breast cancer | (83) |
| USP27X | Snail 1 | Oncogene | Breast and
pancreatic cancer | (75) |
| DUB3 | Snail | Oncogene | Breast cancer | (87) |
| ECM | USP4, USP17 | 6myc-HAS2 | Oncogene | Breast and lung
cells | (91) |
| T-cell
activation | Otub1 | AKT | Tumor
suppressor | B16F10
melanoma | (113) |
| T-cell
immunosuppression | CSN5 | PD-L1 | Oncogene | Breast cancer | (122) |
| USP22 | CSN5, PD-L1 | Oncogene | Non-small cell lung
cancer, liver caner | (123,124) |
| USP9X | PD-L1 | Oncogene | Oral squamous cell
carcinoma | (126) |
| USP7 | Foxp3+,
Tip60 | Oncogene | Colorectal cancer
HCT116, human prostate cancer PC-3 | (129) |
| TAM
polarization | USP24 | β-TrCP, p300 | Oncogene | Lung cancer | (107) |
| A20 | ERα | Oncogene | Endometrial
cancer | (109) |
| DCs | A20 | TRAF6, TRAF2,
cIAP1 | Oncogene | Multiple
myeloma | (140) |
Roles of ubiquitination in the regulation
of hypoxic microenvironments
Hypoxia is a main feature of numerous solid tumors
due to rapid tumor growth. To overcome low oxygen levels, tumor
cells activate a variety of survival pathways and induce
angiogenesis and metastasis. Hypoxia-inducible factor-1 (HIF-1) is
a transcriptional activator that is related to cellular adaptive
responses to hypoxia.
The protein stability of HIFα is mainly regulated by
the oxygen-dependent hydroxylation of two proline residues. Under
normoxic conditions, hydroxylation is carried out by three proline
4-hydroxylase domain enzymes, i.e., PHD1-3 or EGL nine
(Caenorhabditis elegans) homologs (EGLN1-3), that can sense
cellular oxygen. The hydroxylation of proline residues is used to
label the HIFα subunits for ubiquitination and subsequent
proteasome degradation by the von Hippel Lindau (VHL)-containing E3
ligase complex. In addition, HIF-1α and HIF-2α can also be
hydroxylated by HIF at a C-terminal asparagine residue. As a
result, the recruitment of transcriptional coactivator p300 is
blocked, and the transactivation activity of HIF is suppressed. By
contrast, HIFs become stable and active under hypoxic conditions
due to the inactivation of oxygen-dependent hydroxylases.
In the presence of oxygen, the tumor suppressor
protein VHL E3-ubiquitin ligase mediates the degradation of HIFα
subunits via ubiquitination (31). Several DUBs can reverse the action
of VHL on HIFα subunits (32).
For example, USP20 (VDU2) was the first DUB shown to maintain the
stability of HIF-1α and upregulate the expression of HIF-1α target
genes, such as VEGF (33).
Ubiquitin C-terminal hydrolase-L1 (UCH-L1) has been shown to
increase the stability of HIF-1α as a deubiquitinating enzyme and
promote distant tumor metastases under hypoxic conditions in breast
and lung cancers (34).
In the majority of tumors, hypoxia also induces
epithelial-mesenchymal transition (EMT) and this enables tumor
cells to acquire migratory and invasive capabilities.
Ubiquitin-specific protease (USP)47 has been identified in the
process of EMT. In colorectal cancer cells, USP47 is activated by
the hypoxia-mediated Sox9. USP47 subsequently stabilizes Snail by
deubiquitination and promotes EMT and tumor metastasis (35).
Dysfunctional mitochondria are a critical factor for
tumorigenesis, and a low mitochondrial gene expression is
associated with a poor prognosis of patients with breast cancer.
The hypoxia-induced E3 ligase SIAH2 promotes nuclear respiratory
factor 1 (NRF1) degradation through the proteasomal pathway under
hypoxic conditions, thus leading to the reduction of
nuclear-encoded mitochondrial gene expression, metabolic
reprogramming, tumor-associated macrophage polarization and
pro-tumor immune response. Therefore, the inhibition of NRF1
degradation may represent a potential therapeutic strategy against
cancer (36).
Roles of ubiquitination in
angiogenesis
Tumor cells produce angiogenic and proliferation
factors, such as VEGFA, to stimulate the development of new blood
vessels (16). Some
transcriptional factors (e.g., HIF) and signaling pathways (e.g.,
Notch and Wnt) are involved in angiogenesis. Of these, VEGF and its
associated receptors are significant players. It has been shown
that VEGFR2 ubiquitination is required for degradation through the
lysosomes or recycling back to the plasma membrane.
Deubiquitinating enzyme USP8 regulates the stability of VEGFR2,
thus affecting the endothelial cell response and vascular
physiology (37).
As a transcriptional regulator of VEGF, HIF-1α plays
a crucial role in angiogenesis. Its stability and functionality are
generally affected by ubiquitination. The deubiquitinating enzymes
USP20, USP8 and UCHL1 can modulate the HIF-1α level and stability,
and regulate VEGF expression (32). Lysine-specific demethylase 1
(LSD1), a flavin adenine dinucleotide dependent demethylase for
lysine (K) 4 and 9 of histone H3, has been shown to demethylate
non-histone proteins. LSD1 can also demethylate HIF-1α at lysine
(K) 391 and suppress PHD2-induced HIF-1α hydroxylation, thereby
preventing HIF-1α from ubiquitin-mediated protein degradation
(38). The transcription factor
TAp73 opposes HIF-1 activity by recruiting mouse double minute 2
homolog (MDM2) and promotes HIF-1α polyubiquitination and
degradation, thereby inhibiting angiogenesis and tumor progression
(39).
COX-2 also plays a role in angiogenesis and its
expression often upregulated in various types of cancer. COX-2
increases the production of VEGF via the extracellular
signal-regulated kinase (ERK)/HIF-1α/VEGFA pathway. The centromere
protein U, a centromere component for mitosis, is involved in the
tumorigenesis of multiple cancer types. This protein has recently
been shown to promote angiogenesis by inhibiting the
ubiquitin-proteasomal degradation of COX-2 (40).
Cullin-RING ligase is a multi-complex E3 ubiquitin
ligase and controls a number of critical biological processes. As a
RING protein, sensitive to apoptosis gene (SAG) is critical for the
activity of Cullin-RING ligase. SAG is upregulated by the
transcription factors AP-1 and HIF-1 in response to reactive oxygen
species, mitogen and hypoxia in several cancer types; these factors
are associated with a poor prognosis. Stress-inducible SAG can
recruit other components of Cullin-RING E3s to promote the
ubiquitination and degradation of various substrates (41). Tan et al (42) found that SAG is necessary for
endothelial cell migration and tube formation in vitro and
tumor angiogenesis in vivo in a murine B16F10 melanoma
model. Furthermore, the endothelial deletion of SAG E3 ubiquitin
ligase was shown to block tumor angiogenesis (42).
Endothelial Notch signaling limits angiogenesis by
preventing budding and catheter formation (43). Tip cells secrete the Notch ligand
Dll4 which binds to the Notch receptor on stalk cells. The
activation of Notch leads to VEGFR2 transcriptional inhibition and
subsequently suppresses endothelial cell proliferation (44). Ubiquitin ligase F-box and WD
repeat domain-containing 7 (FBW7) plays a positive role in
angiogenesis by inducing Notch ubiquitination and degradation
(45).
The Wnt signaling pathway participates in multiple
aspects of development and is necessary for appropriate vascular
growth in mammals (46). This
pathway promotes the accumulation of β-catenin and enhances the
transcription of pro-angiogenic genes, such as VEGF and IL-8
(47). The stability of β-catenin
can also be regulated by ubiquitination. A previous study revealed
that c-CBL induced the ubiquitination of nuclear β-catenin and
promoted its degradation, thereby negatively regulating
angiogenesis (48). In addition,
LUBAC induces linear ubiquitin on β-catenin, facilitates its
proteasomal degradation, and DUB OTULIN/gumby interacts with LUBAC
to remove linear ubiquitin chains and stabilize β-catenin.
Moreover, gumby interacts with disheveled 2 and promotes Wnt
signaling during angiogenesis. The loss of the DUB activity of
gumby can comprise angiogenesis (49,50).
The process of angiogenesis includes the generation
of new vessels, vascular basement membrane degradation, blood
vessel ECM remodeling, endothelial cell migration, proliferation
and the new generation of matrix components. MMPs play key roles in
tumor rupture, tumor neovascularization and subsequent metastasis
(51). MMP2, a member of the MMPs
family, has been proven to be overexpressed in gastric cancer and
promote the proliferation, angiogenesis, and migration of gastric
cancer cells. Tripartite motif containing (TRIM)25, an E3 ubiquitin
ligase, promotes the ubiquitination of transcription factor SP1 at
K610, further suppressing the expression of MMP2 and inhibiting
angiogenesis in gastric cancer (52).
Roles of ubiquitination in chronic
inflammation-mediated tumor formation
Chronic inflammation is a significant risk factor
for cancer development and ~20% of human cancers are related to
chronic inflammation (53).
Cancer usually occurs in inflammatory tissues, thus indicating the
roles of local inflammation in the initiation and progression of
cancers. Ubiquitination plays an intrinsic role in the chronic
inflammatory TME by regulating transcriptional factors and
cytokines (54).
Forkhead box O (FOXO) transcription factors play a
crucial role in immune homeostasis, particularly FOXO1 and FOXO3a,
which regulate the development and differentiation of lymphocytes,
along with the quiescence of primary T-cells. A number of chronic
inflammatory processes appear to be related to the loss of activity
of these transcription factors (55). There is increasing evidence to
suggest that FOXOs function as tumor suppressors and participate in
a variety of biological processes, including apoptosis,
inflammation regulation, cell cycle checkpoint, oxidative stress
resistance and DNA repair (56).
The dysregulation of FOXO proteins is usually related to the
carcinogenesis, progression and chemoresistance of several human
tumors (57). S-phase
kinase-associated protein-2 (Skp2), an Skp-Cul1-F-box (SCF) UPS
protein subunit, is responsible for the ubiquitination/degradation
of FOXO1 in hepatoma and prostate cancer cells, as well as a mouse
lymphoma model (58). In
addition, MDM2 is a critical universal ubiquitin E3 ligase for the
proteasomal degradation of FOXO family members (59).
Cytokines also mediate the crosstalk between
malignant cells and surrounding cells in the TME. Pro-inflammatory
cytokines, such as TNF-α, IL-1β, IL-6, TGF-β and the
anti-inflammatory cytokine, IL-10, have been shown to be involved
in the occurrence and maintenance of cancer (54). The ubiquitin mechanism is
triggered under infection-inflammation conditions and promotes the
formation of cancer by regulating multiple cytokines. For example,
the SAG ubiquitin-proteasome system is significantly upregulated in
infected tissues and participates in chronic inflammation-induced
cancer via the ubiquitination of key apoptotic factors
(pro-apoptotic SARM and Noxa) and by controlling the ratio of
pro-and anti-apoptosis factors (60,61).
The melanoma differentiation-associated gene-7
(MDA-7)/IL-24 belongs to the anti-inflammatory IL-10 family and
exhibits anticancer effects by inducing apoptosis, inhibiting
angiogenesis and stimulating an immune response (62). The MDA-7/IL-24 protein is
ubiquitinated, and its degradation is controlled by the 26S
proteasome in ovarian and lung cancer cells (63). Inhibiting the degradation of
MDA-7/IL-24 can enhance antitumor activity.
The E2 promoter binding factor (E2F) family of
transcription factors is an essential mediator of cell
proliferation and DNA-damage-induced apoptosis and can exhibit both
tumorigenic and tumor suppressor activity. E2F is overexpressed in
nasopharyngeal carcinoma (NPC) cells and activates the expression
of IL-6, upregulates inflammatory signaling, and promotes malignant
proliferation and tumorigenesis in NPC cells (64). The UPS plays a key role in
regulating E2F function. In the viral HBx microenvironment, HBx can
compete with E2F1 to bind Skp2 E3 ligase, thus resulting in the
accumulation of E2F1 and histone methyltransferase mixed-lineage
leukemia 1. E2F1 stimulates cell proliferation in early-stage
tumors and triggers apoptosis in late-stage tumors. Notably, the
differential promoter occupancy of mixed lineage leukemia 1 (with a
co-activator or co-repressor) appears to specify the paradoxical
functions of E2F1 during the early and late stages of tumor
development (65).
Inflammation-related cancers are also characterized
by mutagenic DNA lesions (66).
Innate immune surveillance systems can recognize cytosolic DNA
through the cyclic GMP-AMP synthase (cGAS)/stimulator of IFN genes
(STING) pathway and activate downstream non-canonical NF-κB
signaling to monitor cell damage and fight against pathogen
infection (67). Aberrant DNA
fragmentation is common in cancer cells due to abnormal chromosome
structure and genome instability (68). Therefore, avoiding this monitoring
process is crucial for tumorigenesis. Recently, Wu et al
(69) revealed that receptor
tyrosine kinase human epidermal growth factor receptor 2 (HER2)
effectively inhibited cGAS/STING signaling and prevented cancer
cells from undergoing apoptosis. HER2 is closely related to STING
and recruits AKT1 (also known as PKB) to directly phosphorylate
TANK-binding kinase 1 (TBK1), thus preventing the TBK1-STING
association and TBK1 K63-linked ubiquitination, thereby weakening
the STING signal (69).
Chronic inflammation and NF-κB activation are
related to the development and progression of cancer. The linear
ubiquitination of the NF-κB essential modulator mediated by LUBAC
is required for NF-κB activation (70). Thus, the dysregulation of linear
ubiquitin signaling is associated with cancer development. For
example, increased LUBAC expression enhances NF-κB activation, thus
accelerating the accumulation of somatic mutations and
lymphomagenesis (71). Enhanced
linear ubiquitination drives breast cancer development in mice
(72). Consistently, DUB OTULIN
(gumby) negatively regulates linear ubiquitin signaling and its
deficiency in hepatocytes causes the development of hepatocellular
carcinoma (73). These results
suggest that LUBAC inhibition may be an effective treatment
strategy for certain types of cancer.
Roles of ubiquitination in the crosstalk
between CAFs and tumor cells
CAFs are the most abundant stromal cells in the TME
and regulate tumor cells and other stromal cells via cell-cell
contacts and secrete regulatory factors, notably TGF-β, IL-6 and
CC-chemokine ligand (CCL)2; they can also synthesize and remodel
the ECM. Thus, CAFs are closely related to cancer progression.
TGF-β, a pleiotropic cytokine, plays essential roles
in tumor cell EMT, migration and invasion. EMT is a
transdifferentiation process from polarized epithelial cells to
motile mesenchymal cells; this is associated with the
downregulation of E-cadherin and the upregulation of the
mesenchymal protein vimentin. TGF-β binding leads to TGF-β type I
and type II receptor (TGF-βRI and TGF-βRII) heterocomplex
activation. SMAD2/3 are then phosphorylated and shuttled to the
nucleus with SMAD4 to regulate the expression of EMT transcription
factors (74). EMT is driven by
Snail, zinc-finger E-box-binding and basic helix-loop-helix (e.g.,
Twist) transcription factors; this inhibits epithelial marker genes
and activates genes related to the mesenchymal phenotype. Due to
the continuous ubiquitination of several F-box-specific E3 ligases,
Snail1 has a short lifespan in normal epithelial cells; however,
its degradation is prevented in cancer cells and activated
fibroblasts. Snail1 is required for the activation of CAFs
(75).
Studies have reported that several USP family
members are involved in mediating TGF-β-induced EMT. For instance,
USP3 expression is upregulated in gastric cancer cells induced by
TGF-β. USP3 interacts with and stabilizes the suppressor of zeste
12 protein homolog via deubiquitination, thus enhancing tumor cell
EMT and metastasis (76). Snail
and Twist are important transcriptional factors in EMT. USP11 is
upregulated and promotes EMT by deubiquitinating Snail in ovarian
cancer (77). USP11 also
regulates TGF-β-induced epithelial-mesenchymal plasticity and
promotes human breast cancer metastasis by stabilizing TGF-βRII
(78). USP26 has been found to
negatively regulate the TGF-β signaling pathway by deubiquitination
and stabilizing SMAD7 in glioblastoma (79). USP15 enhances the tumorigenic
effects of TGF-β in glioblastoma (80), while USP4 promotes TGF-β-induced
EMT and cell migration in breast cancer. Both of these maintain the
stability of TGF-βRI (81). Of
note, Jang et al (82)
found that UCH-L1 was specifically and highly expressed in the
metastatic DU145 prostate cancer cell line, but not in the benign
or weakly metastatic prostate cancer cells. Furthermore, UCH-L1 was
found to promote prostate cancer metastasis via EMT induction
(82). Recently, A20 was found to
be overexpressed in human basal-like breast cancers and mediates
TGFβ1-induced EMT in breast cancers by monoubiquitylating Snail1.
The knockdown of A20 reduced lung cancer metastasis in mouse
engrafts and orthotopic breast cancer models, thus suggesting roles
for A20 and monoubiquitylated Snail1 in metastasis (83). Deubiquitinase USP27X has been
shown to be upregulated by TGF-β during EMT and to maintain Snail1
stability in breast and pancreatic cancer. The inhibition of USP27X
leads to Snail1 destabilization, suppresses EMT and renders tumor
cells sensitive to chemotherapy (75). Collectively, these results
indicate that these deubiquitinases regulate EMT and may serve as a
target for the inhibition of CMF-induced tumor invasion and
chemoresistance.
TGF-α, produced by CAFs, can induce EMT and promote
tumor growth and metastasis via EGFR. Recently, a novel E3 ligase,
RNF5, has been identified to regulate the crosstalk of pancreatic
CAFs and pancreatic ductal adenocarcinoma. The ablation of the
Hedgehog signaling gene Smoothened in pancreatic CAFs can activate
Glycogen synthase kinase 3β and lead to PTEN phosphorylation. RNF5
has been found to ubiquitinate PTEN and induce PTEN degradation. As
a result, AKT and the TGF-α promoter are activated. TGF-α produced
by CAFs is known to undergo crosstalk with adjacent tumor cells and
to promote tumor growth via the activation of EGFR (84).
Recently, IL-6 was also identified as a potential
mediator of the crosstalk between tumor cells and CAFs in
esophageal cancer (85). Chronic
inflammation leads to the activation of normal fibroblasts which
become CAFs. CAFs produce more pro-tumorigenic cytokines, including
IL-6. The direct interaction of CAFs enables tumor cells to secrete
a higher level of IL-6. IL-6 activates its receptor on tumor cells
and CAFs to promote tumor cell proliferation and invasion, and by
suppressing apoptosis via the STAT3 and ERK1/2 signaling pathways.
Thus, IL-6 signaling has become a therapeutic target for several
types of cancer. Wu et al (86) suggested that IL-6 secreted by CAFs
promoted EMT and the metastasis of gastric cancer via the
JAK2/STAT3 signaling pathway. Recently, DUB3 has been identified as
a deubiquitinase of Snail; furthermore, IL-6 promotes tumor cell
EMT by inducing DUB3 expression and by stabilizing Snail in breast
cancer (87).
Chemokines are secreted proteins and participate in
inflammatory and immunoregulatory processes. The increased
concentration of chemokines is associated with a poor prognosis
(88). Liu et al (89) demonstrated that CAFs produce
higher levels of CCL2, CCL5, CCL7 and CXCL16 than peri-tumor
fibroblasts in hepatocellular carcinoma. CCL2 and CCL5 promoted
cell migration via the Hedgehog pathway; CCL7 and CXCL16 enhanced
the migration and invasion of hepatocellular carcinoma cells via
the TGF-β pathway (89).
Oncosuppressor protein Fbxw7 is a substrate receptor for the SCF
ubiquitin ligase complex, and Fbxw7 deficiency has been shown to
give rise to various tumors in mice. The loss of Fbxw7 in bone
marrow-derived stromal cells promotes cancer metastasis by
impairing the degradation of Notch1 and increasing the production
of chemokine CCL2. As the precursors of CAFs, bone marrow-derived
stromal cells are major components of the TME and represent the
sources of CCL2 output (90).
The ECM in tumors changes dynamically and CAFs are
the main contributors to ECM stiffness and degradation. The
production and degradation of the ECM are regulated by
ubiquitination. Hyaluronan, a ubiquitous glycosaminoglycan that is
highly expressed in the ECM, accumulates in rapidly remodeling
tissues, such as breast cancer. Hyaluronan-synthesizing enzyme 2
(HAS2) catalyzes the production of hyaluronan. USP4 and USP17 have
been identified to reduce the ubiquitination of 6myc-HAS2 and
modulate the stability and activity of HAS2. USP4 and USP17 are
upregulated in malignant lines. USP17 maintains the level of
6myc-HAS2 protein by reducing the polyubiquitination of 6myc-HAS2.
USP4 preferentially removes the monoubiquitination of 6myc-HAS2 and
regulates the activity of HAS2 (91).
MMPs are a family of zinc-dependent endopeptidases
that selectively degrade the components of the ECM (92). MMP-2 is upregulated in a number of
cancer types and can reduce cell-cell adhesion, promote tumor
invasion and EMT, and can contribute to tumor aggressiveness and a
poor prognosis (93-95). Skp2 is usually overexpressed in
human cancers and increases the expression of MMP-2 by enhancing
transcription factor Sp1 activity (96,97). The expression of MMP-2 can also be
regulated by epigenetic mechanisms and aberrant epigenetic
regulations of MMP-2 are known to be involved in cancer progression
(98). For example, CpG
methylation of the MMP-2 promoter is associated with the invasive
phenotype. Chromobox 6 is a component of the polycomb repressive
complex. As a transcriptional repressor, Chromobox 6 inhibits the
expression of MMP-2 in non-invasive cells. The enhanced
ubiquitination and degradation of chromo box 6 increases the
expression of MMP-2 and enhances the invasion of mesothelioma
(99). USP2 is overexpressed in
various human cancers and has been regarded as a therapeutic target
in cancer (100). Previously,
USP2 was revealed to enhance tumor migration and invasion by
increasing MMP-2 activity in metastatic triple-negative breast
cancer (101).
Membrane type 1-matrix metalloproteinase (MT1-MMP)
mediates cancer cell invasion by degrading the basement membrane
and ECM, as well as by inducing cell migration. The
MT1-MMP-mediated activation of TGF-β signaling enables the
autocrine and paracrine-mediated induction of EMT (102). Eisenach et al (103) revealed that the ubiquitin E3
ligase NEDD4-1 mediated monoubiquitination within the MT1-MMP
intracellular domain and was involved in MT1-MMP trafficking and
modulated cellular invasion through type I collagen matrices.
Roles of ubiquitination in the crosstalk
between TAM and tumor cells
During tumorigenesis, TAMs are usually regulated by
environmental factors to present the M2 state; this inhibits the
cytotoxic function of immune cells and impairs antitumor immunity
(104). Macrophage M2
polarization involves the activation of STAT6 and STAT6
acetylation, which is mediated by the acetyltransferase
CREB-binding protein (CBP), to suppress macrophage M2 polarization.
CBP-associated E3 ligase TRIM24 induces CBP ubiquitination, which
facilitates the recruitment of CBP to STAT6, promotes STAT6
acetylation, and inhibits macrophage M2 polarization (105). M2 macrophages promote cancer
cell metastasis by secreting growth factors and cytokines, such as
IL-6 (106). Recently, Wang
et al (107) reported
that USP24 was highly expressed in the late stages of lung cancer
and promoted lung cancer metastasis by inducing IL-6 expression.
USP24 can stabilize β-TrCP, promote IκB degradation in lung cancer
cells, and upregulate NF-κB in M2 macrophages, thus leading to the
increased expression of IL-6. Furthermore, USP24 stabilizes p300
and reduces the levels of DNMT1, thereby increasing IL-6
transcription in M2 macrophages and lung cancer cells (107).
In estrogen-driven endometrial cancer, the dominant
M2 macrophages in endometrial cancer lesions are CD163+
macrophages which play essential roles in carcinogenesis (108). CD163+ macrophages can
upregulate the ubiquitin-editing enzyme A20 via cytokines, such as
IL-1α, IL-17A and TNF-α in endometrial lesions. A20 maintains
estrogen receptor α protein level and enhances endometrial cancer
cell proliferation by its deubiquitinase activity (109). Thus, CD163+
macrophages sensitize endometrial cancer cells to estrogen via
A20.
Ubiquitin protein ligase E3 component n-recognin 5
(UBR5, also known as EDD) is overexpressed in multiple cancer
types, especially breast cancer and ovarian cancer, and is
associated with a poor prognosis (110). UBR5 mediates tumor-associated
macrophage recruitment and activation via chemokines and cytokines.
In addition, UBR5 promotes ovarian tumor cell adhesion and spheroid
formation and suppresses apoptosis by regulating p53 levels.
Targeting tumor-derived UBR5 impairs TAM recruitment and TME
immunosuppression and enhances the therapeutic effects of
immunotherapy (111).
Roles of ubiquitination in the crosstalk
between T-cells and tumor cells
Ubiquitination is a crucial mechanism that regulates
the crosstalk between T-cells and tumor cells, including T-cell
immune responses against cancer and tumor immunosuppression.
CD8+ T-cells are primary cytotoxic effector cells of the
immune system against cancer, which specifically depend on
IL-15-mediated AKT signaling for homeostasis (112). DUB Otub1 deubiquitinates AKT and
inhibits the IL-15-stimulated activation of AKT, negatively
regulating the homeostasis and activation of CD8+
T-cells in immune responses. As previously demonstrated in a mouse
model, Otub1 deficiency sensitized CD8+ T-cells and
enhanced tumor rejection (113).
The linker for activation of T-cells (LAT) is a transmembrane
molecule and the key for T-cell activation (114). The ubiquitylation-resistant form
of LAT is more stable than that of wild-type LAT in cells and
enhances T-cell signaling. Therefore, blocking LAT ubiquitination
may be a promising strategy with which to enhance the function of
effector T-cells (115).
Promyelocytic leukemia (PML), a pleiotropic tumor
suppressor, is usually downregulated in a number of cancer types.
Ubiquitin-mediated degradation is a key mechanism for PML
downregulation in tumors (116).
WD repeat 4-containing CUL-RING ubiquitin ligase 4 is responsible
for PML destruction in lung cancer. The WDR4/PML axis induces
several oncogenes and promotes lung cancer progression. It also
elevates intratumoral Tregs and M2-like macrophages, reduces
CD8+ T-cell numbers and induces tumor immunosuppression
(117).
Surface PD-1 molecules can be ubiquitinated and
degraded by proteasomes in activated T-cells. E3 ubiquitin ligase
FBXO38 interacts with PD-1 directly and mediates PD-1
ubiquitination and proteasome degradation. Thus, FBXO38 regulates
T-cell antitumor immunity (118). The immunosuppressive activity of
PD-L1 can also be modulated by ubiquitination and N-glycosylation.
Glycogen synthase kinase (GSK)3β phosphorylates PD-L1 and
facilities the proteasome degradation of PD-L1 by β-TrCP.
N-glycosylation influences the structure and function of PD-L1 and
antagonizes GSK3β binding; thus, only non-glycosylated PD-L1 forms
a complex with GSK3β and β-TrCP and undergoes fast protein
degradation (119). In human
prostate cancer and kidney cancer, cullin 3-speckle-type pox virus
and zinc finger protein has been reported to target PD-L1 for
ubiquitination and degradation (120,121).
The TME can induce cancer immunosuppression by
upregulating PD-L1 protein expression. DUB COP9 signalosome 5
(CSN5) inhibits the ubiquitination and degradation of PD-L1,
subsequently enhancing its interaction with PD-L1 to escape T cell
immune surveillance (122).
USP22 is a novel regulator of PD-L1, which deubiquitinates CSN5 and
regulates the protein levels of PD-L1 via the USP22-CSN5-PD-L1
axis. CSN5 can also directly deubiquitinate PD-L1 and maintains its
stability in human non-small cell lung cancer and liver cancers
(123,124). USP22 is overexpressed and
promotes tumor progression in multiple tumor types; the deletion of
USP22 in pancreatic tumor cells promotes the infiltration of
T-cells and NK cells, thus improving the response to combined
immunotherapy (125). Emerging
data have demonstrated that USP9X is highly expressed in oral
squamous cell carcinoma tissues and stabilizes PD-L1 by
deubiquitination. Reducing USP9X expression also blocks oral
squamous cell growth (126).
Another study demonstrated that USP9X-deficient T-cells were
hyperproliferative and caused spontaneous lupus-like autoimmunity
and lymphoproliferative diseases in aged mice. Moreover, the mRNA
expression of PD-1 was markedly upregulated in cells in which USP9X
was knocked down. As such, mice with USP9X depletion have an
increased number of PD-1 expressing T-cells (127).
Foxp3+ Tregs play an indispensable role
in immunosuppression and lead to tumor immune evasion (128). The Treg lineage-specific
transcription factor Foxp3+, and the histone
acetyltransferase Tip60, are pivotal to the development and
maintenance of the Treg lineage. USP7 maintains Treg functions by
stabilizing the expression of Tip60 and Foxp3 (129). Glucocorticoid-induced TNF
receptor (GITR), a member of the TNF receptor superfamily, is
highly expressed in Tregs, whereas it is expressed at low levels in
naive and memory T-cells (130).
Its agonist antibody DTA-1 can enhance the CD8+ effector
T-cell to Treg ratio by depleting Tregs, as well as activating
CD8+ effector T-cells by promoting GITR oligomerization
(131). The E3 ligase NEDD4 can
also mediate the ubiquitination and degradation of GITR; this
protein is overexpressed in metastatic melanoma and inhibits T-cell
antitumor immunity. Therefore, NEDD4 can be regarded as a novel
prognostic biomarker and therapeutic target for melanoma (132).
Roles of ubiquitination in the crosstalk
between MDSCs and tumor cells
MDSCs are crucial immunosuppressive cells and
inhibit T-cell proliferation and antitumor responses in the TME.
The expansion and function of MDSCs are modulated by the
transcription factor, STAT3; the activation of STAT3 is controlled
by post-translational modifications, such as ubiquitination and
phosphorylation. E3 ligase TNF receptor associated factor (TRAF)6
promotes STAT3 phosphorylation and activation by inducing the
K63-linked polyubiquitination of STAT3 (133). Thus, TRAF6 plays a critical role
in regulating the function of MDSCs. Song et al (134) reported that TRAF6 was highly
expressed in the MDSCs of patients with lung cancer and TRAF6
knockdown impaired the immunosuppressive effects of MDSCs in
vitro and in Lewis xenograft mice.
On the other hand, STING has been shown to suppress
the differentiation of MDSCs by inhibiting STAT3 activation in the
NPC. The underlying mechanism is that STING promotes suppressor of
cytokine signaling 1 (SOCS1) expression via the STING/TBK1/IRF3
axis and SOCS1 prevents STAT3 phosphorylation by binding to its SH2
domain in NPC cells and MDSCs (135). Galectins are a family of lectins
that mediate a variety of biological functions in tumor progression
and immune surveillance (136).
Galectin-9 is usually upregulated in tumor tissues and recruits E3
ligase TRIM29 to mediate the ubiquitination and degradation of
STING in malignant NPC cells. As a result, galectin-9 activates
STAT3, enhances the production of suppressive cytokines and
chemokines including IL-1β and IL-6, and promotes MDSC
differentiation and expansion in patients with NPC (137).
Roles of ubiquitination in the crosstalk
between DCs and tumor cells
DCs are potent antigen-presenting cells with a
number of different subtypes, such as monocyte DCs, conventional
DCs and plasmacytoid DCs (pDCs) (138). DCs can specifically recognize,
process and present cancer antigens, activate CD8+ and
CD4+ T-cells, and regulate immune responses. Thus, DCs
are key players in tumor immunity (139). Human pDCs selectively express
intracellular TLR7 and TLR9 to sense viral stimulation and produce
a large number of IFNs. Thus, pDCs play vital roles in antiviral
responses. In multiple myeloma, tumor cells educate pDCs to induce
tumorigenesis and escape immune surveillance via the E-cadherin
(CDH1) pathway. The homophilic interaction between myeloma cells
and pDCs activates CDH1 and then upregulates ubiquitin-editing
enzyme A20 expression. A20 negatively regulates NF-κB signaling by
inhibiting E3 ligases TRAF6, TRAF2 and cIAP1, and suppresses TLR
pathways and the production of IFN-α in pDCs (140).
To escape immune surveillance, malignant cells may
develop multiple mechanisms, such as reducing antigenicity and
establishing an immunosuppressive microenvironment. Long-non-coding
RNA long intergenic non-coding RNA for kinase activation (LINK-A)
is upregulated in triple-negative breast cancers and is associated
with a poor prognosis of patients with breast cancer. LINK-A
inhibits protein kinase A (PKA) phosphorylation and the
PKA-mediated phosphorylation of the E3 ubiquitin ligase TRIM71. As
a result, LINK-A enhances TRIM71-induced ubiquitination and the
degradation of peptide-loading complex, thereby downregulating
antigen presentation and innate tumor suppression (141).
Roles of ubiquitination in the crosstalk
between NK cells and tumor cells
NK cells play key roles in the innate antitumor
immune response, and ubiquitination is involved in regulating its
function. NK cells express the tyrosine kinase receptors, Tyro3,
Mer and Axl, which play essential roles in NK cell functional
maturation (142). E3 ligase
Casitas B-lineage lymphoma proto oncogene-b (Cbl-b), also expressed
in NK cells, participates in controlling NK cell antitumor
responses via the ubiquitination of Tyro3, Mer and Axl. The
suppression of Cbl-b promotes NK cells to reject melanoma and
breast metastases (143,144).
It is well known that IL-15 contributes to NK cell
survival and function. It was recently demonstrated by Wang et
al (145) that
deubiquitination is involved in the regulation of events downstream
of IL-15. IL-15 activates AKT, which maintains the stability of
X-box binding protein 1 through deubiquitination. Accumulated X-box
binding protein 1 in the nucleus recruits the transcription factor
T-bet and increases the expression of granzyme B and IFN-γ
expression, thus enhancing NK cell function (145). On the other hand, DUB Otub1
induces AKT deubiquitination, attenuating its binding to
phosphatidylinositol 3,4,5-trisphosphate, thereby inhibiting AKT
activation. Thus, Otub1 controls the NK cell activation induced by
IL-15 (113).
IL-12 and IL-18 induce NK cell activation; TGF-β
activated kinase 1 binding protein 2 (TAB2) is known to be involved
in the process as a key adaptor protein. Dou et al (146) reported that E3 ligase TRIM29 was
upregulated in activated NK cells and negatively regulated IFN-γ
production by ubiquitinating and degrading TAB2. NK cell functions
can be enhanced upon IL-12 and IL-18 stimulation when TRIM29 is
deficient. Thus, TRIM29 plays a key role in regulating NK cell
activity.
4. Application of UPS modulators in the TME
in tumor therapy
Ubiquitination is involved in multiple processes in
the crosstalk between a tumor and the TME. Therefore, this process
may provide potential targets for cancer treatment (147). Accumulating evidence indicates
that modulating the ubiquitination of substrates in the TME may
provide a promising strategy for the development of anticancer
drugs. Some UPS modulators and their targets are presented in
Table III.
 | Table IIIAnticancer compounds in clinical
trials modulating the ubiquitination of substrates in the TME. |
Table III
Anticancer compounds in clinical
trials modulating the ubiquitination of substrates in the TME.
| Drug/compound | Substrate | Target | Cancer type | Research stage | (Refs.) |
|---|
| Bortezomib | Proteasome | | Multiple
myeloma | Approved | (148) |
| Delanzomib | Proteasome | | Multiple
myeloma | Approved | (149) |
| RA190 | Proteasome | | Multiple myeloma,
ovarian cancer, hepatocellular carcinoma, intrahepatic
cholangiocarcinoma | Research | (150-155) |
| Thymoquinone | HIF-1α | HIF-1α | Renal cancer | Phase II | (157) |
| Decursin | HIF-1α | HIF-1α | Lung cancer, colon
cancer | Approved | (158) |
| JP3 | TRIM25 | MMP-2 | Gastric cancer | Research | (159) |
| MLN4924 | NEDD8-activating
enzyme | IκBα | Chronic lymphocytic
leukemia B-cells | Phase II | (161) |
| IQGAP1 | SMURF1 | TGF-βRII | Hepatic stellate
cell | Research | (164) |
| Gefitinib | EGF signaling | PD-L1 | Breast cancer | Approved | (119) |
| Dinaciclib | CDK1, 2, 5, 9 | PD-L1 | Lung
adenocarcinoma | Phase II | (166) |
| Berberine | CSN5 | PD-L1 | Non-small cell lung
cancer | Approved | (167) |
| MLN4924 | NEDD8-activiting
enzyme | IκBα | Chronic lymphocytic
leukemia B-cells | Phase II | (161) |
| P217564 | Tip60, Foxp3 | USP7 | Multiple myeloma,
B-cell leukemia, neuroblastoma | Research | (129) |
| HOIPIN-8 | HOIP | NF-κB | Lung carcinoma
cells | Research | (163) |
Proteasome inhibitors
As a vital component of the UPS, the proteasome has
been successfully used as a target for cancer treatment. Bortezomib
and Dalanzomib are proteasome inhibitors approved for recurrent
refractory multiple myeloma (148,149). These drugs induce the apoptosis
of refractory multiple myeloma cells, suppress NF-kB activation and
the production of cytokines (e.g., IL-6, insulin-like growth factor
1and VEGF), and inhibit angiogenesis in the TME. The clinical
success of existing proteasome inhibitors encourages great efforts
to discover more UPS inhibitors.
The bis-benzylidine piperidone RA190 is a novel
proteasome inhibitor, targeting 19S proteasome-associated ubiquitin
receptor RPN13 [also known as adhesion-regulating molecule-1
(ADRM1)] and exerts anticancer effects in multiple types of cancer
(150). For example, RA190
induces the apoptosis and inhibits the proliferation of multiple
myeloma cells in vitro and in vivo by exerting
anti-myeloma activity (151).
STAT3 plays a key role in modulating MDSC immunosuppression and its
activation can be inhibited by endoplasmic reticulum stress
(152). RA190 induces
endoplasmic reticulum stress by accumulating polyubiquitinated
proteins. As a result, RA190 reduces STAT3 expression, and reduces
the levels of arginase, inducible nitric oxide synthase, and IL-10
in MDSCs, suppresses MDSC immunosuppression, and kills ovarian
cancer cells in mice bearing syngeneic ovarian tumors (153). ADRM1 is overexpressed in
intrahepatic cholangiocarcinoma and is associated with a poor
prognosis of patients with intrahepatic cholangiocarcinoma. RA190
inhibits the cell cycle and induces apoptosis in intrahepatic
cholangiocarcinoma by targeting ADRM1 (154). Recently, Soong et al
(155) reported that RA190 could
kill hepatocellular carcinoma by blocking IκBα degradation and
inhibiting NF-κB-mediated inflammation in vitro and in
vivo. Moreover, RA190 overcomes bortezomib resistance and
exhibits synergistic anticancer activity with bortezomib,
lenalidomide or pomalidomide in the treatment of multiple myeloma,
or with sorafenib in the treatment of hepatocellular carcinoma
(155).
HIF-1α inhibitors
Increased levels of HIF-1α in tumors are associated
with angiogenesis, aggressive tumor growth and a poor prognosis
(156). Lee et al
(157) identified thymoquinone
as a HIF-1α inhibitor from a 502 natural compound library.
Thymoquinone promoted HIF-1α degradation by suppressing
HSP90-mediated stabilization, altering the proteasome-dependent
degradation pathway, and blocking anaerobic glycolysis, and
selectively induced tumor cell apoptosis in hypoxic renal cancer
(157).
Decursin, an active compound extracted from the
roots of Angelica gigas, can enhance hydroxylation and
ubiquitination and consequently promote HIF-1α degradation under
hypoxic conditions. As a result, decursin reduces the expression of
HIF-1α target genes, such as VEGF and CXCR4, inhibits cell
proliferation and invasion, and induces the apoptosis of A549 human
lung cancer and HCT116 human colon cancer cells. Moreover, decursin
reduces PD-L1 expression, increases the number of
tumor-infiltrating T-lymphocytes (CD3+ and
CD8+ cells), suppresses Tregs and MDSCs, and improves
immune responses in a Lewis lung carcinoma allograft mouse model
(158). Decursin is a novel
HIF-1α inhibitor exhibiting anticancer and anti-angiogenic
activities.
Targeting MMP-2
MMP-2 is closely related to invasion, angiogenesis
and metastasis in several types of cancer. Therefore, MMP-2 is a
promising target for tumor treatment. It is known that E3 ubiquitin
ligase TRIM25 can promote the ubiquitination and degradation of
SP1, further suppressing MMP-2 expression and inhibiting
angiogenesis in gastric cancer. Recently, Chen et al
(159,160) developed an MMP2-targeted
peptide, JP3, based on the functional fragment from JWA protein, a
microtubule-binding protein with inhibitory activity against SP1.
JP3 maintains TRIM25 stability by phosphorylation at Ser12 and
reducing its ubiquitination. Consequently, JP3 suppresses gastric
cancer angiogenesis, growth and metastasis in vivo without
observable toxic side-effects (159).
Targeting the NF-κB pathway
The activation of the NF-κB pathway leads to cell
survival and chemoresistance in chronic lymphocytic leukemia
B-cells. The ubiquitination of IκBα may partially account for the
constitutive activation of the NF-κB pathway. The small-molecule
inhibitor of NEDD8-activating enzyme, MLN4924, blocks Cullin-RING
ubiquitin ligase activity, thus leading to the accumulation of IκBα
and the inactivation of the NF-κB canonical pathway. Therefore,
treatment with MLN4924 could disrupt NF-κB activation, induce the
chronic apoptosis of lymphocytic leukemia B-cells and prevent
stroma-mediated resistance (161). In addition, the ubiquitin ligase
complex LUBAC consists of three subunits: HOIL-1L, HOIP and
SHARPIN. Katsuya et al (162) developed a small molecule
inhibitor of HOIP, JTP-0819958 (HOIPIN-1), which inhibits LUBAC
activity and suppresses NF-κB activation in vitro. These
authors also synthesized seven derivatives (HOIPIN-2-8) of HOIPIN-1
and found that HOIPIN-8 could suppress LUBAC- and TNF-α-induced
NF-κB activation with high efficiency and without cytotoxicity
(163).
Targeting the TGF-β pathway
In the TME, TGF-β induces the activation of
pericytes and other mesenchymal stromal cells into tumor-associated
myofibroblasts by activating the TGF-β receptor heterocomplex. IQ
motif containing GTPase activating protein 1has been shown to
suppress this process in the liver by recruiting the E3 ligase
SMURF1 to ubiquitinate and induce the degradation of the TGF-β
receptor II (TGF-βRII) in hepatic stellate cells (164).
Modulating immunotherapy
The PD-1/PD-L1 axis is a key determinant of
physiological immune homeostasis. Inhibition of PD-1/PD-L1 is a
promising therapeutic strategy which restores the immune system in
cancer patients. EGF stabilizes PD-L1 via GSK3β inactivation in
basal-like breast cancer. The inhibition of EGF signaling by
gefitinib destabilizes PD-L1, enhances antitumor T-cell immunity,
and improves the therapeutic efficacy of PD-1 blockade in syngeneic
mouse models (119).
Cyclin-dependent kinase 5 (CDK5) plays a critical role in driving
tumor formation and development and is highly expressed in various
lung cancer cells (165). CDK5
depletion promotes E3 ligase TRIM21-mediated PD-L1 ubiquitination
and degradation, and enhances CD8+ T-cell-mediated
immune responses in lung adenocarcinoma. A specific inhibitor of
CDK5 has yet to be developed. Dinaciclib is a potent inhibitor of
CDK1, 2, 5 and 9, and can be downregulated by the PD-L1 protein in
lung adenocarcinoma cells (166). Berberine, an anti-inflammatory
drug from Chinese medicine, has been found to induce PD-L1
degradation by binding to and inhibiting CSN5 deubiquitination
activity in the non-small cell lung cancer lines, H460, H1975, H358
and HCC827. Consequently, berberine activates tumor-infiltrating
T-cells, decreases the number of Tregs and MSDCs, and inhibits
tumor growth in Lewis xenograft mice. Therefore, berberine can be
regarded as a tumor immunotherapeutic agent (167).
The accumulation of Foxp3+ Tregs in the
TME is related to tumor immune evasion and a poor prognosis in
several solid tumors. The current strategies used to block Treg
function are not Treg-specific and exhibit limited and transient
efficacy. It has been demonstrated that USP7 is essential for Treg
function by stabilizing the expression of Tip60 and Foxp3.
Therefore, the pharmacological inhibition of USP7 is a promising
strategy which may be used to inhibit Treg function and promote
antitumor immunity. The P5091 series of USP7 inhibitors are known
to exhibit direct anti-tumor activity in vivo using
xenograft models, although the mechanism involved is unclear
(168). The second-generation
USP7 inhibitor, P217564, selectively modifies the active site of
USP7 and exerts a durable inhibitory effect on USP7. As a result,
P217564 promotes the ubiquitination and degradation of Foxp3 and
Tip60 and impairs Treg functions. Moreover, USP7 regulates a number
of other tumor-associated proteins, such as E3 ligase HDM2, FOXO4,
PTEN, claspin and UHRF1 (169);
thus, inhibiting USP7 has the potential for direct antitumor
activity. In this regard, the inhibition of USP7 may provide dual
antitumor activities: direct tumoricidal effects and
immune-mediated tumor elimination. USP7 selective inhibitors have
been found to significantly repress the growth of multiple tumor
xenograft models in immunodeficient mice, including multiple
myeloma, B-cell leukemia and neuroblastoma (129).
It has been reported that cisplatin can promote the
immunotherapeutic effects of NK cells on HCC by reducing the
expression of the androgen receptor (AR) and by upregulating the
expression of AR-UL16-binding protein 2 (ULBP2). ULBP2 is a major
natural killer group 2 member D ligand, which facilitates the
activation of NK cells. AR binds to the ULBP2 promoter and
negatively regulates the expression of ULBP2. Cisplatin reduces the
levels of AR by suppressing AR expression through increased
mir-34a-5p expression, or by inducing ubiquitination-mediated AR
degradation, thereby increasing ULBP2 expression (170). This finding may provide a
potential method for the combination of traditional chemotherapy
and immunotherapy to control liver cancer.
5. Conclusions and future perspectives
Ubiquitination plays a vital role in the crosstalk
between a tumor and the TME to regulate hypoxic environments,
angiogenesis, chronic inflammation-mediated tumor formation, and
control the functions of CAFs and infiltrating immune cells. The
dysregulation of ubiquitylation is closely associated with the
development of various human cancers. Therefore, the UPS has been
regarded as a promising therapeutic target for novel anti-cancer
drugs. Currently, some ubiquitination modulators in the TME have
been identified for anticancer treatment.
It remains a significant challenge to identify
specific small molecule modulators for different targets. The
development of more specific inhibitors (e.g., CDK5 and USP22
inhibitors) is desirable. Apart from inhibitors, other strategies
may also provide opportunities. For example, PROTAC technologies
provide a strategy with which to target a number of non-druggable
proteins (171). In addition,
the crosstalk of other post-translational modifications, such as
glycosylation and phosphorylation, can affect the ubiquitination
and degradation of proteins (119,159). Thus, ubiquitination can be
modulated by altering the glycosylation or phosphorylation of
target proteins. Furthermore, epigenetic modulators of target
proteins can also be regulated by ubiquitination to control their
expression (99,172). In this regard, epigenetic
modulators, such as CBX6 and CBX4, can also be regarded as
potential targets. A deeper understanding of ubiquitination in the
TME may further provide a larger therapeutic window for tumor
treatment.
Availability of data and materials
Not applicable.
Authors' contributions
XZ wrote and drafted the manuscript. TM revised the
manuscript critically. SC and DL created the figures, and were
involved in the literature search. QP and PW revised and completed
the final version of the manuscript. All authors have read and
approved the final manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Professor Fengtang
Yang, Shandong University of Technology for providing valuable
comments regarding the writing of the manuscript.
Funding
The present study was supported by the Natural Science
Foundation of Shandong Province (grant no. ZR2016CM46) and the
National Natural Science Foundation of China (grant nos. 82173168
and 81702659).
Abbreviations:
|
AR
|
androgen receptor
|
|
CAFs
|
cancer-associated fibroblasts
|
|
Cbl-b
|
Casitas B-lineage lymphoma
proto-oncogene-b
|
|
CBP
|
CREB-binding protein
|
|
CSN5
|
COP9 signalosome 5
|
|
DUB
|
deubiquitinase
|
|
E2F
|
E2 promoter binding factor
|
|
MDA-7
|
melanoma differentiation-associated
gene-7
|
|
MDM2
|
mouse double minute 2 homolog
|
|
MDSCs
|
myeloid-derived suppressor cells
|
|
PD-1
|
programmed cell death protein-1
|
|
PD-L1
|
programmed cell death-ligand 1
|
|
pDCs
|
plasmacytoid DCs
|
|
SCF
|
Skp-Cul1-F-box
|
|
Skp2
|
S-phase kinase-associated
protein-2
|
|
TAMs
|
tumor-associated macrophages
|
|
TME
|
tumor microenvironment
|
|
TNF-α
|
tumor necrosis factor α
|
|
UCH-L1
|
ubiquitin C-terminal hydrolase-L1
|
|
UPS
|
ubiquitination proteasome system
|
|
USPs
|
ubiquitin-specific proteases
|
References
|
1
|
Casey SC, Amedei A, Aquilano K, Azmi AS,
Benencia F, Bhakta D, Bilsland AE, Boosani CS, Chen S, Ciriolo MR,
et al: Cancer prevention and therapy through the modulation of the
tumor microenvironment. Semin Cancer Biol. 35(Suppl): S199–S223.
2015. View Article : Google Scholar
|
|
2
|
Ribas A and Wolchok JD: Cancer
immunotherapy using checkpoint blockade. Science. 359:1350–1355.
2018. View Article : Google Scholar
|
|
3
|
Meng T, Huang R, Jin J, Gao J, Liu F, Wei
Z, Xu X, Chang Z, Lin J, Ta N, et al: A comparative integrated
multi-omics analysis identifies CA2 as a novel target for chordoma.
Neuro Oncol. 23:1709–1722. 2021. View Article : Google Scholar
|
|
4
|
Suryadinata R, Roesley SN, Yang G and
Sarcevic B: Mechanisms of generating polyubiquitin chains of
different topology. Cells. 3:674–689. 2014. View Article : Google Scholar
|
|
5
|
Haglund K, Di Fiore PP and Dikic I:
Distinct monoubiquitin signals in receptor endocytosis. Trends
Biochem Sci. 28:598–603. 2003. View Article : Google Scholar
|
|
6
|
Erpapazoglou Z, Walker O and
Haguenauer-Tsapis R: Versatile roles of k63-linked ubiquitin chains
in trafficking. Cells. 3:1027–1088. 2014. View Article : Google Scholar
|
|
7
|
Gerlach B, Cordier SM, Schmukle AC,
Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H,
Wong WW, et al: Linear ubiquitination prevents inflammation and
regulates immune signalling. Nature. 471:591–596. 2011. View Article : Google Scholar
|
|
8
|
Jahan AS, Elbæk CR and Damgaard RB:
Met1-linked ubiquitin signalling in health and disease:
Inflammation, immunity, cancer, and beyond. Cell Death Differ.
28:473–492. 2021. View Article : Google Scholar
|
|
9
|
Bhattacharya S and Ghosh MK: Cell death
and deubiquitinases: Perspectives in cancer. Biomed Res Int.
2014:4351972014. View Article : Google Scholar
|
|
10
|
Senft D, Qi J and Ronai ZA: Ubiquitin
ligases in oncogenic transformation and cancer therapy. Nat Rev
Cancer. 18:69–88. 2018. View Article : Google Scholar
|
|
11
|
Morrow JK, Lin HK, Sun SC and Zhang S:
Targeting ubiquitination for cancer therapies. Future Med Chem.
7:2333–2350. 2015. View Article : Google Scholar
|
|
12
|
Wang D, Ma L, Wang B, Liu J and Wei W: E3
ubiquitin ligases in cancer and implications for therapies. Cancer
Metastasis Rev. 36:683–702. 2017. View Article : Google Scholar
|
|
13
|
Wei R, Liu X, Yu W, Yang T, Cai W, Liu J,
Huang X, Xu GT, Zhao S, Yang J and Liu S: Deubiquitinases in
cancer. Oncotarget. 6:12872–12889. 2015. View Article : Google Scholar
|
|
14
|
Ishii G, Ochiai A and Neri S: Phenotypic
and functional heterogeneity of cancer-associated fibroblast within
the tumor microenvironment. Adv Drug Deliv Rev. 99(Pt B): 186–196.
2016. View Article : Google Scholar
|
|
15
|
Vosseler S, Lederle W, Airola K,
Obermueller E, Fusenig NE and Mueller MM: Distinct
progression-associated expression of tumor and stromal MMPs in
HaCaT skin SCCs correlates with onset of invasion. Int J Cancer.
125:2296–2306. 2009. View Article : Google Scholar
|
|
16
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar
|
|
17
|
Griffioen AW, Damen CA, Blijham GH and
Groenewegen G: Tumor angiogenesis is accompanied by a decreased
inflammatory response of tumor-associated endothelium. Blood.
88:667–673. 1996. View Article : Google Scholar
|
|
18
|
Rodig N, Ryan T, Allen JA, Pang H, Grabie
N, Chernova T, Greenfield EA, Liang SC, Sharpe AH, Lichtman AH and
Freeman GJ: Endothelial expression of PD-L1 and PD-L2
down-regulates CD8+ T cell activation and cytolysis. Eur
J Immunol. 33:3117–3126. 2003. View Article : Google Scholar
|
|
19
|
Mulligan JK, Day TA, Gillespie MB,
Rosenzweig SA and Young MRI: Secretion of vascular endothelial
growth factor by oral squamous cell carcinoma cells skews
endothelial cells to suppress T-cell functions. Hum Immunol.
70:375–382. 2009. View Article : Google Scholar
|
|
20
|
Mulligan JK and Young MR: Tumors induce
the formation of suppressor endothelial cells in vivo. Cancer
Immunol Immunother. 59:267–277. 2010. View Article : Google Scholar
|
|
21
|
Talmadge JE and Gabrilovich DI: History of
myeloid-derived suppressor cells. Nat Rev Cancer. 13:739–752. 2013.
View Article : Google Scholar
|
|
22
|
Wang J, Li D, Cang H and Guo B: Crosstalk
between cancer and immune cells: Role of tumor-associated
macrophages in the tumor microenvironment. Cancer Med. 8:4709–4721.
2019. View Article : Google Scholar
|
|
23
|
Jayasingam SD, Citartan M, Thang TH, Mat
Zin AA, Ang KC and Ch'ng ES: Evaluating the polarization of
tumor-associated macrophages into M1 and M2 phenotypes in human
cancer tissue: Technicalities and challenges in routine clinical
practice. Front Oncol. 9:15122020. View Article : Google Scholar
|
|
24
|
Laviron M and Boissonnas A: Ontogeny of
tumor-associated macrophages. Front Immunol. 10:17992019.
View Article : Google Scholar
|
|
25
|
Jiang S and Yan W: T-cell immunometabolism
against cancer. Cancer Lett. 382:255–258. 2016. View Article : Google Scholar
|
|
26
|
Gabrilovich DI: Myeloid-derived suppressor
cells. Cancer Immunol Res. 5:3–8. 2017. View Article : Google Scholar
|
|
27
|
Oya Y, Hayakawa Y and Koike K: Tumor
microenvironment in gastric cancers. Cancer Sci. 111:2696–2707.
2020. View Article : Google Scholar
|
|
28
|
Tran Janco JM, Lamichhane P, Karyampudi L
and Knutson KL: Tumor-infiltrating dendritic cells in cancer
pathogenesis. J Immunol. 194:2985–2991. 2015. View Article : Google Scholar
|
|
29
|
Wu SY, Fu T, Jiang YZ and Shao ZM: Natural
killer cells in cancer biology and therapy. Mol Cancer. 19:1202020.
View Article : Google Scholar
|
|
30
|
Wu T and Dai Y: Tumor microenvironment and
therapeutic response. Cancer Lett. 387:61–68. 2017. View Article : Google Scholar
|
|
31
|
Jaakkola P, Mole DR, Tian YM, Wilson MI,
Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji
M, Schofield CJ, et al: Targeting of HIF-alpha to the von
Hippel-Lindau ubiquitylation complex by O2-regulated prolyl
hydroxylation. Science. 292:468–472. 2001. View Article : Google Scholar
|
|
32
|
Mennerich D, Kubaichuk K and Kietzmann T:
DUBs, hypoxia, and cancer. Trends Cancer. 5:632–653. 2019.
View Article : Google Scholar
|
|
33
|
Li Z, Wang D, Messing EM and Wu G: VHL
protein-interacting deubiquitinating enzyme 2 deubiquitinates and
stabilizes HIF-1alpha. EMBO Rep. 6:373–378. 2005. View Article : Google Scholar
|
|
34
|
Goto Y, Zeng L, Yeom CJ, Zhu Y, Morinibu
A, Shinomiya K, Kobayashi M, Hirota K, Itasaka S, Yoshimura M, et
al: UCHL1 provides diagnostic and antimetastatic strategies due to
its deubiquitinating effect on HIF-1α. Nat Commun. 6:61532015.
View Article : Google Scholar
|
|
35
|
Choi BJ, Park SA, Lee SY, Cha YN and Surh
YJ: Hypoxia induces epithelial-mesenchymal transition in colorectal
cancer cells through ubiquitin-specific protease 47-mediated
stabilization of Snail: A potential role of Sox9. Sci Rep.
7:159182017. View Article : Google Scholar
|
|
36
|
Ma B, Cheng H, Mu C, Geng G, Zhao T, Luo
Q, Ma K, Chang R, Liu Q, Gao R, et al: The SIAH2-NRF1 axis
spatially regulates tumor microenvironment remodeling for tumor
progression. Nat Commun. 10:10342019. View Article : Google Scholar
|
|
37
|
Smith GA, Fearnley GW, Abdul-Zani I,
Wheatcroft SB, Tomlinson DC, Harrison MA and Ponnambalam S: VEGFR2
trafficking, signaling and proteolysis is regulated by the
ubiquitin isopeptidase USP8. Traffic. 17:53–65. 2016. View Article : Google Scholar
|
|
38
|
Lee JY, Park JH, Choi HJ, Won HY, Joo HS,
Shin DH, Park MK, Han B, Kim KP, Lee TJ, et al: LSD1 demethylates
HIF1α to inhibit hydroxylation and ubiquitin-mediated degradation
in tumor angiogenesis. Oncogene. 36:5512–5521. 2017. View Article : Google Scholar
|
|
39
|
Amelio I, Inoue S, Markert EK, Levine AJ,
Knight RA, Mak TW and Melino G: TAp73 opposes tumor angiogenesis by
promoting hypoxia-inducible factor 1α degradation. Proc Natl Acad
Sci USA. 112:226–231. 2015. View Article : Google Scholar
|
|
40
|
Pan T, Zhou D, Shi Z, Qiu Y, Zhou G, Liu
J, Yang Q, Cao L and Zhang J: Centromere protein U (CENPU) enhances
angiogenesis in triple-negative breast cancer by inhibiting
ubiquitin-proteasomal degradation of COX-2. Cancer Lett.
482:102–111. 2020. View Article : Google Scholar
|
|
41
|
Sun Y and Li H: Functional
characterization of SAG/RBX2/ROC2/RNF7, an antioxidant protein and
an E3 ubiquitin ligase. Protein Cell. 4:103–116. 2013. View Article : Google Scholar
|
|
42
|
Tan M, Li H and Sun Y: Endothelial
deletion of Sag/Rbx2/Roc2 E3 ubiquitin ligase causes embryonic
lethality and blocks tumor angiogenesis. Oncogene. 33:5211–5220.
2014. View Article : Google Scholar
|
|
43
|
Hasan SS, Tsaryk R, Lange M, Wisniewski L,
Moore JC, Lawson ND, Wojciechowska K, Schnittler H and Siekmann AF:
Endothelial Notch signalling limits angiogenesis via control of
artery formation. Nat Cell Biol. 19:928–940. 2017. View Article : Google Scholar
|
|
44
|
Rabellino A, Andreani C and Scaglioni PP:
Roles of ubiquitination and SUMOylation in the regulation of
angiogenesis. Curr Issues Mol Biol. 35:109–126. 2020. View Article : Google Scholar
|
|
45
|
Tsunematsu R, Nakayama K, Oike Y,
Nishiyama M, Ishida N, Hatakeyama S, Bessho Y, Kageyama R, Suda T
and Nakayama KI: Mouse Fbw7/Sel-10/Cdc4 is required for notch
degradation during vascular development. J Biol Chem.
279:9417–9423. 2004. View Article : Google Scholar
|
|
46
|
Zerlin M, Julius MA and Kitajewski J:
Wnt/Frizzled signaling in angiogenesis. Angiogenesis. 11:63–69.
2008. View Article : Google Scholar
|
|
47
|
MacDonald BT, Tamai K and He X:
Wnt/beta-catenin signaling: Components, mechanisms, and diseases.
Dev Cell. 17:9–26. 2009. View Article : Google Scholar
|
|
48
|
Shivanna S, Harrold I, Shashar M, Meyer R,
Kiang C, Francis J, Zhao Q, Feng H, Edelman ER, Rahimi N and
Chitalia VC: The c-Cbl ubiquitin ligase regulates nuclear β-catenin
and angiogenesis by its tyrosine phosphorylation mediated through
the Wnt signaling pathway. J Biol Chem. 290:12537–12546. 2015.
View Article : Google Scholar
|
|
49
|
Rivkin E, Almeida SM, Ceccarelli DF, Juang
YC, MacLean TA, Srikumar T, Huang H, Dunham WH, Fukumura R, Xie G,
et al: The linear ubiquitin-specific deubiquitinase gumby regulates
angiogenesis. Nature. 498:318–324. 2013. View Article : Google Scholar
|
|
50
|
Wang W, Li M, Ponnusamy S, Chi Y, Xue J,
Fahmy B, Fan M, Miranda-Carboni GA, Narayanan R, Wu J and Wu ZH:
ABL1-dependent OTULIN phosphorylation promotes genotoxic
Wnt/β-catenin activation to enhance drug resistance in breast
cancers. Nat Commun. 11:39652020. View Article : Google Scholar
|
|
51
|
Quintero-Fabián S, Arreola R,
Becerril-Villanueva E, Torres-Romero JC, Arana-Argáez V,
Lara-Riegos J, Ramírez-Camacho MA and Alvarez-Sánchez ME: Role of
matrix metalloproteinases in angiogenesis and cancer. Front Oncol.
9:13702019. View Article : Google Scholar
|
|
52
|
Murray GI, Duncan ME, Arbuckle E, Melvin
WT and Fothergill JE: Matrix metalloproteinases and their
inhibitors in gastric cancer. Gut. 43:791–797. 1998. View Article : Google Scholar
|
|
53
|
Pikarsky E, Porat RM, Stein I, Abramovitch
R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E and
Ben-Neriah Y: NF-kappaB functions as a tumour promoter in
inflammation-associated cancer. Nature. 431:461–466. 2004.
View Article : Google Scholar
|
|
54
|
Chang SC and Ding JL: Ubiquitination and
SUMOylation in the chronic inflammatory tumor microenvironment.
Biochim Biophys Acta Rev Cancer. 1870:165–175. 2018. View Article : Google Scholar
|
|
55
|
Peng SL: Forkhead transcription factors in
chronic inflammation. Int J Biochem Cell Biol. 42:482–485. 2010.
View Article : Google Scholar
|
|
56
|
Huang H and Tindall DJ: Dynamic FoxO
transcription factors. J Cell Sci. 120(Pt 15): 2479–2487. 2007.
View Article : Google Scholar
|
|
57
|
Ramezani A, Nikravesh H and Faghihloo E:
The roles of FOX proteins in virus-associated cancers. J Cell
Physiol. 234:3347–3361. 2019. View Article : Google Scholar
|
|
58
|
Huang H, Regan KM, Wang F, Wang D, Smith
DI, van Deursen JM and Tindall DJ: Skp2 inhibits FOXO1 in tumor
suppression through ubiquitin-mediated degradation. Proc Natl Acad
Sci USA. 102:1649–1654. 2005. View Article : Google Scholar
|
|
59
|
Fu W, Ma Q, Chen L, Li P, Zhang M,
Ramamoorthy S, Nawaz Z, Shimojima T, Wang H, Yang Y, et al: MDM2
acts downstream of p53 as an E3 ligase to promote FOXO
ubiquitination and degradation. J Biol Chem. 284:13987–14000. 2009.
View Article : Google Scholar
|
|
60
|
Chang SC and Ding JL: Ubiquitination by
SAG regulates macrophage survival/death and immune response during
infection. Cell Death Differ. 21:1388–1398. 2014. View Article : Google Scholar
|
|
61
|
Chang SC, Choo WQ, Toh HC and Ding JL:
SAG-UPS attenuates proapoptotic SARM and Noxa to confer survival
advantage to early hepatocellular carcinoma. Cell Death Discov.
1:150322015. View Article : Google Scholar
|
|
62
|
Chada S, Sutton RB, Ekmekcioglu S,
Ellerhorst J, Mumm JB, Leitner WW, Yang HY, Sahin AA, Hunt KK,
Fuson KL, et al: MDA-7/IL-24 is a unique cytokine-tumor suppressor
in the IL-10 family. Int Immunopharmacol. 4:649–667. 2004.
View Article : Google Scholar
|
|
63
|
Gopalan B, Shanker M, Scott A, Branch CD,
Chada S and Ramesh R: MDA-7/IL-24, a novel tumor
suppressor/cytokine is ubiquitinated and regulated by the
ubiquitin-proteasome system, and inhibition of MDA-7/IL-24
degradation enhances the anti-tumor activity. Cancer Gene Ther.
15:1–8. 2008. View Article : Google Scholar
|
|
64
|
Liu P, Zhang X, Li Z, Wei L, Peng Q, Liu
C, Wu Y, Yan Q and Ma J: A significant role of transcription
factors E2F in inflammation and tumorigenesis of nasopharyngeal
carcinoma. Biochem Biophys Res Commun. 524:816–824. 2020.
View Article : Google Scholar
|
|
65
|
Swarnalatha M, Singh AK and Kumar V:
Promoter occupancy of MLL1 histone methyltransferase seems to
specify the proliferative and apoptotic functions of E2F1 in a
tumour microenvironment. Cell Sci. 126(Pt 20): 4636–4646. 2013.
|
|
66
|
Murata M: Inflammation and cancer. Environ
Health Prev Med. 23:502018. View Article : Google Scholar
|
|
67
|
Sun L, Wu J, Du F, Chen X and Chen ZJ:
Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates
the type I interferon pathway. Science. 339:786–791. 2013.
View Article : Google Scholar
|
|
68
|
Bakhoum SF, Ngo B, Laughney AM, Cavallo
JA, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, et
al: Chromosomal instability drives metastasis through a cytosolic
DNA response. Nature. 553:467–472. 2018. View Article : Google Scholar
|
|
69
|
Wu S, Zhang Q, Zhang F, Meng F, Liu S,
Zhou R, Wu Q, Li X, Shen L, Huang J, et al: HER2 recruits AKT1 to
disrupt STING signalling and suppress antiviral defence and
antitumour immunity. Nat Cell Biol. 21:1027–1040. 2019. View Article : Google Scholar
|
|
70
|
Kensche T, Tokunaga F, Ikeda F, Goto E,
Iwai K and Dikic I: Analysis of nuclear factor-κB (NF-κB) essential
modulator (NEMO) binding to linear and lysine-linked ubiquitin
chains and its role in the activation of NF-κB. J Biol Chem.
287:23626–23634. 2012. View Article : Google Scholar
|
|
71
|
Jo T, Nishikori M, Kogure Y, Arima H,
Sasaki K, Sasaki Y, Nakagawa T, Iwai F, Momose S, Shiraishi A, et
al: LUBAC accelerates B-cell lymphomagenesis by conferring
resistance to genotoxic stress on B cells. Blood. 136:684–697.
2020. View Article : Google Scholar
|
|
72
|
Song K, Cai X, Dong Y, Wu H, Wei Y,
Shankavaram UT, Cui K, Lee Y, Zhu B, Bhattacharjee S, et al: Epsins
1 and 2 promote NEMO linear ubiquitination via LUBAC to drive
breast cancer development. J Clin Invest. 131:e1293742021.
View Article : Google Scholar
|
|
73
|
Damgaard RB, Jolin HE, Allison MED, Davies
SE, Titheradge HL, McKenzie ANJ and Komander D: OTULIN protects the
liver against cell death, inflammation, fibrosis, and cancer. Cell
Death Differ. 27:1457–1474. 2020. View Article : Google Scholar
|
|
74
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar
|
|
75
|
Lambies G, Miceli M, Martínez-Guillamon C,
Olivera-Salguero R, Peña R, Frías CP, Calderó I, Atanassov BS, Dent
SYR, Arribas J, et al: TGFβ-Activated USP27X deubiquitinase
regulates cell migration and chemoresistance via stabilization of
snail1. Cancer Res. 79:33–46. 2019. View Article : Google Scholar
|
|
76
|
Wu X, Liu M, Zhu H, Wang J, Dai W, Li J,
Zhu D, Tang W, Xiao Y, Lin J, et al: Ubiquitin-specific protease 3
promotes cell migration and invasion by interacting with and
deubiquitinating SUZ12 in gastric cancer. J Exp Clin Cancer Res.
38:2772019. View Article : Google Scholar
|
|
77
|
Wang W, Wang J, Yan H, Zhang K and Liu Y:
Upregulation of USP11 promotes epithelial-to-mesenchymal transition
by deubiquitinating Snail in ovarian cancer. Oncol Rep.
41:1739–1748. 2019.
|
|
78
|
Garcia DA, Baek C, Estrada MV, Tysl T,
Bennett EJ, Yang J and Chang JT: USP11 enhances TGFβ-Induced
epithelial-mesenchymal plasticity and human breast cancer
metastasis. Mol Cancer Res. 16:1172–1184. 2018. View Article : Google Scholar
|
|
79
|
Kit Leng Lui S, Iyengar PV, Jaynes P, Isa
ZFBA, Pang B, Tan TZ and Eichhorn PJA: USP26 regulates TGF-β
signaling by deubiquitinating and stabilizing SMAD7. EMBO Rep.
18:797–808. 2017. View Article : Google Scholar
|
|
80
|
Eichhorn PJ, Rodó L, Gonzàlez-Juncà A,
Dirac A, Gili M, Martínez-Sáez E, Aura C, Barba I, Peg V, Prat A,
et al: USP15 stabilizes TGF-β receptor I and promotes oncogenesis
through the activation of TGF-β signaling in glioblastoma. Nat Med.
18:429–435. 2012. View Article : Google Scholar
|
|
81
|
Zhang L, Zhou F, Drabsch Y, Gao R,
Snaar-Jagalska BE, Mickanin C, Huang H, Sheppard KA, Porter JA, Lu
CX and ten Dijke P: USP4 is regulated by AKT phosphorylation and
directly deubiquitylates TGF-β type I receptor. Nat Cell Biol.
14:717–726. 2012. View Article : Google Scholar
|
|
82
|
Jang MJ, Baek SH and Kim JH: UCH-L1
promotes cancer metastasis in prostate cancer cells through EMT
induction. Cancer Lett. 302:128–135. 2011. View Article : Google Scholar
|
|
83
|
Lee JH, Jung SM, Yang KM, Bae E, Ahn SG,
Park JS, Seo D, Kim M, Ha J, Lee J, et al: A20 promotes metastasis
of aggressive basal-like breast cancers through
multi-monoubiquitylation of Snail1. Nat Cell Biol. 19:1260–1273.
2017. View Article : Google Scholar
|
|
84
|
Pitarresi JR, Liu X, Avendano A, Thies KA,
Sizemore GM, Hammer AM, Hildreth BE III, Wang DJ, Steck SA, Donohue
S, et al: Disruption of stromal hedgehog signaling initiates
RNF5-mediated proteasomal degradation of PTEN and accelerates
pancreatic tumor growth. Life Sci Alliance. 1:e2018001902018.
View Article : Google Scholar
|
|
85
|
Karakasheva TA, Lin EW, Tang Q, Qiao E,
Waldron TJ, Soni M, Klein-Szanto AJ, Sahu V, Basu D, Ohashi S, et
al: IL-6 mediates cross-talk between tumor cells and activated
fibroblasts in the tumor microenvironment. Cancer Res.
78:4957–4970. 2018. View Article : Google Scholar
|
|
86
|
Wu X, Tao P, Zhou Q, Li J, Yu Z, Wang X,
Li J, Li C, Yan M, Zhu Z, et al: IL-6 secreted by cancer-associated
fibroblasts promotes epithelial-mesenchymal transition and
metastasis of gastric cancer via JAK2/STAT3 signaling pathway.
Oncotarget. 8:20741–20750. 2017. View Article : Google Scholar
|
|
87
|
Wu Y, Wang Y, Lin Y, Liu Y, Wang Y, Jia J,
Singh P, Chi YI, Wang C, Dong C, et al: Dub3 inhibition suppresses
breast cancer invasion and metastasis by promoting Snail1
degradation. Nat Commun. 8:142282017. View Article : Google Scholar
|
|
88
|
Borsig L, Wolf MJ, Roblek M, Lorentzen A
and Heikenwalder M: Inflammatory chemokines and metastasis-tracing
the accessory. Oncogene. 33:3217–3224. 2014. View Article : Google Scholar
|
|
89
|
Liu J, Chen S, Wang W, Ning BF, Chen F,
Shen W, Ding J, Chen W, Xie WF and Zhang X: Cancer-associated
fibroblasts promote hepatocellular carcinoma metastasis through
chemokine-activated hedgehog and TGF-β pathways. Cancer Lett.
379:49–59. 2016. View Article : Google Scholar
|
|
90
|
Yumimoto K and Nakayama KI: Fbxw7
suppresses cancer metastasis by inhibiting niche formation.
Oncoimmunology. 4:e10223082015. View Article : Google Scholar
|
|
91
|
Mehić M, de Sa VK, Hebestreit S, Heldin CH
and Heldin P: The deubiquitinating enzymes USP4 and USP17 target
hyaluronan synthase 2 and differentially affect its function.
Oncogenesis. 6:e3482017. View Article : Google Scholar
|
|
92
|
Stetler-Stevenson WG and Yu AE: Proteases
in invasion: Matrix metalloproteinases. Semin Cancer Biol.
11:143–152. 2001. View Article : Google Scholar
|
|
93
|
Stefanidakis M and Koivunen E:
Cell-surface association between matrix metalloproteinases and
integrins: Role of the complexes in leukocyte migration and cancer
progression. Blood. 108:1441–1450. 2006. View Article : Google Scholar
|
|
94
|
Gontero P, Banisadr S, Frea B and Brausi
M: Metastasis markers in bladder cancer: A review of the literature
and clinical considerations. Eur Urol. 46:296–311. 2004. View Article : Google Scholar
|
|
95
|
Li S and Luo W: Matrix metalloproteinase 2
contributes to aggressive phenotype, epithelial-mesenchymal
transition and poor outcome in nasopharyngeal carcinoma. Onco
Targets Ther. 12:5701–5711. 2019. View Article : Google Scholar
|
|
96
|
Yamada S, Yanamoto S, Naruse T, Matsushita
Y, Takahashi H, Umeda M, Nemoto TK and Kurita H: Skp2 regulates the
expression of MMP-2 and MMP-9, and enhances the invasion potential
of oral squamous cell carcinoma. Pathol Oncol Res. 22:625–632.
2016. View Article : Google Scholar
|
|
97
|
Hung WC, Tseng WL, Shiea J and Chang HC:
Skp2 overexpression increases the expression of MMP-2 and MMP-9 and
invasion of lung cancer cells. Cancer Lett. 288:156–161. 2010.
View Article : Google Scholar
|
|
98
|
Chernov AV, Sounni NE, Remacle AG and
Strongin AY: Epigenetic control of the invasion-promoting
MT1-MMP/MMP-2/TIMP-2 axis in cancer cells. J Biol Chem.
284:12727–12734. 2009. View Article : Google Scholar
|
|
99
|
Sakai K, Nishiuchi T, Tange S, Suzuki Y,
Yano S, Terashima M, Suzuki T and Matsumoto K: Proteasomal
degradation of polycomb-group protein CBX6 confers MMP-2 expression
essential for mesothelioma invasion. Sci Rep. 10:166782020.
View Article : Google Scholar
|
|
100
|
Priolo C, Tang D, Brahamandan M, Benassi
B, Sicinska E, Ogino S, Farsetti A, Porrello A, Finn S, Zimmermann
J, et al: The isopeptidase USP2a protects human prostate cancer
from apoptosis. Cancer Res. 66:8625–8632. 2006. View Article : Google Scholar
|
|
101
|
Qu Q, Mao Y, Xiao G, Fei X, Wang J, Zhang
Y, Liu J, Cheng G, Chen X, Wang J and Shen K: USP2 promotes cell
migration and invasion in triple negative breast cancer cell lines.
Tumour Biol. 36:5415–5423. 2015. View Article : Google Scholar
|
|
102
|
Nguyen HL, Kadam P, Helkin A, Cao K, Wu S,
Samara GJ, Zhang Q, Zucker S and Cao J: MT1-MMP Activation of TGF-β
signaling enables intercellular activation of an
epithelial-mesenchymal transition program in cancer. Curr Cancer
Drug Targets. 16:618–630. 2016. View Article : Google Scholar
|
|
103
|
Eisenach PA, de Sampaio PC, Murphy G and
Roghi C: Membrane type 1 matrix metalloproteinase (MT1-MMP)
ubiquitination at Lys581 increases cellular invasion through type I
collagen. J Biol Chem. 287:11533–11545. 2012. View Article : Google Scholar
|
|
104
|
Noy R and Pollard JW: Tumor-associated
macrophages: From mechanisms to therapy. Immunity. 41:49–61. 2014.
View Article : Google Scholar
|
|
105
|
Yu T, Gan S, Zhu Q, Dai D, Li N, Wang H,
Chen X, Hou D, Wang Y, Pan Q, et al: Modulation of M2 macrophage
polarization by the crosstalk between Stat6 and Trim24. Nat Commun.
10:43532019. View Article : Google Scholar
|
|
106
|
Rőszer T: Understanding the Mysterious M2
Macrophage through activation markers and effector mechanisms.
Mediators Inflamm. 2015:8164602015. View Article : Google Scholar
|
|
107
|
Wang YC, Wu YS, Hung CY, Wang SA, Young
MJ, Hsu TI and Hung JJ: USP24 induces IL-6 in tumor-associated
microenvironment by stabilizing p300 and β-TrCP and promotes cancer
malignancy. Nat Commun. 9:39962018. View Article : Google Scholar
|
|
108
|
Ning C, Xie B, Zhang L, Li C, Shan W, Yang
B, Luo X, Gu C, He Q, Jin H, et al: Infiltrating Macrophages Induce
ERα Expression through an IL17A-mediated epigenetic mechanism to
sensitize endometrial cancer cells to estrogen. Cancer Res.
76:1354–1366. 2016. View Article : Google Scholar
|
|
109
|
Lv Q, Xie L, Cheng Y, Shi Y, Shan W, Ning
C, Xie B, Yang B, Luo X, He Q, et al: A20-mediated deubiquitination
of ERα in the microenvironment of CD163+ macrophages sensitizes
endometrial cancer cells to estrogen. Cancer Lett. 442:137–147.
2019. View Article : Google Scholar
|
|
110
|
Clancy JL, Henderson MJ, Russell AJ,
Anderson DW, Bova RJ, Campbell IG, Choong DY, Macdonald GA, Mann
GJ, Nolan T, et al: EDD, the human orthologue of the hyperplastic
discs tumour suppressor gene, is amplified and overexpressed in
cancer. Oncogene. 22:5070–5081. 2003. View Article : Google Scholar
|
|
111
|
Song M, Yeku OO, Rafiq S, Purdon T, Dong
X, Zhu L, Zhang T, Wang H, Yu Z, Mai J, et al: Tumor derived UBR5
promotes ovarian cancer growth and metastasis through inducing
immunosuppressive macrophages. Nat Commun. 11:62982020. View Article : Google Scholar
|
|
112
|
Surh CD and Sprent J: Homeostasis of naive
and memory T cells. Immunity. 29:848–862. 2008. View Article : Google Scholar
|
|
113
|
Zhou X, Yu J, Cheng X, Zhao B, Manyam GC,
Zhang L, Schluns K, Li P, Wang J and Sun SC: The deubiquitinase
Otub1 controls the activation of CD8+ T cells and NK cells by
regulating IL-15-mediated priming. Nat Immunol. 20:879–889. 2019.
View Article : Google Scholar
|
|
114
|
Zhang W, Sloan-Lancaster J, Kitchen J,
Trible RP and Samelson LE: LAT: The ZAP-70 tyrosine kinase
substrate that links T cell receptor to cellular activation. Cell.
92:83–92. 1998. View Article : Google Scholar
|
|
115
|
Kunii N, Zhao Y, Jiang S, Liu X, Scholler
J, Balagopalan L, Samelson LE, Milone MC and June CH: Enhanced
function of redirected human T cells expressing linker for
activation of T cells that is resistant to ubiquitylation. Hum Gene
Ther. 24:27–37. 2013. View Article : Google Scholar
|
|
116
|
Chen RH, Lee YR and Yuan WC: The role of
PML ubiquitination in human malignancies. J Biomed Sci. 19:812012.
View Article : Google Scholar
|
|
117
|
Wang YT, Chen J, Chang CW, Jen J, Huang
TY, Chen CM, Shen R, Liang SY, Cheng IC, Yang SC, et al:
Ubiquitination of tumor suppressor PML regulates prometastatic and
immunosuppressive tumor microenvironment. J Clin Invest.
127:2982–2997. 2017. View Article : Google Scholar
|
|
118
|
Meng X, Liu X, Guo X, Jiang S, Chen T, Hu
Z, Liu H, Bai Y, Xue M, Hu R, et al: FBXO38 mediates PD-1
ubiquitination and regulates anti-tumour immunity of T cells.
Nature. 564:130–135. 2018. View Article : Google Scholar
|
|
119
|
Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo
CW, Khoo KH, Chang SS, Cha JH, Kim T, et al: Glycosylation and
stabilization of programmed death ligand-1 suppresses T-cell
activity. Nat Commun. 7:126322016. View Article : Google Scholar
|
|
120
|
Zhang J, Bu X, Wang H, Zhu Y, Geng Y,
Nihira NT, Tan Y, Ci Y, Wu F, Dai X, et al: Cyclin D-CDK4 kinase
destabilizes PD-L1 via cullin 3-SPOP to control cancer immune
surveillance. Nature. 553:91–95. 2018. View Article : Google Scholar
|
|
121
|
Song Y, Xu Y, Pan C, Yan L, Wang ZW and
Zhu X: The emerging role of SPOP protein in tumorigenesis and
cancer therapy. Mol Cancer. 19:22020. View Article : Google Scholar
|
|
122
|
Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu
Y, Chang SS, Lin WC, Hsu JM, Hsu YH, et al: Deubiquitination and
Stabilization of PD-L1 by CSN5. Cancer Cell. 30:925–939. 2016.
View Article : Google Scholar
|
|
123
|
Wang Y, Sun Q, Mu N, Sun X, Wang Y, Fan S,
Su L and Liu X: The deubiquitinase USP22 regulates PD-L1
degradation in human cancer cells. Cell Commun Signal. 18:1122020.
View Article : Google Scholar
|
|
124
|
Huang X, Zhang Q, Lou Y, Wang J, Zhao X,
Wang L, Zhang X, Li S, Zhao Y, Chen Q, et al: USP22 Deubiquitinates
CD274 to Suppress Anticancer Immunity. Cancer Immunol Res.
7:1580–1590. 2019. View Article : Google Scholar
|
|
125
|
Li J, Yuan S, Norgard RJ, Yan F, Yamazoe
T, Blanco A and Stanger BZ: Tumor Cell-Intrinsic USP22 suppresses
antitumor immunity in pancreatic cancer. Cancer Immunol Res.
8:282–291. 2020. View Article : Google Scholar
|
|
126
|
Jingjing W, Wenzheng G, Donghua W, Guangyu
H, Aiping Z and Wenjuan W: Deubiquitination and stabilization of
programmed cell death ligand 1 by ubiquitin-specific peptidase 9,
X-linked in oral squamous cell carcinoma. Cancer Med. 7:4004–4011.
2018. View Article : Google Scholar
|
|
127
|
Naik E, Webster JD, DeVoss J, Liu J,
Suriben R and Dixit VM: Regulation of proximal T cell receptor
signaling and tolerance induction by deubiquitinase Usp9X. J Exp
Med. 211:1947–1955. 2014. View Article : Google Scholar
|
|
128
|
Sakaguchi S, Yamaguchi T, Nomura T and Ono
M: Regulatory T cells and immune tolerance. Cell. 133:775–787.
2008. View Article : Google Scholar
|
|
129
|
Wang F, Wang L, Wu J, Sokirniy I, Nguyen
P, Bregnard T, Weinstock J, Mattern M, Bezsonova I, Hancock WW and
Kumar S: Active site-targeted covalent irreversible inhibitors of
USP7 impair the functions of Foxp3+ T-regulatory cells by promoting
ubiquitination of Tip60. PLoS One. 12:e01897442017. View Article : Google Scholar
|
|
130
|
McHugh RS, Whitters MJ, Piccirillo CA,
Young DA, Shevach EM, Collins M and Byrne MC:
CD4(+)CD25(+) immunoregulatory T cells: Gene
expression analysis reveals a functional role for the
glucocorticoid-induced TNF receptor. Immunity. 16:311–323. 2002.
View Article : Google Scholar
|
|
131
|
Knee DA, Hewes B and Brogdon JL: Rationale
for anti-GITR cancer immunotherapy. Eur J Cancer. 67:1–10. 2016.
View Article : Google Scholar
|
|
132
|
Guo Y, Yang L, Lei S, Tan W and Long J:
NEDD4 Negatively Regulates GITR via ubiquitination in immune
microenvironment of melanoma. Onco Targets Ther. 12:10629–10637.
2019. View Article : Google Scholar
|
|
133
|
Trovato R, Fiore A, Sartori S, Canè S,
Giugno R, Cascione L, Paiella S, Salvia R, De Sanctis F, Poffe O,
et al: Immunosuppression by monocytic myeloid-derived suppressor
cells in patients with pancreatic ductal carcinoma is orchestrated
by STAT3. J Immunother Cancer. 7:2552019. View Article : Google Scholar
|
|
134
|
Song G, Zhang Y, Tian J, Ma J, Yin K, Xu H
and Wang S: TRAF6 regulates the immunosuppressive effects of
myeloid-derived suppressor cells in tumor-bearing host. Front
Immunol. 12:6490202021. View Article : Google Scholar
|
|
135
|
Zhang CX, Ye SB, Ni JJ, Cai TT, Liu YN,
Huang DJ, Mai HQ, Chen QY, He J, Zhang XS, et al: STING signaling
remodels the tumor microenvironment by antagonizing myeloid-derived
suppressor cell expansion. Cell Death Differ. 26:2314–2328. 2019.
View Article : Google Scholar
|
|
136
|
Chou FC, Chen HY, Kuo CC and Sytwu HK:
Role of galectins in tumors and in clinical immunotherapy. Int J
Mol Sci. 19:4302018. View Article : Google Scholar
|
|
137
|
Zhang CX, Huang DJ, Baloche V, Zhang L, Xu
JX, Li BW, Zhao XR, He J, Mai HQ, Chen QY, et al: Galectin-9
promotes a suppressive microenvironment in human cancer by
enhancing STING degradation. Oncogenesis. 9:652020. View Article : Google Scholar
|
|
138
|
Fang P, Li X, Dai J, Cole L, Camacho JA,
Zhang Y, Ji Y, Wang J, Yang XF and Wang H: Immune cell subset
differentiation and tissue inflammation. J Hematol Oncol.
11:972018. View Article : Google Scholar
|
|
139
|
Wang Y, Xiang Y, Xin VW, Wang XW, Peng XC,
Liu XQ, Wang D, Li N, Cheng JT, Lyv YN, et al: Dendritic cell
biology and its role in tumor immunotherapy. J Hematol Oncol.
13:1072020. View Article : Google Scholar
|
|
140
|
Bi E, Li R, Bover LC, Li H, Su P, Ma X,
Huang C, Wang Q, Liu L, Yang M, et al: E-cadherin expression on
multiple myeloma cells activates tumor-promoting properties in
plasmacytoid DCs. J Clin Invest. 128:4821–4831. 2018. View Article : Google Scholar
|
|
141
|
Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A,
Egranov SD, Zhang Y, Xia W, Gong J, et al: Oncogenic lncRNA
down-regulates cancer cell antigen presentation and intrinsic tumor
suppression. Nat Immunol. 20:835–851. 2019. View Article : Google Scholar
|
|
142
|
Caraux A, Lu Q, Fernandez N, Riou S, Di
Santo JP, Raulet DH, Lemke G and Roth C: Natural killer cell
differentiation driven by Tyro3 receptor tyrosine kinases. Nat
Immunol. 7:747–754. 2006. View
Article : Google Scholar
|
|
143
|
Paolino M, Choidas A, Wallner S, Pranjic
B, Uribesalgo I, Loeser S, Jamieson AM, Langdon WY, Ikeda F, Fededa
JP, et al: The E3 ligase Cbl-b and TAM receptors regulate cancer
metastasis via natural killer cells. Nature. 507:508–512. 2014.
View Article : Google Scholar
|
|
144
|
Haglund K and Dikic I: The role of
ubiquitylation in receptor endocytosis and endosomal sorting. J
Cell Sci. 125(Pt 2): 265–275. 2012. View Article : Google Scholar
|
|
145
|
Wang Y, Zhang Y, Yi P, Dong W, Nalin AP,
Zhang J, Zhu Z, Chen L, Benson DM, Mundy-Bosse BL, et al: The
IL-15-AKT-XBP1s signaling pathway contributes to effector functions
and survival in human NK cells. Nat Immunol. 20:10–17. 2019.
View Article : Google Scholar
|
|
146
|
Dou Y, Xing J, Kong G, Wang G, Lou X, Xiao
X, Vivier E, Li XC and Zhang Z: Identification of the E3 Ligase
TRIM29 as a critical checkpoint regulator of NK cell functions. J
Immunol. 203:873–880. 2019. View Article : Google Scholar
|
|
147
|
Deng L, Meng T, Chen L, Wei W and Wang P:
The role of ubiquitination in tumorigenesis and targeted drug
discovery. Signal Transduct Target Ther. 5:112020. View Article : Google Scholar
|
|
148
|
Richardson PG, Hideshima T and Anderson
KC: Bortezomib (PS-341): A novel, first-in-class proteasome
inhibitor for the treatment of multiple myeloma and other cancers.
Cancer Control. 10:361–369. 2003. View Article : Google Scholar
|
|
149
|
Piva R, Ruggeri B, Williams M, Costa G,
Tamagno I, Ferrero D, Giai V, Coscia M, Peola S, Massaia M, et al:
CEP-18770: A novel, orally active proteasome inhibitor with a
tumor-selective pharmacologic profile competitive with bortezomib.
Blood. 111:2765–2775. 2008. View Article : Google Scholar
|
|
150
|
Anchoori RK, Karanam B, Peng S, Wang JW,
Jiang R, Tanno T, Orlowski RZ, Matsui W, Zhao M, Rudek MA, et al: A
bis-benzylidine piperidone targeting proteasome ubiquitin receptor
RPN13/ADRM1 as a therapy for cancer. Cancer Cell. 24:791–805. 2013.
View Article : Google Scholar
|
|
151
|
Song Y, Ray A, Li S, Das DS, Tai YT,
Carrasco RD, Chauhan D and Anderson KC: Targeting proteasome
ubiquitin receptor Rpn13 in multiple myeloma. Leukemia.
30:1877–1886. 2016. View Article : Google Scholar
|
|
152
|
Kimura K, Yamada T, Matsumoto M, Kido Y,
Hosooka T, Asahara S, Matsuda T, Ota T, Watanabe H, Sai Y, et al:
Endoplasmic reticulum stress inhibits STAT3-dependent suppression
of hepatic gluconeogenesis via dephosphorylation and deacetylation.
Diabetes. 61:61–73. 2012. View Article : Google Scholar
|
|
153
|
Soong RS, Anchoori RK, Yang B, Yang A,
Tseng SH, He L, Tsai YC, Roden RB and Hung CF: RPN13/ADRM1
inhibitor reverses immunosuppression by myeloid-derived suppressor
cells. Oncotarget. 7:68489–68502. 2016. View Article : Google Scholar
|
|
154
|
Yu GY, Wang X, Zheng SS, Gao XM, Jia QA,
Zhu WW, Lu L, Jia HL, Chen JH, Dong QZ, et al: RA190, a proteasome
subunit ADRM1 inhibitor, suppresses intrahepatic cholangiocarcinoma
by inducing NF-KB-Mediated cell apoptosis. Cell Physiol Biochem.
47:1152–1166. 2018. View Article : Google Scholar
|
|
155
|
Soong RS, Anchoori RK, Roden RBS, Cho RL,
Chen YC, Tseng SC, Huang YL, Liao PC and Shyu YC: Bis-benzylidine
Piperidone RA190 treatment of hepatocellular carcinoma via binding
RPN13 and inhibiting NF-κB signaling. BMC Cancer. 20:3862020.
View Article : Google Scholar
|
|
156
|
Powis G and Kirkpatrick L: Hypoxia
inducible factor-1alpha as a cancer drug target. Mol Cancer Ther.
3:647–654. 2004.
|
|
157
|
Lee YM, Kim GH, Park EJ, Oh TI, Lee S, Kan
SY, Kang H, Kim BM, Kim JH and Lim JH: Thymoquinone selectively
kills hypoxic renal cancer cells by suppressing HIF-1α-mediated
glycolysis. Int J Mol Sci. 20:10922019. View Article : Google Scholar
|
|
158
|
Ge Y, Yoon SH, Jang H, Jeong JH and Lee
YM: Decursin promotes HIF-1α proteasomal degradation and immune
responses in hypoxic tumour microenvironment. Phytomedicine.
78:1533182020. View Article : Google Scholar
|
|
159
|
Chen JJ, Ren YL, Shu CJ, Zhang Y, Chen MJ,
Xu J, Li J, Li AP, Chen DY, He JD, et al: JP3, an antiangiogenic
peptide, inhibits growth and metastasis of gastric cancer through
TRIM25/SP1/MMP2 axis. J Exp Clin Cancer Res. 39:1182020. View Article : Google Scholar
|
|
160
|
Chen Y, Huang Y, Huang Y, Xia X, Zhang J,
Zhou Y, Tan Y, He S, Qiang F, Li A, et al: JWA suppresses tumor
angiogenesis via Sp1-activated matrix metalloproteinase-2 and its
prognostic significance in human gastric cancer. Carcinogenesis.
35:442–451. 2014. View Article : Google Scholar
|
|
161
|
Godbersen JC, Humphries LA, Danilova OV,
Kebbekus PE, Brown JR, Eastman A and Danilov AV: The
Nedd8-activating enzyme inhibitor MLN4924 thwarts
microenvironment-driven NF-κB activation and induces apoptosis in
chronic lymphocytic leukemia B cells. Clin Cancer Res.
20:1576–1589. 2014. View Article : Google Scholar
|
|
162
|
Katsuya K, Hori Y, Oikawa D, Yamamoto T,
Umetani K, Urashima T, Kinoshita T, Ayukawa K, Tokunaga F and
Tamaru M: High-Throughput screening for linear ubiquitin chain
assembly complex (LUBAC) selective inhibitors using homogenous
time-resolved fluorescence (HTRF)-based assay system. SLAS Discov.
23:1018–1029. 2018. View Article : Google Scholar
|
|
163
|
Katsuya K, Oikawa D, Iio K, Obika S, Hori
Y, Urashima T, Ayukawa K and Tokunaga F: Small-molecule inhibitors
of linear ubiquitin chain assembly complex (LUBAC), HOIPINs,
suppress NF-κB signaling. Biochem Biophys Res Commun. 509:700–706.
2019. View Article : Google Scholar
|
|
164
|
Liu C, Billadeau DD, Abdelhakim H, Leof E,
Kaibuchi K, Bernabeu C, Bloom GS, Yang L, Boardman L, Shah VH and
Kang N: IQGAP1 suppresses TβRII-mediated myofibroblastic activation
and metastatic growth in liver. J Clin Invest. 123:1138–1156. 2013.
View Article : Google Scholar
|
|
165
|
Liu JL, Wang XY, Huang BX, Zhu F, Zhang RG
and Wu G: Expression of CDK5/p35 in resected patients with
non-small cell lung cancer: Relation to prognosis. Med Oncol.
28:673–678. 2011. View Article : Google Scholar
|
|
166
|
Gao L, Xia L, Ji W, Zhang Y, Xia W and Lu
S: Knockdown of CDK5 down-regulates PD-L1 via the
ubiquitination-proteasome pathway and improves antitumor immunity
in lung adenocarcinoma. Transl Oncol. 14:1011482021. View Article : Google Scholar
|
|
167
|
Liu Y, Liu X, Zhang N, Yin M, Dong J, Zeng
Q, Mao G, Song D, Liu L and Deng H: Berberine diminishes cancer
cell PD-L1 expression and facilitates antitumor immunity inhibiting
the deubiquitination activity of CSN5. Acta Pharm Sin B.
10:2299–2312. 2020. View Article : Google Scholar
|
|
168
|
Chauhan D, Tian Z, Nicholson B, Kumar KG,
Zhou B, Carrasco R, McDermott JL, Leach CA, Fulcinniti M, Kodrasov
MP, et al: A small molecule inhibitor of ubiquitin-specific
protease-7 induces apoptosis in multiple myeloma cells and
overcomes bortezomib resistance. Cancer Cell. 22:345–358. 2012.
View Article : Google Scholar
|
|
169
|
Nicholson B and Suresh Kumar KG: The
multifaceted roles of USP7: New therapeutic opportunities. Cell
Biochem Biophys. 60:61–68. 2011. View Article : Google Scholar
|
|
170
|
Shi L, Lin H, Li G, Sun Y, Shen J, Xu J,
Lin C, Yeh S, Cai X and Chang C: Cisplatin enhances NK cells
immunotherapy efficacy to suppress HCC progression via altering the
androgen receptor (AR)-ULBP2 signals. Cancer Lett. 373:45–56. 2016.
View Article : Google Scholar
|
|
171
|
Zhang X, Meng T, Cui S, Feng L, Liu D,
Pang Q and Wang P: Ubiquitination of nonhistone proteins in cancer
development and treatment. Front Oncol. 10:6212942021. View Article : Google Scholar
|
|
172
|
Ning B, Zhao W, Qian C, Liu P, Li Q, Li W
and Wang RF: USP26 functions as a negative regulator of cellular
reprogramming by stabilising PRC1 complex components. Nat Commun.
8:3492017. View Article : Google Scholar
|